Abstract
Mutations in Rhodopsin (RHO) gene commonly cause autosomal dominant retinitis pigmentosa (adRP) without effective therapeutic treatment so far. Compared with genomic DNA-targeting CRISPR-Cas9 system, Cas13 edits RNA for therapeutic applications, avoiding the risk of causing permanent changes in the genome. In particular, a compact and high-fidelity Cas13X (hfCas13X) recently has been developed to degrade targeted RNA with minimal collateral effects and could also be packaged in a single adeno-associated virus for efficient in vivo delivery. In this study, we engineered single-guide RNA for hfCas13X to specifically knock down human mutant Rhodopsin transcripts RHO-P23H with minimal effect on wild-type transcripts. Moreover, treatment with hfCas13X alleviated the adRP progression in both RHO-P23H overexpression-induced and humanized hRHOP23H/WT mouse models. Our study indicates the potential of hfCas13X in treating adRP caused by RHO mutations and other genetic diseases.
Keywords: MT: RNA/DNA editing, hfCas13X, rhodopsin, P23H mutation, retinitis pigmentosa, adRP
Graphical abstract
Yang and colleagues demonstrated that high-fidelity Cas13X (hfCas13X)-mediated specific degradation of Rhodopsin mutant transcripts, RHO-P23H, can delay photoreceptor death and retard the disease progression in a humanized hRHOP23H/WT mouse model of retinitis pigmentosa, providing an alternative for treatment of retinal degeneration caused by dominant mutations.
Introduction
Retinitis pigmentosa (RP) is a primary cause of inherited blindness characterized by the progressive loss of vision due to the degeneration of photoreceptors including rod and cone cells, affecting approximately 1 in 4,000 people worldwide.1 RP is genetically heterogeneous, showing autosomal recessive, autosomal dominant, and X-linked inheritance patterns. Rhodopsin (RHO) gene mutations are a prominent cause of autosomal dominant RP (adRP), accounting for 25%–30% of adRP cases. There are over 150 mutations in human RHO gene with the RHO-P23H mutation (c.68C>A; p.Pro23His) being the most frequent one.2,3,4 RHO is a G protein-coupled receptor protein with seven transmembrane domains located in the outer segments of rods, facilitating vision in dim light.5,6 The pathological mechanisms underlying RHO mutations are complex. It has been reported that RHO-P23H mutation induces gain-of-function pathological effects, leading to protein misfolding and aggregation, activating endoplasmic reticulum stress and cell death pathway, eventually resulting in retinal photoreceptor degeneration.7,8,9
Clinical observations suggest that patients with RHO-P23H-dominant mutation carrying a wild-type (WT) allele show milder pathological process, and their photoreceptors have the potential to be rescued, thereby alleviating retinal degeneration.10 Because of the dominant-negative mechanism, it is necessary to suppress the toxic protein instead of simply augmenting the expression of normal RHO gene. Various efforts have been made, such as mutant-independent interference coupled with replacement gene therapy,11,12,13,14 allele-specific knockout or knockdown,15,16,17 and pharmacological strategies to stabilize the functional or clear the toxic rhodopsin.18,19,20 However, there are currently no effective drugs available for RHO-P23H mutation-related adRP treatment.
Traditional gene editing based on the CRISPR-Cas9 system has recently been shown to confer therapeutic effect for several genetic diseases, including RHO-related adRP. However, the direct targeting of RHO-P23H allele by Cas9 showed heterogeneous disruption outcomes with modest editing efficiency in mouse retina.17,21 Notably, Cas9-mediated DNA cleavage has been found to randomly cause large DNA insertion, deletion, and even chromosome rearrangements, posing huge unpredictable safety risk for therapeutic use.22,23 In contrast, CRISPR-Cas13, a class 2 type VI RNA endonuclease, can bind and cleave single-stranded RNA guided by CRISPR RNA (crRNA) to avoid the risk of introducing permanent genomic DNA alterations, which would be a safe alternative for gene therapies.24,25,26,27 Recently, several studies have reported significant therapeutic effects of Cas13 on the treatment of COVID-19 or neurodegenerative diseases in mouse models.28,29,30,31 In particular, recent development of compact and high-fidelity Cas13X (hfCas13X) makes it safer and more suitable for therapeutic applications, as it keeps high on-target activity for RNA degradation with markedly lower collateral side effect. In addition, the compact size of hfCas13X facilitated its package in single adeno-associated virus (AAV) for efficient in vivo delivery.32,33,34 To investigate the therapeutic effect of hfCas13X, we herein used two different RHO-P23H mutation-related adRP mouse models via AAV-RHO(P23H) overexpression and genetically humanized hRHOP23H/WT knockin. We engineered sgRNAs for hfCas13X to specifically degrade human mutant Rhodopsin transcripts RHO-P23H without affecting WT transcripts. Moreover, administration of AAV-hfCas13X targeting human RHO-P23H significantly increased survival of photoreceptors and delayed adRP progression in mouse models. Our study indicated the therapeutic potential of hfCas13X in alleviating adRP and other genetic disorders.
Results
In vitro screening of engineered sgRNA for specifically targeting RHO-P23H mutant transcripts
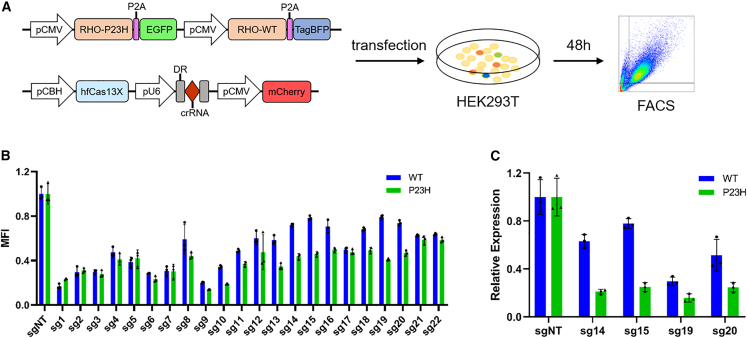
Cas13X-mediated RNA degradation can tolerate some mismatches in the single-guide RNA (sgRNA) without obviously affecting the knockdown efficiency.33 Therefore, we attempted to engineer the hfCas13X-based sgRNA to specifically knock down the human RHO-P23H mutant transcripts, while minimally affecting the WT form. We engineered a series of sgRNAs by substituting or inserting certain bases near the 3′ end adjacent to the scaffold (Table 1). In order to screen for the suitable sgRNAs in vitro, we constructed a single reporter vector encoding human RHO(P23H)-P2A-EGFP (green fluorescent protein) and RHO(WT)-P2A-TagBFP (blue fluorescent protein) simultaneously, and we co-transfected it with a vector expressing sgRNAs-hfCas13X-mCherry into HEK293T cells. After 48 h, we analyzed the mean fluorescence intensity (MFI) of EGFP and BFP by flow cytometry (Figure 1A). We found that some sgRNAs could efficiently reduce the RHO(P23H)-P2A-EGFP expression but had a weaker effect on RHO(WT)-P2A-TagBFP expression (Figure 1B). Furthermore, we conducted real-time quantitative PCR (qPCR) based on the expression of fluorescent tag (EGFP or TagBFP), and we confirmed that sg14 and sg15 were more suitable than other sgRNAs for further research (Figure 1C).
Table 1.
Engineered hfCas13X-based sgRNAs
 |
The blue letters “T”, C > A mutation targeting; the red letters, substitution; the green lowercase letters, insertion
Figure 1.
Screening of engineered hfCas13X-mediated sgRNAs in vitro
(A) Schematic diagram of strategy for experiments of sgRNA screening in vitro. FACS, fluorescence-activated cell sorting. (B) Mean fluorescence intensity (MFI) analysis of EGFP and TagBFP from mCherry + populations. MFI was relative to the sgNT group. (C) Relative degradation of RHO(WT)-TagBFP and RHO(P23H)-EGFP transcripts induced by hfCas13X with engineered sg14, sg15, sg19, and sg20 by qPCR. Gene expression was relative to the sgNT group.
Decrease of photoreceptor death by hfCas13X-mediated specific knockdown of human RHO-P23H transcripts in RHO-P23H overexpression-induced adRP mouse models
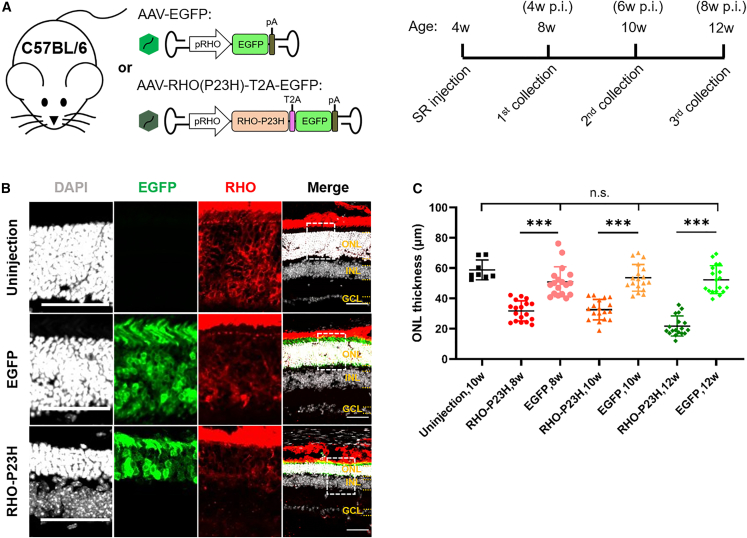
In light of the pathological mechanism of RHO-P23H-mediated adRP, it is possible to mimic the adRP symptoms by overexpressing the toxic RHO-P23H protein in normal retina, causing photoreceptor death and thinning of the outer nuclear layer (ONL). In order to restrict the RHO-P23H protein expression in photoreceptors, human RHO-specific promoter (pRHO) was used.35 Then, we constructed a vector expressing human RHO-P23H linked with EGFP by a T2A peptide, followed by AAV packaging. Meanwhile, EGFP expression only was used as the control. Afterward, we performed subretinal injection in 4-week-old C57BL/6 mice with AAV-EGFP or AAV-RHO(P23H)-T2A-EGFP, and we collected retina tissues at age of 8, 10, and 12 weeks, respectively (Figure 2A). After immunofluorescence (IF) staining, we found that from 8 weeks on, the ONL of retina expressing RHO-P23H became prominently thinner than those uninjected or injected with AAV-EGFP groups (Figures 2B, S1A, and S1B). This was also confirmed by the quantification of ONL thickness (Figure 2C). These results demonstrate that overexpression of human RHO-P23H in normal mouse retina causes photoreceptor death similar to the major phenotypes observed in human adRP.
Figure 2.
Photoreceptor degeneration by overexpression of human RHO-P23H in C57BL/6 mice
(A) Schematic diagram of strategy for experiments of human RHO-P23H overexpression in C57BL/6 mice. SR, subretinal; p.i., post injection. (B) Representative images of 8-week-old C57BL/6 retinas uninjected or injected with AAV-EGFP and AAV-RHO(P23H)-T2A-EGFP, respectively. Individual channels zoomed in from the dashed boxes in the corresponding merged images. Scale bar, 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (C) ONL thickness statistics of 8-, 10- and 12-week-old C57BL/6 retinas uninjected or injected with AAV-EGFP and AAV-RHO(P23H)-T2A-EGFP, respectively. ∗∗∗p < 0.001; n.s., not significant; one-way ANOVA test. Four retinas for uninjection group and six retinas for injection groups.
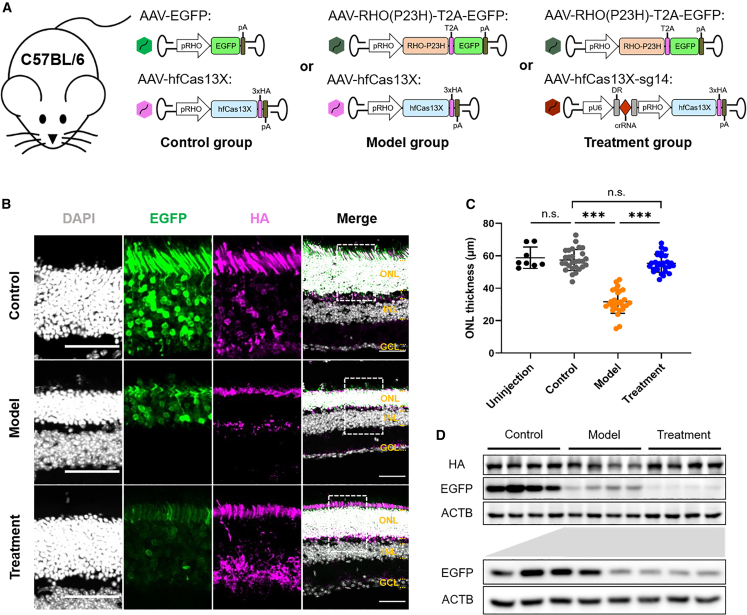
Next, based on adRP models induced by the AAV-RHO(P23H) overexpression, we attempted to evaluate the therapeutic efficacy for hfCas13X-mediated knockdown of human RHO-P23H in preventing photoreceptors degeneration. We generated AAV vector carrying pRHO-driven hfCas13X fused with 3×HA tag and U6-driven sg14. Meanwhile, the AAV vector only expressing hfCas13X was used as control. To minimize the immunogenicity and secondary damage to the retina, we chose to subretinally inject 4-week-old C57BL/6 mice with dual AAVs at the same time (Figure 3A): AAV-EGFP and AAV-hfCas13X (control group), AAV-RHO(P23H)-T2A-EGFP and AAV-hfCas13X (model group), or AAV-RHO(P23H)-T2A-EGFP and AAV-hfCas13X-sg14 (treatment group). At age of 12 weeks, retina tissues were collected and subjected to IF staining. We found that retinas from the treatment group with hfCas13X-sg14 injection had thicker ONL than the model group, remaining similar to the control group (Figures 3B and 3C). These observations suggest the protective role of hfCas13X-mediated targeting in photoreceptor degeneration.
Figure 3.
Decrease of photoreceptor death by specific knockdown of human RHO-P23H with hfCas13X in C57BL/6 mice
(A) Schematic diagram of strategy for experiments of human RHO-P23H-specific knockdown in C57BL/6 mice. (B) Representative images of 12-week-old C57BL/6 retinas injected with AAVs of control, model, and treatment groups, respectively. Individual channels zoomed in from the dashed boxes in the corresponding merged images. Scale bar, 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (C) ONL thickness statistics of 12-week-old C57BL/6 retinas uninjected or injected with AAVs of control, model, and treatment groups, respectively. ∗∗∗p < 0.001; n.s., not significant; one-way ANOVA test. Four retinas for uninjection group and seven retinas for injection groups. (D) Western blot of 8-week-old C57BL/6 retinas injected with AAVs of control, model, and treatment groups, respectively. The part below is western blot of retinas injected with AAVs of model and treatment groups, respectively. Each group presented four retinas.
To detect whether RHO-P23H expression was knocked down by hfCas13X, we performed western blot assay of 8-week-old mouse retinas. The results showed that the expression level of RHO-P23H (EGFP tag) in the treatment group was reduced compared to that of the model group (Figure 3D). Furthermore, in order to examine the knockdown effect of hfCas13X on WT RHO in vivo, 4-week-old C57BL/6 mice were subretinally injected with AAV-RHO(WT)-T2A-EGFP and AAV-hfCas13X (control [WT] group) or AAV-RHO(WT)-T2A-EGFP and AAV-hfCas13X-sg14 (treatment [WT] group) (Figure S2A). In the same way, western blot of 8-week-old mouse retinas showed that the expression level of RHO-WT (EGFP tag) was not reduced by hfCas13X-sg14 in the treatment (WT) group (Figure S2B). Overall, these results indicate that RHO-P23H can be specifically knocked down in vivo by hfCas13X-sg14 without obviously affecting WT RHO, and it decreased the photoreceptor death.
Treatment of adRP by hfCas13X in humanized hRHOP23H/WT mice models
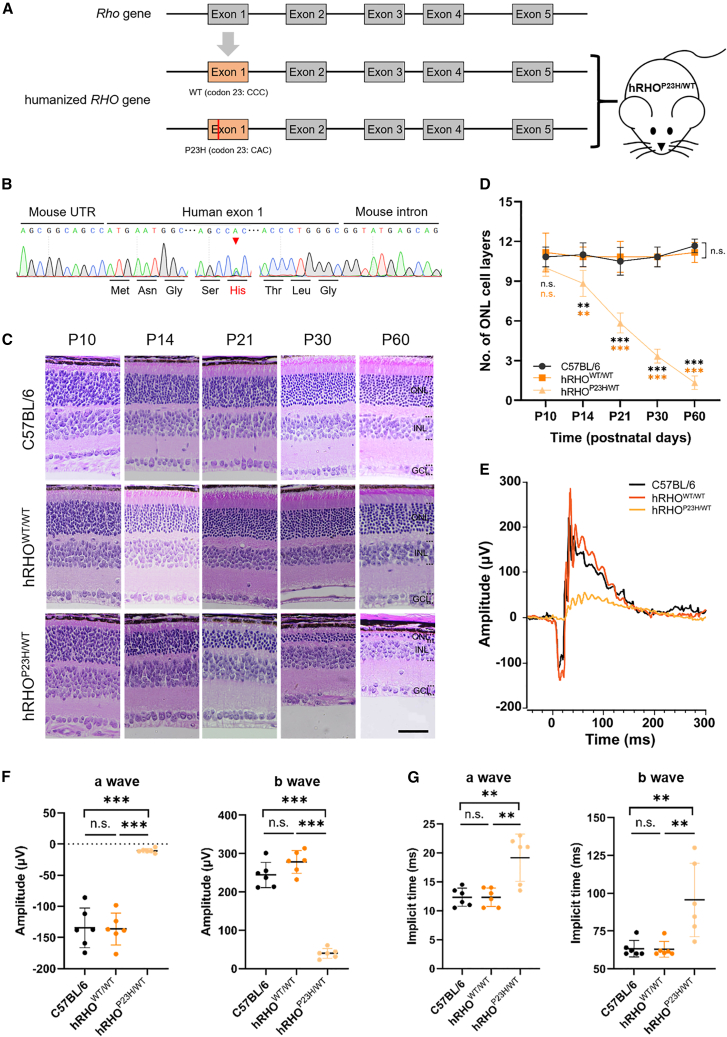
Humanized RHO-mutation transgenic mice have been widely used to study the pathogenesis and therapeutic strategies of adRP.11,36,37 To better simulate natural pathological condition, in this study, we generated a humanized adRP mouse model by replacing mouse Rho exon 1 with the corresponding human exon carrying the WT alleles (hRHOWT/WT) or a P23H mutation (hRHOP23H/WT) (Figures 4A and 4B). In contrast to WT C57BL/6 or hRHOWT/WT mice, the photoreceptors of the hRHOP23H/WT mouse model degenerated gradually over time after birth (Figures 4C and 4D). Moreover, we also performed scotopic electroretinography (ERG) recordings to detect the retinal function at the age of 1 month. The results showed that in hRHOP23H/WT mice, the retinal response decreased notably with reduced amplitudes and increased implicit times of a- and b-wave compared to WT mice (Figures 4E–4G). These results suggest the phenotypes of hRHOP23H/WT mouse model are similar to the major photoreceptor degeneration and dysfunction symptoms of human adRP.
Figure 4.
Construction and characterization of a humanized hRHOP23H/WT mouse model
(A) Schematic diagram of strategy for construction of humanized RHO-WT and -P23H knockin mice. The exon 1 of mouse Rho gene (in gray) was replaced with corresponding normal (WT) or c.68C>A mutation (P23H) exon (in orange) of human RHO gene. (B) Genotyping validation of hRHOP23H/WT mouse by sequencing. Red arrow indicates the double peaks of C and A base. (C) Representative images of HE staining of C57BL/6, hRHOWT/WT, and hRHOP23H/WT retinas at age of postnatal 10, 14, 21, 30, and 60 days, respectively. Scale bar, 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (D) ONL cell layer numbers statistics of C57BL/6, hRHOWT/WT, and hRHOP23H/WT retinas at age of postnatal 10, 14, 21, 30, and 60 days, respectively. ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., not significant; two-way ANOVA test. Three retinas for each group. (E) Representative ERG histograms of C57BL/6, hRHOWT/WT, and hRHOP23H/WT retinas at age of postnatal 30 days, respectively. (F) a- and b-wave amplitude statistics of C57BL/6, hRHOWT/WT, and hRHOP23H/WT retinas at age of postnatal 30 days, respectively. ∗∗∗p < 0.001; n.s., not significant; one-way ANOVA test. Six retinas for each group. (G) a- and b-wave implicit time statistics of C57BL/6, hRHOWT/WT, and hRHOP23H/WT retinas at age of postnatal 30 days, respectively. ∗∗p < 0.01; n.s., not significant; one-way ANOVA test. Six retinas for each group.
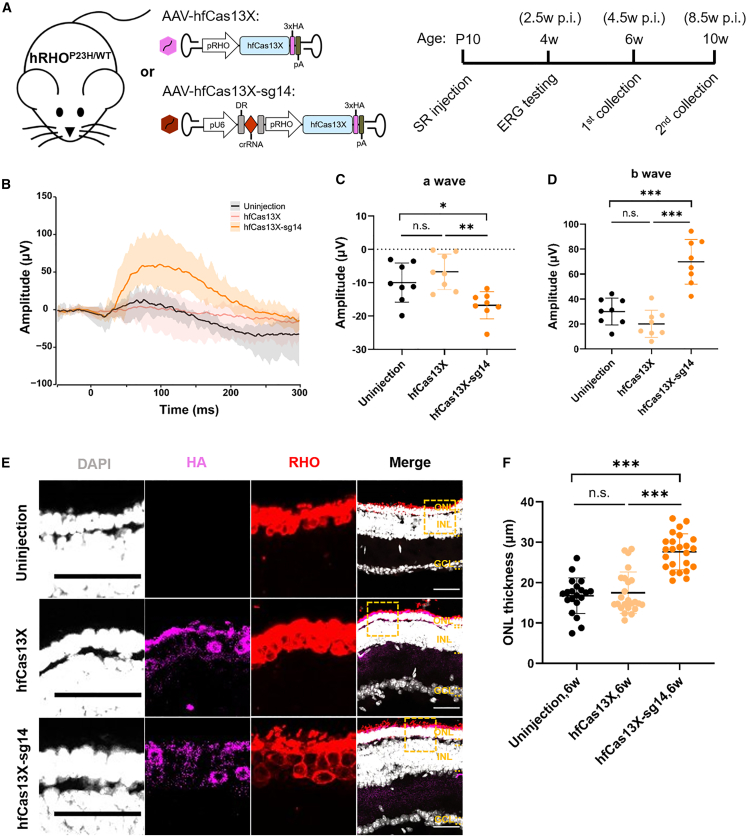
Then, we evaluated the therapeutic efficacy of hfCas13X targeting RHO-P23H in hRHOP23H/WT mice. To minimize the potential damage by AAV injection to newborn mice retinas, we opted for subretinal injection in postnatal 10-day-old (P10) hRHOP23H/WT mice using AAV-hfCas13X-sg14 or AAV-hfCas13X (Figure 5A). At the age of 4 weeks, we assessed the photoreceptor function through ERG test. The group injected with AAV-hfCas13X-sg14 showed a notable light response compared to the groups uninjected or injected with AAV-hfCas13X, with obvious a-wave and b-wave amplitudes (Figures 5B–5D). However, the implicit time did not show obvious changes after treatment (Figures S3A and S3B). At the age of 6 and 10 weeks, we collected the retina tissues to examine the ONL thickness. Our IF and quantification results showed that the ONL from AAV-hfCas13X-sg14-injected mice were thicker than that of uninjected or AAV-hfCas13X-injected mice, suggesting the protective role of AAV-hfCas13X-sg14 treatment for photoreceptor degeneration caused by RHO-P23H mutation (Figures 5E, 5F, S3C, and S3D).
Figure 5.
Treatment of adRP in hRHOP23H/WT mice with hfCas13X
(A) Schematic diagram of strategy for experiments of adRP treatment in hRHOP23H/WT mice. SR, subretinal; ERG, electroretinography; p.i., post injection. (B) Average ERG histograms of 4-week-old hRHOP23H/WT retinas uninjected or injected with AAV-hfCas13X and AAV-hfCas13X-sg14, respectively. Eight retinas for each group. (C) a-wave amplitude statistics of 4-week-old hRHOP23H/WT retinas uninjected or injected with AAV-hfCas13X and AAV-hfCas13X-sg14, respectively. ∗p < 0.05; ∗∗p < 0.01; n.s., not significant; one-way ANOVA test. Eight retinas for each group. (D) b-wave amplitude statistics of 4-week-old hRHOP23H/WT retinas uninjected or injected with AAV-hfCas13X and AAV-hfCas13X-sg14, respectively. ∗∗∗p < 0.001; n.s., not significant; one-way ANOVA test. Eight retinas for each group. (E) Representative images of 6-week-old hRHOP23H/WT retinas uninjected or injected with AAV-hfCas13X and AAV-hfCas13X-sg14, respectively. Individual channels zoomed in from the dashed boxes in the corresponding merged images. Scale bar, 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (F) ONL thickness statistics of 6-week-old hRHOP23H/WT retinas uninjected or injected with AAV-hfCas13X and AAV-hfCas13X-sg14, respectively. ∗∗∗p < 0.001; n.s., not significant; one-way ANOVA test. Six retinas for each group.
Furthermore, we also detected the knockdown effects of hfCas13X on RHO transcripts in humanized mouse models. Considering the difficulty of distinguishing between RHO-P23H and RHO-WT transcripts in heterozygous hRHOP23H/WT mice, we subretinally injected AAV-hfCas13X-sg14 or AAV-hfCas13X in hRHOP23H/WT and hRHOWT/WT mice aged P10, respectively. At P18, the retina tissues were then collected for qPCR, and the results showed that the expression level of total RHO in the hRHOP23H/WT mice injected with AAV-hfCas13X-sg14 was reduced notably, while the expression level in the hRHOWT/WT mice remained unchanged (Figures S3E and S3F). In general, these results demonstrated that hfCas13X-mediated specific knockdown of RHO-P23H transcripts could alleviate the progression of adRP in a humanized mouse model.
Discussion
In this study, we demonstrate that engineered hfCas13X-mediated specific degradation of RHO-P23H transcripts delays photoreceptor death and retards the retina degeneration in vivo. Our findings provide a promising alternative for treating adRP caused by RHO mutations.
Previous studies have reported that genome DNA editing with CRISPR-Cas9 in an allele-dependent or -independent manner can treat RHO mutation-mediated adRP in animal models.16,17,38 EDIT-103, a dual AAV-CRISPR-Cas9-based “knockout and replace” therapeutic strategy for RHO-adRP, has achieved remarkable results in mouse models and non-human primates.39 However, the inherent risk of DNA break hinders its clinical applications. In addition, RNA interference (RNAi) strategy is also used to knock down the RHO-P23H and -WT mRNA indiscriminately, coupled with complement of functional RHO.13,14,40 But, the balance of expression between RNAi and RHO should be well controlled, because of the possible toxicity of overloaded RHO protein in photoreceptors. Recently, a meganuclease Rho1-2 has also been reported to effectively rejuvenate rod structure and function in pig models of adRP with an expanded therapeutic window to late stage.41,42
Our study highlights the potential of using the RNA editing tool, CRISPR-Cas13, to specifically knock down single-base mutant transcripts for treating dominant genetic disorders. The therapeutic efficacy is more sustainable by AAV delivery compared to siRNA or ASO requiring multiple dosing, but still with suboptimal therapeutic effect in hRHOP23H/WT mice, including the unimproved implicit time of ERG and slow degeneration of photoreceptors over time. The reasons may be the limitation of single-site injections in mice, as well as the insufficient dose of AAV and compromised efficiency of on-target cleavage. In this study, for the two kinds of transcripts, RHO-WT and -P23H, with only one base difference, some mismatches were introduced into the hfCas13X-based sgRNAs to improve the specificity of on-target binding. This certainly will compromise the knockdown efficiency, and the photoreceptors will be in a state of slow degeneration with remaining mutant proteins. Further research is required to optimize the administration method and given dosage. Moreover, dual AAV therapy with mutation-independent knockdown of RHO plus overexpression of normal RHO may be another promising therapeutic approach.
More than 150 mutations in RHO gene cause adRP, but CRISPR-Cas13-mediated mutation-specific knockdown can target only one mutation with a single sgRNA, which may limit the scope of clinical application. In this study, the WT and P23H-mutation isoforms of RHO are nearly identical in sequence, differing by only one base (C > A mutation). Nevertheless, as shown in Figure 1, engineered sgRNAs with different designs have varying knockdown efficiency against the RHO-P23H and -WT transcripts, which may indicate the suboptimal on-target effects of hfCas13X. On the other hand, even if there is one different base in the targeted transcripts, the designed sgRNAs can target the better match with higher efficiency, which also indicates the high specificity of hfCas13X. Long-term examination of adRP mice treated by hfCas13X should be carried out in the future to check potential off-target effects. Moreover, further engineering of high-fidelity Cas13X system would also be needed to make this treatment safer and more effective.
Materials and methods
Cell culture, vector transfection, and flow cytometry
HEK293T cell lines were obtained from the cell bank of Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Chinese Academy of Sciences (CAS). They were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1× penicillin/streptomycin (100 U/ml penicillin and 100 μg/mL streptomycin, Gibco) in an incubator at 37°C and 5% CO2. Human rhodopsin gene (RHO) was obtained by joint PCR products of exon fragments from HEK293T genome. P23H mutation (RHO-P23H) was introduced by special primer design. A total 1 μg vectors were transiently transfected with polyethylenimine (PEI) into cultured cells in 24-well clusters (vector:PEI = 1:3). The vectors expressing RHO-WT/RHO-P23H were co-transfected with vectors expressing hfCas13X-sgRNAs (molar ratio 1:1.5). 48 h after transfection, cells were harvested for flow cytometry by BD LSRFortessa X-20 or fluorescence-activated cell sorting (FACS) by BD FACS Aria III. Flow cytometry results were analyzed with FlowJo v10, and mCherry-positive cells were used as input for MFI analysis.
RNA extraction and real-time quantitative PCR (qPCR)
For in vitro screening, 48 h after vector transfection, mCherry-positive cells (top 20%) were collected by FACS. For retina tissues, the EGFP-expressing regions were dissected under a stereo fluorescence microscope and then were fully grinded in TRIzol Reagent (Ambion). RNA was extracted with TRIzol. cDNA was obtained with HiScript Q RT SuperMix for qPCR Kit (Vazyme). Quantitative PCR (qPCR) was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) and LightCycler 480 II (Roche). Results were obtained after normalization to the expression level of the housekeeping gene β-actin.
Adeno-associated virus package and preparation
AAVs used in this study were PHP.eB serotype, in consideration of its extremely efficient infection of photoreceptors in mouse retina.43 AAVs were packaged in the Gene Editing Core Facility in the Institute of Neuroscience (ION), CAS. Viral particles of AAV-PHP.eB were packaged in HEK293T cells co-transfected with the other two plasmids: pAAV-Rep-Cap and pAAV-Helper. Viral particles were harvested and purified with a heparin column (GE Healthcare Biosciences). Then, viral particles were concentrated with an Ultra-4 centrifugal filter unit (Amicon, 100,000 molecular weight cutoff) to reach the titer >1E13 vg/ml quantified by qPCR.
Mice and AAV subretinal injection
C57BL/6 mice were obtained from Vital River Laboratory Animal Technology Co. hRHOP23H/WT mice with C57BL/6 background were obtained by crossing humanized hRHOWT/WT and humanized hRHOP23H/P23H mice from HuidaGene Therapeutics Co., Shanghai, China. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of HuidaGene Therapeutics Co.
Mice were anesthetized with a mixture of xylazine (10 mg/kg) and Zoletil (60 mg/kg, equal mixture of teletamine and zolazepam) by intraperitoneal injection. For subretinal injection, an incision was made using a 35G needle followed by injection using a Hamilton syringe. AAVs were injected into the subretinal space bilaterally. After injection, antibiotics eye ointment was applied to prevent infection. In C57BL/6 mice, AAV-RHO(P23H)-T2A-EGFP was injected for simulating the adRP models and injected together with AAV-hfCas13X-sg14 for treatment in vivo. In hRHOP23H/WT mice, AAV-hfCas13X-sg14 was injected for treatment and AAV-hfCas13X was as control. AAV delivery is summarized (Table 2).
Table 2.
Summary of AAV injection
| Figures | Groups | Mice genotype, age | AAV vectors | Titer (vg/ml) | Volume (μL) | Collection |
|---|---|---|---|---|---|---|
| Figure 2 | EGFP | C57BL/6, 4 w | ITR-pRHO-EGFP-polyA-ITR | 1.0E+12 | 1.0 | 8 w, 10 w, and 12 w (for IF) |
| RHO-P23H | C57BL/6, 4 w | ITR-pRHO-RHO(P23H)-T2A-EGFP-polyA-ITR | 1.0E+12 | 1.0 | ||
| Figure 3 | control | C57BL/6, 4 w | ITR-pRHO-EGFP-polyA-ITR | 1.0E+12 | 1.0 | 8 w (for WB), 12 w (for IF) |
| ITR-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | |||||
| model | C57BL/6, 4 w | ITR-pRHO-RHO(P23H)-T2A-EGFP-polyA-ITR | 1.0E+12 | 1.0 | ||
| ITR-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | |||||
| treatment | C57BL/6, 4 w | ITR-pRHO-RHO(P23H)-T2A-EGFP-polyA-ITR | 1.0E+12 | 1.0 | ||
| ITR-U6-sg14-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | |||||
| Figure S2 | control (WT) | C57BL/6, 4 w | ITR-pRHO-RHO(WT)-T2A-EGFP-polyA-ITR | 1.0E+12 | 1.0 | 8 w (for WB) |
| ITR-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | |||||
| treatment (WT) | C57BL/6, 4 w | ITR-pRHO-RHO(WT)-T2A-EGFP-polyA-ITR | 1.0E+12 | 1.0 | ||
| ITR-U6-sg14-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | |||||
| Figure 5 | hfCas13X | hRHOP23H/WT, P10 | ITR-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | 0.5 | 4 w (for ERG), 6 w and 10 w (for IF) |
| hfCas13X-sg14 | hRHOP23H/WT, P10 | ITR-U6-sg14-pRHO-hfCas13X-3×HA-polyA-ITR | 5.0E+12 | 0.5 |
4 w, 4 weeks old; P10, postnatal 10 days old; AAV, adeno-associated virus; ITR, inverted terminal repeat sequence; RHO, rhodopsin; pRHO, RHO promoter; WT, wild type; vg, virus genome; WB, western blot; ERG, electroretinography; IF, immunofluorescence.
Western blot of retina tissues
The whole retinas were isolated completely and ground fully in RIPA buffer containing proteinase inhibitor, and then the lysis was incubated at 95°C for 15 min with 1× SDS-PAGE loading buffer. The primary antibodies were as follows: anti-HA tag (1:1,000, CST, 3724), anti-EGFP (1:2000, Invitrogen, A11122), and anti-β actin (1:10,000, proteintech, HRP-60008). HRP-conjunction secondary antibodies (Beyotime) and Chemiluminescent HRP Substrate (Vazyme) were used for the detection. Images were analyzed using ImageJ.
Hematoxylin and eosin staining
Mouse eyeballs were collected after being perfused transcardially with PBS, followed by dehydration with gradient concentrations of ethanol. The tissues were sequentially soaked in xylene and paraffin. Sections were performed by a microtome at the thickness of 4 μm (RM2255, Leica). Tissue sections were unfolded and attached to slides and dried for 90 min at 65°C incubator, followed by dewaxing with xylene and gradient concentrations of ethanol. Tissue sections were stained with hematoxylin and rinsed with tap water and then stained with eosin and rinsed with tap water, followed by rinsing with ethanol and xylene.
Immunofluorescent staining and imaging
Mouse eyeballs were collected after being perfused transcardially with PBS and following 4% paraformaldehyde (PFA). Then, the tissues were post-fixed for an hour in 4% PFA at room temperature after removing the cornea, followed by dissection and dehydration overnight at 4°C within 30% (w/v) sucrose in PBS and embedded in O.C.T. compound (Sakura). Sections were performed by a cryostat at the thickness of 20 μm (HM525 NX, Thermo Fisher Scientific).
Before staining, sections were dried for 1 h in 65°C incubator and then were washed in PBS three times for 5 min. Antigen recovery was performed using heated sodium citrate for 20 min. After washing with PBS, the sections were incubated with blocking buffer containing 10% goat serum and 0.1% Triton X-100 in PBS for 1 h at room temperature, followed by incubation at 4°C overnight with primary antibodies diluted in dilution buffer (Beyotime). Sections were then washed three times for 5 min with PBS before incubation in secondary antibodies at 4°C overnight in the dark. Sections were washed three times for 5 min with PBS containing 2% Tween 20 and then mounted with Fluoromount-G (SouthernBiotech). Cell nuclei were counterstained with DAPI (1:2,000, Beyotime). Primary antibodies used were mouse anti-rhodopsin (1:1,000, Millipore, MAB5356) and rabbit anti-HA tag (1:1,000, CST, 3724). Secondary antibodies used were Fluorescein (FITC) AffiniPure goat anti-mouse/rabbit IgG (H + L) (1:1,000, Jackson ImmunoResearch, 111-095-003/115-095-003) and Cy3 AffiniPure goat anti-mouse/rabbit IgG (H + L) (1:1,000, Jackson ImmunoResearch, 115-165-003/111-165-003).
Confocal images were captured using an Olympus FV3000 microscope under a 20× objective lens and analyzed with software ImageJ. To obtain statistics on ONL thickness, one to two sections from each retina were taken as images, and two different sites were measured on each image. The images were scrambled, and the method was consistent for measurement.
Scotopic electroretinography
Before testing, the mice were adapted to darkness more than 5 h. Under dark red light, we dilated pupils of mice with eye drops of 1.0% tropicamide. Local anesthesia of mouse cornea was with eye drops of 5% proparacaine. Mice flash ERGs were recorded by using Diagnosys Celeris next-generation rodent ERG testing. Before testing, the mice’s eyes were cleaned with saline. The visual stimuli for the scotopic ERGs were white-light (6,500 k) flash of 3.00 cd s/m2 (15-s delay, 10 repeats). ERG amplitudes and peak times were measured and analyzed by supporting software. In detail, the amplitude of a-wave was measured from the baseline to the negative trough of the a-wave, and the amplitude of b-wave was measured from the trough of the a-wave to the following peak of the b-wave. The implicit time of a-wave was from flash onset to the trough of the a-wave, and the implicit time of b-wave was from flash onset to the peak of the b-wave.
Data statistics
The data are presented as mean ± standard deviation (SD). For data statistics consisting of more than two groups, varying in a single factor, one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test were used. Software GraphPad Prism 8.0 was used for statistical analysis and was also used to draw figures. Differences in means were considered statistically significant when they reached p value less than 0.05. Significance levels are ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data and code availability
All the data can be accessed online.
Acknowledgments
We thank the members from H. Yang lab for the help of experiments and discussions, G. Geng from the Gene Editing Core Facility of ION for AAV preparation, and Y. Wang, Y. Zhang, and Q. Hu from the Optical Imaging facility of ION for capturing images. This work was supported by the National Science and Technology Innovation 2030 Major Program (2021ZD0200900), Chinese National Science and Technology major project R&D Program of China (2017YFC1001302 and 2018YFC2000101), Strategic Priority Research Program of the Chinese Academy of Science (XDB32060000), National Natural Science Foundation of China (31871502, 31901047, 82021001 and 31922048), Basic Frontier Scientific Research Program of Chinese Academy of Sciences From 0 to 1 original innovation project (ZDBS-LY-SM001), Lingang Laboratory (LG202106-01-02), International Partnership Program of Chinese Academy of Sciences (153D31KYSB20170059), Project of Shanghai Municipal Science and Technology Commission (20MC1920400), and Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2021-AGIS to E.Z.).
Author contributions
Z.Y., E.Z., and H.Y. conceived the project. Z.Y. and L.L. designed and conducted the experiments assisted by Y.Y., L.C., H.Z., S.Z., Q.X., and X.W. Z.Y., E.Z., J.W., C.X., and H.Y. analyzed the data and wrote the manuscript. E.Z. and H.Y. supervised the project and acquired the funds.
Declaration of interests
H.Y. is a founder of HuidaGene Therapeutics Co., Ltd., Shanghai.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.08.002.
Contributor Information
Erwei Zuo, Email: zuoerwei@caas.cn.
Chunlong Xu, Email: xucl@lglab.ac.cn.
Jihong Wu, Email: jihongwu@fudan.edu.cn.
Hui Yang, Email: huiyang@huidagene.com.
Supplemental information
References
- 1.Tsang S.H., Sharma T. Retinitis Pigmentosa (Non-syndromic) Adv. Exp. Med. Biol. 2018;1085:125–130. doi: 10.1007/978-3-319-95046-4_25. [DOI] [PubMed] [Google Scholar]
- 2.Dryja T.P., McGee T.L., Reichel E., Hahn L.B., Cowley G.S., Yandell D.W., Sandberg M.A., Berson E.L. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 3.Dryja T.P., McGee T.L., Hahn L.B., Cowley G.S., Olsson J.E., Reichel E., Sandberg M.A., Berson E.L. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N. Engl. J. Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 4.Athanasiou D., Aguila M., Bellingham J., Li W., McCulley C., Reeves P.J., Cheetham M.E. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 2018;62:1–23. doi: 10.1016/j.preteyeres.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathans J., Hogness D.S. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc. Natl. Acad. Sci. USA. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathans J., Piantanida T.P., Eddy R.L., Shows T.B., Hogness D.S. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 7.Illing M.E., Rajan R.S., Bence N.F., Kopito R.R. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J. Biol. Chem. 2002;277:34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- 8.Tam B.M., Moritz O.L. Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2006;47:3234–3241. doi: 10.1167/iovs.06-0213. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson S.G., Kemp C.M., Sung C.H., Nathans J. Retinal function and rhodopsin levels in autosomal dominant retinitis pigmentosa with rhodopsin mutations. Am. J. Ophthalmol. 1991;112:256–271. doi: 10.1016/s0002-9394(14)76726-1. [DOI] [PubMed] [Google Scholar]
- 11.Chadderton N., Millington-Ward S., Palfi A., O'Reilly M., Tuohy G., Humphries M.M., Li T., Humphries P., Kenna P.F., Farrar G.J. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol. Ther. 2009;17:593–599. doi: 10.1038/mt.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cideciyan A.V., Sudharsan R., Dufour V.L., Massengill M.T., Iwabe S., Swider M., Lisi B., Sumaroka A., Marinho L.F., Appelbaum T., et al. Mutation-independent rhodopsin gene therapy by knockdown and replacement with a single AAV vector. Proc. Natl. Acad. Sci. USA. 2018;115:E8547–E8556. doi: 10.1073/pnas.1805055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlans H.O., McClements M.E., Barnard A.R., Martinez-Fernandez de la Camara C., MacLaren R.E. Mirtron-mediated RNA knockdown/replacement therapy for the treatment of dominant retinitis pigmentosa. Nat. Commun. 2021;12:4934. doi: 10.1038/s41467-021-25204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed C.M., Massengill M.T., Ildefonso C.J., Jalligampala A., Zhu P., Li H., Patel A.P., McCall M.A., Lewin A.S. Binocular benefit following monocular subretinal AAV injection in a mouse model of autosomal dominant retinitis pigmentosa (adRP) Vis. Res. 2023;206:108189. doi: 10.1016/j.visres.2023.108189. [DOI] [PubMed] [Google Scholar]
- 15.Murray S.F., Jazayeri A., Matthes M.T., Yasumura D., Yang H., Peralta R., Watt A., Freier S., Hung G., Adamson P.S., et al. Allele-Specific Inhibition of Rhodopsin With an Antisense Oligonucleotide Slows Photoreceptor Cell Degeneration. Invest. Ophthalmol. Vis. Sci. 2015;56:6362–6375. doi: 10.1167/iovs.15-16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latella M.C., Di Salvo M.T., Cocchiarella F., Benati D., Grisendi G., Comitato A., Marigo V., Recchia A. In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina. Mol. Ther. Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannelli S.G., Luoni M., Castoldi V., Massimino L., Cabassi T., Angeloni D., Demontis G.C., Leocani L., Andreazzoli M., Broccoli V. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 2018;27:761–779. doi: 10.1093/hmg/ddx438. [DOI] [PubMed] [Google Scholar]
- 18.Aguilà M., Bellingham J., Athanasiou D., Bevilacqua D., Duran Y., Maswood R., Parfitt D.A., Iwawaki T., Spyrou G., Smith A.J., et al. AAV-mediated ERdj5 overexpression protects against P23H rhodopsin toxicity. Hum. Mol. Genet. 2020;29:1310–1318. doi: 10.1093/hmg/ddaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Lian L., Liu B., Chen Y., Yang J., Jian S., Zhou J., Xu Y., Ma X., Qu J., Hou L. KIT ligand protects against both light-induced and genetic photoreceptor degeneration. Elife. 2020;9:e51698. doi: 10.7554/eLife.51698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Feng B., Vats A., Tang H., Seibel W., Swaroop M., Tawa G., Zheng W., Byrne L., Schurdak M., Chen Y. Pharmacological clearance of misfolded rhodopsin for the treatment of RHO-associated retinitis pigmentosa. Faseb. J. 2020;34:10146–10167. doi: 10.1096/fj.202000282R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., Kleinstiver B.P., Leon M.Y., Prew M.S., Navarro-Gomez D., Greenwald S.H., Pierce E.A., Joung J.K., Liu Q. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 2018;1:55–64. doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeballos C.M., Gaj T. Next-Generation CRISPR Technologies and Their Applications in Gene and Cell Therapy. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lotfi M., Rezaei N. CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomed. Pharmacother. 2020;131:110738. doi: 10.1016/j.biopha.2020.110738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C., Hu X., Tang C., Liu W., Wang S., Zhou Y., Zhao Q., Bo Q., Shi L., Sun X., et al. CasRx-mediated RNA targeting prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Natl. Sci. Rev. 2020;7:835–837. doi: 10.1093/nsr/nwaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morelli K.H., Wu Q., Gosztyla M.L., Liu H., Yao M., Zhang C., Chen J., Marina R.J., Lee K., Jones K.L., et al. An RNA-targeting CRISPR-Cas13d system alleviates disease-related phenotypes in Huntington's disease models. Nat. Neurosci. 2023;26:27–38. doi: 10.1038/s41593-022-01207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Shen Z., Liu Y., Yan Z., Liu Y., Lin X., Tang J., Lv R., Geng G., Xiong Z.Q., et al. A high-fidelity RNA-targeting Cas13 restores paternal Ube3a expression and improves motor functions in Angelman Syndrome mice. Mol. Ther. 2023;31:2286–2295. doi: 10.1016/j.ymthe.2023.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y., et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods. 2021;18:499–506. doi: 10.1038/s41592-021-01124-4. [DOI] [PubMed] [Google Scholar]
- 34.Tong H., Huang J., Xiao Q., He B., Dong X., Liu Y., Yang X., Han D., Wang Z., Wang X., et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 2023;41:108–119. doi: 10.1038/s41587-022-01419-7. [DOI] [PubMed] [Google Scholar]
- 35.Allocca M., Mussolino C., Garcia-Hoyos M., Sanges D., Iodice C., Petrillo M., Vandenberghe L.H., Wilson J.M., Marigo V., Surace E.M., Auricchio A. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J. Virol. 2007;81:11372–11380. doi: 10.1128/JVI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roof D.J., Adamian M., Hayes A. Rhodopsin accumulation at abnormal sites in retinas of mice with a human P23H rhodopsin transgene. Invest. Ophthalmol. Vis. Sci. 1994;35:4049–4062. [PubMed] [Google Scholar]
- 37.McNally N., Kenna P., Humphries M.M., Hobson A.H., Khan N.W., Bush R.A., Sieving P.A., Humphries P., Farrar G.J. Structural and functional rescue of murine rod photoreceptors by human rhodopsin transgene. Hum. Mol. Genet. 1999;8:1309–1312. doi: 10.1093/hmg/8.7.1309. [DOI] [PubMed] [Google Scholar]
- 38.Wu W.H., Tsai Y.T., Huang I.W., Cheng C.H., Hsu C.W., Cui X., Ryu J., Quinn P.M.J., Caruso S.M., Lin C.S., Tsang S.H. CRISPR genome surgery in a novel humanized model for autosomal dominant retinitis pigmentosa. Mol. Ther. 2022;30:1407–1420. doi: 10.1016/j.ymthe.2022.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C.H., Wolf P., Dong R., Huang Y., Tabbaa D., Marco E., Duke B., Pinilla A., Pant A., D’Souza R., et al. ASGCT Annual Meeting; 2022. A Mutation-Independent CRISPR/Cas9-Based ‘Knockout and Replace’ Strategy to Treat Rhodopsin-Associated Autosomal Dominant Retinitis Pigmentosa (RHO-adRP) [Google Scholar]
- 40.Mao H., Gorbatyuk M.S., Hauswirth W.W., Lewin A.S. Gene delivery of wild-type rhodopsin rescues retinal function in an autosomal dominant retinitis pigmentosa mouse model. Adv. Exp. Med. Biol. 2012;723:199–205. doi: 10.1007/978-1-4614-0631-0_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jalligampala A., Noel J., Fransen J.W., Wang W., Jabbar M., Hasan N., Pangeni G., Sahu B., Lewis W., Smith J., et al. Rho 1-2 meganuclease, an allele-specific gene-editing therapy, rejuvenates rod photoreceptor structure and function in a pig model of autosomal dominant Retinitis Pigmentosa (adRP) Invest. Ophthalmol. Vis. Sci. 2021;62:1478. [Google Scholar]
- 42.Jalligampala A., Noel J.M., Jacobs O.N., Bradley A.L., Wang W., Jabbar M.H., Hasan N., Prestigiacomo J., Smith J., Bartsevich V., et al. Successful late-stage disease treatment of P23H human RHO (hRHO) using ARCUS nuclease gene editing in a pig model of Autosomal Dominant Retinitis Pigmentosa (adRP) Invest. Ophthalmol. Vis. Sci. 2022;63:1107. [Google Scholar]
- 43.Palfi A., Chadderton N., Millington-Ward S., Post I., Humphries P., Kenna P.F., Farrar G.J. AAV-PHP.eB transduces both the inner and outer retina with high efficacy in mice. Mol. Ther. Methods Clin. Dev. 2022;25:236–249. doi: 10.1016/j.omtm.2022.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data can be accessed online.