Abstract
Clonal organisms are particularly useful to investigate the contribution of epigenetics to phenotypic plasticity, because confounding effects of genetic variation are negligible. In the last decade, the apomictic parthenogenetic marbled crayfish, Procambarus virginalis, has been developed as a model to investigate the relationships between phenotypic plasticity and genetic and epigenetic diversity in detail. This crayfish originated about 30 years ago by autotriploidy from a single slough crayfish Procambarus fallax. As the result of human releases and active spreading, marbled crayfish has established numerous populations in very diverse habitats in 22 countries from the tropics to cold temperate regions. Studies in the laboratory and field revealed considerable plasticity in coloration, spination, morphometric parameters, growth, food preference, population structure, trophic position, and niche width. Illumina and PacBio whole-genome sequencing of marbled crayfish from representatives of 19 populations in Europe and Madagascar demonstrated extremely low genetic diversity within and among populations, indicating that the observed phenotypic diversity and ability to live in strikingly different environments are not due to adaptation by selection on genetic variation. In contrast, considerable differences were found between populations in the DNA methylation patterns of hundreds of genes, suggesting that the environmentally induced phenotypic plasticity is mediated by epigenetic mechanisms and corresponding changes in gene expression. Specific DNA methylation fingerprints persisted in local populations over successive years indicating the existence of epigenetic ecotypes, but there is presently no information as to whether these epigenetic signatures are transgenerationally inherited or established anew in each generation and whether the recorded phenotypic plasticity is adaptive or nonadaptive.
Keywords: DNA methylation, environmental adaptation, epigenetics, epigenetic ecotypes, marbled crayfish, phenotypic plasticity, whole-genome sequencing
All animal and plant populations are phenotypically diverse, some more, some less. In sexually reproducing species, this phenomenon is explained as the result of genetic variation plus phenotypic plasticity. Genetic variation of a trait comes from the presence of multiple genotypes (different DNA sequences) in populations that encode different phenotypes. The phenotype includes all characteristics of an organism other than its genes. Phenotypic plasticity is the property of organisms to produce different phenotypes from the same genome in response to different environmental cues or inputs (Schlichting 1986; West-Eberhard 1989; DeWitt and Scheiner 2004; Fusco and Minelli 2010; Sommer 2020; Pfennig 2021; Vogt 2022a). The plastic response may involve changes in morphology, physiology, life-history traits, behavior, trophic position, and niche width, and may be adaptive or nonadaptive (Ghalambor et al. 2007). Phenotypic plasticity always involves a change in gene expression or gene-product use.
Interestingly, asexually reproducing animals and plants often inhabit diverse habitats and geographical regions as well (Massicotte and Angers 2012; Thorson et al. 2017; Shi et al. 2019), but in this case, genetic variation is often very small and cannot contribute much to phenotypic diversity. Therefore, such clonal organisms are particularly suitable to investigate the contribution of environmentally induced epigenetic changes to phenotypic diversity and to uncover its molecular underpinning. Epigenetic mechanisms such as DNA methylation, histone modifications, and noncoding RNAs are prime candidates for mediating phenotypic plasticity, because they can produce different phenotypes from the same genome by changing gene expression, either stochastically or in response to environmental cues (Gibney and Nolan 2010; Feinberg and Irizarry 2010; Duncan et al. 2014; Verhoeven and Preite 2014; Richards et al. 2017; Vogt 2017, 2020a; Johannes and Schmitz 2019; Angers et al. 2020; Biwer et al. 2020).
The marbled crayfish Procambarus virginalis, which is at the center of this paper, appeared on the scene in 1995 (Scholtz et al. 2003). Its particular suitability for research on the relationships of phenotypic plasticity, environmental adaptation, and epigenetics was first propagated in 2008 because of obligatory parthenogenetic reproduction, possession of a broad spectrum of phenotypic traits easy to analyze, genetic uniformity, intense methylation of the genome, and ease of rearing and handling in the laboratory (Vogt 2008; Vogt et al. 2008). In 2013, we have launched a project at the German Cancer Research Center (DKFZ, Heidelberg) to sequence the marbled crayfish genome and establish a genome-wide methylome at single base resolution, which are important requirements to investigate the relationships between phenotypic plasticity and genetic and epigenetic diversity in depth (Vogt et al. 2015; Gatzmann et al. 2018; Gutekunst et al. 2018). Since then, many papers have been published on genetics, epigenetic, phenotypic diversity, and environmental acclimatization of the marbled crayfish (e.g., Martin et al. 2007, 2010a, 2016; Vogt 2008, 2018, 2020a, 2020b, 2022b, 2022c; Vogt et al. 2008, 2015, 2018, 2019; Chucholl et al. 2012; Gatzmann et al. 2018; Gutekunst et al. 2018; Lipták et al. 2019; Andriantsoa et al. 2019; Linzmaier et al. 2020; Legrand et al. 2021; Maiakovska et al. 2021; Tönges et al. 2021a, 2021b; Veselý et al. 2021).
In the last 20 years, marbled crayfish has established numerous populations in very diverse habitats in 22 countries from the tropics to cold temperate regions as the result of human releases and active spreading (reviewed in Chucholl et al. 2012; Vogt 2020b; and Maiakovska et al. 2021). In the following, I will use the term “acclimatization” instead of “adaptation” to refer to this phenomenon to avoid confusion with the restricted use of the latter term in evolutionary biology. In biological literature, the word adaptation is often used in a more general meaning, for example, short-term adaptation of humans to higher temperature in summer, but in evolutionary biology, it is restricted to the alteration of phenotypic traits in response to the environment to increase survival and transmission of the genes, requiring genetic variation, fitness enhancement, selection, and heredity.
This article summarizes the papers on genetics, epigenetics, phenotypic variation, and environmental acclimatization of marbled crayfish and examines the associations between the 4 parameters. The main goal was to test if epigenetic mechanisms could serve as potential molecular mediators of environmentally induced phenotypic plasticity. Based on the marbled crayfish case study, a scheme is proposed on how environmental cues, epigenetic mechanisms, and genes could interact to change gene expression and phenotypic traits.
Epigenetic Mechanisms and Their Responsiveness to Environmental Cues
The best investigated epigenetic mechanisms are DNA methylation, histone modifications, and noncoding RNAs, which usually act together (Jaenisch and Bird 2003; Lennartsson and Ekwall 2009; Du et al. 2015; Moutinho and Esteller 2017). DNA methylation occurs in the vast majority of animals but has been evolutionarily lost in some species and groups (Raddatz et al. 2013; Provataris et al. 2018; Vogt 2022b). The methylation marks are mostly at the cytosines of CpG dinucleotides and occur in intergenic and genic regions including promoters, gene bodies, and repeats (Jaenisch and Bird 2003; Jones 2012; Schübeler 2015). Hypermethylation of promoters and repeats usually results in transcriptional repression, whereas gene body methylation modulates gene expression and seems to reduce transcriptional noise (Schübeler 2015; Neri et al. 2017; Gatzmann et al. 2018). The DNA methylation marks are established by DNA methyltransferases (DNMTs) and erased by ten-eleven-translocation enzymes (TETs) (Wu and Zhang 2017; Lyko 2018).
The histones of the nucleosomes greatly influence DNA transcription by either shielding the DNA or allowing the binding of transcription factors to the DNA. The N-terminal tails of the histones carry post-translational modifications like methylation, acetylation, phosphorylation, and ubiquitination, which help regulate the chromatin structure. Histone acetylation often stimulates gene expression, whereas histone methylation often represses gene expression depending on their location (Bannister and Kouzarides 2011; Allis and Jenuwein 2016). The chemical modifications on the histones are dynamically regulated by numerous enzymes (Marmorstein and Zhou 2014; Morgan and Shilatifard 2020). For example, histone acetylation marks are written by histone acetyltransferases (HATs) and read by bromodomain-containing proteins (BrDs).
Small to long noncoding RNAs (ncRNAs) regulate gene expression at the transcriptional and post-transcriptional levels contributing further to the production of phenotypic variation (Frias-Laserre and Villagra 2017; Long et al. 2017). For example, microRNAs inhibit translation or cause mRNA degradation (Moutinho and Esteller 2017; O’Brien et al. 2018). Small interfering RNAs can regulate gene transcription through transposable element silencing and the interaction with DNA methylation and histone modifications (Holoch and Moazed 2015). Long ncRNAs are involved in transcriptional regulation, dosage compensation, and genomic imprinting (Li et al. 2019).
Further potential epigenetic contributors to phenotypic plasticity are Polycomb group (PcG) and Trithorax group (TrxG) proteins that are involved in the mitotic and meiotic inheritance of epigenetically mediated phenotypes by sustaining silent and active gene expression states, respectively (Steffen and Ringrose 2014). This epigenetic memory maintains gene expression states in the absence of the initiating signals and without a change in DNA sequence (Ciabrelli et al. 2017).
There is multiple evidence from animals, plants, and microorganisms that environmental cues can change gene-regulating epigenetic marks and gene expression. Stronger and longer lasting environmental signals such as temperature, drought, light, oxygen level, food, predator odors, and toxicants seem to be particularly effective (Skinner 2014; Kim et al. 2015; Xue and Acar 2018; Liu and He 2020).
The Study System: Biology, Ecology, and Geographic Distribution of the Marbled Crayfish
The parthenogenetic all-female marbled crayfish Procambarus virginalisLyko, 2017 (Figure 1A, B), was detected in 1995 in the German aquarium trade (Scholtz et al. 2003; Vogt 2020b). Morphological and genetic studies revealed that it is an autotriploid descendant of a single female of slough crayfish, Procambarus fallax Hagen, 1870, which is native to peninsular Florida and southern Georgia, USA (Martin et al. 2007, 2010a, 2016; Vogt et al. 2008, 2015, 2018; Gutekunst et al. 2018, 2021). Searching in museum collections of Floridian crayfish revealed no evidence of marbled crayfish, suggesting that it is a phylogenetically very young species (Vogt 2019a). Time-resolved analysis of its genome evolution dated its origin to a time window between 1947 and 1996 (Legrand et al. 2021). Marbled crayfish was described as a separate species (Lyko 2017) because it represents a distinct reproductive unit and is characterized by significantly enhanced fitness traits (body size, fecundity, and longevity) compared with its parent species (Vogt et al. 2015, 2018, 2019).
Figure 1.
Phenotypic variation of monoclonal marbled crayfish Procambarus virginalis exemplified by the habitus and coloration of specimens from Lake Moosweiher, Germany (A) and the laboratory (B). ct, cephalothorax; p, pleon (A from Vogt et al. 2018; B from Vogt 2017).
Although marbled crayfish is monoclonal, it shows a broad spectrum of variable phenotypic traits including coloration, morphological characters, life-history features, and behaviors (Vogt et al. 2008; Vogt 2022c). These phenotypic variants can arise either by developmental stochasticity (“developmental noise”) or environmental induction as revealed by the rearing of clutchmates in the same or different environments (Vogt 2020b). The coloration pattern is particularly diverse and identifies each specimen unambiguously (Figure 1A,B). The maximum body size (anterior tip of cephalothorax to end of pleon) is about 13 cm and the generation time is often 6–7 months (Vogt et al. 2004; Seitz et al. 2005), but can be shorter than 5 months under favorable conditions (Kouba et al. 2021). The average life span is 2–3 years, reaching a maximum of 4.5 years (Vogt 2010).
Clutch size is positively correlated with the body size of the mother and varies between ~20 and 730 eggs. Hossain et al. (2019) recorded a mean pleopodal fecundity at the first reproduction of 89.72 ± 56.9 eggs. The maximum number of successfully hatched juveniles per clutch was 427 (Vogt 2010; Vogt et al. 2019). In my laboratory, most females spawned twice around the spring and autumn equinoxes (Vogt 2015). Two potential reproductive peaks per year were also recorded in a wild population in Croatia (Dobrović et al. 2021). The maximum number of spawnings per lifetime was 7.
Marbled crayfish can be mass cultured in simple containers or commercially available aquarium systems (Vogt 2008; Jimenez and Faulkes 2010). Maximum growth in the laboratory was obtained at 25 °C and maximum survival at 20 °C (Seitz et al. 2005). Temperatures below 5 °C and above 30 °C can be survived for many weeks, but mortality increases and reproduction decreases under such conditions (Seitz et al. 2005; Veselý et al. 2015; Kaldre et al. 2016). Marbled crayfish are omnivorous generalist consumers (Linzmaier et al. 2020; Veselý et al. 2021). In a food choice experiment, the most preferred mobile food items were chironomids and gammarids, whereas snails were consumed less. Of the nonmobile food items, dead fish were significantly preferred over mussels, macrophytes, and decaying leaves (Linzmaier et al. 2020).
Due to high robustness against handling stress and fluctuations of abiotic factors, marbled crayfish can easily be kept in the laboratory and exposed to very different conditions enabling experiments on phenotypic plasticity. Moreover, in the last 20 years, this species has invaded numerous strikingly diverse habitats on 3 continents, mainly as the result of human releases (reviewed in Chucholl et al. 2012; Vogt 2020b; Maiakovska et al. 2021). These populations make it possible to study the relationships between phenotypic plasticity and genetic and epigenetic diversity under natural conditions.
Marbled crayfish is triploid, showing an AA’B genotype with a high level of heterozygosity and has a haploid chromosome number of 92 (Vogt et al. 2015; Martin et al. 2016; Gutekunst et al. 2018). The haploid genome size is ~3.7 Gb (Gutekunst et al. 2021). The DNA of marbled crayfish is well methylated (Vogt et al. 2008, 2015; Gatzmann et al. 2018) providing a good target for investigating epigenetic diversity. Global DNA methylation, the percentage of methylated cytosines per total cytosines in the genome, is about 2.4% (Vogt et al. 2015), corresponding to half of the value of laboratory mice and humans. Marbled crayfish possess an effective DNA methylation and demethylation toolkit comprising single copies of the DNA methytransferases DNMT1 and DNMT3 and the ten-eleven-translocation methylcytosine dioxygenase TET (Gatzmann et al. 2018).
Marbled crayfish was kept in several research laboratories and exposed to various abiotic and biotic conditions (e.g., Seitz et al. 2005; Vogt 2007; Veselý et al. 2015; Kouba et al. 2021; Tönges et al. 2021b; Tresnakova et al. 2022). These experiments demonstrated that it is very robust and plastic and can cope with a broad spectrum of conditions. The high plasticity is confirmed by numerous wild populations from very different habitats and geographical regions.
Wild populations have been found in 17 European countries (Austria, Belgium, Croatia, Czech Republic, Denmark, Estonia, France, Germany, Hungary, Italy, Malta, the Netherlands, Poland, Romania, Slovakia, Sweden, and Ukraine), 4 Asian countries (China, Israel, Japan, and Taiwan), and 1 African country (Madagascar) (references and coordinates in Vogt 2020b, Maiakovska et al. 2021; https://sites.google.com/view/marmorkrebs/). Most populations by far live in Madagascar, followed by Germany. In Madagascar, marbled crayfish has spread from an initial introduction near the capital Antananarivo around 2005 over more than 100,000 km2 in 2017, mostly by human dispersal (Gutekunst et al. 2018; Andriantsoa et al. 2019). In Germany, the country of its first appearance, marbled crayfish was found at 23 sites in ponds, lakes, and rivers (Vogt et al. 2020b).
Marbled crayfish occur in a broad spectrum of habitats, including rivers, ponds, oligotrophic to eutrophic lakes, rice fields, and cold, thermal, acidic, and polluted waters (Figure 2A–F) (Andriantsoa et al. 2019; Vogt 2020b; Maiakovska et al. 2021). The coldest habitat is River Märstaån in Sweden in which marbled crayfish were seen crawling in 2 °C cold water (Bohman et al. 2013). This population is meanwhile extinct (Patrik Bohman, personal communication). The warmest water bodies inhabited by marbled crayfish are a rice field at Anjingilo, Madagascar (Figure 2F), with 37 °C supplied by thermal water (Andriantsoa et al. 2019), thermal Lake Hévíz in Hungary with summer temperatures of 38 °C (Lőkkös et al. 2016), and Városliget pond in Hungary with summer temperatures of 38.4 °C (Weiperth et al. 2020).
Figure 2.
Examples of strikingly different environments of marbled crayfish including a laboratory setup (A) and pristine, polluted, acidic, cool, and warm natural habitats (B–F) (A and B from Vogt et al. 2018; C from Tönges et al. 2021a; D and E from Tönges et al. 2021b; F from Andriantsoa et al. 2019).
Marbled crayfish even populated acidic water bodies, which is unusual for crayfish. Examples are the recultivated lignite mining sites Lake Murner See (Figure 2C) and Lake Singliser See in Germany with pH 3.9–4.2 (Dümpelmann and Bonacker 2012; Tönges et al. 2021a, 2021b). An example of a heavily polluted marbled crayfish habitat is Ihosy River in Madagascar (Figure 2E) which has high aluminum levels of ~4,800 µg/L due to nearby mining activities (Andriantsoa et al. 2019).
Plasticity of Phenotypic and Ecological Traits in Marbled Crayfish
The acclimatization or adaptation of a species to different habitats generally depends on phenotypic variation, which can be produced by genetic variation, phenotypic plasticity, or a mixture of both. In the marbled crayfish, several phenotypic traits have already been investigated with respect to plasticity including coloration, spination, body proportions, life-history features, and behaviors. However, marbled crayfish are also plastic for food choice, population structure, trophic position, and niche width.
The marmoration pattern (Figure 1A,B) is the most plastic trait in marbled crayfish. Variation is so high that there are no two identical specimens. Because identically raised clutchmates differ from each other and their mother, the high diversity in marmoration is obviously caused by stochastic epimutations and downstream effects and further developmental stochasticity generators such as reaction–diffusion mechanisms (Vogt et al. 2008). However, background tanning between the marbled spots seems to depend on the environment, because it usually differs between laboratory-raised and wild marbled crayfish. Wild specimens mostly display a greenish-brown tanning (Figure 1A), whilst laboratory-raised specimens show a broad variety of colors including reddish (Figure 1B), brown, ochre, or bluish. The dorsal side of wild specimens is very dark (Figure 3A), probably serving as camouflage against predatory fish, otters, and water birds, whereas the dorsal side of laboratory-raised specimens is variably stained and often speckled (Figure 3A).
Figure 3.
Phenotypic plasticity of morphological characters in marbled crayfish adapted to the laboratory and Lake Moosweiher (Germany). (A) Maximum body size of laboratory-raised and wild specimens showing 30% bigger total length (TL) in the lake. Staining of the dorsal side is generally dark brownish in wild specimens but very variable in laboratory-raised individuals. CL, carapace length; CW, carapace width (original from author). (B) Spination of chelipeds in laboratory-raised and wild crayfish, showing much bigger and sharper spines (arrows) in the wild specimen (from Vogt et al. 2018). (C) Body proportions of marbled crayfish from the laboratory (L) and Lake Moosweiher (M) showing significant differences in total length/carapace length ratios (TL/CL) and carapace length/carapace width ratios (CL/CW). The adult offspring of a female that was transferred from the lake to the laboratory (M→L) and reproduced there had TL/CL ratios similar to the wild population but CL/CW ratios similar to the laboratory population. Figures in columns give numbers of specimens investigated; *** significantly different (P < 0.001); ns, not significantly different (based on Vogt 2021, modified).
Lake specimens have some prominent sharp spines on their carapaces and chelipeds, but laboratory-raised specimens of the same size lack such spines and have only small blunt knobs instead (Figure 3B). This difference in spination was repeatedly observed by comparison of specimens from different laboratories with specimens from a small brook in Saxony (Martin et al. 2010b), Lake Singliser See (Dümpelmann and Bonacker 2012), and Lake Moosweiher (Vogt et al. 2018), all located in Germany. Specimens transferred from Lake Moosweiher to the laboratory maintained their spines through several molts until the end of life, but in the F1 progeny, the spines were considerably reduced, resembling members of the laboratory colony (Vogt et al. 2018). Spination of wild marbled crayfish is probably predator driven as observed in other aquatic invertebrates like Daphnia water fleas (cyclomorphosis) and fishes (Tollrian 1995; Price et al. 2015). Augusto et al. (2021) reported that in Daphnia pulex, the presence of predator stimuli induced profound reorganization of the chromatin in parallel to the morphological changes (elongation of head and terminal spine), suggesting the involvement of epigenetic mechanisms like histone modifications in cyclomorphosis.
The laboratory specimens of marbled crayfish also had significantly different body proportions (relatively longer pleons and broader carapaces) when compared to equal-sized specimens from Lake Moosweiher (Figure 3C) (Vogt 2021). Interestingly, the adult offspring of a specimen that was transferred from Lake Moosweiher to the laboratory and reproduced there 1 year later had total length/carapace length ratios similar to the wild population and their mother, but carapace length/carapace width ratios more similar to the laboratory population (Figure 3C) suggesting partial acclimatization to the new conditions.
A comparison of marbled crayfish from my laboratory and Lake Moosweiher also revealed considerable differences in maximal body size. Specimens raised under stringent laboratory conditions and fed with a single pellet food for many generations reproduced well and grew old (4.5 years), but reached maximum total lengths of only 9 cm and weights of 18 g, whereas their relatives from Lake Moosweiher that can utilize a much broader spectrum of food grew to ~12 cm and 40 g (Figure 3A).
Behavioral plasticity is particularly useful for acclimatization or adaptation to the specific conditions of habitats. Examples are food choice (see below) and the use of shelters. Marbled crayfish often hide under stones and water plants, as observed in Lake Moosweiher and other sites, but when such shelters are scarce or absent, they hide in the mud or in burrows. For example, in Lake Murner See, a former lignite mining site, they build burrows as deep as 1 m (Figure 2C) (Frank Lenich, personal communication). In dried-out ponds in Madagascar, they were found buried deep in the mud (Jones et al. 2009). Marbled crayfish usually stay in a suitable water body, but single individuals occasionally leave such habitats and migrate overland, perhaps to search for new habitats. This potential dispersal behavior was repeatedly observed (Chucholl et al. 2012; Herrmann et al. 2018). An example is Lake Epplesee (Germany) where individuals were found more than 100 m away from the shoreline.
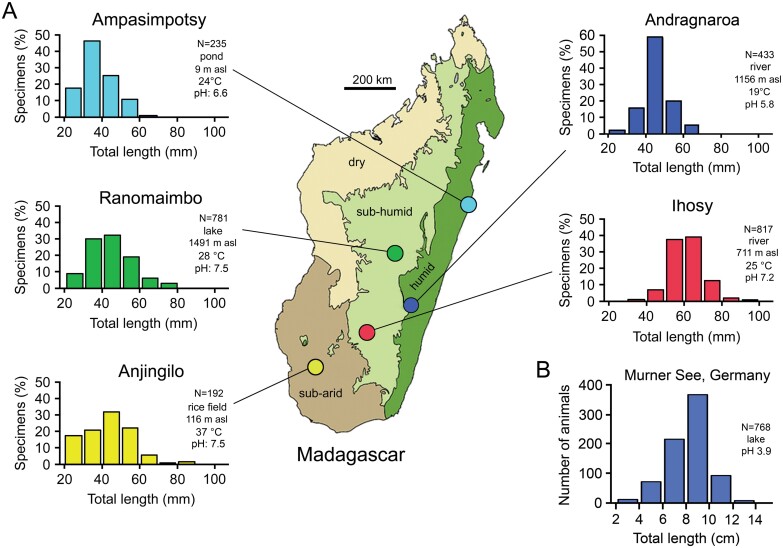
Andriantsoa et al. (2019) analyzed 5 Malagasy populations from different lentic and lotic habitats in 3 bioclimatic regions and revealed marked differences in size–frequency distribution between populations (Figure 4A). The lotic populations live in a mountain river in the Ranomafana National Park rainforest (Andragnaroa River) and a slow-flowing, turbid and highly polluted lowland river near a large city in south-central Madagascar (Ihosy River). The lentic populations include a lake in the highlands in the center of a big city impacted by human activities (Ranomaimbo), a rice field supplied by thermal water in a relatively isolated area in the south (Anjingilo), and a pond in a village on the east coast (Ampasimpotsy). Interestingly, the polluted Ihosy River harbors the highest density of marbled crayfish and significantly bigger specimens (P < 0.05) than the other sites (Andriantsoa et al. 2019).
Figure 4.
Differences in size–frequency distribution among populations of marbled crayfish from diverse habitats. (A) Comparison of Malagasy populations from 2 rivers, a lake, a pond, and a rice field in humid, sub-humid, and sub-arid bioclimatic regions. Values on right side of graphs give number of specimens analyzed, water body type, altitude above sea level, water temperature measured at time of sampling in 10 cm water depth, and pH value (based on Andriantsoa et al. 2019, modified). (B) Population from Lake Murner See in Germany showing considerably larger animals when compared to the Malagasy populations (based on Tönges et al. 2021a, modified).
Analysis of size–frequency distribution in mesotrophic Lake Moosweiher (N = 175) and acidic Lake Murner See (N = 768), both in Germany, revealed the highest percentages of animals in the size ranges of 6.5–8.5 and 8–10 cm TL (Figure 4B), respectively. These values are considerably higher than those of the Malagasy population. The shift toward bigger animals is likely the effect of latitude, among other factors, which may be deduced from the example of the con-generic Procambarus clarkii (red swamp crayfish). This species is of subtropical origin like the ancestor of marbled crayfish and demonstrates a gradual and significant increase in body size with latitude (Chucholl 2011).
Acidic Lake Murner See harbors the population with the largest specimens ever found (Tönges et al. 2021a), which appears odd at first glance, because the abiotic conditions in this lake are certainly not optimal for crayfish. However, the absence of predatory fish (Frank Lenich, personal communication) may have favored this particular population structure. In Lake Moosweiher and most other German marbled crayfish habitats, fish species that prey on adult crayfish are abundant (Chucholl and Pfeiffer 2010).
Analyses of stable carbon 13C and nitrogen 15N isotopes of field samples from a Slovakian gravel pit, 3 German lakes, and the upper and lower stretches of a Hungarian stream revealed that marbled crayfish are also plastic with respect to food utilization, trophic position, and niche width (Lipták et al. 2019; Linzmaier et al. 2020; Veselý et al. 2021). Lipták et al. (2019) studied the food–web interactions of marbled crayfish in an oligotrophic gravel pit in Leopoldov (south-western Slovak Republic) and found that allochthonous detritus (29%), algae (25%), and autochthonous detritus (21%) were the most important food sources, while macrophytes (15%) and zoobenthos (9%) were less frequently used. Moreover, the marbled crayfish were found to be an important food source for top fish predators, but marginal for omnivorous fish.
Linzmaier et al. (2020) investigated the diet composition of marbled crayfish in the gravel pits Lake Moosweiher and Lake Meitzer See and natural Lake Krumme Lanke, all located in Germany. They revealed average proportions of invertebrates, plant-based material, and fish of 62%, 21%, and 16%, respectively, which is in sharp contrast to the Slovakian gravel pit. However, these proportions varied to some degree between habitats as shown in Table 1. Lake Moosweiher and Lake Krumme Lanke are small, mesotrophic, and dimictic lakes with several tree species growing along the seashore. The littoral of both lakes is covered with macrophytes, woody debris on sediments of coarse gravel, and some soft-bottomed areas. Lake Meitzer See is more oligotrophic and larger and deeper (Linzmaier et al. 2020), suggesting that the lakes may considerably vary in the food supply.
Table 1.
Differences in food consumption by marbled crayfish in 3 German lakes (from Linzmaier et al. 2020)
| Food (%) | Moosweiher | Krumme Lanke | Meitzer See |
|---|---|---|---|
| Fish | 14.4 | 16.1 | 16.4 |
| Mussels | 9.9 | 21.8 | 20.6 |
| Snails | 14.1 | 19.3 | 18.7 |
| Arthropods | 32.9 | 18.3 | 16.8 |
| Macrophytes | 4.9 | 4.5 | 5.6 |
| Detritus | 8.9 | 13.1 | 12.2 |
Veselý et al. (2021) investigated the trophic position and niche width of marbled crayfish in the upper and lower sections of a thermal tributary of Barát stream in Budapest, Hungary. In the upper section, marbled crayfish coexist with spiny cheek crayfish, Faxonius limosus, and in the lower section with spiny cheek crayfish and red swamp crayfish. The authors revealed that in the upper section, marbled crayfish feed mainly on detritus (31%) and algae (30%) followed by zoobenthos (24%) and macrophytes (15%). In the lower section, these proportions differ to some degree (detritus 36%, algae 28%, zoobenthos 18%, and macrophytes 17%).
Linzmaier et al. (2020) found that marbled crayfish can occupy a large range of trophic positions from 1.7 to 4.1 in the lakes (Figure 5A,B), meaning that they can act from primary consumers or detritivores (low values) up to predators (high values). This wide span of trophic positions puts marbled crayfish in a key position for energy transfer between trophic levels. Niche width also varied between the 3 lakes (Linzmaier et al. 2020). Veselý et al. (2021) confirmed for the Hungarian stream that marbled crayfish can occupy different trophic niches. In the lower section, the presence of the more carnivorous red swamp crayfish restricted the niches of marbled crayfish and spiny cheek crayfish (Figure 5C,D).
Figure 5.
Differences of trophic position and niche width between marbled crayfish from different habitats. (A) Trophic positions in mesotrophic Lake Moosweiher and oligotrophic Lake Meitzer See, represented by standard ellipse areas. In Lake Moosweiher, marbled crayfish has a higher trophic position than in Lake Meitzer See. The smaller specimens in Lake Moosweiher use more animal food than the larger individuals (based on Linzmaier et al. 2020, modified). (B) Trophic positions and niche widths in the upper and lower sections of the Barát thermal tributary in the presence of 1 (left) or 2 (right) crayfish competitors. The presence of the more carnivorous Procambarus clarkii influences the trophic positions and niche widths of marbled crayfish and Faxonius limosus (based on Veselý et al. 2021, modified).
The studies on food use, trophic position, and niche width demonstrate that marbled crayfish can flexibly cope with different abiotic and biotic conditions in natural environments. This feature may have helped this species to successfully establish populations in many highly diverse habitats and geographical regions and explain, aside from parthenogenetic reproduction, why it has a much higher invasive potential than its parent species P. fallax. Chucholl (2016) calculated an almost double Freshwater Invertebrate Invasiveness Scoring Kit for marbled crayfish when compared to its parent species. By the way, the evolution of marbled crayfish from slough crayfish by autotriploidization and transition from gonochoristic to parthenogenetic reproduction was associated with a considerable enhancement of the fitness traits fecundity, body size, and longevity (Vogt et al. 2019).
Genetic Diversity in Marbled Crayfish
According to prevailing theory, members of monoclonal apomictic parthenogenetic populations are considered genetically identical (except for random mutations), whilst automictic parthenogenetic populations are genetically diverse (Mirzaghaderi and Hörandl 2016). Previous comparison of the mitochondrial cytochrome c oxidase subunit I (COI) gene in marbled crayfish revealed the identity of all marbled crayfish from laboratory colonies and wild populations from different European countries, Madagascar and Japan (Martin et al. 2010a, 2010b; Bohman et al. 2013; Vojkovská et al. 2014; Vogt et al. 2015, 2018; Lőkkös et al. 2016; Usio et al. 2017; Gutekunst et al. 2018; Ercoli et al. 2019).
Comparison of 3 nuclear microsatellite loci between laboratory-raised mothers and their progeny and representatives of different populations confirmed the genetic identity of all marbled crayfish (e.g., Martin et al. 2007; Vogt et al. 2008, 2015, 2018; Parvulescu et al. 2017), suggesting that they are apomictic parthenogens constituting a monoclonal meta-population. Genetic fingerprinting by highly variable microsatellites is a very efficient and highly sensitive method to identify individuals and to study parentage and the genetic diversity and structure of animal populations (Flanagan and Jones 2019; Svishcheva et al. 2020). However, because microsatellites cover only a very short sequence of the genome and because only 3 microsatellites were analyzed, there remained some residual doubts on the genetic identity of marbled crayfish on the whole-genome scale.
To dispel these doubts, the whole genome of marbled crayfish was sequenced, the first time by paired-end lllumina sequencing (Gutekunst et al. 2018) and a second time by long-read PacBio sequencing (Gutekunst et al. 2021). Assembly of the Illumina data revealed a genome of 3.5 Gb and correctly positioned 52.5% of bases including the sequences of the ~22,000 coding genes (Gutekunst et al. 2018). PacBio drastically reduced the number of gaps, which mainly concerned repetitive sequences. The newly assembled sequence has a size of 3.7 Gb and covers 82.1% of the genome, which is a very good value when compared with other animal genomes (Gutekunst et al. 2021).
Gutekunst et al. (2018) compared ca. 20% of the whole-genome sequences of specimens from different German laboratory lineages and wild populations in Germany and Madagascar with the reference genome. The investigated populations are separated from each other for about 20–40 generations. The analysis revealed a total of 416 single nucleotide variants (SNVs) and only small differences of 128–219 SNVs between the reference genome and wild individuals (Figure 6A). The vast majority of these SNVs were silent mutations. The maximum number of nonsynonymous SNVs that can change amino acids in proteins was only 4 between samples.
Figure 6.
Genetic diversity between differently adapted marbled crayfish populations. (A) Phylogenetic tree of marbled crayfish from diverse laboratory (lab) and field sources in Germany (G) and Madagascar based on the comparison of ca. 20% of the Illumina-sequenced genomes. SNV, single nucleotide variant (based on Gutekunst et al. 2018, modified). (B) Whole-genome differences between representatives from several European populations determined by Illumina sequencing and improved bioinformatics approaches. A descendant of the oldest known marbled crayfish population was used as a reference (based on Maiakovska et al. 2021, modified).
More recent whole-genome analysis with improved bioinformatics tools of 14 specimens from 10 populations in 8 European countries and 5 specimens from 5 populations in Madagascar revealed a total of 16,563 SNVs in the 3.7 Gb genome, 74% of them in intergenic regions, and only 4% in the coding regions (Maiakovska et al. 2021). These results indicate a very low degree of evolution of the marbled crayfish genome across its distribution range since its first appearance in 1995, which was solely driven by its natural mutation rate of 3.5 × 10−8 per nucleotide and year (Legrand et al. 2021). Genetic recombination was absent and genome-wide analysis of variant allele frequency did not reveal any evidence for loss of heterozygosity, which is consistent with the notion that marbled crayfish reproduce by apomictic parthenogenesis (Maiakovska et al. 2021). A distance tree showed high genetic similarity among the wild European populations and a more distant relationship to the ancestral aquarium lineage that was used to establish the reference genome (Figure 6B).
The 2 genetic analyses of representatives of multiple populations on the whole-genome scale demonstrate that the total number of SNVs, particularly nonsynonymous variants, in coding regions is extremely small. These data suggest that the extensive phenotypic variation of marbled crayfish cannot be caused by genetic variation and that the occupation of highly diverse habitats is not the result of selection on genetic variation.
Epigenetic Diversity in Marbled Crayfish
If genetic diversity is not sufficient to explain the phenotypic diversity and environmental acclimatization in marbled crayfish, what else could be the underlying mechanism? A promising candidate is epigenetic diversity, because epigenetic mechanism like DNA methylation, histone modifications, and noncoding RNAs can produce multiple phenotypes from the same DNA sequence by changing gene expression either stochastically or in response to environmental inputs (Gibney and Nolan 2010; Duncan et al. 2014; Vogt 2020a).
To get a first idea on whether there are methylation differences between differently acclimatized populations, we measured global DNA methylation by mass spectrometry in laboratory-raised and wild specimens from Lake Moosweiher. We obtained a relatively small difference between the 2 conditions (2.38% in the laboratory vs. 2.27% in the lake) (Vogt 2018). Since global DNA methylation changes reflect the sum of DNA methylation gains in some genes and DNA methylation losses in others, this parameter is obviously not sensitive enough for measuring epigenetic diversity within and between populations. Therefore, the methylation of individual genes was chosen as a new target.
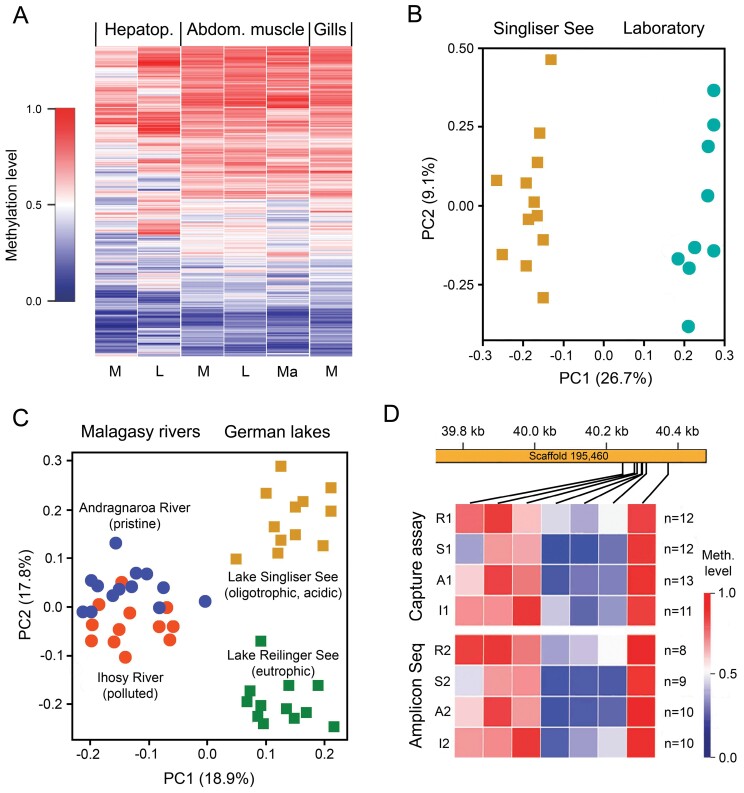
Comparison of whole-genome bisulfite sequencing datasets from selected pure tissues (hepatopancreas and abdominal muscle) of 8 marbled crayfish identified a subset of 697 highly variably methylated genes (Gatzmann et al. 2018) that proved particularly suitable for further research on intra- and interpopulation epigenetic diversity. There were striking differences in the methylation pattern of these genes between samples from the laboratory and Lake Moosweiher (Figure 7A), particularly in the hepatopancreas, the central organ of metabolism in crayfish (Vogt 2019b). There were also considerable differences between tissues, suggesting that the epigenetic data used for within- and between-population comparisons must be obtained from the same tissue.
Figure 7.
Epigenetic diversity among differently adapted marbled crayfish populations. (A) Comparative analysis of 697 variably methylated genes in the hepatopancreas, abdominal muscle, and gills of specimens from the laboratory (L), Lake Moosweiher, Germany (M), and a rice field in Moramanga, Madagascar (Ma). The heatmap shows differences in methylation patterns between individuals, particularly in the hepatopancreas (based on Tönges et al. 2021b, modified). (B) Principal component analysis based on the average methylation of 361 variably methylated genes, showing a clear separation of individuals from Lake Singliser See, Germany, and the laboratory (based on Tönges et al. 2021b, modified). (C) Principal component analysis of cytosine methylation of 122 genes separating four populations living in pristine and polluted rivers in Madagascar and oligotrophic and eutrophic lakes in Germany (based on Tönges et al. 2021b, modified). (D) Persistent DNA methylation fingerprints of populations from Andragnaroa River (A), Ihosy River (I), Lake Reilinger See (R), and Lake Singliser See (S) in consecutive years (1 and 2), exemplified for a small genic region of the hepatopancreatic DNA. The samples were collected at intervals of 12–21 months and analyzed with 2 different methods (based on Tönges et al. 2021b, modified).
To investigate the impact of environmental influences on DNA methylation in more detail, Tönges et al. (2021b) used 3 approaches: (1) comparison of populations from the laboratory and a natural habitat, (2) comparison of wild populations from strikingly different habitats, and (3) experimental exposure of laboratory specimens to strikingly different temperatures. For these comparisons they applied a capture-based subgenome bisulfite sequencing approach that covered variably methylated portions of the marbled crayfish genome.
Principal component analysis based on the average methylation of 361 variably methylated genes from hepatopancreas samples that were enriched for housekeeping gene functions showed a clear separation of individuals from the Heidelberg laboratory colony and Lake Singliser See (Figure 7B). Moreover, the methylation patterns of laboratory specimens kept at 10 °C for 6 months differed markedly from samples kept in the same setup at 20 °C and the complete set of investigated wild samples, providing experimental evidence for environmentally induced methylation changes within a single generation (Tönges et al. 2021b).
Tönges et al. (2021b) then compared the methylation patterns of 122 variably methylated genes by a capture-based subgenome bisulfite sequencing approach in the hepatopancreas of 48 specimens from 4 highly diverse habitats in Madagascar and Germany. The Andragnaroa population lives in a mountain river (1,156 m altitude) with soft water (0.3 dH) flowing through a forest area. The Ihosy population inhabits a river with highly turbid and polluted water including high levels of aluminum (4,800 μg/L) and iron (2,200 μg/L) from nearby mining activities. Lake Reilinger See (Germany) is a 9-ha-large and 16-m-deep eutrophic lake in an environmentally protected area with clean, slightly alkaline (pH 8.4) and hard (18.9 dH) water and numerous predatory fishes. Lake Singliser See is a 74-ha-large and 30-m-deep oligotrophic lake within a renaturalized brown coal mining area with acidic water (pH 4.2) and high levels of sulfur (700 mg/L) and manganese (2.900 μg/L). Fishes are absent. The results revealed specific and highly localized DNA methylation signatures for each population (Figure 7C). Similar differences between populations were also obtained with a smaller dataset from the abdominal muscles (N = 27) of the same animals covering 23 differentially methylated genes, 21 of which overlapped with the hepatopancreas.
Gene ontology analysis of the variably methylated genes in the hepatopancreas and abdominal muscle revealed a significant enrichment of GTP-binding proteins, which transmit signals from the external world to the cells regulating various cellular processes. Enrichment was also recorded for proteins involved in the regulation of transcription and translation, RNA metabolism, response to stress, and immune response (Tönges et al. 2021b).
To validate the location-specific methylation patterns, Tönges et al. (2021b) collected a second set of 37 specimens from the same locations 1–2 years after the initial sampling and analyzed them with a different technique (Amplicon Seq). They identified differentially methylated regions (DMRs) within the set of 361 variably methylated genes and designed PCR assays for targeted bisulfite sequencing. Deep sequencing of the PCR amplicons principally confirmed the capture-based subgenome sequencing results. The methylation ratios of individual CpGs of samples from the same site were highly similar between the initial dataset and the validation dataset from subsequent years, as was shown for several DMRs. An example is given in Figure 7D.
SNVs were absent from all differentially methylated genes investigated, and therefore, the findings discard genetic variants as underlying factors of the observed epigenetic differences and demonstrate that the population-specific methylation signatures were purely epigenetic. The results also suggest that different environments induce different epigenetic fingerprints that remain rather stable over some time.
The observed establishment of “epigenetic ecotypes” in marbled crayfish could either result from the repeated de novo production of identical, environmentally induced epigenetic patterns and phenotypes in each generation or from the transgenerational inheritance and selection of adaptive, epigenetically determined phenotypes. At present, this question is still open. There is increasing evidence in the literature for the transgenerational inheritance of epigenetic signatures in animals and there are ideas on how they might contribute to environmental adaptation and evolution (Jablonka and Raz 2009; Pfennig and Servedio 2013; Jablonka 2017; Anastasiadi et al. 2021; Skinner and Nilsson 2021; Feiner et al. 2022). Common garden experiments, cross-transplantation experiments, and condition and location-specific monitoring of epigenetic fingerprints for many generations are required to uncover, which of the 2 alternatives of environmental acclimatization in marbled crayfish is realized. Such experiments could also help answer the question of whether the observed phenotypic plasticity is adaptive or not and whether it contributes to evolutionary change (Thompson 1991; Price et al. 2003; Crispo 2007).
Scenario on the Generation of Environmentally Induced Phenotypic Diversity by Epigenetic Mechanisms
There is convincing empirical evidence that the production of phenotypic variation from the same genome via epigenetic mechanisms can be triggered by strong environmental cues like temperature, salinity, food, predator cues, toxicants, or disease agents (e.g., Skinner 2014; Guillette et al. 2016; Logan and Storey 2021). This process requires (1) signal transmission from the external world to the nuclei of the target cells, (2) environment-sensitive molecules involved in gene regulation, (3) readers, writers, and editors of epigenetic marks, and (4) molecules that recruit the epigenetic modifiers to specific regions of the DNA and chromatin. Usually, various epigenetic mechanisms like DNA methylation, histone modifications, and ncRNAs cooperate in this process (Du et al. 2015). Figure 8 shows a hypothetical scenario of the production of diverse phenotypes from the same genome in response to environmental signals via epigenetic mechanisms.
Figure 8.
Hypothetical scenario of environmentally induced changes of gene and phenotype expression by epigenetic mechanisms on the example of the invasion of a new environment. Strong environmental signals in the new habitat can trigger gene expression change in the nucleus of the target cells by a cascade of signal-transmitting hormones, second messengers, and an environment-sensitive molecular complex that comprises DNA methylation-modifying-enzymes (DME) and histone modifying enzymes (HME). DNA methylation readers (DMRe), histone modification readers (HMRe), and transcription factors recruit the DMEs and HMEs to specific sites in the chromatin and DNA. Acetylation (filled squares) and deacetylation (open squares) of histones modify the chromatin structure and regulate access to the DNA, and methylation (filled circles) and demethylation (open circles) of CpG dinucleotides in the promoter region and gene bodies of the DNA change gene expression, finally resulting in phenotypic change.
Environmental cues can directly act on the target cells, for example, fatty acids from the food, but mostly they are perceived by sense organs and translated to neurohormonal signals that are conveyed to the target cells. The hormonal signals then elicit cellular signals (second messengers), which finally regulate molecules involved in chromatin remodeling, gene expression, and processing of the transcripts. Serotonin is a good example of an environmental signal-transmitting hormone. In locust polyphenism, the density-dependent change of morphological and behavioral phases, serotonin transmits information on the density of individuals, and initiates the regulation of density-responsive genes via epigenetic mechanisms (Foquet et al. 2021).
A considerable number of the enzymes participating in the regulation of chromatin architecture and gene expression are apparently susceptible to activation and inhibition by environmental cues. Examples are the DNA demethylating TET, which can be up- or downregulated by several environmental factors, including ethanol, food ingredients, air pollution, and radiation (Zhu et al. 2020), proteins of the Polycomb group that are sensitive to the environmental temperature (Voigt and Kost 2021), and transcription factors of the TCP family that mediate environmental signals into growth responses in plants (Danisman et al. 2016). To change gene expression, such environment-sensitive proteins must crosstalk with readers and modifiers of the epigenetic marks and with molecules that recruit them to specific sites of the DNA and chromatin.
The writers, erasers, and editors of the DNA methylation marks include the methylating DNMTs and the demethylating TETs (Wu and Zhang 2017; Lyko 2018). These enzymes form complexes with readers of the DNA methylation marks like proteins of the methyl-CpG-binding domain family (MBDs) and transcription factors to exert their functions (Huang et al. 2018; Ravichandran et al. 2018). The MBDs bind methylated CpG dinucleotides and act as translators between DNA methylation and histone modifications (Du et al. 2015). Transcription factors with different sequence specificity can guide the methylation-modifying complex to specific sites of the DNA (Kribelbauer et al. 2020). Each animal possesses hundreds of such transcription factors.
Environmentally induced changes of the DNA methylation marks and histone modifications are thought to cause changes in gene expression via on–off switching of genes (e.g., via promoter methylation) or modulation of their activity (e.g., via gene body methylation). These changes lead to alterations in corresponding gene networks and eventually to changes of the phenotypic traits that are regulated by these networks.
What Can We Learn from the Marbled Crayfish Case Study About Phenotypic Plasticity?
The marbled crayfish is the first clonal animal in which whole-genome genetic and epigenetic data were used to compare genetic and epigenetic diversity with plasticity in phenotypic and ecological traits (coloration, spination, body proportions, growth, food preference, population structure, trophic position, and niche width) in statistically significant numbers of specimens. The studies revealed that phenotypic diversity and environmental acclimatization in this species are associated with epigenetic diversity but not with genetic diversity, suggesting that epigenetic mechanisms are crucially involved in the production of phenotypic plasticity.
Other studies with clonal animals that were performed with less sophisticated genetic and epigenetic approaches principally support this view. For example, Liew et al. (2018) exposed genetically identical genets of the coral Stylophora pistillata to long-term pH stress in the laboratory and measured associated changes in phenotypic traits and DNA methylation. Exposure to stressful pH 7.2 significantly enhanced cell size and polyp size resulting in more porous skeletons when compared to pH 8.0. It also increased mean methylation levels and caused methylation changes in genes regulating cell cycle and body size.
Another good example from the field is the populations of the New Zealand mud snail Potamopyrgus antipodarum in the western USA. These populations originate from a single clone that was introduced about 35 years ago, and therefore, they are genetically largely identical (Dybdahl and Drown 2011). Thorson et al. (2017) investigated populations from different sites and measured habitat-specific differences in shell shape, which were correlated with water current speed. Using methylated DNA immunoprecipitation (MeDIP) and partial Illumina sequencing of foot pad tissue, the authors also measured significant genome-wide DNA methylation differences between lakes and rivers. Thorson et al. (2019) then compared populations from a rural lake and 2 polluted urban lakes and determined differences in shell shape and allometric growth. These differences were associated with different methylation of several DNA regions. The data suggest that environmentally induced phenotypic plasticity is closely related to epigenetic variation supporting the results with marbled crayfish.
Exposure of marbled crayfish and other clonal animals to different environmental conditions demonstrates that the epigenetic layer above the uniform DNA sequence is the first level of biological organization in which diversity is manifested. This epigenetic diversity and further downstream events are thought to finally lead to the phenotypic plasticity of traits. Strong and longer lasting environmental cues are considered the ultimate cause of phenotypic plasticity and epigenetic mechanisms are likely the proximal cause or mediators.
The studies with the marbled crayfish and other obligately parthenogenetic species suggest that epigenetic mechanisms enable genetically near-identical species and lineages to inhabit very broad ranges of habitats and geographical regions, which is contrary to expectation. Epigenetic mechanisms seem to be particularly important for generating phenotypic diversity in asexually reproducing species and lineages and genetically depauperate populations and invasive groups, but it is likely that they contribute to phenotypic plasticity in sexually reproducing species as well, which represent the vast majority of animals.
The work with clonal animals has revealed a close association between phenotypic diversity and epigenetic diversity, but the detailed cause–effect relationships between the 2 remain to be established. Although associations are not as meaningful as well-established cause–effect relationships, they are the first important step toward the identification of the molecular mechanisms underlying phenotypic plasticity. This knowledge may encourage researchers to proceed with more targeted cause–effect studies, for example, by performing epigenetic and phenotypic association studies (Campagna et al. 2021) or by editing specific epigenetic marks and genes of the epigenetic machinery by CRISPR/Cas (Xu et al. 2020; Nakamura et al. 2021).
The ability to be plastic must anyhow be encoded in the genome and chromatin. Kronholm et al. (2016) investigated phenotypic plasticity in the filamentous fungus Neurospora crassa and found that this phenomenon is not governed by general plasticity genes. It is rather context dependent and regulated by epigenetic mechanisms. For example, histone methylation at H3K36 contributed to the plastic response to high temperature and methylation at H3K4 to the response to pH alteration. This study suggests that plastic and less plastic organisms differ in the possession of modifiable marks on the DNA and histones and in genes that encode the DNA methylation machinery, histone modification machinery, ncRNAs, and further epigenetic systems like Polycomb/Trithorax.
Comparison between triploid marbled crayfish and its diploid parent species P. fallax demonstrated that marbled crayfish is phenotypically more plastic and more invasive (Chucholl 2016; Vogt et al. 2019), which is probably the consequence of polyploidy. Therefore, the marbled crayfish and slough crayfish pair may be a suitable model system to investigate how the genetic make-up influences phenotypic plasticity.
Conclusions
The studies with the monoclonal marbled crayfish revealed a pronounced association between phenotypic diversity and epigenetic diversity but not between phenotypic diversity and genetic diversity, suggesting that epigenetic mechanisms are potent molecular players in producing phenotypic plasticity. The epigenome is apparently the interface at which the environment and genome interact. The studies further demonstrate that the production of different phenotypes from the same genotype with the help of epigenetic mechanisms in response to environmental signals is relevant under natural conditions.
Presently, it is unknown whether the observed phenotypic plasticity in marbled crayfish is adaptive or nonadaptive and whether the plastic changes are related to fitness changes. Experiments, in which specimens from differently acclimatized populations are raised in a common environment, could answer these questions. Such experiments could also reveal how fast and to what degree epigenetic marks are erased and if there are epigenetic signatures and related phenotypes that persist, are inherited, and are genetically fixed in the long term, providing raw material for selection and adaptive evolution.
Future research with the marbled crayfish model should be directed toward identifying cause–effect relationships by performing association studies between phenotypes and epigenetic variants and by editing genes and epigenetic marks by CRISPR/Cas. Future experiments should also be extended to other epigenetic mechanisms such as histone modifications and noncoding RNAs.
Conflict of Interest
The author declares no conflict of interest.
References
- Allis CD, Jenuwein T, 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17:487–500. [DOI] [PubMed] [Google Scholar]
- Anastasiadi D, Venney CJ, Bernatchez L, Wellenreuther M, 2021. Epigenetic inheritance and reproductive mode in plants and animals. Trends Ecol Evol 36:1124–1140. [DOI] [PubMed] [Google Scholar]
- Andriantsoa R, Tönges S, Panteleit J, Theissinger K, Coutinho Carneiro Vet al. , 2019. Ecological plasticity and commercial impact of invasive marbled crayfish populations in Madagascar. BMC Ecol 19:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers B, Perez M, Menicucci T, Leung C, 2020. Sources of epigenetic variation and their applications in natural populations. Evol Applicat 13:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto RC, Minod A, Rey O, Chaparro C, Vidal-Dupiol Jet al. , 2021. A simple ATAC-seq protocol for population epigenetics, version 2. Wellcome Open Res 5:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A, Kouzarides T, 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biwer C, Kawam B, Chapelle V, Silvestre F, 2020. The role of stochasticity in the origin of epigenetic variation in animal populations. Integr Comp Biol 60:1544–1557. [DOI] [PubMed] [Google Scholar]
- Bohman P, Edsman L, Martin P, Scholtz G, 2013. The first Marmorkrebs (Decapoda: Astacida: Cambaridae) in Scandinavia. BioInvasions Rec 2:227–232. [Google Scholar]
- Campagna MP, Xavier A, Lechner-Scott J, Maltby V, Scott RJet al. , 2021. Epigenome-wide association studies: Current knowledge, strategies and recommendations. Clin Epigenet 13:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chucholl C, 2011. Population ecology of an alien “warm water” crayfish Procambarus clarkii in a new cold habitat. Knowl Manag Aquat Ecosyst 401:29. [Google Scholar]
- Chucholl C, 2016. Marbled crayfish gaining ground in Europe: The role of the pet trade as invasion pathway. In: Kawai T, Faulkes Z, Scholtz G, editors. Freshwater Crayfish: Global Overview. Boca Raton (FL): CRC Press, 83–114. [Google Scholar]
- Chucholl C, Morawetz K, Groß H, 2012. The clones are coming: Strong increase in Marmorkrebs [Procambarus fallax (Hagen, 1870) f. virginalis] records from Europe. Aquat Invasions 7:511–519. [Google Scholar]
- Chucholl C, Pfeiffer M, 2010. First evidence for an established Marmorkrebs (Decapoda, Astacida, Cambaridae) population in Southwestern Germany, in syntopic occurrence with Orconectes limosus (Rafinesque, 1817). Aquat Invasions 5:05e412. [Google Scholar]
- Ciabrelli F, Comoglio F, Fellous S, Bonev B, Ninova Met al. , 2017. Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nat Genet 49:876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispo E, 2007. The Baldwin effect and genetic assimilation: Revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 61:2469–2479. [DOI] [PubMed] [Google Scholar]
- Danisman S, 2016. TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front Plant Sci 7:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt T, Scheiner SM, 2004. Phenotypic Plasticity: Functional and Conceptual Approaches. New York (NY): Oxford University Press. [Google Scholar]
- Dobrović A, Maguire I, Boban M, Grbin D, Hudina S, 2021. Reproduction dynamics of the marbled crayfish Procambarus virginalis Lyko, 2017 from an anthropogenic lake in northern Croatia. Aquat Invasions 16:482–498. [Google Scholar]
- Du Q, Luu PL, Stirzaker C, Clark SJ, 2015. Methyl-CpG-binding domain proteins: Readers of the epigenome. Epigenomics 7:1051–1073. [DOI] [PubMed] [Google Scholar]
- Dümpelmann C, Bonacker F, 2012. Erstnachweis des Marmorkrebses Procambarus fallax f. virginalis (Decapoda: Cambaridae) in Hessen. Forum Flusskrebse 18:3–12. [Google Scholar]
- Duncan EJ, Gluckman PD, Dearden PK, 2014. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J Exp Zool B Mol Dev Evol 322:208–220. [DOI] [PubMed] [Google Scholar]
- Dybdahl MF, Drown DM, 2011. The absence of genotypic diversity in a successful parthenogenetic invader. Biol Invasions 13:1663–1672. [Google Scholar]
- Ercoli F, Kaldre K, Paaver T, Groos R, 2019. First record of an established marbled crayfish Procambarus virginalis (Lyko, 2017) population in Estonia. BioInvasions Rec 8:675–683. [Google Scholar]
- Feinberg AP, Irizarry RA, 2010. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA:1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner N, Radersma R, Vasquez L, Ringnér M, Nystedt Bet al. , 2022. Environmentally-induced DNA methylation is inherited across generations in an aquatic keystone species Daphnia magna. iScience 25:104303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SP, Jones AG, 2019. The future of parentage analysis: From microsatellites to SNPs and beyond. Mol Ecol 28:544–567. [DOI] [PubMed] [Google Scholar]
- Foquet B, Castellanos AA, Song H, 2021. Comparative analysis of phenotypic plasticity sheds light on the evolution and molecular underpinnings of locust phase polyphenism. Sci Rep 11:11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Laserre D, Villagra CA, 2017. The importance of ncRNAs as epigenetic mechanisms in phenotypic variation and organic evolution. Front Microbiol 8:2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco G, Minelli A, 2010. Phenotypic plasticity in development and evolution: Facts and concepts. Philos Trans R Soc Lond B 365:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzmann F, Falckenhayn C, Gutekunst J, Hanna K, Raddatz Get al. , 2018. The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenet Chromatin 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN, 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol 21:394–407. [Google Scholar]
- Gibney ER, Nolan CM, 2010. Epigenetics and gene expression. Heredity 105:4–13. [DOI] [PubMed] [Google Scholar]
- Guillette LJ Jr, Parrott BB, Nilsson E, Haque MM, Skinner MK, 2016. Epigenetic programming alterations in alligators from environmentally contaminated lakes. Gen Comp Endocrinol 238:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst J, Andriantsoa R, Falckenhayn C, Hanna K, Stein Wet al. , 2018. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat Ecol Evol 2:567–573. [DOI] [PubMed] [Google Scholar]
- Gutekunst J, Maiakovska O, Hanna K, Provataris P, Horn Het al. , 2021. Phylogeographic reconstruction of the marbled crayfish origin. Commun Biol 4:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A, Schnabler A, Martens A, 2018. Phenology of overland dispersal in the invasive crayfish Faxonius immunis (Hagen) at the Upper Rhine River area. Knowl Manag Aquat Ecosyst 419:30. [Google Scholar]
- Holoch D, Moazed D, 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Kouba A, Buřič M, 2019. Morphometry, size at maturity, and fecundity of marbled crayfish Procambarus virginalis. Zool Anz 281:69–75. [Google Scholar]
- Huang Q, Ma C, Chen L, Luo D, Chen Ret al. , 2018. Mechanistic insights into the interaction between transcription factors and epigenetic modifications and the contribution to the development of obesity. Front Endocrinol 9:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, 2017. The evolutionary implications of epigenetic inheritance. Interface Focus 7:20160135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Raz G, 2009. Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Quart Rev Biol 84:131–176. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A, 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33:245–254. [DOI] [PubMed] [Google Scholar]
- Jimenez SA, Faulkes Z, 2010. Establishment and care of a colony of parthenogenetic marbled crayfish, Marmorkrebs. Invertebr Rearing 1:10–18. [Google Scholar]
- Johannes F, Schmitz RJ, 2019. Spontaneous epimutations in plants. New Phytol 221:1253–1259. [DOI] [PubMed] [Google Scholar]
- Jones PA, 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. [DOI] [PubMed] [Google Scholar]
- Jones JPG, Rasamy JR, Harvey A, Toon A, Oidtmann Bet al. , 2009. The perfect invader: A parthenogenic crayfish poses a new threat to Madagascar’s freshwater biodiversity. Biol Invasions 11:1475–1482. [Google Scholar]
- Kaldre K, Meženin A, Paaver T, Kawai T, 2016. A preliminary study on the tolerance of marble crayfish Procambarus fallax f. virginalis to low temperature in Nordic climate. In: Kawai T, Faulkes Z, Scholtz G, editors. Freshwater Crayfish: A Global Overview. Boca Raton (FL): CRC Press, 54–62. [Google Scholar]
- Kim J-M, Sasaki T, Ueda M, Sako K, Seki M, 2015. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci 6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba A, Lipták B, Kubec J, Bláha M, Veselý Let al. , 2021. Survival, growth, and reproduction: Comparison of marbled crayfish with four prominent crayfish invaders. Biology 10:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kribelbauer JF, Lu CJ, Rohs R, Mann RS, Bussemaker HJ, 2020. Toward a mechanistic understanding of DNA methylation readout by transcription factors. J Mol Biol 432:1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronholm I, Johannesson H, Ketola T, 2016. Epigenetic control of phenotypic plasticity in the filamentous fungus Neurospora crassa. G3 Genes Genomes Genet 6:4009–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand C, Andriantsoa R, Lichter P, Lyko F, 2021. Time-resolved, integrated analysis of clonal genome evolution in parthenogenetic animals and in cancer. bioRxiv: preprint. [Google Scholar]
- Lennartsson A, Ekwall K, 2009. Histone modification patterns and epigenetic codes. Biochim Biophys Acta 1790:863–868. [DOI] [PubMed] [Google Scholar]
- Li MZ, Xiao HM, He K, Li F, 2019. Progress and prospects of noncoding RNAs in insects. J Integr Agric 18:729–747. [Google Scholar]
- Liew YJ, Zoccola D, Li Y, Tambutté E, Venn AAet al. , 2018. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci Adv 4:eaar8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzmaier SM, Musseau C, Matern S, Jeschke JM, 2020. Trophic ecology of invasive marbled and spiny-cheek crayfish populations. Biol Invasions 22:3339–3356. [Google Scholar]
- Lipták B, Veselý L, Ercoli F, Bláha M, Buřič Met al. , 2019. Trophic role of marbled crayfish in a lentic freshwater ecosystem. Aquat Invasions 14:299–309. [Google Scholar]
- Liu J, He Z, 2020. Small DNA methylation, big player in plant abiotic stress responses and memory. Front Plant Sci 11:95603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SM, Storey KB, 2021. MicroRNA expression patterns in the brown fat of hibernating 13-lined ground squirrels. Genomics 113:769–781. [DOI] [PubMed] [Google Scholar]
- Lőkkös A, Müller T, Kovács K, Várkonyi L, Specziár Aet al. , 2016. The alien, parthenogenetic marbled crayfish (Decapoda: Cambaridae) is entering Kis-Balaton (Hungary), one of Europe’s most important wetland biotopes. Knowl Manag Aquat Ecosyst 417:16. [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR, 2017. How do lncRNAs regulate transcription? Sci Adv 3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, 2017. The marbled crayfish (Decapoda: Cambaridae) represents an independent new species. Zootaxa 4363:544–552. [DOI] [PubMed] [Google Scholar]
- Lyko F, 2018. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev Genet 19:81–92. [DOI] [PubMed] [Google Scholar]
- Maiakovska O, Andriantsoa R, Tönges S, Hanna K, Pârvulescu Let al. , 2021. Genome analysis of the monoclonal marbled crayfish reveals genetic separation over a short evolutionary timescale. Commun Biol 4:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM, 2014. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6:a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Dorn NJ, Kawai T, van der Heiden C, Scholtz G, 2010a. The enigmatic Marmorkrebs (marbled crayfish) is the parthenogenetic form of Procambarus fallax (Hagen, 1870). Contrib Zool 79:107–118. [Google Scholar]
- Martin P, Kohlmann K, Scholtz G, 2007. The parthenogenetic Marmorkrebs (marbled crayfish) produces genetically uniform offspring. Naturwiss 94:843–846. [DOI] [PubMed] [Google Scholar]
- Martin P, Shen H, Füllner G, Scholtz G, 2010b. The first record of the parthenogenetic Marmorkrebs (Decapoda, Astacida, Cambaridae) in the wild in Saxony (Germany) raises the question of its actual threat to European freshwater ecosystems. Aquat Invasions 5:397–403. [Google Scholar]
- Martin P, Thonagel S, Scholtz G, 2016. The parthenogenetic Marmorkrebs (Malacostraca: Decapoda: Cambaridae) is a triploid organism. J Zool Syst Evol Res 54:13–21. [Google Scholar]
- Massicotte R, Angers B, 2012. General-purpose genotype or how epigenetics extend the flexibility of a genotype. Genet Res Internat 2012:317175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaghaderi G, Hörandl E, 2016. The evolution of meiotic sex and its alternatives. Proc Biol Sci 283:20161221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MAJ, Shilatifard A, 2020. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat Genet 52:1271–1281. [DOI] [PubMed] [Google Scholar]
- Moutinho C, Esteller M, 2017. MicroRNAs and epigenetics. Adv Cancer Res 135:189–220. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Gao Y, Dominguez AA, Lei S, 2021. CRISPR technologies for precise epigenome editing. Nat Cell Biol 23:11–22. [DOI] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato Cet al. , 2017. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543:72–77. [DOI] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y, Peng C, 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pârvulescu L, Togor A, Lele S-F, Scheu S, Șinca Det al. , 2017. First established population of marbled crayfish Procambarus fallax (Hagen, 1870) f. virginalis (Decapoda, Cambaridae) in Romania. BioInvasions Rec 6:357–362. [Google Scholar]
- Pfennig DW, 2021. Phenotypic Plasticity and Evolution. Causes, Consequences, Controversies. Boca Raton (FL): CRC Press. [Google Scholar]
- Pfennig DW, Servedio MR, 2013. The role of transgenerational epigenetic inheritance in diversification and speciation. Non-Genet Inherit 1:17–26. [Google Scholar]
- Price SA, Friedman ST, Wainwright PC, 2015. How predation shaped fish: The impact of fin spines on body form evolution across teleosts. Proc R Soc B 282:20151428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD, Qvarnström A, Irwin DE, 2003. The role of phenotypic plasticity in driving genetic evolution. Proc R Soc Lond B 270:1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provataris P, Meusemann K, Niehuis O, Grath S, Misof B, 2018. Signatures of DNA methylation across insects suggest reduced DNA methylation levels in Holometabola. Genome Biol Evol 10:1185–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz G, Guzzardo PM, Olova N, Rosado Fantappié M, Rampp Met al. , 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci USA 110:8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran M, Jurkowska RZ, Jurkowski TP, 2018. Target specificity of mammalian DNA methylation and demethylation machinery. Org Biomol Chem 16:1419. [DOI] [PubMed] [Google Scholar]
- Richards CL, Alonso C, Becker C, Bossdorf O, Bucher Eet al. , 2017. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol Lett 20:1576–1590. [DOI] [PubMed] [Google Scholar]
- Schlichting CD, 1986. The evolution of phenotypic plasticity in plants. Ann Rev Ecol Syst 17:667–693. [Google Scholar]
- Scholtz G, Braband A, Tolley L, Reimann A, Mittmann Bet al. , 2003. Parthenogenesis in an outsider crayfish. Nature 421:806. [DOI] [PubMed] [Google Scholar]
- Schübeler D, 2015. Function and information content of DNA methylation. Nature 517:321–326. [DOI] [PubMed] [Google Scholar]
- Seitz R, Vilpoux K, Hopp U, Harzsch S, Maier G, 2005. Ontogeny of the Marmorkrebs (marbled crayfish): A parthenogenetic crayfish with unknown origin and phylogenetic position. J Exp Zool A 303:393–405. [DOI] [PubMed] [Google Scholar]
- Shi W, Chen X, Gao L, Xu CY, Ou Xet al. , 2019. Transient stability of epigenetic population differentiation in a clonal invader. Front Plant Sci 9:1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, 2014. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Nilsson EE, 2021. Role of environmentally induced epigenetic transgenerational inheritance in evolutionary biology: Unified evolution theory. Environ Epigenet 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, 2020. Phenotypic plasticity: From theory and genetics to current and future challenges. Genetics 215:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Ringrose L, 2014. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 15:340–356. [DOI] [PubMed] [Google Scholar]
- Svishcheva G, Babayan O, Lkhasaranov B, Tsendsuren B, Abdurasulov Aet al. , 2020. Microsatellite diversity and phylogenetic relationships among East Eurasian Bos taurus breeds with an emphasis on rare and ancient local cattle. Animals 10:1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, 1991. Phenotypic plasticity as a component of evolutionary change. Trends Ecol Evol 6:246–249. [DOI] [PubMed] [Google Scholar]
- Thorson JLM, Smithson M, Beck D, Sadler-Riggleman I, Nilsson Eet al. , 2017. Epigenetics and adaptive phenotypic variation between habitats in an asexual snail. Sci Rep 7:14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorson JLM, Smithson M, Sadler-Riggleman I, Beck D, Dybdahl Met al. , 2019. Regional epigenetic variation in asexual snail populations among urban and rural lakes. Environ Epigenet 5:dvz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollrian R, 1995. Predator-induced morphological defenses: Costs, life history shifts, and maternal effects in Daphnia pulex. Ecology 76:1691–1705. [Google Scholar]
- Tönges S, Masagounder K, Lenich F, Gutekunst J, Tönges Met al. , 2021a. Evaluating invasive marbled crayfish as a potential livestock for sustainable aquaculture. Front Ecol Evol 9:651981. [Google Scholar]
- Tönges S, Venkatesh G, Andriantsoa R, Hanna K, Gatzmann Fet al. , 2021b. Location-dependent DNA methylation signatures in a clonal invasive crayfish. Front Cell Dev Biol 9:794506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnakova N, Kubec J, Stara A, Zuskova E, Faggio Cet al. , 2022. Chronic toxicity of primary metabolites of chloroacetamide and glyphosate to early life stages of marbled crayfish Procambarus virginalis. Biology 11:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usio N, Azuma N, Sasaki S, Oka T, Inoue M, 2017. New record of Marmorkrebs from western Japan and its potential threats to freshwater ecosystems. Cancer 26:5–11. [Google Scholar]
- Verhoeven KJF, Preite V, 2014. Epigenetic variation in asexual reproducing organisms. Evolution 68:644–655. [DOI] [PubMed] [Google Scholar]
- Veselý L, Buřič M, Kouba A, 2015. Hardy exotics species in temperate zone: Can “warm water” crayfish invaders establish regardless of low temperatures? Sci Rep 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselý L, Ruokonen TJ, Weiperth A, Kubec J, Szajbert Bet al. , 2021. Trophic niches of three sympatric invasive crayfish of EU concern. Hydrobiologia 848:727–737. [Google Scholar]
- Vogt G, 2007. Exposure of the eggs to 17α-methyl testosterone reduced hatching success and growth and elicited teratogenic effects in postembryonic life stages of crayfish. Aquat Toxicol 85:291–296. [DOI] [PubMed] [Google Scholar]
- Vogt G, 2008. The marbled crayfish: A new model organism for research on development, epigenetics and evolutionary biology. J Zool 276:1–13. [Google Scholar]
- Vogt G, 2010. Suitability of the clonal marbled crayfish for biogerontological research: A review and perspective, with remarks on some further crustaceans. Biogerontol 11:643–669. [DOI] [PubMed] [Google Scholar]
- Vogt G, 2015. Bimodal annual reproduction pattern in laboratory-reared marbled crayfish. Invert Reprod Dev 59:218–223. [Google Scholar]
- Vogt G, 2017. Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: Insights from clonal, invasive, polyploid, and domesticated animals. Environ Epigenet 3:dvx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, 2018. Investigating the genetic and epigenetic basis of big biological questions with the parthenogenetic marbled crayfish: A review and perspectives. J Biosci 43:189–223. [PubMed] [Google Scholar]
- Vogt G, 2019a. Estimating the young evolutionary age of marbled crayfish from museum samples. J Nat Hist 53:2353–2363. [Google Scholar]
- Vogt G, 2019b. Functional cytology of the hepatopancreas of decapod crustaceans. J Morphol 280:1405–1444. [DOI] [PubMed] [Google Scholar]
- Vogt G, 2020a. Disentangling the environmentally induced and stochastic developmental components of phenotypic variation. In: Levine H, Jolly MK, Kulkarni P, Nanjundiah V, editors. Phenotypic Switching: Implications in Biology and Medicine. San Diego (CA): Academic Press, 207–251. [Google Scholar]
- Vogt G, 2020b. Biology, ecology, evolution, systematics and utilization of the parthenogenetic marbled crayfish Procambarus virginalis. In: Ribeiro FB, editor. Crayfish: Evolution, Habitat and Conservation Strategies. Hauppauge (NY): Nova Science Publishers, 137–227. [Google Scholar]
- Vogt G, 2021. Epigenetic variation in animal populations: Sources, extent, phenotypic implications, and ecological and evolutionary relevance. J Biosci 46:24. [PubMed] [Google Scholar]
- Vogt G, 2022a. Epigenetics and phenotypic plasticity in animals. In: Vaschetto LM, editor. Epigenetics, Development, Ecology and Evolution. Cham: Springer, 35–108. [Google Scholar]
- Vogt G, 2022b. Evolution, functions and dynamics of epigenetic mechanisms in animals. In: Tollefsbol T, editor. Handbook of Epigenetics: The New Molecular and Medical Genetics. 3rd edn. Cambridge: Academic Press, 521–549. [Google Scholar]
- Vogt G, 2022c. Studying phenotypic variation and DNA methylation across development, ecology and evolution in the clonal marbled crayfish: A paradigm for investigating epigenotype-phenotype relationships in macro-invertebrates. Sci Nat 109:16. [DOI] [PubMed] [Google Scholar]
- Vogt G, Dorn NJ, Pfeiffer M, Lukhaup C, Williams BWet al. , 2019. The dimension of biological change caused by autotriploidy: A meta-analysis with the triploid Procambarus virginalis and its diploid parent Procambarus fallax. Zool Anz 281:53–67. [Google Scholar]
- Vogt G, Falckenhayn C, Schrimpf A, Schmid K, Hanna Ket al. , 2015. The marbled crayfish as a paradigm for saltational speciation by autopolyploidy and parthenogenesis in animals. Biol Open 4:1583–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJet al. , 2008. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol 211:510–523. [DOI] [PubMed] [Google Scholar]
- Vogt G, Lukhaup C, Pfeiffer M, Dorn NJ, Williams BWet al. , 2018. Morphological and genetic characterization of the marbled crayfish, including a determination key. Zootaxa 4524:329–350. [DOI] [PubMed] [Google Scholar]
- Vogt G, Tolley L, Scholtz G, 2004. Life stages and reproductive components of the Marmorkrebs (marbled crayfish), the first parthenogenetic decapod crustacean. J Morphol 261:286–311. [DOI] [PubMed] [Google Scholar]
- Voigt S, Kost L, 2021. Differences in temperature-sensitive expression of PcG regulated genes among natural populations of Drosophila melanogaster. G3 Genes Genomes Genet 11:jkab237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojkovská R, Horká I, Tricarico E, Ďuriš Z, 2014. New record of the parthenogenetic marbled crayfish Procambarus fallax f. virginalis from Italy. Crustaceana 87:1386–1392. [Google Scholar]
- Weiperth A, Bláha M, Szajbert B, Seprős R, Bányai Zet al. , 2020. Hungary: A European hotspot of non-native crayfish biodiversity. Knowl Manag Aquat Ecosyst 421:43. [Google Scholar]
- West-Eberhard MJ, 1989. Phenotypic plasticity and the origins of diversity. Ann Rev Ecol Syst 20:249–278. [Google Scholar]
- Wu H, Zhang Y, 2017. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat Rev Genet 18:517–534. [DOI] [PubMed] [Google Scholar]
- Xu S, Pham T, Neupane S, 2020. Delivery methods for CRISPR/Cas9 gene editing in crustaceans. Mar Life Sci Technol 2:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Acar M, 2018. Mechanisms for the epigenetic inheritance of stress response in single cells. Curr Genet 64:1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Brown AP, Ji H, 2020. The emerging role of ten-eleven translocation 1 in epigenetic responses to environmental exposures. Epigenetics Insights 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]