Abstract
The genetic characteristics of rectal neuroendocrine tumors (R-NETs) were poorly understood. Depicting the genetic characteristics may provide a biological basis for prognosis prediction and novel treatment development. Tissues of 18 R-NET patients were analyzed using whole-exome sequencing. The median tumor mutation burden (TMB) and microsatellite instability (MSI) were 1.15 Muts/MB (range, 0.03–23.28) and 0.36 (range, 0.00–10.97), respectively. Genes involved in P53 signaling, PI3K-AKT signaling, DNA damage repair, WNT signaling, etc. were frequently altered. Higher TMB (P = 0.078), higher CNV (P = 0.110), somatic mutation of CCDC168 (P = 0.049), HMCN1 (P = 0.040), MYO10 (P = 0.007), and amplification of ZC3H13 (P < 0.001) were associated with shorter OS. Potentially targetable gene alterations (PTGAs) were seen in 72% of the patients. FGFR1 amplification (22%) was the most common PTGA followed by BARD1 and BRCA2 mutation (each 17%). As for gene variations associated with the efficacy of immune checkpoint blockade (ICB), FAT1 alteration (39%) and PTEN depletion (28%) were commonly observed. In conclusion, frequently altered oncogenic pathways might contribute to the development and progression of R-NETs. Gene alterations significantly associated with prognosis might be potential novel targets. Targeted therapy might be a promising strategy as targetable alterations were prevalent in R-NETs. FAT1 alteration and PTEN depletion might be the main genetic alterations influencing the response to ICB besides overall low TMB and MSI in R-NETs.
Keywords: rectal neuroendocrine tumors, whole-exome sequencing, prognosis, targeted therapy, immune checkpoint blockade
Introduction
Neuroendocrine neoplasms (NENs) are a group of rare malignancies with strong heterogeneity, which arise from neuroendocrine cells distributed throughout the body and produce peptide hormone and biogenic amine (Klöppel 2017). NENs can affect all parts of the gastrointestinal tract. Rectal neuroendocrine neoplasms (R-NENs) are the most prevalent NENs and rank second for the incidence rate of gastrointestinal NENs, with a rate of 1.04/100,000 (Dasari et al. 2017). According to the differentiation, they can be classified as well-differentiated rectal neuroendocrine tumors (R-NETs) and poorly differentiated neuroendocrine carcinomas (R-NECs). In recent years, several drugs or treatment strategies have been developed for the treatment of R-NETs, such as somatostatin analogs (SSAs) (Rinke et al. 2009), mTOR inhibitors (Everolimus) (Yao et al. 2016), vascular-targeting agents (Surufatinib) (Xu et al. 2020), and peptide receptor radionuclide therapy (PRRT) (Strosberg et al. 2017). Nevertheless, the treatment options for R-NETs are still limited, especially for advanced patients with drug resistance after a multi-line of treatment. Advanced patients usually suffer from liver metastasis and bear a heavy financial burden with low quality of life. Thus, it is necessary to develop novel drugs for these patients.
So far, the genomic characteristics of R-NETs are not well understood. The current genomic research for NENs mainly focused on the pancreas (Jiao et al. 2011, Scarpa et al. 2017), small intestine (Banck et al. 2013, Francis et al. 2013), and lung (Gabriel et al. 2020), while the genomic data based on whole-exome sequencing in R-NETs was absent. Evidence has shown that the sensitivity to mTOR inhibitors of R-NETs is higher than that of NETs from other primary sites (Singh et al. 2018), suggesting the unique molecular characteristics of R-NETs. However, most of the previous genomic studies have integrated the genomic data of gastroenteropancreatic NENs (Mafficini & Scarpa 2019, Puccini et al. 2020, Venizelos et al. 2021), while genomic studies analyzing R-NETs alone are rare, which leads to a lack of understanding of the unique genomic characteristics of R-NETs and further limits the drug development for individualized treatment. In this study, we performed whole-exome sequencing for 18 Chinese R-NET patients to comprehensively depict the genetic characteristics based on somatic gene mutation and copy number variation (CNV) in Chinese R-NETs. We investigated frequently altered signaling pathways, variations potentially affecting the prognosis, potentially targetable gene alterations (PTGAs), and gene alterations potentially influencing the efficacy of immune checkpoint blockade (ICB).
Materials and methods
Patient selection
Between 2015 and 2021, a total of 66 patients with R-NETs at China-Japan Friendship Hospital in Beijing underwent endoscopic or surgical resection. After excluding patients without sufficient tumor samples or complete clinical/follow-up information, a total of 18 R-NET patients were finally included in this study. Before the commencement of the study, all experimental protocols have been approved by the Ethics Committee of China-Japan Friendship hospital (2019-24-K18-1), and all patients have signed informed consent before collecting tissues or blood. The study was conducted following the declaration of Helsinki.
Clinicopathological information collection and follow-up
All hematoxylin and eosin-stained tumor sections were examined by gastrointestinal pathologists to confirm the diagnosis and evaluate histopathological features, including tumor size, mitotic count, tumor grade, depth of invasion, lymphatic invasion, vascular invasion, perineural invasion, and status of the resection margin. Clinical information, including age, gender, tumor grade, tumor stage, lymph node involvement, treatment, and overall survival (OS) were collected. The patients were followed up by outpatient visits, ward hospitalization, or telephone inquiry until death occurred or the end of the study in May 2022.
Sample collection
Eighteen primary R-NET samples including 3 fresh tumor tissues and 15 formalin-fixed paraffin embedding (FFPE) specimens and corresponding blood samples were collected. FFPE or fresh tumor samples were used for whole-exome sequencing with corresponding adjacent normal tissues or peripheral blood lymphocytes used as control. Tumor samples with more than 10% of tumor content were eligible for sequencing, while patients with tumor content of less than 10% were asked for resampling. Two samples were collected after treatment (sample 9 was collected from a patient who had received capecitabine, and sample 12 was from a patient who had received surufatinib). Other samples were all collected before systemic medical treatment.
Sample processing and whole-exome sequencing
For FFPE samples, genomic DNA was extracted from ten tumor sections of 5 μm using QIAamp DNA FFPE Tissue Kit (Qiagen GmbH). For blood samples, DNA was extracted from peripheral blood lymphocytes (PBL) using a blood genomic DNA Extraction Kit (centrifugal column method) (Tiangen Biotech (Beijing) Co., Ltd). For fresh tumor tissues, DNA was extracted using MagPure FFPE DNA LQ Kit F (Magen Biotechnology Co., Ltd). DNA was quantified using the Qubit 4.0 fluorometer and Qubit dsDNA HS Analysis Kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. 5× WGS fragmentation mix, 10× WGS fragmentation buffer, T4 DNA Ligase, 5× rapid ligation buffer (Tiangen Biotech (Beijing) Co., Ltd.), KAPA HiFi HotStart Readymix, KAPA Library Amplification Primer Mix (Roche molecular systems, Inc.) were used for library construction. Then, the library was purified by KAPA HyperPure Beads (Roche molecular systems, Inc.). Hybridization capture was carried out using the combined probe of KAPA HyperExplore Max and KAPA HyperExome (Roche molecular systems, Inc.). PCR amplification was carried out for seven to eight cycles according to the input amount of the hybridization library. DNA sequencing was performed on Illumina Novaseq 6000 (Illumina, Inc.) system with an average depth of 650×.
Genomic data processing and analysis
The Fastp software (v0.23.0) (https://github.com/OpenGene/fastp) was used to filter the raw data. The filtered reads were further aligned with the reference genome hg19 (GRCh37) using BWA MEM (v0.7.15-rll40) (http://bio-bwa.sourceforge.net/). Sambamba (V 0.6.6) (https://github.com/lomereiter/sambamba/issues) was used to remove duplicated results after alignment. The Baserecalibrator program in gatk (v4.1.1.0) was used to calculate the error distribution of base mass. Applybqsr program in gatk (v4.1.1.0) was used to correct the base mass. Mutect2 tool in gatk (v4.1.1.0) was used for somatic mutation (SNVs, Indels) analysis. The default parameter was used for filtering SNVs and Indels. Cnvkit (v0.9.6) was used for CNV analysis (https://cnvkit.readthedocs.io/en/stable/). Further annotation of mutation sites was performed by Annovar (https://annovar.openbioinformatics.org/en/latest/). Msisensor (v0.5) (https://github.com/ding-lab/msisensor) is used for microsatellite instability analysis which counted the number of known microsatellite loci that were altered by somatic insertion or deletion. TMB was calculated by counting the number of nonsynonymous somatic mutations within an average of 1 million bases in the tumor genome. TMB-high (TMB-H) was defined as TMB more than or equal to ten mutations/MB. MSI-high (MSI-H) was defined as the MSI index more than or equal to ten. R package deconstructSigs was used for the analysis of mutational signatures based on the signatures reported by Alexandrov et al. (Alexandrov et al. 2013). Mutation and CNV profiles were drawn by the R package Complexheatmap. Five data sources: Cancer Gene Census (https://cancer.sanger.ac.uk/census/), Integrative Onco Genomics (https://www.intogen.org/search), publication by Bert Vogelstein (Vogelstein et al. 2013), significantly mutated genes from the The Cancer Genome Atlas (TCGA) Pan-cancer analysis (Kandoth et al. 2013), and driver genes derived from integrated analysis (Tamborero et al. 2013) were used to screen out known driver genes. Hallmark gene set in Molecular signatures database (Msigdb) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for signaling pathway analysis. The Gene ontology (GO) database was interrogated for functional enrichment analysis by R package clusterProfiler. Potentially targetable gene alterations were interrogated from Oncology Knowledge Base (OncoKB) (https://www.oncokb.org) (Chakravarty et al. 2017).
Statistical analysis and data visualization
Statistical analysis and data visualization were performed using R software (v 4.1.2). For continuous normal distribution variables, the mean ± s.d. was calculated with the Student’s t-test applied to show the significance of the difference. For categorical variables, the percentage was calculated with Fisher’s exact test applied to determine the significance of the difference. Survival analysis was performed by R package survival and survminer. The Kaplan–Meier (KM) method was used to estimate the probability of OS, with the log-rank test performed to evaluate significance. All tests were two-sided. P < 0.05 was considered statistically significant, while P < 0.1 was regarded as a trend.
Results
Patient characteristics
Eighteen R-NET patients were included in this study. The clinical information of the included patient is summarized in Table 1. There were 4 females and 14 males. The mean age at the time of sampling was 57.56 (range, 37–78) years (Table 1). There were seven patients in grade 1, ten patients in grade 2, and one patient in grade 3 (Table 1). Most tumors (12/18, 66.67%) had sizes more than 1 cm in the maximum diameters. Six patients (33.3%) had localized or regional disease, while 12 patients (66.7%) presented distant metastasis. Three patients (16.7%) received endoscopy, six patients received (33.3%) radical surgery, and nine patients (50%) received systemic therapy (including one patient treated with capecitabine, five patients with SSAs, three patients with anti-angiogenesis tyrosine kinase inhibitors) as the first treatment, respectively (Table 1).
Table 1.
Clinical characteristics of 18 RNET patients.
| Characteristics | NET | |
|---|---|---|
| (n = 18) | ||
| Sex (%) | Female | 4 (22.2) |
| Male | 14 (77.8) | |
| Age (mean (s.d.)) | 57.56 (11.55) | |
| Tumor size (median (IQR)) | 1.70 (1.30, 2.25) | |
| Ki67 (median (IQR)) | 3.00 (1.25, 8.75) | |
| T stage (%) | T1 | 6 (33.3) |
| T2 | 3 (16.7) | |
| T3 | 0 (0.0) | |
| T4 | 4 (22.2) | |
| Tx | 5 (27.8) | |
| N stage (%) | N0 | 6 (33.3) |
| N1 | 11 (61.1) | |
| Nx | 1 (5.6) | |
| M stage (%) | M0 | 6 (33.3) |
| M1 | 12 (66.7) | |
| Stage (%) | I | 3 (16.7) |
| III | 3 (16.7) | |
| IV | 12 (66.7) | |
| Grade (%) | G1 | 7 (38.9) |
| G2 | 10 (55.6) | |
| G3 | 1 (5.6) | |
| Treatment (%) | Chemotherapy | 1 (5.6) |
| Endoscopy | 3 (16.7) | |
| SSA | 5 (27.8) | |
| Surgery | 6 (33.3) | |
| TKI | 3 (16.7) |
R-NET, rectal neuroendocrine tumor; SSA, somatostatin analog; TKI, tyrosine kinase inhibitor.
Somatic mutation characteristics
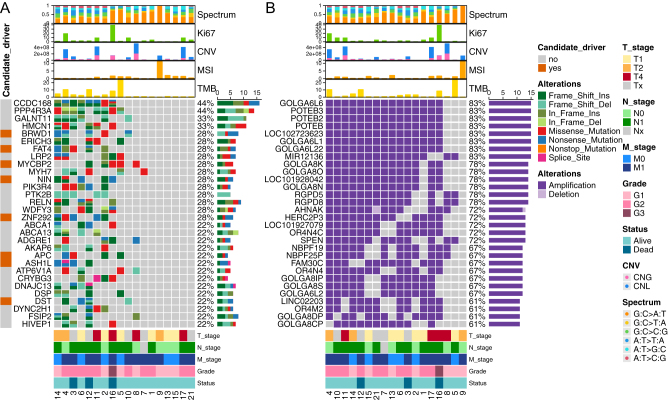
The whole-exome sequencing of 18 R-NETs found a median of 45 single nucleotide variants (SNVs) and 19 small insertion deletions (Indels) for somatic nonsynonymous mutation. Figure 1A showed the top 30 high-frequency gene mutations. CCDC168 and PPP4R3A mutation were the somatic mutations with the highest frequency which were both detected in eight (44.4%) patients. GALNT11 and HMCN1 mutations were detected in six (33.3%) patients. Other high-frequency gene mutations included BRWD1, ERICH3, FAT4, LRP2, MYCBP2, MYH7, NIN, PIK3R4, PTK2B, RELN, WDFY3, and ZNF292 (for each gene, n = 5, 27.8%). Notably, APC, the well-recognized frequently mutated genes in colorectal adenocarcinoma (COREAD) and colorectal NEC (CR-NEC) was mutated in four patients (22.2%) in our R-NET cohort (Fig. 1A). The most common mutational spectrum was G: C>A: T transition (Fig. 1, Supplementary Fig. 1B, see section on supplementary materials given at the end of this article). For mutational signature, signature 1A was the most prevalent (13/18, 72.2%) followed by signature 3 (9/18, 50%) and signature 15 (6/18, 33.3%). Patients were clustered into four groups mainly based on the signature 1A, signature 3, and signature 15. The fourth cluster exhibited few of these three signatures (Supplementary Fig. 1C). All patients had mutations on the chromosome (Chr) 12 and Chr 19 (Supplementary Fig. 1A). For the identification of tumor driver genes, we interrogated five data sources (see methods) to screen out known driver genes. Top mutated candidate driver genes documented in at least one of the data sources including BRWD1, FAT4, MYCBP2, NIN, ZNF292 (each 28.8%) etc. were annotated in Fig. 1A. APC was the candidate driver gene documented in all of the five data sources which had the highest mutation rates in the R-NET samples (4/18, 22.2%), followed by BRCA2 (3/18, 16.7%) (Supplementary Table 1). The total identified driver genes are provided in the Supplementary Table 1. The median TMB was 1.15 Muts/MB (range, 0.03–23.28) which was lower than the TMB of colorectum NEC (median: 5.18 Muts/MB) (Chen et al. 2020). TMB of three samples (16.7%) was more than 10. The median MSI was 0.36 (range, 0.00–10.97). Only one patient (5.6%) presented as MSI-H (MSI>10). For CNV, we found that regions of Chr 15q11.2 which covered the location of GOLGA6L1, GOLGA6L22, GOLGA6L6, POTEB, POTEB2, and POTEB3 were amplified in a large proportion (83.3%) of patients, and regions of Chr 15q13.3 (covered location of GOLGA8K, GOLGA8N, and GOLGA8O), Chr 2q13 (covered RGPD5), and Chr 2q14.1 (covered RGPD8) were amplified in 77.8% of patients. Other common regions of amplification (13/18, 72.2%) included Chr 11q12.3 (covered AHNAK), Chr 15q11.1-q11.2 (covered HERC2P3, OR4N4C), Chr 1p36.21-p36.13 (covered SPEN), and etc. (Fig. 1B). CNV and copy number amplification (CNA) are most frequently found on Chr 19 and Chr 1 with incidence rates of 100% (18/18). Copy number loss (CNL) is most frequently observed on Chr 10 with an incidence rate of 61.1% (11/18) (Supplementary Fig. 1A). Besides, we detected no gene fusion events.
Figure 1.
The genomic landscape of the 18 R-NETs shows the top 30 genes with somatic mutation (A) and the top 30 genes with copy number variation (B). Genes were ordered by their frequency in 18 R-NET patients. Clinicopathologic and genetic parameters were annotated in the top bar plots and bottom colored bars. Identified candidate drive genes were annotated in the left colored bar (A). A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Functional enrichment and signaling pathway analysis for frequently altered genes
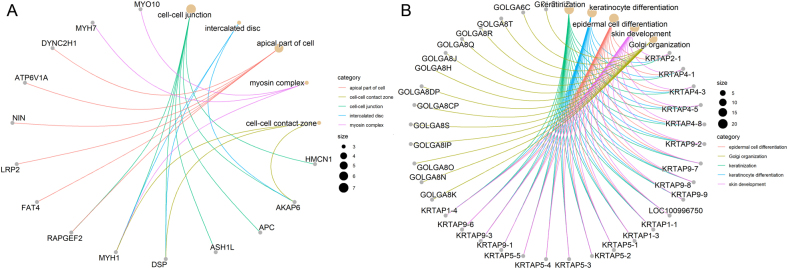
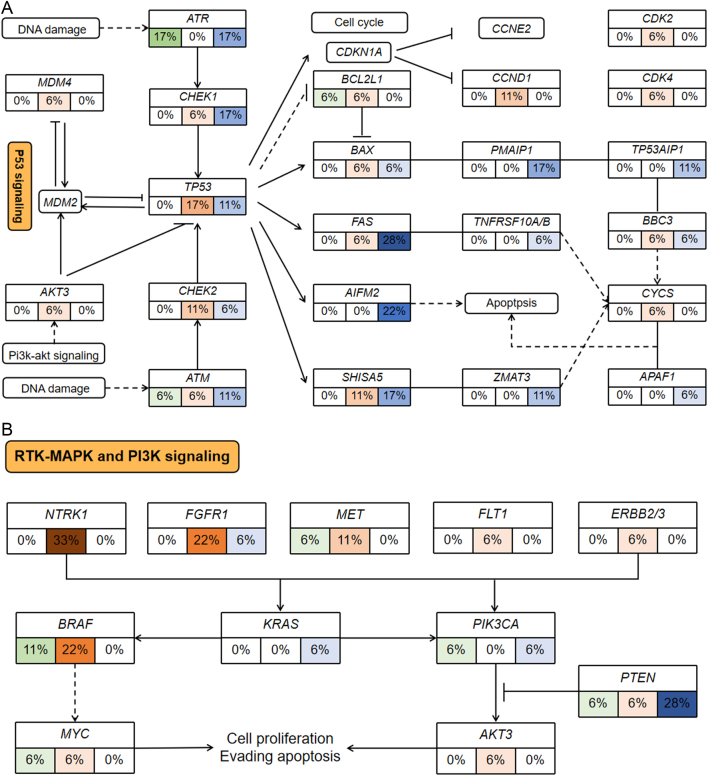
To investigate the biological function of frequently altered genes, we performed GO enrichment analysis for genes with mutational rates more than or equal to 16.7% (n = 169) and genes with CNV rates more than or equal to 44.4% (n = 183), respectively. Top enriched GO terms for frequently mutated genes included microtubule-based movement, regulation of cytokinesis, cell–cell junction, microtubule, the extrinsic component of membrane, ATPase activity, etc. (Supplementary Fig. 2A). Top enriched GO terms for genes with high-frequency CNV included keratinization, keratinocyte differentiation, epidermal cell differentiation, Golgi organization, endomembrane system organization, keratin filament, intermediate filament cytoskeleton, endopeptidase activity, etc. (Supplementary Fig. 2B). Enriched functions of top altered genes with mutation rates of more than 16.7% and genes with CNV of rates more than 44.4% and the corresponding matched genes were shown in Fig. 2. Next, we analyzed the gene alteration ratio and frequency of patients with gene alterations in each signaling pathway based on the Hallmark gene set in the Msigdb. We found all patients had alterations in the P53 pathway, mitotic spindle, estrogen response, E2F targets, and PI3K-AKT-mTOR signaling pathway (Supplementary Fig. 3A). Highly affected pathways in terms of altered gene ratio included mitotic spindle (with 75.9% of genes in this pathway altered), MYC targets v2 (altered gene ratio: 75.9%), reactive oxygen species (altered gene ratio: 75.5%), Notch signaling (altered gene ratio: 75.0%), apoptosis (altered gene ratio: 74.5%), etc. (Supplementary Fig. 3B). We further annotated the main altered genes with variation type and frequency in the P53 pathway, receptor tyrosine kinase (RTK)-MAPK, and PI3K signaling based on the signaling pathway maps in KEGG (Fig. 3). Other related frequently altered signaling pathways including WNT signaling, cell cycle, apoptosis, DNA repair (base excision repair, nucleotide excision repair, mismatch repair, homologous recombination repair, and non-homologous end-joining repair) were also mapped (Supplementary Fig. 4, 5, 6, 7, 8, 9, 10 and 11).
Figure 2.
The enriched function of frequently altered genes. Enriched functions of top altered genes with mutation rates of more than 16.7% (A) and genes with CNV rates of more than 44.4% (B) were shown. Functional enrichment was based on Gene Ontology (GO). Enriched functions and the corresponding matched genes were linked with specific colored lines. A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Figure 3.
Part of highly altered signaling pathways in R-NETs. The frequency of somatic mutations and copy number variations is shown for key genes in the P53 signaling pathway (A), MAPK, and PI3K-AKT signaling pathway (B). Different colors represent different gene alterations. Green represents mutation, red represents copy number amplification, and blue represents copy number deletion. The darker the color, the higher the frequency. A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Genetic alterations and survival
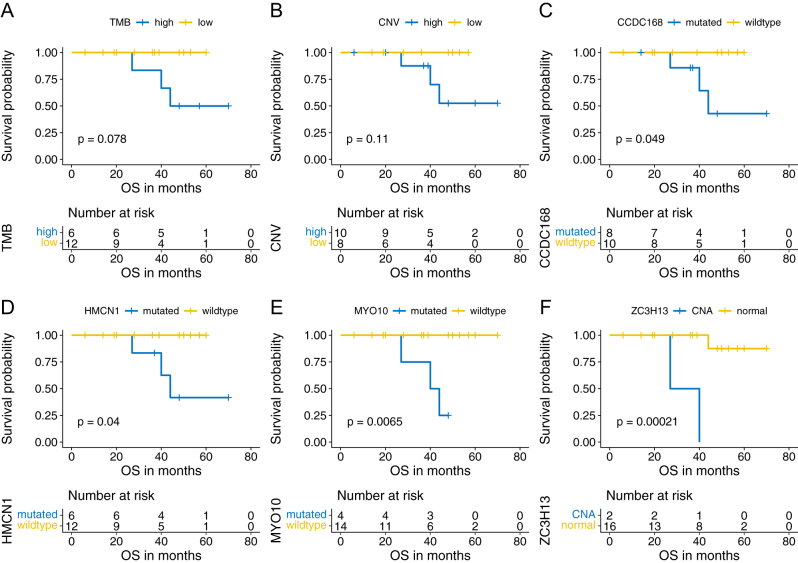
To explore the potential correlation of gene alterations with OS, we followed up on these 18 patients. In the whole cohort, the median follow-up time was 39.5 months. Three patients (3/18, 16.7%) died at the last follow-up. According to the optimal cut-off value of TMB, CNV, and MSI (TMB: 3.08/MB; CNV: 4.30E+7, MSI: 0.15) derived from the maximally selected rank statistics by the R package survminer, patients were divided into two groups respectively. Patients with higher TMB or CNV tended to have shorter OS (TMB: P = 0.078; CNV: P = 0.11, Fig. 4A and B), while MSI did not have a prognostic significance (MSI: P = 0.26). For somatic mutation, we found significant correlation of CCDC168, HMCN1, MYO10 mutation with worse OS (CCDC168: P = 0.049; HMCN1: P = 0.040; MYO10: P = 0.007, Fig. 4C, D, and E). For CNV, we found a significant correlation of ZC3H13 CNA with worse OS (P < 0.001, Fig. 4F). We also investigated the correlation of gene variation involved in the P53 signaling pathway and PI3K-AKT-pathway with prognosis. We found patients with mutation of ITGB8 (P = 0.032), BRCA1 (P = 0.014), SGK1 (P = 0.014) and CNL of APAF1 (P < 0.001) had significantly shorter OS (Supplementary Fig. 12A, B, C and D).
Figure 4.
Genetic variations with significant correlation with overall survival (OS). Survival analysis indicated that patients with higher TMB (P = 0.078) (A), higher CNV (P = 0.110) (B) tended to have worse OS. Patients with mutation of CCDC168 (P = 0.049) (C), HMCN1 (P = 0.040) (D), MYO10 (P = 0.007) (E), and copy number amplification (CNA) of ZC3H13 (P < 0.001) (F) had significantly worse OS in R-NETs. A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Pathway alterations and survival
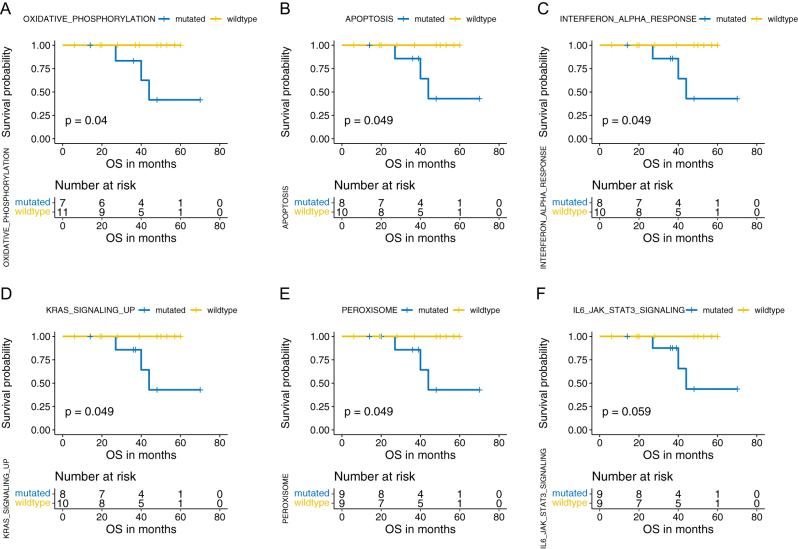
The correlation of pathway alterations with survival was investigated based on the Hallmark gene set. In total, we found five pathways with gene mutations significantly correlated with worse OS: oxidative phosphorylation (P = 0.040), apoptosis (P = 0.049), interferon alpha response (P = 0.049), KRAS signaling up (P = 0.049), and peroxisome (P = 0.049) (Fig. 5A, B, C, D and E). Besides, patients with gene mutations in L6-JAK-STAT3 signaling tended to have shorter OS (P = 0.059) (Fig. 5F).
Figure 5.
Signaling pathway alteration with significant correlation with overall survival (OS). Survival analysis indicated that gene mutations in pathways of oxidative phosphorylation (P = 0.040) (A), apoptosis (P = 0.049) (B), interferon alpha response (P = 0.049) (C), KRAS signaling up (P = 0.049) (D), and peroxisome (P = 0.049) were significantly correlated with worse OS in R-NETs. Gene mutations in the IL6-JAK-STAT3 signaling pathway tended to associate with worse OS with a P value of 0.059. Included signaling pathways and corresponding gene sets were derived from Hallmark gene sets in the Molecular Signatures Database (Msigdb). A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Potentially targetable gene alterations for targeted therapy
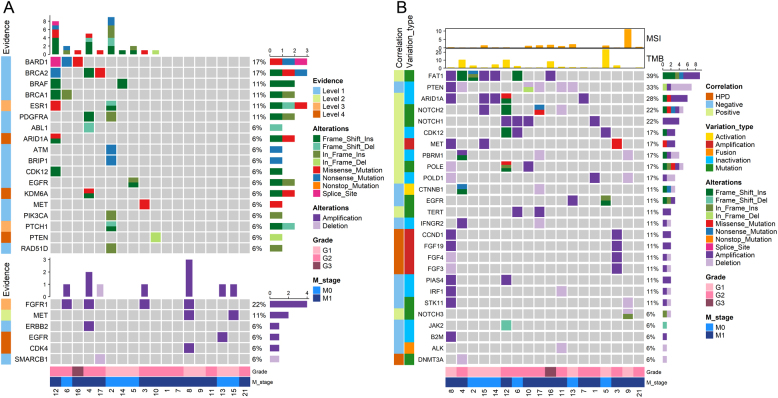
To find potentially targetable gene alterations (PTGAs) for R-NETs, the frequency of gene alterations annotated by OncoKB was counted in our cohort. In total, 13/18 (72.2%) of the patients had at least one gene alteration for all-level targets documented in the OncoKB database, which included 10/18 (55.6%) of patients who harbored at least one targetable gene mutation and 7/18 (38.9%) of patients who harbored at least one targetable gene CNV. Of these PTGAs, targetable gene mutations with top frequency included BARD1, BRCA2 (each n = 3, 16.7%), BRAF, BRCA1, ESR1, and PDGFRA (each n = 2, 11.1%) (Fig. 6A). Besides, mutation of ABL1, ARID1A, ATM, BRIP1, CDK12, EGFR, KDM6A, MET, PIK3CA, PTCH1, PTEN, and RAD51D was each found in one patient (each n = 1, 5.5%). For potentially targetable CNVs, FGFR1 amplification was the most prevalent CNV with a frequency of 22.2% (n = 4), followed by MET amplification (n = 2, 11.1%). Other potentially targetable CNVs included amplification of CDK4, EGFR, ERBB2, and deletion of SMARCB1 (each n = 1, 5.5%) (Fig. 6A). The total targetable gene list and corresponding drugs were shown in Supplementary Table 2.
Figure 6.
Gene alterations associated with therapy. Potentially actionable gene alterations for targeted therapy (A) and potential gene alterations with impact on the efficacy of immune checkpoint blockade (ICB) (B). Gene lists for targeted therapy and ICB were selected based on the Oncology KnowledgeBase (oncoKB) database and literature reports respectively. Clinicopathologic and genetic parameters were annotated in the top bar plots and bottom colored bars. A full-colour version of this figure can be found at https://doi.org/10.1530/ERC-22-0257.
Gene alterations potentially influencing immune checkpoint blockade
In addition to targeted therapy, we also evaluated patients carrying gene alterations associated with the efficacy of ICB. Among 33 genes reported to be correlated with response to ICB, 26 genes (78.8%) had variation in our cohort (the total gene list was shown in Supplementary Table 3). We found that 17/18 (94.4%) of the patients carried these gene alterations potentially influencing the efficacy of ICB (Fig. 6B). FAT1 gene alterations were the most common gene alterations potentially influencing the activity of ICB (n = 7, 38.9%, including three mutations and four amplifications, Fig. 6B). In total, the negative genetic predictors of ICB were found in eight patients (44.4%). PTEN inactivation was the most common gene alteration found to negatively influence the response to ICB (n = 5, 27.8%, including one patient with concurrent PTEN mutation and deletion and four patients with PTEN deletion, Fig. 6B). Besides, mutation of CTNNB1, EGFR, and JAK2 and deletion of IFNGR2, IRF1, and STK11 each were found in one patient (5.5%). In addition, gene alterations associated with hyperprogression (HPD) included amplification of CCND1 (n = 2, 11.1%), FGF3 (n = 1, 5.5%), FGF4 (n = 1, 5.5%), and FGF19 (n = 2, 11.1%) and deletion of DNMT3A (n = 1, 5.5%) were found in total three patients (16.7%).
Discussion
To our knowledge, this study is the first to exclusively depict the unique genomic characteristics of R-NETs using whole-exome sequencing in the Chinese population. We found the top mutated genes were significantly distinct from the results using the targeted gene panel. More genetic variations were detected by whole-exome sequencing. Genes like CCDC168, PPP4R3A, GALNT11, and HMCN1 were frequently mutated which have not been reported in previous studies in R-NENs (Chen et al. 2020, 2021). Functional enrichment analysis showed these frequently altered genes are mainly involved in microtubule-based movement, regulation of cytokinesis, cell–cell junction, microtubule, the extrinsic component of membrane, ATPase activity, etc. Besides, a large number of high-frequency CNV events were found, like the amplification of GOLGA6L6, POTEB3, POTEB2, POTEB, LOC102723623, GOLGA6L1, and GOLGA6L22. These genes mainly function in keratinization, keratinocyte differentiation, epidermal cell differentiation, Golgi organization, endomembrane system organization, keratin filament, intermediate filament cytoskeleton, endopeptidase activity, etc. However, further functional studies are needed to validate the biological effects of these genes in R-NETs. Signature 1A and G: C>A: T were the most prevalent mutation characteristic in this cohort. Signature 1A was related to the elevated rate of spontaneous deamination of 5-methyl-cytosine which results in C>T transitions and showed a strong association with age (Alexandrov et al. 2013). The predominant of signature 1A might reflect that the etiology of R-NET was mainly related to age but lack of carcinogen exposures. Besides, signature 3 is found in half of the patients which is associated with BRCA1 and BRCA2 mutation, implicating that the deficiency of homologous recombination repair might contribute to the pathogenesis of R-NET. Previous studies investigating the CNVs of NEN were mainly limited to the gastrointestinal tract and pancreas (Puccini et al. 2020). Our study might provide unique insight for understanding the genetic characteristics of NET in the rectum.
Previous genomic studies of R-NENs were mainly performed in CR-NECs, with frequently mutated genes including TP53 (24–65.5%), RB1 (4–16.7%), APC (16–59.5%), KRAS (24–36.9%), BRAF (8.3–20.2%), etc. (Chen et al. 2020, 2021, Venizelos et al. 2021). In general, these gene alterations were scarcely observed in our R-NETs cohort, except for APC, which had a mutation rate of 22.2% (4/18) in our R-NET cohort (Supplementary Fig. 13). APC was also frequently mutated in COREAD (55–96%) (Conlin et al. 2005, Chen et al. 2020). However, previous studies had found APC had a lower frequency of mutation in pancreatic NENs (1–15%) (Puccini et al. 2020, Venizelos et al. 2021), esophagus NEC (11%) (Venizelos et al. 2021), and gastric NEC (6%) (Venizelos et al. 2021). This indicated that APC mutation was tumor-site specific but not dependent of tumor grade and differentiation. For TP53, we found no mutation but two deletions. For RB1, one patient had the insertional mutation and the other one patient had RB1 deletion. All patients with TP53 and/or RB1 alterations were in low grade in this study. This suggests that TP53 and RB1 inactivation can occur in low-grade NETs but had a lower frequency compared with NECs. We found no KRAS mutations, indicating KRAS is likely to mainly function in the CR-NECs instead of R-NETs. However, we found four insertional mutations of BRAF in two R-NET patients (11.1%) and four additional BRAF amplifications (22.2%). Besides, three BRCA mutations (16.7%), five PTEN deletions, and one PTEN mutation (27.8%) were found in our cohort which might be the characteristic molecular alteration in R-NET (Supplementary Fig. 13).
In this study, we found that all patients had at least one gene alteration in the P53 signaling pathway, indicating the pivotal role of the P53 signaling pathway in R-NETs. The P53 signaling pathway was involved in the regulation of DNA damage repair, cell cycle, and apoptosis. We found that key genes promoting apoptosis, such as FAS (28%), PRF1 (28%), CASP7 (22%), AIFM2 (22%), TNFSF10 (22%), and MAPK8 (22%) had high-frequency copy number deletion (CND), while NTRK1 (33%), FGFR1 (22%), MET (11%), BRAF (22%) in the RTK-MAPK pathway which promoted proliferation had high-frequency CNA. In addition, PTEN (28%), the key negative regulator of the PI3K-AKT-mTOR pathway had frequent depletion. These alterations might lead to abnormal activation of the downstream cell proliferation signal. We speculated that the frequent variation in MAPK and PI3K-AKT signaling pathway might contribute to the relatively high sensitivity of R-NETs to the mTOR inhibitor Everolimus. In addition, we found that the DNA damage repair pathway also had frequent alterations. The DNA damage sensors, ATM (mutation: 6%, deletion: 11%) and ATR (mutation: 17%, deletion: 17%), were frequently mutated and depleted. Among various DNA damage repair mechanisms, genes involved in homologous recombination and nucleotide excision repair had relatively higher mutation rates, such as BRCA1 (11%), BRCA2 (17%), BARD1 (17%), and ERCC6 (17%). While genes involved in non-homologous end-joining repair had relatively higher rates of CNL, such as POLL (22%) and DNTT (28%). We found that the key genes involved in mismatch repair, such as MLH1, MSH2, and MSH3, mainly had CNV but rarely mutated. In addition, genes like APC (33%), WNT5B (11%), and AXIN2 (11%) in the WNT signaling pathway that regulates cell proliferation and differentiation frequently exhibited CNA. Simultaneously, BIRC5 (11%), MYC (6%), and CCND1 (11%), the downstream molecules of WNT signaling, also showed CNA, suggesting activation of the WNT-β catenin signaling pathway in a subset of patients.
We found higher TMB was associated with poor prognosis which was consistent with the previous report (Chen et al. 2020). For single gene variation, we found a mutation of CCDC168, HMCN1, MYO10, and CNA of ZC3H13 significantly correlated with worse OS. CCDC168 is a protein-coding gene with a poorly understood function known to be mutated in several cancer types (Wang et al. 2018). CCDC168 is reported to have a frequent mutation in pediatric renal cell carcinomas (Beck et al. 2022), malignant pleural mesothelioma (Yu et al. 2021), and colon adenocarcinoma (Cortes-Ciriano et al. 2017). Our study identified CCDC168 as the top mutant gene and associated with poor OS in R-NETs. HMCN1 encodes a large extracellular member of the immunoglobulin superfamily. HMCN1 is frequently mutated in prostate cancer (Zhao et al. 2019) and gastric and colorectal cancer (Lee et al. 2015). Mutation of HMCN1 is associated with poor prognosis in clear cell renal carcinoma (Gong et al. 2022) and breast cancer (Kikutake et al. 2018). Besides, it was found to be associated with higher TMB in ovarian cancer (Lu et al. 2021). In our study, we also found R-NETs with HMCN1 mutation exhibited higher TMB (median: 6.39 vs 0.60, P = 0.010). The underlying mechanism of HMCN1 mutation to potentially cause high TMB and the potential link between HMCN1 mutation and response to ICB are interesting to be further studied. MYO10 encodes a member of the myosin superfamily which functions as an actin-based molecular motor and plays a role in the integration of F-actin and microtubule cytoskeletons during meiosis. MYO10 is found to be upregulated in many humor cancers and correlated with poor prognosis, such as lung squamous cell carcinoma (Dvornikov et al. 2018) and cervical cancer (He et al. 2020). High expression of MYO10 contributes to the tumor invasion and migration of breast cancer (Cao et al. 2014), prostate cancer (Makowska et al. 2015), and colorectal cancer (Ou et al. 2022). Our study first reported that MYO10 mutation is associated with poor prognosis in R-NETs. Evidence has shown MYO10 promotes tumor progression by inducing genomic instability which in turn creates an immunogenic environment for ICB (Mayca Pozo et al. 2021). In this study, we found patients with MYO10 mutations had higher TMB (median: 6.39 vs 0.65 Muts/MB, P = 0.029), supporting the role of MYO10 in the regulation of genomic stability. However, the correlation of MYO10 mutation with response to ICB remains to be further elucidated in R-NETs. ZC3H13 was a part of the RNA N6-methyladenosine methyltransferase complex involved in RNA epigenetic modification, which was reported to be a tumor suppressor in colorectal cancer, breast cancer, and endometrial cancer (Zhu et al. 2019, Gong et al. 2020, Ma et al. 2021). However, in our study, it seemed to be an unfavorable molecule since patients with ZC3H13 CNA had worse OS. These gene alterations might play important roles in the development and progression of R-NETs and might be potential therapeutic targets that deserved to be further investigated.
Our results showed that up to 13/18 (72.2%) of the R-NETs had at least one PTGA, suggesting the promising prospect of targeted therapy in R-NETs. Our study showed up to 22% of the R-NETs harbored FGFR1 amplification which was the most common PTGA, followed by BARD1 mutation and BRCA2 mutation (each 17%). Drugs targeting the FGFR1 amplification included infigratinib, Debio1347, and AZD4547, all of which were FGFR1-3 inhibitors. Infigratinib and AZD4547 had shown activity in lung squamous cell carcinoma with FGFR1 amplification (Zhang et al. 2012, Nogova et al. 2017, Paik et al. 2017). Debio 1347 had also demonstrated encouraging efficacy across multiple solid tumor types with FGFR1 amplification (Voss et al. 2019). However, these drugs had not been attempted in neuroendocrine tumors. Further preclinical studies and clinical trials are needed to test the efficacy of the FGFR inhibitors in R-NETs. Besides, our study suggested the wide range of adaptability of olaparib in R-NETs since 33.3% (6/18) of the patients harbored the targetable alterations for olaparib (including mutation of BARD1, BRCA2 (each 16.7%), BRCA1 (11.1%), ATM, BRIP1, CDK12, and RAD51D (each 5.6%)). However, olaparib had not been used in R-NETs. Besides, we analyzed all the variation types of potentially targetable genes validated in other malignancies. We found NTRK1 amplification in six (33.3%) patients. ROS1 was mutated in two patients and was amplified in four additional patients. However, drugs targeting these specific variation types mostly have not been developed yet. It remained to see whether the currently approved drugs targeting NTRK1 rearrangements and ROS1 fusions, entrectinib also had effectiveness in R-NETs. BRAF was not only mutated in two patients but also amplified in an additional four patients. However, currently approved drugs only target BRAF V600 mutations like dabrafenib and vemurafenib. The efficacy of these drugs for patients with BRAF amplification is unsure and specific drugs targeting the BRAF amplification are needed. In sum, these frequently altered genes are mostly involved in RTK-MAPK and PI3K-AKT signaling indicating that targeting these signaling pathways might be promising therapeutic strategies for R-NETs. This was supported by the exceptional efficacy of the mTOR inhibitor, Everolimus, implicated in advanced G1-2 R-NENs (Yao et al. 2016). Moreover, Surufatinib, the novel approved multitargeted receptor tyrosine kinase inhibitor (RTKI) targeting VEGFR1-3, FGFR1, and CSF1R has also demonstrated benefit for patients with advanced extra-pancreatic NETs in China (Xu et al. 2020). Other novel RTKIs like cabozantinib, lenvatinib, and pazopanib have been developed and shown promising efficacy (Das & Dasari 2021). With a relatively higher ORR of 15% for extra-pancreatic NETs and well tolerance observed in the phase II study (Chan et al. 2014), the randomized phase III CABINET trial of cabozantinib for advanced extra-pancreatic NETs has been initiated (NCT03375320), which may bring better efficacy and safety in R-NET patients.
ICB represented a promising strategy for cancer treatment in a broad range of malignancies. However, the immune checkpoint inhibitors (ICIs) targeting PD-(L)1 and/or CTLA-4 have not seen clinical meaningful anti-tumor effects in gastroenteropancreatic (GEP) -NETs, except for the rare subsets of patients with MSI-H or TMB-H (Mehnert et al. 2020, Patel et al. 2020, Chan et al. 2021). In our study, the overall TMB and MSI were extremely low with only three patients having TMB-H and one patient having MSI-H, which was concordant with the previous reports (Mitsuhashi et al. 2015, Chen et al. 2020, Venizelos et al. 2021). The overall low TMB and MSI in GEP-NETs might contribute to the overall poor response to immunotherapy. Nevertheless, ICB might serve as an alternative option for selected patients with TMB-H given that patients with higher TMB suffered from worse OS. Besides the widely accepted predictive biomarkers, TMB and MSI, we investigated other genetic predictors of response to ICB reported in previous literature. We found 44.4% of the patients harbored at least one negative genetic predictor of ICB. PTEN depletion (n = 5, 27.8%) was the most common gene alteration found to potentially negatively influence the response to ICB. PTEN depletion was found to be correlated with immune resistance and tumor immune evasion and predicted poor response to ICB in a variety of malignancies (Peng et al. 2016, Vidotto et al. 2020, Lin et al. 2021). Besides, FAT1 was also frequently altered with four patients harboring CNAs and three patients harboring insertional mutations. Previous studies had indicated a positive correlation of FAT1 mutation with the durable clinical benefit of ICB in non-small cell lung cancer (Fang et al. 2019, Feng et al. 2022). However, conflicting results were shown in another data-mining study (Zhu et al. 2021). Our study showed that FAT1 was not only mutated but also had relatively high-frequency CNA in R-NETs. Further studies are needed to validate the potential effects of FAT1 multi-type variations on response to ICB. Other genetic negative predictors found in our cohort but with a low frequency included mutation of CTNNB1, EGFR, and JAK2 and deletion of IFNGR2, IRF1, and STK11 (each n = 1). Notably, gene alterations associated with HPD were found in a total of three patients (16.7%), which underscores the need for real-time imaging monitoring for R-NET with these alterations when receiving ICB and the value of genetic testing upfront of the therapy. Oncogenic pathway activation was found as a critical tumor-intrinsic mechanism for resistance to ICB (Kalbasi & Ribas 2020). Given frequent gene alterations in the MAPK, PI3K-AKT, and WNT signaling pathway observed in this study, further combinational strategies concurrently targeting these pathways and negative immune checkpoints are deserved to be tested in R-NETs.
There were some limitations in this work. First, this study was performed in a single center and the retrospective nature may lead to some bias. Secondly, the sample size was limited. However, given the rarity of R-NETs, including a large number of patients was difficult. Thirdly, two samples were collected after treatment which might influence the sequencing results. Fourthly, sample types are heterogeneous and this might further impact the results to a certain degree. However, given the complexity of the realistic situations, the specimen types can be diverse. Since only 3 fresh tumor tissues were included, and all of the remaining 15 samples are FFPE tissues. We believed that this might have a much smaller impact on the heterogeneity of the results than the inherent heterogeneity between tumor tissues. Further genomic studies with larger sample sizes and samples of homogeneity are needed.
In summary, multiple oncogenic pathways were frequently altered potentially leading to the development and progression of R-NETs. Several gene alterations were found to be significantly associated with worse OS which might be potential therapeutic targets. Targeted therapy might be a promising strategy as targetable gene alterations were prevalent in a high fraction of R-NETs. FAT1 alteration and PTEN depletion might be the main genetic alterations influencing the response to ICB besides overall low TMB and MSI in R-NETs.
Supplementary Materials
Declaration of interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2019YFB1309704).
Data availability
Sequencing data are available at the request of the corresponding authors.
Author contribution statement
LYL, CZX, TC, CYY, and SF collected the patients and samples; CZX, YFH, and CRA collected the clinicopathological information; SYF and LJ performed pathological review, GYY and ZSH performed the bioinformatic data analysis; LYL, GYY, and WZZ drafted the manuscript; THY and LJ directed the project and revised the manuscript.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale ALet al. 2013Signatures of mutational processes in human cancer. Nature 500415–421. ( 10.1038/nature12477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banck MS, Kanwar R, Kulkarni AA, Boora GK, Metge F, Kipp BR, Zhang L, Thorland EC, Minn KT, Tentu Ret al. 2013The genomic landscape of small intestine neuroendocrine tumors. Journal of Clinical Investigation 1232502–2508. ( 10.1172/JCI67963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck P, Selle B, Madenach L, Jones DTW, Vokuhl C, Gopisetty A, Nabbi A, Brecht IB, Ebinger M, Wegert Jet al. 2022The genomic landscape of pediatric renal cell carcinomas. iScience 25 104167. ( 10.1016/j.isci.2022.104167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Chen J, Zhang X, Zhai Y, Qing X, Xing W, Zhang L, Malik YS, Yu H, Zhu X.2014Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. British Journal of Cancer 111539–550. ( 10.1038/bjc.2014.298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MHet al. 2017OncoKB: A precision oncology knowledge base. JCO Precision Oncology 1. ( 10.1200/PO.17.00011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Faris JE, Murphy JE, Blaszkowsky LS, Ryan DP.2014A phase II trial of cabozantinib in patients with carcinoid and pancreatic neuroendocrine tumors 35228–228. ( 10.1200/JCO.2017.35.4_suppl.228) [DOI] [Google Scholar]
- Chan JA, Raj NP, Aggarwal RR, Calabrese S, Demore A, Dhawan MS, Fattah D, Fong L, Grabowsky J, Hope TAet al. 2021Phase II study of pembrolizumab-based therapy in previously treated extrapulmonary poorly differentiated neuroendocrine carcinomas: results of Part B (pembrolizumab + chemotherapy). Journal of Clinical Oncology 394148–4148. ( 10.1200/JCO.2021.39.15_suppl.4148) [DOI] [Google Scholar]
- Chen D, Bao X, Zhang R, Ding Y, Zhang M, Li B, Zhang H, Li X, Tong Z, Liu Let al. 2020Depiction of the genomic and genetic landscape identifies CCL5 as a protective factor in colorectal neuroendocrine carcinoma. British Journal of Cancer 125994–1002. ( 10.1038/s41416-021-01501-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu M, Zhang Y, Guo Y, Chen MH, Chen J.2021Genetic characteristics of colorectal neuroendocrine carcinoma: more similar to colorectal adenocarcinoma. Clinical Colorectal Cancer 20177–185.e13. ( 10.1016/j.clcc.2020.09.001) [DOI] [PubMed] [Google Scholar]
- Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ.2005The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 541283–1286. ( 10.1136/gut.2005.066514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ.2017A molecular portrait of microsatellite instability across multiple cancers. Nature Communications 8 15180. ( 10.1038/ncomms15180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Dasari A.2021Novel therapeutics for patients with well-differentiated gastroenteropancreatic neuroendocrine tumors. Therapeutic Advances in Medical Oncology 1317588359211018047. ( 10.1177/17588359211018047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T, Yao JC.2017Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncology 31335–1342. ( 10.1001/jamaoncol.2017.0589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvornikov D, Schneider MA, Ohse S, Szczygieł M, Titkova I, Rosenblatt M, Muley T, Warth A, Herth FJ, Dienemann Het al. 2018Expression ratio of the TGFβ-inducible gene MYO10 is prognostic for overall survival of squamous cell lung cancer patients and predicts chemotherapy response. Scientific Reports 8 9517. ( 10.1038/s41598-018-27912-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, Wang F, Bao H, Wu X, Yang Yet al. 2019Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clinical Cancer Research 255015–5026. ( 10.1158/1078-0432.CCR-19-0585) [DOI] [PubMed] [Google Scholar]
- Feng Z, Yin Y, Liu B, Zheng Y, Shi D, Zhang H, Qin J.2022Prognostic and immunological role of FAT family genes in non-small cell lung cancer. Cancer Control 2910732748221076682. ( 10.1177/10732748221076682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, Banck MS, Kanwar R, Kulkarni AA, Karpathakis Aet al. 2013Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nature Genetics 451483–1486. ( 10.1038/ng.2821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel AAG, Mathian E, Mangiante L, Voegele C, Cahais V, Ghantous A, McKay JD, Alcala N, Fernandez-Cuesta L, Foll M.2020A molecular map of lung neuroendocrine neoplasms. GigaScience 9 giaa112. ( 10.1093/gigascience/giaa112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong PJ, Shao YC, Yang Y, Song WJ, He X, Zeng YF, Huang SR, Wei L, Zhang JW.2020Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer. Frontiers in Oncology 10 578963. ( 10.3389/fonc.2020.578963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Wu X, Guo Q, Du HZ, Zhan FH, Kong Y.2022Comprehensive analysis of HMCN1 somatic mutation in clear cell renal cell carcinoma. Genes 13 1282. ( 10.3390/genes13071282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JH, Chen JG, Zhang B, Chen J, You KL, Hu JM, Xu JW, Chen L.2020Elevated MYO10 predicts poor prognosis and its deletion hampers proliferation and migration potentials of cells through rewiring PI3K/Akt signaling in cervical cancer. Technology in Cancer Research and Treatment 191533033820936773. ( 10.1177/1533033820936773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MAet al. 2011DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science (New York, NY) 3311199–1203. ( 10.1126/science.1200609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbasi A, Ribas A.2020Tumour-intrinsic resistance to immune checkpoint blockade. Nature Reviews. Immunology 2025–39. ( 10.1038/s41577-019-0218-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MAet al. 2013Mutational landscape and significance across 12 major cancer types. Nature 502333–339. ( 10.1038/nature12634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikutake C, Yoshihara M, Sato T, Saito D, Suyama M.2018Intratumor heterogeneity of HMCN1 mutant alleles associated with poor prognosis in patients with breast cancer. Oncotarget 933337–33347. ( 10.18632/oncotarget.26071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel G.2017Neuroendocrine neoplasms: dichotomy, origin and classifications. Visceral Medicine 33324–330. ( 10.1159/000481390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Je EM, Yoo NJ, Lee SH.2015HMCN1, a cell polarity-related gene, is somatically mutated in gastric and colorectal cancers. Pathology Oncology Research 21847–848. ( 10.1007/s12253-014-9809-3) [DOI] [PubMed] [Google Scholar]
- Lin Z, Huang L, Li SL, Gu J, Cui X, Zhou Y.2021PTEN loss correlates with T cell exclusion across human cancers. BMC Cancer 21 429. ( 10.1186/s12885-021-08114-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Liu J, Xu M, Liang JQ, Wang YC, Wu ZP, Xing Y, Diao FY.2021CSMD3 is associated with tumor mutation burden and immune infiltration in ovarian cancer patients. International Journal of General Medicine 147647–7657. ( 10.2147/IJGM.S335592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yang D, Ma XX.2021Immune infiltration-related N6-methyladenosine RNA methylation regulators influence the malignancy and prognosis of endometrial cancer. Aging (Albany NY) 1316287–16315. ( 10.18632/aging.203157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafficini A, Scarpa A.2019Genetics and epigenetics of gastroenteropancreatic neuroendocrine neoplasms. Endocrine Reviews 40506–536. ( 10.1210/er.2018-00160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowska KA, Hughes RE, White KJ, Wells CM, Peckham M.2015Specific myosins control actin organization, cell morphology, and migration in prostate cancer cells. Cell Reports 132118–2125. ( 10.1016/j.celrep.2015.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayca Pozo F, Geng X, Tamagno I, Jackson MW, Heimsath EG, Hammer JA, Cheney RE, Zhang YW.2021MYO10 drives genomic instability and inflammation in cancer. Science Advances 7 eabg6908. ( 10.1126/sciadv.abg6908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert JM, Bergsland E, O'Neil BH, Santoro A, Schellens JHM, Cohen RB, Doi T, Ott PA, Pishvaian MJ, Puzanov Iet al. 2020Pembrolizumab for the treatment of programmed death-ligand 1-positive advanced carcinoid or pancreatic neuroendocrine tumors: results from the KEYNOTE-028 study. Cancer 1263021–3030. ( 10.1002/cncr.32883) [DOI] [PubMed] [Google Scholar]
- Mitsuhashi K, Yamamoto I, Kurihara H, Kanno S, Ito M, Igarashi H, Ishigami K, Sukawa Y, Tachibana M, Takahashi Het al. 2015Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget 622114–22125 ( 10.18632/oncotarget.4294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogova L, Sequist LV, Perez Garcia JM, Andre F, Delord JP, Hidalgo M, Schellens JH, Cassier PA, Camidge DR, Schuler Met al. 2017Evaluation of BGJ398, a fibroblast growth factor Receptor 1–3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global Phase I, dose-escalation and dose-expansion study. Journal of Clinical Oncology 35157–165. ( 10.1200/JCO.2016.67.2048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou H, Wang L, Xi Z, Shen H, Jiang YF, Zhou FX, Liu Y, Zhou YF.2022MYO10 contributes to the malignant phenotypes of colorectal cancer via RACK1 by activating integrin/Src/FAK signaling. Cancer Science 1133838–3851. ( 10.1111/cas.15519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik PK, Shen R, Berger MF, Ferry D, Soria JC, Mathewson A, Rooney C, Smith NR, Cullberg M, Kilgour Eet al. 2017A phase Ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clinical Cancer Research 235366–5373. ( 10.1158/1078-0432.CCR-17-0645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SP, Othus M, Chae YK, Giles FJ, Hansel DE, Singh PP, Fontaine A, Shah MH, Kasi A, Baghdadi TAet al. 2020A Phase II basket trial of dual anti-CTLA-4 and anti-PD-1 blockade in rare tumors (DART SWOG 1609) in patients with nonpancreatic neuroendocrine tumors. Clinical Cancer Research 262290–2296. ( 10.1158/1078-0432.CCR-19-3356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang Xet al. 2016Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discovery 6202–216. ( 10.1158/2159-8290.CD-15-0283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini A, Poorman K, Salem ME, Soldato D, Seeber A, Goldberg RM, Shields AF, Xiu J, Battaglin F, Berger MDet al. 2020Comprehensive genomic profiling of gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs). Clinical Cancer Research 265943–5951. ( 10.1158/1078-0432.CCR-20-1804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke A, Muller HH, Schade-Brittinger C, Klose KJ, Barth P, Wied M, Mayer C, Aminossadati B, Pape UF, Blaker Met al. 2009Placebo-controlled, double-blind, prospective, randomized study on the effect of octreotide LAR in the control of tumor growth in patients with metastatic neuroendocrine midgut tumors: a report from the PROMID Study Group. Journal of Clinical Oncology 274656–4663 ( 10.1200/JCO.2009.22.8510) [DOI] [PubMed] [Google Scholar]
- Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini Aet al. 2017Whole-genome landscape of pancreatic neuroendocrine tumors. Nature 54365–71. ( 10.1038/nature21063) [DOI] [PubMed] [Google Scholar]
- Singh S, Carnaghi C, Buzzoni R, Pommier RF, Raderer M, Tomasek J, Lahner H, Valle JW, Voi M, Bubuteishvili-Pacaud Let al. 2018Everolimus in neuroendocrine tumors of the gastrointestinal tract and unknown primary. Neuroendocrinology 106211–220. ( 10.1159/000477585) [DOI] [PubMed] [Google Scholar]
- Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, Mittra E, Kunz PL, Kulke MH, Jacene Het al. 2017Phase 3 trial of (177) Lu-Dotatate for midgut neuroendocrine tumors. New England Journal of Medicine 376125–135 ( 10.1056/NEJMoa1607427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, Lawrence MS, Getz G, Bader GD, Ding Let al. 2013Comprehensive identification of mutational cancer driver genes across 12 tumor types. Scientific Reports 3 2650. ( 10.1038/srep02650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venizelos A, Elvebakken H, Perren A, Nikolaienko O, Deng W, Lothe IMB, Couvelard A, Hjortland GO, Sundlöv A, Svensson Jet al. 2021The molecular characteristics of high-grade gastroenteropancreatic neuroendocrine neoplasms. Endocrine-Related Cancer 291–14. ( 10.1530/ERC-21-0152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidotto T, Melo CM, Castelli E, Koti M, Dos Reis RB, Squire JA.2020Emerging role of PTEN loss in evasion of the immune response to tumours. British Journal of Cancer 1221732–1743. ( 10.1038/s41416-020-0834-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW.2013Cancer genome landscapes. Science 3391546–1558. ( 10.1126/science.1235122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss MH, Hierro C, Heist RS, Cleary JM, Meric-Bernstam F, Tabernero J, Janku F, Gandhi L, Iafrate AJ, Borger DRet al. 2019A Phase I, open-label, multicenter, dose-escalation study of the oral selective FGFR inhibitor Debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clinical Cancer Research 252699–2707. ( 10.1158/1078-0432.CCR-18-1959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VG, Kim H, Chuang JH.2018Whole-exome sequencing capture kit biases yield false negative mutation calls in TCGA cohorts. PLoS One 13 e0204912. ( 10.1371/journal.pone.0204912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shen L, Zhou Z, Li J, Bai C, Chi Y, Li Z, Xu N, Li E, Liu Tet al. 2020Surufatinib in advanced extrapancreatic neuroendocrine tumours (SANET-ep): a randomised, double-blind, placebo-controlled, phase 3 study. Lancet. Oncology 211500–1512. ( 10.1016/S1470-2045(2030496-4) [DOI] [PubMed] [Google Scholar]
- Yao JC, Fazio N, Singh S, Buzzoni R, Carnaghi C, Wolin E, Tomasek J, Raderer M, Lahner H, Voi Met al. 2016Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet 387968–977. ( 10.1016/S0140-6736(1500817-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Yu M, Zhu LJ, Yuan XY, Zhang X.2021Expression and clinical significance of SETD2 in malignant pleural mesothelioma. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 3991–98. ( 10.3760/cma.j.cn121094-20200831-00505) [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, Fan S, Yin X, Xu Y, Liu Ket al. 2012Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clinical Cancer Research 186658–6667. ( 10.1158/1078-0432.CCR-12-2694) [DOI] [PubMed] [Google Scholar]
- Zhao X, Lei Y, Li G, Cheng Y, Yang HF, Xie LB, Long H, Jiang R.2019Integrative analysis of cancer driver genes in prostate adenocarcinoma. Molecular Medicine Reports 192707–2715 ( 10.3892/mmr.2019.9902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Zhou J, Zhao J, Jiang G, Zhang X, Zhang Y, Dong M.2019ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. Journal of Cellular Physiology 2348899–8907. ( 10.1002/jcp.27551) [DOI] [PubMed] [Google Scholar]
- Zhu G, Ren D, Lei X, Shi R, Zhu S, Zhou N, Zu L, Mello RA, Chen J, Xu S.2021Mutations associated with no durable clinical benefit to immune checkpoint blockade in non-S-cell lung cancer. Cancers 13 1397. ( 10.3390/cancers13061397) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at the request of the corresponding authors.



 This work is licensed under a
This work is licensed under a