Abstract
The DNA adduct 6-oxo-M1dG, (3-(2′-deoxy-β-D-erythro-pentofuranosyl)-6-oxo-pyrimido(1,2alpha)purin-10(3H)-one) is formed in the genome via oxidation of the peroxidation-derived adduct M1dG. However, the effect of 6-oxo-M1dG adducts on subsequent DNA replication is unclear. Here we investigated the ability of the human Y-family polymerase hPol η to bypass 6-oxo-M1dG. Using steady-state kinetics and analysis of DNA extension products by liquid chromatography–tandem mass spectrometry, we found hPol η preferentially inserts a dAMP or dGMP nucleotide into primer–templates across from the 6-oxo-M1dG adduct, with dGMP being slightly preferred. We also show primer–templates with a 3′-terminal dGMP or dAMP across from 6-oxo-M1dG were extended to a greater degree than primers with a dCMP or dTMP across from the adduct. In addition, we explored the structural basis for bypass of 6-oxo-M1dG by hPol η using X-ray crystallography of both an insertion-stage and an extension-stage complex. In the insertion-stage complex, we observed that the incoming dCTP opposite 6-oxo-M1dG, although present during crystallization, was not present in the active site. We found the adduct does not interact with residues in the hPol η active site but rather forms stacking interactions with the base pair immediately 3′ to the adduct. In the extension-stage complex, we observed the 3′ hydroxyl group of the primer strand dGMP across from 6-oxo-M1dG is not positioned correctly to form a phosphodiester bond with the incoming dCTP. Taken together, these results indicate 6-oxo-M1dG forms a strong block to DNA replication by hPol η and provide a structural basis for its blocking ability.
Keywords: DNA damage, DNA repair, DNA polymerase, X-ray crystallography, mutagenesis mechanism, M1dG, 6-oxo-M1dG, translesion synthesis, DNA adduct, Oxidative damage
DNA modification can arise by a variety of mechanisms from both foreign as well as endogenously produced agents (1, 2, 3). The DNA adduct M1dG (3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido(1,2-alpha)purin-10(3H)-one)is produced from dG by reaction with base propenal or malondialdehyde (4, 5, 6). M1dG has been detected in various human tissues, including liver, pancreas, breast, leukocytes, and lymphocytes, with levels ranging from 1 to 120 adducts/108 nucleotides (7, 8, 9, 10, 11, 12). The amount of M1dG is 50 to 100 times higher in mitochondrial DNA than in genomic DNA (13). M1dG has been shown to be mutagenic in bacterial and mammalian cells and induces both base pair substitutions and frameshifts (14, 15).
Cells have devised many mechanisms of damage surveillance and repair. M1dG is removed from the genome by nucleotide excision repair (16) and can be bypassed by translesion synthesis (TLS) (17, 18, 19). Humans express five translesion DNA polymerases with four belonging to the Y-family (20, 21) and one from the B-family of polymerases (Pol ) (22). Translesion polymerases contain larger active sites than other replicative polymerases. This allows them to accommodate bulky DNA lesions; however, they lack the fidelity of other replicative polymerases and are thus error prone (23, 24). M1dG has been shown to be bypassed by , Rev1, and the Sulfolobus sulfataricus enzyme Dpo4. HPol , hPol , and Rev1 preferentially insert a dCMP opposite M1dG during in vitro bypass (18); dAMP is the favored nucleotide inserted across from M1dG by hPol and Dpo4 (17, 19).
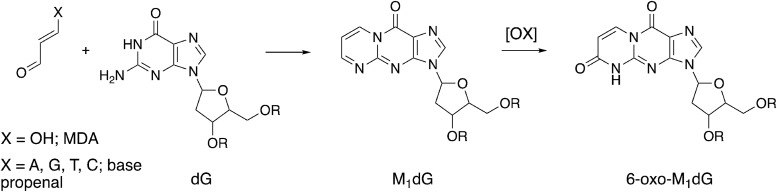
M1dG in the genome of human cells is oxidized to 6-oxo-M1dG (3-(2′-deoxy-β-D-erythro-pentofuranosyl)-6-oxo-pyrimido(1,2-alpha)purin-10(3H)-one) in a process that occurs more rapidly than removal of M1dG by nucleotide excision repair (Fig. 1) (25). 6-Oxo-M1dG differs from M1dG in that it has an added carbonyl oxygen at carbon 6 in the exocyclic ring. Little is known about the bypass or mutagenicity of 6-oxo-M1dG. There are two steps for successful bypass of an adduct. The first step is insertion of a nucleotide across from the adduct, and the second step is extension by the addition of nucleotides beyond this point. We have previously reported that recombinant human hPol ι is able to catalyze the insertion step by incorporating either dCMP or dTMP across from 6-oxo-M1dG in a template–primer duplex in vitro, but it is not able to extend beyond this single nucleotide addition (26). In the present study, we chose to focus on the bypass of 6-oxo-M1dG by hPol . This translesion polymerase has been shown previously to catalyze both the insertion and extension steps across from M1dG (19). Furthermore, hPol can accommodate bulky adducts in its active site (27, 28, 29). We investigated the bypass of 6-oxo-M1dG by human hPol using a primer–template duplex containing a single adduct. In addition, we determined crystal structures of hPol in complex with an oligonucleotide primer–template duplex containing a single 6-oxo-M1dG. Structures of both an insertion-stage complex and an extension-stage complex were obtained. The results indicate that 6-oxo-M1dG is a powerful block to replication by hPol and provide a structural basis for the inability of this polymerase to bypass the lesion.
Figure 1.
Formation of 6-oxo-M1dG. M1dG is formed from the lipid peroxidation product malondialdehyde or from the DNA oxidation product base propenal. Oxidation by an unknown enzyme forms 6-oxo-M1dG.
Results
Nucleotide insertion and extension past the 6-oxo-M1dG adduct by hPol
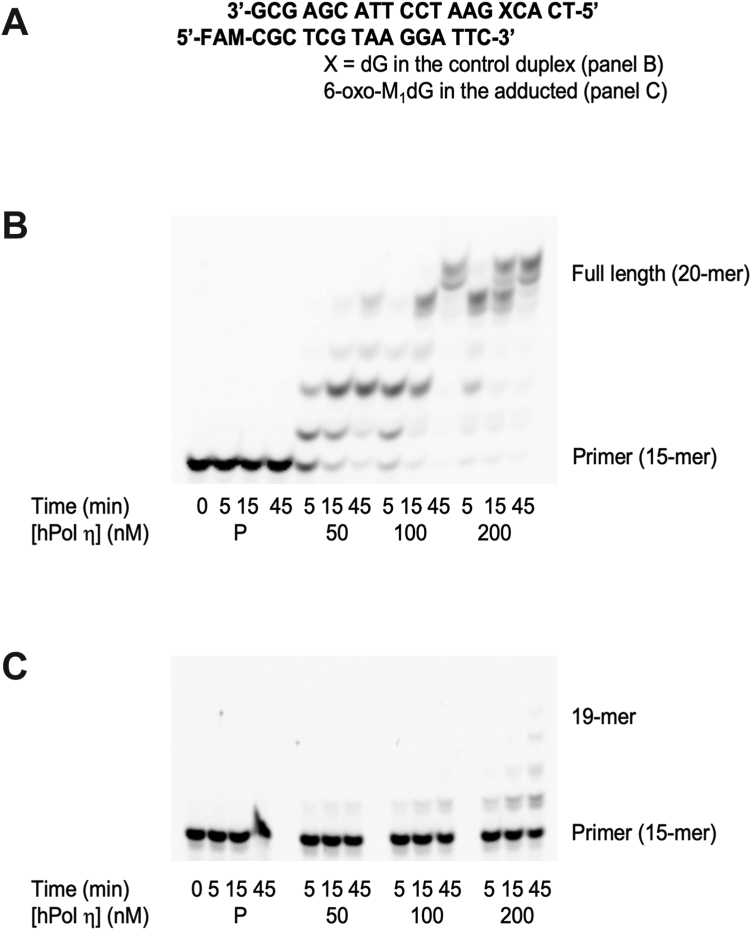
A 20-mer 6-oxo-M1dG-containing template was synthesized as described in Experimental Procedures and annealed to a complementary 15-mer (standing start) primer with a terminus immediately 3′ to the adduct (Fig. 2A). In addition, a control 20-mer template containing dG in place of 6-oxo-M1dG was annealed to the complementary 15-mer primer. A time course of insertion and extension of these primers in the presence of all four dNTPs and increasing amounts of hPol was performed. With sufficient time and/or hPol concentration, the 15-mer primer was extended almost completely to 20 nucleotides when it was annealed to the control template (Fig. 2B). Densitometry analysis of the gel confirmed 98% product formation at the 45 min time point for both the 100 nM and 200 nM concentrations of hPol . In contrast, hPol displayed noticeably less primer insertion across from the template containing 6-oxo-M1dG. At the highest enzyme concentration (200 nM) and longest incubation time (45 min), hPol was able to insert a nucleotide across from the adduct in 34% of the primer–template duplex and a small amount was further extended to form 17-mers and 18-mers, which in total corresponded to approximately 10% of the starting amount of template–primer duplex. (Fig. 2C). In addition, the gel of the primer insertion reaction revealed a doublet of bands for the 16-nucleotide product. This suggested the incorporation of two different nucleotides across from 6-oxo-M1dG in the insertion step. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis determined that dAMP (37% of total) and dGMP (58% of total) were incorporated across from the 6-oxo-M1dG adduct in the 16-mer products (30). Very little dCMP (4.3% of total) and no dTMP incorporation were seen (Fig. S1).
Figure 2.
Incorporation of nucleotides opposite 6-oxo-M1dG by hPolη. Sequence of the DNA duplex with the standing start primer (A). Time course of nucleotide incorporation into the standing start primer opposite the control oligonucleotide template (B) or the 6-oxo-M1dG oligonucleotide template (C). Increasing concentrations of hPol (50–200 nM) were incubated with 5 μM DNA duplex and a 500 μM mix of all four dNTPs except for the primer input (P), which was incubated with 200 nM hPol , 5 μM DNA duplex, and an equal volume of water instead of dNTPs. The reaction was stopped at 5, 15, and 45 min.
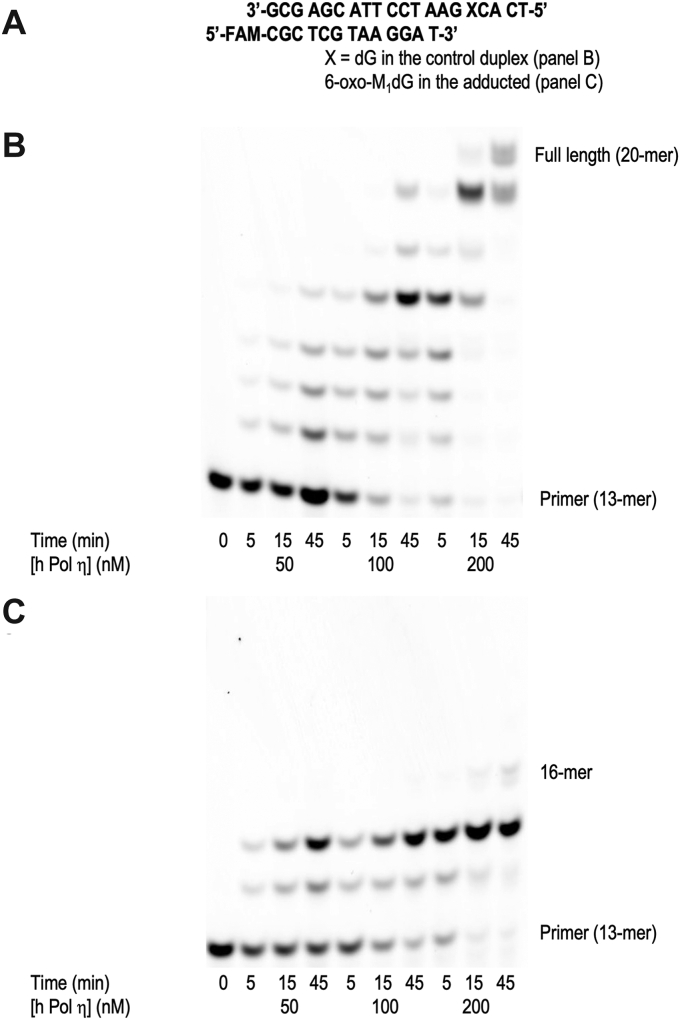
The primer extension reaction was also conducted with a 13-mer (running start) primer annealed to the control template or the 6-oxo-M1dG template (Fig. 3A). With the 6-oxo-M1dG template, hPol was able to efficiently extend the primer to 15 nucleotides, but it formed very little 16-mer product, which corresponded to incorporation of a nucleotide opposite 6-oxo-M1dG. Densitometry analysis of the 45 min time point with 200 nM hPol revealed insertion of the nucleotide across from 6-oxo-M1dG in only 10% of the primer–template duplex. Furthermore, extension occurred in less than 1% of the duplex. A faint band was seen corresponding to the 17-mer product. With the control template, hPol was able to completely extend either to a 19-nucleotide product or the full length 20-nucleotide product (Fig. 3, B and C).
Figure 3.
Incorporation of nucleotides opposite 6-oxo-M1dG by hPolη. Sequence of the DNA duplex with the running start primer (A). Time course of nucleotide incorporation into the running start primer using the control template (B) and the 6-oxo-M1dG template (C). Increasing concentrations of hPol (50–200 nM) were incubated with 5 μM DNA duplex and a 500 μM mix of all four dNTPs except for the zero time point, which was incubated with an equal volume of water. The reaction was stopped at 5, 15, and 45 min.
Steady-state kinetics analysis of dNTP insertion across from dG and 6-oxo-M1dG by hPol
Control or 6-oxo-M1dG oligonucleotide primer–templates were incubated with hPol and increasing concentrations of single dNTPs to determine the kinetics of insertion for each dNTP. The specificity constants (kcat/Km) were calculated to determine the catalytic efficiency of each insertion reaction by hPol and to compare kinetics among the individual dNTPs. As shown in Table 1, all four dNTPs were poorly incorporated across from 6-oxo-M1dG by hPol . The specificity constants for dGTP and dATP were the highest, at 0.039 and 0.034 μM−1 min−1, respectively. These results matched the LC-MS/MS data from the primer extension experiments that showed essentially all but a minor amount of extension products contained dGMP or dAMP across from 6-oxo-M1dG. The specificity constants for dCTP and dTTP were approximately 9-fold and 13-fold lower than those of dGTP, respectively. With the control duplex, dCTP was the preferred substrate for hPol incorporation across from dG with a specificity constant of 7.1 μM−1 min−1. The remaining three nucleotides were all poorly utilized compared with dCTP. The specificity constants were 100- to 3500-fold lower and ranged from 0.0020 to 0.067 μM−1 min−1.
Table 1.
Steady-state kinetics parameters for insertion of single nucleotides opposite dG and 6-oxo-M1dG by hPol
| Nucleotide | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) | Ki (μM) |
|---|---|---|---|---|
| dATP | 1.9 ± 0.3 | 47 ± 17 | 0.040 | ND |
| dCTP | 10 ± 2 | 1.4 ± 0.3 | 7.1 | 650 ± 225 |
| dGTP | 1.8 ± 0.1 | 27 ± 7 | 0.067 | 6600 ± 2100 |
| dTTP | 1.9 ± 0.9 | 950 ± 540 | 0.0020 | 2200 ± 1900 |
| Control oligonucleotide | ||||
| dATP | 0.91 ± 0.12 | 27 ± 10 | 0.034 | 2400 ± 1300 |
| dCTP | 0.27 ± 0.05 | 65 ± 33 | 0.0042 | 1800 ± 950 |
| dGTP | 0.51 ± 0.03 | 13 ± 3 | 0.039 | 5020 ± 1400 |
| dTTP | 1.1 ± 0.3 | 370 ± 190 | 0.0030 | 5000 ± 4200 |
| 6-Oxo-M1dG oligonucleotide |
Abbreviation: ND, not detectable.
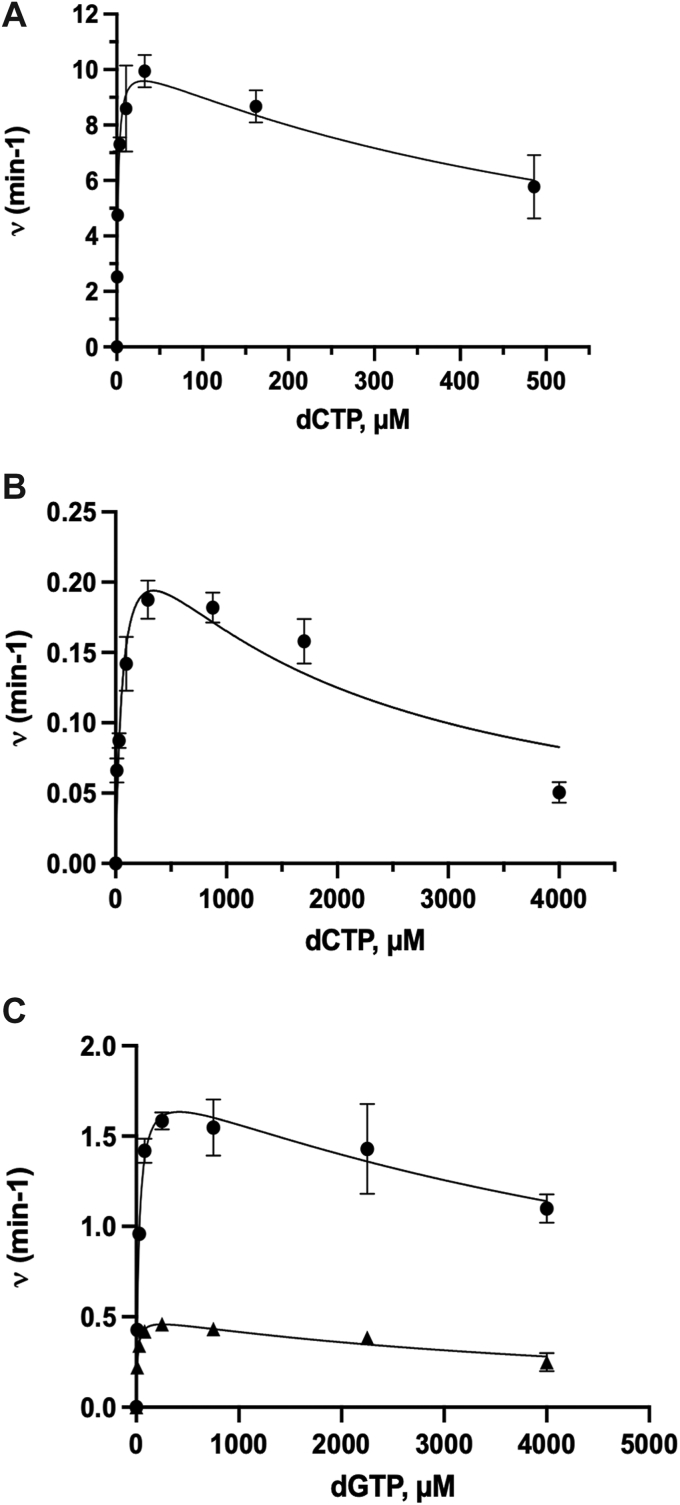
The Michaelis–Menten plots for incorporation of dCTP and dGTP are shown in Figure 4 and those for dATP and dTTP in Fig. S2. Interestingly, substrate inhibition is seen to varying degrees with all of the nucleotides. The data were analyzed in Prism using the substrate inhibition model, and Ki values were also measured for all nucleotides as shown in Table 1. The Ki values for substrate inhibition were lowest for dCTP, at 650 μM for the control oligonucleotide duplex and 1800 μM with the 6-oxo-M1dG oligonucleotide duplex. The Ki values were substantially higher for the other nucleotides regardless of the primer–template used. Gel electrophoresis data used to derive these plots are shown in Figs S3–S6.
Figure 4.
Steady-state kinetics for dCTP and dGTP incorporation opposite dG or 6-oxo-M1dG by hPolη. Control (A) and 6-oxo-M1dG (B) oligonucleotide duplexes (5 μM) were incubated with 10 nM or 200 nM hPol , respectively, and increasing concentrations of dCTP for 6 min for control duplex or 10 min for 6-oxo-M1dG duplex. C, control (•) and 6-oxo-M1dG (▲) oligonucleotide duplexes (5 μM) were incubated with 50 nM or 200 nM hPol , respectively, and increasing concentrations of dGTP for 10 min. Each point represents the mean and standard deviation of triplicate determinations. Data were analyzed using the substrate inhibition model in GraphPad Prism.
Substrate inhibition was not observed during steady-state kinetics analysis of M1dG bypass by hPol (19). There were two main differences between the experiments previously used to determine steady-state kinetics parameters for M1dG and those used here for 6-oxo-M1dG: a higher concentration of enzyme and primer–template duplex for 6-oxo-M1dG and also higher nucleotide concentrations were employed in the present work. To determine if substrate inhibition was an artifact of using a higher concentration of enzyme and substrate, we repeated the experiment for steady-state kinetics analysis of dCTP incorporation into the control primer–template duplex at a much lower enzyme (1.6 nM) and primer–template duplex (80 nM) concentration. A 1-min incubation period ensured linear enzyme activity under these conditions. We analyzed product formation both by gel electrophoresis and LC-MS/MS analysis (26). As seen in Figs S7 and S8, substrate inhibition also occurred with the lower enzyme and primer–duplex concentrations, suggesting that hPol activity decreased with increasing dCTP starting at high micromolar to low millimolar concentrations, suggesting that substrate inhibition is not dependent on the amount of enzyme and primer–template used.
Extension of primers containing a nucleotide across from 6-oxo-M1dG by hPol
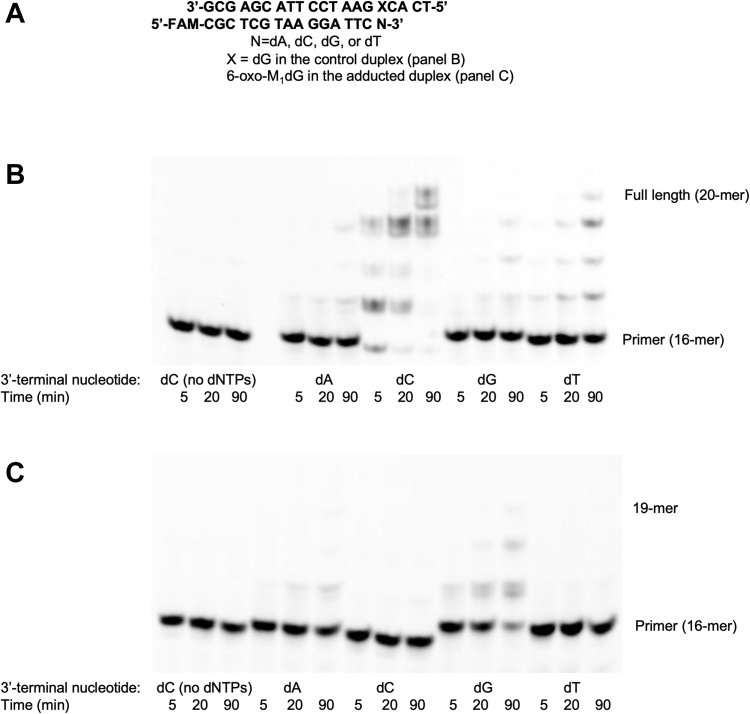
We investigated extension of 16-nucleotide primers containing a 3′-terminal dA, dC, dG, or dT annealed to the 6-oxo-M1dG oligonucleotide or the control oligonucleotide. The 6-oxo-M1dG adduct or dG in the control template was placed directly across from the 3′-terminal nucleotide in the complementary 16-mer primer (Fig. 5A). With the 6-oxo-M1dG primer–template, the 16-mers with a 3′-terminal dA or dG displayed the most extension, at 16% or 58% by the 90 min time point, respectively (Fig. 5B). Primers with a 3′-terminal dC or dT exhibited extension of 3% of the primer–template duplex. Thus, not only are dAMP and dGMP the most common nucleotides inserted across from 6-oxo-M1dG, they are more likely to be further extended past this point. With the control template primer duplex, the 16-mer with a 3′-terminal dC showed the greatest amount of extension. Over 90% of the template–primer duplex was extended either to 19 or 20 nucleotides, consistent with the expected correct Watson–Crick base pairing of the terminal dC with the dG in the control oligonucleotide. The amount of extension seen with the primers containing a 3′-terminal dA, dG, or dT was noticeably less (Fig. 5C).
Figure 5.
Extension of 6-oxo-M1dG-containing and control oligonucleotide duplexes by hPolη.A, sequence of the duplexes used to evaluate extension past 6-oxo-M1dG. HPol was incubated separately or together with a 500 μM mix of all four dNTPs and the indicated control (B) or 6-oxo-M1dG-containing (C) oligonucleotide duplexes. The polymerase concentrations of hPol and the oligonucleotide complexes were 200 nM and 5 μM, respectively, and the reactions were stopped after the indicated times. The first three lanes for both the 6-oxo-M1dG oligonucleotide duplex and control oligonucleotide duplex contained an equal volume of water in place of the added dNTPs.
Crystal structures of hPol insertion- and extension-stage complexes
The results of the in vitro insertion and extension experiments indicate that 6-oxo-M1dG is a strong block to replication by hPol . The results with hPol are particularly surprising because this polymerase has been demonstrated to bypass a range of DNA adducts including some that are quite large (27, 28, 29). To get insight into the basis for the observed replication block, we attempted to determine the structures of 6-oxo-M1dG-containing template–primers bound to hPol during insertion or extension steps.
For crystallization experiments intended to produce ternary complexes composed of hPol , DNA template–primer duplex, and incoming nucleoside triphosphate, we prepared two template strands with incorporated 6-oxo-M1dG (X): 5′-d(CAT XAT GAC GCT)-3′ and 5′-d(CAT GXT GAC GCT)-3′. The first 12-mer was combined with the primer 3′-d(TA CTG CGA)-5′ and, separately, with dATP, dCTP, dGTP, or dTTP, to capture an insertion-stage polymerase complex. The second template strand was combined, separately, with one of the four primers 3′-d(NA CTG CGA)-5′ (N = A, C, G, T) and dCTP to capture an extension-stage polymerase complex. Thus, 6-oxo-M1dG was placed opposite dA, dC, dG, or dT in separate binary hPol –DNA complexes, and the incoming dCTP was anticipated to pair with template G that was located 5′-adjacent to the adduct. Crystallization conditions were screened for hPol insertion and extension complexes with these combinations of template and primer strands as well as incoming nucleotides in the presence of Ca2+. Crystals obtained in various droplets were mounted in loops, cryoprotected, and stored in liquid nitrogen prior to testing them for diffraction at the Advanced Photon Source.
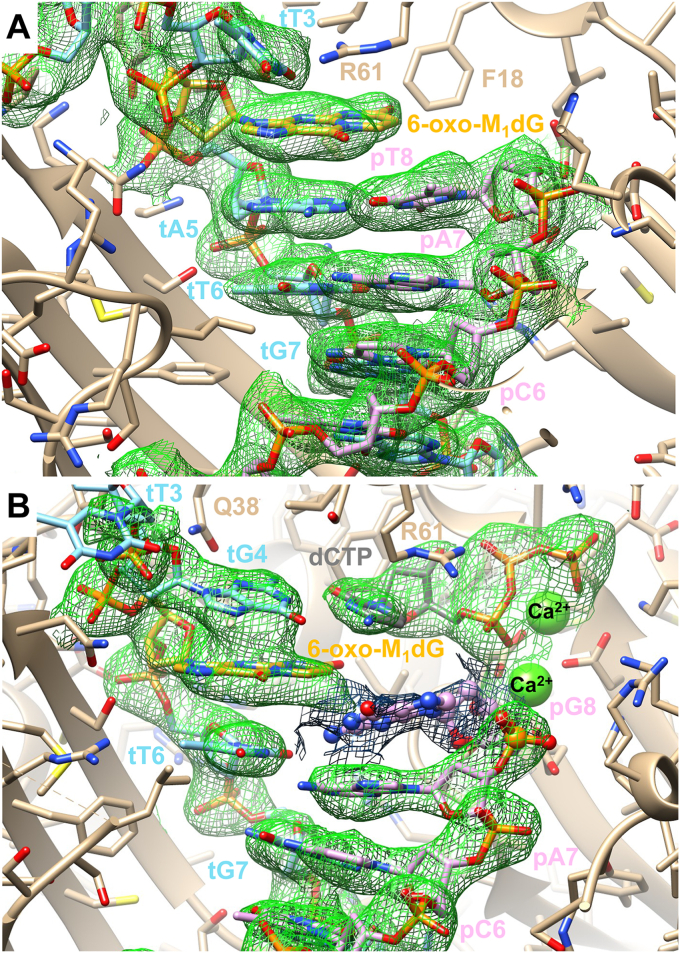
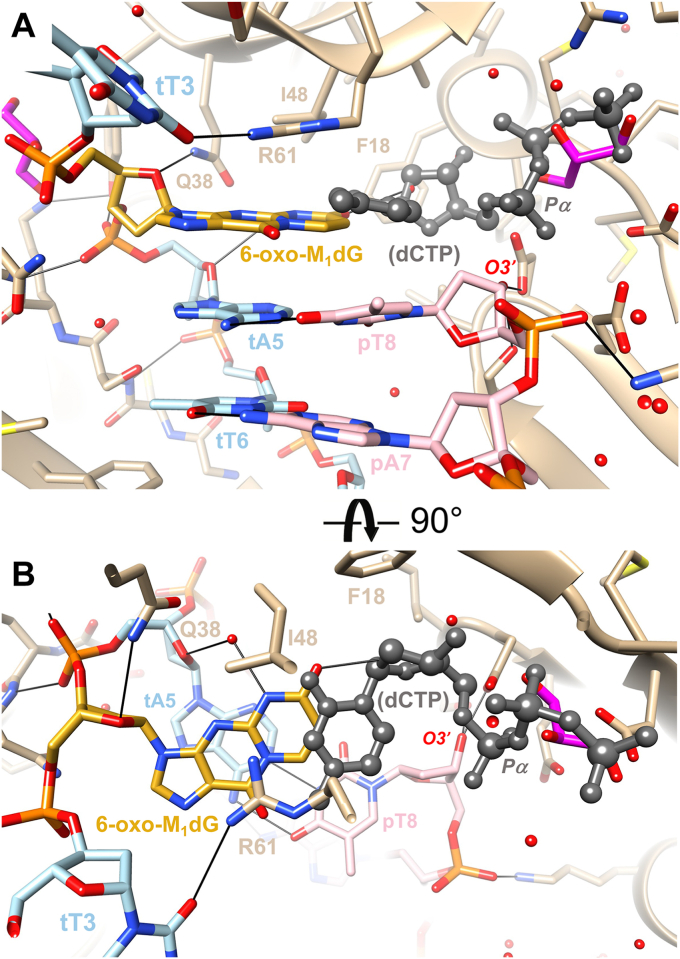
We identified two viable crystals. The first comprised hPol crystalized in the presence of the insertion-stage oligonucleotide duplex and dCTP. This putative insertion complex diffracted to 2.09-Å resolution. The second comprised hPol crystalized in the presence of the extension-stage oligonucleotide duplex containing dG at the 3′-terminus of the primer strand and dCTP. This extension complex diffracted to 2.61-Å resolution. Selected crystal data, data collection statistics, and refinement parameters are listed in Table 2, and illustrations depicting the quality of the final electron density in the active site region of the insertion and extension-stage complexes are shown in Figure 6, A and B, respectively. Nucleotides in the template (t) strand are numbered 1 to 12 from the 5′- to 3′-end, and nucleotides in the primer (p) strand are numbered 1 to 8 from the 5′- to 3′-end. The crystal structure of the insertion-stage complex revealed that the adduct is in an anti-conformation at the active site that was devoid of an incoming nucleotide (Fig. 6A). The 6-oxo-M1dG base moiety does not engage in direct interactions with side- or main-chain atoms of polymerase residues (Fig. 7) and instead forms extensive stacking interactions with the dA5:pT8 base pair (Fig. 7B).
Table 2.
Selected crystal data, X-ray data collection statistics, and refinement parametersa
| Complex | Insertion | Extension |
|---|---|---|
| Data Collection | ||
| Space group | P61 | P61 |
| Unit cell constants: a, b, c [Å] | 98.99, 98.99, 81.47 | 98.67, 98.67, 81.74 |
| Unit cell constants: α, β, γ [°] | 90, 90, 120 | 90, 90, 120 |
| Resolution [Å] | 32.40–2.35 (2.41–2.35) | 30.00–2.87 (2.92–2.87) |
| Wavelength [Å] | 1.12723 | 1.12723 |
| No. of unique reflections | 18,885 (1397) | 10,308 (536) |
| Completeness [%] | 99.50 (99.50) | 98.50 (99.30) |
| R-merge | 0.087 (0.674) | 0.106 (0.697) |
| R-pim | 0.064 (0.499) | 0.049 (0.392) |
| I/σ(I) | 10.30 (2.10) | 15.68 (1.98) |
| Redundancy | 5.30 (5.30) | 5.40 (4.10) |
| Refinement | ||
| No. of protein molecules/DNA duplexes per a.u. | 1/1 | 1/1 |
| Resolution [Å] | 32.40–2.35 (2.47–2.35) | 29.53–2.87 (3.28–2.87) |
| Number of reflections | 18,867 (2559) | 10,282 (3295) |
| R-work | 0.189 (0.230) | 0.204 (0.262) |
| R-free | 0.244 (0.292) | 0.251 (0.331) |
| No. of protein/nucleotide atoms | 3327/393 | 3225/374 |
| No. of waters/ions/ligands | 90/3/7 | 23/2/1 |
| R.m.s. deviations bonds [Å] | 0.008 | 0.004 |
| R.m.s. deviations angles [°] | 1.00 | 0.75 |
| Avg. B-factor, protein/nucleotide atoms [Å2] | 48.82/49.69 | 52.81/53.35 |
| Avg. B-factor, H2O/ions/ligands [Å2] | 48.05/67.64/53.90 | 48.77/50.07/43.20 |
| PDB entry code | 8EVE | 8EVF |
Numbers in parentheses refer to the outermost shell.
Figure 6.
Final Fourier 2Fo-Fcsum electron density around the DNA template–primer duplex in the active site region.Green depicts the 1.1 threshold, and gray, the 0.7 σ threshold. A, active site of the pol •DNA insertion-stage complex viewed from the major groove side of the DNA duplex. Carbon atoms of hPol , template, primer, and the 6-oxo-M1dG residue are colored in tan, light blue, pink, and golden rod, respectively. B, active site of the pol •DNA•dCTP extension-stage complex viewed from the major groove side of the DNA duplex. The color code is the same as in A; Ca2+ ions are shown as green spheres and carbon atoms of the incoming dCTP are colored in gray. The 3′-terminal pG8 in a partially occupied orientation is depicted in ball-and-stick mode.
Figure 7.
Crystal structure of an hPolη•DNA insertion-stage complex. The polymerase active site (A) viewed from the major groove side of the DNA template–primer duplex and (B) rotated around the horizonal axis by 90°, viewed from the ceiling and along the base stack. Carbon atoms of hPol , template, primer, and the 6-oxo-M1dG residue are colored in tan, light blue, pink, and golden rod, respectively. Selected residues and atoms are labeled, H-bonds are thin solid lines, and water molecules are small red spheres. The structure of the complex did not reveal an incoming dCTP, and the nucleotide depicted in ball-and-stick mode is from the superimposed structure of the extension-stage complex to demonstrate a clash between incoming residue and the adduct in an anti-conformation. Two glycerol molecules located in the vicinity of the active site are shown with carbon atoms colored in magenta.
When the extension-stage complex was superimposed on the insertion-stage complex, the incoming dCTP in the former clearly clashed with the adduct lodged at the active site of the latter (gray dCTP in ball-and-stick mode in Fig. 7). Thus, the Watson–Crick edge of the cytosine base is shifted over atoms of the outer six-membered ring of the adduct in this hypothetical scenario (Fig. 7B). The only way to accommodate dCTP would require a staggered arrangement between adduct and incoming nucleoside triphosphate, with suboptimal stacking on the adjacent template–primer base pair. Instead, at the concentrations used for crystallization of the complex, dCTP was not inserted, and the adduct occupies most of the hPol active site. Arg-61 that protrudes from the ceiling of the active site is stacked on 6-oxo-M1dG and turned away from the α- and β-phosphates of the incoming nucleotide with which it normally interacts. No calcium ions were observed at the active site, and a glycerol used to cryoprotect the crystal is instead trapped in the region normally occupied by the β- and γ-phosphate moieties of the incoming dCTP (Fig. 7).
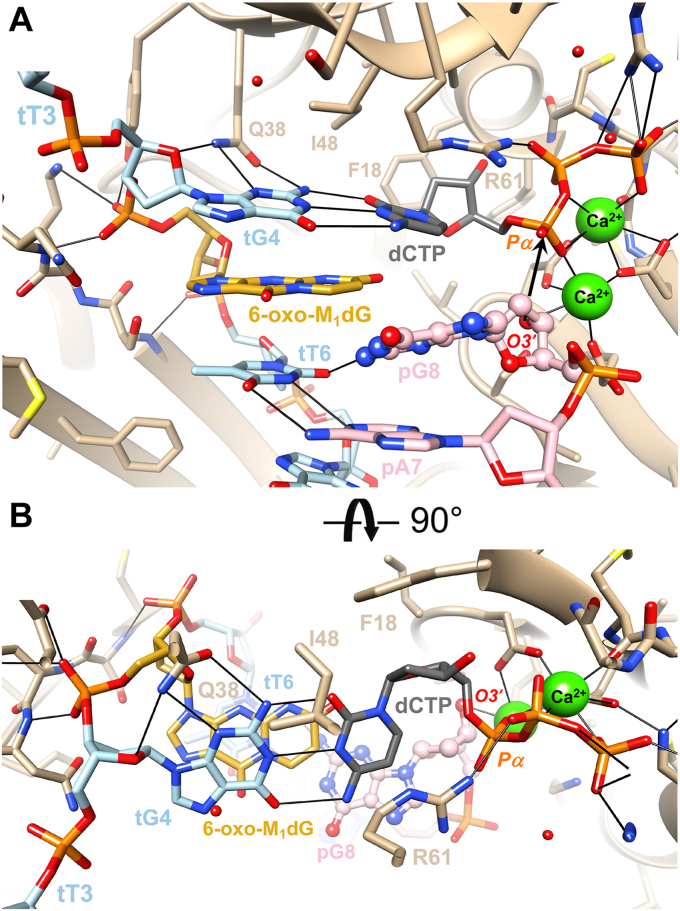
In the extension-stage complex, the adduct maintains an anti-conformation and forms a stack with G4 and T6 of the template strand (Figs. 6B and 8). The former is engaged in a canonical base pair with the incoming dCTP, whereby Gln-38 forms H-bonds with the minor groove edge of guanine (N2 and N3) as well as O4′ of the deoxyribose. Arg-61 is now directing its guanidino moiety toward the α- and β-phosphates of the incoming nucleotide, and the active site harbors two calcium ions (Fig. 8). The position of the 3′-terminal residue of the primer, G8, is only partially occupied judging from the relatively weak electron density (Fig. 6B). In the refined model, it assumes a staggered orientation vis-à-vis the adduct, and the N2 amino group of guanine is engaged in an H-bond (2.58 Å) with O2 of template T6 underneath 6-oxo-M1dG. As a result, the primer residue opposite template T6, A7, is pushed down, and distances of 3.36 Å between O4 and N6 and 3.43 Å between N3 and N1 suggest a weakened T:A pair (Fig. 8A). As in the case of the insertion-stage complex, no H-bond interactions are observed between hPol side chains and the 6-oxo-M1dG base moiety. Interestingly, the wedge-like arrangement of the tG4:dCTP and tT6•pG8 pairs that wrap around the adduct base places O3′ of the 3′-terminal primer G8 at 4.27 Å from P of the incoming dCTP (Fig. 8A). The near in-line orientation of O3′(tG8) and P-O3A(dCTP) together with the O3′-P distance in the refined model is compatible in principle with an extension of the primer by hPol in the presence of the 6-oxo-M1dG adduct (Fig. 8). However, the partial occupancy of the base and deoxyribose of the terminal primer residue indicate a dynamic makeup of the active site and argue against a conformationally preorganized O3′-nucleophile⋅⋅⋅α-phosphate pair for facile extension.
Figure 8.
Crystal structure of a ternary polη•DNA•dCTP extension-stage complex. The polymerase active site (A) viewed from the major groove side of the DNA template–primer duplex and (B) rotated around the horizonal axis by 90°, viewed from the ceiling and along the base stack. The color code is identical to that in Figure 6. Nucleobase and sugar atoms of the 3′-terminal nucleotide of the primer strand, pG8, are highlighted in ball-and-stick mode to indicate partial occupancy of this residue. Calcium ions are shown as green spheres, and the putative nucleophilic attack of O3′(pG8) at the α-phosphate group is indicated with an arrow (A).
Discussion
Previous studies had shown that a member of the Y-family of polymerases, hPol ι, can insert dCMP or dTMP across from 6-oxo-M1dG but is unable to catalyze further extension (26). HPol differs in that it inserts a dAMP or dGMP across from the adduct and is able to catalyze a marginal amount of extension past this point with high polymerase concentrations and extended times. Steady-state kinetics analysis of nucleotide incorporation into a primer–template in which the primer terminus is one nucleotide short of the adduct site on the template (standing start primer) demonstrated that dGMP had the highest specificity constant for insertion across from 6-oxo-M1dG (0.039 μM−1 min−1) and that for dAMP was slightly lower (0.034 μM−1 min−1). The specificity constants for dCMP and dTMP were 8.1- and 11-fold less than that for dAMP. LC-MS/MS analysis of the 16-nucleotide products from nucleotide extension reactions was consistent with the steady-state kinetics results and revealed 58% of the products contained dGMP and 37% of the products contained dAMP across from the adduct. The use of a standing start primer provides a maximal opportunity for polymerase-catalyzed insertion and extension. Indeed, when the primer strand terminates three nucleotides short of the adduct position on the template (running start primer), hPol catalyzes primer extension to the site of the adduct but barely catalyzes synthesis beyond that point (Fig. 3C). Thus, experiments with both a standing start and a running start primer indicate 6-oxo-M1dG is a strong block to replication by hPol .
In the steady-state kinetics experiments, substrate inhibition was seen at high nucleotide concentrations. This phenomenon had not been seen with M1dG, and a literature survey did not uncover any mention of it in other in vitro replication studies. Data from most prior investigations of steady-state kinetics values for hPol have been reported in tabular form, making it impossible to evaluate the presence of substrate inhibition in those studies. The investigations of hPol activity that presented results as graphs of nucleotide concentration versus enzyme activity did not use a high enough nucleoside triphosphate concentration to observe substrate inhibition as observed in the present work; however, in a few of these articles a slight decrease in activity at the highest nucleotide concentration was observed (31, 32). It is possible that activity would have continued to decrease with increasing concentrations of nucleotides in those assays. Notably, the Ki values reported in Table 1 are all over 600 μM, whereas the intracellular concentrations of nucleotides are reported to be below 50 μM (33). Thus, the observed substrate inhibition reported here is unlikely to be of great physiological significance.
Contrary to the previous results with hPol ι, hPol displayed a small amount of extension past the adduct in standing start experiments. With the highest enzyme concentration and the longest time point in the standing start primer extension experiment, hPol inserted a nucleotide across from the adduct in approximately 35% of the template–primer duplex and was able to extend approximately 12% of the total template–primer duplex. Although dAMP is a comparable substrate to dGMP for the insertion reaction, a 3′-terminal dGMP across from the adduct (dG•6-oxo-M1dG pair) appears to be the most favorable nucleotide for extension, with 58% of the total primer–duplex displaying some amount of further nucleotide incorporation compared with 16% when the 3′-terminal nucleotide is dAMP (dA•6-oxo-M1dG pair). When a template that contains a dG residue is paired with a primer containing dA or dG at the 16th position, hPol extends the primer very poorly. This contrasts sharply with the results described above for 6-oxo-M1dG and suggests that the presence of the adduct actually facilitates extension from a mismatched terminus. Thus, 6-oxo-M1dG is capable of inducing an initial mutagenic insertion by hPol opposite the adduct and then “fixing” it by enabling extension of the mutagenic primer terminus.
6-Oxo-M1dG is an in vivo intragenomic metabolite of M1dG that arises by introduction of an oxygen atom at the 6-position of the exocyclic ring followed by tautomerization to an amide functionality. This introduces a small amount of additional steric bulk in the minor grove that leads to a dramatic difference in replication outcome compared with that of M1dG. Previous experiments indicate that hPol efficiently bypasses M1dG with a marked preference for incorporation of dATP opposite the adduct. Using template–primers with identical sequence contexts in the vicinity of the adduct reveals that the misincorporation frequency (relative to dCTP opposite dG) for dATP opposite M1dG is 0.12, whereas for dATP opposite 6-oxo-M1dG, the misincorporation frequency is 0.0047. The misincorporation frequencies for dGTP opposite M1dG and 6-oxo-M1dG are 0.007 and 0.0055, respectively. With primers containing a 3′-terminal nucleotide paired with the M1dG adduct, a 3′-terminal dCMP had the highest activity ratio for extension with a single nucleoside triphosphate followed by dAMP. The activity ratio for a dG•dC pair was similar in catalytic efficiency to that of a M1dG•dC pair. In contrast, hPol preferentially extended primers with dGMP opposite 6-oxo-M1dG followed by dAMP. Extension of a primer–template with dCMP opposite 6-oxo-M1dG was ∼20-fold less efficient than extension of dGMP.
Crystallizations were conducted to generate ternary complexes of hPol , a 6-oxo-M1dG-containing template–primer and an incoming dNTP. Crystals were obtained from one template–primer representing an insertion-stage complex and one template–primer representing an extension-stage complex, and structures were obtained via X-ray diffraction. The crystal structure of hPol at the insertion stage with template 6-oxo-M1dG lodged at the active site does not reveal an incoming dCTP, although it was in the crystallization mix. The isolation of only a binary hPol -template–primer complex is consistent with the adduct’s ability to act as a strong replicative block (Figs. 6A and 7). In the crystal structure of a ternary hPol •DNA•dCTP complex at the extension stage, the 3′-terminal G of the primer strand is accommodated in a staggered orientation opposite the adduct in the anti-conformation. This results in a base triple involving template T6, the 3′-terminal primer G, and the penultimate A from the primer. The wedge-like arrangement between tG:dCTP above the adduct and tT:pG below the adduct places O3′ of the 3′-terminal primer G at ca. 4.3 Å from the Pα phosphate moiety (Fig. 8). However, the 3′-terminal nucleotide of the primer is only partially ordered in the crystal structure (Fig. 6B), consistent with increased mobility of this residue at the active site of the extension-stage complex. The relative orientation of the primer O3′ and the dCTP P-O bond in the refined model with partially occupied positions of tG8 base and sugar atoms does not preclude an extension of the primer by hPol but suggests it could be greatly slowed. Thus, the structural data provide explanations for the fact that 6-oxo-M1dG strongly retards extension.
The anti-conformation of 6-oxo-M1dG in the binary and ternary complexes introduces significant steric hindrance to the hPol active site. Modeling the incoming dCTP of the extension-stage complex to the structure of the insertion-stage complex reveals multiple potential steric clashes between the pyrimidine ring and the exocyclic ring of 6-oxo-M1dG. These would likely preclude facile dNTP binding and dramatically slow the insertion process. The anti-conformation is analogous to the anti-conformation observed in complexes like the archeal Y-family polymerase Dpo4 with M1dG (17).
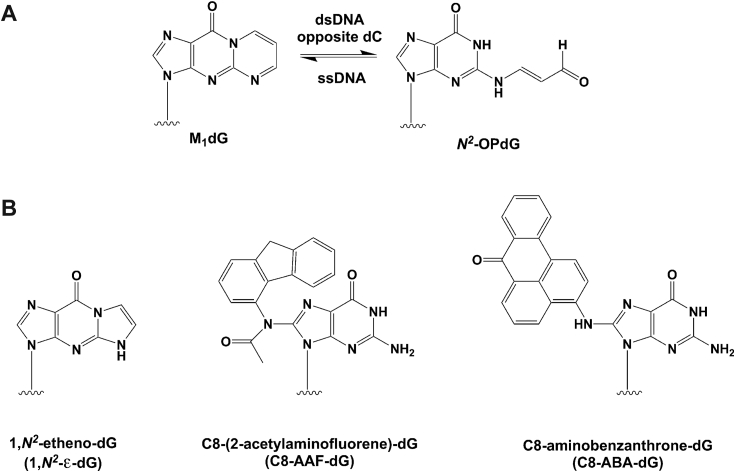
The significant differences in bypass efficiency and outcome in the reactions of hPol with 6-oxo-M1dG-versus M1dG-containing template–primers constitute a dramatic example of the impact a single atom—in this case oxygen—can have on DNA replication. What is the basis for this difference? An attractive hypothesis is based on the chemical stability and behavior of M1dG and 6-oxo-M1dG in duplex DNA. M1dG is unstable to base above pH 9 and ring-opens to N2-oxopropenyl-dG (N2-OPdG) (Fig. 9A). When M1dG is placed in duplex DNA opposite dC at neutral pH, it ring-opens to N2-OPdG in a process that is catalyzed by DNA and is fully reversible on denaturing the duplex DNA (34). N2-OPdG base-pairs to dC, is bypassed more efficiently than M1dG, and is much less mutagenic (35). In contrast, 6-oxo-M1dG does not ring-open when exposed to base or when positioned in duplex DNA opposite dC. Ring-opening of M1dG to N2-OPdG in the active site of hPol could explain its more facile bypass and lower mutagenicity than 6-oxo-M1dG. Despite the attractiveness of this hypothesis, there is currently no evidence to support it. The structures of complexes of Dpo4 bound to template–primers containing M1dG with an incoming dGTP or with a dCMP at the end of the primer strand directly opposite the adduct reveal that M1dG exists only in the closed exocylic form (17). Importantly, the bypass efficiency and outcome of Dpo4 replication of M1dG-containing template–primers are very similar to that of hPol in that the lesion is readily bypassed and dATP is the preferred dNTP incorporated. Thus, it seems likely that M1dG at the active site of hPol adopts the anti-conformation seen in Dpo4.
Figure 9.
Ring-opening of M1dG and guanine adducts repaired by hPol η. Ring-opening of M1dG (A) and guanine adducts repaired by hPol η (B).
As far as bypass by hPol is concerned, 6-oxo-M1dG stands out among exocyclic dG lesions in terms of its strong blockage of insertion and extension. 6-Oxo-M1dG also constitutes a more serious obstacle to hPol than several other bulky adducts of dG. Thus, hPol can bypass 1,N2-etheno-dG (1,N2--dG) (Fig. 9B) by mostly incorporating purines opposite the adduct. Single nucleotide incorporation assays with hPol opposite 1,N2--dG revealed the following order of insertion: dGTP > dATP > dTTP > dCTP (27). When full-length extension products were analyzed, dG accounted for 85% of nucleotides inserted opposite the lesion and 63% of nucleotides in the first extension step. Extension from a misincorporated dA was also possible, but extension from the correct dC was not observed. The crystal structure of the ternary hPol •DNA•dAMPPnP (dAMPPnP is a nonhydrolyzable analogue of dATP) showed the adenine base staggered between 1,N2--dG in a syn orientation and the 5′-adjacent template dT. This arrangement was stabilized by a bifurcated H-bond between N6(H)2 of A and O6 of 1,N2--dG and O4 of dT. The 3′-terminal primer dC was coplanar with syn 1,N2--dG in the crystal structure of the ternary complex with formation of a single H-bond between N4(H)2 of C and O6 of 1,N2--dG. The syn orientation of 1,N2--dG seen in the structures accounts for miscoding combined with limited blockage, as the two exocyclic carbon atoms of the lesion are rotated into the major groove, thereby resulting in the Hoogsteen edge of G facing the incoming base.
Two other bulky dG adducts that can be bypassed by Pol are C8-(2-acetylaminofluorene-dG) (C8-AAF-dG) and C8-aminobenzanthrone-dG (C8-ABA-dG) (Fig. 9B). In both cases, Pol correctly inserts dC opposite the adduct and then extends from there. With C8-AAF-dG, the crystal structure of a first complex between Saccharomyces cerevisiae Pol and an adducted primer–template duplex revealed that the acetylaminofluorene moiety is stacked on the previous Watson–Crick base pair with guanine lodged in the major groove (29). In the second complex, dG has moved toward the templating position in a syn orientation, thereby partially freeing the active site and pushing the AAF moiety into the minor groove. A model of the correct bypass scenario envisions guanine in the syn orientation and formation of a single H-bond between O6 and N4(H)2 of the incoming dCTP (29). Although constituting a considerable TLS block, the C8-AAF-dG adduct can thus be bypassed in a largely error-free manner.
The same is true for the C8-ABA-dG adduct (Fig. 9B), although the crystal structure of the ternary hPol •DNA•C8-ABA-dG complex reveals that the basis underlying the correct bypass by this polymerase here is quite different from that seen in the case of C8-AAF-dG (28). Specifically, guanine is accommodated at the hPol active site in the standard anti orientation opposite the incoming dCTP. The aminobenzanthrone moiety is wedged into a mostly hydrophobic pocket to the side of the hPol active site. In this fashion, it is kept out of the way and precluded from stacking interactions with the previous template–primer base pair, thereby blocking the incoming nucleotide and TLS. While individual dG adducts have different consequences for bypass, e.g., reduced TLS efficiency paired with miscoding/frameshifting or slower bypass but mostly error-free insertion and extension, none appears to hamper TLS polymerases to the extent that 6-oxo-M1dG does. It will be exciting to evaluate the mutagenicity and replication blockade by 6-oxo-M1dG in vivo.
Experimental Procedures
Reagents
All DNA oligonucleotides with the exception of the 6-oxo-M1dG oligonucleotide were from Integrated DNA Technologies (IDT), Inc. Methanol, acetonitrile, and water were HPLC grade and were purchased from Fisher. Formic acid, triethylamine, and hexafluoroisopropanol were from Sigma. All chemicals were the best available quality. PreScission protease was from Apex bio.
Purification of hPol η
The codon-optimized cDNA clone encoding hPol (residues 1–432 in a pET28a plasmid) developed in the lab of Wei Yang (National Institute of Diabetes and Digestive and Kidney Diseases) was a gift from Dr F.P. Guengerich, Vanderbilt University. The encoded protein contains a 6x-histidine tag and a PreScission protease recognition site (LEVLFQGP) at its N-terminal end. The plasmid was transformed into E. coli BL21 (DE3) pLysS cells, which were then grown at 37 ˚C and 250 rpm until A600 reached 0.6. At that time, 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added, and the cells were grown at 30 ˚C and 250 rpm for 3 h. The cells were centrifuged, and the pellet was resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4; 0.5 M NaCl; 10% glycerol; 5 mM β-mercaptoethanol, 1 mg/ml lysozyme, and a protease inhibitor cocktail [Roche]) and incubated on ice for 30 min. The cells were disrupted by sonication, and the lysate was cleared by centrifugation at 16,000 rpm for 30 min. The soluble portion was added to a 10-ml column of Ni-NTA resin equilibrated with 50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and 10 mM imidazole. The resin was washed using the same buffer with the imidazole concentration increased to 30 mM. HPol was eluted from the column by adding 50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and 400 mM imidazole. Fractions (1 ml) were collected, and those that contained hPol (as determined by SDS-PAGE) were pooled and concentrated using an Amicon 30 concentrator (Millipore). The concentrated sample was treated by PreScission protease for 16 h at 4 °C. Protease-treated hPol was added to a 120-ml exclusion column (HiLoad 16/600 Superdex 200 pg) equilibrated with 25 mM Hepes (pH 7.5), 0.1 M NaCl, 5 mM β-mercaptoethanol, and 10% glycerol. Fractions (5 ml) were collected. Those containing hPol were concentrated using an Amicon-30 concentrator, and the concentrated sample was diluted 2-fold with 25 mM Hepes (pH 7.5), 0.1 M NaCl, 5 mM β-mercaptoethanol, and 50% glycerol. The enzyme was divided into 20-μl aliquots, and these were flash frozen and stored at −80 °C.
Nucleotide insertion and extension past the 6-oxo-M1dG adduct
Generation of the primer–template DNA duplex for in vitro assays
A 20-mer oligonucleotide (5′-TC ACX GAA TCC TTA CGA GCG-3′, X = site of 6-oxo-M1dG incorporation) was synthesized and characterized as described (26). Control oligonucleotide (5′-TC ACG GAA TCC TTA CGA GCG-3′) or the 6-oxo-M1dG oligonucleotide was mixed with an equimolar amount of 5′ FAM (fluorescein amidite)-labeled complementary primer (5′-FAM-CGC TCG TAA GGA TTC-3′). The mixture was heated at 95 °C for 5 min and allowed to cool slowly overnight.
Primer extension assays
Primer extension assays were conducted at 37 °C with 5 mM dithiothreitol, 0.05% bovine serum albumin, 5% glycerol, 5 mM MgCl2, 50 mM NaCl, 40 mM Tris-HCl (pH 7.5), 5 μM primer–template complex, and 250 to 500 μM dNTP mix (all four dNTPs) or 500 μM single dNTPs (dATP, dCTP, dGTP, or dTTP). The concentration of hPol varied from 25 to 250 nM. The total reaction volume was 10 μl. The reactions were stopped at time points from 0 to 90 min by removing 2 μl from the total reaction volume and heating at 95 °C for 5 min. Water (8 μl) was added to each sample, and 1 μl of this was combined with quench solution (95% formamide, 20 mM ethylaminediaminetetraacetic acid, 0.1% bromophenol blue, and 0.1% xylene cyanol). The samples were then separated by electrophoresis on a 20% (w/v) denaturing polyacrylamide gel with 7 M urea. The gels were imaged on a Typhoon Trio Mode Imager in fluorescence mode with the green (532 nm) laser in conjunction with the 526-nm short-pass filter.
Mass spectrometry analysis of oligonucleotides from primer extension assays
A portion of the quenched reaction volume (7 μl) was added to 50 μl of water, and 20 μl of the total was analyzed by LC-MS/MS. All LC-MS/MS analyses were done on a Shimadzu Nexera X2 system in-line with a SCIEX 6500 QTrap mass spectrometer. Oligonucleotides were chromatographed on a reverse-phase system using an Acquity BEH C8 column (10 × 0.2 cm, 1.7 μm) held at 60 °C under a gradient elution scheme. Mobile phase component A was water with 15 mM triethylamine and 100 mM hexafluoroisopropanol plus 1% (v:v) methanol. Component B was 1:1 (v:v) methanol:acetonitrile with 15 mM triethylamine and 100 mM hexafluoroisopropanol. A typical gradient was 4% B for 0 to 0.5 min, followed by a linear increase to 19% B over 6 min followed by a 2-min hold at 19% B. The column was equilibrated for 2 min before each injection. The SCIEX Analyst software package was used to collect and process data. The predicted mass of each oligonucleotide was calculated using the Mongo Oligo Mass Calculator from Prof Jef Rozenski (ORCID 0000-0001-9624-5536) (http://mass.rega.kuleuven.be/mass/mongo.htm).
Steady-state kinetics analysis
Steady-state kinetics reactions were performed using a duplex of control template annealed to the complementary FAM-labeled primer or a duplex of 6-oxo-M1dG template annealed to the same complementary FAM-labeled primer. The reaction time was optimized for initial velocities, and reactions were carried out at 37 °C for 5 to 10 min. The reaction mixture was the same as listed for primer extension assays except dNTP concentrations varied from 0 to 4000 μM. Reactions were terminated, and products were electrophoresed and visualized as described for primer extension and single nucleotide insertion assays. Bands were quantified using Image J (downloaded from nih.gov), and kinetic parameters were determined as described (30).
Statistical analyses
From the gel electrophoresis data analyzed using Image J, the ratio product/(product + substrate) was calculated (Rp). The turnover value (ν in min−1) was calculated according to the equation below, with Di as the initial oligonucleotide duplex concentration, E as the enzyme concentration, and t as time in min:
Using GraphPad Prism, the turnover values were plotted for each nucleotide concentration. The graphs were analyzed by nonlinear regression using the substrate inhibition model to determine kcat, Km, and Ki. The results in Table 1 are from a representative experiment done in triplicate for each dNTP. Values are presented as mean ± standard deviation.
Crystal structures of ternary hPol •DNA•dNTP complexes
Crystallization experiments
Solutions of a 6-oxo-M1dG-modified template and the complementary primer DNAs were mixed and annealed at a 1:1 molar ratio in 10 mM sodium Hepes buffer (pH 8.0), 0.1 mM ethylaminediaminetetraacetic acid, and 50 mM NaCl at 85 °C for 5 to 10 min, followed by slow cooling to room temperature. HPol was mixed with DNA duplexes in a 1:1.2 molar ratio in 50 mM Tris-HCl, pH 7.5, containing 450 mM KCl, and 3 mM dithiothreitol, followed by the addition of 5 μl of 100 mM CaCl2. Using a spin concentrator with an Amicon cutoff filter (Millipore), solutions of complexes were reduced to a final concentration of ∼2 mg/ml. dNTPs were added to the concentrated mixtures containing Ca2+. The ternary complex solutions were mixed with an equal volume of reservoir solution containing 0.1 M Mes (pH 5.5), 5 mM MgCl2, and 15 to 22% (w/v) polyethyleneglycol 2000 monomethyl ether and equilibrated against 500 μl of reservoir solution. Crystals were obtained by the hanging drop vapor diffusion technique at 18 °C. Crystals typically appeared within a few days and were allowed to grow for a few weeks. They were then transferred to cryoprotectant solution containing reservoir solution mixed with 25% glycerol (v/v) and flash frozen in liquid nitrogen for data collection.
X-ray diffraction data collection, structure determination and refinement
X-ray diffraction data were collected at 100 K on the 21-ID-D beamline of the Life Sciences Collaborative Access Team at the Advanced Photon Source, Argonne National Laboratory. All data were processed with the program XDS (36) and scaled using AIMLESS (37). Data collection statistics are summarized in Table 2. Both structures were determined by molecular replacement using the program MOLREP (38, 39), with the coordinates of the complex between hPol and DNA (PDB ID code 5L1I) (40) serving as the search model. Structures were initially refined using Refmac (38, 41) and later refined in the PHENIX suite10 using phenix.refine (42). Model building and inspection were carried out in COOT (43). Structural illustrations were generated with the program UCSF Chimera (44).
Data availability
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 8EVE and 8EVF.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Amritraj Patra for help with crystallization experiments and Dr Zdzislaw Wawrzak for help with diffraction data collection and processing. LC-MS/MS analyses were carried out at the Vanderbilt Mass Spectrometry Core Facility. The Vanderbilt-Ingram Cancer Center was funded by NIH grant P30 CA-068485. Vanderbilt University and the Vanderbilt Center for Structural Biology assisted with the purchase of in-house crystallographic instrumentation. Crystallographic data were collected on the 21-ID-D beamline of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Supporting institutions may be found at http://ls-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31109-Eng-38.
Author contributions
R. R.-J., P. P., P. J. K., and N. K. investigation; R. R.-J., P. J. K., and P. P. formal analysis; R. R.-J. writing - original draft; L. J. M. and M. E. conceptualization; L. J. M. and M. E. writing - review and editing.
Funding and additional information
This work was supported by the National Institutes of Health (P01 CA-160032 to M. E. and R01 CA-87819 to L. J. M.). Funding for open access charge: National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Supporting information
References
- 1.Marnett L.J., Plastaras J.P. Endogenous DNA damage and mutation. Trends Genet. 2001;17:214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 2.Migliore L., Coppede F. Genetic and environmental factors in cancer and neurodegenerative diseases. Mutat. Res. 2002;512:135–153. doi: 10.1016/s1383-5742(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Olinski R., Gackowski D., Foksinski M., Rozalski R., Roszkowski K., Jaruga P. Oxidative DNA damage: assessment of the role in carcinogenesis, atherosclerosis, and acquired immunodeficiency syndrome. Free Radic. Biol. Med. 2002;33:192–200. doi: 10.1016/s0891-5849(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 4.Basu A.K., O'Hara S.M., Valladier P., Stone K., Mols O., Marnett L.J. Identification of adducts formed by reaction of guanine nucleosides with malondialdehyde and structurally related aldehydes. Chem. Res. Toxicol. 1988;1:53–59. doi: 10.1021/tx00001a010. [DOI] [PubMed] [Google Scholar]
- 5.Dedon P.C., Plastaras J.P., Rouzer C.A., Marnett L.J. Indirect mutagenesis by oxidative DNA damage: formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plastaras J.P., Riggins J.N., Otteneder M., Marnett L.J. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: base propenals, malondialdehyde, and n(epsilon)-oxopropenyllysine. Chem. Res. Toxicol. 2000;13:1235–1242. doi: 10.1021/tx0001631. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhary A.K., Nokubo M., Reddy G.R., Yeola S.N., Morrow J.D., Blair I.A., et al. Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 8.Leuratti C., Singh R., Lagneau C., Farmer P.B., Plastaras J.P., Marnett L.J., et al. Determination of malondialdehyde-induced DNA damage in human tissues using an immunoslot blot assay. Carcinogenesis. 1998;19:1919–1924. doi: 10.1093/carcin/19.11.1919. [DOI] [PubMed] [Google Scholar]
- 9.Ma B., Villalta P.W., Balbo S., Stepanov I. Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem. Res. Toxicol. 2014;27:1829–1836. doi: 10.1021/tx5002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marnett L.J. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 11.Nair U., Bartsch H., Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radic. Biol. Med. 2007;43:1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Wang M., Dhingra K., Hittelman W.N., Liehr J.G., de Andrade M., Li D. Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol. Biomarkers Prev. 1996;5:705–710. [PubMed] [Google Scholar]
- 13.Wauchope O.R., Mitchener M.M., Beavers W.N., Galligan J.J., Camarillo J.M., Sanders W.D., et al. Oxidative stress increases m1dg, a major peroxidation-derived DNA adduct, in mitochondrial DNA. Nucleic Acids Res. 2018;46:3458–3467. doi: 10.1093/nar/gky089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fink S.P., Reddy G.R., Marnett L.J. Mutagenicity in escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanderVeen L.A., Hashim M.F., Shyr Y., Marnett L.J. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14247–14252. doi: 10.1073/pnas.2332176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson K.A., Fink S.P., Marnett L.J. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J. Biol. Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 17.Eoff R.L., Stafford J.B., Szekely J., Rizzo C.J., Egli M., Guengerich F.P., et al. Structural and functional analysis of sulfolobus solfataricus y-family DNA polymerase dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct. Biochemistry. 2009;48:7079–7088. doi: 10.1021/bi9003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddukuri L., Eoff R.L., Choi J.Y., Rizzo C.J., Guengerich F.P., Marnett L.J. In vitro bypass of the major malondialdehyde- and base propenal-derived DNA adduct by human y-family DNA polymerases kappa, iota, and rev1. Biochemistry. 2010;49:8415–8424. doi: 10.1021/bi1009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stafford J.B., Eoff R.L., Kozekova A., Rizzo C.J., Guengerich F.P., Marnett L.J. Translesion DNA synthesis by human DNA polymerase eta on templates containing a pyrimidopurinone deoxyguanosine adduct, 3-(2'-deoxy-beta-d-erythro-pentofuranosyl)pyrimido-[1,2-a]purin-10(3h)-one. Biochemistry. 2009;48:471–480. doi: 10.1021/bi801591a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 21.Ohmori H., Friedberg E.C., Fuchs R.P., Goodman M.F., Hanaoka F., Hinkle D., et al. The y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 22.Nelson J.R., Lawrence C.W., Hinkle D.C. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann A.R. Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 24.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 25.Wauchope O.R., Beavers W.N., Galligan J.J., Mitchener M.M., Kingsley P.J., Marnett L.J. Nuclear oxidation of a major peroxidation DNA adduct, m1dg, in the genome. Chem. Res. Toxicol. 2015;28:2334–2342. doi: 10.1021/acs.chemrestox.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christov P.P., Richie-Jannetta R., Kingsley P.J., Vemulapalli A., Kim K., Sulikowski G.A., et al. Site-specific synthesis of oligonucleotides containing 6-oxo-m1dg, the genomic metabolite of m1dg, and liquid chromatography-tandem mass spectrometry analysis of its in vitro bypass by human polymerase iota. Chem. Res. Toxicol. 2021;34:2567–2578. doi: 10.1021/acs.chemrestox.1c00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghodke P.P., Mali J.R., Patra A., Rizzo C.J., Guengerich F.P., Egli M. Enzymatic bypass and the structural basis of miscoding opposite the DNA adduct 1,n(2)-ethenodeoxyguanosine by human DNA translesion polymerase eta. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patra A., Politica D.A., Chatterjee A., Tokarsky E.J., Suo Z., Basu A.K., et al. Mechanism of error-free bypass of the environmental carcinogen n-(2'-deoxyguanosin-8-yl)-3-aminobenzanthrone adduct by human DNA polymerase eta. Chembiochem. 2016;17:2033–2037. doi: 10.1002/cbic.201600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schorr S., Schneider S., Lammens K., Hopfner K.P., Carell T. Mechanism of replication blocking and bypass of y-family polymerase eta by bulky acetylaminofluorene DNA adducts. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20720–20725. doi: 10.1073/pnas.1008894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Flaherty D.K., Guengerich F.P. Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products. Curr. Protoc. Nucleic Acid Chem. 2014;59:7.21.21–13. doi: 10.1002/0471142700.nc0721s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njuma O.J., Su Y., Guengerich F.P. The abundant DNA adduct n(7)-methyl deoxyguanosine contributes to miscoding during replication by human DNA polymerase eta. J. Biol. Chem. 2019;294:10253–10265. doi: 10.1074/jbc.RA119.008986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomforde J., Fu I., Rodriguez F., Pujari S.S., Broyde S., Tretyakova N. Translesion synthesis past 5-formylcytosine-mediated DNA-peptide cross-links by hpoleta is dependent on the local DNA sequence. Biochemistry. 2021;60:1797–1807. doi: 10.1021/acs.biochem.1c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 34.Mao H., Schnetz-Boutaud N.C., Weisenseel J.P., Marnett L.J., Stone M.P. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashim M.F., Riggins J.N., Schnetz-Boutaud N., Voehler M., Stone M.P., Marnett L.J. In vitro bypass of malondialdehyde-deoxyguanosine adducts: differential base selection during extension by the klenow fragment of DNA polymerase i is the critical determinant of replication outcome. Biochemistry. 2004;43:11828–11835. doi: 10.1021/bi049360f. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project N. The ccp4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Vagin A., Teplyakov A. Molecular replacement with molrep. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 40.Patra A., Zhang Q., Guengerich F.P., Egli M. Mechanisms of insertion of dctp and dttp opposite the DNA lesion o6-methyl-2'-deoxyguanosine by human DNA polymerase eta. J. Biol. Chem. 2016;291:24304–24313. doi: 10.1074/jbc.M116.755462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., et al. Refmac5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. Ucsf chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 8EVE and 8EVF.