Summary
The production and release of small phospholipid membrane vesicles, or extracellular vesicles (EVs), is a trait of most prokaryotic and eukaryotic cells. EVs display heterogeneity in content, size, biogenesis, activity, and function. B cells uniquely express immunoglobulin and produce EVs; however, the relationship between these entities has not been clarified. Here, we used several methodologies to isolate large (11,000 × g) and small (110,000 × g) EVs and evaluate their IgM content, characteristics and activity. We found that B cells from multiple cell lines and primary B cells produce EVs that display monomeric IgM on the surface and contain encapsulated monomeric IgM, which is independent of secreted pentameric IgM. Our data indicate EV IgM can bind antigen specifically, and EV IgM can be incorporated intracellularly into secondary cells. These results suggest immunological activities different from secreted pentameric IgM that may constitute a separate and distinct antibody distribution system.
Subject areas: Immunology, Cell biology
Graphical abstract
Highlights
-
•
B cells release two sizes of extracellular vesicles (EVs) containing IgM
-
•
EV IgM is located on the EV surface and inside
-
•
EV IgM can be transferred to and enter secondary cells
-
•
EV IgM can bind antigen specifically
Immunology; Cell biology
Introduction
The most ancient precursors to our adaptive immune system appeared with the rise of jawed vertebrates, forming a diverse, antigen-specific defense that could be layered with the established components of innate immunity.1 Mammalian B cell populations and secreted immunoglobulin (antibody), generated over time through molecular and cellular adaptation, reflect further expansion in immune diversity.2,3,4,5 Predating adaptive immunity in phylogenetic origin is the cellular capacity to manufacture and release extracellular vesicles (EVs). Identified in all branches of life, from bacteria to plants to humans, and produced by cells from divergent developmental pathways, accumulating evidence indicates that EVs are a fundamental mode for intercellular communication, existing in parallel with soluble bioactive molecules secreted through the conventional endoplasmic reticulum-to-golgi cell secretory pathway.6,7,8,9,10,11 EV-mediated interactions can be observed in normal organismal development, immunity, aging, and in disease states that reflect dysfunctional cellular behavior.12,13,14,15,16,17,18,19,20 This is also evident across species, from helminth parasitism to the gut microbiome, and in natural environments, where for example marine bacteria may evade bacteriophage infection by releasing EV decoys.21,22,23,24 In all biological contexts, EVs are characterized by a phospholipid construction that generates distinct compartments, encapsulated interior and membrane exterior, and are notable for their heterogeneity, that manifests in molecular content (solutes, nucleic acid, lipids, proteins), biogenesis (endosomal, plasma membrane), and interaction with secondary cells (receptor-mediated endocytosis, phagocytosis).25,26 Although dissimilar in physical form, EV-associated and secreted soluble molecules share the ability to impart biological activity at a distance, independent of cell-to-cell contact.27,28,29,30,31,32,33,34,35,36 There has been an increasing appreciation for the combined contribution of extracellular factors via the conventional secretory and EV pathways.
Although millions of years have shaped B cell and EV biology, experimental investigation into B cell-derived EVs and associated immunoglobulin is surprisingly limited. Early seminal work relating to MHC-II endosomal trafficking and EV-mediated immune activation revealed that B cells could activate T cells through the release of small EVs that contained MHC-II complexed antigen, and other reports provided insight into B cell EV protein and lipid content.37,38,39,40 Additional evaluation using transformed B cells and murine primary B cells identified factors that regulate small EV production and have better characterized EV protein content, and more recent work demonstrated EV-mediated protein transfer between B cell lines.41,42,43,44,45,46,47,48,49 As demonstrated for EV produced by other cell types, a link has been established between B cell EV and disease, including cancer and parasitic diseases.50,51,52,53,54 Although informative and essential in establishing that B cell EVs exist and harbor functional attributes, these and other studies in this area have neglected to thoroughly evaluate EV immunoglobulin content, across multiple B cell populations and multiple EV populations based on size. Both points are pertinent, the former given the basic concept that antibody expression is singular to B-lineage cells and comprises the foundational element in humoral immunity, and the latter given the growing evidence that unique EV populations are functionally distinct.55,56,57,58,59,60 Additionally, B cells can secrete large amounts of antibody at certain developmental stages and in response to appropriate external stimulation; however, how this relates to regulated EV release and immunoglobulin expression is unknown. A complete account of extracellular antibody, and its functional implications, would thus need to include elucidation and characterization of EV-associated immunoglobulin.
To address these points, we combined multiple EV isolation techniques and analytical methodologies to first characterize immunoglobulin M (IgM) expression, molecular form, and topography in small (also referred to as exosomes) and large (also referred to as microvesicles) EVs isolated from murine B lymphoma cell lines and murine splenic B cells. We then evaluated IgM associated with peritoneal cavity EVs, in both EVs isolated from washout fluid and EVs produced by sorted peritoneal B cell populations. Lastly, we assessed the antigen binding capacity of EV IgM and transfer potential to secondary cells. Our results indicate that IgM is differentially expressed in multiple EV populations, that EV IgM expression occurs independently from secreted IgM, that IgM can be both sequestered within the EV cavity and presented on the EV surface, and that it is present under homeostatic conditions in peritoneal cavity EVs and is present in EVs released by both B1 and B2 cells. We further show using phosphatidylcholine (PtC)-specific CH12 B lymphoma cells, that express IgM in both small and large EVs, that EVs can bind antigen specifically. Finally, we provide evidence that IgM associated with large EVs can be internalized by secondary cells and persist intact. Our results suggest that EV IgM holds immunoregulatory potential, perhaps unique by comparison to secreted IgM given the differences in antibody presentation, both biological (soluble vs. lipid encapsulated) and molecular (pentameric vs. monomeric), between the conventional and EV cellular release mechanisms. We therefore propose that IgM packaging into EV is a fundamental cellular process, performed across multiple B cell populations alongside conventional IgM secretion.
Results
Multiple extracellular vesicle (EV) populations produced by WEHI-231 cells contain immunoglobulin M
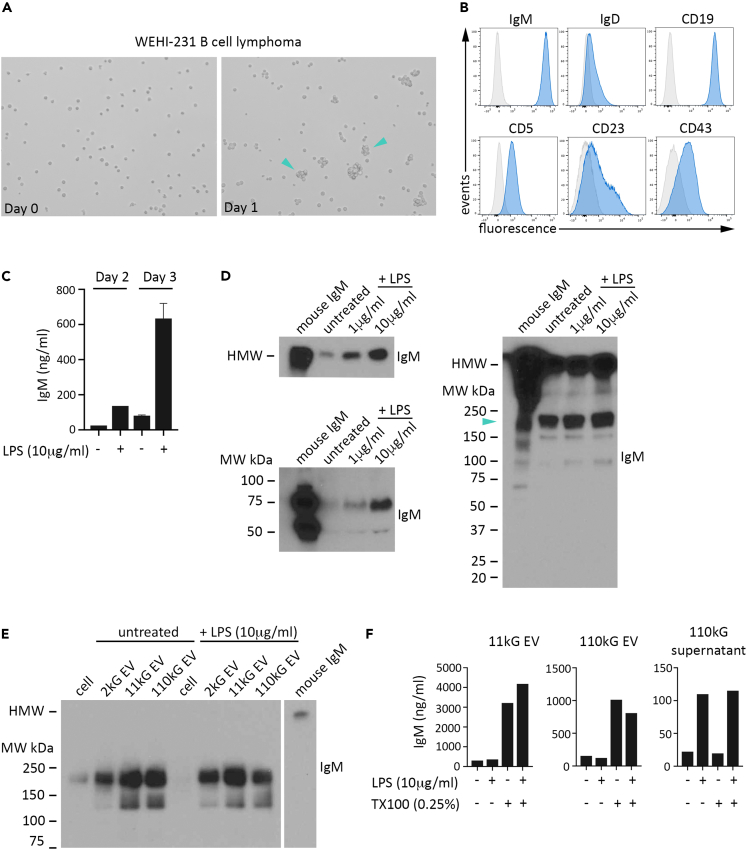
EVs, like cells, display exceptional diversity in molecular composition and physical size. Reports documenting immunoglobulin M (IgM) expression in B cell-derived EVs, albeit limited, have focused on small EVs, or exosomes, thus leaving a large void in our basic understanding of IgM content across different B cell EV populations. Differential ultracentrifugation (DUC) has persisted over decades as a primary EV isolation methodology, revealing that segregation by sedimentation velocity, or size, can reflect unique EV biogenesis pathways, biological cargo signatures, and functional profiles.61,62,63,64 We therefore utilized DUC to isolate EV populations and then synergized immunoblot, ELISA, and flow cytometry analyses to investigate both EV IgM expression and EV IgM topography. We initially capitalized on several characteristics inherent to the B cell lymphoma line WEHI-231, including an extensive literature record of LPS-regulated IgM secretion, evidence placing IgM in WEHI-231 exosomes, consistent cell doubling time and high cell viability, and relatively robust EV production.45,65 It has been documented that WEHI-231 variants exist, so we first evaluated our WEHI-231 line for established hallmark features.66 Microscopic analysis of unstimulated low-density cultures after one day proliferation showed the expected appearance of small cell clusters in suspension (Figure 1A). Flow cytometry analysis revealed a cell surface antigen profile of IgM+IgD−/lowCD19+CD5+CD23-/lowCD43-/low, in line with what has been reported (Figure 1B). ELISA analysis of conditioned media from LPS-treated cells showed the expected increase in secreted IgM (Figure 1C), and a concomitant dose-dependent loss in IgM cell surface expression, as determined by flow cytometry (Figure S1A, left graph). Cell viability was comparable for all conditions (right graph). Combined, these results indicated a phenotype and LPS response consistent with WEHI-231 cells as previously described.
Figure 1.
Multiple extracellular vesicle populations produced by WEHI-231 cells contain immunoglobulin M (IgM)
(A) WEHI-231 cells were imaged by bright field microscopy at the time of plating (day 0, left) and after 24 h (day 1, right) in complete media. Cell clusters, interspersed with single cells, are indicated with a turquoise arrowhead.
(B) WEHI-231 cells were evaluated by flow cytometry for expression of IgM, IgD, CD19, CD5, CD23, and CD43 (blue) compared to fluorescence minus one (FMO) stained control cells (gray). Data are representative of two independent experiments.
(C) IgM in 2000 × g supernatant (2kG supernatant) from untreated or LPS (10 μg/mL) treated WEHI-231 cells at day 2 (columns 1,2) and at day 3 (columns 3,4) was determined by anti-mouse IgM ELISA. Data are from one (day 2) and two (day 3) independent experiments. Data are represented as mean ± SEM.
(D) Conditioned media from untreated WEHI-231 cells, or WEHI-231 cells treated with 1 or 10 μg/mL LPS, was centrifuged at 2000 × g and IgM expression in the resulting supernatant (8 μL/well) was determined by SDS/PAGE and immunoblot. Presented are immunoblots for IgM in samples prepared with unreduced (left, upper) or reduced (left, lower) sample buffer. Right image displays a longer membrane exposure of the unreduced samples, with low molecular weight IgM indicated by turquoise arrowhead. Purified mouse IgM (25 ng) was run as a positive control.
(E) Lysate preparations of whole cells, and extracellular vesicle (EV) populations isolated by differential ultracentrifugation (2 kG, 11,000 × g (11 kG), 110,000 × g (110kG)), from untreated or LPS (10 μg/mL) treated WEHI-231 cells were evaluated for IgM by unreduced immunoblot. Total protein was 1 μg for all samples. Purified mouse IgM (3.1 ng) was run as a positive control in a non-contiguous lane and is presented at the far right in both images.
(F) IgM in 11 kG EV (left), 110 kG EV (center), and 110kG supernatant (right) was determined by ELISA under buffer only or buffer +0.25% TX100 conditions, as indicated.
Data in (D) and (E) are representative of at least two independent experiments. See also Figure S1.
IgM protein is translated as either one of two species, a monomeric form that constitutes in part the plasma membrane B cell receptor (BCR), or a version modified in amino acid sequence at the carboxy terminus that assembles primarily as five bound monomers (pentameric structure with J chain) and is secreted.67 Each species functions uniquely due to these differences in molecular structure and physical location, and thus distinguishing IgM monomer (denoted herein as Low Molecular Weight IgM (LMW IgM)) from IgM pentamer (denoted herein as High Molecular Weight IgM (HMW IgM)) in assayed experimental samples provides essential knowledge about a key characteristic that is problematic to ascertain by standard ELISA and flow cytometry analyses and is often unaddressed. We exploited the quality of dissimilar molecular weights and performed SDS/PAGE and immunoblot analysis under reducing and non-reducing conditions on equivalent volumes of conditioned media (CM) from untreated or LPS stimulated WEHI-231 cells (Figure S1B, total protein for unreduced samples). Using purified murine IgM as a comparative molecular weight control, we detected elevated IgM with LPS treatment, which was in accordance with the ELISA results above (Figure 1D; unreduced conditions upper, reduced conditions lower). However, in addition to HMW IgM (migration distance similar to purified IgM control; ∼980 kDa), we observed LMW IgM (turquoise arrowhead; migration distance similar to monomeric IgM; ∼180 kDa) upon longer membrane exposure of the unreduced samples (Figure 1D, right image). It has been established that IgM is secreted from the cell by the conventional secretory pathway as a high molecular species only, suggesting that the LMW IgM species we detected was EV IgM. Subsequent EV isolation from WEHI-231 CM by DUC and immunoblot analysis of equivalent protein from cell and EV preparations revealed that LMW IgM is greatly enriched in three populations of EVs (2000 × g (2 kG EV), 11000 × g (11 kG EV), 110000 × g (110 kG EV) we evaluated (Figure S1C, total protein; Figure 1E, IgM immunoblot; lanes 1–4 untreated, lanes 5–8 + LPS (10 μg/mL)). We did not observe a notable increase in LMW EV IgM under LPS conditions. Analysis by ELISA supported our immunoblot results (Figure 1F). When completed under standard buffer conditions, IgM detection in isolated 11kG and 110 kG EV was similar (left and center graphs, columns 1 and 2), whereas 110kG supernatant IgM, or secreted IgM, was greater in the LPS treated sample (right graph, columns 1 and 2). Most striking was the influence of detergent treatment (0.25% Triton X-100; TX100) prior to ELISA, an established methodology to solubilize the EV phospholipid membrane and thus expose sequestered, intravesicular molecules for interrogation.68,69 Under solubilized conditions, we observed a large increase in detectable IgM above standard conditions in both EV populations (columns 1 vs. 3 and 2 vs. 4, respectively), but not in the 110kG supernatant, providing additional support that we had in fact isolated detergent-sensitive secreted particles (EVs) of different sizes that harbor IgM in biological form different than that of secreted IgM.
Monomeric IgM is enriched in WEHI-231 EV, expressed on the EV surface, and sequestered in EVs
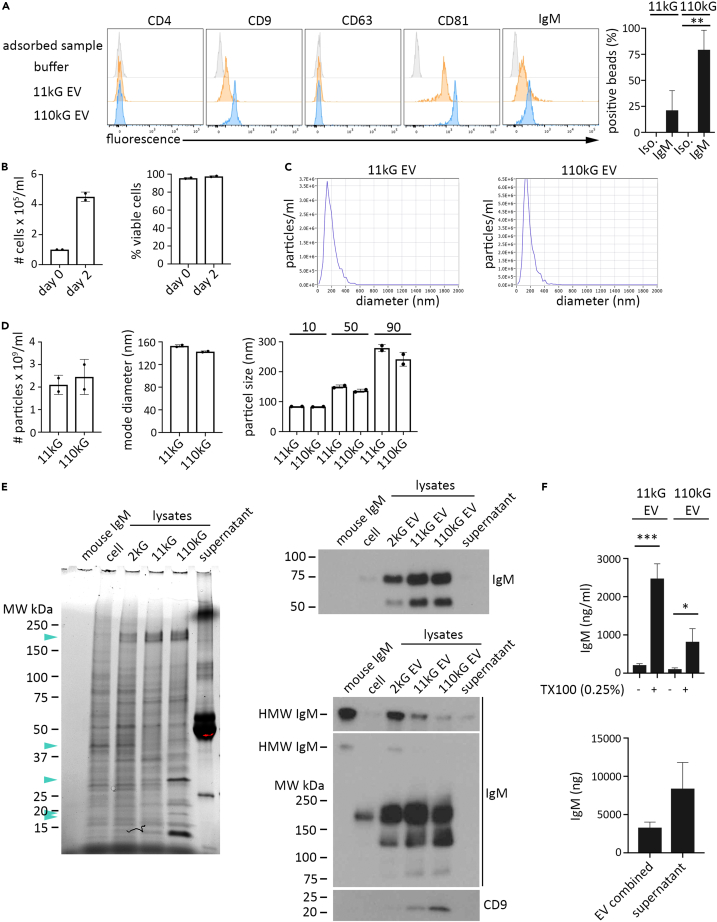
Although recent phenotyping advancements have refined EV classification, the search for definitive molecular EV population markers is ongoing. Proteins of the tetraspanin family, notably CD9, CD63, and CD81, persist as classical markers due to their enrichment in EV derived from a broad spectrum of cell lines and ex vivo tissue samples.37,70,71 Further, the evidence indicates that their relative expression can reflect divergent EV biogenesis pathways. We utilized these features to help validate our isolation techniques and provide a more thorough characterization of WEHI-231 EVs. First, we determined the tetraspanin profile of WEHI 231 cells and found robust surface expression of CD9 and CD81 compared to CD63 (Figures S2A and S2B). Following EV isolation and subsequent adsorption to microbeads, flow cytometry analysis revealed that both 11kG and 110 kG EV express surface CD9 and CD81, but not CD63, in addition to surface IgM (Figure 2A; Figures S3A and S3B). Nanoparticle tracking analysis of isolated EVs from viable WEHI 231 resulted in a histogram profile consistent with previous reports for large and small vesicles, with an average mode diameter of 152.8 nm and 142.7 nm, respectively (Figures 2B–2D). SDS/PAGE and immunoblot analysis of equivalent total protein from whole cell, 2kG, 11kG, and 110 kG EV lysate preparations showed not only discordant protein expression visible by eye (Figure 2E; left, turquoise arrowheads), but also EV enrichment of IgM and CD9 in relation to cells (Figure 2E; center, upper image, reduced conditions; center, lower image, unreduced conditions), thus providing support for our isolation methodology. An enrichment of the ER-associated protein calnexin and actin in WEHI 231 cell lysate preparations compared to WEHI 231 EV samples was observed as well (Figure S2C). LMW IgM was the dominant EV IgM species, as the HMW IgM band was visualized only upon a long membrane exposure (unreduced conditions, top). We then substantiated our preliminary results (see Figure 1F) and evaluated by ELISA the proportional contribution of EV IgM and secreted IgM to the post-2kG WEHI-231 secretome (Figure 2F). Not only was IgM significantly increased in 11kG and 110 kG EV with TX100 solubilization (top graph; compare bars 1 and 2, 3 and 4), with ∼2.4 more IgM contained in 11kG compared to 110 kG EV, but total EV IgM constituted a substantial amount of total extracellular IgM, as is evident by comparison with IgM levels in the supernatant (bottom graph). Effective lysis by TX100 was confirmed by sedimentation analysis and comparison to PBS and RIPA lysis buffer treated EV preparations (Figure S3D). These results in total indicate that IgM released from WEHI-231 cells exists in two predominant forms, a pentameric secreted species and a monomeric EV-associated species. Further, the data suggest that a large portion of 11kG and 110 kG EV IgM is encapsulated or membrane-sequestered, as detergent-mediated solubilization resulted in a large increase in detectable IgM.
Figure 2.
Monomeric IgM is enriched in WEHI-231 EV and is detected at a greater level following detergent solubilization
(A) Equivalent amounts of 11kG (upper row) and 110kG (lower row) EVs isolated by differential ultracentrifugation were titrated and adsorbed onto aldehyde sulfate microbeads, stained for surface expression of IgM (or isotype control), canonical EV marker proteins, and negative control protein (CD4), and analyzed by flow cytometry. Beads incubated with buffer alone were included as a control. Histogram overlays display the fluorescence for each antigen (CD4, CD9, CD63, CD81, or IgM). The graph indicates the percentage of beads positive for IgM compared to beads stained with isotype control antibody. Data are representative of at least three independent experiments.
(B) WEHI 231 cells (16 × 10 cm plates; 0.1 × 106 cells/mL media) were cultured for two days, after which EV were isolated by DUC, resuspended in 1.1 mL PBS and serially titrated at 1:10, and subsequently analyzed by nanoparticle tracking analysis. Displayed are the starting and ending cell number (left) and cell viability (right).
(C) Histogram plots for particle diameters of 11kG (left) and 110kG (right) samples are presented.
(D) Displayed are the particle concentration (left), mode diameter (center), and percentile distribution by particle size (right) of 11kG and 110kG samples. N = 2 independent experiments.
(E) Whole cell and isolated EV (2kG, 11kG, 110kG) lysates (2.3 μg protein/well) were evaluated for IgM and CD9 expression by SDS/PAGE and immunoblot. Total protein (left) and immunoblot detection of IgM and CD9 under reduced (right, top) or unreduced (right, bottom) conditions are presented. A longer (upper) and shorter (lower) exposure are presented for the unreduced immunoblot. Turquoise arrowheads (total protein gel) indicate visible discrepancies in expression of protein species across the four lysate preparations. Purified IgM (2.5 ng) and 110kG supernatant (10.5 μL) were run as control samples. N = 2 independent experiments.
(F) The amount of IgM in 11kG and 110 kG EV populations under buffer only (no TX100) and buffer +0.25% TX100 conditions (top graph) and the total IgM in 11kG plus 110 kG EV populations combined as compared to 110kG supernatant (bottom graph), determined by anti-mouse IgM ELISA, are presented.
Data are representative of three independent experiments. ANOVA and Tukey’s post-hoc analysis was applied to data presented in (A) and (F) prior to direct comparison. ∗ = p < 0.05; ∗∗ = p < 0.005; ∗∗∗ = p < 0.001. Data are represented as mean ± SEM. See also Figures S2 and S3.
WEHI-231 EV populations isolated by gradient fractionation display coordinated expression of IgM and CD81
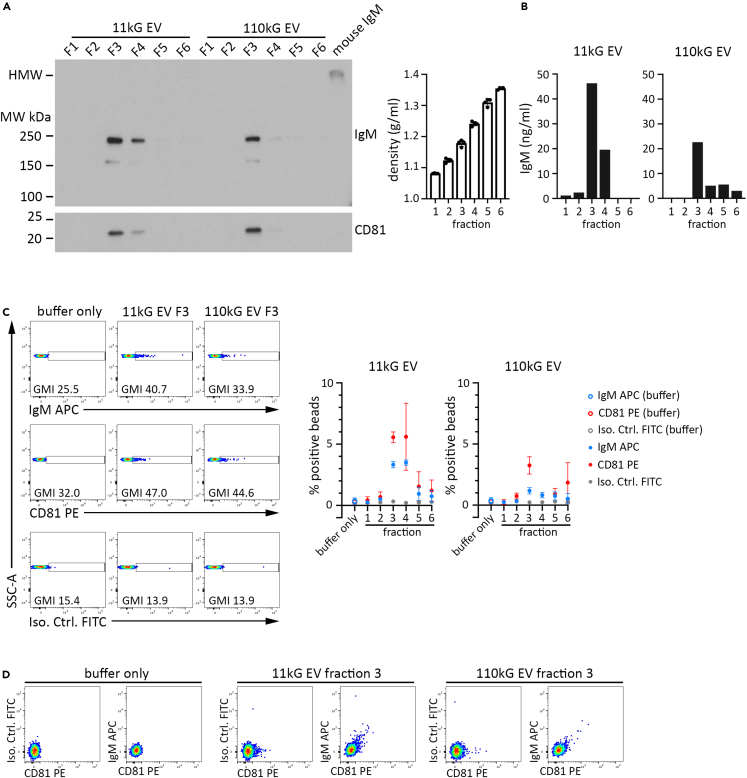
Density gradient centrifugation provides an additional level of EV isolation stringency based on vesicle density. We subjected 11 kG and 110 kG EV preparations to sucrose gradient centrifugation and resuspended each isolated fraction in an equivalent volume of buffer. An equivalent proportion of each fraction was then evaluated by immunoblot, ELISA, and flow cytometry for IgM and CD81 protein expression. Immunoblot analysis (Figure 3A, left) of fraction 1 (F1) least dense to fraction 6 (F6) most dense (Figure 3A, right) showed that CD81 was highly enriched in fraction 3 (F3) for both 11 kG and 110 kG EV samples, indicating that EVs separated primarily to this layer. Importantly, monomeric IgM tracked with CD81, indicating not only that IgM was associated with EVs but also that the parent EV samples isolated by differential centrifugation were largely devoid of non-EV or contaminating IgM. These data were corroborated by ELISA analysis, where IgM was concentrated in F3 (Figure 3B; left graph, 11 kG EV and right graph, 110 kG EV). We then adsorbed each fraction onto microbeads, stained for IgM and CD81, and analyzed the samples by flow cytometry. There was sufficient IgM and CD81 surface expression on the F3 EV populations to observe a distinct PE+ bead population, as compared to beads incubated in buffer only (Figure 3C; IgM top row, CD81 center, isotype control lower; buffer only left column, 11kG F3 center, 110kG F3 right). A comparison of bead fluorescence for all fractions demonstrated that F3, and F4 to a degree, had elevated IgM and CD81 surface expression above the other fractions, supporting the immunoblot and ELISA results (Figure 3C, right). Biparametric analysis of fraction 3 samples indicated a positive correlation for IgM and CD81, providing additional support that 11 kG and 110 kG EVs express surface IgM (Figure 3D).
Figure 3.
WEHI-231 11kG and 110 kG EV populations fractionated by sucrose gradient centrifugation reveal correlated expression of IgM and CD81
EV were first isolated by differential ultracentrifugation and then subjected to density gradient fractionation by overlay of sucrose/PBS layers with decreasing density. Fractions were collected (F1 least dense to F6 most dense) and washed with buffer. An equivalent proportion of each resuspended fraction was subsequently utilized for downstream analyses.
(A) Density gradient fractions of EV were subjected to PAGE and immunoblotted for IgM and CD81, as indicated. Purified mouse IgM was run as a positive control. The measured density (g/mL) of each fraction are shown (right graph). N = 2 independent experiments.
(B) Density gradient fractions of EV were tested by anti-mouse IgM ELISA under buffer +0.25% TX100 conditions for 11kG (left) and 110kG (right) EV populations.
(C) Density gradient fractions were adsorbed onto aldehyde sulfate microbeads, stained for IgM and CD81, and analyzed by flow cytometry. Dot plots for IgM (top), CD81 (middle), and isotype control (bottom) of fraction 3 are shown, with gates indicating the positive bead populations. The graphs (bottom) show the percentage of positive beads for all 11 kG EV (left) and 110 kG EV (right) fractions.
(D) Biparametric dot plots of buffer (left), fraction 3 11 kG EV (center), and fraction 3 110 kG EV (right) samples displaying the expression of isotype control/CD81 or IgM/CD81 are presented.
Data presented in (A)–(D) are representative of at least two independent experiments. Data are represented as mean ± SEM.
Mouse splenocytes from WT and μsKO mice release 11 kG and 110 kG EV that contain IgM
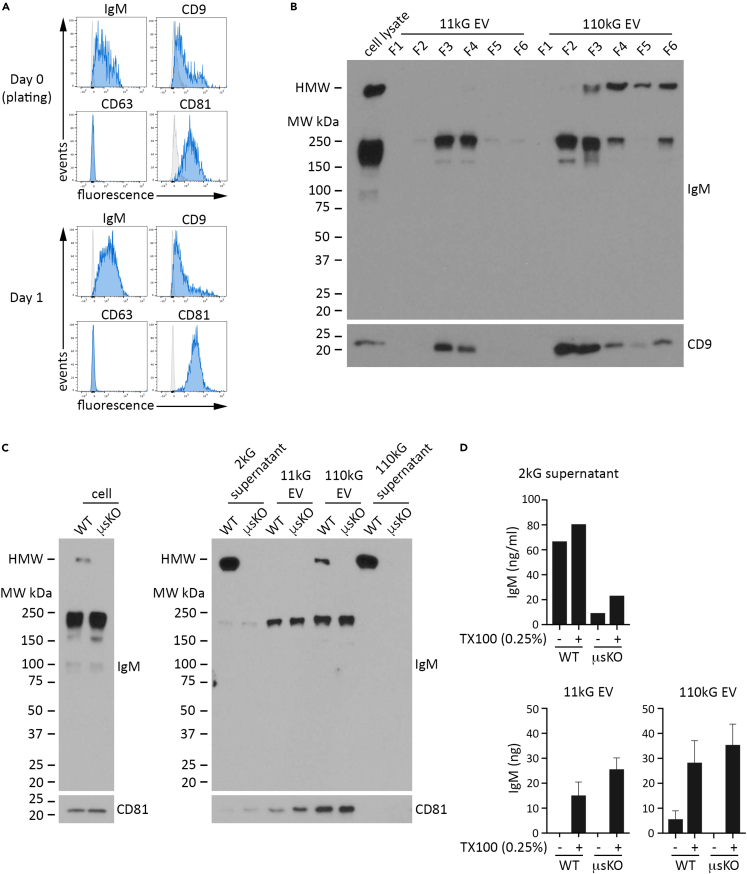
Our results from WEHI-231 B lymphoma cells demonstrated that we had in place a system to isolate EVs, by vesicle size (differential centrifugation) and vesicle density (gradient centrifugation), and measure protein expression using several methodologies. We next applied this approach to evaluate IgM content in EVs produced by primary cells, focusing initially on isolated splenic B cells (immunocapture magnetic bead negative selection) from WT mice. Flow cytometry analysis indicated that ex vivo unstimulated B cells at the time of harvest and after one day in culture express the tetraspanins CD9 and CD81, but not CD63, a profile similar to what we observed for EVs from WEHI-231 cells (Figure 4A; upper histograms, day 0 and lower histograms, day 1). Immunoblot analysis of gradient fractionated 11kG and 110 kG EV samples isolated from day one conditioned media (CM) showed a strong correlation of CD9 and LMW IgM, indicating that IgM is contained in both smaller and larger EVs produced by primary B cells (Figure 4B).
Figure 4.
Splenic B cells from WT and μsKO mice release 11kG and 110 kG EV that contain low molecular weight IgM
WT mouse splenic B cells were isolated by immunocapture magnetic bead negative selection and cultured unstimulated for 24 h, after which conditioned media was harvested for EV isolation and analysis.
(A) Cell surface expression of IgM, CD9, CD63, and CD81 was determined by flow cytometry at the time of plating (day 0) and at harvest (day 1). Histogram overlays of cells stained for antigen (blue) and corresponding FMO stained cells (gray) are presented.
(B) EV (11kG and 110kG) in day 1 conditioned media were isolated by differential ultracentrifugation (DUC) and then fractionated by sucrose gradient centrifugation. Following buffer wash, each fraction was resuspended in an equivalent volume and then resolved by SDS/PAGE for immunoblot detection of IgM and CD9, as indicated. A sample of day 1 WT splenic B cell lysate (3.75 μg protein) was run as a positive control. Data are representative of at least two independent experiments.
(C) WT and μsKO splenic B cells were isolated by immunocapture magnetic bead negative selection and cultured unstimulated for 24 h, after which conditioned media was collected and cell lysates were prepared. EV were isolated from conditioned media by DUC and resuspended in an equivalent volume of buffer. Equivalent protein (cell lysates; 2 μg) or volume (2kG supernatant, 11 kG EV, 110 kG EV, post-110kG supernatant) for WT and μsKO samples was resolved by SDS/PAGE under unreduced conditions, followed by immunoblot detection of IgM and CD81.
(D) The amount of IgM, as determined by anti-mouse IgM ELISA under buffer and buffer +0.25% TX100 conditions, is shown for 2kG supernatant (top), 11 kG EV (lower left), and 110 kG EV (lower right).
Data in (C) and (D) are representative of two independent experiments. Data are represented as mean ± SEM. See also Figure S4.
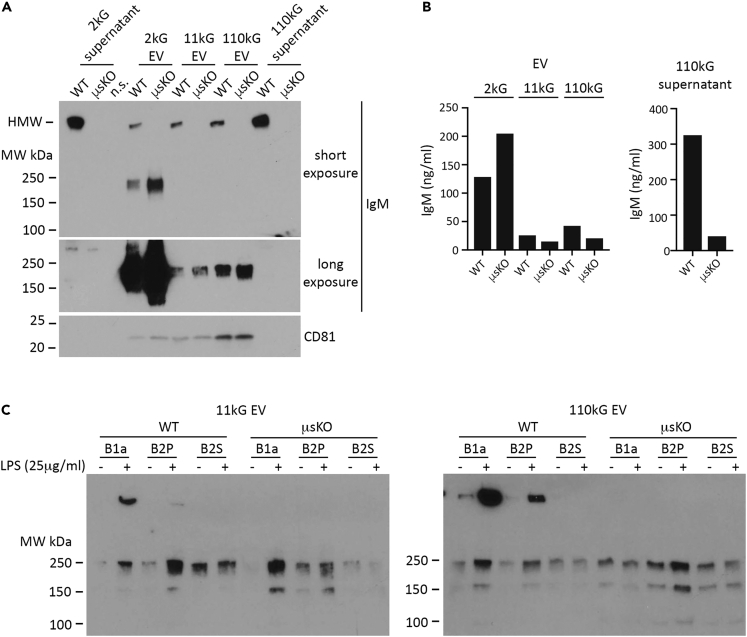
Although our data strongly suggested that EV IgM expression is not dependent on traditional IgM secretion, an alternative explanation for the presence of EV LMW IgM could be that secreted IgM pentamer undergoes molecular degradation and association with EVs. We addressed this through direct comparison of B cell-derived EV from C57BL/6 WT and μsKO mice, the latter model genetically modified to translate IgM monomer only, and thus does not secrete IgM pentamer.72 Sorted splenic B cells from WT and μsKO mice were cultured unstimulated for one day, followed by CM harvest and isolation of the 11kG and 110 kG EV populations. Immunoblot analysis showed expression of monomer LMW IgM in both WT and μsKO cells, but pentameric HMW IgM only in the WT (Figure 4C; left image). In the 2kG supernatant, we again observed HMW IgM in WT only, however there was also a much less pronounced but distinct LMW IgM band of similar intensity in WT and μsKO samples (Figure 4C; right image, lanes 1 and 2). Following isolation of the 11kG and 110 kG EV populations, this band became dominant by comparison in both WT and μsKO EVs (Figure 4C; right image, lanes 3–6), indicating not only the expected enrichment of monomeric IgM in EVs, but also demonstrating that EV IgM can exist independently of secreted IgM. Additionally, CD81 expression tracked well with the EV samples and monomeric IgM (Figure 4C; compare lanes 3–6 (EV samples) to lanes 7 and 8 (110kG supernatant)). Our ELISA results supported these data, as we detected substantially more IgM in WT 2kG supernatant, with solubilization increasing detection in both samples, especially for μsKO, presumably due to IgM contained in EVs (Figure 4D, top). This was reflected upon isolation of the EV populations, where the most appreciable amount of IgM in both WT and μsKO EV was observed under solubilized conditions (Figure 4D, bottom). WT and μsKO 110 kG EV preparations were then subjected to density gradient fractionation and immunoblot analysis (Figure S4). We observed that the vast majority of IgM had floated to fractions 2 and 3 for both WT and μsKO and was coordinately expressed with CD81 for both, thus providing additional support that EV IgM is produced from both WT and μsKO B cells.
Peritoneal cavity B cell populations from WT and μsKO mice produce EVs that contain IgM
Our analyses to this point had focused on EVs produced by splenic B cells, however, it is well documented that the mouse peritoneal cavity, an area surrounding the abdominal organs, houses multiple immune cell populations, including B cells, and is intimately involved in immunoregulation. Although most commonly studied in mouse gastric cancer models and implicated in metastatic dissemination, peritoneal cavity extracellular vesicle populations to our knowledge have not been well characterized and remain largely unexplored.73,74 We therefore wanted to determine if peritoneal cavity EVs contain IgM. To address this, murine peritoneal cavity fluid from healthy age-matched WT and μsKO mice was subjected to the same differential ultracentrifugation protocol utilized for WEHI-231 and splenic B cell conditioned media and then evaluated by immunoblot and ELISA for IgM. Samples from each cohort were processed identically to permit a direct comparison. Analysis of 2kG supernatant showed that HMW IgM was readily detected in peritoneal fluid from WT mice but not μsKO mice, and that it was in excess compared to LMW IgM (Figure 5A, lanes 1 and 2; upper image, short exposure; lower image, long exposure). However, upon EV isolation we observed LMW IgM in both 11kG and 110 kG EV from WT and μsKO mice, and relatively robust expression in 2 kG EV (lanes 4–9), indicating that at least a portion of the isolated EV populations were in fact produced by peritoneal cavity B cells. We also detected HMW IgM in all three WT EV samples, which was, in comparison to LMW IgM, proportionally much greater here than what we observed in the EVs isolated from the conditioned media of cultured B cells (WEHI-231 cells and splenic B cells). This phenomenon has been reported in the blood, where HMW IgM was shown to occur in association with reticulocyte exosomes.75 We observed a similar expression profile in 110 kG EV isolated from mouse blood plasma and then gradient fractionated, where although we could detect LMW IgM in EVs, expression was comparatively much lower than HMW IgM (Figure S5). As expected in peritoneal fluid, the WT 110kG supernatant contained the major proportion of HMW IgM and lacked any detectable amount of the LMW species (Figure 5A; lanes 10 and 11). CD81 expression tracked with EVs and not supernatant, indicating effective EV isolation (Figure 5A; lower). IgM quantitation by ELISA supported our immunoblot results, where we detected IgM in all EV samples, with 2 kG EVs showing the highest IgM levels, and IgM in large excess in WT 110kG supernatant (Figure 5B; EV left graph, supernatant right graph). In total, using mice that lack the ability to express HMW IgM, we demonstrated that EV IgM is a naturally occurring form of extracellular IgM that is produced in healthy mice without dependence on expression or secretion of HMW IgM, and that EV populations of different sizes contain LMW IgM.
Figure 5.
EV isolated from WT and μsKO mouse peritoneal cavity fluid and peritoneal B cell cultures contain low molecular weight IgM
(A) Peritoneal cavity washout fluid was obtained from WT and μsKO mice and subjected to DUC to isolate 2kG, 11kG, and 110 kG EV populations, which were then resuspended in an equivalent volume of buffer, resolved by SDS/PAGE under unreduced conditions, and immunoblotted for IgM and CD81, along with 2kG and 110kG supernatant samples. Data are representative of two independent experiments.
(B) IgM expression in the peritoneal cavity samples described in (A) was determined by anti-mouse IgM ELISA under buffer +0.25% TX100 conditions. Results are shown for the EV samples (left) and 110kG supernatant (right).
(C) Peritoneal B-1a cells (CD5+B220loCD43+CD23−CD19+), peritoneal B2 (B2P) cells (CD5-B220highCD43−CD23+CD19+), and splenic B2 (B2 S) cells (CD5-B220highCD43−CD23highCD21lo/−CD19+) were sorted from WT and μsKO, as detailed in materials and methods. The sorted B-1a, B2P, and B2 S cells were cultured in media alone or media + LPS (25 μg/mL) for 24-h, after which conditioned media was harvested and the 11kG and 110 kG EV populations were subsequently isolated by differential ultracentrifugation. EV samples (11 kG EV, left and 110 kG EV, right) were resuspended in an equivalent volume of buffer, resolved by SDS/PAGE under non-reducing conditions, and immunoblotted for IgM.
Data are representative of at least two independent experiments. See also Figures S5 and S6.
The peritoneal cavity contains two primary B cell populations that express IgM, innate-type B1 cells (natural antibody) and traditional B2 lineage cells (adaptive antibody). Accurate cell-of-origin determination in tissue-derived EV preparations is problematic due to cell heterogeneity, therefore, we FACS sorted B1a (the dominant peritoneal B1 sub-type) and B2 peritoneal (B2P) cells from WT and μsKO mice and determined if these isolated cells express EV IgM (Figures S6A and S6B). Splenic B2 cells (B2 S) were FACS sorted as well for comparison (Figure S6C). Prior reports have shown that peritoneal B cells rapidly respond to LPS stimulation and increase IgM secretion, so we cultured the three populations in media alone or with LPS (25 μg/mL) for one day followed by EV isolation from the conditioned media. Sample preparation was identical for all samples to permit direct comparison. Immunoblots revealed that in response to LPS, both peritoneal B cell populations from WT and μsKO mice increased LMW IgM expression in 11 kG EV (Figure 5C; left image). The same trend held for 110 kG EV, except for those isolated from μsKO B1a cells (Figure 5C; right image). We did not detect any appreciable increase in IgM following LPS treatment in either EV population from splenic B2 cells, an expected result given the short exposure to LPS. In summary, our data indicate that EVs produced by peritoneal B1a and B2 cells contain LMW IgM and that expression is not dependent on secreted IgM.
IgM contained in B cell-derived EVs can bind antigen
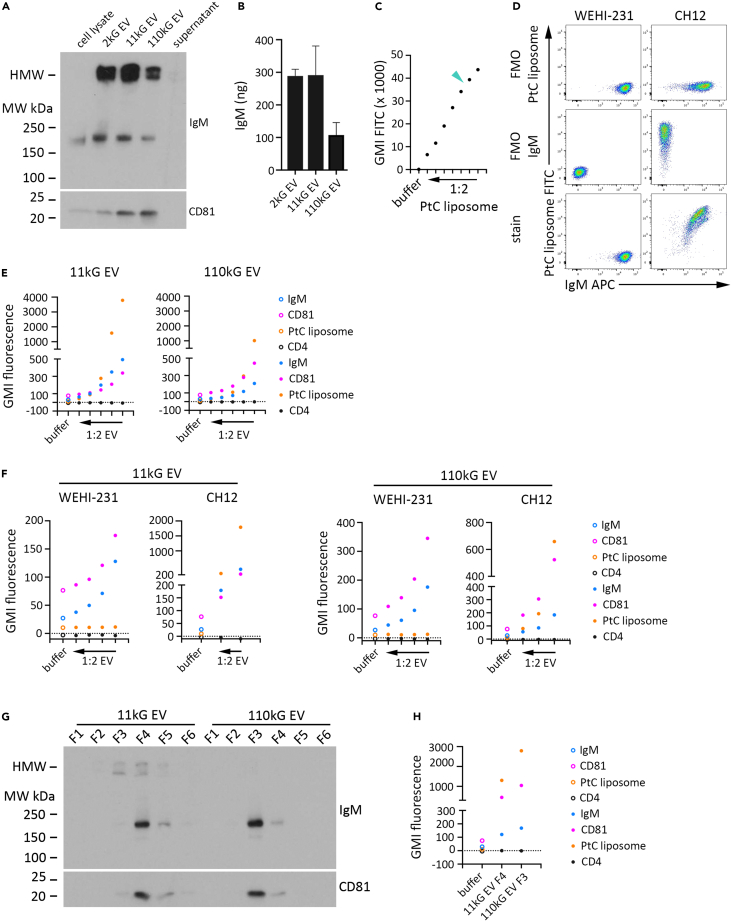
Antigen binding is a critical step during antibody-mediated immune responses. Not only must the two molecules be in close proximity for this interaction to occur, but antibody orientation and molecular integrity must be maintained and sufficient. Although we had revealed the presence of IgM in EVs produced by the WEHI-231 cell line and by mouse primary B cells, and in EVs isolated directly from the peritoneal cavity, a significant question remained as to whether EV IgMs can bind antigen. To address this, we utilized the CH12 murine B lymphoma cell line, a model that expresses anti-phosphatidylcholine (PtC) antibodies that are IgM isotype.76,77,78 We first isolated EVs from CH12 conditioned media by DUC and then evaluated IgM and CD81 expression in cell lysates, EV preparations, and 110kG supernatant by immunoblot (Figure 6A). Similar to WEHI-231 cells (see Figure 2B), LMW IgM was detected in CH12 cell lysates (lane 1) and all three EV populations (lanes 2–4). The presence of CD81 in the EV preparations, and lack thereof in the 110kG supernatant (lane 5), indicated a high level of EV enrichment. We then analyzed a proportionally equivalent amount of sample by ELISA under solubilization conditions (0.25% TX100) and observed that the data tracked well with immunoblot, as the 2kG and 11 kG EVs contained approximately two times more IgM than the 110 kG EV (Figure 6B). These data demonstrated that multiple EV populations produced by CH12 cells contain IgM, indicating we had identified a suitable model to test antigen specific EV IgM binding.
Figure 6.
IgM contained in B cell-derived EV can bind antigen
CH12 cells were cultured in media alone for two days, after which the conditioned media was subjected to differential ultracentrifugation (DUC) to isolate EV. The EV samples (2kG, 11kG, and 110kG) were resuspended in an equivalent volume of buffer for subsequent analysis by immunoblot and ELISA.
(A) An equivalent proportion of each sample was resolved by SDS/PAGE under unreduced conditions, and immunoblotted for IgM and CD81. CH12 cell lysate (lane 1; 7.5 μg) and 110kG supernatant (lane 5), were run as controls.
(B) The amount of IgM in each EV sample under buffer +0.25% TX100 conditions was determined by anti-mouse IgM ELISA. For (A) and (B), data are representative of two independent experiments.
(C) CH12 cells were incubated with PtC liposomes for 30 min on ice at 1:2000 stock and at sequential 1:2 dilutions thereof, after which FITC fluorescence was determined by flow cytometry and GMI determined. The extreme left data point is the GMI FITC for CH12 cells incubated in buffer only. The turquoise arrowhead indicates the reagent concentration (1:5000 stock) utilized for subsequent experiments.
(D) WEHI 231 and CH12 cells were stained with IgM APC and PtC liposome FITC reagent for flow cytometry analysis. Dot plots indicate FMO PtC liposome (first row), FMO IgM APC (second row), and complete stain (third row) fluorescence profiles of single, viable WEHI-231 (left column) and CH12 (right column) cells.
(E) EV samples (11kG and 110kG) were isolated by DUC from CH12 conditioned media, resuspended in an equivalent volume of buffer, sequentially titrated 1:2, incubated with an identical volume of aldehyde sulfate microbeads, and subsequently stained with a fluorescent panel of IgM APC, CD81 BV241, CD4 PE/Cy5 and PtC liposome FITC reagent for flow cytometry analysis. GMI for each fluorophore is shown for the most concentrated EV preparation (extreme right data point) and each titrated sample (progressing left). GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point. Data are representative of two independent experiments.
(F) EV (11kG and 110kG) were isolated from WEHI-231 and CH12 conditioned media by differential ultracentrifugation and then titrated 1:2, adsorbed onto an equivalent volume of aldehyde sulfate microbeads, and then stained with the reagent panel described above for flow cytometry analysis (11 kG EV, left graphs; 110kG right graphs). GMI for each fluorophore is shown for the most concentrated EV preparation (extreme right data point) and each titrated sample (progressing left). GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point.
(G) EV (11kG and 110kG) were isolated from CH12 conditioned media by DUC and subjected to sucrose gradient fractionation. Equivalent volumes of resultant fractions were subjected to SDS/PAGE under non-reducing conditions and immunoblotted for IgM and CD81. Data are representative of two independent experiments.
(H) Fraction 4 from 11 kG EV and fraction 3 from 110 kG EV were analyzed by flow cytometry following adsorption to aldehyde sulfate microbeads and staining with the reagent panel described above. GMI fluorescence for each fluorophore is shown. GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point. Data are represented as mean ± SEM.
PtC-constituted liposomes formulated with encapsulated fluorescein is a commonly applied reagent to identify B cell populations that express anti-PtC antibodies. We first verified anti-PtC antibody expression using CH12 and WEHI-231 cells, the later serving as a control that expresses surface IgM but of unknown (non-anti-PtC) antigen specificity. Incubation of titrated PtC liposomes with CH12 cells indicated a suitable reagent dilution at 1:5000, which was utilized for all subsequent experiments (Figure 6C; turquoise arrowhead). Flow cytometry analysis of WEHI-231 and CH12 cell lines demonstrated that although both lines express surface IgM, only CH12 cells bind PtC liposomes, indicating both expression of intact, functional IgM by CH12 cells and absence of non-specific liposome adsorption (Figure 6D; WEHI-231 left column, CH12 right column; FMO rows one and two, complete stain row three). We then isolated CH12 11kG and 110 kG EV by DUC and adsorbed titrated EV preparations to beads, followed by flow cytometry analysis for surface IgM, CD81, CD4 expression, and PtC liposome binding. We observed correlated fluorescence intensity for IgM, PtC liposomes, and CD81 in both EV populations across the titrated samples, demonstrating association of the three elements, along with the expected absence of CD4 detection, which is not expressed by B cells (Figure 6E; left 11 kG EV, right 110 kG EV). These data indicated that, like CH12 cells, EVs produced by CH12 cells bind PtC. We then completed a similar experiment by directly comparing titrated 11kG and 110 kG EV from WEHI-231 and CH12 cells, and again observed association of IgM, CD81, and PtC in CH12 EV, but only IgM and CD81 in WEHI-231 EV, which recapitulated the cell phenotypes and further supported that PtC liposome binding was specific (Figure 6F; left 11 kG EV, right 110 kG EV). Sucrose gradient fractionated 11kG and 110 kG EV populations showed overlapping IgM and CD81 expression and enrichment in 11kG fraction 4 (F4) and 110kG fraction 3 (F3) (Figure 6G; 11kG lanes 1–6 and 110kG lanes 7–12). Bead adsorption and subsequent flow cytometry analysis of these fractions for IgM, CD81, and PtC showed that all three are expressed together in 11 kG EV fraction F4 and 110 kG EV fraction F3, providing additional evidence that PtC binding was through IgM monomer present on the surface of CH12 EVs (Figure 6H; column 1 buffer alone, column 2 11kG fraction 4 (F4), column 3 110kG fraction 3 (F3)).
IgM contained in B cell-derived EVs can accumulate intracellularly in secondary cells
Characterization of EVs as secreted bioactive agents that can interact with secondary cells and impart functional consequences is extensive. For B cell-derived EVs, examples of this relationship include the early discovery that MHCII-presented antigen on the EV surface could be recognized by T cells, to more recent results showing the transfer of IgM from one B cell to the surface of another.38,45 Given that we had identified IgM as a component of different size EV populations from an array of B cell lines, primary B cells, and tissues enriched in B cell populations, we sought to determine whether IgM present in EVs could persist intact extracellularly and be acquired by adjacent cells. Our initial experiments exploited fundamental differences between the IgM-positive mouse WEHI-231 B cell lymphoma and IgM-negative adherent human HeLa cervical cancer cell line as EV donor and EV acceptor cell, respectively. We further utilized complementary methodologies of immunoblot and confocal microscopy to assess IgM transfer and subcellular location following transfer. A short transfer time course was first conducted using the fluorescently conjugated lectin wheat germ agglutinin Alexa Fluor 488 (WGA-AF488) to non-specifically label membrane peptidoglycans in WEHI-231 EV preparations prior to application (11kG and 110kG) (Figure S7A; left to right, 30 min to 4 h). Following EV isolation and labeling, we observed fluorescence in HeLa monolayers as early as 1-h post-EV application. The WGA-AF488 signal coordinated predominantly with cells (defined by actin) and not tissue culture substrate, and the signal appeared to be non-randomly distributed throughout the cell, providing evidence against a simple blanketing effect by settling EVs. This was much more apparent at 24 h, as the signal generated following transfer of either 11kG or 110 kG EV showed strong peri-nuclear orientation (Figure S7B; left 11 kG EV, center 110 kG EV, right vehicle only).
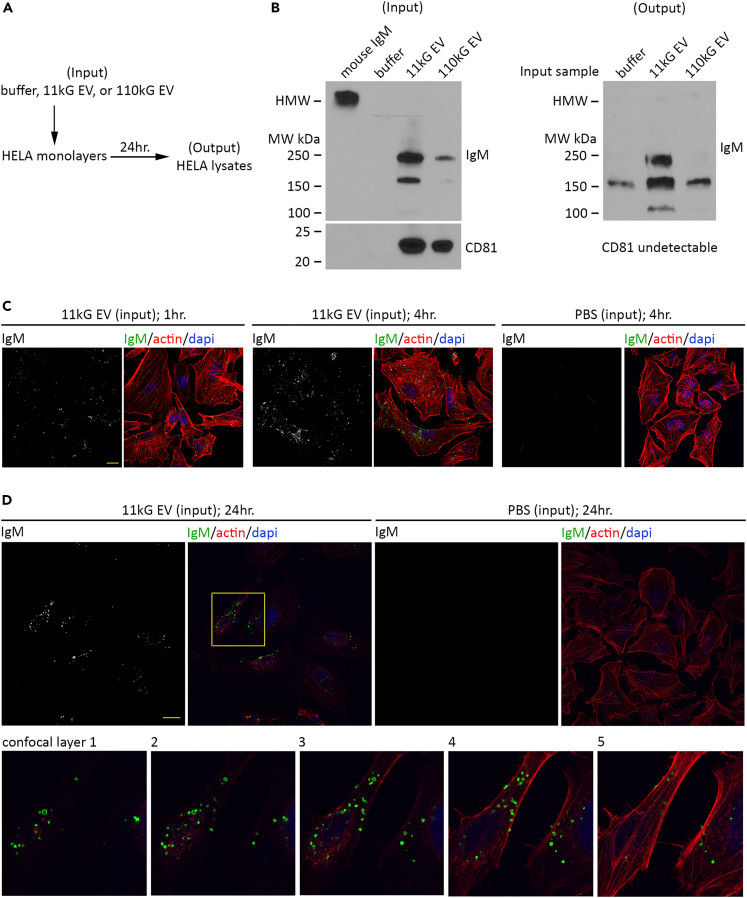
Although these data suggested B cell EV transfer to HeLa cells and cellular uptake, they did not specifically indicate IgM transfer. To address this, we performed immunoblot detection of IgM on HeLa whole cell lysates 24 h post-application of 11kG or 110kG WEHI-231 EVs (Figure 7A; schematic). Prior to resuspension in lysis buffer, the adherent cells were washed, trypsinized, pelleted, washed, and pelleted with the intent to remove loosely associated, unbound EVs. While we could detect IgM and CD81 in both EV preparations prior to transfer (Figure 7B; left immunoblot (Input), lanes 3 and 4), a distinct band indicating LMW IgM was present only in HeLa lysates that had been treated with 11 kG EV (right immunoblot (Output), lane 2). We were unable to detect CD81. We subsequently focused on 11 kG EV and evaluated IgM presence and sub-cellular localization by anti-IgM immunofluorescence staining and confocal microscopy. Similar to the indirect method presented above using WGA-AF488, we observed distinct fluorescence in HeLa monolayers at one and 4 h after 11 kG EV transfer (Figure 7C; left 1 h, center 4 h, right buffer only 4 h; Figures S8A and S8B). IgM appeared to be associated with cells, but not uniformly distributed throughout the cell, and additional evaluation of confocal microscopic sections at 24 h post-transfer indicated co-detection of actin and IgM, providing support that IgM was intracellular (Figure 7D, top row; Figures S8C and S8D). At higher magnification, a punctate, peri-nuclear IgM staining pattern was clearly visible across consecutive layers, some of which also contained a substantial level of actin (Figure 7D, bottom row 1–5; consecutive high magnification sections of yellow-outlined region in top row). Combined with the immunoblot results, these data indicate that not only do B cell EV associate with secondary cells, but that IgM contained in EVs is incorporated within the target cell. We then performed immunoblot and confocal microscopy analyses following transfer of EVs isolated from the S9.6 hybridoma cell line (monoclonal IgG2a isotype) and observed similar results, suggesting that cellular uptake of EV immunoglobulin can occur for multiple immunoglobulin isotypes (Figures S9A–S9C).
Figure 7.
IgM contained in B cell-derived EV can accumulate intracellularly in secondary cells
EV (11kG and 110kG) were isolated from WEHI-231 cell culture supernatant and subsequently applied to HeLa cell monolayers, followed by HeLa cell lysate preparation and immunoblot analysis or immunofluorescent confocal microscopy of intact monolayers.
(A) Schematic showing the experimental procedure, and designation of buffer, 11 kG EV, or 110kG (EV) as (input) and HeLa lysates as (output).
(B) Immunoblot for IgM and CD81 in isolated EV prior to application (left, (input)) and in HeLa lysate preparations generated 24 h post-EV application (right, (output)). Mouse IgM was run as a positive control. Data are representative of two independent experiments.
(C) Representative confocal microscopy images of HeLa monolayers 1 h (left set) and 4 h (center set) after application of 11 kG EV. Images show IgM alone (anti-mouse IgM AF488), and pseudocolored IgM (green) with actin (phalloidin rhodamine, red) and nuclei (dapi, blue). HeLa monolayers 4 h after application of vehicle (PBS) (right set) that were subjected to the identical staining reagents utilized above are shown.
(D) Representative microscopy image of one confocal layer (0.75 μm) 24 h post-application of 11 kG EV (left set). A confocal microscopy image of HeLa treated with buffer only, that were subjected to the identical staining reagents utilized above, is presented (right set). A region of interest (yellow box) from five sequential layers of HeLa treated with the 11 kG EV was magnified and is presented in the lower panel (1–5).
Data in (C) and (D) are representative of two independent experiments. Scale bar indicates 20 μm. See also Figures S7–S9.
Discussion
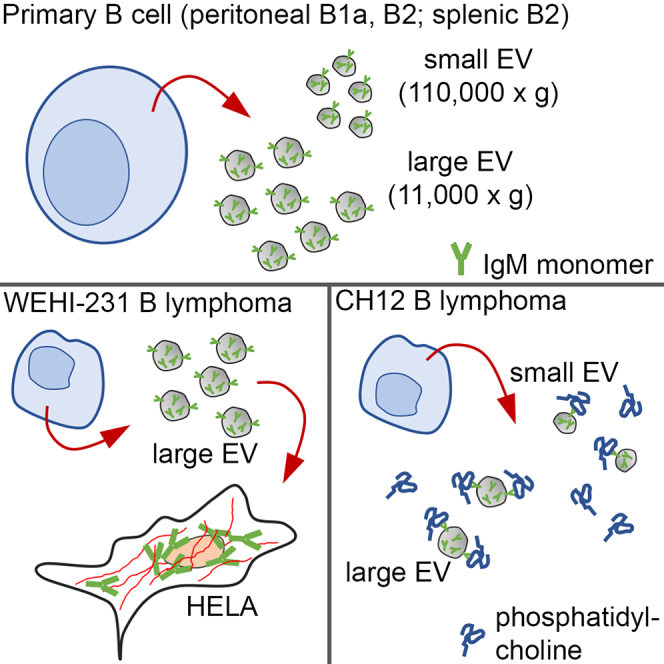
In this report, we provide evidence that the representation, and biological presentation, of extracellular immunoglobulin M extends beyond the secreted pentameric species. Using multiple cell lines (WEHI-231, CH12), sorted mouse primary B cell populations (peritoneal B1, B2; splenic total B cells; splenic B2), and mouse tissues (peritoneal cavity fluid, blood plasma), we found that IgM exists extracellularly as a low molecular weight species associated with EVs. IgM was present in large and small EV populations, isolated by both differential centrifugation and density gradient centrifugation, and this EV IgM aligned with EV-associated tetraspanins CD9 and CD81. We observed that the amount of secreted IgM pentamer or EV IgM monomer varied proportionally depending on the cell population or tissue sample that was analyzed. Detection of EV-associated IgM monomer in a broad panel of unstimulated cell cultures and in healthy mice led us to conclude that IgM packaging into EVs is an inherent, controlled cellular process, and not a phenomenon due to selective sampling, experimental conditions, or method of analysis.
There are several points worth noting that place our results in the larger picture of B cell immunity. The first is that a direct comparison of IgM expression in both large (11kG) and small (110kG) EV, across multiple cell populations and tissues, has not been reported to our knowledge. It has been suggested that EV segregation into different sized populations reflects differences in biogenesis pathways, with large forming from plasma membrane and small forming in the endosome, but this has not been established.79,80,81,82 It was evident, however, that observable protein (Figures S1C and 2E) and IgM levels (Figures 2E, 2F, 4C, 4D, 5A, 5B, 6A, and 6B) in each EV population were not equivalent, suggesting that at some level the mechanisms pertaining to EV formation and IgM packaging are differentially regulated. The second relates to IgM topography, meaning location within the EVs. It became clear, across multiple cellular sources (Figures 1F, 2F, and 4D) and in both 11kG and 110kG populations, that EV treatment with a solubilizing agent (0.25% TX-100) resulted in substantially more detectable IgM. As this approach is utilized to quantitate EV-encapsulated bioactive molecules, our data suggest that a portion of EV-associated IgM is sequestered inside EVs.68 Other results presented here, and supported by prior reports, place immunoglobulin on the EV surface as well.45,47,48 IgM presentation, whether encapsulated or surface, could have implications regarding antibody stability, cellular uptake and processing, and immune function. Although not tested here, encapsulation may provide a mechanism for IgM to be released but remain inactive, or hidden to antigen, until conditions arise that trigger EV membrane solubilization and intraluminal content release. A third point is that low molecular weight EV IgM cannot be viewed as a byproduct of secreted pentameric IgM on the basis of studies with secretory IgM (μs) knockout mice. A direct comparison of cell culture supernatant (Figures 4C, 4D, S4, and S5C) and peritoneal cavity fluid (Figures 5A and 5B) showed the expected lack of secreted IgM in μsKO 110kG supernatant samples, but approximately equivalent monomeric IgM levels in EV samples from WT and μsKO mice. This indicates that the mechanisms regulating EV IgM packaging and release can occur independently of those required for secretion, and demonstrate that extracellular EV IgM may accumulate in the absence of IgM pentamer. Further experimentation with different stimuli will help elucidate the signaling cascades that regulate these processes. It should be noted as well that we directed our investigation to IgM, for several reasons including that IgM is expressed first phylogenetically (most ancient) and during B cell development, that IgM is the primary isotype involved in innate-type protection, that the membrane and secreted form differ substantially in structure and molecular weight, and the availability of the μs knockout mouse model, which allowed us to directly test the dependence of EV LMW IgM expression on pentameric HMW IgM.72,83,84,85 To our knowledge expression and topography of EV IgA, IgD, IgE, or IgG have not been thoroughly evaluated, and more investigation in this area may identify a broader role for B cell EV-mediated immunity.
Although we could detect EV IgM under the above conditions, we questioned if EV IgM could bind antigen, as this would indicate a potential immune function. Our results showed that small and large EV harvested from the PtC-specific CH12 B cell line could bind to PtC-formulated liposomes (Figures 6E–6H), which did not occur with WEHI-231-produced EVs. Although not directly translatable to in vivo conditions, it does open the intriguing possibility that association between EV-IgM and antigen does occur, under homeostatic and/or disease conditions. Although not abundant, there are reports of lower molecular weight IgM in serum collected from autoimmune patients, and in the sera of infants.86,87,88,89 It was concluded in the former that this was not a byproduct of secreted IgM; however, the biological process responsible for the observation was never delineated, raising the possibility that IgM-associated EV played a role.90,91 Biochemical analysis of isolated, serum EV, under healthy and disease states, would help answer this. Perhaps a lack of evidence for extracellular IgM monomer is more reflective of methodology than biology. Future experimentation and tissue immunoglobulin characterization should utilize approaches (immunoblot) that would help distinguish IgM species. If low molecular weight IgM is more broadly expressed in tissues than previously recognized, this would raise additional questions as to IgM monomer binding properties (affinity) and functional capabilities, alone and by comparison to pentameric IgM. Furthermore, it is not known if encapsulated EV IgM monomer can bind antigen, and if so whether the properties (antibody affinity) are similar to its membrane counterpart.
If EV-associated LMW IgM release occurs in vivo, as it appears from our results from peritoneal fluid and mouse plasma, one would then have to consider EV IgM interaction with other cells. Our results under controlled, in vitro conditions (Figures 7 and S7–S9) indicate that EV-associated immunoglobulin can be internalized following transfer, as determined by two immunodetection methods (immunoblot and immunofluorescent confocal microscopy). Two EV-associated isotypes (IgM and IgG2a) were evaluated, and both showed a distinct staining pattern, largely peri-nuclear, after overnight transfer. Intracellular accumulation of EV-associated antibody was a property of 11 kG EV rather than 110 kG EV, providing evidence that these differently sized EV function differently, and emphasizing the importance of further study directed at EV that are larger than exosomes and that have been less studied to date. The mechanism of uptake and functional output were not evaluated, but our results do place EV-associated antibody inside a target cell for a substantial period of time. Thus, it may be that EV association can provide an (alternate) route to cell entry distinct from soluble HMW IgM. This could be relevant given the functional consequences of EV-mediated intercellular communication observed for other proteins in other cell types.56 Additional studies will need to determine if molecular interactions exist between B cell-derived EV and receptor cell that confer specificity. Our use of HeLa, however, indicates that EV-mediated intracellular antibody-antigen interactions may be possible in cancer cells, and perhaps other cell lineages as well.
In conclusion, we have provided evidence that EV IgM contributes to the pool of extracellular IgM, in monomer form on the EV surface and encapsulated inside the EV, and is functional in antigen binding and cellular targeting. Given the differences in molecular weight and biological presentation as compared to pentameric IgM, we propose that consideration should be given to EV IgM under all experimental conditions and as a potential source of novel immune related activities.
Limitations of the study
B cell populations are heterogeneous in form and function, and we utilized several methodologies to isolate the populations evaluated herein. We acknowledge that this is only a subset of the complete, known B cell repertoire, and future studies will uncover EV production by additional B cell populations. Along this line, the criteria that define EV populations is evolving, along with the methodologies to isolate specific populations. Continued refinement in both areas will likely reveal new information regarding B cell EVs and immunoglobulin production, as they relate to both homeostatic processes and disease. Advancement in EV isolation and sorting techniques, along with surface antigen expression, will also permit better cellular source-of-origin for tissue isolated EVs. Although we carefully evaluated B cell EV immunoglobulin, B cell EV size, and EV surface antigens, further study is needed to fully characterize the contents of large and small B cell-derived IgM-containing EV. We followed several established methodologies for detergent solubilization of EV. As it has been shown that EV lipid content and EV structural integrity can vary considerably under different physical, environmental, and stress conditions, we acknowledge that additional experimentation is needed to more thoroughly characterize EV IgM topography and orientation (inside vs. outside, Fab and Fc domain accessibility). While our use of the μsKO mouse model revealed that expression of low molecular weight (monomeric) IgM in EV is not dependent on pentameric IgM expression, we acknowledge that our analyses by immunoblot, ELISA, and flow cytometry did not address the precise molecular and structural characteristics of EV IgM, features that by comparison are well known for surface IgM contained in the B cell receptor complex. To perform our EV transfer studies, we utilized HeLa cells to establish proof-of-principle, however we acknowledge this model is limited in scope, and the mechanism underlying perinuclear localization of large EV in target cells remains to be determined. Experimentation with additional immunoglobulin-containing EV source material and secondary cells is needed to better characterize EV IgM trafficking and function.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-B220 FITC, clone RA3-6B2 | BD | Cat.# 553088; RRID:AB_394618 |
| anti-CD5 BB700, clone 53-7.3 | BD | Cat.# 742083; RRID:AB_2871364 |
| anti-CD19 BV421, clone 1D3 | BD | Cat.# 562701; RRID:AB_2737731 |
| anti-CD21 APC, clone 7G6 | BD | Cat.# 561770; RRID:AB_10892818 |
| anti-CD43 PE, clone S7 | BD | Cat.# 553271; RRID:AB_394748 |
| anti-CD23 PE-Cy7, clone B3B4 | BioLegend | Cat.# 101614; RRID:AB_2103036 |
| anti-CD16/32 | BD | Cat.# 553142; RRID:AB_394657 |
| anti-IgD PE, clone 11 -26c.2a | BD | Cat.# 558597; RRID:AB_647211 |
| anti-CD19 PE-CF594, clone 1D3 | BD | Cat.# 562329; RRID:AB_11154580 |
| anti-IgM APC, clone RMM-1 | BioLegend | Cat.# 406509; RRID:AB_315059 |
| anti-CD43 APC-Cy7, clone 1B11 | Biolegend | Cat.# 121220; RRID:AB_2194192 |
| anti-CD5 PerCP-Cy5.5, clone 53-7.3 | ebioscience | Cat.# 45-0051-82; RRID:AB_914334 |
| biotinylated anti-CD9, clone KMC8 | BD | Cat.# 558749; RRID:AB_397103 |
| anti-CD63 PE, clone NVG-2 | BD | Cat.# 564222; RRID:AB_2738678 |
| biotinylated anti-CD81, clone Eat2 | BD | Cat.# 559518; RRID:AB_397259 |
| anti-IgM HRP | SouthernBiotech | Cat.# 1020-05; RRID:AB_2794201 |
| anti-IgG2a HRP | SouthernBiotech | Cat.# 1080-05; RRID:AB_2734756 |
| anti-CD81 BV421, clone Eat2 | BD | Cat.# 740060; RRID:AB_2739825 |
| rat IgG2a FITC isotype control, clone R35-95 | BD | Cat.# 554688; RRID:AB_10053774 |
| rat IgG2a APC isotype control, clone RTK2758 | Biolegend | Cat.# 400512; RRID:AB_2814702 |
| anti-CD4 PE-Cy5, clone GK1.5 | ebioscience | Cat.# 15-0041-82; RRID:AB_468695 |
| anti-IgG AF488 | ThermoFisher Scientific | Cat.# A11055; RRID:AB_2534102 |
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharide | Sigma | Cat.# L6511 |
| PtC liposomes FITC | Dr. Aaron Kantor, Kantor Bioscience Consulting | |
| streptavidin PE | ebioscience | Cat.# 12-4317-87 |
| RBC lysis buffer | Lonza | Cat.# 10-548E |
| LIVE/DEAD fixable cell stain | Invitrogen | Cat.# L34966 |
| RIPA lysis buffer | ThermoFisher Scientific | Cat.# 89900 |
| Halt protease inhibitor cocktail | ThermoFisher Scientific | Cat.# 1862209 |
| Halt phosphatase inhibitor cocktail | ThermoFisher Scientific | Cat.# 1862495 |
| Triton X-100 | Sigma | Cat.# X100 |
| Tween 20 | Sigma | Cat.# 9416 |
| Laemmli SDS sample buffer | Bio-Rad | Cat.# 1610747 |
| streptavidin HRP | Pierce | Cat.# 21130 |
| purified mouse IgM | BD | Cat.# 557275 |
| wheat germ agglutinin AF488 | Invitrogen | Cat.# W11261 |
| phalloidin rhodamine | ThermoFisher Scientific | Cat.# R415 |
| Prolong Gold/DAPI | ThermoFisher Scientific | Cat.# P10144 |
| Critical commercial assays | ||
| Miltenyi gentleMACS Octo dissociator | Miltenyi | Cat.# 130-095-937 |
| Miltenyi pan B cell isolation kit II | Miltenyi | Cat.# 130-104-443 |
| bicinchoninic acid (BCA) protein assay kit | Pierce | Cat# 23227 |
| 4-15% tris-glycine gels | Bio-Rad | Cat# 4568086 |
| Aldehyde sulfate beads (4 μm); Aldehyde sulfate beads (5μm) | ThermoFisher Scientific | Cat.# A37304; Cat.# A37306 |
| imperial protein stain | ThermoFisher Scientific | Cat# 24615 |
| anti-mouse IgM ELISA kit | Bethyl | Cat.# E90-101 |
| round glass coverslips, #1.5, 12mm | Thomas Scientific | Cat.# 1217N79 |
| Immun-Blot PVDF membrane | Bio-Rad | Cat# 1620177 |
| SuperSignal West Pico PLUS ECL reagents | ThermoFisher Scientific | Cat# 34580 |
| Lionheart FX automated live cell imager | Biotek | |
| AR1+ confocal microscope | Nikon | |
| Fortessa flow cytometer | BD | |
| Influx cell sorter | BD | |
| Experimental models: Cell lines | ||
| WEHI-231 mouse B cell lymphoma | frozen stock | |
| S9.6 hybridoma | American Type Culture Collection (ATCC) | Cat.# HB-8730; lot 63476317 |
| CH-12 mouse B cell lymphoma | frozen stock | |
| HELA human cervical cancer | frozen stock | |
| Experimental models: Organisms/strains | ||
| C57BL/6 mouse | The Jackson Laboratory | https://www.jax.org |
| Ighm knock-out mice (B6; 129S4-Ighmtm1che/J) | Dr. Kishore Alugupalli | |
| Software and algorithms | ||
| Flowjo, v10.6.2 | BD | https://www.flowjo.com |
| Image J, version 1.51 | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Prism, version 8.1.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Photoshop 2018, 2021 | Adobe | https://www.adobe.com/products/photoshop.html |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| Gen5 software (Lionheart microscope) | Biotek | |
| NIS-Elements C (Nikon confocal microscope) | Nikon | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael Gutknecht (michael.gutknecht@wmed.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mouse model
The Ighm knock-out mice (B6; 129S4-Ighmtm1che/J) originally created by Dr. Jianzhu Chen were kindly provided by Dr. Kishore Alugupalli. We backcrossed this line onto a pure C57BL/6 background within our facility and then bred and housed wild type, heterozygous, and homozygous animals. Both male and female mice were used for experimentation. The mice were housed at 5 mice per cage with a 12-h light/12-h dark cycle and ad libitum access to water and food. Mice were cared for and handled in accordance with the Guide for the Care and Use of Laboratory Animals, National Institutes of Health, and institutional guidelines. All animal studies were approved by the Western Michigan University Homer Stryker M.D. School of Medicine institutional IACUC committee.
Method details
Cell culture and preparation of vesicle production media
The WEHI-231 B cell lymphoma line was cultured in DMEM (4.5 g/ml glucose) supplemented with 10% fetal bovine serum (FBS), penicillin (100 Units/ml), streptomycin (100 μg/ml), L-glutamine (2 mM), 2-mercaptoethanol at 37°C/5% CO2 upon thaw of frozen stock. The cells were passaged every 2–3 days at 0.075 - 0.1 × 106 cells/ml and maintained >90% viability. S9.6 hybridoma cells, obtained from ATCC (lot 63476317) and cultured under the same conditions, were passaged every two days at 0.125 × 106 cells/ml and maintained >95% viability. CH12 cells were cultured in RPMI 1640 supplemented with the reagents listed above upon thaw of frozen stock, and passaged at 0.15 × 106 cells/ml every two days. Viability was maintained at >90%. HELA cells were maintained in DMEM supplemented with the reagents listed above, and passaged prior to confluency. To deplete FBS-derived vesicles in media for cell culture experiments involving extracellular vesicle analysis, supplemented culture media was prepared at 20% FBS, vacuum filtered (0.2 μm), and centrifuged overnight at 110,000 × g (4°C). The supernatant was then combined with serum free supplemented culture media at 1:1 and filtered (0.2 μm) a second time prior to use.63 Where indicated, LPS (Sigma, from Salmonella enterica serotype typhimurium) was added to cultures at the time of plating.
Mouse tissue harvest and cell purification (FACS)
Peritoneal lavage and spleen removals were performed on euthanized mice. Spleens were homogenized using the rough ends of glass slides or the Miltenyi gentleMACS dissociator and then passed through a 70-μm cell strainer. All samples were treated with RBC lysis buffer for 2 min (Lonza), subsequently diluted with HBSS with 2.5% FBS, and then centrifuged at 1200 rpm for 10 min. For cell sorting, the cells were resuspended in HBSS with 2.5% FBS, stained with immunofluorescent antibodies, and then sorted on the Influx cell sorter (BD Biosciences) with gating on live cells by forward side scatter and/or Aqua Live/Dead stain (Invitrogen). The following antibodies were obtained from BD Pharmingen: anti-B220 FITC or BB700 (clone RA3-6B2), anti-CD5 BB700 or BV421 (clone 53-7.3), anti-CD19 BV421 or BUV395 (clone 1D3), anti-CD21 APC (clone 7G6), anti-CD43 PE (clone S7); from Biolegend: anti-CD23 PE-Cy7 (clone B3B4). Gated peritoneal B-1a cells (CD5+B220loCD43+CD23−CD19+), peritoneal B2 (B2P) cells (CD5-B220highCD43−CD23+CD19+), and splenic B2 (B2 S) cells (CD5-B220highCD43−CD23highCD21lo/−CD19+) were collected, centrifuged, enumerated, and cultured. Post-sort re-analysis of B cell subsets showed them to be >98% pure.
For splenic B cell isolation using immunocapture magnetic beads, upon removal the spleens were placed in MACS buffer (0.5% BSA/2 mM EDTA/PBS), disrupted on a 70 μm filter by gentle pressure with a syringe plunger, treated with RBC lysis buffer (4 min at room temperature), and resuspended in MACS buffer. Splenic B cells were isolated by negative selection using the Miltenyi B cell isolation kit II according to the manufacturer’s instructions, with reagent volumes adjusted according to cell number. The resultant flow through (B cell population; viability >95%) was resuspended in MACS buffer for subsequent analysis or media for culture.
Flow cytometry
All washes and antibody incubations for flow cytometry staining described below were completed in MACS buffer at 4°C, unless otherwise indicated. For WEHI-231 phenotyping, cells were aliquoted in 96 well conical plates (0.5–1.0 × 106 cells/well), washed, incubated with viability dye (Invitrogen), washed, blocked with anti-CD16/32 (BD), and incubated with the following antibody panel. From BD, anti-IgD PE (clone 11-26 c.2a), anti-CD19 PE-CF594 (clone 1D3); from Biolegend, anti-IgM APC (clone RMM-1), anti-CD23 PE-Cy7 (clone B3B4), anti-CD43 APC-Cy7 (clone 1B11); from ebioscience, anti-CD5 PerCP-Cy5.5 (clone 53-7.3). After washes, the cells were resuspended in MACS buffer for flow cytometry acquisition (BD LSR Fortessa). For assessment of PtC liposome binding to CH12 or WEHI-231 cells, washed cells were aliquoted at 0.5 × 106 cells/tube and subjected to the viability and block protocol described above. The cells were then stained with FITC PtC liposomes (obtained from Dr. Aaron Kantor, Kantor Bioscience Consulting, Redwood City, CA) and anti-IgM APC (clone RMM-1) for 30 min on ice, washed, and acquired on the flow cytometer. The composition of the PtC liposomes used is DSPC:DSPG:Chol (Molar ratio: 45:5:50). For splenic B cell phenotyping, cells were prepared in 96 well plates (0.225 × 106 - 0.5 × 106 cells/well), subjected to the viability and block protocol described above, and incubated in the following single stain antibody preparations: anti-IgM APC (clone RMM-1), anti-CD9 biotin (clone KMC8), anti-CD63 PE (clone NVG-2), anti-CD81 biotin (clone Eat2). Anti-CD19 BV 421 (clone 1D3) was included in each stain to assess B cell purity. The cells were washed, incubated with streptavidin PE (ebioscience) where necessary, washed, and acquired on the cytometer. Images were constructed with FlowJo v10.6.2 Software (BD).
Extracellular vesicle isolation by differential ultracentrifugation (DUC)
Following removal of intact cells (300 × g at 4°C, 10 min), extracellular vesicle (EV) populations in buffer or media solutions were isolated by differential centrifugation.63 Solutions were first centrifuged at 2000 × g (4°C, 20 min), and the supernatant was collected by pipet and centrifuged at 11,000 × g (4°C, 40 min). Supernatant was again collected by pipet and subjected to 110,000 × g (4°C, 100 min). The remaining supernatant was decanted and saved as the EV depleted solution (labeled herein 110kG supernatant). The resultant pellets after each centrifugation step were washed in PBS, centrifuged, and resuspended in buffer for downstream assays. Isolated EVs were maintained at 4°C (or on ice) until analyzed by the methodological approaches utilized herein. At no time was conditioned media prior to EV isolation or isolated EVs frozen.
Extracellular vesicle isolation by sucrose gradient fractionation
The washed EV populations following DUC were again centrifuged, the supernatant was removed, and the pellet was resuspended in 1.5 mL 2.5 M sucrose/PBS, which was then overlaid with graded sucrose/PBS solutions (0.75 mL each: 2.0 M, 1.5 M, 1.25 M, 1.0 M, 0.50 M, 0.25 M). Total solution volume was then brought to 11 mL with 0.25 M sucrose, and the samples were centrifuged at 200,000 × g (4°C, acceleration slow, ∼16 hrs at speed, deceleration no brake). Sample was then collected by pipet down to 5 mL volume, upon which collection proceeded in 1 mL fractions. Care was taken to limit disruption to the adjacent fraction during removal by pipet. The collected fractions (labeled F1 (least dense) to F6 (most dense)) were brought to 11 mL with PBS, mixed by pipet, and centrifuged. The supernatant was then decanted, and the pellets were resuspended in buffer for downstream assays.
Polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis
Isolated vesicles were either lysed in RIPA buffer/inhibitor cocktail (ThermoFisher) or solubilized directly in Laemmli SDS sample buffer (BioRad). RIPA-mediated lysis occurred for 20 min on ice, with occasional vortex, followed by centrifugation at 20,000 × g for 20 min to pellet cellular debris. Protein concentration of the supernatant was determined by the BCA method (Pierce). Samples were resolved by Tris/glycine PAGE, transferred to PVDF membranes (0.2 μm pore), washed (TBS/0.1% Tween 20 (TBS/T20)), blocked (TBS/T20/5% non-fat dry milk), and incubated with primary antibody overnight (in blocking buffer). The membranes were then incubated with streptavidin HRP (in blocking buffer) if required, washed, and developed by enhanced chemiluminescence. Antibodies and reagents utilized for immunoblot analysis included: from Southern Biotech, anti-IgM HRP, anti-IgG2a HRP; from BD, anti-CD9 biotin (clone KMC8), anti-CD81 biotin (clone Eat2), control purified mouse IgM; from Pierce, streptavidin HRP.
Extracellular vesicle adsorption to microbeads
Isolated vesicles in PBS were incubated with 4 μm or 5 μm aldehyde sulfate beads (ThermoFisher) to allow adsorption (overnight 4°C or 120 min room temperature). Samples were prepared at either a ratio of 0.5 μg protein:1.0ul bead stock, or an equivalent volume of isolated EV for direct comparative analyses, and then titrated to confirm the measured fluorescence was in the linear range of detection. Beads incubated with buffer alone were included as a control. After adsorption, the beads were blocked sequentially with 100 mM glycine and PBS/0.5%BSA/2 mM EDTA (blocking buffer).63,92 The beads were incubated with primary antibody (in blocking buffer) or isotype control, washed, and if necessary, incubated with streptavidin PE (in blocking buffer), washed, and resuspended in blocking buffer for flow cytometry acquisition. Antibodies utilized for staining included: from BD, anti-IgM APC (clone RMM-1), anti-IgM FITC (clone R6-60.2), anti-CD9 biotin (clone KMC8), anti-CD63 PE (clone NVG-2), anti-CD81 biotin (clone Eat2), anti-CD81 BV421 (clone Eat2), rat IgG2a FITC (clone R35-95); from Biolegend, rat IgG2a APC (clone RTK2758); from ebioscience, anti-CD4 PE-Cy5 (clone GK1.5), anti-CD4 eFluor 660 (clone GK1.5); and FITC PtC liposomes.
ELISA analysis of isolated extracellular vesicles
Samples were prepared and diluted in either buffer alone (TBS/1.0% BSA/0.05% Tween 20) or buffer +0.25% TX100 followed by a 20s vortex pulse to disrupt the EV lipid membrane. Quantitation of IgM in intact and solubilized vesicles was completed using an anti-mouse IgM specific ELISA kit (Bethyl), according to the manufacturer’s instructions.
Nanoparticle tracking analysis (NTA) of isolated extracellular vesicles
Freshly prepared WEHI 231 11kG and 110 kG EV samples in PBS (4°C) were serially titrated 1:10 in PBS. Acquisition and analysis were completed on a ZetaView Twin NTA instrument (Particle Metrix) at the University of Michigan Biointerfaces Institute under the guidance of the research staff. The instrument was calibrated using polystyrene beads. Size distribution was measured using the 488 nm laser at 11 positions for sample dilution 1:100. Analysis parameters for all samples were: Max. area 1000, Min. area 5, Min. brightness 20.
Harvest and preparation of mouse plasma
Plasma was prepared according to Jeppesen et al., with modification.93 Microcentrifuge collection tubes (1.5 mL) and syringe + needle (1.0 mL syringe/27G 0.5 inch needle) were first rinsed with 0.5 M EDTA. Blood from WT and μsKO mice was harvested by cardiac puncture and deposited (800 μL/tube) into tubes pre-loaded with 0.5 M EDTA (8 μL), then centrifuged at 3000 × g for 15 min. The supernatant was collected and centrifuged at 3000 × g for an additional 15 min. Equivalent volumes of WT and μsKO supernatant (cell and platelet depleted plasma) were then diluted in PBS and centrifuged at 15,000 × g for 40 min. The supernatant was recovered, and subjected to the DUC protocol described above to isolate the 110 kG EV population. The samples were then fractionated by sucrose gradient, followed by SDS/PAGE and immunoblot analysis, as described above. At each step, samples from WT and μsKO mice were prepared in an equivalent volume of buffer.
Extracellular vesicle transfer to HELA cells
To test in vitro transfer of isolated EV populations to HELA monolayers, HELA cells were plated at either 0.025 × 106 cells/mL (24 well cluster plate with glass coverslips for microscopy; 1 mL/well) or 0.075 × 106 cells/mL (6 well plate for generation of whole cell lysates; 2 mL/well) and cultured for at least one day prior to EV addition to allow for adherence. Plating density was determined to permit sufficient cell number for analysis but remain below cell confluence. Extracellular vesicles (EV) were isolated by differential centrifugation as described above and resuspended in PBS. For antibody-mediated detection of EV following transfer, the isolated EV were prepared in vesicle production media and added directly to HELA cells for the indicated period of time before staining. For direct fluorescent labeling of EV, wheat germ agglutinin AF488 (Invitrogen) prepared in PBS was added to the EV suspensions [5 μg/mL]f, which were then incubated for 15 min at 37°C, diluted with PBS, and placed on ice. Following a series of two PBS washes, the labeled EV were resuspended in vesicle production media and added to the HELA cultures. To analyze cells by confocal microscopy, the coverslips were washed twice with PBS, fixed with 4% paraformaldehyde (15 min), washed twice with PBS, permeabilized with 0.1% Triton X-100 for 4 min, washed twice with PBS, blocked with 5% BSA/PBS (30 min), and then incubated with primary antibody (goat IgG anti-mouse IgM (Southern Biotech); goat IgG anti-mouse IgG2a (Southern Biotech). After washes, the cells were incubated with phalloidin rhodamine (ThermoFisher) and secondary antibody (ThermoFisher, donkey anti-goat IgG AF488). The coverslips were then washed, removed, and mounted with Prolong Gold/DAPI (ThermoFisher) on glass microscope slides for confocal microscopy imaging. To generate whole cell lysates, HELA monolayers were washed twice with PBS, trypsinized (0.25% porcine trypsin/0.1% EDTA, 10 min), quenched with complete media, centrifuged, and then washed with PBS. The pellets were subsequently lysed in RIPA buffer and clarified by centrifugation, followed by protein quantification, PAGE, and immunoblot, as described above.
Quantification and statistical analysis
Prism software (version 8.1.1; GraphPad) was utilized to generate graphs and for statistical analyses. One-way ANOVA was performed for comparison of three or more groups, followed by Tukey’s multiple comparisons post-hoc test. Statistically significant differences between groups (p < 0.05) were determined by unpaired two-tailed t test. For ELISA, the standard curve was established using sigmoidal 4PL parameters. Brightfield microscopy images were acquired using a Lionheart FX automated live cell imager (Biotek) with Gen5 software (Biotek). Confocal microscopy was conducted on a Nikon AR1+ confocal microscope integrated with NIS-Elements C control software (Nikon). Sequential sections were imaged every 0.75 μm, where indicated. Established laser settings remained unchanged across evaluated samples. PAGE gels were imaged on the chemidoc imaging system (Bio-Rad) and processed with ImageLab software (Bio-Rad). Film for enhanced chemiluminescent immunoblot analysis was processed and developed on the AFP Imaging mini-medical X-ray film processor. All adjustments and transformations during image preparation in Image J (National Institutes of Health) were applied equivalently to whole images, and across multiple images subjected to comparative analysis. Final figure layouts were completed using Photoshop 2018 and 2021 (Adobe).
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Numbers R01AI142004 (TLR) and R01AI154539 (NEH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support was provided by Western Michigan University Homer Stryker M.D. School of Medicine as well. We would also like to acknowledge the Western Michigan University Homer Stryker M.D. School of Medicine Flow Cytometry and Imaging Core and Michael Clemente for assistance with EV analysis by flow cytometry. Additionally, we acknowledge the University of Michigan Biointerfaces Institute for use of NTA instrumentation, and Dr. Sarah Spanninga for her expertise and guidance in EV acquisition and analysis.
Author contributions
Conceptualization, M.F.G., T.L.R., and N.E.H.; methodology, M.F.G. and T.L.R.; investigation, M.F.G. and N.E.H.; Visualization, M.F.G., T.L.R., and N.E.H.; writing – original draft, M.F.G. and T.L.R.; writing – review and editing, M.F.G., T.L.R., and N.E.H.; supervision, T.L.R. and N.E.H.; funding acquisition, T.L.R. and N.E.H.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107526.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Flajnik M.F., Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briney B., Inderbitzin A., Joyce C., Burton D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cyster J.G., Allen C.D.C. B cell responses: Cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parra D., Takizawa F., Sunyer J.O. Evolution of B cell immunity. Annu. Rev. Anim. Biosci. 2013;1:65–97. doi: 10.1146/annurev-animal-031412-103651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunyer J.O. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets. 2012;12:200–212. doi: 10.2174/187152612800564419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deatherage B.L., Cookson B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012;80:1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill S., Catchpole R., Forterre P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019;43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janda M., Robatzek S. Extracellular vesicles from phytobacteria: Properties, functions and uses. Biotechnol. Adv. 2022;58 doi: 10.1016/j.biotechadv.2022.107934. [DOI] [PubMed] [Google Scholar]
- 9.Kameli N., Dragojlovic-Kerkache A., Savelkoul P., Stassen F.R. Plant-derived extracellular vesicles: Current findings, challenges, and future applications. Membranes. 2021;11:411. doi: 10.3390/membranes11060411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira D.L., Nakayasu E.S., Joffe L.S., Guimarães A.J., Sobreira T.J.P., Nosanchuk J.D., Cordero R.J.B., Frases S., Casadevall A., Almeida I.C., et al. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutter B.D., Innes R.W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017;173:728–741. doi: 10.1104/pp.16.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capello M., Vykoukal J.V., Katayama H., Bantis L.E., Wang H., Kundnani D.L., Aguilar-Bonavides C., Aguilar M., Tripathi S.C., Dhillon D.S., et al. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat. Commun. 2019;10:254. doi: 10.1038/s41467-018-08109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Anca M., Fenoglio C., Serpente M., Arosio B., Cesari M., Scarpini E.A., Galimberti D. Exosome determinants of physiological aging and age-related neurodegenerative diseases. Front. Aging Neurosci. 2019;11:232. doi: 10.3389/fnagi.2019.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitan E., Green J., Bodogai M., Mode N.A., Bæk R., Jørgensen M.M., Freeman D.W., Witwer K.W., Zonderman A.B., Biragyn A., et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci. Rep. 2017;7:1342. doi: 10.1038/s41598-017-01386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C., Fisher K.P., Hammer S.S., Navitskaya S., Blanchard G.J., Busik J.V. Plasma exosomes contribute to microvascular damage in diabetic retinopathy by activating the classical complement pathway. Diabetes. 2018;67:1639–1649. doi: 10.2337/db17-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGough I.J., Vincent J.P. Exosomes in developmental signalling. Development. 2016;143:2482–2493. doi: 10.1242/dev.126516. [DOI] [PubMed] [Google Scholar]
- 17.Stahl P.D., Raposo G. Extracellular vesicles: Exosomes and microvesicles, integrators of homeostasis. Physiology. 2019;34:169–177. doi: 10.1152/physiol.00045.2018. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi T., Suzuki M., Fujikake N., Popiel H.A., Kikuchi H., Futaki S., Wada K., Nagai Y. Intercellular chaperone transmission via exosomes contributes to maintenance of protein homeostasis at the organismal level. Proc. Natl. Acad. Sci. USA. 2015;112:E2497–E2506. doi: 10.1073/pnas.1412651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: Communication from a distance. Dev. Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Kim M.S., Jia B., Yan J., Zuniga-Hertz J.P., Han C., Cai D. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548:52–57. doi: 10.1038/nature23282. Erratum in: Nature. 2018 Aug;560:E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biller S.J., Schubotz F., Roggensack S.E., Thompson A.W., Summons R.E., Chisholm S.W. Bacterial vesicles in marine ecosystems. Science. 2014;343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 22.Chávez A.S.O., O'Neal A.J., Santambrogio L., Kotsyfakis M., Pedra J.H.F. Message in a vesicle - trans-kingdom intercommunication at the vector-host interface. J. Cell Sci. 2019;132 doi: 10.1242/jcs.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortés A., Sotillo J., Rinaldi G., Cantacessi C. Gut-microbiota-derived extracellular vesicles: Overlooked mediators in host-helminth interactions? Trends Parasitol. 2021;37:690–693. doi: 10.1016/j.pt.2021.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Díaz-Garrido N., Badia J., Baldomà L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles. 2021;10 doi: 10.1002/jev2.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 26.Yáñez-Mó M., Siljander P.R.M., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4 doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escrevente C., Keller S., Altevogt P., Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi B.S., de Beer M.A., Giepmans B.N.G., Zuhorn I.S. Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano. 2020;14:4444–4455. doi: 10.1021/acsnano.9b10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanada M., Bachmann M.H., Hardy J.W., Frimannson D.O., Bronsart L., Wang A., Sylvester M.D., Schmidt T.L., Kaspar R.L., Butte M.J., et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA. 2015;112:E1433–E1442. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maas S.L.N., Breakefield X.O., Weaver A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathieu M., Martin-Jaular L., Lavieu G., Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 33.Mir B., Goettsch C. Extracellular vesicles as delivery vehicles of specific cellular cargo. Cells. 2020;9:1601. doi: 10.3390/cells9071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulcahy L.A., Pink R.C., Carter D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raposo G., Stahl P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019;20:509–510. doi: 10.1038/s41580-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 36.Théry C., Ostrowski M., Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 37.Escola J.M., Kleijmeer M.J., Stoorvogel W., Griffith J.M., Yoshie O., Geuze H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 38.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West M.A., Lucocq J.M., Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369:147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 40.Wubbolts R., Leckie R.S., Veenhuizen P.T.M., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 41.Ayre D.C., Elstner M., Smith N.C., Moores E.S., Hogan A.M., Christian S.L. Dynamic regulation of CD24 expression and release of CD24-containing microvesicles in immature B cells in response to CD24 engagement. Immunology. 2015;146:217–233. doi: 10.1111/imm.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayre D.C., Chute I.C., Joy A.P., Barnett D.A., Hogan A.M., Grüll M.P., Peña-Castillo L., Lang A.S., Lewis S.M., Christian S.L. CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo. Sci. Rep. 2017;7:8642. doi: 10.1038/s41598-017-08094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clayton A., Turkes A., Dewitt S., Steadman R., Mason M.D., Hallett M.B. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18:977–979. doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- 44.Clayton A., Turkes A., Navabi H., Mason M.D., Tabi Z. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 2005;118:3631–3638. doi: 10.1242/jcs.02494. [DOI] [PubMed] [Google Scholar]
- 45.Phan H.D., Longjohn M.N., Gormley D.J.B., Smith R.H., Dang-Lawson M., Matsuuchi L., Gold M.R., Christian S.L. CD24 and IgM stimulation of B cells triggers transfer of functional B cell receptor to B cell recipients via extracellular vesicles. J. Immunol. 2021;207:3004–3015. doi: 10.4049/jimmunol.2100025. [DOI] [PubMed] [Google Scholar]
- 46.Rialland P., Lankar D., Raposo G., Bonnerot C., Hubert P. BCR-bound antigen is targeted to exosomes in human follicular lymphoma B-cells. Biol. Cell. 2006;98:491–501. doi: 10.1042/BC20060027. [DOI] [PubMed] [Google Scholar]
- 47.Saunderson S.C., Schuberth P.C., Dunn A.C., Miller L., Hock B.D., MacKay P.A., Koch N., Jack R.W., McLellan A.D. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J. Immunol. 2008;180:8146–8152. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 48.Saunderson S.C., Dunn A.C., Crocker P.R., McLellan A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood. 2014;123:208–216. doi: 10.1182/blood-2013-03-489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunderson S.C., McLellan A.D. Role of lymphocyte subsets in the immune response to primary B cell-derived exosomes. J. Immunol. 2017;199:2225–2235. doi: 10.4049/jimmunol.1601537. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh A.K., Secreto C.R., Knox T.R., Ding W., Mukhopadhyay D., Kay N.E. Circulating microvesicles in B-cell chronic lymphocytic leukemia can stimulate marrow stromal cells: Implications for disease progression. Blood. 2010;115:1755–1764. doi: 10.1182/blood-2009-09-242719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato T., Fahrmann J.F., Hanash S.M., Vykoukal J. Extracellular vesicles mediate B cell immune response and are a potential target for cancer therapy. Cells. 2020;9:1518. doi: 10.3390/cells9061518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pegtel D.M., Cosmopoulos K., Thorley-Lawson D.A., van Eijndhoven M.A.J., Hopmans E.S., Lindenberg J.L., de Gruijl T.D., Würdinger T., Middeldorp J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toledo M.D.S., Cronemberger-Andrade A., Barbosa F.M.C., Reis N.F.d.C., Dupin T.V., Soares R.P., Torrecilhas A.C., Xander P. Effects of extracellular vesicles released by peritoneal B-1 cells on experimental Leishmania (Leishmania) amazonensis infection. J. Leukoc. Biol. 2020;108:1803–1814. doi: 10.1002/JLB.3MA0220-464RR. [DOI] [PubMed] [Google Scholar]
- 54.Zhang F., Li R., Yang Y., Shi C., Shen Y., Lu C., Chen Y., Zhou W., Lin A., Yu L., et al. Specific decrease in B-cell-derived extracellular vesicles enhances post-chemotherapeutic CD8+ T cell responses. Immunity. 2019;50:738–750.e7. doi: 10.1016/j.immuni.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 55.Abou Karam P., Rosenhek-Goldian I., Ziv T., Ben Ami Pilo H., Azuri I., Rivkin A., Kiper E., Rotkopf R., Cohen S.R., Torrecilhas A.C., et al. Malaria parasites release vesicle subpopulations with signatures of different destinations. EMBO Rep. 2022;23 doi: 10.15252/embr.202254755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lässer C., Jang S.C., Lötvall J. Subpopulations of extracellular vesicles and their therapeutic potential. Mol. Aspects Med. 2018;60:1–14. doi: 10.1016/j.mam.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Tkach M., Kowal J., Zucchetti A.E., Enserink L., Jouve M., Lankar D., Saitakis M., Martin-Jaular L., Théry C. Qualitative differences in T-cell activation by dendritic cell-derived extracellular vesicle subtypes. EMBO J. 2017;36:3012–3028. doi: 10.15252/embj.201696003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tkach M., Kowal J., Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373 doi: 10.1098/rstb.2016.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wahlund C.J.E., Güclüler G., Hiltbrunner S., Veerman R.E., Näslund T.I., Gabrielsson S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willms E., Johansson H.J., Mäger I., Lee Y., Blomberg K.E.M., Sadik M., Alaarg A., Smith C.I.E., Lehtiö J., El Andaloussi S., et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016;6 doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 64.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boyd A.W., Goding J.W., Schrader J.W. The regulation of growth and differentiation of a murine B cell lymphoma. I. Lipopolysaccharide-induced differentiation. J. Immunol. 1981;126:2461–2465. [PubMed] [Google Scholar]
- 66.Spillmann F.J.X., Beck-Engeser G., Wabl M. Differentiation and Ig-allele switch in cell line WEHI-231. J. Immunol. 2007;179:6395–6402. doi: 10.4049/jimmunol.179.10.6395. [DOI] [PubMed] [Google Scholar]
- 67.Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980;20:303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- 68.Fitzgerald W., Freeman M.L., Lederman M.M., Vasilieva E., Romero R., Margolis L. A system of cytokines encapsulated in extracellular vesicles. Sci. Rep. 2018;8:8973. doi: 10.1038/s41598-018-27190-x. Erratum in: Sci Rep. 2020 Oct 29;10:18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osteikoetxea X., Sódar B., Németh A., Szabó-Taylor K., Pálóczi K., Vukman K.V., Tamási V., Balogh A., Kittel Á., Pállinger É., Buzás E.I. Differential detergent sensitivity of extracellular vesicle subpopulations. Org. Biomol. Chem. 2015;13:9775–9782. doi: 10.1039/c5ob01451d. [DOI] [PubMed] [Google Scholar]
- 70.Andreu Z., Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou F., Wang X., Han X., Rothschild G., Zheng S.G., Basu U., Sun J. Expression and function of tetraspanins and their interacting partners in B cells. Front. Immunol. 2018;9:1606. doi: 10.3389/fimmu.2018.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boes M., Esau C., Fischer M.B., Schmidt T., Carroll M., Chen J. Enhanced B-1 cell development, but impaired IgG antibody responses in mice deficient in secreted IgM. J. Immunol. 1998;160:4776–4787. [PubMed] [Google Scholar]
- 73.Wu H., Fu M., Liu J., Chong W., Fang Z., Du F., Liu Y., Shang L., Li L. The role and application of small extracellular vesicles in gastric cancer. Mol. Cancer. 2021;20:71. doi: 10.1186/s12943-021-01365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang F., Guo C., Cao X., Yan Y., Zhang J., Lv S. Gastric cancer cell-derived extracellular vesicles elevate E2F7 expression and activate the MAPK/ERK signaling to promote peritoneal metastasis through the delivery of SNHG12. Cell Death Discov. 2022;8:164. doi: 10.1038/s41420-022-00925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanc L., Barres C., Bette-Bobillo P., Vidal M. Reticulocyte-secreted exosomes bind natural IgM antibodies: Involvement of a ROS-activatable endosomal phospholipase iPLA2. Blood. 2007;110:3407–3416. doi: 10.1182/blood-2007-04-085845. [DOI] [PubMed] [Google Scholar]
- 76.Arnold L.W., LoCascio N.J., Lutz P.M., Pennell C.A., Klapper D., Haughton G. Antigen-induced lymphomagenesis: Identification of a murine B cell lymphoma with known antigen specificity. J. Immunol. 1983;131:2064–2068. [PubMed] [Google Scholar]
- 77.Lanier L.L., Arnold L.W., Raybourne R.B., Russell S., Lynes M.A., Warner N.L., Haughton G. Transplantable B-cell lymphomas in B10. H-2aH-4bp/Wts mice. Immunogenetics. 1982;16:367–371. doi: 10.1007/BF00372309. [DOI] [PubMed] [Google Scholar]
- 78.Mercolino T.J., Arnold L.W., Haughton G. Phosphatidyl choline is recognized by a series of Ly-1+ murine B cell lymphomas specific for erythrocyte membranes. J. Exp. Med. 1986;163:155–165. doi: 10.1084/jem.163.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bobrie A., Colombo M., Krumeich S., Raposo G., Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 81.de la Torre-Escudero E., Bennett A.P.S., Clarke A., Brennan G.P., Robinson M.W. Extracellular vesicle biogenesis in helminths: More than one route to the surface? Trends Parasitol. 2016;32:921–929. doi: 10.1016/j.pt.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Fordjour F.K., Guo C., Ai Y., Daaboul G.G., Gould S.J. A shared, stochastic pathway mediates exosome protein budding along plasma and endosome membranes. J. Biol. Chem. 2022;298:102394. doi: 10.1016/j.jbc.2022.102394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blandino R., Baumgarth N. Secreted IgM: New tricks for an old molecule. J. Leukoc. Biol. 2019;106:1021–1034. doi: 10.1002/JLB.3RI0519-161R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flajnik M.F. Comparative analyses of immunoglobulin genes: surprises and portents. Nat. Rev. Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 85.Grönwall C., Vas J., Silverman G.J. Protective roles of natural IgM antibodies. Front. Immunol. 2012;3:66. doi: 10.3389/fimmu.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harisdangkul V., McDougal J.S., Knapp M., Christian C.L. Naturally occurring low molecular weight IgM in patients with rheumatoid arthritis, systemic lupus erythematosus and macroglobulinemia. I. Purification and immunologic studies. J. Immunol. 1975;115:216–222. [PubMed] [Google Scholar]
- 87.Hunter A., Feinstein A., Coombs R.R. Immunoglobulin class of antibodies to cow's milk casein in infant sera and evidence for low molecular weight IgM antibodies. Immunology. 1968;15:381–388. [PMC free article] [PubMed] [Google Scholar]
- 88.McDougal J.S., Harisdangkul V., Christian C.L. Naturally-ocurring low molecular weight IgM in patients with rheumatoid arthritis, systemic lupus erythematosus, and macroglobulinemia. II. Structural studies and comparison of some physicochemical properties of reduced and alkylated IgM, and low molecular weight IgM. J. Immunol. 1975;115:223–229. [PubMed] [Google Scholar]
- 89.Xu H., Geddes R., Roberts-Thomson P.J. Low molecular weight IgM and CD5 B lymphocytes in rheumatoid arthritis. Ann. Rheum. Dis. 1994;53:383–390. doi: 10.1136/ard.53.6.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maione F., Cappellano G., Bellan M., Raineri D., Chiocchetti A. Chicken-or-egg question: Which came first, extracellular vesicles or autoimmune diseases? J. Leukoc. Biol. 2020;108:601–616. doi: 10.1002/JLB.3MR0120-232R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Solomon A., McLaughlin C.L. Biosynthesis of low molecular weight (7S) and high molecular weight (19S) immunoglobulin M. J. Clin. Invest. 1970;49:150–160. doi: 10.1172/JCI106214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gutknecht M.F., Kaku H., Rothstein T.L. Microparticle immunocapture assay for quantitation of protein multimer amount and size. Cell Rep. Methods. 2022;2 doi: 10.1016/j.crmeth.2022.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.