Abstract
Background
Accumulating evidence has indicated the role of gut microbiota in remodeling host immune signatures, but various interplays underlying colorectal cancers (CRC) with deficient DNA mismatch repair (dMMR) and proficient DNA mismatch repair (pMMR) remain poorly understood. This study aims to decipher the gut microbiome-host immune interactions between dMMR and pMMR CRC.
Method
We performed metagenomic sequencing and metabolomic analysis of fecal samples from a cohort encompassing 455 participants, including 21 dMMR CRC, 207 pMMR CRC, and 227 healthy controls. Among them, 50 tumor samples collected from 5 dMMR CRC and 45 pMMR CRC were conducted bulk RNA sequencing.
Results
Pronounced microbiota and metabolic heterogeneity were identified with 211 dMMR-enriched species, such as Fusobacterium nucleatum and Akkermansia muciniphila, 2 dMMR-depleted species, such as Flavonifractor plautii, 13 dMMR-enriched metabolites, such as retinoic acid, and 77 dMMR-depleted metabolites, such as lactic acid, succinic acid, and 2,3-dihydroxyvaleric acid. F. plautii was enriched in pMMR CRC and it was positively associated with fatty acid degradation, which might account for the accumulation of dMMR-depleted metabolites classified as short chain organic acid (lactic acid, succinic acid, and 2,3-dihydroxyvaleric acid) in pMMR CRC. The microbial-metabolic association analysis revealed the characterization of pMMR CRC as the accumulation of lactate induced by the depletion of specific gut microbiota which was negatively associated with antitumor immune, whereas the nucleotide metabolism and peptide degradation mediated by dMMR-enriched species characterized dMMR CRC. MMR-specific metabolic landscapes were related to distinctive immune features, such as CD8+ T cells, dendritic cells and M2-like macrophages.
Conclusions
Our mutiomics results delineate a heterogeneous landscape of microbiome-host immune interactions within dMMR and pMMR CRC from aspects of bacterial communities, metabolic features, and correlation with immunocyte compartment, which infers the underlying mechanism of heterogeneous immune responses.
Keywords: gastrointestinal neoplasms
WHAT IS ALREADY KNOWN ON THIS TOPIC
Gut microbiota regulates the host immune and impacts the response to immunotherapy.
The gut microbial-metabolic interplay and microbial-host immune interactions between proficient DNA mismatch repair (pMMR) and deficient DNA mismatch repair (dMMR) colorectal cancer (CRC) have not yet been studied.
WHAT THIS STUDY ADDS
An increased alpha diversity and enriched microbial communities were identified in dMMR CRC compared with pMMR CRC.
In dMMR CRC, activated nucleotide metabolism and peptide degradation were correlated with dMMR-enriched species, such as Akkermansia muciniphila.
In pMMR CRC, the depletion of gut microbiota contributed to the accumulation of short chain organic acids, including lactic acid and propanoic acid, therefore leading to an immunosuppressive tumor microenvironment.
Identifying distinctive gut microbiome-host immune interactions provides insight into the response of immunotherapy from the perspective of the microbiome.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Targeting gut microbiota may be a promising intervention to improve the immunotherapeutic efficacy for CRC, especially pMMR CRC.
Introduction
Colorectal cancer (CRC) is a highly heterogeneous disease with escalating incidence and mortality globally followed in the step of westernization.1 As central hallmarks of cancer development, genetic instability and mutability serially fuel the malignant behavior in most CRC carcinogenesis.2 DNA mismatch repair (MMR) deficiency (dMMR), mainly defined by particular mutations within short tandem repetitive DNA sequences and evaluated by the molecular fingerprint of microsatellite instability (MSI), is observed in approximately 10%–15% of CRCs.3 The MMR deficiency derives from functional germline mutations in MMR-related genes such as MLH1, MSH2, MSH6, PMS1, and EPCAM, leading to a highly heritable Lynch syndrome4 or results from epigenetic abnormality including 5' cytosine-phosphate-guanine-3' (CpG) island methylator phenotype and pathogenic BRAF mutations, which is more commonly seen in sporadic CRC.5 Molecular and immunological features of dMMR CRC have been elucidated by preliminary epidemiologic, mechanistic and clinical studies. Compared with MSI-low/proficient DNA mismatch repair (pMMR), dMMR CRC is characterized by an increased mutational burden with abundant neoantigens, ensuring a more robust immune response.6 7 Clinically, dMMR CRC is predisposed to a better prognosis in early-stage patients with a lower incidence of distant metastasis8 and being less sensitive to adjuvant chemotherapy but more responsive to immune checkpoint inhibitor (ICI) therapy.9–13
As the most prominent commensal resident in the human ecosystem, enteric microorganisms along with metabolites play an intriguing role with multilayer crosstalk in carcinogenesis, immunosurveillance, and tumor microenvironment.14–18 Some studies have provided an initial demonstration of the intimate association between influential bacterium with specific MSI status. For example, Fusobacterium nucleatum has a higher relative abundance in MSI-high (MSI-H) CRCs and is negatively correlated with tumor-infiltrating lymphocytes.19 In patients with head and neck squamous cell carcinoma, F. nucleatum induced epigenetic alteration via suppressing MMR-related gene expression.20 Emerging evidence emphatically points to the significance of gut microbiota and its metabolite in influencing and predicting the clinical benefit of cancer therapy.21 22 Association between gut microbiota composition and therapeutic efficacy of immunotherapy in preclinical models and human cohorts of non-small-cell lung cancer, melanoma, and other tumors treated with ICI therapy highlights their potential of being novel predictive biomarkers and therapeutic targets.23–27 Bacteroidales has shown immunostimulatory effects of augmenting anti-cytotoxic T-lymphocyte associated protein 4 (CTLA-4) therapeutic efficacy and is associated with immune-related adverse event.27 A recent study suggested that fecal Akkermansia muciniphila was found to be closely associated with the clinical benefit of ICI therapy in patients with non-small-cell lung cancer.28 In addition, a favorable enteric microbiotype of Lachnospiraceae and Actinobacteria in patients with melanoma treated with anti-programmed cell death protein 1 (PD-1) is shown to exhibit better clinical outcome.29 More importantly, fecal microbiota transplantation (FMT) from patients responsive to ICI makes the first appearance to foster a protective immune signature and reverse anti-PD-1 non-responsiveness by reprogramming the tumor microenvironment.26 27 30
Metagenomics and metabolomic analysis make it possible to decode gut microbial communities, define the unique composition and infer the complex interplay among microbiota, metabolites and CRC in the context of various risk factors and clinical phenotype.31 32 However, these studies did not incorporate particular genetic subtypes such as MMR status. Thus, more delicate features or multilayer crosstalk of fecal microbial load and metabolites abundance in microsatellite stability-specific CRCs remain cloaked in mystery. High-quality observation with large-scale sample analysis and omics-based profiling of particular cohorts is sorely needed to delineate and stratify the dMMR and pMMR CRC population from the perspective of microbiota.
In this study, we devise a cohort of 455 samples of carefully curated dMMR and pMMR patients and carry out comprehensive multi-omics analyses with metagenomic sequencing and metabolomic analysis of fecal samples and bulk RNA sequencing of tumor tissues. We interrogate the heterogeneity of microbial communities along with metabolites and identify the host-microbial interplay underlying dMMR and pMMR CRC. Depicting the MMR-specific landscape of microbial communities in patients with CRC would aid a more nuanced understanding of microbiome-host immune interactions and provide a fundamental basis for future improvement of ICI therapy.
Results
Participant information
A total of 455 fecal samples and 50 tumor tissue samples were included in the study (figure 1A). The dMMR group and pMMR group were similar in clinical characteristics, including age, gender, body mass index (BMI), differentiation, tumor node metastasis (TNM) stage, KRAS/NRAS/BRAF mutation, and CA199, whereas tumor size, differentiation, the rate of nerve invasion and vascular invasion, and carcinoembryonic antigen (CEA) were significantly different between dMMR and pMMR CRC. Detailed demographic, clinical, and biochemical profiles of the cohort were provided in online supplemental table S1.
Figure 1.
Study design and bacterial diversity of the fecal microbiota associated with mismatch repair status. (A) Study design and flow diagram. A total of 455 comprising 228 CRC and 227 CTRL were included in the study. The fecal metagenome was analyzed from 21 dMMR CRC, 207 pMMR CRC, and 227 CTRL. The fecal metabolome was analyzed from 14 dMMR CRC, 177 pMMR CRC, and 227 CTRL. Bulk RNA sequencing of tumor tissue samples was analyzed from 5 dMMR CRC and 45 pMMR CRC. (B) Alpha diversity measured by the Shannon Index of patients with CRC and health control in different groups. The p value (Student’s t-test) is calculated and shown in the figure. Data are delivered via the IQRs with the median as a black horizontal line and the whiskers extending up to the most extreme points within 1.5× the IQR. (C–D) PCoA of microbiota at the genus level (C) and metabolites (D) calculated by Bray-Curtis distance between dMMR, pMMR, and CTRL. The p values are calculated by the pairwise adonis test and adjusted by Bonferroni correction and shown in the figure. CRC, colorectal cancer; CTRL, control; dMMR, deficient DNA mismatch repair; PCoA, principal coordinates analysis; pMMR, proficient DNA mismatch repair; RNA-seq, RNA sequencing.
jitc-2023-007420supp001.pdf (146.4KB, pdf)
jitc-2023-007420supp006.pdf (47.7KB, pdf)
Decreased alpha diversity in pMMR CRC and altered overall microbial composition in dMMR and pMMR CRC
Fecal alpha diversity estimated by the Shannon Index was significantly decreased in pMMR CRC compared with healthy control (CTRL) or dMMR CRC (p=0.00081 and 0.048, respectively, Student’s t-test). Interestingly, the alpha diversity was similar between CTRL and dMMR (figure 1B). Principal coordinates analysis (PCoA) demonstrating beta diversity calculated by Bray-Curtis distance showed a significantly diverse distribution of fecal bacteria between dMMR and CTRL and between pMMR and CTRL (p=0.038 and p=0.0005, respectively, pairwise adonis test, figure 1C), while no significant differences between dMMR and pMMR (p=0.6582, pairwise adonis test). Similar results were seen in the PCoA of metabolites (figure 1D). Our data suggested that dMMR CRC possessed a higher alpha diversity but shared similar beta diversity when compared with pMMR CRC, indicating the potential function of microbiota affecting the host in an MMR-specific manner.
Taxonomic signatures reveal heterogeneity of microbiota in dMMR and pMMR CRC
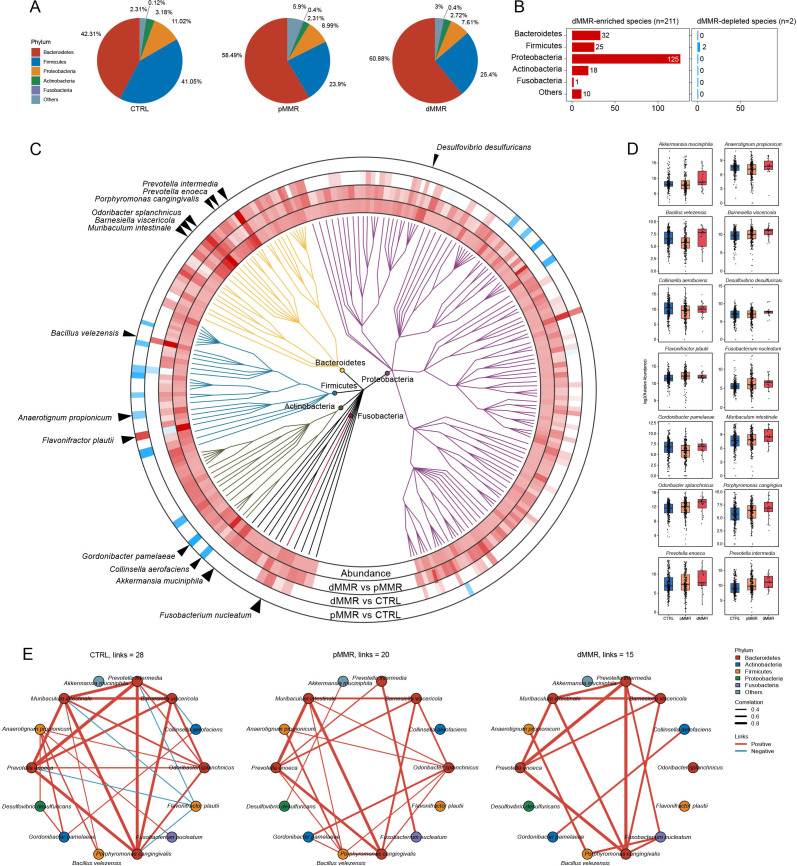
To better discern the dissimilarities of the microbial community, we first checked the microbial proportion at the phylum level in CTRL, dMMR CRC, and pMMR CRC (figure 2A). Next, we identified a total of 213 differentially abundant species between dMMR and pMMR CRC in each phylum, with 211 species enriched in dMMR CRC and 2 species depleted in dMMR CRC (figure 2B). A phylogenic tree of the differentially abundant species between dMMR and pMMR CRC was drawn (figure 2C, online supplemental table S2). The top 30 abundant differential species of 455 experimental subjects were then calculated, showing a more predominant distribution of dMMR-enriched species (online supplemental figure S1). Among them, 14 differential species were notably selected, including dMMR-enriched A. muciniphila, F. nucleatum, Odoribacter splanchnicus, Prevotella intermedia and pMMR-enriched Flavonifractor plautii (figure 2D). The probiotic A. muciniphila and the tumorigenic bacteria F. nucleatum have been reported to modulate tumor immune microenvironment through different mechanisms and manifest the potential to improve programmed cell death ligand 1 (PD-L1) inhibitor efficacy.33–36 F. plautii, a flavonoid degrading bacteria, is reported to be enriched in early-onset CRC and present the suppressive immune responses of Th2.37 To assess the possible microbial associations, interaction networks of the 14 selected microbial species were drawn and significant correlation with p<0.05 were displayed within CTRL, pMMR CRC, and dMMR CRC, respectively (figure 2). In the CTRL group, the pMMR-enriched bacteria F. plautiii was negatively correlated with Bacteroidetes, including P. intermedia, Prevotella enoeca, Muribaculum intestinale, and Porphyromonas cangingivalis, while the correlations with F. plautii diminished in the pMMR and dMMR CRC. In pMMR CRC, the dMMR-enriched bacteria O. splanchnicus was positively correlated with Barnesiella viscericola, M. intestinale, Anaerotignum propionicum, Gordonibacter pamelaeae, Bacillus velezensis, and P. cangingivalis, whereas the correlations vanished in dMMR CRC. These findings revealed distinct composition and interactions of gut microbiota in dMMR and pMMR CRC, which may be associated with differences in tumor clinicopathological characteristics and ICI responses.
Figure 2.
Taxonomic microbial signatures of dMMR CRC and pMMR CRC. (A) Pie charts of microbial proportions at the phylum level in CTRL, pMMR, and dMMR CRC. (B) Bar chart of the number of dMMR enriched and depleted species in each phylum, including Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, and Others. (C) Two hundred and thirteen differentially abundant bacteria species between dMMR and pMMR CRC are shown in the phylogenetic tree, grouped in the phyla Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Fusobacteria. The innermost circle shows species log10 relative abundances averaged over all samples. In the outer rings, species are marked for significant (p<0.05; multivariable regression model with age, sex, and BMI as covariates) elevation (red) or depletion (blue). (D) Box plots show the log2 relative abundance of 14 species marked by arrows in (A). Data are delivered via the IQRs, with the median as a black horizontal line and the whiskers extending up to the most extreme points within 1.5× the IQR. (E) Co-occurrence relationships among species in dMMR CRC, pMMR CRC, and CTRL calculated by Spearman’s correlation. Only significant (p < 0.05) correlations are shown. The red lines indicate positive species interactions; the blue lines indicate negative interactions. The colors of the species represent the phylum. CRC, colorectal cancer; CTRL, control; dMMR, deficient DNA mismatch repair; pMMR, proficient DNA mismatch repair; BMI, body mass index.
jitc-2023-007420supp007.xlsx (22.2KB, xlsx)
jitc-2023-007420supp002.pdf (172.1KB, pdf)
Fecal metabolomic alterations in dMMR and pMMR CRC
To investigate how gut microbiota affects metabolism in dMMR and pMMR CRC, the non-targeted metabolomics was carried out on fecal samples (14 dMMR, 177 pMMR, and 227 CTRL). A total of 90 differential metabolites, including 13 metabolites enriched and 77 metabolites depleted in dMMR CRC, was shown in figure 3A and online supplemental table S3. A broad overview of differential metabolomic expression levels categorized by the compound superclass was shown in online supplemental figure S2. The metabolites enrichment overview based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database showed significantly enriched metabolic pathways, including purine metabolism, pyrimidine metabolism, starch and sucrose metabolism, sphingolipid metabolism, and propanoate metabolism (figure 3B). Specifically, metabolites involved in propanoate metabolism, including propionic acid and lactic acid, as well as other short chain organic acids, such as succinic acid (succinate) and 2,3-dihydroxyvaleric acid, were decreased in dMMR CRC (figure 3C). Lactate and succinate as key metabolites involved in glycolysis and tricarboxylic acid cycle modulating non-metabolic immune responses may impact antitumor responses to immunotherapy via different pathways in distinct subgroups of immune cells.38–40 Organic acids and diverse peptides had immunomodulation properties and exerted biological activity in the immune cell signaling pathway. Among the total 25 organic acids and derivatives, amino acid D-alanine, dipeptides such as glutamylvaline and Phe-Val, and polypeptides such as Arg-Pro-Pro and Arg-Glu-Asp-Val were depleted in dMMR CRC, whereas only hydroxyprolyl-proline was enriched in dMMR CRC, reflecting the attenuated proteolysis activity and possible inflammatory (figure 3C).
Figure 3.
Fecal metabolome changes between dMMR and pMMR CRC. (A) Volcano plot of differential metabolites between dMMR and pMMR CRC (p value<0.05 and absolute log2 fold change > 0.3; multivariable regression model with age, sex, and BMI as covariates). (B) Bar plot shows the over-representation analysis of differential metabolites based on the metabolites set from the KEGG database. Significantly enriched metabolism pathways are annotated with asterisks (p value<0.05). (C) Box plots show the log2 abundance of metabolites. Data are delivered via the IQRs, with the median as a black horizontal line and the whiskers extending up to the most extreme points within 1.5× the IQR. (D) A heatmap of Spearman’s correlation between differential species and metabolites. *p<0.05; **p<0.01; ***p<0.001. CRC, colorectal cancer; CTRL, control; dMMR, deficient DNA mismatch repair; pMMR, proficient DNA mismatch repair; BMI, body mass index; KEGG, Kyoto Encyclopedia of Genes and Genomes; TCA, tricarboxylic acid.
jitc-2023-007420supp008.xlsx (18.1KB, xlsx)
jitc-2023-007420supp003.pdf (324.1KB, pdf)
Lipids and long-chain fatty acids (LCFAs) also presented diverse alteration in fecal metabolites, comprising dMMR-enriched retinoic acid (RA), dMMR-depleted LCFAs, including trans-vaccenic acid, 9,10-epoxyoctadecenoic acid, and xi-10-hydroxyoctadecanoic acid (figure 3C). The function of lipids and long-chain fatty acids in the microbiota-host immune landscape required in-depth research. The dMMR-enriched D-alpha-tocopherol succinate, the most effective analog of vitamin E, presented multiple biological functions, including chemopreventative and anticancer activity.41 To explore the role of microbiota in the metabolic process, we performed a Spearman’s correlation of differential microbiota and metabolites (figure 3D). Notably, the dMMR-enriched RA was mainly correlated with B. velezensis while the dMMR-depleted lactate was negatively correlated with most differential species. Moreover, the dMMR-enriched F. nucleatum presented positive associations with carboxylic acid, including succinic acid and propionic acid. On the contrary, the pMMR-enriched F. plautii was negatively correlated with most of the differential metabolites. The above results suggested that fecal metabolic composition was heterogeneous between dMMR and pMMR CRC, which might alter the bioavailability of metabolites and reconstruct tumor microenvironment. We speculated that metabolic factors might well play a requisite role in microbiota affecting host immunometabolism and antitumor immune response.
The alteration of microbial KEGG Orthology genes and KEGG pathway from dMMR and pMMR CRC
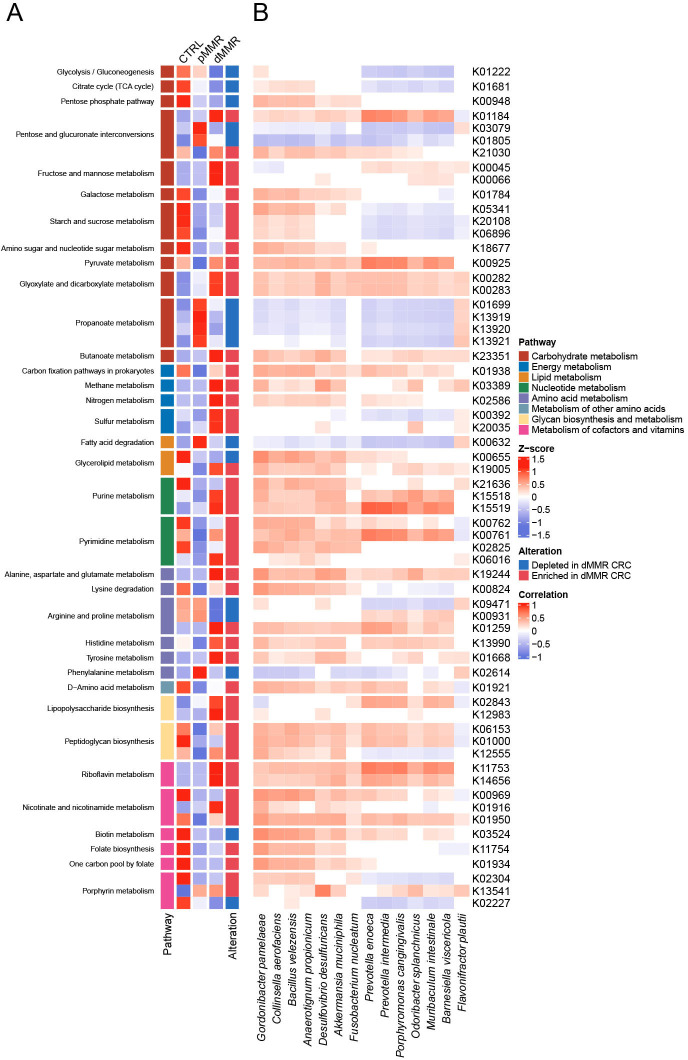
To further interrogate the metabolic programs of microbiota in dMMR and pMMR CRC, we compared the microbial genes annotated from the KEGG Orthology (KO) database, categorized by KEGG metabolic pathways. A total of 111 differential KO genes, comprising 85 upregulated genes and 26 downregulated genes in dMMR CRC, were summarized in figure 4A and online supplemental table S4. Most differential genes related to energy metabolism, nucleotide metabolism, glycan biosynthesis and metabolism, and metabolic cofactors and vitamins were upregulated, while genes involved in lipid metabolism, such as fatty acid degradation and glycerophospholipid metabolism, were downregulated. Furthermore, we dived deeply into the specific metabolic pathways of the differential genes. Interestingly, four KO genes involved in the propanoate metabolism, including pduC, pduD, and pduE, constructing the subunit of propanediol dehydratase, and pduQ (1-propanol dehydrogenase), were significantly depleted in dMMR CRC, which might reduce the production of propanal and propanol, therefore contributing to the depletion of propanoic acid as demonstrated in the above metabolomic data. In the nucleotide metabolism pathway, genes participating in the purine metabolism and pyrimidine metabolism (nrdD, dgk, dck, pyrE, upp, pyrR, and pydC) were significantly enriched in dMMR CRC, indicating a reinforced conversion of nucleotides, which was concordant with the depletion of nucleotide, purine, and pyrimidine in dMMR CRC including uridine, deoxyinosine, deoxyguanosine, thymine, hypoxanthine, guanine, and uracil (online supplemental figure S2). Next, we conducted the Spearman’s correlation analysis to further investigate the association between differential microbiota and the KO genes (figure 4B). The species under phylum Bacteroidetes, including P. enoeca, P. intermedia, P. cangingivalis, O. splanchnicus, M. intestinale, B. viscericola, showed similar patterns of correlations with KO genes, indicating their identical metabolic function in CRC. The positive correlation between dMMR-enriched species and purine and pyrimidine metabolism that augmented in dMMR CRC suggested the potential disturbance in nucleotide metabolism of gut microbiota affected by the host MMR status. Consistent with metabolomic analysis, these identified differential KO genes may bridge the alteration in metabolic processes catalyzed or affected by gut microbiota in dMMR CRC and pMMR CRC.
Figure 4.
The mismatch repair status-associated microbial genes are categorized into KO genes and KEGG pathways. (A) The heatmap shows the significantly altered KO gene expression from each pathway in CTRL, dMMR, and pMMR. The cell color and intensity represent the relative abundance of KO genes. The first row of cells annotated the KEGG pathways, including carbohydrate metabolism, energy metabolism, lipid metabolism, nucleotide metabolism, amino acid metabolism, metabolism of other amino acids, glycan biosynthesis and metabolism, and metabolism of cofactors and vitamins. The split cells are annotated with the pathway names. The last row of cells marked the alteration of significant differential KO genes between dMMR and pMMR, with red representing enriched in dMMR CRC and blue representing depleted in dMMR CRC (p< 0.05, multivariable regression model with age, sex, and body mass index as covariates). (B) The heatmap represents Spearman’s correlations between 14 selected species and KO genes (adjusted p value<0.05). CRC, colorectal cancer; CTRL, control; dMMR, deficient DNA mismatch repair; KO, KEGG Orthology; pMMR, proficient DNA mismatch repair; KEGG, Kyoto Encyclopedia of Genes and Genomes; TCA, tricarboxylic acid.
jitc-2023-007420supp009.xlsx (26.3KB, xlsx)
Microbiota-driven metabolic reprogramming in dMMR CRC and pMMR CRC
To further interpret the microbial-metabolic interplay in CRC with different MMR status, we conducted Spearman’s correlation analysis among differential species, metabolites, and KO genes and identified 462, 456, and 195 links in CTRL, dMMR CRC, and pMMR CRC, respectively (correlation coefficients >0.2 and p<0.05) (figure 5A–C; online supplemental table S5). A. muciniphila was positively correlated with K00282 (gcvPA) and K00283 (gcvPB), encoding the glycine dehydrogenase subunit 1 and 2, respectively, and K19244 (ala) encoding the alanine dehydrogenase in CTRL and pMMR CRC, suggesting its potential influence in amino acid degradation. Of note, the dMMR-depleted species F. plautiii was positively correlated with KO gene K00632 (fadA), while other dMMR-enriched species were negatively correlated with fadA. FadA encodes the key enzyme acetyl-CoA acyltransferase participating in beta-oxidation that affects alpha−linolenic acid metabolism, fatty acid degradation and valine, leucine and isoleucine degradation. Short chain hydroxy fatty acids, such as lactic acid and 2,3-dihydroxyvaleric acid, were negatively correlated with the dMMR-enriched species O. splanchnicus, Collinsella aerofaciens, B. viscericola, P. intermedia, M. intestinale, P. enoeca, and P. cangingivalis in CTRL and pMMR CRC, suggesting they might be the potential consumer of these acids and the depletion of these species contribute to the accumulation of these dMMR-depleted acids. Overall, more differential metabolites with a higher proportion of dipeptide and polypeptide were observed in the microbial-metabolic interplay in dMMR CRC, indicating the diverse metabolic process in dMMR CRC. Our results profiled distinctive microbial-metabolic interaction within dMMR and pMMR CRC, which may improve our knowledge of the role of gut microbiota with regard to MMR status in patients with CRC.
Figure 5.
Microbial-metabolic remodeling in CTRL, pMMR CRC, and dMMR CRC. (A–C) The chord diagram among species, KO genes, and metabolites in CTRL (A), pMMR CRC (B), and dMMR CRC (C). Only significant (p < 0.05, Spearman’s correlation) correlations are shown. The colors of nodes indicate the group of features. The colors of each grid showed the feature groups (red, species; blue, metabolites; orange, KO genes). The colors of the lines indicate the type of correlation (red, positive interactions; blue, negative interactions). CRC, colorectal cancer; CTRL, control; dMMR, deficient DNA mismatch repair; pMMR, proficient DNA mismatch repair; KEGG, Kyoto Encyclopedia of Genes and Genomes Orthology; KO, KEGG Orthology.
jitc-2023-007420supp010.xlsx (41.9KB, xlsx)
Microbial-metabolic-host immune interplay revealed by integrative multiomics signatures of dMMR CRC and pMMR CRC
Given the fact that dMMR CRC responds to ICI therapy with a frequent and durable effect other than pMMR CRC and gut microbial implication in systemic and local immunity, we raised the core question of how gut microbiota specifically remodel immune response.42 43 Bulk RNA sequencing of the tumor samples from 5 dMMR CRC and 45 pMMR CRC was conducted and analyzed to depict transcriptomic signatures. A total of 374 differential expressed genes (p value corrected by Bonferroni <0.05 and log2 fold change (log2FC) >1.5 were observed, with 94 upregulated and 280 downregulated in dMMR CRC (online supplemental figure S3A and table S6). Next, we used CIBERSORTx to determine the immune landscape and identified differential immune cells composition. There was a higher proportion of M1 macrophages compartment in dMMR versus pMMR tumors. Among the component enriched in pMMR tumors were B cells naïve, B cells memory, plasma cells, natural killer (NK) cells activated, and dendritic cell (DC) resting (online supplemental figure S3B; Student’s t-test, p<0.05).44 Then, we conducted Spearman’s correlation analysis between differential species and genes, and between differential species and immune cells in dMMR CRC and pMMR CRC. A total of 36 and 72 links were shown in pMMR and dMMR, respectively (p<0.05, figure 6A,B). As known, dMMR tumors are characterized by a large number of neoantigens and thus enhanced immunogenicity, which is tightly associated with immune infiltration in tumor microenvironment and the solid foundation of success in immunotherapy.45 In consequence, the probability of microbial species interacting with local immune compartments is dramatically increased and potentially contributed to greater correlation links within dMMR CRC. In pMMR CRC, F. plautii was negatively correlated with monocytes and positively correlated with macrophages M2, MIR31HG, and LYG1. Among T-cell subgroups, T cells CD4 memory activated was positively correlated with F. nucleatum, A. propionicum, and Desulfovibrio desulfuricans, while T cells CD4 memory resting was negatively correlated with P. enoeca, P. cangingivalis, O. splanchnicus, B. viscericola. In dMMR CRC, F. nucleatum was positively correlated with DC activated, and negatively correlated with macrophages M1. P. intermedia was negatively correlated with macrophages M0 and M1, and positively correlated with T cells CD4 memory resting and neutrophils, presenting diverse multicellular interactions. NK cells resting and DC activated positively correlated with A. muciniphila, O. splanchnicus, G. pamelaeae. The microbial-metabolic-host immune interplay showed a negative correlation between pMMR-enriched propionic acid and T cells CD8, which might explain the poor response to immunotherapy in pMMR CRC (online supplemental figure S4A). The pMMR-enriched KO genes, including K01699, K13919 and K13920 encoding pduC, pduD, pduE, respectively, and comprising the subunit of propanediol dehydratase participating involved in propanoate metabolism, presented a negative correlation with monocytes and a positive correlation with macrophages M2, indicating the role of gut microbiota in monocyte differentiation toward macrophages M2 (online supplemental figure S4B). Collectively, our analyses identified differential genes potentially regulated by gut microbiota and revealed the interactions between microbes and host immune cells infiltration in tumor microenvironment in dMMR and pMMR CRC, which may impact the immune response to ICI therapy. Targeting gut microbiota, such as oral supplementation of dMMR-enriched species and FMT, is a promising adjuvant strategy to improve immunotherapy.
Figure 6.
Microbial-host immune crosstalk in pMMR CRC and dMMR CRC. The integrated network among species, host genes, and immunocytes in pMMR CRC and dMMR CRC. Only significant (p<0.05, Spearman’s correlation) correlations are shown. The colors of nodes indicate the group of features (red, species; blue, RNA; orange, immunocytes). The colors of the lines indicate the type of correlation (red, positive interactions; blue, negative interactions). CRC, colorectal cancer; dMMR, deficient DNA mismatch repair; pMMR, proficient DNA mismatch repair.
jitc-2023-007420supp004.pdf (308.7KB, pdf)
jitc-2023-007420supp011.xlsx (209.6KB, xlsx)
jitc-2023-007420supp005.pdf (864.7KB, pdf)
Discussion
Our study combined metagenomic sequencing and metabolomic analysis of fecal samples with bulk RNA sequencing of tumor samples from dMMR CRC and pMMR CRC, revealing the 211 dMMR-enriched species, such as F. nucleatum, A. muciniphila, and O. splanchnicus, and only 2 dMMR-depleted species, such as F. plautii, dMMR-enriched metabolites RA and dMMR-depleted metabolites lactic acid and 2,3-dihydroxyvaleric acid. We further gained resolution on the microbial-metabolic interplay in dMMR CRC and pMMR CRC and speculated that the accumulation of lactic acid induced by the depletion of gut microbiota characterized the pMMR CRC, whereas the activated nucleotide metabolism and peptide degradation mediated by dMMR-enriched species characterized the dMMR CRC. We also analyzed the microbial-host immune interactions and revealed the association between species and immunocytes in the tumor microenvironment, which might account for the differences in the immune response to ICI therapy mediated by gut microbiota.
Our results showed that gut microbiota in dMMR CRC had a higher alpha diversity than pMMR CRC, and the differential species were mostly enriched in dMMR CRC. Considering that the impact of antibiotics on the depletion of gut microbiota contributed to the poor response to immunotherapy, the enrichment of microbial species such as A. muciniphila in dMMR CRC corroborated the crucial role of gut microbiota in the effective response of dMMR patients to immunotherapy.46–48 These data indicated that the poor response of pMMR CRC to immunotherapy was partly attributed to the depletion of gut microbiota. FMT is one of the therapeutic approaches to alter the gut microbiota in patients. A preliminary report of a phase I clinical trial demonstrated the safety of FMT to 20 patients with advanced melanoma and its efficacy of improving immune response to ICI therapy. Another approach is oral supplementation of dMMR-enriched species that might be associated with the immune response, but the mechanism of how these microbes impact the host immune response has not been fully interpreted yet. Therefore, combining ICI therapy and FMT of fecal samples from dMMR CRC to pMMR CRC might be a promising approach to treat pMMR CRC.
The crosstalk among commensal species and immunocytes reflected the heterogeneity of tumor microenvironment between dMMR CRC and pMMR CRC. Basically, dMMR tumors are characterized by accumulation of abnormal proteins and thus higher levels of neoantigens, enabling a greater recruitment and infiltration of immune cells. Additionally, our correlation analysis of differential gut microbial species revealed numerous bacteria enriched in dMMR CRC compared with pMMR CRC. The heightened immunogenicity and microbial immunomodulatory capacity in dMMR may provide the foundation for the extensive associations observed between gut bacteria and immune components. The elevated immunization threshold in dMMR appears to foster intricate microbial-immune interplay, whereby resident microbes may remodel antitumor immunity through diverse mechanisms influencing various immune cell subsets and pathways. The dMMR-enriched species F. nucleatum was reported to induce PD-L1 expression and augment the immune response to ICI therapy via STING signaling and recruitment of CD8+ T cells.34 F. nucleatum was reported to exhibit a proinflammatory signature and recruit tumor infiltrated myeloid cells including conventional DCs, classical myeloid DCs and CD103+ regulatory DCs, partly concordant with our result that F. nucleatum positively correlated with activated DCs in dMMR CRC. F. nucleatum is present in both dMMR and pMMR CRC, yet exhibits distinct correlations with immune cell subsets. We postulated several factors that may contribute to the differential immunomodulation by F. nucleatum, including the divergent mutational landscapes of dMMR versus pMMR tumors and discrepancies between fecal microbiota and intratumoral microbiota, where additional tumor-associated microbial community members and immune microenvironment may shape the immune impacts of F. nucleatum. Our result also suggested a positive correlation between A. muciniphila and activated DCs in dMMR CRC. The probiotic A. muciniphila exhibited benefits in multiple diseases, including obesity, diabetes, and improvement of responses to immunotherapy.49 A. muciniphila was also reported to induce the production of cytokines involved in the immunogenicity of PD-1 blockade from DCs and promote antigen-specific T-cell responses.36 48 50 Although the exact mechanisms of such species that remodeling immunotherapy efficacy responses were not amply interpreted, our analyses established a hypothetical bridge between the gut microbiota and tumor microenvironment.
Alterations of the metabolic landscape also make a great difference to immune cell function and tumor microenvironment. Increasing evidence has shown that short-chain fatty acids (SCFAs), including propionic acid and butyric acid, acted as novel regulators of host homeostasis and tumor immunity.51 Lactic acid has been shown to exert an immunosuppressive effect by inducing anti-inflammatory genes transcription, promoting M2-like tumor-associated macrophage phenotype52 and impairing cytotoxic T-cell function.53 Clinical supplement of propionic acid contributed to the upregulation of interleukin-10 and suppressive immunologic function of CD25+ Foxp3+ regulatory T cells, indicating the immunomodulatory property of propionic acid through a metabolite-immune interaction.54 Interestingly, our metabolome exactly demonstrated that pMMR CRC was characterized by a higher level of both propionic acid and lactic acid, with a negative correlation between propionic acid and CD8+ T cells, which might account for a relatively immunosuppressive tumor microenvironment in pMMR CRC and the subsequent inefficacy in ICI therapy. Besides, lactic acid was negatively correlated with most dMMR-enriched species, which referred to a relative depletion of these species in the pMMR group. The possible underlying connection of specific microorganism might account for the accumulation of lactic acid in the local tumor niche, which contributes to a relatively immunosuppressive tumor microenvironment in pMMR CRC and the subsequent inefficacy in ICI therapy. Apart from SCFAs and lactic acid, metabolic reprogramming of LCFAs in cancer cells or immunocytes also plays a pivotal role in affecting tumor-related inflammatory responses by means of mediated-immune phenotypes or lipotoxicity. For instance, the accumulation of specific LCFAs dampened the cytotoxic T cells’ ability to repress tumor progression.55 RA catalyzed from vitamin A-derivative retinol in intestinal epithelial cell is considered as a key regulator of local immunity influenced by gut microbiota, which is involved in many physiological properties of immune cells. RA has been found to accumulate in many kinds of cancers and promotes antitumor immunity in CRC. Specifically, it elevated the expression of major histocompatibility complex I on CRC tumor cells and promoted a robust CD8+ T cells response.56 The supplement of RA in CRC reduced tumor burden by elevating the cytotoxic CD8+ T cells frequency and promoted PD-L1 blockade efficacy by recruiting inflammatory macrophages. In addition, suppression of RA synthesis restored the balance of T-cell subset ratio and modulated the activity of cytokines such as interleukin-22.57 58 On the contrary, tumor-derived RA was found to inhibit anti-PD-1 therapy by promoting intratumoral monocyte differentiation toward immunosuppressive macrophages other than DCs.59 Given the critical influence of RA on intestinal immunity, we speculated that enriched RA in dMMR CRC have substantial implication for immunosuppressive tumor microenvironment. The 2,3-dihydroxyvaleric acid belongs to the class of hydroxy fatty acids. Notably, it is negatively correlated with most of the dMMR-enriched species based on our former analysis. However, there is a limited number of existing research on this metabolite and its role in immune response. Further research is needed to determine its precise interaction with microbial species and underlying mediation in tumor-associated immunologic function. The heterogeneity of metabolic landscape in CRC with different MMR status provided clues for variant immune microenvironment. Since metabolites can be derived from or modeled by gut microbiota and host cells under different circumstances, our data preliminarily threw light on the underlying microbial-host immune interaction.
This study has some inherent limitations. The small dMMR CRC cohort constrains the conclusions that can be drawn. Some patients underwent neoadjuvant therapy prior to specimen collection, which could potentially confound microbiome and metabolic profiles. The limited sample size coupled with variability in fecal sampling timing and neoadjuvant interventions restricts interpretability and generalizability of the findings. Several factors may contribute to microbiome compositional differences between our control cohort and those in other studies. Subject characteristics like ethnicity, geography, diet, and lifestyle can influence the microbiota and often vary between control groups. Additionally, technical factors like sample preservation, DNA extraction and sequencing platforms used across studies could impact the analytic results. Moving forward, larger prospective studies controlling for potential confounders including medical interventions will be essential to derive meaningful conclusions. What is more, the analysis combining metagenome and host transcriptome provides proof of concept that gut microbiota is associated with immunocytes in the tumor microenvironment. However, the fecal metagenomic and metabolomic profiles do not fully represent the intratumoral microbiota and tumor-associated metabolism. Also, whether certain metabolites serve as an intermediate connecting microbiota with heterogeneous antitumor immunity such as immunocytes compartment or a pro/anti-inflammatory tumor microenvironment and the exact mechanism remain unsolved. The intratumoral microbiome can be identified from the whole genome, bulk RNA transcriptome, and single-cell RNA transcriptome sequencing, decoding tumor-specific microbiome, bacteria-associated gene expression, immune activity, and prognosis.60 61 However, sample contamination from environmental microorganisms and experimental operation, and bioinformatic noise obscure the accurate identification of intratumor species. The decontaminated bioinformatic pipelines filtered contaminant species stringently to ensure quality control, leading to reduced microbial information derived from the sequencing data, which restricted the study of the intratumoral microbiome. Recently, new approaches, such as spatial transcriptomics sequencing and invasion–adhesion-directed expression sequencing (INVADEseq), have been developed to visualize the intratumoral microbiota and map in situ the host-bacterial interactions.62 Therefore, further experiments focusing on the interaction between microbial species and immunocytes in situ, are required to corroborate the results from the bioinformatic analysis and confirm the hypothesis.
Altogether, our study identified dMMR and pMMR-specific gut microbiota and metabolites and provided an integrative multiomics analysis of the microbial-metabolic interplay and microbial-host immune interactions. These analyses paved the road for targeting gut microbiota to improve the immune response in CRC via species supplementation and FMT.
Methods
Study subjects and sample collection
A total of 455 fecal samples and 50 CRC tumor tissue samples were collected from the Fudan University Shanghai Cancer Center, Shanghai, China, and the Second Hospital of Shandong University, Shandong, China, from the year 2018 to 2021. Fecal samples were collected before surgery and stored at −80°C before metagenomic sequencing and metabolomic analysis. Tumor tissue samples were collected after surgery and stored at −80°C before RNA bulk sequencing. No patients received treatment with ICIs during the study period or sample collection. The tumor stage was evaluated based on the TNM staging system. MMR status was determined by four major MMR proteins (MLH1, MSH2, MSH6, and PMS2) via immunohistochemistry staining. The healthy control group encompassed volunteers with no gastrointestinal tumors confirmed by colonoscopy screening. Participants’ clinicopathological characteristics, including age, gender, BMI, tumor location, tumor size, differentiation, stage, nerve invasion, vascular invasion, KRAS/NRAS/BRAF mutation, CEA, CA199, and patients received neoadjuvant therapy were collected from the electronic medical record system. Written informed consent was provided by all subjects before sampling.
Metagenomic sequencing and data analysis
DNA extraction, library construction, and metagenomic sequencing were manipulated as previously described.63 The taxonomy profile at the phylum, genus, and species level and the KO genes profile were selected for further analysis. MaAsLin264 was used to identify differential microbial features (p value<0.05) and evaluate the multivariable association between phenotypes (age, gender, and BMI) and microbial features, including taxonomy abundance and KO gene profile.
Non-targeted metabolomic sequencing and data analysis
Fecal metabolite extraction and liquid chromatography tandem mass spectrometry were manipulated as previously described.63 MaAsLin264 was used to identify differential metabolites between dMMR CRC and pMMR CRC (p value<0.05, absolute log2FC > 0.3), with age, gender and BMI as covariates.
Bulk RNA sequencing and data analysis
RNA was isolated from tumor tissue as described above. The quality of FASTQ files was assessed using FastQC V.0.10.165 and RSeQC V.2.6.4.66 Reads were aligned by HISAT2 V.2.0.67 Principal component analysis was used to identify outliers, and then differential expression was performed using R package edgeR.68 Genes with an adjusted p value<0.05 and absolute log2FC>1.5 were considered differentially expressed. CIBERSORTx was used for the immune cell analysis of the bulk RNA sequencing data, running with the following parameters: FPKM values of the gene expression, LM22 signature gene file, 1000 permutations, and quantile normalization disabled.44
Statistical analyses
Statistical analyses were performed using R V.4.1.2 (R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/). Continuous variables were compared using the two-sided Student’s t-test or Mann-Whitney U test. Categorical variables were compared using Pearson’s χ2 test or Fisher’s exact test. Spearman’s correlation analysis was performed to analyze the correlation among taxa, metabolites, KO genes, RNA expression, and immune cells.
Acknowledgments
The authors take this opportunity to thank all of the participating patients and healthy volunteers for supporting this study by donating the precious samples used in this research.
Footnotes
JL, YG and JL contributed equally.
Contributors: JLi, YG, JLiu and YM designed the experiments. YY, XL, JLiu and YM provided the clinical samples and performed the experiments. JLi and YG analyzed the data. JLi, YG, JL, FG and YM wrote the manuscript. YM is responsible for the overall content as guarantor. All authors edited the manuscript.
Funding: National Natural Science Foundation of China (No. 81920108026); Program of Shanghai Academic Research Leader (No. 20XD1421200 to YM); CSCO-Roche Tumor Research Fund (No. Y-2019Roche-079 to YM).
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Further information and requests for resources and reagents should be directed to the lead contact Yanlei Ma (yanleima@fudan.edu.cn).
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
Ethical approval was obtained from the Institutional Review Board of Fudan University Shanghai Cancer Center (ID: 050432-4-1911D). Participants gave informed consent to participate in the study before taking part.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–64. 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 2006;7:335–46. 10.1038/nrm1907 [DOI] [PubMed] [Google Scholar]
- 4.Lynch HT, Snyder CL, Shaw TG, et al. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer 2015;15:181–94. 10.1038/nrc3878 [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Bañobre J, Goel A. Dna mismatch repair deficiency and immune checkpoint inhibitors in gastrointestinal cancers. Gastroenterology 2019;156:890–903. 10.1053/j.gastro.2018.11.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloor M, von Knebel Doeberitz M. The immune biology of Microsatellite-unstable cancer. Trends Cancer 2016;2:121–33. 10.1016/j.trecan.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 7.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer—the stable evidence. Nat Rev Clin Oncol 2010;7:153–62. 10.1038/nrclinonc.2009.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch Repair-Deficient/Microsatellite Instability-High metastatic colorectal cancer. J Clin Oncol 2018;36:773–9. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 10.André T, Lonardi S, Wong KYM, et al. Nivolumab plus low-dose ipilimumab in previously treated patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: 4-year follow-up from CheckMate 142. Ann Oncol 2022;33:1052–60. 10.1016/j.annonc.2022.06.008 [DOI] [PubMed] [Google Scholar]
- 11.Chalabi M, Fanchi LF, Dijkstra KK, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med 2020;26:566–76. 10.1038/s41591-020-0805-8 [DOI] [PubMed] [Google Scholar]
- 12.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le DT, Uram JN, Wang H, et al. Pd-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohaumilitzky L, Kluck K, Hüneburg R, et al. The different immune profiles of normal Colonic mucosa in cancer-free Lynch syndrome carriers and Lynch syndrome colorectal cancer patients. Gastroenterology 2022;162:907–19. 10.1053/j.gastro.2021.11.029 [DOI] [PubMed] [Google Scholar]
- 15.Ballhausen A, Przybilla MJ, Jendrusch M, et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat Commun 2020;11:4740. 10.1038/s41467-020-18514-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremonesi E, Governa V, Garzon JFG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018;67:1984–94. 10.1136/gutjnl-2016-313498 [DOI] [PubMed] [Google Scholar]
- 17.Ma C, Han M, Heinrich B, et al. Gut Microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018;360:eaan5931. 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam KC, Araya RE, Huang A, et al. Microbiota triggers STING-type I IFN-dependent monocyte Reprogramming of the tumor Microenvironment. Cell 2021;184:5338–56. 10.1016/j.cell.2021.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsueh C-Y, Lau H-C, Huang Q, et al. Fusobacterium Nucleatum impairs DNA mismatch repair and stability in patients with squamous cell carcinoma of the head and neck. Cancer 2022;128:3170–84. 10.1002/cncr.34338 [DOI] [PubMed] [Google Scholar]
- 21.Allen-Vercoe E, Coburn B. A Microbiota-Derived metabolite augments cancer immunotherapy responses in mice. Cancer Cell 2020;38:452–3. 10.1016/j.ccell.2020.09.005 [DOI] [PubMed] [Google Scholar]
- 22.Yu T, Guo F, Yu Y, et al. Fusobacterium Nucleatum promotes Chemoresistance to colorectal cancer by Modulating Autophagy. Cell 2017;170:548–63. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KA, Thomas AM, Bolte LA, et al. Cross-Cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat Med 2022;28:535–44. 10.1038/s41591-022-01695-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 25.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Annals of Oncology 2017;28:1368–79. 10.1093/annonc/mdx108 [DOI] [PubMed] [Google Scholar]
- 26.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derosa L, Routy B, Thomas AM, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med 2022;28:315–24. 10.1038/s41591-021-01655-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCulloch JA, Davar D, Rodrigues RR, et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat Med 2022;28:545–56. 10.1038/s41591-022-01698-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371:595–602. 10.1126/science.abf3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long X, Wong CC, Tong L, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol 2019;4:2319–30. 10.1038/s41564-019-0541-3 [DOI] [PubMed] [Google Scholar]
- 32.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713–32. 10.1038/s41575-019-0189-8 [DOI] [PubMed] [Google Scholar]
- 33.Yang X, Guo Y, Chen C, et al. Interaction between intestinal microbiota and tumour immunity in the tumour microenvironment. Immunology 2021;164:476–93. 10.1111/imm.13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Bi D, Xie R, et al. Fusobacterium Nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct Target Ther 2021;6:398. 10.1038/s41392-021-00795-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostic AD, Chun E, Robertson L, et al. Fusobacterium Nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune Microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansaldo E, Slayden LC, Ching KL, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019;364:1179–84. 10.1126/science.aaw7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang G, Hong S, Yang P, et al. Discovery of an ene-reductase for initiating Flavone and Flavonol catabolism in gut bacteria. Nat Commun 2021;12:790. 10.1038/s41467-021-20974-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Certo M, Tsai C-H, Pucino V, et al. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol 2021;21:151–61. 10.1038/s41577-020-0406-2 [DOI] [PubMed] [Google Scholar]
- 39.O'Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016;16:553–65. 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumagai S, Koyama S, Itahashi K, et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell 2022;40:201–18. 10.1016/j.ccell.2022.01.001 [DOI] [PubMed] [Google Scholar]
- 41.Majima D, Mitsuhashi R, Fukuta T, et al. Biological functions of α-tocopheryl succinate. J Nutr Sci Vitaminol 2019;65:S104–8. 10.3177/jnsv.65.S104 [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Yuan X, Wang M, et al. Gut Microbiota influence Immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol 2022;15:47. 10.1186/s13045-022-01273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Sinicrope FA. Mismatch repair-deficient colorectal cancer: building on checkpoint blockade. J Clin Oncol 2022;40:2735–50. 10.1200/JCO.21.02691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cercek A, Lumish M, Sinopoli J, et al. Pd-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med 2022;386:2363–76. 10.1056/NEJMoa2201445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y, Zheng P, Gao L, et al. Effects of antibiotic use on outcomes in cancer patients treated using immune checkpoint inhibitors: a systematic review and meta-analysis. J Immunother 2021;44:76–85. 10.1097/CJI.0000000000000346 [DOI] [PubMed] [Google Scholar]
- 47.Elkrief A, Derosa L, Kroemer G, et al. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: a new independent prognostic factor? Annals of Oncology 2019;30:1572–9. 10.1093/annonc/mdz206 [DOI] [PubMed] [Google Scholar]
- 48.Routy B, Le Chatelier E, Derosa L, et al. Gut Microbiome influences efficacy of PD-1-based Immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 49.Cani PD, Depommier C, Derrien M, et al. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol 2022;19:625–37. 10.1038/s41575-022-00631-9 [DOI] [PubMed] [Google Scholar]
- 50.Bae M, Cassilly CD, Liu X, et al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature 2022;608:168–73. 10.1038/s41586-022-04985-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koh A, De Vadder F, Kovatcheva-Datchary P, et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 52.Ivashkiv LB. The hypoxia–lactate axis tempers inflammation. Nat Rev Immunol 2020;20:85–6. 10.1038/s41577-019-0259-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Angelin A, Gil-de-Gómez L, Dahiya S, et al. Foxp3 Reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab 2017;25:1282–93. 10.1016/j.cmet.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duscha A, Gisevius B, Hirschberg S, et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 2020;180:1067–80. 10.1016/j.cell.2020.02.035 [DOI] [PubMed] [Google Scholar]
- 55.Manzo T, Prentice BM, Anderson KG, et al. Accumulation of long-chain fatty acids in the tumor Microenvironment drives dysfunction in Intrapancreatic Cd8+ T cells. J Exp Med 2020;217:e20191920. 10.1084/jem.20191920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharya N, Yuan R, Prestwood TR, et al. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8 + T cell-mediated immunity in colorectal cancer. Immunity 2016;45:641–55. 10.1016/j.immuni.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grizotte-Lake M, Zhong G, Duncan K, et al. Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate interleukin-22 activity and prevent microbial dysbiosis. Immunity 2018;49:1103–15. 10.1016/j.immuni.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins CB, Aherne CM, Kominsky D, et al. Retinoic acid attenuates Ileitis by restoring the balance between T-helper 17 and T regulatory cells. Gastroenterology 2011;141:1821–31. 10.1053/j.gastro.2011.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devalaraja S, To TKJ, Folkert IW, et al. Tumor-derived retinoic acid regulates Intratumoral monocyte differentiation to promote immune suppression. Cell 2020;180:1098–114. 10.1016/j.cell.2020.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Ghaddar B, Biswas A, Harris C, et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 2022;40:1240–53. 10.1016/j.ccell.2022.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galeano Niño JL, Wu H, LaCourse KD, et al. Effect of the Intratumoral Microbiota on spatial and cellular heterogeneity in cancer. Nature 2022;611:810–7. 10.1038/s41586-022-05435-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kong C, Liang L, Liu G, et al. Integrated Metagenomic and Metabolomic analysis reveals distinct gut-Microbiome-derived phenotypes in early-onset colorectal cancer. Gut 2023;72:1129–42. 10.1136/gutjnl-2022-327156 [DOI] [PubMed] [Google Scholar]
- 64.Mallick H, Rahnavard A, McIver LJ, et al. Multivariable Association discovery in population-scale meta-Omics studies. PLOS Comput Biol 2021;17:e1009442. 10.1371/journal.pcbi.1009442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data, 2010. [Google Scholar]
- 66.Wang L, Wang S, Li W. RSeQC: quality control of RNA-Seq experiments. Bioinformatics 2012;28:2184–5. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 67.Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–15. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of Digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007420supp001.pdf (146.4KB, pdf)
jitc-2023-007420supp006.pdf (47.7KB, pdf)
jitc-2023-007420supp007.xlsx (22.2KB, xlsx)
jitc-2023-007420supp002.pdf (172.1KB, pdf)
jitc-2023-007420supp008.xlsx (18.1KB, xlsx)
jitc-2023-007420supp003.pdf (324.1KB, pdf)
jitc-2023-007420supp009.xlsx (26.3KB, xlsx)
jitc-2023-007420supp010.xlsx (41.9KB, xlsx)
jitc-2023-007420supp004.pdf (308.7KB, pdf)
jitc-2023-007420supp011.xlsx (209.6KB, xlsx)
jitc-2023-007420supp005.pdf (864.7KB, pdf)
Data Availability Statement
Data are available on reasonable request. Further information and requests for resources and reagents should be directed to the lead contact Yanlei Ma (yanleima@fudan.edu.cn).