Abstract
Retroviruses require both spliced and unspliced RNA for replication. Accumulation of unspliced Rous sarcoma virus RNA is facilitated in part by a negative cis element in the gag region, termed the negative regulator of splicing (NRS), which serves to repress splicing of viral RNA but can also block splicing of heterologous introns. The NRS binds components of the splicing machinery including SR proteins, U1 and U2, small nuclear ribonucleoproteins (snRNPs) of the major splicing pathway, and U11 snRNP of the minor pathway, yet splicing does not normally occur from the NRS. A mutation that abolishes U11 binding (RG11) also abrogates NRS splicing inhibition, indicating that U11 is functionally important for NRS activity and suggesting that the NRS is recognized as a minor-class 5′ splice site (5′ ss). We show here, using specific NRS mutations to disrupt U11 binding and coexpression of U11 snRNA genes harboring compensatory mutations, that the NRS U11 site is functional when paired with a minor-class 3′ ss from the human P120 gene. Surprisingly, the expectation that the same NRS mutants would be defective for splicing inhibition proved false; splicing inhibition was as good as, if not better than, that for the wild-type NRS. Comparison of these new mutations with RG11 indicated that the latter may disrupt binding of a factor(s) other than U11. Our data suggest that this factor is U1 snRNP and that a U1 binding site that overlaps the U11 site is also disrupted by RG11. Analysis of mutations which selectively disrupted U1 or U11 binding indicated that splicing inhibition by the NRS correlates most strongly with U1 snRNP. Additionally, we show that U1 binding is facilitated by SR proteins that bind to the 5′ half of the NRS, confirming an earlier proposal that this region is involved in recruiting snRNPs to the NRS. These data indicate a functional role for U1 in NRS-mediated splicing inhibition.
Most protein-encoding genes in metazoans are interrupted by introns which must be accurately spliced out of the pre-mRNA to generate mRNA. The excision process involves two transesterification reactions and takes place in a large macromolecular complex, called the spliceosome, that is composed of several small nuclear ribonucleoprotein (snRNP) particles and a large number of non-snRNP factors (28). While there is considerable understanding of the splicing reaction itself, much less is known about how splice sites are appropriately paired. The problem becomes more complicated in the many instances of alternative splicing, where different combinations of splice sites are utilized to ultimately generate different proteins from a common pre-mRNA (38). Many viruses that infect eukaryotic cells have evolved to exploit RNA splicing as a means to expand the coding capacity of their typically small genomes and as a mechanism of posttranscriptional gene regulation. As an extreme example, human immunodeficiency virus (HIV) utilizes numerous 5′ and 3′ splice sites (5′ and 3′ ss) in a temporal manner to generate an estimated 40 different mRNAs in an infected cell (32, 35).
In addition to alternative splicing observed in some viruses, incomplete RNA splicing is a feature common to all retroviruses. The primary transcript serves as an mRNA for the Gag-Pol proteins or as genome for progeny virions, but a population must also be spliced to subgenomic mRNAs, which in the simplest cases encode the Env protein (6). In HIV, it appears that the 5′ ss are efficiently recognized and that regulation occurs at the 3′ ss (31); both positive and negative elements that contribute the HIV 3′ ss control have been described (1, 36, 39). In Rous sarcoma virus (RSV), splicing control is achieved through the action of several cis elements, two of which represent the env and src 3′ ss themselves, which are maintained in suboptimal forms (18, 49). Another element that is located upstream of the src 3′ ss controls src splicing specifically and also has a mild inhibitory effect on heterologous introns (2, 4, 25, 48). Elements required for cytoplasmic accumulation of retroviral RNA, the Rev/Rev response element system in HIV (10) and constitutive transport elements in simian and avian retroviruses (5, 30, 37), might also influence the unspliced/spliced RNA ratio by modulating the availability of the unspliced RNA pool for splicing. Efficient replication of RSV requires a precise balance between spliced and unspliced RNA since viruses that display even modest increases in splicing efficiency exhibit replication defects (17, 49).
Besides the elements that are coincident with or reside near splice sites, RSV contains a novel negative element in the gag gene that is remote from the splice sites yet acts in a global manner to regulate splicing. Deletions in this element, termed the negative regulator of splicing (NRS), result in a substantial increase in splicing at both the env and src 3′ ss (3, 40). The NRS can also block splicing of heterologous introns in vivo and in vitro (14, 26), indicating that NRS inhibition is not unique to the viral context but rather results from a general effect on splicing. The importance of accumulating large amounts of unspliced viral RNA is underscored by the presence of multiple and diverse control elements, all of which are required to achieve the proper ratio of unspliced to spliced RNA.
While maintenance of suboptimal 3′ ss and splice site-specific inhibitory elements are common features of in retrovirus splicing control, the NRS element has been described only for avian retroviruses. Efforts to understand how the NRS inhibits splicing have suggested a major role for components of the splicing machinery itself. Mapping studies demonstrated that two subregions of a ∼227-base RNA fragment are required for splicing inhibition (26). The downstream region is necessary but not sufficient for splicing inhibition and harbors sequences with similarity to 5′ and 3′ ss. It was shown that U1 and U2 snRNP particles, abundant splicing factors that interact with 5′ and 3′ ss, respectively, bind the NRS in vitro (14). The same study revealed binding of a third and lower-abundance snRNP, U11, to the downstream region. The U11 snRNP interaction appears important for function since NRS mutations that abolish U11 binding also substantially reduce splicing inhibition activity. Mutations that specifically abolish U1 and U2 binding have yet to be identified, and the significance of their binding has been ambiguous. The upstream region of the NRS contains a ∼35-nucleotide (nt) purine-rich region that is also necessary but not sufficient for inhibition and to which the SR protein splicing factor SF2/ASF binds (24). Members of the SR protein family of splicing factors have several activities, including facilitating the entry of snRNP particles into the assembling spliceosome and mediating the function of RNA splicing enhancers (13, 22, 44). These two aspects of SR protein function are likely to be important for NRS action because the purine-rich NRS 5′ region (NRS5′) has enhancer activity and heterologous enhancers can substitute for NRS5′ and support splicing inhibition (23).
U11 snRNP was recently shown to be the U1 counterpart for 5′ ss recognition in a spliceosome that excises a minor class of introns that contain noncanonical splice sites (15, 20, 42, 43). These introns are often referred to as minor-class introns. Rather than having /GT (the slash denotes the splice site) and AG/ terminal dinucleotides that are characteristic of a loosely conserved consensus associated with most nuclear pre-mRNA introns (5′ ss AG/GTRAGT), the minor introns were originally described as being bounded by /AT and AC/ but also adhering to a much stricter 5′ consensus sequence, /ATATCCTT. The /AT-AC/ nature of the junctions also led to the term “attack” introns (29). More recently, minor-class introns containing /GT and AG/ termini have been identified, but the remainder of the sequences clearly resemble the minor-class consensus (11). Since the terminal dinucleotides no longer specify which splicing pathway a particular intron will use, the major and minor introns are also called U2-dependent and U12-dependent introns, respectively (11). Like the major spliceosome, a minor spliceosome assembles in a stepwise fashion through the sequential addition of other unique snRNPs (U12, U4atac, and U6atac) that serve functions analogous to the major pathway counterparts (U2, U4, and U6); U5 snRNP is present in both spliceosomes (16, 20, 41, 42). Significantly, the short sequence in the NRS 3′ region that is required for U11 binding (/GTATCCTT) matches the highly conserved minor-class 5′ ss consensus sequence.
In vitro, the NRS assembles into an RNP complex that is dependent on SR proteins and U1 and U11 snRNPs (but not U2), and whose characteristics most closely resemble complexes that assemble on a U1-dependent 5′ ss (7, 9). Despite binding SR proteins and U1/U2 snRNPs from the major splicing pathway, our efforts to force splicing from the NRS have been unsuccessful, even when it is paired with 5′ or 3′ ss that are normally used efficiently. This observation, the general lack of data supporting a functional role for U1 binding, and the apparent importance of U11 snRNP for inhibition have led us to propose that the NRS is primarily recognized as a minor-class 5′ ss that elicits splicing inhibition when placed within major-class introns (23).
In this work, we tested the notion that the NRS might serve as a minor-class 5′ ss in the appropriate context and found (i) that splicing did occur to an authentic minor-class (U12-dependent) 3′ ss from the human P120 gene in transfected cells and (ii) that U11 snRNP was required for splicing. Surprisingly, an NRS mutation that abolished U11 binding in vitro and minor pathway splicing in vivo still blocked splicing of a U2-dependent heterologous intron, suggesting that a factor other than U11 was responsible for splicing inhibition. Data presented here indicate that binding of U1 snRNP to a sequence that overlaps the U11 site most strongly correlates with splicing inhibition by the NRS. The U1 site must be suboptimal, since splicing was activated when the site was mutated to match the U1 consensus. We further show that the upstream purine-rich element, through SR proteins, is required for efficient U1 snRNP binding in vitro. Thus, while U1 was known to bind the NRS, our data now suggest that binding is an important event in splicing inhibition.
MATERIALS AND METHODS
Plasmid constructs.
RSV fragments were from the Prague C strain (27); sequence coordinates are as specified by Schwartz et al. (34). Plasmids harboring human P120 genomic fragments encompassing wild-type exons 5 to 8, or containing the CT67GA and TT78AA mutations at the minor-class 5′ ss, and U11 snRNA gene expression plasmids were generously provided by R. Padgett (Cleveland Clinic) (16, 20). pP120 contains P120 exon 6, intron F (a minor-class intron), and most of exon 7 inserted into the HindIII-BamHI sites of pRSV2 (26). The P120 fragment, generated by PCR (all primer sequences available upon request), contained nt 1 of exon 6 through nt 180 of exon 7 and had HindIII and BglII sites appended to the 5′ and 3′ ends, respectively. The chimera pNRS-P120 was created by replacing the HindIII-Bsu36I fragment of pP120 (exon 6 through position 61 of the 99-nt intron) with an NRS PCR fragment harboring nt 714 to 979 (to which HindIII and Bsu36I restriction sites were appended to the 5′ and 3′ ends, respectively). Mutations depicted in Fig. 1A were introduced into pNRS-P120 either by replacement of P120 sequences with PCR fragments derived from mutant NRS DNA or by site-directed mutagenesis of NRS sequences in pNRS-P120 by the U.S.E. (unique site elimination) method (Pharmacia Biotech).
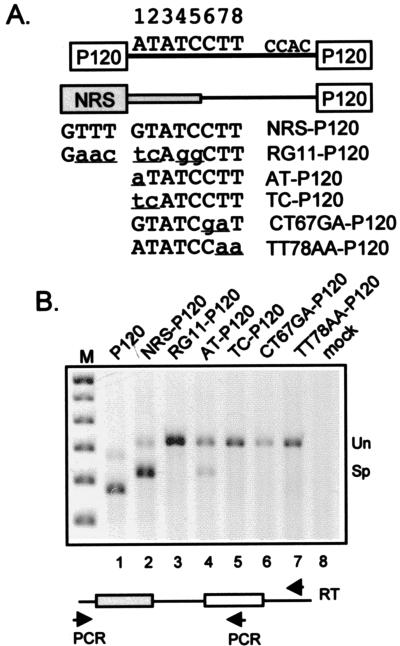
FIG. 1.
Splicing from the NRS to a U12-dependent 3′ ss. (A) Diagram of P120 and NRS-P120 chimeric constructs. A two-exon construct containing the human P120 gene exons 6 and 7 (open boxes) and intervening minor class intron (line) were expressed from the RSV promoter (not shown). The minor-class 5′ and 3′ ss sequences are shown, and the nucleotide positions at the 5′ ss are numbered. The NRS portions of the chimeric constructs are in gray, with the thick and thin regions representing sequences upstream and downstream, respectively, of the U11 site. The sequences of the NRS and mutant U11 sites are shown, with the changes in lowercase and underlined. The diagram is not to scale. (B) RT-PCR of RNA from transfected 293 cells. Below the gel is a schematic of the RT and PCR primers. Total RNA (2 μg) was reverse transcribed with a primer to common vector sequences, cDNA was subjected to PCR with an upstream vector primer and a downstream P120 exon 7 primer (see Materials and Methods), and the PCR products were resolved in an agarose gel and stained with ethidium bromide. The image was captured electronically, and the pixels were inverted. The names above the lanes refer to the constructs in panel A. Mock, transfection with an empty expression vector; M, 100-bp markers. Bands corresponding to unspliced (Un) and spliced (Sp) are indicated on the right.
To test for splicing inhibition activity, NRS fragments were inserted into the SacII intron position of the myc intron of pRSVNeo-int (21), using the KpnI-XbaI fragment shuttling approach described previously (23). All fragments consisted of nt 701 to 1011 and had KpnI and XbaI sites appended to the 5′ and 3′ ends by PCR. Fragments to be used to generate RNA for affinity selections were also inserted into pGEM-3Z. NRS mutations were created either with a U.S.E. kit (Pharmacia Biotech) or by recombinant PCR (33). The mutations are indicated in the figures. All sequences were verified by DNA sequencing.
Transfection of 293 cells and analysis of RNA.
293 cells were grown in minimal essential medium supplemented with 10% fetal calf serum and penicillin-streptomycin. Cells grown to about 40 to 60% confluence in 6-cm-diameter dishes were transfected with 2 to 3 μg of DNA by the calcium phosphate method (Pharmacia Biotech), and total RNA harvested 40 h later was isolated with Qiagen RNAeasy columns according to the manufacturer’s instructions. For reverse transcription (RT)-PCR, 1 μg of total RNA was reverse transcribed with an antisense primer directed to pRSV2 vector sequences downstream of the transcription unit (GCAGACACTCTATGCCTGTGTGG) and common to all RNAs in 20 μl, using 200 U of reverse transcriptase (GibcoBRL) and the manufacturer’s recommended reaction conditions. Two microliters of the RT reaction was subjected to 28 cycles of PCR using a 5′ vector primer directed upstream of the transcription unit (CACCACATTGGTGTGC) and a 3′ primer to sequences in P120 exon 6. Thus, the same primer pair was used for all constructs. RNase protection assays were as described elsewhere (23), using 5 μg of RNA. A plasmid for generating the 3′ ss probe was made by inserting a blunt-ended AflII-PstI fragment from pRSVNeo-int that spans the 3′ ss into the pGEM-3Z SmaI site such that T7 RNA polymerase and HindIII-cut DNA generated an antisense probe. Quantitation was done with a Molecular Dynamics Storm 860 PhosphorImager.
Affinity selection.
Affinity selection was performed essentially as described previously (9). Briefly, RNA transcribed in vitro in the presence of biotin-11-UTP (20% of total UTP) from pGEM constructs linearized with XbaI was incubated under splicing conditions with ATP in HeLa cell nuclear extract (12) for 30 min at 30°C. Streptavidin-agarose beads were added and mixed at 300 mM KCl for 1 h at 4°C and then washed extensively at 300 mM KCl. Bound nucleic acids were released by proteinase K digestion, phenol extracted, precipitated with ethanol, and subjected to electrophoresis in a 8 M urea–8% polyacrylamide gel. RNA was electroblotted to a ZetaProbe membrane (Bio-Rad) and hybridized with riboprobes to U1 and U11 snRNA; the membrane was then subjected to autoradiography.
RESULTS
Splicing from the NRS to a U12-dependent 3′ ss requires U11 snRNP.
Given that the NRS interacts with a number of splicing factors, one might expect the NRS to be productively used as a splice site under certain conditions, yet our efforts to detect splicing to or from the NRS in a variety of contexts have been unsuccessful. The description of U11 snRNP as the component of the minor splicing pathway that recognizes the 5′ ss of U12-dependent introns (20, 46) prompted us to investigate whether our failures to detect NRS splicing stemmed from its inappropriate pairing with major-class splice sites. Might the NRS function in splicing if paired with a minor-class, U12-dependent 3′ ss? To examine this possibility, we made a number of chimeras utilizing the RSV promoter by fusing the NRS to a human P120 gene fragment encompassing part of intron F and exon 7 that contains a U12-dependent 3′ ss (Fig. 1A). The parental vector contained most of the P120 exon 6, the entire intron F, and most of exon 7. A similar construct containing P120 exons 5 through 8 is known to splice accurately in vivo (16), and exon 6 is spliced to exon 7 accurately in vitro (11, 42). For the NRS-P120 chimeras, the P120 exon 6 and 61 nt of the 99-nt intron were replaced with NRS sequences. The NRS fragment contained nt 719 to 979 and so contained 61 nt downstream of the putative U11 5′ ss such that if the site were active, the intron would be the natural size of P120 intron F, 99 nt. As controls, additional constructs contained the RG11 NRS mutation that abolishes U11 binding and splicing inhibition (14) and would be expected to abrogate minor-class splicing; a G-to-A change at the first nucleotide of the U11 consensus to give an AT dinucleotide (AT), which should not affect U11 binding or function (11); a GT-to-TC change at positions 1 and 2 (TC) which would abolish splicing but would not be predicted to significantly affect U11 binding; and CT67GA and TT78AA mutations that are known to abolish P120 splicing (20). The constructs were transfected into 293 cells, and the extent of splicing was assessed by subjecting the harvested RNA to RT-PCR with an RT primer to downstream vector sequences, and PCR primers to upstream vector sequences and downstream P120 exon 7, sequences common to all constructs. Control reactions which lacked RT showed that the PCR signals were due to expression of transfected plasmids rather than endogenous P120 mRNA or DNA (data not shown).
As expected for the P120 RNA, RT-PCR products of the size expected for unspliced and spliced RNA were observed, with the spliced product predominating (Fig. 1B, lane 1). When P120 exon 6 and the upstream two-thirds of the intron were replaced with the NRS, a band whose size was consistent with utilization of the NRS U11 5′ and P120 3′ splice sites was observed (lane 2), and the splicing efficiency of the NRS-P120 chimera was similar to that for the P120 construct. Thus, pairing the NRS with a minor-class 3′ ss activated splicing from the NRS for the first time in our hands. Southern blotting with intron and exon probes confirmed the conclusion that the bands were derived from unspliced and spliced RNA (data not shown). Significantly, the putative spliced product was not observed with the RG11 mutant that fails to bind U11 snRNP in vitro and that would be expected to abolish minor pathway splicing (lane 3). Further support for the idea that splicing occurred from the NRS U11 site stemmed from the result with the A-to-T mutation. If the observed splice had occurred by aberrant U1 recognition of the GT dinucleotide associated with the NRS U11 site, the AT mutation might be expected to abolish splicing, whereas an A at the +1 position should have little effect on minor-class splicing. Consistent with the latter possibility, splicing was still observed with the AT mutation (lane 4). In addition, three mutations expected to inactivate the NRS U11 site, two of which should be highly specific to U11 (CT67GA and TT78GA) (20), completely abolished the spliced band. These data are most compatible with the NRS splice occurring via the minor pathway at the predicted U11 site.
Still, the presence of several U1-like 5′ ss sequences and the previously observed U1 snRNP binding to the NRS (14), coupled with the presence of a /GT rather than an /AT in the minor 5′ consensus, left open the possibility that the splicing observed with NRS-P120 chimera resulted from the major pathway via U1 snRNP, perhaps to one of several AG/ dinucleotides surrounding the P120 U12-dependent 3′ ss. To address this, a number of independent cDNAs representing the NRS-P120 spliced RNA were cloned and sequenced to determine the splice junctions. Sequencing of the control P120 mRNA showed accurate splicing (/AT to AC/), and the predicted NRS /GT 5′ ss was always used in NRS-P120 mRNA, but in no instance was the authentic P120 AC/ 3′ ss used (data not shown; see Discussion). Rather, in the majority of cases the splice junction was shifted two nucleotides downstream to an AT/ dinucleotide (22 of 27 sequences), with five cDNAs showing an AG/ junction at +8. Interestingly, this apparent anomaly might have been expected since this is exactly what is observed when the 5′ ss is changed from AT to GT in P120 splicing; however, the GT-to-AT and GT-to-AG splicing was still by the minor pathway (11). As expected, sequencing of the NRS AT-P120 cDNA showed accurate minor class splicing (AT to AC [data not shown]). Thus, the sequencing data were highly suggestive but not conclusive enough to unambiguously assign the splicing observed with NRS-P120 to the minor pathway.
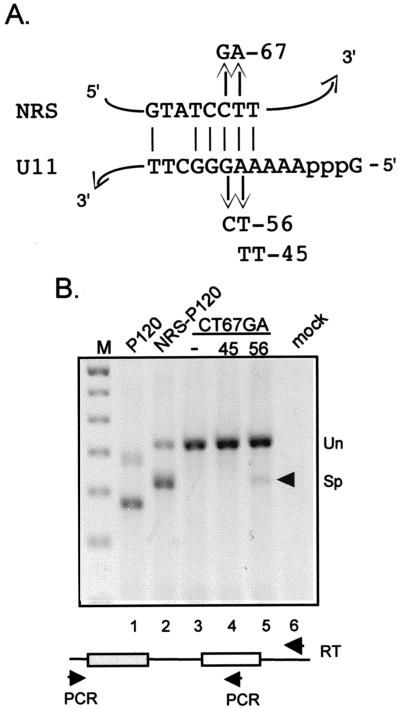
Recently, a genetic approach was taken to demonstrate that U11 functionally interacts with the P120 5′ ss (20). Specifically, the splicing defect of the CT67GA mutant 5′ ss was suppressed by coexpression of a U11 snRNA gene harboring a compensatory mutation (AG56TC) predicted to reestablish base pairing with the substrate. The base pairing potential of U11 snRNA with the NRS is shown in Fig. 2A. To definitively show that the minor pathway was utilized for NRS-P120 splicing, we similarly coexpressed mutant U11 genes with the CT67AG NRS-P120 chimera. As shown in Fig. 2B, a modest level of splicing was restored to the CT67GA mutant chimera when the U11 AG56TC gene (lane 5) was coexpressed. However, no splicing was observed when an inappropriate U11 allele was used (lane 4), indicating specific suppression of the NRS mutation by the altered U11 snRNA. These data are strong evidence that the splicing from the NRS in the chimera was by the minor pathway.
FIG. 2.
Allele-specific suppression of the CT67GA mutant splicing defect by expression of compensatory U11 snRNA. (A) Potential base pairing interaction between the NRS and U11 snRNA. The NRS CT67GA mutation and the U11 snRNA compensatory mutations (AG56CT and AA45TT) at the 5′ end of U11 are shown. (B) RT-PCR of RNA from transfected 293 cells was performed as described in the legend to Fig. 1. Lanes 3 to 5 are reactions from the cells transfected with the NRS CT67GA mutant. Indicated below is cotransfection with an empty vector (−), or the AA45TT (45) or AG56CT (56) U11 snRNA expression plasmid. The arrowhead indicates the spliced band restored by the compensatory AG56CT U11 construct.
NRS U11 snRNP binding mutant (CT67GA) still blocks splicing.
Having shown genetically that the U11 5′ ss associated with the NRS can function the correct context, we set out to demonstrate that splicing inhibition was also mediated by U11 snRNP. NRS inhibition activity was assessed by monitoring the splicing efficiency in transfected 293 cells of a heterologous intron into which the NRS is inserted, as shown in Fig. 3A (26). The distance of the insertion site from the 5′ ss is similar to the natural location of the NRS in RSV. Our expectation was that the NRS CT67GA mutant would no longer inhibit splicing and that splicing inhibition would be restored by coexpression of the compensatory U11 snRNA. As expected, the NRS elicited splicing inhibition in an orientation-specific manner (Fig. 3, lanes 3 and 4), and the RG11 mutation eliminated splicing inhibition (lane 5), consistent with an elimination of U11 binding. Surprisingly, the CT67GA mutation had no effect on splicing inhibition and sometimes resulted in slightly more unspliced RNA. A similar result was obtained with the TT78AA mutant (data not shown). Thus, the same mutations that eliminated minor-class splicing in the NRS-P120 chimera had no effect on splicing inhibition.
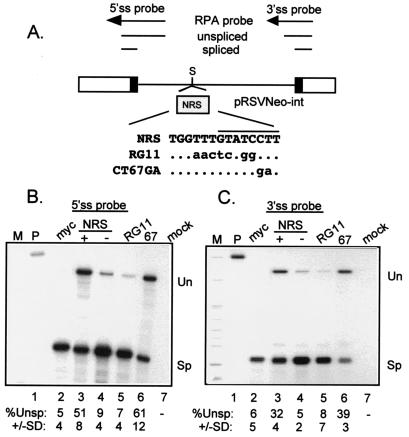
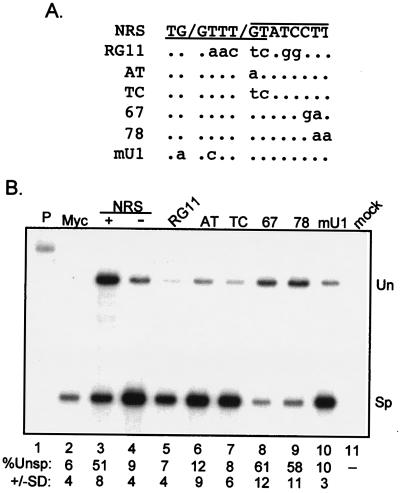
FIG. 3.
The CT67GA NRS mutation does not impair NRS splicing inhibition. (A) Diagram of the pRSVNeo-int construct and RNase protection probes used to measure NRS activity. Open boxes indicate Neo sequences, black boxes represent the small portions of myc exons, and the line denotes the myc intron. Above is a schematic of the RNase protection probes and protected fragments. The shaded box represents NRS fragments that were inserted into the SacII site (S) of the myc intron of pRSVNeo-int. The sequence surrounding the U11 site of the mutant and wild-type NRS fragments is shown. The dots indicate unchanged bases; changes are in lowercase. The U11 site is overlined. (B and C) RNase protection assays on RNA from transfected 293 cells. Total RNA from 293 cells transfected with the indicated constructs was used for RNase protection with 5′ ss (B) and 3′ ss (C) probes (see Materials and Methods), and protected fragments were run on a 4% (B) or 6% (C) denaturing polyacrylamide gels and visualized by autoradiography. M, markers; P, unprocessed probe; myc, pRSVNeo-int with no insert; + and −, sense and antisense orientations of the NRS; RG11 and CT67GA (67) NRS mutants; mock, RNA from mock-transfected cells; Un and Sp, positions of bands representing unspliced and spliced RNA. Below the lanes is the quantitated percent unspliced RNA as determined by PhosphorImager analysis; the data are representative of at least three separate experiments.
It was possible that the lack of inhibition observed with NRS CT67GA was an artifact of using a probe to the 5′ ss to assay splicing of the test intron. For example, the specific elimination of U11 binding may have activated major-class splicing from the NRS to the normal test intron 3′ ss such that the probe would then be hybridizing to upstream exon sequences; the full-length protected probe would no longer be diagnostic for splicing. This was addressed by subjecting the same RNA to RNase protection with a probe to the 3′ ss, the results from which should differ from the 5′ ss probe data if cryptic splicing is activated within the NRS. The results with the 3′ ss probe mirrored those with the 5′ ss probe (Fig. 3C), favoring the conclusion that the CT67GA mutation failed to abolish splicing inhibition. One interpretation is that the CT67GA mutation may have had no effect on U11 binding and splicing inhibition but selectively abolished the splicing potential of the chimeras used for Fig. 1 and 2. Alternatively, a factor other than or in addition to U11 snRNP may be responsible for splicing inhibition by the NRS.
snRNP binding to the NRS.
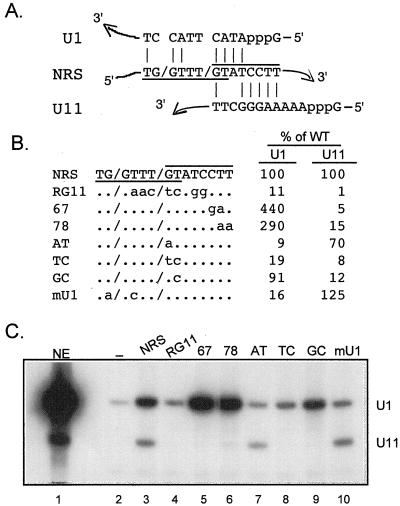
In addition to having four changes in the U11 binding sequence rather than the two changes present in CT67GA, the original U11 binding mutation, RG11, contains three other changes immediately upstream of the U11 site (Fig. 1A) which could be the source of the different inhibition activities observed above for the two mutations. Interestingly, this sequence is similar to a U1-type 5′ ss (TG/GTTTGT versus the AG/GTRAGT consensus), and the NRS is known to bind U1 (14) although the binding site(s) has not been identified. Thus, the RG11 mutation might simultaneously affect U11 and U1 binding sites. The base pairing potential of U1 and U11 is shown in Fig. 4A. This possibility was directly addressed by examining the effect of RG11 on U11 and U1 snRNP binding. Biotinylated RNA transcripts of the NRS and a number of mutants (Fig. 4B) were incubated in nuclear extract and affinity selected with streptavidin agarose, and the associated snRNPs were identified by Northern blotting of extracted RNAs. A representative result is shown in Fig. 4C, and the percentage of U1 and U11 binding of the mutants relative to the wild type is presented in Fig. 4B. We recently reported that U11 snRNP is poorly selected by NRS nt 701 to 932 (9), but as shown in Fig. 4C, U11 binding to NRS nt 701 to 1011 is much more efficient (lane 3 and data not shown). When RG11 was used, the U11 signal was virtually eliminated, as expected, but the U1 signal was also decreased by about 90% (lane 4). In contrast, the U11-specific mutations CT67GA and TT78AA also eliminated U11 binding but resulted in more efficient U1 binding (lanes 5 and 6). These results suggest that a U1 binding site is very close to the U11 site. Also, the increased U1 binding to the U11 mutants suggests that the two snRNPs compete for binding to the same region and perhaps to overlapping sequences.
FIG. 4.
A U1 snRNP binding site overlaps the U11 site. (A) Diagram of potential base pairing interactions (vertical lines) of U1 and U11 snRNA with the NRS. The NRS U1 site is underlined; the U11 site is overlined; slashes indicate where splicing would be predicted to occur if the sites were functional. (B) Sequences of NRS and mutants used for affinity selection. To the right, bands in panel C were quantitated with a PhosphorImager, and the intensity of each of the U1 and U11 signals generated by the mutants was compared to that for wild-type (WT) NRS RNA, which was set to 100%. (C) Affinity selection. Equal moles of the indicated biotinylated RNAs were incubated in nuclear extract and affinity selected with streptavidin-agarose beads, and snRNA components of snRNPs associated with the NRS were extracted with phenol-chloroform (see Materials and Methods). The extracted RNA was resolved in a denaturing polyacrylamide gel, electroblotted to a nylon membrane, and hybridized with U1 and U11 antisense riboprobes. Background binding to the beads was determined with nonbiotinylated RNA (−). NE, U1 and U11 snRNA markers extracted from 3 μl of nuclear extract. The positions of U1 and U11 are indicated on the right.
One prediction of the overlapping binding site model is that U11 binding would be unaffected by the AT mutation but that since the mutated G is at the important +5 position for U1 (28), U1 binding would be affected. As predicted, U1 binding to AT was greatly reduced whereas U11 binding was largely unaffected (Fig. 4C, lane 7). Additionally, the TC and GC mutations had severe and mild effects on U1 binding, as expected (lanes 8 and 9). Surprisingly, these two mutations also eliminated U11 binding, indicating that changing the second position in the U11 consensus, which is not predicted to contribute to base pairing interactions with U11, nonetheless has a significant impact on U11 binding in this assay. The mU1 mutant, which should specifically reduce U1 binding, nearly eliminated U1 binding and resulted in a small but reproducible increase in U11 binding (lane 10). Thus, mutations designed to impact binding of both snRNPs, and likewise mutations targeted to one or the other snRNP, had the predicted effects. These data support the view that U1 and U11 binding sites overlap, sharing a central GT. Therefore, it is possible that the effect of previous mutations on NRS function might have been through disruption of U1 binding rather than U11, or both.
U1 snRNP binding correlates with splicing inhibition.
Having shown that the CT67GA mutation retains inhibitory activity (Fig. 3) and that the RG11 mutation affects both U11 and U1 snRNP binding (Fig. 4), we used selected mutants to determine the effect of mutating the putative U1 site while preserving U11 binding on NRS splicing inhibition (Fig. 5A). The wild-type NRS and RG11 served as positive and negative controls. As shown in Fig. 5B, compared to the wild type (lane 3), unspliced RNA accumulation was decreased substantially when the RG11, AT, TC, and mU1 mutants were inserted into the test intron at the SacII site (lanes 5 to 7 and 10). This finding is consistent with the loss of U1 binding for each (Fig. 4C) but would not expected for AT and mU1 if U11 contributes to splicing inhibition. The level of unspliced RNA with the U11-specific CT67GA and TT78AA mutants was similar to or slightly higher than the wild-type level (lanes 8 and 9), again suggesting that U11 binding in not required for splicing inhibition. These results establish a correlation between splicing inhibition and U1 binding and indicate that U11 binding is of less importance.
FIG. 5.
Selective mutation of the U1 site severely reduces NRS splicing inhibition. (A) The sequence of the U1 and U11 sites of the NRS is shown at the top. Slashes indicate where splicing would occur if the sites were used. Below are sequences of the mutants, with mutations in lowercase and unchanged bases indicated as dots. The U1 site is underlined; the U11 site is overlined. The fragments were inserted into the SacII intron site of pRSVNeo-int. (B) RNase protection assay on RNA from transfected 293 cells. Total RNA from 293 cells transfected with the indicated constructs was used for RNase protection with the 5′ ss probe as for Fig. 3B. Un and Sp, positions of bands representing unspliced and spliced RNA. Below the lanes is the percent unspliced RNA as determined by PhosphorImager analysis; the data are representative of at least three separate experiments.
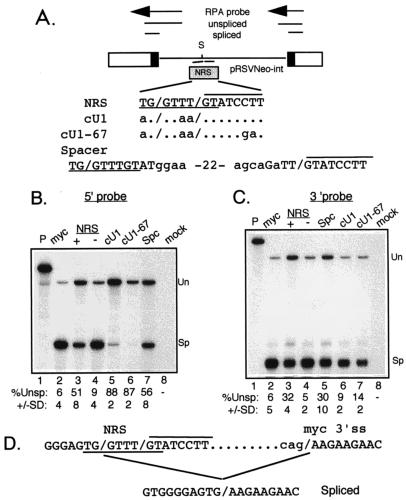
A consensus U1 site activates splicing from the NRS.
The NRS U1 site deviates from the consensus U1 5′ ss at positions −2, +3, and +4. Since the above results indicated that U1 binding is important for splicing inhibition, the constructs in Fig. 6A were used to determine if altering the NRS U1 site to match the consensus would result in more potent splicing inhibition. Both a mutant containing a consensus U1 site and the normal overlapping U11 site (cU1) and a construct harboring the U11 CT67GA mutation and the improved U1 site (cU1-67) appeared to inhibit splicing more efficiently than wild type when assayed with the 5′ ss probe (Fig. 6B; compare lanes 5 and 6 to lane 3). The U11 site was also changed to the U1 consensus in the cU1 background, generating two overlapping U1 sites. Again, a higher than normal level of inhibition was observed (data not shown). The U1 and U11 sites were also separated by 22 nt to relieve competition, but this construct (Spc) produced normal levels of unspliced RNA. Thus, it appeared that a stronger U1 site resulted in increased splicing inhibition efficiency. However, as in Fig. 3, the result with the 5′ ss probe would be artifactual if the consensus U1 sites were actually used for splicing. When the same RNAs were used in RNase protection assays with the 3′ ss probe to control for this possibility (Fig. 6C), roughly 90% of the RNA appeared to be spliced when a consensus U1 site was present (lanes 6 and 7). These data are consistent with splicing occurring from the NRS in the mutants containing a consensus U1 site. This possibility was confirmed by sequencing RT-PCR products generated from the same RNA. The size of the products was consistent with splicing from the NRS to the myc 3′ ss (data not shown) and the sequence indicated splicing from the consensus U1 sites to the normal myc 3′ ss of the test intron (Fig. 6D). We conclude that the NRS U1 site must be suboptimal to inhibit splicing; consensus sites are productively used.
FIG. 6.
Productive splicing from a consensus NRS U1 site. (A) Schematic of pRSVNeo-int, 5′ and 3′ ss probes, and NRS insertion site. Relevant sequences of the NRS (U1 site underlined, U11 site overlined) and mutants inserted into the SacII site (S) of the pRSVNeo-int test intron are shown. Dots indicate unchanged bases; changes are in lowercase; slashes (/) indicate where splicing would be predicted to occur if the U1 or U11 sites were functional. For the spacer (Spc) construct, the U1 and U11 sites were separated by 22 nt. (B and C) Results of RNase protection assays on RNA from transfected 293 cells obtained with the 5′ ss and 3′ ss probes, respectively (note the difference in lane order). Below the lanes is the quantitated percent unspliced RNA as determined by PhosphorImager analysis; the data are representative of at least three separate experiments. Un and Sp, positions of bands representing unspliced and spliced RNA. (D) Sequence of cDNA from spliced RNA. The cut RNA samples used for panels B and C were subjected to RT-PCR, and the PCR products corresponding to spliced RNA were sequenced. The relevant sequence of the NRS and myc 3′ ss is shown. Intron sequences are represented by dots, the terminal myc nucleotides are in lowercase, and the myc exon sequence is in uppercase.
The NRS purine region and SR proteins promote snRNP binding to the NRS.
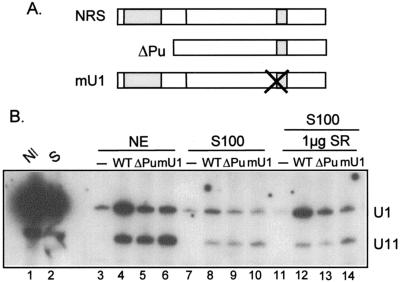
The purine-rich NRS5′ binds SR proteins, possesses RNA splicing enhancer activity, and is required for splicing inhibition. We previously proposed that the purine region might function by promoting snRNP binding to the NRS (23). However, in nuclear extract only a small positive effect of NRS5′ on U11 binding has been observed. The above data indicated a primary role for U1 in splicing inhibition and led us to examine the effect of the purine region on U1 binding, using the affinity selection assay and the RNAs shown in Fig. 7A. As shown above, U1 and U11 were efficiently selected by the NRS (Fig. 7B, lane 4), whereas the mU1 mutation substantially reduced U1 binding while modestly increasing U11 binding (lane 6). The NRS lacking the purine region (ΔPu) again selected U11 with only slightly reduced efficiency, but U1 binding was strongly affected (lane 5). These results suggest that U1 binding is more strongly influenced by the purine region, and by extension SR protein binding, than U11.
FIG. 7.
The NRS purine-rich region and SR proteins promote U1 snRNP binding to the NRS. (A) Diagram of the constructs used in biotin-streptavidin affinity selection experiments. The larger and smaller shaded regions indicate the NRS purine-rich region and snRNP binding sites, respectively. (B) Affinity selection experiment. The indicated biotinylated RNAs (−, nonbiotinylated NRS RNA) were incubated in nuclear extract (NE; lanes 3 to 6), S100 (lanes 7 to 10), or S100 supplemented with SR proteins (lanes 11 to 14), and the complexes assembled on the RNA were affinity selected with streptavidin-agarose. Bound nucleic acids were released by treatment with proteinase K and phenol extracted, and RNA was electroblotted to a nylon membrane and hybridized with U1 and U11 antisense riboprobes. snRNA markers were extracted directly from nuclear extract or S100 (lanes 1 and 2). The positions of U1 and U11 are indicated to the right.
We directly investigated the role of SR proteins in U1 and U11 binding with an S100 extract that lacks SR proteins (47). Binding of both snRNPs was strongly reduced in S100, regardless of the substrate used (Fig. 7B, lanes 8 to 10), which suggests but does not prove that SR proteins promote binding of U1 and U11. When 1 μg of total SR proteins purified from HeLa cells was added to S100, binding of U1 was rescued, but only in the presence of the purine region and an intact U1 site (lanes 12 to 14). In contrast, U11 binding was not restored in S100, even with the addition of up to 8 μg of SR proteins (data not shown). It appears that in addition to SR proteins, U11 binding requires another factor(s) that is not present in S100. Given that U11 binding is less dependent on the purine region and that NRS activity in vivo requires the purines, these results are most consistent with a primary role for U1 in splicing inhibition.
DISCUSSION
Three snRNPs have been shown to interact with the NRS: U1 and U2 snRNPs of the major splicing pathway, and U11 snRNP of the minor pathway. Deciphering which snRNPs are required for splicing inhibition is important for understanding the mechanism by which the NRS ultimately blocks splicing. Several observations have suggested a primary role for U11 snRNP in NRS-mediated splicing inhibition. First, U11 binding to the NRS is dependent on a critical sequence that matches the minor class 5′ ss consensus to which U11 is known to bind, and mutation of this sequence abolished splicing inhibition of a heterologous intron in vivo (14). Further, assembly of an RNP complex on the NRS in vitro, NRS-C, is partially dependent on an intact U11 site (7). Less certain were the roles for U1 and/or U2 snRNP, since their binding sites within the NRS had not been identified and thus mutational studies had not been possible. U2 snRNP is not required for NRS-C assembly in vitro (9), which can be taken as evidence against a role for U2 in NRS activity. In contrast, disruption or sequestration of U1 snRNP results in a large decrease in NRS-C assembly (9). This result suggested a role for U1 in NRS activity; however, without functional data, it was possible that in vitro binding to the NRS reflected the high abundance of U1 in nuclear extract. Surprisingly, the results reported here provide strong evidence that a U1-type 5′ ss overlaps the U11 site and that U1 snRNP is, in fact, more important than U11 for splicing inhibition.
We speculated that our inability to force splicing from the NRS in numerous contexts, despite its binding of splicing factors, stemmed from the fact that it was always paired with major-class splice sites which are thought to be incompatible with minor-class splicing (19). Our results indicate that minor-class splicing can occur efficiently from the NRS via U11 snRNP provided that a U12-dependent 3′ ss is supplied. Sequencing of chimeric cDNAs showed that the /GT of the NRS U11 site was used but the natural P120 AC/ 3′ ss was not. Rather, a cryptic TG/ 2 nt downstream from the 3′ ss was preferentially used along with a nearby AG/ dinucleotide that is characteristic of the major pathway. Dietrich et al. (11) recently showed that mutation of the P120 gene U11 5′ ss to GT, which matches the NRS sequence, results in cryptic splicing to the very same 3′ TG/ and AG/ dinucleotides and that splicing was by the minor pathway. This work, our demonstration that NRS CT67GA-P120 splicing could be restored in an allele-specific manner by expression of a compensatory U11 snRNA, and accurate minor-class splicing from the AT-P120 construct provide strong evidence that the NRS splice was by the minor pathway. Interestingly, Weldon and Wills (45) detected a splice from the /GT of the NRS U11 site to cytochrome c sequences in an expression vector where the yeast cytochrome c gene replaced PR in the RSV gag gene. The 3′ ss junction was an AG/ dinucleotide characteristic of a major class splice, but 8 nt upstream is a close match to the U12-dependent branch point sequence, suggesting that this splice was also by the minor pathway and that the cyt gene sequences fortuitously provided a U12 site. We generalize these observations and suggest that splicing from the NRS does not normally occur unless a U12-dependent 3′ ss is provided. We are unaware of any instance where a major class splice has occurred from NRS, despite the many configurations that have provided a U2-dependent 3′ ss. Until this study, these results were consistent with previous models of a role for U11 snRNP in NRS-mediated splicing inhibition.
Based on the results with the NRS-P120 chimera, we fully expected the CT67GA mutation to abolish splicing inhibition, just as it had abolished splicing of the chimera. Surprisingly, CT67GA had no effect on splicing inhibition. The demonstration that the mutation rendered U11 binding almost undetectable indicates that U11 is not required for, and perhaps is not involved in, NRS-mediated splicing inhibition. This suggested that binding of a factor in addition to U11 is disrupted by the original RG11 mutation, and that factor appears to be U1 snRNP. Four observations suggest a major role for U1 snRNP binding in splicing inhibition by the NRS. First, the mutations that selectively disrupted U1 severely reduced splicing inhibition and slightly increased U11 binding. This finding is in conflict with a predominant role for U11. Second, mutations that disrupted U11 binding resulted in an increase in U1 binding, and these mutants often showed enhanced splicing inhibition. Third, we previously showed that NRS5′ binds SR proteins (24) and harbors splicing enhancer activity (23) and proposed that this region is responsible for recruiting snRNPs to the NRS. Our findings here that the NRS purine-rich region and SR proteins are required for efficient U1 binding, but less so for U11, is consistent with a principal role for U1 snRNP in inhibition. Fourth, incapacitation of U1 has a dramatic affect on assembly of the NRS complex in vitro (9), and selective mutation of the U1 site, but not U11, abolished an in vitro interaction with a U2-dependent 3′ ss (see below) (8).
While our data suggest that U1 is of primary importance for the NRS, it is still possible that U11 snRNP can inhibit major pathway splicing to a lesser degree. Mutations which disrupt binding of both U1 and U11 abolished residual inhibitory activity often seen with the U1 binding mutant, indicating that a low level inhibition could be due to U11. Also, a moderate level of inhibition was observed when the U1 binding mutants were inserted at a proximal intron position (24a). Additionally, the authentic P120 minor 5′ ss region inhibits splicing of this construct to roughly the same degree as the mU1 mutant, but again only when inserted at the intron proximal site (24a). In these latter cases, it remains to be determined if the U11 site is required or if the inhibition can be attributed to secondary U1 sites. The very weak activity of U1 binding mutants when placed in the test intron at a position similar to the native NRS location also calls into question the significance of U11 binding for the virus. We are currently incorporating specific U1 and U11 site mutations into the virus in order to assess their effect on viral replication.
How might U1 snRNP bring about splicing inhibition when bound to the NRS? Put another way, why, once it is bound, does splicing not occur from the NRS to U2-dependent 3′ ss? While these questions remain unanswered, it does appear that the NRS U1 site must be in a suboptimal form, since converting it to a consensus 5′ ss activated splicing. There is also no evidence for collaboration with U11, since in its absence inhibition was efficient. We previously proposed that the NRS might block splicing by the formation of a nonproductive complex between factors bound to the NRS and the 3′ ss and hypothesized that U11 snRNP would be involved (23). The results of Cook and McNally (8) indicate that an interaction does occur but that it is not through U11, at least not in vitro. Rather, they showed that U1 engages in an early interaction with factors associated with the branch point/pyrimidine tract of an adenovirus U2-dependent 3′ ss. Inhibition might take place at this early stage directly through U1, which could arrest spliceosome assembly. Alternatively, the observation that an adenovirus splicing substrate harboring the NRS in the intron failed to splice in vitro yet assembled abnormally large RNP complexes with full spliceosomal snRNP representation argues that inhibition may take place at subsequent spliceosome assembly steps, perhaps through other snRNPs and/or non-snRNP factors. These issues are currently being investigated in our laboratory.
ACKNOWLEDGMENTS
We thank Richard Padgett for P120 and U11 plasmids and Brent Fogel for some of the NRS-P120 chimeric plasmids. We thank members of the McNally lab for critical reviews of the manuscript.
This research was supported by a Public Health Service grant R29 CA63348 from the National Cancer Institute.
REFERENCES
- 1.Amendt B A, Hesslein D, Chang L J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt B A, Simpson S B, Stoltzfus C M. Inhibition of RNA splicing at the Rous sarcoma virus src 3′ splice site is mediated by an interaction between a negative cis element and a chicken embryo fibroblast nuclear factor. J Virol. 1995;69:5068–5076. doi: 10.1128/jvi.69.8.5068-5076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berberich S L, Stoltzfus C M. Mutations in the regions of the Rous sarcoma virus 3′ splice sites: implications for regulation of alternative splicing. J Virol. 1991;65:2640–2646. doi: 10.1128/jvi.65.5.2640-2646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1767–1847. [Google Scholar]
- 7.Cook C R, McNally M T. Characterization of an RNP complex that assembles on the Rous sarcoma virus negative regulator of splicing element. Nucleic Acids Res. 1996;24:4962–4968. doi: 10.1093/nar/24.24.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook C R, McNally M T. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J Virol. 1999;73:2394–2400. doi: 10.1128/jvi.73.3.2394-2400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook C R, McNally M T. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R. Regulation of human immunodeficiency virus replication. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich R C, Incorvaia R, Padgett R A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R D. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 14.Gontarek R R, McNally M T, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 15.Hall S L, Padgett R A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 16.Hall S L, Padgett R A. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 17.Katz R A, Kotler M, Skalka A M. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988;62:2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohrman D C, Harris J B, Meisler M H. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J Biol Chem. 1996;271:17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- 20.Kolossova I, Padgett R A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 21.Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987;49:93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- 22.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 23.McNally L M, McNally M T. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNally L M, McNally M T. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.McNally, M. T. Unpublished data.
- 25.McNally M T, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally M T, Gontarek R R, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 27.Meric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. In: Gesteland R, Atkins J, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 29.Mount S M. AT-AC introns; an ATtACk on dogma. Science. 1996;271:1690–1691. doi: 10.1126/science.271.5256.1690. [DOI] [PubMed] [Google Scholar]
- 30.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Reilly M M, McNally M T, Beemon K L. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 32.Purcell D F, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saiki R K, Gelfand D H, Stoffel S, Scharf S, Higuchi R H, Horn G T, Mulli K B, Erlich H A. Primer-directed enzymatic amplification with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz S, Felber B K, Benko D M, Fenyo E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si Z H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith C W J, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 39.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoltzfus C M, Fogarty S J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989;63:1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarn W Y, Steitz J A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 42.Tarn W Y, Steitz J A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 43.Tarn W Y, Steitz J A. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 44.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 45.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y T, Steitz J A. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc Natl Acad Sci USA. 1997;94:6030–6035. doi: 10.1073/pnas.94.12.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Simpson S B, Stoltzfus C M. Selection and characterization of replication-competent revertants of a Rous sarcoma virus src gene oversplicing mutant. J Virol. 1996;70:3636–3644. doi: 10.1128/jvi.70.6.3636-3644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Stoltzfus C M. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206:1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]