Abstract
Tissues are heterogeneous with respect to cellular and non-cellular components and in the dynamic interactions between these elements. To study the behaviour and fate of individual cells in these complex tissues, intravital microscopy (IVM) techniques such as multiphoton microscopy have been developed to visualize intact and live tissues at cellular and subcellular resolution. IVM experiments have revealed unique insights into the dynamic interplay between different cell types and their local environment, and how this drives morphogenesis and homeostasis of tissues, inflammation and immune responses, and the development of various diseases. This Primer introduces researchers to IVM technologies, with a focus on multiphoton microscopy of rodents, and discusses challenges, solutions and practical tips on how to perform IVM. To illustrate the unique potential of IVM, several examples of results are highlighted. Finally, we discuss data reproducibility and how to handle big imaging data sets.
Intravital microscopy (IVM) provides an unrivalled ability to appreciate the principles regulating tissue and organ function at cellular and subcellular levels by capturing cell behaviours in situ and in real time. IVM is, broadly, any kind of microscopy performed on any kind of living organism. In this Primer, we focus on the high-resolution imaging of individual cells in living rodents with techniques such as multiphoton microscopy. In the last two decades, these high-resolution microscopy technologies in rodents have rapidly become popular in the fields of cell biology and medicine, as they shed new light on physiological and pathological processes. Cellular behaviour is driven by the tissue (micro) environment, which is often incomplete and oversimplified in the in vitro context. Indeed, fundamental differences in cell behaviour have been observed between in vivo and in vitro conditions. For example, IVM in tumour-bearing mice showed that the mode of action of taxanes, an important class of chemotherapeutics, is different between in vitro and in vivo conditions1. Although cancer cells undergo mitotic cell death in vitro upon treatment with taxanes, in vivo the majority of the same cancer cells die without any signs of mitotic defects, underscoring the importance of in vivo validation of chemotherapeutic agents. IVM studies not only can confirm or dispute hypotheses generated through in vitro or histological studies but have also enabled the discovery of biological principles that were otherwise unexpected.
In this Primer, we do not describe the principles of microscopy techniques in detail, which have been extensively described elsewhere2–5. We aim to generate a guide that helps researchers to set up or optimize IVM in rodents in their own laboratory. For this, we discuss microscopy set-ups, optimizing an IVM experiment and the type of analyses that can be performed. We present some examples of the use of advanced microscopy techniques — such as photobleaching, photoactivation, photo-conversion, Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) — where the in situ and real-time nature of IVM expands the degree of information that can be achieved with these approaches. IVM researchers face challenges based on the nature of the specimens and the need to maintain physiology in anaesthetized animals. We outline how to overcome these challenges, including procedures on how to correct for movement aberrations due to breathing and heartbeat and guidelines on how to implement techniques to excite and detect fluorophores deep within tissues and organs that are most often difficult to access. Finally, we provide some considerations on the complex relationship between IVM and the principles of the 3Rs of animal work: although IVM procedures are often complex and more invasive for the animals, they often enable reduction of the overall number of animals used and are irreplaceable in terms of providing unique information. Indeed, IVM in rodents generates rich data sets that can be analysed deeply and uncover unexpected and otherwise untestable biological mechanisms in healthy and diseased tissues. For example, IVM has revealed the cooperation of multiple cell types to enable the intravasation of immune cells during inflammation6 or metastatic cells leading to tumour spread7, and the unexpected migratory behaviour of multiple stem cells8–11 and chemoresistant tumour cells12,13.
Experimentation
Microscopy modalities
Modern IVM platforms consist of a laser, a scan head and detectors, which enable either point, line or multifocal scanning, or light sheet detection of intact tissue in living animals14,15 (TABLE 1). Wide-field and confocal microscopes detect fluorophores by single-photon excitation with light in the visible wavelength range (<700 nm). Single-photon excitation achieves high resolution (down to 200 nm) and imaging speed (multiple frames per second) within around 100 μm from the tissue surface, including precise detection of cell organelles, membrane trafficking and cytoskeletal dynamics16,17. Excitation by visible light suffers from strong scattering by tissue structures, leading to loss of signal and resolution when focusing deep inside tissues18 (FIG. 1a). For fast imaging of superficial tissue layers, wide-field microscopy enables frame rates up to 1,000 s−1, which is sufficient for monitoring fast calcium currents and arterial blood flow.
Table 1 |.
Microscopy techniques used for intravital microscopy
| Microscope | Strength for IVM | Weakness for IVM |
|---|---|---|
| Wide-field | Very sensitive Fast imaging Commonly available microscope Cost-efficient |
Imaging depth is limited to tens of micrometres in non-cleared tissues Poor z-resolution |
| Light sheet | Very fast imaging of large areas in three dimensions Minimum photodamage and fluorophore bleaching Optimally suited for embryos, brain and fixed tissue after clearing |
Very large data sets Imaging depth is limited to tens of micrometres in non-cleared tissues Cannot be applied to rodents Requires specialized microscopes and expertise Susceptible to shadowing artefacts |
| Single-point scanning confocal | Thin optical section Commonly available microscope |
Imaging depth is limited to 100 μm in non-cleared tissues Slow imaging |
| Spinning disc confocal | Thin optical section Fast imaging of single planes |
Imaging depth is limited to tens of micrometres in non-cleared tissues Requires bright samples due to large light losses |
| Multiphoton | Improved imaging depth with hundreds of micrometres for three-photon and four-photon excitation in non-cleared tissues Thin optical sections Enables higher harmonics of tissue structures Up to seven channels simultaneously detected |
Requires specialized microscopes and expertise Costly |
Fig. 1 |. Intravital microscopy by multiphoton excitation.
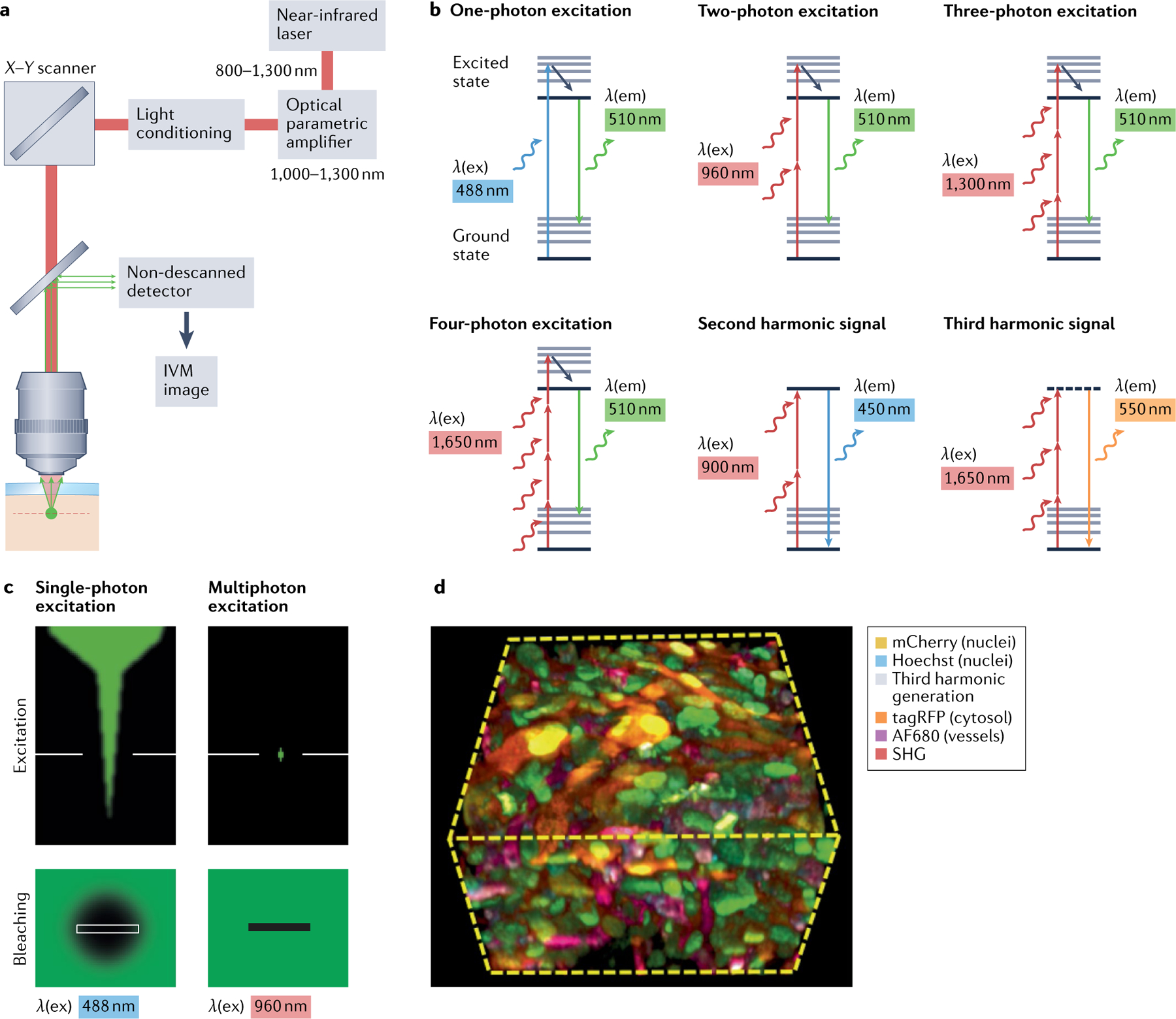
a | Multiphoton microscopy platform. A pulsed pump laser is introduced into a tunable oscillator or amplifier to generate high-power laser light in the range of 1,000–1,700 nm. Multiphoton-excited signal in the sample is detected without the need of a pinhole. b | Jablonski diagrams of one-photon, two-photon, three-photon and four-photon-excited fluorescence and higher harmonic generation. Arrows indicate individual photons causing excitation (upward arrows) and emission (downward arrows). c | Single-photon excitation excites all fluorophores potentially causing photodamage in and out of the imaging focus; two-photon and multiphoton excitation occurs only in the imaging focus and, thereby, minimizes photodamage in out-of-focus planes. d | Example of a multicolour intravital microscopy (IVM) image including second harmonic generation (SHG), third harmonic generation and multiple colour channels. Displayed tissue volume in xyz represents 210 × 210 × 110 μm3. Part d adapted from REF.21, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Multiphoton imaging.
To excite deeper layers and allow 3D imaging in complex in vivo environments, multi-photon imaging is the method of choice. Near-infrared (700–1,000 nm) or infrared (1,000–1,700 nm) laser light is introduced into the sample by pulsed lasers, causing high photon density in the focus and the excitation of fluorophores by multiple photons18,19 (FIG. 1a). In contrast to single-photon excitation, two-photon imaging fluorophores are excited by simultaneous absorbance of two photons with half the excitation energy18,19 (FIG. 1b). More recently, infrared pulsed light sources can achieve three-photon and four-photon excitation of tissue-intrinsic fluorescent molecules and exogenous fluorescent reporters, as well as generate high harmonic light scattering20,21 (FIG. 1b). Multiphoton excitation allows penetration up to 1 mm in some tissues and causes less photodamage due to a lack of excitation in out-of-focus regions, rendering this approach suitable for deep tissue microscopy in small animals22 (FIG. 1c). As a recent development, increasing the pulse energy of the laser light but reducing the pulse repetition rate (from 80 MHz down to 1–2 MHz) allows a dramatic increase in photon density within the laser focus, without increasing photodamage to the sample21,23,24. This high photon density enables three-photon excitation at an excitation wavelength of 1,300 nm, and at 1,650–1,700 nm a combination of three-photon excitation of red fluorophores together with four-photon excitation of green and blue fluorophores (FIG. 1b). Up to twofold increased tissue penetration is achieved in the brain and peripheral tissues when using three-photon and four-photon microscopy compared with conventional two-photon microscopy, due to increased pulse energies, due to improved signal-to-noise ratio and because 1,300 nm and 1,650–1,700 nm both reside in a region of low water and biomolecule absorption, known as the near-infrared window21,25,26. Using an integrated platform design, it is now possible to combine multicolour confocal microscopy with multiphoton imaging in the near-infrared and infrared excitation spectrum27–29 (FIG. 1d).
Commercial microscope vendors have made great strides in microscope hardware, incorporating features such as FLIM electronics30, optical parametric oscillators27,29, group velocity dispersion compensators31, multicolour detection32 or video-rate acquisition33, all of which were once only research endeavours and of limited accessibility. To allow for further customization of multiphoton microscopy systems, companies offer kits and publish open-source designs34 to reduce the complexity of system design and remove technical barriers. Novel systems are still pushing the envelope of system capabilities, incorporating new features such as multispectral/hyperspectral imaging35,36, unique light sources21,37 or mesoscale imaging38. Software for system control and data acquisition is intimately tied to the hardware chosen. Commercial systems, or those that build upon them, must use the software provided by the company. Kits and home-built systems provide the option either to write software from scratch or to employ one of several open-source initiatives39–43. Image analysis can be performed in various different commercial or open-source platforms.
Upright and inverted configurations.
The two major microscope configurations used for IVM are upright and inverted microscopes. For inverted IVM, the sample is immobilized on the stage above the objective. Conversely, for upright IVM the objective is positioned above the sample and can be dipped into surgically exposed samples or positioned above a coverslip, which helps to immobilize and flatten the sample. Although the majority of tissues can be imaged using both configurations (FIG. 2a), intravital brain microscopy is commonly performed on upright microscopes (FIG. 2a), which allows immobilization/stabilization of the animal head using stereotactic head holders44–46.
Fig. 2 |. Intravital microscopy of tissues through optical imaging windows.
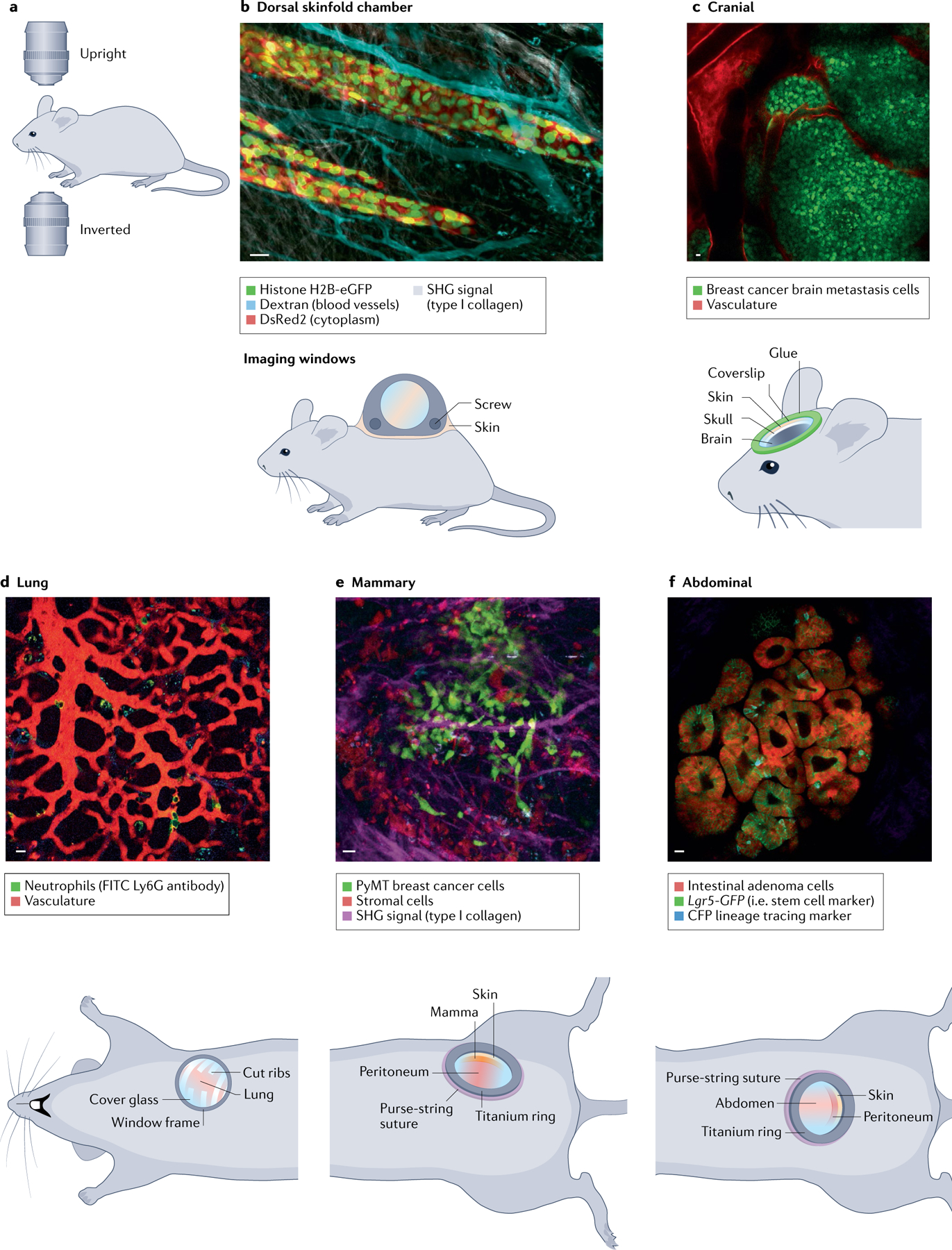
Murine models can be imaged with upright or inverted microscopes (part a). Examples of chamber types (cartoons) and representative example multicolour intravital microscopy (IVM) images taken through the imaging windows of the dorsal skinfold chamber (part b), cranial window (part c), lung window (part d), mammary imaging window (part e) and abdominal imaging window (part f). Scale bars represent 20 μm. eGFP, enhanced green fluorescent protein; SHG, second harmonic generation. Part b (IVM image) reprinted with permission from REF.384, Rockefeller University Press.
Gaining optical access
Several vertebrate animal models, including zebrafish, are naturally transparent during early developmental phases, and therefore well suited to perform IVM. In small rodents, including commonly used mouse models, a few organs present optically non-turbid surfaces — for example, skin, eyes and the oral cavity — that allow direct access for in vivo microscopy. Gaining optical access at a cellular or subcellular resolution at much deeper sites is essential for many experiments.
Microscope access to most tissues and organs requires surgical intervention, either by ad hoc surgery to expose the tissue for direct coupling to the optics or by implantation of an optical window (FIG. 2b–f). Ad hoc approaches restrict imaging to limited periods of time (a few hours) due to the risk of side effects caused by extended use of anaesthetics and by limitations in maintaining the physiological conditions of exposed organs. Ad hoc approaches are therefore mostly used to visualize short-term cellular dynamics such as cell migration or invasion47–49. Optical windows protect the tissue, reduce motion artefacts and enable longitudinal imaging over days to months50–54 (FIG. 2b–f). A multitude of imaging windows have been developed over the past years providing optical access to nearly all organs in the mammalian body, including brain, breast and abdominal organs50. Windows were initially fabricated using passivated stainless steel, titanium or selected bio-compatible polymers. Recent advances include the development of flexible silicon windows to image subcutaneous organs such as muscles and mammary tissue55,56; the imaging window with a replaceable lid, which provides physical access to the tissue of interest over the entire duration of the imaging experiment57; as well as the development of an imaging window to follow embryonic development in utero at a high resolution58. Other windows that are not frequently used but can improve imaging depth beyond the limits imposed by the scattering property of the tissue include gradient-index lenses and prisms59–61. Gradient-index lenses are based on either a single lens or a series of thin lenses (diameter 1–2 mm) inserted into soft tissues (primarily the brain) to reach the desired depth. Prisms are inserted with the hypotenuses coated with a reflective material, enabling the translation of the rastering pattern of the excitation beam from an x–y plane to an x–z plane.
Animal maintenance
In order to limit sample movement, rodents typically need to be anaesthetized for IVM. Gaseous isoflurane is commonly used for induction and maintenance of anaesthesia (5% or 0.75–3% anaesthesia in oxygen or air, respectively, at an oxygen flow rate of 1–2 l min−1). Imaging through chronic imaging windows is not invasive and therefore requires a minimal level of anaesthesia, allowing rapid recovery of the animal, even after extended periods of time (>8 h)50,62–64. For more invasive procedures where the tissue of interest is surgically exposed for IVM (such as skin flap surgery or acute imaging windows)64–66, analgesia is required for pain relief in combination with anaesthetic agents. For example, a combination of zoletil or ketamine and xylazine has been used for non-recovery IVM of subcutaneous tumours exposed via skin flap surgery, which involves euthanizing the animal after imaging65–67. Other combinations involving the use of midazolam, fluanisone and fentanyl have also been described for acute non-recovery IVM68,69. In order to avoid a too light or too deep plane of anaesthesia, the animal can be monitored by checking reflexes (such as in response to a toe pinch), by observing respiration or by monitoring the heartbeat, respiration and blood oxygenation57,65,67,70.
Although anaesthetics and analgesics can help to limit animal movement and reduce pain sensation, they may also interfere with physiological processes. For example, it has been shown that different anaesthetic and analgesic agents can distinctly affect cellular activity, behaviour and function71–73 compared with the awake state. Care should therefore be taken to choose an anaesthetic and analgesic regimen which does not affect the biological processes to be observed by IVM and to design complementary (non-imaging) experiments to further support the experimental hypothesis.
Keeping the sample happy should be the hallmark of in vivo imaging74 (BOX 1). During IVM in rodents, the body temperature needs to be maintained near 37 °C to ensure physiological cell and tissue function and vascular flow (which regulates tissue oxygenation, nutrient supply/metabolism as well as the behaviour of circulating cells such as cancer cells)75. This is essential as anaesthetized animals lose the ability to maintain body temperature independently. Temperature fluctuations have been shown to affect IVM readouts, such as fluorescence lifetime measurements76, highlighting the importance of a physiological environment throughout the IVM session. In anaesthetized rodents, heated environmental enclosures, heating blankets and/or a heated stage are commonly used to maintain body temperature, which can be monitored using a rectal probe. The use of heated immersion objectives or dry objectives may help to maintain temperature in the tissue of interest, counteracting heat transfer away from the sample via a cooler immersion medium77. Where possible, temperature maintenance near the tissue of interest can be monitored using a small probe close to the tissue and coverslip. IVM using a microscope set-up that is fully integrated into an enclosed incubating chamber can further help to maintain animal/sample temperature while also reducing temperature fluctuations of the IVM equipment.
Box 1 | Responsible use of intravital microscopy: beyond the 3Rs.
Intravital microscopy (IVM) touches upon the public and ethical debate on animal testing. This requires careful reflection on the application of IVM.
There are legal frameworks that provide guidance in the context of IVM, such as EU Directive 2010/63. Although the technique is used on a global level, legal frameworks often function only for a specific region or country. A mere reference to a legal framework will not suffice for the responsible use of IVM in general. There exist diverse views on the value of animals in research, and there are differences between regions and between animal species. For instance, the use of dogs in research often raises more public discussion than the use of mice, and replacing mice with zebrafish in IVM is sometimes considered a preferred alternative, even though these three animal species have similar capacity to experience pain and can be harmed by the testing procedures. Finally, the 3R principles (replacement, reduction and refinement)385 have been presented as an essential step to more responsible practice in animal research, with the aim to decrease animal use and reduce the impact of the procedures for the involved animals. Although these principles are important for responsible animal research, a simple reference to these principles is not sufficient, and the principles can be in conflict with each other. For instance, the tension between using a small number of animals for an invasive procedure where the tissue of interest is surgically exposed for IVM against a design that is less invasive but requires ten times more animals. The 3R principles provide no direct guidance in this case and often overlook the underlying reasons for or against the use of IVM. These principles function as guidance to improve the system of animal testing, but as such do not provide the ingredients for the societal and ethical justification385,386.
The applications of IVM should be in line with recent views on responsible animal research. At the experimental design stage, this entails a testing strategy that reduces in vivo experiments. A comparison of non-invasive IVM methods with in vivo results is an important step to find ways to apply IVM techniques that do not harm animals or that no longer require the use of animals. Exchange between research groups and consorted action are required to adhere to the international landscape standards. This exchange may also contribute to harmonization of guidelines on an international level. The harms and benefits of IVM should be carefully assessed, including the application of the 3R principles. Establishment and maintenance of a culture of care387 is also needed for all who are involved in the use of animals for IVM, including an attitude of care for the animals, careful compliance with protocols and guidelines, and professional development.
Ethical reflection should be an integrated part of IVM to provide unique insights into the in situ behaviour of cells, and to solve fundamental biological and medical problems.
For long imaging sessions (>3 h), the animal should also be supplemented intraperitoneally, subcutaneously or via intravenous catheters with fluids, such as saline, in order to prevent animal dehydration. Dehydration of the tissue of interest itself is another challenge in IVM, which can lead to tissue shrinkage as well as motion artefacts during imaging. Whereas optical imaging windows above the tissue of interest can prevent tissue dehydration, surgically exposed tissues require a moist environment, such as constant flushing with warm Tyrode’s salt solution or saline78,79 or applications of selected gels80, to protect from dehydration. Incubating chambers enclosing the microscope set-up can help to circulate humidified air around the sample to maintain humidity/hydration.
Fluorescent labelling
In order to visualize molecular and cellular behaviour in vivo, many IVM approaches rely on the use of fluorescent protein tags to provide spatial and temporal information on a protein of interest. In particular, red-shifted fluorescent proteins are well suited for IVM. Red-shifted emission light has long wavelengths leading to less scattering in tissues, thereby permitting increased imaging depth in tissues and reducing the contribution of endogenous tissue fluorescence to the overall signal. However, red and far-red fluorescent proteins often suffer from poor quantum yield and low photostability, which limits the available photon budget per sample, reduces the signal-to-noise ratio and impedes long-term imaging. Furthermore, many red-shifted fluorescent proteins present as oligomers, which can impose steric hindrance if tagged to a protein of interest and lead to biological artefacts. The recent generation of monomeric red and far-red fluorescent proteins with improved brightness and photostability81,82, or the genetically engineered red-shifting of existing green fluorescent proteins83, has helped to overcome these limitations for IVM studies.
Labelling strategies for IVM may include the implantation of fluorescent cells into animals, for example in tumour xenograft settings84–86, studies of immune cell behaviour87,88 or to visualize exchange of molecules between cells carried by extracellular vesicles89–91. Where cells and tissues can readily be accessed, in vivo electroporation may be used for delivery of fluorescent protein-encoding genes as demonstrated for IVM of embryos58, postnatal tissues, such as the brain92, retina93 and skin94, or tumours95. Viral gene delivery can also be achieved in vivo, for example via viral inhalation to label the lung96 or systemic administration to label abdominal organs such as the liver97. Other labelling agents may include fluorescent dextrans or quantum dots for IVM of the vasculature7,69, fluorescent antibodies for affinity labelling of specific cell-surface proteins (intravital immunofluorescence)98,99, probes for intracellular pH changes100, reactive oxygen species generation100,101, protease activity102 or dyes to label fatty acids103, lipid droplets104 or vesicles105.
For specific and permanent labelling of cell types and tissues, genetically engineered mouse models may be used, where fluorescently labelled proteins are expressed from ubiquitous gene loci, such as Rosa26 or Hprt, or from endogenous gene loci to recapitulate native expression patterns. Knock-in of fluorescent proteins in the endogenous locus can be used to visualize cell state changes in real time using IVM. For example, endogenous labelling of CDH1 with a fluorescent protein was used to monitor real-time CDH1 localization and expression levels as a readout for epithelial-to-mesenchymal transition during cancer progression106,107. Similarly, using a mouse model in which enhanced green fluorescent protein (eGFP) is knocked-in behind the Lgr5 gene, in vivo stem cell plasticity was visualized over time in homeostatic and tumorigenic conditions108–110. To follow cell cycle progression using IVM, transgenic fluorescence ubiquitination-based cell cycle indicator (FUCCI) reporter mice were generated wherein a cyclic change in colour from green over yellow to red can provide information on the cell cycle phase in which an individual cell resides111. This has, for example, been used to demonstrate that myogenic cells migrate faster during S/G2/M phase during muscle regeneration112, whereas invadopodia formation predominantly occurred during G1 phase in a mouse model of breast cancer113. FUCCI IVM has also been used to optimize response to standard-of-care chemotherapies in preclinical cancer models84. Furthermore, transgenic mice have been generated using fusion proteins of fluorescent timers, where a change in spectral properties occurs following synthesis over the course of fluorescent protein maturation. For example, a fluorescent timer fusion to histone H2B knocked into the Hprt locus was used to determine/map individual cell cycle length in vivo114. Similarly, fusion of the fluorescent timer to the TCR downstream gene Nr4a3 or the transcription factor Foxp3 was used to characterize the temporal dynamics of TCR signalling and transcription factors115,116.
Cre-inducible multicolour reporter mouse models can be used to study long-term cellular dynamics and lineage tracing. For example, transgenic Brainbow mice express a Cre-inducible brainbow cassette consisting of up to four fluorescent proteins under the control of the neuron-specific Thy1 promoter. Here, Cre recombination allows for stochastic fluorescent protein expression to individually label Thy1-expressing cells with an arbitrary combination of fluorescent proteins in order to map neuronal circuits in the brain117,118. This concept was expanded upon by inserting brainbow into the Rosa26 locus (Confetti mice)119 or the β-actin locus (Rainbow mice) to induce fluorescent protein expression in any tissue or cell type of interest using Cre driver lines, which allowed live lineage tracing in developing embryos120 and the mammary gland66,121 or the in vivo tracking of stem cell fate in intestinal crypts108,119, the mammary gland121,122 and tumours.
Label-free techniques
The inherent fluorescent properties of endogenous molecules can also be used to provide a label-free context to IVM data. Label-free strategies have the advantage that they provide environmental information without the need for genetic modification or in vivo labelling of tissues. Label-free strategies exploit the excitation of select endogenous molecules by either linear or non-linear methods. For example, NADH, flavonoids and other small molecules are extensively used to visualize cellular architecture and metabolism by using either confocal or multiphoton microscopy123. Less commonly used techniques that are technically challenging, such as coherent anti-Stokes Raman scattering and stimulated Raman scattering, and label-free ultrasound and opto-acoustic imaging, have been used to image, respectively, lipids and myelin, or DNA88,124,125 and the vasculature or tissue oxygenation126,127.
A commonly used label-free imaging method in IVM is higher harmonic generation, a light scattering process that arises when light interacts with electrons bound to molecules via non-linear restorative forces, causing the emission of photons with frequencies that are integer multiples of the illumination light128. Substantial higher harmonic generation is only possible either with high-illumination intensities (such as occurring with high peak power ultrashort femtosecond pulses that simultaneously reduce the risk of photodamage and phototoxicity), highly non-linear restorative forces or in polymeric molecules where the monomers are spaced such that emitted photons from each monomer can efficiently transfer their momentum along the macromolecule to generate large aggregate signals129. Second harmonic generation (SHG) results in the frequency doubling of photons. Biological structures efficient in generating SHG include fibrillar collagen, actomyosin and biomaterials15,130. Third harmonic generation results in the frequency tripling of light and can originate from cell and tissue interfaces with a mismatch of refractive index, such as tissues that contain water–lipid and protein–lipid interfaces131,132 (such as cell membranes or lipid droplets) or endogenous compounds such as metabolites132,133.
Lastly, the visualization of endogenous fluorophores, such as the intracellular metabolites NAD(P)H and FAD, can be used as a label-free imaging technique to map cells and their metabolic state in live tissues133. Metabolite IVM has, for example, previously shown promise for the detection or delineation of diseased and healthy tissues due to the differential expression and abundance of endogenous fluorophores134. This ability for label-free FLIM imaging may also open up opportunities for applications in human volunteers and patients to diagnose disease tissue or the disease stage using IVM and endomicroscopy, demonstrating the bench to bedside capacity of in vivo imaging135,136. Closely related to label-free imaging of endogenous molecules is the use of autofluorescence generated by molecules introduced into a biological system for other reasons. For example, some anti-cancer therapeutics are themselves fluorescent and this property can be used to track drug distribution and uptake137,138.
Advanced techniques
Intravital photobleaching, photoactivation and photoconversion applications.
Whereas fluorescent labels can provide spatial information on a cell or protein of interest, tracking this signal over time can provide insights into molecular and cellular dynamics and behaviour in vivo139,140.
Photobleaching techniques, such as fluorescence recovery after photobleaching (FRAP) (FIG. 3A) or fluorescence loss in photobleaching (FIG. 3B), can be used to assess the mobility of fluorescently labelled proteins or agents. FRAP involves the laser-mediated bleaching of the fluorophore in a defined region of interest followed by tracking of fluorescence recovery over time (FIG. 3A). This approach provides information on the mobile and immobile fractions of a molecule of interest, its half-time of recovery and the mode of movement — for example, lateral diffusion versus cytoplasmic exchange for membrane proteins. Fluorescence loss in photobleaching serves as a complementary approach where the loss of fluorescence adjacent to a bleached region is monitored over time (FIG. 3B). Intravital FRAP and fluorescence loss in photobleaching imaging have been used to assess cell–cell junction dynamics during cancer progression68,141, subcellular protein dynamics in actomyosin cytoskeleton networks of the salivary gland16,140,142, granule trafficking in dendrites of epidermal T cells143, glutamate receptor dynamics in dendritic spines144 or the dynamics of protein aggregation in the brain, which can often underlie neurodegenerative diseases145. Similarly, intravital FRAP using fluorescent dyes can also be used to characterize changes in vessel leakiness upon wound healing146, lymphatic viscosity and flow in a mouse model of arthritis147, cell–cell communication via gap junctions148–150 or molecule diffusion and interstitial connectivity in extracellular spaces151,152.
Fig. 3 |. Advanced imaging techniques.
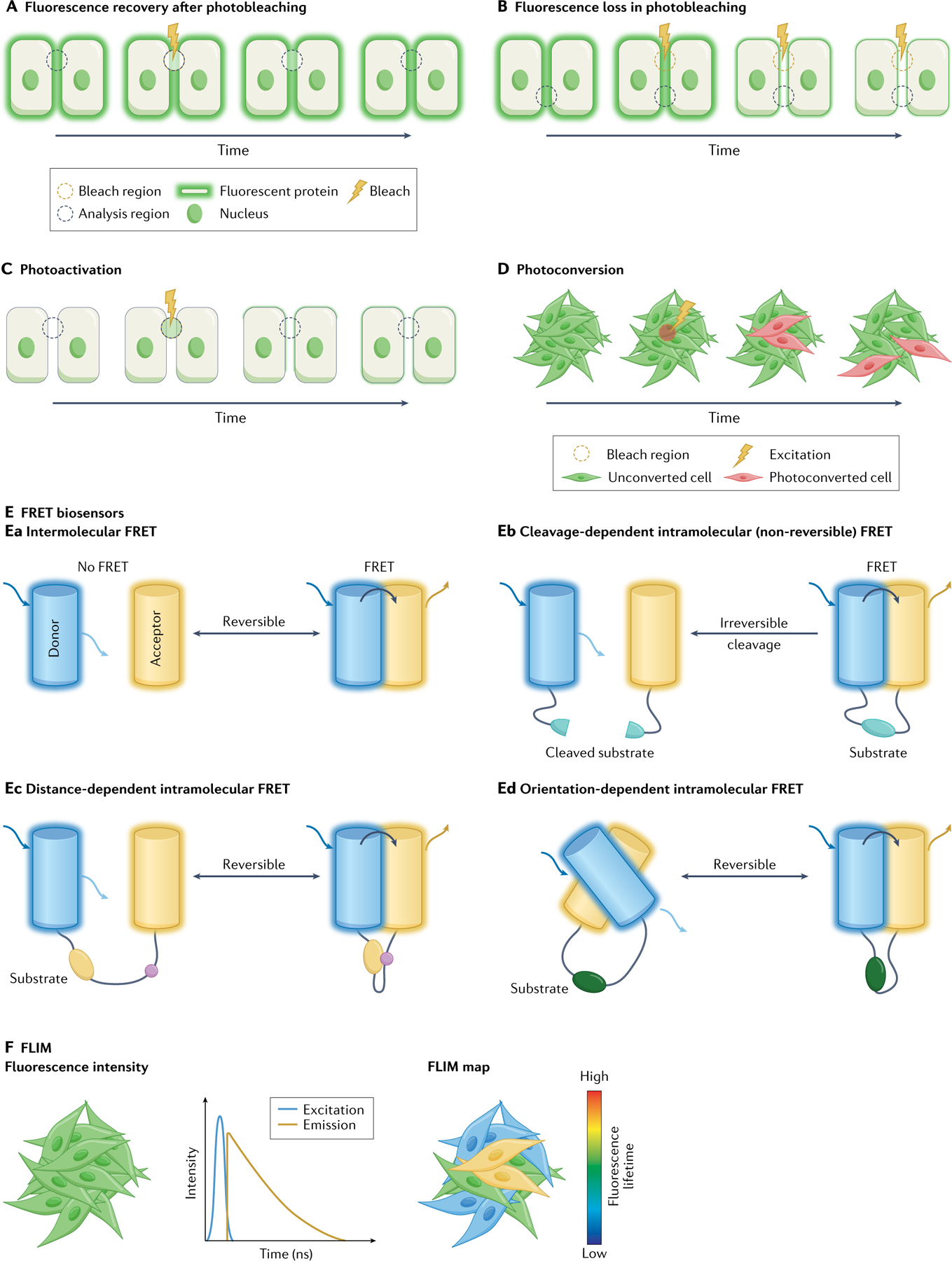
A | Fluorescence recovery after photobleaching (FRAP) imaging involves the laser-mediated bleaching of a fluorophore in a region of interest (yellow dashed circle) followed by monitoring of fluorescence recovery into the same region of interest (black dashed circle). B | During fluorescence loss in photobleaching imaging, the fluorophore in a region of interest is continuously bleached (yellow dashed circle) while observing loss of fluorescence in adjacent/connected regions (black dashed circle). C | Photoactivation studies use caged (non-fluorescent) fluorophores, which can be uncaged and activated by a laser pulse to emit a fluorescent signal (yellow dashed circle) followed by monitoring the loss of fluorescence in the same region of interest (black dashed circle). D | During photoconversion the spectral properties of a fluorophore are permanently changed, which can, for example, allow/facilitate the tracking of individual cells in large populations. E | Examples of Förster resonance energy transfer (FRET) biosensors: intermolecular FRET from the donor fluorophore (cyan) to an unlinked receptor fluorophore (yellow) (part Ea); cleavage-dependent FRET biosensors whereby an irreversible cleavage of the substrate peptide leads to irreversible loss of/decrease in FRET (part Eb); distance-dependent FRET biosensors whereby a reversible change in the linker region leads to a reversible modification in the distance between donor and acceptor fluorophores, thus affecting FRET efficiency (part Ec); and orientation-dependent FRET biosensors whereby a reversible modification in the linker region leads to a reversible change in the alignment between donor and acceptor fluorophores, thus affecting FRET efficiency (part Ed). F | Fluorescence lifetime imaging microscopy (FLIM) is used to quantify the fluorescence lifetime, which is the time a fluorophore spends in the excited state prior to emission of a photon, an inherent feature of each fluorophore, which also depends on environmental conditions. Parts A–C and E adapted from REF.139, CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
Photobleaching imaging can be complemented by photoactivation studies, wherein an initially caged (non-fluorescent) agent is activated via a laser pulse to emit a fluorescent signal (FIG. 3C). The expansion of this localized pool of activated fluorescent proteins (or loss of fluorescence in the activated region of interest) can be tracked over time (FIG. 3C) to provide information on protein mobility, movement and kinetics, such as tracking protein dynamics in live tumours141, the bile flux mechanisms in the live liver153 or transcription factor dynamics in living embryos154. For further details on the set-up of photobleaching imaging studies, we refer the reader to two publications155,156.
Photoconvertible agents are another excellent tool for IVM studies that involve tracking of dynamic events, such as cell fate mapping51,157, the movement of cancer cells70,158–163 or immune cells164–166, or the dynamics of subcellular compartments167. Upon excitation at a specific wavelength, photoconvertible proteins, such as the green to red converting proteins Kikume, Kaede or Dendra2, for example, undergo an irreversible switch in their spectral properties due to a light-induced peptide cleavage (FIG. 3D). This can be explored to precisely label and track individual cells (FIG. 3D) with the aforementioned advantage over photoactivation that a fluorescent signal is already obtained prior to photoconversion.
FRET imaging and biosensors.
FRET involves the transfer of energy from a donor fluorophore to an acceptor fluorophore provided that the donor and acceptor display sufficient spectral overlap, proximity and alignment to each other (FIG. 3E). FRET can be quantified using ratiometric microscopy or FLIM.
Intermolecular FRET involves the energy transfer between unlinked donor and acceptor fluorophores, which can be used to assess heterotypic or homotypic protein–protein interactions168 (FIG. 3Ea). Intramolecular FRET in FRET biosensors occurs between donor and acceptor fluorophores that are physically connected to each other via a linker substrate, and can be used to study molecular activity, such as proteolytic cleavage or phosphorylation by protein kinases. For example, cleavage-dependent FRET biosensors contain a linker sequence that can serve as a protease substrate, where protease-mediated cleavage of the linker irreversibly separates donor and acceptor fluorophores from each other, decreasing FRET efficiency (FIG. 3Eb). Examples of cleavage-dependent FRET biosensors include biosensors to assess caspase activity (which coordinates cell apoptosis)1 or matrix metalloproteinase activity (which regulates remodelling and turnover of extracellular matrix)169.
Proximity-dependent and orientation-dependent FRET sensors contain a linker that serves as a substrate for post-translational modification, thereby affecting the proximity and alignment of donor and acceptor and, subsequently, FRET efficacy (FIG. 3Ec,Ed). These reversible FRET biosensors include reporters for kinase activity such as ERK170–174, AKT/PKB175,176, SRC69,177,178, FAK84,179,180 or ROCK181,182 and can provide information on drug target inactivation dynamics in vivo to see whether, for how long and to what extent a given drug target can be inhibited. This information can be used to fine-tune drug treatment regimens in vivo for maximized drug target engagement while minimizing overtreatment, which is often associated with side effects and toxicities69,84,135,159,177,183,184.
Furthermore, proteins themselves can serve as a linker where changes in protein domain binding or protein folding and configuration determine FRET efficiency. Examples include the Raichu FRET biosensor series of small GTPases where the small GTPase binds to or dissociates from the binding domain dependent on GDP (inactive) or GTP (active) loading, thereby affecting FRET efficacy to quantify rapid changes in these notoriously difficult to assess signalling events in live tissue159,184,185. Similarly, IVM of a glucose FRET biosensor, which contains the bacterial MglB protein as a substrate for reversible glucose binding, was used to reveal single-cell heterogeneity of breast cancer cell metabolism in vivo with higher levels of intracellular glucose detected at the tumour borders186. Lastly, FRET biosensors of microenvironmental conditions have also been described, where changes in oxygenation or pH can lead to altered fluorescent protein folding of biosensor configuration, which consequently affects FRET efficacy187,188.
Exciting new developments allow multiplexed imaging of several FRET biosensors to simultaneously assess several biological or molecular processes at the same time. Especially, the generation of new red-shifted and far-red-shifted FRET pairs facilitated the simultaneous imaging of several FRET biosensors81,83,189,190 with application for deep in vivo imaging as well as multiplexing with optogenetic tools to switch on distinct signalling pathways190,191. Recent live cell imaging studies have further advanced the field and highlight what may be possible in IVM in future by specifically interrogating protein activity at a distinct/endogenous subcellular location using split fluorescent proteins and split FRET biosensors192, mapping of signalling molecule gradients using FRET-based nanorulers193 or the tracking of entire signalling networks using biosensor barcoding in combination with deep learning-based analysis approaches194. For a detailed overview on FRET biosensors, the reader is referred to the following review articles180,195,196.
Fluorescence lifetime imaging microscopy.
FLIM can be used to quantify the fluorescence lifetime, which is the time a fluorescent molecule spends in the excited state before emitting a photon and returning to the ground state (FIG. 3F). The fluorescence lifetime is an inherent property of each molecule which varies depending upon the molecule’s environment. The fluorescence lifetime can therefore be used to identify individual fluorescent species (FIG. 3F). FLIM imaging can help to resolve or separate signals from overlapping spectra or be used to quantify changes in FRET efficiency, for example in FRET biosensors. This has been used to assess many key aspects of in vivo biology such as calcium dynamics in B cells in vivo197, real-time signalling dynamics during cancer cell extravasation into the live liver178, to map endogenous fluorophores such as the intracellular metabolites NAD(P)H and FAD, in live tissues133,198 or for drug target engagement in a model of ovarian cancer metastasis using intravital endomicroscopy199.
Image acquisition
Microscope acquisition parameters such as the frame rate, resolution and magnification are dictated by the biological processes of interest, but must also consider limitations of the microscope, the signal source and the tissue properties as well. In principle, aiming for the Nyquist condition — where acquisitions are performed at more than twice the highest spatial or temporal frequency component in each of these parameters — is ideal. However, the interplay between the frame rate, magnification and light exposure means that this ideal is not always attainable. For example, higher magnification often means a laser beam scans over a tissue at a slower rate, increasing the amount of light exposure, phototoxicity and photodamage. Attempting to capture rapid dynamics requires high frame rates, but high frame rates increase the overall exposure of the tissue to light and may lead to photodamage. Optimal conditions attempt to maximize tissue exposure while minimizing photodamage and phototoxicity. Although this is highly dependent on the microscope and experiment, there are some guidelines that can be provided. Resolution is limited by either the optical resolution (laser spot size determined by the numerical aperture of the objective lens) or the pixel size (determined by the scan angle and magnification of the objective). Scan angles and line scan speeds determine the pixel dwell time (how long a laser spends illuminating a pixel). Longer pixel dwell times increase exposure, but acquisition electronics typically do not capture this increased signal29. In addition, continuous excitation may induce photodamage more rapidly than allowing molecules a period of rest. Thus, it is better to increase the signal-to-noise ratio by averaging multiple scans, rather than increasing the illumination intensity. When averaging, line averages can accommodate motion artefacts better than frame averages. Phototoxicity and photodamage can be assessed by recording a large number of images in succession and plotting the signal intensity over time, and by observing the tissue behaviour in real time (for example, with cell stress reporter activity) or cell death and inflammation after imaging. The highest laser power that does not cause cell stress and/or photodamage at a given magnification and scan angle should be used.
Overcoming physiological motion
Physiological motion, such as from respiration, heartbeat, peristalsis or muscle and vascular tone, can affect tissue geometry over longitudinal IVM sessions and lead to blurred images200, and is one of the major challenges of IVM regardless of the type of IVM imaging platform. Motion correction and compensation strategies include gating approaches to synchronize image acquisition to the cardiac and/or respiratory cycles in order to obtain IVM data when physiological motion is minimal. However, it should be noted that this approach can result in a trade-off for temporal resolution as IVM only occurs at dedicated time points. Many studies have focused on suppressing sample motion during IVM. This often poses a trade-off or compromise between stabilizing the sample without introducing artefacts that might affect the biology to be observed. In order to reduce motion of surgically exposed tissues or readily accessible tissues, such as the tongue, during IVM, custom-made holders can be used with care to not interfere with the blood circulation between the exposed tissue and the animal16,201,202. Optical imaging windows can also be surgically inserted above the tissue of interest with adhesives between the tissue and the window frame providing stability50,52,53,70,203. Additional stability for IVM through optical imaging windows can be achieved if the window can be fitted into a custom-made window stage, which can be readily added to an inverted IVM microscope set-up. Similarly, stereotactic head holders have been glued to surgically exposed skull bone to stably assess the interactions between osteoblasts and osteoclasts during bone turnover204,205. Extremely slow, non-periodic movements such as tissue growth can also occur which do not necessarily impede IVM data acquisition itself but might have to be accommodated for during longitudinal IVM studies. Here, the suture-less insertion of silicone-based imaging windows above the tissue of interest for up to 35 days can prevent growth restriction55,56.
Results
Correcting for physiological motion
In addition to physical restraint, real-time or post-IVM correction approaches have also been used. For example, an off-path light beam was used to measure motion displacement followed by adjustment of the objective position in order to correct for axial motion206,207. In another example, a feedback system was used that is based on anatomical 3D landmarks, which can be obtained from the SHG signal or blood vessels, in combination with high-speed computing to correct for sample drift208. Furthermore, the ImageJ macro Intravital_Microscopy_Toolbox can be used to automatically remove severely distorted images if they exceed a dissimilarity score to a reference frame209. The software tool IMART achieves image stabilization using alignment and similarity to a reference frame by rigid registration of images to correct for overall movement followed by non-rigid registration to correct for small-scale aberrations/movements210. StackReg (recently modified as HyperstackReg to work on 4D hyperstack data) is a sub-pixel registration algorithm for Fiji that minimizes the mean square difference of intensities between images211–213. Similarly, Galene was developed to correct for physiological motion in FLIM data, for instance when performing FRET measurements in intestinal crypts, which are subject to peristaltic movement178. FIGURE 4 shows the Galene workflow for correction of FLIM–FRET data correction, which can also be followed to correct motion in other IVM data sets.
Fig. 4 |. Overcoming tissue motion.
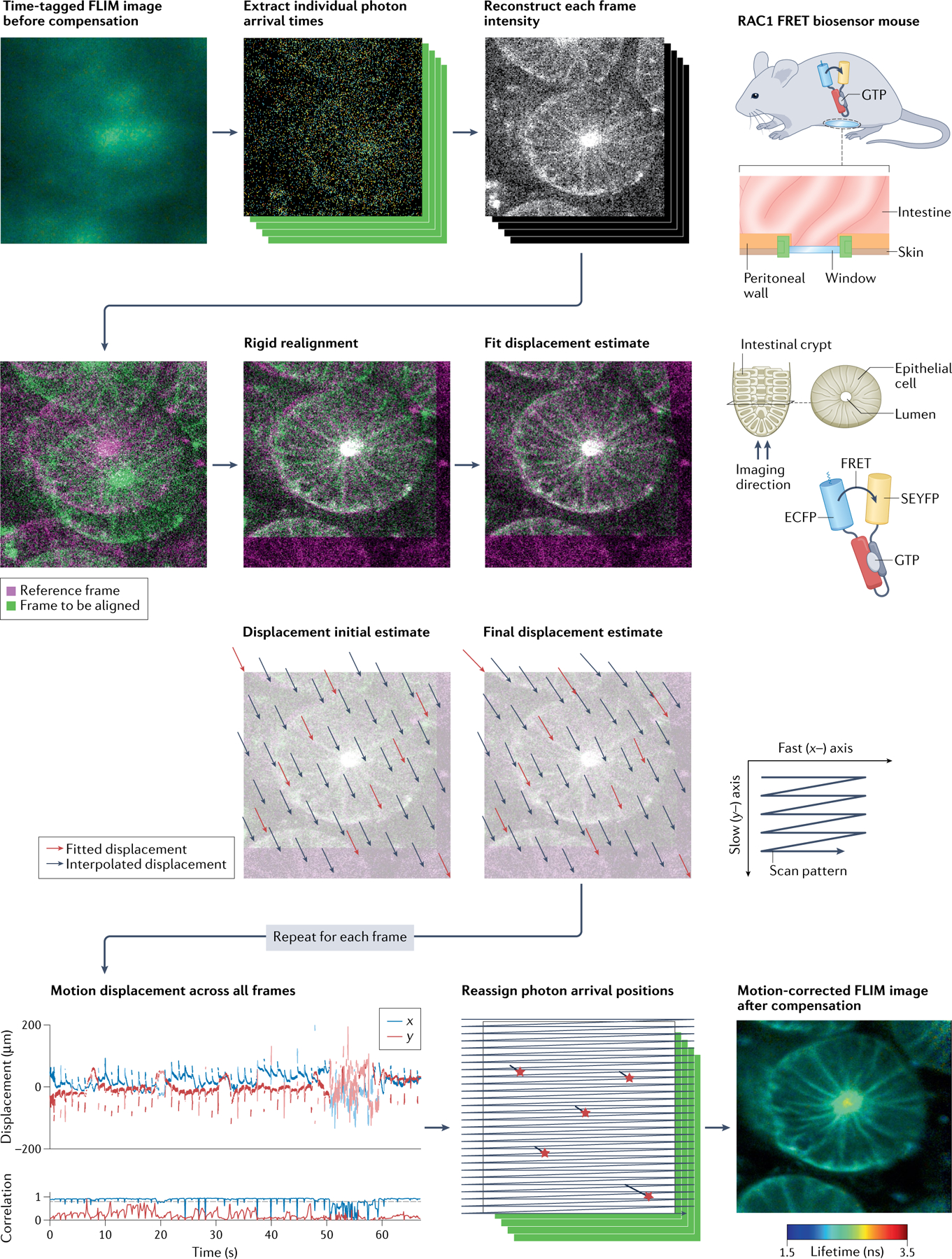
General workflow for motion correction of intravital fluorescence lifetime imaging microscopy (FLIM)–Förster resonance energy transfer (FRET) data using the software Galene. Physiological motion, such as respiration, heartbeat, peristalsis or muscle and vascular tone, can render intravital microscopy (IVM) data unintelligible as shown here for intravital FLIM–FRET in the intestine as an example. After extracting the photon arrival times, individual frames are reconstructed and aligned against a reference frame based on fluorescence intensity information. Following alignment, the individual photon arrival positions are reassigned for each frame to allow quantification of fluorescence lifetimes per pixel. Similarly, Galene can also be used to correct for motion in other IVM data including fluorescence time series or endogenous fluorescence data. ECFP, enhanced cyan fluorescent protein; SEYFP, super enhanced yellow fluorescent protein. Adapted from REF.178, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Segmentation and quantification
IVM can provide highly complex information and many routinely encountered challenges, including moving image position, low signal and inconsistent signal strength between channels, can be mitigated by using image processing, such as broadly used open-access software (ImageJ/FIJI, Cell Profiler). Segmentation can help to select those data for subsequent quantification which are of relevance to the experiment, such as specific fluorescence channels and spectra, individual cells or cell populations, or subcellular structures. This can, for example, be achieved by manual selection, fluorescence intensity thresholding as well as supervised, semi-supervised or automatic data segmentation. Tracking tools, such as the TrackMate214–216 plug-in for ImageJ/FIJI or the Icy plug-in Spot Tracker217, can be used to segment cells in an image and track their dynamics during an acquired time series. This information can then be used for track visualization and quantification of dynamic cell behaviours, for example, cell migratory parameters, such as cell displacement, migration speed or directionality. Similarly, surface segmentation — provided in the commercial software Imarisor Amira — can be used to delineate the boundaries of cells or other structures in volumetric data for subsequent quantification of parameters such as cell volume, shape or position. To improve the speed, objectivity and reproducibility of IVM data analysis, deep learning methods have been developed. These approaches, which are based on the training of computer algorithms or artificial neural networks, are not mainstream yet but are progressively integrated into segmentation of IVM data. For machine learning assisted data segmentation, the user can initially train a model by manually labelling areas containing a signal compared with areas containing no signal, from which the neural networks can then extract thresholds to develop a model. If the model was sufficiently trained, it can segment all remaining data. This concept was used in an IVM database of leukocyte movement, a highly heterogeneous and plastic cell type. Manually annotated tracks for different leukocyte populations upon exposure to a range of stimuli served as a training and validation data set for machine learning approaches to automate cell migration and tracking analysis218.
Furthermore, a cell segmentation approach based on a trainable clustering algorithm was developed, which separates highly heterogeneous immune cell populations into individual cells219. Similarly, a semi-supervised learning approach assisted by pixel classification was used to segment and track immune cell dynamics in the lymph node in vivo220 as well as fluorescently labelled synapses in the cortical volume of mice upon whisker stimulation144. In addition to cell segmentation algorithms for specific data sets, U-Net, a plug-in for ImageJ/Fiji, although pre-trained for cell segmentation and detection, allows the assignment of new tasks221, and has recently been used to track cells in the live retina from phase contrast data222 or to segment cells for subsequent ratiometric FRET analysis of small GTPase activity223. In order to improve accessibility to deep learning tools, several platforms have been released, providing a range of pre-trained machine learning-based models for image processing and analysis without requiring extensive coding skills, including the ImageJ/Fiji plug-in DeepImageJ224 which is compatible with BioImage Model Zoo, an open repository for deep learning models developed by and for the scientific community, and ZeroCostDL4Mic, a platform which allows users to train and run deep learning models on free to access cloud spaces, thus reducing the amount of computing hardware required for deep learning assisted analysis225. For a detailed discussion on best practices in the use of deep learning models for image analysis, we refer the reader to the following excellent article226.
Quantification
Depending on the biological question, quantification of IVM data following segmentation can include quantification of fluorescence intensity, to analyse changes in gene expression (if the fluorophore is coupled to a protein); quantification of size or shape, to assess changes in cellular or subcellular morphology; and quantification of x–y–z coordinates/location over time, to track subcellular dynamics, migration of individual cells or cell populations, as well as cell–cell or cell–environment interactions. Furthermore, quantification of microenvironmental parameters, such as the extracellular matrix, can reveal dynamic changes in matrix deposition or remodelling, which can lead to fibrosis under pathological conditions and accompanies diseases, such as cancer or cardiovascular disease227,228. Similarly, quantification of vascular features can be performed to monitor vascular structure, flow, perfusion and patency. Analysis of fluorescently labelled drugs can be used to assess drug uptake, distribution or metabolism to provide valuable information about drug pharmacokinetics and pharmacodynamics in vivo in a preclinical context, which may inform future clinical applications or mechanisms of action135,136,183.
Available software
Paralleling the vast range of potential IVM readouts, there is an abundant source of software tools available to the scientific community to facilitate data analysis. Many software packages have been developed, which are bundle solutions to image data correction (motion/drift correction post IVM) and segmentation/quantification/visualization (cell tracking, quantification of cellular/subcellular/molecular features). Proprietary software packages include IMARIS, Amira, MetaMorph, ImagePro Plus and Arivis Vision 4D, whereas open-source applications include FIJI, QuPath, Ilastik and Cell Profiler. Supplementary Table 1 presents a non-exhaustive list of image analysis tools that were made available to the scientific community over recent years. In an effort to incorporate user-driven need and collaborative effort, ImageJ is an open-source image analysis software that readily allows addition of new features or analysis workflows, which are distributed and added to ImageJ via plug-ins and macros229,230. In order to expand upon its applicability, ImageJ2 was released, which for example allows the import, handling and analysis of multidimensional data, data types and size231. Fiji serves as a bundled release of ImageJ2 with pre-selected plug-ins, thus presenting a more complete installation package232. A comprehensive overview of the diverse applications of ImageJ and its derivatives is presented elsewhere233,234. Similarly, the platform TAPAS (Towards an Automated Processing and Analysis System) has recently been published with an enhanced focus on image data organization and the establishment and sharing of workflows/protocols for automated image processing in order to improve data standardization and reproducibility235.
Applications
IVM has provided seminal insights into the dynamics of biology across many different research fields, ranging from neurobiology and stem cell biology to cancer biology. As it is impossible to highlight all applications of IVM, we provide a few examples in the field of expertise of the authors.
Imaging immunity
The ability of IVM to visualize the immune response in an ever-broadening array of tissues and physiological situations has dramatically expanded — and fundamentally altered — our understanding of inflammation and immunity. IVM has allowed the mapping of the multistep leukocyte recruitment cascade, defining specific, sequential and regulated stages of leukocyte tethering, rolling, activation, adhesion and transmigration, identifying the role of selectins, integrins and chemokines as well as interaction with tumour cells followed by cytotoxic effector function236–239. Subsequent application of fluorescently labelled antibodies and reporters, and of confocal and multiphoton microscopy, further refined this understanding, allowing differential labelling of leukocyte subsets and improved resolution over conventional light microscopy240–242 (FIG. 5). Although these early studies were limited, they were ground-breaking in the adoption of fluorescent reporters241, the adaptation of laser-scanning confocal microscopy240 and the use of charge-coupled device (CCD) cameras to achieve high imaging frame rates242. Recently, IVM recordings of immune cells were used to introduce a new framework whereby a large number of motility and shape parameters can be quantified and analysed in an automated way, using computational tools typically employed for transcriptional and proteomic profiling. These behavioural profiles can then be used as a metric to determine the impact of individual genes and proteins on the behavioural state of individual cells243. Moreover, adaptation of intravital approaches to additional vascular beds, including the application of epifluorescence to the mouse brain244, spinning disc imaging to the liver245 and two-photon imaging to the breathing lung246, has identified tissue-specific exceptions to this central leukocyte recruitment cascade244,246,247, further refining our understanding of inflammation and immunity.
Fig. 5 |. Bone marrow imaging in situ in the leukaemia mouse model.
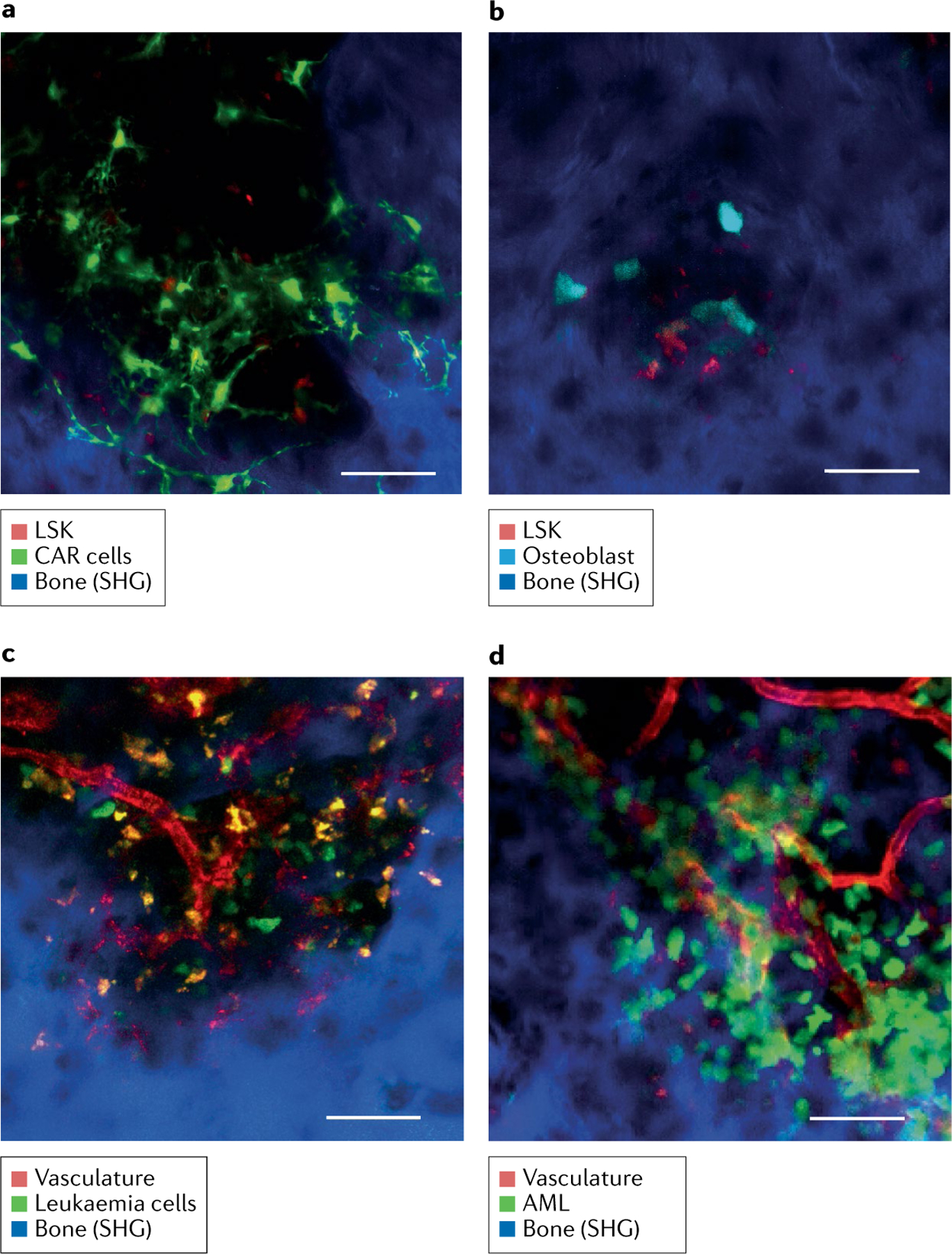
Representative intravital two-photon skull images. Bone tissue was detected by second harmonic generation (SHG), which captures collagen molecules without staining (represented in blue). Scale bar: 50 μm. a,b | Images of mouse LIN−SCA1+KIT− (LSK) cells within the bone marrow niche: mouse LSK-tdTomato cells transplanted into Cxcl12-GFP recipient mouse (green, CXCL12-abundant reticular (CAR) cells; red, LSK cells) (part a); and mouse LSK-tdTomato cells transplanted into Col1a1 2.3-CFP mouse (cyan, osteoblasts; red, LSK cells) (part b). c | Intravital image of a murine chronic myeloid leukaemia model made by transplantation of leukaemia cells (BCR-ABL-ires-GFP transfected LIN−SCA1+KIT+ cells). Green, leukaemia cells; red, blood vessels. d | Intravital image of a murine acute myeloid leukaemia (AML) model made by transplantation of leukaemia cell line (C1498-eGFP). Green, leukaemia cells; red, blood vessels.
IVM has also been transformative in our understanding of cell–cell and cell–pathogen interactions. Using models of bacterial infection and spinning disc confocal microscopy, the function of tissues such as the liver in pathogen clearance has been elucidated, identifying roles for both antibody-mediated and independent bacterial capture248,249. Other elegant studies have employed multiphoton imaging approaches in animals with cells expressing fluorescent protein reporter molecules and labelled antigens to define mechanisms involved in immune-complex drainage through lymphatics and antigen trafficking within lymph nodes250–252, lymphocyte entry, trafficking and retention within secondary lymphoid tissues253, the initiation of neutrophil swarming within sites of infection and/or damage254 and graft rejection following transplant255. Wide-field microscopy has helped to define the role of high endothelial vessels in regulating cellular entry to lymph nodes and to tumours256–258, whereas other work using CCD-based epifluorescence has been critical in defining shear-dependent alterations to neutrophil behaviour259. Additionally, the balance achieved between image resolution and the acquisition rates possible with spinning disc confocal imaging has been essential to our evolving understanding of numerous immune effector mechanisms including the formation and function of neutrophil extracellular traps260,261, and understanding how viral infection reprogrammes the host immune response262. Within the context of viral infection, the higher resolution achieved with resonant scanning confocal IVM has allowed for the direct visualization of viral delivery to tissues263,264, and has allowed for the mapping of the innate immune response to acute viral infection265. Over the past decade, IVM has also brought the role of the platelet in the host immune response to the fore. Imaging of direct platelet–neutrophil interactions, with a range of platforms from CCD-camera based epifluorescence to two-photon microscopy, has identified roles for platelets in neutrophil recruitment and activation266–268, helping to facilitate lymphocyte adhesion and antigen recognition within the virally infected liver269 and as key contributors to immunothrombosis102,270. More recently, the imaging depth possible with two-photon microscopy has allowed us to observe the function of the immune system within previously inaccessible tissues such as the bone marrow, providing single-cell resolution analysis of engineered T cell therapies in murine models of cancer271,272.
Stem cell behaviour in solid tissues
Adult stem cells self-renew and give rise to all specialized cells in the body, and are therefore the key driver cells for tissue homeostasis, regeneration and diseases such as cancer. Our knowledge on stem cells was predominantly derived from static genetic and molecular data that defined stem cells as a homeostatic pool of quiescent cells that only occasionally give rise to transient amplifying cells, which in their turn give rise to a burst of specialized offspring. However, pioneering IVM studies in hair follicle, intestine and pubertal mammary epithelia drew a completely different picture, and showed that adult stem cells can be highly migratory and proliferative and that their behaviour and fate are dynamic, heterogeneous and compartmentalized108,121,273,274. Stem cells are identified in IVM experiments directly by a fluorophore that is expressed by a stem cell promoter, or indirectly by their self-renewal behaviour and potential using fluorescent lineage-tracing approaches. The latter requires multi-day imaging of the same cells and is therefore often combined with optical windows. One particular aspect which has been elucidated using multi-day IVM is the high degree of plasticity in their behaviour and fate. In the skin, IVM experiments revealed that stem cell fate decisions (that is, symmetric or asymmetric division) are directed by the behaviour of the neighbouring cells275 and the location of the stem cells within the niche273. In the hair follicle, IVM showed that stem cells move along the niche during the hair follicle growth cycle, and, depending on their position, these stem cells generate distinct differentiated cell types of the hair274. Multi-day imaging of the pubertal mammary gland showed that stem cell movement enables stem cells to enter various positions and niches so that they all have equal behaviour and fate121,122. IVM also showed that cell movement is important for the fate decisions of intestinal tissues276,277. Differentiated cells have been shown to move away from the stem cell niche277 and migration in the reverse direction has also been shown to enable cells to enter or stay in the stem cell niche and, therefore, function as stem cells regardless of their molecular profile276. Lastly, multi-day multiphoton microscopy directly visualized the competition between wild-type intestinal stem cells and intestinal stem cells that acquired a (oncogenic) mutation278. Interestingly, imaging every few weeks the same area of surgically exposed intestine with multiphoton microscopy showed that caloric restriction increases the number of functional stem cells per stem cell niche, leading to slower but stronger competition and a more frequent loss of mutations279,280 (FIG. 6). Together, these IVM studies highlight that stem cell fate and differentiation potential are highly plastic and dependent on environmental cues.
Fig. 6 |. Intravital microscopy of calorie restriction reveals slower and stronger stem cell competition.
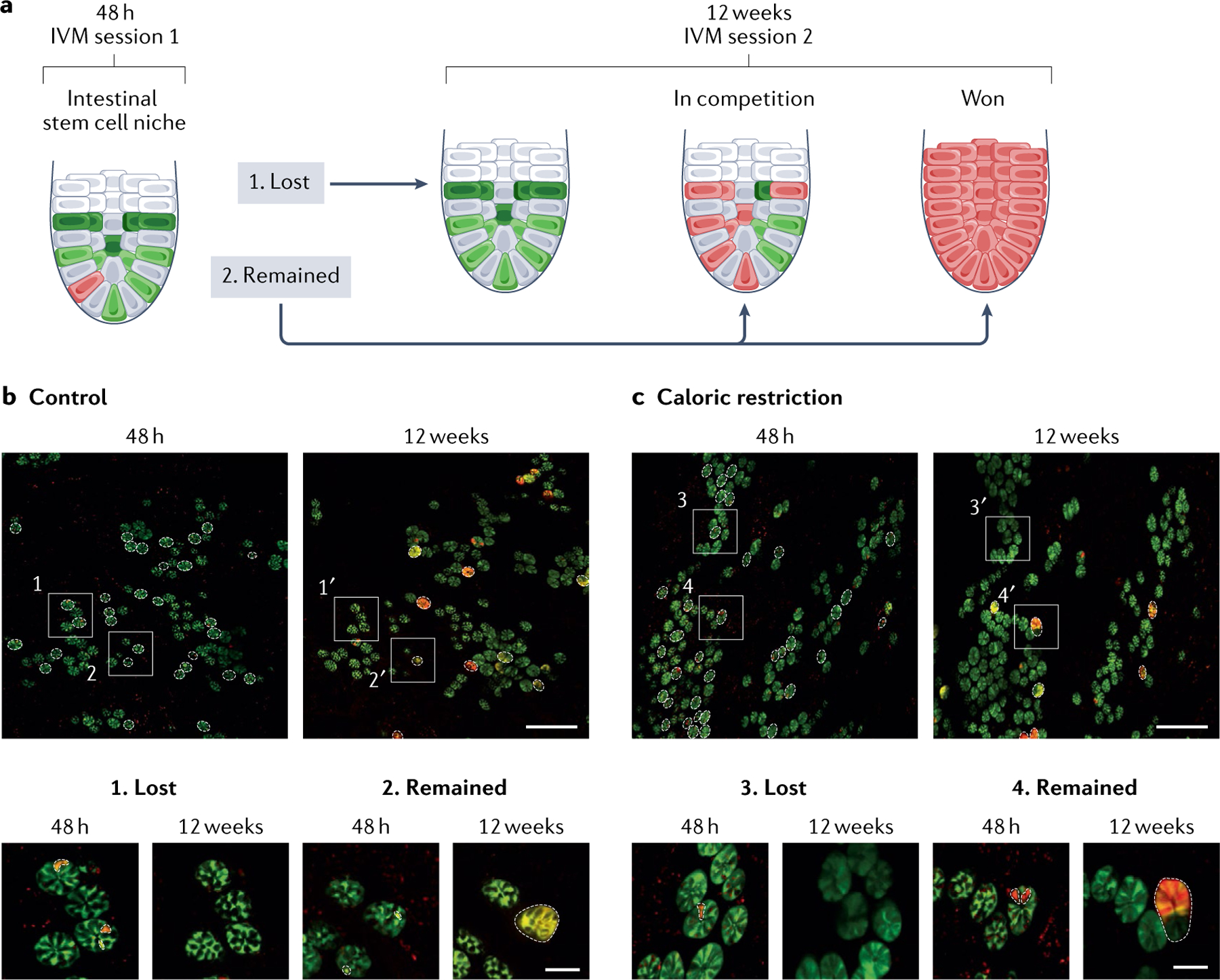
a | By repeated intravital microscopy (IVM) of the same crypts 48 h and 12 weeks after fluorescent labelling of the stem cells, stem cell competition can be visualized. Stem cell clones are either lost or remain within the crypts, represented by the presence of red cells. b,c | IVM images of the same intestinal areas in control conditions (part b) and upon caloric restriction (part c). Dotted lines highlight the crypts in which labelled stem cell clones were present at 48 h and 12 weeks after stochastic fluorescent labelling. Zoomed-in images (bottom panels) show examples of clones that were lost (1 and 3) or remained (2 and 4) in both conditions. Scale bars represent 250 μm (top panels) or 50 μm (bottom panels). Adapted with permission from REF.279, Elsevier.
Imaging haematopoietic and leukaemic stem cell behaviour and dynamics.
IVM has contributed to our understanding of the behaviour and dynamics of haematopoieticstem cells (HSCs) and leukaemic stem cells (LSCs) that reside within the bone marrow through minimally invasive surgery to expose the skull calvarium or tibia and imaging of the bone marrow cavities using time-lapse multiphoton microscopy (FIG. 5). It has been important to reveal spatio-temporal interactions of HSCs or LSCs with their bone marrow niche, which supports HSC maintenance and haematopoiesis or which is used by LSCs to escape from immune cell attacks.
Transplantation of stained HSCs into mouse calvaria bone marrow revealed that steady-state HSCs engrafted near the endosteum and resided within the endothelial niche11,281–283. In addition, differentiating progeny of HSCs reside within their specialized bone marrow niches, which can regulate the dynamics, localization and proliferation of stem and progenitor cells284. To precisely analyse the behaviour of HSCs in vivo, transgenic mouse strains with fluorescently labelled HSCs are key tools, such as the α-catulin-GFP and Fgd5-mCherry strains, in which HSCs and vascular endothelial cells are labelled285, and the Hoxb5-tri-mCherry (REF.286), Mds1GFP/+Flt3Cre (REF.10) and Pdzk1ip1 (REF.9) strains, in which only some of the long-term HSCs are labelled. Even though these strains are useful for HSC imaging, there is still no ideal HCS-specific reporter mouse strain that labels all phenotypically identifiable HSCs with bright fluorescence, and it is essential to develop new reporter mouse strains for further studies. Imaging analyses of leukaemic cells in mouse bone marrow have also advanced in recent years using time-lapse multiphoton microscopy. IVM studies showed that B cell acute lymphoid leukaemia cells resided within an endothelial bone marrow niche283, and interact with the vascular endothelium287. Moreover, T cell acute lymphoid leukaemia cells were shown to be highly migratory, especially following chemotherapy treatment13. By contrast, several studies reported localization of acute myeloid leukaemia (AML) cells around the endosteum. Bone marrow imaging of MLL-AF9-driven murine AML showed that remodelling of the endosteal region by local AML cells resulted in a reduced supporting capacity of the endosteal niche for HSCs288, and that a subset of AML cells are more migratory than the rest289, potentially taking a role akin to that of invasive tumour cells. In addition, IVM showed that the chemotherapy drug cytarabine suppressed cell motility of the transplanted mouse AML cell line via the RHO–MRTF–SRF pathway, suggesting a relationship between residual AML cell dynamics and chemo-resistance290. Recently, it has become possible to visualize the function of cells with their dynamics in living bone marrow using fluorescent probes. For example, by applying a metabolic sensor to human AML cells, it was shown that high-glycolysis LSCs prefer to reside within the endosteal niche and proliferate to maintain their leukemogenic activities291. Also, dynamic measurements of a real-time indicator of mTORC1 activity in other mouse AML models showed that the mTORC1 activity of AML cells varied according to their distance to the bone and AML cells had high mTORC1 activity under cytarabine chemotherapy292.
Progression of solid tumours
IVM has been important to reveal dynamic processes that drive several distinct progression steps of solid tumours of epithelial origin (such as breast and pancreas tumours). At the very early stage of tumour development, solid tumours are confined within a basement membrane and are identifiable by the presence of tumour cells filling up and expanding individual ducts, acini or lobules. The process of cellular expansion within these spatially well-defined units has been visualized through mammary imaging windows by time-lapse multiphoton microscopy121. It was found that during the in situ stage, the observed cellular movement is predominantly passive and occurs as a consequence of increased proliferation of cells located near the basement membrane, which push luminal cells further towards the centre of the unit (FIG. 7). It is widely thought that dissemination to distant sites requires cancer cells to actively degrade this basement membrane. IVM through organ-specific imaging windows and time-lapse multiphoton microscopy has demonstrated that interstitial tissue invasion is mediated by single-cell dissemination as well as collective invasion, which can be visualized using genetically encoded reporters for cytoplasm or membrane markers and nucleus293,294. By co-registration of tumour cells with SHG and third harmonic generation, the guiding tissue structures and tracks of least resistance can be detected, including collagen bundles, perfused blood vessels, myofibres, nerves and adipocytes131.
Fig. 7 |. Intravital microscopy of the ductal carcinoma in situ in the PyMT mouse model.
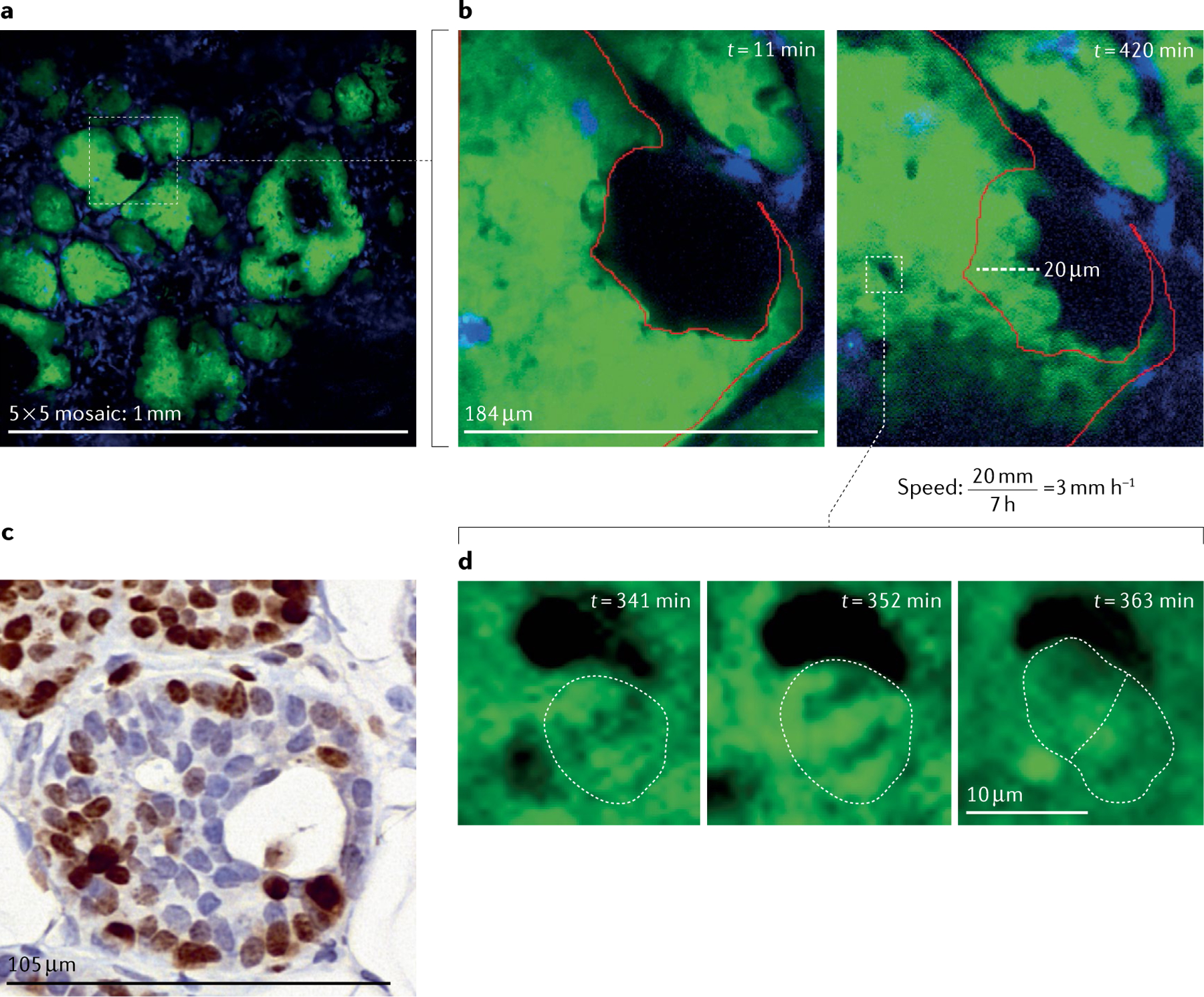
a | Large-volume high-resolution intravital microscopy (IVM) of the ducts provides an overview of the tissue and allows identification of the tissue stage and of incompletely filled ducts (white box). A 5 × 5 mosaic of the mammary gland imaged through a mammary imaging window implanted in a transgenic mouse model that expresses the PyMT oncogene and the fluorescent proteins Dendra2 in the mammary epithelium (green) and CFP in macrophages (blue). b | Stills from a continuous time-lapse movie of the duct showing migration of the cells over 7 h imaging duration. Red outline indicates position of cells at time t = 0 min. After only a brief imaging period (left), displacement of the cells is not obvious. However, after 7 h of imaging (right), cells have moved appreciably. Dashed line indicates the distance the cells have moved and can be used to calculate the speed of cell movement during imaging. c | Ki67 staining (brown) of fixed tissues taken from another mouse shows ~60% of cells are actively proliferating in similar structures. d | High-resolution stills from the IVM movie captures the division of a single cell (white dashed outline drawn by hand indicates the boundary of the dividing cell and the daughter cells), as evidenced by chromosomal separation over ~30 min.
IVM has further led to a paradigm-shifting observation demonstrating that cancer cells can intravasate during the in situ stage295. This observation explained the finding of disseminated tumour cells (DTCs) in the bone marrow of patients with in situ breast cancer296. Using IVM, the patterns of invasive cancer cell migration, single cell7,297 and collective294,298–300, as well as the effect of cancer cell interaction with non-cancerous cells within the tumour microenvironment on cancer cell phenotype109,212,260,261,301 have been described in detail. For example, IVM studies indicate that whereas intravasation of cancer cells into lymphatics occurs by collective cell migration298, intravasation of cancer cells into blood vessels occurs by single cancer cells7,106,212,302. IVM showed that in breast cancer, invasive cancer cells intravasate through tumour microenvironment of metastasis doorways, stable structures on blood vessels composed of a pro-angiogenic macrophage, a cancer cell overexpressing the actin regulator Mena and an endothelial cell. Tumour microenvironment of metastasis doorways are sites where intravasation of single cancer cells has been observed in real time7,212,302,303.
The latest stages of solid tumour progression, such as extravasation of DTCs and their outgrowth at the secondary sites, has been studied by multiphoton microscopy through specific imaging windows in several models. IVM studies revealed the fate of DTCs immediately upon extravasation into the brain304, liver53,305,306, bone283 and, most recently, lungs301. For example, IVM demonstrated that the primary and metastatic tumour microenvironment induces changes in the cancer cell states (including stemness109, epithelial-to-mesenchymal transition106 or a dormant phenotype260,301), which in turn drives cancer cell dissemination by providing these cells with a survival advantage within the metastatic site, and the potential to grow into metastatic nodules when conditions become suitable. Furthermore, IVM determined that cancer cells can re-disseminate haematogenously from already formed metastatic nodules in the lymph nodes307,308 and lungs52,161, potentially increasing the tumour burden.
Imaging haemostasis and thrombosis
Thrombosis, leading to the occlusion of vessels and, subsequently, to strokes and heart attacks, is a major cause of morbidity and mortality worldwide. To study this lethal process, several thrombosis models for IVM have been developed309, leading to the discovery of unexpected mechanisms which fostered drug development to target these major human diseases. For example, a ferric chloride-injured thrombosis model in mesenteric arterioles has been developed310 and, later, used to visualize the formation of platelet plugs, thereby revealing a role for fibronectin in this process311–313. An alternative laser-induced endothelial injury model was also developed, and confocal and wide-field microscopes were used to image in real time the formation of a thrombus. The authors combined multiple fluorescent channels with a bright-field channel, and visualized the deposition of platelets, accumulation of tissue factor and generation of fibrin314. Furthermore, IVM showed that fully activated platelets were overlaid with unstable shell and less-activated platelets315. Additional studies have made use of activatable protein substrates to reveal protease activity in living animals, defining where and when thrombin, the terminal enzyme in the clotting cascade, is active in the blood vessels102,316,317. These studies have been critical in knowledge linking infection, inflammation and coagulation265,318–320. Moreover, these types of IVM experiments have helped to find new potential ways to prevent thrombosis. For example, an IVM study has found a potential link between apoA-IV and platelet activity, suggesting that raising this plasma protein, either upon infusion of this protein or upon increased synthesis on consumption of unsaturated fat, may act as a strategy to prevent thrombosis and strokes321. Other studies have attempted to uncouple infection and coagulation, allowing us to prevent pathological immunothrombosis while preserving the critical functions of haemostasis322,323.
Imaging subcellular processes
High-resolution IVM depends upon procedures minimizing motion artefacts caused by the heartbeat and respiration, improved high numerical aperture optics, high scan speed64 and rapid engineering of mouse models expressing suitable bright reporters. These advances allow the study of in vivo cell biology at a level of subcellular detail previously reserved to in vitro microscopy, thus providing the unique opportunity of unravelling how intracellular events are controlled by the tissue environment and how they contribute to tissue pathophysiology. Although confocal and spinning disc microscopy are an ideal choice for subcellular imaging, their use is restricted to a maximum of 50–75 μm from the tissue surface. Multiphoton microscopy is the obligatory choice for deeper penetration in tissues. For example, seminal studies of transcytosis of albumin and folate receptor carried out by two-photon microscopy in the kidney324,325 led to the discovery that a significant fraction of proteins is recaptured in the kidney proximal tubules. This finding revealed an additional mechanism aside from the well-established glomerular filtration, with significant consequences for the understanding of kidney diseases and the development of novel therapies326,327. Work in various secretory organs, in immune cells and cancer models, has revealed how unique combinations of extracellular cues are transduced into cascades of intracellular signals that trigger the assembly and activation of force-generating structures to remodel intracellular membranes16,79,90,105,201,328. For example, stimulation of β-adrenergic signalling in exocrine glands elicits the assembly of a novel actomyosin structure composed of geodesic cages, revealed by spinning disc microscopy, which drive the regulated exocytosis of secretory vesicles16, a process that is coordinated with the activation of mitochondrial dynamics and bioenergetics329. Another study performed by two-photon microscopy revealed that during neutrophil extravasation the autocrine/paracrine release of chemokine signalling activates myosin II to control integrin trafficking to drive cell adhesion on the endothelium and to generate the forces required for trans-endothelial migration330. Notably, these unique structures and processes have not been described in cell or organ cultures, further highlighting the power of this approach in bridging cell biology with organ pathophysiology16,17,105,328.
Reproducibility and data deposition
Regardless of the chosen imaging modality, IVM data are typically voluminous compared with conventional digital imaging data, as they contain multidimensional spatial information, temporal information by time-lapse imaging and multichannel information for multiple fluorescent labels. The raw data can be of multiple terabytes in size, and often researchers can only save processed data after analysis, which requires systematic and efficient management of processing, communication and information security to prevent mistakes. Building a system to deposit and back up IVM data in laboratories is therefore necessary for proper data management, and managing the data on the computer controlling the microscope is not recommended. Connecting the computer to the network poses a risk of data leakage and storing high volumes of raw data on microscope-controlling computers can interfere with the control of the microscope. Raw data should therefore be transferred to and stored on external devices and network-attached storage connected to a closed network. When performing analyses on other platforms or computers, the required data can be requested from the network-attached storage, and after processing the processed data are also stored and managed on the network-attached storage. Backing up the data is even more important because it requires much effort and time to acquire a single image in IVM experiments. Even if the requested data are lost, they can be recovered if the raw data remain on the external server. Sufficient data storage capacities and net bandwidth for exchanging data of multiple terabytes in size are essential.
Biological image management systems that manage imaging data together with metadata related to microscope information and imaging conditions are available online. OMERO (Open Microscopy Environment Remote Object)331 and Bisque (BioImage Semantic Query User Environment)332 are frequently used and good examples of web-based open-source repositories. OMERO can be used to further improve/streamline the workflow from image processing to data visualization, which also allows deposition of image data in a secure repository and can help to improve the organization, analysis and sharing of big IVM data331,333. OMERO supports more than 150 image file formats and converts them to an OME-based model that any software can use, and the platform and related toolkits are frequently updated334,335. Bisque supports more than 100 image file formats, and all related data have a unique Uniform Resource Locator (URL) to access them via a web browser. The repository also provides a 3D multi-fluorophore image viewer. The RIKEN Center for Advanced Photonics in Japan also released a cloud-based system for sharing, processing and visualizing 3D biomedical images336. These repositories are useful when applying previous analyses to other projects or data. However, sharing and comparing data remain difficult due to the complexity of imaging technology. Details on the experimental animal (including genotype, sex and age) and imaging details (including excitation and emission wavelengths, fluorophores and used objectives) should be included as metadata. Full recommendations for the standardization of life science imaging data formats and the improvement of data sharing were announced in 2021 to advance open science by creating an environment in which large amounts of imaging data can be shared appropriately337. In the future, data management in a standardized format may become a global standard.
Reproducibility in live imaging results
The generation of each image is extremely complex, and therefore all imaging settings need to be well annotated. A high number of samples is required for IVM because of large individual differences among animal models, and even if the imaging conditions are the same, the appearance may change depending on the conditions of the microscope system. Furthermore, due to the limited observation range, it takes an appreciable amount of time and effort to generate one image. Imaging data are not only used for qualitative analyses but also for image processing and quantitative biological measurements to demonstrate that the data are reproducible. Although many tools have been developed for quantitative analysis, detailed methods and parameters must be chosen subjectively. These details are often not reported in papers given their word restrictions and that no unified method can be described. However, even relatively small differences in unreported processing methods can lead to conclusions with inconsistent results and many differences. Therefore, future establishment of standardized reporting guidelines to accurately and completely describe all image processing methods can help to improve the reproducibility of IVM studies.
Limitations and optimizations
The main limitations of IVM are linked to resolution, throughput and the use of animals both ethically and as a model for human disease (BOX 1). Although important differences exist between mice and humans, when appropriately designed, murine models can be strikingly similar in physiology and pathology with respect to disease initiation and progression including the driving genetic alterations and factors such as inflammation. Yet the applicability of murine models to human disease needs to be evaluated for each research question. Mice remain an important animal model to obtain fundamental knowledge on stimuli and processes that drive normal physiology and alterations that lead to disease338. Although a wealth of studies have provided striking data sets of tissue dynamics in murine models at cellular resolution, subcellular resolution is harder to achieve, especially when cells deep within tissues are observed. In mature tissue, high cell density, fibrillar collagen, fat tissue, blood vessels and mineralized structures scatter the incident light and decrease the signal intensity with scanning depth21.
The development of imaging protocols that can gradually increase laser power and detector gain in deeper sections or 3D stacks, and the choice of multiphoton over confocal microscopy, improve resolution at depth. However, in most cases, we are limited to imaging the 100–200 μm of tissues that are closest to the objective lens in locations that are surgically exposed, optically accessible through imaging windows or directly physically accessible by the objective. Still, subcellular resolution has been achieved64 and workflows including FLIM and FRET can be used to identify interactions between specific molecules133,178,196.
Biological tissues can display a very complex, multilayered structure with variations in the refractive index of the sample introducing aberrations and blurring, which are increased further with increasing imaging depth, obscuring image quality and limiting resolution and readouts. Mismatches between the refractive index of the sample and the immersion medium can also contribute to aberrations. Once light enters a sample, these aberrations can distort the wavefront, leading to aberrations and reducing image quality. Although still not commercialized, adaptive optics has been developed to artificially introduce a compensatory aberration to the light wavefront of equal but opposite magnitude to the sample aberration, correcting the wavefront and improving image quality and resolution at depth. For an in-depth discussion on adaptive optics, we refer the reader to recently published, comprehensive review articles339,340. Initially developed to fix light aberrations in telescopes used to observe remote stars, adaptive optics promise to expand the next frontier of deep tissue microscopy.
An alternative imaging approach is provided by the use of endoscopy-based approaches. Multimode optical fibres have been used to reach deep regions of the brain with minimal tissue damage341. In general, these approaches are still very much at the prototype stage, but it is hoped that further collaborations between physicists and engineers will lead to widely applicable solutions.
Currently, the most difficult limitation to overcome is the low throughput of IVM. Because real-time biological processes that occur over several hours or days can only be tracked in only one or a very few animals at a time, IVM is typically time-intensive and labour-intensive. Sometimes very rare cells are observed (for example, HSCs)9,11, although even when the cells and structures observed are more abundant (such as in the case of intestinal crypts or hair follicles) it is challenging to monitor several fields of view at a time, particularly if 3D data sets are generated. The speed of acquisition is a still an unbroken limiting step in determining the throughput of acquisition. When working with confocal and two-photon microscopes, alternative scanners that allow faster acquisition rates are still under-used. At the low end of temporal scales, scanning imaging systems, such as confocal and multiphoton microscopes, have employed various means for rapid scanning including spinning polygons342, acousto-optic deflectors343 and resonant galvanometers344, although these advances have yet to become commercialized and, thus, widely available. In the cases where samples are limited in the number of available fluorescent molecules, fast-scanning techniques may be of limited utility. However, although it is commonly thought that short dwell times do not allow sufficient time for an appreciable amount of fluorescence to be generated, this is actually not a limitation to fast scanning. As acquisition electronics only sample a very small portion of light emitted during a pixel’s dwell time, the signal collected by most systems is rather independent of scan speed29. Indeed, many video-rate confocal and multiphoton systems have been successfully developed and used. For example, video-rate acquisition of 2D images using an opto-acoustic scanner has been demonstrated282, and an acousto-optic deflector-based multiphoton systems was developed to visualize neurotransmitter release in the intact brain using a pH-sensitive GFP variant fused to a synaptic vesicle membrane protein343. Ideally, microscope arrays that would allow monitoring cells in two to four animals simultaneously would increase the number of cells observed in each session; however, at the present time the cost of such set-ups, even if multiple microscopes could share single and multiphoton lasers, remains prohibitive. Attempts to widen the spatial scale of acquisitions have taken the form of mosaicked imaging (large-volume high-resolution intravital imaging)64,202, or optical systems specifically designed for large field of view, mesoscopic imaging345, which have been used to solve problems where rare and stochastic events are captured and followed over time to determine their fate. These include spontaneous tumour initiation202 and DTC arrest in the lungs301.
More recently, new techniques have focused on reducing scan times by the selective illumination of subregions of interest346. These techniques are particularly useful for imaging immobile but functionally active structures such as neurons. Because neural networks comprise multiple neurons connected to the same functional circuit, but potentially separated by millimetres of distance, labelling the cells with calcium indicators can allow them to be probed together. Innovative new techniques that are focused either on unique scan patterns347 or on combining multiphoton with light sheet microscopy348 are still being developed.
A further limitation is posed by the duration that an animal can be kept anaesthetized while maintaining its physiology, and therefore the length of each imaging session. Although implanted windows have not been found to cause inflammation or a major alteration in the physiology50,53, the accumulated effects of multiple procedures (window, anaesthesia, imaging and so on) on physiology over extended periods of time can and should never be excluded. Thus, it is crucial that proper control experiments be designed to ensure that an observed effect is due to the phenomenon being studied, and not a response to the technical approach. Although there are examples of tissues being constantly monitored for up to 72 h (REFs.8,51,108,349,350), the challenge of maintaining animal physiology increases as the time of anaesthesia is extended beyond a few hours. Electrophysiology and imaging set-ups where animals are awake while recordings are taking place are not common but have been des cribed24,72,347,351, although sessions remain relatively short; time limits in these cases are linked to the amount of time an animal can be kept restrained without affecting its physiology. Miniaturized microscope devices would revolutionize our ability to observe cells in situ over long periods of time352,353. Recently, a lightweight two-photon miniscope was developed to perform calcium imaging in awake non-restrained mice over time spans of weeks354, but significant technological developments will be required for this to happen in other organs.
Finally, image analysis is a major bottleneck in the generation of data from IVM sessions. Traditional segmentation pipelines do not deal well with depth-linked changes in signal and customized approaches are needed355. Artificial intelligence approaches hold the promise to increase our ability to recognize, label and measure cells and structures efficiently, and will undoubtedly enable a wide implementation of multispectral imaging approaches, so that we will eventually be able to follow several — if not all — different cell types found in a certain tissue area and truly grasp the cellular dynamics underpinning health and disease225,226.
Outlook
Future perspectives on the 3Rs
Although recent advances in cell culture techniques allow for live imaging of cell dynamics and behaviour of many different cell types, these in vitro systems can still not fully replace in vivo biology. Therefore, the observed behaviour can significantly differ for cells in the in vitro and in vivo settings. In the absence of in vitro systems to study cell behaviour and dynamics in their near-native environment, IVM will remain an important technology to provide unique insights into the in situ behaviour of cells. In this light, it will be important to consider and reflect on the ethics regarding animal use for experimentation and further develop the 3Rs (replacement, reduction and refinement) for IVM (BOX 1).
Replacement.
Before designing an IVM study, it should always be considered whether animal models are necessary to address the research questions. For example, human samples can be analysed with histopathology or sequencing approaches to identify cell types and gene alterations, and many organoid models (mini-organs) have been developed to study cell behaviour in a 3D environment, and to enable the study of dynamics of cell–cell interactions356,357. However, histopathology and sequencing only provide a snapshot of highly dynamic processes and tissues, and organoids are a simplification of the full in situ environment. Therefore, it is still inevitable that animal models will be used to address key biological and medical questions.
Reduction.
The past decade has seen a wide variety of implantable imaging windows for various organs358,359. The introduction of windows has helped to reduce the number of required animals because it allows measurements over time in the same animal rather than at different time points in multiple animals. In addition to the increase of data obtained from a single animal, the number of mice required for a statistically significant result is reduced as paired statistics can be performed. Another important development to reduce the use of animals is new approaches to cure data after, for example, movement distortions. These technologies reduce the number of experiments that fail, and therefore the number of required animals.
Refinement.
In addition to reducing the number of required mice, imaging windows are important refinement tools as they dramatically reduce the invasiveness and expand the accessibility of tissues for in vivo investigation even to the point that imaging can be performed for some tissues in freely moving, awake mice360. Combined with development in anaesthetics and analgesics, IVM experiments can be optimally refined.
Rodent models are typically considered the gold standard of biological research due to the high level of similarity in genetic, physiological and pathological features to humans361. However, non-mammalian models can, potentially, be a refinement, such as nematodes362, Drosophila363 and zebrafish364. Zebrafish are likely the most common model after mice and have been utilized for a diverse range of investigations that span cancer cell metastasis365, mitophagy366 and sensory hair-cell development and regeneration367, to name just a few. Their small size and high level of transparency make them suitable for imaging with a wide range of optical microscopy techniques364.
Priorities for the next decade
IVM could be advanced to decode cause and effect relationships of paracrine and juxtacrine communications between cells during specific physiological and pathological processes, as well as the effect of pharmacological interventions on tissue microenvironments. These capabilities would enhance our understanding of the natural progression of various diseases and the ways current therapies could be modified to achieve maximum benefit. In pursuit of this goal, IVM has seen several waves of innovation that have built upon each other. Advances in microscope technology enabled better visualization of cells and non-cellular components within tissues and drove the development of better methods of visualizing cellular dynamics. Advances in fluorescent proteins, and techniques for their insertion into somatic and germ-line cells in a cell type-specific manner, drove the need for developments in surgical engineering. Some of the newest advances have been the use of fluorescent reporters and biosensors to go beyond the identification of cell type and location, venturing into the cell state, lineage tracing368 and fate mapping70,110. Reporters for stemness109,212, phase of the cell cycle369 and exposure to hypoxia370 are just a few examples.
The next frontier that is ripe to be addressed is that of functional imaging, where the natural progression of diseases and the effect of pharmacological interventions can be tested within the context of intact tissues. This can take several approaches: microfluidic-containing implantable devices for the controlled release of biological or chemical agents, where the first proof-of-principle forays have already begun371,372 but only a very few applications have been realized373,374; or optogenetics375, a discipline where genes encoding light-sensitive proteins control protein expression376,377, activity378,379 or function380. To date, the predominant use of optogenetic tools has been in the discipline of neuroscience381, with limited applications in cardiovascular382 and developmental research383. IVM is envisioned to provide advanced understanding of biological processes within intact healthy and diseased tissues which would ultimately impact patient care.
Conclusion
Taken together, IVM is a powerful technology that has the unique potential to reveal cell and tissue dynamics in the unperturbed tissue context. IVM has significantly contributed to our understanding of in vivo cell behaviour and plasticity. We foresee that in the next years, technical advances will even further push the boundaries of in vivo imaging depth, resolution, throughput and complexity to explore the unseen worlds of tissue biology.
Supplementary Material
Fluorescence lifetime imaging microscopy (FLIM).
An imaging technique capable of assessing the chemical environment of a fluorophore by detecting the characteristic time from excitation to emission for a fluorophore rather than the number of photons emitted.
Intravasation
The entry of a cell or material from tissues into the lumen of a blood or lymph vessel.
Pulse repetition rate
The frequency at which high-energy laser light is emitted, typically 1–2 MHz for low-repetition rate and 80 MHz for high-repetition rate lasers.
Optical parametric oscillators
Femtosecond pulsed light sources capable of emitting light in the range of 1,100–2,000 nm, increasing the wavelength of a pulsed femtosecond laser beyond 1,000 nm.
Group velocity dispersion compensators
Optical devices capable of maintaining the duration of femtosecond light pulses in the presence of dispersive optical elements such as objective lenses.
Optical window
A surgically implantable device that creates an optically clear aperture through which deeper tissues (such as internal organs) may be viewed.
Gradient-index lenses
Very small diameter lenses (~1–2 mm) made of a material of non-uniform index of refraction that can be positioned at physical sites that cannot be accessed by standard lenses.
Quantum yield
The ratio of photons emitted to photons observed, informing on the efficiency of fluorescence emission.
Photon budget
The number of emitted photons per molecule before becoming non-fluorescent, which can depend on the quantum yield and photostability of a fluorophore.
Invadopodia
Plasma membrane protrusions with proteolytic activity to break down extracellular matrix proteins, which can be used by cancer cells to locally invade into adjacent tissues.
Calvarium
The top part of the skull. In the mouse, this includes the frontal bones, with the central suture joining them.
Endosteum
The interface between bone and bone marrow, typically lined by osteoblasts and osteoclasts.
MLL-AF9-driven murine AML
A murine model of poor-prognosis acute myeloid leukaemia (AML). The model can be based on the doxycycline-induced expression of the oncogene MLL-AF9 or on retroviral transduction of MLL-AF9 into haematopoietic progenitor cells. The different variants have some differences, for example, in the expression levels of the oncogene and latency of disease, but all develop into AML.
Acini
A cluster of cells that form a lobed structure that looks similar to a cluster of grapes.
apoA-IV
A plasma protein that shares structural features with other apolipoproteins.
Juxtacrine
signalling that requires cell–cell or cell–matrix contact.
Surgical engineering
The combination of novel surgical protocols and engineering designs to expose these tissues for short-term and longitudinal microscopic analysis.
Acknowledgements
C.L.G.J.S. was supported by a European Molecular Biology Organization (EMBO) long-term postdoctoral fellowship (ALTF 1035–2020), a Federation of the European Biochemical Societies (FEBS) excellence award and an Excellence of Science (EOS) grant (project ID: 40007532) of Fonds Wetenschappelijk Onderzoek — Le Fonds de la Recherche Scientifique (FWO-FNRS). P.T. was supported by the Len Ainsworth Fellowship in Pancreatic Cancer Research and is a National Health and Medical Research Council (NHMRC) Senior Research Fellow. D.H. was supported by a Cancer Institute NSW (CINSW) Early Career Research Fellowship. R.W. was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) Center for Cancer Research Intramural Research Program (ZIA BC 011682). C.N.J. was supported by the Canada Research Chairs Program. P.F. was supported by the NIH U54 CA261694–01 and ERC-2021-ADG 101054921. J.v.R. was supported by the VICI (09150182110004) of ZonMW of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) and the Doctor Josef Steiner Foundation. J.v.R. and P.F. were also funded by the CancerGenomics.nl (Netherlands Organization for Scientific Research) programme. D.E. and M.H.O. are supported by the NCI (CA255153) and The Gruss-Lipper Biophotonics Center and its associated Integrated Imaging Program, and by Jane A. and Myles P. Dempsey.
RELATED LINKS
Fiji: https://fiji.sc/
ImageJ: https://imagej.nih.gov/ij/
Intravital_Microscopy_Toolbox: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0053942
Principles of the 3Rs: https://nc3rs.org.uk/who-we-are/3rs
Footnotes
Competing interests
P.T. receives reagents from Kadmon, InxMed (also consultant), Redx Pharma, Équilibre Biopharmaceuticals and Amplia Therapeutics. Under a licensing agreement between Amplia Therapeutics and Garvan Institute of Medical Research, D.H. and P.T. (consultant) are entitled to milestone payments. All other authors declare no competing interests.
Peer review information
Nature Reviews Methods Primers thanks Heyu Ni, Helene Moreau and the other, anonymous, reviewer for their contribution to the peer review of this work.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s43586-022-00168-w.
References
- 1.Janssen A, Beerling E, Medema R & van Rheenen J Intravital FRET imaging of tumor cell viability and mitosis during chemotherapy. PLoS ONE 8, e64029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy DB & Davidson MW Fundamentals of Light Microscopy and Electronic Imaging 2nd edn (Wiley, 2012). [Google Scholar]
- 3.Montero Llopis P et al. Best practices and tools for reporting reproducible fluorescence microscopy methods. Nat. Methods 18, 1463–1476 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Wollman AJM, Nudd R, Hedlund EG & Leake MC From Animaculum to single molecules: 300 years of the light microscope. Open Biol 5, 150019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawley JB Handbook of Biological Confocal Microscopy 3rd edn (Springer, 2006). [Google Scholar]
- 6.Rius C & Sanz MJ Intravital microscopy in the cremaster muscle microcirculation for endothelial dysfunction studies. Methods Mol. Biol 1339, 357–366 (2015). [DOI] [PubMed] [Google Scholar]
- 7. Harney AS et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov 5, 932–943 (2015). This study uses IVM to show that blood vessel opening and breast cancer cell intravasation are coordinated events that happen only at tumour microenvironment of metastasis doorways: sites within tumour nests where TIE2hi macrophages, Mena-expressing tumour cells and endothelial cells make direct contact.
- 8.Kitadate Y et al. Competition for mitogens regulates spermatogenic stem cell homeostasis in an open niche. Cell Stem Cell 24, 79–92.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Upadhaya S et al. Intravital imaging reveals motility of adult hematopoietic stem cells in the bone marrow niche. Cell Stem Cell 27, 336–345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christodoulou C et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashidi NM et al. In vivo time-lapse imaging shows diverse niche engagement by quiescent and naturally activated hematopoietic stem cells. Blood 124, 79–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duarte D et al. Defining the in vivo characteristics of acute myeloid leukemia cells behavior by intravital imaging. Immunol. Cell Biol 97, 229–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins ED et al. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538, 518–522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pittet MJ & Weissleder R Intravital imaging. Cell 147, 983–991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondossola E, Friedl P, Dondossola E & Friedl P Host responses to implants revealed by intravital microscopy. Nat. Rev. Mater 7, 6–22 (2022). [Google Scholar]
- 16.Ebrahim S et al. Dynamic polyhedral actomyosin lattices remodel micron-scale curved membranes during exocytosis in live mice. Nat. Cell Biol 21, 933–939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porat-Shliom N et al. In vivo tissue-wide synchronization of mitochondrial metabolic oscillations. Cell Rep 9, 514–521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theer P & Denk W On the fundamental imaging-depth limit in two-photon microscopy. J. Opt. Soc. Am. A Opt Image Sci. Vis 23, 3139 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Denk W, Strickler JH & Webb WW Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990). [DOI] [PubMed] [Google Scholar]
- 20.You S et al. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun 9, 2125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakker GJ et al. Intravital deep-tumor single-beam 3-photon, 4-photon, and harmonic microscopy. eLife 11, e63776 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andresen V et al. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr. Opin. Biotechnol 20, 54–62 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Kiepas A, Voorand E, Mubaid F, Siegel PM & Brown CM Optimizing live-cell fluorescence imaging conditions to minimize phototoxicity. J. Cell Sci 133, jcs242834 (2020). [DOI] [PubMed] [Google Scholar]
- 24. Wang T et al. Three-photon imaging of mouse brain structure and function through the intact skull. Nat. Methods 15, 789–792 (2018). This work presents three-photon IVM through the intact skull of vascular and calcium dynamics in the live brain of awake mice at imaging depths of up to ~500 μm.
- 25. Scholkmann F et al. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. Neuroimage 85, 6–27 (2014). This paper, although not focused on IVM, reviews the current state of instrumentation and methodology of near-infrared imaging, and its discussion on the selection of optimum wavelengths gives an excellent description of the ‘near-infrared window’ that is crucial to the success of multiphoton IVM.
- 26.Jansen K, Wu M, Van der Steen AFW & Van Soest G Photoacoustic imaging of human coronary atherosclerosis in two spectral bands. Photoacoustics 2, 12–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herz J et al. Expanding two-photon intravital microscopy to the infrared by means of optical parametric oscillator. Biophys. J 98, 715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paddock S Over the rainbow: 25 years of confocal imaging. Biotechniques 44, 643–648 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Entenberg D et al. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat. Protoc 6, 1500–1520 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerritsen HC, Vroom JM & De Grauw CJ Combining two-photon excitation with fluorescence lifetime imaging. IEEE Eng. Med. Biol. Mag 18, 31–36 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Koester HJ, Baur D, Uhl R & Hell SW Ca2+ fluorescence imaging with pico- and femtosecond two-photon excitation: signal and photodamage. Biophys. J 77, 2226–2236 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M et al. Dual-color fluorescence imaging distinguishes tumor cells from induced host angiogenic vessels and stromal cells. Proc. Natl Acad. Sci. USA 100, 14259–14262 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan GY et al. Video-rate scanning two-photon excitation fluorescence microscopy and ratio imaging with cameleons. Biophys. J 76, 2412–2420 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenegger DG, Tran CHT, LeDue J, Zhou N & Gordon GR A high performance, cost-effective, open-source microscope for scanning two-photon microscopy that is modular and readily adaptable. PLoS ONE 9, e110475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rakhymzhan A et al. Method for multiplexed dynamic intravital multiphoton imaging. Methods Mol. Biol 2350, 145–156 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Bares AJ et al. Hyperspectral multiphoton microscopy for in vivo visualization of multiple, spectrally overlapped fluorescent labels. Optica 7, 1587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang S, Liu J, Krasieva TB, Chen Z & Tromberg BJ Developing compact multiphoton systems using femtosecond fiber lasers. J. Biomed. Opt 14, 030508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clough M et al. Flexible simultaneous mesoscale two-photon imaging of neural activity at high speeds. Nat. Commun 12, 6638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno XC, Al-Kadhimi S, Alvelid J, Bodén A & Testa I ImSwitch: generalizing microscope control in Python. J. Open Source Softw 6, 3394 (2021). [Google Scholar]
- 40.Chhetri R et al. Software for microscopy workshop white paper. Preprint at arXiv 10.48550/arxiv.2005.00082 (2020). [DOI]
- 41.Nguyen QT, Tsai PS & Kleinfeld D MPScope: a versatile software suite for multiphoton microscopy. J. Neurosci. Methods 156, 351–359 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Nguyen Q-T, Driscoll J, Dolnick EM & Kleinfeld D in In Vivo Optical Imaging of Brain Function 2nd edn Ch. 4 (Frostig RD) 133–158 (CRC, 2009). [Google Scholar]
- 43.Pinkard H et al. Pycro-Manager: open-source software for customized and reproducible microscope control. Nat. Methods 18, 226–228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardin JA, Crair MC & Higley MJ Mesoscopic imaging: shining a wide light on large-scale neural dynamics. Neuron 108, 33–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tehranian C et al. The PI3K/Akt/mTOR pathway as a preventive target in melanoma brain metastasis. Neuro Oncol 24, 213–225 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw K et al. Neurovascular coupling and oxygenation are decreased in hippocampus compared to neocortex because of microvascular differences. Nat. Commun 12, 3190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson AJ et al. Multiscale imaging of basal cell dynamics in the functionally mature mammary gland. Proc. Natl Acad. Sci. USA 117, 26822–26832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Headley MB et al. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 531, 513–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawkins ED et al. Intravital imaging of cardiac function at the single-cell level. Nature 111, 11257–11262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alieva M, Ritsma L, Giedt RJ, Weissleder R & van Rheenen J Imaging windows for long-term intravital imaging: general overview and technical insights. Intravital 3, e29917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messal HA, van Rheenen J & Scheele CLGJ An intravital microscopy toolbox to study mammary gland dynamics from cellular level to organ scale. J. Mammary Gland Biol. Neoplasia 26, 9–27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Entenberg D et al. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat. Methods 15, 73–80 (2018). This article uses surgical engineering to develop and validate an implantable optical window for serially visualizing the murine lung with single cell-resolution IVM over a period of weeks. This is used to visualize each step of the metastatic cascade (arrival, lodging, survival, extravasation and growth) in the lung.
- 53.Ritsma L et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci. Transl Med 4, 158ra145 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Cramer SW et al. Through the looking glass: a review of cranial window technology for optical access to the brain. J. Neurosci. Methods 354, 109100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maiorino L et al. Longitudinal intravital imaging through clear silicone windows. J. Vis. Exp 10.3791/62757 (2022). [DOI] [PMC free article] [PubMed]
- 56.Jacquemin G et al. Longitudinal high-resolution imaging through a flexible intravital imaging window. Sci. Adv 7, eabg7663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mourao L, Ciwinska M, Rheenen, Jvan & Scheele CLGJ Longitudinal intravital microscopy using a mammary imaging window with replaceable lid. J. Vis. Exp 10.3791/63326 (2022). [DOI] [PubMed]
- 58. Huang Q et al. Intravital imaging of mouse embryos. Science 368, 181–186 (2020). This work presents a removable imaging window implanted above the uterus allowing IVM of live embryonic development in utero as well as embryonic manipulation for IVM studies.
- 59.Okano F, Arai J & Okui M Amplified optical window for three-dimensional images. Opt. Lett 31, 1842–1844 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Yang Y et al. A two-step GRIN lens coating for in vivo brain imaging. Neurosci. Bull 35, 419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L et al. Unit title: miniscope GRIN lens system for calcium imaging of neuronal activity from deep brain structures in behaving animals. Curr. Protoc. Neurosci 86, e56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rakhilin N et al. An intravital window to image the colon in real time. Nat. Commun 10, 5647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heo C et al. A soft, transparent, freely accessible cranial window for chronic imaging and electrophysiology. Sci. Rep 6, 27818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Entenberg D et al. Time-lapsed, large-volume, high-resolution intravital imaging for tissue-wide analysis of single cell dynamics. Methods 128, 65–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ewald AJ, Werb Z & Egeblad M Preparation of mice for long-term intravital imaging of the mammary gland. Cold Spring Harb. Protoc 2011, pdb.prot5562 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson CA, Mueller SN, Lindeman GJ, Rios AC & Visvader JE Intravital microscopy of dynamic single-cell behavior in mouse mammary tissue. Nat. Protoc 16, 1907–1935 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Ewald AJ, Werb Z & Egeblad M Monitoring of vital signs for long-term survival of mice under anesthesia. Cold Spring Harb. Protoc 2011, pdb. prot5563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erami Z et al. Intravital FRAP imaging using an E-cadherin–GFP mouse reveals disease- and drug-dependent dynamic regulation of cell–cell junctions in live tissue. Cell Rep 14, 152–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nobis M et al. Intravital FLIM–FRET imaging reveals dasatinib-induced spatial control of Src in pancreatic cancer. Cancer Res 73, 4674–4686 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Kedrin D et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods 5, 1019–1021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun W et al. In vivo two-photon imaging of anesthesia-specific alterations in microglial surveillance and photodamage-directed motility in mouse cortex. Front. Neurosci 13, 421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen C et al. Long-term imaging of dorsal root ganglia in awake behaving mice. Nat. Commun 10, 3087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W et al. Anesthetics fragment hippocampal network activity, alter spine dynamics, and affect memory consolidation. PLoS Biol 19, e3001146 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scherf N & Huisken J The smart and gentle microscope. Nat. Biotechnol 33, 815–818 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Follain G et al. Fluids and their mechanics in tumour transit: shaping metastasis. Nat. Rev. Cancer 20, 107–124 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Laine R et al. Fluorescence lifetime readouts of Troponin-C-based calcium FRET sensors: a quantitative comparison of CFP and mTFP1 as donor fluorophores. PLoS ONE 7, e49200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roche M et al. In vivo imaging with a water immersion objective affects brain temperature, blood flow and oxygenation. eLife 8, e47324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barkaway A et al. Age-related changes in the local milieu of inflamed tissues cause aberrant neutrophil trafficking and subsequent remote organ damage. Immunity 54, 1494–1510.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Milberg O et al. Concerted actions of distinct nonmuscle myosin II isoforms drive intracellular membrane remodeling in live animals. J. Cell Biol 216, 1925–1936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masedunskas A & Weigert R Intravital two-photon microscopy for studying the uptake and trafficking of fluorescently conjugated molecules in live rodents. Traffic 9, 1801–1810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mo GCH, Posner C, Rodriguez EA, Sun T & Zhang J A rationally enhanced red fluorescent protein expands the utility of FRET biosensors. Nat. Commun 11, 1848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wannier TM et al. Monomerization of far-red fluorescent proteins. Proc. Natl Acad. Sci. USA 115, E11294–E11301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang S & Ai HW A general strategy to red-shift green fluorescent protein-based biosensors. Nat. Chem. Biol 16, 1434–1439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Murphy KJ et al. Intravital imaging technology guides FAK-mediated priming in pancreatic cancer precision medicine according to Merlin status. Sci. Adv 7, eabh0363 (2021). This work on IVM of metastatic pancreatic cancer demonstrates that short-term FAK inhibition reduces tissue fibrosis and improves standard-of-care chemotherapy performance at primary and secondary cancer sites leading to a significant extension in pancreatic cancer survival.
- 85.Chishima T et al. Cancer invasion and micrometastasis visualized in live tissue by green fluorescent protein expression. Cancer Res 57, 2042–2047 (1997). [PubMed] [Google Scholar]
- 86.Farina KL et al. Cell motility of tumor cells visualized in living intact primary tumors using green fluorescent protein. Cancer Res 58, 2528–2532 (1998). [PubMed] [Google Scholar]
- 87.Stolp B et al. Salivary gland macrophages and tissue-resident CD8+ T cells cooperate for homeostatic organ surveillance. Sci. Immunol 5, eaaz4371 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Pittet MJ et al. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. Cell 11, 1100–1111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steenbeek SC et al. Cancer cells copy migratory behavior and exchange signaling networks via extracellular vesicles. EMBO J 37, e98357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zomer A et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lai CP et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun 6, 7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barnabe-Heider F et al. Genetic manipulation of adult mouse neurogenic niches by in vivo electroporation. Nat. Methods 5, 189–196 (2008). [DOI] [PubMed] [Google Scholar]
- 93.Matsuda T & Cepko CL Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA 101, 16–22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maruyama H et al. Skin-targeted gene transfer using in vivo electroporation. Gene Ther 8, 1808–1812 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Aung W et al. Visualization of in vivo electroporation-mediated transgene expression in experimental tumors by optical and magnetic resonance imaging. Gene Ther 16, 830–839 (2009). [DOI] [PubMed] [Google Scholar]
- 96.Buckley SM et al. Lentiviral transduction of the murine lung provides efficient pseudotype and developmental stage-dependent cell-specific transgene expression. Gene Ther 15, 1167–1175 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Reetz J et al. Development of adenoviral delivery systems to target hepatic stellate cells in vivo. PLoS ONE 8, e67091 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thanabalasuriar A, Neupane AS, Wang J, Krummel MF & Kubes P iNKT cell emigration out of the lung vasculature requires neutrophils and monocyte-derived dendritic cells in inflammation. Cell Rep 16, 3260–3272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dawson CA et al. Tissue-resident ductal macrophages survey the mammary epithelium and facilitate tissue remodelling. Nat. Cell Biol 22, 546–558 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Surewaard BGJ et al. Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med 213, 1141–1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park I et al. Neutrophils disturb pulmonary microcirculation in sepsis-induced acute lung injury. Eur. Respir. J 53, 1800786 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McDonald B et al. Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood 129, 1357–1367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choe K et al. Intravital imaging of intestinal lacteals unveils lipid drainage through contractility. J. Clin. Invest 125, 4042–4052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moon J et al. Intravital two-photon imaging and quantification of hepatic steatosis and fibrosis in a live small animal model. Biomed. Opt. Express 12, 7918–7927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Masedunskas A et al. Kinetics of milk lipid droplet transport, growth, and secretion revealed by intravital imaging: lipid droplet release is intermittently stimulated by oxytocin. Mol. Biol. Cell 28, 935–946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beerling E et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep 14, 2281–2288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bornes L et al. Fsp1-mediated lineage tracing fails to detect the majority of disseminating cells undergoing EMT. Cell Rep 29, 2565–2569.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ritsma L et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fumagalli A et al. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell 26, 569–578.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zomer A et al. Brief report: intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cell 31, 602–606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakaue-Sawano A et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 112.Konagaya Y et al. Intravital imaging reveals cell cycle-dependent myogenic cell migration during muscle regeneration. Cell Cycle 19, 3167–3181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bayarmagnai B et al. Invadopodia-mediated ECM degradation is enhanced in the G1 phase of the cell cycle. J. Cell Sci 132, jcs227116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eastman AE et al. Resolving cell cycle speed in one snapshot with a live-cell fluorescent reporter. Cell Rep 31, 107804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jennings E et al. Nr4a1 and Nr4a3 reporter mice are differentially sensitive to T cell receptor signal strength and duration. Cell Rep 33, 108328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bending D et al. A timer for analyzing temporally dynamic changes in transcription during differentiation in vivo. J. Cell Biol 217, 2931–2950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Livet J et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Cai D, Cohen KB, Luo T, Lichtman JW & Sanes JR Improved tools for the Brainbow toolbox. Nat. Methods 10, 540–547 (2013). [PubMed] [Google Scholar]
- 119.Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 120.Tabansky I et al. Developmental bias in cleavage-stage mouse blastomeres. Curr. Biol 23, 21–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scheele CLGJ et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542, 313–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corominas-Murtra B et al. Stem cell lineage survival as a noisy competition for niche access. Proc. Natl Acad. Sci. USA 117, 201921205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shirshin EA et al. Label-free multiphoton microscopy: the origin of fluorophores and capabilities for analyzing biochemical processes. Biochemistry. 84, 69–88 (2019). [DOI] [PubMed] [Google Scholar]
- 124.El Waly B, Buttigieg E, Karakus C, Brustlein S & Debarbieux F Longitudinal intravital microscopy reveals axon degeneration concomitant with inflammatory cell infiltration in an LPC model of demyelination. Front. Cell. Neurosci 14, 165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Feizpour A, Marstrand T, Bastholm L, Eirefelt S & Evans CL Label-free quantification of pharmacokinetics in skin with stimulated raman scattering microscopy and deep learning. J. Invest. Dermatol 141, 395–403 (2021). [DOI] [PubMed] [Google Scholar]
- 126.Estrada H et al. Intravital optoacoustic and ultrasound bio-microscopy reveal radiation-inhibited skull angiogenesis. Bone 133, 115251 (2020). [DOI] [PubMed] [Google Scholar]
- 127.Ron A, Dean-Ben XL, Gottschalk S & Razansky D Volumetric optoacoustic imaging unveils high-resolution patterns of acute and cyclic hypoxia in a murine model of breast cancer. Cancer Res 79, 4767–4775 (2019). [DOI] [PubMed] [Google Scholar]
- 128.Mazumder N et al. Label-free non-linear multimodal optical microscopy — basics, development, and applications. Front. Phys 7, 170 (2019). [Google Scholar]
- 129.Tian L et al. Microscopic second-harmonic generation emission direction in fibrillous collagen type I by quasi-phase-matching theory. J. Appl. Phys 108, 054701 (2010). [Google Scholar]
- 130.Chen X, Nadiarynkh O, Plotnikov S & Campagnola PJ Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc 7, 654–669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weigelin B, Bakker GJ & Friedl P Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci 129, 245–255 (2016). [DOI] [PubMed] [Google Scholar]
- 132.You S et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proc. Natl Acad. Sci. USA 116, 24012–24018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winfree S, Hato T & Day RN Intravital microscopy of biosensor activities and intrinsic metabolic states. Methods 128, 95–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zurauskas M et al. Assessing the severity of psoriasis through multivariate analysis of optical images from non-lesional skin. Sci. Rep 10, 9154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Conway JRW et al. Developments in preclinical cancer imaging: innovating the discovery of therapeutics. Nat. Rev. Cancer 14, 314–328 (2014). [DOI] [PubMed] [Google Scholar]
- 136.Miller MA & Weissleder R Imaging of anticancer drug action in single cells. Nat. Rev. Cancer 17, 399–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nakasone ES et al. Imaging tumor–stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 21, 488–503 (2012). This study uses IVM to visualize the effect of the chemotherapy on breast tumours and finds that chemotherapy induces a CCR2-mediated infiltration of myeloid cells into the tumour, which when blocked leads to better responses to treatment.
- 138.Timpson P, Mcghee EJ & Anderson KI Imaging molecular dynamics in vivo-from cell biology to animal models. J. Cell Sci 124, 2877–2890 (2011). [DOI] [PubMed] [Google Scholar]
- 139.Nobis M et al. Molecular mobility and activity in an intravital imaging setting — implications for cancer progression and targeting. J. Cell Sci 131, jcs206995 (2018). [DOI] [PubMed] [Google Scholar]
- 140.Ebrahim S & Weigert R Intravital microscopy in mammalian multicellular organisms. Curr. Opin. Cell Biol 59, 97–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Serrels A et al. Real-time study of E-cadherin and membrane dynamics in living animals: implications for disease modeling and drug development. Cancer Res 69, 2714–2719 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Appaduray MA et al. Recruitment kinetics of tropomyosin Tpm3.1 to actin filament bundles in the cytoskeleton is independent of actin filament kinetics. PLoS ONE 11, e0168203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chodaczek G, Toporkiewicz M, Zal MA & Zal T Epidermal T cell dendrites serve as conduits for bidirectional trafficking of granular cargo. Front. Immunol 9, 1430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Graves AR et al. Visualizing synaptic plasticity in vivo by large-scale imaging of endogenous AMPA receptors. eLife 10, e66809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Spinelli KJ et al. Presynaptic α-synuclein aggregation in a mouse model of Parkinson’s disease. J. Neurosci 34, 2037–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Machado MJ & Mitchell CA Temporal changes in microvessel leakiness during wound healing discriminated by in vivo fluorescence recovery after photobleaching. J. Physiol 589, 4681–4696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bouta EM et al. In vivo quantification of lymph viscosity and pressure in lymphatic vessels and draining lymph nodes of arthritic joints in mice. J. Physiol 592, 1213–1223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yi C, Teillon J, Koulakoff A, Berry H & Giaume C Monitoring gap junctional communication in astrocytes from acute adult mouse brain slices using the gap-FRAP technique. J. Neurosci. Methods 303, 103–113 (2018). [DOI] [PubMed] [Google Scholar]
- 149.Abbaci M et al. Gap junctional intercellular communication capacity by gap-FRAP technique: a comparative study. Biotechnol. J 2, 50–61 (2007). [DOI] [PubMed] [Google Scholar]
- 150.Farnsworth NL, Hemmati A, Pozzoli M & Benninger RK Fluorescence recovery after photobleaching reveals regulation and distribution of connexin36 gap junction coupling within mouse islets of Langerhans. J. Physiol 592, 4431–4446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pluen A et al. Role of tumor–host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc. Natl Acad. Sci. USA 98, 4628–4633 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Netti PA, Berk DA, Swartz MA, Grodzinsky AJ & Jain RK Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res 60, 2497–2503 (2000). [PubMed] [Google Scholar]
- 153.Vartak N et al. Intravital dynamic and correlative imaging of mouse livers reveals diffusion-dominated canalicular and flow-augmented ductular bile flux. Hepatology 73, 1531–1550 (2021). [DOI] [PubMed] [Google Scholar]
- 154.Kaur G et al. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nat. Commun 4, 1637 (2013). [DOI] [PubMed] [Google Scholar]
- 155.Sullivan KD, Majewska AK & Brown EB Single- and two-photon fluorescence recovery after photobleaching. Cold Spring Harb. Protoc 2015, pdb top083519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fritzsche M & Charras G Dissecting protein reaction dynamics in living cells by fluorescence recovery after photobleaching. Nat. Protoc 10, 660–680 (2015). [DOI] [PubMed] [Google Scholar]
- 157.Moran I & Phan TG Fate mapping and transcript profiling of germinal center cells by two-photon photoconversion. Methods Mol. Biol 1623, 59–72 (2017). [DOI] [PubMed] [Google Scholar]
- 158.Fumagalli A et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl Acad. Sci. USA 114, E2357–E2364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Floerchinger A et al. Optimizing metastatic-cascade-dependent Rac1 targeting in breast cancer: guidance using optical window intravital FRET imaging. Cell Rep 36, 109689 (2021). [DOI] [PubMed] [Google Scholar]
- 160.Alieva M et al. Preventing inflammation inhibits biopsy-mediated changes in tumor cell behavior. Sci. Rep 7, 7529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Borriello L, Condeelis J, Entenberg D & Oktay MH Breast cancer cell re-dissemination from lung metastases — a mechanism for enhancing metastatic burden. J. Clin. Med 10, 2340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fluegen G et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat. Cell Biol 19, 120–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alieva M et al. Intravital imaging of glioma border morphology reveals distinctive cellular dynamics and contribution to tumor cell invasion. Sci. Rep 9, 2054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kitano M et al. Imaging of the cross-presenting dendritic cell subsets in the skin-draining lymph node. Proc. Natl Acad. Sci. USA 113, 1044–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Torcellan T et al. In vivo photolabeling of tumor-infiltrating cells reveals highly regulated egress of T-cell subsets from tumors. Proc. Natl Acad. Sci. USA 114, 5677–5682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Suan D et al. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity 42, 704–718 (2015). [DOI] [PubMed] [Google Scholar]
- 167.Amornphimoltham P et al. Rab25 regulates invasion and metastasis in head and neck cancer. Clin. Cancer Res 19, 1375–1388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Holmes BB et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl Acad. Sci. USA 111, E4376–E4385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ouyang M et al. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J. Biol. Chem 283, 17740–17748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Harvey CD et al. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl Acad. Sci. USA 105, 19264–19269 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ishii M, Tateya T, Matsuda M & Hirashima T Retrograde ERK activation waves drive base-to-apex multicellular flow in murine cochlear duct morphogenesis. eLife 10, e61092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kinjo T et al. FRET-assisted photoactivation of flavoproteins for in vivo two-photon optogenetics. Nat. Methods 16, 1029–1036 (2019). This work develops FRET-mediated photoactivation and genetically engineered mouse models to allow single-cell two-photon excitation of optogenetic proteins for IVM.
- 173.Hiratsuka T et al. Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. eLife 4, e05178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hirata E et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 27, 574–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yan C et al. Peripheral-specific Y1 receptor antagonism increases thermogenesis and protects against diet-induced obesity. Nat. Commun 12, 2622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yoshizaki H, Mochizuki N, Gotoh Y & Matsuda M Akt–PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell 18, 119–128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Vennin C et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl Med 9, eaai8504 (2017). This study uses IVM to demonstrate that fine-tuned manipulation of tissue tension and vascular patency in pancreatic cancer via ROCK inhibition can improve chemotherapy performance in the primary tumour and secondary metastases leading to a significant improvement in pancreatic cancer outcome.
- 178.Warren SC et al. Removing physiological motion from intravital and clinical functional imaging data. eLife 7, e35800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seong J et al. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat. Commun 2, 406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Murphy KJ, Reed DA, Trpceski M, Herrmann D & Timpson P Quantifying and visualising the nuances of cellular dynamics in vivo using intravital imaging. Curr. Opin. Cell Biol 72, 41–53 (2021). [DOI] [PubMed] [Google Scholar]
- 181.Li C et al. A FRET biosensor for ROCK based on a consensus substrate sequence identified by KISS technology. Cell Struct. Funct 42, 1–13 (2017). [DOI] [PubMed] [Google Scholar]
- 182.Imanishi A et al. Visualization of spatially-controlled vasospasm by sympathetic nerve-mediated ROCK activation. Am. J. Pathol 191, 194–203 (2021). [DOI] [PubMed] [Google Scholar]
- 183.Ng TSC, Garlin MA, Weissleder R & Miller MA Improving nanotherapy delivery and action through image-guided systems pharmacology. Theranostics 10, 968–997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nobis M et al. A RhoA-FRET biosensor mouse for intravital imaging in normal tissue homeostasis and disease contexts. Cell Rep 21, 274–288 (2017). [DOI] [PubMed] [Google Scholar]
- 185.Johnsson AE et al. The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Rep 6, 1153–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kondo H et al. Single-cell resolved imaging reveals intra-tumor heterogeneity in glycolysis, transitions between metabolic states, and their regulatory mechanisms. Cell Rep 34, 108750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Potzkei J et al. Real-time determination of intracellular oxygen in bacteria using a genetically encoded FRET-based biosensor. BMC Biol 10, 28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Awaji T, Hirasawa A, Shirakawa H, Tsujimoto G & Miyazaki S Novel green fluorescent protein-based ratiometric indicators for monitoring pH in defined intracellular microdomains. Biochem. Biophys. Res. Commun 289, 457–462 (2001). [DOI] [PubMed] [Google Scholar]
- 189.Watabe T, Terai K, Sumiyama K & Matsuda M Booster, a red-shifted genetically encoded Förster resonance energy transfer (FRET) biosensor compatible with cyan fluorescent protein/yellow fluorescent protein-based FRET biosensors and blue light-responsive optogenetic tools. ACS Sens 5, 719–730 (2020). [DOI] [PubMed] [Google Scholar]
- 190.Shcherbakova DM, Cox Cammer N, Huisman TM, Verkhusha VV & Hodgson L Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat. Chem. Biol 14, 591–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shcherbakova DM, Stepanenko OV, Turoverov KK & Verkhusha VV Near-infrared fluorescent proteins: multiplexing and optogenetics across scales. Trends Biotechnol 36, 1230–1243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang JZ et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 182, 1531–1544.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bock A et al. Optical mapping of cAMP signaling at the nanometer scale. Cell 182, 1519–1530.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang JM et al. Deciphering cell signaling networks with massively multiplexed biosensor barcoding. Cell 184, 6193–6206.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ni Q, Mehta S & Zhang J Live-cell imaging of cell signaling using genetically encoded fluorescent reporters. FEBS J. 285, 203–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Conway JRW, Warren SC & Timpson P Context-dependent intravital imaging of therapeutic response using intramolecular FRET biosensors. Methods 128, 78–94 (2017). [DOI] [PubMed] [Google Scholar]
- 197.Ulbricht C et al. Intravital quantification reveals dynamic calcium concentration changes across B cell differentiation stages. eLife 10, e56020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Skala MC et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl Acad. Sci. USA 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sparks H et al. Heterogeneity in tumor chromatin-doxorubicin binding revealed by in vivo fluorescence lifetime imaging confocal endomicroscopy. Nat. Commun 9, 2662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Soulet D, Lamontagne-Proulx J, Aube B & Davalos D Multiphoton intravital microscopy in small animals: motion artefact challenges and technical solutions. J. Microsc 278, 3–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Masedunskas A et al. Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy. Proc. Natl Acad. Sci. USA 108, 13552–13557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Amornphimoltham P, Thompson J, Melis N & Weigert R Non-invasive intravital imaging of head and neck squamous cell carcinomas in live mice. Methods 128, 3–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ritsma L et al. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat. Protoc 8, 583–594 (2013). [DOI] [PubMed] [Google Scholar]
- 204.Morimoto A et al. SLPI is a critical mediator that controls PTH-induced bone formation. Nat. Commun 12, 2136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Furuya M et al. Direct cell–cell contact between mature osteoblasts and osteoclasts dynamically controls their functions in vivo. Nat. Commun 9, 300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sekiguchi KJ et al. Imaging large-scale cellular activity in spinal cord of freely behaving mice. Nat. Commun 7, 11450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Laffray S et al. Adaptive movement compensation for in vivo imaging of fast cellular dynamics within a moving tissue. PLoS ONE 6, e19928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Vladymyrov M, Haghayegh Jahromi N, Kaba E, Engelhardt B & Ariga A VivoFollow 2: distortion-free multiphoton intravital imaging. Front. Phys 7, 222 (2020). [Google Scholar]
- 209.Soulet D, Pare A, Coste J & Lacroix S Automated filtering of intrinsic movement artifacts during two-photon intravital microscopy. PLoS ONE 8, e53942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dunn KW, Lorenz KS, Salama P & Delp EJ IMART software for correction of motion artifacts in images collected in intravital microscopy. Intravital 3, e28210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 212. Sharma VP et al. Live tumor imaging shows macrophage induction and TMEM-mediated enrichment of cancer stem cells during metastatic dissemination. Nat. Commun 12, 7300 (2021). This article uses IVM of a fluorescent reporter for stemness to determine that tumour-associated macrophages induce programmes of stemness in disseminating breast cancer cells as they intravasate, and that this stemness phenotype is carried with the cells to the secondary site of the lung.
- 213.Sharma V ImageJ plugin HyperStackReg V5.6. Zenodo https://zenodo.org/record/2252521#.YzagunZBz-g (2018).
- 214.Ershov D et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022). [DOI] [PubMed] [Google Scholar]
- 215.Tinevez JY et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 216.Jaqaman K et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chenouard N, Bloch I & Olivo-Marin JC Multiple hypothesis tracking for cluttered biological image sequences. IEEE Trans. Pattern Anal. Mach. Intell 35, 2736–2750 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Pizzagalli DU et al. Leukocyte tracking database, a collection of immune cell tracks from intravital 2-photon microscopy videos. Sci. Data 5, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pizzagalli DU, Gonzalez SF & Krause R A trainable clustering algorithm based on shortest paths from density peaks. Sci. Adv 5, eaax3770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pizzagalli DU, Thelen M, Gonzalez SF & Krause R Semi-supervised machine learning facilitates cell colocalization and tracking in intravital microscopy. Preprint at bioRxiv 10.1101/829838 (2019). [DOI]
- 221.Falk T et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 222.Joseph A, Chu CJ, Feng G, Dholakia K & Schallek J Label-free imaging of immune cell dynamics in the living retina using adaptive optics. eLife 9, e60547 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cheung BCH, Hodgson L, Segall JE & Wu M Spatial and temporal dynamics of RhoA activities of single breast tumor cells in a 3D environment revealed by a machine learning-assisted FRET technique. Exp. Cell Res 410, 112939 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gomez-de-Mariscal E et al. DeepImageJ: a user-friendly environment to run deep learning models in ImageJ. Nat. Methods 18, 1192–1195 (2021). [DOI] [PubMed] [Google Scholar]
- 225.von Chamier L et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun 12, 2276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Laine RF, Arganda-Carreras I, Henriques R & Jacquemet G Avoiding a replication crisis in deep-learning-based bioimage analysis. Nat. Methods 18, 1136–1144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lopez B et al. Diffuse myocardial fibrosis: mechanisms, diagnosis and therapeutic approaches. Nat. Rev. Cardiol 18, 479–498 (2021). [DOI] [PubMed] [Google Scholar]
- 228.Cox TR The matrix in cancer. Nat. Rev. Cancer 21, 217–238 (2021). [DOI] [PubMed] [Google Scholar]
- 229.Schneider CA, Rasband WS & Eliceiri KW NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Abràmoff MD, Magalhães PJ & Ram SJ Image processing with ImageJ. Biophotonics Int 11, 36–42 (2004). [Google Scholar]
- 231.Rueden CT et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Collins TJ ImageJ for microscopy. Biotechniques 43, 25–30 (2007). [DOI] [PubMed] [Google Scholar]
- 234.Schroeder AB et al. The ImageJ ecosystem: open-source software for image visualization, processing, and analysis. Protein Sci 30, 234–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gilles JF & Boudier T TAPAS: towards automated processing and analysis of multi-dimensional bioimage data. F1000Res 9, 1278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Springer TA Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76, 301–314 (1994). [DOI] [PubMed] [Google Scholar]
- 237.Butcher EC Leukocyte–endothelial cell recognition: three (or more) steps to specificity and diversity. Cell 67, 1033–1036 (1991). [DOI] [PubMed] [Google Scholar]
- 238.Ley K, Laudanna C, Cybulsky MI & Nourshargh S Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol 7, 678–689 (2007). [DOI] [PubMed] [Google Scholar]
- 239.Weigelin B et al. Cytotoxic T cells are able to efficiently eliminate cancer cells by additive cytotoxicity. Nat. Commun 12, 5217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suzuki T, Yanagi K, Ookawa K, Hatakeyama K & Ohshima N Flow visualization of microcirculation in solid tumor tissues: intravital microscopic observation of blood circulation by use of a confocal laser scanning microscope. Front. Med. Biol. Eng 7, 253–263 (1996). [PubMed] [Google Scholar]
- 241.Potter SM, Wang CM, Garrity PA & Fraser SE Intravital imaging of green fluorescent protein using two-photon laser-scanning microscopy. Gene 173, 25–31 (1996). [DOI] [PubMed] [Google Scholar]
- 242.Bonder CS et al. Rules of recruitment for TH1 and TH2 lymphocytes in inflamed liver: a role for α−4 integrin and vascular adhesion protein-1. Immunity 23, 153–163 (2005). [DOI] [PubMed] [Google Scholar]
- 243. Crainiciuc G et al. Behavioural immune landscapes of inflammation. Nature 601, 415–421 (2022). This study uses large-scale IVM of leukocytes during inflammation to profile dynamic immune cell behaviour in vivo mirroring orthogonal transcriptomic and proteomic approaches.
- 244.Carvalho-Tavares J et al. A role for platelets and endothelial selectins in tumor necrosis factor-α-induced leukocyte recruitment in the brain microvasculature. Circ. Res 87, 1141–1148 (2000). [DOI] [PubMed] [Google Scholar]
- 245.Menezes GB et al. Selective down-regulation of neutrophil Mac-1 in endotoxemic hepatic microcirculation via IL-10. J. Immunol 183, 7557–7568 (2009). [DOI] [PubMed] [Google Scholar]
- 246.Kreisel D et al. In vivo two-photon imaging reveals monocyte-dependent neutrophil extravasation during pulmonary inflammation. Proc. Natl Acad. Sci. USA 107, 18073–18078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lee WY & Kubes P Leukocyte adhesion in the liver: distinct adhesion paradigm from other organs. J. Hepatol 48, 504–512 (2008). [DOI] [PubMed] [Google Scholar]
- 248.Zeng Z et al. Sex-hormone-driven innate antibodies protect females and infants against EPEC infection. Nat. Immunol 19, 1100–1111 (2018). [DOI] [PubMed] [Google Scholar]
- 249.Zeng Z et al. CRIg functions as a macrophage pattern recognition receptor to directly bind and capture blood-borne Gram-positive bacteria. Cell Host Microbe 20, 99–106 (2016). [DOI] [PubMed] [Google Scholar]
- 250.Allen CD, Okada T, Tang HL & Cyster JG Imaging of germinal center selection events during affinity maturation. Science 315, 528–531 (2007). [DOI] [PubMed] [Google Scholar]
- 251.Phan TG, Green JA, Gray EE, Xu Y & Cyster JG Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat.Immunol 10, 786–793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Choe K et al. Intravital three-photon microscopy allows visualization over the entire depth of mouse lymph nodes. Nat. Immunol 23, 330–340 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Breart B & Bousso P S1P 1 downregulation tailors CD8+ T-cell residence time in lymph nodes to the strength of the antigenic stimulation. Eur. J. Immunol 46, 2730–2736 (2016). [DOI] [PubMed] [Google Scholar]
- 254.Lammermann T et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Celli S, Albert ML & Bousso P Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat. Med 17, 744–749 (2011). [DOI] [PubMed] [Google Scholar]
- 256.Moussion C & Girard JP Dendritic cells control lymphocyte entry to lymph nodes through high endothelial venules. Nature 479, 542–546 (2011). [DOI] [PubMed] [Google Scholar]
- 257.Lafouresse F et al. L-Selectin controls trafficking of chronic lymphocytic leukemia cells in lymph node high endothelial venules in vivo. Blood 126, 1336–1345 (2015). [DOI] [PubMed] [Google Scholar]
- 258.Asrir A et al. Tumor-associated high endothelial venules mediate lymphocyte entry into tumors and predict response to PD-1 plus CTLA-4 combination immunotherapy. Cancer Cell 40, 318–334.e9 (2022). [DOI] [PubMed] [Google Scholar]
- 259.Sundd P et al. Slings enable neutrophil rolling at high shear. Nature 488, 399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Albrengues J et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361, eaao4227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Park J et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci. Transl Med 8, 361ra138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Neupane AS et al. Patrolling alveolar macrophages conceal bacteria from the immune system to maintain homeostasis. Cell 183, 110–125.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 263.Naumenko V et al. Visualizing oncolytic virus–host interactions in live mice using intravital microscopy. Mol. Ther. Oncolytics 10, 14–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Carestia A et al. Modulation of the liver immune microenvironment by the adeno-associated virus serotype 8 gene therapy vector. Mol. Ther. Methods Clin. Dev 20, 95–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kim SJ et al. Platelet-mediated NET release amplifies coagulopathy and drives lung pathology during severe influenza infection. Front. Immunol 12, 772859 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Devi S et al. Platelet recruitment to the inflamed glomerulus occurs via an alphaIIbbeta3/GPVI-dependentαIIbβ3/GPVI-dependent pathway. Am. J. Pathol 177, 1131–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lefrancais E, Mallavia B, Zhuo H, Calfee CS & Looney MR Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight 3, e98178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Riffo-Vasquez Y et al. A non-anticoagulant fraction of heparin inhibits leukocyte diapedesis into the lung by an effect on platelets. Am. J. Respir. Cell Mol. Biol 55, 554–563 (2016). [DOI] [PubMed] [Google Scholar]
- 269.Guidotti LG et al. Immunosurveillance of the liver by intravascular effector CD8+ T cells. Cell 161, 486–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Surewaard BGJJ et al. alpha-toxinα-Toxin induces platelet aggregation and liver injury during Staphylococcus aureus sepsis. Cell Host Microbe 24, 271–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cazaux M et al. Single-cell imaging of CAR T cell activity in vivo reveals extensive functional and anatomical heterogeneity. J. Exp. Med 216, 1038–1049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Boulch M et al. A cross-talk between CAR T cell subsets and the tumor microenvironment is essential for sustained cytotoxic activity. Sci. Immunol 6, eabd4344 (2021). [DOI] [PubMed] [Google Scholar]
- 273.Rompolas P, Mesa KR & Greco V Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xin T, Gonzalez D, Rompolas P & Greco V Flexible fate determination ensures robust differentiation in the hair follicle. Nat. Cell Biol 20, 1361–1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mesa KR et al. Homeostatic epidermal stem cell self-renewal is driven by local differentiation. Cell Stem Cell 23, 677–686.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Azkanaz M et al. Retrograde movements determine effective stem cell numbers in the intestine. Nature 607, 548–554 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Krndija D et al. Active cell migration is critical for steady-state epithelial turnover in the gut. Science 365, 705–710 (2019). [DOI] [PubMed] [Google Scholar]
- 278.Huels DJ et al. Wnt ligands influence tumour initiation by controlling the number of intestinal stem cells. Nat. Commun 9, 1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bruens L et al. Calorie restriction increases the number of competing stem cells and decreases mutation retention in the intestine. Cell Rep 32, 107937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Van Rheenen J, Bruens L, Lotte B & Van Rheenen J Cellular protection mechanisms that minimise accumulation of mutations in intestinal tissue. Swiss Med. Wkly 147, w14539 (2017). [DOI] [PubMed] [Google Scholar]
- 281.Fujisaki J et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature 474, 216–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lo Celso C et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Sipkins DA et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature 435, 969–973 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Koechlein CS et al. High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo. Nat. Commun 7, 12169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Acar M et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Bajaj J et al. CD98-mediated adhesive signaling enables the establishment and propagation of acute myelogenous leukemia. Cancer Cell 30, 792–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Duarte D et al. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell 22, 64–77.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pirillo C et al. Metalloproteinase inhibition reduces AML growth, prevents stem cell loss, and improves chemotherapy effectiveness. Blood Adv 6, 3126–3141 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Morimatsu M et al. Migration arrest of chemoresistant leukemia cells mediated by MRTF–SRF pathway. Inflamm. Regen 40, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hao X et al. Metabolic imaging reveals a unique preference of symmetric cell division and homing of leukemia-initiating cells in an endosteal niche. Cell Metab 29, 950–965.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 292.Oki T et al. Imaging dynamic mTORC1 pathway activity in vivo reveals marked shifts that support time-specific inhibitor therapy in AML. Nat. Commun 12, 245 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Khalil AA et al. Collective invasion induced by an autocrine purinergic loop through connexin-43 hemichannels. J. Cell Biol 219, e201911120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ilina O et al. Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion. Nat. Cell Biol 22, 1103–1115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Harper KL et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 588–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Sanger N et al. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int. J. Cancer 129, 2522–2526 (2011). [DOI] [PubMed] [Google Scholar]
- 297.Beerling E, Oosterom I, Voest E, Lolkema M & Rheenen J Intravital characterization of tumor cell migration in pancreatic cancer. Intravital 5, e1261773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Giampieri S et al. Localized and reversible TGFβ signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol 11, 1287–1296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ilina O et al. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model. Mech 11, dmm034330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Alexander S, Koehl GE, Hirschberg M, Geissler EK & Friedl P Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem. Cell Biol 130, 1147–1154 (2008). [DOI] [PubMed] [Google Scholar]
- 301. Borriello L et al. Primary tumor associated macrophages activate programs of invasion and dormancy in disseminating tumor cells. Nat. Commun 13, 626 (2022). This paper uses serial IVM to track the fate of DTCs in the lung, showing that primary tumour-associated macrophages induce in DTCs a triple-threat phenotype (invasive, stem and dormant) that is carried to the lung and confers to them survival and extravasation advantages.
- 302.Liu X et al. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov 9, 96–113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Karagiannis GS et al. Assessing tumor microenvironment of metastasis doorway-mediated vascular permeability associated with cancer cell dissemination using intravital imaging and fixed tissue analysis. J. Vis. Exp 10.3791/59633 (2019). [DOI] [PMC free article] [PubMed]
- 304.Kienast Y et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med 16, 116–122 (2010). [DOI] [PubMed] [Google Scholar]
- 305.Bagge U, Skolnik G & Ericson LE The arrest of circulating tumor cells in the liver microcirculation: a vital fluorescence microscopic, electron microscopic and isotope study in the rat. J. Cancer Res. Clin. Oncol 105, 134–140 (1983). [DOI] [PubMed] [Google Scholar]
- 306.Scherbarth S & Orr FW Intravital videomicroscopic evidence for regulation of metastasis by the hepatic microvasculature: effects of interleukin-1α on metastasis and the location of B16F1 melanoma cell arrest. Cancer Res 57, 4105–4110 (1997). [PubMed] [Google Scholar]
- 307.Padera TP et al. Lymphatic metastasis in the absence of functional intratumor lymphatics. Science 296, 1883–1886 (2002). [DOI] [PubMed] [Google Scholar]
- 308.Brown M et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 359, 1408–1411 (2018). [DOI] [PubMed] [Google Scholar]
- 309.Lei X, MacKeigan DT & Ni H Control of data variations in intravital microscopy thrombosis models. J. Thromb. Haemost 18, 2823–2825 (2020). [DOI] [PubMed] [Google Scholar]
- 310.Denis C et al. A mouse model of severe von Willebrand disease: defects in hemostasis and thrombosis. Proc. Natl Acad. Sci. USA 95, 9524–9529 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ni H et al. Persistence of platelet thrombus formation in arterioles of mice lacking both von Willebrand factor and fibrinogen. J. Clin. Invest 106, 385–392 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ni H et al. Plasma fibronectin promotes thrombus growth and stability in injured arterioles. Proc. Natl Acad. Sci. USA 100, 2415–2419 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Wang Y et al. Plasma fibronectin supports hemostasis and regulates thrombosis. J. Clin. Invest 124, 4281–4293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Falati S, Gross P, Merrill-skoloff G, Furie BC & Furie B Real-time in vivo imaging of platelets, tissue factor and fibrin during arterial thrombus formation in the mouse. Nat. Med 8, 1175–1180 (2002). [DOI] [PubMed] [Google Scholar]
- 315.Stalker TJ et al. Hierarchical organization in the hemostatic response and its relationship to the platelet-signaling network. Blood 121, 1875–1885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Olson ES et al. In vivo fluorescence imaging of atherosclerotic plaques with activatable cell-penetrating peptides targeting thrombin activity. Integr. Biol 4, 595–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Jaffer FA, Tung CH, Gerszten RE & Weissleder R In vivo imaging of thrombin activity in experimental thrombi with thrombin-sensitive near-infrared molecular probe. Arterioscler. Thromb. Vasc. Biol 22, 1929–1935 (2002). [DOI] [PubMed] [Google Scholar]
- 318.Campos J et al. Neutrophil extracellular traps and inflammasomes cooperatively promote venous thrombosis in mice. Blood Adv 5, 2319–2324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Massberg S et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat. Med 16, 887–896 (2010). [DOI] [PubMed] [Google Scholar]
- 320.von Brühl ML et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J. Exp. Med 209, 819–835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Xu XR et al. Apolipoprotein A-IV binds αIIbβ3 integrin and inhibits thrombosis. Nat. Commun 9, 3608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Carestia A, Davis RP, Grosjean H, Lau MW & Jenne CN Acetylsalicylic acid inhibits intravascular coagulation during Staphylococcus aureus-induced sepsis in mice. Blood 135, 1281–1286 (2020). [DOI] [PubMed] [Google Scholar]
- 323.Chauhan AK et al. ADAMTS13: a new link between thrombosis and inflammation. J. Exp. Med 205, 2065–2074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Dunn KW et al. Functional studies of the kidney of living animals using multicolor two-photon microscopy. Am. J. Physiol. Cell. Physiol 283, C905–C916 (2002). [DOI] [PubMed] [Google Scholar]
- 325.Sandoval RM, Kennedy MD, Low PS & Molitoris BA Uptake and trafficking of fluorescent conjugates of folic acid in intact kidney determined using intravital two-photon microscopy. Am. J. Physiol. Cell Physiol 287, C517–C526 (2004). [DOI] [PubMed] [Google Scholar]
- 326.Russo LM et al. The normal kidney filters nephrotic levels of albumin retrieved by proximal tubule cells: retrieval is disrupted in nephrotic states. Kidney Int 71, 504–513 (2007). [DOI] [PubMed] [Google Scholar]
- 327.Russo LM, Sandoval RM, Brown D, Molitoris BA & Comper WD Controversies in nephrology: response to ‘renal albumin handling, facts, and artifacts’. Kidney Int 72, 1195–1197 (2007). [DOI] [PubMed] [Google Scholar]
- 328.Meyer K et al. A Predictive 3D multi-scale model of biliary fluid dynamics in the liver lobule. Cell Syst 4, 277–290.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Porat-Shliom N, Harding OJ, Malec L, Narayan K & Weigert R Mitochondrial populations exhibit differential dynamic responses to increased energy demand during exocytosis in vivo. iScience 11, 440–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Subramanian BC et al. The LTB4–BLT1 axis regulates actomyosin and β2-integrin dynamics during neutrophil extravasation. J. Cell Biol 219, e201910215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Allan C et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Kvilekval K, Fedorov D, Obara B, Singh A & Manjunath BS Bisque: a platform for bioimage analysis and management. Bioinformatics 26, 544–552 (2010). [DOI] [PubMed] [Google Scholar]
- 333.Swedlow JR et al. Informatics and quantitative analysis in biological imaging. Science 300, 100–102 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Burel J-M et al. Publishing and sharing multi-dimensional image data with OMERO. Mamm. Genome 26, 441–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Hay J et al. PyOmeroUpload: a Python toolkit for uploading images and metadata to OMERO. Wellcome Open Res 5, 96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Morita M et al. Communication platform for image analysis and sharing in biology. Int. J. Netw. Comput 4, 369–391 (2014). [Google Scholar]
- 337.Swedlow JR et al. A global view of standards for open image data formats and repositories. Nat. Methods 18, 1440–1446 (2021). [DOI] [PubMed] [Google Scholar]
- 338.Genzel L et al. How the COVID-19 pandemic highlights the necessity of animal research. Curr. Biol 30, 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ji N Adaptive optical fluorescence microscopy. Nat. Methods 14, 374–380 (2017). [DOI] [PubMed] [Google Scholar]
- 340.Hampson KM et al. Adaptive optics for high-resolution imaging. Nat. Rev. Methods Primers 1, 68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Vasquez-Lopez SA et al. Subcellular spatial resolution achieved for deep-brain imaging in vivo using a minimally invasive multimode fiber. Light Sci. Appl 7, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Entenberg D, Roorda RD & Toledo-Crow R Non-linear microscope for imaging of the neural systems in live Drosophila. Biomed. Top. Meet 10.1364/BIO.2004.ThE4 (2004). [DOI]
- 343.Roorda RD, Hohl TM, Toledo-Crow R & Miesenbock G Video-rate nonlinear microscopy of neuronal membrane dynamics with genetically encoded probes. J. Neurophysiol 92, 609–621 (2004). [DOI] [PubMed] [Google Scholar]
- 344.Kirkpatrick ND et al. Video-rate resonant scanning multiphoton microscopy: an emerging technique for intravital imaging of the tumor microenvironment. Intravital 1, intv.21557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tsai PS et al. Ultra-large field-of-view two-photon microscopy. Opt. Express 23, 13833–13847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Li B, Wu C, Wang M, Charan K & Xu C An adaptive excitation source for high-speed multiphoton microscopy. Nat. Methods 17, 163–166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Schuck R et al. Multiphoton minimal inertia scanning for fast acquisition of neural activity signals. J. Neural. Eng 15, 025003 (2018). [DOI] [PubMed] [Google Scholar]
- 348.Maioli V et al. Fast in vivo multiphoton light-sheet microscopy with optimal pulse frequency. Biomed. Opt. Express 11, 6012–6026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hara K et al. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14, 658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Bornes L, Windoffer R, Leube RE, Morgner J & Van Rheenen J Scratch-induced partial skin wounds re-epithelialize by sheets of independently migrating keratinocytes. Life Sci. Alliance 4, e202000765 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Villette V et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608.e23 (2019). This study presents an ultrafast local volume excitation approach for rapid scanning at up to 15 kHz deep in the brain of awake mice to monitor changes in electrical activity using a genetically encoded voltage sensor.
- 352.Ziv Y & Ghosh KK Miniature microscopes for large-scale imaging of neuronal activity in freely behaving rodents. Curr. Opin. Neurobiol 32, 141–147 (2015). [DOI] [PubMed] [Google Scholar]
- 353.Chen S et al. Miniature fluorescence microscopy for imaging brain activity in freely-behaving animals. Neurosci. Bull 36, 1182–1190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Zong W et al. Large-scale two-photon calcium imaging in freely moving mice. Cell 185, 1240–1256.e30 (2022). This work develops a lightweight two-photon miniaturized microscope to allow for almost unrestricted animal movement and behaviour during large-scale IVM deep in the brain.
- 355.Khorshed RA et al. Automated identification and localization of hematopoietic stem cells in 3D intravital microscopy data. Stem Cell Rep 5, 139–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Kim J, Koo BK & Knoblich JA Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol 21, 571–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Clevers H Modeling development and disease with organoids. Cell 165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 358.Vaghela R, Arkudas A, Horch RE & Hessenauer M Actually seeing what is going on — intravital microscopy in tissue engineering. Front. Bioeng. Biotechnol 9, 627462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Prunier C, Chen N, Ritsma L & Vrisekoop N Procedures and applications of long-term intravital microscopy. Methods 128, 52–64 (2017). [DOI] [PubMed] [Google Scholar]
- 360.Zong W et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017). [DOI] [PubMed] [Google Scholar]
- 361.Rosenthal N & Brown S The mouse ascending: perspectives for human-disease models. Nat. Cell Biol 9, 993–999 (2007). [DOI] [PubMed] [Google Scholar]
- 362.Kerr RA Imaging the activity of neurons and muscles. WormBook 10.1895/wormbook.1.113.1 (2006). [DOI] [PMC free article] [PubMed]
- 363.Supatto W, McMahon A, Fraser SE & Stathopoulos A Quantitative imaging of collective cell migration during Drosophila gastrulation: multiphoton microscopy and computational analysis. Nat. Protoc 4, 1397–1412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Abu-Siniyeh A & Al-Zyoud W Highlights on selected microscopy techniques to study zebrafish developmental biology. Lab. Anim. Res 36, 12 (2020). This review discusses several of the common non-invasive microscopy techniques that are utilized for investigating zebrafish embryo and larvae in developmental biology.
- 365.Benjamin DC & Hynes RO Intravital imaging of metastasis in adult zebrafish. BMC Cancer 17, 660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Wrighton PJ et al. Quantitative intravital imaging in zebrafish reveals in vivo dynamics of physiological-stress-induced mitophagy. J. Cell Sci 134, jcs256255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Pinto-Teixeira F et al. Intravital imaging of hair-cell development and regeneration in the zebrafish. Front. Neuroanat 7, 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Buckingham ME & Meilhac SM Tracing cells for tracking cell lineage and clonal behavior. Dev. Cell 21, 394–409 (2011). [DOI] [PubMed] [Google Scholar]
- 369.Yano S et al. Spatial-temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle 13, 2110–2119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Wang Y et al. Direct visualization of the phenotype of hypoxic tumor cells at single cell resolution in vivo using a new hypoxia probe. Intravital 5, e1187803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Takehara H et al. Lab-on-a-brain: implantable micro-optical fluidic devices for neural cell analysis in vivo. Sci. Rep 4, 6721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Williams JK et al. Validation of a device for the active manipulation of the tumor microenvironment during intravital imaging. Intravital 5, e1182271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Szulczewski JM et al. In vivo visualization of stromal macrophages via label-free FLIM-based metabolite imaging. Sci. Rep 6, 25086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Myneni P, Terekhov A, Wright G, Hofmeister W & Janetopoulos C Intravital microfluidic windows for delivery of chemicals, drugs and probes. Microsc. Microanal 20, 1352–1353 (2014). [Google Scholar]
- 375. Deisseroth K et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci 26, 10380–10386 (2006). This seminal article, which coins the term ‘optogenetics’, reviews the (at the time) emerging confluence of optics, genetics and bioengineering for studies of intact neural circuits.
- 376.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Huang Z et al. Engineering light-controllable CAR T cells for cancer immunotherapy. Sci. Adv 6, eaay9209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Benedetti L Optogenetic tools for manipulating protein subcellular localization and intracellular signaling at organelle contact sites. Curr Protoc 1, e71 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Weis D & Di Ventura B Optogenetic control of nucleocytoplasmic protein transport. Methods Mol. Biol 2173, 127–136 (2020). [DOI] [PubMed] [Google Scholar]
- 380.Gasser C et al. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl Acad. Sci. USA 111, 8803–8808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rost BR, Schneider-Warme F, Schmitz D & Hegemann P Optogenetic tools for subcellular applications in neuroscience. Neuron 96, 572–603 (2017). [DOI] [PubMed] [Google Scholar]
- 382. Joshi J, Rubart M & Zhu W Optogenetics: background, methodological advances and potential applications for cardiovascular research and medicine. Front. Bioeng. Biotechnol 7, 466 (2020). This review article covers the development and use of optogenetic tools in cardiovascular medicine and provides an excellent summary of the different methods for introducing optogenetic tools into cells, including plasmids, adenoviruses and lentiviruses.
- 383.Krueger D et al. Principles and applications of optogenetics in developmental biology. Development 146, dev175067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Haeger A et al. Collective cancer invasion forms an integrin-dependent radioresistant niche. J. Exp. Med 217, e20181184 1(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Graham ML & Prescott MJ The multifactorial role of the 3Rs in shifting the harm–benefit analysis in animal models of disease. Eur. J. Pharmacol 759, 19–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Grimm H, Olsson IAS & Sandøe P Harm–benefit analysis — what is the added value? A review of alternative strategies for weighing harms and benefits as part of the assessment of animal research. Lab. Anim 53, 17–27 (2019). [DOI] [PubMed] [Google Scholar]
- 387.Davies GF et al. Developing a collaborative agenda for humanities and social scientific research on laboratory animal science and welfare. PLoS ONE 11, e0158791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


