Abstract
PURPOSE
Given regulatory approval of immune checkpoint inhibitors in patients with mismatch repair–deficient (MMR-D) cancers agnostic to tumor type, it has become important to characterize occurrence of MMR-D and develop cost-effective screening approaches. Using a next-generation sequencing (NGS) panel (OncoPanel), we developed an algorithm to identify MMR-D frequency in tumor samples and applied it in a clinical setting with pathologist review.
METHODS
To predict MMR-D, we adapted methods described previously for use in NGS panels, which assess patterns of single base-pair insertion or deletion events occurring in homopolymer regions. Tumors assayed with OncoPanel between July 2013 and July 2018 were included. For tumors tested after June 2017, sequencing results were presented to pathologists in real time for clinical MMR determination, in the context of tumor mutation burden, other mutational signatures, and clinical data.
RESULTS
Of 20,301 tumors sequenced, 2.7% (553) were retrospectively classified as MMR-D by the algorithm. Of 4,404 samples with pathologist sign-out of MMR status, the algorithm classified 147 (3.3%) as MMR-D: in 116 cases, MMR-D was confirmed by a pathologist, five cases were overruled by the pathologist, and 26 were assessed as indeterminate. Overall, the highest frequencies of OncoPanel-inferred MMR-D were in endometrial (21%; 152/723), colorectal (9.7%; 169/1,744), and small bowel (9.3%; 9/97) cancers. When algorithm predictions were compared with historical MMR immunohistochemistry or polymerase chain reaction results in a set of 325 tumors sequenced before initiation of pathologist assessment, the overall sensitivity and specificity of the algorithm were 91.1% and 98.2%, respectively.
CONCLUSION
We show that targeted, tumor-only NGS can be leveraged to determine MMR signatures across tumor types, suggesting that broader biomarker screening approaches may have clinical value.
INTRODUCTION
As regulatory approvals for immunotherapy agents expand across tumor types, efforts to identify predictive biomarkers of response or resistance to treatment are similarly expanding. Comprehensive analysis of colorectal cancers has shown that tumors with defects in mismatch repair (MMR), resulting in microsatellite instability (MSI), have elevated lymphocytic infiltration.1 In addition, tumors with MSI demonstrate upregulated expression of inhibitory immune checkpoints, including PD-L1, on tumor-infiltrating lymphocytes and myeloid cells. These developments and others led to clinical trials evaluating the efficacy of immune checkpoint blockade on the basis of MMR status. On the basis of data from 149 patients with mismatch repair deficient (MMR-D) or MSI-high (MSI-H) tumors in five single-arm trials, the PD-1 inhibitor pembrolizumab was approved in 2017 by the US Food and Drug Administration (FDA) for patients with unresectable or metastatic MMR-D or MSI-H solid tumors of any tissue type.2,3 This was the first tumor-agnostic, biomarker-based drug approval, with approvals for larotrectinib and entrectinib for neurotrophic tyrosine receptor kinase fusion-positive tumors following shortly thereafter.4,5 These breakthroughs underscore the need for broadly applicable, cost-effective methods of identifying tumors with MMR-D and other potential biomarkers.
CONTEXT
Key Objective
To develop a bioinformatics algorithm to infer mismatch repair–deficient (MMR-D) cancers using tumor-only next-generation sequencing (NGS) data and describe the prevalence of MMR-D across cancer types.
Knowledge Generated
We characterized MMR status in a cohort of 20,301 tumor samples across a wide range of adult and pediatric malignancies, including cancer types not commonly assayed for MMR-D as part of routine clinical practice. Overall, the algorithm classified 2.7% of samples as MMR-D.
Relevance
We show the value of using a routine NGS panel to identify new, clinically relevant biomarkers that might otherwise not be assessed because of low prevalence in certain tumor types and to identify potential candidates for immunotherapy.
Patients with colorectal and endometrial tumors undergo routine clinical testing for MSI and/or MMR-D using polymerase chain reaction (PCR) assays by assessing loci within the Bethesda panel or related biomarkers or by assessing gene expression in the MMR pathway via immunohistochemistry (IHC).6 However, given the lower prevalence of MMR-D outside colorectal and endometrial cancers, MSI/MMR testing using IHC or PCR is not routinely pursued in clinical practice in other tumor types, despite National Comprehensive Cancer Network testing recommendations for the following cancers: cervical, colon, rectal, endometrial, esophageal, gastroesophageal junction, gastric carcinoma, gallbladder, intrahepatic cholangiocarcinoma, extrahepatic cholangiocarcinoma, neuroendocrine, ovarian, pancreatic, prostate, testicular, and vulvar. Broad implementation of a targeted, tumor-only next-generation sequencing (NGS) assay (OncoPanel) for patients at Dana-Farber Cancer Institute (DFCI), Brigham and Women’s Hospital (BWH), and Boston Children’s Hospital (BCH) allowed us to assess whether bioinformatic analysis of OncoPanel7 data could characterize the prevalence of MMR-D/MSI-H (hereafter MMR-D) across tumors and increase the number of patients who could be considered for immunotherapy.
Methods such as mSINGS,8 MSI-Sensor9 and cell-free DNA10 have previously been reported for detection of MMR-D. However, given the wide implementation and relatively low cost of tumor-only NGS, it is important to develop approaches suited for this paradigm and understand potential differences in results. It is also critical to better characterize the frequency of MMR-D across tumors to inform selective use of IHC, PCR, or NGS assays in clinical settings where NGS panels and MMR testing are used less frequently. This study expands on previous work to develop OncoPanel for clinical assessment of MMR-D using IHC and PCR for validation11 and to characterize MMR status in 645 upper GI tract cancers12 and 304 sarcomas.13 Here we describe a bioinformatics algorithm that infers MMR status using an institution-wide NGS panel and report the prevalence of MMR-D across > 20,000 adult and pediatric tumor specimens.
METHODS
Patient Population
Patients treated for a cancer diagnosis at DFCI, BWH, or BCH were offered participation in the research study PROFILE (Dana-Farber/Harvard Cancer Center Institutional Review Board [IRB] protocol #11-104/17-000) as previously described.14 Tumor samples from patients who consented to participate in PROFILE or from patients with cancers eligible for clinical NGS were tested using a tumor-only targeted DNA sequencing panel (OncoPanel).14 Data acquisition, bioinformatic analysis, chart review, and orthogonal MSI testing via PCR were performed under DFCI IRB protocol #17-465.
We included patients who provided written informed consent and underwent successful targeted OncoPanel sequencing of a primary or metastatic tumor between August 2013 (OncoPanel study initiation) and July 2018 to allow adequate time for patient follow-up. As of the study cutoff date, 20,301 tumors had OncoPanel results and could be assessed for MMR-D via the bioinformatics algorithm described below. Of these tumors, 4,488 were sequenced after the implementation on June 27, 2017 of a process for pathologist sign-out of NGS-based, MMR-D assessment, described in more detail below. A total of 4,404 of these 4,488 tumors met criteria for MMR assessment via the OncoPanel algorithm, which measures insertion or deletion (indel) events within homopolymeric microsatellite loci to infer MMR status, and were presented for final interpretation by a molecular pathologist. Signed-out OncoPanel reports, including MMR-D calls, were returned to physicians for clinical use.
Patients from the aforementioned study of 304 sarcomas13 are also included in the summary data here, although with different subclassifications. Here, we used the original pathologists’ diagnoses as recorded at OncoPanel sign-out, rather than more detailed subclassification of sarcomas using chart review.
Sequencing
From August 2013 to July 2014, patient samples were tested with OncoPanel Version 1, which covers 0.753334 Mb and the full coding regions of 275 genes, plus selected intronic regions of 30 genes for rearrangement detection. OncoPanel Version 2 began August 2014, covering 0.826167 Mb, with full coding regions of 300 cancer genes plus selected intronic regions of 35 genes. OncoPanel Version 3 activated in October 2016, covering 1.315078 Mb, 447 cancer genes, and 191 regions across 60 genes for rearrangement detection. At the time of clinical pathology assessment for MMR deficiency (on or after June 27, 2017), all samples were being sequenced using OncoPanel Version 3.14 Variants observed in public genome databases at a frequency of > 0.1% were removed, unless observed more than twice in the Catalogue of Somatic Mutations in Cancer,15 in which case they were presented for manual review. Finally, variants were tiered and reported based on clinical significance by a molecular pathologist using methods previously described.16 All testing was conducted in a Clinical Laboratory Improvement Amendments–certified environment.
MMR Algorithm
On the basis of methods described previously for identifying MMR-D in colorectal cancer using OncoPanel Versions 1-2,11 an internally developed algorithm was adjusted to predict MMR status across all tumor types. For this study, a homopolymer was defined as a genomic region with more than four repeat nucleotides. Tumors harboring ≥ 1.5 single base-pair indels/Mb were called MMR-D. Only patterns of single base-pair indels in homopolymers were considered (with a minimum threshold of 1.5/Mb), and tumor mutational burden (TMB) was ignored.
Possible values resulting from the OncoPanel algorithm are proficient (MMR-P), deficient (MMR-D), and cannot assess (for tumors with < 12 variants), although a pathologist may override algorithmic calls or mark a tumor “indeterminate.” For the 4,488 samples where MMR results were communicated to the ordering physician, a pathologist manually reviewed the algorithmic MMR status to assign final status. Pathologists interpreted the algorithmic MMR determination in the context of TMB, other mutational signatures validated on the platform (UV [ultraviolet], tobacco, APOBEC [apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like], temozolomide, and POLE [DNA polymerase epsilon catalytic subunit]), and existing clinical data such as MMR protein expression, when available. Recognizing that the positive and negative predictive values of the MMR algorithm are lower near the defined cutoff, and that patterns of MMR-D are not well defined for tumors outside of colon and endometrium, the pathologist could elect to override the algorithmic classification. Possible reasons for overriding the classification included homopolymer indel rates near the cutoff, low TMB for that tumor type, or no MMR gene alterations in the sequencing data. The treating clinician was also encouraged to seek confirmatory testing with IHC or PCR in this setting when clinically relevant.
TMB
TMB was calculated by including all nonsynonymous mutations in coding regions of the genome covered by OncoPanel. The sum of such mutations was divided by the size of the bait set to determine mutations per megabase (mut/Mb). TMB results were compared with other tumors sequenced with the same panel version as well as with other tumors of the same cancer type. We reported three TMB metrics for each tumor: mutations per megabase, percentile of TMB across all tumors sequenced in the current OncoPanel version, and percentile of TMB across tumors of each cancer type.
Orthogonal Testing
Twelve of the 26 tumor samples with discrepant calls between algorithm and pathologist, specifically those with algorithm-predicted MMR-D, a pathologist call of indeterminate, and sufficient leftover DNA, underwent orthogonal, tumor-only PCR testing. MSI PCR was performed using a five-marker microsatellite panel and capillary electrophoresis as previously described.11 Results were interpreted by the pathologist according to revised Bethesda guidelines.17
RESULTS
Frequencies of MMR-D
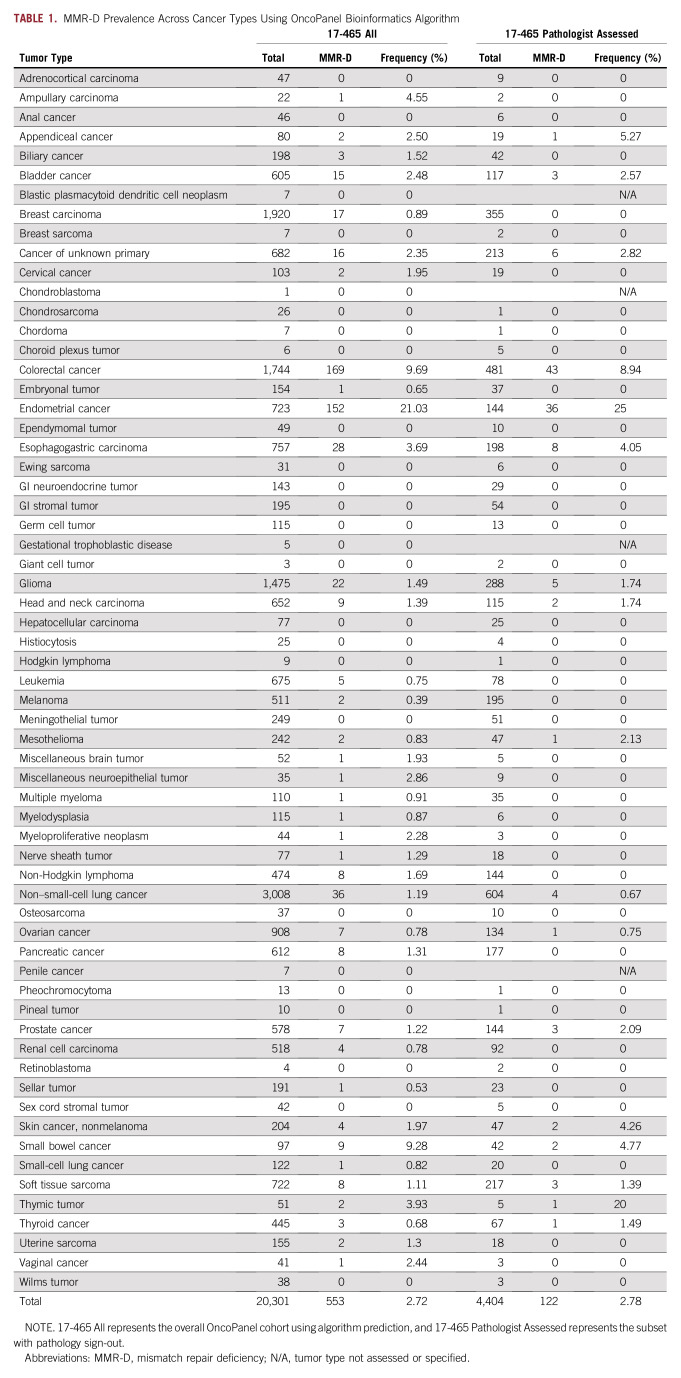
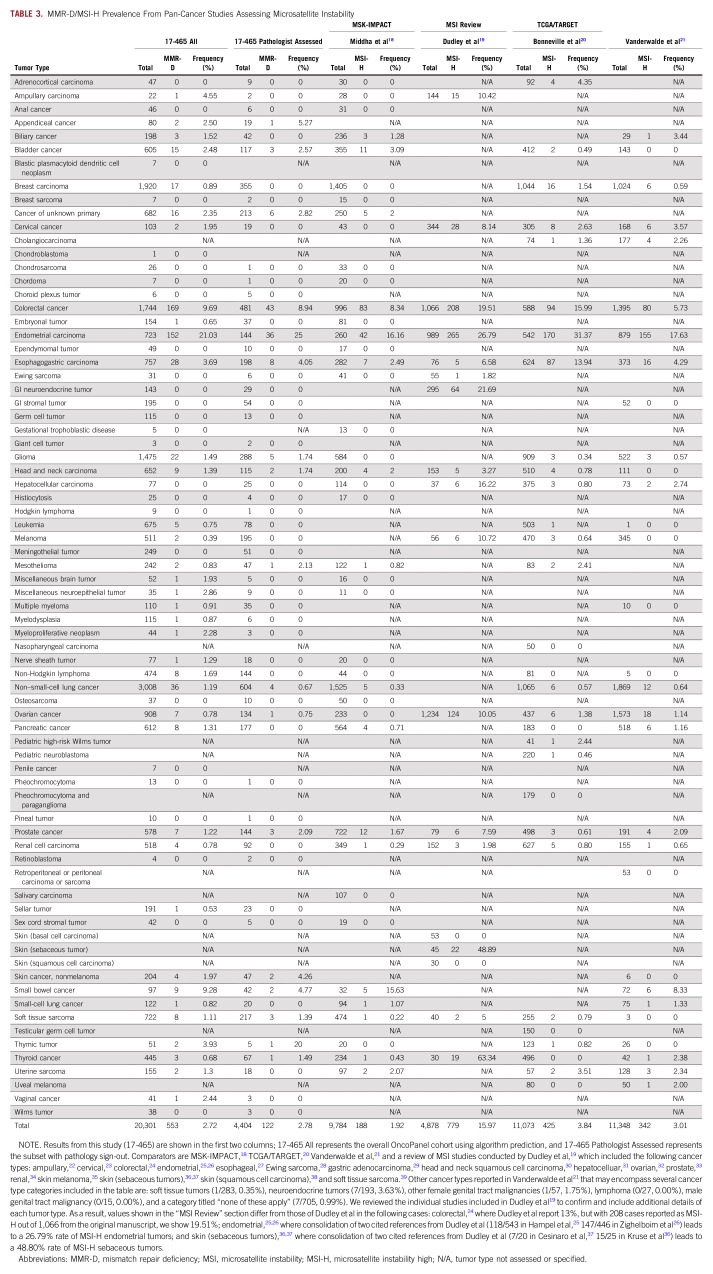
We measured MMR-D frequencies detected using the OncoPanel algorithm in 20,301 samples across tumor types (Tables 1 and 2) and compared them to historical reports of MMR-D prevalence (Tables 1 and 3, Fig 1).18-39 The OncoPanel cohort comprised 13,542 primary biopsies, 4,734 metastatic recurrences, 655 local recurrences, and 1,370 tumors with unspecified biopsy site.
TABLE 1.
MMR-D Prevalence Across Cancer Types Using OncoPanel Bioinformatics Algorithm
TABLE 2.
Mapping of Tumor Types From Sources
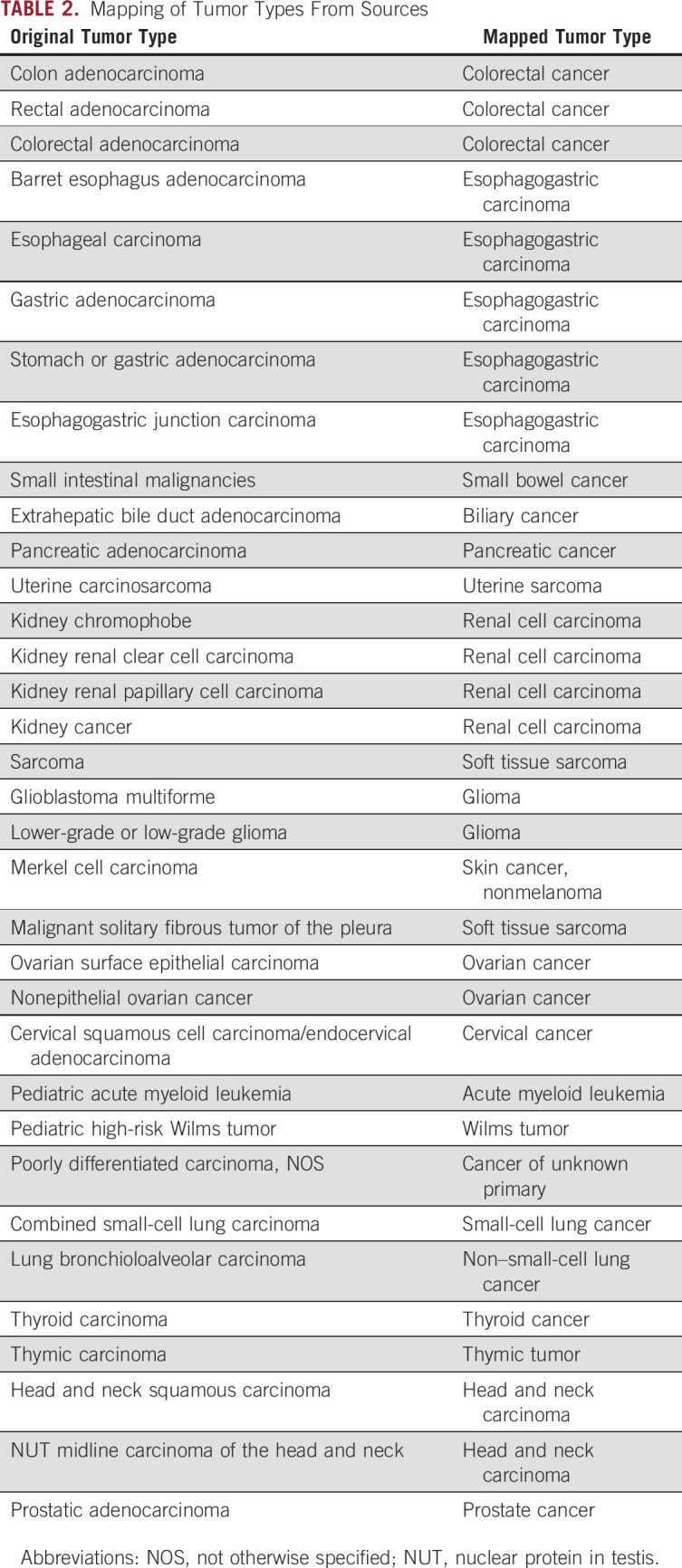
TABLE 3.
MMR-D/MSI-H Prevalence From Pan-Cancer Studies Assessing Microsatellite Instability
FIG 1.
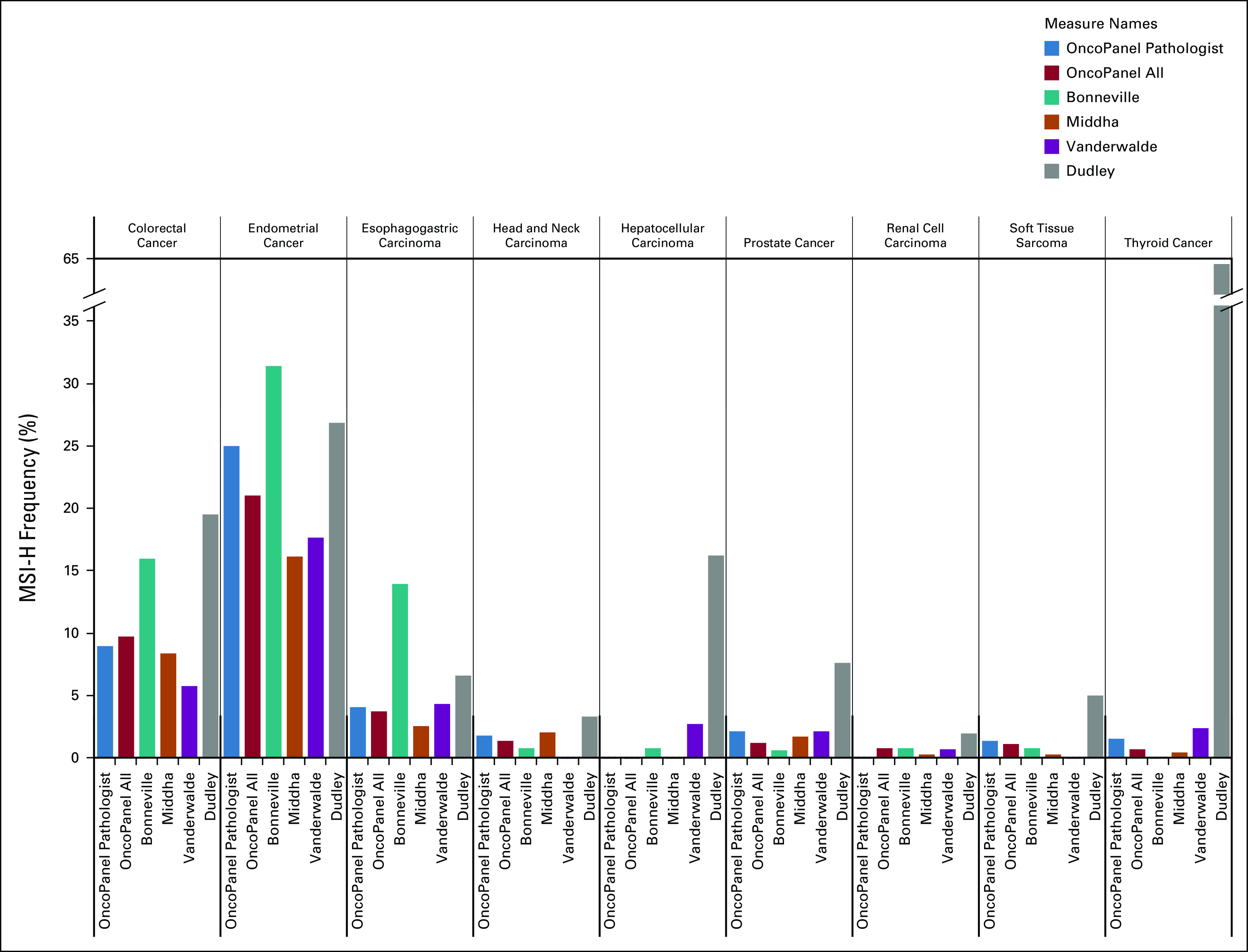
Comparison of mismatch repair–deficient (MMR-D)/microsatellite instability–high (MSI-H) patterns across tumor types. Across all studies described in Table 3,18-39 the frequency of MMR-D or MSI-H was reported across all six cohorts in nine tumor types: colorectal, endometrial, esophagogastric, head and neck, hepatocellular, prostate, renal cell carcinoma, thyroid cancer, and soft tissue sarcoma. Hepatocellular carcinoma and thyroid cancer had considerably higher MSI-H rates reported in the review by Dudley et al.19
One of the main goals of this work was to characterize prevalence of MMR-D outside the tumor types where it is commonly assayed in routine clinical practice. Overall, among 3,779 noncolorectal, nonendometrial samples for which OncoPanel results were returned to clinicians, 43 (1.14%) tumors were detected as MSI-H/MMR-D, and results were reported to the patients’ treating clinicians. Without another indication for clinical MSI/MMR testing, these 43 patients are unlikely to have been tested outside of the OncoPanel program.
Across the 816 pediatric tumors profiled, eight were predicted as MMR-D algorithmically. Of these 816, 208 were also assessed for MMR status by a pathologist for the clinical OncoPanel report. One case of pediatric small bowel cancer was reported as MMR-D algorithmically, with concordant pathologist sign-out. One pediatric embryonal tumor was predicted algorithmically as MMR-D but ruled indeterminate by the pathologist. PCR-based orthogonal validation was performed on the small bowel case to confirm MSI-H status, and MSI was detected for three of four microsatellite markers.
We identified two adult appendiceal tumors with predicted MMR-D: one predicted by the algorithm in the historical cohort and the other identified by a pathologist during routine clinical sign-out. To the best of our knowledge, MMR-D has not been reported previously in appendiceal cancer. In addition, we found one thymic MMR-D tumor among five cases (20%), indicating that thymic cancers too can be MMR-D, corroborating a recent report.20
The Cancer Genome Atlas (TCGA) and Therapeutically Applicable Research to Generate Effective Treatments (TARGET) projects20 reported MSI-H status in 3.8% of tumors, using paired tumor-normal sequencing across cancer types and the algorithm MANTIS,40 and identified MSI in 27 cancer types (see Discussion). This rate is consistent with our algorithmic predictions of MMR-D in clinically signed-out tumors (3.33%), although only 2.78% were pathologist confirmed.
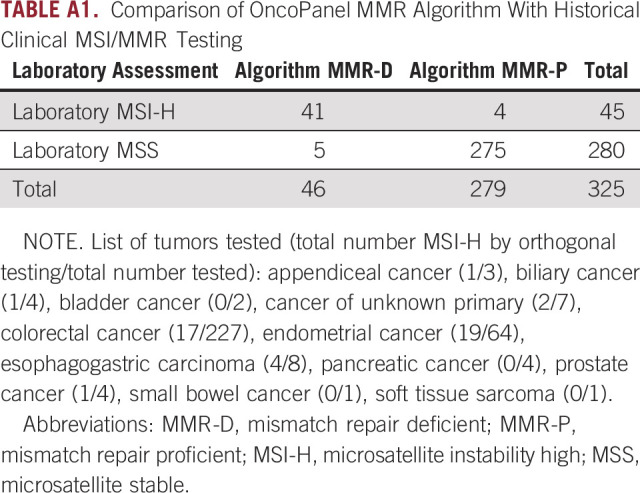
We also measured algorithm sensitivity and specificity using a historical set of 325 tumors tested with IHC or PCR. This yielded a sensitivity and specificity of 91% and 98%, respectively (Appendix Table A1).
Concordance of OncoPanel MMR Algorithm With Pathologist Assessment
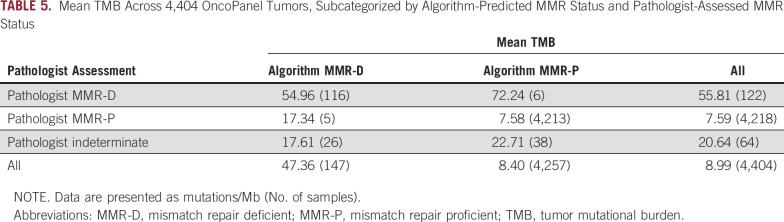
Concordance was assessed between the purely algorithmic MMR/OncoPanel calls and pathologist calls. For the 4,404 clinical cases with MMR status signed out by pathologists, there was substantial agreement (Cohen’s κ coefficient = 0.75) between algorithmic and pathologist calls (Table 4). To better understand discordant cases, we selected 14 tumors for orthogonal MSI validation via PCR that were algorithmically predicted to be MMR-D and also had sufficient sample material. One of the samples was confirmed to be MMR-D: a pediatric embryonal tumor. None of the other samples were MSI-H by PCR, supporting the additional value of manual pathologist review. One factor among many that could play a role in discordance between algorithmic prediction and pathologist assessment (Table 5) is TMB, which was relatively high in concordant MMR-D cases (mean TMB, 55.8 mut/Mb). Concordant MMR-P cases had a mean TMB of 7.59 mut/Mb, and indeterminate cases yielded a mean TMB of 20.6 mut/Mb.
TABLE 4.
Concordance Between Algorithmic Prediction and Final Pathology Assessment for MMR Status
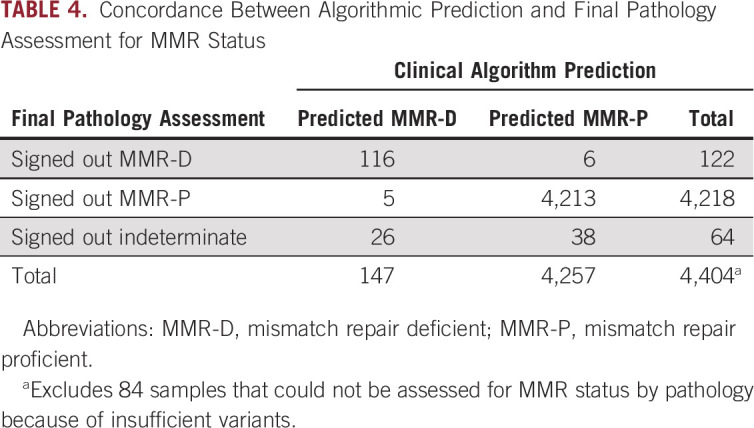
TABLE 5.
Mean TMB Across 4,404 OncoPanel Tumors, Subcategorized by Algorithm-Predicted MMR Status and Pathologist-Assessed MMR Status
DISCUSSION
We found 2.72% of 20,301 tumor samples to be MMR-D across a wide range of adult and pediatric malignancies using a bioinformatics algorithm to process tumor-only NGS data. In a 4,404 sample subset, we report substantial agreement between purely algorithmic calls and formal pathologist classification. It should be noted that these two measures are not independent, because the pathologists included the algorithmic calls as a part of their assessment, along with TMB, results of other mutational signatures, and existing clinical data. Orthogonal laboratory validation of tumors predicted to be MMR-D but signed out as indeterminate demonstrated the value of manual pathologist review as a final step in the MMR-D classification process and suggests that inclusion of other data types could improve future algorithms.
Confirming prior clinical observations, the highest frequencies of predicted MMR-D were in endometrial and colorectal cancers, but MMR-D was also observed in many different cancer types not commonly assessed for MMR-D in routine clinical practice, including appendiceal cancer and nonmelanoma skin cancer.
Further manual review of predicted MMR-D tumors signed out as indeterminate highlighted the tendency for tumors with a pattern of indels in homopolymer regions but low TMB to be called indeterminate. Orthogonal PCR validation of 14 discordant cases confirmed only one as MSI-H. It could thus be useful to incorporate TMB explicitly in future MMR-D prediction algorithms. Our results suggest confirmatory laboratory testing should continue to be used, especially when borderline NGS results are obtained or MMR-D is predicted in a tumor with low MMR-D prevalence. Correlation between high TMB and MSI-H has been reported in other NGS studies, including in prostate cancer, where 71% of 111 tumors with high TMB (≥ 20 mut/Mb) were MSI-H,41 and in adrenocortical carcinoma (n = 92) and cervical squamous cell and cervical adenocarcinoma (n = 305).20 In a larger NGS panel study (n = 11,000), only 27% of tumors with high TMB (defined as ≥ 17 mut/Mb) were MSI-H (≥ 46 altered microsatellite loci/Mb), whereas 70% of MSI-H cases had high TMB.21 As exemplified in these studies, however, there is significant variability in the definitions of “high TMB” and the methodologies used to measure it.42 Recent FDA approval of pembrolizumab for high-TMB tumors defined high TMB as ≥ 10 mut/Mb, with a targeted gene panel as a companion diagnostic. Although TMB and MSI are both now broadly relevant biomarkers for immunotherapy, they reflect different aspects of tumor biology. Thus, assessing MMR independently from TMB allows clinicians to evaluate these data independently when creating patient treatment plans.43
Certain cancers have considerable variability in reported MMR-D frequencies, including hepatocellular carcinoma (0.0%-16.2%), ampullary carcinoma (0.0%-10.4%), GI neuroendocrine tumors (0.0%-20.7%), ovarian cancer (0.0%-10.0%), and thyroid cancer (0%-63.3%). Potential reasons include variation in sample sizes and detection methods, and it also remains unknown if MMR-D differs based on the site of disease (primary v metastatic) or timing (treatment-naïve v treatment-exposed).
It is encouraging, however, that MMR-D prevalence in most tumor types across independent studies was generally consistent (Fig 1), especially for other NGS-based assessments,18,20,21 although more traditional MSI testing performed via PCR or IHC showed greater variability in some tumor types.19 For the TCGA and TARGET data sets,18 MSI-H status was observed using tumor-normal paired NGS in 27 different tumor types at an overall frequency of 3.8%.37 Our predicted MMR-D frequency using tumor-only NGS is similar for clinically signed-out cases (3.3%), although only 2.8% were pathologist confirmed. Coupled with the high sensitivity and specificity we observed in comparison with manual review, this suggests tumor-only NGS detection of MMR-D supported by simple computational predictions may be a useful and cost-effective screening tool for research and clinical use. In this paradigm, computationally predicted MMR-D tumors should, in many cases, still undergo validation with a clinical test (eg, PCR or IHC) before being used for clinical care. PCR testing has been considered the gold standard for the determination of microsatellite stability, although the five-marker Bethesda panel44 and updated MSI Analysis System45 were originally optimized and validated in colorectal cancer. Across tumor types, it remains unclear which methodology to determine MMR status provides greater accuracy, and NGS computational algorithms may improve identification of these tumors compared with TMB, PCR, or IHC testing. It will also be important for future studies to measure the impact of MMR-D assessment on treatment decisions and patient outcomes.
Given the low overall frequency of MMR-D across most noncolorectal, nonendometrial tumors, we believe our algorithm provides a cost-effective initial screening tool to identify patients for PCR- or IHC-based testing. From a clinical implementation standpoint, rather than testing all patients with cancer with PCR or IHC to identify the < 3% of patients who may be candidates for immune checkpoint inhibitors on the basis of MMR-D, implementing the algorithm in the context of an existing, multipurpose NGS tumor-only panel, can provide an efficient initial screen and allow clinicians to select only those tumors most likely to be MMR-D for further testing. Biomarker studies in patients with MMR-D tumors treated with immune checkpoint inhibition will provide further insight.
In the near future, combined testing of many biomarkers across a large panel of genes as described here may be more efficient in cost, time, and sample material than sequential or more selective biomarker testing, and it also offers the ability to quickly incorporate new biomarker discoveries into clinical practice and bring effective treatments to patients who would otherwise not benefit from these discoveries. Efforts to evaluate the clinical utility of NGS-inferred MMR-D, determined by the impact of physician notification of MMR/MSI status on treatment decision making, are ongoing and may provide further support for the implementation of targeted NGS panels in routine clinical practice.
ACKNOWLEDGMENT
We thank Kaitlyn T. Bifolck, BA, for her editorial support.
Appendix
TABLE A1.
Comparison of OncoPanel MMR Algorithm With Historical Clinical MSI/MMR Testing
SUPPORT
Supported by a research grant from Merck Sharp & Dohme, a subsidiary of Merck, Kenilworth, NJ.
EQUAL CONTRIBUTION
A.A. and A.C.G.-C. contributed equally to this work.N.U.L. and J.M.J. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Adem Albayrak, Ana C. Garrido-Castro, Kai-Li Liaw, Mayur Amonkar, Katherine A. Janeway, Lynette Sholl, Nancy U. Lin, Jason Johnson
Financial support: Bruce E. Johnson, Kai-Li Liaw, Nancy U. Lin
Administrative support: Bruce E. Johnson
Provision of study material or patients: Marios Giannakis, Elizabeth H. Stover, Bruce E. Johnson, Katherine A. Janeway, Jonathan A. Nowak, Lynette Sholl, Nancy U. Lin
Collection and assembly of data: Adem Albayrak, Ana C. Garrido-Castro, Marios Giannakis, Monica Devi Manam, Alanna J. Church, Katherine A. Janeway, Lynette Sholl, Nancy U. Lin, Jason Johnson
Data analysis and interpretation: Adem Albayrak, Ana C. Garrido-Castro, Marios Giannakis, Renato Umeton, Monica Devi Manam, Elizabeth H. Stover, Rebecca L. Porter, Bruce E. Johnson, Kai-Li Liaw, Alanna J. Church, Katherine A. Janeway, Jonathan A. Nowak, Lynette Sholl, Nancy U. Lin, Jason Johnson
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Adem Albayrak
Employment: Health Catalyst
Stock and Other Ownership Interests: Health Catalyst
Honoraria: BC Platforms
Research Funding: Merck (Inst)
Travel, Accommodations, Expenses: Health Catalyst
Other Relationship: Guidepoint
Marios Giannakis
Research Funding: Bristol Myers Squibb, Merck
Renato Umeton
Consulting or Advisory Role: Health Catalyst
Patents, Royalties, Other Intellectual Property: Patent: Portable medical device and method for quantitative retinal image analysis through a smartphone; Patent: Epstein Barr virus genotypic variants and uses thereof as risk predictors, biomarkers, and therapeutic targets of multiple sclerosis
Elizabeth H. Stover
Patents, Royalties, Other Intellectual Property: Patents pending: “Dissecting treatment resistance in patients with ovarian cancer and PDX models using single-cell RNA-sequencing” and “Compositions and methods for treating ovarian tumors,” unrelated to the current work.
Bruce E. Johnson
Consulting or Advisory Role: Novartis, Foundation Medicine, Hengrui Therapeutics, Daiichi Sankyo, Chugai Pharmaceuticals, Lilly, Checkpoint Therapeutics
Research Funding: Toshiba (Inst), Novartis (Inst), Novartis
Patents, Royalties, Other Intellectual Property: Dana-Farber Cancer Institute
Kai-Li Liaw
Employment: Merck Sharp & Dohme
Stock and Other Ownership Interests: Merck Sharp & Dohme
Research Funding: Merck Sharp & Dohme
Travel, Accommodations, Expenses: Merck Sharp & Dohme
Mayur Amonkar
Employment: Merck
Stock and Other Ownership Interests: Merck
Alanna J. Church
Honoraria: BioRad Laboratories, Jackson Laboratories
Consulting or Advisory Role: AlphaSights, Samba Scientific, Perceptive Advisors, Jackson Laboratories, Bayer
Travel, Accommodations, Expenses: Jackson Laboratories, Bayer
Katherine A. Janeway
Honoraria: Foundation Medicine
Consulting or Advisory Role: Bayer, Ipsen
Travel, Accommodations, Expenses: Bayer
Jonathan A. Nowak
Patents, Royalties, Other Intellectual Property: Provisional patent application involving novel methods for characterizing immune cell distributions in solid tumors.
Lynette Sholl
Honoraria: AstraZeneca
Consulting or Advisory Role: LOXO Oncology, EMD Serono
Research Funding: Roche/Genentech
Nancy U. Lin
Consulting or Advisory Role: Seattle Genetics, Puma Biotechnology, Daiichi Sankyo, California Institute for Regenerative Medicine, Denali Therapeutics
Research Funding: Genentech, Pfizer, Seattle Genetics, Merck
Patents, Royalties, Other Intellectual Property: Royalties for chapter in Up-to-Date regarding management of breast cancer brain metastases; Royalties, Jones & Bartlett
Jason M. Johnson
Consulting or Advisory Role: UCB, PatientsLikeMe
Research Funding: Merck
Travel, Accommodations, Expenses: UCB
No other potential conflicts of interest were reported.
REFERENCES
- 1.Llosa NJ, Cruise M, Tam A, et al. : The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 5:43-51, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.US Food and Drug Administration : FDA approves first cancer treatment for any solid tumor with a specific genetic feature. https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm.
- 3.Le DT, Durham JN, Smith KN, et al. : Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357:409-413, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott LJ: Larotrectinib: First global approval. Drugs 79:201-206, 2019 [DOI] [PubMed] [Google Scholar]
- 5.Al-Salama ZT, Keam SJ: Entrectinib: First global approval. Drugs 79:1477-1483, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Murphy KM, Zhang S, Geiger T, et al. : Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn 8:305-311, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Peng Y, Peng G: Mismatch repair-based stratification for immune checkpoint blockade therapy. Am J Cancer Res 8:1977-1988, 2018 [PMC free article] [PubMed] [Google Scholar]
- 8.Salipante SJ, Scroggins SM, Hampel HL, et al. : Microsatellite instability detection by next generation sequencing. Clin Chem 60:1192-1199, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Niu B, Ye K, Zhang Q, et al. : MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30:1015-1016, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willis J, Lefterova MI, Artyomenko A, et al. : Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 25:7035-7045, 2019 [DOI] [PubMed] [Google Scholar]
- 11.Nowak JA, Yurgelun MB, Bruce JL, et al. : Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn 19:84-91, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christakis AG, Papke DJ, Nowak JA, et al. : Targeted cancer next-generation sequencing as a primary screening tool for microsatellite instability and Lynch syndrome in upper gastrointestinal tract cancers. Cancer Epidemiol Biomarkers Prev 28:1246-1251, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Doyle LA, Nowak JA, Nathenson MJ, et al. : Characteristics of mismatch repair deficiency in sarcomas. Mod Pathol 32:977-987, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Sholl LM, Do K, Shivdasani P, et al. : Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1:e87062, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tate JG, Bamford S, Jubb HC, et al. : COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 47:D941-D947, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MM, Datto M, Duncavage EJ, et al. : Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19:4-23, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umar A, Boland CR, Terdiman JP, et al. : Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96:261-268, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1200/PO.17.00084. Middha S, Zhang L, Nafa K, et al: Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol 10.1200/PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley JC, Lin MT, Le DT, et al. : Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res 22:813-820, 2016 [DOI] [PubMed] [Google Scholar]
- 20. doi: 10.1200/PO.17.00073. Bonneville R, Krook MA, Kautto EA, et al: Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 10.1200/PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanderwalde A, Spetzler D, Xiao N, et al. : Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 7:746-756, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruemmele P, Dietmaier W, Terracciano L, et al. : Histopathologic features and microsatellite instability of cancers of the papilla of vater and their precursor lesions. Am J Surg Pathol 33:691-704, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Lazo PA: The molecular genetics of cervical carcinoma. Br J Cancer 80:2008-2018, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampel H, Frankel WL, Martin E, et al. : Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851-1860, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Hampel H, Frankel W, Panescu J, et al. : Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res 66:7810-7817, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Zighelboim I, Goodfellow PJ, Gao F, et al. : Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol 25:2042-2048, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Farris AB III, Demicco EG, Le LP, et al. : Clinicopathologic and molecular profiles of microsatellite unstable Barrett esophagus-associated adenocarcinoma. Am J Surg Pathol 35:647-655, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Alldinger I, Schaefer KL, Goedde D, et al. : Microsatellite instability in Ewing tumor is not associated with loss of mismatch repair protein expression. J Cancer Res Clin Oncol 133:749-759, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network : Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202-209, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glavac D, Volavsek M, Potocnik U, et al. : Low microsatellite instability and high loss of heterozygosity rates indicate dominant role of the suppressor pathway in squamous cell carcinoma of head and neck and loss of heterozygosity of 11q14.3 correlates with tumor grade. Cancer Genet Cytogenet 146:27-32, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Chiappini F, Gross-Goupil M, Saffroy R, et al. : Microsatellite instability mutator phenotype in hepatocellular carcinoma in non-alcoholic and non-virally infected normal livers. Carcinogenesis 25:541-547, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Murphy MA, Wentzensen N: Frequency of mismatch repair deficiency in ovarian cancer: A systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int J Cancer 129:1914-1922, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burger M, Denzinger S, Hammerschmied CG, et al. : Elevated microsatellite alterations at selected tetranucleotides (EMAST) and mismatch repair gene expression in prostate cancer. J Mol Med (Berl) 84:833-841, 2006 [DOI] [PubMed] [Google Scholar]
- 34. Hammerschmied C, Stoehr R, Walter B, et al: The role of elevated microsatellite alterations at selected tetranucleotides (EMAST) in renal cell carcinoma (RCC). Cancer Res 67, 2007 (abstr 1934) [Google Scholar]
- 35.Palmieri G, Ascierto PA, Cossu A, et al. : Assessment of genetic instability in melanocytic skin lesions through microsatellite analysis of benign naevi, dysplastic naevi, and primary melanomas and their metastases. Melanoma Res 13:167-170, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Kruse R, Rütten A, Schweiger N, et al. : Frequency of microsatellite instability in unselected sebaceous gland neoplasias and hyperplasias. J Invest Dermatol 120:858-864, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Cesinaro AM, Ubiali A, Sighinolfi P, et al. : Mismatch repair proteins expression and microsatellite instability in skin lesions with sebaceous differentiation: A study in different clinical subgroups with and without extracutaneous cancer. Am J Dermatopathol 29:351-358, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Reuschenbach M, Sommerer C, Hartschuh W, et al. : Absence of mismatch repair deficiency-related microsatellite instability in non-melanoma skin cancer. J Invest Dermatol 132:491-493, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Kawaguchi K, Oda Y, Takahira T, et al. : Microsatellite instability and hMLH1 and hMSH2 expression analysis in soft tissue sarcomas. Oncol Rep 13:241-246, 2005 [PubMed] [Google Scholar]
- 40.Kautto EA, Bonneville R, Miya J, et al. : Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 8:7452-7463, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. doi: 10.1200/PO.18.00283. Chung JH, Dewal N, Sokol E, et al: Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis Oncol 10.1200/PO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. doi: 10.1200/PO.19.00171. Vokes NI, Liu D, Ricciuti B, et al: Harmonization of tumor mutational burden quantification and association with response to immune checkpoint blockade in non-small-cell lung cancer. JCO Precis Oncol 10.1200/PO.19.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.US Food and Drug Administration : FDA approves pembrolizumab for adults and children with TMB-H solid tumors. https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors
- 44.Boland CR, Thibodeau SN, Hamilton SR, et al. : A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248-5257, 1998 [PubMed] [Google Scholar]
- 45.Bacher JW, Flanagan LA, Smalley RL, et al. : Development of a fluorescent multiplex assay for detection of MSI-high tumors. Dis Markers 20:237-250, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]