Abstract
RBM10 is a nuclear RNA-binding protein (RBP) that regulates the alternative splicing of primary transcripts. Recently, research on RBM10 has become increasingly active owing to its clinical importance, as indicated by studies on RBM0 mutations that cause TARP syndrome, an X-linked congenital pleiotropic developmental anomaly, and various cancers such as lung adenocarcinoma in adults. Herein, the molecular biology of RBM10 and its significance in medicine are reviewed, focusing on the gene and protein structures of RBM10, its cell biology, molecular functions and regulation, relationship with the paralogous protein RBM5, and the mutations of RBM10 and their associated diseases. Finally, the challenges in future studies of RBM10 are discussed in the concluding remarks.
Keywords: Alternative splicing, Splicing network, Regulation of RBM10, X-inactivation, RBM5, RBM10 mutations and diseases, Antithetical effects of RBM10
1. Introduction
Owing to the association of RBM10 mutations with various cancers, research on RBM10 has been rapidly expanding in recent years; in the 25 years since its discovery, approximately half of the articles on RBM10 indexed in PubMed have been published in the last 3 years. RBM10 is a regulator of alternative splicing (AS) of various genes (Wang et al., 2013; Bechara et al., 2013; Zheng et al., 2013; Inoue et al., 2014; Sutherland et al., 2017; Rodor et al., 2017; Sun et al., 2017; Collins et al., 2017). It inhibits cell proliferation (Mueller et al., 2009; Bechara et al., 2013; Hernandez et al., 2016; Zhao et al., 2017; Han et al., 2018; Jin et al., ´ 2019; Jung et al., 2020) and promotes apoptosis (Sutherland et al., 2005; Mueller et al., 2009; Wang et al., 2012; Han et al., 2018; Jung et al., 2020); hence, it is generally regarded as a tumor suppressor (Hernández et al., 2016; Zhao et al., 2017; Han et al., 2018; Jin et al., 2019; Jung et al., 2020).
RBM10 is ubiquitously expressed in growing as well as quiescent cells. In general, RBM10 is more strongly expressed in actively transcribing cells (Inoue et al., 2008). There are several mechanisms controlling the cellular levels of RBM10, both transcriptionally and post-transcriptionally (Coleman et al., 1996; Goto and Kimura, 2009; Loiselle et al., 2017; Sun et al., 2017; Loiselle and Sutherland, 2018; Wang L. et al., 2019).
In the AS reaction, RBM10 promotes the exclusion of cassette exons from target pre-mRNAs, and less frequently, other AS events (Wang et al., 2013; Bechara et al., 2013; Zheng et al., 2013; Inoue et al., 2014). It is also involved in reactions other than AS, such as 3′ end processing of some pre-mRNAs (Mueller et al., 2009; Mohan et al., 2018), p53 stabilization (Jung et al., 2020), cell cycle arrest (Guan et al., 2017; Kunimoto et al., 2020), and anti-viral reactions (Pozzi et al., 2020).
RBM5 is similar to RBM10, functioning as a splicing regulator (Fushimi et al., 2008; Bonnal et al., 2008; Bechara et al., 2013) and as a tumor suppressor (M.-Maarabouni and Williams, 2002; Oh et al., 2006; Fushimi et al., 2008; Sutherland et al., 2010; Bechara et al., 2013; Jamsai et al., 2017). There is a cross-regulation between RBM5 and RBM10 (Sun et al., 2017; Loiselle et al., 2017). In addition, RBM10 influences the splicing or expression of multiple other splicing regulators (Wang et al., 2013; Sun et al., 2017), and RBM10 pre-mRNA itself is subjected to AS events. Thus, RBM10, RBM5 and other splicing regulators constitute a splicing network(s). However, its biological significance is not well understood.
Defects in splicing regulation have been associated with various human diseases (Cooper et al., 2009; Cieply and Carstens, 2015; Seiler et al., 2018; Urbanski et al., 2018; Coomer et al., 2019). In fact, mutations in RBM10 cause diseases such as TARP syndrome (Gorlin et al., 1970; Johnston et al., 2010) and various cancers (Imielinski et al., 2012; Seiler et al., 2018). In these cancers, RBM10 has been shown to lose its tumor suppressive activity. However, in some cases, it oppositely functions as a tumor promoter or growth enhancer (Witkiewicz et al., 2015; Rodor et al., 2017; Loiselle et al., 2017; Loiselle and Sutherland, 2018; Sun et al., 2019). Downstream effectors of RBM10 have been studied, and NUMB has been the most studied. RBM10 mutations that nullify or reduce its AS activity lead to the generation of a cell proliferative NUMB isoform, which increases proliferation of various cancers through the Notch signaling pathway (M.-Ali et al., 2011; Bechara et al., 2013; Cieply and Carstens, 2015; Hernández et al., 2016 ). Further studies on unelucidated RBM10 target genes and RBM10-related pathways are expected to promote deeper understanding of the pathogenesis and progression of diseases caused by RBM10 mutations, as well as the antithetical actions of RBM10 as a tumor suppressor and tumor promoter.
In the following sections, the above-mentioned studies and other topics are reviewed in detail.
2. RBM10: Gene and protein
2.1. RBM10 gene
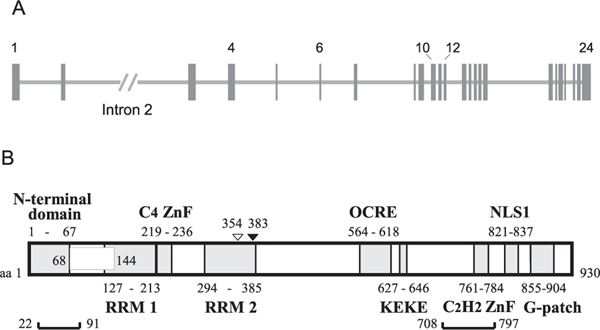
The RBM10 gene is located at p11.3 on the X chromosome; it spans ~41.6 kb and contains 24 exons (Fig. 1A). In a series of projects aiming to accumulate information on unidentified human genes, Nagase et al. (1995) sequenced 40 randomly selected cDNA clones from a myeloid cell line library. One of the cDNAs, KIAA0122, was an RBM10 clone. This polypeptide comprised 929 residues (counted from the first Met), corresponding to isoform 4 of RBM10. The sequence contained an RNP1 motif often found in RBPs (see section 2.2.1.). They also demonstrated that the gene is located on the X chromosome (Nagase et al., 1995). During the same period, Coleman et al. (1996) identified DXS8237E (RBM10) in a human brain cDNA library in an attempt to identify genes in the vicinity of UBE1, which escapes X-inactivation, the mechanism by which most of the genes on one of the two X chromosomes in female cells are transcriptionally silenced. RBM10 was the closest mapped gene, and was shown to be subjected to X-inactivation (Coleman et al., 1996). Subsequently, while cloning hnRNP S1 proteins B2, C1 and D1 (currently identified as AUF1 (hnRNPD) isoforms), Inoue et al. (1996) isolated a cDNA clone, S1–1, from a rat liver library by virtue of the weak cross-reactivity of RBM10 with an anti-S1 protein antibody used in the cloning. S1–1 encoded an RBP of 852 aa residues (Inoue et al., 1996), corresponding to isoform 2 of RBM10. Later, Thiselton et al. (2002) constructed and analyzed a ~2.7-Mb physical map of chromosome Xp11.3–Xp11.23, which harbors several disease-related genes, and identified RBM10 as one of the genes on the map. The RBM10 gene spanned ~41.6 kb (NCBI Reference Sequence: NG_012548.1), consisting of 23 exons (exon 1 was not counted), and its transcript encoded a protein of 930 residues (Thiselton et al., 2002), corresponding to isoform 1 of RBM10 (Table 1).
Fig. 1.
Gene and Domain Structure of RBM10. (A) RBM10 gene structure (NCBI Reference Sequence: NG_012548.1). The RBM10 gene contains 24 exons, indicated by vertical closed boxes, with some of these numbered. Horizontal lines between exons indicate introns. Intron 2 is indicated with its relative length shortened by two-sevenths. (B) Schematic domain structure of RBM10. The structure of isoform 1 (930 aa residues) is shown. RBM10 has two RNA-binding domains in the N-terminal region: the RRM1-C4 ZnF di-domain and RRM2 domain. RBM10 isoforms 1 – 4 are determined by the inclusion or exclusion of the region encoded by exon 4 (box at residues 68–144) and of a Val residue at 354 (open triangle) (see also Fig. 3). The brackets at the bottom (residues 22–91 and 709–797) indicate the regions where phosphorylation occurs at multiple sites. K383 (closed triangle) undergoes acetylation/ubiquitylation. Note that the beginning and ending residue-numbers of the domains may not be strictly defined.
Table 1.
Basic properties of the RBM10 gene and its protein product.
| Human | Mouse | |
|---|---|---|
| Gene symbol | RBM10 | Rbm10 |
| NCBI Gene ID | 8241 | 236732 |
| Chromosome | Xp11.3 | X A1.3 |
| Uniprot ID | P98175 | Q99KG3 |
| Ensembl ID | ENSG00000182872 | ENSMUSG00000031060 |
| Gene length | 41593 bp | 33532 bp |
| (24 exons and 23 introns) | (24 exons and 23 introns) | |
| Protein length: residue number and mass (Da) of isoform 1 |
930 aa (103533 Da) | 930 aa (103492 Da) |
The recent NCBI data on the human RBM10 gene (as of Dec. 1, 2020) indicate the presence of an additional exon between exons 9 and 10, and thereby a total of 25 exons on the RBM10 gene. Based on automated computational analysis using the Gnomon gene prediction tool (NCBI Gene), the additional exon was indicated as the 5′ untranslated exon 1 of the predicted RBM10 mRNA variant X14 (XM_024452465.1), which encodes the predicted isoform X13 (XP_024308233.1). In this review, the conventional number of 24 exons was adopted to follow the literature until the assignment of 25 exons is verified to be generally accepted.
2.2. RBM10 protein and domains
RBM proteins constitute a large family of more than 50 proteins, which generally have one to several RNA-binding domains called RNA recognition motifs (RRMs). The term “RBM” was derived from “RNA binding motif” (note that in addition to these RBM proteins, there are > 160 other RRM-containing proteins with their respective symbol names. HGNC: HUGO Gene Nomenclature Committee). The name “RBM10” was coined after its paralog, LUCA-15 or H37, was renamed as RBM5 (Timmer et al., 1999). RBM10 is highly conserved among mammals; the human RBM10 protein shares 96% and 97% aa sequence homology with that of mice and rats, respectively. This high homology indicates that the human, mouse, and rat RBM10 proteins perform essentially the same molecular functions. The main domains that constitute RBM10 are shown in Fig. 1B.
2.2.1. RNA recognition motif and zinc fingers
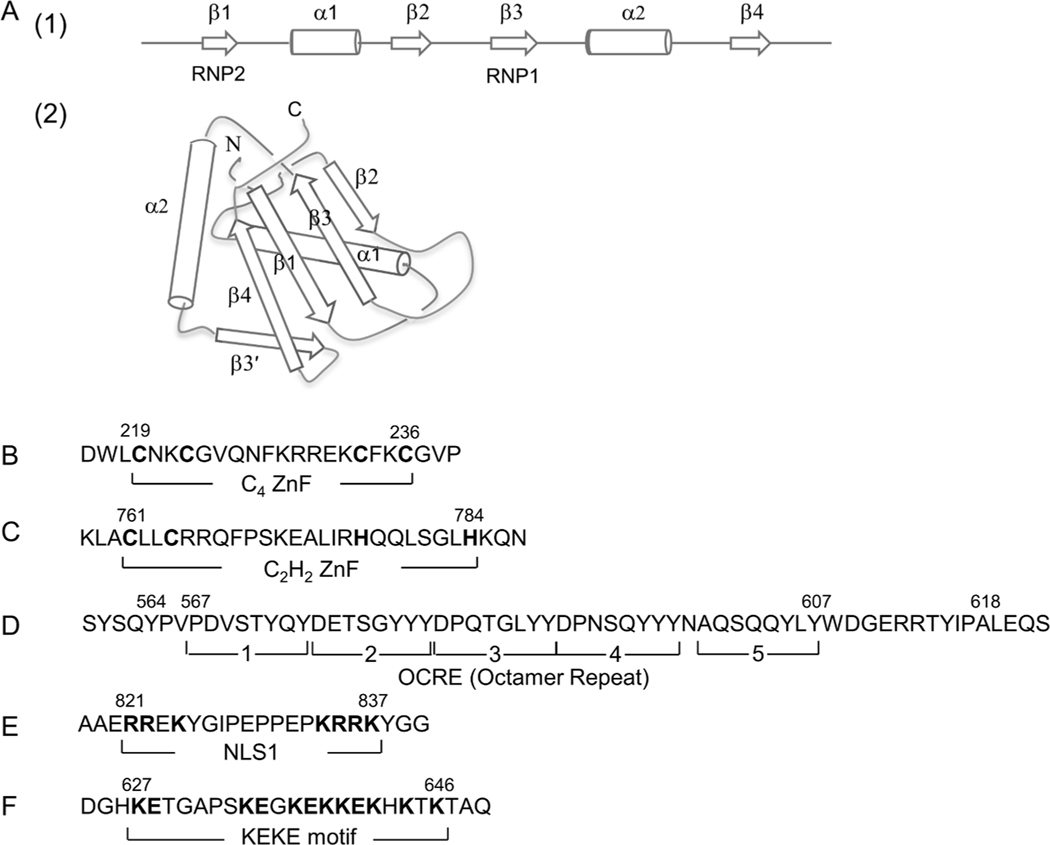
The RRM domain, also known as the ribonucleoprotein (RNP) domain, comprises ~75–90 aa residues that form four anti-parallel β-sheets and two α-helices arranged in a β1–α1–β2–β3–α2–β4 fold (Wittekind et al., 1992; Gorlach et al., 1992; Birney et al., 1993) [Fig. 2A (1)]. The sub-motifs of the 8-aa RNP1 and N-terminal 6-aa RNP2 are conserved sequences within RRM (Dreyfuss et al., 1988), which lie side-by-side on the two central β3- and β1-strands of antiparallel β-sheets backed by two α-helices (α1 and α2) and form a surface to interact with RNA (Oubridge et al., 1994) [Fig. 2A (2)]. RBM10 contains two RRMs, RRM1 and RRM2, at residues 127–213 and 294–385, respectively (Fig. 1B). RRM1 exhibits a variation in the canonical RRM structure, with an additional β strand (β3ʹ) between α2 and β4 [Fig. 2A (2)].
Fig. 2.
Domain structures of RBM10. (A) RNA recognition motifs (RRMs). (1) Schematic representation of a canonical RRM. RRM is composed of four β-sheets and two α-helices arranged in the order β1–α1–β2–β3–α2–β4. (2) 3D structure of RRM1. The figure schematically shows the RRM1 structure of RBM10 determined by NMR (Serrano et al., 2018; Protein Data Bank: 2LX1). The β3- and β1-strands containing RNP1 and RNP2 shown in panel (1) form an RNA-binding surface. (B and C) C4 ZnF and C2H2 ZnF. The cysteine and histidine residues critical for the C4- and C2H2-type ZnF structures and functions are highlighted in boldface. (D) OCRE (Octamer repeat). Brackets 1–5 represent five repeating octamers (8 aa) originally assigned based on hydrophilicity/hydrophobicity profiles (Inoue et al., 1996). The extended aa 564–618 forms a globular fold of 6 anti-parallel β-sheets (Martin et al., 2016). (E) NLS1. Basic aa residues in the bipartite sequence are highlighted. (F) KEKE region. Positive and negative aa residues characterizing the KEKE region are highlighted.
RBM10 has two zinc fingers (ZnFs), which are motifs containing a Zn2+ ion coordinated by 4 cysteine residues or 2 cysteine and 2 histidine residues. The C4-type ZnF at residues 219–236 is next to RRM1 in the N-terminal region (Fig. 1B and 2B). It is a member of a family of RanBP2-type ZnFs that recognize single-stranded RNA (Nguyen et al., 2011). Together with RRM1, it likely acts as a two-domain RNA-binding unit (Collins et al., 2017). Accordingly, RBM10 has an RNA-binding region composed of this RRM1–C4 ZnF di-domain and the RRM2 domain in the N-terminal region of the molecule. These domains act cooperatively to increase the binding affinity for target RNA, as compared with that of the individual motifs (Serrano et al., 2018).
The C2H2-type ZnF, which is present in the C-terminal region at residues 761–784 (Fig. 2C), is essential for the AS function of RBM10 (Zhao et al., 2017; Wang L. et al., 2019). This ZnF also acts to localize/sequester RBM10 in the nuclear domains (S1–1 NBs) when cellular transcription decreases (Wang L. et al., 2019).
2.2.2. Octamer repeat
An OCRE domain is composed of a tandem repeat of octamers, each containing a complete or incomplete Tyr triplet in an archetypical 8-aa sequence of DP–S(Q/G)YYY (Inoue et al., 1996) (Fig. 2D). NMR structure determination has shown that the OCRE in RBM10 forms a globular structure of 55 residues (residues 564–618), consisting of an anti-parallel arrangement of six β-strands, with the first five strands containing complete or incomplete Tyr triplets (Martin et al., 2016). An OCRE motif is also present in the paralogous protein, RBM5, an AS regulator similar to RBM10 (see Section 6). The OCRE in RBM5 has been shown to interact with spliceosomal snRNP proteins, SmN/B/B’, and is suggested to interfere with spliceosome assembly around the target/cassette exons and thereby to promote cassette exon skipping (Bonnal et al., 2008; Mourão et al., 2016). The OCRE of RBM10 is expected to have a similar function.
2.2.3. G-patch
The G-patch is a glycine-rich domain found in certain RNA-processing proteins. A G-patch motif is present in the C-terminal region (residues 855–904, Fig. 1B) of RBM10 (Aravind and Koonin, 1999). RBM10 has been shown to interact with a spliceosomal RNA helicase, DHX15/PRP43 (Hegele et al., 2012). Niu et al. demonstrated that the G-patch domain of RBM5 also interacts with DHX15 and stimulates its helicase activity (Niu et al., 2012). These findings suggest a mechanistic participation of RBM5 in the structural modulation of target RNAs in pre-mRNA AS, and that the G-patch domain of RBM10 likely exhibits a similar function.
2.2.4. Nuclear localization signals
RBM10 is localized in the nucleus; it contains three NLSs (NLS1, NLS2, and NLS3) (Inoue et al., 1996; Xiao et al., 2013). NLS1 in the C-terminal region exhibits a strong nuclear localization activity. It is a typical bipartite NLS with an RREK–(aa)9–KRRK sequence (two basic aa clusters separated by a 9-aa spacer, Fig. 2E). The RRM1 and OCRE in RBM10 exhibit additional NLS activity, and are referred to as NLS2 and NLS3, respectively. The multiple NLSs act cooperatively, even though NLS1 alone is nearly sufficient to localize RBM10 to the nucleus (Xiao et al., 2013). The NLS activity of RRM motifs has also been found in other RBPs (Cassola et al., 2010).
2.2.5. KEKE motif
The KEKE motif is characterized by a stretch greater than 12 residues in length, containing more than 60% K and E/D residues, but lacking 5 K or E/D residues in a row, and is thought to be involved in the association between proteins (Realini et al., 1994). RBM10 contains a KEKE motif (Inoue et al., 1996) at residues 627–646 (Fig. 2F). Together with the C2H2-type ZnF, the KEKE motif promotes RBM10 targeting toward S1–1 NBs, wherein RBM10 is sequestered when cellular transcription declines (Wang L. et al., 2019).
2.2.6. Tyrosine-rich hydrophilic N-terminal region
The N-terminus (residues 1–67) of RBM10 is hydrophilic and highly charged; 69% of its residues are hydrophilic, and it contains 19 acidic and 14 basic residues that constitute approximately half of the region. This region also contains as many as 7 tyrosine residues and many potential phosphorylation sites such as Ser22, 50, 60, 61, and 65, and Tyr36, 41, 48, and 57 (Dephoure et al., 2008; UniphoSite Plus). In addition, it contains 3 RS dipeptides and 1 SR dipeptide in residues 21–43, which likely act as a clustered dipeptide RS domain (see Section 4) used in the interaction with other splicing factors (Long and Caceres, 2009). Therefore, this hydrophilic N-terminal region is likely exposed and involved in inter-molecular interactions that are controlled by phosphorylation. However, the functional significance of this region is yet to be examined.
2.2.7. Region between RRM2 and OCRE
The middle 171-aa region of the molecule at residues 391–561 (Fig. 1B) has a characteristically low sequence homology (84.3%) between human and mouse proteins, in contrast to the 98.5% and 98.9% homology of the N-terminal (residues 1–390) and C-terminal (residues 562–930) regions, respectively. This middle region may be involved in molecular interactions in a species-specific manner.
2.3. Isoforms of RBM10
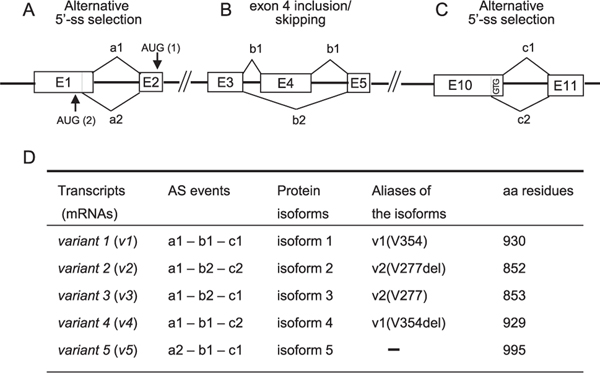
RBM10 has multiple protein isoforms produced by AS of RBM10 pre-mRNA. These events are either an inclusion or a skipping of exon 4 that encodes a 77-aa sequence (Figs. 1B and 3B), and alternative 5′-splice site selection that generates variants with or without a GTG (Val) codon at the end of exon 10 (Figs. 1B and 3C), thereby producing 4 transcript variants, v1–v4, which give rise to protein isoforms 1–4, respectively [Fig. 3D; note that the isoforms have aliases (Tessier et al., 2015)]. RBM10 has another isoform translated from transcript v5, which is generated by an internal 5′-splice site selection in exon 1 (Fig. 3A, a2), and is translated from a start codon in exon 1 through the same reading frame as that of transcripts v1–v4. Accordingly, isoform 5 (995 residues) has a longer 65-aa N-terminus compared with that of isoform 1 (930 residues) (Fig. 3D). As isoform 1 is expressed predominantly and isoform 5 is rarely in human cells, and because isoform 1 is the most extensively studied RBM10 isoform, the aa residue numbers of RBM10 are indicated using those of isoform 1 in this review. The residue numbers of other isoforms are specified when necessary.
Fig. 3.
Transcript variants and protein isoforms of RBM10. Alternative splicing events in the primary transcript of RBM10 (A ~ C) generate mRNA variants v1–v5, which give rise to protein isoforms 1–5 (D). Exons, introns, and splice choices are illustrated using boxes, thick lines, and angled lines, respectively. The exons and introns are not shown to scale. Translation of v1–v4 and v5 starts from an initiation codon AUG (1) in exon 2 and an AUG codon (2) in exon 1, respectively. The isoforms have aliases (D), whose V354 and V277 correspond to the GTG (Val) codon at the end of exon 10 shown in C. The aliases in italic stand for their corresponding transcripts (not shown). The RefSeq numbers of each transcript and their protein isoforms as well as their sequences are found in the gene section of NCBI under RBM10.
The amino acid sequence of exon 4 (residues 68–144) is characteristic: it contains stretches of a basic poly-Arg (R6-H-R-H) sequence at residues 80–88 and an acidic poly-Glu (E7-D-E5) sequence at residues 113–125, as well as the RNP2 submotif of RRM1 at residues 130–136 ( Figs. 1B and 2). Therefore, the RRM1 domain of isoforms 2 and 3, which do not contain the exon 4 sequence, is an imperfect RRM, and the RBM10 isoforms with or without the exon 4 sequence are thought to have different specificity in their interactions with other molecules, including target RNAs. In this respect, it should be noted that the binding preference of RBM10 for poly (U) and poly (G) >poly (C) >poly (A) was determined with isoform 2 (Inoue et al., 1996). However, the significance of exon 4 is not fully understood. The expression of the exon 4-minus RBM10 variant is associated with high proliferation of breast cancers (M.-Arribas et al., 2006). On the other hand, its expression is positively correlated with apoptosis as shown by proapoptotic Bax gene expression (M.-Arribas et al., 2006) and apoptotic caspase-3 expression (M.-Garabato et al., 2008) in breast cancers, as well as by TNF-related apoptosis-inducing ligand (TRAIL) upregulation in MCF-7 breast cancer cells (Wang et al., 2012).
Human cells generally express isoform 1 predominantly, whereas rodent cells generally express more isoform 2 than isoform 1 (Inoue et al., 2008). The biological rationale for this difference in isoform expression between humans and rodents is unknown.
As mentioned above, RBM10 either contains V354 or not. This valine is located in the middle of RRM2 (Fig. 1B), and its functional significance has been reported differently (Loiselle and Sutherland, 2018). Hernandez et al. showed that, irrespective of the presence of V354, RBM10 isoforms displayed similar AS activity in exon 9 skipping of the NUMB (proliferation regulatory protein) transcript (Hernández et al., 2016). In addition, Serrano et al. demonstrated that the binding affinity of RRM2 for the Fas exon 6 splice site was almost the same with or without V354 (Serrano et al., 2018). Conversely, Tessier et al. showed that RBM10 containing V354 enhanced the NUMB exon 9 skipping (exon 11 in their description), whereas V354-minus RBM10 enhanced its inclusion, thereby inhibiting or promoting cell proliferation, respectively, through the Notch signaling pathway (Tessier et al., 2015).
Other RBM10 isoforms called RBM10 isoforms X1–X13 have been indicated, which were derived via automated computational analysis using the gene prediction tool, Gnomon (NCBI, human RBM10 in Gene database). The occurrence and biological significance of these possible RBM10 isoforms remain to be verified.
3. Cell biology of RBM10: Expression and subcellular localization
Coleman et al. (1996) first showed that RBM10 (DXS8237E) is widely expressed in human cell lines and mouse tissues (Coleman et al., 1996). In fact, RBM10 is expressed in almost every cell type, both growing as well as quiescent, and notably, its expression levels are diverse [UniProtKB-P98175 (human) and Q99KG3 (mouse); The Human Protein Atlas]. In general, RBM10 is more strongly expressed in actively transcribing cells (Inoue et al., 2008).
Among the 22,283 sequences examined on microarrays, RBM10 in the THP-1 monocytic leukemia cell line was one of the genes most strongly influenced by the exposure of cells to a moderately low temperature (32 °C for 24 h), wherein the expression of RBM10 decreased by 74% as compared to that of the control (Sonna et al., 2006). Moreover, the major target RNAs bound by RBM10 indicate that it is involved in various metabolic processes such as oxidative phosphorylation, and in the pathways linked to cell proliferation, apoptosis, cell adhesion, actin/cytoskeleton reorganization, and various diseases such as cancers and neurodegenerative disorders (Bechara et al., 2013; Loiselle et al., 2017). These data, together with the ubiquitous expression of RBM10, suggest that it is a general cellular component functioning in various biological processes including homeostasis, and that its functional aberrations bring about diseases.
RBM10 localizes to the nucleoplasm, where transcription and splicing occur, as well as in membrane-less nuclear compartments called S1–1 NBs. There are ~10–40 S1–1 NBs per interphase nucleus. Their numbers and sizes (about 0.5 μm) vary with cell type and cellular conditions. When the transcription of RNA polymerase II decreases, RBM10 in the nucleoplasm is sequestered in S1–1 NBs; it returns to its initial state according to elevated cellular transcription (Inoue et al., 2008; Wang L. et al., 2019). S1–1 NBs often overlap with nuclear speckles (also known as splicing speckles or interchromatin granule clusters) (Inoue et al., 2008; Salichs et al., 2009), which function as sites for the storage of splicing factors and the modification and metabolism of RNAs (Galganski et al., 2017). Like S1–1 NBs, the speckles become larger and spherical upon inhibition of polymerase II transcription (Zeng et al., 1997). These alterations and their frequent side-by-side orientations indicate a close functional relationship between the nuclear speckles and S1–1 NBs.
Although RBM10 is generally present in the nucleus, its cytoplasmic localization has also been observed under specific conditions. Typical examples are shown in neutrophils (Xiao et al., 2013) and human melanoma A7 cells (Yamada et al., 2016). It is unknown how and why RBM10 localizes outside the nucleus in neutrophils. In A7 cells, forced expression of the Src family tyrosine kinase, Fyn, induces translocation of RBM10 from the nucleus to the cell periphery and results in co-localization of RBM10 with FilGAP at small cell protrusions. FilGAP is a GTPase-activating protein that suppresses lamellar formation and cell spreading. Although RBM10 and FilGAP seem to interact indirectly and the phosphorylated protein(s) responsible for the above phenomena have not been identified, the suppression of cell spreading by FilGAP does not occur in the absence of RBM10 (Yamada et al., 2016). It would be intriguing if the results reflect a tumor suppressive function of RBM10.
Interestingly, in cultured fibroblasts from patients with impaired metabolism of vitamin B12, RBM10 and other RBPs, viz., HuR and hnRNPA1, are localized in the cytoplasm, and their cytoplasmic mislocalization is reversed upon incubation of the cells with vitamin B12 (a cofactor of methionine synthesis), S-adenosylmethionine, or SRT1720, an activator of NAD+-dependent deacetylase sirtuin-1 (SIRT1) that protects cells from endoplasmic reticulum (ER) stress (Prola et al., 2017). Ghemrawi et al. concluded that cellular stress induced by defects in vitamin B12 metabolism (Ghemrawi et al., 2013) causes cytoplasmic mislocalization of RBM10, HuR, and hnRNPA1 (Ghemrawi et al., 2019).
4. Functions of RBM10
4.1. Splicing regulations
4.1.1. Regulation of alternative splicing
Nearly all eukaryotes have introns, and > 95% of human multi-exon genes undergo AS events (Black, 2003; Pan et al., 2008). There are seven (Matlin et al., 2005; Scotti and Swanson, 2016) or eight (Wang et al., 2008; Chen and Weiss, 2015) types of AS events. AS generates variant mRNAs from a single gene, producing multiple protein isoforms with different biological activities or antagonistic functions, and thereby creating cellular and functional diversity and complexity. These AS events play critical roles in both normal and disease contexts (Chen and Weiss, 2015; Scotti and Swanson, 2016; Yang et al., 2016).
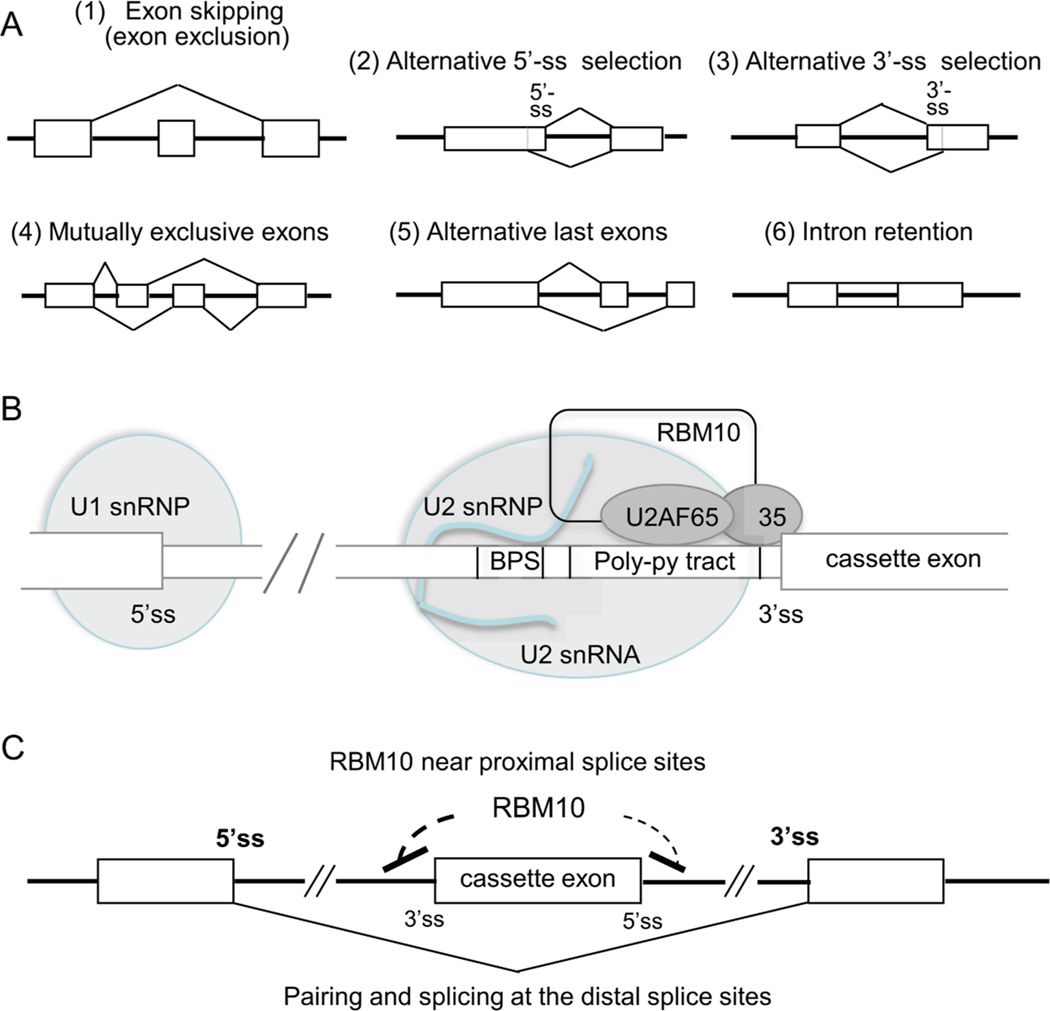
In the AS of pre-mRNAs, RBM10 promotes exclusion of specific exons called alternative or cassette exons from target transcripts (Wang et al., 2013; Bechara et al., 2013; Zheng et al., 2013; Inoue et al., 2014; Rodor et al., 2017; Fig. 4A (1)). Wang et al. (2013) demonstrated that RBM10 regulates the AS of several hundred genes in human embryonic kidney (HEK)-293 cells. The majority (74%) of splicing changes induced by RBM10 overexpression and knockdown were RBM10-enhanced exon skipping events, which were confirmed in 21 candidate transcripts such as DNA methyltransferase 3 beta (DNMT3B), DNA polymerase delta-interacting protein 3 (POLDIP3), spermidine/spermine acetyltransferase 1 (SAT1), RBM5, poly(U)-binding splicing factor (PUF60), cell fate-determining protein NUMB, CREB-binding protein (CREBBP), and dual specificity protein kinase CLK2 (Wang et al., 2013). Exon skipping by RBM10 was also shown with other transcripts such as death receptor Fas (Inoue et al., 2014), pro-apoptotic Bcl-2 member BID, growth arrest and DNA damage-inducible protein GADD4A (Bechara et al., 2013); RBM10 (Sun et al., 2017), survival of motor neuron 2 (SMN2), RAS signaling pathway-associated genes (Sutherland et al., 2017), microRNA pathway-related TNRC6A, histone deacetylase HDAC1, heat shock factor protein HSF2 (Collins et al., 2017), Ser/Thr protein kinase WNK1, and CDKN1A-interacting ZnF protein 1 (CIZ1) (Rodor et al., 2017). Such a diversity of target RNAs indicates that RBM10 participates in a variety of biological pathways and processes (Loiselle et al., 2017).
Fig. 4.
Action model of RBM10. (A) AS events promoted by RBM10. RBM10 promotes skipping of target exons in substrate pre-mRNAs (1). In addition, it may promote other AS events (2) – (6), albeit less frequently. Exons, introns, and splice choices are illustrated using boxes, thick lines, and angled lines, respectively. (B) Association of RBM10 with the vicinity of the 3′-splice site (ss) of a target cassette exon in the spliceosome A complex. RBM10 brings about AS (skipping) of the cassette exon in association with the 35- and 65-kDa subunits of splicing factor U2AF and U2 snRNA of U2 snRNP. (C) Model of RBM10-mediated skipping of a cassette exon. RBM10 is believed to interfere with the 3′- and 5′-ss recognition and/or splice site pairing of the cassette exon, and to enhance the pairing of the distal 5′- and 3′-ss (in boldface). Exons, introns, and exon skipping by pairing of distal splice sites are indicated using boxes, thick horizontal lines, and an angled line, respectively.
Depending on the target exons, RBM10 may promote, albeit less frequently, other AS events such as alternative 5′- or 3′-splice site selection, mutually exclusive exons, alternative last exons, or intron retention [Fig. 4A, (2)–(6)] (Wang et al., 2013; Bechara et al., 2013; Inoue et al., 2014; Rodor et al., 2017). In this respect, it is worth noting that the AS events detected by RBM10 knockdown or overexpression represent not only the direct RBM10-targeted splicing events but also the secondary effects of other splicing regulators whose expression and/or splicing is changed/regulated by RBM10 (section 6.3 and Table 2) (Wang et al., 2013).
Table 2.
Splicing regulators affected by RBM10
| Expression | PPIH, PTBP2, APEH, PRPF39, U2AF2, RBM10 |
| Splicing | ZRANB2, PTBP2, CELF1, PCBP2, HNRNPM, RALY, THOC5, RBM5 |
The splicing regulators whose expression or splicing is altered by RBM10 perturbation (overexpression or knockdown) are indicated. PPIH is downregulated by RBM10 knockdown. PTBP2, PRPF39 and U2AF2 are upregulated and APEH is downregulated by RBM10 overexpression (Supporting information in Ref. Wang et al. (2013)). RBM10 is decreased by auto-regulation via AS-NMD (Sun et al., 2017).
The type of AS promoted by RBM10 is believed to be dependent on its binding position around the exons (Ule and Blencowe, 2019) and other interacting trans-acting splicing factors, e.g., SR proteins (a family of RBPs composed of RRM(s) and a serine-arginine-rich domain referred to as the RS domain (Manley and Krainer, 2010), hnRNP proteins (a group of RBPs with various RNA-binding domains (Geuens et al., 2016), and originally identified as the proteins binding to heterogeneous nuclear RNAs or pre-mRNAs (Dreyfuss et al., 1993); their binding sites are usually in introns of pre-mRNAs), and/or other splicing regulatory RBPs such as SF1 and U2AF (U2 auxiliary factor) (Fu and Ares, 2014; Lee and Rio, 2015; Rahman et al., 2020). Cis-elements on pre-mRNAs act as splicing enhancer or silencer sequences; SR proteins generally bind to splicing enhancer elements and promote splicing, whereas hnRNP proteins generally bind to silencer elements and suppress splicing (Wahl et al., 2009; Cieply and Carstens, 2015).
4.1.2. Mechanistic scheme of RBM10 action in AS: Exon-skipping
The presence of RBM10 in spliceosomes was first shown in 2002 (Rappsilber et al., 2002); later, RBM10 was found in the spliceosomal A (Behzadnia et al., 2007; Kuhn et al., 2009) and B complexes (Deckert et al., 2006; Kuhn et al., 2009). The major spliceosome comprises U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs), each containing proteins and respective uridine-rich small nuclear RNA (i.e., U1–U6 snRNA). Assembly of a spliceosome on a pre-mRNA strictly depends on the presence of the 5′-splice site, branchpoint sequence (BPS), and 3′-splice site. It begins with the formation of the E complex, wherein U1 snRNP binds to the 5′-splice site of an intron, splicing factor SF1 binds to the conserved adenosine-containing branchpoint sequence (BPS) in the 20–50 nt intron region upstream of the 3′-splice site, and the 35 and 65 kDa subunits of U2AF bind to the 3′-splice site and to polypyrimidine tract just downstream of the BPS, respectively. U2AF then promotes the binding of U2 snRNP to the BPS, replacing SF1, to produce the A complex (Fig. 4B). Further conformational and compositional rearrangements proceed from the A complex to B complex, which includes U4/U6.U5 tri-snRNP, to form the catalytically active B* complex (Wahl et al., 2009).
RBM10 interacts with SF1 and U2AF (Hegele et al., 2012), as well as U2 snRNA (Makarov et al., 2011; Wang et al., 2013; Rodor et al., 2017). In accordance with these associations, the RBM10 binding sites on pre-mRNAs are highly enriched within the BPS-containing ~70–100 nt intronic region upstream of cassette exons (Fig. 4B). The binding sites are also detected within an ~100 nt intronic region downstream of cassette exons, as well as in the exons (Wang et al., 2013; Bechara et al., 2013; Rodor et al., 2017; Collins et al., 2017). The presence of RBM10 near the 5′-splice site in the downstream intron was experimentally shown to enhance cassette-exon skipping (Wang et al., 2013). These data suggest that in the splicing reaction of target pre-mRNAs, RBM10 binds close to the splice sites of cassette exons and interferes with the 3′- and 5′-splice site recognition and/or splice site pairing, thereby enhancing the pairing of the 5′- and 3′-splice sites distal to the cassette exons and ultimately leading to the exclusion of the cassette exons and the flanking upstream and downstream introns (Wang et al., 2013; Bechara et al., 2013; Zheng et al., 2013) (Fig. 4C). This mechanism is consistent with the expected function of the OCRE domain of RBM10 (see Section 2). The possible simultaneous interference of the 3′- and 5′-splice sites at cassette exons by RBM10 may be brought about by the formation of its dimer (Hegele et al., 2012) or an exon loop using its two RNA-binding domains (Fig. 1B).
4.1.3. RBM10 target sequences
In the process of AS, RBM10 recognizes and binds to target pre-mRNAs. RBM10 contains two RNA-binding domains in the N-terminal region: the RRM1–C4-type ZnF di-domain, which recognizes a GGA-centered motif, and the RRM2 domain, which recognizes a C-rich sequence (Collins et al., 2017). However, the target sequences identified using CLIP-Seq, PAR-CLIP, and iCLIP (Wang et al., 2013; Bechara et al., 2013; Maaskola and Rajewsky, 2014; Rodor et al., 2017), or in vitro assays (Nguyen et al., 2011; Collins et al., 2017) show a considerably broad spectrum (Loiselle and Sutherland, 2018). For instance, the most enriched motifs are CUCUGAACUC and GAUCCC(U/A) in HeLa cells (Bechara et al., 2013), and UCCAA, CCAAA, and CCCCA in mouse mandibular MEPA cells (Rodor et al., 2017). The reason for such low consensus or diversity in the binding sequences of RBM10 is unclear. However, it is noteworthy that the RNA-binding domains of RBPs generally recognize short (i.e., 3–4 nt) motifs, and that many RBPs interact cooperatively to achieve the regulation of AS (Ule and Blencowe, 2019).
4.2. Splicing-independent functions of RBM10
4.2.1. RBM10 regulates transcription
RBM10 influences the expression of many genes as shown in HEK-293 cells (Wang et al., 2013; Mohan et al., 2018), HeLa cells (Bechara et al., 2013), lung cancer cells (Loiselle et al., 2017; Guan et al., 2017), and mouse mandibular cell lines (Rodor et al., 2017). For example, RBM10 knockdown in HEK cells upregulates 801 genes and downregulates 1166 genes (Mohan et al., 2018), and overexpression of RBM10 in lung adenocarcinoma (LUAD) cells upregulates 304 genes and downregulates 386 genes (Guan et al., 2017).
Thus, RBM10 regulates the transcription of many genes. However, in most cases, this regulation is likely brought about indirectly via its AS function. For example, RBM10 regulates the AS of DNA (cytosine-5)-methyltransferase 3b (DNMT3B). Under RBM10 deficiency, the level of the active DNMT3B2 isoform increases, which in turn increases DNA methylation of the promoters of NF-κB–responsive genes, resulting in the suppression of NF-κB-mediated transcription (Atsumi et al., 2017). Similarly, overexpression of RBM10 has been shown to enhance the transcription of TNF-α in the human breast adenocarcinoma cell line, MCF-7, the Jurkat human T lymphoblastic leukemia cell line (Wang et al., 2012), and in osteosarcoma cells (Han et al., 2018). The increase in TNF-α levels correlated with an increase in apoptosis, presumably through the TNF–TNFR signaling pathway. It was suggested that the regulation of TNF-α transcription occurred indirectly through an AS event promoted by RBM10 (Wang et al., 2012).
Histone H2A deubiquitinase (2A-DUB) forms a complex with approximately 10 proteins such as histone acetyltransferase p300/CBP-associated factor (p/CAF; presently, PCAF), thyroid hormone receptor-interacting protein 5 (Trip5) and RBM10 (Zhu et al., 2007). The 2A-DUB complex epigenetically regulates transcriptional initiation by coordinating histone acetylation and de-ubiquitination and by destabilizing the association of linker histone H1 with nucleosomes (Zhu et al., 2007). As frequently observed in transcriptional regulation by co-association between transcription factors and RBPs at gene promoters (Xiao et al., 2019), RBM10 may be involved in the transcriptional regulation by 2A-DUB.
4.2.2. Splicing-independent RNA-binding reactions
RBM10 also participates in splicing-independent RNA-binding events. Mueller et al. showed that S1–1 (RBM10) stabilizes angiotensin II receptor (AT1) mRNA by binding to its 3ʹ-UTR in vascular smooth muscle cells (Mueller et al., 2009). Mohan et al. demonstrated that RBM10 mediates anti-hypertrophy of the heart by acting as a co-regulator of Star-PAP (TUT1). Star-PAP is a non-canonical poly(A) polymerase that exhibits polyadenylation and uridylation activity. The RRM2 domain of RBM10 binds to the catalytic domain of Star-PAP and stimulates Star-PAP activity for polyadenylation (but not uridylation). Along with Star-PAP, RBM10 binds to the 3′-UTRs of various cardiac pre-mRNAs of anti-hypertrophy regulators and participates in the production of their mature mRNAs (Mohan et al., 2018). These data indicate that RBM10 controls the 3′ end processing of distinct subsets of pre-mRNAs, a splicing-independent regulatory mechanism of RBM10 for protein expression and function (Li et al., 2017; Mohan et al., 2018).
MicroRNAs (miRNAs) regulate gene expression by reducing target mRNAs or their translational efficiency. RBM10 binds to a hairpin structure of the let-7 g miRNA precursor and likely participates in regulating the biogenesis of let-7 g miRNA (Treiber et al., 2017), whose major function appears to be the promotion of terminal differentiation in development and tumor suppression (E.-Kerscher and Slack, 2006). Intriguingly, binding of RBM10 to a variety of non-coding RNAs has been also observed (Rodor et al., 2017).
4.2.3. p53 stabilization, cell cycle arrest, and anti-viral function
RBM10 exhibits another function through protein–protein interactions. In a study on the role of RBM10 in tumor suppression, Jung et al. showed that overexpressed RBM10 stabilizes p53 by binding to MDM2. MDM2 is a major negative regulator of p53, acting as an E3 ubiquitin ligase to trigger the degradation of p53 via a proteasome pathway. They concluded that by interfering with the p53–MDM2 interaction, RBM10 suppresses the ubiquitination and degradation of p53, and thereby enhances the p53-dependent apoptosis and inhibition of cell proliferation (Jung et al., 2020).
Kunimoto et al. reported that overexpressed RBM10 or its C-terminal half (residues 401–930) causes monopolar spindle formation and cell cycle arrest in the M phase in HepG2 cells. Overexpressed RBM10 forms large nuclear domains, which sequester PLK4, STIL, and SAS6 (the regulatory proteins involved in centriole duplication), resulting in centrioles without PLK4 and STIL and impairment of centriole duplication. In contrast, RBM10 knockdown increases the cytoplasmic PLK4 levels and causes abnormal cells with more than 4 centrioles in HepG2 cells, but not in A549 cells. This is an example to indicate that the cellular levels of RBM10 should be securely controlled (Kunimoto et al., 2020). Guan et al. also reported cell cycle arrest due to overexpressed RBM10 in the G0/G1 phase in A549 cells and in the S phase in H1299 cells (Guan et al., 2017). These different phases in cell cycle arrest by overexpressed RBM10 seem to result from different cellular conditions and/or contexts.
RBM10 performs an anti-viral task (Pozzi et al., 2020). SAT1 (spermidine/spermine acetyltransferase 1) is known for its anti-viral action, where it decreases the reservoir of cellular polyamines and thereby limits viral replication (Li and MacDonald, 2016). In dengue virus-infected A549 cells, RBM10 is a target of the dengue virus polymerase, NS5, which binds to RBM10 and mediates its proteasomal degradation. The decrease in RBM10 levels alters the AS of SAT1 pre-mRNA and generates the exon 4-containing SAT1 transcript, which contains a premature termination codon (PTC) and is degraded by nonsense mediated mRNA decay (NMD). Hence, the anti-viral SAT1 is downregulated in dengue virus-infected cells. Conversely, RBM10 overexpression in infected cells prevents SAT1 splicing changes and limits viral replication. In addition, RBM10 interacts with viral RNA and RIG-I (retinoic acid-inducible gene I), which is an essential protein in the innate immune system for sensing viral RNA. Furthermore, RBM10 depletion attenuates the expression of interferon and pro-inflammatory cytokines. These results suggest that RBM10 fulfills diverse pro-inflammatory anti-viral tasks in addition to AS regulation (Pozzi et al., 2020).
5. Regulation of RBM10
5.1. Transcriptional regulation of RBM10 gene
In female somatic cells, one of the two X chromosomes undergoes X-inactivation. Approximately 85% (Balaton et al., 2015) of the genes in the inactive X chromosome (Xi) are transcriptionally silenced by heterochromatin formation, which is brought about by progressive coating of the chromosome with Xist long noncoding RNA. In 1996, Coleman et al. isolated a cDNA clone, DXS8237E (presently, RBM10), which was the closest gene to that of ubiquitin-activating enzyme E (UBE1, currently called ubiquitin-like modifier activating enzyme [UBA1]) in Xp11.23 (Xp11.3, to be exact) and showed that these genes had discordant X-inactivation statuses; RBM10 is subjected to X-inactivation, whereas UBE1 escapes it (Coleman et al., 1996). The 5′ region of the active UBE1/UBA1 gene on Xi is unmethylated and enriched with acetylated H3K9, indicating its escape from gene inactivation (Carrel et al., 1996), whereas the exon of the adjacent RBM10 gene and its proximal intergenic region have high levels of inactive heterochromatin markers, viz., tri-methylated histone H3 at lysine 9 (H3K9me3) and tri- methylated H4 at lysine 20 (H4K20me3) (Chadwick and Willard, 2004; Goto and Kimura, 2009). In addition, the insulator protein CTCF (also known as CCCTC-binding factor) (Cuddapah et al., 2009) and site-specific CpG hypomethylation are distributed between UBE1 and RBM10 genes, constituting the chromatin boundary that prevents the spread of transcriptional silencing (Goto and Kimura, 2009).
The transcriptional control of RBM10 in active X chromosome has also been reported. The expression of RBM10 was repressed in endometrial tumor cells, and this decrease in expression correlated with low H3K27 acetylation and an increase in CpG island methylation in the gene promoter (Dou et al., 2020). In addition, RBM10 expression is shown to decrease strongly under hypothermal conditions (Sonna et al., 2006; see section 3).
5.2. Autoregulation of RBM10
As described above, most genes on one of the two X chromosomes in female cells are transcriptionally inactivated, and RBM10 is one of the genes subjected to this X-inactivation (Coleman et al., 1996; Thiselton et al., 2002; Goto and Kimura, 2009). This implies that a double dose of RBM10 can be unfavorable to cells. In fact, cellular levels of RBM10 are regulated by several mechanisms.
Sun et al. found that the RNA and protein levels of endogenous RBM10 were considerably reduced by the overexpression of exogenous RBM10–EGFP in HEK cells; hence, a feedback-type autoregulation likely operated in RBM10 expression. In fact, overexpressed RBM10 binds to its own pre-mRNA and brings about AS to exclude exon 6 or 12 (see Fig. 4A (1) for the exon-exclusion/skipping reaction). The resulting transcripts contain a PTC and undergo degradation via NMD, ultimately downregulating both the RBM10 mRNA and protein levels (Sun et al., 2017). Multiple skipping events at exons 6 and 12 are expected to reduce the cellular concentration of RBM10 efficiently. Sun et al. further suggested that auto-regulation by AS-coupled NMD (AS-NMD) would operate similarly in other splicing regulators to maintain transcriptome homeostasis, as has been shown with polypyrimidine tract-binding proteins (PTBPs) (Wollerton et al., 2004; Sun et al., 2017).
In addition, another self-regulatory mechanism of RBM10 was reported. In RBM5-null GLC cells, RBM10 (most likely, isoform 1) binds to RBM10 mRNAs (the smaller v2 variant in particular), which leads to downregulation of RBM10 expression, seemingly through translational self-regulation (Loiselle et al., 2017; Loiselle and Sutherland, 2018).
RBM10 further undergoes a different autoregulation. When transcription by RNA polymerase II decreases, RBM10 sequesters itself in nuclear domains called S1–1 NBs. This process is reversible, and RBM10 returns to its initial state when cellular transcription is restored (Inoue et al., 2008). RBM10 has a C2H2-type ZnF, which is not only essential for AS regulatory function but also for targeting the RBM10 molecule toward S1–1 NBs (Zhao et al., 2017; Wang L. et al., 2019). The NB-targeting activity of the ZnF is likely exerted when it becomes free from AS under decreased transcription. The multiple auto-regulation mechanisms of RBM10 described above suggest that the cellular levels of RBM10 should be controlled not to exceed necessary levels.
RBM10 also undergoes downregulation by microRNA-335, as shown in endometrial tumors. This microRNA is upregulated in the tumor and targets the 3′-UTR of RBM10 mRNA, resulting in a reduction in RBM10 protein levels. This decrease in RBM10 levels brings about the AS of NUMB transcripts to produce the cell-proliferative NUMB isoform, which promotes tumor growth by activating the Notch signaling pathway (Dou et al., 2020).
5.3. Post-translational modifications of RBM10
5.3.1. Phosphorylation
RBM10 undergoes various post-translational modifications. It is phosphorylated at serine, threonine, and tyrosine residues, and more than 50 phosphorylation sites have been identified (UniProtKB-P98175; PhosphoSitePlus RBM10). This large number suggests that the activity and function of RBM10 are regulated by phosphorylation in response to various cellular conditions or stimuli. A significantly high number of modification sites are found in the N-terminal region (residues 22–91) upstream of RRM1 and in the C-terminal region (residues 708–797) (Fig. 1B). In the M/G1 phase, RBM10 is phosphorylated at S723, S733, S736, and S738, and in the G1 phase, at Y48 and Y57, as well as S60, S61, S65, and S797 (Dephoure et al., 2008). Stimulation with epidermal growth factor (EGF) results in phosphorylation at Y732, S733, S736, and S738 (Olsen et al., 2006), and overexpression of c-Src kinase or stimulation with platelet-derived growth factor (PDGF) leads to tyrosine phosphorylation at Y16, Y435, and Y906 (Amanchy et al., 2008). Phosphorylation at S517, S736, and S738 has also been observed in the absence of stimulation (Beausoleil et al., 2004). In addition, S60, S288, and S517 are phosphorylated in the DNA damage response caused by ionizing radiation (Matsuoka et al., 2007). As above, the phosphorylation sites on RBM10 have been identified; however, how these modification events (1) regulate the activity and function of RBM10 as well as the RBM10-related cellular processes remains to be elucidated.
5.3.2. Ubiquitylation and acetylation
RBM10 undergoes ubiquitylation at K126, K246, K335, K383, K834, K880, and K881 (Stes et al., 2014; Akimov et al., 2018). The lysine residue at position 383 undergoes acetylation as well (Choudhary et al., 2009). A cross-talk between these modifications at K383 may control the cellular levels of RBM10 via proteasome-mediated degradation of RBM10, by ubiquitylation, and its prevention, by acetylation.
5.3.3. Methylation
Guo et al. identified over 1000 and ~160 arginine and lysine methylation sites, respectively, in the human colon cancer cell line, HCT116. They showed that RBM10 isoform 2 was modified by monomethylation at K803, R817, and R824. Interestingly, R824 methylation occurs in a tissue-specific manner; it is detected in a mouse embryo, but not in the mouse brain (Guo et al., 2014). However, the biological significance of these methylation events is not known.
As discussed above, the cellular levels and activity of RBM10 are controlled by X-inactivation and several post-transcriptional self-regulations, and most likely by post-translational modifications as well. These multiple mechanisms of RBM10 regulation indicate that proper levels or activity of RBM10 are important for the cellular processes in which RBM10 is involved (Sun et al., 2017). Hence, experimental data obtained by overexpression of exogenously introduced RBM10 need to be carefully interpreted, and its expression should be controlled to avoid possible biological artifacts caused by its overproduction. To this end, more physiological experiments, such as RBM10 depletion, may be required to support the data obtained by RBM10 overexpression.
6. Relationship between RBM5, RBM6, and RBM10
RBM5, RBM6, and RBM10 are paralogs that arose from gene duplications during genome evolution. RBM5 was first identified in 1996 (Wei et al., 1996), and called LUCA-15 (M.-Maarabouni and Williams, 2002) and H37 (Oh et al., 1999). RBM6 was identified as NY-LU-12 in 1998, a partial sequence of which had been called g16 (Güre et al., 1998), and subsequently as DEF-3 (Drabkin et al., 1999). The names “RBM5” and “RBM6” were coined by Timmer et al. in 1999 (Timmer et al., 1999), and RBM10 was so called afterwards, by replacing the previous name S1–1 (chronologically, RBM10 was discovered before RBM5). The RBM5 and RBM6 genes are separated by ~11 kb and tandemly map to 3p21.31 (Timmer et al., 1999), a tumor suppressor region that frequently shows loss of heterozygosity in various cancers (for a review, see Ref. Ji et al. (2005)). In fact, RBM5 and RBM6 are tumor suppressors (Oh et al., 2006; Sutherland et al., 2010; Jamsai et al., 2017; Wang Q. et al., 2019). They regulate cell growth and cell cycle progression (M.-Maarabouni and Williams, 2002; Oh et al., 2006), promote apoptosis (Sutherland et al., 2005; Fushimi et al., 2008; Bonnal et al., 2008), and suppress transformation-associated processes (Jamsai et al., 2017; Wang Q. et al., 2019). In general, RBM5 retards cell growth (e.g., see Refs. Sutherland et al. (2010) and Shao et al. (2012)). As with RBM10, RBM5 and RBM6 are found in spliceosome complexes (Deckert et al., 2006; Hegele et al., 2012; Papasaikas et al., 2015), and function as AS regulators of various genes (Fushimi et al., 2008; Bonnal et al., 2008; Bechara et al., 2013).
6.1. Origin of genes
There are different views on which gene among RBM5 and RBM10 evolved first and produced the other via gene duplication. RBM5 is predicted to be the progenitor gene, based on shared synteny indicating preserved co-localization of genes around USP (deubiquitinating enzymes) genes on chromosomes (Vlasschaert et al., 2015; Loiselle and Sutherland, 2018). RBM10 (DXS8237E) has been predicted to be so, based on gene orientations and distances with respect to nearby UBE genes (ubiquitin-like modifier activating enzymes) (Timmer et al., 1999). Although the data suggest that RBM5 is likely the progenitor gene, further analysis is required to clarify this issue. On chromosome 3, RBM6 is present next to RBM5, and is thought to have arisen by gene duplication from RBM5 and then undergone considerable sequence alterations (Timmer et al., 1999).
6.2. Comparison: Sequences and functions of RBM5, RBM6, and RBM10
RBM5, RBM6, and RBM10 commonly contain two RRMs, two ZnFs, an OCRE, a bipartite NLS (NLS1), and a G-patch domain (Sutherland et al., 2005; Bechara et al., 2013) (Fig. 1B). The protein sequences of RBM5 (815 residues) and RBM10 isoform 1 (930 residues) or isoform 3 (853 residues) show 52% identity. The sequence identity of RBM5 and RBM6 (1123 residues) is 24%, and that of RBM6 and RBM10 (isoform 1) is 25%. Their C-terminal halves containing the C2H2 ZnF, OCRE, NLS1 and G-patch domains are more homologous than the N-terminal regions: the sequence identity is 68% in the C-terminal two fifths of the RBM5 and RBM10 molecules, in contrast to 41% in the remaining N-terminal regions including the RNA-binding domains. The corresponding RBM6 N-terminal region shows very low homologies with those of RBM5 and RBM10. In AS, RBM5 and RBM6 preferentially bind to exon regions in target RNAs, whereas RBM10 binding sites are enriched in the intronic regions close to splice sites (Bechara et al., 2013; Rodor et al., 2017). In some cases, these RBM proteins likely bind to the same target RNAs; about 10% of the RNAs immunoprecipitated with an anti-RBM10 antibody from small-cell lung cancer cell line extracts were also precipitated with an anti-RBM5 antibody (Loiselle et al., 2017). In fact, in the AS events in HeLa cells, about 20% of target transcripts regulated by RBM5, RBM6, or RBM10 are controlled by more than one RBM protein, suggesting also that these RBM proteins act synergistically or antagonistically on a subset of their targets (Bechara et al., 2013).
The antagonistic actions of RBM6 and RBM10 during cell proliferation have been observed in HeLa cells (Bechara et al., 2013). RBM10 knockdown enhances cell proliferation and colony formation, whereas RBM6 knockdown decreases these processes. Their antagonistic effects on cell proliferation are explained by their opposite AS actions on NUMB: RBM6 enhances NUMB exon 9 inclusion, whereas RBM10 enhances NUMB exon 9 skipping (Bechara et al., 2013). Similarly, RBM5 knockdown in neuronal cells decreases staurosporine-induced caspase-9 activation, whereas RBM10 knockdown increases it (Jackson et al., 2015). In addition, their effects can overlap. Both RBM5 and RBM10 similarly enhance the AS of exon 6 skipping in Fas receptor pre-mRNA (Bonnal et al., 2008; Inoue et al., 2014). In many cases, however, they do not correlate to each other. For instance, RBM5 enhances the skipping of exon 9 of caspase 2 pre-mRNA (Fushimi et al., 2008) and exon 7 of c-FLIP (a regulator of the Fas signaling pathway) pre-mRNA (Bonnal et al., 2008), RBM10 is hardly involved directly in these reactions (Inoue et al., 2014).
6.3. Cross-regulation and networks of splicing regulation
RBM10 negatively cross-regulates RBM5. Overexpression of RBM10 enhances the skipping of exons 6 and 16 of RBM5 transcripts, and introduces PTCs, thereby causing AS-coupled NMD and ultimately reducing RBM5 expression (Sun et al., 2017). In addition, although RBM5 does not influence the transcription or AS of RBM10, overexpressed RBM5 specifically binds to RBM10 v2 transcript and significantly increases its protein levels, presumably by promoting RBM10v2 translation (Loiselle et al., 2017).
As described above, RBM5 and RBM10 act on the same subset of target RNAs (Bechara et al., 2013) and there are cross-regulations between RBM5 and RBM10 (Sun et al., 2017; Loiselle et al., 2017). Furthermore, RBM10 perturbation (knockdown or overexpression) alters the splicing of eight splicing regulators, including RBM5, and significantly influences the expression of six others, including RBM10 (Table 2) (Wang et al., 2013; Sun et al., 2017). In addition, RBM10 itself undergoes multiple AS events, producing its isoforms by the action of unidentified splicing regulators (see section 2). These data suggest that RBM10 constitutes a splicing network(s), which presumably functions in transcriptomic homeostasis or in organized cellular processes/pathways (Jangi and Sharp, 2014; Ule and Blencowe, 2019).
7. RBM10 mutations and associated diseases
Mutations in splicing regulators and abnormal splicing of RNA targets are associated with various human diseases (Cooper et al., 2009; Cieply and Carstens, 2015; Seiler et al., 2018; Urbanski et al., 2018; Coomer et al., 2019). RBM10 regulates many genes, and the phenotypes caused by its mutations differ by the stages of development and tissues. Examples include TARP syndrome (Gorlin et al., 1970; Johnston et al., 2010), a rare X-linked congenital disorder with various phenotypes in male neonates, and cancers such as lung adenocarcinoma (LUAD) (Imielinski et al., 2012) and bladder carcinoma (BLCA) in adults (Seiler et al., 2018). Probably because RBM10 is located on the X chromosome, its mutations are found differently in males and females, as observed in TARP syndrome and LUAD, which are more common in men than in women (Cancer Genome Atlas Research Network, 2014; Yuan et al., 2016; Yin et al., 2018; Seiler et al., 2018). RBM10 generally functions as a tumor suppressor; however, it may exert an opposite oncogenic function. The antithetical effects of RBM10 in cell proliferation and apoptosis are shown below, as well as in the review by Loiselle and Sutherland (2018).
7.1. TARP syndrome
TARP syndrome is characterized by pleiotropic defective development and malformations (Johnston et al., 2010; Schneider et al., 2011; Gripp et al., 2011). The term “TARP” comes from Talipes equinovarus (clubfoot), Atrial septal defect, Robin sequence [micrognathia (undersized jaw), glossoptosis (tongue displacement or retraction), and cleft palate], and Persistent left superior vena cava. In 2010, Johnston et al. identified a frameshift and a nonsense mutation in RBM10 in two families of individuals afflicted with TARP syndrome (Johnston et al., 2010), one of which was originally described by Gorlin et al. (1970). They also showed that RBM10 was highly expressed in the branchial arches and limbs of mid-gestation mouse embryos, consistent with the human malformation sites observed in TARP syndrome (Johnston et al., 2010). Although TARP syndrome is pre- or postnatally lethal and affected males generally do not survive, patients with TARP aged 11, 14, and 28 years have been reported (Wang et al., 2013; Højland et al., 2018; Niceta et al., 2019). Their phenotype is variable and includes expanded TARP syndrome phenotypes, some of which are low-set posteriorly angulated ears, upslanting palpebral fissures, various structural abnormalities of the brain, imperforate anus, cryptorchidism (absence of one or both testes in the scrotum), scoliosis (curved spine), multiple hemivertebrae, aplasia of thumbs and first toes, and intellectual disability. Hence, abnormal features may be observed in the ears, eyes, oral region, head, heart, spine, hands, and feet, and in the cardiovascular, skeletal, genitourinary, and nervous systems (Gripp et al., 2011; Johnston et al., 2014; Powis et al., 2017; Højland et al., 2018).
In this disorder, mutations are found throughout the RBM10 gene (Johnston et al., 2010, 2014; Gripp et al., 2011; Wang et al., 2013; Powis et al., 2017; Højland et al., 2018; Niceta et al., 2019), and most of them are null or highly likely to be null, owing to PTCs that cause NMD, and thus, loss of RBM10 function. The first patient to have survived to adulthood (28 years old) had a mutation, c.273_283delinsA, in exon 4 (Højland et al., 2018) (Fig. 1A and B) that caused a frameshift and produced a PTC in the transcript (p.G92AfsX39). It has been suggested that such transcripts undergo AS to skip exon 4 and generate normal RBM10 isoforms 2 and 3 (Fig. 3), resulting in a milder phenotype and survival into adulthood (Højland et al., 2018). The mutation identified in a 14-year-old patient with intellectual disability and multiple congenital malformations as well as the main pathological phenotypes was an in-frame deletion of residues 651–889, which removed the C-terminal region spanning 6 exons (Wang et al., 2013). The lost region included the second ZnF (C2H2 ZnF) domain and an NLS (NLS1) of RBM10 (Fig. 1B). Although these domains are important for the AS function of RBM10 in the nucleus (Wang Y. et al., 2013; Wang L. et al., 2019), its molecular function(s) seemed to have arisen partly.
7.2. Lung adenocarcinoma
In 2012, Imielinski et al. identified somatic mutations in RBM10 in LUAD samples (Imielinski et al., 2012). RBM10 mutations in LUAD occur throughout the RBM10 coding region rather than at the same recurrent aa positions (Imielinski et al., 2012; Zhao et al., 2017). In addition, many RBM10 mutations are protein-truncating variants, including nonsense, frameshift, and splice site mutations, which often cause loss of function. Therefore, the mutational spectra of these RBM10 mutations are similar to those of tumor suppressor genes (Vogelstein et al., 2013; Zhao et al., 2017); hence, RBM10 is often classified as a tumor suppressor (Imielinski et al., 2012; Vinayanuwattikun et al., 2016; Zhao et al., 2017; Seiler et al., 2018). In fact, overexpression of RBM10 mutants identified in LUAD or knockdown of wild-type RBM10 results in enhanced cell proliferation, reduced apoptosis, and malignant cancer cell behavior in various cultured or xenografted cells (Bechara et al., 2013; Hernandez et al., 2016; Guan et al., 2017; Yin et al., 2018; ´ Jin et al., 2019). In addition, a decrease in the expression of wild type RBM10 was observed in the tissues of patients with LUAD and in patients with poor prognosis (Guan et al., 2017; Ji et al., 2017; Yin et al., 2018). Conversely, overexpression of wild-type RBM10 reduces cell growth and enhances apoptosis (Guan et al., 2017; Ji et al., 2017; Zhao et al., 2017; Yin et al., 2018; Han et al., 2018; Jin et al., 2019; Jung et al., 2020), and high levels of wild-type RBM10 expression correlates with longer overall survival of patients with LUAD (Zhao et al., 2017). Thus, these data reinforce the notion that RBM10 is a tumor suppressor.
The frequency of RBM10 mutation in LUAD has been reported as 7% (Imielinski et al., 2012); 8% (Cancer Genome Atlas Research Network, 2014); 21% (Vinayanuwattikun et al., 2016), and 22% (Yin et al., 2018), and the mutation frequency increases to 22% in the advanced tumor stage of LUAD (Yin et al., 2018).
RBM10 regulates AS of NUMB. NUMB is the most studied downstream effector of RBM10, and misregulation of NUMB AS is often found in lung cancer (M.-Ali et al., 2011). Wild-type RBM10 promotes skipping of exon 9 of NUMB, generating a NUMB isoform that causes ubiquitination followed by proteasomal degradation of the Notch receptor, thereby inhibiting the Notch signaling cell-proliferation pathway (M.-Ali et al., 2011; Bechara et al., 2013; Cieply and Carstens, 2015). An RBM10 mutation, V354E, found in the lung cancer cell line A549, for example, inhibits the AS activity of RBM10 and produces an exon 9-including NUMB isoform. This isoform promotes cell proliferation and LUAD pathogenesis (Bechara et al., 2013; Hernández et al., 2016). Zhao et al. also showed that an increase in the proliferation rate of LUAD cells correlates with a decrease in the AS activity of RBM10 caused by missense mutations, and suggested that RBM10 mutations contribute to LUAD pathogenesis by deregulating NUMB splicing (Zhao et al., 2017). Notably, the same group recently reported that not NUMB but eukaryotic translation initiation factor 4H (EIF4H) is the main effector of RBM10 in suppressing lung cancer progression; loss of RBM10 promotes the expression of exon 5-containing EIF4H isoform (named EIF4H-L) that is critical for LUAD progression (Zhang et al., 2020).
7.3. Renal cell carcinoma
Translocation or inversion of the TFE3 gene is associated with some renal cell carcinomas (RCCs). The gene encodes a transcription factor and maps at Xp11.2. In Xp11 translocation RCCs, TFE3 is fused with various genes present on other chromosomes such as chromosome 1 or 17 or on the same X chromosome (Gotoh et al., 2014; Just et al., 2016). The RBM10 gene at Xp11.3 and the TFE3 gene have opposite transcriptional orientations. Their fusion produces the RBM10–TFE3 inversion variant, which appears to be a recurrent molecular event in Xp11.2 RCCs (Xia et al., 2017; Argani et al., 2017; Kato et al., 2019). The inversion occurs between RBM10 exon 17 and TFE3 exon 5, resulting in a chimeric gene that produces a transcript composed of RBM10 exons 1–17 and TFE3 exons 5–10, with their reading frames conserved (Gotoh et al., 2014; Just et al., 2016). The fusion likely activates or dysregulates TFE3 activity (Pei et al., 2019) and promotes the expression of a wide variety of genes involved in cell proliferation or may inactivate the tumor suppressor activity of RBM10.
7.4. Other RBM10-related cancers
RBM10 mutations or reduced expression of RBM10 has also been demonstrated in various other cancers (Loiselle and Sutherland, 2018) such as bladder carcinoma (Seiler et al., 2018), pancreatic cancers (Furukawa et al., 2011; Witkiewicz et al., 2015), colorectal cancers (Lawrence et al., 2014; Giannakis et al., 2016), thyroid cancers (Ibrahimpasic et al., 2017, 2019; Antonello et al., 2017), breast cancers (Kan et al., 2010), prostate cancer (Kan et al., 2010), bile duct cancers (Tian et al., 2020; Schwab et al., 2020), and hepatocellular carcinoma (Zhao et al., 2020), as well as in brain tumor meningiomas (Juratli et al., 2018) and astroblastomas (Majd et al., 2019).
7.5. Pathways involving RBM10 in cancers
RBM10 regulates the splicing of hundreds of genes (Wang et al., 2013; Bechara et al., 2013; Sutherland et al., 2017; Sun et al., 2017; Collins et al., 2017; Guan et al., 2017). Accordingly, it is thought to be involved in various pathways, as suggested or shown in the EGFR/KRAS/MAPK (Imielinski et al., 2012; Guan et al., 2017), NRAS/PIK3CA (Imielinski et al., 2012; Ibrahimpasic et al., 2017) and RAP1A/AKT/CREB (Guan et al., 2017; Jin et al., 2019) pathways. Jin et al. demonstrated that RBM10 suppresses the RAP1/AKT/CREB signaling pathway (Jin et al., 2019). In this pathway, RAP1 (a small GTPase that acts as a cellular switch in signal transduction) activates AKT (also known as protein kinase B), which in turn phosphorylates and activates CREB (a transcription factor that induces cell proliferation and reduces apoptosis). Overexpression of RBM10 in LUAD A549 cells decreases RAP1 activation, which reduces the phosphorylation/activation of AKT and CREB, and suppresses the proliferation of cells. Thus, these results suggest that RBM10 suppresses LUAD cell proliferation by inhibiting the RAP1/AKT/CREB signaling pathway (Guan et al., 2017; Jin et al., 2019). Further studies on various RBM10 target genes and RBM10-related pathways are expected to provide new clues/strategies for the diagnosis and treatment of cancers caused by RBM10 mutations.
7.6. Antithetical effects of RBM10
With respect to cell proliferation and apoptosis, most cancer studies on RBM10 have suggested that it functions as a tumor suppressor (Wang et al., 2012; Bechara et al., 2013; Hernandez et al., 2016; Ji et al., 2017; ´ Zhao et al., 2017; Han et al., 2018; Jin et al., 2019; Jung et al., 2020). However, in certain cases, RBM10 acts as a tumor promoter or growth enhancer (Loiselle and Sutherland, 2018). For instance, RBM10 expression can reduce p53 expression, inhibit apoptosis, and promote cell growth as well as other transformation-associated processes as shown in human lung cancer cell lines A549, p53-deficient H1299 (Sun et al., 2019), and RBM5-null GLC20 (Loiselle et al., 2017). In addition, RBM10 depletion leads to proliferation defects in mouse ES cells (Rodor et al., 2017). Furthermore, patients with PDAC (pancreatic ductal adenocarcinoma) harboring RBM10 mutations exhibit a survival rate remarkably higher than the general 5-year PDAC survival rate of less than 7–8% (Witkiewicz et al., 2015; Balachandran et al., 2017; Siegel et al., 2018). These studies are in contrast with those showing RBM10 as a tumor suppressor. As discussed by Rodor et al. (2017) and Loiselle et al. (2017), these antithetical effects of RBM10 are thought to arise from different experimental conditions or cellular contexts composed of different responsible constituents or biological pathways. For instance, Loiselle and Sutherland showed that in an RBM5-suppressed environment, RBM10 promotes cell transformation (Loiselle and Sutherland, 2018). Further elucidation of RBM10-mediated biological pathways will help understand the antithetical outcomes of RBM10 actions.
8. Concluding remarks
Progress has been made in understanding the molecular and cellular biology of RBM10 and diseases caused by its mutations. However, many aspects remain unclear and need to be studied further. These include (1) the molecular structure of RBM10, (2) the functional significance of RBM10 domains such as the region encoded by exon 4 and the tyrosine-rich hydrophilic N-terminal region, (3) the molecular mechanisms of RBM10-mediated AS events, and the basis for the broad spectrum of RNA sequence motifs recognized by RBM10, (4) the cellular processes and pathways in which RBM10 is involved based on the fact that RBM10 regulates the splicing of hundreds of genes, (5) the regulation and mechanisms of RBM10 gene expression, (6) the effects of various post-translational modifications on the activity and functions of RBM10, (7) the biological significance of AS networks organized by RBM10, RBM5, and other splicing regulators, (8) the mechanisms of antithetical RBM10 actions in cancers as a tumor suppressor and tumor promoter, and (9) further understanding of the disease pathogenesis and progression caused by RBM10 mutations.
Studies on these aspects are mutually correlated and will promote the development of strategies to treat the diseases caused by RBM10 mutations.
Acknowledgements
This review and the corresponding Gene Wiki article were written as part of the Gene Wiki Review Series—a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by the National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE.
The author would like to thank Dr. Kishiko Sunami, Department of Otolaryngology, Head and Neck Surgery, Osaka City University Graduate School of Medicine, Japan, for her support. The corresponding Gene Wiki entry for this review can be found here: <<https://en.wikipedia.org/wiki/RBM10>>.
Funding
This work was supported by a Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (Grant number: 19K08378).
Abbreviations:
- RBP
RNA binding protein
- RRM
RNA recognition motif
- aa
amino acid
- OCRE
octamer repeat
- NLS
nuclear localization signal
- ZnF
zinc finger
- NB
nuclear body
- AS
alternative splicing
- NMD
nonsense mediated mRNA decay
- AS-NMD
AS-coupled NMD
- PTC
premature termination codon
- BPS
branchpoint sequence
- LUAD
lung adenocarcinoma
Footnotes
Declaration of Competing Interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Some post-translational modification sites are shown in the supplemental data of the references.
References
- Akimov V, B. -Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Jensen B, D B, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, et al. , 2018. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640. 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- Amanchy R, Zhong J, Molina H, Chaerkady R, Iwahori A, Kalume DE, Grønborg M, Joore J, Cope L, Pandey A, 2008. Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J. Proteome Res. 7, 3900–3910. 10.1021/pr800198w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonello ZA, Hsu N, Bhasin M, Roti G, Joshi M, Van Hummelen P, Ye E, Lo AS, Karumanchi SA, Bryke CR, Nucera C, 2017. Vemurafenib-resistance via de novo RBM genes mutations and chromosome 5 aberrations is overcome by combined therapy with palbociclib in thyroid carcinoma with BRAFV600E. Oncotarget 8, 84743–84760. 10.18632/oncotarget.21262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV, 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24, 342–344. 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR, 2017. RBM10-TFE3 renal cell carcinoma: A potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am. J. Surg. Pathol. 41, 655–662. 10.1097/PAS.0000000000000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T, Suzuki H, Jiang JJ, Okuyama Y, Nakagawa I, Ota M, Tanaka Y, Ohki T, Katsunuma K, Nakajima K, et al. , 2017. Rbm10 regulates inflammation development via alternative splicing of Dnmt3b. Int. Immunol. 29, 581–591. 10.1093/intimm/dxx067. [DOI] [PubMed] [Google Scholar]
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, et al. , 2017. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaton BP, Cotton AM, Brown CJ, 2015. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differences 6: 35. 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li JX, Cohn MA, Cantley LC, Gygi SP, 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135. 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J, 2013. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell. 52, 720–733. 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, et al. , 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26, 1737–1748. 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR, 1993. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucl. Acids Res. 21, 5803–5816. 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL, 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336. 10.1146/annurev.biochem.72.121801.161720. PMID 12626338. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Martínez C, Förch P, Bachi A, Wilm M, Valcárcel J, 2008. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell 32, 81–95. 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. 10.1038/nature13385. Author correction: Nature 2018, 559(7715):E12. . [DOI] [PubMed] [Google Scholar]
- Carrel L, Clemson CM, Dunn JM, Miller AP, Hunt PA, Lawrence JB, Willard HF, 1996. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Human Mol. Genet. 5, 391–401. 10.1093/hmg/5.3.391. [DOI] [PubMed] [Google Scholar]
- Cassola A, Noé G, Frasch AC, 2010. RNA recognition motifs involved in nuclear import of RNA-binding proteins. RNA Biol. 7, 339–344. 10.4161/rna.7.3.12087. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF, 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. USA 101, 17450–17455. 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Weiss WA, 2015. Alternative splicing in cancer: implications for biology and therapy. Oncogene 34, 1–14. 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M, 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cieply B, Carstens RP, 2015. Functional roles of alternative splicing factors in human disease. WIREs RNA 6, 311–326. 10.1002/wrna.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, Ambrose HJ, Carrel L, Nemeth AH, Willard HF, Davies KE, 1996. A novel gene, DXS8237E, lies within 20 kb upstream of UBE1 in Xp11.23 and has a different X inactivation status. Genomics 31, 135–138. 10.1006/geno.1996.0022. [DOI] [PubMed] [Google Scholar]
- Collins KM, Kainov YA, Christodolou E, Ray D, Morris Q, Hughes T, Taylor IA, Makeyev EV, Ramos A, 2017. An RRM-ZnF RNA recognition module targets RBM10 to exonic sequences to promote exon exclusion. Nucl. Acids Res. 45, 6761–6774. 10.1093/nar/gkx225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomer AO, Black F, Greystoke A, Munkley J, Elliott DJ, 2019. Alternative splicing in lung cancer. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194388. 10.1016/j.bbagrm.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G, 2009. RNA and disease. Cell 136, 777–793. 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K, 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32. 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, et al. , 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP, 2008. A quantitative atlas of mitotic phosphorylation. PNAS 105, 10762–10767. 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou XQ, Chen XJ, Wen MX, Zhang SZ, Zhou Q, Zhang SQ, 2020. Alternative splicing of VEGFA is regulated by RBM10 in endometrial cancer. Kaohsiung J. Med. Sci. 36, 13–19. 10.1002/kjm2.12127. [DOI] [PubMed] [Google Scholar]
- Dou XQ, Chen XJ, Zhou Q, Wen MX, Zhang SZ, Zhang SQ, 2020. miR-335 modulates Numb alternative splicing via targeting RBM10 in endometrial cancer. Kaohsiung J. Med. Sci. 36, 171–177. 10.1002/kjm2.12149. [DOI] [PubMed] [Google Scholar]
- Drabkin HA, West JD, Hotfilder M, Heng YM, Erickson P, Calvo R, Dalmau J, Gemmill RM, Sablitzky F, 1999. DEF-3(g16/NY-LU-12), an RNA binding protein from the 3p21.3 homozygous deletion region in SCLC. Oncogene 18, 2589–2597. 10.1038/sj.onc.1202601. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Swanson MS, Pinol-Roma S, 1988. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 13, 86–91. 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG, 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62, 289–321. 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- KerscherA E, , Slack FJ, 2006. Oncomirs–microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fu XD, Ares M, 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701. 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, Yamamoto M, Shibata N, Shimizu K, Kamatani N, Shiratori K, 2011. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep. 1, 161. 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Ray P, Kar A, Wang L, Sutherland LC, Wu JY, 2008. Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc. Natl. Acad. Sci. USA 105, 15708–15713. 10.1073/pnas.0805569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galganski L, Urbanek MO, Krzyzosiak WJ, 2017. Nuclear speckles: molecular organization, biological function and role in disease. Nucl. Acids Res. 45, 10350–10368. 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V, 2016. The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghemrawi R, Arnold C, Hsu B, Pourié SF, Trinh G, Bassila I, Rashka C, Wiedemann C, Flayac A, Robert J, Dreumont A, Feillet N, Guéant F, Gueant JL Coelho D, 2019. SIRT1 activation rescues the mislocalization of RNA-binding proteins and cognitive defects induced by inherited cobalamin disorders. Metabolism 101:153992 10.1016/j.metabol.2019.153992. [DOI] [PubMed] [Google Scholar]
- Ghemrawi R, Pooya S, Lorentz S, Gauchotte G, Arnold C, Gueant J-L, B.-Hsu S-F, 2013. Decreased vitamin B12 availability induces ER stress through impaired SIRT1-deacetylation of HSF1. Cell Death Dis. 4:e553 10.1038/cddis.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Mansour A, A., Yamauchi, M., et al. , 2016. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 15, 857–865 10.1016/j.celrep.2016.03.075. Erratum in: Cell Rep. 17, 1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G, 1992. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 11, 3289–3295. 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cervenka J, Anderson RC, Sauk JJ, Bevis WD, 1970. Robin’s syndrome: A probably X-linked recessive subvariety exhibiting persistence of left superior vena cava and atrial septal defect. Am. J. Dis. Child. 119, 176–178. 10.1001/archpedi.1970.02100050178020. [DOI] [PubMed] [Google Scholar]
- Goto Y,. Kimura H, 2009. Inactive X chromosome-specific histone H3 modifications and CpG hypomethylation flank a chromatin boundary between an X-inactivated and an escape gene. Nucl. Acids Res. 37, 7416–7428. 10.1093/nar/gkp860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh M, Ichikawa H, Arai E, Chiku S, Sakamoto H, Fujimoto H, Hiramoto M, Nammo T, Yasuda K, Yoshida T, Kanai Y, 2014. Comprehensive exploration of novel chimeric transcripts in clear cell renal cell carcinomas using whole transcriptome analysis. Genes Chromosomes Cancer 53, 1018–1032. 10.1002/gcc.22211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Hopkins E, Johnston JJ, Krause C, Dobyns WB, Biesecker LG, 2011. Long-term survival in TARP syndrome and confirmation of RBM10 as the disease-causing gene. Am. J. Med. Genet. A 155A, 2516–2520. 10.1002/ajmg.a.34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan G, Li R, Tang W, Liu T, Su Z, Wang Y, Tan J, Jiang S, Wang K, 2017. Expression of RNA-binding motif 10 is associated with advanced tumor stage and malignant behaviors of lung adenocarcinoma cancer cells. Tumour Biol. 39: 1010428317691740. 10.1177/1010428317691740. [DOI] [PubMed] [Google Scholar]
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Comb MJ, et al. , 2014. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics 13, 372–387. 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güre AO, Altorki NK, Stockert E, Scanlan MJ, Old LJ Chen Y-T, 1998. Human lung cancer antigens recognized by autologous antibodies: Definition of a novel cDNA derived from the tumor suppressor gene locus on chromosome 3p21.3. Cancer Res. 58, 1034–1041. [PubMed] [Google Scholar]
- Han LP, Wang CP, Han SL, 2018. Overexpression of RBM10 induces osteosarcoma cell apoptosis and inhibits cell proliferation and migration. Med Sci (Paris). 34 (Focus issue F1), 81–86 10.1051/medsci/201834f114. [DOI] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U, 2012. Dynamic protein–protein interaction wiring of the human spliceosome. Mol. Cell. 45, 567–580 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Hernández J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcárcel J, 2016. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 13, 466–472. 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højland AT, Lolas I, Okkels H, Lautrup CK, Diness BR, Petersen MB, Nielsen IK, 2018. First reported adult patient with TARP syndrome: A case report. Am. J. Med. Genet. A 176, 2915–2918. 10.1002/ajmg.a.40638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, Seshan V, Deraje S, Carlson DL, Migliacci J, Knauf JA, et al. , 2017. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: Identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin. Cancer Res. 23, 5970–5980. 10.1158/1078-0432.CCR-17-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimpasic T, Ghossein R, Shah JP, Ganly I, 2019. Poorly differentiated carcinoma of the thyroid gland: Current status and future prospects. Thyroid 29, 311–321. 10.1089/thy.2018.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. , 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Takahashi KP, Kimura M, Watanabe T, Morisawa S, 1996. Molecular cloning of a RNA binding protein, S1–1. Nucl. Acids Res. 24, 2990–2997. 10.1093/nar/24.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Tsugawa K, Tokunaga K, Takahashi KP, Uni S, Kimura M, Nishio K, Yamamoto N, Honda K, Watanabe T, et al. , 2008. S1–1 nuclear domains: characterization and dynamics as a function of transcriptional activity. Biol. Cell 100, 523–535. 10.1042/BC20070142. [DOI] [PubMed] [Google Scholar]
- Inoue A, Yamamoto N, Kimura M, Nishio K, Yamane H, Nakajima K, 2014. RBM10 regulates alternative splicing. FEBS Lett. 588, 942–947. 10.1016/j.febslet.2014.01.052. [DOI] [PubMed] [Google Scholar]
- Jackson TC, Du L, Feldman JK, Vagni VA, Dezfulian C, Poloyac SM, Jackson EK, Clark RS, Kochanek PM, 2015. The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J Cereb. Blood Flow Metab. 35, 655–666 10.1038/jcbfm.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsai D, Watkins DN, O’Connor AE, Merriner DJ, Gursoy S, Bird AD, Kumar B, Miller A, Cole TJ, Jenkins BJ, O’Bryan MK, 2017. In vivo evidence that RBM5 is a tumour suppressor in the lung. Sci. Rep. 7, 16323. 10.1038/s41598-017-15874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Sharp PA, 2014. Building robust transcriptomes with master splicing factors. Cell 159, 487–498. 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Minna JD, Roth JA, 2005. 3p21.3 tumor suppressor cluster: prospects for translational applications. Future Oncol. 1, 79–92. 10.1517/14796694.1.1.79. [DOI] [PubMed] [Google Scholar]
- Ji Y, Xie S, Jiang L, Liu L, Li L, Luo L, Chen Y, Zhang J, Yu L, Zhang Y, Tang N, Liu B, 2017. Increased cell apoptosis in human lung adenocarcinoma and in vivo tumor growth inhibition by RBM10, a tumor suppressor gene. Oncol Lett. 14, 4663–4669. 10.3892/ol.2017.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Di X, Wang R, Ma H, Tian C, Zhao M, Cong S, Liu J, Li R, Wang K, 2019. RBM10 inhibits cell proliferation of lung adenocarcinoma via RAP1/AKT/CREB signalling pathway. J Cell Mol Med. 23, 3897–3904. 10.1111/jcmm.14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, NIH Intramural Sequencing Center (NISC), Chong K, Mullikin JC, Biesecker LG, 2010. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 86, 743–748. 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Sapp JC, Curry C, Horton M, Leon E, Ozog CK, , Dobyns WB, Hudgins L, Zackai E, Biesecker LG, 2014. Expansion of the TARP syndrome phenotype associated with de novo mutations and mosaicism. Am. J. Med. Genet. A 164A, 120–128 10.1002/ajmg.a.36212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Lee H, Cao B, Liao P, Zeng SX, Lu H, 2020. RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. Oncogene 39, 1031–1040. 10.1038/s41388-019-1034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juratli TA, McCabe D, Nayyar N, Williams EA, Silverman IM, Tummala SS, Fink AL, Baig A, M.-Lage M, Selig MK, et al. , 2018. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 136, 779–792. 10.1007/s00401-018-1899-7. [DOI] [PubMed] [Google Scholar]
- Just PA, Letourneur F, Pouliquen C, Dome F, Audebourg A, Biquet P, Vidaud M, Terris B, Sibony M, Pasmant E, 2016. Identification by FFPE RNA-Seq of a new recurrent inversion leading to RBM10-TFE3 fusion in renal cell carcinoma with subtle TFE3 break-apart FISH pattern. Genes Chromosomes Cancer 55, 541–548. 10.1002/gcc.22356. [DOI] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, et al. , 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873. 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Kato I, Furuya M, Baba M, Kameda Y, Yasuda M, Nishimoto K, Oyama M, Yamasaki T, Ogawa O, Niino H, et al. , 2019. RBM10-TFE3 renal cell carcinoma characterised by paracentric inversion with consistent closely split signals in break-apart fluorescence in-situ hybridisation: study of 10 cases and a literature review. Histopathology 75, 254–265. 10.1111/his.13866. [DOI] [PubMed] [Google Scholar]
- Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Lührmann R, 2009. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 15, 153–175. 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto H, Inoue A, Kojima H, Yang J, Zhao H, Tsuruta D, Nakajima K, 2020. RBM10 regulates centriole duplication in HepG2 cells by ectopically assembling PLK4-STIL complexes in the nucleus. Genes Cells 25, 100–110. 10.1111/gtc.12741. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G, 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC, 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323. 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li W, Laishram RS, Hoque M, Ji Z, Tian B, Anderson RA, 2017. Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases. Nucl. Acids Res. 45, 8930–8942. 10.1093/nar/gkx560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MMH, MacDonald MR, 2016. Polyamines: Small molecules with a big role in promoting virus infection. Cell Host Microbe 20, 123–124. 10.1016/j.chom.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Loiselle JJ, Roy JG, Sutherland LC, 2017. RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5. PLoS One 12, e0180258. 10.1371/journal.pone.0180258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselle JJ, Sutherland LC, 2018. RBM10: Harmful or helpful-many factors to consider. J. Cell Biochem. 119, 3809–3818. 10.1002/jcb.26644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF, 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 417, 15–27. 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- M.-Ali CM, Cheng E, O’Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ, 2011. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol. Cell Biol. 31, 138–150. 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M.-Arribas F, Agudo D, Pollán M, Gómez-Esquer F, Díaz-Gil G, Lucas R, Schneider J, 2006. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J. Cell Biochem. 97, 1275–1282. 10.1002/jcb.20725. [DOI] [PubMed] [Google Scholar]
- M.-Garabato E, M.-Arribas F, Pollán M, Lucas AR, Sánchez J, Schneider J, 2008. The small variant of the apoptosis-associated X-chromosome RBM10 gene is co-expressed with caspase-3 in breast cancer. Cancer Genomics Proteomics. 5, 169–173. [PubMed] [Google Scholar]
- M.-Maarabouni M, ., Williams GT, 2002. RBM5/LUCA-15 —Tumour Suppression by Control of Apoptosis and the Cell Cycle? Sci. World J. 2, 1885–1890. 10.1100/tsw.2002.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaskola J, Rajewsky N, 2014. Binding site discovery from nucleic acid sequences by discriminative learning of hidden Markov models. Nucl. Acids Res. 42, 12995–13011. 10.1093/nar/gku1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd NK, Metrus NR, S.-Pinheiro F, Trevino CR, Fuller GN, Huse JT, Chung C, Ketonen L, Anderson MD, P.-Prado M, 2019. RBM10 truncation in astroblastoma in a patient with history of mandibular ameloblastoma: A case report. Cancer Genet. 231–232, 41–45 10.1016/j.cancergen.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Makarov EM, Owen N, Bottril l.A., Makarova OV, 2011. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucl. Acids Res. 40, 2639–2652. 10.1093/nar/gkr1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Krainer AR, 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074. 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BT, Serrano P, Geralt M, Wüthrich K, 2016. Nuclear magnetic resonance structure of a novel globular domain in RBM10 containing OCRE, the octamer repeat sequence motif. Structure 24, 158–164. 10.1016/j.str.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CWJ, 2005. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398. 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. , 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166. 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mohan N, Kumar V, Kandala DT, Kartha CC, Laishram RS, 2018. A splicing-independent function of RBM10 controls specific 3’ UTR processing to regulate cardiac hypertrophy. Cell Rep. 24, 3539–3553. 10.1016/j.celrep.2018.08.077. [DOI] [PubMed] [Google Scholar]
- Mourão A, Bonnal S, Soni1 K, Warner L, Bordonné R, Valcárcel J, Sattler M, 2016. Structural basis for the recognition of spliceosomal SmN/B/B’ proteins by the RBM5 OCRE domain in splicing regulation. eLife 5, e14707. 10.7554/eLife.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CF, Berger A, Zimmer S, Tiyerili V, Nickenig G, 2009. The heterogenous nuclear riboprotein S1–1 regulates AT1 receptor gene expression via transcriptional and posttranscriptional mechanisms. Arch Biochem Biophys. 488, 76–82. 10.1016/j.abb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N, 1995. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 2, 167–174. 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- Nguyen CD, Mansfield RE, Leung W, Vaz PM, Loughlin FE, Grant RP, Mackay JP, 2011. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 407, 273–283. 10.1016/j.jmb.2010.12.041. [DOI] [PubMed] [Google Scholar]
- Niceta M, Barresi S, Pantaleoni F, Capolino R, Dentici ML, Ciolfi A, Pizzi S, Bartuli A, Dallapiccola B, Tartaglia M, Digilio MC, 2019. TARP syndrome: Long-term survival, anatomic patterns of congenital heart defects, differential diagnosis and pathogenetic considerations. Eur. J. Med. Genet. 62:103534 10.1016/j.ejmg.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Niu Z, Jin W, Zhang L, Li X, 2012. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 586, 977–983. 10.1016/j.febslet.2012.02.052. [DOI] [PubMed] [Google Scholar]
- Oh JJ, Grosshans DR, Wong SG, Slamon DJ, 1999. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucl. Acids Res. 27, 4008–4017. 10.1093/nar/27.20.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JJ, Razfar A, Delgado I, Reed RA, Malkina A, Boctor B, Slamon DJ, 2006. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 66, 3419–3427. 10.1158/0008-5472.CAN-05-1667. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M, 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648. 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K, 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438. 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ, 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415. 10.1038/ng.259. PMID 18978789. [DOI] [PubMed] [Google Scholar]
- Papasaikas P, Tejedor JR, Vigevani L, Valcárcel L, 2015. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell 57, 7–22. 10.1016/j.molcel.2014.10.030. [DOI] [PubMed] [Google Scholar]
- Pei J, Cooper H, Flieder DB, Talarchek JN, Al-Saleem T, Uzzo RG, Dulaimi E, Patchefsky AS, Testa JR, Wei S, 2019. NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma. Mod. Pathol. 32, 710–716. 10.1038/s41379-018-0191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis Z, Hart A, Cherny S, Petrik I, Palmaer E, Tang S, Jones C, 2017. Clinical diagnostic exome evaluation for an infant with a lethal disorder: genetic diagnosis of TARP syndrome and expansion of the phenotype in a patient with a newly reported RBM10 alteration. BMC Med. Genet. 18, 60. 10.1186/s12881-017-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi B, Bragado L, Mammi P, Torti MF, Gaioli N, Gebhard LG, Solá MEG, Vaz-Drago R, Iglesias NG, García CC, Gamarnik AV, Srebrow A, 2020. Dengue virus targets RBM10 deregulating host cell splicing and innate immune response. Nucl. Acids Res. 48, 6824–6838. 10.1093/nar/gkaa340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prola A, Da Silva JP, Guilbert A, Lecru L, Piquereau J, Ribeiro M, Mateo P, Gressette M, Fortin D, Boursier C, et al. , 2017. SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation. Cell Death Differ. 24, 343–356. 10.1038/cdd.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Krainer AR, Abdel-Wahab O, 2020. SnapShot: Splicing alterations in cancer. Cell 180, 208–208.e1. 10.1016/j.cell.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M, 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245. 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini C, Rogers SW, Rechsteiner M, 1994. KEKE motifs: Proposed roles in protein-protein association and presentation of peptides by MHC Class I receptors. FEBS Lett. 348, 109–113. 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- Rodor J, FitzPatrick DR, Eyras E, Caceres JF, 2017. The RNA-binding landscape of ´ RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 14, 45–57. 10.1080/15476286.2016.1247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichs E, Ledda A, Mularoni L, Albà MM, de la Luna S, 2009. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLOS Genet. 5:e1000397 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Gómez-Esquer F, Díaz-Gil G, Torrejón R, Pollán M, 2011. mRNA expression of the putative antimetastatic gene BRMS1 and of apoptosis-related genes in breast cancer. Cancer Genomics Proteomics. 8, 195–197. [PubMed] [Google Scholar]
- Schwab ME, Song H, Mattis A, Phelps A, Vu LT, Huang FW, Nijagal A, 2020. De novo somatic mutations and KRAS amplification are associated with cholangiocarcinoma in a patient with a history of choledochal cyst. J. Pediatr. Surg. 55, 2657–2661. 10.1016/j.jpedsurg.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS, 2016. RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32. 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, Smith PG; Cancer Genome Atlas Research Network, Buonamici S, Yu L, 2018. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.e4 10.1016/j.celrep.2018.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P, Hammond JA, Geralt M, Wüthrich K, 2018. Splicing site recognition by synergy of three domains in splicing factor RBM10. Biochem. 57, 1563–1567. 10.1021/acs.biochem.7b01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Zhao L, Wang K, Xu W, Zhang J, Yang B, 2012. The tumor suppressor gene RBM5 inhibits lung adenocarcinoma cell growth and induces apoptosis. World J. Surg. Oncol. 10:160 10.1186/1477-7819-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2018. Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Kuhlmeier MM, Carter HC, Hasday JD, Lilly CM, Fairchild KD, 2006. Effect of moderate hypothermia on gene expression by THP-1 cells: a DNA microarray study. Physiol. Genomics 26, 91–98. 10.1152/physiolgenomics.00296.2005. [DOI] [PubMed] [Google Scholar]
- Stes E, Laga M, Walton A, Samyn N, Timmerman E, De Smet I, Goormachtig S, Gevaert K, 2014. A COFRADIC protocol to study protein ubiquitination. J. Proteome Res. 136, 3107–3113. 10.1021/pr4012443. [DOI] [PubMed] [Google Scholar]
- Sun Y, Bao Y, Han W, Song F, Shen X, Zhao J, Zuo J, Saffen D, Chen W, Wang Z, You X, Wang Y, 2017. Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay. Nucl. Acids Res. 45, 8524–8540. 10.1093/nar/gkx508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jia M, Sun W, Feng L, Gu C, Wu T, 2019. Functional role of RBM10 in lung adenocarcinoma proliferation. Int. J. Oncol. 54, 467–478. 10.3892/ijo.2018.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland LC, Rintala-Maki ND, White RD, Morin CD, 2005. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J. Cell Biochem. 94, 5–24. 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- Sutherland LC, Wang K, Robinson AG, 2010. RBM5 as a putative tumor suppressor gene for lung cancer. J. Thorac. Oncol. 5, 294–298. 10.1097/JTO.0b013e3181c6e330. [DOI] [PubMed] [Google Scholar]
- Sutherland LC, Thibault P, Durand M, Lapointe E, Knee JM, Beauvais A, Kalatskaya I, Hunt SC, Loiselle JJ, Roy JG, et al. , 2017. Splicing arrays reveal novel RBM10 targets, including SMN2 pre-mRNA. BMC Mol. Biol. 18, 19. 10.1186/s12867-017-0096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier SJ, Loiselle JJ, McBain A, Pullen C, Koenderink BW, Roy JG, Sutherland LC, 2015. Insight into the role of alternative splicing within the RBM10v1 exon 10 tandem donor site. BMC Res. Notes 8, 46. 10.1186/s13104-015-0983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiselton DL, McDowall J, Brandau O, Ramser J, d’Esposito F, Bhattacharya SS, Ross MT, Hardcastle AJ, Meindl A, 2002. An integrated, functionally annotated gene map of the DXS8026-ELK1 interval on human Xp11.3-Xp11.23: potential hotspot for neurogenetic disorders. Genomics. 79, 560–572. Erratum in: Genomics 79, 891. 10.1006/geno.2002.6733. [DOI] [PubMed] [Google Scholar]
- Tian W, Hu W, Shi X, Liu P, Ma X, Zhao W, Qu L, Zhang S, Shi W, Liu A, Cao J, 2020. Comprehensive genomic profile of cholangiocarcinomas in China. Oncol Lett. 19, 3101–3110. 10.3892/ol.2020.11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer T, Terpstra P, van den Berg A, Veldhuis PM, Ter Elst A, Voutsinas G, Hulsbeek MM, Draaijers TG, Looman MW, Kok K, et al. , 1999. A comparison of genomic structures and expression patterns of two closely related flanking genes in a critical lung cancer region at 3p21.3. Eur. J. Hum. Genet. 7, 478–486. 10.1038/sj.ejhg.5200334. [DOI] [PubMed] [Google Scholar]
- Timmer T, Terpstra P, van den Berg A, Veldhuis PM, Ter Elst A, van der Veen AY, Kok K, Naylor SL, Buys CH, 1999. An evolutionary rearrangement of the Xp11.3–11.23 region in 3p21.3, a region frequently deleted in a variety of cancers. Genomics 60, 238–240. 10.1006/geno.1999.5878. PMID 10486212. [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiss JL, Eichner N, Lehmann G, Schall K, Urlaub H, Meister G, 2017. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol. Cell 66, 270–284. 10.1016/j.molcel.2017.03.014. [DOI] [PubMed] [Google Scholar]
- Ule J, Blencowe BJ, 2019. Alternative splicing regulatory networks: Functions, mechanisms, and evolution. Mol. Cell 76, 329–345. 10.1016/j.molcel.2019.09.017. [DOI] [PubMed] [Google Scholar]
- Urbanski LM, Leclair N, Anczuków O, 2018. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev. RNA 9, e1476. 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayanuwattikun C, Le Calvez-Kelm F, Abedi-Ardekani B, Zaridze D, Mukeria A, Voegele C, Vallée M, Purnomosari D, Forey N, Durand G, et al. , 2016. Elucidating genomic characteristics of lung cancer progression from in situ to invasive adenocarcinoma. Sci. Rep. 6, 31628. 10.1038/srep31628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasschaert C, Xia X, Coulombe J, Gray DA, 2015. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 15: 230 10.1186/s12862-015-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW, 2013. Cancer genome landscapes. Science 339, 1546–1558. 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R, 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718. 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB, 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Bacon ML, Tessier JJ, Rintala-MakI ND, Tang V, Sutherland LC, 2012. RBM10 modulates apoptosis and influences TNF-α gene expression. J. Cell Death. 5, 1–19. 10.4137/JCD.S9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gogol-Döring A, Hu H, Fröhler S, Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, et al. , 2013. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol. Med. 5, 1431–1442. 10.1002/emmm.201302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xiao S, Kunimoto H, Tokunaga K, Kojima H, Kimura M, Yamamoto T, Yamamoto N, Hong Z, Nishio K, et al. , 2019. Targeting of RBM10 to S1–1 nuclear bodies: Targeting sequences and its biological significance. bioRxiv, 10.1101/516831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang F, Zhong W, Ling H, Wang J, Cui J, Xie T, Wen S, Chen J, 2019. RNA-binding protein RBM6 as a tumor suppressor gene represses the growth and progression in laryngocarcinoma. Gene 697, 26–34. 10.1016/j.gene.2019.02.025. [DOI] [PubMed] [Google Scholar]
- Wei MH, Latif F, Bader S, Kashuba V, Chen JY, Duh FM, Sekido Y, Lee CC, Geil L, Kuzmin I, et al. , 1996. Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3: progress toward the isolation of a lung cancer TSG. Cancer Res. 56, 1487–1492. [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. , 2015. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744. 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M, Gorlach M, Friedrichs M, Dreyfuss G, Mueller L, 1992. 1H, 13C, and 15N NMR assignments and global folding pattern of the RNA-binding domain of the human hnRNP C proteins. Biochemistry 31, 6254–6265. 10.1021/bi00142a013. [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, G.-Blanco MA, , Smith CWJ, 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell, 13, 91–100. 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Xia QY, Wang XT, Zhan XM, Tan X, Chen H, Liu Y, Shi SS, Wang X, Wei X, Ye SB, et al. , 2017. Xp11 translocation renal cell carcinomas (RCCs) with RBM10-TFE3 gene fusion demonstrating melanotic features and overlapping morphology with t(6;11) RCC: Interest and diagnostic pitfall in detecting a paracentric inversion of TFE3. Am. J. Surg. Pathol. 41, 663–676. 10.1097/PAS.0000000000000837. [DOI] [PubMed] [Google Scholar]
- Xiao R, Chen J-Y, Liang Z, Luo D, Chen G, Lu ZJ, Chen Y, Zhou B, Li H, Du X, et al. , 2019. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121. 10.1016/j.cell.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SJ, Wang LY, Kimura M, Kojima H, Kunimoto H, Nishiumi F, Yamamoto N, Nishio K, Fujimoto S, Kato T, Kitagawa S, Yamane H, Nakajima K, Inoue A, 2013. S1–1/RBM10: multiplicity and cooperativity of nuclear localisation domains. Biol Cell. 105, 162–174. 10.1111/boc.201200068. [DOI] [PubMed] [Google Scholar]
- Yamada H, Tsutsumi K, Nakazawa Y, Shibagaki Y, Hattori S, Ohta Y, 2016. Src family tyrosine kinase signaling regulates FilGAP through association with RBM10. PLoS One 11:e0146593. 10.1371/journal.pone.0146593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, C.-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, et al. , 2016. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805–817. 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin LL, Wen XM, Li M, Xu YM, Zhao XF, Li J, Wang XW, 2018. A gene mutation in RNA-binding protein 10 is associated with lung adenocarcinoma progression and poor prognosis. Oncol. Lett. 16, 6283–6292. 10.3892/ol.2018.9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, Li J, Mills GB, Shu Y, Li L, Liang H, 2016. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell 29, 711–722. 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Kim E, Warren SL, Berget SM, 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16, 1401–1412. 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bao Y, Shen X, Pan Y, Sun Y, Xiao M, Chen K, Wei H, Zuo J, Saffen D, et al. , 2020. RNA binding motif protein 10 suppresses lung cancer progression by controlling alternative splicing of eukaryotic translation initiation factor 4H. EBioMedicine 61, 103067. 10.1016/j.ebiom.2020.103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun Y, Huang Y, Song F, Huang Z, Bao Y, Zuo J, Saffen D, Shao Z, Liu W, Wang Y, 2017. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci. Rep. 7, 40488. 10.1038/srep40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Li J, Shen F, 2020. Protective effect of the RNA-binding protein RBM10 in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 24, 6005–6013. 10.26355/eurrev_202006_21494. [DOI] [PubMed] [Google Scholar]
- Zheng S, Damoiseaux R, Chen L, Black DL, 2013. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res. 23, 998–1007. 10.1101/gr.147546.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG, 2007. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621. 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]