Abstract
Upon transient expression in cell culture, the ie-2 gene of Autographa californica nuclear polyhedrosis virus (AcMNPV) displays three functions: trans activation of viral promoters, direct or indirect stimulation of virus origin-specific DNA replication, and arrest of the cell cycle. The ability of IE2 to trans stimulate DNA replication and coupled late gene expression is observed in a cell line derived from Spodoptera frugiperda but not in a cell line derived from Trichoplusia ni. This finding suggested that IE-2 may exert cell line-specific or host-specific effects. To examine the role of ie-2 in the context of infection and its possible influence on the host range, we constructed recombinants of AcMNPV containing deletions of different functional regions within ie-2 and characterized them in cell lines and larvae of S. frugiperda and T. ni. The ie-2 mutant viruses exhibited delays in viral DNA synthesis, late gene expression, budded virus production, and occlusion body formation in SF-21 cells but not in TN-5B1-4 cells. In TN-5B1-4 cells, the ie-2 mutants produced more budded virus and fewer occlusion bodies but the infection proceeded without delay. Examination of the effects of ie-2 and the respective mutants on immediate-early viral promoters in transient expression assays revealed striking differences in the relative levels of expression and differences in responses to ie-2 and its mutant forms in different cell lines. In T. ni and S. frugiperda larvae, the infectivities of the occluded form of ie-2 mutant viruses by the normal oral route of infection was 100- and 1,000-fold lower, respectively, than that of wild-type AcMNPV. The reduction in oral infectivity was traced to the absence of virions within the occlusion bodies. The infectivity of the budded form of ie-2 mutants by hemocoelic injection was similar to that of wild-type virus in both species. Thus, ie-2 mutants are viable but exhibit cell line-specific effects on temporal regulation of the infection process. Due to its effect on virion occlusion, mutants of IE-2 were essentially noninfectious by the normal route of infection in both species tested. However, since budded viruses exhibited normal infectivity upon hemocoelic injection, we conclude that ie-2 does not affect host range per se. The possibility that IE-2 exerts tissue-specific effects has not been ruled out.
Four genes of the baculovirus Autographa californica nuclear polyhedrosis virus (AcMNPV) are known to trans activate early baculovirus promoters in transient expression assays: ie-0 (5, 17), ie-1 (11, 12), ie-2 (3, 4), and pe-38 (18, 20). IE-1, the product of ie-1, is the most thoroughly characterized trans regulator of AcMNPV and is thought to play a central role in the regulation of early gene expression and possibly DNA replication (8). IE-1 is required for AcMNPV replication (32), and in transient expression assays, it strongly stimulates transcription from several early viral promoters, including the promoters of the 39K and p35 genes (11, 12, 25). The effect of IE-1 is strongest when a homologous repeat (hr) sequence, comprised of repeated imperfect palindromic sequences to which IE-1 binds, is cis linked to the early promoter (11–13, 25, 33). IE-0 is identical to IE-1 except that it has an additional 54 amino acids at its N terminus, since it is derived from a spliced transcript which initiates from a separate promoter approximately 4 kb upstream of the ie-1 promoter (5). IE-2 was originally identified as augmenting IE-1 activation of the 39K gene promoter (3) and was subsequently found to activate transcription from the ie-1 promoter approximately 2.5-fold in transient expression assays, although it does not appear to be able to bind DNA directly (38, 39). PE-38 stimulates expression from the early p143 gene promoter (20).
In addition to its ability to trans activate expression from the ie-1 promoter in SF-21 cells, IE-2 exhibits two additional activities in transient assays. (i) IE-2 blocks the progression of the cell cycle in a variety of cell lines, including those derived from Spodoptera frugiperda and Trichoplusia ni (30). The cycle appears to arrest in late S phase since transfected cells accumulate a greater than 4N complement of DNA with no evidence of mitotic spindle formation. This activity of IE-2 requires a functional RING finger motif, whereas its ability to trans activate the ie-1 promoter is not affected by alterations within the RING finger motif. (ii) IE-2, in the presence of IE-1 and six other AcMNPV genes, augments the replication and stability of reporter plasmids containing hr sequences (16, 21) and also augments expression from cis-linked late promoters in transient expression assays (27). It is possible that the role of IE-2 in these assays is simply to stimulate ie-1 expression or expression from other early genes required for plasmid replication or stability. The stimulatory effect of IE-2 on hr-containing plasmid DNA replication or stability and coupled late gene expression is observed in SF-21 cells, a cell line derived from the fall armyworm S. frugiperda, but not in TN-368 cells or in TN-5B1-4 cells (22), derived from the cabbage looper T. ni.
Mutants of AcMNPV defective in ie-2 have not been previously described, but the cell line-specific effects of ie-2 on DNA replication and late gene expression prompted us to attempt to construct viruses with ie-2 mutations in order to define the function of IE-2 and its possible role in host range. We have previously described the effects of two site-specific deletion mutations on the properties of ie-2 in transient expression assays (30). These mutations selectively affect either ie-1 trans activation or cell cycle arrest. In this study, we show that AcMNPV ie-2 mutants are viable and describe the properties of viruses containing mutations which lack the RING finger required for cell cycle arrest and/or a region required for transcriptional activation.
MATERIALS AND METHODS
Cell lines and insects.
S. frugiperda IPBL-SF-21 (SF-21) (35) and T. ni BTI-TN-5B1-4 (TN-5B1-4) (36) and TN-368 (14) cells were cultured at 27°C in TC 100 medium (GIBCO, Gaithersburg, Md.) supplemented with 10% fetal bovine serum and 0.26% tryptose broth, as described previously (26). S. frugiperda and T. ni eggs were provided by W. Deryck Perkins (Agricultural Research Service, U.S. Department of Agriculture, Tifton, Ga.) and Mark Harmon (Abbott Laboratories, Chicago, Ill.), respectively. Larvae were reared in individual cups of artificial diet (25) at 27°C under a 14 h–10 h light-dark cycle.
Reporter plasmids and plasmid constructs.
Reporter plasmids phcIE1, phcIE2, phcIE0, and pCAPCAT, containing the chloramphenicol acetyltransferase (CAT) gene under the transcriptional control of the AcMNPV ie-1, ie-2, ie-0, and late vp39 promoters, respectively, have been described previously (24). Plasmid pBs-PstN contains the AcMNPV PstI N fragment from 97.0 to 98.9 map units (27). Plasmid pBs-PstNfs contains a frameshift mutation of ie-2 within pBs-PstN (30).
To generate ie-2 mutant viruses containing deletions of different regions of ie-2, we constructed a plasmid, pBs-PstNlacZ, that has an insertion of the Escherichia coli lacZ gene under control of the Drosophila melanogaster hsp70 promoter within ie-2. To construct pBs-PstNlacZ, the BamHI fragment containing the E. coli lacZ gene under hsp70 promoter control from pHSP70lacZ (24) was ligated into the BglII site located in 148 to 150 codons within the ie-2 coding sequences in pBs-PstN (27). The resulting plasmid, pBs-PstNlacZ, contains lacZ inserted into the BglII site of ie-2. Plasmids pBs-PstNd(94-173) and pBs-PstNd(215-274) with in-frame deletions in ie-2 were described previously (30). To construct pBs-PstNd(94-274), plasmid pBs-PstN was digested with a high concentration of HpaI to digest HpaI sites and an HpaI-like site (HpaI* [30]) and then religated. The expected in-frame deletion of the 543 bp HpaI-HpaI* fragment in pBs-PstNd(94-274) was confirmed by sequence analysis.
Transfection, CAT assays, and DNA replication assays.
Plasmid DNA was introduced into SF-21, TN-368, or TN-5B1-4 cells (106 cells per 60-mm-diameter dish) by lipofectin-mediated transfection (26). At 24 h posttransfection, 2.0 × 106 cells of different cell cultures were pelleted by centrifugation at 6,000 × g for 2 min, and cell extracts were prepared as described previously (10, 31). The same amount of each cell extract was used to determine CAT activity.
Construction of ie-2 mutant viruses.
The L1 strain of wild-type (wt) AcMNPV (19) and ie-2 mutant viruses vie2Z, vie2d(94-274), vie2d(94-173), and vie2d(215-274) were propagated and titers were determined in TN-5B1-4 cells.
vie2Z, containing the lacZ gene within ie-2 coding sequences, was constructed by lipofectin-mediated transfection of pBs-PstNlacZ with wt AcMNPV DNA in TN-5B1-4 cells. Supernatant fluids were harvested at 96 h posttransfection and screened for viruses having a blue plaque phenotype in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Three independent isolates of vie2Z were obtained, and all exhibited a defect in occlusion body (OB) production. A single isolate was chosen for further work. A marker-rescued revertant of this vie2Z mutant was isolated and was found to have a phenotype similar, if not identical, to that of wt AcMNPV. The ie-2 mutant viruses vie2d(94-274), vie2d(94-173), and vie2d(215-274) were generated by cotransfection of pBs-PstNd(94-274), pBs-PstNd(94-173), or pBs-PstNd(215-274) DNA with vie2Z DNA and screened as viruses which formed white plaques in the presence of X-Gal. All recombinant viruses were plaque purified four times on TN-5B1-4 cells. The structures of vie2d(94-274), vie2d(94-173), and vie2d(215-274) were confirmed by restriction enzyme analysis of the viral DNA as well as by PCR analysis.
Metabolic labeling and protein analysis.
Monolayers of SF-21 or TN-5B1-4 cells (106 cells) were infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274) at a multiplicity of infection (MOI) of 20 PFU/cell. After 1.5 h, inocula were replaced with complete TC 100 medium. Two hours before designated time points, TC 100 medium was exchanged for TC 100 medium deficient in methionine and cysteine (incomplete TC 100 medium). After 1 h of incubation, 25 μCi of Trans 35S label (New England Nuclear, Boston, Mass.) per plate was added. Cells were labeled for 1 h, washed twice with TC 100 medium and resuspended in 100 μl of sodium dodecyl sulfate (SDS) loading buffer as previously described (26). Labeled proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and the radioisotope distribution was detected by fluorography.
Immunoblot analysis.
SF-21 or TN-5B1-4 cells (106 cells) were infected with either wt AcMNPV or ie-2 mutant viruses at an MOI of 20 PFU/cell. At different times postinfection (p.i.), cells were harvested and lysed in SDS loading buffer as previously described (26). Proteins from lysates of the infected cells were separated on SDS–12% polyacrylamide gels and transferred to nylon membranes (Amersham). To detect major capsid protein VP39, rabbit polyclonal anti-VP39 antiserum was diluted 1:8,000 in Tris-buffered saline (pH 7.6). Polyhedrin antiserum was diluted 1:20,000. In both cases, mouse anti-rabbit antibody conjugated with horseradish peroxidase (Promega) was used as a secondary antibody at a 1:25,000 dilution. Immunoblots were visualized by the Amersham enhanced chemiluminescence system.
OBs used in Western immunoblot analysis were purified from wt- and ie-2 mutant-infected T. ni larvae by isopycnic centrifugation on 40 to 65% sucrose step gradient as described previously (26). A total of 3 × 108 OBs were resuspended in 100 μl of 2 × SDS loading buffer and analyzed as described above.
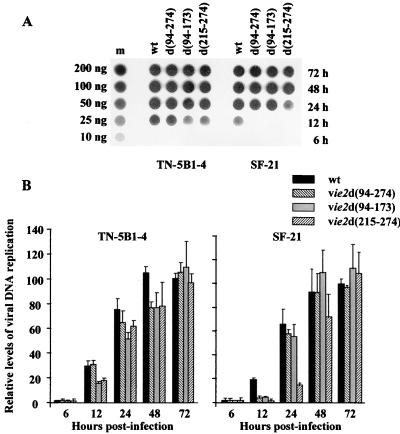
Dot blot analysis of viral DNA replication.
SF-21 or TN-5B1-4 cells were infected with wt AcMNPV or ie-2 mutant virus at an MOI of 20 PFU/cell. At different times p.i., cells were pelleted by centrifugation at 10,000 × g for 10 min. The cell pellet was resuspended in 500 μl of 0.4 M NaOH–10 mM EDTA solution, incubated at 100°C for 10 min, and blotted onto a nylon membrane using a dot blot apparatus with a vacuum as described previously (23). Samples were hybridized to radiolabeled AcMNPV DNA. The blot was visualized by autoradiography, and the bound probe was quantified with a PhosphorImager 4000 (Molecular Dynamics, Sunnyvale, Calif.).
Characterization of BV production.
SF-21 or TN-5B1-4 cells (2.0 × 106/60-mm-diameter dish) were infected with either wt AcMNPV or ie-2 mutant virus at an MOI of 20 PFU/cell. After 1.5 h of incubation, viral inoculum was removed. Infected cells were washed twice with TC 100 medium and 4 ml of TC 100 medium was added (0 h p.i.). Culture medium (500 μl) was harvested at the designated times. Budded virus (BV) production was determined by plaque assay (26) on TN-5B1-4 cells.
Larval bioassays.
OBs were prepared by infection of fifth-instar T. ni larvae by hemocoelic injection with 2 × 105 PFU of BV, as determined by plaque assay in TN-5B1-4 cells, from wt AcMNPV or ie-2 mutant virus. The LC50s (concentrations of OBs required to reach 50% larval mortality) of wt AcMNPV and ie-2 mutant viruses were determined in neonates of S. frugiperda or T. ni larvae as previously described (34). T. ni or S. frugiperda neonates were infected with wt AcMNPV by feeding on diet containing 0, 104, 2.5 × 104, 5.0 × 104, and 105 OBs/ml and 0, 5.0 × 104, 2.0 × 105, 5.0 × 105, and 106 OBs/ml, respectively. To obtain the LC50 for ie-2 mutant viruses, T. ni or S. frugiperda neonates were fed diet containing vie2d(94-274), vie2d(94-173), or vie2d(215-274) in concentrations ranging from 3 × 105 to 5.0 × 107 OBs/ml for T. ni and from 2.0 × 107 to 1.9 × 109 OBs/ml for S. frugiperda neonates. The LC50 was quantified with the aid of probit analysis.
The 50% lethal dose (LD50) of BV was determined with larvae in their penultimate larval instar (fourth instar for T. ni and fifth instar for S. frugiperda) by hemocoelic injection with different doses of BV (produced and titered in TN-5B1-4 cells) from either wt AcMNPV or ie-2 mutants diluted in incomplete TC 100 medium. Thirty insects per dose were tested. Thirty insects injected with TC 100 medium alone were used as a control. Total mortality was recorded when all larvae were dead or had pupated.
RESULTS
Construction of ie-2 mutant viruses.
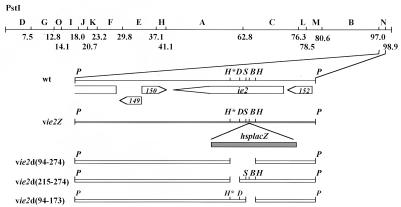
To examine the role of the AcMNPV ie-2 gene during viral infection, we constructed ie-2 mutant viruses with deletions of different functional regions of IE-2. Since ie-2 is not essential for transient trans activation of the late vp39 promoter in TN-368 cells (22), all ie-2 mutant viruses were constructed by using a cell line derived from T. ni cells. The first of these recombinant viruses, vie2Z, contained an insertion of the E. coli lacZ gene within the ie-2 coding sequences (Fig. 1). The mutant, vie2Z, formed blue occlusion-positive plaques on both TN-368 and TN-5B1-4 cells and was used to construct the mutant viruses vie2d(94-173), vie2d(215-274), and vie2d(94-274). Mutant vie2d(215-274) has an in-frame deletion within ie-2 resulting in the deletion of amino acids 215 to 274 including the majority of the RING finger motif of IE-2. In vie2d(94-173), amino acids 94 to 173 encompassing the region required for trans-regulatory activity of IE-2 were deleted in frame. Mutant vie2d(94-274) contains an in-frame deletion of amino acids 94 to 274 including both the RING finger motif and transcriptional activation region (Fig. 1).
FIG. 1.
Locations and nature of ie-2 mutants of AcMNPV. A linear representation of the PstI restriction map of the AcMNPV genome is at the top, with map units indicated below the line. Positions and orientations of the open reading frames in the PstI N fragment from 97.0 to 98.9 map units (1) are indicated by open arrows. The location of the lacZ gene under hsp70 promoter control in vIE2Z and deletions of the HpaI-HpaI*, DraI-HpaI*, and HpaI-SnaBI fragments in vie2d(94-274), vie2d(215-274), and vie2d(94-173), respectively, are shown. Abbreviations for restriction sites: P, PstI; H, HpaI; H*, HpaI*; D, DraI; S, SnaBI; B, BglII.
OB production in ie-2 mutant-infected cells.
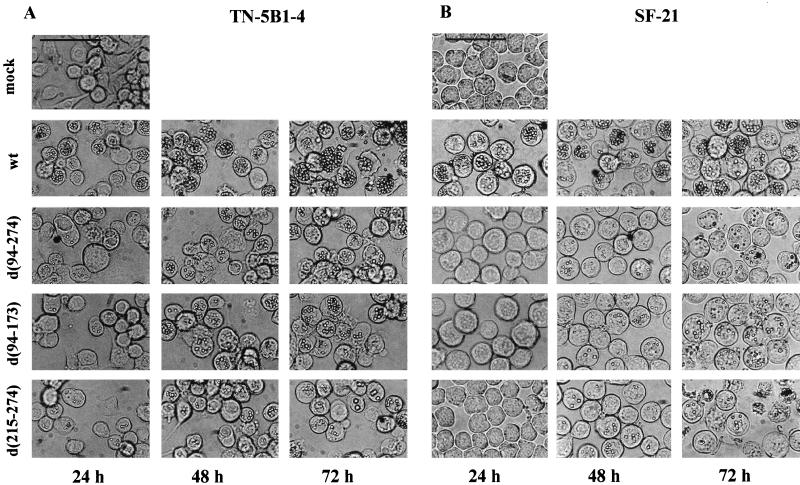
In TN-5B1-4 cells, all of the ie-2 mutants produced fewer OBs than wt AcMNPV throughout the very late phase of infection; even at 72 h p.i., fewer OBs were present in nuclei of cells infected with vie2Z (data not shown), vie2d(94-274), vie2d(94-173), and vie2d(215-274) than in wt AcMNPV-infected cells (Fig. 2A). vie2d(215-274) was the most severely impaired of the four ie-2 mutants tested, with very few OBs produced per cell even at 72 h p.i. However, the temporal regulation of infection appeared to be unaltered, as the initial appearance of cytopathic effects and OB production was similar to that for wt virus in ie-2 mutant-infected TN-5B1-4 cells.
FIG. 2.
Effects of ie-2 deletion mutants on OB production in SF-21 and TN-5B1-4 cells evidenced in light micrographs of TN-5B1-4 (A) and SF-21 (B) cells infected with wt, vie2d(94-274), vie2d(94-173), or vie2d(215-274). Panels on the left, middle, and right correspond to 24, 48, and 72 h p.i., respectively. Mock-infected SF-21 and TN-5B1-4 cells are shown in the upper left panels. Bar, 50 μm.
Mutants of ie-2 exhibited a more severe phenotype in SF-21 cells which included a striking delay in the infection process (Fig. 2B). At 24 h p.i., most wt-infected cells contained OBs (Fig. 2B) but the majority of cells infected with ie-2 mutants contained no OBs. Cells infected with vie2d(94-274) or vie2d(94-173) showed cytopathic effects including the rounding and enlargement of cells at this time (Fig. 2B). SF-21 cells infected with vie2d(215-274), a mutant lacking only the RING finger motif of IE-2, appeared to be uninfected and were similar in appearance to mock-infected cells at 24 h p.i. (Fig. 2B). This mutant also caused the premature disintegration of the cells. At 72 h p.i., only 50% of vie2d(215-274)-infected cells had intact cellular membranes compared to wt-infected cells or cells infected with other ie-2 mutant viruses (Fig. 2B). Thus, the vie2d(94-274) mutant in which both the RING finger motif and transcription activation region were deleted seemed to have a less severe defect than vie2d(215-274) lacking only the RING finger. Since we have not isolated and characterized a revertant of vie2d(215-274), we cannot eliminate the possibility that this more severe defect is due to a second mutation elsewhere in the genome.
By 48 and 72 h p.i., a large proportion of SF-21 cells infected with vie2Z (data not shown), vie2d(94-274), vie2d(94-173), or vie2d(215-274) contained OBs (Fig. 2B). However, cells infected with ie-2 mutants had significantly fewer OBs than did wt-infected cells. Thus, mutations of the ie-2 gene resulted in decreased numbers of OBs in both SF-21 and TN-5B1-4 cell lines and caused a delay in the onset of OB formation in SF-21 cells but not in TN-5B1-4 cells.
Temporal expression of viral polypeptides in SF-21 and TN-5B1-4 cells infected with wt AcMNPV and ie-2 mutant viruses.
We next studied the kinetics of protein synthesis in both SF-21 and TN-5B1-4 cells infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274) (Fig. 3). In SF-21 cells at 12 h p.i., the rate of synthesis of several virus-induced proteins was lower in mutant- than in wt-infected cells (Fig. 3A). This probably reflects differences in the rate of onset of the late phase of infection but may also reflect differences in the overall regulation of early and late gene expression. The shutoff of host protein synthesis occurred between 12 and 24 h p.i. for all ie-2 mutants as well as wt AcMNPV (Fig. 3A). Polyhedrin synthesis was observed in SF-21 cells infected with wt virus at 24 h p.i. but not in cells infected with vie2d(94-274), vie2d(94-173), or vie2d(215-274), suggesting a delay in the onset of the very late phase of infection as well (Fig. 3A). Polyhedrin synthesis was observed in ie-2 mutant-infected cells at 48 h p.i., but at a rate lower than that in wt-infected cells.
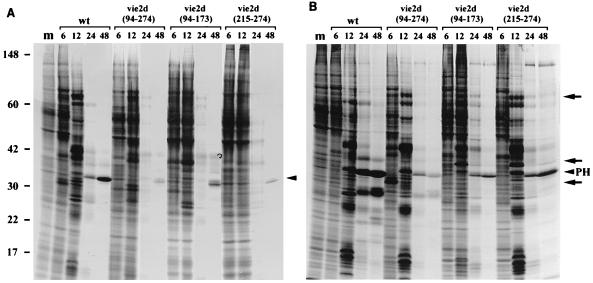
FIG. 3.
Kinetics of protein synthesis in SF-21 and TN-5B1-4 cells infected with wt AcMNPV or ie-2 mutant virus. TN-5B1-4 and SF-21 cells were infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274). Cells were pulse-labeled with a mixture of [35S]methionine and [35S]cysteine at the time indicated above each lane, harvested, and lysed. Total cellular proteins were resolved by SDS-PAGE using 12% acrylamide and visualized by autoradiography. Mock-infected cells (lanes m) were treated in parallel as a control. The sizes (in kilodaltons) and positions of the protein standards are indicated on the left. The position of polyhedrin (PH) in the gels is indicated on the right. Arrows point to the proteins of 75, 37, and 33 kDa which are differentially expressed in wt- or ie-2 mutant-infected cells.
In TN-5B1-4 cells, infection proceeded more rapidly than in SF-21 cells, and polyhedrin synthesis was detected in both wt- and mutant-infected cells at 24 h p.i. (Fig. 3B). However, the overall rate of polyhedrin synthesized at 24 and 48 h p.i. in ie-2 mutant-infected cells was lower than in wt-infected TN-5B1-4 cells (Fig. 3B). Thus, there was no evidence of a delay in the onset of the very late phase of infection in mutant-infected TN-5B1-4 cells, but there was a defect in the level at which polyhedrin synthesis was sustained.
Analysis of the protein profiles in TN-5B1-4 cells infected with ie-2 mutant viruses or with wt AcMNPV showed differences in the rate of synthesis of three other proteins. A 75-kDa protein was synthesized at 12 and 24 h p.i. in cells infected with ie-2 mutants, vie2d(94-274), vie2d(94-173), and vie2d(215-274) but not in wt-infected cells (Fig. 3B). The synthesis of this protein may be normally down-regulated by IE2 in this cell line. A 33-kDa protein was rapidly synthesized in vie2d(94-274)-infected cells, but not in cells infected with wt AcMNPV and other ie-2 mutant viruses, at 6 h p.i., whereas a 37-kDa polypeptide was synthesized in cells infected with vie2d(94-173) and vie2d(215-274) but not in wt-infected cells or in cells infected with vie2d(94-274). These proteins may be products of the ie-2 gene itself, although they migrate faster than expected for the sizes of the products predicted for the mutant ie-2 genes.
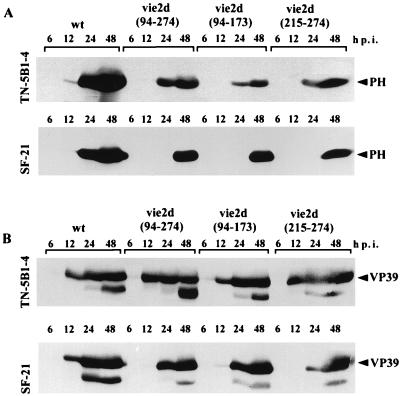
For both SF-21 and TN-5B1-4 cells, reduced steady-state levels of polyhedrin were demonstrated by Western immunoblot analysis (Fig. 4A). In TN-5B1-4 cells infected with wt AcMNPV, polyhedrin could be detected at 12 h p.i.; by 48 h p.i., abundant levels of this protein were observed (Fig. 4A, upper panel). In ie-2 mutant-infected cells, polyhedrin was observed at 24 h p.i. and continued to accumulate through 48 h p.i. but at lower than wt levels. In SF-21 cells, a 24-h delay in polyhedrin production was observed in mutant-infected cells (Fig. 4A, bottom panel). This correlates with the observed delay in OB appearance in mutant-infected SF-21 cells.
FIG. 4.
Western blot analysis of the levels of polyhedrin and VP39 major capsid protein in SF-21 and TN-5B1-4 cells infected with wt AcMNPV or ie-2 mutant virus. (A) Cell lysates from SF-21 and TN-5B1-4 cells infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274) were prepared at indicated times p.i., subjected to Western blot analysis, and probed with polyhedrin (PH) immune serum. (B) The blots in panel A were stripped and reprobed with VP39 immune serum.
To further assess the effect of ie-2 on late viral protein synthesis, the same blots used to detect polyhedrin were stripped and then reprobed with an antibody raised to the major viral capsid protein, VP39 (Fig. 4B). No differences in amount or temporal expression pattern of VP39 were observed in TN-5B1-4 cells infected with wt AcMNPV or with ie-2 mutants, confirming that mutations of ie-2 do not affect the regulation of early and late phases of expression in these cells (Fig. 4B, upper panel). In SF-21 cells infected with wt AcMNPV, VP39 was detected at 12 h p.i., and stable levels of this protein were maintained through 48 h p.i. (Fig. 4B, bottom panel). The production of VP39 was delayed in SF-21 cells infected with ie-2 mutants. No detectable levels of VP39 were observed in vie2d(94-274)-, vie2d(94-173)-, or vie2d(215-274)-infected cells at 12 h p.i., but by 24 h p.i., VP39 was expressed in all mutant-infected cells. At 24 h p.i., less VP39 was observed in vie2d(215-274)-infected cells than in cells infected with vie2d(94-274) and vie2d(94-173), consistent with the more severe phenotype of this mutant. Similar levels of VP39 production were observed in SF-21 cells infected with wt AcMNPV or with ie-2 mutants by 48 h p.i. Collectively, the results show that mutations of ie-2 cause a delay in late protein synthesis in SF-21 cells.
Effects of ie-2 mutations on viral DNA replication.
To determine if the delay in late protein synthesis in ie-2 mutant-infected SF-21 cells is correlated with a delay in viral DNA synthesis, we examined viral DNA replication in both SF-21 and TN-5B1-4 cell lines infected with wt or ie-2 mutant viruses. The level of viral DNA was determined during infection by dot blot analysis using AcMNPV genomic DNA as a probe (Fig. 5). In TN-5B1-4 cells, the levels of viral DNA were similar (not more than twofold different) for wt AcMNPV and all three ie-2 mutants throughout infection (Fig. 5A, left panels; Fig. 5B). In contrast, viral DNA synthesis was delayed approximately 6 to 12 h in SF-21 cells infected with vie2d(94-274) and vie2d(94-173) compared to wt-infected cells (Fig. 5A, right panels; Fig. 5B). By 24 h p.i., similar levels of DNA synthesis were observed for wt, vie2d(94-274), and vie2d(94-173). A longer delay was observed for vie2d(215-274)-infected SF-21 cells, with a fivefold reduction in viral DNA accumulation at 24 h p.i. By 48 h p.i., levels of DNA synthesis in SF-21 cells infected with vie2d(215-274) were comparable to those in wt-, vie2d(94-274)-, and vie2d(94-173)-infected cells. This result is consistent with the more severe phenotype of this mutant.
FIG. 5.
Dot blot analysis of wt and mutant viral DNA replication in two different cell lines. (A) Total cellular DNA was isolated from SF-21 and TN-5B1-4 cells infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274) at the indicated times p.i. (shown on the right), dot blotted, and hybridized to radiolabeled AcMNPV DNA. Dots on lane m represent purified AcMNPV DNA (amounts are shown on the left) used as a standard. (B) Graphic representation of the data quantified by PhosphorImager reading of each dot. Levels of viral DNA replication relative to that from wt-infected cells at 72 h p.i. (100%) are presented. The data represent the results of two or more experiments, and standard errors are indicated.
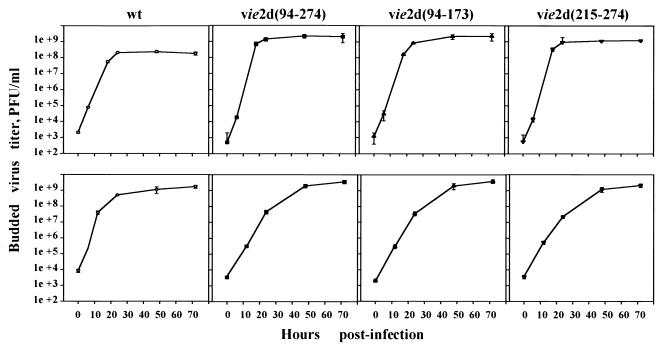
Effect of ie-2 mutations on BV production.
We next examined the effect of ie-2 mutations on BV production. SF-21 and TN-5B1-4 cells were infected with ie-2 mutant viruses or with wt, and yields of BV were determined on TN-5B1-4 cells. In TN-5B1-4 cells, the rate of BV release from ie-2 mutant-infected cells seemed to be similar to, if not higher than, than that observed for wt-infected cells (Fig. 6, top panels). Furthermore, all of the ie-2 mutant viruses produced similar amounts of BV at each time point; by 24 h p.i., BV levels reached plateaus of 2.0 × 109, 1.2 × 109, and 1.8 × 109 PFU/ml, respectively. By 24 h p.i., wt AcMNPV produced a maximum titer which was 10-fold lower than those obtained for ie-2 mutants.
FIG. 6.
BV production of wt and ie-2 mutant viruses in two different cell lines. SF-21 and TN-5B1-4 cells were infected with wt AcMNPV (open circles), vie2d(94-274), vie2d(94-173), or vie2d(215-274) (closed symbols) and cultured at 27°C. At 0, 6, 12, 24, 48, and 72 h p.i., 500 μl of culture medium was collected and subjected to plaque assay on TN-5B1-4 cells to determine virus titer. The results represent the average of two independent titers.
In SF-21 cells, ie-2 mutants exhibited a retarded rate of BV production throughout infection (Fig. 6, bottom panels). Similar BV titers were observed for vie2d(94-274), vie2d(94-173), and vie2d(215-274), but the levels reached plateaus 24 h later (e.g., 48 h p.i.) than for wt AcMNPV, indicating that mutations of ie-2 caused a delay of BV production in SF-21 cells. The final titers of wt and ie-2 mutant BVs were similar by 48 h p.i. in this cell line.
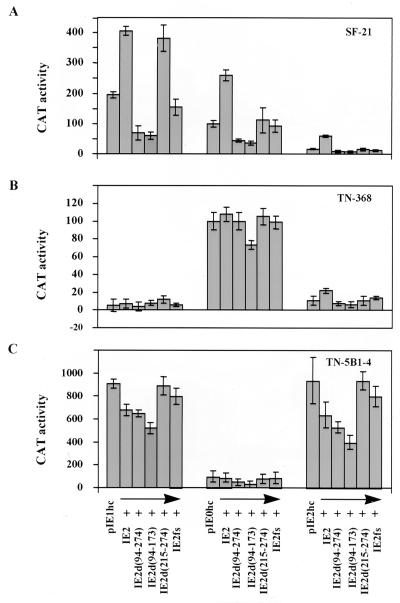
Effects of ie-2 mutants on AcMNPV immediate-early promoters in transient expression assays.
It was of interest to know whether the phenotypes of the ie-2 mutants correlated with their effects on expression from early promoters observed in transient expression assays in the different cell lines examined. However, IE-2 trans regulation of early promoters of AcMNPV has been studied only in cell lines derived from S. frugiperda. We therefore examined the ability of wt and mutant forms of ie-2 to trans activate the ie-1, ie-0, and ie-2 promoters in transient expression assays in SF-21, TN-368, and TN-5B1-4 cells (Fig. 7). Plasmids phcIE1, phcIE0, and phcIE2 were used as reporter plasmids and contained the CAT gene under ie-1, ie-0, and ie-2 promoter control, respectively. Plasmid pBs-PstNfs, which contained a frameshift mutation in ie-2 at codon 148, was considered as a null mutant in these experiments. As previously shown (30), IE2d(215-274), which lacks the RING finger motif, does not affect the ability of IE2 to trans activate expression from the ie-1 promoter in SF-21 cells (Fig. 7A). In contrast, IE2d(94-274) and IE2d(94-173) lacking the trans-regulatory region were unable to trans activate the ie-1 promoter and exerted a negative effect on expression from this promoter, exhibiting approximately two- to threefold decreases in CAT gene expression compared to that observed in cells transfected with the reporter plasmid phcIE1 alone. Similar results were observed for the reporter plasmids phcIE0 and phcIE2: deletion of residues 94 to 173 or 94 to 274 completely eliminated the ability of ie-2 to trans activate these promoters in SF-21 cells. Moreover, the same negative effect of these ie-2 mutants, resulting in two- to threefold decreases of CAT activity from ie-2 and ie-0 promoters, was observed. IE2d(215-274), which retained the ability to trans activate the ie-1 promoter, did not activate the ie-2 and ie-0 promoters and showed levels of CAT gene expression similar to those for IE2fs in SF-21 cells. Thus, deletion of the RING finger of IE-2 affected its ability to trans regulate two of three promoters tested. The IE2d(215-274) mutant has a similar but more severe phenotype than IE2d(94-173) and IE2d(94-274), and so there is no obvious correlation between trans activation of the ie-0, ie-1, or ie-2 promoter with mutant phenotype.
FIG. 7.
Analysis of the trans-regulatory activity of ie-2 deletion mutants in different cell lines. SF-21 (A), TN-368 (B), and TN-5B1-4 (C) cells (106/60-mm-diameter dish) were transfected with the reporter plasmids phcIE-1, phcIE-0, and phcIE-2 alone or in combination with pBs-PstN, pBs-PstNd(94-274), pBs-PstNd(94-173), or pBs-PstNd(215-274), expressing the intact ie-2 gene, ie-2 deletion mutants, or a frameshift mutant of ie-2. Transfected cells were harvested at 24 h posttransfection, and CAT activity was determined by enzyme assays (see Materials and Methods). CAT activity relative to that obtained from the reporter plasmid phcIE0 (100%) in each cell line was determined. The results represent the average of two independent experiments, and standard errors are indicated.
In TN-5B1-4 and TN-368 cells, ie-2 did not show any significant trans activation of the ie-0 or ie-1 promoter in transient expression assay, indicating this trans-regulatory ability of ie-2 is cell line dependent. A weak trans activation of expression from the ie-2 promoter was observed for wt but not mutant IE2s in TN-368 cells but not in TN-5B1-4 cells, indicating cell line-specific effects. Mutants IE2d(94-173) and IE2d(94-274) lacking the transcriptional activation region exerted a negative influence on expression from some of the promoters in one or both T. ni-derived cell lines.
Striking differences in the relative levels of activity of the ie-1, ie-0, and ie-2 promoters were observed in SF-21, TN-368, and TN-5B1-4 cells transfected with the reporter plasmids alone (Fig. 7). The most notable feature was that the ie-1 promoter appeared to have little or no activity in the TN-368 cell line, whereas the ie-0 promoter was highly active (Fig. 7B). The level of expression from the ie-1 promoter in phcIE1 was significantly increased in TN-368 cells when phcIE1 was cotransfected with pBs-IE1/HC expressing the AcMNPV ie-1 gene, but coexpression with ie-2, even in combination with ie-1, did not increase the levels of CAT gene expression from the ie-1 promoter in this cell line (data not shown).
Effect of ie-2 deletions on the virulence and infectivity of AcMNPV in insect larvae.
To determine whether ie-2 mutants affect the infectivity of AcMNPV at the organism level, we determined the LC50 of orally administered OBs. In T. ni, the LC50s of the ie-2 mutant viruses vie2d(94-274), vie2d(94-173), and vie2d(215-274) were similar within the experimental error limits and were approximately 100-fold higher than the LC50 obtained for wt AcMNPV (Table 1). An even larger difference between the LC50s of wt and ie-2 mutants was observed in S. frugiperda larvae, a species which is generally less susceptible to AcMNPV infection than T. ni. Mutants vie2d(94-274) and vie2d(94-173) had LC50s approximately 1,000-fold higher than the wt value, and the LC50 of vie2d(215-274) was 5,000-fold higher than that of wt AcMNPV in S. frugiperda. Thus, mutations in ie-2 reduced the infectivity of AcMNPV OBs in both S. frugiperda and T. ni larvae, although the effect was more pronounced in S. frugiperda.
TABLE 1.
Dose-mortality responses of neonate S. frugiperda and T. ni larvae infected orally with OBs of wt AcMNPV or ie-2 mutant virusesa
| Host and virus | LC50b (OBs/ml) | 95% Fiducial limit
|
Slope (mean ± SE) | Heterogeneity factorc | |
|---|---|---|---|---|---|
| Upper | Lower | ||||
| S. frugiperda | |||||
| wt AcMNPV | 2.6 × 105 | 4.2 × 105 | 1.5 × 105 | 1.148 ± 0.177 | 0.31 |
| vie2d(94-274) | 3.9 × 108 | 9.9 × 108 | 2.5 × 108 | 1.131 ± 0.224 | 0.34 |
| vie2d(94-173) | 3.1 × 108 | 5.2 × 108 | 2.0 × 108 | 1.142 ± 0.193 | 0.30 |
| vie2d(215-274) | 1.2 × 109 | 1.9 × 109 | 0.5 × 109 | 0.756 ± 0.210 | 0.12 |
| T. ni | |||||
| wt AcMNPV | 1.4 × 104 | 1.8 × 104 | 1.0 × 104 | 2.941 ± 0.514 | 0.40 |
| vie2d(94-274) | 1.3 × 106 | 1.9 × 106 | 0.9 × 106 | 1.554 ± 0.225 | 0.42 |
| vie2d(94-173) | 1.8 × 106 | 2.8 × 106 | 0.8 × 106 | 1.715 ± 0.386 | 0.79 |
| vie2d(215-274) | 3.1 × 106 | 4.2 × 106 | 2.3 × 106 | 1.926 ± 0.249 | 0.16 |
Six groups of neonates (less than 12 h old) were allowed to feed on diet contaminated with various concentrations of OBs per ml for 24 h and then transferred to uncontaminated diet. The number of dead larvae at 10 days p.i. was used to determine the LC50.
Values were calculated by probit analysis.
χ2 divided by the degrees of freedom (POLO-PC; LeOra Software).
The infectivity of the budded form of wt AcMNPV and ie-2 mutants was assessed by determining their LD50s in S. frugiperda and T. ni larvae (Table 2). Insects in the penultimate larval instar were injected hemocoelically with selected doses of BV and monitored for mortality. No differences were observed among LD50s of the viruses in T. ni or S. frugiperda larvae. Thus, the infectivity of the budded form of ie-2 mutants was normal in both insect species.
TABLE 2.
Dose-mortality responses of S. frugiperda and T. ni penultimate larval instars to the budded form of wt AcMNPV or ie-2 mutant viruses following hemocoelic injectiona
| Host and virus | LD50 (PFU/insect) | 95% Fiducial limit
|
Slope (mean ± SE) | Heterogeneity factor | |
|---|---|---|---|---|---|
| Upper | Lower | ||||
| S. frugiperda | |||||
| wt AcMNPV | 0.6 × 104 | 1.4 × 104 | 0.3 × 104 | 0.988 ± 0.152 | 0.69 |
| vie2d(94-274) | 0.4 × 105 | 1.0 × 105 | 0.1 × 105 | 0.781 ± 0.127 | 0.70 |
| vie2d(94-173) | 0.9 × 104 | 2.7 × 104 | 0.4 × 104 | 0.697 ± 0.102 | 0.59 |
| vie2d(215-274) | 0.6 × 104 | 1.4 × 104 | 0.3 × 104 | 0.921 ± 0.140 | 0.31 |
| T. ni | |||||
| wt AcMNPV | 0.9 × 102 | 2.1 × 102 | 0.3 × 102 | 0.968 ± 0.196 | 0.62 |
| vie2d(94-274) | 1.0 × 102 | 2.6 × 102 | 0.3 × 102 | 0.862 ± 0.177 | 0.89 |
| vie2d(94-173) | 0.9 × 102 | 2.7 × 102 | 0.2 × 102 | 0.558 ± 0.106 | 0.73 |
| vie2d(215-274) | 3.1 × 102 | 9.6 × 102 | 0.9 × 102 | 0.484 ± 0.086 | 0.66 |
Six groups of S. frugiperda and T. ni penultimate larval instars were injected hemocoelically with various doses of BV and then observed for mortality. For other details, see Table 1, footnotes b and c.
Deficiency of virions in the OBs of ie-2 mutants.
The greatly reduced infectivity of OBs in both T. ni and S. frugiperda larvae suggested that the OBs of ie-2 mutants might be defective. To determine if a normal number of virions were embedded in the OBs, we determined the relative amounts of the major capsid protein, VP39, in wt and mutant OBs purified from infected T. ni larvae (Fig. 8). The same number of OBs (3 × 108) of each virus tested was purified and subjected to SDS-PAGE followed by Western immunoblot analysis using an antibody against VP39. No significant differences were observed in the size of occlusion bodies of the ie-2 mutant viruses compared to wt OBs (data not shown). Levels of VP39 were much lower for all ie-2 mutants than for wt OBs (Fig. 8A). Although the levels were not quantified, it appeared that wt OBs had over 100 times more VP39 than ie-2 mutant OBs, which would account for their lack of infectivity. To confirm that equivalent numbers of OBs for ie-2 mutants and wt were used, the blot used to detect VP39 protein was stripped and then reprobed with polyclonal immune serum to polyhedrin. The amounts of polyhedrin in the OBs obtained from the ie-2 mutant- or wt-infected larvae were similar (Fig. 8B).
FIG. 8.
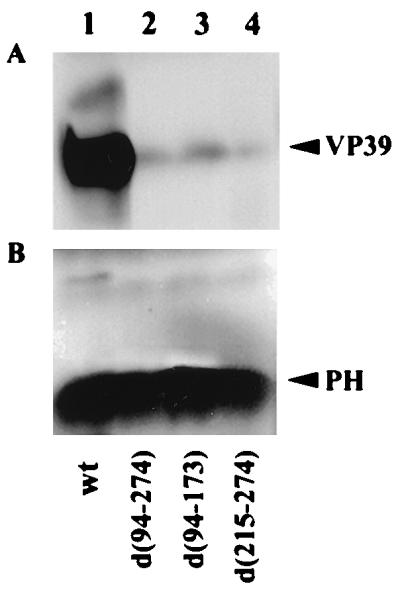
Western blot analysis of the levels of VP39 major capsid protein and polyhedrin in OBs obtained from T. ni larvae infected with wt AcMNPV or ie-2 mutant viruses. (A) A total of 3 × 108 OBs purified from insect infected with wt AcMNPV, vie2d(94-274), vie2d(94-173), or vie2d(215-274) were boiled in SDS loading buffer and subjected to Western blot analysis using a VP39 polyclonal antibody. (B) The blots in panel A, stripped and reprobed with a polyhedrin (PH) polyclonal antibody.
DISCUSSION
We have isolated mutants of AcMNPV defective in ie-2; these mutants replicate in both SF-21 and TN-5B1-4 cell lines even though ie-2 is known from transient expression assays to trans activate viral gene expression and DNA replication in SF-21 cells and block cell cycle progression in both cell lines. The mutant viruses do, however, display mutant phenotypes in both cell lines, with the SF-21 cell line displaying the more severe phenotype. In TN-5B1-4 cells, the ie-2 mutants produce less polyhedrin, fewer OBs per cell, and more BV. In SF-21 cells, there is a substantial delay in viral DNA replication, late gene expression, BV production, and polyhedrin production in addition to a reduction in the number of OBs per cell. The delay in viral DNA replication in SF-21 but not TN5-B1-4 cells is consistent with a role of IE-2 in trans activation of gene expression from the ie-0, ie-1, and ie-2 promoters in SF-21 cells but not TN5-B1-4 cells. However, all three ie-2 mutants which we constructed displayed a delayed-replication phenotype in SF-21 cells, and one of these mutants, vie2d(215-274), lacking only the RING finger motif, retained ie-1 promoter trans-activating ability but exhibited the strongest phenotype with regard to delay in DNA replication and later events. This finding suggests that IE-2 activation of the ie-1 promoter is not directly correlated with the delayed phenotype of the ie-2 mutants in the SF-21 cell line. It is possible that IE-2 trans regulates expression of another early viral gene yet to be identified or is involved in regulation of some cellular factors which respond differently to the IE-2 mutants. We have not examined the effect of the mutant viruses on cell cycle progression owing to the interfering effects of virus DNA replication on DNA content and cellular integrity.
There are at least two other differences between SF-21 and TN-5B1-4 cells which might affect the phenotypes of the ie-2 mutants. In TN-5B1-4 cells, all three mutants exhibited elevated rates of synthesis of a 75-kDa protein; altered levels of expression of this protein were not observed in SF-21 cells. Furthermore, all three mutants displayed elevated levels of expression of either a 34- or a 37-kDa protein. The 34- and 37-kDa proteins might be products of the mutant ie-2 genes, but why they are overexpressed in TN-5B1-4 cells and not in SF-21 cells is not clear. We have previously shown by Western blot analysis that the ie-2 mutants are well expressed in SF-21 cells upon transient expression (30) as well as in transfected cells additionally infected with vie2Z (data not shown). The identity of the 75-kDa protein is not known, but given its relatively large size, only a few viral proteins would qualify as candidates. The overexpression of this protein in all ie-2 mutant-infected TN 5B1-4 cells suggests that IE-2 is involved in down-regulating the expression of this gene in this cell line. We apparently have much to learn concerning the normal molecular role of IE-2 in the infection process, and neither the known trans-activation function nor the cell cycle regulation function of IE-2 can fully account for the phenotypes of the mutants.
Two other AcMNPV genes, p35 and lef-7, were previously shown to activate transient expression of a late reporter gene in SF-21 cells but have little or no effect in equivalent transient expression assays in TN-368 cells (22). Mutant viruses defective in p35 or lef-7 have also been successfully isolated and propagated in T. ni cells (6, 7). The severity of the phenotypes of these mutants in SF-21 cells correlates well with the relative contribution of the genes (p35 > lef-7 > ie-2) to plasmid replication and/or stability in this cell line (22).
One of the unexpected results from our study of IE-2 trans activation of the ie-1, ie-0, and ie-2 promoters was the enormous differences in the relative levels of expression from the three different promoters in the three cell lines tested. The most dramatic difference observed was in the relative levels of expression from the ie-1 and ie-0 promoters in SF-21 and TN-368 cells. Whereas expression levels from these two promoters are similar in SF-21 cells, the ie-1 promoter is 20-fold less active than the ie-0 promoter in TN-368 cells. Expression from the ie-1 promoter was trans activated by ie-1 (data not shown), but ie-2 was unable to trans activate it either in the presence or in the absence of ie-1 (Fig. 8 and data not shown). Thus, the ie-1 promoter appears to be dependent on IE-1 or IE-0 for its expression in this cell line. The effect is cell line related rather than a species effect, since basal levels of expression from the ie-1 promoter are higher than ie-0 in TN-5B1-4 cells. This finding suggests that the relative contributions of IE-0 and IE-1 in the infection process may be tissue dependent in the insect.
The infectivity of OBs of the ie-2 mutants was drastically reduced in both S. frugiperda and T. ni larvae. The reduced infectivity of the mutant OBs correlated with a reduction in the level of the major capsid protein VP39 within the OBs, indicating a deficiency of occluded virions. Noninfectious OBs lacking virions are also formed by a class of mutants known as FP (few polyhedra) mutants (28, 29, 37). Like FP mutants, the OBs of ie-2 mutants have very low infectivity upon oral infection of larvae (28). It may also be noteworthy that, like FP mutants, the ie-2 mutants produce approximately 10-fold more BV and fewer OBs in T. ni-derived cells. The genetic defect of a number of FP mutants has been traced to another gene known as the 25K or FP25 gene (2). The 25K and ie-2 genes may independently affect the embedding of virions within OBs, or ie-2 may regulate 25K gene expression.
The BVs of ie-2 mutants exhibited the same infectivity (LD50) as wt virus, demonstrating that ie-2 is not required for the virus to disseminate in larvae and cause mortality. The pathology of the larval infections initiated by BVs was not examined at the tissue level, and so it remains to be determined whether ie-2 affects the tissue tropism of the virus. IE-2, however, does not appear to be a host range determinant per se, although it clearly influences the infectivity of OBs in both species by its effect on virion occlusion.
During the course of this work, an ie-2 mutant of the closely related Bombyx mori nuclear polyhedrosis virus (BmNPV) was described (9). The mutant was viable in BmN-4 cells but showed a delay in DNA replication. However, IE-2 of BmNPV has not been characterized with respect to either trans activation or cell cycle arrest functions, nor have its cell line- or species-specific effects been examined. Since ie-2 of AcMNPV was known to exert cell line-specific trans activation, it was important to determine if AcMNPV ie-2 mutants were viable and if they had altered host range properties. Furthermore, with mutations specifically affecting trans activation or cell cycle arrest, we were interested in determining if we could distinguish different phenotypes for the different alleles.
Like the BmNPV ie-2 mutant, a lacZ insertion mutant, all AcMNPV ie-2 mutants exhibited delayed DNA replication, but this delay was cell line dependent since it was pronounced in SF-21 cells but was not detectable in TN-5B-1 cells. Whereas the growth rate of the BmNPV ie-2 mutant appears to be unaffected in BmN-4 cells (9), growth of the AcMNPV ie-2 mutants was severely retarded in SF-21 cells. Although the growth rates of the AcMNPV ie-2 mutants appear to be almost normal in TN-5B-1 cells, the mutants produced approximately 10-fold more BV than wt, and there was a defect in occlusion since occlusion bodies were deficient in virions. Thus, ie-2 mutants of AcMNPV are viable, although impaired, in cell culture but are effectively incapable of infection through the normal route of infection in the insect. Since ie-2 alleles which differentially affect cell cycle arrest and transactivation had generally similar phenotypes with regard to virus growth rate, BV production, and infectivity in cell cultures and in two insect species, it is not clear which, if any, of these transient expression properties of ie-2 are directly relevant to the role that ie-2 plays in virus infection. A comparison of the effects of ie-2 on transient gene expression in different cell lines revealed a complex pattern of gene activity and gene activation in these cell lines. Our results clearly establish that ie-2 of AcMNPV confers a growth advantage in a cell line-specific manner and is essential for proper OB formation and oral infectivity.
ACKNOWLEDGMENTS
We thank Grigori Prikhod’ko for technical advice and assistance in bioassays. We are grateful to Jeanne McLachlin for technical advice.
This research was supported in part by Public Health Service grant AI 23719 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Beames B, Summers M D. Location and nucleotide sequence of the 25K protein missing from baculovirus few polyhedra (FP) mutants. Virology. 1989;168:344–353. doi: 10.1016/0042-6822(89)90275-4. [DOI] [PubMed] [Google Scholar]
- 3.Carson D D, Guarino L A, Summers M D. Functional mapping of an AcNPV immediately early gene which augments expression of the ie-1 trans-activated 39k gene. Virology. 1988;162:444–451. doi: 10.1016/0042-6822(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 4.Carson D D, Summers M D, Guarino L A. Molecular analysis of a baculovirus regulatory gene. Virology. 1991;182:279–286. doi: 10.1016/0042-6822(91)90671-w. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm G E, Henner D J. Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosis virus ie-1 gene. J Virol. 1988;62:3193–3200. doi: 10.1128/jvi.62.9.3193-3200.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C J, Thiem S M. Differential infectivity of two Autographa californica nucleopolyhedrovirus mutants on three permissive cell lines is the result of lef-7 deletion. Virology. 1997;227:88–95. doi: 10.1006/viro.1996.8341. [DOI] [PubMed] [Google Scholar]
- 7.Clem R J, Miller L K. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J Virol. 1993;67:3730–3738. doi: 10.1128/jvi.67.7.3730-3738.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen P D. Regulation of baculovirus early gene expression. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 141–170. [Google Scholar]
- 9.Gomi S, Zhoi C E, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230:35–47. doi: 10.1006/viro.1997.8457. [DOI] [PubMed] [Google Scholar]
- 10.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarino L A, Summers M D. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol. 1986;57:565–571. doi: 10.1128/jvi.57.2.563-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarino L A, Summers M D. Nucleotide sequence and temporal expression of a baculovirus regulatory gene. J Virol. 1987;61:2091–2099. doi: 10.1128/jvi.61.7.2091-2099.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino L A, Dong W. Functional dissection of the Autographa californica nuclear polyhedrosis virus enhancer element hr5. Virology. 1994;200:328–335. doi: 10.1006/viro.1994.1197. [DOI] [PubMed] [Google Scholar]
- 14.Hink W F. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 15.Hughes P R. ViStat: statistical package for the analysis of baculovirus bioassay data. Ithaca, N.Y: Boyce Thompson Institute at Cornell University; 1990. [Google Scholar]
- 16.Kool M, Ahrens C, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacs G R, Guarino L A, Summers M D. Novel regulatory properties of the IE1 and IE0 transactivators encoded by the baculovirus Autographa californica multicapsid nuclear polyhedrosis virus. J Virol. 1991;65:5281–5288. doi: 10.1128/jvi.65.10.5281-5288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krappa R, Knebel-Mörsdorf D. Identification of the very early transcribed baculovirus gene pe-38. J Virol. 1991;65:805–812. doi: 10.1128/jvi.65.2.805-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H H, Miller L K. Isolation of genotypic variants of Autographa californica nuclear polyhedrosis virus. J Virol. 1978;27:754–767. doi: 10.1128/jvi.27.3.754-767.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu A, Carsten E B. Immediately-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology. 1993;195:710–718. doi: 10.1006/viro.1993.1422. [DOI] [PubMed] [Google Scholar]
- 21.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu A, Miller L K. Differential requirements for baculovirus late expression factor genes in two cell lines. J Virol. 1995;69:6265–6272. doi: 10.1128/jvi.69.10.6265-6272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu A, Miller L K. Species-specific effects of the hcf-1 gene on baculovirus virulence. J Virol. 1996;70:5123–5130. doi: 10.1128/jvi.70.8.5123-5130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris T D, Miller L K. Promoter influence of baculovirus-mediated gene expression in permissive and nonpermissive insect cell lines. J Virol. 1992;66:7397–7405. doi: 10.1128/jvi.66.12.7397-7405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen M S, Friesen P D. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J Virol. 1989;63:493–503. doi: 10.1128/jvi.63.2.493-503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Reilly D A, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 27.Passarelli A L, Miller L K. Three baculovirus genes involved in late and very late gene expression: ie-1, ie-n, and lef-2. J Virol. 1993;67:2149–2158. doi: 10.1128/jvi.67.4.2149-2158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potter K N, Faulkner P, MacKinnon E A. Strain selection during serial passage of Trichoplusia ni nuclear polyhedrosis virus. J Virol. 1976;18:1040–1050. doi: 10.1128/jvi.18.3.1040-1050.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potter K N, Jacques R P, Faulkner P. Modification of Trichoplusia ni nuclear polyhedrosis virus passaged in vivo. Intervirology. 1978;9:76–85. doi: 10.1159/000148925. [DOI] [PubMed] [Google Scholar]
- 30.Prikhodko E A, Miller L K. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J Virol. 1998;72:684–692. doi: 10.1128/jvi.72.1.684-692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin C, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene. 1988;70:39–50. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro B, Hutchinson K, Miller L K. A mutant baculovirus with a temperature-sensitive IE-1 transregulatory protein. J Virol. 1994;68:1075–1084. doi: 10.1128/jvi.68.2.1075-1084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodems S M, Friesen P D. Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J Virol. 1995;69:5368–5375. doi: 10.1128/jvi.69.9.5368-5375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomalski M D, Miller L K. Expression of a paralytic neurotoxin gene to improve insect baculoviruses as biopesticides. Bio/Technology. 1992;10:545–549. [Google Scholar]
- 35.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 36.Wichman T J, Davis T T, Granados R R, Shuler M L, Wood H A. Screening insect cell lines for the production of recombinant virus in the baculovirus expression system. Biotechnol Prog. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 37.Wood H A. Isolation and replication of an occlusion body-deficient mutant of the Autographa californica nuclear polyhedrosis virus. Virology. 1980;105:338–344. doi: 10.1016/0042-6822(80)90035-5. [DOI] [PubMed] [Google Scholar]
- 38.Yoo S, Guarino L A. Functional dissection of the ie2 gene product of the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:164–172. doi: 10.1006/viro.1994.1332. [DOI] [PubMed] [Google Scholar]
- 39.Yoo S, Guarino L A. The Autographa californica nuclear polyhedrosis virus ie-2 gene encodes a transcriptional regulator. Virology. 1994;202:746–753. doi: 10.1006/viro.1994.1396. [DOI] [PubMed] [Google Scholar]