Abstract
The US Food and Drug Administration (FDA) regulatory process often involves several reviewers who focus on sets of information related to their respective areas of review. Accordingly, manufacturers that provide submission packages to regulatory agencies are instructed to organize the contents using a structure that enables the information to be easily allocated, retrieved, and reviewed. However, this practice is not always followed correctly; as such, some documents are not well structured, with similar information spreading across different sections, hindering the efficient access and review of all of the relevant data as a whole. To improve this common situation, we evaluated an artificial intelligence (AI)-based natural language processing (NLP) methodology, called Bidirectional Encoder Representations from Transformers (BERT), to automatically classify free-text information into standardized sections, supporting a holistic review of drug safety and efficacy. Specifically, FDA labeling documents were used in this study as a proof of concept, where the labeling section structure defined by the Physician Label Rule (PLR) was used to classify labels in the development of the model. The model was subsequently evaluated on texts from both well-structured labeling documents (i.e., PLR-based labeling) and less- or differently structured documents (i.e., non-PLR and Summary of Product Characteristic [SmPC] labeling.) In the training process, the model yielded 96% and 88% accuracy for binary and multiclass tasks, respectively. The testing accuracies observed for the PLR, non-PLR, and SmPC testing data sets for the binary model were 95%, 88%, and 88%, and for the multiclass model were 82%, 73%, and 68%, respectively. Our study demonstrated that automatically classifying free texts into standardized sections with AI language models could be an advanced regulatory science approach for supporting the review process by effectively processing unformatted documents.
Introduction
Regulatory documents are typically large and cover a broad range of information. Individual reviewers usually focus on specific sets of information, such as safety or efficacy, in accordance with their review assignments. Therefore, regulatory documents need to be organized by using a structure within which the information can be easily allocated, retrieved, and reviewed. Unfortunately, this is not always the case, although structured documents are recognized as crucial to an improved regulatory review process. For example, the structure and information in FDA labeling documents have changed over the past 40 years. In 2005, the FDA published “Guidance for Industry: Providing Regulatory Submissions in Electronic Format – Content of Labeling”, which provided guidelines for regulatory submissions in the Structured Product Labeling (SPL) format.1 With the SPL format, texts are preannotated into specific labeling sections, making it easier for FDA researchers and reviewers to retrieve and analyze the documents’ textual information.
Nevertheless, many labeling documents are still not well-formatted, which hinders access to and use of the information they contain. In addition, it would not be unusual if, in the future, the structure of labeling documents is again revised to enhance the clarity and organization of information. This situation raises a question about how to normalize information from a previous version of labeling with future formats to provide a holistic view of drug safety and efficacy, particularly when reviewing drugs from the same therapeutic area or pharmaceutical class. Moreover, different countries have their own formats for structuring information; to expand the FDA knowledge base, their regulatory documents must be converted to the format used in the FDA review process.
Text classification is one of the principal tasks in NLP, which has significantly advanced with transformer-based language models.2−4 While text classification models often focus on sentiment analysis,5−7 several studies have demonstrated NLP application in classifying free (unstructured) texts into predefined categories.8,9 For instance, Dernoncourt and Lee8 classified sentences into sections, (i.e., background, objective, method, result, or conclusion), for approximately 200,000 medical abstracts with more than 90% accuracy. Moreover, in an attempt to distinguish between sentences with and without propaganda,9 a BERT-based classification model achieved 55–80%+ accuracy for the various classification tasks in accordance with 18 different propaganda techniques and categories.
Several studies have aimed to classify or group drugs and/or drug labeling documents based on patterns within their texts.10,11 In 2011, Bisgin et al.10 categorized drugs with similar safety concerns, therapeutic uses, or both by applying the unsupervised text mining method of topic modeling to labeling documents for 794 FDA-approved drugs. Furthermore, in 2019, Wu et al.11 grouped drug labeling documents by conducting a hierarchical cluster analysis to uncover similar patterns among the MedDRA (Medical Dictionary for Regulatory Activities) -preferred terms and adverse drug reactions (ADRs) within the boxed warning sections of 367 single-ingredient drugs. These studies demonstrated that hidden patterns exist within the texts of drug labeling documents, enabling these documents and their sections to be grouped with computer-aided or machine learning technologies.
Our study expands on these concepts, and as such, we developed a language model to automatically classify free-text information from FDA drug labeling documents into defined and standardized sections. We set the PLR labeling format as the standard in constructing the language model. Given the diversity of the drug labeling sections, we used various categorical configurations when training the classification model to examine how performance is impacted by specific circumstances such as how results may change when the number of categories is increased. The model was evaluated with texts from PLR-formatted labeling documents and subsequently applied to non-PLR-formatted labeling documents, the format usually found in older labeling documents. We also applied the model to the classification of UK drug labeling documents with the SmPC format.
Materials and Methods
US FDA Drug Labeling
Based on the PLR, FDA prescription drug labeling documents generally can have one of two formats. The PLR format, first published by the FDA in 2006, is the gold standard for prescription drug labeling formats, as all prescription drug labeling submitted after June 2001 is required to conform to it (documents submitted between June 2001 and 2006 were required to retroactively update to the PLR format). On the other hand, nonprescription drug labeling, such as those for over-the-counter (OTC) drugs, and prescription labeling documents approved before 2001 are not required to use PLR format and are considered non-PLR formatted documents.
It is important to note the differences between the PLR and non-PLR format. The FDA posits that the PLR format “enhances the safe and effective use of human prescription drugs ... and reduces the number of adverse reactions resulting from medication errors due to misunderstood or incorrectly applied drug information”.12 Additionally, the PLR format is important for making prescription information more accessible to healthcare practitioners, patients, and researchers. The FDA asserts that the PLR format’s modern approach to communicating accurate drug use information makes prescription information “more accessible for use with electronic prescribing tools and other electronic information resources’.13 Despite the benefits of using the PLR format vs the non-PLR format, some older labeling documents remained in the non-PLR format; only new drug applications (NDAs) and biologics license applications (BLAs) approved from June 2001 to 2006 being retroactively updated to be PLR compliant.12
With the PLR format being the preferred standard for retrieving and using information from drug labeling documents, this study aimed to organize all types of drug labeling into appropriate PLR-formatted sections. To begin, 45,626 prescription drug labeling documents were obtained and processed from DailyMed’s full release of human prescription labeling (retrieved February 28, 2022).14 Of these documents, 29,709 (65%) were in the PLR format, while 15,917 (35%) were in the non-PLR format. A total of 17,453,802 sentences were extracted using Python and Natural Language Toolkit (NLTK) libraries.15 These sentences were further mapped with logical observation identifiers, names, and codes (LOINC),16 which are the official codes used to determine the located sections in the labeling documents. PLR and non-PLR labeling documents had to be processed separately, because they have different LOINC codes.
Figure 1 provides examples of the contents included within various major sections within FDA prescription drug labeling documents. From a drug labeling document for the adalimumab-atto injection (which this study is not associated with), Figure 1A is an excerpt from the “indications and usage” section, Figure 1B is an excerpt from the “warnings and precautions” section, and Figure 1C is an excerpt from the “adverse reactions” section. Together, these figures provide a general overview of what information can be expected within these key drug labeling sections.
Figure 1.
Excerpts from drug labeling document. (A) Indications and usage. (B) Warnings and precautions. (C) Adverse reactions.
UK Drug Labeling
In the UK, the primary drug labeling documents are SmPCs. These provide vital information to healthcare professionals, such as how to use and prescribe medicines.17 SmPCs are written and updated by pharmaceutical companies based on their research, and are checked and approved by the UK or European medicines licensing agencies. They are akin to FDA-regulated prescription drug labeling in that each document contains labeling sections comparable to those found in the PLR or non-PLR formats. For instance, the SmPC section “Therapeutic Indications” contains similar information to the FDA labeling section “Indications and Usage.”
To determine if the language model produced in this study could be applicable to external drug labeling documents, a collection of 9580 SmPCs was obtained from the UK medicine database Electronic Medicines Compendium (retrieved June 26, 2022).17 Using similar data processing techniques, we collected 2,180,388 sentences.
Summary of Data Sets
Table 1 summarizes the three data sets used in this study. Overall, we collected over 55,000 labeling documents and over 19 million sentences from (1) PLR, (2) non-PLR, and (3) SmPC formatted documents.
Table 1. Summary of Datasets.
| Data set | Origin | No. Documents | No. Sentences |
|---|---|---|---|
| 1. PLR | US – DailyMed | 29,709 | 14,072,802 |
| 2. non-PLR | US – DailyMed | 15,917 | 3,380,819 |
| US Total | 45,626 | 17,453,802 | |
| 3. SmPC | UK – EMC | 9580 | 2,180,388 |
| Overall Total | 55,206 | 19,634,190 |
Modeling Algorithms
In the primary task, we used BERT to train the sentence classification model. BERT is a state-of-the-art language model popularly used for a wide variety of NLP tasks, including text classification, question answering, and next-sentence prediction.18 As its full name implies, BERT is a multilayer encoder with a transformer architecture, or an attention-based model.19 It was pretrained on BooksCorpus (800 million words) and Wikipedia (2500 million words). Given its self-attention mechanism, the trained model could be further fine-tuned for a multitude of tasks by training different heads on top of the model architecture.
Besides the basic BERT model (BERT-base), there are many different BERT models. To explore the model’s impact on the results, several alternative BERT models, ALBERT,20 DistilBERT,21 and RoBERTa,22 were fine-tuned and tested on the same data. These models were selected due to their unique and proven capabilities. ALBERT implements parameter-reduction techniques to lower memory consumption and increase the training speed of BERT while limiting the loss of language understanding. DistilBERT leverages knowledge distillation during the pretraining phase to reduce model size and increase training speed while retaining most of its language understanding capabilities. RoBERTa implements a longer training phase with more data and dynamically changes the masking pattern applied to the training data, enabling it to perform equally well or better than models published after BERT. Random forest (RF)23 and support vector machine (SVM)24 models from the scikit-learn package25 were used as a baseline. For more information about the models used in this study, see Table 2 for a brief overview of each model.
Table 2. Overview of the Selected Models.
| Model | Description |
|---|---|
| RF23 | A random forest (RF) classifier is a machine learning algorithm that combines the output of multiple decision trees to produce a single result. In more detail, it fits a number of decision tree classifiers on a collection of subsamples from the data set and uses averaging to improve predictive accuracy and control overfitting. |
| SVM24 | A support vector machine (SVM) is a supervised machine learning algorithm that is primarily used for classification problems. In this algorithm, each data item is plotted as a point in n-dimensional space, representing each feature as a coordinate. Then, the algorithm finds the optimal decision boundary (i.e., the hyperplane) to separate the various classes by using the extreme points/vectors (i.e., the support vectors). |
| BERT18 | BERT, which stands for Bidirectional Encoder Representations from Transformers, is a general-purpose language model that was pretrained on the BookCorpus (800 M words) and English Wikipedia (2500M) data sets. By employing the use of self-attention mechanisms, BERT can be fine-tuned with additional layers to complete new tasks with new data, making it a foundation for many transformer-based language models. At the time of its release, BERT achieved state-of-the-art performance for several tasks of the General Language Understanding Evaluation (GLUE) benchmark. The base model is composed of approximately 100 million parameters. |
| ALBERT20 | ALBERT, or A Lite BERT, addresses the problems of memory limitation and lengthy training times by modifying BERT through the incorporation of the parameter reduction techniques of factorized embedding parametrization and cross-later parameter sharing. Despite having much fewer parameters (i.e., only 12 million parameters in the base model), ALBERT experiences minimal loss in language understanding, performing almost equally to BERT for the GLUE benchmark. |
| DistilBERT21 | DistilBERT, a distilled version of BERT, addresses concerns about the computational efficiency of large transformer-based language models by applying knowledge distillation during BERT’s pretraining phase. This method reduced the size of BERT by 40%, while retaining 97% of its language understanding capabilities and being 60% faster. |
| RoBERTa22 | RoBERTa, a robustly optimized BERT pretraining approach, modifies BERT by training the model longer and with more data, training on longer sequences, and dynamically changing the masking pattern applied to the training data. In more detail, this model’s training data is composed of the BookCorpus and English Wikipedia (16GB), CC-News (76GB), OpenWebText (38GB), and Stories (31GB) data sets. With these modifications, RoBERTa improved on the results obtained by BERT, achieving several state-of-the-art results. The base model is composed of approximately 110 million parameters. |
Fine-Tuning the Model
To explore the model’s ability to predict which section a given drug labeling sentence belonged to, a series of binary and multiclass classification tasks were developed, with the focus on several key PLR sections: (1) “Indications and Usage,” (2) “Warnings and Precautions,” and (3) “Adverse Reactions”. For the primary binary classification task, the end points were “Indications and Usage” and “Warnings and Precautions”; and since texts from these sections are easily discernible, this was expected to provide a solid baseline of the model’s language understanding capabilities. Conversely, the primary multiclass task included these end points plus “Adverse Reactions” and “Other/Unknown” (including all the remaining drug labeling sections), giving the model a much more difficult task and providing a measure of its ability to differentiate among a multitude of drug labeling sections. From here, the training and testing data sets were prepared. For each classification modeling task, 10,000 sentences were obtained for each of the end points present in each data set, and as such, the data sets have balanced classes.
For the BERT-based models, these data sets were tokenized using their respective HuggingFace autotokenizer. The processed and tokenized training data sets were split into 80% for training and 20% for validation. For each BERT-based model, their respective HuggingFace model was fine-tuned using a PLR-formatted training data set. More specifically, each model was fine-tuned over the course of 10 epochs using the model’s default parameters and the “Accuracy” metric. The models were fine-tuned for only 10 epochs, as it was noted that improvements in performance plateaued before or around this stage. Finally, each model was evaluated using PLR-, non-PLR-, and SmPC-formatted testing data sets, each containing 10,000 sentences per end point that were new to or unseen by the model.
On the other hand, for the RF and SVM models, the data sets were tokenized and processed using the NLTK package15 for Python. In more detail, the “word_tokenizer” function was used to tokenize the sentences and “WordNetLemmatizer” was used to lemmatize each word. The sentences were then vectorized using the scikit-learn25 “TfidfVectorizer” tool. As before, the training data sets were split into 80% for training and 20% for validation. For the RF and SVM models, their respective scikit-learn25 model was trained using a PLR-formatted data set. Finally, each model was evaluated with PLR-, non-PLR-, and SmPC-formatted testing data sets, using the “accuracy_score” function from scikit-learn.25
Model Explainability Analysis
After obtaining results from the aforementioned models, Shapley additive explanations (SHAPs)26 were calculated to determine which words had the largest influence for the various drug labeling sections found within the three drug labeling document formats. Mean Shapley values provide the average relative impact of a particular word on models’ predictions for the section end point of a given sentence. Furthermore, SHAPs can be plotted to visualize the parts of a sentence that lead to a certain prediction and those that do not.
Results
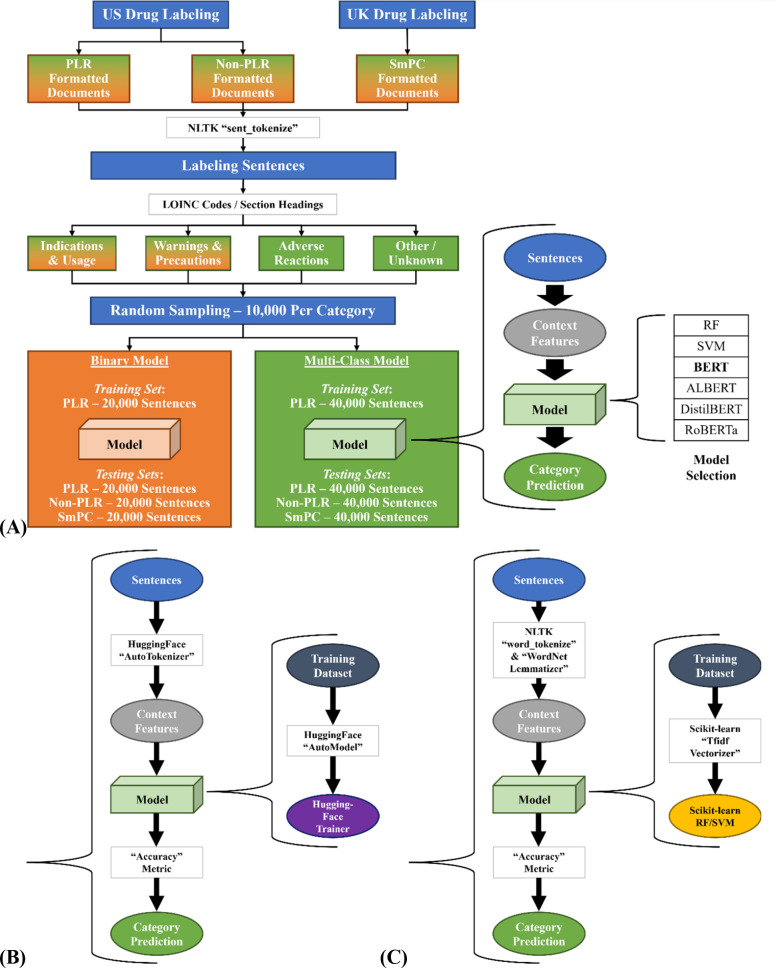
Model Development Flow
Figure 2A depicts the overall workflow of this study, with Figure 2B expanding the modeling procedure for the BERT-based models and Figure 2C expanding the modeling procedure for the RF and SVM models. The labeling data were collected from US and UK drug labeling resources, and each sentence was categorized based on its derived labeling sections. After data collection and processing, two classification models were developed. One was a binary model developed to separate texts of “Indications and Usage” and “Warnings and Precautions.” The other was a multiclass model consisting of four end points: “Indications and Usage”, “Warnings and Precautions”, “Adverse Reactions”, and “Other/Unknown”. Both classification models were developed based on PLR-formatted labeling texts and then tested on PLR, non-PLR, and SmPC labeling texts. The texts were first transformed into context features by tokenization and encoding representation approaches. Next, six modeling algorithms, RF, SVM, BERT-base, ALBERT, DistilBERT, and RoBERTa, were applied for model development. For more detailed information, please see the Materials and Methods section.
Figure 2.
Study workflow. (A) Overall workflow. (B) Model procedure for BERT-based models. (C) Model procedure for RF and SVM models.
Model Testing Results
In the training of both the binary and multiclass models, the evaluation accuracy was saturated after 10 epochs; therefore, we ended training at that point to avoid overtraining. The results reported within this section are the average achieved accuracy of ten testing samples (each containing 10,000 randomly selected records per end point), with the standard deviation of these results provided in parentheses. Table 3 shows the results obtained from the BERT binary classification model. As expected, the results obtained for the PLR testing data set are highest since the model was fine-tuned with PLR-formatted documents. However, note that the accuracies and precisions for the non-PLR and SmPC data sets were very similar, demonstrating that the model works well for all types of external testing data sets. Furthermore, with a training validation accuracy of 0.9635 and average testing accuracies of 0.9486, 0.8756, and 0.8827 for the PLR, non-PLR, and SmPC testing data sets, respectively, this model excelled at differentiating sentences in these two categories.
Table 3. Binary Classification Model Results, in Predictive Accuracy.
| Overall | Indications and Usage | Warnings and Precautions | ||
|---|---|---|---|---|
| Val. | PLR | 0.9635 | ||
| Testing with Avg (stdev) | PLR | 0.9486 (0.0010) | 0.9313 (0.0013) | 0.9659 (0.0016) |
| non-PLR | 0.8756 (0.0019) | 0.8564 (0.0013) | 0.8947 (0.0029) | |
| SmPC | 0.8827 (0.0018) | 0.8809 (0.0013) | 0.8846 (0.0031) |
Table 4 shows the results obtained from the BERT multiclass classification model. Again, note that accuracies and precisions for the non-PLR and SmPC data sets were very similar. Moreover, with a training validation accuracy of 0.8798 and testing accuracies of 0.8194, 0.7302, and 0.6846 for the PLR, non-PLR, and SmPC testing data sets, respectively, this model efficiently differentiated sentences from these four categories. However, it should be noted that the addition of the two new categories significantly decreased the model’s prediction precision for the “Warnings and Precautions” section. This may be due to the inherent similarities between this section and the “Adverse Reactions” section, or perhaps, the variability of the “Other/Unknown” section led to the model’s incorrect predictions.
Table 4. Multiclass Classification Model results in predictive accuracy.
| Overall | Indications and Usage | Warnings and Precautions | Adverse Reactions | Other/Unknown | ||
|---|---|---|---|---|---|---|
| Val. | PLR | 0.8798 | ||||
| Testing with Avg (stdev) | PLR | 0.8194 (0.0019) | 0.9040 (0.0017) | 0.9044 (0.0023) | 0.8166 (0.0038) | 0.6525 (0.0039) |
| non-PLR | 0.7302 (0.0019) | 0.8061 (0.0021) | 0.5982 (0.0045) | 0.7812 (0.0038) | 0.7351 (0.0018) | |
| SmPC | 0.6846 (0.0012) | 0.8538 (0.0015) | 0.6554 (0.0036) | 0.6513 (0.0045) | 0.5781 (0.0038) |
Comparing Different Modeling Algorithms
Many different BERT models can be used for the text classification task; therefore, the performance of a collection of models for the binary and multiclass classification tasks was compared with one set of training and testing samples. As shown in Table 5, BERT-based models outperformed and had lower error rates than did the baseline models, (i.e., for the PLR testing binary classification task, the RF and SVM models had error rates of 7–8%, while BERT had an error rate of 5%, or a ∼50% decrease in errors.) This, along with the “black box” nature of the RF and SVM models, showed the advantages of using BERT with SHAPs to interpret model predictions.
Table 5. Model Comparison, in Predictive Accuracy.
| Validation |
Testing |
|||||||
|---|---|---|---|---|---|---|---|---|
| PLR |
PLR |
non-PLR |
SmPC |
|||||
| Binary | Multiclass | Binary | Multiclass | Binary | Multiclass | Binary | Multiclass | |
| RF | 0.94 | 0.81 | 0.92 | 0.81 | 0.88 | 0.73 | 0.85 | 0.66 |
| SVM | 0.95 | 0.85 | 0.93 | 0.81 | 0.88 | 0.74 | 0.85 | 0.66 |
| BERT | 0.96 | 0.88 | 0.95 | 0.84 | 0.89 | 0.74 | 0.89 | 0.67 |
| ALBERT | 0.96 | 0.87 | 0.95 | 0.84 | 0.89 | 0.72 | 0.87 | 0.66 |
| DistilBERT | 0.96 | 0.88 | 0.94 | 0.83 | 0.88 | 0.74 | 0.89 | 0.66 |
| RoBERTa | 0.97 | 0.88 | 0.95 | 0.83 | 0.89 | 0.74 | 0.90 | 0.66 |
For the most part, each BERT-based model performed at the same level; however, different models had slight edges in some areas. For instance, the RoBERTa model, which has a slightly higher training validation accuracy, performed the binary classification task particularly well on the SmPC data set. Thus, this model may be preferable to others if time is not a consideration, since it has a longer training phase. Furthermore, it is noteworthy that the ALBERT and DistilBERT models, while significantly smaller and faster than the BERT model, have minimal losses in accuracy. This finding showed that these models might be preferable to other BERT-based models, when there are potential time constraints. Altogether, this analysis provided more insight into the various strengths and weaknesses of the selected BERT-based models, which might be useful in future studies.
Figure 3 compares the training and evaluation loss for the four BERT-based models over the ten epochs of which they were trained. Figure 3A compares the training loss of the models, while Figure 3B compares the evaluation loss. Based on these graphs, it is revealed that each model follows a very similar pattern, with training loss gradually decreasing over time and evaluation loss gradually increasing over time. Fine-tuning was cut off after ten epochs to prevent the evaluation loss from getting too high.
Figure 3.
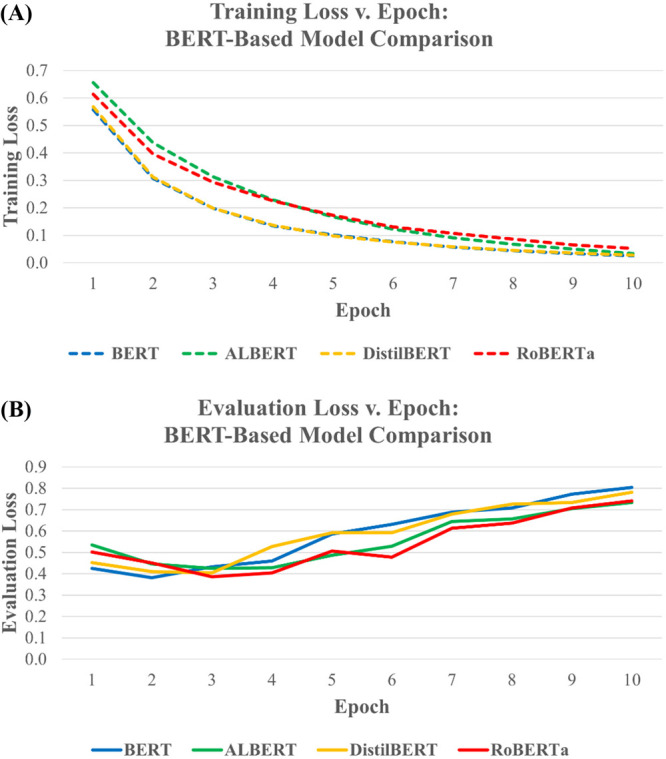
Loss plots for the BERT-based models. (A) Training loss over 10 epochs. (B) Evaluation loss over 10 epochs.
Keywords Most Influential to Predictions
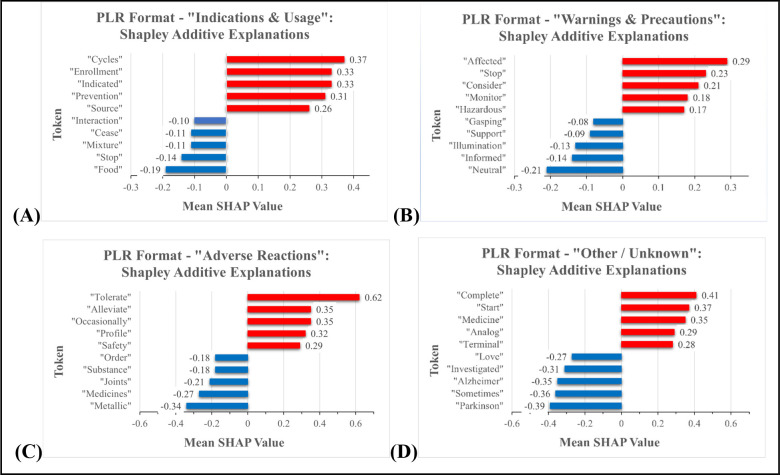
To determine the words with the largest influence in various drug labeling sections, SHAPs were calculated for each end point in the three drug labeling document formats, using the fine-tuned BERT model. For each format, 1000 records per end point were used for these calculations. Figure 4 shows the words with the greatest influence in PLR-formatted documents. The recorded mean Shapley values provided the average relative impact of a particular word on the model’s prediction for the section end point of a given sentence. Values with a positive correlation are colored red, while those with a negative relationship are displayed in blue. The top five positive and negative values are shown. See the Supporting Information document for the model explainability analysis of non-PLR and SmPC-formatted documents.
Figure 4.
PLR format Shapley additive explanations: (A) Indications and Usage; (B) Warnings and Precautions; (C) Adverse Reactions; and (D) Other/Unknown. Values with a positive correlation are shown in red, while those with a negative relationship are displayed in blue.
Based on the results, the words “Indicated” and “Prevention” are among the most influential in the “Indications and Usage” section (Figure 4A), while the words “Affected”, “Stop”, and “Consider” are key to the “Warnings and Precautions” section (Figure 4B). Furthermore, the “Adverse Reactions” section (Figure 4C) was largely influenced by the words “Tolerate”, “Alleviate”, and “Occasionally”. For the most part, the words that dominated these three sections made sense; their usages largely coincided with the information covered in each particular section. However, the tokens selected for the “Other/Unknown” category (Figure 4D) seem random, likely due to this category’s coverage of a broad variety of drug labeling sections. Altogether, this analysis provided more insight into the token words that were most important for discerning among the PLR-formatted sections.
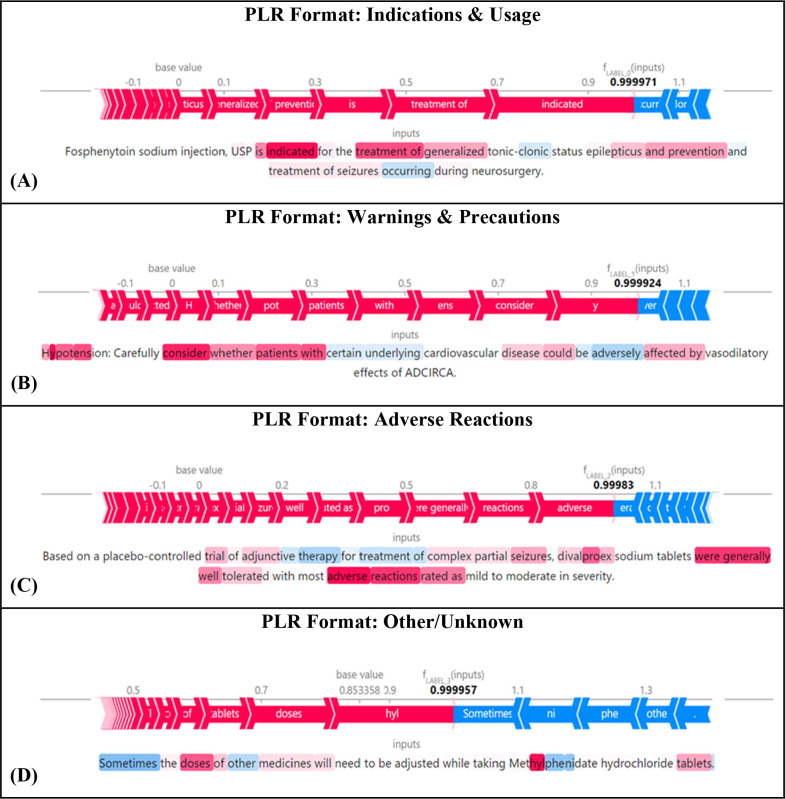
How the Model Weighted Sentences for Prediction
To demonstrate the impacts of certain tokens, a collection of sentences was plotted using the SHAP library and the fine-tuned BERT model. Figure 5 shows the text plots for four sentences retrieved from PLR-formatted documents (one for each of the four categorizations). The tokens highlighted in blue are negatively correlated with the sentence’s section end point, while those highlighted in red are positively correlated. The SHAPs illustrated within these diagrams show which tokens within the individual sentences had the largest influence and led to their classifications. For example, in Figure 5A, which includes a sentence from the “Indications and Usage” section, the tokens “Indicated” and “Treatment [of]” played the biggest roles in this sentence’s categorization. Furthermore, in Figure 5B, which includes a sentence from the “Warnings and Precautions” section, the tokens “Consider” (positive impact) and “Adversely” (negative impact) largely led to this sentence’s classification. Overall, this analysis helped reveal the key factors that led to the language models’ classification of sentences into organized drug labeling sections.
Figure 5.
PLR format Shapley text plots: (A) Indications and Usage; (B) Warnings and Precautions; (C) Adverse Reactions; and (D) Other/Unknown. The tokens highlighted in blue are negatively correlated with the sentence’s section end point, while those highlighted in red are positively correlated.
Discussion
Using Paragraphs or Sentences as Inputs
We further examined how different input levels would affect the results. To accomplish this, the experiments were conducted again, but with paragraphs rather than sentences as inputs. Table 6 compares the results obtained for the BERT binary classification model. For this task, the paragraph-input model outperforms the sentence-input model in each metric: its training validation accuracy, overall testing accuracies, and individual end point prediction precisions were all superior. Since paragraphs inherently provided the model with more context or information than did sentences, the model could make more accurate predictions.
Table 6. Sentence vs Paragraph-Input Binary Classification Models, in Predictive Accuracy.
| Overall | Indications and Usage | Warnings and Precautions | |||
|---|---|---|---|---|---|
| Sentence Input | Val. | PLR | 0.96 | ||
| Testing | PLR | 0.95 | 0.94 | 0.96 | |
| non-PLR | 0.89 | 0.91 | 0.88 | ||
| Paragraph Input | Val. | PLR | 0.98 | ||
| Testing | PLR | 0.97 | 0.96 | 0.98 | |
| non-PLR | 0.92 | 0.92 | 0.92 |
Overall, these findings showed that, in comparison to sentence-input models, paragraph-input models produced better results for core drug labeling sections, suggesting that this may be a promising path to explore in future studies. Nonetheless, there are several disadvantages regarding the use of paragraphs over sentences. For instance, training and testing of the paragraph-input models took much longer than for the sentence-input models, which could pose problems for reviewers faced with time constraints. Additionally, and perhaps most importantly, the categorization of individual sentences might be more useful for researchers and reviewers, so sentence-level predictions might be more convenient for future projects. Overall, even though the paragraph-input models produced slightly better results, the sentence-input models were preferred for the task at hand due to their more balanced and accessible predictions.
Limitations and Future Directions
For the current study, we used only a limited number of section end points to train and test the models. Specifically, the multiclassification model only analyzed three drug labeling sections: “Indications and Usage,” “Warnings and Precautions,” and “Adverse Reactions,” with all the remaining section end points grouped together to form an “Other/Unknown” category. Many of these grouped section end points also provided essential or valuable information for researchers or reviewers. Thus, future research in this area should focus on developing models with different end point configurations, which could potentially lead to unique or novel findings. Furthermore, future studies could involve utilizing more data during the fine-tuning of the model, as performance has been observed to increase with more data points. We noted an increase in performance from using 10,000—from an original 1000—records per end point. Nonetheless, the resulting models provide a foundation for future research related to organizing unformatted regulatory documents into structured data sets for efficient regulatory use.
This case study aimed to evaluate the ability of transformer-based language models to categorize free-texts into appropriate drug labeling sections. As such, an encoder-style language model (i.e., BERT) was selected due to the understanding that this architecture outperforms others for tasks such as sentiment analysis and text classification. However, decoder-style language models, such as those of the GPT series, would potentially be useful for applying a text format or structure to drug labeling texts. Thus, these models should be explored in future studies. Nonetheless, this case study provides evidence that deep learning neural networks are capable of connecting and grouping texts from different formats of drug labeling documents into standardized categories.
Implications for Regulatory Science
Overall, this study uniquely contributes to the field of regulatory science with several broad applications. First, with the knowledge gained and the developed language model, novel techniques for automatically structuring regulatory submissions could emerge, streamlining the submission process for regulatory documents. Next, this research could lead to more understandable, safety-oriented prescription drug labeling resulting from well-structured documents (i.e., PLR vs non-PLR formats). In the future, the language model developed in this study could potentially be applied in the processing of other unformatted, (i.e., scanned or photographed) documents and their contents, adding to the regulatory knowledge base.
Conclusion
In this study, to make unstructured text information more accessible to regulatory reviewers and researchers, we developed a language model that could classify texts or sentences into defined or standardized drug labeling sections. By employing BERT-based models, automatically classifying free text into appropriate drug labeling sections is possible, to a notable extent. Thus, this project paves a pathway for future regulatory science endeavors.
Acknowledgments
This project was supported in part by an appointment to the Research Participation Program at the National Center for Toxicological Research, FDA, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA. We would like to thank Joanne Berger, FDA Library, for manuscript editing assistance.
Glossary
Abbreviations
- ADR
adverse drug reaction
- AI
artificial intelligence
- BERT
Bidirectional Encoder Representations from Transformers
- BLA
Biologics License Application
- FDA
US Food and Drug Administration
- LOINC
Logical Observation Identifiers, Names, and Codes
- MedDRA
Medical Dictionary for Regulatory Activities
- NDA
new drug applications
- NLP
natural language processing
- NLTK
natural language toolkit
- OTC
over-the-counter
- PLR
Physician Label Rule
- RF
random forest
- SHAP
Shapley additive explanation
- SmPC
Summary of Product Characteristic
- SPL
structured product labeling
- SVM
support vector machine
Data Availability Statement
The code and data sets utilized in this project have been made available on GitHub at the following address: https://github.com/magnusgray1/drug_label.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.3c00028.
Shapley additive explanations and analysis for non-PLR- and SmPC-formatted drug labeling documents (PDF)
Author Contributions
CRediT: Magnus Gray data curation, formal analysis, methodology, visualization, writing-original draft; Joshua Xu conceptualization, project administration, validation, writing-review & editing; Weida Tong project administration, supervision, writing-review & editing; Leihong Wu conceptualization, data curation, funding acquisition, investigation, methodology, project administration, supervision, validation, visualization, writing-review & editing.
The views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification and is not intended as an endorsement.
The authors declare no competing financial interest.
Supplementary Material
References
- U.S. Food & Drug Administration . Structured product labeling resources [Internet], 2022.https://www.fda.gov/industry/fda-data-standards-advisory-board/structured-product-labeling-resources (accessed Jan 30, 2023).
- Kowsari K.; Jafari Meimandi K.; Heidarysafa M.; Mendu S.; Barnes L.; Brown D. Text classification algorithms: a survey. Information. 2019, 10 (4), 150. 10.3390/info10040150. [DOI] [Google Scholar]
- Minaee S.; Kalchbrenner N.; Cambria E.; Nikzad N.; Chenaghlu M.; Gao J. Deep learning based text classification: a comprehensive review. ACM computing surveys (CSUR). 2022, 54 (3), 1–40. 10.1145/3439726. [DOI] [Google Scholar]
- Kadhim A. I. Survey on supervised machine learning techniques for automatic text classification. Artificial intelligence review. 2019, 52 (1), 273–92. 10.1007/s10462-018-09677-1. [DOI] [Google Scholar]
- Zhang L.; Wang S.; Liu B. Deep learning for sentiment analysis: a survey. Wiley interdisciplinary reviews: data mining and knowledge discovery 2018, 8 (4), e1253. 10.1002/widm.1253. [DOI] [Google Scholar]
- Yadav A.; Vishwakarma D. K. Sentiment analysis using deep learning architectures: a review. Artificial intelligence review. 2020, 53 (6), 4335–85. 10.1007/s10462-019-09794-5. [DOI] [Google Scholar]
- Dang N. C.; Moreno-García M. N.; De la Prieta F. Sentiment analysis based on deep learning: a comparative study. Electronics. 2020, 9 (3), 483. 10.3390/electronics9030483. [DOI] [Google Scholar]
- Dernoncourt F.; Lee J. Y.. PubMed 200k RCT: a dataset for sequential sentence classification in medical abstracts. arXiv (Computation and Language), 1710.06071, October, 2017, ver. 1. https://arxiv.org/abs/1710.06071 (accessed Jan 30, 2023).
- Madabushi H. T.; Kochkina E.; Castelle M.. Cost-sensitive BERT for generalisable sentence classification with imbalanced data. arXiv (Computation and Language), 2003.11563, March, 2020, ver. 1. https://arxiv.org/abs/2003.11563 (accessed Jan 30, 2023).
- Bisgin H.; Liu Z.; Fang H.; Xu X.; Tong W. Mining FDA drug labels using an unsupervised learning technique-topic modeling. BMC bioinformatics. 2011, 12 (10), 1–8. 10.1186/1471-2105-12-S10-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Ingle T.; Liu Z.; Zhao-Wong A.; Harris S.; Thakkar S.; Zhou G.; Yang J.; Xu J.; Mehta D.; Ge W.; et al. Study of serious adverse drug reactions using FDA-approved drug labeling and MedDRA. BMC bioinformatics. 2019, 20 (2), 129–39. 10.1186/s12859-019-2628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food & Drug Administration . Prescription labeling rule requirements [Internet], 2022. https://www.fda.gov/drugs/laws-acts-and-rules/prescription-drug-labeling-resources (accessed Jan 30, 2023).
- Sohrabi F.Converting labeling for older drugs from the old Format to the PLR format [PowerPoint Presentation]. Center for Drug Evaluation and Research (CDER), 2017.https://www.fda.gov/media/109318/download (accessed Jan 30, 2023).
- DailyMed. SPL resources: download all drug labels [Internet]. U.S. National Library of Medicine. National Institutes of Health, 2022. https://dailymed.nlm.nih.gov/dailymed/spl-resources-all-drug-labels.cfm (accessed Jan 30, 2023).
- NLTK Project . Documentation: nltk.tokenize package [Internet], 2022. https://www.nltk.org/api/nltk.tokenize.html (accessed Jan 30, 2023).
- U.S. Food & Drug Administration . Section headings (LOINC) [Internet], 2018. https://www.fda.gov/industry/structured-product-labeling-resources/section-headings-loinc (accessed Jan 30, 2023).
- Datapharm. Latest Medicine Updates [Internet]. Electronic Medicines Compendium (EMC). https://www.medicines.org.uk/emc/ (accessed Jan 30, 2023).
- Devlin J.; Chang M. W.; Lee K.; Toutanova K.. BERT: pre-training of deep bidirectional transformers for language understanding. arXiv (Computation and Language), 1810.04805, October, 2018, ver. 1. https://arxiv.org/abs/1810.04805 (accessed Jan 30, 2023).
- Vaswani A.; Shazeer N.; Parmar N.; Uszkoreit J.; Jones L.; Gomez A. N.; Kaiser Ł; Polosukhin I.. Attention is all you need. Proceedings from the 31st Conference on Neural Information Processing Systems, December 4–9, 2017, Long Beach, CA; ACM, 2017. https://proceedings.neurips.cc/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf.
- Lan Z.; Chen M.; Goodman S.; Gimpel K.; Sharma P.; Soricut R.. ALBERT: a lite BERT for self-supervised learning of language representations. arXiv (Computation and Language), 1909.11942, September, 2019, ver. 1. https://arxiv.org/abs/1909.11942 (accessed Jan 30, 2023).
- Sanh V.; Debut L.; Chaumond J.; Wolf T.. DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. arXiv (Computation and Language), 1910.01108, October, 2019, ver. 1. https://arxiv.org/abs/1910.01108 (accessed Jan 30, 2023).
- Liu Y.; Ott M.; Goyal N.; Du J.; Joshi M.; Chen D.; Levy O.; Lewis M.; Zettlemoyer L.; Stoyanov V.. RoBERTa: a robustly optimized BERT pretraining approach. arXiv (Computation and Language), 1907.11692, July 2019, ver. 1. https://arxiv.org/abs/1907.11692 (accessed Jan 30, 2023).
- Breiman L. Random forests. Machine learning. 2001, 45, 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Cortes C.; Vapnik V. Support-vector networks. Machine learning 1995, 20, 273–97. 10.1007/BF00994018. [DOI] [Google Scholar]
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; Vanderplas J. Scikit-learn: machine learning in Python. journal of machine learning research 2011, 12, 2825–2830. [Google Scholar]
- Lundberg S.Welcome to the SHAP documentation [Internet]. SHAP, 2018. https://shap.readthedocs.io/en/latest/index.html (accessed Jan 30, 2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code and data sets utilized in this project have been made available on GitHub at the following address: https://github.com/magnusgray1/drug_label.