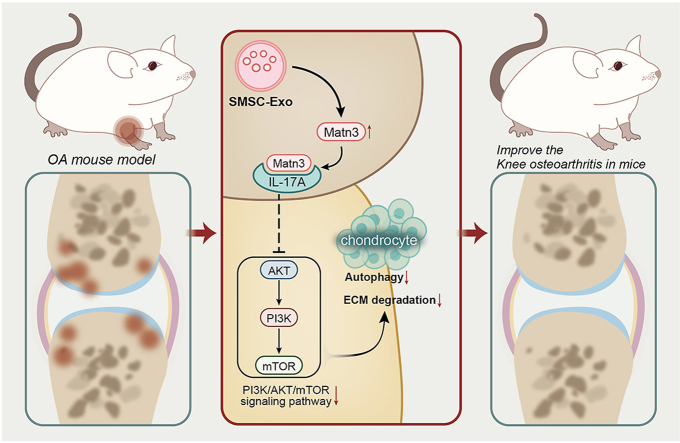
Abstract
Background
Synovial mesenchymal stem cell (SMSC) exerts chondroprotective effects in osteoarthritis (OA) clinical models. However, the regulatory potentials of SMSC-derived exosomes (SMSC-Exo) in OA still need to be discovered, which attracted our attention.
Methods
The destabilization of the medial meniscus surgery was performed on the knee joints of a mouse OA model, followed by injection of SMSC-Exo. In addition, SMSC-Exo was administrated to mouse chondrocytes to observe the functional and molecular alterations.
Results
Both of SMSC-Exo and overexpression of Matrilin-3 (MATN3) alleviated cartilage destruction and suppressed degradation of extracellular matrix (ECM) in the OA rat model. In addition, assays concerning the in vitro OA model induced by IL-1β showed that SMSC-Exo could promote chondrocyte viability and inhibit autophagy defects. Furthermore, SMSC-Exo achieved the chondroprotective effects through the delivery of MATN3/IL-17A, and MATN3 could suppress the activation of PI3K/AKT/mTOR signaling through IL-17A.
Conclusion
SMSC-Exo exerts beneficial therapeutic effects on OA by preventing ECM degradation and autophagy defects by delivering MATN3/IL-17A.
The Translational Potential of this Article
The translational potential of this study is not only limited to the treatment of knee osteoarthritis but also provides new insights for the treatment of other joint diseases by exploring the mechanism of MATN3. In addition, SMSCExo, as a novel drug carrier, has great potential for treating and diagnosing other diseases. With further research, these findings will provide new directions for developing personalized and innovative treatment options.
Keywords: Knee osteoarthritis, Synovial mesenchymal stem cells, Exosomes, MATN3, IL-17A, PI3K/AKT/mTOR signaling pathway
Graphical abstract
1. Introduction
Osteoarthritis (OA) is the most common disease worldwide and has become a major medical burden on society as more and more people suffer from OA as they age [1]. The knee and hip are two commonly affected joints, and approximately 30% of people over 45 years old show radiographic evidence of knee OA [2]. Characterized by cartilage degeneration and synovial inflammation, OA is often associated with the destruction of extracellular matrix (ECM) and reduced autophagy [[3], [4], [5]]. Therefore, expounding the mechanism that could affect the degradation of ECM, inflammatory response, and autophagy may provide a new direction for studying OA pathogenesis [6,7].
A recent study has shown that mesenchymal stem cells (MSC) have been widely used in the treatment of OA and that MSC-derived exosome (Exo) can suppress the development of OA [[8], [9], [10]]. Moreover, it has been illustrated by a prior study that MSC-Exo could be taken as a new therapy for OA since it boosts cartilage regeneration and thus repairs cartilage tissues [11,12]. In an established mouse model, extracellular vesicles derived from lipopolysaccharide-preconditioned human synovial MSCs Synovial mesenchymal stem cells (SMSCs) can suppress ECM degradation and knee OA [13]. Furthermore, previous studies have shown that exosomes derived from bone marrow mesenchymal stem cells (MSC-Exos) carry complex components, including proteins, lipids, and nucleic acids, which play a pivotal role in intercellular communication [14]. dECM-BMSC-Exo can promote the synthetic metabolism and migration of chondrocytes and inhibit chondrocyte apoptosis by up-regulating miR-3473b targeting PTEN, thereby promoting the relief of OA [15]. In addition, BMSCs-Exo can maintain the ultrastructure of ACL-deficient mice, inhibit abnormal vascularization of subchondral bone, and produce a protective effect against pain and bone absorption induced by OA [16]. In addition, exosomes derived from human SMSCs can alleviate IL-1β-induced OA by targeting HMGB1 through shuttling miR-129-5p [17].
From in silico analysis in our study, we identified that SMSC-derived exosomes (SMSC-Exo) might improve OA by carrying Matrilin-3 (MATN3) to chondrocytes. According to Guo Z et al., Exo from uroderived stem cells could deliver MATN3 to repair and regenerate tissues, thus relieving disc degeneration [18]. Evidence demonstrates that Matrilins, acting as the bridge between collagen II and proteoglycan networks, are adaptor proteins of the cartilage ECM, and MATN3 is the third member belonging to the family of ECM proteins [19]. Moreover, MATN3 is reported as a critical cartilage matrix protein released by chondrocytes and may suppress premature chondrocyte hypertrophy through blockage of BMP-2/Smad1 activity [20]. Our bioinformatics prediction results also revealed a possible interaction between MATN3 and interleukin-17A (IL-17A) in OA. The IL-17 family of cytokines is widely validated as a contributor to OA pathogenesis, and IL-17A has been documented in an increased level in serum and synovial fluid from OA patients (relative to healthy controls), clarifying positive correlations with disease severity scores [21]. IL-6 and TNF-α are representative inflammatory markers that characterize changes in inflammatory levels in OA disease. These two common inflammatory factors are closely related to OA disease [[22], [23], [24], [25]]. Additionally, they are often used as the detection factors for changes in inflammatory levels [[26], [27], [28], [29], [30], [31], [32]].
Baicalin alleviates chondrocyte injury by activating mitochondrial autophagy via PINK1/Parkin and PINK1/Drp-1 pathways and inhibiting PI3K/AKT/mTOR pathway [33]. Orexin B has a chondroprotective effect by inhibiting the PI3K/AKT/mTOR signaling pathway, enhancing the autophagic process, and exerting anti-inflammatory effects to alleviate OA [34]. The downregulation of ciRS-7 or upregulation of miR-7 exacerbates cartilage degradation and autophagy defects in IL-17A-mediated PI3K/AKT/mTOR activation in OA [35]. Nonivamide effectively inhibits the IL-17A and Akt/mTOR pathways, promotes keratinocyte differentiation, activates autophagy, and relieves imiquimod-induced psoriasis-like diseases in mice [36]. Notably, IL-17A is involved in the progression of OA, which can activate the PI3K/AKT/mTOR pathway to affect cartilage degradation and autophagy defection [35]. However, the functional mechanism in OA remains to be supplemented.
In this study, we assumed that SMSC-Exo might affect OA in association with MATN3, IL-17A, and the PI3K/AKT/mTOR pathway. Therefore, we further tested whether SMSC-Exo could carry MATN3 to affect knee OA in a manner that can help explain its function on ECM degradation and autophagy defects. Therefore, we performed a series of experiments in OA-modeled mice to illustrate the mechanism as a potential target for OA treatment.
2. Materials and methods
2.1. Bioinformatics analysis
The OA-related microarrays GSE26475 and GSE104793 were downloaded from the Gene Expression Omnibus (GEO) database. In the GSE26475 dataset, OA was induced in mice with destabilization of the medial meniscus (DMM). The gene expression in cartilage tissues was detected at 3 d and 7 d after DMM (sham-3 d: n = 3, DMM-3 d: n = 3; sham-7d: n = 3, DMM-7 d: n = 3). Meanwhile, GSE104793 showed gene expression in mouse primary chondrocytes after IL-1β induction (control: n = 3; IL-1β: n = 3).
Differential gene analysis was performed by the R software “limma” package for screening out the differentially expressed genes (DEGs) in the GSE26475 with | logFoldChange (FC) | > 1 and p < 0.05 as the screening condition. Heatmaps for DEGs were drawn using the R software “heatmap” package, and the intersection of DEGs was made using the tool jvenn. Boxplots for the expression of key genes were visualized using the R software “ggpubr” package. After protein–protein interaction (PPI) analysis of key factors using the Genemania database to obtain their interacting proteins, these proteins were subjected to functional enrichment analysis using the “ClusterProfiler” package in the R software, and the Fisher test was applied to identify significantly enriched KEGG pathways, with p < 0.05 considered statistically significant.
2.2. Laboratory animals
Wild-type C57BL/6J mice (male; 8 weeks of age, body weight: about 20 g–25 g) were purchased from Vital River Laboratory Animals (Beijing, China; 219). Mice were caged separately with 60%–65% humidity at 22–25 °C and had free access to food and water under a 12 h/12 h light and dark cycle. Mice were acclimated for a week before the experiment. The Animal Ethics Committee of our hospital approved these experimental procedures and animal use protocol.
2.3. Isolation of SMSC-Exo by ultracentrifugation
Mouse joint sites were exposed under a dissecting microscope, and synovial tissues were dissected from the joint and placed in a petri dish containing phosphate buffer saline (PBS) and 1% penicillin-streptomycin (10378016). Scratches were made at the bottom of the 12-well plate to enhance tissue transfer and promote adhesion. Tissue samples were transferred to 12-well plates which contained SMSC amplification medium replenished with Dulbecco's modified eagle medium (DMEM)/F-12 (11320033), 10% fetal bovine serum (FBS; 10099141C), 1% nonessential amino acids (11140050), 1% penicillin-streptomycin, and 0.2% β-mercaptoethanol (21985023). Cells were then placed in an incubator in 2% O2, and 5% CO2 at 37 °C, with the medium renewed every 2–3 d. When cells were adherent, tissue slices were removed, and the SMSCs were passaged into a T25 culture flask, followed by medium renewal every 2–3 d. The above reagents were all purchased from ThermoFisher (Waltham, MA, USA).
The surface antigens of isolated SMSCs were analyzed by flow cytometry. First, SMSCs were incubated with monoclonal antibodies of CD34 (567304; BD Biosciences, San Diego, CA, USA), CD45 (567451, BD Biosciences), CD90 (553016, BD Biosciences), and CD105 (562761, BD Biosciences). Cells were then washed to remove the unbound antibodies, and a flow cytometer (Millipore, Billerica, MA, USA) was used to analyze these surface antigens.
Exo was isolated using the Ribo™ Exo isolation reagent (C10130-1; Ribobio, Beijing, China). After the cells were cultured in Exo-free medium for 72 h, the medium supernatant was collected and centrifuged at 2000 g for 30 min at 4 °C to remove floating cells, cell debris, and unwanted proteins. The cell supernatant was then transferred to a new tube and placed on ice, followed by the addition of a third volume of Ribo ™ Exo isolation reagent. Subsequently, the samples were placed in a 4 °C refrigerator overnight. The next day, a total of 2 mL mixture was pipetted to a centrifuge tube and then centrifuged at 1500 g at 4 °C for 30 min to remove the supernatant and collect the precipitation. This centrifugation step was repeated thrice until all samples were transferred. The Exo was then eluted, and the Exo precipitation was resuspended in 1 × PBS.
2.4. Transmission electron microscopy (TEM), dynamic light scattering (DLA), and Western blot
Exo was identified by TEM (Talos F200X S/TEM, Opton, Portland). DLA detected the diameter of Exo. The experiment used the Zetasizer Nano-ZS90 instrument (Malvin, UK), with the excitation light wavelength λ = 532 nm.
EVs surface-labeled proteins were identified by Western blot with the help of CD63 (ab216130, 1:1000, Abcam, Cambridge, UK), HSP70 (ab2787, 1:1000, Abcam), and endoplasmic reticulum marker protein Calnexin (ab22595, 1:1000, Abcam).
2.5. Isolation of mouse chondrocytes
Under sterile conditions, the knee joint cartilage of C57BL/6J mice (n = 24) was placed in the culture dish, rinsed with PBS, cut to 1 × mm × 1 mm × 1 mm, and transferred to another culture dish. The tissue slices were detached using 0.2% type II collagenase (171010105) and 0.25% trypsin (25200072) in an incubator at 37 °C for 3 h. After digestion, the cell suspension was filtered in a 150-mesh stainless steel filter and centrifuged at 1000 r/min for 3–5 min, with the supernatant discarded. Next, the suspension was added to the DMEM (11965092) with 10% FBS (10099141C) and 1% penicillin-streptomycin (10378016). After counting, culture dishes containing chondrocytes were placed in an incubator with saturation humidity and a volume fraction of 5% CO2. The medium was changed 24 h depending on the cell adherent condition and once every 2 d after that. Cell growth was observed under an inverted microscope, and when the confluence of chondrocytes reached 90%, they were passaged using 2.5 g/L trypsin +0.02% ethylenediamine tetraacetic acid. Chondrocytes at the 3rd to 6th passages were used for subsequent experiments. When the confluence of cells seeded in 10 cm dishes reached 80%, chondrocytes were treated with 10 ng/mL of IL-1β (HY-P7073, MedChemExpress, Princeton, NJ, USA) for 24 h to construct an in vitro OA model [37]. The reagents described above were all purchased from ThermoFisher.
Mouse chondrocytes were identified by type II collagen immunofluorescent cell staining. Specifically, the primary and passaged mouse chondrocytes were seeded on the sterile cover slide and cultured. The prepared cell slides were washed in PBS, fixed by 40 g/L paraformaldehyde for 20 min, and blocked by blocking solution for 2 h. Cell slides were incubated with primary antibodies against Collagen II (ab34712, 1:100, Abcam) at room temperature for 3–5 h, followed by incubation with fluorescent secondary antibodies in a dark chamber. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI), while chondrocytes were observed under the fluorescence microscope. PBS was a negative control (NC; Supplementary Fig. 2A).
2.6. Uptake assay
The isolated SMSC-Exo was incubated with the red fluorescent dye PKH26 (PKH26GL-1 KT, Sigma-Aldrich, St.Louis, MO, USA) for 5 min at room temperature. SMSC-Exo was washed with PBS and centrifuged at 100,000 g for 90 min, suspended in a basal medium. SMSC-Exo with a concentration of 10 μg/mL was incubated with chondrocytes at 37 °C for 24 h and then fixed by 4% paraformaldehyde. Nuclei were stained with 10 μg/mL of Hoechst 33342 (C1025, Beyotime, Nantong, China) for 10 min, and the cytoskeleton was stained with a microfilament green fluorescent probe (C1033, Beyotime) for 30 min. Chondrocytes were placed under a confocal laser scanning microscope (Zeiss LSM710, Germany) to observe the uptake of SMSC-Exo by chondrocytes.
2.7. Cell infection
Cells were overexpressed or silenced by lentiviral infection, and lentiviral packaging services were provided by Sangon Bioengineering (Shanghai, China). Plasmids of pHAGE-puro, pSPAX2, pMD2.G, pSuper-retro-puro, gag/pol, and VSVG were co-transfected into 293T cells, which were cultured for 48 h. Then, the supernatant was collected, filtered with a 0.45 μm filter, and centrifuged to collect the virus. After 72 h, the supernatant was concentrated by centrifugation, and the obtained virus was mixed to determine the titer. Cells were digested with trypsin when the chondrocytes or SMSC were in the logarithmic phase. Cells were seeded in a 6-well plate at 1 × 105 cells/well density. After routine culture for 24 h, the cell confluence reached about 75%, an appropriate amount of packaged lentivirus (MOI = 10, working titer of about 5 × 106 TU/mL), and 5 μg/mL polybrene (Merck, TR-1003, USA) were added to infect the culture media. After 4-h infection, an equal amount of medium was added to dilute polybrene, and fresh medium was replaced 24 h after infection.
SMSCs were infected with MATN3 overexpressing lentiviral vector (or-MATN3), silencing MATN3 lentivirus 1 (sh-MATN3-1), sh-MATN3-2, or the relevant NC (oe-NC and sh-NC). The sequences of the silenced lentiviruses are shown in Supplementary Table 1.
Chondrocytes were treated with PBS (control), 10 ng/mL IL-1β, lentivirus of oe-MATN3, sh-MATN3, oe-NC, sh-NC, IL-17A (10 ng/mL; HY-P73174, MedChemExpress), DMSO (treated for 48 h), mTORC1 inhibitor rapamycin (rapa; 25 μM, S1039, Selleck, Shenzhen, China), an activator of PI3K/AKT pathway LY294002 (50 μM, S1105, Selleck), alone or in combination.
IL-1β-induced chondrocytes were co-cultured with SMSC-Exo (at density of 10 μg/mL) for 24 h and assigned into IL-1β + PBS, IL-1β + SMSC-Exo, IL-1β + SMSC-Exo-oe-NC, IL-1β + SMSC-Exo-oe-MATN3, IL-1β + SMSC-Exo-sh-NC, and IL-1β + SMSC-Exo-sh-MATN3 groups.
2.8. Establishment of OA mouse model
88 C57BL/6J mice were used for this study, of which 80 mice underwent DMM surgery, and the remaining 8 underwent sham surgery as control. The DMM operation was performed as previously described [38]. Four weeks after DMM molding, 10 μL lentivirus (titer of 1 × 1011 PFU/mL, Ribobio) or 10 μL SMSC-Exo (100 μg/mL) was injected into the joint cavity for three consecutive weeks (weekly injection). Four weeks later, mice were euthanized, and knee bone and joint tissues were removed for subsequent experiments. Mice were subjected to different treatments (8 mice in each treatment) with the sham operation and DMM operation, while mice received DMM operation were subjected to intra-articular injection of lentivirus of oe-NC, oe-MATN3, sh-NC, sh-MATN3, PBS (10 μL), and SMSC-Exo (10 μL).
2.9. Hematoxylin-eosin (H&E) staining for pathological changes
Cartilage tissue sections of mouse knee were dried at room temperature, fixed at room temperature for 30 s, stained with hematoxylin for 60 s, differentiated with 1% Hydrol alcohol differentiation solution for 3 s, and stained with eosin for 3 min. Finally, under the microscope (BX63, Olympus, Japan), the morphological structure of cartilage tissues of mouse knee joints was observed, with at least three visual fields randomly selected per sample. The synovitis score was identified by H&E staining and evaluated according to the principles published in previous studies [39,40].
Immunohistochemistry.
Immunohistochemistry was performed as a previously published method [23] with primary antibody MATN3 (1:100, PA5-112604, ThermoFisher Scientific).
2.10. Toluidine blue staining for glycosaminoglycan content
Cartilage tissues from the knee joint of differently-treated mice were dehydrated with conventional gradient alcohol (70%, 80%, 95%, 95% ethanol, absolute ethanol), washed with xylene and deionized water, then stained with 0.5% toluidine blue for 30 min. Soaked in 0.5% glacial acetic acid for 5s, the sections were finally dehydrated, transparentized, and mounted. The morphological structure of the cartilage tissues of the knee was observed under a light microscope, and at least three fields were randomly selected per sample for detection. The glycosaminoglycan content in cartilage tissues was quantified by observing the blue color in toluidine blue staining.
2.11. Safranin O green staining for degradation of ECM
Paraffin-embedded sections of cartilage tissues were conventionally dewaxed to water and stain. The sections were stained with hematoxylin for about 10–30 min, washed with running water for 15 min, and faded with 1% hydrochloric acid ethanol solution for about 2–10 s (till the sections turned red, the color was lighter.) Sections were further soaked in 50%, 70%, and 80% ethanol for 5 min. Subsequently, the sections were put in 95% ethanol to wash off the excess red and in absolute ethanol for 5 min. After sealing, the sections were examined with microscopy. Images were collected and analyzed, with at least three visual fields randomly selected per sample. The OA Research Society International (OARSI) score was performed according to the OARSI score rules [41].
2.12. Autophagosomes observed under TEM
Mouse knee cartilage tissues were fixed with 2.5% glutaraldehyde, washed three times in PBS at pH 7.2, fixed in 1% osmium tetroxide, dehydrated in gradient ethanol and acetone solution, and embedded in epoxy resin. The sections were subsequently stained under the supervision of a microscope. Autophagosomes in cells from different samples were observed and imaged under TEM.
2.13. Immunofluorescence
The chondrocytes to be measured were counted and cultured in an immunofluorescence chamber with a density of 2 × 105 cells per well. When the cell confluence reached about 90%, cells were washed 3 times with pre-cooled PBS and fixed with 1 mL of 4% paraformaldehyde. Following standing for 15 min at room temperature, cells were triturated for 10 min with 0.2% Triton and then blocked by 5% BSA. Cells were then incubated with antibodies of rabbit anti-MATN3 (2.5 μg/mL, PA5-20727, Invitrogen) and mouse anti-IL-17A (1:50, sc-374218, Santa Cruz, USA) at 4 °C overnight. After 3 washes in PBS, cells were then incubated in the dark with secondary antibodies of goat anti-rabbit immunoglobulin G (IgG) H&L (Alexa Fluor® 488; ab150077, Abcam) and goat anti-mouse IgG H&L (Alexa Fluor® 647; ab150115, Abcam) for 1 h at room temperature. After 3 washes in PBS, DAPI (D9542, Sigma-Aldrich) was added into cells for staining in the void of light for 15 min. The co-localization of cells was observed and photographed under a fluorescent microscope (Olympus, Japan).
2.14. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted by Trizol (16096020, Thermo Fisher Scientific), and the RNA concentration and purity were determined by using the NanoDrop One/OneC trace nucleic acid protein concentration tester (Thermo Fisher Scientific), which showed that A260/A280 = 2.0, and the concentration was >5 μg/μL. The RNA was synthesized using the complementary DNA first-strand synthesis kit (D7168L, Beyotime, Shanghai).
The RT-qPCR experiment was performed with the kit (Q511-02, Vazyme Biotech, Nanjing, China). PCR amplification was performed on a Bio-rad real-time quantitative PCR instrument CFX96. Primer sequences (Supplementary Table 2) were designed and provided by Sangon Biotech (Shanghai, China). With glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference, the mRNA levels were determined using the 2−ΔΔCt method.
2.15. Western blot analysis
Total proteins were extracted from tissues and cells by the radioimmunoprecipitation analysis (RIPA) lysis buffer (P0013B, Beyotime) containing 1% phenylmethanesulfonyl fluoride (PMSF), and the total protein was extracted by following the instructions of protein extraction kit (P0028, Beyotime). The total protein concentration of each sample was determined using the BCA kit (P0011, Beyotime). According to the size of the target protein band, 8%–12% sodium dodecyl sulfate gels were prepared, and the protein samples were added to each lane for electrophoresis separation. The protein from the gel was transferred to a polyvinylidene fluoride membrane (1620177, BIO-RAD), and the membrane was blocked by 5% skim milk or 5% BSA for 1 h at room temperature. The primary antibodies shown in Supplementary Table 3 were added to the membrane and incubated overnight at 4 °C. After washing, the membrane was incubated with horseradish peroxide-labeled goat anti-rabbit IgG (ab6721, 1:2000, Abcam) or goat anti-mouse (ab6789, 1:2000, Abcam) secondary antibodies for l h at room temperature. The membrane was immersed in the enhanced chemiluminescence reaction solution (1705062, Bio-Rad) at room temperature for 1 min. The protein band was exposed and imaged on an Image Quant LAS 4000C gel imager (GE Appliances, Louisville, KY, USA). Total cellular and cytoplasmic proteins were determined with GAPDH as an internal reference.
2.16. Co-immunoprecipitation (Co-IP) for the interaction of MATN3 and IL-17A
Co-IP detected the interaction between MATN3 and IL-17A. Mouse chondrocytes at the logarithmic growth phase were harvested and lysed by 300 μL of cell lysis buffer (R0100, Solarbio) containing protease inhibitor (A8260, Solarbio). The supernatant was collected, 50 μL of which was taken as Input, and the remaining supernatant was supplemented with IL-17A (# 13838, 1:200, CST, Danvers, MA, USA) or IgG (# 3423, 1:20, CST) antibodies and incubated overnight at 4 °C. Next, after adding 20 μL of protein A/G-agarose pellets (36403ES03, Yeasen, USA) and shaking at 4 °C for 3 h, cells were collected, lysed, and washed 3 times, and then precipitated by boiling with 40 μL of sample loading buffer for 5 min. The pulled MATN3 protein levels were analyzed by Western blot analysis.
2.17. Enzyme-linked immunosorbent assay (ELISA) for IL-6 and TNF-α levels in cells or serum
The supernatant from chondrocyte culture medium or mouse serum was collected. The IL-6 (Mouse IL-6 ELISA Kit, PI326, Beyotime) and tumor necrosis factor-α (TNF-α; Mouse TNF-α ELISA Kit, PT512, Beyotime) levels were detected, respectively, using the ELISA kits.
2.18. Cell counting kit-8 (CCK-8) assay for mouse chondrocyte viability
The effect of IL-1β and SMSC-Exo on the viability of mouse chondrocytes was assessed using the CCK-8 kit (K1018, Apexbio, Houston, TX, USA). Cells were seeded at a density of 1 × 103 cells/well into 96-well culture plates containing 100 μL of medium replenished with 10% FBS. Cells were cultured for 48 h, then 10 μL CCK-8 solution was added to each plate well and incubated at 37 °C for an additional 4 h. The optical density (OD) values were measured at 450 nm, and the absorbance values of the cells cultured for 0 h, 12 h, 24 h, 36 h, and 48 h were recorded. Five parallel wells were set for each experiment, and the experiment was repeated three times independently. The proliferation rates were relative to the control were used to plot cell growth curves and measured using a one-way analysis of variance (ANOVA).
2.19. Statistical analysis
Statistical analyses of this study were performed using SPSS 21.0 (IBM, Armonk, NY, USA) statistical software. Measurement data were expressed as mean ± standard deviation, followed by normality and homogeneity of variance tests. For data conforming to a normal distribution and homogeneous variance, the comparison between groups was made using an unpaired t-test, while multi-group comparisons were made by one-way ANOVA with Tukey's post-hoc test. Data between groups at different time points were compared by repeated measures ANOVA, and Tukey's was used for the post hoc test. A p < 0.05 indicates a statistically significant difference.
3. Results
3.1. Bioinformatics prediction found that SMSC-Exo could deliver MATN3 and regulate the IL-17 signaling pathway to participate in OA development
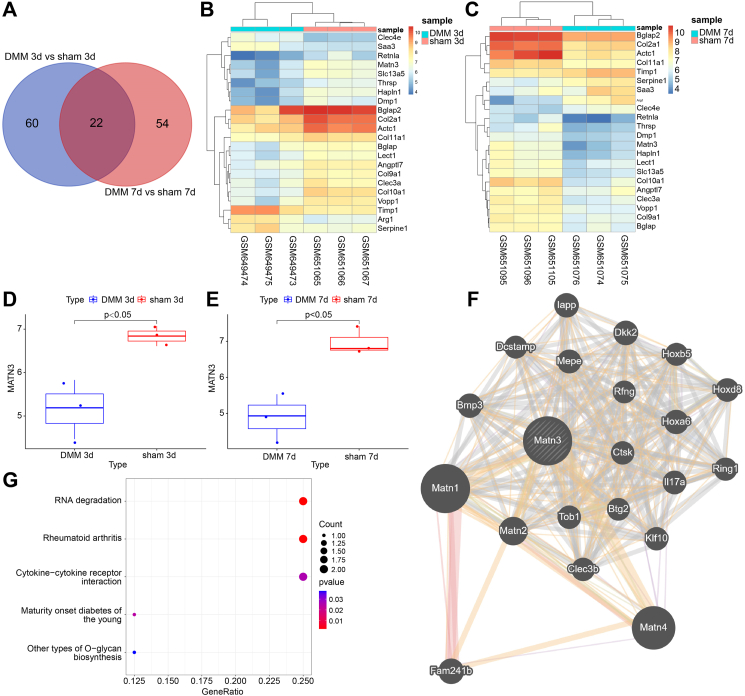
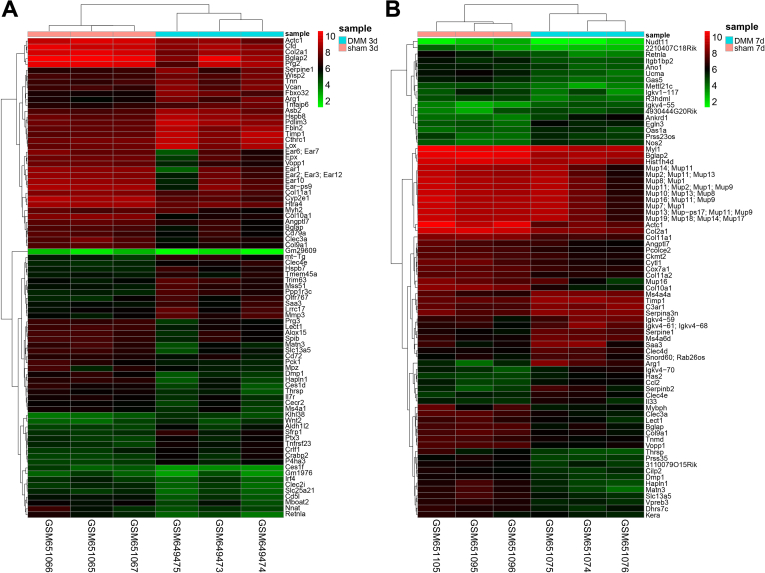
The GEO database was searched to explore the specific molecular mechanism of SMSC-Exo ameliorating OA, with the mouse OA-related microarray GSE26475 found. In that microarray, OA was induced through DMM. We then detected DEGs in cartilage tissues at 3 d and 7 d post-sham operation and DMM operation, respectively, as shown in Supplementary Figs. 1A–B, the heatmap of the DEGs at 3 d and 7 d after DMM was plotted. The intersection of DEGs yielded 22 key genes (Fig. 1A). The heatmap of these 22 intersected genes is shown in Fig. 1B and C. In addition, we found that Col10a1, Col11a1, Col2a1, Actc1, Col9a1, Clec3a, MATN3, Hapln1, Bglap, Vopp1, Retnla, Lect1, Bglap2, Thrsp, Slc13a5, Dmp1, and Angptl7 were significantly downregulated at both 3 d and 7 d after DMM operation.
Figure 1.
Bioinformatics prediction of the molecular mechanisms involving SMSC-Exo in the occurrence and development of OA. A, Venn diagram showing the intersection genes between 3 d after sham and DMM operations and 7 d after sham and DMM operations. B, heatmap showing the expression of the 22 intersection genes 3 d after the sham and DMM operations (sham 3 d n = 3, DMM 3 d n = 3). C, heatmap showing the expression of the 22 intersection genes 7 d after the sham and DMM operations (sham 7 d n = 3, DMM 7 d n = 3). D-E, Expression of MATN3 in GSE26475 at 3 d and 7 d after DMM operation. F, PPI analysis of MATN3 in the Genemania database yielded 22 interacting proteins. G, KEGG enrichment analysis of 22 interacting proteins (bubble color indicates p-value, The darker the color, the smaller the p-value; Size indicates the number of genes). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Further analysis of GSE26475 found that MATN3 expression was significantly reduced in the cartilage tissues of OA mice 3 d and 7 d after DMM (Fig. 1D and E). As existing literature documented, mice lacking MATN3 can develop mild skeletal abnormalities and are prone to age-related OA [32]. Therefore, we hypothesized that SMSC-Exo carrying MATN3 might ameliorate OA.
To further explore the specific molecular mechanism of MATN3, we used the Genemania database for PPI analysis of MATN3 with 22 interacting proteins obtained (Fig. 1F). Following KEGG enrichment analysis of these 22 proteins identified, two proteins interacting with MATN3, namely, CTSK and IL17A, were enriched in the Rheumatoid arthritis pathway (Fig. 1G). As an inflammatory factor, IL-17A is involved in the inflammatory response and promotes the development of OA [42]. Therefore, based on the above bioinformatics analysis, we speculated that SMSC-Exo could deliver MATN3 to regulate the IL-17 signaling pathway, thus participating in OA development.
3.2. Overexpression of MATN3 alleviates OA
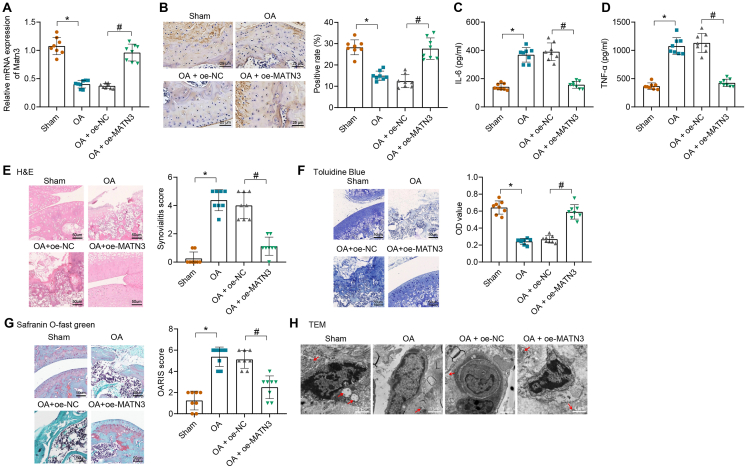
OA was induced by DMM operation, and modeled mice were injected with lentivirus with MATN3 overexpression into the joint cavity. MATN3 expression in cartilage tissues of differently-treated mice was detected by RT-qPCR, which showed that MATN3 expression was significantly reduced in OA mice compared with sham-operated mice. However, it was distinctly enhanced in OA mice injected with lentivirus overexpressing MATN3 (Fig. 2A). Immunohistochemistry revealed that OA mice showed reduced MATN3 expression while further on-MATN3 treatment led to increased one (Fig. 2B). Furthermore, the levels of inflammatory factors IL-6 and TNF-α were accumulated in the serum of OA mice compared with that of the sham-operated mice, which then significantly lessened in the serum of OA mice injected with lentivirus overexpressing MATN3 (Fig. 2C and D). In addition, H&E staining observed that sham-operated mice had smooth cartilage surfaces, healthy articular synovial membranes, neat articular cartilage edges, and well-aligned chondrocytes without the formation of a free body. However, OA mice had rough and dry cartilage surfaces, thick synovial surfaces, various cartilage cracks, poor articular cartilage boundary, messy arrangement, and loss of chondrocytes, while overexpression of MATN3 effectively alleviated these symptoms (Fig. 2E).
Figure 2.
MATN3 overexpression suppresses ECM degradation and autophagy defects in cartilage tissues to relieve OA in mice. An RT-qPCR for MATN3 expression in the cartilage tissues of each group of mice (n = 8). B, The expression of MATN3 in the cartilage tissues of mice in each group was detected by immunohistochemistry (bar = 25 μm). C-D, ELISA for IL-6 and TNF-α contents in mouse serum. E, H&E staining for the pathology of the cartilage tissues (bar = 50 μm) and the synovitis score. F, Toluidine blue staining for aggrecan content (bar = 50 μm) and semi-quantitative analysis in the cartilage tissue of mice. G, Safranin O green staining for ECM degradation (bar = 50 μm) and OARSI score of mouse cartilage tissue. H, TEM for the autophagosome in the chondrocytes of mice (bar = 1 μm). ∗ indicates p < 0.05, compared with the sham group; # indicates p < 0.05 compared with the OA + oe-NC group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, aggrecan content was reduced in OA mice, along with seriously degraded ECM and increased OARSI score. However, after MATN3 overexpression, aggrecan content was increased, along with relieved ECM degradation and decreased OARSI score (Fig. 2F and G). It is well known that autophagy increases in OA chondrocytes to protect chondrocytes from adaptive responses to various environmental changes, but autophagy decreases as the cartilage gradually degenerates [43]. Therefore, we further observed the autophagosomes in the mouse chondrocytes using TEM, which showed that the autophagosomes were decreased in OA mice compared with the sham-operated mice, while overexpression of MATN3 increased the number of autophagosomes (Fig. 2H). This histological experiment showed that MATN3 overexpression effectively inhibited the progression of OA in mice.
The above results indicate that overexpression of MATN3 can alleviate the OA progression.
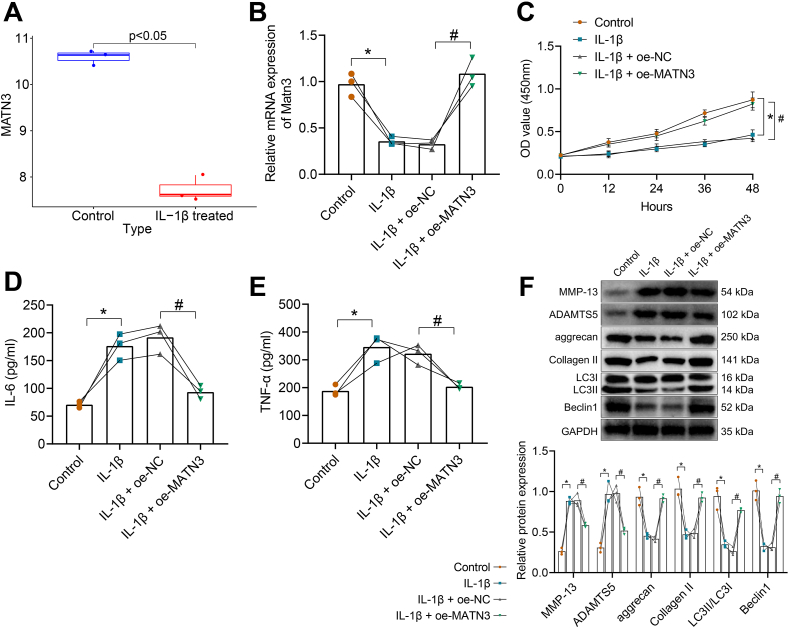
Overexpression of MATN3 inhibits IL-1β-induced ECM degradation and autophagy defects in chondrocytes.
We also found that MATN3 expression was significantly reduced in IL-1β-induced chondrocytes in the microarray GSE104793 (Fig. 3A). To further explore the role of MATN3 in OA, we isolated mouse chondrocytes (Supplementary Fig. 2A) and induced them with 10 ng/mL of IL-1β to simulate OA in vitro [37]. We subsequently overexpressed MATN3 in chondrocytes. MATN3 expression was reduced after treatment with IL-1β and was increased significantly after MATN3 overexpression (Fig. 3B). Then, CCK-8 found that the cell viability was weakened significantly after IL-1β induction, but overexpression of MATN3 led to facilitated cell viability (Fig. 3C). The expression of inflammatory factors IL-6 and TNF-α was significantly elevated in the supernatants of mouse chondrocytes after IL-1β stimulation, while they were decreased after MATN3 overexpression (Fig. 3D and E). Additionally, MMP-13 and ADAMTS5 expression was increased in the chondrocytes induced by IL-1β, but aggrecan, Collagen II, LC3II/LC3I, and Beclin1 expression was decreased, while overexpression of MATN3 led to opposite trends (Fig. 3F).
Figure 3.
MATN3 overexpression inhibits IL-1β-induced ECM degradation and autophagy defects in chondrocytes. A, Analysis of MATN3 expression in IL-1β-induced chondrocytes in microarray GSE104793 (control sample n = 3, IL-1β treated sample n = 3). B, RT-qPCR for MATN3 expression in chondrocytes. C, CCK-8 for the viability of chondrocytes in each group. D-E, ELISA for IL-6 and TNF-α levels in the supernatant of chondrocytes. F, Western blot for expression of MMP-13, ADAMTS5, aggrecan, Collagen II, LC3II/LC3I, and Beclin1 in chondrocytes. ∗ indicates p < 0.05 compared with the control group; # indicates p < 0.05 compared with the IL-1β + oe-NC group. Cell experiments were performed in triplicate.
Thus, overexpression of MATN3 inhibits IL-1β-induced ECM degradation, autophagy defects, and inflammatory responses in chondrocytes.
3.3. Mouse SMSC-Exo delivers MATN3 to repair IL-1β-induced chondrocyte damage and relieve OA
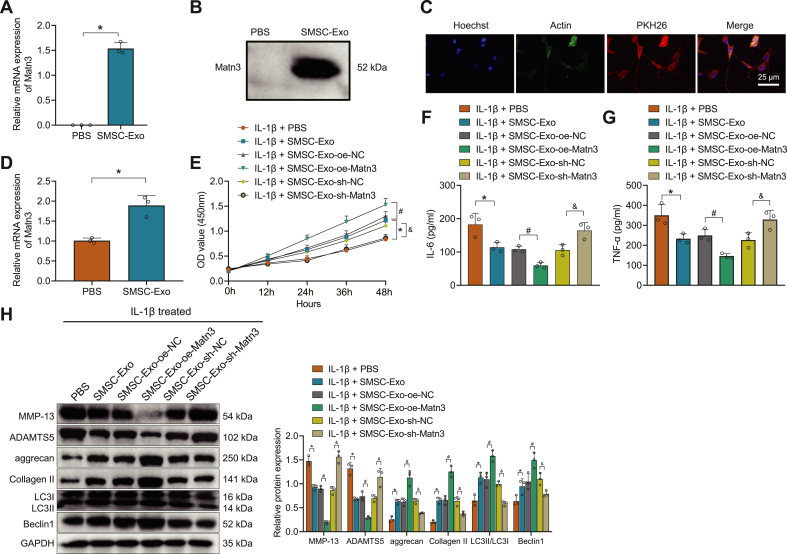
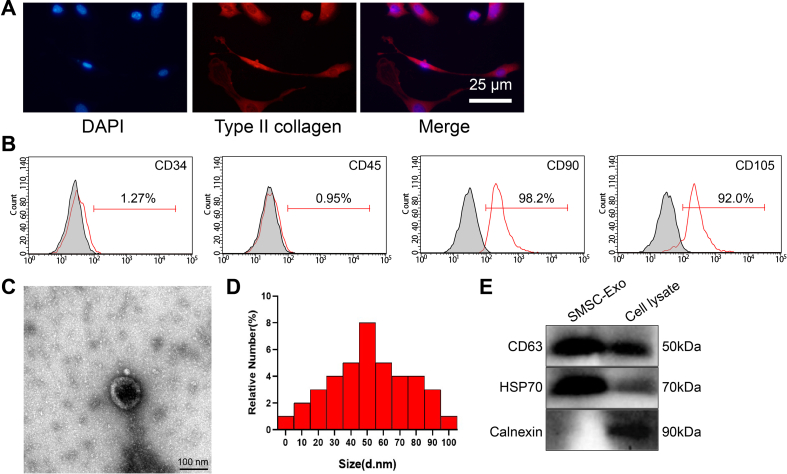
We isolated the SMSC-Exo from the mouse to observe whether it could carry MATN3 to the chondrocytes to ameliorate OA. First, we detected the surface antigen by flow cytometry, which showed positive for CD90 and CD105 but negative for CD34 and CD45, indicating the successful isolation of SMSC (Supplementary Fig. 2B). Next, SMSC-Exo was further isolated, and through TEM, DLS, and Western blot detection, SMSC-Exo had a double-layer membrane structure and showed a tea-ware tray shape, with a diameter between 30 and 100 nm, and Exo highly expressed CD63 and HSP70 but no Calnexin (Supplementary Figs. 2C–E). Detection of the RNA and protein levels of MATN3 showed that a higher expression of MATN3 was found in SMSC-Exo (Fig. 4A and B). After co-culture of PKH26-labeled SMSC-Exo with chondrocytes for 24 h, SMSC-Exo could be internalized by chondrocytes (Fig. 4C). MATN3 expression was further studied in the chondrocytes after co-culture with SMSC-Exo, and the results showed that MATN3 expression increased significantly in the chondrocytes after the co-culture (Fig. 4D).
Figure 4.
SMSC-Exo delivers MATN3 to repair IL-1β-induced chondrocyte injury to relieve OA. A, RT-qPCR for MATN3 expression in isolated SMSC-Exo. B, Western blot for the protein level of MATN3 in the isolated SMSC-Exo. C, Fluorescence microscopy for SMSC-Exo labeled by PKH26 to observe the internalization of chondrocytes (bar = 25 μm). D, RT-qPCR for MATN3 expression in chondrocytes co-cultured with SMSC-Exo. E, CCK-8 for the viability of chondrocytes. F-G, ELISA for IL-6 and TNF-α contents in chondrocytes after co-culture. H, Western blot for the expression of MMP-13, ADAMTS5, aggrecan, Collagen II, LC3II/LC3I, and Beclin1 in the chondrocytes. ∗ indicates p < 0.05 compared with the IL-1β + PBS group; # indicates p < 0.05 compared with the IL-1β + SMSC-Exo-oe-NC group; & indicates p < 0.05 compared with the IL-1β + SMSC-Exo-sh-NC group. ∗ indicates p < 0.05 compared with the PBS group. Cell experiments were performed in triplicate.
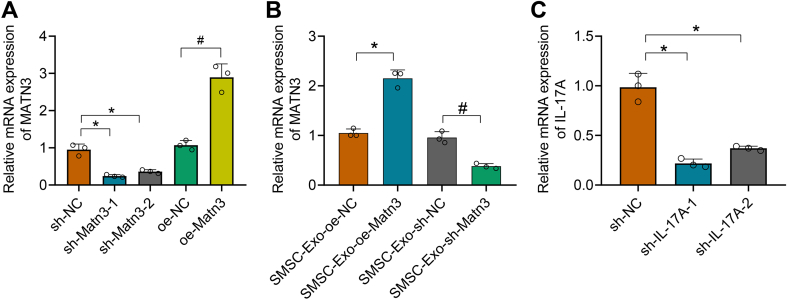
To further determine that SMSC-Exo acts by delivering MATN3 to chondrocytes, we infected SMSC with lentivirus of sh-MATN3 or oe-MATN3 for knockdown or overexpression. As shown in Supplementary Fig. 3A, sh-MATN3-1 showed the best silencing effect, which was chosen for further experiments. Meanwhile, Exo was extracted from differently-treated SMSC, and expression of MATN3 in Exo was detected (Supplementary Fig. 3B).
After co-culture of SMSC-Exo with chondrocytes, IL-1β was further added. Through CCK-8 (Fig. 4E), ELISA (Fig. 4F and G), and Western blot (Fig. 4H), we found that the chondrocyte viability was significantly strengthened after the addition of SMSC-Exo, while the release of IL-6 and TNF-α was significantly reduced, along with downregulated expression of ECM proteins MMP-13 and ADAMTS5, as well as up-regulated expression of aggrecan, Collagen II, LC3II/LC3I, and Beclin1. Moreover, after IL-1β + SMSC-Exo-or-MATN3, the chondrocyte viability was further strengthened, along with lower levels of IL-6 and TNF-α, MMP-13, and ADAMTS5, but higher levels of aggrecan, Collagen II, LC3II/LC3I, and Beclin1; however, IL-1β + SMSC-Exo-sh-MATN3 led to opposite trends. The above experiments demonstrate that SMSC-Exo can play a role in OA by delivering MATN3 to the chondrocytes.
3.3.1. SMSC-Exo delivers MATN3 to improve OA symptoms
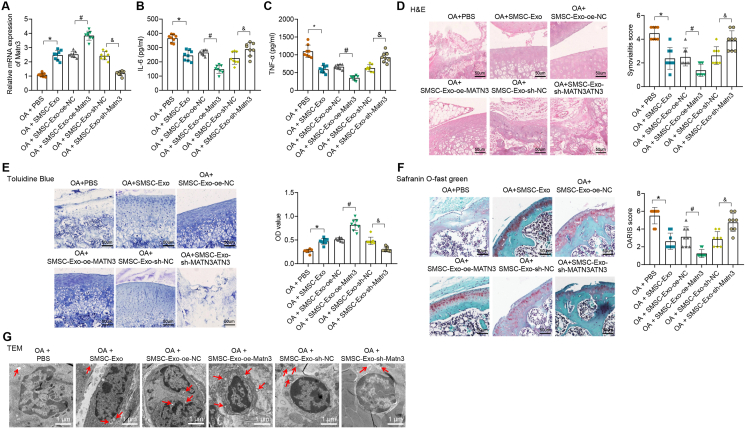
To further validate our findings, we injected the differently-treated SMSC-Exo into the joint cavity of the OA mice to observe the effect of SMSC-Exo-MATN3 on OA. MATN3 expression was increased in the OA mice treated with SMSC-Exo, which was then significantly reduced after SMSC-Exo-sh-MATN3 (Fig. 5A). In addition, levels of IL-6 and TNF-α were decreased in the serum of OA mice treated with SMSC-Exo, which were further decreased after SMSC-Exo-or-MATN3, but then increased after SMSC-Exo-sh-MATN3 (Fig. 5B and C).
Figure 5.
MATN3 delivered by mouse SMSC-Exo relieves OA symptoms in mice. A, RT-qPCR for MATN3 expression in the cartilage tissues of differently-treated mice (n = 8). B-C, ELISA for inflammatory factor IL-6 and TNF-α contents in mouse serum. D, H&E staining for the pathology of the cartilage tissue (bar = 50 μm) and the synovitis score. E, Toluidine blue staining for aggrecan content (bar = 50 μm) and semi-quantitative analysis in the cartilage tissues of mice. F, Safranin O green staining for ECM degradation (bar = 50 μm) and the OARSI score. G, TEM for autophagosome (bar = 1 μm). ∗ indicates p < 0.05 compared with the OA + PBS group; # indicates p < 0.05 compared with the OA + SMSC-Exo-oe-NC group; & indicates p < 0.05 compared with the OA + SMSC-Exo-sh-NC group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The subsequent H&E (Fig. 5D), toluidine blue (Fig. 5E), and Safranin O green (Fig. 5F) staining results showed that after SMSC-Exo injection, the histopathological conditions of cartilages in mice were bettered. Moreover, in OA mice injected with SMSC-Exo-or-MATN3, aggrecan content was increased, accompanied by relieved ECM degradation, which showed the best treatment effect for OA. However, in OA mice injected with SMSC-Exo-sh-MATN3, aggrecan content was lessened, while ECM was severely degraded, which showed the worst treatment effect for OA. The observation of autophagosomes by TEM showed that the number of autophagosomes in cartilage tissues increased in OA mice injected with SMSC-Exo, which was further increased after injection of SMSC-Exo-or-MATN3. However, the number of autophagosomes in cartilage tissues was reduced in OA mice injected with SMSC-Exo-sh-MATN3 (Fig. 5G).
These experiments show that mouse SMSC-Exo can repair IL-1β-induced chondrocyte damage and relieve OA by delivering MATN3.
3.4. MATN3 inhibits the release of inflammatory cytokines, ECM degradation, and autophagy defects by interacting with IL-17A
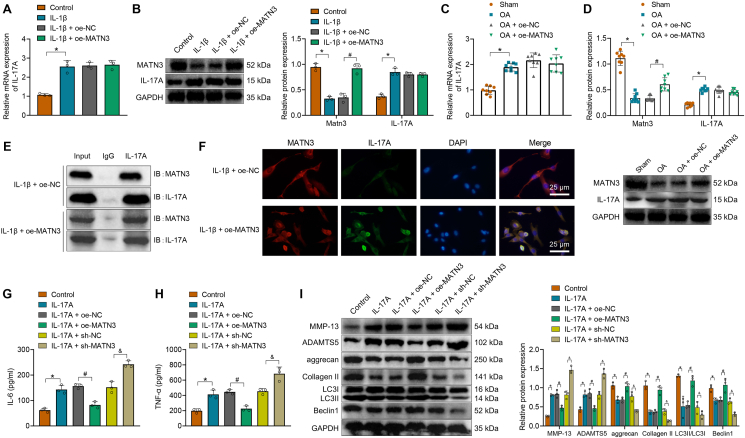
Previously, our bioinformatics studies showed that MATN3 might participate in the occurrence and development of OA through interaction with IL-17A. Therefore, we first examined IL-17A expression in chondrocytes and found that IL-17A expression was elevated in chondrocytes induced by IL-1β, which did not significantly change after adding lentivirus of oe-MATN3 (Fig. 6A and B). In addition, IL-17A expression in the cartilage tissues of mice also increased compared with sham-operated mice, which also did not significantly change after injection of lentivirus of oe-MATN3 (Fig. 6C and D).
Figure 6.
MATN3 inhibits IL-1β-induced ECM degradation and autophagy defects in chondrocytes through interaction with IL-17A. A, RT-qPCR for IL-17A expression in chondrocytes. B, Western blot for IL-17A and MATN3 protein levels in chondrocytes. C, RT-qPCR for mRNA levels of IL-17A in the cartilage tissues of mice. D, Western blot for IL-17A and MATN3 protein levels in the cartilage tissue of mice. E, Co-IP experiment for the interaction between IL-17A and MATN3. F, Immunofluorescence for the co-localization of IL-17 and MATN3 in chondrocytes (bar = 25 μm). G-H, ELISA for the IL-6 and TNF-α content in the supernatants of the chondrocytes. I, Western blot for expression of MMP-13, ADAMTS5, aggrecan, Collagen II, LC3II/LC3I, and Beclin1 in chondrocytes. ∗ indicates p < 0.05 compared with the control group/sham; # indicates p < 0.05 compared with the IL-17A + oe-NC/OA + oe-NC group; & indicates p < 0.05 compared with the IL-17A + sh-NC group. Cell experiments were performed in triplicate.
Therefore, we further observed whether there was an interaction between MATN3 and IL-17A through Co-IP experiments. The results showed that after stimulating chondrocytes with IL-1β, a large amount of MATN3 was pulled down by IL-17A antibody when MATN3 was overexpressed, indicating a significant interaction between the MATN3 and IL-17A (Fig. 6E). Immunofluorescence further revealed a significant co-localization between MATN3 and IL-17A, which was more apparent in chondrocytes treated with IL-1β and lentivirus of oe-MATN3 (Fig. 6F).
To determine whether MATN3 alleviated OA by interacting with IL-17A, chondrocytes were induced with 10 ng/mL IL-17A. Downstream IL-6 and TNF-α expression were significantly increased, while overexpression of MATN3 could reduce IL-6 and TNF-α expression. The silencing of MATN3 caused opposite trends (Fig. 6G and H). In addition, ECM and autophagy-related proteins in chondrocytes were determined. Results showed that MMP-13 and ADAMTS5 levels were significantly increased while aggrecan, Collagen II, LC3II/LC3I, and Beclin1 levels were decreased in chondrocytes induced with IL-17A. However, after overexpression of MATN3, MMP-13, and ADAMTS5, levels were decreased while aggrecan, Collagen II, LC3II/LC3I, and Beclin1 levels were increased. Silencing of MATN3 enhanced levels of MMP-13 and ADAMTS5 but decreased levels of aggrecan, Collagen II, LC3II/LC3I, and Beclin1 (Fig. 6I).
The above experiments show that MATN3 inhibits the release of downstream inflammatory cytokines, ECM degradation, and autophagy defects by interacting with IL-17A.
3.5. MATN3 inhibits IL-17A-induced activation of the PI3K/AKT/mTOR signaling axis, thereby inhibiting IL-1β-induced ECM degradation and autophagy defects in chondrocytes
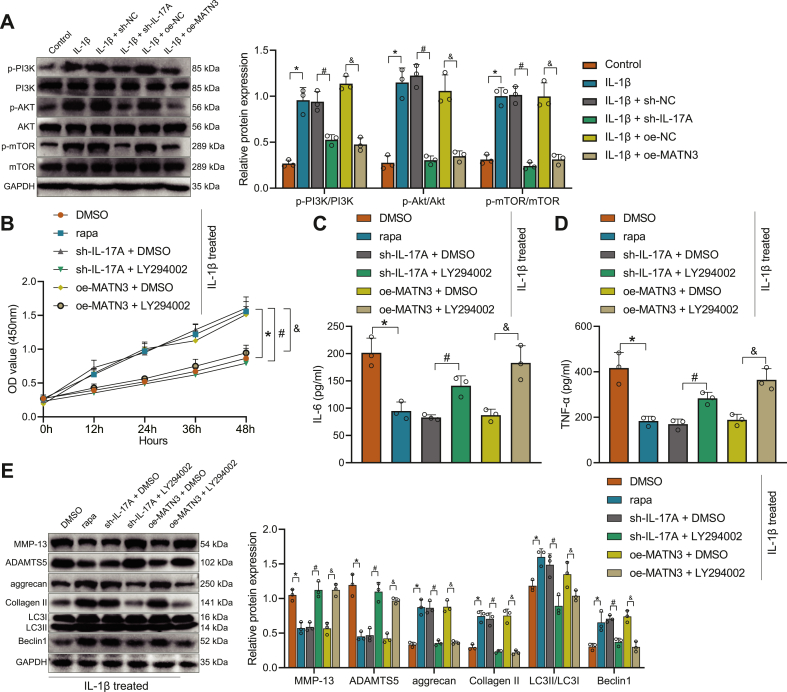
IL-17A can activate the PI3K/AKT/mTOR pathway to inhibit autophagy [44], so we next investigated whether MATN3 can regulate the activation of the PI3K/AKT/mTOR signaling axis by IL-17A to promote autophagy in OA chondrocytes. We transfected chondrocytes with lentivirus of sh-IL-17A to achieve the effect of silencing IL-17A expression and performed subsequent experiments with the sh-IL-17A-1 sequence (Supplementary Fig. 3C). As shown in Fig. 7A, the phosphorylation levels of PI3K, Akt, and mTOR were distinctively increased after IL-1β induction, while treatment with sh-IL-17A or oe-MATN3 significantly inhibited the phosphorylation of PI3K/AKT/mTOR pathway induced by IL-1β.
Figure 7.
MATN3 suppresses IL-1β-induced ECM degradation and autophagy defects in chondrocytes via the IL-17A/PI3K/AKT/mTOR axis. A Western blot for the protein levels of PI3K, AKT, and mTOR and phosphorylation levels of PI3K, AKT, and mTOR in cells. B, CCK-8 for the viability of chondrocytes. C-D, ELISA for IL-6 and TNF-α contents in the supernatants of chondrocytes. E, Western blot for the expression levels of MMP-13, ADAMTS5, aggrecan, Collagen II, LC3II/LC3I, and Beclin1 in chondrocytes. ∗ indicates p < 0.05 compared with control/DMSO group; # indicates p < 0.05 compared with the IL-1β + sh-NC/IL-1β + sh-IL-17A + DMSO group; & indicates p < 0.05 compared with the IL-1β + oe-NC/IL-1β + oe-MATN3 + DMSO group. Cell experiments were performed in triplicate.
Subsequently, we treated chondrocytes with LY294002 and rapa. CCK-8 (Fig. 7B), ELISA (Fig. 7C and D), and Western blot (Fig. 7E) experiments found that chondrocyte viability was strengthened by treatment of IL-1β and rapa, while IL-6 and TNF-α content was reduced, along with reduced expression of MMP-13 and ADAMTS5 but increased expression of aggrecan, Collagen II, LC3II/LC3I, and Beclin1. However, relative to IL-1β-induced chondrocytes treated with sh-IL-17A or oe-MATN3, further LY294002 treatment weakened cell viability, elevated IL-6 and TNF-α content, and expression of MMP-13 and ADAMTS5 but decreased expression of aggrecan, Collagen II, LC3II/LC3I, and Beclin1.
Collectively, MATN3 can inhibit the activation of the PI3K/AKT/mTOR signaling axis induced by IL-17A, thus inhibiting IL-1β-induced ECM degradation and autophagy defects in chondrocytes.
4. Discussion
SMSCs show high multi-lineage differentiation and self-renewal potentials, a good source for stem cell treatment applications on diseases, including OA [45,46]. Moreover, the significance of MSC-based therapies has been highly reported, and MSC exosome as a cell-free MSC therapy for OA also possesses great potential [47]. The current study was conducted with the central objective of the function of SMSC-Exo in chondrocytes during OA and the molecular mechanism. Following in vitro and in vivo models, our study demonstrated that SMSC-Exo-MATN3 prevented ECM degradation and autophagy defects from inhibiting OA progression associated with the IL-17A/PI3K/AKT/mTOR axis.
Our bioinformatics analyses found that MATN3 was involved in the mechanism by which SMSC-Exo affects OA. The promising potential of SMSC-Exos in preventing OA through the delivery of miR-140-5p has been proved previously [48]. We further verified the effects of MATN3 on OA and found that MATN3 was poorly expressed in OA-modeled mice, which led to the assumption that the absence of MATN3 may cause cartilage destruction and aggravate OA. In line with our findings, Jayasuriya CT et al. proposed that MATN3 possessed chondroprotective properties and that deletion of MATN3 could result in the onset of OA [49]. Further experiments revealed that overexpressed MATN3 decreased the content of IL-6 and TNF-α in serum, increased the expression of aggrecan, and inhibited autophagy defects. Autophagy defects are an essential cellular homeostasis mechanism observed in OA articular cartilage, and autophagosome formation is linked to autophagy defects [50]. MATN3, a non-collagenous, cartilage-specific ECM protein, can facilitate the microenvironment of the nanofibrous poly(l-lactic acid) scaffold for cartilage regeneration and keep cartilage property [51].
Similarly, recombinant human MATN3 protein increased gene expression of ACAN to exert anti-inflammatory and chondroprotective potentials in OA [49]. Conforming to our prediction, a previous study reported that MATN3 was an ECM protein during the chondrogenesis of mesenchymal fibroblasts from synovium [52]. Moreover, urine-derived stem cell-Exo was rich in MATN3 protein, and exosomal MATN3 was responsible for Exo-triggered ECM synthesis and treatment of intervertebral disc degeneration [18]. However, limited prior work detected the delivery of MATN3 by SMSC-Exo, which was one of the highlights of our current study. Our further bioinformatics analyses predicted that MATN3 might exert protective effects on cartilage by interacting with IL-17A. The most important factor controlling OA is inflammatory cytokines, including IL-17 [27]. A prior work stated that IL-17A was highly expressed in the serum of patients with knee OA [53], which aligns with our experimental result.
Additionally, IL-17 can induce fibroblasts and chondrocytes in OA to accelerate the release of cytokines such as TNF-α, IL-6, and IL-1β and further induce the generation of cartilage-destroying factors and facilitate synovial infiltration [54]. The alleviatory role of MATN3 in ECM degradation of nucleus pulposus cells through modulation of IL-1 receptor antagonist has been validated [55], while its interaction with IL-17A remains further investigated. Moreover, the regulatory effects of MATN3 interacting with IL-17A on OA were achieved through the PI3K/AKT/mTOR signaling. It has been reported that IL-17A can activate PI3K/AKT/mTOR pathway to inhibit autophagy [44]. As recently highlighted, the ciRS-7/miR-7 axis may relieve cartilage degradation and the defection of autophagy by IL-17A-mediated PI3K/AKT/mTOR activation in IL-1β-induced chondrocytes [35], which was partly in line with our finding.
Based on the in vitro and in vivo OA models, our study demonstrated that injection of SMSC-Exo-MATN3 reduced inflammation, and relieved cartilage damage. Furthermore, it prevented ECM degradation and autophagy defects, thus inhibiting OA progression in association with the IL-17A/PI3K/AKT/mTOR pathway. These results further suggested that SMSC-Exo is a candidate therapy for the treatment of OA, and MATN3 aided in the effects of relieving OA mediated by SMSC-Exo.
The limitations of this study mainly include the following aspects. Firstly, in exploring the mechanism by which SMSC-Exo improves the occurrence and development of mouse knee OA by transferring MATN3, this study did not focus on two important signal pathways and influencing factors, PI3K/AKT/mTOR and autophagy, in OA. Secondly, in the in vivo experiments, this study used DMM surgery to construct an OA mouse model. Furthermore, it injected SMSC-Exo for treatment but did not include the corresponding control group, which lacks certain comparisons and verification. Thus, the results need to be further confirmed through future studies. In addition, in the cell experiments, this study used IL-1β to induce mouse chondrocytes to construct an in vitro OA model and treated them with SMSC-Exo, but did not explore the effect of SMSC-Exo on OA models caused by other factors. Finally, although this study found that the interaction of MATN3 and IL-17A can inhibit the release of downstream factors and inhibit their activation of the PI3K/AKT/mTOR signaling axis, the specific details of this interaction mechanism were not further elucidated. Therefore, these limitations must be fully considered and further confirmed in future studies.
Author contributions
LL and YXS designed the study. GYZ, YC and FL collated the data, carried out data analyses and produced the initial draft of the manuscript. LL, YXS and HW contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Funding
Not applicable.
Ethics statement
The animal experimental processes were approved by the Ethnic Committee of the Second Affiliated Hospital of Soochow University and conducted in strict accordance to the standard of the Guide for the Care and Use of Laboratory Animals published by the Ministry of Science and Technology of the People‘s Republic of China in 2006.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2023.06.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1.
Heatmap of the DEGs in microarray GSE26475.
A, Heatmap of DEGs in cartilage tissue 3 d after DMM and sham operation in GSE26475 (the upper right histogram is color scale, Red color indicates the high expression, while Green indicates low expression) (sham 3 d n = 3, DMM 3 d n = 3). B, Heatmap of DEGs in cartilage tissues 7 d after DMM and sham operations in GSE26475 (sham 7 d n = 3, DMM 7 d n = 3).
Supplementary Figure 2.
Identification of chondrocytes, SMSC, and SMSC-Exo.
A, expression of chondrocyte collagen in primary cultured mice. The collagen secreted by cultured chondrocytes is red under a fluorescence microscope and blue in the nucleus, showing typical chondrocyte morphology (bar = 25 μm). B, Flow cytometry for SMSC surface antigen. C, TEM for SMSC-Exo morphology. D, DLS for SMSC-Exo particle diameter. E, Western blot for CD63, HSP70, and Calnexin expression in SMSC-Exo. Cell experiments were performed in triplicate.
Supplementary Figure 3.
Test results for the efficiency of lentiviral infection.
A-B, RT-qPCR for mRNA levels of MATN3 in each group of SMSC and SMSC-Exo. C, RT-qPCR for mRNA level of IL-17A in each group of chondrocytes.∗ indicates p < 0.05 compared with the sh-NC group; # indicates p < 0.05 compared with the oe-NC group. ∗∗ indicates p < 0.01. Cell experiments were performed in triplicate.
References
- 1.van den Bosch M.H.J. Osteoarthritis year in review 2020: biology. Osteoarthritis Cartilage. 2021;29(2):143–150. doi: 10.1016/j.joca.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Katz J.N., Arant K.R., Loeser R.F. Diagnosis and treatment of hip and knee osteoarthritis: a review. JAMA. 2021;325(6):568–578. doi: 10.1001/jama.2020.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang S., Lee K., Ju J.H. Recent updates of diagnosis, pathophysiology, and treatment on osteoarthritis of the knee. Int J Mol Sci. 2021;22(5) doi: 10.3390/ijms22052619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng Z., Sun H., Bunpetch V., Koh Y., Wen Y., Wu D., et al. The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration. Biomaterials. 2021 doi: 10.1016/j.biomaterials.2020.120555. [DOI] [PubMed] [Google Scholar]
- 5.Huang R.Z., Zheng J., Liu F.L., Li Q.L., Huang W.H., Zhang D.M., et al. A novel autophagy-related marker for improved differential diagnosis of rheumatoid arthritis and osteoarthritis. Front Genet. 2021 doi: 10.3389/fgene.2021.743560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong H., Wang X.Q., Zhang X.A. Exercise for osteoarthritis: a literature review of pathology and mechanism. Front Aging Neurosci. 2022 doi: 10.3389/fnagi.2022.854026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Zheng J.J.Y., Li G., Yuan J., Ebert J.R., Li H., et al. Pathogenesis and clinical management of obesity-related knee osteoarthritis: impact of mechanical loading. J. Orthop. Translat. 2020;24:66–75. doi: 10.1016/j.jot.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mianehsaz E., Mirzaei H.R., Mahjoubin-Tehran M., Rezaee A., Sahebnasagh R., Pourhanifeh M.H., et al. Mesenchymal stem cell-derived exosomes: a new therapeutic approach to osteoarthritis? Stem Cell Res Ther. 2019;10(1):340. doi: 10.1186/s13287-019-1445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y., Zhang J., Xu H., Lin Z., Chang H., Liu W., et al. Mesenchymal stem cells in knee osteoarthritis treatment: a systematic review and meta-analysis. J Orthop Translat. 2020;24:121–130. doi: 10.1016/j.jot.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu R., Li H., Sun C., Liu J., Chen D., Yu H., et al. Exosome-based strategy for degenerative disease in orthopedics: recent progress and perspectives. J Orthop Translat. 2022;36:8–17. doi: 10.1016/j.jot.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vonk L.A., van Dooremalen S.F.J., Liv N., Klumperman J., Coffer P.J., Saris D.B.F., et al. Mesenchymal stromal/stem cell-derived extracellular vesicles promote human cartilage regeneration in vitro. Theranostics. 2018;8(4):906–920. doi: 10.7150/thno.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L., Liu G., Wu X. Exosomes derived from umbilical cord mesenchymal stem cells in mechanical environment show improved osteochondral activity via upregulation of LncRNA H19. J Orthop Translat. 2021;26:111–120. doi: 10.1016/j.jot.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan A., Shen K., Li B., Li C., Zhou H., Kong R., et al. Extracellular vesicles derived from LPS-preconditioned human synovial mesenchymal stem cells inhibit extracellular matrix degradation and prevent osteoarthritis of the knee in a mouse model. Stem Cell Res Ther. 2021;12(1):427. doi: 10.1186/s13287-021-02507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T.S., Lai R.C., Lee M.M., Choo A.B., Lee C.N., Lim S.K. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38(1):215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Qi G., Yan Y., Wang C., Wang Z., Jiang C., et al. Exosomes derived from bone marrow mesenchymal stem cells pretreated with decellularized extracellular matrix enhance the alleviation of osteoarthritis through miR-3473b/phosphatase and tensin homolog axis. J Gene Med. 2023 doi: 10.1002/jgm.3510. [DOI] [PubMed] [Google Scholar]
- 16.Wang R., Xu B. TGFbeta1-modified MSC-derived exosome attenuates osteoarthritis by inhibiting PDGF-BB secretion and H-type vessel activity in the subchondral bone. Acta Histochem. 2022;124(7) doi: 10.1016/j.acthis.2022.151933. [DOI] [PubMed] [Google Scholar]
- 17.Qiu M., Liu D., Fu Q. MiR-129-5p shuttled by human synovial mesenchymal stem cell-derived exosomes relieves IL-1beta induced osteoarthritis via targeting HMGB1. Life Sci. 2021;269 doi: 10.1016/j.lfs.2020.118987. [DOI] [PubMed] [Google Scholar]
- 18.Guo Z., Su W., Zhou R., Zhang G., Yang S., Wu X., et al. Exosomal MATN3 of urine-derived stem cells ameliorates intervertebral disc degeneration by antisenescence effects and promotes NPC proliferation and ECM synthesis by activating TGF-beta. Oxid Med Cell Longev. 2021 doi: 10.1155/2021/5542241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P., Fleischhauer L., Nicolae C., Prein C., Farkas Z., Saller M.M., et al. Mice lacking the Matrilin family of extracellular matrix proteins develop mild skeletal abnormalities and are susceptible to age-associated osteoarthritis. Int J Mol Sci. 2020;21(2) doi: 10.3390/ijms21020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Trehan S.K., Guan Y., Sun C., Moore D.C., Jayasuriya C.T., et al. Matrilin-3 inhibits chondrocyte hypertrophy as a bone morphogenetic protein-2 antagonist. J Biol Chem. 2014;289(50):34768–34779. doi: 10.1074/jbc.M114.583104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimpen J.Y., Baldwin M.J., Cribbs A.P., Philpott M., Carr A.J., Dakin S.G., et al. Interleukin-17A causes osteoarthritis-like transcriptional changes in human osteoarthritis-derived chondrocytes and synovial fibroblasts in vitro. Front Immunol. 2021 doi: 10.3389/fimmu.2021.676173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldring M.B., Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M., Chen C., Wang K., Chen Y., Xia J. Astilbin influences the progression of osteoarthritis in rats by down-regulation of PGE-2 expression via the NF-kappaB pathway. Ann Transl Med. 2020;8(12):766. doi: 10.21037/atm-20-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pozgan U., Caglic D., Rozman B., Nagase H., Turk V., Turk B. Expression and activity profiling of selected cysteine cathepsins and matrix metalloproteinases in synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Biol Chem. 2010;391(5):571–579. doi: 10.1515/BC.2010.035. [DOI] [PubMed] [Google Scholar]
- 25.Shen J., Abu-Amer Y., O'Keefe R.J., McAlinden A. Inflammation and epigenetic regulation in osteoarthritis. Connect Tissue Res. 2017;58(1):49–63. doi: 10.1080/03008207.2016.1208655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene M.A., Loeser R.F. Aging-related inflammation in osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1966–1971. doi: 10.1016/j.joca.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wojdasiewicz P., Poniatowski L.A., Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediat Inflamm. 2014;2014 doi: 10.1155/2014/561459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari M.Y., Ahmad N., Haqqi T.M. Oxidative stress and inflammation in osteoarthritis pathogenesis: role of polyphenols. Biomed Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Rong Y., Luo C., Cui W. Bone marrow mesenchymal stem cell-derived exosomes prevent osteoarthritis by regulating synovial macrophage polarization. Aging (Albany NY) 2020;12(24):25138–25152. doi: 10.18632/aging.104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu Y.S., Bamodu O.A., Fong I.H., Lee W.H., Lin C.C., Lu C.H., et al. The JAK inhibitor Tofacitinib inhibits structural damage in osteoarthritis by modulating JAK1/TNF-alpha/IL-6 signaling through Mir-149-5p. Bone. 2021;151 doi: 10.1016/j.bone.2021.116024. [DOI] [PubMed] [Google Scholar]
- 31.Yang X., Guo H., Ye W., Yang L., He C. Pulsed electromagnetic field attenuates osteoarthritis progression in a murine destabilization-induced model through inhibition of TNF-alpha and IL-6 signaling. Cartilage. 2021;13(2_suppl):1665S. doi: 10.1177/19476035211049561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P. Diagnostic value of combined serum IL-6, TNF-alpha, and leptin levels in patients with post-traumatic osteoarthritis. Clin Lab. 2020;66(10) doi: 10.7754/Clin.Lab.2020.200229. [DOI] [PubMed] [Google Scholar]
- 33.He J., He J. Baicalin mitigated IL-1beta-Induced osteoarthritis chondrocytes damage through activating mitophagy. Chem Biol Drug Des. 2023;101(6):1322–1334. doi: 10.1111/cbdd.14215. [DOI] [PubMed] [Google Scholar]
- 34.Lu R., He Z., Zhang W., Wang Y., Cheng P., Lv Z., et al. Oroxin B alleviates osteoarthritis through anti-inflammation and inhibition of PI3K/AKT/mTOR signaling pathway and enhancement of autophagy. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.1060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., Li J., Zhou Y., Yang Z., Yang H., Li D., et al. Down-regulated ciRS-7/up-regulated miR-7 axis aggravated cartilage degradation and autophagy defection by PI3K/AKT/mTOR activation mediated by IL-17A in osteoarthritis. Aging (Albany NY) 2020;12(20):20163–20183. doi: 10.18632/aging.103731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy T., Banang-Mbeumi S., Boateng S.T., Ruiz E.M., Chamcheu R.N., Kang L., et al. Dual targeting of mTOR/IL-17A and autophagy by fisetin alleviates psoriasis-like skin inflammation. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.1075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L., He T., Xing J., Zhou Q., Fan L., Liu C., et al. Bone marrow mesenchymal stem cell-derived exosomes protect cartilage damage and relieve knee osteoarthritis pain in a rat model of osteoarthritis. Stem Cell Res Ther. 2020;11(1):276. doi: 10.1186/s13287-020-01781-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Li Z., Pi Y., Chen Y., Mei L., Luo Y., et al. MicroRNA-375 exacerbates knee osteoarthritis by repressing chondrocyte autophagy by targeting ATG2B. Aging (Albany NY) 2020;12(8):7248–7261. doi: 10.18632/aging.103073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Q., Kong N., Liu X., Tian R., Jiao M., Li Y., et al. Pirfenidone attenuates synovial fibrosis and postpones the progression of osteoarthritis by anti-fibrotic and anti-inflammatory properties in vivo and in vitro. J Transl Med. 2021;19(1):157. doi: 10.1186/s12967-021-02823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krenn V., Morawietz L., Burmester G.R., Kinne R.W., Mueller-Ladner U., Muller B., et al. Synovitis score: discrimination between chronic low-grade and high-grade synovitis. Histopathology. 2006;49(4):358–364. doi: 10.1111/j.1365-2559.2006.02508.x. [DOI] [PubMed] [Google Scholar]
- 41.Glasson S.S., Chambers M.G., Van Den Berg W.B., Little C.B. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage. 2010;18(Suppl):17–23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 42.Huang T., Wang J., Zhou Y., Zhao Y., Hang D., Cao Y. LncRNA CASC2 is up-regulated in osteoarthritis and participates in the regulation of IL-17 expression and chondrocyte proliferation and apoptosis. Biosci Rep. 2019;39(5) doi: 10.1042/BSR20182454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y.S., Zhang F.J., Zeng C., Luo W., Xiao W.F., Gao S.G., et al. Autophagy in osteoarthritis. Joint Bone Spine. 2016;83(2):143–148. doi: 10.1016/j.jbspin.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Varshney P., Saini N. PI3K/AKT/mTOR activation and autophagy inhibition plays a key role in increased cholesterol during IL-17A mediated inflammatory response in psoriasis. Biochim Biophys Acta, Mol Basis Dis. 2018;1864(5 Pt A):1795–1803. doi: 10.1016/j.bbadis.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima H., Uchida S., Hatakeyama A., Murata Y., Yamanaka Y., Tsukamoto M., et al. Isolation and characterization of synovial mesenchymal stem cells derived from patients with chronic lateral ankle instability: a comparative analysis of synovial fluid, adipose synovium, and fibrous synovium of the ankle joint. Orthop J Sports Med. 2022;10(5) doi: 10.1177/23259671221094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaguchi Y., Sekiya I., Yagishita K., Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52(8):2521–2529. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- 47.Toh W.S., Lai R.C., Hui J.H.P., Lim S.K. MSC exosome as a cell-free MSC therapy for cartilage regeneration: implications for osteoarthritis treatment. Semin Cell Dev Biol. 2017:6756–6764. doi: 10.1016/j.semcdb.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Tao S.C., Yuan T., Zhang Y.L., Yin W.J., Guo S.C., Zhang C.Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7(1):180–195. doi: 10.7150/thno.17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayasuriya C.T., Goldring M.B., Terek R., Chen Q. Matrilin-3 induction of IL-1 receptor antagonist is required for up-regulating collagen II and aggrecan and down-regulating ADAMTS-5 gene expression. Arthritis Res Ther. 2012;14(5):R197. doi: 10.1186/ar4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez de Figueroa P., Lotz M.K., Blanco F.J., Carames B. Autophagy activation and protection from mitochondrial dysfunction in human chondrocytes. Arthritis Rheumatol. 2015;67(4):966–976. doi: 10.1002/art.39025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Q., Wang J., Chen Y., Zhang Z., Saunders L., Schipani E., et al. Suppressing mesenchymal stem cell hypertrophy and endochondral ossification in 3D cartilage regeneration with nanofibrous poly(l-lactic acid) scaffold and matriline-3. Acta Biomater. 2018:7629–7638. doi: 10.1016/j.actbio.2018.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pei M., Luo J., Chen Q. Enhancing and maintaining chondrogenesis of synovial fibroblasts by cartilage extracellular matrix protein matrilines. Osteoarthritis Cartilage. 2008;16(9):1110–1117. doi: 10.1016/j.joca.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamel S., Khalaf R., Moness H., Ahmed S. Serum and synovial fluid levels of interleukin-17a in primary knee osteoarthritis patients: correlations with functional status, pain, and disease severity. Arch Rheumatol. 2022;37(2):187–194. doi: 10.46497/ArchRheumatol.2022.7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Na H.S., Park J.S., Cho K.H., Kwon J.Y., Choi J., Jhun J., et al. Interleukin-1-Interleukin-17 signaling Axis induces cartilage destruction and promotes experimental osteoarthritis. Front Immunol. 2020 doi: 10.3389/fimmu.2020.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X.D., Liu Y.R., Zhang Z.Y. Matrilin-3 alleviates extracellular matrix degradation of nucleus pulposus cells via induction of IL-1 receptor antagonist. Eur Rev Med Pharmacol Sci. 2020;24(10):5231–5241. doi: 10.26355/eurrev_202005_21305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.