Abstract
SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, has had an enduring impact on global public health. However, SARS-CoV-2 is only one of multiple pathogenic human coronaviruses (CoVs) to have emerged since the turn of the century. CoVs encode for several nonstructural proteins (nsps) that are essential for viral replication and pathogenesis. Among them is nsp15, a uridine-specific viral endonuclease that is important in evading the host immune response and promoting viral replication. Despite the established endonuclease function of nsp15, little is known about other determinants of its cleavage specificity. In this study we investigate the role of RNA secondary structure in SARS-CoV-2 nsp15 endonuclease activity. Using a series of in vitro endonuclease assays, we observed that thermodynamically stable RNA structures were protected from nsp15 cleavage relative to RNAs lacking stable structure. We leveraged the s2m RNA from the SARS-CoV-1 3’UTR as a model for our structural studies as it adopts a well-defined structure with several uridines, two of which are unpaired and thus highly probable targets for nsp15 cleavage. We found that SARS-CoV-2 nsp15 specifically cleaves s2m at the unpaired uridine within the GNRNA pentaloop of the RNA. Further investigation revealed that the position of uridine within the pentaloop also impacted nsp15 cleavage efficiency suggesting that positioning within the pentaloop is necessary for optimal presentation of the scissile uridine and alignment within the nsp15 catalytic pocket. Our findings indicate that RNA secondary structure is an important determinant of nsp15 cleavage and provides insight into the molecular mechanisms of RNA recognition by nsp15.
Introduction
Coronaviruses (CoVs) are a diverse group of enveloped, positive sense single-stranded RNA (+ssRNA) viruses within the Nidovirales order. CoVs infect a wide range of host species and are associated with mild to severe disease in livestock and humans [1–3]. The Coronaviridae family is composed of four genera: Alpha-, Beta-, Gamma-, and Deltacoronaviruses [4]. Since the turn of the century, several pathogenic CoVs have emerged in the human population including SARS-CoV-1, MERS-CoV, and SARS-CoV-2 with the latter being the causative agent of the COVID-19 pandemic [1, 2].
Replication of the large (~30kb) CoV genome is facilitated by multiple nonstructural proteins (nsp1-16) which are produced from two open reading frames encoded by the first two thirds of the viral genome. These proteins encode several key functions necessary for synthesis and post-translational processing of the viral genome, including RNA-dependent RNA polymerase (nsp12), helicase (nsp13), RNA M7 and 2ʹ-O-methyltransferase (nsp14, nsp16/nsp10), protease (nsp3, nsp5), and exonuclease (nsp14) activities. Structural proteins which assemble to form the virion, as well as accessory proteins, are encoded in a set of nested subgenomic RNAs which are coterminal with the 3’ end of the genome.
The viral replication/transcription complex (RTC) is housed within membrane structures derived from the host endoplasmic reticulum (ER) which are induced by expression of nsp3, 4, and 6 [5–12]. Like other +ssRNA viruses, these double-membrane vesicles (DMVs) provide a microenvironment that supports efficient viral replication and sequester double-stranded RNA (dsRNA) replication intermediates from detection by host pathogen recognition receptors (PRRs) [13, 14]. Previous studies have demonstrated a key role for the interferon (IFN) response in restriction of CoVs. Accordingly, CoVs have evolved several mechanisms to evade antiviral immunity (e.g., 2ʹ-O-methylation) and encode several proteins which antagonize the IFN response (nsp1, ORF6) [15–17]. Nsp15 is a uridine-specific endonuclease (endoU) encoded by CoVs and other Nidoviruses which has been implicated in evasion of host-mediated RNA sensing and IFN activation [18, 19]. While nsp15 has been shown to be non-essential for viral replication of several CoVs, absence of nsp15 endoU activity (Δnsp15) leads to greater accumulation of dsRNA replication intermediates which trigger IFN activation in immune competent cells such as macrophages [18–20]. During CoV infection, dsRNA sensing occurs primarily via MDA-5, though other RNA sensors (PKR) have also been implicated [18, 21].
While originally described as a uridine-specific endonuclease, mouse hepatitis virus (MHV) nsp15 has been shown to preferentially cleave pyrimidine-adenine dinucleotides (U↓A or C↓A) [22]. More recently, viral targets of MHV nsp15 have been identified in both the polyU tract of 5’ negative-sense RNA, as well as at several sites throughout the CoV genome [19, 22]. Although multiple nsp15 cleavage sites have been mapped throughout the MHV genome, no other sequence determinants have yet been identified. Despite the abundance of pyrimidine-adenine dinucleotides across the MHV genome, only a fraction of these sites are cleaved by nsp15 suggesting that other RNA determinants likely affect cleavage specificity of viral RNA. Earlier in vitro studies have shown that SARS-CoV-1 nsp15 cleaves a single unpaired uridine in the stem-loop II motif (s2m) RNA structural element found in the 3’-UTR of the viral genome suggesting that the structural context in which the scissile uridine is presented may play a role in determining nsp15 cleavage specificity [23]. In this study, we use SARS-CoV-1 s2m as a model RNA to explore the secondary structure requirements for SARS-CoV-2 nsp15 cleavage. Our findings reveal that thermodynamically stable RNAs are protected from SARS-CoV-2 nsp15 cleavage and cleaved less efficiently. We show for the first time that the thermodynamic stability of RNA structure as well as positioning and sequence context of the scissile uridine play a role in determining SARS-CoV-2 nsp15 cleavage specificity. This is consistent with previous studies of SARS-CoV-1 nsp15, suggesting that SARS-CoV-2, and CoVs more broadly, may use RNA structure to regulate nsp15 cleavage specificity. These data shed further light on requirements for viral endoU target recognition and demonstrate an underappreciated role for viral RNA structure in evasion of innate immunity.
Materials and methods
Generation of SARS-CoV-2 RNA segments
100nt segments of SARS-CoV-2 genome were amplified using the primers listed in Table 1. Amplicons were purified using Mag-Bind Total Pure NGS beads (Omega Bio-Tek). 1.5x bead volume relative to amplification reactions was used and purified as per manufacturer’s instructions. Purified amplicons were subcloned into a vector containing the HDV ribozyme [24]. These constructs were linearized and purified prior to transcription.
Table 1. Primers used for generation of SARS-CoV-2 RNA segments.
| RNA | Forward Primer | Reverse primer |
|---|---|---|
| RNA 1 | CATGAGCTTA AGTAATACGA CTCACTATAG GCTACTAACA ATGCCAT | CATGAGGCTA GCAAGATTGT GTCCGCTTAA AA |
| RNA 2 | CATGAGCTTA AGTAATACGA CTCACTATAG ACCTGAGCAT AGTCTTG | CATGAGGCTA GCGAGAACAC ACAGCCTC |
| RNA 3 | CATGAGCTTA AGTAATACGA CTCACTATAG TGAAGTGCTG TCTGAC | CATGAGGCTA GCGTTACACG ATAACCAGTA AAG |
Primers used to amplify RNAs 1–3 prior to subcloning as described above.
Approximately 5 pmol of purified DNA was used as a template for T7 transcription reactions. RNAs were transcribed using the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs). Reactions were set up as per manufacturer’s instructions with the addition of 4 units of Murine RNase Inhibitor (NEB) and 5% DMSO. Transcriptions were incubated at 42°C overnight after which, the reactions were treated with DNaseI (NEB) as per manufacturer’s instructions. DNase-treated transcriptions were then purified using Mag-Bind Total Pure NGS beads as described above.
Transcriptions were gel purified by running samples on urea gels. 15% TBE-Urea gels were made using SequaGel UreaGel 29:1 Denaturing Gel System (National Diagnostics). Urea was added to the samples to a final concentration of 4M before running on the gels at 200V for 90 minutes. Gels were briefly stained with 0.02% Methylene Blue in TBE until ladder was visible. Gels were imaged on Gel Doc XR+ (Bio-Rad), and bands corresponding to 100nt fragments were cut out and shredded by centrifugation. RNAs were eluted overnight on rotator in 3x volume of elution buffer (10mM Tris-HCl pH 7.5, 1mM EDTA, 300mM NaCl, 0.1% SDS) relative to gel weight. Gel slurries were applied to cellulose acetate columns and frozen at -80°C for 10 minutes. Samples were centrifuged at 10,000x g for 3 minutes. Eluate was adjusted up to 600μL with nuclease-free water and purified by acid phenol-chloroform extraction.
All other RNAs (s2m and ∆s2m and corresponding mutant RNAs) were synthesized by Integrated DNA Technologies. RNA sequences can be found in S1 Table.
Expression and purification of recombinant nsp15
Recombinant nsp15 was expressed and purified as previously described [25]. Briefly, a pET28a(+) construct consisting of a codon-optimized SARS-CoV-2 nsp15 gene sequence with a N-terminal hexahistidine tag was expressed in Rosetta BL21(DE3) E. coli competent cells using autoinduction methods. The K290A mutant was generated using a Q5 site-directed mutagenesis kit (NEB) and the forward and reverse primers: CGGCAGCAGCGCATGCGTGTGC and GTCTGCGCATCGGTAATAAAATAG. Recombinant protein was purified using 5 mL HisTrap FF immobilized metal ion affinity chromatography columns (GE Healthcare) and fractionated using a Superdex 200 size-exclusion chromatography column and an NGC Chromatography System (Bio-Rad). Fractions containing purified nsp15 were flash frozen in liquid nitrogen and stored at -80°C.
Endonuclease assay
RNAs were diluted in folding buffer (50mM Tris-HCl pH 7.5, 50mM KCl, 5mM MgCl2, 5mM DTT). RNAs were folded by incubating at 95°C for 10 minutes then cooled to 25°C at a rate of 0.1°C/sec (S1 Fig). Purified nsp15 was diluted in assay buffer (50mM Tris-HCl pH 7.5, 50mM KCl, 5mM MnCl2, 5mM MgCl2, 5mM DTT) to the appropriate concentration for indicated nsp15 hexamer:RNA ratios. To denature nsp15, the protein was incubated at 95°C for 5 minutes. Denatured wt protein showed no cleavage when compared to the nsp15 catalytic mutant protein (S2 Fig). Folded RNA was aliquoted into individual tubes, and diluted nsp15 was mixed in equal amounts to each RNA simultaneously. Reactions were sampled and arrested at indicated timepoints with a final concentration of 20mM EDTA before flash freezing in liquid nitrogen. Denatured nsp15 reactions were incubated for 240 minutes.
TBE-Urea gels were made using SequaGel UreaGel 29:1 Denaturing Gel System (National Diagnostics). 15% or 22.5% gels were poured as indicated in each experiment. RNA samples were thawed and mixed with an equal volume of 2x RNA Loading Dye (NEB) and incubated at 70°C for 10 minutes prior to loading onto the gels. Gels were run at 200V for 1 hour in 1x TBE. Gels were stained with SYBR Green II (ThermoFisher) diluted 1:5000 in TBE for 30 minutes while shaking and imaged on Gel Doc XR+ (Bio-Rad). Full-length bands at each time point were calculated by densitometry using Image Lab software (Bio-Rad). Density of bands were normalized to denatured nsp15 control for each reaction.
Differential Radial Capillary Action of Ligand Assay (DRaCALA)
DRaCALAs were performed as described previously [26]. Briefly, RNAs were dephosphorylated and 5’ radiolabeled with 30μCi ATP. Radiolabeled RNAs were set up in reactions with the following components: DRaCALA buffer (50mM HEPES pH 7.0, 100mM KCl), 5mM MgCl2, 50ng/μL yeast tRNA, 1mM DTT, 5% glycerol. RNAs were heated to 95°C for 2 minutes and cooled at RT for 10 minutes. Serial dilutions of catalytic mutant nsp15 K290A were added to the reactions which were incubated for 15 minutes at RT and spotted onto a 0.45μm nitrocellulose membrane. Blots were exposed to a phosphor screen overnight and imaged on Sapphire Molecular Imager (Azure Biosystems). Binding was measured by densitometry as previously described [27] using Fiji.
Statistical analysis
For quantification of band density in endonuclease assays, the area under the curve (AUC) was calculated for each replicate using GraphPad Prism software. Statistical analysis of the AUC was performed using one-way ANOVA with multiple comparisons, whereby the mean of each column was compared to the mean of all other columns. p<0.05 (*), p<0.01 (**), p<0.001 (***), p<0.0001(****) For statistical analysis of DRaCALA, the dissociation constant (KD) was calculated for each RNA and a one-way ANOVA with multiple comparisons was used to calculate p-values for each comparison.
Results
RNA secondary structure modulates nsp15 cleavage efficiency
Nsp15 cleavage sites have previously been mapped in the MHV genome following viral infection [19, 22]. Despite the abundance of pyrimidine-adenine dinucleotides throughout the genome, MHV nsp15 was found to cleave only a fraction of these sites suggesting that other determinants play a role in CoV nsp15 cleavage specificity. Using the MEME-suite XSTREME motif analysis and discovery tool [28] we analyzed 200 nt sequences corresponding to 100 nt upstream and downstream of the MHV nsp15 cleavage sites identified by Ancar et al. to identify shared motifs that might contribute to specific recognition or cleavage of these sequences by nsp15. Although some motifs were found to be enriched in sequences surrounding the MHV nsp15 cleavage sites, they were not uniformly conserved across all cleavage sites and were also found at other sites not previously identified as nsp15 targets.
As RNA structure has previously been implicated in nsp15 cleavage specificity [23], we next compared the underlying predicted secondary structure of the same 200 nt fragments using LocRNA software [29, 30]. We did not identify any conserved structural elements at nsp15 cleavage sites in MHV nor did we identify conserved structural elements upstream or downstream of these sites. Despite the absence of any conserved sequence or structural motifs, we hypothesized that the presence of thermodynamically stable RNA elements would prevent cleavage of RNA by nsp15.
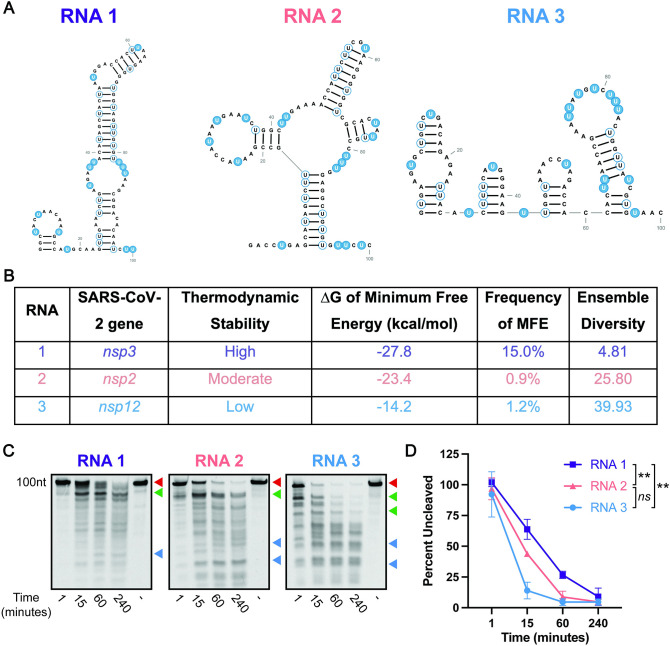
To test this hypothesis, we used RNAfold [31] to identify regions across the SARS-CoV-2 genome predicted to have varying degrees of thermodynamic stability (RNA 1, RNA 2, RNA 3; Fig 1A, Table 2) [32, 33]. We identified representative regions of high, moderate, and low thermodynamic stability as indicated by their relative ΔG values (Fig 1B) with more stable predicted structures having lower ΔG values [34]. Additionally, we compared the predicted RNA structures to previously published SHAPE-MaP data (S3 Fig) [35]. RNA 1, the most thermodynamically stable RNA, shared significant similarity to these data. However, we observed less correlation between the RNAfold predictions and SHAPE-MaP data for RNA 2 and particularly RNA 3. These two RNAs were predicted to have higher ensemble diversities (Fig 1B) and, as such, are predicted to be more structurally variable (adopt a larger number of conformers than RNA 1). Therefore, it follows that less correlation was observed between predicted and experimentally determined structures. 100 nt RNAs corresponding to these regions were synthesized in vitro and cleavage efficiency of these RNAs was assessed using a SARS-CoV-2 nsp15 endonuclease assay [25] (Fig 1C and 1D). Folded RNAs were incubated with recombinant SARS-CoV-2 nsp15 for various times (1, 15, 60, and 240 minutes) and cleavage products were analyzed by denaturing polyacrylamide gel electrophoresis (Fig 1C). Cleavage of full-length RNA was additionally quantified by densitometry by comparing the abundance of full-length RNA at each time point to a denatured nsp15 control (no cleavage, (-) far-right lane) (Fig 1D). As we hypothesized, we observed that RNAs with relatively moderate or low predicted thermodynamic stability were cleaved more rapidly than RNA 1 with relatively high thermodynamically stability (Fig 1C, compare abundance of full-length transcript at 1, 15 and 60 min [red arrow]; Fig 1D compare percentage uncleaved RNA). Notably, RNA with relatively low thermodynamic stability showed rapid cleavage as early as 1 min following the addition of nsp15. While each RNA yielded different cleavage products owing to the differences in RNA sequence, we observed the appearance of distinct cleavage products that either decreased (green arrows) or increased (blue arrows) in abundance over time (Fig 1C). These likely represent cleavage intermediates or end products respectively and suggest that other structural features or specific sequences also likely contribute to the higher efficiency of nsp15 cleavage of some RNA sequences over others. Collectively, these data suggest that RNA secondary structure impacts nsp15-dependent RNA cleavage.
Fig 1. Secondary structure protects RNA from cleavage by SARS-CoV-2 nsp15.
A. Predicted minimum free energy structures (RNAfold) of 100 nt RNAs from the SARS-CoV-2 genome of varying thermodynamic stability (high, medium, and low). Unpaired uridines are highlighted in blue, and paired uridines are outlined in blue. All RNA structures were predicted in RNAfold and designed in RNA2Drawer [34, 36]. B. The locations within the SARS-CoV-2 genome are indicated for each structure, and thermodynamic stability is indicated as -ΔG. Frequency of MFE and ensemble diversity is listed as predicted by RNAfold. C. Endonuclease assays of RNAs from (A) at a nsp15 hexamer:RNA ratio of 1:600. Full-length (uncut) RNAs are indicated by the red arrows. Diminishing cleavage products (cleavage intermediates) are indicated by green arrows. Accumulating cleavage products (cleavage end products) indicated by blue arrows. RNAs were run on a 15% TBE-Urea Gel. Representative images from one of three total experiments are shown. D. Quantitation of data from (C). The percentage of full-length RNA remaining was measured by densitometry. Most thermodynamically stable RNA (RNA 1) was cleaved least rapidly relative to other RNAs. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). Area under the curve (AUC) was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. Data represents three independent experiments.
Table 2. Sequences of RNAs 1–3 from SARS-CoV-2.
| RNA | Sequence |
|---|---|
| RNA 1 (3496–3595) | GGCUACUAAC AAUGCCAUGC AAGUUGAAUC UGAUGAUUAC AUAGCUACUA AUGGACCACU UAAAGUGGGU GGUAGUUGUG UUUUAAGCGG ACACAAUCUU |
| RNA 2 (1401–1500) | GACCUGAGCA UAGUCUUGCC GAAUACCAUA AUGAAUCUGG CUUGAAAACC AUUCUUCGUA AGGGUGGUCG CACUAUUGCC UUUGGAGGCU GUGUGUUCUC |
| RNA 3 (16700–16799) | GUGAAGUGCU GUCUGACAGA GAAUUACAUC UUUCAUGGGA AGUUGGUAAA CCUAGACCAC CACUUAACCG AAAUUAUGUC UUUACUGGUU AUCGUGUAAC |
SARS-CoV-2 nsp15 cleaves unpaired bases in structured RNAs
In vitro biochemical studies by Bhardwaj et al. previously showed that SARS-CoV-1 nsp15 specifically cleaves 3’ of unpaired uridines in s2m, a conserved structural element found in the 3’UTR of the SARS-CoV-1 genome and some other related CoVs [23, 37, 38]. In addition to our results described above (Fig 1), this study suggests that the context in which the cleaved uridine is presented may be important for determining the cleavage specificity of nsp15. In the case of s2m, the two unpaired uridines (U25 and U30) are located in two unstructured loops (Fig 2A, highlighted in blue). Structural resolution of s2m has shown that U25, which is located within the GNRNA sequence of the pentaloop of s2m, is highly disordered relative to other nucleotides, and that U30 is also moderately disordered [37]. Interestingly, while there is significant homology between s2m from SARS-CoV-1 and SARS-CoV-2, SARS-CoV-2 s2m has two single nucleotide polymorphisms (SNPs) that change the structure of the RNA (S4 Fig). Based on previous findings and the availability of structural information, we posited that SARS-CoV-1 s2m would be a tractable model RNA for studying the determinants of cleavage specificity of SARS-CoV-2 nsp15.
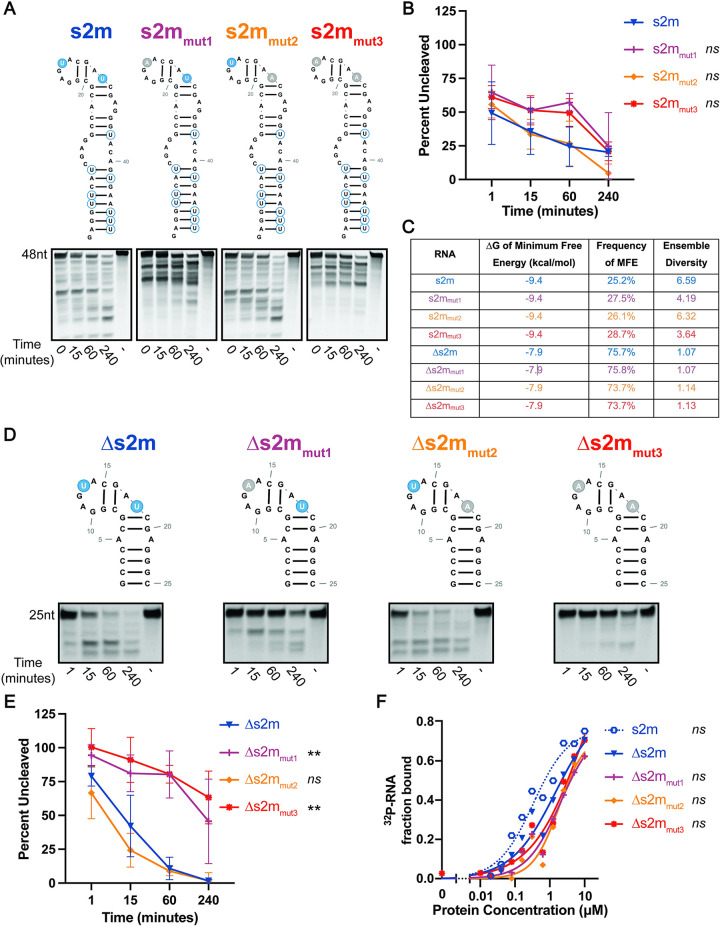
Fig 2. SARS-CoV-2 nsp15 cleaves structured RNAs at specific sites.
A. Endonuclease assays of full-length s2m mutant RNAs at a nsp15 hexamer:RNA ratio of 1:30. RNAs were run on a 15% TBE-Urea Gel. Full-length bands indicated at the top of the gel with cleavage products below. Representative images from one experiment. B. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (-). RNAs with uridines present in the pentaloop (s2m and s2mmut2) cleaved more efficiently than s2mmut1 and s2mmut3. AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. AUC of s2m mutant RNAs are compared to wt s2m. Data represents three independent experiments. C. Thermodynamic stability, frequency of MFE, and ensemble diversity of s2m and mutant RNAs indicated as predicted by RNAfold. D. Endonuclease assays of modified s2m RNAs at a nsp15 hexamer:RNA ratio of 1:6. RNAs were run on a 22.5% TBE-Urea Gel. Full-length bands indicated at the top of the gel with cleavage products below. Representative images from one experiment. E. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (-). RNAs with uridines present in the pentaloop (∆s2m and ∆s2mmut2) were cleaved more efficiently than ∆s2mmut1 and ∆s2mmut3. AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. AUC of ∆s2m mutant RNAs are compared to wt ∆s2m. Data represents three independent experiments. F. Nsp15 binding curves of Δs2m RNAs as compared to wt s2m. ∆s2m RNA binding curves overlap with each other while wt s2m curve is shifted left indicating improved binding with nsp15. Dissociation constant (KD) was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the KD. AUC of wt full-length s2m and ∆s2m mutant RNAs are compared to wt ∆s2m. Data represents three independent experiments.
We first sought to determine whether SARS-CoV-2 nsp15 exhibits cleavage specificity for unpaired uridines in s2m similar to what was previously observed for SARS-CoV-1 nsp15 [23]. To identify whether nsp15 preferentially cleaves at U25, U30, or both, we designed the wild type (wt) SARS-CoV-1 s2m RNA, as well as single and double s2m mutants containing U-to-A substitutions at U25 and U30 (Fig 2A) and compared cleavage efficiency of these RNAs via endonuclease assay (Fig 2A and 2B). In contrast to wt s2m, cleavage of the double mutant (s2mmut3) was noticeably impaired consistent with previous studies showing nsp15 cleavage preference for unpaired uridines. However, in contrast to previous findings with SARS-CoV-1 nsp15, we observed that mutation of U30 (U30A; s2mmut2) did not impact RNA cleavage, whereas mutation of U25 (U25A; s2mmut1) did. Although quantitative differences in full-length RNA cleavage were not found to be statistically significant (Fig 2B), accumulation of cleavage products in the U25A (s2mmut1) and double mutant (s2mmut3) was clearly impaired relative to wt and U30A (s2mmut2) RNAs.
While we would expect to see the accumulation of cleavage products over time, interestingly, we also observe that certain cleavage products diminish throughout the time course (Fig 2A). These diminishing products may indicate that step wise cleavage of RNA and RNA intermediates is altered in the absence of the scissile U25. However, while mutation of U25 was not predicted to impact the structure of s2m using RNAfold analysis (Fig 2C), we cannot rule out the possibility that mutation of U25 leads to changes in s2m RNA structure which impact nsp15-mediated cleavage.
Due to the presence of multiple uridines in s2m, accurate identification of cleavage products and intermediates was challenging. In order to eliminate extraneous cleavage products to clarify interpretation of endonuclease data and further dissect RNA structural determinants of cleavage efficiency, we designed truncated s2m RNAs (Δs2m) which lack the base of the stem and all uridines except U25 and U30 (Fig 2D). Similar to the experiments described above, we also engineered Δs2m mutants with U-to-A substitutions at U13 (Δs2mmut1) and U18 (Δs2mmut2) (equivalent to U25 and U30 in wt s2m), or both U13 and U18 (Δs2mmut3). As with the full-length s2m RNAs, the free energies and ensemble diversities of the ∆s2m mutant RNAs were similar to each other (Fig 2C). We analyzed cleavage of wt and mutant Δs2m RNAs over time by endonuclease assay as described above (Fig 2D and 2E). In keeping with experiments with full-length s2m RNA, we observed rapid accumulation of cleavage products in wt Δs2m and Δs2mmut2 RNAs. In contrast, mutation of both U13 and U18 (U25 and U30 equivalent), or U13 (U25 equivalent) alone, resulted in delayed and reduced nsp15 cleavage of RNA. Interestingly, we still observed some modest cleavage of Δs2mmut1 suggesting that while U13 is the predominant cleavage site in the Δs2m RNA, nsp15 is also able to recognize and cleave at other sites. While mutation of both U13 and U18 led to a further reduction in nsp15-mediated cleavage, some cleavage products were still detected in the double mutant lending further support to the hypothesis that nsp15 can also cleave non-uridine bases, albeit at lower efficiency. Indeed, while nsp15 displays a strong preference of cleavage 3’ of uridines, previous work has shown that cleavage at other bases (especially cytosine) also occurs [39].
We hypothesized that RNA secondary structure contributes to nsp15 cleavage specificity by presenting the scissile uridine in a structural context favorable for nsp15 cleavage. However, an alternative explanation could be that RNA structure serves to sterically hinder or promote nsp15-RNA binding. To address whether the differences we observed in cleavage of wt and mutant Δs2m RNAs was due to differences in protein-RNA binding alone, we compared nsp15-RNA binding affinities using differential radial capillary action of ligand assay (DRaCALA) [26, 27]. Here, 5’-end radiolabeled Δs2m RNAs were incubated with increasing concentrations of recombinant mutant nsp15 (nsp15 (K290A)), complexes applied to nitrocellulose membrane, and the fraction of bound vs unbound RNA quantified (Fig 2F). Nsp15 (K290A) was used to eliminate endonuclease activity and contains a mutation of one of the conserved catalytic residues necessary for endoU activity [23, 40–42]. We compared binding of wt and mutant Δs2m RNA, in addition to full-length wt s2m. The latter was chosen as a control, as it is presently unknown what the minimum length requirement is for RNA recognition and binding by nsp15. We observed binding of nsp15 (K290A) to wt Δs2m (KD = 1.5μM). When we compared binding of K290A to each mutant Δs2m RNA, we observed only slight decreases in binding affinity (approximately 1.3-fold; Δs2mmut1 KD = 2.0μM, Δs2mmut2 KD = 1.9μM, Δs2mmut3 KD = 8.5μM), which were not found to be statistically significant. Notably, we observed a modest increase in binding affinity of K290A for the full-length s2m RNA (approximately 4-fold; s2m KD = 0.3μM) suggesting that other RNA-protein contacts in longer RNAs may modestly contribute to binding affinity. As nsp15 exhibited similar binding affinities for all Δs2m RNAs we concluded that differences in cleavage efficiency of Δs2m RNAs is not due to differences in protein-RNA binding but likely driven by positioning of the scissile uridine in these differently structured RNAs.
Flexible uridine nucleotides in structured pentaloops are susceptible to SARS-CoV-2 nsp15 cleavage
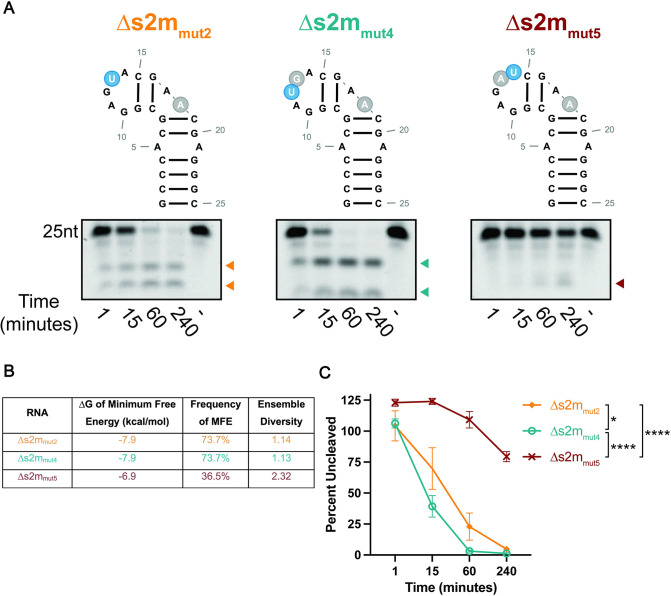
Recent structural studies have suggested that SARS-CoV-2 nsp15 uses a base flipping mechanism to position the scissile uridine in the catalytic pocket, thus enabling cleavage of dsRNA [43]. The crystal structure of s2m shows that U25 in the GNRNA pentaloop adopts a similar flipped out orientation [37] suggesting that this mechanism could also account for the specificity of nsp15 cleavage for U25 in s2m. RNA pentaloops adopt a characteristic structure which consists of a sheared G-A base pair that closes the pentaloop and induces base stacking of first N, R, and A nucleotides. Notably, this conformation induces extrusion (or ‘flipping’) of the second N (GNRNA) in the pentaloop [44, 45]. Thus, we postulated that the position of the scissile uridine in the pentaloop (GAGUA) might be necessary to confer the correct orientation required for nsp15 cleavage. To test whether the relative position of U25 is necessary for cleavage specificity, we constructed two additional Δs2m mutants in which the position of the scissile uridine has been altered (Fig 3A, top), and compared cleavage efficiency in the endoU assay (Fig 3A, bottom). In Δs2mmut4 the adjacent guanidine and uridine in the pentaloop were swapped (GAUGA), such that the scissile uridine now sits at the apex of the pentaloop. In Δs2mmut5 the scissile uridine was positioned downstream at the 3’ end of the pentaloop and adjacent to the closing GC base pair of the helix (GAGAU). The second uridine in the bulge of Δs2m (U18) was also mutated in both RNAs to assess the impact of U13 (U25 equivalent in full-length s2m) cleavage only. We predicted that in Δs2mmut4 positioning of the scissile uridine at the apex would have minimal effect on cleavage but that placement of uridine adjacent to the helix would place structural constraints on this base such that it would be unable to adopt the necessary conformation (flipped out) required for positioning within the catalytic pocket of nsp15. When we compared cleavage of each mutant, we observed that Δs2mmut4 (GAUGA) was cleaved to completion earlier than both Δs2mmut2 (control) and Δs2mmut5 (GAGAU) (Fig 3B). We also designed ∆s2mmut6 and ∆s2mmut7 with a uridine in the second position to assess cleavage of these RNAs by nsp15 (S6A Fig). We observed a slight, but consistent increase in nsp15 cleavage activity in ∆s2mmut6 and ∆s2mmut7 relative to ∆s2mmut2. In contrast, we observed almost no cleavage of Δs2mmut5 supporting the hypothesis that the scissile uridine might be more structurally constrained at this position and unable to engage with the catalytic pocket of nsp15. From these data, we concluded that uridine position within the pentaloop influences nsp15 cleavage activity, likely through structural constraints.
Fig 3. Uracil position in loop structures impacts SARS-CoV-2 nsp15 cleavage efficiency.
A. Endonuclease assays of s2m pentaloop mutant RNAs. Reactions were run at a nsp15 hexamer:RNA ratio of 1:6 run on a 22.5% TBE-Urea Gel. Full-length bands indicated at the top of the gel with cleavage products below. Representative images from one experiment. Cleavage products that accumulate over time are indicated by arrows. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of ∆s2m mutant RNAs indicated as predicted by RNAfold. C. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). RNAs with uridines positioned closer to the apex of the pentaloop cleaved more rapidly than ∆s2mmut5 in which the uridine is at the end of the pentaloop. AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. Data represents three independent experiments.
We also tested further RNAs from the SARS-CoV-2 genome that adopt a similar structure to s2m RNAs. These s2m-like RNAs shared features with s2m including comparable length, a loop size of 5–7 nucleotides, a similar loop sequence to the GNRUA pentaloop of s2m, and internal bulges (S7A Fig). Additionally, only RNAs which fulfilled the above criteria and had high confidence SHAPE-MaP structures were chosen [35]. We also chose a control RNA based on the SHAPE-MaP that lacks the s2m pentaloop as well as the dinucleotide context that has been shown to be important for nsp15 cleavage. We found that both s2m-like RNAs in the SARS-CoV-2 genome were cleaved while the control RNA was not (S7C Fig). These results provide further evidence of structural determinants of RNA substrates playing a role in nsp15 cleavage activity.
Discussion
Nsp15 is a viral endonuclease conserved across the Nidovirales and plays a crucial role in CoV evasion of host innate immunity. The crystal structure of several CoV nsp15 proteins has been solved to date, including SARS-CoV-2 [40, 42, 46–50]. Structural and biochemical analyses have revealed key aspects of nsp15 function, including the requirement of Mn2+ for endonuclease activity at neutral pH as well as formation of higher order oligomers (hexamers) which is also necessary for endoU function [23, 40, 41, 46, 50, 51]. Nsp15 primarily functions to evade innate immune responses by cleaving dsRNA replicative intermediates and preventing activation of MDA-5 and possibly other RNA sensors in the host cell [18–22, 52–55]. Some studies also suggest that CoV nsp15 may play a role in viral transcriptional regulation during viral replication [22] and that endoU-independent activities additionally contribute to modulation of host responses [54, 56–59].
At present, the RNA targets of nsp15 endoU from across the Coronaviridae are poorly defined. This can be attributed in part to the fact that in vitro studies have used different RNA species (dsRNA vs ssRNA) and different RNA sequences and lengths, making cross-comparison of these data challenging. Nsp15 has been shown to cleave 3’ of pyrimidines, in particular uridine, and exhibit a preference for weak bases (A or U) 3’ of the scissile nucleotide. However, to date no specific nsp15 recognition motifs have been described. In the absence of a sequence motif, we postulate that RNA secondary structure may function to regulate both RNA binding and cleavage efficiency of nsp15. Thus, we sought to determine the role of RNA structure in the regulation of SARS-CoV-2 nsp15-mediated cleavage and define specific structural features of RNAs that contribute to this. We demonstrated that increased thermodynamic stability of RNA is associated with decreased nsp15 cleavage efficiency (Fig 1) and postulated that stable secondary structure may contribute to nsp15 cleavage by two modes: 1) steric hinderance and 2) inducing optimal positioning of the scissile uridine in the nsp15-RNA complex. To test this second hypothesis, we used s2m as a model RNA to explore the structural determinants of nsp15 cleavage specificity.
Using an in vitro biochemical approach, we first demonstrated that nsp15 exhibits specificity for cleavage of the uridine nucleotide located in the GNRNA pentaloop of s2m (Fig 2). This contrasts with previous data from Bhardwaj et al., in which the authors observed preferential cleavage of U30 located in the bulge sequence downstream of the pentaloop [23]. It should be noted that these previous studies were performed with SARS-CoV-1 nsp15, which shares 88% identity with SARS-CoV-2 nsp15. One explanation for the discrepancy in our data could be structural differences between these proteins which lead to altered cleavage specificity.
We also tested s2m-like RNAs that shared a similar loop size and sequence to wt s2m (S6A Fig) and found that the s2m-like RNAs were cleaved far more rapidly that a control RNA (S6C Fig). However, we cannot rule out that other variables may be playing a role in this difference including the positioning and the number of uridines within the loops of these RNAs. Furthermore, the control RNA contains a tetraloop as opposed to pentaloop or heptaloop (S6A Fig). As we have not thoroughly tested the influence of loop size on nsp15 cleavage, it is possible that nsp15 cleaves tetraloops with a lower efficiency than pentaloops.
Notably, the specificity for cleavage of the pentaloop uridine (GAGUA) was not due to differences in nsp15-RNA binding (Fig 2F), thus we postulated that the sequence context of the scissile uridine might be important for this specificity. The characteristic structure adopted by GNRNA pentaloops induces extrusion, or base flipping, of the scissile uridine away from the body of the RNA [44, 45], and this base flipping mechanism has been suggested to be important for correct positioning of uridine in the catalytic pocket of nsp15 [43]. Pentaloop motifs have also been implicated in base editing mediated by Adenosine Deaminases that Act on RNA 2 (ADAR2) [60], though in this instance the GCUMA pentaloop is located downstream of the edited A and is implicated in recruitment of ADAR2 via interactions between the dsRNA binding motif of ADAR2 and the target RNA. We speculated that if structural conformation of the scissile uridine was important for base recognition and cleavage that altering the position (and conformation) of the uridine base would lead to altered cleavage efficiency. Indeed, repositioning of the scissile uridine to the apex of the pentaloop (Δs2mmut4) slightly enhanced cleavage efficiency, whereas repositioning adjacent to the closing G-C base pair (Δs2mmut5) significantly reduced RNA cleavage (Fig 3). We postulate that the marked decrease in Δs2mmut5 cleavage is likely due to altered positioning of the scissile uridine which we predict is no longer extruded from the pentaloop, as well as decreased flexibility of this base which is now structurally constrained by the neighboring G-C base pair. However, RNAfold predictions indicate a potential wobble base pairing between the G-U nucleotides in the pentaloop of ∆s2mmut5. Thus, it is also possible that occlusion of the scissile uridine may be contributing to the lack of cleavage of ∆s2mmut5 (Fig 3). Interestingly, based on the anticipated cleavage sites of ∆s2mmut2 and ∆s2mmut4, we expected to see a single nucleotide difference in the bands between these RNAs, However, there appears to be a larger size discrepancy between these products (S7 Fig). These may represent distinct cleavage products produced from each RNA, suggesting that additional cleavage at noncanonical sites may occur. Further studies are needed to confirm the identity of these cleavage products and the specific cleavage sites in ∆s2mmut2 and ∆s2mmut4 RNAs.
Another possibility is that cleavage efficiency is impacted by altering the sequence context of the scissile uridine. Nsp15 has been shown to preferentially cleave uridines flanked by a weak base (A/U) which is found in wt Δs2m and Δs2mmut2 (U↓A) but is changed to a strong base in Δs2mmut5 (U↓C). However, we observed a modest increase in cleavage efficiency of Δs2mmut4 which also has a strong base at the 3’ position (U↓G) and lends support to the hypothesis that the structural context of the scissile uridine, not sequence context, may be more relevant in dictating cleavage specificity and efficiency. To further address the importance of uridine position within the s2m pentaloop, we designed ∆s2mmut6 and ∆s2mmut7 RNAs which contained the uridine in the second position of the pentaloop. These RNAs were cleaved slightly but consistently more rapidly than ∆s2mmut2 (S5C Fig). Previous work has shown that trinucleotide sequence context is also important for nsp15 activity and that G-U↓A trinucleotides were cleaved most efficiently [39]. Interestingly, ∆s2mmut6 (G-U↓G) and ∆s2mmut7 (G-U↓A) were cleaved at similar rates (S5C Fig) whereas ∆s2mmut7 and ∆s2mmut2 both share the same sequence context (G-U↓A) but were cleaved at slightly different rates. This lends further support to the hypothesis that both sequence (trinucleotide) context as well as RNA structure are necessary for optimal cleavage of target RNAs. Future combinatorial studies which examine the impact of both sequence and structural context will help to further elucidate determinants of nsp15 substrate specificity.
The studies described herein were based largely on RNA structure prediction. While RNA structure prediction favors the formation of the most thermodynamically stable conformer (MFE), the MFE structure is not necessarily the predominant, or most biologically relevant, conformer. Furthermore, RNAs are dynamic molecules that can adopt and transition between multiple conformations, thus interpretation of data using structurally diverse RNAs can be challenging. To ameliorate this issue, we compared the 100nt RNAs from SARS-CoV-2 to established SHAPE-MaP data and found that the most thermodynamically stable RNA highly correlated with published structural data (S3 Fig) [35]. For the same reason, we chose to use s2m as a model RNA for our studies because it has an established crystal structure [37]. Nonetheless, it is important to note that these substrates are likely undergoing conformational changes throughout the course of the reaction. Despite this, we show large differences in cleavage of RNAs that have even single nucleotide changes (Figs 2 and 3) thus indicating that nsp15 preferentially acts on certain substrates. Furthermore, in our endonuclease assays, we used several RNAs with loop structures. While loops are commonly thought to be regions lacking structure, unpaired nucleotides in loops can form tertiary structures through non-Watson-Crick base pairing interactions with each other [61]. While prediction of tertiary structures can be challenging, prediction software is accurate when a large percentage of the queried RNA contain traditional Watson-Crick base pairing [61]. Because the structure of s2m has been verified by x-ray crystallography [37], and ∆s2m RNAs are based off the full-length s2m and have a high percentage of base pairing in the stem, we are confident in the predicted structure of these RNAs.
Collectively, our data suggests that RNA secondary structure plays two distinct roles in determining nsp15 cleavage specificity. First, thermodynamically stable structures likely sterically hinder nsp15-RNA interactions, thus preventing engagement and cleavage of some RNA sequences. Second, susceptible bases are presented in a structural context which facilitates optimal positioning of the scissile nucleotide in the catalytic pocket of nsp15 (e.g., extrusion of the scissile U25 in the GNRNA pentaloop). However, several questions remain unanswered, most notably, what are the biologically relevant targets (host and viral) of nsp15 cleavage during CoV infection and are viral targets conserved across different CoVs? Two studies to date have identified cleavage sites within the positive-sense genomic RNA and 3’ polyU tails of the negative-sense RNA of MHV [19, 22]. Despite this, no recognition motif or other regulatory element associated with nsp15 cleavage has been identified. While the in vitro assays performed here and in other studies provide a tractable system for dissecting the molecular mechanism of nsp15-RNA interactions and cleavage determinants, a recent computational model suggests that nsp15 is part of a larger replication-transcription complex (RTC) which contains other viral proteins and host factors. How nsp15 protein-protein interactions and RNA interactions with other replicase proteins (e.g., nsp12, nsp14, nsp16) impacts nsp15 cleavage of RNA remains unsolved. Indeed, recent models of the RTC suggest the hexameric structure of nsp15 provides the central structure and arrangement of a hexameric RTC [62]. In this model where nsp15 forms part of a multimeric RTC, it is possible there may be differences in its cleavage of viral RNAs due to its 3-D interaction with the nascent RNA as opposed to when nsp15 is free from the RTC as was the case in this study. The importance of examining nsp15-RNA interactions in their relevant biological context is further highlighted by recent studies showing that the requirement for Mn2+ and the structural conformation of nsp15 is highly dependent on the pH of the environment [63]. Given that the redox state of the viral RTC of +ssRNA viruses is altered during replication, the conditions under which nsp15 endoU activity is examined is highly relevant. To address this our future studies will explore the impact of pH and redox state on nsp15 structure and function in vitro.
Overall, our findings suggest a role for RNA secondary structure in regulation of nsp15 cleavage. Identification of bona fide nsp15 cleavage sites in SARS-CoV-2 RNA during replication and examination of how perturbation of these sites impacts the outcome of replication and pathogenesis will provide key insights into the broader roles for nsp15 endoU activity during infection (e.g., viral transcriptional/translational regulation, host translation). Furthermore, defining key structural features of RNAs which are resistant or susceptible to nsp15 cleavage may provide avenues for development of antiviral therapeutics.
Supporting information
Δs2mmut4 and Δs2mmut7 RNAs were denatured and folded as outlined in the materials and methods prior to being electrophoresed on a native gel. Denatured RNA was heated in 2x RNA Loading Dye (NEB) at 70°C for 10 minutes. Folded RNAs show the presence of conformers as indicated by arrows.
(TIF)
∆s2mmut7 RNA was incubated with wt nsp15, catalytically inactive nsp15 (K290A), or denatured wt or K290A nsp15. For denatured samples, nsp15 was denatured by heating recombinant protein for 5 minutes at 95°C prior to the addition of folded RNA. Endonuclease reactions were incubated for 15 minutes then terminated with the addition of EDTA to a final concentration of 20mM and flash frozen (see Materials and Methods).
(TIF)
A. Minimum free energy structures of RNAs 1, 2, and 3 as predicted by RNAfold. B. Published structures of RNA 1, 2, and 3 as resolved by SHAPE-MaP (Huston et al. Molecular Cell 2021). Nucleotides highlighted in blue match with SHAPE data while bases highlighted in purple and red do not. Purple nucleotides are paired with a different nucleotide whereas red nucleotides are unpaired in SHAPE structures.
(TIF)
A. Minimum free energy structures of SARS-CoV-1 and SARS-CoV-2 s2m. The U25 (pentaloop) and U30 (bulge) nucleotides are highlighted in blue. Two nucleotide changes between the RNAs in the stem (highlighted in red) change the MFE structure. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of s2m from each virus. Increased ∆G and ensemble diversity in SARS-CoV-2 s2m indicate that it does not maintain the highly conserved structure of SARS-CoV-1 s2m. C. Alignment of s2m from both viruses with relevant polymorphisms in SARS-CoV-2 s2m indicated by red arrows.
(TIF)
Endonuclease assays of full-length s2m mutant RNAs as shown in Fig 2. Full-length bands indicated at the top of the gel with cleavage products below. Selected cleavage products are numbered and indicated by arrows. Representative images from one experiment. B. Fold change in each cleavage product relative to the amount present at the one-minute time point. Cleavage products above the dashed lines indicate accumulating products while those below are diminishing over the course of the reaction. Data represents three independent experiments.
(TIF)
A. Endonuclease assays of additional ∆s2m mutant RNAs. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:6. RNAs were run on a 22.5% TBE-Urea Gel. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of ∆s2m mutant RNAs indicated as predicted by RNAfold. C. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). RNAs with uridines in the second position of the pentaloop (∆s2mmut6/7) were consistently cleaved more rapidly than ∆s2mmut2 in which the uridine is in the fourth position. P-values were calculated using one-way ANOVA with multiple comparisons analysis of the AUC. Data represents three independent experiments.
(TIF)
A. Minimum free energy structures of s2m-like and control RNAs. Representative images from endonuclease assays are indicated below each RNA. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:120. RNAs were run on a 22.5% TBE-Urea Gel. B. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. Data represents three independent experiments.
(TIF)
Time course of ∆s2mmut2, ∆s2mmut4, ∆s2mmut5, RNAs (from Fig 3) run on a single gel. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:6. RNAs were run on a 22.5% TBE-Urea Gel. Cleavage products run side by side show distinct banding patterns despite the presumed cleavage sites in each RNA being only one nucleotide apart indicating potential alternative cleavage sites.
(TIF)
(XLSX)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No.: 75N93022C00036. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. Epub 2020/02/06. doi: 10.1038/s41586-020-2012-7 ; PubMed Central PMCID: PMC7095418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J Clin Med. 2020;9(4). Epub 2020/04/30. doi: 10.3390/jcm9041225 ; PubMed Central PMCID: PMC7230578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635–64. Epub 2005/12/13. doi: 10.1128/MMBR.69.4.635-664.2005 ; PubMed Central PMCID: PMC1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. Epub 2019/03/08. doi: 10.1016/j.meegid.2019.03.001 ; PubMed Central PMCID: PMC7106202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricciardi S, Guarino AM, Giaquinto L, Polishchuk EV, Santoro M, Di Tullio G, et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 2022;606(7915):761–8. Epub 2022/05/14. doi: 10.1038/s41586-022-04835-6 ; PubMed Central PMCID: PMC7612910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oudshoorn D, Rijs K, Limpens R, Groen K, Koster AJ, Snijder EJ, et al. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio. 2017;8(6). Epub 2017/11/23. doi: 10.1128/mBio.01658-17 ; PubMed Central PMCID: PMC5698553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundin A, Dijkman R, Bergstrom T, Kann N, Adamiak B, Hannoun C, et al. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10(5):e1004166. Epub 2014/05/31. doi: 10.1371/journal.ppat.1004166 ; PubMed Central PMCID: PMC4038610 our adherence to all PLOS Pathogens policies on sharing data and materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4). Epub 2013/08/15. doi: 10.1128/mBio.00524-13 ; PubMed Central PMCID: PMC3747587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76(8):3697–708. Epub 2002/03/22. doi: 10.1128/jvi.76.8.3697-3708.2002 ; PubMed Central PMCID: PMC136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Meer Y, Snijder EJ, Dobbe JC, Schleich S, Denison MR, Spaan WJ, et al. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73(9):7641–57. Epub 1999/08/10. doi: 10.1128/JVI.73.9.7641-7657.1999 ; PubMed Central PMCID: PMC104292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi ST, Schiller JJ, Kanjanahaluethai A, Baker SC, Oh JW, Lai MM. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J Virol. 1999;73(7):5957–69. Epub 1999/06/11. doi: 10.1128/JVI.73.7.5957-5969.1999 ; PubMed Central PMCID: PMC112657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279(11):10136–41. Epub 2003/12/31. doi: 10.1074/jbc.M306124200 ; PubMed Central PMCID: PMC7957857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overby AK, Popov VL, Niedrig M, Weber F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J Virol. 2010;84(17):8470–83. Epub 2010/06/18. doi: 10.1128/JVI.00176-10 ; PubMed Central PMCID: PMC2919015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. Epub 2008/09/19. doi: 10.1371/journal.pbio.0060226 ; PubMed Central PMCID: PMC2535663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006;103(34):12885–90. Epub 2006/08/17. doi: 10.1073/pnas.0603144103 ; PubMed Central PMCID: PMC1568942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A. 2020;117(45):28344–54. Epub 2020/10/25. doi: 10.1073/pnas.2016650117 ; PubMed Central PMCID: PMC7668094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3(8):e109. Epub 2007/08/19. doi: 10.1371/journal.ppat.0030109 ; PubMed Central PMCID: PMC1941747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Hackbart M, Mettelman RC, O’Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114(21):E4251–E60. Epub 2017/05/10. doi: 10.1073/pnas.1618310114 ; PubMed Central PMCID: PMC5448190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117(14):8094–103. Epub 2020/03/22. doi: 10.1073/pnas.1921485117 ; PubMed Central PMCID: PMC7149396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volk A, Hackbart M, Deng X, Cruz-Pulido Y, O’Brien A, Baker SC. Coronavirus Endoribonuclease and Deubiquitinating Interferon Antagonists Differentially Modulate the Host Response during Replication in Macrophages. J Virol. 2020;94(11). Epub 2020/03/20. doi: 10.1128/JVI.00178-20 ; PubMed Central PMCID: PMC7269425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2):e1006195. Epub 2017/02/06. doi: 10.1371/journal.ppat.1006195 ; PubMed Central PMCID: PMC5310923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ancar R, Li Y, Kindler E, Cooper DA, Ransom M, Thiel V, et al. Physiologic RNA targets and refined sequence specificity of coronavirus EndoU. RNA. 2020;26(12):1976–99. Epub 2020/09/30. doi: 10.1261/rna.076604.120 ; PubMed Central PMCID: PMC7668261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361(2):243–56. Epub 2006/07/11. doi: 10.1016/j.jmb.2006.06.021 ; PubMed Central PMCID: PMC7118729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avis JM, Conn GL, Walker SC. Cis-acting ribozymes for the production of RNA in vitro transcripts with defined 5’ and 3’ ends. Methods Mol Biol. 2012;941:83–98. Epub 2012/10/16. doi: 10.1007/978-1-62703-113-4_7 . [DOI] [PubMed] [Google Scholar]
- 25.Choi R, Zhou M, Shek R, Wilson JW, Tillery L, Craig JK, et al. High-throughput screening of the ReFRAME, Pandemic Box, and COVID Box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLoS One. 2021;16(4):e0250019. Epub 2021/04/23. doi: 10.1371/journal.pone.0250019 ; PubMed Central PMCID: PMC8062000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel DK, Gebbie MP, Lee VT. Assessing RNA interactions with proteins by DRaCALA. Methods Enzymol. 2014;549:489–512. Epub 2014/11/30. doi: 10.1016/B978-0-12-801122-5.00021-0 . [DOI] [PubMed] [Google Scholar]
- 27.Orr MW, Lee VT. Differential Radial Capillary Action of Ligand Assay (DRaCALA) for High-Throughput Detection of Protein-Metabolite Interactions in Bacteria. Methods Mol Biol. 2017;1535:25–41. Epub 2016/12/04. doi: 10.1007/978-1-4939-6673-8_3 . [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43(W1):W39–49. Epub 2015/05/09. doi: 10.1093/nar/gkv416 ; PubMed Central PMCID: PMC4489269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. Rna. 2012;18(5):900–14. doi: 10.1261/rna.029041.111 WOS:000302909000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raden M, Ali SM, Alkhnbashi OS, Busch A, Costa F, Davis JA, et al. Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Research. 2018;46(W1):W25–W9. doi: 10.1093/nar/gky329 WOS:000438374100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–31. Epub 2003/06/26. doi: 10.1093/nar/gkg599 ; PubMed Central PMCID: PMC169005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavares RCA, Mahadeshwar G, Wan H, Huston NC, Pyle AM. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. J Virol. 2021;95(5). Epub 2020/12/04. doi: 10.1128/JVI.02190-20 ; PubMed Central PMCID: PMC8092842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews RJ, O’Leary CA, Tompkins VS, Peterson JM, Haniff HS, Williams C, et al. A map of the SARS-CoV-2 RNA structurome. NAR Genom Bioinform. 2021;3(2):lqab043. Epub 2021/05/29. doi: 10.1093/nargab/lqab043 ; PubMed Central PMCID: PMC8140738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–4. Epub 2008/04/22. doi: 10.1093/nar/gkn188 ; PubMed Central PMCID: PMC2447809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huston NC, Wan H, Strine MS, de Cesaris Araujo Tavares R, Wilen CB, Pyle AM. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol Cell. 2021;81(3):584–98 e5. Epub 2021/01/15. doi: 10.1016/j.molcel.2020.12.041 ; PubMed Central PMCID: PMC7775661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson PZ, Kasprzak WK, Shapiro BA, Simon AE. RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 2019;16(12):1667–71. Epub 2019/08/24. doi: 10.1080/15476286.2019.1659081 ; PubMed Central PMCID: PMC6844559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson MP, Igel H, Baertsch R, Haussler D, Ares M Jr., Scott WG . The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3(1):e5. Epub 2005/01/05. doi: 10.1371/journal.pbio.0030005 ; PubMed Central PMCID: PMC539059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tengs T, Kristoffersen AB, Bachvaroff TR, Jonassen CM. A mobile genetic element with unknown function found in distantly related viruses. Virol J. 2013;10:132. Epub 2013/04/27. doi: 10.1186/1743-422X-10-132 ; PubMed Central PMCID: PMC3653767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazier MN, Dillard LB, Krahn JM, Perera L, Williams JG, Wilson IM, et al. Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 2021;49(17):10136–49. Epub 2021/08/18. doi: 10.1093/nar/gkab719 ; PubMed Central PMCID: PMC8385992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhardwaj K, Palaninathan S, Alcantara JM, Yi LL, Guarino L, Sacchettini JC, et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283(6):3655–64. Epub 2007/11/30. doi: 10.1074/jbc.M708375200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci U S A. 2006;103(32):11892–7. Epub 2006/08/03. doi: 10.1073/pnas.0601708103 ; PubMed Central PMCID: PMC2131687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X, Zhai Y, Sun F, Lou Z, Su D, Xu Y, et al. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol. 2006;80(16):7909–17. Epub 2006/07/29. doi: 10.1128/JVI.00525-06 ; PubMed Central PMCID: PMC1563835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazier MN, Wilson IM, Krahn JM, Butay KJ, Dillard LB, Borgnia MJ, et al. Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease. Nucleic Acids Res. 2022. Epub 2022/07/09. doi: 10.1093/nar/gkac589 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scharpf M, Sticht H, Schweimer K, Boehm M, Hoffmann S, Rosch P. Antitermination in bacteriophage lambda. The structure of the N36 peptide-boxB RNA complex. Eur J Biochem. 2000;267(8):2397–408. Epub 2000/04/12. doi: 10.1046/j.1432-1327.2000.01251.x . [DOI] [PubMed] [Google Scholar]
- 45.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93(2):289–99. Epub 1998/05/06. doi: 10.1016/s0092-8674(00)81579-2 . [DOI] [PubMed] [Google Scholar]
- 46.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, et al. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol. 2007;81(12):6700–8. Epub 2007/04/06. doi: 10.1128/JVI.02817-06 ; PubMed Central PMCID: PMC1900129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Li L, Yan L, Ming Z, Jia Z, Lou Z, et al. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. J Virol. 2018;92(22). Epub 2018/08/24. doi: 10.1128/JVI.00893-18 ; PubMed Central PMCID: PMC6206473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng A, Shi Y, Shen Z, Wang G, Shi J, Xiong Q, et al. Insight into the evolution of nidovirus endoribonuclease based on the finding that nsp15 from porcine Deltacoronavirus functions as a dimer. J Biol Chem. 2018;293(31):12054–67. Epub 2018/06/12. doi: 10.1074/jbc.RA118.003756 ; PubMed Central PMCID: PMC6078464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–605. Epub 2020/04/19. doi: 10.1002/pro.3873 ; PubMed Central PMCID: PMC7264519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pillon MC, Frazier MN, Dillard LB, Williams JG, Kocaman S, Krahn JM, et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun. 2021;12(1):636. Epub 2021/01/29. doi: 10.1038/s41467-020-20608-z ; PubMed Central PMCID: PMC7840905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol. 2005;353(5):1106–17. Epub 2005/10/12. doi: 10.1016/j.jmb.2005.09.007 ; PubMed Central PMCID: PMC7094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Sun L, Zhao Y, Feng D, Cheng J, Zhang G. Coronavirus Endoribonuclease Ensures Efficient Viral Replication and Prevents Protein Kinase R Activation. J Virol. 2020. Epub 2020/12/29. doi: 10.1128/JVI.02103-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng X, Buckley AC, Pillatzki A, Lager KM, Faaberg KS, Baker SC. Inactivating Three Interferon Antagonists Attenuates Pathogenesis of an Enteric Coronavirus. J Virol. 2020;94(17). Epub 2020/06/20. doi: 10.1128/JVI.00565-20 ; PubMed Central PMCID: PMC7431798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Fang P, Fang L, Hong Y, Zhu X, Wang D, et al. Porcine deltacoronavirus nsp15 antagonizes interferon-beta production independently of its endoribonuclease activity. Mol Immunol. 2019;114:100–7. Epub 2019/07/28. doi: 10.1016/j.molimm.2019.07.003 ; PubMed Central PMCID: PMC7112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol. 2019;234(3):2143–51. Epub 2018/08/28. doi: 10.1002/jcp.27155 ; PubMed Central PMCID: PMC7166610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei Y, Moore CB, Liesman RM, O’Connor BP, Bergstralh DT, Chen ZJ, et al. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4(5):e5466. Epub 2009/05/01. doi: 10.1371/journal.pone.0005466 ; PubMed Central PMCID: PMC2674933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhardwaj K, Liu P, Leibowitz JL, Kao CC. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J Virol. 2012;86(8):4294–304. Epub 2012/02/04. doi: 10.1128/JVI.07012-11 ; PubMed Central PMCID: PMC3318636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Athmer J, Fehr AR, Grunewald ME, Qu W, Wheeler DL, Graepel KW, et al. Selective Packaging in Murine Coronavirus Promotes Virulence by Limiting Type I Interferon Responses. mBio. 2018;9(3). Epub 2018/05/03. doi: 10.1128/mBio.00272-18 ; PubMed Central PMCID: PMC5930304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Y, Zhang H, Shi Z, Chen J, Li M, Shi H, et al. Porcine Epidemic Diarrhea Virus nsp15 Antagonizes Interferon Signaling by RNA Degradation of TBK1 and IRF3. Viruses. 2020;12(6). Epub 2020/06/04. doi: 10.3390/v12060599 ; PubMed Central PMCID: PMC7354440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stefl R, Allain FH. A novel RNA pentaloop fold involved in targeting ADAR2. RNA. 2005;11(5):592–7. Epub 2005/04/21. doi: 10.1261/rna.7276805 ; PubMed Central PMCID: PMC1370747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vicens Q, Kieft JS. Thoughts on how to think (and talk) about RNA structure. Proc Natl Acad Sci U S A. 2022;119(17):e2112677119. Epub 2022/04/20. doi: 10.1073/pnas.2112677119 ; PubMed Central PMCID: PMC9169933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry JK, Appleby TC, Bilello JP, Feng JY, Schmitz U, Campbell EA. An atomistic model of the coronavirus replication-transcription complex as a hexamer assembled around nsp15. J Biol Chem. 2021;297(4):101218. Epub 2021/09/26. doi: 10.1016/j.jbc.2021.101218 ; PubMed Central PMCID: PMC8494237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Godoy AS, Nakamura AM, Douangamath A, Song Y, Noske GD, Gawriljuk VO, et al. Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease. Nucleic Acids Res. 2023. Epub 2023/04/28. doi: 10.1093/nar/gkad314 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Δs2mmut4 and Δs2mmut7 RNAs were denatured and folded as outlined in the materials and methods prior to being electrophoresed on a native gel. Denatured RNA was heated in 2x RNA Loading Dye (NEB) at 70°C for 10 minutes. Folded RNAs show the presence of conformers as indicated by arrows.
(TIF)
∆s2mmut7 RNA was incubated with wt nsp15, catalytically inactive nsp15 (K290A), or denatured wt or K290A nsp15. For denatured samples, nsp15 was denatured by heating recombinant protein for 5 minutes at 95°C prior to the addition of folded RNA. Endonuclease reactions were incubated for 15 minutes then terminated with the addition of EDTA to a final concentration of 20mM and flash frozen (see Materials and Methods).
(TIF)
A. Minimum free energy structures of RNAs 1, 2, and 3 as predicted by RNAfold. B. Published structures of RNA 1, 2, and 3 as resolved by SHAPE-MaP (Huston et al. Molecular Cell 2021). Nucleotides highlighted in blue match with SHAPE data while bases highlighted in purple and red do not. Purple nucleotides are paired with a different nucleotide whereas red nucleotides are unpaired in SHAPE structures.
(TIF)
A. Minimum free energy structures of SARS-CoV-1 and SARS-CoV-2 s2m. The U25 (pentaloop) and U30 (bulge) nucleotides are highlighted in blue. Two nucleotide changes between the RNAs in the stem (highlighted in red) change the MFE structure. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of s2m from each virus. Increased ∆G and ensemble diversity in SARS-CoV-2 s2m indicate that it does not maintain the highly conserved structure of SARS-CoV-1 s2m. C. Alignment of s2m from both viruses with relevant polymorphisms in SARS-CoV-2 s2m indicated by red arrows.
(TIF)
Endonuclease assays of full-length s2m mutant RNAs as shown in Fig 2. Full-length bands indicated at the top of the gel with cleavage products below. Selected cleavage products are numbered and indicated by arrows. Representative images from one experiment. B. Fold change in each cleavage product relative to the amount present at the one-minute time point. Cleavage products above the dashed lines indicate accumulating products while those below are diminishing over the course of the reaction. Data represents three independent experiments.
(TIF)
A. Endonuclease assays of additional ∆s2m mutant RNAs. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:6. RNAs were run on a 22.5% TBE-Urea Gel. B. Thermodynamic stability, frequency of MFE, and ensemble diversity of ∆s2m mutant RNAs indicated as predicted by RNAfold. C. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). RNAs with uridines in the second position of the pentaloop (∆s2mmut6/7) were consistently cleaved more rapidly than ∆s2mmut2 in which the uridine is in the fourth position. P-values were calculated using one-way ANOVA with multiple comparisons analysis of the AUC. Data represents three independent experiments.
(TIF)
A. Minimum free energy structures of s2m-like and control RNAs. Representative images from endonuclease assays are indicated below each RNA. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:120. RNAs were run on a 22.5% TBE-Urea Gel. B. Percent of full-length RNA remaining as measured by densitometry. Percentage of uncut RNA was calculated by normalizing to a denatured nsp15 control (indicated by (-) in the far-right lane). AUC was calculated for each RNA, and one-way ANOVA with multiple comparisons was performed on the AUC. Data represents three independent experiments.
(TIF)
Time course of ∆s2mmut2, ∆s2mmut4, ∆s2mmut5, RNAs (from Fig 3) run on a single gel. Reactions were carried out at a nsp15 hexamer:RNA ratio of 1:6. RNAs were run on a 22.5% TBE-Urea Gel. Cleavage products run side by side show distinct banding patterns despite the presumed cleavage sites in each RNA being only one nucleotide apart indicating potential alternative cleavage sites.
(TIF)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.