Summary
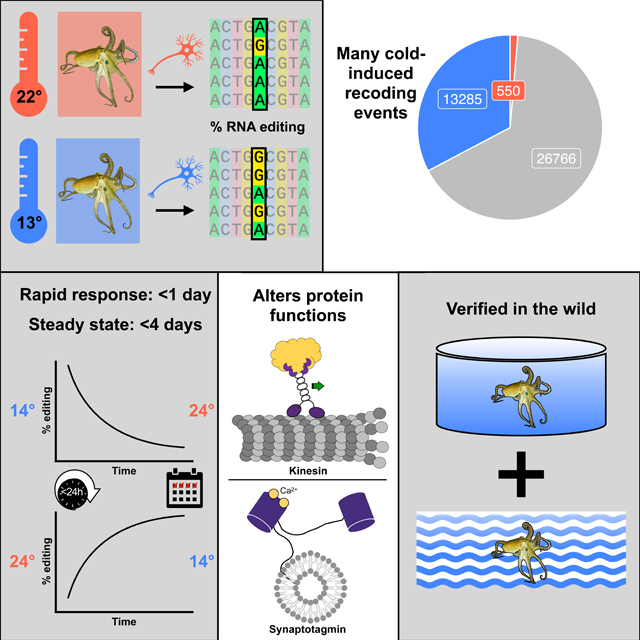
In poikilotherms, temperature changes challenge the integration of physiological function. Within the complex nervous systems of the behaviorally sophisticated coleoid cephalopods, these problems are substantial. RNA editing by adenosine deamination is a well-positioned mechanism for environmental acclimation. We report that the neural proteome of Octopus bimaculoides undergoes massive reconfigurations via RNA editing following a temperature challenge. Over 13,000 codons are affected, and many alter proteins that are vital for neural processes. For two highly temperature sensitive examples, recoding tunes protein function. For synaptotagmin, a key component of Ca2+-dependent neurotransmitter release, crystal structures and supporting experiments show that editing alters Ca2+ binding. For kinesin-1, a motor protein driving axonal transport, editing regulates transport velocity down microtubules. Seasonal sampling of wild-caught specimens indicates that temperature-dependent editing occurs in the field as well. These data show that A-to-I editing tunes neurophysiological function in response to temperature in octopus and likely other coleoids.
Keywords: ADAR, cephalopod, kinesin, synaptotagmin, acclimation, RNA editing, temperature
In brief:
Octopuses utilize RNA editing to rapidly respond to environmental temperature changes by altering protein function.
Graphical Abstract
Introduction
The temperatures that marine organisms experience can vary drastically both spatially and temporally, due to environmental factors such as tides, thermoclines, and seasons. Because of the high thermal conductivity of water, these changes pose physiological challenges to poikilotherms, particularly in their nervous systems where a variety of molecular and physiological processes must be properly integrated. Excitability provides a good example, where the resting membrane potential and the individual components of the action potential can have different temperature dependencies1–11. To underscore the challenges associated with integrating these complex processes, even modest acute temperature changes can result in death, brought about by a failure of the nervous system12. Accordingly, molecular and physiological temperature acclimation is a key driver of organismal success.
Due to its transient nature, genetic information within mRNA provides an ideal target for acclimation. Many studies have identified changes in RNA expression, localization, or splicing in response to temperature (reviewed by Somero13). RNA editing by adenosine deamination provides a potentially powerful, highly specific alternative mechanism for acclimation because it can directly alter what a messenger RNA encodes. Catalyzed by the ADAR (Adenosine Deaminases that Act on RNA) family of enzymes, specific adenosines are converted to inosine, a mimic for guanosine during translation and other biological processes14,15. When editing occurs at a non-synonymous position of a codon within an mRNA, the codon is recoded to another amino acid. Thus, editing has the potential to change amino acids both spatially and temporally in response to environmental change. Unlike changes within DNA, RNA edits are not binary, and can occur with variable penetrance across the population of RNAs. Because of its transient, specific and highly versatile ability to alter genetic information, RNA editing is well-positioned as a mechanism for acclimation. Few data, however, support the idea that it is used for this purpose.
RNA editing is rarely used for protein recoding in most organisms. There are millions of editing sites in human mRNAs16 and thousands have been identified in mouse17–19, but the vast majority lie within double-stranded RNA structures formed by inverted repetitive elements in non-coding portions19–23, and their role is to prevent an aberrant innate immune response24–26. Recoding sites are far less abundant. Only ~3% of human messages harbor a recoding site, and most are only weakly edited27. Furthermore, only a few dozen recoding sites are known to be conserved throughout the mammalian lineage27,28. Although there are functionally important editing sites in mammals, many of the editing sites are likely to provide no adaptive advantage27,29. RNA editing has not been broadly assessed across invertebrates, however data from a small number of taxa indicate that, as in mammals, recoding is infrequent. It has been studied most extensively in Drosophila, where ~1300 recoding sites have been identified in ~4% of Drosophila messages30–38. Many of these sites are conserved across Drosophila lineages and are thought to be under positive selection37–39. Some recoding sites were shown to alter protein function40–46, and it has been suggested that the primary role of RNA editing in Drosophila is to fine-tune nervous function38,43. Thus, the process is used, but to a limited extent.
The coleoid cephalopods (octopuses, squids, and cuttlefishes) are a clear exception to this pattern. Over 60% of squid brain transcripts (Doryteuthis pealeii) have at least one recoding site, and many are edited at multiple sites47. Editing is enriched in transcripts encoding proteins involved in neuronal processes. Similar levels of editing occur in other squids (Euprymna scolopes and Sepioloidea lineolata), cuttlefish (Sepia officinalis), and two species of octopuses (Octopus vulgaris and Octopus bimaculoides), but not in nautilus (Nautilus pompilius) or in Aplysia californica, leading to the conclusion that high-level recoding is a coleoid cephalopod innovation48,49. Furthermore, highly edited sites tend to recode, and a large number of these, along with the structures that drives editing, are conserved across coleoid taxa47,48. These data support the idea that cephalopod editing is under positive selection and leads to phenotypic advantage48–50. Individual recoding sites can directly affect protein function, particularly that of ion channels and ion transporters48,51–53. Because cephalopods inhabit exceptionally varied marine environments, a central question is whether their abundant recoding is generally used to respond to environmental changes.
Past studies have hinted that editing may be used for temperature acclimation. For example, a study that examined orthologous voltage dependent K+ channel messages from diverse octopus species demonstrated that positional editing frequencies correlated well with the thermal environments from which the specimens were sampled51. Furthermore, a specific recoding event in one of the channel’s transmembrane spans accelerated the channel’s closing rate, and the editing frequency at this position increased with decreasing water temperature where the animals were captured. In this previous study, acute temperature changes were not tested so it was unclear whether the editing changes were an acclimation or an adaptation. In a study using Drosophila melanogaster, populations inhabiting different temperature microclimates showed differences in editing frequencies at a large number of sites54. In related studies, editing in Drosophila melanogaster was shown to respond to acute temperature changes. However, these studies were either limited to just a handful of transcripts55,56, detected a rather small number (<50) of temperature-sensitive recoding sites30,57 or found very small changes in editing37.
In this study, we explore the effects of temperature on recoding across the neural transcriptome of Octopus bimaculoides, taking advantage of the extensive number of RNA editing sites in this species and its varied thermal environment. Our data show that temperature affects ~33% of all recoding sites and that editing changes occur within hours. In the vast majority of cases, editing frequencies are negatively correlated with temperature, and we identified over 20,000 sites that are edited more extensively in the cold. Furthermore, we demonstrate that some sites affect the function of proteins that are key to neurophysiology. A single, highly temperature-sensitive editing site in messages encoding kinesin affects the motor’s transport velocity and run length along microtubules, two key metrics for axonal transport. By solving the structure and measuring ion binding of edited and unedited versions of synaptotagmin, we show that another highly temperature-sensitive editing site regulates Ca2+ binding, a key physiological property for synaptic transmission. Lastly, we demonstrate that the editing changes observed in the lab also take place in the field in the face of complex and dynamic environments.
Results
Cold-induced RNA editing is abundant in Octopus bimaculoides
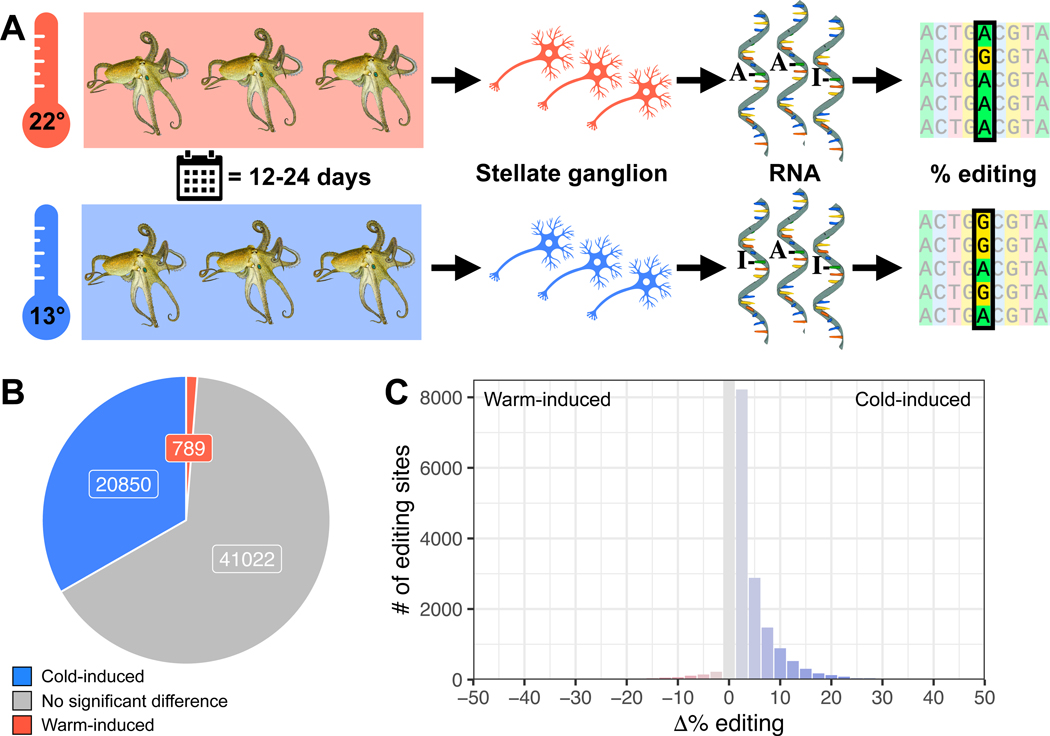
Octopus bimaculoides are found in the Pacific Ocean in nearshore waters off southern California and northern Baja California58. They are an ideal species for this study because they experience relatively large seasonal temperature changes, have a high-quality sequenced genome59, and a comprehensive map of editing sites across their neural transcriptome has been constructed48. To assess the temperature-dependence of RNA editing, adult wild-caught O. bimaculoides were transferred to the Marine Biological Laboratory and allowed to equilibrate to ambient conditions for 2–3 weeks in temperature-controlled aquaria. At the onset of the experiment, the temperature was gradually shifted to either ~13 or 22°C (Figure 1A). Temperature data loggers were placed in each tank and the time-course of the temperature change was monitored (Figure S1). The cold temperature shifts were largely complete in 10–12 days. The warm temperature shifts were much faster because the final temperature was close to the ambient tank temperature at the onset. The target temperatures were then maintained for 12–24 days. At the end of the acclimation period, animals were sacrificed. RNA extracted from the stellate ganglia, a motor center in the peripheral nervous system that is known to edit at high levels48,59 was sequenced to assess RNA editing levels across the neural transcriptome at all previously-mapped editing sites48. These data revealed that the editing frequencies at a large number of sites are temperature-sensitive. Across the 62,661 editing sites with sufficient coverage, ~33% (20,850) had significantly higher editing levels at 13°C compared to 22°C (cold-induced), while only ~1% (789) had higher editing levels at 22°C than at 13°C (warm-induced) (Figure 1B, Tables S1,2). Furthermore, thousands of these temperature-sensitive sites showed robust changes in editing percentages (>5%, up to 51%, Figure 1C). Results were similar among edits that recoded a codon − 33% (13,285) of sites were cold-induced and only 1% (550) were warm-induced (Figure 2A). Cold-induced protein recoding via RNA editing was abundant across the transcriptome but also appeared in distinctive patterns.
Figure 1: Octopuses exposed to cold temperatures exhibit stronger RNA editing activity.
Panel A: Octopus bimaculoides (n=6 per temperature) were kept at 13 or 22°C for 12–24 days before dissecting stellate ganglia to measure A-to-I editing levels. Panel B: A large proportion of the O. bimaculoides editome exhibits increased editing at colder temperature (blue), but only 789 sites show a significant increase in warm samples (red). Panel C: Cold-induced increases in editing levels were both more common and higher in magnitude than warm-induced increases. See also Figure S1. Octopus drawings are reproduced, with permission, from Roger Hall.
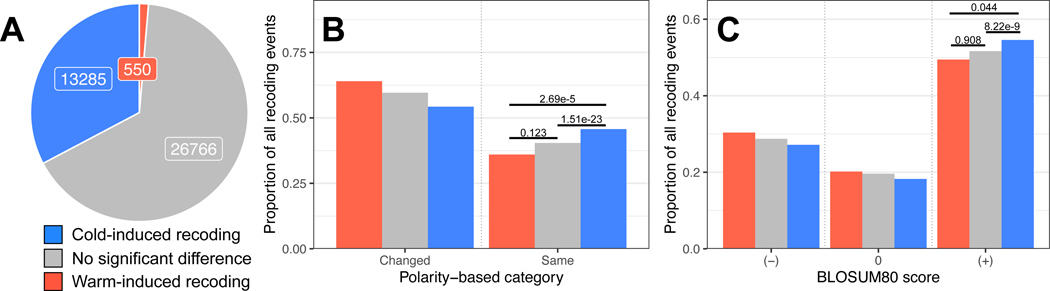
Figure 2: Cold-induced RNA editing sites are enriched in subtle, common amino acid substitutions.
Panel A: the majority of recoding editing sites do not show a statistically-significant temperature-sensitivity (grey) but a large proportion (33%, blue) are cold-induced and a small proportion are warm-induced (1%, red). Panel B: The fraction of recoding sites where amino acid substitutions stayed within the same polarity category is higher in cold-induced sites than warm-induced sites or sites with insignificant temperature sensitivity. The Bonferroni adjusted p-values from pairwise χ2 tests are shown above each comparison. Panel C: The fraction of recoding sites resulting in evolutionarily common amino acid substitutions (positive BLOSUM scores) is higher in cold-induced sites than sites with insignificant temperature sensitivity. The Bonferroni adjusted p-values from pairwise t-tests on the raw BLOSUM scores are shown above each comparison. See also Figure S2.
Temperature-sensitive recoding sites were not randomly distributed across messages. Firstly, transcripts with recoding sites showing ≥10% cold-induced change in editing tended to encode specific classes of proteins; membrane proteins, especially synaptic proteins, calcium binding or dependent proteins, and autophagous proteins were particularly enriched (Table S3). Amongst the membrane-associated proteins, cold induced sites were not found preferentially in any specific protein region (e.g. intracellular, extracellular, or transmembrane spans). Secondly, cold-induced recoding events tended to conserve the polarity of the amino acid (χ2 test, p=1.51e-23, Figure 2B, Figure S2) and generate evolutionarily conserved substitutions (i.e. positive BLOSUM80 scores, t-test, p=8.22e-9, Figure 2C, Figure S2) more so than warm-induced sites or sites without significant temperature-induced changes. These trends suggest that cold-induced editing favors subtle, common amino acid substitutions over rare, drastic changes.
A natural question arising from these results is what controls the temperature-sensitivity which varies across different editing sites? The underlying mechanism is likely complex and may involve both global and site-specific factors. We assessed three of the most likely candidates that could impart temperature-sensitivity on editing. First we tested for temperature-dependent expression of ADARs, the enzymes that catalyze A-to-I RNA editing, and found that both octopus ADAR paralogs60 are expressed similarly in our cold and warm acclimated RNA-Seq datasets (ADAR1: p=0.79, ADAR2: p=0.40, Figure S3A).
A second possible explanation relates to the double-stranded RNA (dsRNA) structures that ADARs recognize. Equilibrium RNA structures are determined by a temperature-dependent balance of energy and entropy, making all structures more stable at lower temperatures. The added stability of structures surrounding temperature-sensitive editing sites in the cold might make them more editable. To test this idea, we followed Avram-Shperling et al.61 and searched in silico for RNA structures surrounding all known editing sites, at 13 and 22°C, to examine whether changes in dsRNA stability can explain the higher observed editing in the cold. As expected, the stability of the dsRNA structures increases in the cold in the vast majority of cases, in agreement with the general trend towards higher editing. However, we could not identify any distinguishable relationship between the shift in free energy due to temperature change and the observed changes in editing levels among cold-induced sites (Figure S3B,C). Nevertheless, it is possible that subtle changes in the structure affect the editing levels in ways that are not detected by our in-silico RNA structure prediction.
A third potential mechanism to explain temperature-sensitive editing would be the temperature-dependent expression of trans-acting proteins that regulate ADAR activity. Site-specific regulation by ADAR interactors has been demonstrated in the nervous system of many organisms62–64, but nothing is known about ADAR interactors in cephalopods. We made a list of 310 proteins that interact with ADARs in humans, and looked at the temperature dependence of their homologs’ expression in octopus. Interestingly, 65 were upregulated in the cold, while only 9 were upregulated at the warmer temperature (Figure S3D, Table S4). Although these data suggest that some of these interactors could play a role in temperature-sensitive editing, in the vast majority of cases it is not known whether these proteins up- or downregulate ADAR activity in humans and nothing is known about the conservation of these roles in cephalopods. Overall, these results raise the possibility of a complex temperature-dependent regulatory network; however, the mechanistic underpinnings of temperature-dependent editing remain an open question.
RNA editing responds to temperature within hours
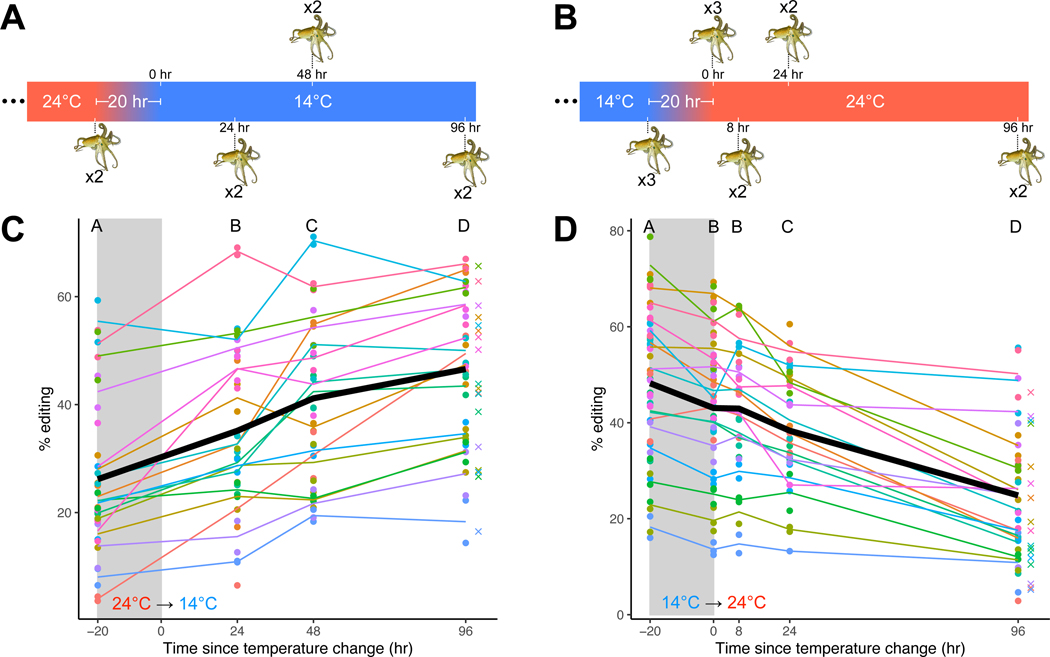
Cephalopods often face dynamic thermal environments where the temperature can change rapidly (e.g. due to thermoclines) or slowly (e.g. due to seasons). Accordingly, the speed at which RNA editing can recode the pool of messages influences the utility of the process. In order to assess how rapidly editing levels change, time series experiments were conducted. In one series, juvenile O. bimaculoides were equilibrated in aquaria to 24°C for one week after which the temperature was reduced to 14°C over the course of 20 hours (Figure 3A). In a reciprocal series, animals were equilibrated to 14°C and then the temperature was raised to 24°C (Figure 3B). These temperatures are reasonable matches for the temperature range experienced by this species in southern California65. The 20-hour acclimation was required to avoid temperature shock induced by an acute temperature change. Animals were sacrificed immediately before the temperature change and from 0 to 96 hours after the temperature change was complete. RNA was extracted and sequenced and editing was quantified at 18 highly temperature sensitive editing sites (see File S1). Cold-induced editing is seen within hours and reaches a steady-state within ~4 days. Statistically significant changes in editing levels were observed both during the 20-hour temperature shift (for the 14°C→24°C series) and at nearly every timepoint afterwards during both warm-to-cold and cold-to-warm experiments (Figure 3C-D, Table S5). The editing frequencies observed in animals 96 hours after the temperature change were indistinguishable from those during the long-term temperature acclimations shown in Figure 1 (paired t-tests, cold: p=0.951, warm: p=0.362), suggesting that a new steady state had been reached within 4 days at both temperatures. These data describe the time-course of temperature-dependent changes in RNA editing and provide a reference for the minimal time that this process could be used for temperature acclimation.
Figure 3: RNA editing changes within hours and reaches a steady state within days of a change in temperature.
Panels A and B: Octopuses were sampled before and at various timepoints after a 10°C transition that occurred over 20 hours. Panel C: Editing levels at 18 selected sites (see File S1) rise during a 10°C fall in temperature and at every timepoint afterwards. Each color represents a different editing site. The black line represents the mean editing level across all sites. The crosses represent the mean editing level from the long-term experiments at the equivalent ending temperature. Successive time points showing a statistically significant difference (see Table S5) are marked by distinct letters. Panel D: Editing levels at 18 selected sites decline during a 10°C rise in temperature and at almost every timepoint afterward. Colors and symbols are the same as described in panel C. Octopus drawings are reproduced, with permission, from Roger Hall.
Cold-induced editing changes kinesin motility
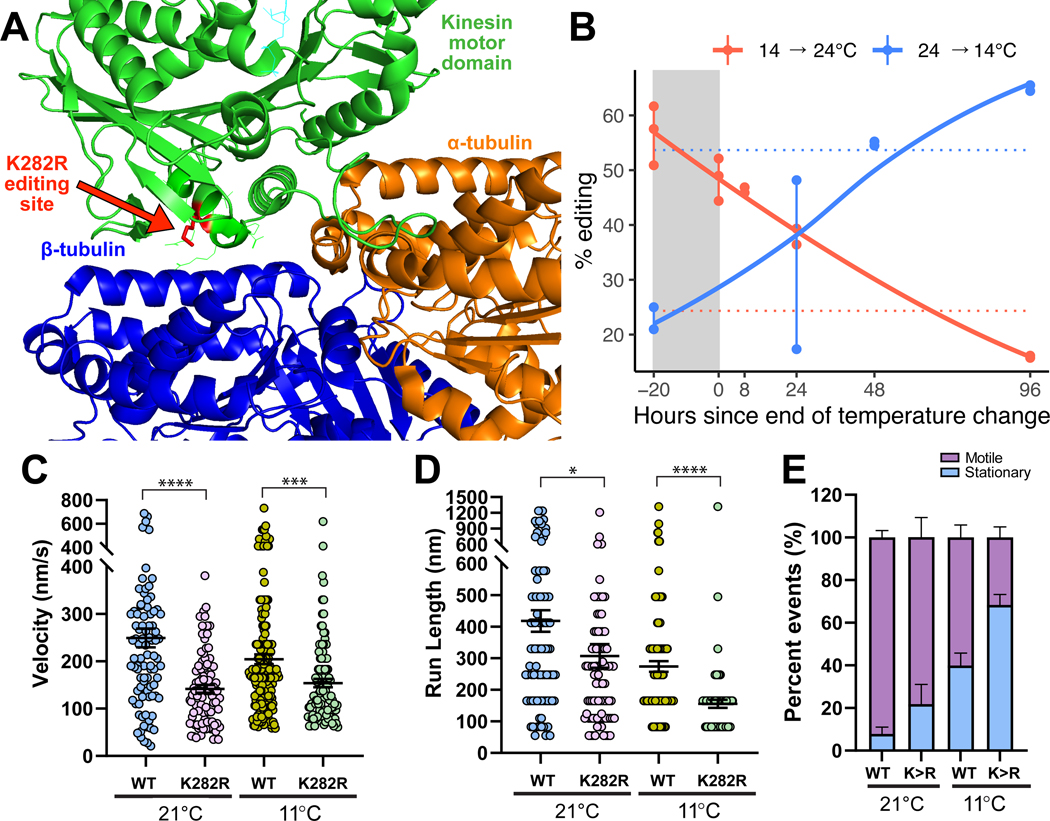
Given that there are >13,000 cold induced recoding sites, a central question is to what extent do these changes alter protein structure and function? To explore this question directly, we focused on two highly temperature-sensitive recoding sites within proteins that are critical for nervous system function. The first site recodes a lysine to an arginine (K282R) in kinesin-1, the primary molecular motor responsible for moving cargo in the anterograde direction down microtubules in axons (see File S1 for coordinates). The genomically encoded lysine at this position is universally conserved across 162 species within 4 phyla (Figure S4A), suggesting purifying selection, and lies in kinesin’s motor domain that faces the microtubule (Figure 4A). We thus predicted that it could influence kinesin’s transport dynamics. Editing at this site undergoes a 30% shift in response to a 10°C change in temperature (Figure 4B). To examine the effects on kinesin-1 motility, we utilized an O. bimaculoides kinesin-1 construct containing the full motor domain and a portion of the dimerization stalk with a HaloTag on the C-terminus (Figure S4B). Edited and unedited versions of the protein were visualized walking on taxol-stabilized microtubules using TIRF microscopy at 21 and 11°C. These experiments showed that the edited version of octopus kinesin-1 has a lower velocity than the wild-type version at both warm (t-test, p=1.7e-6) and cold temperatures (p=0.0007; Figure 4C). Interestingly, the edited version displayed an essentially temperature-invariant velocity, such that velocity was comparable at both 21°C and 11°C (t-test, p=0.312). The edited version also had shorter run lengths than wild-type versions at both temperatures (t-tests, warm p=0.029, cold p=8.9e-7, Figure 4D). Finally, the edited kinesin-1 had a greater propensity to be stationary at both temperatures (Figure 4E). The conservation of the lysine across kinesin-1 proteins suggests that recoding to arginine would have similar effects across species. Indeed, mutation of K283R in rat kinesin-1 (KIF5C) resulted in a similar decrease in velocity and run length (Figure S4B,C). Taken together, these results suggest that recoding of a conserved residue in the Octopus bimaculoides kinesin-1 motor domain alters motility properties, including transport velocity and run length, in a temperature-dependent manner.
Figure 4: An editing site (K282R) on the motor domain of kinesin-1 is highly temperature-sensitive and induces strong changes in motility.
Panel A: Human monomeric kinesin (KIF5B) bound to tubulin (RCSB: 2P4N). Side chains are revealed for editing site and 10 conserved neighboring residues. Panel B: The K282R editing site is highly temperature-sensitive. Point data are shown from amplicon sequencing of a 4-day time-lapse experiment. Dotted horizontal lines represent editing levels during long-term temperature exposures at 13°C (blue) and 22°C (red). Panels C-E: Motility properties of individual wild-type (WT) and edited (K282R) octopus kinesin-1 along taxol-stabilized microtubules was visualized using single-molecule TIRF microscopy. From kymographs, the C) velocity, (D) run length, and (E) proportion of motile and stationary kinesins were determined. Motility properties were compared between wildtype (WT) and edited (K282R) kinesins at 21 and 11°C (*, p<0.05; **, p<0.01; ***, p<0.001; and ****, p<0.0001). Panel E: The proportion of motile and stationary kinesins observed along microtubules were compared between WT and K282R and between temperatures. Error bars represent standard error. See also Figure S4.
Cold-induced editing modifies synaptotagmin structure and Ca2+ binding
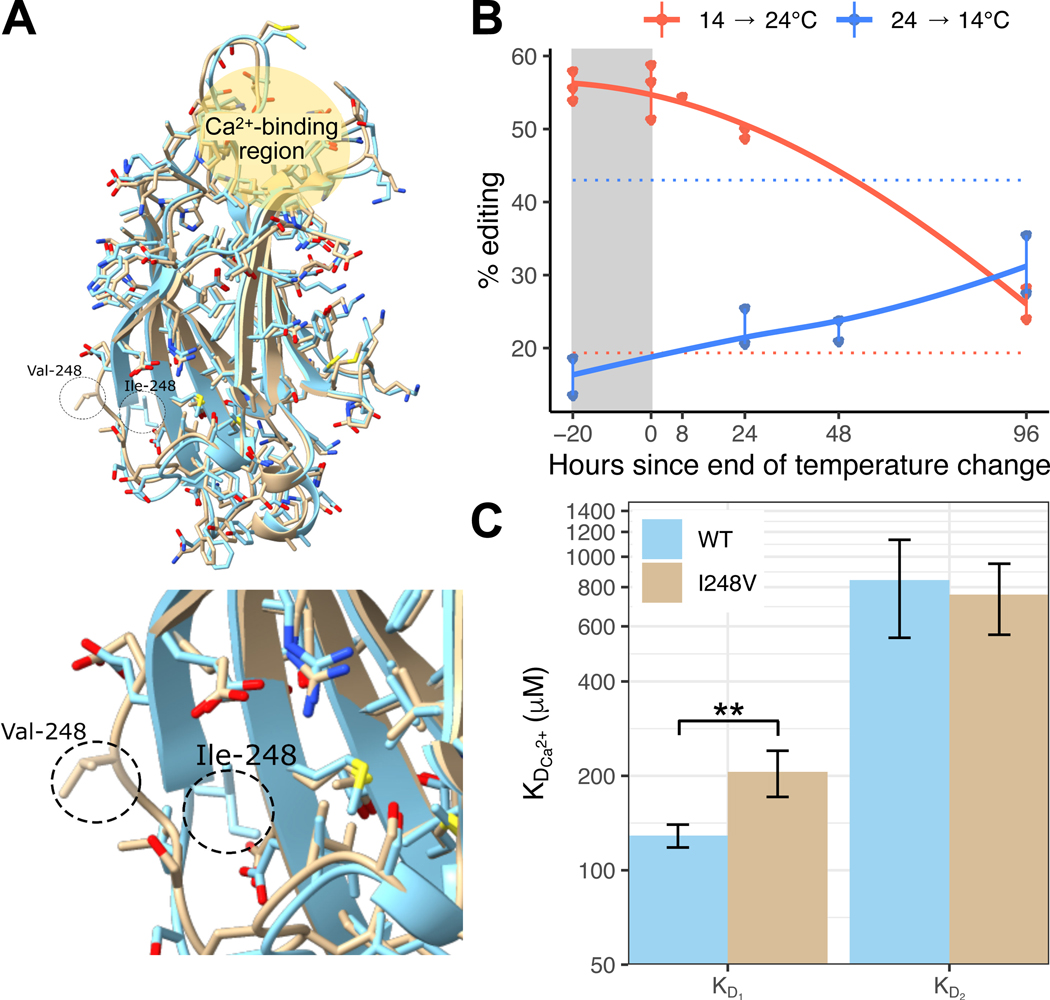
To explore how another temperature-sensitive editing site impacts protein function, we examined an I→V recoding event in synaptotagmin-1 (Syt1), a key protein involved in synaptic transmission (see File S1 for coordinates). The I248V edit is highly temperature sensitive, increasing by 24% in the cold (Figure 5B). Among 32 molluscan sequences, >60% have either I or V at this position, with the remaining sequences mostly containing other nonpolar residues. Synaptotagmin-1 lies at the interface of neurotransmitter-containing presynaptic vesicles and the presynaptic membrane. When the concentration of intracellular Ca2+ rises during presynaptic excitation, Ca2+ ions bind to synaptotagmin and induce a conformational change that promotes the initiation of vesicle docking to the presynaptic membrane. Synaptotagmin is composed of an N-terminal transmembrane domain embedded in the synaptic vesicle and two calcium binding domains (C2A and C2B). Each C2 domain is capable of binding at least two Ca2+ ions as well as phospholipid membranes66. The C2A domain is composed of 8 beta-sheets with neighboring high-affinity and low-affinity Ca2+ ion-binding sites. Phospholipid-binding, which is promoted by the bound Ca2+ ions, occurs at the same location as ion binding, both at one end of the domain67.
Figure 5: A cold-induced editing site (I248V) on the C2A domain of synaptotagmin-1 changes protein conformation to alter Ca2+-binding affinity.
Panel A: Wild-type (WT) Octopus bimaculoides synaptotagmin-1 C2A domain (blue, RCSB: 8FAF) and the I248V edited version (tan, RCSB: 8FAM) superimposed together. Top image shows the entire C2A domain including the Ca2+-binding region. The inset below zooms on residue 248 and the surrounding loop between β-strands 6 and 7, showing the change in conformation caused by the edit. See also Table S6. Panel B: The I248V editing site is highly temperature-sensitive. Point data are shown from amplicon sequencing of a 4-day time-lapse experiment. Dotted horizontal lines represent editing level during long-term temperature exposures at 13°C (blue) and 22°C (red). Panel C: Ca2+-binding affinity of the first (KD1) and second (KD2) Ca2+ ions to bind the C2A domain were determined via ITC for both the WT and I248V domains. **, p<0.01. Data are presented as mean +/− SD.
Based on protein structures from rat, the I248V edit is on the opposite side of the C2A domain from Ca2+ ion binding sites68. To assess the impact of this edit, we first solved the structure of the wild-type and the I248V C2A domains via x-ray crystallography. The high-resolution (1.85Å) crystal structures of Octopus synaptotagmin-1 C2A matched very closely to the rat synaptotagmin-1 C2A domain, composed of 8 beta-sheets mapping to homologous positions of the primary structure (RMSD=0.552Å, Table S6). The position 248 lies within an exposed loop between β-strands 6 and 7, on the opposite side of the domain to the ion binding sites (Figure 5A). In the unedited Syt1 C2A domain structure, I248 is in direct contact with the hydrophobic core of the domain and has a relatively low solvent accessibility surface (SAS) of 25Å2. In the edited version, the side chain is completely exposed to solvent, with an SAS exposure of 103Å2, a change brought about by the elimination of a single methyl group. Furthermore, the β6–7 loop containing this position is fairly rigid. The overall B-factor for the wild-type C2A domain is 45.26Å2 (Table S6). The average B-factor for residues that make up the β6–7 loop of the unedited C2A domain (residues 238–250) is 60.1Å2, indicating that the loop is more flexible, on average, than the macromolecule itself, despite being associated with the hydrophobic core of the domain. In the edited C2A structure, however, the loop possessing the edited residue (V248) is exposed to solvent, yet still relatively rigid. The B-factor for the edited domain overall is 35.2Å2, while the refined B-factor for the residue range around the loop is 35.7Å2. However, despite the relative rigidity in the loops, the β6–7 loop for the wild-type C2A domain is flipped inward, with the I248 residue directly contacting the hydrophobic core, while the same residue in the edited C2A domain (V248) was flipped out. There are no other major structural differences between the wild-type and edited C2A domain structures. Thus, editing generates a very specific change that may be poised to fine-tune function.
We next examined whether this small structural change might affect Ca2+ ion binding, despite its distance from the Ca2+ ion binding sites (Figure 5A). Using isothermal titration calorimetry (ITC), we directly examined the Ca2+ binding affinities of the edited and unedited recombinant proteins. Surprisingly, we found that the I248V edit lowers the binding-affinity of the first bound Ca2+ (KD1) by nearly 60% (p=0.003), while the affinity for the second Ca2+ (KD2) remains unchanged (p=0.59, Figure 5C). Together, these results demonstrate that the removal of a single methyl group on the C2A domain of synaptotagmin-1 changes the protein’s conformation sufficiently to alter Ca2+-binding dynamics in response to temperature.
Wild populations exhibit temperature-sensitive RNA editing
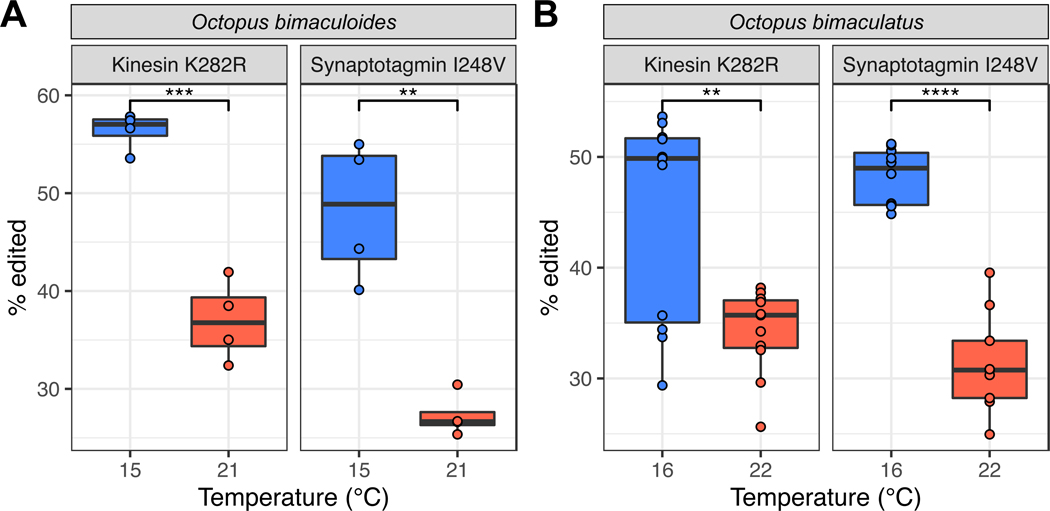
Our data on laboratory-raised animals demonstrated that temperature changes editing levels under tightly controlled conditions: animals were housed under identical conditions outside of temperature. A natural question is whether temperature would induce a similar effect seasonally on wild caught specimens confronting dynamic, varied environments. To address this, we collected adult specimens of O. bimaculoides during the winter and late summer in shallow waters near Long Beach, CA, USA. Prior to collection, temperature data loggers were placed near the octopuses’ dens and the water temperature was recorded for 1–2 months before collection. The water temperature was 21–22°C in the late summer and ~15°C in the winter during the week prior to collection (Figure S5). After collection, the extent of editing for the kinesin-1 K282R and the synaptotagmin I248V sites was determined by PCR followed by direct sequencing. Both sites exhibited robust increases in editing frequencies in the cold (Figure 6A, p=0.0001, p=0.001, respectively), and the magnitudes were strikingly similar to those observed for the lab-reared animals (Figure 4B). We next asked whether temperature-dependent recoding at these sites is evolutionarily conserved. Octopus bimaculatus is a closely related congener of O. bimaculoides that inhabits the same geographical range69. We collected specimens of O. bimaculatus off Santa Catalina Island, CA using SCUBA during the same months that we collected O. bimaculoides. Data loggers for these experiments showed a temperature of ~16°C in the winter and ~22°C in the late summer (Figure S5). Editing at the kinesin-1 K282R and synaptotagmin I248V sites was conserved in this species, and showed a similar temperature-dependence (Figure 6B, p=0.002, p=1.22e-8), as the seasonal values matched closely with those from O. bimaculoides. These data showed that RNA editing for these two temperature-dependent sites is evolutionarily conserved across these two species.
Figure 6: Temperature-sensitive editing sites discovered in the laboratory show comparable temperature-sensitivity in wild-caught animals undergoing seasonal temperature.
Panel A: Kinesin-1 K282R and synaptotagmin-1 I248V editing sites present in O. bimaculoides caught in the wild both exhibit higher editing in February (15°C, blue) than in September (21°C, red), corroborating laboratory experiments. Panel B: The same temperature-dependent pattern is conserved in the sister species Octopus bimaculatus caught in the wild. **, p<0.01; ***, p<0.001; and ****, p<0.0001. See also Figure S5.
Discussion
As a mechanism for environmental acclimation, A→I RNA editing appears ideal. It has the potential to recode single amino acids within proteins transiently, as needed. Data that support this idea, however, are remarkably limited. Studies on Drosophila spp reported 17–47 temperature-sensitive recoding editing sites30,55,56, and a study on hibernating ground squirrels uncovered 12 recoding sites57. Most of these sites showed small changes in editing. A previous study on various octopus species showed that editing at specific sites in a K+ channel message correlated well with the thermal environment from which the animals were sampled, but it was not determined whether these differences were due to acclimation or adaptation51. The physiological effects of the recoding event were only examined in the octopus study. Here, we demonstrate that: (i) temperature-sensitive RNA editing in coleoid cephalopods is abundant (20,850 sites), (ii) the changes in editing occur rapidly following a temperature change (i.e, within hours), (iii) recoding events affect the structure and function of proteins involved in critical neuronal functions and (iv) temperature-dependent recoding is robust in the face of dynamic environments. Therefore, for coleoid cephalopods, and perhaps other poikilotherms, RNA editing appears to be a valuable tool for fine-tuning neurophysiological function in response to temperature.
At the vast majority of temperature-dependent sites, editing increases in the cold. This is true for the sites uncovered in this study, and also for those reported for Drosophila and ground squirrels during hibernation. The mechanism underlying this phenomenon remains unclear. We addressed three possibilities but methodological limitations prevent our reaching firm conclusions. It should be noted that the extent of temperature-dependent changes in editing levels varies considerably across sites (Fig. 1C), thus a global mechanism is unlikely, but it can’t be ruled out. The identification of the factors that drive temperature sensitivity in editing will help us to better understand the evolution of this process.
Is temperature dependent RNA editing used for acclimation, or is it simply a byproduct of the temperature changes? A detailed examination of how recoding events affect protein function can provide critical data to answer this question. At the outset one might expect editing sites used for temperature acclimation to increase reaction rates in order to compensate for the cold. In the limited number of cases where the functional effects of cephalopod edits have been studied, they do accelerate reaction rates. For example, an I→V edit in the squid Na+/K+ ATPase increases the transport rate and the same change in a K+ channel increases the closing rate51,52, consistent with temperature compensation, but it was not determined if their editing frequencies underwent acute changes due to temperature. In this study, we show that a single I→V edit in the C2A domain of synaptotagmin decreases its affinity for one of the two Ca2+ ions that it binds. This might help compensate for increased calcium concentrations caused by longer duration depolarizations of cephalopod presynaptic terminals in the cold6,8.
It is remarkable that the loss of a single methyl group from the side chain of the edited residue so far away from the Ca2+-binding sites would change the binding affinity, but this is not unprecedented. Site-directed mutagenesis has shown that point mutations far away from active or catalytic sites can change temperature-stability and temperature-dependent function of molluscan proteins70. In the synaptotagmin example here, the most likely mechanism is that transient interactions of the valine residue with the core of the domain transmits small reorientations of the residues that make up the hydrophobic core; thereby impacting the Ca2+ binding residues on the other end of the domain. The idea of an intra-domain allosteric communications network transmitted through the hydrophobic core is an under-appreciated concept in altering the activity of C2 domains. Squid synaptotagmin-1 has an I>M editing site at the same locus47,48 and our data have shown that it too is temperature sensitive (unpublished). As most other synaptotagmin C2A domains that have been sequenced utilize a polar residue at this locus, this may be a unique feature of cephalopod synaptotagmin.
As a second example of the functional effects of temperature dependent recoding, we found that a cold-induced editing site in kinesin-1’s motor domain led to a decline in motility. Upon first consideration, the slowing of kinesin under colder temperatures seems counterintuitive, as one might expect it to accelerate to help compensate for the cold. However, we presume that changes in cellular cargo transport rates should optimally match the changes in general cellular processes so that supply matches demand. The rate of most cellular processes increase 2–3x for every 10°C within the temperature range at which an organism is adapted71. Similarly, axoplasmic transport rates measured in a variety of animals also show 2–3x changes over a 10°C change in temperature72–74. The wild-type octopus kinesin measured here, however, showed only a 1.2x difference in velocity over a 10°C gradient, appreciably less temperature-sensitive than expected. Shifting from a relatively fast wild-type kinesin in the warm to a relatively slow edited kinesin in the cold, however, increases its temperature sensitivity, perhaps better matching other cellular processes dependent on cargo transport that are more temperature sensitive. Furthermore, cellular cargo transport rates at a given temperature are dependent on both kinesin and cytoplasmic dynein in a “tug-of-war” pulling the cargo in opposite directions along a microtubule. Additional recoding editing sites on kinesin or dynein, whether temperature-sensitive or not, may also modulate overall transport rates. In the O. bimaculoides editome48, there are 17 recoding editing sites in kinesin-1 heavy chain mRNA and 8 are temperature-sensitive (though none as strongly as K282R). Thus, the K282R edit does not occur in isolation, but rather in concert with other editing events and may exert its function in an epistatic manner. Cytoplasmic dynein-1 heavy chain has 78 recoding editing sites, 39 of which are temperature-sensitive. The ability of these motors to drive axonal transport depends not just on velocity and run length of individual motors, as measured here, but also on their ability to work collectively as teams on a cargo and to withstand opposing forces during transport. Thus, it will be necessary to examine the collective effects of the recoding sites on transport. The sheer number of temperature sensitive and insensitive recoding sites in cephalopod motor molecules is a profound example of the regulatory potential of RNA editing.
Our data show that temperature dependent recoding is widespread across the neural transcriptome. What is it being used to regulate? Phospholipid membranes are one of the most temperature-sensitive cellular structures and cells must actively modulate their fluidity to compensate for temperature75. Proteins embedded within the membranes must also compensate for the viscous drag associated with their movement within these structures76. This is especially critical in physiologically dynamic regions of the membrane such as at the synapse. Therefore, it is unsurprising that membrane-associated proteins, and synaptic proteins in general, possess a disproportionately large number of cold-induced recoding sites. In addition, our results indicate that temperature dependent changes in editing begin within hours, but take 3–4 days to reach a new steady state. This likely is due to the balance between the rates of mRNA synthesis and decay. For the effects of recoding to be realized, the rates of protein synthesis and decay must also be factored in. Therefore, it may be expected for RNA editing-mediated amino acid recoding to take several days to a week to reach a new steady state after a temperature change. In marine environments, temperature can change appreciably over the course of days to months due to factors such as upwelling events, the seasons, or seasonal migrations. RNA editing would not be well suited to accommodate rapid temperature changes due to the tides or crossing thermoclines.
We show two examples of functional effects caused by recoding on keystone neurophysiological properties: axonal transport and synaptic transmission. Due to the extraordinarily large number of temperature sensitive events, we expect that their effects are widespread across neurophysiological processes. Furthermore, it will be interesting to see whether RNA editing can respond to other changes in the physical environment. In the larger picture, the evolutionary pressures that drove the coleoid cephalopods, but not other poikilotherms, to adopt high-level mRNA recoding remain enigmatic and fascinating.
Limitations of the Study
We present robust evidence that temperature-induced RNA editing can alter the performance of key neuronal proteins. However, at present it is not possible to assess the functional impacts of these changes at cellular or organismal scales. Recent advancements in the development of genetic tools for cephalopod species77,78 may provide the means to hardwire edited or unedited codons within transgenic lines in order to study the effects of individual editing sites at higher levels of biological organization.
The mechanism that drives temperature-sensitivity remains unclear. The expression levels of ADAR messages appear similar across temperatures; however, our data do not rule out post-transcriptional mechanisms controlling ADAR protein expression. In addition, we could not identify differences in the thermal stabilities of mRNA structures surrounding temperature-sensitive and insensitive editing sites; however, our folding prediction is limited, and it is possible that small perturbations in the dsRNA structures, which are not seen using bulk in silico folding algorithms, play an important role. Expression of many octopus homologs to known ADAR interactors are upregulated in the cold, but these results must be interpreted with caution because ADAR-interactions are known to be cell-type and developmental stage specific62,63. It is also quite possible that reported ADAR-interactors in human cell lines play different roles in octopus neurons. Furthermore, it is likely that many cephalopod-specific ADAR-interactors remain undescribed and could play a role in temperature-sensitive editing. In this study, the function of single editing sites were studied. It should be noted that many cephalopod transcripts contain multiple editing sites and epistatic interactions between these sites may be important. In general, the mechanism that drives abundant recoding in coleoid cephalopods is poorly understood. A better understanding of the contribution of each ADAR paralog may help us to better understand the process and its temperature-dependence.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joshua Rosenthal (jrosenthal@mbl.edu).
Materials availability
Plasmids generated in this study have been deposited to AddGene. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| DH5α E. coli cells | New England BioLabs | Cat#C2987H |
| Biological samples | ||
| Octopus bimaculoides | Aquatic Research Consultants | fishes4study.com |
| Chemicals, peptides, and recombinant proteins | ||
| Schneider’s Drosophila medium | Gibco | Cat#21720024 |
| Fetal bovine serum | Gibco | Cat#16000044 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat#11960 |
| Fetal Clone III serum | HyClone | Cat#SH30109.03 |
| GlutaMAX supplement | Gibco | Cat#35050061 |
| RNAlater | Invitrogen | Cat#AM7021 |
| TRIzol reagent | Invitrogen | Cat#15596026 |
| GlycoBlue Coprecipitant | Invitrogen | Cat#AM9516 |
| DNase I | New England BioLabs | Cat# M0303S |
| Phusion High-Fidelity DNA Polymerase | New England BioLabs | Cat#M0530L |
| Lipofectamine LTX with PLUS reagent | Invitrogen | Cat#15338100 |
| Janelia Fluor 552 (JF552) Halo ligand | Janelia Farms | Cat#JF552 |
| Trans-IT LT1 | Mirus | Cat#MIR2305 |
| Protease inhibitors | Sigma-Aldrich | Cat#P8340 |
| HiLyte488 tubulin | Cytoskeleton | Cat#TL488M |
| Biotin-labeled tubulin | Cytoskeleton | Cat#T333P |
| HiLyte647 tubulin | Cytoskeleton | Cat#TL670M |
| Taxol | Cytoskeleton | Cat#TXD01 |
| Bovine serum albumin (BSA) | Sigma | Cat#A9647 |
| Casein | Sigma | Cat#C8654 |
| Glucose oxidase | Sigma-Aldrich | Cat#G7141-10KU |
| Catalase | Sigma | Cat#C3515 |
| Aminopropyltrimethoxysilane | Sigma | Cat#A3648 |
| mPEG | Laysan Bio | Cat#MPEG-SVA-5000 |
| Biotin-PEG | Laysan Bio | Cat#BIO-PEG-SVA-5000 |
| Disuccinimidyl tartrate | Soltec Bioscience | Cat#CL108 |
| NeutrAvidin protein | Thermo Scientific | Cat#31000 |
| Restriction enzyme: NdeI | New England BioLabs | Cat#R0111 |
| Restriction enzyme: XhoI | New England BioLabs | Cat#R0146 |
| Restriction enzyme: EcoRI | New England BioLabs | Cat#R0101 |
| IPTG | UBPBio | Cat#P1010-25 |
| His60 Ni Superflow Resin | Takara | Cat#635660 |
| TEV protease | AddGene | Cat#pRK793 |
| SP Sepharose Fast Flow | Cytiva | Cat#17072901 |
| Superdex 75 Prep Grade | Cytiva | Cat#17104402 |
| Critical commercial assays | ||
| RNAqueous Phenol-free total RNA Isolation kit | Invitrogen | Cat#AM1912 |
| TruSeq Stranded mRNA Sample Prep Kit | Illumina | Cat#20020594 |
| AccuScript High Fidelity 1st Strand cDNA Synthesis Kit | Agilent | Cat#200820 |
| Monarch DNA Gel Extraction Kit | New England BioLabs | Cat#T1020L |
| Gibson Assembly Master Mix | New England BioLabs | Cat#E2611L |
| QuikChange II Site-directed Mutagenesis kit | Agilent | Cat#200523 |
| Mix and Go! Transformation kit | Zymogen | Cat#T3001 |
| Deposited data | ||
| Illumina reads | This paper | SRA: PRJNA948369 |
| X-ray crystallography structure of unedited octopus synaptotagmin-1 | This paper | PDB: 8FAF |
| X-ray crystallography structure of edited octopus synaptotagmin-1 | This paper | PDB: 8FAM |
| Experimental models: Cell lines | ||
| Drosophila S2 cells | DGRC | RRID: CVCL_TZ72 |
| COS-7 cells (monkey kidney fibroblast) | ATCC | RRID: CVCL_0224 |
| Oligonucleotides | ||
| See File S1 for amplicon primers | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pAc5.1 V5-HisB | Dr. Patrick Emery, University of Massachusetts | N/A |
| Plasmid: rat kinesin-1 (KIF5C, residues 1–559) appended by 3 tandem mCitrine fluorophores | Cai et al. 2009 | N/A |
| Plasmid: Octopus wild-type kinesin | This paper | AddGene: 201551 |
| Plasmid: Octopus K282R edited kinesin | This paper | AddGene: 201552 |
| Plasmid: Octopus wild-type synaptotagmin | This paper | AddGene: 201553 |
| Plasmid: Octopus I248V edited synaptotagmin | This paper | AddGene: 201554 |
| Software and algorithms | ||
| Nikon Elements | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| Fiji/ImageJ2 | Schindelin et al. 2012 | https://imagej.net/software/fiji |
| HKL | HKL Research | https://hkl-xray.com |
| Phenix | Liebschner et al. 2019 | http://www.phenix-online.org |
| Biopython | Cock et al. 2009 | https://biopython.org |
| nanoAnalyze | TA Instruments | https://www.tainstruments.com/itcrun-dscrun-nanoanalyze-software |
| Bowtie2 (v2.1.0 and 2.3.2) | Langmead and Salzberg 2012 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| REDItools (v1.0.4) | Picardi and Pesole 2013 | https://github.com/BioinfoUNIBA/REDItools |
| Salmon (v0.8.2) | Patro et al. 2017 | https://salmon.readthedocs.io/en/latest |
| R (v4.2.1) | R Core Team 2022 | https://cran.r-project.org |
| R package: Biostrings (v2.64.0) | Pagès et al. 2022 | https://bioconductor.org/packages/release/bioc/html/Biostrings.html |
| DAVID Functional Annotation Tool (v6.8) | Huang et al. 2009 | https://david.ncifcrf.gov |
| DeepTMHMM | Hallgren et al. 2022 | https://dtu.biolib.com/DeepTMHMM |
| BLAST (v2.13.0+) | Camacho et al. 2009 | https://blast.ncbi.nlm.nih.gov/doc/blast-help/downloadblastdata.html |
| Prism (v8.0.0 (224)) | GraphPad | https://www.graphpad.com |
| R package: respirometry (v1.3.0) | Birk | https://cran.r-project.org/package=respirometry |
| Original code to analyze data and create figures | This paper; GitHub; Zenodo | https://github.com/matthewabirk/Temperature-dependent-RNA-editing; https://doi.org/10.5281/zenodo.7874071 |
| R package: DESeq2 (v1.36.0) | Love et al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R package: apeglm (v1.18.0) | Zhu et al. 2019 | https://bioconductor.org/packages/release/bioc/html/apeglm.html |
| RNAStructure | Reuter and Mathews 2010 | https://www.urmc.rochester.edu/rna/ |
| bpRNA | Danaee et al. 2018 | https://github.com/padidehdanaee/bpRNA |
| Other | ||
| Illumina HiSeq2000 sequencing platform | Illumina | N/A |
| #1.5 coverslip for imaging kinesin | Fisher Scientific | Cat#2850-18 |
| Glass slide for imaging kinesin | Fisher Scientific | Cat#12-544-3 |
| Inverted microscope Ti-E/B equipped with perfect focus system, a 100× 1.49 NA oil immersion TIRF objective, and three 20-mW diode lasers (488 nm, 561 nm, and 640 nm) | Nikon | N/A |
| Electron-multiplying charge-coupled device detector | Andor Technology | Cat#iXon X3DU897 |
| TIRF microscope with 60× 1.49NA objective, a 1.6X zoom lens on an Olympus IX81 microscope base with cellTIRF module, equipped with a 50mW 488nm laser and 100mW 561nm laser | Olympus | N/A |
| PID controller | TE Technology | Cat#TC-24-10 |
| Rigaku Screen Machine fitted with a Saturn CCD detector | Rigaku Americas Corporation | Out of production |
| nanoITC | TA Instruments | https://www.tainstruments.com/pdf/brochure/BROCH-MICRO-EN.pdf |
| HOBO pendant temperature data logger | Onset | Cat#UA-002-64 |
Data and code availability
Illumina RNASeq data have been deposited at the NCBI SRA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. X-ray crystallography structures have been deposited at the RCSB PDB and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
All original code has been deposited at GitHub and Zenodo and is publicly available as of the date of publication. URLs and DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Octopus bimaculoides
For the initial 12–24 day temperature trials, adult California two-spot octopuses (Octopus bimaculoides) were collected from Long Beach, CA, USA by a commercial vendor (Aquatic Research Consultants), shipped to the Marine Biological Laboratory (MBL) in Woods Hole, MA, USA, and held in flow-through seawater systems until trials began.
For time course experiments, an adult female with eggs was collected from Long Beach, CA, USA by a commercial vendor (Aquatic Research Consultants), shipped to the Marine Biological Laboratory (MBL) in Woods Hole, MA, USA, and held in a flow-through seawater system. Upon hatching, juvenile O. bimaculoides (n=12; 0.5–4.6 g) were held in isolation in flow-through seawater systems and fed grass shrimp twice daily before and during temperature acclimation trials. Although the use of cephalopods for research is not currently regulated in the USA, the Marine Biological Laboratory has implemented strict internal policies to ensure their ethical and humane treatment. All cephalopod specimens used in this study conformed to the Marine Biological Laboratory’s ‘Policy for the use of cephalopods for research and teaching’. Since February 2022, all cephalopod research at the MBL has required IACUC approval and this work was conducted under the approved protocol number 22–13A to JJCR.
Wild juvenile and adult Octopus bimaculoides specimens were collected from Long Beach, CA, USA by Aquatic Research Consultants in September 2019 (n=4, 80–122 g, 2 female 2 male, T=21°C) and February 2022 (n=4, 69–107 g, all male, T=15°C).
Octopus bimaculatus
Wild juvenile and adult specimens of Octopus bimaculatus were collected by SCUBA diving from Two Harbors, CA on Santa Catalina Island in September 2019 (n=11, 42–283 g, 5 female 6 male, T=22°C) and February/March 2020 (n=11, 106–456 g, 5 female 5 male 1 uncertain sex, T=16°C).
Drosophila S2 cells
Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Gibco) supplemented with 10% (vol/vol) FBS (HyClone) at 26°C.
COS-7 cells
COS-7 cells (monkey kidney fibroblast, ATCC, RRID: CVCL_0224) were cultured in DMEM (Cat #11960; Gibco) with 10% (vol/vol) Fetal Clone III (HyClone) and 1% (vol/vol) GlutaMAX (Gibco) at 37°C with 5% CO2.
Escherichia coli DH5α cells
DH5α E. coli competent cells were ordered from New England BioLabs (C2987H). After transformation, cells were plated on antibiotic agar plates and colonies were cultured in LB media supplemented with antibiotic at 37°C and 250 rpm.
METHOD DETAILS
Octopus temperature acclimation
Adult California two-spot octopuses (Octopus bimaculoides, Pickford and McConnaughey 1949) were acclimated to either 13 or 22°C seawater over approximately two weeks and held at these treatment temperatures for 12–24 days (n=3 for each temperature, Figure S1). At the end of the trial, animals were sacrificed and stellate ganglia were dissected. Stellate ganglion samples destined for RNA extraction were immediately preserved in RNAlater. All samples were then stored at −80°C. This trial was then repeated for a further 6 animals (3 at each temperature). The time course of the temperature changes for each tank was recorded via data-loggers and is presented in Figure S1.
Temperature-sensitive editing site discovery
For the transcriptome-wide assessment of site-specific editing frequencies at different temperatures, total RNA was extracted from stellate ganglia using the RNAqueous solution (Life Technologies, Carlsbad, CA). RNA-Seq libraries were prepared from these samples using the TruSeq Stranded mRNA Sample Prep Kit, as described by the manufacturer (Illumina), and were sequenced using one lane for each sample on an Illumina HiSeq 2000 instrument.
Time course of changes in RNA editing
To assess the time-course of changes in RNA editing levels in response to temperature, juvenile O. bimaculoides (n=12; 0.5–4.6 g) were acclimated in a flow-through seawater system at 14°C for 3+ weeks. As a baseline, three individuals were sacrificed and stellate ganglia were dissected. Immediately thereafter, the seawater was heated at a rate of 0.5°C / hour to 24°C and further samples were collected at times 0, 8, 24, and 96 hours after reaching 24°C (n=2–3 per timepoint). A similar trial was also conducted from 24°C to 14°C with samples collected before the temperature change and 24, 48, and 96 hours after reaching 14°C (n=2 per timepoint). For all experiments, animal euthanasia was conducted by a 5-minute immersion in seawater containing 3% ethanol.
To quantify single editing sites from these animals, tissue samples were immediately ground in 0.5–1.0 mL ice-cold TRIzol Reagent (Invitrogen) and processed following the manufacturer’s instructions for RNA isolation. 3 μL of GlycoBlue Coprecipitant (Invitrogen) were added to facilitate RNA precipitation and RNA pellets were resuspended in 20 μL of DEPC-treated water. Samples were treated with dNase I (NEB) following the manufacturer’s instructions. cDNA was then synthesized using the AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent) following the manufacturer’s instructions using an oligo(dT) primer.
Editing levels were quantified for 18 editing sites within four messages that were shown to be cold-induced from the transcriptome-wide analysis. The four messages were targeted for amplicon sequencing (see File S1 for editing sites and primer sequences), and PCR amplified using Phusion High-Fidelity DNA Polymerase (NEB). Amplicons were purified from agarose gel slices using the Monarch DNA Gel Extraction Kit (NEB) and then directly sequenced by a commercial vendor (Genewiz) using the Sanger protocol. Editing levels were quantified at 18 editing sites by comparing the T and C peak heights within the electropherograms in the strand opposing the target sites. Sequencing the antisense strand improves peak accuracy compared to A or G peaks in the sense strand 79.
Single-molecule motility assays of kinesin
Plasmid constructs
Wild-type and edited (K282R) versions of octopus kinesin-1 (KIF5, residues 1–558) were chemically synthesized with a HaloTag appended on their C-terminus by a G4S linkage (Figure S4B). These products were cloned into the EcoRI site of pAc5.1 V5-HisB plasmid (generously provided by Dr. P. Emery, University of Massachusetts) by Gibson assembly. Plasmids were then transformed into DH5α E. coli cells and the final product was verified by Sanger sequencing. In addition, as a positive control, a plasmid encoding rat kinesin-1 (KIF5C, residues 1–559) appended by 3 tandem mCitrine fluorophores 80 was mutated at the equivalent to position 282 from Octopus to create an arginine (rat K283R) using a QuikChange II site-directed mutagenesis kit (Agilent).
Cell culture, transfection, and cell lysates
Plasmids were transfected into cultured cells for protein production. Octopus kinesin-HaloTag plasmids (wild-type and K282R) were transfected into Drosophila S2 cells using Lipofectamine LTX with PLUS reagent (Invitrogen) according to the manufacturer’s instructions. Meanwhile, the protein was fluorescently labeled by the inclusion of 50 nM JF552 Halo ligand (Janelia Farms) in the growth medium. Rat kinesin-1 KIF5C(1–559)-3xmCit plasmids (wild-type and K283R) were transfected into COS-7 cells using Trans-IT LT1 (Mirus) according to the manufacturer’s instructions.
COS-7 or S2 cells were harvested 24 or 72h after transfection, respectively, by low-speed centrifugation at 4°C. The cell pellet was rinsed twice in 1xPBS buffer and resuspended in ice-cold lysis buffer (25 mM HEPES/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 1% Triton X-100, pH 7.4) freshly supplemented with 1 mM ATP, 1 mM PMSF, 1 mM DTT (for S2 cell lysate) and 1% (vol/vol) protease inhibitors (Sigma-Aldrich). Insoluble material was pelleted by centrifugation at full speed at 4°C in a table-top microcentrifuge. Aliquots of the supernatant were snap-frozen in liquid nitrogen and stored at − 80°C until further use.
Single-molecule motility assay
HiLyte488- and biotin-labeled microtubules or HiLyte647-labeled microtubules were polymerized from purified tubulin and 10% labeled tubulin (Cytoskeleton) in BRB80 buffer (80 mM PIPES/KOH pH 6.8, 1 mM MgCl2, and 1 mM EGTA) supplemented with 1 mM GTP and 2.5 mM MgCl2 at 37°C for 30 min. 20 μM taxol in prewarmed BRB80 buffer was added and incubated at 37°C for an additional 30 min to stabilize microtubules. Microtubules were stored in the dark at room temperature for further use.
For COS-7 cell lysates, a flow cell (~10 μL volume) was assembled by attaching a clean #1.5 coverslip (Fisher Scientific) to a glass slide (Fisher Scientific) with two strips of double-sided tape. Polymerized microtubules were diluted in BRB80 buffer supplemented with 10 μM taxol and then infused into the flow cell and incubated for 5 min at room temperature for nonspecific adsorption to the coverslips. Subsequently, blocking buffer [15 mg/ml BSA and 10 μM taxol in P12 buffer (12 mM Pipes/KOH pH 6.8, 1 mM MgCl2, 1 mM EGTA)] was infused and incubated for 5 min. Finally, 0.5 μL cell lysate expressing the rat 3xmCit-tagged kinesin motor in motility mixture [2 mM ATP, 0.4 mg/ml casein, 6 mg/ml BSA, 10 μM taxol, and oxygen scavengers (1 mM DTT, 1 mM MgCl2, 10 mM glucose, 0.2 mg/ml glucose oxidase, and 0.08 mg/ml catalase) in P12 buffer] was added and the flow cell was sealed with molten paraffin wax. Images were acquired by TIRF microscopy using an inverted microscope Ti-E/B (Nikon) equipped with perfect focus system (Nikon), a 100× 1.49 NA oil immersion TIRF objective (Nikon), three 20-mW diode lasers (488 nm, 561 nm, and 640 nm) and an electron-multiplying charge-coupled device detector (iXon X3DU897; Andor Technology). Image acquisition was controlled using Nikon Elements software and all assays were performed at room temperature (22°C). Images were acquired at 100 ms per frame for 300 frames.
For S2 cell lysates, glass-bottom dishes were plasma-cleaned and then amine-functionalized by incubating 1 hour in a vacuum chamber with 100 uL aminopropyltrimethoxysilane (Sigma A3648). The surface was further functionalized with a solution of mPEG and biotin-PEG (Laysan Bio MPEG-SVA-5000 and BIO-PEG-SVA-5000, respectively) to prevent non-specific protein adsorption while providing a specific attachment for biotinylated microtubules. Unreacted amine groups on the surface were removed by incubation with disuccinimidyl tartrate (Soltec Bioscience CL108). A stainless steel cooler ring was installed on the coated glass surface with vacuum grease. 0.5 mg/ml NeutrAvidin was infused into the cooler ring and incubated for 5 min at room temperature, followed by aspirating the solution. The glass bottom was rinsed twice using blocking buffer (1mg/ml casein in BRB80 buffer). Subsequently, polymerized Hilyte488- and biotin-labeled microtubules were diluted in BRB80 buffer supplemented with 10 μM taxol. The diluted microtubules then were infused into the cooler ring and incubated for 5 min at room temperature for biotin-NeutrAvidin binding, followed by aspirating the solution. The glass bottom was rinsed twice using blocking buffer. Finally, 5 uL cell lysate expressing octopus kinesin motor in motility mixture [2 mM ATP, 0.4 mg/ml casein, 6 mg/ml BSA, 10 μM taxol, and oxygen scavengers (1 mM DTT, 1 mM MgCl2, 10 mM glucose, 0.2 mg/ml glucose oxidase, and 0.08 mg/ml catalase) in P12 buffer] was added into the cooler ring. Images were acquired by TIRF microscopy (60× 1.49NA objective with a 1.6X zoom lens on an Olympus IX81 microscope base with cellTIRF module, equipped with a 50mW 488nm laser and 100mW 561nm laser, imaged with an Andor iXon DU-897U EMCCD camera). The sample was cooled with a bespoke sample chiller composed of an aluminum base cooled by a thermoelectric cooler controlled by a PID controller (TE Technology). The cooler ring fits into a recess in the chilled base, with thermal contact made through water deposited in the glass-bottom outside the cooler ring. Images were acquired at 200 ms per frame for 200 frames at room temperature (21°C), or 100 ms per frame for 400 frames at 11°C.
Crystallization of synaptotagmin C2A domains
Plasmid constructs
Wild-type and edited (I248V) versions of octopus synaptotagmin-1 (Syt1) C2A domain (residues 145–273) were chemically synthesized and cloned between the NdeI and XhoI sites of the p202 plasmid by Gibson assembly. Plasmids were then transformed into DH5α E. coli cells and their sequences verified by Sanger sequencing.
Expression & purification of Syt1 C2A domains
The expression plasmids encoding wt and I248V C2A domains were transformed into BL21(DE3) cells using the Mix and Go kit from Zymogen. Transformants were selected on kanamycin plates (50µg/ml) and inoculated into 10 ml of LB media + kanamycin and grown to saturation. 10 ml of the confluent cells were then inoculated into 1L of Terrific Broth. Cells were grown at 37°C until the OD600 reached ~2.0, then the cells were chilled to 18°C while shaking at 250 rpm. 400 µM of IPTG was added to induce gene expression and the cultures were grown for an additional 18 hours at 18°C. Cells were harvested by centrifugation, frozen in liquid nitrogen and stored at −80°C until needed.
Cell pellets were thawed on ice and resuspended in lysis buffer (100 mM HEPES, pH 7.4, 300 mM NaCl). The cell suspension was then lysed in a Microfluidics fluidizer and centrifuged in an SS-34 rotor for 45 minutes. The supernatant was passed through a Ni2+-NTA affinity resin to select the His6-tagged-maltose-binding protein (MBP) fusion proteins. The column was washed with 250 ml of lysis buffer, then washed with 250 ml of lysis buffer plus 15 mM imidazole to remove any non-specific binding proteins. The His-tagged-MBP-C2A proteins were eluted with lysis buffer plus 250 mM imidazole. The resulting fusion protein was then cleaved overnight at 4°C with 1 mg of TEV protease to separate the MBP component from the C2A domain. After cleavage, the protein solution was purified further by ion exchange over a SAE-Sepharose column (Buffer A: 100 mM HEPES, pH 7.4, Buffer B: 100 mM HEPES, pH 7.4 + 1M NaCl). The MBP eluted in the flow-through fraction while the C2A component eluted as a single peak during a 0 to 1 M salt gradient. The final step of the purification was over an 80 cm gel filtration column (Superdex 75) in lysis buffer plus 300 mM NaCl. All relevant protein peaks were analyzed using PAGE gels and imaged with the BioRad stainfree system.
Crystallization
The purified synaptotagmin C2A domains were concentrated to 20 mg/ml and screened for crystallization condition using the multifactorial approach. Multiple hits were discovered. For structure determination, Octopus Syt1 C2A crystals were grown on 31% PEG 4000 with 100 µM CHES, pH 9.3.
X-ray data collection, structure solution & refinement
Crystals were mounted in quartz capillary tubes and Cu-Kα X-ray data were collected at room temperature with a Rigaku Screen Machine fitted with a Saturn CCD detector. Data were integrated and reduced using the HKL software package (Table S6). The resolution cutoff was selected using the CC1/2 values.
Molecular replacement (MR), as implemented in Phenix, was used to solve the structure. The ultra-high resolution structure of rat synaptotagmin C2A (4wee) was used as the MR template. Multiple rounds of manual model fitting and X-ray refinement in Phenix 81 were used to complete the structure of Octopus synaptotagmin C2A. Multi-conformer, anisotropic refinement was conducted. Riding hydrogens were included. Solvent accessibility surface (SAS) of key residues was determined by the Shrake-Rupley SAS algorithm as implemented in Biopython 82.
Isothermal Titration Calorimetry
The Ca2+ binding affinity of the purified C2A domains was assessed using a nanoITC from TA Instruments/Waters (nWT=4, nI248V=6). To remove any residual Ca2+ from the C2A samples prior to ITC analysis, we added 10 mM EDTA, pH 8.0 to the protein sample and allowed it to equilibrate for 20 min on ice. A PD-10 buffer exchange column was then equilibrated with “Chelexed” buffer (40 mM HEPES pH: 7.4, 150 mM NaCl). The C2A-EDTA solution was then concentrated to 1 ml and applied to the resin bed of the PD-10 column. The desalted C2A domain fractions were collected and pooled. 400 µM purified C2A domain was titrated against 10 mM Ca2+ analyte at 10°C in 25 injections of 2 µl each. The mixer speed was set to 250 rpm. Data were processed using nanoAnalyze.
Temperature-sensitive RNA editing in the wild
Wild O. bimaculoides specimens were collected from Long Beach, CA, USA by Aquatic Research Consultants in September 2019 (n=4, T=21°C) and February 2022 (n=4, T=15°C). Specimens of Octopus bimaculatus were collected by SCUBA diving from Two Harbors, CA on Santa Catalina Island in September 2019 (n=11, T=22°C) and February/March 2020 (n=11, T=16°C). Temperatures were recorded at the capture sites for both species with HOBO pendant temperature data loggers (Onset) deployed for 4–9 weeks preceding animal capture (Figure S5). Species identifications were confirmed by amplicon sequencing of cytochrome c oxidase I (COI) containing species-specific sequences (File S1).
Animals were euthanized by immersion in 2% ethanol for 5 minutes followed by 5% ethanol for 5 minutes. Stellate ganglia were then immediately dissected, stored in RNAlater, and processed identically to those for the “time course of changes in RNA editing” samples.
QUANTIFICATION AND STATISTICAL ANALYSIS
Temperature-sensitive editing site discovery
To search for differential editing between the cold and warm samples from the initial 12-day temperature acclimation experiments, we first aligned the reads to the Octopus bimaculoides genome 60 using Bowtie2 with local alignment configuration and default parameters 83. Editing was quantified for 105,975 previously identified sites within the coding sequence (Liscovitch-Brauer et al. 2017, Supp. Table 5), using the REDItools command REDItoolKnown with the following parameters: -v 0 -n 0.001 -c 0 -t 2 -q −30 -m 40 84. For this analysis, we considered only editing sites covered by at least 100 reads in each of the twelve samples (62,661 sites). Sites with significant differential editing were identified using two one-tailed t-tests with a Benjamini–Hochberg multiple-testing correction (separately for each of the one-tailed tests; false discovery rate (FDR) ≤ 0.1).
Temperature-sensitive editome analyses
Assessments of editome-wide patterns
Depending on their position, many edits are capable of recoding a codon. Among the 16 possible codon substitutions by A-to-I RNA editing (Figure S2A), we assessed whether there was a systemic temperature-induced bias towards different kinds of amino acid recoding. Firstly, amino acids were grouped according to whether they are nonpolar, polar, positively-charged, or negatively-charged as outlined in Figure S2B. Then, changes were categorized as either inter-group (“changed”) or intra-group (“same”). The proportion of these events amongst warm-induced, temperature insensitive or cold-induced sites was compared using pairwise chi-squared tests with Bonferroni-adjusted p-values. Similarly, editing-induced amino acid substitutions were also grouped based on their BLOSUM80 score, as provided by the Biostrings R package 85. To test for statistical significance, we applied pairwise t-tests for the difference in scores between the original and recoded amino acid.
To test whether the differences between the groups may be due to differences in the distribution of editing levels, we constructed matched groups with similar editing level distributions. All significant differences remained significant after controlling for editing levels, except for the comparison of BLOSUM80 score between the cold and warm groups.
In addition, we also determined which protein traits (expressed as Uniprot keywords) were enriched in temperature-sensitive sites. The DAVID Functional Annotation tool v 6.8 86 was used to test for enrichment in transcripts containing a cold-induced recoding editing site with >10% increase in editing (n=571) compared to all transcripts with recoding sites (n=5417). Uniprot Keywords were considered significant when FDR < 0.05. The position of the large temperature-sensitive recoding sites within membrane-associated proteins (n=218) was predicted with DeepTMHMM 87 to assess whether recoding edits are biased towards transmembrane segments.
Potential mechanisms of temperature-sensitivity
One possible mechanism to induce temperature-sensitive RNA editing is a change in expression of ADARs, the catalytic enzymes that induce A-to-I RNA editing. To assess this possibility, RNASeq reads were quasimapped to the octopus exome 60 using Salmon (v0.8.2)88 with default parameters. The expression (TPM) of both catalytic paralogs (ADAR1, Ocbimv22018643m and ADAR2, Ocbimv22009676m) was compared between warm and cold samples using a t-test.
Another popular hypothesis to explain cold-induced RNA editing is a higher stability of dsRNA structure at lower temperature. To assess this possibility, we folded RNA sequences at 13 and 22°C to examine whether changes in dsRNA stability can explain higher observed % editing in the cold. Nucleotide sequences of the editing site flanked by 400 basepairs on each side were compiled from both the genome and exome sequences. Although RNA editing has traditionally been thought to occur mainly in the nucleus during transcription, we assessed both pre-mRNA and mature mRNA because cephalopods have been demonstrated to exhibit abundant editing of mature mRNA in the cytoplasm as well as pre-mRNA in the nucleus89. We found the lowest energy structure for each sequence at 295.15K and 286.15K using Fold from the RNAStructure package90 with default parameters. Then, the substructure surrounding the editing site was extracted using bpRNA91, and for generating a single RNA sequence, we connected its two arms by 7 “N” base-pairs. We recalculated this substructure’s free energy (ΔG) using Fold, taking the most probable structure. Sites for which the editing site didn’t reside in a dsRNA segment in the most probable structure were assigned ΔG = 0.
Lastly, we also examined differential expression of known ADAR-interacting proteins with temperature. RNASeq reads were quasimapped to the octopus transcriptome60 using Salmon (v0.8.2)88 with default parameters. Transcriptome-wide differential expression due to temperature was then analyzed using DESeq2 (v1.36.0)92. Estimated counts were generated from abundance using length-scaled TPM. Estimated log2-fold changes were adjusted using the adaptive Student’s t prior shrinkage estimator from the ‘apeglm’ package93. Significantly differentially-expressed genes were identified as those with adjusted p-values < 0.05. Amongst the differentially-expressed genes, we searched for octopus genes homologous to human genes that are known to interact with human ADAR1 or ADAR2 in human cell lines. Known ADAR-interacting proteins were compiled from BioGRID (v4.4.219)94 utilizing only those with in vivo evidence of interaction (e.g. affinity capture, n=370) and from Freund et al.64 (n=243). To identify homologs, we used BLAST95 and searched for an octopus hit to each human interactor query. If there was one or more octopus hits with a BLAST e-value < 1e-5 and covering >50% of the query gene’s length, the hit with the lowest e-value was defined as the octopus homolog.
Time course of changes in RNA editing
The temporal dynamics of the changes in editing in response to temperature were quantified for the cold-to-warm and warm-to-cold experiments. The difference in editing level from each timepoint to the next was quantified for each of the 18 sites assessed. Paired t-tests were conducted for each timeshift with p-values adjusted for multiple comparisons using the Bonferroni method.
Due to a constraint in the number of individuals, the time-course experiments were completed after 96 hours. To estimate whether the editing levels observed at this point had reached a steady-state, the values were compared to the editing levels at the same sites at the completion of the long-term temperature acclimation study. Because the short- and long-term studies differed in their temperatures by 1–2°C, the editing level values from the short-term studies were “prorated” accordingly based on the ∆editing level observed from the long-term experiment, assuming a linear relationship between ∆T and ∆editing level. The linearity assumption has been verified in squid (data not shown). The “prorated” editing levels were then compared to the long-term experiment values using a paired t-test.
Single molecule motility assays of kinesin
From the movies, maximum-intensity projections were generated to highlight microtubule-based events and kymographs were produced by drawing an ROI along microtubule tracks (width=3 pixels) using Fiji/ImageJ296 (Figure S4D). Motor events that lasted >=4 pixels were included in the analysis. Motile events were defined as events with a change in position over time and motor velocity was calculated as the run length (x axis of the kymograph) divided by the time (y axis of the kymograph). Stationary events were defined as events with no change in position over time.
Statistical analysis was performed and graphs were generated using Prism software (GraphPad). The comparisons between WT and mutant at room temperature and 11°C were made by using a two-tailed t-test. 70–100 motile events were evaluated for octopus kinesin-1 and >400 motile events were evaluated for rat kinesin-1 across 2–3 independent experiments. The temperature sensitivities of WT and mutant were calculated using the Q10 function from the ‘respirometry’ R package97.
Temperature-sensitive RNA editing in the wild
Temperature-sensitivity between winter and summer samples was assessed using t-tests in R98.
Supplementary Material
Related to Figures 1 and 2. Each vertical line represents one day.
Related to Figure 2. Panel A: Amongst the 16 amino acid substitutions possible due to A-to-I RNA editing, some were enriched or depleted amongst warm-induced sites (red), temperature-insensitive (grey), or cold-induced sites (blue). Panel B: Four polarity-based categories of amino acids, used to determine if a recoding event changes the category of the encoded amino acid (Figure 2).
Related to STAR Methods. Panel A: mRNA expression levels (TPM) of ADAR1 and ADAR2, estimated for the long-term acclimation samples, are comparable at cold and warm temperatures. Panels B and C: The free energies of the local dsRNA structures surrounding editing sites decrease in the cold but the decrease is not correlated with the temperature-sensitivity of editing. Panel B: Changes in the free-energies (∆G) of the local pre-mRNA (genomic sequences) dsRNA structure upon changing the temperature from 22°C to 13°C. Panel C: As in Panel B, for transcriptomic (mRNA) sequences. Panel D: Many putative ADAR-interactors exhibited an increased mRNA expression in the cold. Far more transcripts exhibited cold-induced than warm-induced expression. The horizontal dashed line marks adjusted p=0.05.
Related to Figure 4. Panel A: The edited lysine residue (K282; equivalent to K283 in human and rat KIF5) is universally conserved in 162 species across four phyla of kinesin-1 sequences available on NCBI. Panel B: Schematic of the domain organization of octopus and rat kinesin-1 proteins. For motility assays, the autoinhibitory stalk and tail domains were deleted, yielding constructs containing just the motor and dimerizing stalk domains (amino acids 1–558 of octopus kinesin-1 KIF5 and 1–559 of rat kinesin-1 KIF5C). The position of the edited lysine residue is shown. Panel C: Representative kymographs from single-molecule motility assays of WT and edited rat KIF5C(1–559)-3xmCit protein at RT and quantification of the effects on motor velocity and run length. Panel D: Representative kymographs from single-molecule motility assays of octopus KIF5(1–558)-Halo as visualized by TIRF microscopy at both room temperature (RT) and 11°C.
Related to Figure 6. Each x-axis tick mark spans one week.
Related to STAR Methods.
Related to STAR Methods and Figure S3.
Highlights of “Temperature-dependent RNA editing in octopus extensively recodes the neural proteome”.
Octopus bimaculoides increase A-to-I RNA editing at >20,000 sites in the cold.
Editing shifts occur within hours and are observed in wild populations.
As a functional example, one cold-induced site alters kinesin motility.
Another cold-induced site alters Ca2+-binding affinity of synaptotagmin.
Acknowledgements
Octopus bimaculatus individuals sampled on Santa Catalina Island were collected under CA Department of Fish and Wildlife scientific collecting permit SC-191820001. M.A.B. was supported by an NSF Postdoctoral Research Fellowship in Biology (DBI-1907197). K.J.V. was supported by NIH R35GM131744. EE was supported by NSF-BSF (BSF 2020759), BSF 2017262 and BSF 2013094. JJCR was supported by NSF-BSF 2110074, NSF 2220587, BSF 2017262, BSF 2013094, and NSF 1827509. All octopus drawings in figures were created by Roger Hall. We thank the MBL’s cephalopods program for their support in this study.
Inclusion and Diversity:
One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
Footnotes
Statements
Declaration of Interests:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hodgkin AL, and Keynes RD. (1955). Active transport of cations in giant axons from Sepia and Loligo. J. Physiol. 128, 28–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Leary T, and Marder E. (2016). Temperature-Robust Neural Function from Activity-Dependent Ion Channel Regulation. Curr. Biol. 26, 2935–2941. 10.1016/j.cub.2016.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter DO. (1981). Ionic and metabolic bases of neuronal thermosensitivity. Fed. Proc. 40, 2808–2813. [PubMed] [Google Scholar]
- 4.Montgomery JC, and MacDonald JA. (1990). Effects of temperature on nervous system: implications for behavioral performance. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 28, R191–R196. [DOI] [PubMed] [Google Scholar]
- 5.Kukita F. (1982). Properties of sodium and potassium channels of the squid giant axon far below 0°C. J. Membr. Biol. 68, 151–160. 10.1007/BF01872261. [DOI] [PubMed] [Google Scholar]
- 6.Weight FF, and Erulkar SD. (1976). Synaptic transmission and effects of temperature at the squid giant synapse. Nature 261, 720–722. [DOI] [PubMed] [Google Scholar]
- 7.Joyner RW. (1981). Temperature effects on neuronal elements. Fed. Proc. 40, 2814–2818. [PubMed] [Google Scholar]
- 8.Charlton MP, and Atwood HL. (1979). Synaptic transmission: temperature-sensitivity of calcium entry in presynaptic terminals. Brain Res. 170, 543–546. [DOI] [PubMed] [Google Scholar]
- 9.Rosenthal JJC, and Bezanilla F. (2000). Seasonal Variation in Conduction Velocity of Action Potentials in Squid Giant Axons. Biol. Bull. 199, 135–143. [DOI] [PubMed] [Google Scholar]
- 10.Katz B, and Miledi R. (1965). The effect of temperature on the synaptic delay at the neuromuscular junction. J. Physiol. 181, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagerspetz KYH. (1974). Temperature acclimation and the nervous system. Biol. Rev. 49, 477–514. [DOI] [PubMed] [Google Scholar]
- 12.Prosser CL, and Nelson DO. (1981). The role of nervous systems in temperature adaptation of poikilotherms. Annu. Rev. Physiol. 43, 281–300. [DOI] [PubMed] [Google Scholar]
- 13.Somero GN. (2018). RNA thermosensors: how might animals exploit their regulatory potential? J. Exp. Biol. 221, jeb162842. 10.1242/jeb.162842. [DOI] [PubMed] [Google Scholar]
- 14.Nishikura K. (2010). Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349. 10.1146/annurev-biochem-060208-105251.Functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg E, and Levanon EY. (2018). A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490. 10.1038/s41576-018-0006-1. [DOI] [PubMed] [Google Scholar]
- 16.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. (2014). A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376. 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daneck P, Nellaker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, Durbin R, Keane TM, and Adams DJ. (2012). High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 13, R26. 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licht K, Kapoor U, Amman F, Picardi E, Martin D, Bajad P, and Jantsch MF. (2019). A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 29, 1453–1463. 10.1101/gr.242636.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neeman Y, Levanon EY, Jantsch MF, and Eisenberg E. (2006). RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 12, 1802–1809. 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. (2004). Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005. 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 21.Kim DDY, Kim TTY, Walsh T, Kobayashi Y, Matise TC, Buyske S, and Gabriel A. (2004). Widespread RNA Editing of Embedded Alu Elements in the Human Transcriptome. Genome Res. 14, 1719–1725. 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blow M, Futreal PA, Wooster R, and Stratton MR. (2004). A survey of RNA editing in human brain. Genome Res. 14, 2379–2387. 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Athanasiadis A, Rich A, and Maas S. (2004). Widespread A-to-I RNA Editing of Alu-Containing mRNAs in the Human Transcriptome. PLoS Biol. 2, e391. 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, Mclaughlin PJ, et al. (2014). The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA. Cell Rep. 9, 1482–1494. 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, and Walkley CR. (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120. 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, and Stetson DB. (2015). Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 43, 933–944. 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabay O, Shoshan Y, Kopel E, Ben-Zvi U, Mann TD, Bressler N, Cohen‐Fultheim R, Schaffer AA, Roth SH, Tzur Z, et al. (2022). Landscape of adenosine-to-inosine RNA recoding across human tissues. Nat. Commun. 13, 1184. 10.1038/s41467-022-28841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto Y, Cohen HY, and Levanon EY. (2014). Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G, and Zhang J. (2014). Human coding RNA editing is generally nonadaptive. Proc. Natl. Acad. Sci. 111, 3769–3774. 10.1073/pnas.1321745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchumenski I, Bartok O, Ashwal-Fluss R, Pandey V, Porath HT, Levanon EY, and Kadener S. (2017). Dynamic hyper-editing underlies temperature adaptation in Drosophila. PLOS Genet. 13, e1006931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savva YA, Laurent G.St., and Reenan RA. (2016). Genome-Wide Analysis of A-to-I RNA Editing. In Post-Transcriptional Gene Regulation Methods in Molecular Biology., Dassi E, ed. (Springer; New York: ), pp. 255–268. 10.1007/978-1-4939-3067-8_15. [DOI] [PubMed] [Google Scholar]
- 32.Mazloomian A, and Meyer IM. (2015). Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing. RNA Biol. 12, 1391–1401. 10.1080/15476286.2015.1107703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, and Li JB. (2013). Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132. 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St Laurent G, Tackett MR, Nechkin S, Shtokalo D, Antonets D, Savva YA, Maloney R, Kapranov P, Lawrence CE, and Reenan RA. (2013). Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat. Struct. Mol. Biol. 20, 1333–1339. 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
- 36.Sapiro AL, Shmueli A, Henry GL, Li Q, Shalit T, Yaron O, Paas Y, Billy Li J, and Shohat-Ophir G. (2019). Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain. Proc. Natl. Acad. Sci. 116, 2318–2327. 10.1073/pnas.1811768116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan Y, Dou S, Luo S, Zhang H, and Lu J. (2017). Adaptation of A-to-I RNA editing in Drosophila. PLOS Genet. 13, e1006648. 10.1371/journal.pgen.1006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R, Deng P, Jacobson D, and Li JB. (2017). Evolutionary analysis reveals regulatory and functional landscape of coding and non-coding RNA editing. PLOS Genet. 13, e1006563. 10.1371/journal.pgen.1006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Zhou H, Kong Y, Pan B, Chen L, Wang H, Hao P, and Li X. (2016). The Landscape of A-to-I RNA Editome Is Shaped by Both Positive and Purifying Selection. PLOS Genet. 12, e1006191. 10.1371/journal.pgen.1006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckingham SD, Biggin PC, Sattelle BM, Brown LA, and Sattelle DB. (2005). Insect GABA Receptors: Splicing, Editing, and Targeting by Antiparasitics and Insecticides. Mol. Pharmacol. 68, 942–951. 10.1124/mol.105.015313. [DOI] [PubMed] [Google Scholar]
- 41.Jepson JEC, Savva YA, Yokose C, Sugden AU, Sahin A, and Reenan RA. (2011). Engineered Alterations in RNA Editing Modulate Complex Behavior in Drosophila. J. Biol. Chem. 286, 8325–8337. 10.1074/jbc.M110.186817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keegan LP, Brindle J, Gallo A, Leroy A, Reenan RA, and O’Connell MA. (2005). Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 24, 2183–2193. 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palladino MJ, Keegan LP, O’Connell MA, and Reenan RA. (2000). dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6, 1004–1018. 10.1017/S1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan MY, Maloney R, Fineberg JD, Reenan RA, and Horn R. (2012). RNA editing in eag potassium channels: Biophysical consequences of editing a conserved S6 residue. Channels 6, 443–452. 10.4161/chan.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savva YA, Jepson JEC, Sahin A, Sugden AU, Dorsky JS, Alpert L, Lawrence C, and Reenan RA. (2012). Auto-regulatory RNA editing fine-tunes mRNA re-coding and complex behaviour in Drosophila. Nat. Commun. 3, 790. 10.1038/ncomms1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenov EP, and Pak WL. (1999). Diversification of Drosophila Chloride Channel Gene by Multiple Posttranscriptional mRNA Modifications. J. Neurochem. 72, 66–72. 10.1046/j.1471-4159.1999.0720066.x. [DOI] [PubMed] [Google Scholar]
- 47.Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJC, and Eisenberg E. (2015). The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife 4, e05198. 10.7554/eLife.05198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJC, and Eisenberg E. (2017). Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 169, 191–202.e11. 10.1016/j.cell.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoshan Y, Liscovitch-Brauer N, Rosenthal JJC, and Eisenberg E. (2021). Adaptive Proteome Diversification by Nonsynonymous A-to-I RNA Editing in Coleoid Cephalopods. Mol. Biol. Evol. 38, 3775–3788. 10.1093/molbev/msab154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moldovan M. (2020). Adaptive evolution at mRNA editing sites in soft-bodied cephalopods. PeerJ 8, e10456. 10.7717/peerj.10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrett S, and Rosenthal JJC. (2012). RNA Editing Underlies Temperature Adaptation in K+ Channels from Polar Octopuses. Science 335, 848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colina C, Palavicini JP, Srikumar D, Holmgren M, and Rosenthal JJC. (2010). Regulation of Na+/K+ ATPase Transport Velocity by RNA Editing. PLOS Biol. 8, e1000540. 10.1371/journal.pbio.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal JJC, and Bezanilla F. (2002). Extensive Editing of mRNAs for the Squid Delayed Rectifier K+ Channel Regulates Subunit Tetramerization. Neuron 34, 743–757. [DOI] [PubMed] [Google Scholar]
- 54.Yablonovitch AL, Fu J, Li K, Mahato S, Kang L, Rashkovetsky E, Korol AB, Tang H, Michalak P, Zelhof AC, et al. (2017). Regulation of gene expression and RNA editing in Drosophila adapting to divergent microclimates. Nat. Commun. 8, 1570. 10.1038/s41467-017-01658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stocker J, Huang H-W, Wang H-M, Chang H-W, Chiu C-C, Cho C-L, and Tseng C-N. (2013). Reduction of RNA A-to-I editing in Drosophila acclimated to heat shock. Kaohsiung J. Med. Sci. 29, 478–483. 10.1016/j.kjms.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Rieder LE, Savva YA, Reyna MA, Chang Y-J, Dorsky JS, Rezaei A, and Reenan RA. (2015). Dynamic response of RNA editing to temperature in Drosophila. BMC Biol. 13, 1. 10.1186/s12915-014-0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riemondy KA, Gillen AE, White EA, Bogren LK, Hesselberth JR, and Martin SL. (2018). Dynamic temperature-sensitive A-to-I RNA editing in the brain of a heterothermic mammal during hibernation. RNA 24, 1481–1495. 10.1101/288159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jereb P, Roper CFE, Norman MD, and Finn JK eds. (2013). Cephalopods of the world. An annotated and illustrated catalogue of cephalopod species known to date. Volume 3. Octopods and Vampire Squids 4th ed. (FAO; ). [Google Scholar]
- 59.Albertin CB, Medina-Ruiz S, Mitros T, Schmidbaur H, Sanchez G, Wang ZY, Grimwood J, Rosenthal JJC, Ragsdale CW, Simakov O, et al. (2022). Genome and transcriptome mechanisms driving cephalopod evolution. Nat. Commun. 13, 2427. 10.1038/s41467-022-29748-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, and Rokhsar DS. (2015). The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224. 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avram-Shperling A, Kopel E, Twersky I, Gabay O, Ben-David A, Karako-Lampert S, Rosenthal JJC, Levanon EY, Eisenberg E, and Ben-Aroya S. (2023). Identification of exceptionally potent adenosine deaminases RNA editors from high body temperature organisms. PLOS Genet. 19, e1010661. 10.1371/journal.pgen.1010661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinones-Valdez G, Tran SS, Jun H-I, Bahn JH, Yang E-W, Zhan L, Brümmer A, Wei X, Van Nostrand EL, Pratt GA, et al. (2019). Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2, 1–14. 10.1038/s42003-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajendren S, Dhakal A, Vadlamani P, Townsend J, Deffit SN, and Hundley HA. (2021). Profiling neural editomes reveals a molecular mechanism to regulate RNA editing during development. Genome Res. 31, 27–39. 10.1101/gr.267575.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freund EC, Sapiro AL, Li Q, Linder S, Moresco JJ, Yates JR, and Li JB. (2020). Unbiased Identification of trans Regulators of ADAR and A-to-I RNA Editing. Cell Rep. 31. 10.1016/j.celrep.2020.107656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.NOAA NCEI Coastal Water Temperature Guide. https://www.ncei.noaa.gov/access/coastal-water-temperature-guide/all_table.html
- 66.Davletov BA, and Sudhof TC. (1993). A Single C2 Domain from Synaptotagmin I Is Sufficient for High Affinity Ca2+/Phospholipid Binding. J. Biol. Chem. 268, 26386–26390. [PubMed] [Google Scholar]
- 67.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, and Sprang SR. (1995). Structure of the first C2 domain of synaptotagmin I: A novel Ca2+/phospholipid-binding fold. Cell 80, 929–938. 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Q, Lai Y, Bacaj T, Zhao M, Lyubimov AY, Uervirojnangkoorn M, Zeldin OB, Brewster AS, Sauter NK, Cohen AE, et al. (2015). Architecture of the synaptotagmin - SNARE machinery for neuronal exocytosis. Nature 525, 62–67. 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pickford GE, and McConnaughey BH. (1949). The Octopus bimaculatus problem: a study in sibling species. Bull. Bingham Oceanogr. Collect. 12, 1–66. [Google Scholar]
- 70.Liao M, Somero GN, and Dong Y. (2019). Comparing mutagenesis and simulations as tools for identifying functionally important sequence changes for protein thermal adaptation. Proc. Natl. Acad. Sci. 116, 679–688. 10.1073/pnas.1817455116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scholander PF, Flagg W, Walters V, and Irving L. (1953). Climatic Adaptation in Arctic and Tropical Poikilotherms. Physiol. Zool. 26, 67–92. 10.1086/physzool.26.1.30152151. [DOI] [Google Scholar]
- 72.Gross GW. (1973). The effect of temperature on the rapid axoplasmic transport in C-fibers. Brain Res. 56, 359–363. 10.1016/0006-8993(73)90353-3. [DOI] [PubMed] [Google Scholar]
- 73.Cosens B, Thacker D, and Brimijoin S. (1976). Temperature-dependence of rapid axonal transport in sympathetic nerves of the rabbit. J. Neurobiol. 7, 339–354. 10.1002/neu.480070406. [DOI] [PubMed] [Google Scholar]
- 74.Edström A, and Hanson M. (1973). Temperature effects on fast axonal transport of proteinsin vitro in frog sciatic nerves. Brain Res. 58, 345–354. 10.1016/0006-8993(73)90006-1. [DOI] [PubMed] [Google Scholar]
- 75.Cossins AR, and Prosser CL. (1978). Evolutionary adaptation of membranes to temperature. Proc. Natl. Acad. Sci. 75, 2040–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hochachka PW, and Somero GN. (2002). Biochemical Adaptation: Mechanism and Process in Physiological Evolution (Oxford University Press; ). [Google Scholar]
- 77.Crawford K, Diaz Quiroz JF, Koenig KM, Ahuja N, Albertin CB, and Rosenthal JJC. (2020). Highly Efficient Knockout of a Squid Pigmentation Gene. Curr. Biol. 30, 3484–3490. 10.1016/j.cub.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahuja N, Hwaun E, Pungor J, Rafiq R, Nemes S, Sakmar T, Vogt MA, Grasse B, Diaz Quiroz JF, Montague T, et al. (2023). Creation of an Albino Squid Line by CRISPR-Cas9 and Its Application for in vivo Functional Imaging of Neural Activity. 10.2139/ssrn.4369821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rinkevich FD, Schweitzer PA, and Scott JG. (2012). Antisense sequencing improves the accuracy and precision of A-to-I editing measurements using the peak height ratio method. BMC Res. Notes 5, 63. 10.1186/1756-0500-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cai D, McEwen DP, Martens JR, Meyhofer E, and Verhey KJ. (2009). Single Molecule Imaging Reveals Differences in Microtubule Track Selection Between Kinesin Motors. PLoS Biol. 7, e1000216. 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung L-W, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol. 75, 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422– 1423. 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langmead B, and Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–360. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Picardi E, and Pesole G. (2013). REDItools: high-throughput RNA editing detection made easy. Bioinformatics 29, 1813–1814. 10.1093/bioinformatics/btt287. [DOI] [PubMed] [Google Scholar]
- 85.Pagès H, Aboyoun P, Gentleman R, and DebRoy S. (2022). Biostrings: Efficient manipulation of biological strings. R package version 2.64.0. https://bioconductor.org/packages/Biostrings. [Google Scholar]
- 86.Huang DW, Sherman BT, and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 87.Hallgren J, Krogh Anders, Tsirigos Konstantinos D., Pedersen Mads D., Armenteros José Juan Almagro, Marcatili Paolo, Henrik Nielsen, and Winther O. (2022). DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. 10.1101/2022.04.08.487609. [DOI] [Google Scholar]
- 88.Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vallecillo-Viejo IC, Liscovitch-Brauer N, Diaz Quiroz JF, Nemes S, Rangan KJ, Levinson SR, Eisenberg E, and Rosenthal JJC. (2020). Spatially regulated editing of genetic information within a neuron. Nucleic Acids Res, 1–13. 10.1093/nar/gkaa172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reuter JS, and Mathews DH. (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129. 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Danaee P, Rouches M, Wiley M, Deng D, Huang L, and Hendrix D. (2018). bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res. 46, 5381–5394. 10.1093/nar/gky285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu A, Ibrahim JG, and Love MI. (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084– 2092. 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, and Tyers M. (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539. 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL. (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Birk MA. (2021). respirometry: Tools for Conducting and Analyzing Respirometry Experiments. R package version 1.3.0. http://www.cran.r-project.org/package=respirometry. [Google Scholar]
- 98.R Core Team (2022). R: A language and environment for statistical computing. http://www.r-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figures 1 and 2. Each vertical line represents one day.
Related to Figure 2. Panel A: Amongst the 16 amino acid substitutions possible due to A-to-I RNA editing, some were enriched or depleted amongst warm-induced sites (red), temperature-insensitive (grey), or cold-induced sites (blue). Panel B: Four polarity-based categories of amino acids, used to determine if a recoding event changes the category of the encoded amino acid (Figure 2).
Related to STAR Methods. Panel A: mRNA expression levels (TPM) of ADAR1 and ADAR2, estimated for the long-term acclimation samples, are comparable at cold and warm temperatures. Panels B and C: The free energies of the local dsRNA structures surrounding editing sites decrease in the cold but the decrease is not correlated with the temperature-sensitivity of editing. Panel B: Changes in the free-energies (∆G) of the local pre-mRNA (genomic sequences) dsRNA structure upon changing the temperature from 22°C to 13°C. Panel C: As in Panel B, for transcriptomic (mRNA) sequences. Panel D: Many putative ADAR-interactors exhibited an increased mRNA expression in the cold. Far more transcripts exhibited cold-induced than warm-induced expression. The horizontal dashed line marks adjusted p=0.05.
Related to Figure 4. Panel A: The edited lysine residue (K282; equivalent to K283 in human and rat KIF5) is universally conserved in 162 species across four phyla of kinesin-1 sequences available on NCBI. Panel B: Schematic of the domain organization of octopus and rat kinesin-1 proteins. For motility assays, the autoinhibitory stalk and tail domains were deleted, yielding constructs containing just the motor and dimerizing stalk domains (amino acids 1–558 of octopus kinesin-1 KIF5 and 1–559 of rat kinesin-1 KIF5C). The position of the edited lysine residue is shown. Panel C: Representative kymographs from single-molecule motility assays of WT and edited rat KIF5C(1–559)-3xmCit protein at RT and quantification of the effects on motor velocity and run length. Panel D: Representative kymographs from single-molecule motility assays of octopus KIF5(1–558)-Halo as visualized by TIRF microscopy at both room temperature (RT) and 11°C.
Related to Figure 6. Each x-axis tick mark spans one week.
Related to STAR Methods.
Related to STAR Methods and Figure S3.
Data Availability Statement
Illumina RNASeq data have been deposited at the NCBI SRA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. X-ray crystallography structures have been deposited at the RCSB PDB and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
All original code has been deposited at GitHub and Zenodo and is publicly available as of the date of publication. URLs and DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.