Abstract
The adeno-associated virus (AAV) is unique in its ability to target viral DNA integration to a defined region of human chromosome 19 (AAVS1). Since AAVS1 sequences are not conserved in a rodent’s genome, no animal model is currently available to study AAV-mediated site-specific integration. We describe here the generation of transgenic rats and mice that carry the AAVS1 3.5-kb DNA fragment. To test the response of the transgenic animals to Rep-mediated targeting, primary cultures of mouse fibroblasts, rat hepatocytes, and fibroblasts were infected with wild-type wt AAV. PCR amplification of the inverted terminal repeat (ITR)-AAVS1 junction revealed that the AAV genome integrated into the AAVS1 site in fibroblasts and hepatocytes. Integration in rat fibroblasts was also observed upon transfection of a plasmid containing the rep gene under the control of the p5 and p19 promoters and a dicistronic cassette carrying the green fluorescent protein (GFP) and neomycin (neo) resistance gene between the ITRs of AAV. The localization of the GFP-Neo sequence in the AAVS1 region was determined by Southern blot and FISH analysis. Lastly, AAV genomic DNA integration into the AAVS1 site in vivo was assessed by virus injection into the quadriceps muscle of transgenic rats and mice. Rep-mediated targeting to the AAVS1 site was detected in several injected animals. These results indicate that the transgenic lines are proficient for Rep-mediated targeting. These animals should allow further characterization of the molecular aspects of site-specific integration and testing of the efficacy of targeted integration of AAV recombinant vectors designed for human gene therapy.
Adeno-associated virus (AAV) is a human parvovirus with a single-stranded DNA genome of about 4.7 kb. For efficient replication, AAV requires coinfection with a helper virus such as adenovirus (Ad) or herpesvirus; however, a low level of helper-independent AAV replication can occur in cells exposed to genotoxic stress. In the absence of helper functions, AAV integrates its DNA into the host genome, where it is maintained as a latent provirus. Ad superinfection of latently infected cells leads to the rescue of the AAV genome from the host and to production of infectious particles (3). Genetic analysis of latently infected human cell lines has shown that integration of viral DNA occurs preferentially in a region of the human genome mapped to human chromosome 19q13.3-qter called AAVS1 (12, 27, 38). Two AAV elements are required both for viral DNA replication and for integration: the inverted terminal repeats (ITR) and the two larger Rep polypeptides Rep68 and Rep78 (1, 23, 28, 36, 42, 45). The inverted terminal repeats are 145-bp palindromic sequences located at each end of the single-stranded viral genome that are folded in a hairpin structure and function as the origin of DNA replication (39, 47, 49). The rep gene encodes four overlapping polypeptides: Rep78, Rep68, Rep52, and Rep40. These polypeptides are derived by transcription from the p5 (Rep78 and Rep68) and the p19 (Rep52 and Rep40) promoters. Rep78 and Rep52 derive from the unspliced and Rep68 and Rep40 from the spliced forms of the corresponding mRNA (44).
The AAVS1 preintegration site has been cloned from human chromosome 19 as an 8.2-kb EcoRI fragment of which 4 kb at the leftmost 5′ region have been sequenced (26). The sequence shows no clear homology to the AAV genome, supporting the notion that site-specific integration of viral DNA into the host genome occurs via nonhomologous recombination (6, 27, 29). Targeted integration by wild-type (wt) AAV has been demonstrated in numerous human cell lines, ranging from aneuploid 293, Huh-7, HeLa, Detroit 6, and IB3 cells to diploid WI-38, colon, and T cells (13, 23, 29, 33, 35, 38). Analysis of the AAVS1 region in latently infected cells has revealed integration of the AAV genome at multiple points clustered in a hot spot region of about 600 nucleotides (nt) (26, 35, 38, 42).
Although many features of interest were identified in the AAVS1 sequence, including a putative open reading frame, short repeats, and a CpG island, only a small region of AAVS1 appears to be essential for site-specific integration. Indeed, critical sequences for targeted integration have been localized by Berns and coworkers with a transient integration system based on an Epstein-Barr virus (EBV) shuttle vector (12, 31) to 500 bp located in the 5′-end region of AAVS1 (12). Further deletions have narrowed down the region necessary to drive site-specific integration to 33 nt in the 500-bp fragment containing a Rep binding site (RBS) separated by an 8-nt spacer from a sequence called the terminal resolution site (trs) that can act as a substrate for Rep endonucleolytic activity (21, 22, 49). Mutations in either the RBS or the trs elements located within AAVS1 abolish targeted integration (31).
A sequence homologous to the human AAVS1 preintegration site has been found to date only in green monkeys, and it is absent in rodents (38), where integration of AAV DNA is believed to occur randomly. Thus, the lack of an animal model for AAV site-specific integration has hampered the molecular characterization of Rep-mediated targeted insertion in the host chromosome both in primary diploid cells and in vivo. Indeed, the only primary cells in which site-specific integration of wt AAV has been documented so far are human hematopoietic progenitors (13). Although wt AAV DNA has been detected by PCR in peripheral blood leukocytes (15), in female genital tissue, and in samples from spontaneous abortions (10, 17, 46, 48), targeted integration in vivo has never been demonstrated. In this study we report the generation of transgenic rat and mouse lines carrying the AAVS1 site. We demonstrate that site-specific integration of both wt virus and AAV-derived vectors occurs in vitro in primary fibroblasts and hepatocytes, as well as in vivo in muscle cells.
MATERIALS AND METHODS
AAV and Ad virus production and purification.
wt AAV-2 virus was prepared by transfection of Ad5 ΔE1β-gal-infected 293 human embryonic kidney cells with plasmid pSub201 according to published protocols (37, 40). The infectious titer of the AAV preparation was determined by replication center assay (2). Ad contamination was measured by infecting strain 293 cells with an aliquot of the wt AAV stock followed by staining for β-galactosidase expression. Ad ΔE1β-gal was present at 1 transducing unit (t.u.)/5 × 107 wt AAV infectious units (i.u.).
AAV replication capacity in transgenic primary rat hepatocytes and fibroblasts, as well as in mouse embryonic fibroblasts, was assessed by infecting cell monolayers at a multiplicity of infection (MOI) of 100. The cells were also infected with wt Ad5 at an MOI of 100. Low-molecular-weight DNA was isolated according to published protocols (19) from infected cultures 48 h postinfection, and analyzed by Southern blot with a probe specific for the Rep sequence.
Generation and analysis of transgenic rodents.
The AAVS1 fragment used for microinjection was derived from plasmid pRVK (K. Berns, Cornell Medical School, New York, N.Y.) by EcoRI and KpnI digestion and covers nt 1 to 3525 of the original 8.2-kb clone (26). Transgenic Sprague-Dawley rats were prepared by pronuclear microinjection in one-cell-stage embryos by BRL (Basel) as described previously (20). Transgenic mice carrying the 3.5-kb fragment of AAVS1 were prepared by transfection of pluripotent embryonic stem cells with plasmid pLNeo-AAVS1 following selection with geneticin. Plasmid pBsLNeo was produced by inserting the blunt end XbaI/SalI fragment derived from pL2-neo (16), which contains the neo resistance gene under the pMCI promoter and flanked by two loxP sites, into pBluescript II KS linearized with SmaI. The AAVS1 3.5-kb fragment was inserted into the EcoRV/KpnI sites of plasmid pBsLNeo, thus generating pLNeo-AAVS1. To obtain stable cell clones carrying the AAVS1 fragment, 30 μg of plasmid pLNeo-AAVS1, linearized with XbaI, was used to electroporate 8 × 106 E14 cells (240 V, 500 mF). Two days after transfection, cells were seeded in two 10-cm plates and placed under G418 selection (200 mg/ml). Thirty-eight neo-resistant clones were individually picked and expanded, and their genomic DNA was analyzed by Southern blot with the AAVS1 3.5-kb fragment as a probe. Seven clones that carried only one copy of the AAVS1 sequence were identified, and four of these were injected into blastocysts C57BL6 and transplanted into the uterus of foster mothers. Then, 60% and 80% male chimeras were obtained from one clone. They were mated to BALB/c females, and agouti offsprings were screened for the presence of the AAVS1 sequence. Genotype analysis was performed by Southern blot analysis on 10 μg of total genomic DNA extracted from tail biopsies with the AAVS1 3.5-kb fragment as a probe.
Animals and treatments.
Rodents were maintained in standard conditions under a 12-h light-dark cycle and provided with irradiated food (4RF21; Mucedola) and chlorinated water ad libitum.
Intramuscular injection was performed in the left quadriceps of six 3-month-old male rats and eight 1-month-old mice. wt AAV prepared as described above was injected at a dose of 108 i.u./rat and 5 × 107 i.u./mouse in a 100-μl volume with a 28 gauge 3/10-ml insulin syringe (Becton Dickinson, Paramus, N.J.). Muscle genomic DNA was extracted from the injected animals at 2, 6, 30, and 75 days postinfection according to standard procedures (20). Where indicated, DNA from the injected animals was analyzed by PCR amplification and Southern blot analysis to assess for AAV infection and viral DNA integration.
Primary cultures.
Primary fibroblasts were prepared from ear biopsies of transgenic and nontransgenic rats and from 14 p.c. embryonic mice. The rat tissue samples were briefly washed in 70% ethanol, placed in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum (FCS), and then cut into 1-mm3 pieces with a sterile blade. The tissues was dissociated by 1 h of incubation at 37°C in DMEM containing 0.3% collagenase-dispase from Vibrio alginolyticus and Bacillus polymyxa (Boehringer Mannheim). Similarly, mouse embryos were cut into small fragments and incubated several times with trypsin-EDTA for 5 min at 37°C. After incubation, the cells were centrifuged, washed with phosphate-buffered saline (PBS), and maintained at 37°C in DMEM with 10% FCS.
Primary hepatocytes were prepared by performing double-step liver perfusion (4). Heparin (200 μl of a 5,000-U/ml mixture) was injected into the femoral veins of anesthetized transgenic rats, and the portal veins were cannulated. The livers were then perfused in a nonrecirculating fashion after cava and aorta resection. The liver was first perfused for 15 min with HEPES buffer (10 mM HEPES, 137 mM NaCl, 2.7 mM KCl, 0.28 mM Na2HPO4) at a flow rate of 30 ml/min. A second perfusion was performed for 12 min with HEPES buffer plus 0.025% collagenase H (Boehringer) and 0.075% CaCl2 at a flow rate of 15 ml/min. The liver was then removed and rinsed in HEPES buffer, and the parenchyma was subsequently disrupted in Leibowitz-15 medium containing penicillin-streptomycin and 2 mg of bovine serum albumin per ml. The flocculent cell suspension was filtered through a 70-mm nylon filter. After sedimentation the cells were washed three times in HEPES buffer and once in complete medium (William’s E–penicillin-streptomycin–glucose–10% FCS) and centrifuged each time at 72 × g for 1 min. Hepatocytes were isolated by Percoll gradient centrifugation according to the standard protocol. The viability of the hepatocyte preparation was assessed by trypan blue exclusion, and the hepatocytes were typically 90% viable.
Transfection of primary fibroblasts.
Primary adult fibroblasts were transfected with plasmid pITR(GFP-Neo)P5Rep (35). The fibroblasts were plated the day before transfection in a 24-well microtiter plate at a density of 2.5 × 104 cells/well and were transfected with 80-kDa polyethyleneimine (PEI) (Fluka) (5). PEI was used as a 10 mM monomer aqueous stock solution. For each well, a PEI-DNA mixture consisting of 60 μl of PEI stock solution and 2 μg of DNA was incubated separately in 50 μl of 150 mM NaCl. After a 10-min incubation at room temperature, the two solutions were mixed by adding PEI to DNA, followed by immediate vortexing. The PEI-DNA mixture was added to the cell monolayer that had been washed with PBS and then incubated in serum-free medium. The microtiter plate was centrifuged at 380 × g for 5 min. After an incubation of 3 h at 37°C, FCS was added to the cell medium to a final concentration of 10%, and the cells were incubated for an additional 24 h. To isolate stable cell clones, cells were treated with trypsin 48 h after transfection, diluted in selection medium (DMEM, 10% FCS, 600 μg of G418 per ml) and seeded at a density of 6 × 105 cells in 15-cm plates. Neomycin-resistant clones were isolated 11 days after transfection, expanded, and processed for genomic DNA extraction.
Southern blot analysis of transfected clones.
High-molecular-weight DNA was prepared according to standard protocols. Ten micrograms of chromosomal DNA was digested with 40 U of the chosen restriction enzyme in a 0.1-ml volume for 12 h at 37°C. The digested DNA was electrophoresed on a 0.8% agarose gel, processed as previously described (34), and hybridized with random primed 32P-labeled probes. To determine site-specific events, filters were first hybridized with a green fluorescent protein (GFP)- or a neo-specific probe; the hybridized probe was then removed by boiling the filters in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% sodium dodecyl sulfate for 10 min, and the same filters were then washed and hybridized to an AAVS1-specific probe. Filters were exposed to X-ray film with an intensifying screen for 1 week.
In situ hybridization.
A 3.5-kb DNA fragment corresponding to the entire GFP-Neo sequence and an 80-kb AAVS1 DNA (34) were labeled by using the Nick Translation kit (Boehringer Mannheim) according to manufacturer’s instructions and then used as probes in chromosome analysis. The chromosome spreads from selected clones were prepared according to typical cytogenetic techniques (30). Cytogenetic preparations were hybridized to biotin- and digoxigenin-labeled probes as described earlier (34). Images were processed by using Adobe Photoshop on a Power Macintosh computer (Apple).
PCR amplification of the ITR-AAVS1 junction.
Integration of ITR-flanked DNA into AAVS1 site was determined, as previously described (34), by PCR amplification by using nested primer pairs that flank the ITR-AAVS1 junction. The amplified products were run in duplicate on a 1.5% agarose gel and subjected to Southern blot analysis and probed with ITR (nt 4563 to 4670)- and AAVS1 (nt 210 to 1207)-specific probes. For molecular cloning of the amplified DNA fragments, the product of the amplification reaction was purified on a 1.5% agarose gel and subcloned by blunt-end ligation into plasmid pZERO-2.1 (Invitrogen). Sequencing was performed by standard chain termination protocols.
RESULTS
Generation of AAVS1 transgenic rodents.
The observation reported by Berns and coworkers that the signals which direct recombination between the AAV genome and the AAVS1 site are localized within a 33-nt region at the 5′ end of the 8.2-kb AAVS1 DNA (31) prompted us to utilize an AAVS1 fragment of 3.5 kb to generate transgenic rodents. This DNA fragment has been shown to be fully competent for site-specific integration (12) and was thus deemed to be appropriate for preparing transgenic rats and mice for in vivo studies on Rep-mediated site-specific integration.
Four founder rats were obtained from the microinjection of the 3.5-kb DNA fragment in one-cell-stage embryos. However, in three lines the transgene was heavily rearranged mainly at its 5′ end, and these animals were no longer studied (data not shown). Founder rat 15, which did not show any rearrangement of the AAVS1 fragment, transmitted the transgene to its progeny and was subject to further analysis. F1 rats were found to carry approximately seven copies of the transgene arranged in a head-to-tail configuration, as determined by Southern blot analysis of genomic DNA and quantitation by slot blot hybridization (data not shown).
Transgenic mice carrying a single copy of the 3.5-kb AAVS1 DNA fragment were generated by transfection of mouse embryonic stem cells with a construct carrying the neomycin resistance gene and the AAVS1 fragment. These animals were generated with the intent of minimizing potential interfering effects in the analysis of the integration process due to the presence of multiple copies of the target AAVS1 sequence. The neo-mycin-resistant clones were analyzed by dot and Southern blot analysis, and four selected clones were injected into blastocysts of C57BL/6 mice to generate transgenic animals. One of the chimeric animals transmitted the transgene to its progeny. From the pattern of transmission and transgene structure analysis, it was evident that the AAVS1 sequence had integrated into the X chromosome and had not undergone any obvious rearrangements (data not shown).
AAV infection of primary cells.
To verify that the AAVS1 3.5-kb fragment was competent for site-specific integration upon insertion into the genome of the transgenic animals, we chose to analyze AAV targeted insertion into primary cells. Fibroblasts were used in initial experiments on transgenic rats and mice, since human fibroblasts are competent both for infection by wt AAV and for Rep-mediated site-specific integration of AAV-derived vectors (34). Because liver is an important target for gene therapy, primary rat hepatocytes were also tested. Human hepatoma cells Huh-7 are permissive to AAV infection and Rep-mediated integration (35).
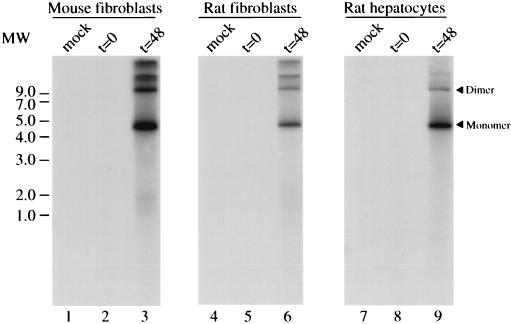
Given the fact that events leading to the integration of the AAV genome may involve the replication of the viral DNA (31), we reasoned that it was important to assess the AAV replication capacity in these cells. The ability of primary fibroblasts and hepatocytes to support AAV replication was assayed by coinfecting the cells with wt AAV and human Ad at an MOI of 100. Hirt supernatants were prepared at 48 h postinfection (19) and fractionated on agarose gel. The DNA was subjected to Southern blot analysis with a probe specific for the p5 region of the AAV genome (Fig. 1). Two bands of approximately 4.7 and 9.6 kb were detected in the infected cells (Fig. 1, lanes 3, 6, and 9) by the Rep-specific probe that correspond to the monomeric and dimeric forms of the AAV genome, respectively. Additional bands of approximately 13 kb or more were detected in the infected mouse and rat fibroblasts which probably correspond to multimeric forms of the AAV genome (Fig. 1, lanes 3 and 6). No viral DNA was detected at t = 0 or in mock-infected cell DNA (Fig. 1, lanes 1, 2, 4, 5, 7, and 8). Although the yield of AAV virus from these infected cells was not determined, the intensity of the bands hybridized by the AAV-specific probe was not dissimilar from that observed upon analysis of human cell lines infected with a similar protocol (data not shown), suggesting that replication of wt AAV in these primary cultures is fairly efficient.
FIG. 1.
Replication of AAV DNA in AAVS1 transgenic primary cultures. Primary fibroblasts and hepatocytes from AAVS1 transgenic rats, as well as mouse embryonic fibroblasts, were infected with wt AAV and at an MOI of 100. After 1 h for virus adsorption (t = 0), the inoculum was removed and the cells were washed and refed with medium. Two days postinfection (t = 48), low-molecular-weight DNA was isolated from uninfected (mock) and infected cells analyzed on a Southern blot with a 32P-labeled probe specific for the Rep coding sequence. The positions of the molecular-weight standards (in kilobases) are indicated.
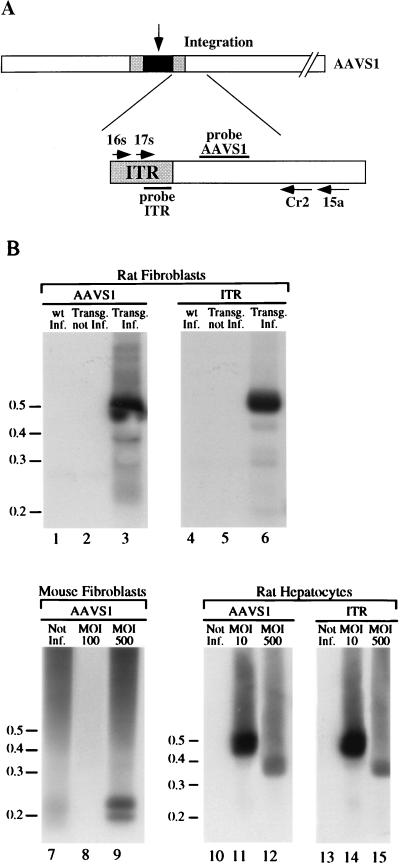
To determine whether the AAV genome integrates into the AAVS1 sequence, fibroblasts from adult transgenics rats and wt littermates were infected with wt AAV at an MOI of 100. The infected cells were passaged four times, and chromosomal DNA was extracted at each passage from an aliquot of the infected cells. Site-specific integration of the viral genome was assessed by PCR amplification of the ITR-AAVS1 junctions by using two sets of nested primers that anneal to the viral ITR and to the AAVS1 region just 3′ of the region, where most of the AAV insertions have been mapped (Fig. 2A) (26, 35, 38, 42). Amplified DNA was loaded onto agarose gel and sequentially analyzed by Southern blotting with AAVS1- and ITR-specific probes. The hybridization pattern of rat fibroblasts infected with AAV and passaged four times is shown in Fig. 2B. A 500-bp band hybridized to both genomic and viral probes (Fig. 2B, lanes 3 and 6), confirming that the amplified DNA corresponds to the viral-chromosomal junction. No DNA fragments corresponding to ITR-AAVS1 junctions were detected in uninfected transgenic fibroblasts (Fig. 2B, lanes 2 and 5) or in infected wt fibroblasts (Fig. 2B, lanes 1 and 4). The same hybridization pattern indicative of integration was detected at each passage of the infected cells (data not shown).
FIG. 2.
PCR amplification of ITR-AAVS1 junctions. (A) Schematic representation of the components of the PCR assay. AAV genome integrated at the AAVS1 is represented by dark box. The gray boxes indicate the AAV ITRs. The open box indicates the AAVS1. The primers used in the PCR analysis are indicated by arrows and are labeled with the associated numbers. Primers 16s and 17s are derived from the AAV ITR. Primers 15a and Cr2 are derived from AAVS1 (34). (B) Site-specific integration of wt AAV genome in transgenic rat and mouse fibroblasts. Rat wt (lanes 1 and 4) and transgenic (lanes 3 and 6) fibroblasts were infected with wt AAV virus at an MOI of 100. Mouse transgenic embryonic fibroblasts were infected with wt AAV at MOIs of 100 (lane 8) and 500 (lane 9). Transgenic rat primary hepatocytes were infected with wt AAV at MOIs of 10 (lanes 11 and 14) and 500 (lanes 12 and 15). Two days postinfection, genomic DNA was prepared and subjected to nested set PCR amplification with AAV-derived and AAVS1-derived primers. As a control, high-molecular-weight DNA from mock-infected rat (lanes 2 and 5) and mouse transgenic embryonic fibroblasts (lane 7) and from rat primary hepatocytes (lanes 10 and 13) were also subjected to the PCR amplification protocol. The products were run on an agarose gel, transferred to nylon membrane, and sequentially hybridized to AAVS1 (lanes 1 to 3 and lanes 7 to 12)- and AAV-specific probes (lanes 4 to 6 and lanes 13 to 15) as described earlier (34). The positions of the molecular-weight standards (in kilobases) are indicated.
Integration of the AAV genome into the AAVS1 site was not limited to the rat but was also observed in transgenic mouse embryonic fibroblasts that had been infected with AAV at an MOI of 500. Two DNA bands of approximately 200 and 220 bp were hybridized by the AAVS1-specific probe (Fig. 2B, lane 9) that were not detected in the mock-infected cells (Fig. 2B, lane 7). In this case, the amplified DNA was not hybridized by the ITR-specific probe, suggesting that partial deletion of the ITR sequence had occurred upon integration (data not shown). Lastly, no amplification of the ITR-AAVS1 junction was obtained upon infection of mouse fibroblasts with an MOI of 100 (Fig. 2B, lane 8).
Finally, rat primary hepatocytes were infected with wt AAV at a MOIs of 10 and 500. Genomic DNA was extracted 48 h postinfection and subjected to PCR amplification of the ITR-AAVS1 junction. Two DNA fragments of approximately 370 and 500 bp were detected with ITR- and AAVS1-specific probes at 48 h postinfection in infected cells with a MOIs of 500 (Fig. 2B, lanes 11 and 14) and 10 (Fig. 2B, lanes 12 and 15), respectively.
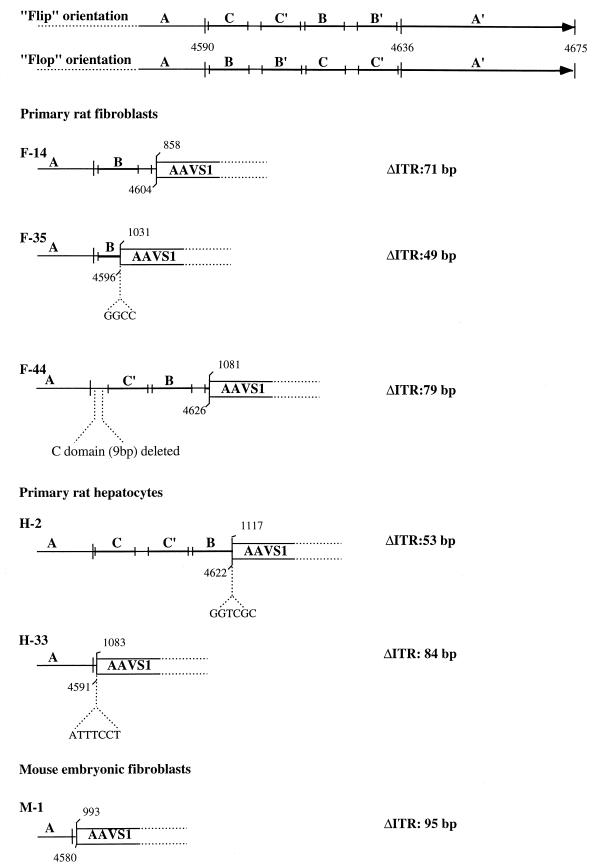
To map the insertions of the AAV genome in the transgenic AAVS1 sequence, some of the PCR products were cloned and sequenced. The junctions obtained from the amplification of DNA from infected rat fibroblasts (clones F-14, F-35, and F-44), hepatocytes (clones H-2 and H-33), and mouse fibroblasts (M-1) are shown in Fig. 3. In clones F-14 and F-35, the insertion of the AAV genome had occurred at nt 858 and 1031 of AAVS1, with concomitant deletions of 71 and 49 nt of the ITR, respectively. In clone F-44 the insertion of the AAV genome was mapped to nt 1081 of AAVS1, with a deletion of 79 nt in the viral terminal repeat. Similarly, the amplified junctions from infected primary hepatocytes showed that the insertion of the AAV genome had occurred at nt 1117 (clone H-2) and 1083 (clone H-33) with deletions of 53 and 84 nt of the ITR, respectively. Interestingly, insertions of 6 and 7 nt were detected at the viral-cellular crossover in junctions H-2 and H33, respectively, whereas an insertion of 4 nt was observed in junction F-35. The cloned junction from infected mouse fibroblasts (clone M-1) indicated that the insertion of the AAV genome had occurred at nt 993 of the AAVS1, with a deletion of 95 nt of the ITR. It is not clear whether some of the deletions detected in the junction sequences could be the result of PCR artifacts or were due to the integration process. Nonetheless, these data indicate that AAV integration can be targeted to the human AAVS1 sequence in mouse and rat cells from transgenic animals. Furthermore, as observed upon AAV infection of human cultured cell lines (25, 26, 27, 35, 38), the AAV genome has integrated in the 5′ region of the AAVS1 site. Thus, these results validate the transgenic lines as a model for site-specific integration.
FIG. 3.
Analysis of PCR-amplified AAV-AAVS1 junctions. Amplified junctions from infected primary fibroblasts (F-14, F-44, and F-35) and hepatocytes (H-2 and H-33) of transgenic rats and from embryonic mouse fibroblasts (M-1) were sequenced and compared with the AAVS1 sequence. The ITR sequence is indicated with the nucleotide numbering for the right terminal repeat. Palindromic sequences within the ITR are indicated by the lettering A, C, C′, B, B′, and A, and the two possible orientations Flip and Flop are indicated (39). For each ITR-AAVS1 crossover sequence analyzed, the amount of viral sequence is indicated by the letters representing the ITR palindromic sequences. The numbers above indicate the nucleotide position on the last identifiable viral and AAVS1 sequence. In smaller letters the amplified sequences that cannot be directly associated with either the ITR or the AAVS1 are shown. The number of deleted bases in each ITR analyzed is indicated at the right.
Site-specific integration of an AAV-derived construct.
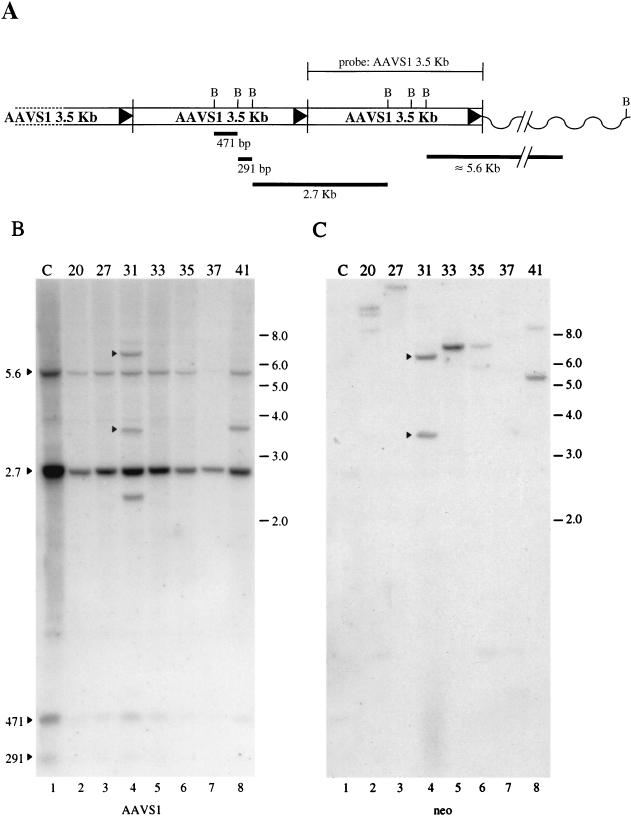
The data reported above do not allow to establish the efficiency of site-specific integration in the transgenic cells. Several investigators have demonstrated that plasmids carrying a selectable marker flanked by the AAV terminal repeats and a Rep-expressing cassette can be targeted to the AAVS1 locus, albeit at a lower frequency than the wt virus (1, 35, 42, 45). Therefore, to further investigate the integration proficiency of the primary cells, rat transgenic fibroblasts were transfected with plasmid pITR(GFP-Neo)P5Rep. This construct carries the GFP gene and the neo gene inserted between the 5′ and 3′ ITRs of AAV, as well as the rep gene under the transcriptional control of the p5 and p19 promoters outside of the two ITRs but next to the 3′ terminal repeat. This plasmid has been shown to drive site-specific integration of the ITR-flanked DNA when transfected into Huh-7 and HeLa cells with integration efficiencies of 12 and 25%, respectively (35). To assess the integration efficiency of pITR(GFP-Neo)P5Rep, rat fibroblasts were transfected with plasmid DNA complexed with PEI. Seven Neor clones were isolated and expanded, and high-molecular-weight DNA was extracted, digested with BamHI, and subjected to Southern blot analysis. BamHI cleaves three times within the AAVS1 sequence (Fig. 4A), but it does not cleave within the ITR-flanked DNA. Thus, the insertion of the ITR-flanked DNA into the AAVS1 site can be easily detected by the altered migration pattern of AAVS1-derived restriction fragments.
FIG. 4.
Southern blot analysis of Neor clones derived from the transfection of transgenic primary rat fibroblasts with plasmid pITR(GFP-Neo)P5Rep. The conditions of transfection and DNA analysis were as described in Materials and Methods. (A) Schematic representation of the predicted head-to-tail configuration of the seven copies of the AAVS1 sequence inserted in rats of line 15. The BamHI restriction sites and the expected restriction fragments are indicated. (B) Hybridization to an AAVS1-specific probe. (C) Same membrane after rehybridization to a neo-specific probe. The bands that are annealed to both probes are considered to be indicative of site-specific integration and are indicated with an arrowhead. Mock-transfected cell DNA sample is indicated with the letter “C”. The number referring to each clone is also shown. The positions of the molecular-weight standards (in kilobases) are indicated.
A diagram of the head-to-tail configuration of the multiple copies of the AAVS1 sequence inserted into rats of line 15 and the size of the expected restriction products is shown in Fig. 4A. The pattern of hybridization of the AAVS1 probe on transgenic rat genomic DNA cut with BamHI consists of four bands (Fig. 4B, lane 1). The smaller bands of 471 and 291 bp correspond to the internal fragments, whereas the 2.7-kb band corresponds to the junction between the 3′ region of one AAVS1 sequence and the 5′ portion of the adjacent AAVS1 copy. The larger, 5.6-kb band represents the fusion between the rightmost AAVS1 monomer and the adjacent mouse genome. The opposite 5′ genomic flanking sequence was not detectable upon digestion with BamHI in this rat cell line. Hybridization of the AAVS1-specific probe to DNA from G418 selected clones shows that the probe hybridizes to the same four DNA fragments (Fig. 4B, lanes 2 to 8). In clone 31, the AAVS1 probe hybridized to three additional bands of approximately 6.0, 3.7, and 2.5 kb (Fig. 4B, lane 4), whereas in clone 41 the AAVS1 probe annealed to an additional band of 3.7-kb (Fig. 4B, lane 8). To verify that these bands were due to site-specific integration, the same blot was hybridized with the neo-specific probe (Fig. 4C). The neo probe hybridized to the 6.0- and 3.7-kb bands, suggesting that in clone 31 the neomycin gene had integrated into the AAVS1 site (Fig. 4C, lane 4). Interestingly, only the 6.0-kb band was detected by the GFP probe (data not shown). In contrast, the new band present in clone 41 was not detected by either the neo- (Fig. 4C, lane 8) or the GFP-specific probes, suggesting that this band corresponds to a rearranged AAVS1 sequence. Additionally, in the other clones analyzed, the neo-specific probe hybridized to one or more bands, indicating that the integration of the neomycin resistance marker had occurred at sites other than AAVS1 (Fig. 4C, lanes 2 to 3 and lanes 5 to 7).
In agreement with previous reports, targeted insertion into the AAVS1 site was limited to the ITR-flanked DNA, with the concomitant exclusion of the remaining part of the transfected plasmid (1, 35). In fact, upon hybridization of the genomic blots with a p5-specific probe, no specific band could be detected. Additionally, PCR amplification of the rep gene with sequence-specific primers also failed to reveal its presence (data not shown).
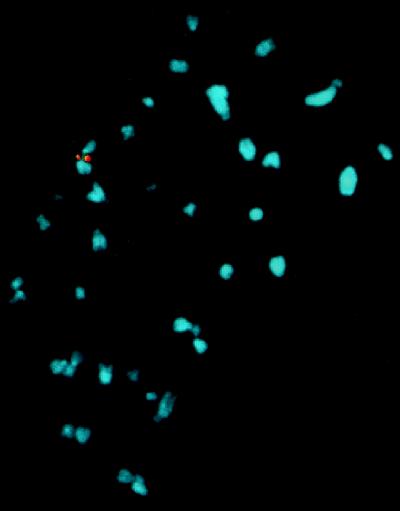
Site-specific integration of the AAV-derived plasmid in AAVS1 was confirmed by fluorescent in situ hybridization (FISH) analysis of cytogenically prepared rat fibroblasts. Metaphase spreads were hybridized with AAVS1- and GFP-Neo-specific probes. Figure 5 demonstrates the presence of the GFP-Neo sequence on each sister chromatid of a single chromosome of fibroblast cell clone 31. Additionally, the same region of the chromosome was hybridized by the AAVS1-specific probe. No GFP-Neo hybridization was detected with mock-transfected cells (data not shown). Thus, the data from the FISH analysis provides further evidence for the targeted insertion of the ITR-GFP-Neo DNA fragment into the AAVS1 site of the transgenic fibroblasts.
FIG. 5.
In situ hybridization of metaphase chromosomes from selected clone 31. Chromosome preparations were hybridized with a GFP-Neo (in red)- or AAVS1 (in yellow)-specific probes as described earlier (34).
The detection of site-specific integration in one of seven transgenic-fibroblast clones analyzed (14%) suggests a frequency similar to that obtained with human hepatoma cells (35). Taken together, these results indicate that the AAVS1 3.5-kb fragment present in transgenic rat line 15 behaves in a manner functionally similar to that of the AAVS1 locus in human cells.
In vivo integration of AAV genomic DNA.
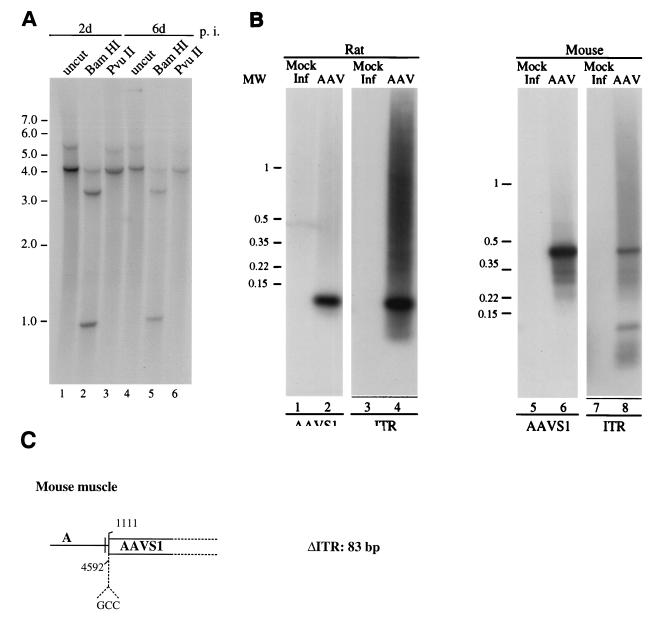
Site-specific integration of AAV genomic DNA in vivo was assessed by injection of wt virus into the left quadriceps muscle of transgenic rats and mice. The quadriceps muscle was chosen as a target in view of the ease of manipulation of this tissue and of the high efficiency of transduction of these cells observed upon injection of recombinant AAV virus (7, 9, 18, 24, 43, 50). Six transgenic rats were injected with 108 i.u. of wt AAV, and eight transgenic mice were injected with a dose of 5 × 107 i.u. To confirm AAV infection of the injected tissue and to assess viral genome integration, high-molecular-weight DNA was extracted at 2, 6, 30, and 75 days postinfection. The presence of AAV genome was confirmed by PCR amplification of the rep gene with rep-specific primers (data not shown). To analyze the structure of the AAV genome in the infected tissue, high-molecular-weight DNA was analyzed by Southern blot. For this purpose, injected muscle DNA was either run on agarose gel undigested or after restriction with BamHI or PvuII and then subjected to Southern blot analysis with an AAV-specific probe. BamHI cleaves once within the AAV genome, generating two bands of 3.7 and 1.0 kb, whereas PvuII does not cut AAV. The latter enzyme was chosen to identify the presence of integrated forms of the AAV DNA. As shown in Fig. 6A, two bands of approximately 3.7 and 1.0 kb, respectively, hybridized with the AAV-specific probe in both samples digested with BamHI (Fig. 6A, lanes 2 and 5). A band of approximately 4.7 kb was detected at both 2 and 6 days postinfection which probably corresponds to undigested AAV genomic monomers, since it was also detected in the PvuII-digested samples. Additionally, a 5.5-kb band, probably corresponding to nonintegrated form of viral DNA, was detected in both 2 and 6 days postinfection in the undigested DNA samples, as well as the PvuII-restricted DNA (Fig. 6A, lanes 1, 3, 4, and 6). No association of AAV sequences with high-molecular-weight DNA was detected, nor were smaller DNA forms consistent with the presence of single-stranded viral DNA detected. Thus, essentially a complete conversion of the single-stranded viral genome to double-stranded DNA had occurred within 48 h of injection. A similar hybridization pattern was detected upon analysis of injected muscle murine DNA, indicating that the wt AAV virus had undergone a similar conversion process upon infection of the transgenic mouse quadriceps muscle (data not shown).
FIG. 6.
Analysis of AAV genomic DNA from the injected muscle of transgenic rodents. (A) Southern blot analysis of total muscle DNA from injected transgenic rats. Total cellular DNA was prepared from quadriceps of injected rats 2 or 6 days postinfection. DNA samples (10 μg) were resolved on agarose gel either uncut or after restriction with BamHI or PvuII. The gel was transferred to a nylon membrane and hybridized with a probe specific for the rep gene. Uninjected muscle DNA did not demonstrate any detectable hybridization (data not shown). (B) PCR amplification of the ITR-AAVS1 junction from injected muscle DNA. Total muscle DNA was prepared from injected transgenic rat and mouse tissue 75 and 6 days postinfection, respectively. DNA was subjected to PCR amplification of the viral-chromosomal junction, and amplified DNA was sequentially hybridized to an AAVS1 (lanes 1, 2, 5, and 6)- and an ITR-specific probe (lanes 3, 4, 7, and 8) as described previously (34). The positions of the molecular-weight standards (in kilobases) are indicated. (C) Schematic representation of the sequence of the ITR-AAVS1 junction derived from injected mouse muscle DNA.
Integration of the AAV genome into the AAVS1 site was analyzed by PCR amplification of ITR-AAVS1 junctions. No bands indicative of integration into the AAVS1 site were detected in the injected rat samples at 6 and 30 days postinfection (data not shown). In contrast, a band of approximately 150 bp amplified from the hindleg muscle DNA extracted at 75 days postinfection from one of the two injected rats hybridized to both the AAVS1- and the ITR-specific probes (Fig. 6B, lanes 2 and 4). The ITR-chromosomal junction was also amplified from injected muscle DNA of one of the two transgenic mice at 6 days postinfection. The amplified 500-bp DNA band hybridized to the AAVS1- and ITR-specific probes (Fig. 6B, lanes 6 and 8). A similar hybridization pattern was detected with muscle DNA at 30 days postinfection (data not shown).
To further confirm that AAV genome had integrated the AAVS1 site, PCR products of injected mice were cloned and sequenced. As shown in Fig. 6C, one of the insertions of the viral DNA into the mice genome had occurred at nt 1111 of AAVS1, with a concomitant deletion of 83 bp in the viral terminal repeat and an insertion of 3 nt (GCC) at the viral-cellular junction. Thus, these results indicate that the AAV genome could integrate into the AAVS1 sequence present in the transgenic rat and mice DNA also upon in vivo infection of muscle cells.
DISCUSSION
We have developed transgenic rats and mice to study Rep-mediated site-specific integration. Targeted integration of the AAV genome occurs in transgenic primary cells with the same molecular characteristics and a similar efficiency as in human cell lines. The results also demonstrate that integration of the AAV genome into the AAVS1 site occurs in vivo upon infection of the transgenic animals.
The AAVS1 3.5-kb DNA fragment rearranged upon generation of various transgenic rat lines. Only one founder animal was obtained that carried several copies of the unrearranged 3.5-kb AAVS1 sequence in a head-to-tail configuration. Similar rearrangements of the AAVS1 DNA fragment were observed upon manipulation of the ES cell clones (data not shown). The recombinogenic instability of the AAVS1 sequence in eukaryotic cells has been described by Berns and coworkers with an episomal EBV shuttle vector. The signal associated with episomal DNA rearrangements was mapped to nt 209 to 326 of AAVS1 (31). This sequence is located in close proximity to the RBS and trs, and it carries a motif, M26, which has been characterized as an enhancer of meiotic recombination in fission yeast (14, 41). It is not clear whether the same region is responsible for the AAVS1 instability observed in the generation of transgenic animals. Nonetheless, it is likely that cellular factors involved in the insertion of genes into the DNA of transgenic animals may interact with the AAVS1 DNA, determining its rearrangement during the transgene integration process. No rearrangement has been observed throughout the amplification of five generations of rat and moose lines, suggesting that the AAVS1 sequence is stabilized once it is inserted into the DNA of transgenic animals.
Analysis of primary cultures infected with wt AAV or transfected with plasmid pITR(GFP-Neo)P5Rep shows that the AAVS1 DNA fragment present in the transgenic cells can be recognized as target for Rep-mediated integration of an ITR-flanked DNA. Integration of AAV genomic DNA into the AAVS1 site of the transgenic cells was mapped by PCR amplification of the ITR-AAVS1 junction to the same locus of integration, i.e., near nt 1000, observed in latently infected and transfected human cells (26, 34, 35, 38, 42). Interestingly, no relevant differences were noted between the junctions obtained from infected mouse and rat fibroblasts, suggesting that the presence of multiple copies of the 3.5-kb DNA fragment in the transgenic rat did not influence the integration of the AAV genome. Additionally, the partial deletion of the ITRs, the insertion of a short stretch of nucleotides in between the ITR-cellular junctions (Fig. 3), and the observation that the transfection of plasmid pITR(GFP-Neo)P5Rep in transgenic rat fibroblasts results in the integration of the ITR-GFP-Neo DNA fragment into the AAVS1 site (Fig. 4) strongly mirrors what has been observed in human cell lines where Rep-mediated integration has been analyzed (35, 51). Lastly, Southern blot analysis of the selected fibroblast clones where several bands hybridized only to the AAVS1 probe indicate that rearrangement of AAVS1 as a consequence of rep expression may have taken place (Fig. 4), much as has been observed upon transfection of AAV-derived plasmids and subsequent selection of neomycin-resistant 293 clones (45).
ITR-AAVS1 junctions could not be detected by PCR in mouse primary fibroblasts infected at an MOI of 100, but they could be detected in primary rat fibroblasts infected an MOI of 100 and in rat hepatocytes infected at a MOIs of 10 and 500. Although it is possible that a certain virus load must be achieved in the infected cell to obtain successful site-specific integration, it is more likely that the lack of detection of integrated virus is due to the sensitivity of the PCR amplification protocol. Nonetheless, this observation is in agreement with results reported by Berns and coworkers showing that the AAVS1 3.5-kb DNA contains the minimal sequence required for integration (12). These results suggest that the AAVS1 DNA fragment is functional for targeted integration, even in cells of nonhuman origin, and that the host factors that are required for Rep-mediated targeting of the ITR-flanked DNA to the AAVS1 site are also present in rodent cells. Finally, the differences in integration efficiency observed between the transgenic primary cells and an EBV shuttle vector carrying the same AAVS1 DNA fragment (12) suggest that the chromatin context in which the 3.5-kb DNA fragment is inserted may play an important role in Rep-mediated targeting.
A proposed model for Rep-mediated site-specific integration requires the RBS and the trs and involves multiple strand switching during novel DNA strand synthesis within a Rep-mediated complex formed between ITR-flanked DNA and the AAVS1 sequence (31). The mechanism of strand switching must be, however, quite precisely regulated, since the viral-AAVS1 junctions are mostly clustered within a 600-bp span (26, 38). The localization of the ITR-AAVS1 junctions in the infected transgenic cells somehow contrasts with the finding that the ITR-AAVS1 junctions are clustered more closely to the RBS around position 400 when the 3.5-kb DNA fragment is located on an episomal shuttle plasmid (11). One possible explanation is that Rep-mediated targeted integration could involve the formation of a synaptic complex comprising several auxiliary factors, as observed for other site-specific recombinases in prokaryotes. The assembly of such a complex with the target DNA could result in a highly organized arrangement of DNA strands only when AAVS1 is in a chromosomal structure and not when it is placed on an episome. It has been recently reported that the high-mobility-group 1 (HMG1) nonhistone chromosomal protein, which is ubiquitous in eukaryotic cells, physically interacts with the Rep polypeptide, promoting the formation of Rep-DNA complexes and stimulating the activity of Rep in strand- and site-specific cleavage of DNA (8). Such interaction could contribute, at least in part, to the correct targeting of the AAV genomic DNA.
We observed that wt viral DNA was readily converted to double-stranded monomer by 2 days after the infection of muscle cells both in transgenic rats and mice (Fig. 6). Thus, wt AAV differs from recombinant AAV viruses, since the latter have been shown to persist for at least 30 days in mouse muscle as single-stranded DNA and are subsequently converted into a high-molecular-weight form without an apparent double-stranded episomal intermediate (24, 32, 50). This difference is very likely due to the presence of the Rep coding sequence which may be expressed upon infection and determine a rapid conversion of the single-stranded DNA into a double-stranded monomer.
We also observed that Rep-mediated integration occurs following AAV injection into muscle cells, although the efficiency was low. In fact, the viral DNA was not detectable as high-molecular-weight species in any of the samples analyzed (Fig. 6); rather, PCR amplification of the ITR-chromosomal junction was necessary in order to obtain evidence of integration. Furthermore, targeted integration was not detected in all of the injected animals. The variability in detection of the viral-AAVS1 junction is not unusual and may be due to the sensitivity of the PCR protocol or to the amount of virus injected. However, it may also reflect variations in the infection efficiency of each animal. We cannot exclude that other factors may have influenced the efficiency of integration into the DNA of injected muscle cells. These may include possible differences in Rep activity in primary cells in vitro and in vivo due to the level of transcription from the p5 promoter and/or to the differential abundance of cellular accessory factors required for integration. Additionally, although we have no evidence of the type of cells where integration has occurred, the observation that targeted insertion can be observed in muscle cells and primary hepatocytes suggests that integration is also possible in quiescent cells. This observation is in agreement with the recent report of Kay and coworkers, who described the random in vivo integration of a recombinant AAV vector in mouse liver (32). We are currently examining the AAV integration proficiency of other organs and, in particular, the liver to assess whether the potential differences exist from one tissue to the other that may influence in vivo the targeted integration mediated by Rep.
A better understanding of the molecular mechanisms that regulate targeted integration of the AAV genome is fundamental for the evaluation of the potential of site-specific integration for human gene therapy. The results presented in this work indicate that the transgenic rodent models we have developed for site-specific integration are functional and thus can be exploited to study the mechanism of Rep-mediated targeting to the AAVS1 site both in vitro and in vivo.
ACKNOWLEDGMENTS
We thank F. Graham, G. Migliaccio, A. Nicosia, and F. Palombo for critical review and helpful suggestions. We also thank T. Alonzi for his involvement in the initial phase of this work and M. Emili for graphics.
REFERENCES
- 1.Balagué C, Kalla M, Zhang W W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett J S, Samulski J R. Methods in molecular medicine. In: Robbins P D, editor. Gene therapy protocols. Totowa, N.J: Humana Press, Inc.; 1997. pp. 25–40. [Google Scholar]
- 3.Berns K I. Parvoviridiae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 4.Berry M, Friend D. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structure study. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boussif O, Zanta M A, Behr J-P. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Ther. 1996;3:1074–1080. [PubMed] [Google Scholar]
- 6.Cheung A K, Hoggan M D, Hauswirth W W, Berns K I. Integration of the adeno-associated virus genome into cellular DNA of latently infected human Detroit 6 cells. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark K R, Sferra T J, Johnson P R. Recombinant adeno-associated viral vectors mediate long-term expression in muscle. Hum Gene Ther. 1997;8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 8.Costello E, Saudman P, Wincour E, Pizer L, Beard P. High-mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity, and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher K J, Joos K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 10.Friedman-Einat M, Grossman Z, Mileguir F, Smetana Z, Ashkenazi M, Barkai G, Varsano N, Glick E, Mendelson E. Detection of adeno-associated virus type 2 sequences in the human genital tract. J Clin Microbiol. 1997;35:71–78. doi: 10.1128/jcm.35.1.71-78.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraud C, Winocour E, Berns K I. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman S, Xiao X, Donahue R E, Moulton A, Miller J, Walsh C, Young N S, Samulski R J, Nienhuis A W. Recombinant adeno-associated virus-mediated gene transfer into hemapoietic progenitor cells. Blood. 1994;84:1492–1500. . (Erratum, 85:862, 1995). [PubMed] [Google Scholar]
- 14.Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;136:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman Z, Mendelson E, Brok-Simoni F, Mileguir Y, Leitner G, Rechavi G, Ramot B. Detection of adeno-associated virus type 2 in human peripheral blood cells. J Gen Virol. 1992;73:961–966. doi: 10.1099/0022-1317-73-4-961. [DOI] [PubMed] [Google Scholar]
- 16.Gu H, Zou Y-R, Rajewski K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 17.Han L, Parmley T H, Keith S, Kozlowski K J, Smith L J, Hermonat P L. High prevalence of adeno-associated virus (AAV) type 2 rep DNA in cervical material: AAV may be sexually transmitted. Virus Genes. 1996;12:47–52. doi: 10.1007/BF00370000. [DOI] [PubMed] [Google Scholar]
- 18.Herzog R W, Hagstrom J N, Kung S-H, Tai S J, Wilson J, Fisher K J, High K. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt B. Selective extraction of polyoma DNA from infected mouse cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 20.Hogan B, Costantini F, Lacey E. Manipulating the mouse embryo. Plainview, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 21.Im D-S, Muzyczka N. The AAV origin-binding protein Rep68 is an ATP dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 22.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep68, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns W G, Afione S A, Fulmer S B, Pang M G, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 24.Kessler P D, Podsakoff F M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotin R M, Berns K I. Organization of adeno-associated virus DNA in latently infected Detroit 6 cell line. Virology. 1989;170:460–467. doi: 10.1016/0042-6822(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 26.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S K, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laughlin C A, Christine B, Cardellichio B, Doon H C. Latent infection of KB cells with adeno-associated virus type 2. J Virol. 1988;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence J B, Villnave C A, Singer R H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 31.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao C H, Snyder R O, Schowalter D B, Patjin G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. [DOI] [PubMed] [Google Scholar]
- 33.Muro-Cacho C A, Samulski R J, Kaplan D. Gene transfer in human lymphocytes using a vector based on adeno-associated virus. J Immunother. 1992;11:231–237. doi: 10.1097/00002371-199205000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G, La Monica N. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieroni L, Fipaldini C, Monciotti A, Cimini D, Sgura A, Fattori E, Epifano O, Cortese R, Palombo F, La Monica N. Targeted integration of AAV derived plasmids in transfected human cells. Virology. 1998;249:249–259. doi: 10.1006/viro.1998.9332. [DOI] [PubMed] [Google Scholar]
- 36.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X-S, Srivastava A. Lack of site-specific integration of the adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 37.Samulski R J, Chang L-S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Gen Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 40.Samulski R J, Berns K I, Tan M, Muzyczka N. Cloning of AAV into pBR322: rescue of intact viruses from the recombinant plasmid into human cells. Proc Natl Acad Sci USA. 1982;109:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuchert P, Langsford M, Kaslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shelling A N, Smith M. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 43.Snyder R O, Spratt S K, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava A, Lubsy E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobiasch E, Rabreau M, Geletneky K, Larue-Charlus S, Severin F, Becker N, Schlehofer J R. Detection of adeno-associated virus DNA in human genital tissue and in material from spontaneous abortion. J Med Virol. 1994;44:215–222. doi: 10.1002/jmv.1890440218. [DOI] [PubMed] [Google Scholar]
- 47.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep proteins. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walz C, Deprez A, Dupressoir T, Durst M, Rabreau M, Schlehofer J R. Interaction of human papillomavirus type 16 and adeno-associated virus type 2 co-infecting human cervical epithelium. J Gen Virol. 1997;78:1441–1452. doi: 10.1099/0022-1317-78-6-1441. [DOI] [PubMed] [Google Scholar]
- 49.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vectors. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]