Abstract
Xbp1 splicing and regulated IRE1-dependent RNA decay (RIDD) are two RNase activities of the ER stress sensor IRE1. While Xbp1 splicing has important roles in stress responses and animal physiology, the physiological role(s) of RIDD remain enigmatic. Genetic evidence in C. elegans connects XBP1-independent IRE1 activity to organismal stress adaptation, but whether this is via RIDD, and what are the targets is yet unknown. We show that cytosolic kinase/RNase domain of C. elegans IRE1 is indeed capable of RIDD in human cells, and that sensory neurons use RIDD to signal environmental stress, by degrading mRNA of TGFβ-like growth factor DAF-7. daf-7 was degraded in human cells by both human and worm IRE1 RNAse activity with same efficiency and specificity as Blos1, confirming daf-7 as RIDD substrate. Surprisingly, daf-7 degradation in vivo was triggered by concentrations of ER stressor tunicamycin too low for xbp-1 splicing. Decrease in DAF-7 normally signals food limitation and harsh environment, triggering adaptive changes to promote population survival. Because C. elegans is a bacteriovore, and tunicamycin, like other common ER stressors, is an antibiotic secreted by Streptomyces spp., we asked whether daf-7 degradation by RIDD could signal pending food deprivation. Indeed, pre-emptive tunicamycin exposure increased survival of C. elegans populations under food limiting/high temperature stress, and this protection was abrogated by overexpression of DAF-7. Thus, C. elegans uses stress-inducing metabolites in its environment as danger signals, and employs IRE1’s RIDD activity to modulate the neuroendocrine signaling for survival of upcoming environmental challenge.
Introduction
Serine/Threonine-Protein Kinase/Endoribonuclease IRE1 (inositol-requiring enzyme 1) is the most ancient and conserved sensor in the unfolded protein response (UPR) pathway. Its main function is to resolve proteostatic stress in the endoplasmic reticulum (ER), or, alternatively, trigger apoptosis when encountering prolonged and severe stress1. IRE1 is a single spanning type I trans-membrane protein2 with a stress-sensing lumenal domain and an unusual combined kinase and RNase domain located in cytosol3,4. Upon ER stress, IRE1 undergoes dimerization and oligomerization5,6, accompanied by a series of conformational changes that initiate from the lumenal domain and proceed to the active conformation of the RNase domain3,7–9. Activated IRE1 has two distinct RNase activities: 1) cleaving of mRNA of the X-box binding protein (XBP1) transcription factor10–12, resulting in removal of a small non-canonical intron and, upon ligation by the tRNA ligase RTCB13,14, production of an active transcription factor Xbp1s (known as ‘splicing activity’), and 2) cleaving a set of transcripts and microRNAs, resulting in their degradation by exonucleases (known as the regulated IRE1 dependent decay of RNA, or RIDD)15. The Xbp1 splicing activity of IRE1 is strongly conserved across evolution, with the fission yeast Schizosaccharomyces pombe being the only identified exception so far16. Importantly, in addition to orchestrating the transcriptional response to acute stress in the ER, the Xbp1 splicing activity functions in multiple aspects of metazoan physiology, such as differentiation of secretory cell types, or lipid metabolism17–21.
In contrast to the splicing activity, the functional importance of IRE1’s RIDD activity remains poorly understood. In cells, activation of RIDD by severe stress has been associated with either pro- or anti-apoptotic outcomes via degradation of specific RIDD targets. For example, RIDD activity increases caspase-2 synthesis by degrading micro RNAs22, but inhibits caspase-8 activation by promoting decay of DR-523. Many findings also indicate that RIDD helps maintain protein homeostasis in the ER under acute stress, but it does so by different means in different cell types and organisms. For example, in fission yeast S. pombe16 and in Drosophila S2 cells24, IRE1 RIDD activity degrades a broad range of mRNAs, decreasing the biosynthetic load in the ER. In contrast, in mammalian cells, this protection mechanism seems more restricted, since both the number of targets and the magnitude of downregulation are lower than in yeast and Drosophila cells24–26. Instead, RIDD protects these cells from ER stress by degrading individual targets. For example, degradation of the 28S rRNA by the human IRE1b decreases protein synthesis under ER stress in intestinal cells27; similarly, translational attenuation may result from RIDD of mRNA for a regulatory subunit of eukaryotic translation initiation factor 2α (eIF2α) phosphatase, by IRE1α28; third, degradation of the Blos1 transcript may assist the clearance of misfolded species and aggregates by reorganizing early endosomes and lysosomes29,30.
At organismal level, ectopic activation of RIDD, either by acute ER stress or by deleting XBP1 (which results in compensatory over-activation of IRE131), can also lead to both protective or detrimental outcomes32. In C. elegans, IRE-1-dependent degradation of flp-6 mRNA, coding for a neuropeptide secreted by the ASE sensory neuron, resulted in abnormal somatic differentiation of germ cells in animals genetically prone to forming germline tumors33. In mice, RIDD may contribute to neuroinflammation under hypoxia–ischemia stress by targeting miR-1734, which in turn activates NLRP3 inflammasome35. On the other hand, activation of RIDD in the liver protected mice from hepatotoxicity36,37.
The physiological in vivo functions of RIDD are even less understood, for three reasons. First, it has been challenging to identify in vivo RIDD targets. Unlike for Xbp1, there is no definitive mRNA structure of a RIDD target that specifies IRE1 recognition and cleavage. The Xbp1-like stem-loops have only been found in some RIDD substrates, including the most canonical substrate Blos125,38. The other RIDD-targeted transcripts have no Xbp1 like stem-loops or cleavage sites, yet are downregulated in stress-dependent manner by IRE122,39,40. Moreover, none of the 13 in vitro - identified RIDD targets that were used to define the consensus recognition/cleavage sequence showed ER stress-dependent decay in the same study41. Thus, identification of in vivo RIDD targets mainly relies on analyzing the transcriptomic changes in IRE1- and ER stress- dependent manner.
Second, it is difficult to uncouple RIDD from Xbp1 splicing mechanistically, or to experimentally activate RIDD without affecting Xbp1 splicing. Yet, RIDD is selectively and physiologically activated in specialized secretory cells that experience high biosynthetic flux, such as insulin-producing pancreatic beta cells, mucin-producing goblet cells, and chylomicrons-producing enterocytes, where it limits the ER overload by targeting the messages of high-flux proteins42–45. That RIDD is protective in these cells indicates that its selective physiological activation may serve as an adaptive strategy.
Finally, there are few identified examples to date where a physiological in vivo function of RIDD is not limited to diminishing the protein folding burden in the ER. In mice, RIDD controls muscle regeneration by degrading mRNA of a TGFβ-family protein myostatin46. During development of the Drosophila eye, RIDD activity is required to increase the synthesis of the rhodopsin, when rhabdomeres are formed47. In C. elegans neurons, IRE1 functions as a controller of morphological change - dendritic branching during development and formation of the stress-resistant dauer larva, in XBP1-independent fashion48–50. Although the latter is suggestive of RIDD, the worm IRE1 has never been tested for its ability to perform the RIDD activity, and the potential targets are unknown. The paucity of molecularly characterized examples makes it hard to determine if RIDD is a commonly used mechanism in animal development or physiology.
We have examined previously published C. elegans transcriptomic data51 and identified several potential RIDD targets, including the growth factor DAF-7, a well-known developmental regulator from the TGFβ superfamily, secreted mainly from the ASI chemosensory neurons52. Transcription of daf-7 is environmentally regulated, and is silenced when neurons sense unfavorable conditions - lack of food, high temperature, and high population density52,53. Interestingly, while secretion of DAF-7 in young larvae signals reproductive development, silencing of daf-7 expression signals dauer transition52,53, a process that is genetically linked to the IRE1’s RIDD activity48,49. Importantly, dauers are exceptionally stress resistant, and decrease in DAF-7 even outside of the dauer signaling leads to increased resistance to heat stress and starvation and extends adult life-span54–56, and increases resistance to pathogens57.
In this study, we first use human cells to confirm that a chimeric IRE1 with the C. elegans cytosolic kinase/RNase domain is capable of performing the RIDD activity, and that daf-7 mRNA is indeed a RIDD target. We then show in vivo that downregulation of daf-7 mRNA by RIDD occurs upon exposure to environmental toxin tunicamycin (Tm), under conditions insufficient for detectable activation of xbp-1 splicing. We find that downregulation of daf-7 by RIDD provides a preemptive protective mechanism against limited food/high temperature environment, and this protection can be reversed by DAF-7 overexpression. Because Tm, like several other ER stressors, is a secreted secondary metabolite of Streptomyces spp., which also secrete numerous antibiotics in order to compete with other bacteria, its presence in the environment may serve as a warning for upcoming depletion of bacteria that C. elegans feeds on. Our data suggest that C. elegans sensory neurons have ‘learned’ to utilize IRE1’s RIDD activity to signal an upcoming environmental challenge and improve the population survival.
Results:
The kinase and RNase domains of C. elegans IRE1 are functional in human cells
Because C. elegans IRE1 has not been demonstrated to have RIDD activity, we first sought to test whether C. elegans IRE1 can cleave a canonical RIDD target, such as the mammalian Blos1 transcript. We chose to use a human leukemia-derived, essentially haploid cell line HAP1 that provides robust and reliable results for both RNase activities58,59. A subclone of this cell line with the endogenous IRE1 knocked-out via CRISPR, IRE1KO, serves as a host for testing the activity of exogenous IRE1s58. The kinase and RNase domains of C. elegans IRE1 are well-conserved relative to their human counterparts, with 79% and 83% similarity, respectively, including the key phosphorylation residues in the active loop of the kinase (marked in red, Suppl. Fig. S1A), while the lumenal and transmembrane (TM) domains are more divergent. We therefore designed a chimeric protein where the entire cytosolic portion of the human IRE1 was replaced by C. elegans sequences (Fig. 1A), reasoning that such configuration would avoid the potential confounding effects of the worm lumenal and TM domains needing to sense misfolded proteins and interact with the mammalian ER membranes and chaperones. The chimeric protein should thus maintain the stress recognition module of the human IRE1, while enabling functional assessment of the worm’s catalytic domains. Finally, to allow visualization of the chimeric protein in live cells, we inserted a superfolder GFP (sfGFP) between the TM and cytosolic domains (Fig. 1A), mimicking the well-characterized and fully functional human IRE1-sfGFP or -mCherry proteins58,59.
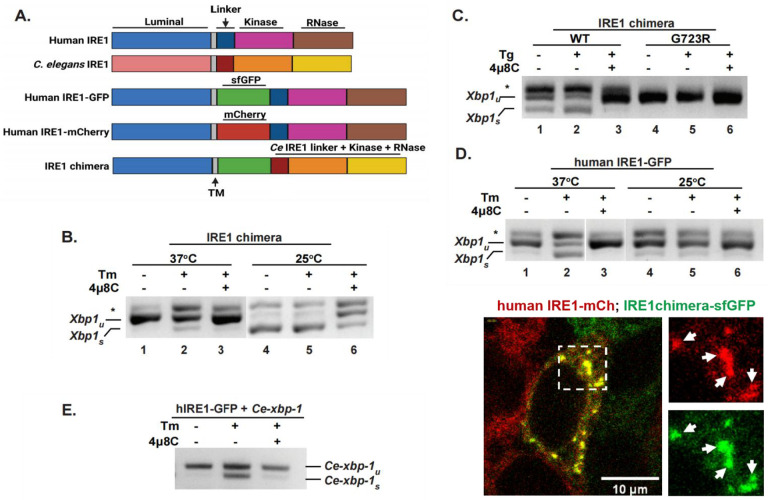
Figure 1. C. elegans cytosolic domains recognize human targets and are catalytically active in human cells.
(A) Schemes of domain organization in the human and C. elegans IRE1 proteins, and the constructs used in this work. The IRE1 chimera is composed of human lumenal and transmembrane (TM) domains (amino acid 1 to 494 of human IRE1α), a super-folder GFP (sfGFP), and C. elegans IRE1 cytosolic portion including the linker region and the kinase/RNase domain (amino acid 456 to 967 of C. elegans IRE1). (B, C) IRE1 chimera is active and kinase-dependent. RT-PCR assay was used to measure Xbp1 splicing in HAP1 IRE1KO cells reconstituted with an IRE1 chimera, either wildtype or G723R kinase-dead mutant. Cells were treated with 4μg/ml tunicamycin (Tm) or 2.5 μM thapsigargin (Tg) for 4hrs, either at 25°C or 37°C, as indicated. IRE1 inhibitor 4μ8C was used at 16 μM. Xbp-1u: unspliced Xbp1; Xbp1s: spliced Xbp1; *, non-specific PCR product, likely a hybrid of spliced and unspliced products. (D) RNase activity of human IRE1 is also temperature-dependent. HAP1 IRE1KO cells stably transfected with human IRE1-GFP protein were treated as in B. Human IRE1 is nearly inactive at 25°C. (E) Human IRE1 recognizes and cleaves C. elegans xbp-1. HAP1 IRE1KO cells stably expressing human IRE1-GFP protein were transfected with C. elegans full-length xbp-1 and treated as in C. (F) IRE1 chimera is able to oligomerize and form clusters with human IRE1 under strong ER stress. HAP1 IREKO cells expressing both the human IRE1-mCherry and IRE1 chimera (containing sfGFP) were treated with SubAB (0.1μg/ml) for 4hrs. Cells were imaged at 25°C, confocal projection of a z-stack is shown for a representative cell containing clusters. Arrows: examples of clusters.
The IRE1 chimera was introduced into HAP1 IRE1KO cells through lentivirus infection and stably expressed in these cells (Fig. S1B, C). To avoid any artifacts of overexpression, we selected cell lines where chimeric proteins were expressed at levels lower than those of the fully functional58,59 human IRE1-GFP protein (Fig. S1C).The functionality of the chimera was first verified by RNase activity towards human Xbp1 RNA (Fig. 1B, C). Treatment of HAP1 IRE1KO cells, expressing the IRE1 chimera, with the ER stressor tunicamycin (Tm) resulted in production of spliced Xbp1; the splicing was inhibited by the selective IRE1 inhibitor 4μ8C60, confirming it was due to the activity of the worm IRE1 domains (Fig. 1B, lanes 1–3). As the splicing activity of the chimeric protein was somewhat limited, we wondered whether worm catalytic domains were better adapted for lower temperatures, since C. elegans lives at the temperature range of 15°C to 25°C, lower than mammalian cells. Indeed, when HAP1 cells expressing chimeric IRE1 were treated with Tm at 25°C, a much stronger Xbp1 splicing activity was detected, with up to 80% spliced Xbp1 observed even without stress treatment (Fig. 1B, lanes 4–6). Importantly, this high splicing activity was still inhibited by 4μ8C. The splicing activity was also induced by a different ER stressor, thapsigargin (Tg)61 (Fig. 1C, lanes 1–3, Fig. S1D). Finally, to test whether the Xbp1 splicing activity of the chimera required auto-phosphorylation, like that of mammalian IRE1, the kinase-inactivating mutation (G723R)11 was introduced into the chimera. The cells with mutant chimera failed to show any splicing activity even under ER stress treatment (Fig 1B). We conclude that the IRE1 chimera is functional, able to recognize and cleave a mammalian substrate, and is activated by the proper progression of kinase-RNase activation.
We asked whether the temperature-dependence of Xbp1 splicing activity is a general property of any IRE1, by testing the activity of the human IRE1 at 25°C. We found that human IRE1-GFP essentially lost its activity and was no longer responsive to ER stress at 25°C (Fig. 1D, lanes 4–6), confirming that temperature-dependence is not a peculiar feature of the worm protein.
Finally, we asked whether cross-species recognition between IRE1 and Xbp1 is reciprocal. We found that the human IRE1 does efficiently cleave C. elegans xbp-1 cDNA under stress (Fig. 1E). These results indicate that human and worm IRE1s recognize and cleave their substrates in the same manner.
Oligomerization of the IRE1 chimera and temperature dependence of IRE1.
Activated yeast and mammalian IRE1 proteins exhibit a distinct manifestation of activation – clustering, reflecting their strong oligomerization58,62. Thus, we asked whether the chimeric IRE1 clustered in human cells, and whether it was able to co-cluster with the human protein. We found no cells with clustered IRE1 chimera under Tm stress at 25°C, when its RNAse is active (n=65). Nonetheless, IRE1 chimera could co-cluster with human IRE1-mCherry at 25°C under treatment with subAB (Fig. 1F), the most robust activator of ER stress58, indicating that the cytosolic domains of C. elegans IRE1 could adopt the conformation required to interact with human IRE1.
Interestingly, similar to the abolished splicing activity, the clustering activity of the human IRE1 protein was severely compromised at 25°C, with only ~7% of cells (10 out of 151 cells scored) exhibiting clusters, compared to ~85% (60 out of 71 cells) at 37°C. Strikingly, the human IRE1 clusters that were formed at 37°C dispersed within 15 mins upon shifting cells to 25°C (Fig. S2). These results suggest that IRE1 activity in both species is temperature-dependent and optimized to the growth temperature range of the host organism. Moreover, strong activation of the chimera splicing activity at 25°C without additional stressor (Fig. 1B) is consistent with a physiological UPR in worms at this temperature63,64, and may indicate that growth at this temperature is already sufficient to activate the enzymatic domains of C. elegans IRE1.
Importance of autologous linker sequence.
Considering the conservation of the enzymatic domains of the two proteins, we next wondered whether the presence of the worm linker sequences in the cytosolic portion of our chimera contributed to either the temperature sensitivity or the constitutive activity of the chimeric IRE1 protein. A linker region is located between the IRE1 transmembrane domain and kinase domain and is required for oligomerization and activation of IRE165. In yeast, the linker region is also required for the recruitment and docking to IRE1 of the Hac1 (yeast Xbp1 homologue) substrate66. The human linker is considerably longer (106 aa) than the C. elegans one (62 aa), with no significant similarities (Fig. S1A). Notably, the mammalian linker has three Gly/Ser-rich sequences that can impart greater polypeptide flexibility, but also more Pro residues compared to C. elegans linker.
Thus, we exchanged the worm linker region with the human one (Fig. S1E). Strikingly, the new chimeric protein with the human linker, h-linker-IRE1 chimera, completely failed to splice Xbp1 at either 25°C or 37°C, even under Tm stress (Fig. S1E). This indicates that an autologous linker region is required to activate the kinase and RNase domains. Thus, only the IRE1 chimera with C. elegans linker region was used in subsequent experiments.
C. elegans IRE1 can perform RIDD
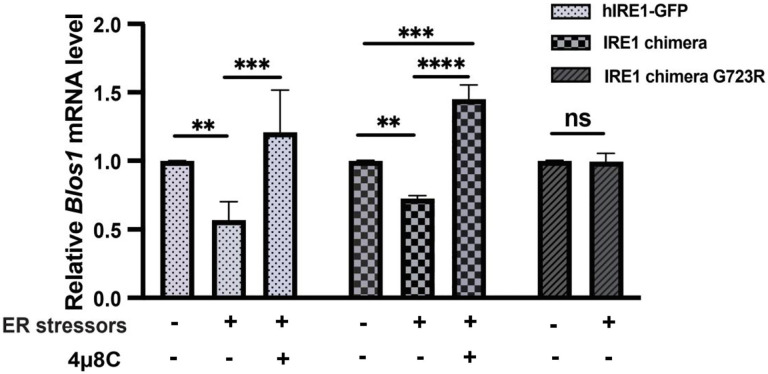
Having shown that the IRE1 chimera is functional and the cytosolic portion of C. elegans IRE1 can recognize and cleave human Xbp1, we tested if it can cleave a known RIDD substrate in HAP1 cells. mRNA of BLOC1S1 (Biogenesis of lysosome-related organelles complex 1 subunit 1) is one of the most conserved RIDD substrates, and its degradation via IRE1 RIDD activity has been shown in multiple cell lines25,38 upon ER stress. Therefore, we asked whether IRE1 chimera can downregulate Blos1 mRNA. At 25°C, Blos1 mRNA was consistently decreased by ~25% upon Tg treatment in chimera-expressing cells, relative to mock treated cells (Fig. 2). The decreased Blos1 reflected activation of the worm IRE1 catalytic RNase activity, since the IRE1 RNase inhibitor 4μ8C not only fully inhibited the downregulation of Blos1 triggered by Tg treatment, but also increased the Blos1 mRNA level by an additional ~50%. This “upregulation” is likely due to inhibition of the spontaneous RNase activity of the chimera that we observed previously in Xbp1 splicing assay (Fig. 1B and C). The kinase-dead mutant (G723R) chimera failed to downregulate Blos1 mRNA (Fig. 2), further confirming the dependence on IRE1 activity. Blos1 downregulation by ER stress was selective, since non-RIDD substrates in mammalian cells, such as Gapdh (Fig. S3) or mCherry (Fig. S4), were unaffected. We conclude that C. elegans IRE1 RNAse domain can recognize and cleave a canonical RIDD substrate.
Figure 2. C. elegans IRE1 can perform RIDD activity.
RT-qPCR was used to quantify mRNA levels of the canonical RIDD substrate Blos1 in HAP1 IRE1KO cells expressing indicated IRE1 proteins. Cells with hIRE1-GFP were treated with Tm at 37°C, cells with IRE1 chimeras were treated with Tg at 25°C. Blos1 mRNA level is normalized to the housekeeping gene Rpl19. 3 independent experiments, error bars: mean+/−SD, Anova with Tukey multiple comparison correction was used for hIRE1-GFP and IRE1 chimera, t-test was used for IRE1 chimera-G723R. Significance as described in Methods.
daf-7 is a RIDD substrate in human cells.
With the indication that worm IRE1 is RIDD-active, we initiated a search for its endogenous RIDD substrates, based on the following expected features: their transcripts should be downregulated under ER stress, and downregulation should require the presence of IRE1 but not XBP1, and be independent of the PERK and ATF-6 branches of the UPR. We examined the previously published microarray data in Shen et al51, using these criteria (see Methods for details). Out of 3538 transcripts with expression changes under ER stress, only 43 candidates met the above criteria (Table 1). As expected for most RIDD substrates32,67, 30 of the candidates were predicted to be either secretory or membrane proteins. Surprisingly, the recently identified RIDD target, flp-633, was not among the selected candidates, as it was not downregulated upon Tm treatment in this microarray (Table 1). However, based on WormBase modENCODE data68, flp-6 is mainly expressed in the early larval and dauer stages; since Shen et al51. used non-dauer larval stage 2 (L2) animals51, our approach may not be sensitive enough to detect downregulation of flp-6. Interestingly, two of the selected candidate genes (lect-2 and daf-7) are involved in dendrite arborization, which, based on the previous genetic data, may depend on RIDD48–50. The muscle-secreted lect-2 contributes to the patterning of all somatosensory dendrite arbors48,49,69,70, while daf-7 is linked to arborization of IL2 neurons specifically during transition into the stress-resistant dauer stage71. Because decrease in DAF-7 expression signals transition to the dauer stage, we focused on daf-7 as a candidate RIDD target.
Table 1:
List of candidate RIDD targets, adapted from Table 5 in Shen et al51.
| Gene | Fold change after Tm (log(2)) | ||||
|---|---|---|---|---|---|
| N2 | Ire1 KO | xbp-1 KO | pek-1 KO | atf-6 KO | |
| clec-265 | −0.46 | 0.82 | −0.52 | −0.98 | −0.71 |
| R05G6.9 | −0.54 | 0.6 | −0.64 | −0.82 | −0.24 |
| srlf-4 | −0.48 | 0.53 | −1.03 | −0.98 | −0.47 |
| nol-58 | −0.46 | 0.49 | −0.36 | −0.39 | −0.33 |
| Eppl-1 | −0.48 | 0.5 | −1.27 | −0.51 | −0.67 |
| mxl-3 | −0.64 | 0.5 | −0.6 | −0.4 | −0.34 |
| T03D8.7 | −0.42 | 0.31 | −0.5 | −0.66 | −0.33 |
| osm-8 | −0.57 | 0.41 | −0.43 | −0.62 | −0.36 |
| daf-7 | −0.53 | 0.31 | −0.62 | −0.62 | −0.2 |
| F42F12.3 | −0.52 | 0.29 | −0.43 | −0.33 | −0.76 |
| osr-1 | −0.56 | 0.28 | −0.76 | −0.9 | −0.36 |
| T07D3.9 | −0.56 | 0.25 | −0.57 | −0.27 | −0.94 |
| K08E7.5 | −0.81 | 0.35 | −0.45 | −0.62 | −0.19 |
| wrt-1 | −0.47 | 0.19 | −1.04 | −1.08 | −0.17 |
| F18C5.5 | −0.55 | 0.21 | −0.62 | −0.66 | −0.59 |
| F33H2.3 | −0.43 | 0.12 | −0.58 | −0.33 | −0.36 |
| ech-7 | −0.48 | 0.13 | −1.61 | −0.9 | −1.06 |
| lect-2 | −1.05 | 0.26 | −0.63 | −1.18 | −0.35 |
| C34C12.8 | −0.5 | 0.12 | −0.96 | −0.24 | −0.79 |
| irg-7 | −0.42 | 0.1 | −0.87 | −0.93 | −0.46 |
| F54D5.4 | −0.85 | 0.21 | −0.39 | −1.01 | −0.65 |
| mrp-5 | −0.55 | 0.13 | −0.77 | −0.8 | −1.15 |
| gcy-11 | −1.08 | 0.25 | −0.66 | −1.32 | −0.96 |
| clec-62 | −1.21 | 0.22 | −0.77 | −0.83 | −0.9 |
| zig-2 | −0.66 | 0.09 | −0.54 | −1.15 | −0.63 |
| tomm-40 | −0.5 | 0.07 | −0.91 | −0.36 | −0.64 |
| ccg-1 | −1.21 | 0.15 | −0.65 | −1.28 | −0.97 |
| ttr-1 | −0.47 | 0.06 | −0.56 | −0.53 | −0.58 |
| D2023.4 | −0.47 | 0.05 | −0.77 | −0.21 | −1.05 |
| F35D11.4 | −0.52 | 0.06 | −0.8 | −0.4 | −0.83 |
| asp-4 | −1.02 | 0.09 | −0.89 | −1.18 | −0.78 |
| F58H1.2 | −0.73 | 0.05 | −0.5 | −0.97 | −0.16 |
| Y41C4A.6 | −0.72 | 0.04 | −0.88 | −0.71 | −0.66 |
| F35E12.6 | −1.8 | 0.1 | −1.14 | −1.69 | −1.63 |
| clec-5 | −1.68 | 0.1 | −1.11 | −0.92 | −1.8 |
| R151.2 | −0.47 | 0.01 | −0.86 | −0.41 | −0.59 |
| F30H5.3 | −0.53 | 0.01 | −1.01 | −0.85 | −0.18 |
| nac-1 | −0.5 | 0.01 | −1.11 | −0.41 | −0.79 |
| scl-24 | −1.71 | −0.02 | −1.43 | −1.3 | −0.24 |
| drd-1 | −0.81 | −0.01 | −0.76 | −0.48 | −1.21 |
| capa-1 | −1.28 | −0.06 | −1.6 | −1.7 | −0.83 |
| F08D12.3 | −0.43 | −0.03 | −0.62 | −0.61 | −0.42 |
| T05B4.8 | −0.63 | −0.05 | −1.16 | −0.58 | −1.29 |
| flp-6 | 0.15 | 0.25 | 0.33 | 0.13 | 0.22 |
Transcripts are sorted by the largest difference in downregulation between wild type (N2) and IRE1-deficient animals (ΔN2-ΔIRE1KO)/ΔN2≥0.9, see Methods). Grey shading indicates transcripts coding for known secreted and membrane proteins, and those with predicted ER signal sequences. daf-7 is shaded in green; flp-6 (shaded blue) did not show downregulation in this microarray.
Since the above data on daf-7 regulation by ER stress is based on genetic deletions, we sought to first independently confirm that daf-7 mRNA can be regulated by the IRE1 RIDD activity. Thus, we compared it with the known RIDD substrate in human HAP1 cells, where the RIDD activity has been characterized26. Since we already showed that human IRE1 can correctly recognize and cleave the worm xbp-1 (Fig. 1E), we expected human IRE1 to also process worm RIDD substrates, if any. We thus introduced daf-7 mRNA into HAP1 cells, as mCherry fusion of DAF-7 protein (Fig. 3A) that we have previously demonstrated in C. elegans to be fully functional72.
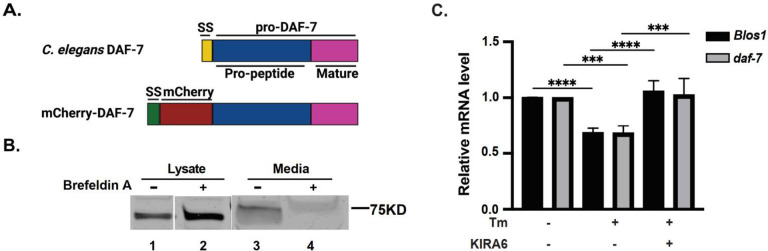
Figure 3. daf-7 is degraded by the human IRE1 as a RIDD substrate.
(A) Scheme of the domain composition of DAF-7 protein and the construct used in this work. C. elegans DAF-7 contains a signal peptide (SS) and pro-DAF-7 (aa. 23 to 350 of DAF-7), including latency-associate peptide (Pro-peptide) and mature DAF-7. The mCherry-DAF-7 construct contains human IGF2 signal peptide (aa. 1 to 24 of IGF2) in place of the original DAF-7 signal peptide, followed by mCherry and pro-DAF-7 (aa. 23 to 350 of DAF-7 protein), and a C-terminal V5/6-His tag. (B) mCherry-DAF-7 is secreted in HAP1 cells. mCherry-DAF-7 was stably expressed in HAP1 WT cells, cells were treated with Brefeldin A (2μg/ml) for 24hrs to block secretion, and analyzed by Western blot with anti-mCherry antibody. (C) daf-7 mRNA is degraded by human IRE1 under ER stress. RT-qPCR analysis of Blos1 and daf-7 mRNA levels in HAP1 cells expressing mCherry-DAF-7. Cells were treated with Tm (4μg/ml) and IRE1-specific inhibitor KIRA6 (1μg/ml) for 6hrs, 3 independent experiments, error bars: mean+/−SD, Anova with Tukey multiple comparison correction, significance as described in Methods.
To ensure that daf-7 mRNA can be recognized by translation elements and targeted to the ER, we replaced DAF-7 signal peptide with that of human IGFII. ER targeting was confirmed by secretion to the culture media, which was sensitive to Brefeldin A (Fig. 3B). We then treated HAP1 cells expressing mCherry::DAF-7 with Tm, and assayed daf-7 mRNA levels. daf-7 message was downregulated to the same extent as the canonical RIDD substrate Blos1 (31+/−2% and 32+/−3% respectively, n=4) (Fig. 3C). Furthermore, the downregulation of both daf-7 and Blos1 was suppressed by IRE1 inhibitors, KIRA6, a type II kinase/IRE1inhibitor73 (Fig. 3C), and 4μ8C. Importantly, the downregulation reflected cleavage of daf-7 sequences within the mCherry::DAF-7-coding transcript, since mCherry sequence was unaffected by either Tm or IRE1 inhibitors treatments (Fig. S4). Thus, daf-7 mRNA is degraded by the human IRE1 like a RIDD substrate.
As mentioned above22,25,32,38–40, RNAs degraded by RIDD activity may or may not have a Xbp1-like spatially conserved CNG↓CNGN IRE1 recognition sequence41 C. elegans xbp-1 message contains two such loops that are specifically cleaved by IRE1 during splicing11 (Fig. S5A). The RNA fold prediction algorithm74 identified a stem-loop in daf-7 mRNA with a potential IRE1 cleavage site (Fig. S5A, right) . The size of the loop is 9nts, larger than the typical 7nts. We asked whether this was the IRE1 recognition site, by mutating conserved residues on both sides of the predicted cleavage site. Incidentally, one of the mutations also eliminates a cysteine residue in the growth factor domain (C314S). Tm treatment still downregulated both mutated daf-7 mRNAs with similar efficiency to the Blos1 (Fig. S5B, C), suggesting either the existence of more than one cleavage sites, or of a different type of a cleavage site(s).
daf-7 is downregulated by IRE1 in vivo at correct developmental stages and under physiological stress levels.
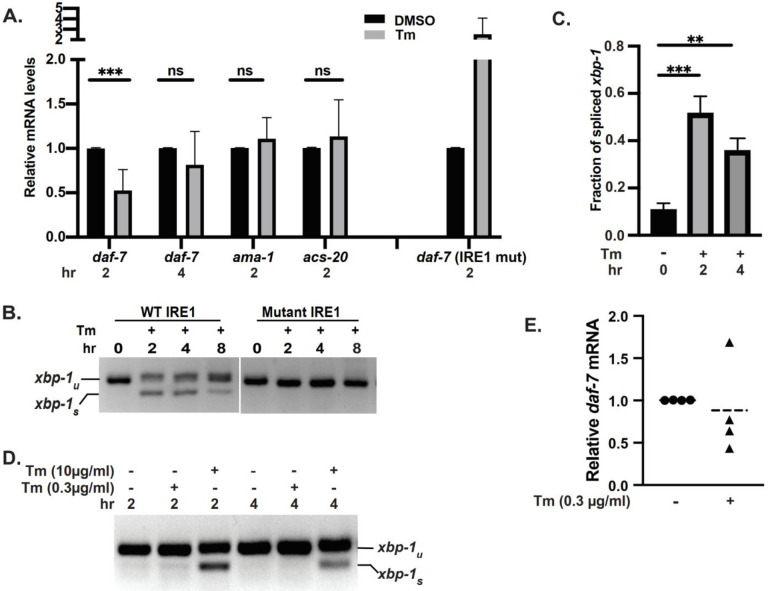
The data so far show that C. elegans IRE1 is capable of performing a RIDD activity (Fig. 2), that daf-7 message is downregulated by Tm stress in mixed populations of C. elegans in a manner consistent with it being a RIDD target51 (Table 1), and that daf-7 mRNA is degraded by a human IRE1 as a RIDD substrate (Fig. 3). To understand the possible physiological significance of daf-7 regulation by RIDD, we considered that decrease in DAF-7 secretion signals the presence of stressful environment specifically in late L1 (first larval stage)/early L2 animals. Thus, we tested whether activation of IRE1 can reduce daf-7 transcript levels at these stages. When synchronized late L1 populations were treated for 2 hours with Tm under conditions that activate robust IRE1-dependent xbp-1 splicing75, the transcript levels of daf-7 were strongly downregulated by 48% +/− 29% (Fig. 4A, n=6). This was not due to non-specific decrease in transcription, since two unrelated housekeeping genes, ama-1 and acs-20 (Fig. 4A), as well as the transcript of another neuro-secretory protein, ins-1 (Fig. S6A), were not affected. As expected for IRE1-dependent process75, the downregulation of daf-7 begun to attenuate at 4hrs Tm treatment (Fig. 4A), paralleling the xbp-1 splicing kinetics in the same animals (Fig. 4B, C). Importantly, the downregulation of daf-7 was dependent on IRE1 activity, as Tm treatment of larvae carrying enzymatically inactive mutant of IRE1 (G723R, zc14 allele)11 failed to downregulate daf-7 transcript, and in fact increased its levels (Fig. 4A). Conversely, the downregulation of daf-7 was still observed in larvae without functional XBP-1 (Fig. S6B). This indicates that daf-7 message is a selective RIDD target of activated IRE1 at the developmental stage when it functions to signal stressful environment.
Figure 4. daf-7 mRNA is a RIDD substrate in vivo.
(A) daf-7 mRNA is selectively downregulated by IRE1 in C. elegans under ER stress. RT-qPCR analysis of daf-7 mRNA levels in synchronized late L1 larvae, normalized to actin. Animals were treated with Tm (10μg/ml) for 2hrs or 4hrs. Mutant IRE1: SJ30(zc14) animals carrying kinase-dead IRE1(G723R). daf-7: 6 (2hrs) or 4 (4hrs) independent experiments; acs-20 and ama-1: 3 experiments. IRE1 mut: 4 experiments. Error bars: means +/− SD, t-test, significance as described in Methods. (B, C) Time-course of UPR induction. RT-PCR xbp-1 splicing assay in late L1 larvae treated with Tm (10μg/ml) for indicated number of hours. C: quantitation in animals with wild type IRE1. 3 independent experiments, error bars: mean+/−SD, Anova with Tukey multiple comparison correction. (D) Marginal activation of xbp-1 splicing at low Tm concentration. Late L1 larvae were treated with high (10μg/ml) or low (0.3μg/ml) Tm concentration for 2 or 4hrs. Representative RT-PCR from 3 (high conc.) and 4 (low conc.) experiments. (E) daf-7 mRNA is still downregulated at low Tm concentration. RT-qPCR analysis of daf-7 mRNA levels in late L1 larvae treated with 0.3μg/ml Tm for 4hrs. 4 independent experiments shown.
We next considered whether degradation of daf-7 message was only triggered under severe ER stress. We treated synchronized late L1 animals with 0.3μg/ml Tm, a 100-fold lower concentration than was used previously to induce ER stress in worms51. Treatment with 0.3μg/ml Tm resulted in xbp-1 splicing at only barely detectable levels at 2 hours, and no detectable splicing at 4 hours (Fig. 4D). Strikingly, even at this low Tm concentration, daf-7 mRNA was downregulated in 3 out of 4 independent experiments (Fig. 4E), to the levels comparable to those triggered by high Tm (Fig. 4A). Downregulation of daf-7 mRNA by the low concentration of Tm was selective, since non-RIDD target ins-1 was unaffected (Fig. S6A). Thus, RIDD of daf-7 is the preferred IRE1 activity under low ER stress.
Downregulation of daf-7 message by Tm is not sufficient to induce dauer transition
The best-defined phenotype expected from reducing DAF-7 levels is dauer larvae formation. Thus, we tested dauer induction by combining a standard treatment of crowding-induced starvation and higher temperature (25°C)76 with an added dose of Tm. In mock-treated (DMSO) wild type animals, dauer larvae started to appear within 48hrs after starvation and their number increased by 96 hrs (Fig. S7, left). Addition of Tm not only failed to increase the number of dauer larvae, but inhibited dauer formation, and this was true even at very low Tm concentration of 0.1μg/ml (Fig. S7). This suggests that downregulating daf-7 by RIDD does not alone induce dauer larvae. Since even at high concentration (10μg/ml), Tm only downregulated daf-7 mRNA by ~50% (Fig. 4A), RIDD activity may not reduce daf-7 levels enough to trigger dauer transition. Alternatively, Tm may prevent dauer transition by interfering with various required secretory events77,78.
Downregulation of daf-7 by RIDD allows for survival and growth of C. elegans populations in limited food/high temperature environments
While performing dauer experiments, we noticed that worm populations under higher temperature grew better when treated with low Tm concentrations compared to non-Tm controls. Thus, we considered environmental conditions under which the animals could encounter Tm. Although Tm is commonly used to experimentally activate the UPR in eukaryotic cells, it is a natural secondary metabolite antibiotic secreted by several Streptomyces spp.79. Moreover, a number of other compounds that are used to trigger stress in eukaryotic cells, including such common UPR inducers as lactacystin, ionomycin, and calcium ionophore, are in fact antimicrobials secreted by Streptomyces. The presence of Streptomyces may thus negatively impact worm populations not only by secreting nematocidal metabolites, but by depleting other bacteria that serves as food source for C. elegans. Moreover, reproduction of C. elegans, and thus population growth, is inhibited at temperatures that are appropriate for Streptomyces (27°C or higher)80,81 (Fig. 5C). Because deletion of daf-7 is known to increase larval survival of starvation and high temperature streses55, we asked if downregulation of daf-7 message by RIDD could be protective under conditions mimicking the presence of Streptomyces - a combination of 27°C and sparse food availability. Embryos were placed sparsely on Tm-containing plates and allowed to grow until plates were crowded with next generation larvae and food became limiting. Strikingly, inclusion of low concentrations of Tm (0.1 and 0.3μg/ml) rescued the population growth defect under these conditions, increasing the population size (Fig. 5A) and the number of laid embryos (Fig. S8A) by 2–3 folds within 72 hours. This rescue continued into the next generation of animals, despite complete depletion of food, since low concentrations of Tm still increased the population size more than two-fold after 96 hours (Fig. S8B).
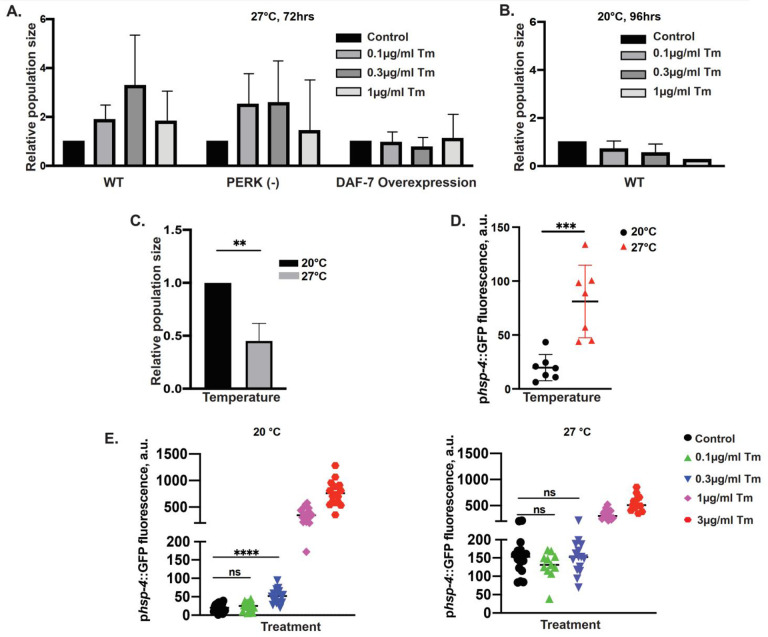
Figure 5. Pre-emptive downregulation of daf-7 by RIDD supports survival of nutritional/temperature stress.
(A) Pretreatment with low Tm concentrations increases population growth under stressful environment through downregulation of DAF-7. Population size assays: animals of indicated genotypes were exposed to Tm or DMSO (control) starting from embryo stage, under low population density (25 embryos per 3cm plate) and plentiful food. Animals were allowed to grow until plates were crowded and food nearly depleted (72hrs) at 27°C. PERK(−): pek-1(ok275); DAF-7 overexpression: animals transgenic for pdaf-7::mcherry::daf-7. 3 independent experiments, error bars: mean+/−SD. (B) Low Tm is not beneficial at normal growth temperature. Wild type embryos were plated as in A and allowed to grow until plates were crowded and food nearly depleted (96hrs) at 20°C. 3 independent experiments, error bars: mean+/−SD. (C) Elevated temperature (27°C) restricts population growth. Embryos were plated as in A and population sizes measured after 72hrs growth at 27°C or 96hrs at 20°C for. 3 independent experiments, error bars: mean+/−SD, t-test, significance as described in Methods. (D) Induction of UPR reporter by growth at elevated temperature (27°C). GFP fluorescence in late L1 animals carrying phsp4::GFP reporter, grown at 20°C or 27°C from embryo stage. error bars: mean+/−SD, t-test. (E) Elevated temperature (27°C) has a dominant effect on UPR reporter induction at low Tm concentrations. Animals were treated (as in D). Comparison of control (DMSO) and low concentration Tm (0.1μg/ml and 0.3μg/ml) treatments: Anova with Tukey multiple comparison correction.
Importantly, overexpression of DAF-7 completely abolished the rescue effect of Tm, indicating that decrease in daf-7 was required for the rescue (Figs. 6A and S9). Notably, the rescue was specific to the growth in these otherwise restrictive conditions, since even low concentrations of Tm were detrimental at the normal growth temperature of 20°C (Fig. 5B). Thus, Tm- and RIDD-mediated decrease in daf-7 can serve as an adaptation strategy to ensure population survival in stressful environments.
Hormesis or PERK activity does not account for the rescue of survival by low Tm.
Unfortunately, we were unable to directly test whether the rescue of population growth by Tm exposure requires IRE1 or XBP1, since animals deficient for either of these proteins arrest early in development at elevated temperatures (ref63 and our observations). Thus, we examined the possible alternative explanations. First, low Tm concentrations at elevated temperature could induce higher levels of general ER stress, in which case the rescue could be due to a hormesis-like effect of UPR82,83, rather than depending on decrease of DAF-7. We measured the fluorescence of the well-characterized phsp-4::GFP UPR reporter11 and found that the temperature alone was sufficient to increase the reporter fluorescence (Fig. 5D), in agreement with the chimeric IRE1 activation in human cells (Fig. 1B,C) and physiological UPR in worms64,84. However, since the temperature effect appeared to be dominant, and the rescuing Tm concentrations (0.1 and 0.3μg/ml) did not increase the reporter fluorescence compared to the DMSO control at 27°C (Fig. 5E), these data are inconsistent with hormesis effect.
Second, we tested the possibility that the related stress sensor, PERK, mediated the rescue by low Tm. Previously, induction of strong ER stress in the ASI neuron in combination with elevated temperature was shown to trigger dauer transition85, which may provide survival advantage under stress. The dauer induction was shown to be due to translational silencing of the ASI neuron via PERK-mediated eIF2α phosphorylation; since the ASI neuron is the main source of DAF-7 for dauer signaling, activation of PERK by Tm could be responsible for our rescue observation. However, when PERK-deficient worms (ok275) were subjected to the same regime as the as the wild type animals, low Tm dose still increased the size of the population at 27°C by more than 2-fold (Fig. 5A). Thus, rescue of population growth by low Tm treatment is independent of PERK. Combined with the inability of Tm to rescue animals that over-express DAF-7, these data argue that activation of RIDD activity of IRE1 in sensory neurons by environmental stress promotes survival of the worm population via degradation of daf-7 message.
Discussion:
The major conclusions from this study are that in addition to the xbp-1 splicing activity, C. elegans IRE1 has a RIDD activity that is similar to that of mammalian IRE1; that daf-7 is a physiological RIDD target; and that downregulation of daf-7 levels by the RIDD activity modulates neuroendocrine output of sensory neurons and thus allows for survival of worm populations under conditions mimicking the presence of Streptomyces spp. capable of killing the bacteria that worms feed on86. This protection is achieved not through the already known mechanisms such as translational silencing of neurons85, XBP-1-dependent neuronal signaling87,88, or hormesis83,89. Previous genetic data implicated IRE1 RIDD activity in morphological remodeling of neurons in context of dauer transition49. We propose that pre-emptive activation of RIDD in environmentally-exposed sensory neurons of C. elegans acts as signaling mechanism for protection against adverse environmental conditions, including anticipated nutrient depletion and temperature stress.
C. elegans IRE1 is capable of RIDD
Since IRE1 RIDD activity is not well-defined in C. elegans, and there is only one verified RIDD target, flp-633, we tested C. elegans IRE1 RIDD using a human/C. elegans chimera in mammalian cells. We show that C. elegans RNase domain can process the heterologous RIDD substrate Blos138, with the same efficiency and specificity as that of the human IRE1. Blos1 often serves as a diagnostic RIDD substrate since, unlike many other mammalian RIDD targets, it is universally degraded by the activated IRE1 in different cells. Similarly, both human and C. elegans IRE1 efficiently and specifically degraded the newly-identified worm RIDD target, daf-7. RIDD of daf-7 can operate in vivo during the time of developmental transition between the larval L1 to L2 stage, which is orchestrated by secretion of DAF-7 protein52, suggesting a physiological role for the worm RIDD activity. During this time, daf-7 exhibits a sharp peak of expression68, making its levels sensitive to tuning either by the well-characterized transcriptional regulation52,53, the poorly understood control of its release, or the newly discovered here regulatory modality - degradation of its mRNA by the RIDD activity.
A strong argument for the physiological relevance of the RIDD activity of C. elegans IRE1 is that it is activated in the environmentally-exposed sensory neurons under conditions of low tunicamycin exposure, prior to the measurable induction of xbp-1 splicing. This is important, because high doses of this toxin cause massive ER stress, which can force artificial enzyme activation. Moreover, tunicamycin inhibits glycosylation in eukaryotic cells, which can in itself interfere with normal signaling by various growth factors90. We observed a protective effect of pre-emptive exposure to 100–300 times lower amounts of tunicamycin that those typically used to induce ER stress. Intriguingly, both the stressful environment by itself and the low tunicamycin in the absence of stressful environment are inhibitory for the growth and survival of the population, while pre-exposure to tunicamycin decreases these inhibitory effects. Thus, the neurons may have ‘subverted’ this cellular homeostatic stress response as stress-signaling modality.
Environmental and structural requirements
In addition to the cross-recognition of each other’s RIDD substrates, the C. elegans IRE1 and associated machinery processed human Xbp1 and conversely, C. elegans xbp-1 was processed by the human machinery. This cross-species recognition has also been observed in vitro between mammalian and yeast91 and between human and C. elegans33 IRE1 and substrates. On the other hand, we observed a complete dependence of enzyme activity on the correspondence between the linker and kinase/RNase domains: the chimera was only functional when the linker was cognate to the catalytic domains, rather than to the rest of the molecule (TM and lumenal domains) or the host cell. The reason for this is unclear at present. The linker region could transfer the conformational change from the lumenal domain, or contain yet unknown functionally important regulatory sites. The different length or amino acid composition may impart distinct biophysical properties to the linkers, which may then affect the formation of a face-to-face kinase interface for dimerization and autophosphorylation65, or a back-to-back interface for oligomerization4.
IRE1 proteins from two species exhibited sharp temperature dependence - the chimera had more robust RNase activity at 25°C compared to 37°C, while the human IRE1 in the same experiment was active at 37°C and inactive at 25°C. Although these data are limited to two species, it is tempting to speculate that IRE1 activation is adapted to the animal’s environment, as is also suggested by the induction of worm’s canonical UPR gene, the inducible BiP/hsp-4, by high growth temperatures64,84. This IRE1 adaptation is different from the classical model of enzyme thermo-sensitivity, in which elevated temperatures increase the rate of reaction until denaturing temperature is reached. Indeed, C. elegans enzymatic domains have much lower optimal temperature, yet remain activatable at the high temperatures. It is more compatible with recently proposed three-state model, where an active and inactive (but activatable) states of an enzyme are in reversible equilibrium92; the sequence variation between species may determine the parameters of such equilibrium. The rapid switching between active and inactive forms that we observed in the temperature shift experiments support the idea of reversible equilibrium.
daf-7 is a physiological RIDD target.
After myostatin, DAF-7 is now a second member of the TGFβ family to be physiologically regulated by RIDD46. To identify candidates in C. elegans, we took advantage of previously performed transcriptional analysis of UPR in strains deficient for individual sensors/effectors51. The candidates conform to expectations of a limited number of strong RIDD substrates (43 out of 3538 transcripts in the array), and of enrichment for ER-targeted transcripts of secretory proteins (~70%). Since two of the strong candidates, daf-7 and lect-2, participate in the morphological remodeling of dendrites during transition into dauer, and we show that RIDD tunes daf-7’s signaling at developmental stage when the dauer decision is made, RIDD activity of IRE1 may have an unappreciated role in regulating the developmental and behavioural decisions at this crucial developmental point.
Downregulation of daf-7 mRNA by RIDD activity is partial, and thus it regulates DAF-7 signaling differently than a complete loss of function or gene deletion. Different degrees of DAF-7 loss lead to distinct phenotypes55,93. The worms without both daf-7 alleles enter only the pre-dauer L2d state if grown at 20°C with plentiful food, while they enter dauer at almost complete penetrance at 25°C52,72; heterozygous animals with one wildtype daf-7 allele show no dauer phenotype even at 25°C. Importantly, DAF-7 deletion mutants exhibit increased lifespan and increased resistance to heat stress; this lifespan extension is not observed in animals treated with daf-7 RNAi, which partially downregulate DAF-755. Therefore, physiological roles of DAF-7 signaling are protein dose-dependent. While the partial loss of DAF-7 due to RIDD is insufficient to initiate dauer transition, we find that it causes a novel, paradoxical phenotype, where treatment with a toxin results in improved animal survival under restrictive high temperature (27°C).
A physiological function of daf-7 RIDD: signaling impending stress
RIDD can promote survival of cells enduring ER stress, either by inhibiting apoptosis23, or by improving stress resistance29. Furthermore, RIDD can be protective even before initiation of UPR, by reducing levels of transcripts of high flux proteins to prevent ER overload. Such anticipatory response is important for differentiation of secretory cells, like the insulin producing or antibody-secreting cells94,95. We argue that RIDD of daf-7 fits into the protective role by preparing C. elegans for the impending temperature/nutritional stresses when encountering predatory bacteria, such as Strepomyces spp.86. These predatory bacteria threaten the food supply of the nematode by secreting antibiotics that kill other bacteria that worms feed on96. We note that secreted antibiotics have been suggested to act as signaling molecules in microbial communities97. As some of these antibiotics, like tunicamycin, lactacystine and ionomycin96,98, are ER stressors99–102, C. elegans can exploit them as warning signals for anticipated starvation.
The presence of such ‘warning signals’ appears to activate IRE1 RIDD and downregulate daf-7 mRNA even at low concentrations that, by themselves, are not sufficient to trigger a global ER stress response. We propose that this preferential activation of IRE1’s RIDD activity in chemosensory neurons that are exposed to the nematode environment serves a signaling function that coordinates protection from upcoming stress. A similar anticipatory protection has been described for neuronal sensing of temperature, which allows induction of the heat-shock response in non-neuronal tissues prior to protein misfolding103, as well as for olfactory detection of pathogenic bacteria, which triggers UPR104 and primes HSF-1105. Worm neurons can detect Streptomyces and trigger avoidance behaviour106; the RIDD activity in sensory neurons modulates their neuroendocrine signaling to coordinate pro-survival strategy.
Material and methods:
Cell culture and viral infection:
HAP1 IRE1α KO (HAP1 IRE1KO) is an engineered HAP1 cells (a near-haploid human cell line) with 8 base pairs deleted in exon 6 of endogenous IRE1α using CRISPR-Cas9 were purchased from Horizon (HZGHC000742c006). HAP1 and HAP1 IRE1KO cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) (Invitrogen) with additional 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA), 1% sodium pyruvate (Corning, Corning, NY, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific).
HAP1 IRE1KO cells were infected with a pTight Tet-On lentiviruses coding for either wild type or mutant IRE1 chimeras, or mCherry-DAF-7. pTight lentiviral vector with human IRE1-GFP was used as the backbone58. Infected HAP1 cells were selected with puromycin, and IRE1 clones were chosen based on the protein expression to avoid overexpression, since it can alone activate IRE1. Expression of transgenes was induced with 1μg/ml doxycycline (dox) (Biochemika) for 18 hours. Tunicamycin, 4μ8C, KIRA6 were from Calbiochem. Thapsigargin was from MP Biomedicals, and Brefeldin A were from MilliporeSigma.
Transfection:
HAP1 IRE1KO expressing human IRE1-GFP cells were transfected with pBud4 plasmid carrying full length (unspliced) C. elegans xbp-1. Transfections were done using Lipofectmine 2000 (Invitrogen), for 6hrs. Cells were incubated overnight, trypsinized, and split into in 6-well plates for pharmacological treatments.
Mutagenesis:
DAF-7-mCherry lentiviral plasmid was used as template to introduce G941C or C942T mutations (nucleotide numbers based on cDNA). Site-directed mutagenesis was performed with Pfu Ultra II Fusion HS polymerase (Agilent). Mutations were confirmed with Sanger sequencing.
Selection of candidate RIDD targets:
We used data from microarray analyses in Shen et al., 200551, which determined transcriptional changes upon Tm treatment in L2 animals of wild type strain and strains with individually deactivated IRE1, XBP1, PERK and ATF6. 43 candidate transcripts from Table 5 in Shen et al. (3538 transcripts total) were selected in the following way: we first chose genes that were downregulated by Tm treatment by at least 25% in the wild type (N2) animals. From these, we next selected transcripts whose downregulation depended on IRE1 ((ΔN2-ΔIRE1KO)/ΔN2≥0.9), resulting in 112 transcripts. Finally we selected from the remaining transcript those that showed no or low dependence on XBP1 (with threshold of 20% downregulation) and PERK or ATF6 (10 % downregulation). Removing duplicates and genes without functional annotation resulted in 43 candidate genes, 30 of which are either know or predicted to be coding for secretory proteins (Table 1).
C. elegans strains:
Nematodes were grown at 20°C on nematode growth medium (NGM) plates seeded with Escherichia. coli OP50108. Animals were synchronized as described for tunicamycin treatment below. Wild type C. elegans strain (Bristol N2) and the following strains: SJ4005(zcIs4[phsp-4::GFP]), SJ30(ire-1(zc14) II), zcIs4[phsp-4::GFP]), RB545(pek-1(ok275) X), SJ17(xbp-1(zc12) III) were obtained from the Caenorhabditis Genetic Center. DAF-7 overexpression strain drxEx20 [pdaf-7::mCherry::daf-7::unc-54 3’UTR] is described in ref72.
Activation of C. elegans IRE1 by tunicamycin treatment:
Care has to be taken throughout the described steps that animals are not crowded and have plentiful food, as even small amounts of nutritional or density stresses affect UPR induction. ~500 embryos were collected from well maintained (for at least three generations) C. elegans plates by scraping, to generate first roughly synchronized population. This population was allowed to grow on fresh 10cm plates for 96 hours, to reach the maximum egg-laying rate. Second roughly synchronized population was generated by washing off larvae and adults, collecting embryos by scraping, and transferring them to new NGM plates. After incubation at 20°C for 96hrs, adults were transferred to fresh NGM plates and allowed to lay eggs for 90 mins (~1000 eggs were laid). For tunicamycin treatments, embryos were allowed to hatch and grow for 28hrs. The hatched (fine synchronized) young larvae then were transferred to assay plates containing defined concentrations of drugs, as needed.
Tm-containing plates for C. elegans treatment:
To prepare Tm-containing plates, Tm stock (10mg/ml in DMSO) was first diluted in M9 buffer (1/200 of total medium for NGM plate) supplemented with 1μl of 3M NaOH to prevent precipitation. This solution of Tm was added to warm NGM-agar solution (65–70°C) to the desired concentration under rapid mixing.
RT-qPCR in human cells:
Total RNA was extracted with Trizol (Life Technologies) following manufacturers protocol. 200ng of extracted RNA was retro-transcribed to cDNA by using oligo (dT)12–18 (Invitrogen) and SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed using SYBR Green reagent (Applied Biosystems) and Applied Biosystems StepOne Plus thermocycler with annealing temperature of 60°C, primer sequences are provided below. Data were analyzed by normalizing to Rpl19 using the ΔΔ-Ct method.
RT-qPCR in C. elegans.
Total RNA was extracted from ~1000 synchronized late L1 animals using Trizol (Life Technologies) following manufacturers standard protocol. 100ng total RNA was converted to cDNA, and analyzed as described for RT-qPCR in human cells. Data were analyzed by normalizing to actin using the ΔΔCt method.
Western blots:
HAP1 cells were lysed using following lysis buffer: 50 mM Tris-HCl pH 8, 150 mM NaCl, 5 mM KCl, 5 mM MgCl2, 1% NP-40, 1X complete protease inhibitors (Roche). Protein concentration was determined by BCA assay (Thermo Fisher). Lysates were resolved on 7 or 10% gels by SDS-PAGE, and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked with milk, probed with primary and secondary antibodies, and scanned with Odyssey Infrared imager (Li-Cor). Primary antibodies used were: anti-N term IRE1 (GeneTex); anti-mCherry (Thermo Fisher Scientific); anti-α tubulin (Sigma). IRDye-conjugated secondary antibodies were from Li-Cor.
daf-7 RNA stem-loop structure prediction
Stem-loop structure was predicted using ViennaRNA Web Services (http://rna.tbi.univie.ac.at/) with minimum free energy (MFE) model. Short RNA sequences (around 30 base pairs) were used as input for the algorithms.
Population growth assay
25 embryos were seeded onto Tm-containing 3cm NGM plates with plentiful food. Embryos were incubated at 27°C or 20°C for 72 hours or 96 hours, the number of offspring larvae were counted in each condition by sampling, and normalized to the number of larvae in NGM plates without Tm.
Dauer induction assay
L4 larvae were transferred to peptone-free NGM plates (two per plate) with concentrated OP50 bacteria, and incubated at 25°C until plates were starved. Dauer animals were counted after 48hrs, 72hrs or 96hrs. Dauers were identified by the resistance to 1% SDS.
Imaging:
Confocal:
Transfected cells were imaged in μ-Dish 35mm culture plates (Ibidi) with Zeiss LSM700 microscope equipped with CO2 and temperature-controlled chamber, at Cell Imaging Center, Drexel University. Cells were imaged with 1.4NA 63x oil objective, and 12 bit confocal stacks were reconstructed in ImageJ as projections.
Stereo:
animals were immobilized with sodium azide on chilled unseeded NGM plates and imaged with Leica M205FA microscope and Hamamatsu Orca R2 camera, keeping magnification and intensity of fluorescent sources (Chroma PhotoFluor 2) constant within experiment. All animals within one experiment were imaged in one session. Fluorescence intensity was quantified with ImageJ.
Statistics:
Unpaired t-test analyses were performed using Prism software (GraphPad, USA). significance levels are indicated in the assays with at least three repeats.
Data were analyzed with t-test or one-way ANOVA with Tukey multiple comparison correction. All analyses were performed using Prism software (GraphPad, USA). α value in all experiemnts was 0.05, and significance levels as follows: ns: P > 0.05; *: P ≤ 0.05; **: P ≤ 0.01; ***: P ≤ 0.001; **** means P ≤ 0.0001.
List of primers used:
| pBud-C. elegans XBP-1 cloning | |
| xbp-1 | Forward: 5’- CTGAGGTACC ATGAGCAACTATCCAAAACG-3’ Reverse: 5’- GTACAGATCTTTAATTTTGGAAGAAATTTAGGTC-3’ |
| cloning IRE1 chimera with C. elegans linker or with human linker | |
| C. elegans IRE1 cytosolic domains | Forward: 5’- TCTGTACAAGCAATGTGGAAGACAATGGGATCAAC-3’ Reverse: 5’-GGATGAGTACCCGAGGTTAACAC-3’ |
| Forward 2: 5’-GTGTTAACCTCGGGTACTCATCC-3’ Reverse 2: 5’-ATACAGAATTCTTAATCGGTGTTCGGATTTGATTTC-3’ | |
| human IRE1 linker | Forward: 5’-TCTGTACAAGACTAGTCAGCTGC-3’ Reverse: 5’-GATCTAGAGGAAATTTTCCCAACTATCACC-3’ |
| C. elegans kinase domain | Forward: 5’-AGTCTAGATATAGTCCATCAGATATTCTTGGAAC-3’ |
| pTight-DAF-7-mCherry cloning | |
| human IGF2 signal peptide | Forward: 5’-TTAGCGGCCGCATGGGAATCCCAATGGGGAAGTCGATGC-3’ Reverse: 5’-TATAGGTACCAGCAATGCAGCACGAGGCGAAGG-3’ |
| mCherry | Forward: 5’-TCATGGTACCGTGAGCAAGGGCGAGGAGGATAAC −3’ Reverse: 5’-TACTAGATCTTCCTTGTACAGCTCGTCCATGCC-3’ |
| DAF-7 propeptide | Forward: 5’-CATGTGTACAAGAACTGCACCAACTCAGGTGTTTG-3’ Reverse: 5’-TACTAGATCTTCTGAGCAACCGCATTTCTTGGCG-3’ |
| Site-directed mutagenesis | |
| DAF-7 C942T | Forward: 5’- CTGAGATCGGATACTGTTGCCATCCAACCGAG −3’ Reverse: 5’- CTCGGTTGGATGGCAACAGTATCCGATCTCAG −3’ |
| DAF-7 G914C | Forward: 5’-CCTGAGATCGGATACTCCTGCCATCCA-3’ Reverse: 5’-TGGATGGCAGGAGTATCCGATCTCAGG-3’ |
| RT-PCR | |
| human Xbp1 | Forward: 5’-AAACAGAGTAGCAGCTCAGACTGC-3’ Reverse: 5’-TCCTTCTGGGTAGACCTCTGGGAG-3’ |
| C. elegans xbp | Forward: 5’-CGTCGTCTACGAAGAAGAAGTCGTC-3’ Reverse: 5’-GATGATAGTTAGATACATATCCACACTG-3’ |
| qPCR primers | |
| Rpl19 | Forward: 5’-AAAACAAGCGGATTCTCATGGA-3’ Reverse: 5’-TGCGTGCTTCCTTGGTCTTAG-3’ |
| Blos1 | Forward: 5’-CAAGGAGCTGCAGGAGAAGA-3’ Reverse: 5’-GCCTGGTTGAAGTTCTCCAC-3’ |
| Gapdh | Forward: 5’-CTGGGCTACACTGAGCAGG-3’ Reverse: 5’-AAGTGGTCGTTGAGGGCAATG-3’ |
| C.elegans actin | Forward: 5’-ATCACCGCTCTTGCCCCATC-3’ Reverse: 5’-GGCCGGACTCGTCGTATTCTTG-3’ |
| daf-7 | Forward: 5’-CGCTACAAAGGGATCCGAAC-3’ Reverse: 5’-CCCGTCGAGCATTGCCTTG-3’ |
| Forward: 5’-GATCTACGATCTGTGCAAAGC-3’ Reverse: 5’-CGAGTACTCGTTCGGCGGT-3’ |
|
| acs-20 | Forward: 5’-TCATAGAGTTGGAGCAAGACTTG-3’ Reverse: 5’-CTGAAGGAGCATCCAT GAT ACC-3’ |
| ins-1 | Forward: 5’-AACCGCTTTCAAACGTTCCG-3’ Reverse: 5’-CGTCCTGATTGCAGCAGAATG-3’ |
Supplementary Material
Acknowledgments:
This work was supported by the NIH grant AG063029 to TG and YA. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References:
- 1.Walter P. & Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086, doi: 10.1126/science.1209038 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Cox J. S., Shamu C. E. & Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206, doi: 10.1016/0092-8674(93)90648-a (1993). [DOI] [PubMed] [Google Scholar]
- 3.Tirasophon W., Welihinda A. A. & Kaufman R. J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12, 1812–1824, doi: 10.1101/gad.12.12.1812 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K. P. et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132, 89–100, doi: 10.1016/j.cell.2007.10.057 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamu C. E. & Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15, 3028–3039 (1996). [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P. & Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2, 326–332, doi: 10.1038/35014014 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M. & Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A 102, 18773–18784, doi: 10.1073/pnas.0509487102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karagoz G. E. et al. An unfolded protein-induced conformational switch activates mammalian IRE1. Elife 6, doi: 10.7554/eLife.30700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagoz G. E., Acosta-Alvear D. & Walter P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol 11, doi: 10.1101/cshperspect.a033886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox J. S. & Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404, doi: 10.1016/s0092-8674(00)81360-4 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Calfon M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96, doi: 10.1038/415092a (2002). [DOI] [PubMed] [Google Scholar]
- 12.Sidrauski C. & Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039, doi: 10.1016/s0092-8674(00)80369-4 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Jurkin J. et al. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 33, 2922–2936, doi: 10.15252/embj.201490332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y., Liang F. X. & Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell 55, 758–770, doi: 10.1016/j.molcel.2014.06.032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollien J. & Weissman J. S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107, doi: 10.1126/science.1129631 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Kimmig P. et al. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife 1, e00048, doi: 10.7554/eLife.00048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reimold A. M. et al. An essential role in liver development for transcription factor XBP-1. Genes Dev 14, 152–157 (2000). [PMC free article] [PubMed] [Google Scholar]
- 18.Todd D. J. et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med 206, 2151–2159, doi: 10.1084/jem.20090738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sriburi R., Jackowski S., Mori K. & Brewer J. W. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167, 35–41, doi: 10.1083/jcb.200406136 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipson K. L. et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab 4, 245–254 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13, 89–102, doi: 10.1038/nrm3270 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Upton J. P. et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338, 818–822, doi: 10.1126/science.1226191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang T. K. et al. Coordination between Two Branches of the Unfolded Protein Response Determines Apoptotic Cell Fate. Mol Cell 71, 629–636 e625, doi: 10.1016/j.molcel.2018.06.038 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Gaddam D., Stevens N. & Hollien J. Comparison of mRNA localization and regulation during endoplasmic reticulum stress in Drosophila cells. Mol Biol Cell 24, 14–20, doi: 10.1091/mbc.E12-06-0491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore K. & Hollien J. Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol Biol Cell 26, 2873–2884, doi: 10.1091/mbc.E15-02-0074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollien J. et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186, 323–331, doi: 10.1083/jcb.200903014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwawaki T. et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol 3, 158–164, doi: 10.1038/35055065 (2001). [DOI] [PubMed] [Google Scholar]
- 28.So J. S., Cho S., Min S. H., Kimball S. R. & Lee A. H. IRE1alpha-Dependent Decay of CReP/Ppp1r15b mRNA Increases Eukaryotic Initiation Factor 2alpha Phosphorylation and Suppresses Protein Synthesis. Mol Cell Biol 35, 2761–2770, doi: 10.1128/MCB.00215-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae D., Moore K. A., Mella J. M., Hayashi S. Y. & Hollien J. Degradation of Blos1 mRNA by IRE1 repositions lysosomes and protects cells from stress. J Cell Biol 218, 1118–1127, doi: 10.1083/jcb.201809027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae D. et al. Regulation of Blos1 by IRE1 prevents the accumulation of Huntingtin protein aggregates. Mol Biol Cell 33, ar125, doi: 10.1091/mbc.E22-07-0281 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee A. H., Scapa E. F., Cohen D. E. & Glimcher L. H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496, doi: 10.1126/science.1158042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurel M., Chevet E., Tavernier J. & Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci 39, 245–254, doi: 10.1016/j.tibs.2014.02.008 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Levi-Ferber M. et al. Neuronal regulated ire-1-dependent mRNA decay controls germline differentiation in Caenorhabditis elegans. Elife 10, doi: 10.7554/eLife.65644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D. et al. IRE1alpha inhibition decreased TXNIP/NLRP3 inflammasome activation through miR-17–5p after neonatal hypoxic-ischemic brain injury in rats. J Neuroinflammation 15, 32, doi: 10.1186/s12974-018-1077-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerner A. G. et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 16, 250–264, doi:S1550-4131(12)00284-7 [pii] 10.1016/j.cmet.2012.07.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hur K. Y. et al. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med 209, 307–318, doi: 10.1084/jem.20111298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So J. S. et al. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab 16, 487–499, doi: 10.1016/j.cmet.2012.09.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bright M. D., Itzhak D. N., Wardell C. P., Morgan G. J. & Davies F. E. Cleavage of BLOC1S1 mRNA by IRE1 Is Sequence Specific, Temporally Separate from XBP1 Splicing, and Dispensable for Cell Viability under Acute Endoplasmic Reticulum Stress. Mol Cell Biol 35, 2186–2202, doi: 10.1128/MCB.00013-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quwaider D. et al. RNA sequencing identifies novel regulated IRE1-dependent decay targets that affect multiple myeloma survival and proliferation. Exp Hematol Oncol 11, 18, doi: 10.1186/s40164-022-00271-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Thomas A. et al. Decoding non-canonical mRNA decay by the endoplasmic-reticulum stress sensor IRE1alpha. Nat Commun 12, 7310, doi: 10.1038/s41467-021-27597-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oikawa D., Tokuda M., Hosoda A. & Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res 38, 6265–6273, doi: 10.1093/nar/gkq452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipson K. L., Ghosh R. & Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS One 3, e1648, doi: 10.1371/journal.pone.0001648 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee A. H., Heidtman K., Hotamisligil G. S. & Glimcher L. H. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A 108, 8885–8890, doi: 10.1073/pnas.1105564108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuru A. et al. Negative feedback by IRE1beta optimizes mucin production in goblet cells. Proc Natl Acad Sci U S A 110, 2864–2869, doi: 10.1073/pnas.1212484110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iqbal J. et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab 7, 445–455, doi: 10.1016/j.cmet.2008.03.005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He S. et al. IRE1alpha regulates skeletal muscle regeneration through Myostatin mRNA decay. J Clin Invest 131, doi: 10.1172/JCI143737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coelho D. S. et al. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep 5, 791–801, doi: 10.1016/j.celrep.2013.09.046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei X. et al. The unfolded protein response is required for dendrite morphogenesis. Elife 4, e06963, doi: 10.7554/eLife.06963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Androwski R. J. et al. Mutually exclusive dendritic arbors in C. elegans neurons share a common architecture and convergent molecular cues. PLoS Genet 16, e1009029, doi: 10.1371/journal.pgen.1009029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salzberg Y. et al. Reduced Insulin/Insulin-Like Growth Factor Receptor Signaling Mitigates Defective Dendrite Morphogenesis in Mutants of the ER Stress Sensor IRE-1. PLoS Genet 13, e1006579, doi: 10.1371/journal.pgen.1006579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X., Ellis R. E., Sakaki K. & Kaufman R. J. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet 1, e37, doi: 10.1371/journal.pgen.0010037 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren P. et al. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389–1391, doi: 10.1126/science.274.5291.1389 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Schackwitz W. S., Inoue T. & Thomas J. H. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719–728, doi: 10.1016/s0896-6273(00)80203-2 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Munoz M. J. & Riddle D. L. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163, 171–180, doi: 10.1093/genetics/163.1.171 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw W. M., Luo S., Landis J., Ashraf J. & Murphy C. T. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol 17, 1635–1645, doi: 10.1016/j.cub.2007.08.058 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lithgow G. J., White T. M., Melov S. & Johnson T. E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A 92, 7540–7544, doi: 10.1073/pnas.92.16.7540 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiffer J. A. et al. Caenorhabditis elegans processes sensory information to choose between freeloading and self-defense strategies. Elife 9, doi: 10.7554/eLife.56186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ricci D. et al. Clustering of IRE1alpha depends on sensing ER stress but not on its RNase activity. FASEB J 33, 9811–9827, doi: 10.1096/fj.201801240RR (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricci D. et al. An interdomain helix in IRE1alpha mediates the conformational change required for the sensor’s activation. J Biol Chem 296, 100781, doi: 10.1016/j.jbc.2021.100781 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cross B. C. et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A 109, E869–878, doi: 10.1073/pnas.1115623109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lytton J., Westlin M. & Hanley M. R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J Biol Chem 266, 17067–17071 (1991). [PubMed] [Google Scholar]
- 62.Kimata Y. et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol 179, 75–86, doi: 10.1083/jcb.200704166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richardson C. E., Kinkel S. & Kim D. H. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet 7, e1002391, doi: 10.1371/journal.pgen.1002391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pispa J., Matilainen O. & Holmberg C. I. Tissue-specific effects of temperature on proteasome function. Cell Stress Chaperones 25, 563–572, doi: 10.1007/s12192-020-01107-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korennykh A. V. et al. The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693, doi: 10.1038/nature07661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Anken E. et al. Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1. Elife 3, e05031, doi: 10.7554/eLife.05031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guydosh N. R., Kimmig P., Walter P. & Green R. Regulated Ire1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. Elife 6, doi: 10.7554/eLife.29216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grove C. et al. Using WormBase: A Genome Biology Resource for Caenorhabditis elegans and Related Nematodes. Methods Mol Biol 1757, 399–470, doi: 10.1007/978-1-4939-7737-6_14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diaz-Balzac C. A. et al. Muscle- and Skin-Derived Cues Jointly Orchestrate Patterning of Somatosensory Dendrites. Curr Biol 26, 2397, doi: 10.1016/j.cub.2016.07.078 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Zou W. et al. A multi-protein receptor-ligand complex underlies combinatorial dendrite guidance choices in C. elegans. Elife 5, doi: 10.7554/eLife.18345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schroeder N. E. et al. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr Biol 23, 1527–1535, doi: 10.1016/j.cub.2013.06.058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klabonski L., Zha J., Senthilkumar L. & Gidalevitz T. A Bystander Mechanism Explains the Specific Phenotype of a Broadly Expressed Misfolded Protein. PLoS Genet 12, e1006450, doi: 10.1371/journal.pgen.1006450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghosh R. et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548, doi: 10.1016/j.cell.2014.07.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kerpedjiev P., Hammer S. & Hofacker I. L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379, doi: 10.1093/bioinformatics/btv372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsialikas J. & Argon Y. xbp-1 mRNA splicing is attenuated under prolonged exposure to ER stress. MicroPubl Biol 2017, doi: 10.17912/W2707X (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Golden J. W. & Riddle D. L. A pheromone-induced developmental switch in Caenorhabditis elegans: Temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc Natl Acad Sci U S A 81, 819–823, doi: 10.1073/pnas.81.3.819 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flatt K. M. et al. Epidermal Remodeling in Caenorhabditis elegans Dauers Requires the Nidogen Domain Protein DEX-1. Genetics 211, 169–183, doi: 10.1534/genetics.118.301557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zha J., Ying M., Alexander-Floyd J. & Gidalevitz T. HSP-4/BiP expression in secretory cells is regulated by a developmental program and not by the unfolded protein response. PLoS Biol 17, e3000196, doi: 10.1371/journal.pbio.3000196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takatsuki A., Arima K. & Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 24, 215–223, doi: 10.7164/antibiotics.24.215 (1971). [DOI] [PubMed] [Google Scholar]
- 80.Shepherd M. D., Kharel M. K., Bosserman M. A. & Rohr J. Laboratory maintenance of Streptomyces species. Curr Protoc Microbiol Chapter 10, Unit 10E 11, doi: 10.1002/9780471729259.mc10e01s18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McMullen P. D. et al. Macro-level modeling of the response of C. elegans reproduction to chronic heat stress. PLoS Comput Biol 8, e1002338, doi: 10.1371/journal.pcbi.1002338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cypser J. R. & Johnson T. E. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci 57, B109–114, doi: 10.1093/gerona/57.3.b109 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Matai L. et al. Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc Natl Acad Sci U S A 116, 17383–17392, doi: 10.1073/pnas.1900055116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vakkayil K. L. & Hoppe T. Temperature-Dependent Regulation of Proteostasis and Longevity. Front Aging 3, 853588, doi: 10.3389/fragi.2022.853588 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kulalert W. & Kim D. H. The unfolded protein response in a pair of sensory neurons promotes entry of C. elegans into dauer diapause. Curr Biol 23, 2540–2545, doi: 10.1016/j.cub.2013.10.058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumbhar C., Mudliar P., Bhatia L., Kshirsagar A. & Watve M. Widespread predatory abilities in the genus Streptomyces. Arch Microbiol 196, 235–248, doi: 10.1007/s00203-014-0961-7 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Taylor R. C. & Dillin A. XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell 153, 1435–1447, doi: 10.1016/j.cell.2013.05.042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Imanikia S., Sheng M., Castro C., Griffin J. L. & Taylor R. C. XBP-1 Remodels Lipid Metabolism to Extend Longevity. Cell Rep 28, 581–589 e584, doi: 10.1016/j.celrep.2019.06.057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eisermann D. J., Wenzel U. & Fitzenberger E. PEK-1 is crucial for hormesis induced by inhibition of the IRE-1/XBP-1 pathway in the Caenorhabditis elegans mev-1 mutant. Biochem Biophys Res Commun 473, 1052–1057, doi: 10.1016/j.bbrc.2016.04.014 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Dricu A., Carlberg M., Wang M. & Larsson O. Inhibition of N-linked glycosylation using tunicamycin causes cell death in malignant cells: role of down-regulation of the insulin-like growth factor 1 receptor in induction of apoptosis. Cancer Res 57, 543–548 (1997). [PubMed] [Google Scholar]
- 91.Wiseman R. L. et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell 38, 291–304, doi: 10.1016/j.molcel.2010.04.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peterson M. E., Daniel R. M., Danson M. J. & Eisenthal R. The dependence of enzyme activity on temperature: determination and validation of parameters. Biochem J 402, 331–337, doi: 10.1042/BJ20061143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crook M. & Grant W. N. Dominant negative mutations of Caenorhabditis elegans daf-7 confer a novel developmental phenotype. Dev Dyn 242, 654–664, doi: 10.1002/dvdy.23963 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Edghill E. L. et al. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes 57, 1034–1042, doi: 10.2337/db07-1405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eletto D., Eletto D., Boyle S. & Argon Y. PDIA6 regulates insulin secretion by selectively inhibiting the RIDD activity of IRE1. FASEB J 30, 653–665, doi: 10.1096/fj.15-275883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Procopio R. E., Silva I. R., Martins M. K., Azevedo J. L. & Araujo J. M. Antibiotics produced by Streptomyces. Braz J Infect Dis 16, 466–471, doi: 10.1016/j.bjid.2012.08.014 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Davies J., Spiegelman G. B. & Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9, 445–453, doi: 10.1016/j.mib.2006.08.006 (2006). [DOI] [PubMed] [Google Scholar]
- 98.Liu W. C. et al. Ionomycin, a new polyether antibiotic. J Antibiot (Tokyo) 31, 815–819, doi: 10.7164/antibiotics.31.815 (1978). [DOI] [PubMed] [Google Scholar]
- 99.Xu Y. et al. Proteasome inhibitor lactacystin enhances cisplatin cytotoxicity by increasing endoplasmic reticulum stress-associated apoptosis in HeLa cells. Mol Med Rep 11, 189–195, doi: 10.3892/mmr.2014.2683 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Werno C., Zhou J. & Brune B. A23187, ionomycin and thapsigargin upregulate mRNA of HIF-1alpha via endoplasmic reticulum stress rather than a rise in intracellular calcium. J Cell Physiol 215, 708–714, doi: 10.1002/jcp.21351 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Wang X. Z. et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J 17, 5708–5717, doi: 10.1093/emboj/17.19.5708 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kitiphongspattana K., Mathews C. E., Leiter E. H. & Gaskins H. R. Proteasome inhibition alters glucose-stimulated (pro)insulin secretion and turnover in pancreatic beta-cells. J Biol Chem 280, 15727–15734, doi: 10.1074/jbc.M410876200 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Prahlad V., Cornelius T. & Morimoto R. I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814, doi:320/5877/811 [pii] 10.1126/science.1156093 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De-Souza E. A., Thompson M. A. & Taylor R. C. Olfactory chemosensation extends lifespan through TGF-beta signaling and UPR activation. Nat Aging, doi: 10.1038/s43587-023-00467-1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ooi F. K. & Prahlad V. Olfactory experience primes the heat shock transcription factor HSF-1 to enhance the expression of molecular chaperones in C. elegans. Sci Signal 10, doi: 10.1126/scisignal.aan4893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tran A. et al. C. elegans avoids toxin-producing Streptomyces using a seven transmembrane domain chemosensory receptor. Elife 6, doi: 10.7554/eLife.23770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sievers F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539, doi: 10.1038/msb.2011.75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94, doi: 10.1093/genetics/77.1.71 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.