Significance
Understanding the molecular basis of competence acquisition by the lineage-specific progenitor cells provides insights into tissue development and regeneration. The sensory epithelium of the inner ear represents a convenient model to study this process, as only two cell types—the mechanosensory hair cells and their associated supporting cells—are specified from a single pool of progenitors in this lineage. In the present manuscript, we uncover some of the mechanisms by which competence for mechanosensory receptor differentiation is acquired in the early organ of Corti progenitor cells. Specifically, we show that the two SoxC family members, Sox 4 and Sox11, establish a permissive chromatin landscape that allows the hair cell gene regulatory network to be activated upon differentiation cues.
Keywords: inner ear, hair cell, competence, chromatin remodeling, auditory sensory epithelium
Abstract
The auditory organ of Corti is comprised of only two major cell types—the mechanosensory hair cells and their associated supporting cells—both specified from a single pool of prosensory progenitors in the cochlear duct. Here, we show that competence to respond to Atoh1, a transcriptional master regulator necessary and sufficient for induction of mechanosensory hair cells, is established in the prosensory progenitors between E12.0 and 13.5. The transition to the competent state is rapid and is associated with extensive remodeling of the epigenetic landscape controlled by the SoxC group of transcription factors. Conditional loss of Sox4 and Sox11—the two homologous family members transiently expressed in the inner ear at the time of competence establishment—blocks the ability of prosensory progenitors to differentiate as hair cells. Mechanistically, we show that Sox4 binds to and establishes accessibility of early sensory lineage–specific regulatory elements, including ones associated with Atoh1 and its direct downstream targets. Consistent with these observations, overexpression of Sox4 or Sox11 prior to developmental establishment of competence precociously induces hair cell differentiation in the cochlear progenitors. Further, reintroducing Sox4 or Sox11 expression restores the ability of postnatal supporting cells to differentiate as hair cells in vitro and in vivo. Our findings demonstrate the pivotal role of SoxC family members as agents of epigenetic and transcriptional changes necessary for establishing competence for sensory receptor differentiation in the inner ear.
During embryogenesis, timely specification of the differentiated cell types relies on acquisition of competence by the lineage-specific progenitor cells. In the central nervous system (CNS), neurogenesis starts from a pool of proliferative neuroepithelial stem cells that transform into radial glia and acquire competence to differentiate as diverse neuronal and glial cell types (1, 2). Similarly, the differentiation of major neuronal and photoreceptor populations in the retina depends on both spatial and temporal cues that modulate the competence of progenitor cells to differentiate sequentially into particular retinal derivatives (3). Yet, how the responsiveness to differentiation cues is acquired by the progenitor cells in these systems remains incompletely understood, partly due to their rich cellular diversity and also because self-renewing progenitors, committed progenitors, and differentiated cells coexist in these tissues over a prolonged developmental time window.
In the organ of Corti, the auditory sensory epithelium of the inner ear, induction of sensory differentiation is spatially and temporally uncoupled from the cell cycle exit making it a unique system to study the molecular basis of competence establishment and lineage commitment (4–10). The organ of Corti originates from a Sox2-positive domain established in the early cochlear duct, where cells undergo active mitosis until embryonic day 12.0 (E12.0) (4, 8–10). Between E12.5 and E14.5, a narrow strip of cells within the broader Sox2-positive domain exit the cell cycle in a wave, spreading from the apex toward the base of the cochlea. Organ of Corti progenitors in this undifferentiated but postmitotic prosensory domain express high level of the cyclin-dependent kinase inhibitor p27Kip1 (5, 6, 10). After proliferation ceases, a wave of sensory differentiation is initiated by upregulation of the basic helix-loop-helix transcription factor Atoh1, which occurs in an opposing base-to-apex gradient starting at E14.5. Atoh1 expression in progenitor cells is refined by Notch-mediated lateral inhibition into a stereotyped mosaic of Atoh1-positive hair cells and Atoh1-negative supporting cells – the only two major cell types in the organ of Corti (7, 10–12). Construction of a functional auditory system depends on this strict regulation of the timing for cell cycle exit and differentiation (6). In p27Kip1-deficient mice, where the cell cycle exit and sensory differentiation become coupled in the organ of Corti similar to vestibular sensory organs, retina, and the CNS, patterning defects arise, leading to the profound hearing deficits (5, 6, 13).
In nonmammalian vertebrates, dying hair cells can be replenished by the residual supporting cells, either directly through transdifferentiation, or after undergoing mitosis (14–16). During embryonic and neonatal development in mammals, supporting cells retain the capacity for hair cell differentiation (17–23). However, this plasticity is lost during the first week after birth, rendering the organ of Corti incapable of hair cell regeneration by postnatal day 6 (P6) (24–30). Because hair cells are limited in number and can be easily damaged by noise or ototoxic drugs (24, 25, 31), the inability of supporting cells to give rise to new hair cells in mammals leads to irreversible hearing loss and balance disorders.
In this study, we characterized the competence of cochlear prosensory progenitor cells to differentiate as hair cells in response to Atoh1. We demonstrated that competence is rapidly established in the developing cochlea between E12.0 and E13.5, immediately following but independent of the cell cycle exit. Using unbiased genome-wide analyses, we identified a group of genomic loci made accessible de novo during competence acquisition. We demonstrated that two SoxC transcription factors, Sox4 and Sox11, are required to establish accessibility of these putative regulatory elements. Using single-cell RNA-sequencing (scRNA-seq), we showed that conditional loss of SoxC genes blocks both spontaneous and Atoh1-induced hair cell differentiation in the organ of Corti without affecting either establishment of the prosensory domain or the timing of cell cycle exit. Moreover, overexpression of SoxC genes prior to the onset of differentiation promoted hair cell fate in the noncompetent progenitor cells in vitro. Finally, we showed that at later stages of postnatal development, SoxC genes are down-regulated as supporting cells lose the ability to transdifferentiate into hair cells and that ectopic expression of either Sox4 or Sox11 restored this ability.
Together our data demonstrate that SoxC transcription factors induce competence to respond to Atoh1 by setting up a permissive chromatin state in cochlear progenitor cells. This primed state is maintained in neonatal supporting cells that retain SoxC expression and the ability to transdifferentiate, and is lost postnatally. These data suggest that SoxC genes are necessary for both establishment and maintenance of competence for hair cell differentiation in the inner ear.
Results
The Organ of Corti Progenitor Cells Acquire Competence to Differentiate as Hair Cells and Supporting Cells upon Transition to the Postmitotic State.
Prior to differentiating as hair cells and supporting cells, Sox2-positive progenitors in the prosensory domain of the cochlear duct rapidly up-regulate p27Kip1 and exit the cell cycle between E12.5 and E14.5 (Fig. 1A) (5, 6, 10). It has been previously shown that p27Kip1-positive postmitotic progenitor cells have the capacity for sensory differentiation that they retain in cochlear explants (32, 33). However, hair cell differentiation is disrupted when cochlear cultures are initiated at E12.0 (32).
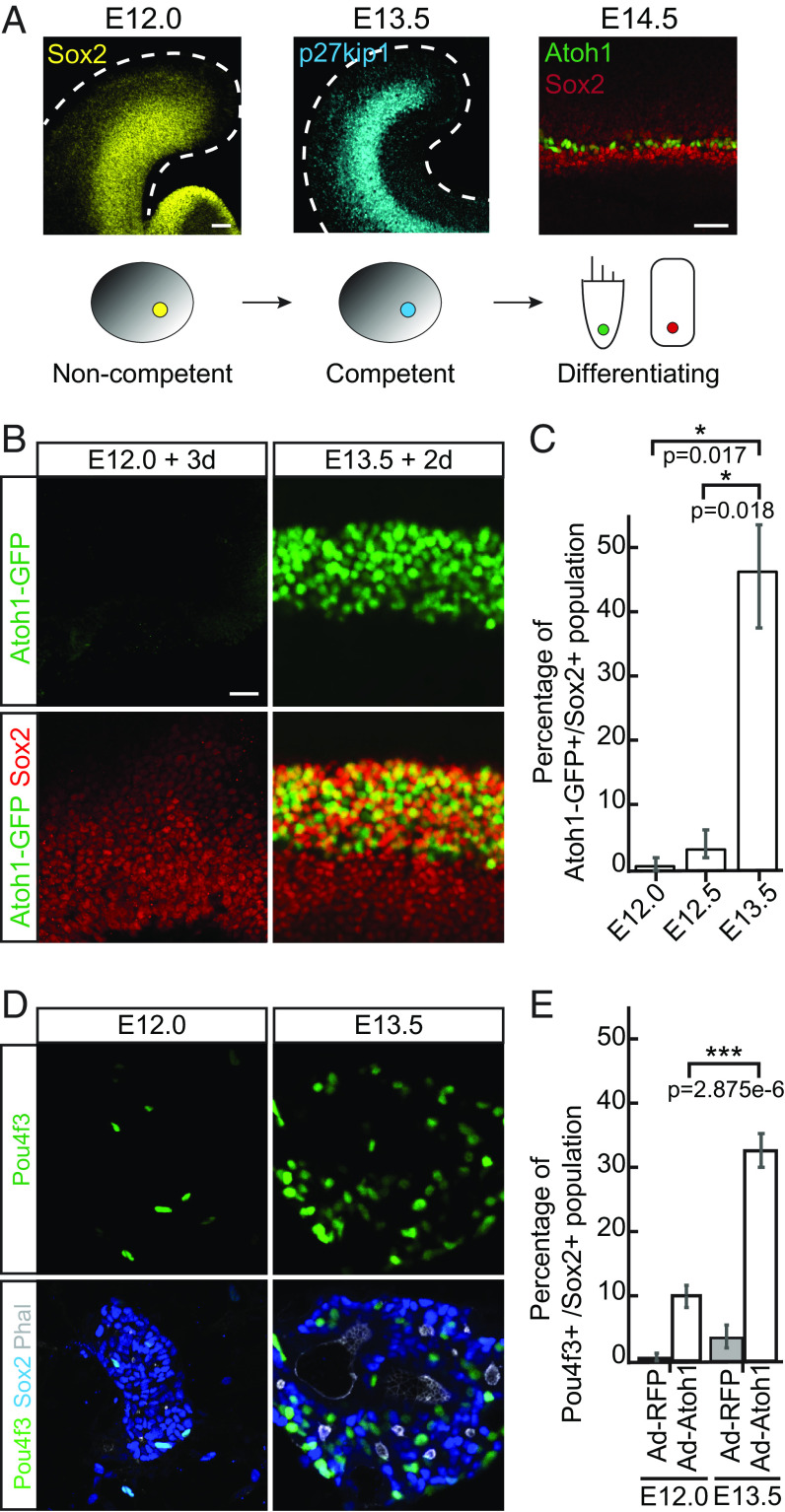
Fig. 1.
Prosensory progenitors in the cochlear duct acquire competence to differentiate as hair cells and supporting cells between E12.0 and E13.5. (A) Diagram demonstrates early embryonic development of the organ of Corti. At E12.0, Sox2-positive proliferating progenitors (yellow) start to exit cell cycle in a wave spreading from the apex toward the base of the cochlea. By E13.5, most progenitors become postmitotic and express high levels of p27kip1 (cyan). At E14.5, Atoh1-positive hair cells (HCs, green) and the surrounding Sox2-positive supporting cells (SCs, red) are specified. (Scale bar, 50 μm.) (B) Representative immunofluorescent images of the whole cochleae isolated from E12.0 and E13.5 Atoh1-GFP transgenic reporter animals and harvested for characterization after 3-d or 2-d in culture, respectively. GFP-positive hair cells (green) and Sox2-positive supporting cells (red) are labeled. (Scale bar, 20 μm.) (C) Quantitative analysis of the cultures in B demonstrates activation of Atoh1-GFP reporter in E12.0 explants (0.8%; n = 6), E12.5 explants (3.6%; n = 3) and E13.5 explants (47%; n = 3). (D) Representative immunofluorescent images show the progenitor cells isolated from the cochlear ducts at E12.0 and E13.5, infected with Ad-RFP-control or Ad-Atoh1-RFP virus, and maintained in culture for 3 d. Note that only in E13.5 cultures Atoh1 overexpression results in formation of the sensory rosettes with a small lumen formed by the actin-rich (Phalloidin, white) apical surfaces of the polarized Pou4f3–positive hair cells (green) and surrounding Sox2-positive supporting cells (blue). (Scale bar, 20 μm.) (E) Bar graph shows the quantitative analysis of cultures in D. Compared to other conditions, a significant increase in the percentage of Pou4f3-positive hair cells is observed in the E13.5 cultures where Atoh1 was overexpressed (n = 3 for each condition).
To confirm these observations, and to test if loss of sensory differentiation in the cochlear explants is caused by the lack of paracrine signals that may be present in vivo, we used three-dimensional organotypic cultures that preserve near-innate anatomy of the cochlear duct and all surrounding tissues (34). The auditory organs were explanted from embryos either prior to initiation of the wave of progenitor cell cycle exit (E12.0 and E12.5), or after p27Kip1-positive postmitotic domain establishment (E13.5) (10). The explants were embedded in collagen matrix and cultured for either 3 d (E12.0 and E12.5) or 2 d (E13.5), before harvesting for subsequent analysis. Initiation of sensory differentiation was assessed using reporter mice, which express green fluorescent protein (GFP) under the control of the Atoh1 3′ autoregulatory enhancer to visualize differentiating hair cells (Atoh1-GFP reporter) (35). After 3 d in culture, fewer than one percent of Sox2-positive progenitors differentiated as GFP-positive hair cells in E12.0 explants and 3.6% in E12.5 explants (n = 6 for E12.0 and n = 3 for E12.5; Fig. 1 B and C and SI Appendix, Fig. S1). In stark contrast, 47% of Sox2-positvie progenitors converted to the hair cells in E13.5 explants cultured for 2 d (n = 3; P < 0.05; Fig. 1 B and C and SI Appendix, Fig. S1).
It was shown that Atoh1 is both necessary and sufficient to initiate sensory hair cell differentiation in the organ of Corti. Atoh1 directly up-regulates a number of key hair cell–specific genes, including transcription factor Pou4f3 (36, 37). To directly compare the competence of E12.0 and E13.5 progenitor cells to respond to Atoh1 induction, we ectopically up-regulated this transcription factor in vitro. Cochlear ducts from E12.0 or E13.5 wild-type (WT) mice were dissociated and infected with adenoviruses expressing either Atoh1 (Ad-Atoh1-RFP) or a control RFP-reporter construct (Ad-RFP). We used this dissociated culture system as, unlike intact organ cultures, dissociated cochlear cells have only a limited capacity to spontaneously differentiate as hair cell (38). Consistent with these earlier observations, only a few Pou4f3-positive hair cells were detected in either E12.0 or E13.5 RFP-controls after 3 d in culture, despite 80 to 90% infection efficiency (Fig. 1 D and E and SI Appendix, Fig. S2). Atoh1 overexpression dramatically increased the number of E13.5 progenitor cells differentiated as Pou4f3-positive hair cells (32% ± 0.8%; Fig. 1 D and E). In these E13.5 Atoh1-overexpression cultures, we also observed formation of the sensory rosettes, where hair cells and supporting cells were polarized with their actin-rich apical surfaces pointing to a small lumen. In contrast, Atoh1 induced significantly fewer Pou4f3-positive hair cells scattered throughout disorganized monolayered cultures at E12.0 (10% ± 1.4%; n = 3, P < 0.01; Fig. 1 D and E), resembling RFP-controls (SI Appendix, Fig. S2).
Together, these two experiments strongly suggested that the competence to form hair cells, as determined by spontaneous and Atoh1-induced differentiation, is acquired by the organ of Corti progenitor cells between E12.0 and E13.5.
Competence for Hair Cell Differentiation is Associated with the Emergence of Newly Established Accessible Chromatin Regions in the E13.5 Progenitor Cells.
Despite its necessity for hair cell differentiation, Atoh1 lacks pioneer factor activity necessary to facilitate chromatin remodeling needed for hair cell maturation (29, 30). We hypothesized that a permissive chromatin state may be established prior to Atoh1 upregulation and is required in the progenitor cells to respond to Atoh1 as part of the competence for sensory differentiation. To test this hypothesis, we used previously published assay for transposase-accessible chromatin with sequencing (ATAC-seq) data to directly compare the chromatin states in E12.0 and E13.5 progenitor cells (39), focusing on the loci that gained accessibility during the transition to the competent state. We identified 13,352 of such loci and demonstrated that, consistent with their potential role in hair cell fate acquisition, these were maintained in immature hair cells and supporting cells at E17.5 (Fig. 2A). The same regions lost accessibility in P6 supporting cells no longer capable of transdifferentiation as sensory hair cells (Fig. 2A). Further suggesting their role in establishment of competence in the organ of Corti, almost a quarter of the chromatin regions that gained accessibility at E13.5 overlapped with previously characterized putative regulatory elements bound by Atoh1 in E17.5 hair cells (24%; 3,163 of 13,352; Fig. 2A) (29).
Fig. 2.
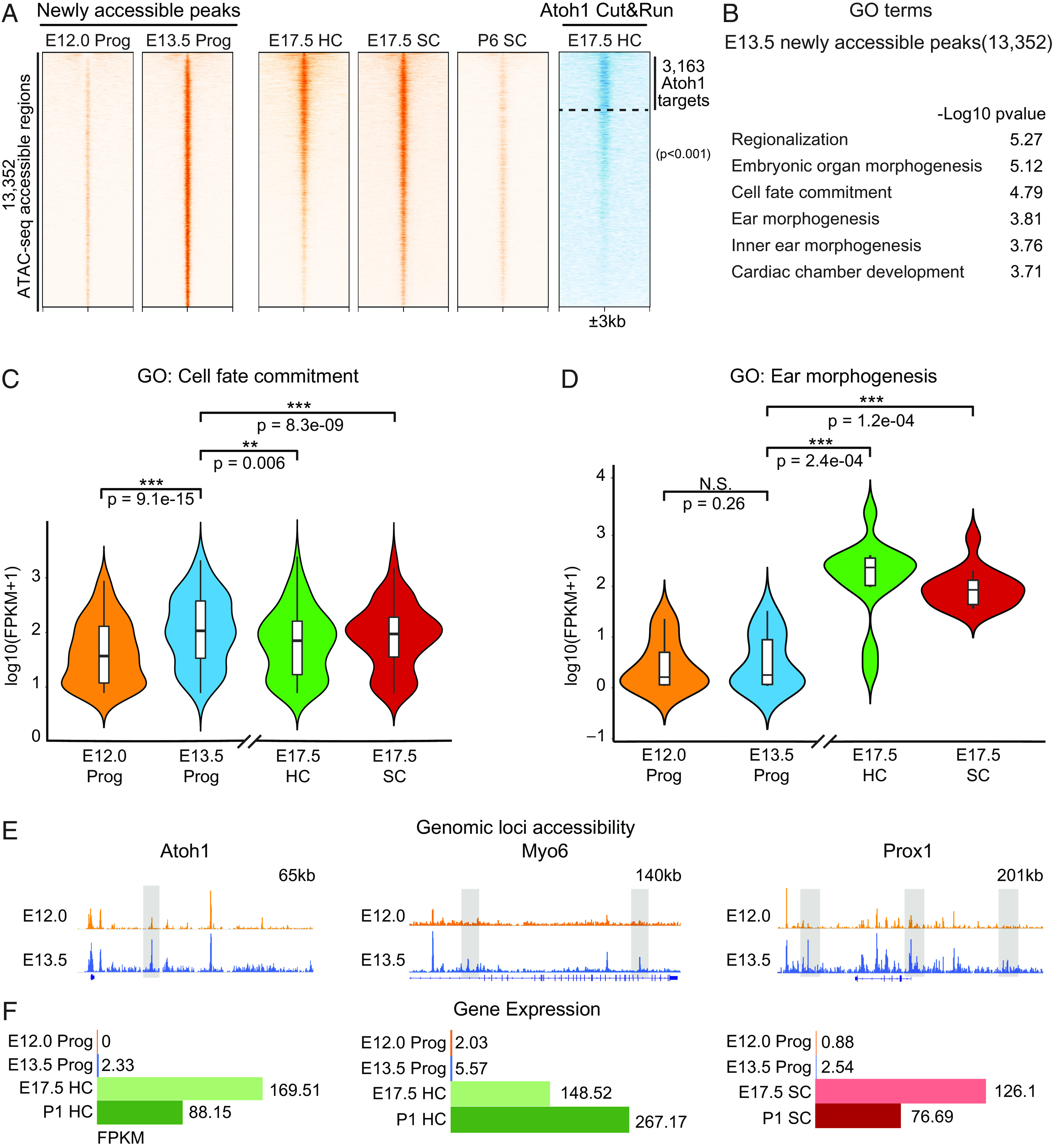
Progenitor cell competence for sensory differentiation is associated with emergence of newly established accessible chromatin regions. (A) A Heatmap demonstrates that most of the ATAC-seq accessible genomic regions (orange) newly emerged during the transition from E12.0 to E13.5 (13,352 peaks) remain open in E17.5 HC and supporting cells (SC), and gradually lose accessibility in the postnatal supporting cells (P6 SC). Out of 13,352, 3,163 genomic loci are bound by Atoh1 in E17.5 HC (Atoh1 Cut&Run, blue). Scale of each sample column is ±3 kb from the center the peaks. (B) GO analysis using GREAT shows the top six most-enriched biological process terms for E13.5 newly accessible peaks. (C) Violin plots show that expression [Log10(FPKM+1)] of the genes in the cell fate commitment GO term is significantly up-regulated in E13.5 progenitors and then down-regulated in E17.5 supporting cells and hair cells (Wilcoxon signed-rank test). (D) The same analysis shows that expression of hair cell–specific and supporting cell–specific genes in the inner ear morphogenesis GO term is unchanged at E13.5 but is significantly up-regulated upon differentiation (Wilcoxon signed-rank test). (E) Integrative Genomics Viewer (IGV) tracks show ATAC-seq profiles of representative genomic loci of hair cell–specific (Atoh1, Myo6) and supporting cell–specific genes (Hes5, Prox1) at E12.0 and E13.5. Their putative enhancers that gain accessibility at E13.5 are highlighted in gray boxes. (F) The expression of the hair cell– and supporting cell–specific genes shown in E is shown in E12.0 and E13.5 progenitor cells (Prog) and in E17.5 and P1 HC or supporting cells (SC).
We analyzed the potential regulatory functions of the 13,352 newly emerged E13.5 peaks using genomic regions enrichment of annotations tool (GREAT) (40) and identified that genes involved in cell fate commitment [Gene ontology (GO): cell fate commitment, P < 0.001] were strongly associated with these genomic loci (Fig. 2B). These included many established prosensory lineage genes, such as Isl1, Eya1, and Fgf20, highly up-regulated at E13.5 (n = 85; P < 0.001; Fig. 2C). In addition, the E13.5 de novo sites were also associated with genes involved in inner ear morphogenesis (GO: Ear morphogenesis, P < 0.001; GO: Inner ear morphogenesis, P < 0.001; Fig. 2B), including both hair cell (HC)– and supporting cell (SC)–specific genes not yet expressed at E13.5 [n = 14; P < 0.01 (HC) and P < 0.01 (SC); Fig. 2D]. These included the key regulators and markers of sensory hair cell lineage differentiation such as Atoh1, Myo6, and Prox1 (Fig. 2 E and F).
Together these observations suggested that the genomic loci that became accessible at E13.5 may be required for Atoh1-induced differentiation and, more broadly, encompass a gene regulatory network that is necessary to establish the competence for sensory hair cell differentiation in the cochlea.
SoxC Genes Are Required for Sensory Hair Cell Differentiation but Are Dispensable for Establishment of the Postmitotic Prosensory Domain in the Cochlea.
To identify transcription factors that may establish chromatin accessibility as part of acquisition of competence for sensory differentiation, we analyzed the newly accessible E13.5 regulatory elements for enriched DNA motifs (HOMER)(41). This analysis identified several motifs associated with transcription factors previously shown to play roles in the inner ear development, most prominently represented by the Sox, Six, and Gata families (Fig. 3A) (42–47). We noted that Sox DNA–binding motifs were the most enriched and represented in 32.31% of the peaks (P < 0.001; Fig. 3A).
Fig. 3.
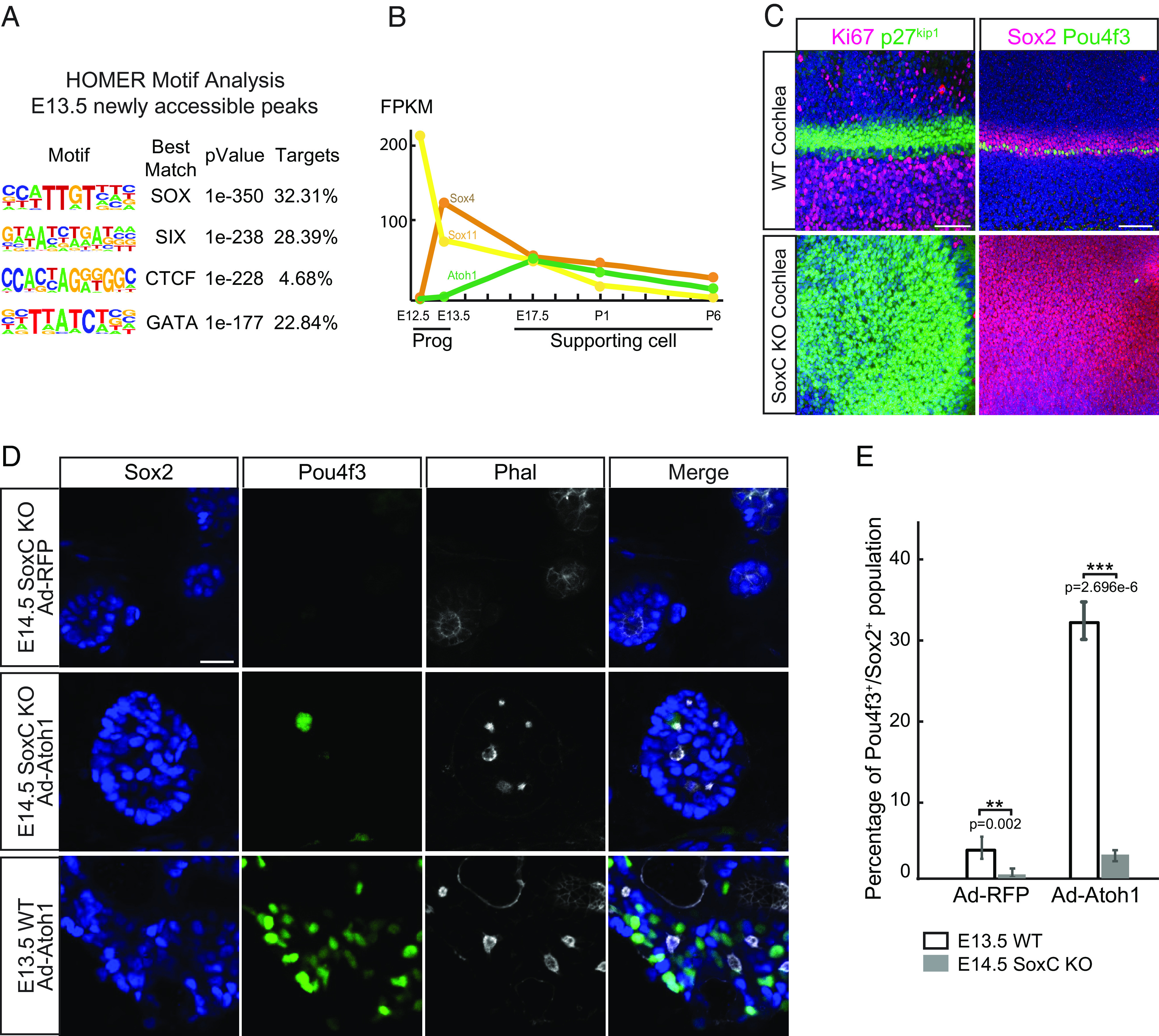
SoxC transcription factors are essential for initiation of sensory differentiation during organ of Corti development. (A) HOMER motif analysis shows top four most enriched DNA-binding motifs in E13.5 newly accessible chromatin regions. (B) Gene expression in fragments per kilobase of transcript per million mapped reads (FPKM) for Sox4, Sox11, and Atoh1 is demonstrated in E12.0 and E13.5 progenitors (Prog) and E17.5-P6 supporting cell (SC) (n = 2 for Progs, n = 3 for SCs). Note that Sox4 expression is significantly up-regulated in E13.5 progenitors prior to the onset of Atoh1 expression. (C) Representative immunofluorescent images show the whole cochleae isolated at E14.5 from the WT and SoxC cKO littermate embryos. Ki67-positive (magenta) proliferating cells and the p27kip1-positive (green) postmitotic progenitor cells (Left) are labeled. Note that progenitor cell cycle exit is not affected in the SoxC cKO organs. Sox2-positive supporting cells (magenta) and one row of Pou4f3-positive inner hair cells (green) are detected in the WT but not in the SoxC cKO organs. Cell nuclei are labeled with DAPI (blue). (Scale bar, 100 μm.) (D) Representative immunofluorescent images show dissociated cochlea cells isolated at E14.5 from the SoxC cKO and WT mice littermates, infected with Ad-RFP control or Ad-Atoh1-RFP virus, and maintained in culture for 3 d. Actin is labeled by Phalloidin (white), hair cells are labeled with Pou4f3 (green), and supporting cells are labeled by Sox2 (blue). Note that although the cells are organized into the sensory rosettes, Atoh1 overexpression fails to induce Pou4f3-positive hair cells (green) in absence of SoxC. (Scale bar, 20 μm.) (E) Bar graph shows quantitative analysis of the cultures in D. Compared to the WT cultures, the percentage of hair cells is significantly decreased in the SoxC cKO cultures transduced with either Ad-RFP or Ad-Atoh1-RFP virus (n = 3 for each condition).
Suggestive of their role in establishing competence for sensory differentiation in the inner ear, the Sox family of transcription factors is well documented to contain regulators of cell fate commitment and sensory specification (48). To identify the members of the family that may be responsible for de novo chromatin remodeling at E13.5, we analyzed previously published RNA-seq data for E12.0 and E13.5 progenitor cells (39), as well as E17.5, P1, and P6 supporting cells (29). Sox2, Sox4, Sox10, and Sox13 expression was significantly up-regulated in E13.5 progenitors as compared to E12.0 (Sox2: fold = 5, P < 0.001; Sox4: fold = 32, P < 0.001; Sox10: fold = 5, P < 0.001; Sox13: fold = 5, P < 0.001). However, expression of only Sox4 was also lost in the P6 supporting cells (Fig. 3B), temporally correlating with the loss of competence for hair cell differentiation and loss of chromatin accessibility at the 13,352 competence-related loci (Fig. 2A).
Sox4 and its close homologue Sox11 belong to the SoxC family of transcription factors (49–51) and loss of both genes disrupts hair cell formation via an uncharacterized molecular mechanism (52). Using conventional in situ hybridization, we have previously reported that in the developing utricle, Sox11 expression is largely restricted to the marginal regions of the growing sensory epithelium, while Sox4 is expressed more broadly throughout the macula (52). We confirmed this pattern of expression in the E15.5 utricle using RNAScope (SI Appendix, Fig. S3). We then investigated if the pattern of Sox4 and Sox11 expression in the developing organ of Corti was as predicted by the RNA-seq results (Fig. 3B). While Sox11 was most highly expressed in the cochlear duct and the spiral ganglion at E12.0, its expression remained in the same structures but decreased at E13.5 and E17.5 (SI Appendix, Fig. S3). Sox4 expression was also most prominent in the cochlear duct and the spiral ganglion and increased from E12.0 to E13.5, with the strong signal remaining in the immature hair cells and spiral ganglion neurons at E17.5 (SI Appendix, Fig. S3).
We have previously reported that conditional inner ear–specific ablation of the two SoxC genes using Pax2Cre (53) and Sox4 fl/fl Sox11fl/fl (54) transgenic animals resulted in formation of stunted sensory epithelia that completely lack sensory hair cells (52). In particular, we showed that cochlear outgrowth was affected and that at E18.5 Sox2-positive cells in, what appeared to be, an undifferentiated prosensory domain remained in multiple nuclear layers similar to that in the WT E12.0 cochlea (52). To test their role in establishing competence for sensory hair cell differentiation in the organ of Corti, we assessed the effects of Sox4 and Sox11 ablation early during cochlear development in the same transgenic animals. Signifying the initiation of sensory differentiation at E14.5, a single row of Pou4f3-positive inner hair cells was detected in the mid-base region of the WT cochlea (Fig. 3C). However, although the p27Kip1-positive postmitotic prosensory domain was established in the knockout littermates, initiation of hair cell differentiation failed in absence of Sox4 and Sox11. To exclude the possibility that hair cell survival rather than differentiation was affected, we also assessed for activated Caspase-3 activity. This analysis revealed no difference in the rate of apoptosis in the cochlear epithelia between the WT and the knockout organs (SI Appendix, Fig. S3).
To directly test if loss of SoxC rendered organ of Corti progenitor cells incapable of sensory differentiation, we overexpressed Atoh1 in E14.5 dissociated progenitor cells isolated from SoxC double knockout or control littermate cochleae (Fig. 3 D and E). Strikingly, Atoh1 overexpression failed to induce hair cell differentiation in absence of SoxC genes, as only 2.6% of Sox2-positive infected progenitor cells up-regulated Pou4f3. This rate of differentiation was over 12-fold lower than that in the E13.5 WT cells (n = 3; P < 0.001; Fig. 3 D and E).
Together, these experiments demonstrated that SoxC transcription factors are necessary for establishment of competence for sensory hair cell differentiation and that these factors likely act upstream of Atoh1. Interestingly, these data also revealed that sensory cell fate commitment is established independently of cell cycle exit, as loss of Sox4 and Sox11 did not affect formation of the postmitotic prosensory domain or the timing of p27Kip1 upregulation in the cochlea.
SoxC Transcription Factors Control Expression of the Key Sensory Lineage Genes.
To identify downstream targets of SoxC in the developing organ of Corti, we performed scRNA-seq analysis of knockout cochlear duct epithelia prior to the onset of hair cell differentiation (E13.5). For a comparative analysis between WT and knockout organs, we integrated the two datasets using the canonical correlation analysis (CCA) algorithm (Fig. 4A). Cells were visualized using uniform manifold approximation and projection (UMAP) plots after clustering and dimension reduction. After excluding nonepithelial cell types (e.g., mesenchyme – cluster 9), the same 10 clusters were identified in both WT and knockout datasets, suggesting that loss of Sox4 and Sox11 did not affect the overall cellular composition of the cochlear duct (Fig. 4A). Established markers for known cochlear cell populations were used to assign identity to each cluster (Fig. 4C and SI Appendix, Fig. S4 and Dataset S1). Postmitotic sensory progenitor cells grouped distinctly as cluster 2 in WT and knockout organ, characterized by combined expression of Cdkn1b (6), Sox2 (8), and Hey2 (Fig. 4 B and C and Dataset S1) (55). In addition, other cell populations such as the Otx2-positive cochlear roof domain (56), Bmp4-positive outer sulcus (57), as well as Sox2- and Jag1-positive Kölliker's organ (58, 59) were identified in both WT and KO cochlear ducts (SI Appendix, Fig. S4 and Dataset S1).
Fig. 4.
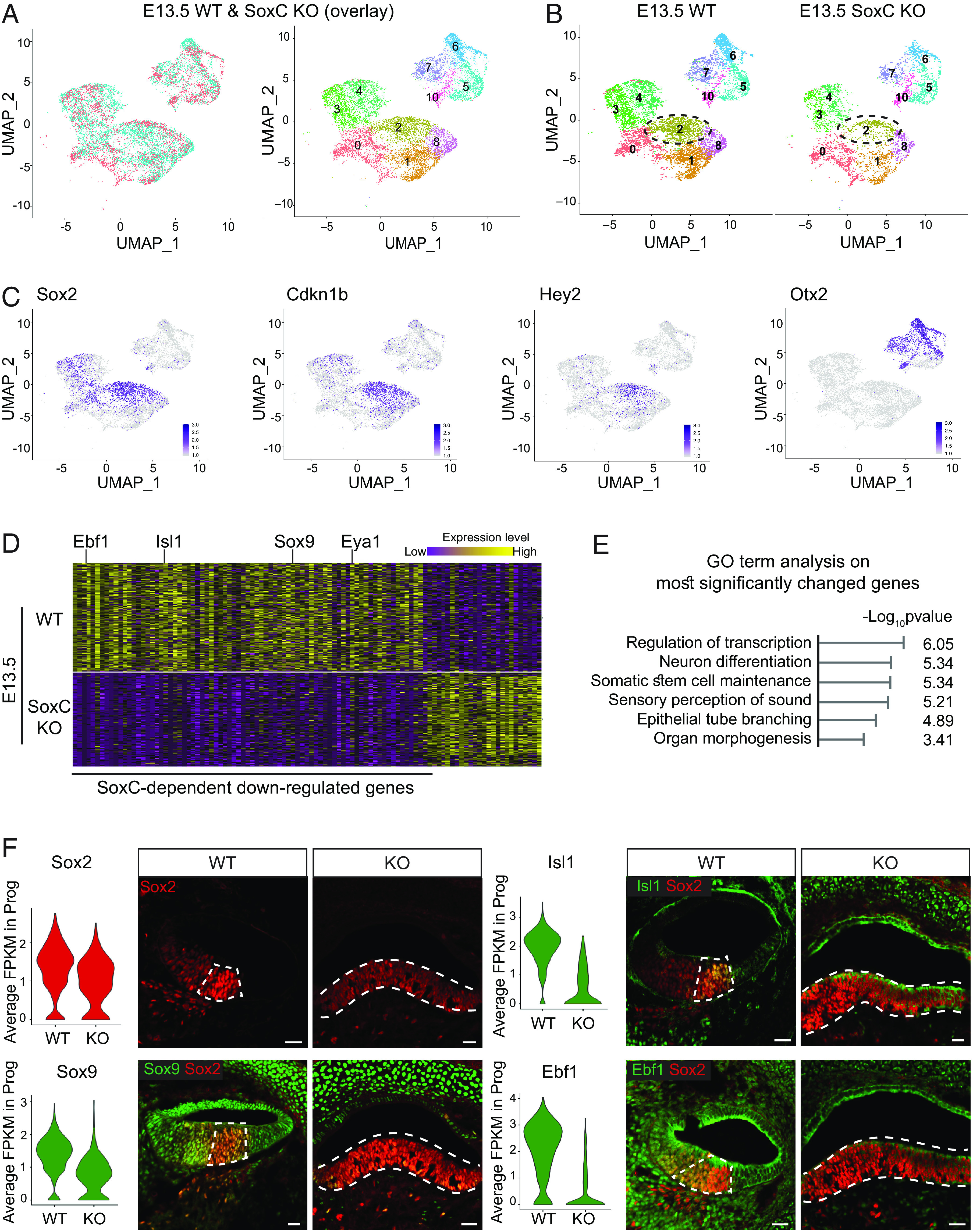
scRNA-seq indicates that SoxC transcription factors promote hair cell differentiation by regulating expression of the key sensory lineage genes. (A) UMAP plots of 7,365 cells collected from WT cochlea duct epithelium and 5,693 cells collected from SoxC cKO cochlea duct epithelium at E13.5 generated by integration using Seurat CCA are shown. The same 11 cell types are identified in the WT and SoxC cKO organs, and the cells within each cell type group together regardless on the genotype. Note that cluster 9, representing mesenchyme, is removed. (B) UMAP visualization of WT and SoxC cKO cochlea cell populations plotted based on the genotype. Cluster 0 represents nondividing Kölliker's organ cells, Clusters 1 and 8 are nondividing outer sulcus cells, cluster 2 represents postmitotic organ of Corti progenitor cells, and Cluster 5, 6, and 10 are nondividing cells of the cochlea roof domain. Proliferating cells of the Kölliker's organ and organ of Corti (cluster 3), outer sulcus (cluster 4), and roof (cluster 7) are also identified. Note that cluster 9, representing mesenchyme, is removed. (C) Feature plots demonstrate transcript levels for cell markers specific to the known cochlear cell types. The organ of Corti progenitor cell population is identified based on the Cdkn1b, Sox2, and Hey2 expression. The roof domain of the cochlear duct is identified by high level of Otx2 expression. A full list of Top 20 genes for each cluster can be found in Dataset S1. (D) Heatmap visualizes the 109 genes most significantly differentially expressed between SoxC cKO and WT postmitotic organ of Corti progenitor cell clusters [Log2(Fold change) > 1.5, P < 0.001]. Key sensory lineage genes, including Isl1, Sox9, and Eya1, are highlighted. High expression level is shown in yellow, low expression level is depicted in purple. A full list of genes can be found in Dataset S2. (E) GO analysis performed using the database for annotation, visualization and integrated discovery shows top six biological process terms enriched in the genes most significantly down-regulated in SoxC cKO progenitor cells compared to WT control. (F) Violin plots show the average expression (FPKM) of the representative genes significantly down-regulated in SoxC cKO progenitor cells compared to WT control. Accompanying immunofluorescent images of the sections through E14.5 cochlear ducts validate decreased protein levels for the same genes. Sox2-positive progenitor cells (red) are counterstained in each section for the investigated protein (Ebf1, Sox9, Isl1; green). (Scale bar, 20 μm.)
To investigate the prosensory role of SoxC transcription factors, we assessed the gene expression changes associated with loss of Sox4 and Sox11 in the postmitotic progenitor cells (cluster 2) (Fig. 4D and SI Appendix, Fig. S4 and Dataset S2). Suggesting that SoxC function as transcriptional activators, two-thirds of the most significantly differentially expressed genes were down-regulated in the knockouts [Log2(Fold change) > 1.5, P < 0.01; Fig. 4D]. These genes included many crucial regulators of sensory lineage specification, such as Isl1 (60), Eya1 (61), Sox9 (62–64), and Fgf20 (65). Among potential targets of SoxC, we also identified genes with no known function in the inner, but shown to be essential for neuronal lineage specification, such as Ebf1 (66) and Sall3 (67). We confirmed downregulation of these key sensory lineage factors in the knockouts using immunohistochemistry (Fig. 4F), asserting the role of SoxC transcription factors as crucial regulators of hair cell fate initiation.
SoxC Transcription Factors Regulate Progenitor Cell Chromatin Accessibility to Promote Hair Cell Differentiation.
To understand how SoxC factors establish competence in the organ of Corti progenitor cells, we profiled chromatin accessibility changes in E13.5 cochlear epithelium after ablation of Sox4 and Sox11 genes. This analysis demonstrated that accessibility of 18% of the genomic regions (42,725 out of 266,580) was dependent on SoxC and was lost in the knockouts (SI Appendix, Fig. S5). GREAT analysis demonstrated that genes associated with regulation of timing of cell differentiation and auditory receptor cell fate commitment were associated with these SoxC-dependent regions (SI Appendix, Fig. S5). Of the peaks that lost accessibility in the knockout cochlear duct, 4,374 overlapped with the progenitor-specific peaks found either at E12.0 or E13.5. Strikingly, 61% of these genomic regions corresponded to the loci that newly gained accessibility at E13.5 (Fig. 5A). Conversely, of the genomic regions that gained accessibility in the knockout cochlea epithelium, 6,246 were open in the progenitor cells and 70% of those regions were specific to E12.0 (Fig. 5A). These data strongly suggested that loss of SoxC genes reverted the chromatin accessibility in the postmitotic E13.5 progenitor cells to that largely resembling E12.0 noncompetent state.
Fig. 5.
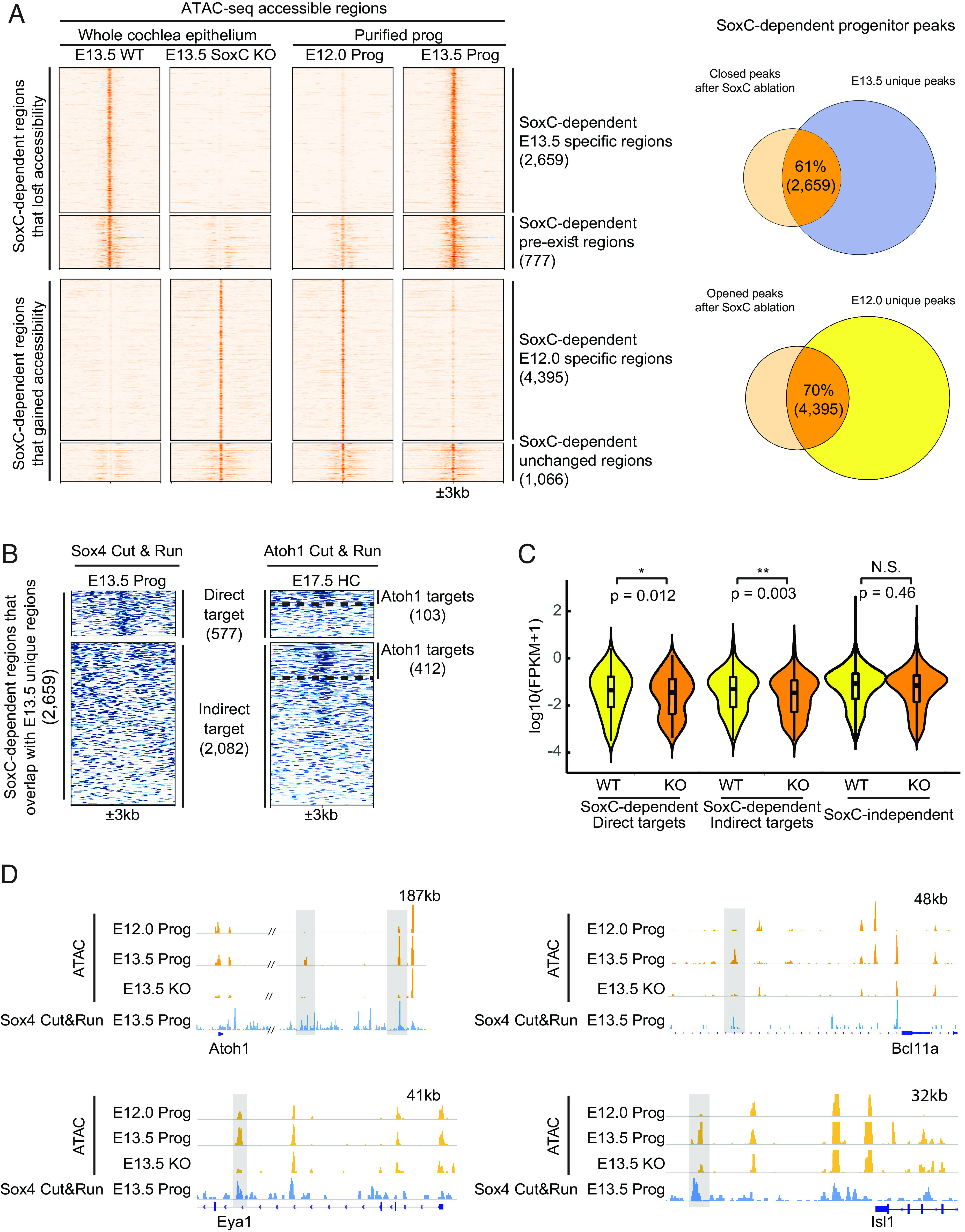
SoxC transcription factors control chromatin accessibility to promote hair cell differentiation in the E13.5 organ of Corti. (A) Heatmap shows accessible genomic regions identified by ATAC-seq in the SoxC cKO cochlea at E13.5 in relation to the E12.0 and E13.5 progenitor cell peaks. Among all the SoxC-dependent peaks, 2,659 that lose accessibility in the SoxC cKO cochlea overlap with E13.5 newly emerged regions; 4,395 peaks that gain accessibility in the SoxC cKO overlap with E12.0 progenitor–specific regions that are closed at E13.5. Scale of each sample column is ±3 kb from the center of the peaks. Venn diagrams on the right show numbers and percentages of the overlap between SoxC-dependent genomic regions and E13.5 (Upper) or E12.0 (Lower) unique progenitor peaks. (B) Heatmap shows direct and indirect Sox4 targets identified using ChIP-seq (Sox4 Cut&Run) in E13.5 progenitors. Twenty-two percent (577) of the E13.5 newly opened SoxC-dependent progenitor peaks (2,659) are direct targets of Sox4. Atoh1 Cut&Run on the right demonstrates the genomic loci bound by Atoh1 in E17.5 hair cells (HC): 103 SoxC-dependent peaks are both Sox4 and Atoh1 direct targets, 412 indirect Sox4 targets are directly bound by Atoh1. Dotted lines separate direct/indirect targets of Atoh1. (C) Violin plots show the expression changes [log10(FPKM+1)] of genes associated with SoxC-dependent (577 direct targets and 2,082 indirect targets demonstrated in panel B) and SoxC-independent (88,674 peaks common in between WT and KO) regulatory elements in WT and SoxC cKO progenitor cells (scRNA-seq datasets). Genes associated with SoxC-dependent regulatory elements are significantly down-regulated, while genes associated with SoxC-independent regulatory elements are not significantly changed upon SoxC ablation. (D) IGV tracks show ATAC-seq profiles (orange) and Sox4 Cut&Run profile (blue) of representative genomic loci for key sensory lineage genes (Atoh1, Bcl11a, Eya1 and Isl1). The putative enhancers highlighted in gray boxes, gain accessibility in E13.5 progenitors (Prog), lose their accessibility upon SoxC ablation, and are bound by Sox4 transcription factor.
To assess if SoxC transcription factors directly control the accessibility of the competency-related regulatory elements, we profiled Sox4 targetome in the E13.5 progenitor cells using CUT&RUN (Fig. 5B). This analysis demonstrated that 21% of the genomic regions that gained accessibility between E12.0 and E13.5 and lost accessibility in absence of SoxC represented direct targets of Sox4 (Dataset S3). Notably, around 20% (515 out of 2,659) of SoxC-dependent regions also contained Atoh1-binding sites (Fig. 5B). To determine if the chromatin accessibility changes were responsible for the changes in gene expression observed in the knockout sensory epithelia, we associated the SoxC-dependent genomic regions with the closest genes and analyzed their expression in our scRNA-seq dataset (Fig. 5C). This analysis demonstrated that both direct (Sox4-occupied) and indirect putative downstream target genes were significantly down-regulated in SoxC-deficient progenitor cells (direct targets, P < 0.05; indirect targets, P < 0.01), while the expression of genes associated with SoxC-independent genomic regions remained unchanged (P > 0.05). HOMER analysis revealed that, in addition to Sox transcription factors, Gata and Isl motifs were significantly enriched in the SoxC-dependent peaks lost in the knockouts, suggesting these families of transcription factors may also be involved in setting up the permissive chromatin state established at E13.5 in conjunction with or downstream of SoxC (SI Appendix, Fig. S5). Further, by correlating the chromatin accessibility (ATAC-seq) in the WT and SoxC knockout progenitors and Sox4 DNA-binding (CUT&RUN), we have identified a number of confident direct targets of Sox4 (Dataset S3). These included key transcription factors previously shown to control sensory hair cell fate, such as Atoh1, Bcl11a, Eya1, and Isl1 (Fig. 5D) (17, 60, 68). In particular, we showed that Sox4 occupies several putative distal enhancer elements. For two of these, located around 130 and 180 kb downstream of the Atoh1 transcription start site, accessibility was established at E13.5 and lost in SoxC cKO, coinciding with the loss of Atoh1 upregulation in the cochlear prosensory domain (Fig. 5D).
Collectively these data strongly suggested that SoxC transcription factors establish competence for sensory receptor differentiation by directly binding to and establishing accessibility of the regulatory elements to initiate expression of the key sensory lineage genes, including master regulator Atoh1.
SoxC Transcription Factors Induce Hair Cell Formation In Vitro and In Vivo.
To directly test if SoxC transcription factors can promote Atoh1 expression and sensory differentiation, we overexpressed either Sox4 or Sox11 in the dissociated E12.5 progenitor cells not capable of differentiating toward hair cells spontaneously (Fig. 1) (38). Isolated from the Atoh1-GFP reporter mice, the E12.5 cochlea progenitor cells were infected with adenoviruses containing RFP, Sox2-RFP, Sox4-RFP, or Sox11-RFP coding sequences, and hair cell formation was analyzed 3 d later. While RFP- or Sox2-control overexpression failed to promote GFP reporter activation (SI Appendix, Fig. S6), either Sox4 or Sox11 potently induced formation of the GFP-positive hair cells (sixfold increase in the percentage of Atoh1-GFP-positive hair cells; n = 3, P < 0.01; Fig. 6 A–C). SoxC also induced supporting cells and hair cells to organize as the sensory rosettes similar to ones induced by Atoh1 at E13.5 (Figs. 1C and 6A). Together with the loss-of-function data, these results demonstrate that Sox4 and Sox11 transcription factors are both necessary and sufficient for sensory cell fate initiation in the organ of Corti progenitor cells.
Fig. 6.
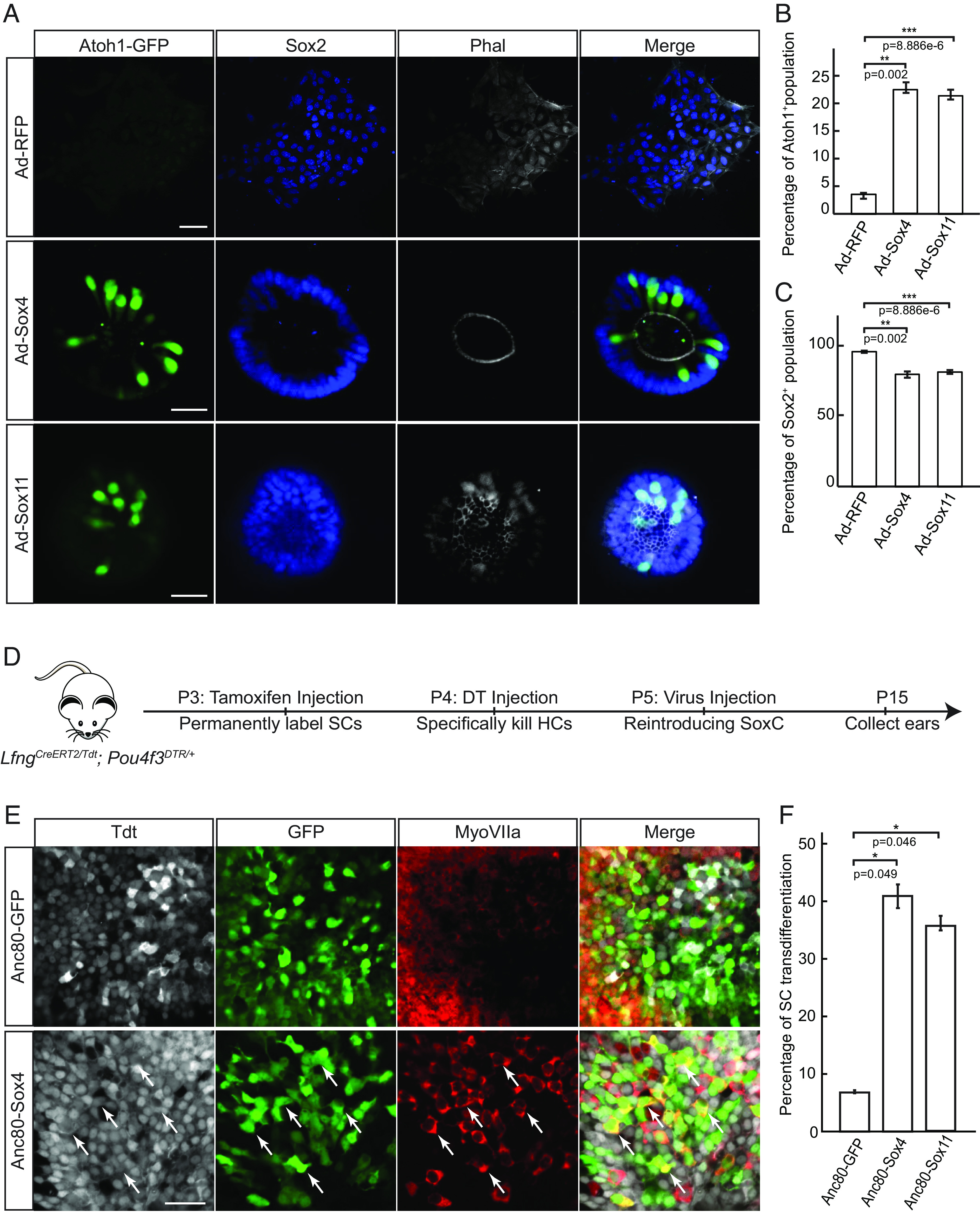
SoxC transcription factors are sufficient to trigger new hair cell generation in the utricles and the organ of Corti in vitro and in vivo. (A) Representative immunofluorescent images demonstrate that overexpression of SoxC promotes sensory differentiation in E12.5 cochlear progenitor cells isolated from the Atoh1-GFP reporter mice. Atoh1-GFP-positive hair cells (green) are detected in the Sox4-RFP and Sox11-RFP overexpressing cochlea cultures, where these sensory receptors and surrounding Sox2-positive supporting cells (blue) are organized into the sensory rosettes. Actin is labeled in white (phalloidin). (Scale bar, 20 μm.) (B and C) Bar graphs show quantitative analysis of the cultures in A. (B) A significant increase in the percentage of Atoh1-GFP-positive hair cell and (C) a corresponding decrease in the percentage of Atoh1-GFP-negative supporting cells is observed within the population of infected Sox2-positive cells in Sox4-RFP and Sox11-RFP overexpression conditions compared to the RFP-control (n = 3 for each condition). (D) Schematic representation shows the experimental design for in vivo overexpression of Sox4 and Sox11 in LfngCreER Pou4f3DTR mice after hair cell damage. Tamoxifen was administrated at P3 to permanently label supporting cells (SCs). Diphtheria toxin (DT) was injected at P4 to ablate Pou4f3-positive hair cell (HC) and the Anc80-AAV viruses containing each construct (GFP-control, Sox4-GFP, and Sox11-GFP) were injected at P5 through posterior semicircular canal. Sensory organs were collected at P15 for analysis. (E) Representative images show immunohistochemical labeling of the whole-mount utricles isolated at P15 from the GFP-control or Sox4-GFP overexpression conditions described in detail in D. tdTomato-labeled supporting cells are shown in gray, Anc80-infected cells expressing GFP-reporter are labeled in green, hair cells are labeled with MyoVIIa in red. White arrows indicate transdifferentiated triple labeled supporting cells. (Scale bar, 20 μm.) (F) Quantification analysis of the rate of supporting cell transdifferentiation in the utricles in E is shown. An increase in numbers of transdifferentiated supporting cells is detected in Sox4-GFP and Sox11-GFP overexpression conditions compared to GFP-controls (n = 3 for each condition).
In mammals, organ of Corti maturation is associated with reduction in transdifferentiation potential of supporting cell and is culminated in loss of capacity for hair cell regeneration by P6 (24–30). Concomitantly, both expression of Sox4 and Sox11 genes (Fig. 3B) and the accessibility of the competence-related chromatin loci (Fig. 2A) are lost. To test if SoxC transcription factors can reestablish competence for sensory differentiation, we overexpressed Sox4 or Sox11 in postnatal supporting cells. LfngGFP transgenic mice were used to purify supporting cells via fluorescence-activated cell sorting (FACS) from P6 cochlea and transfected with adenoviruses carrying RFP-control, Sox4-RFP, or Sox11-RFP (SI Appendix, Fig. S7). As demonstrated by Myo7a-positivity, Sox4 or Sox11 overexpression significantly increased the percentage of Lfng-GFP-positive infected supporting cells that converted to hair cells compared to RFP-control (eightfold for Sox4, fivefold for Sox11; n = 3; P < 0.05; SI Appendix, Fig. S7). We also noted the presence of GFP-positive neuron-like cells in Sox11 overexpression condition.
Because these results suggested that SoxC overexpression enhances supporting cell transdifferentiation potential in culture, we then tested whether Sox4 or Sox11 overexpression may stimulate hair cell regeneration after injury in vivo. By crossing the LfngCreERT2 (69), Rosa26tdTomato reporter (70), and the Pou4f3DTR/+ (71) mice, we generated transgenic animals in which supporting cells can be permanently labeled with tdTomato prior to induction of hair cell damage. After tamoxifen administration at P3, diphtheria toxin was injected to ablate hair cells a day later. At P5, we introduced GFP-control, Sox4-GFP, or Sox11-GFP overexpression vectors using a synthetic adeno-associated virus, Anc80 (72). We described previously that DT-induced hair cell damage allows this viral vector to transduce inner ear supporting cells (39). Because of the infection efficiency, we focused this in vivo analysis largely on the vestibular sensory organ – the utricle. By assessing the proportion of transduced supporting cells that converted to Myo7a-positive hair cells 10 d after adeno associated virus (AAV) administration, we first determined the spontaneous rate of transdifferentiation in GFP-controls to be about 6% in the utricle (Fig. 6 D–F), corresponding closely to the previously published data (71). Introduction of either ectopic Sox4 or Sox11 dramatically increased the proportion of transdifferentiating supporting cells to near 40% (Sox4: n = 3, P < 0.05; Sox11: n = 3, P < 0.05; Fig. 6 D–F). Although the rate of supporting cell infection was reduced in the cochlea impeding the analysis, several newly formed hair cells were detected after Sox4- or Sox11-overexpression, in contrast to the GFP-controls (SI Appendix, Fig. S7). These observations suggested that reintroduction of SoxC transcription factors promotes transdifferentiation of postnatal supporting cells in the inner ear.
Discussion
In this study, we have identified a molecular mechanism of competence establishment in the embryonic inner ear. We demonstrated that the window of plasticity during which organ of Corti progenitor cells can differentiate as hair cells is governed by the SoxC transcription factors, Sox4 and Sox11. Mechanistically, we show that SoxC factors control the ability of Atoh1 to up-regulate sensory receptor genes by making them epigenetically accessible. Conditional loss of Sox4 and Sox11 disrupts sensory progenitor cell differentiation, while ectopic expression of either gene outside the window of developmental plasticity is sufficient to promote hair cell fate in the inner ear. Our study builds a model of sensory fate establishment and provides insights to understanding failure of regeneration in the mammalian inner ear sensory organs.
Conserved Role for SoxC in Establishing Competence in Lineage-Specific Progenitor Cells.
SoxC transcription factors are shown to be critical for neuronal differentiation in other systems (51, 73, 74). Ablation of Sox4 and Sox11 in chick spinal cord and mouse hippocampus leads to failure of neuronal differentiation, while overexpression of the transcription factors promotes neurogenesis by targeting neuron-specific genes, such as Tuj1 and Map2 (51, 75, 76). Similarly, SoxC genes are necessary and sufficient to promote retinal ganglion cell differentiation and expression of lineage-specific genes like Isl1 (74). In these systems, the sensory fate commitment is thought to be tightly coupled with the cell cycle exit (3, 77–79). However, we observe that in absence of Sox4 and Sox11 expression, organ of Corti progenitor cells exit the cell cycle in a timely manner and are paused in a postmitotic yet undifferentiated state. Further, loss of SoxC transcription factor activity prevents sensory progenitor differentiation even when the master regulator of hair cell fate—Atoh1—is ectopically expressed. These data demonstrate that sensory cell fate commitment is not directly dependent on the cell cycle exit, suggesting that SoxC-dependent chromatin remodeling in postmitotic progenitor cells may be a common molecular mechanism for competence establishment in a number of cell lineages.
Beyond regulation of gene expression, SoxC transcription factors are known to cooperate with chromatin remodelers to alter epigenetic states (80–82). For example, Sox4 recruits SWI/SNF subunits to remodel chromatin during human MRC-5 fibroblast to neuron conversion and knockdown of the gene is sufficient to block reprogramming due to loss of around 60% of accessible chromatin regions (81). Mechanistically, chromatin immunoprecipitation assays with sequencing (ChIP-seq) results reveal that Sox4 binding on the regulatory elements of key neuronal lineage genes induces acetylation of the lysine 27 on histone 3 (H3K27ac)—the active histone mark that promotes gene expression (81). However, Sox4 was also shown to up-regulate expression of the epigenetic modifier Ezh2, a functional enzymatic component within the Polycomb Repressive Complex 2 (PRC2) that establishes repressive chromatin structure by introducing trimethylation of lysine 27 on histone 3 (H3K27me3) (82). These data are consistent with our results, as ATAC-seq experiments revealed both gain and loss of chromatin accessibility in SoxC cKO organ of Corti at E13.5, where chromatin structure largely reverted to that of E12.0 noncommitted progenitor cells. Further, scRNA-seq revealed that Ezh2 expression is largely down-regulated in SoxC cKO. Whether SoxC transcription factors cooperate with chromatin remodelers to establish histone modifications in the otic lineage requires further characterization.
SoxC Transcription Factors Likely Act Downstream of Sox2 but Upstream of Atoh1 to Promote Sensory Receptor Lineage Specification in the Inner Ear.
The master regulator Atoh1 is required and sufficient for hair cell differentiation and survival in the inner ear (7). However, what transcription factors initiate Atoh1 expression at the onset of sensory differentiation remains largely unknown. Sox2, the SoxB1 family member, was shown previously to bind the Atoh1 3′ enhancer to up-regulate its expression (8, 83–85). In fact, conditional loss of Sox2 prevents inner ear sensory lineage specification (8, 85, 86). Yet, Sox2 is expressed in the sensory progenitor cells prior to establishment of competence for sensory differentiation and its expression in supporting cells remains high after the loss of transdifferentiation potential. Further, Sox2 haploinsufficiency was shown to promote sensory differentiation (58), highlighting its complex role in the inner ear (8, 58, 83, 84, 86, 87).
We show that expression of Sox4 is initiated at E13.5, immediately prior to the onset of Atoh1 upregulation, while Sox2 (SoxB1) transcription factor is expressed from initiation of the otic lineage (88). Further, unlike Sox2, overexpression of Sox4 or Sox11 strongly promoted Atoh1 expression in undifferentiated organ of Corti progenitor cells. These data demonstrate that SoxC transcription factors likely act upstream of Atoh1 and may directly up-regulate its expression. Mechanistically, CUT&RUN data showed that Sox4 does not bind the proximal preestablished autoregulatory 3′ enhancer of Atoh1 occupied by Sox2. However, in Atoh1-GFP transgenic mice where reporter expression is driven by this enhancer, GFP expression lags the endogenous onset of Atoh1 upregulation in the prosensory domain (35, 89). These data suggest that the initial upregulation of Atoh1 relies on other regulatory elements, and we noted several putative enhancers in the Atoh1 locus. Two of such elements, located around 130 kb and 180 kb downstream of the Atoh1 transcription start site, are established during the transition to the competent state, occupied by Sox4, and disappear in SoxC cKO. It will be important to test if Sox4-mediated accessibility of this putative enhancer directly regulates initial Atoh1 upregulation to drive sensory differentiation in the inner ear.
SoxC Transcription Factors and Hair Cell Regeneration.
Previous studies concluded that reintroducing Atoh1 alone in the mammalian cochlea after P6 is not sufficient to trigger hair cell regeneration (20, 21), likely due to loss of chromatic accessibility and decommissioning of hair cell–specific enhancers (29, 30). Our data further suggest that the normal downregulation of SoxC expression and the consequent decrease in accessibility of the competence-related genomic regions in the postnatal supporting cells may hamper their transdifferentiation potential. By reintroducing SoxC gene expression, we were able to promote supporting cell transdifferentiation in the postnatal ear after hair cell damage. Additionally, both Sox4 and Sox11 genes are significantly up-regulated during direct reprogramming of mouse embryonic fibroblasts of induced hair cells (90). Further, sox4a is one of the first genes up-regulated just prior to atoh1a during hair cell regeneration in the zebrafish lateral line (91). Together these data suggest that expression of SoxC in supporting cells after hair cell damage may promote plasticity and stimulate sensory regeneration. Further experiments, using coexpression of Atoh1, SoxC, and other key hair cell lineage genes may achieve better transdifferentiation rates and promote hair cell recovery in the mature inner ear in mammals.
Materials and Methods
All experiments were conducted according to the policies of the Institutional Animal Care and Use Committees of the Keck School of Medicine at the University of Southern California. p27Kip1-GFP mice were previously described in our laboratory (10). Atoh1-GFP mice were obtained from Jane Johnson at University of Texas Southwestern (89). Sox2GFP mice were obtained from the Jackson laboratory and were described previously (92). Pax2Cre (49, 53) mice were provided by Groves, Baylor College of Medicine. To create conditional double knockouts, Sox4fl/fl Sox11fl/fl Sox12−/− mice were obtained from Veronique Lefebvre (49) bred to Pax2Cre line. LfngCreERt2 mice were obtained from Andrew Groves, Baylor College of Medicine (69). Rosa26tdTomato were obtained from the Jackson laboratory, and Pou4f3DTR/+ mice were obtained from Jennifer Stone, University of Washington (69, 71). The three alleles were combined to obtain transgenic mice used for lineage-tracing of supporting cells after hair cell damage.
Detailed protocols for primary cochlear progenitor and supporting cell isolation and culture, immunohistochemistry, RNA in situ hybridization, viral production, and sequencing data acquisition and analysis can be found in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (PDF)
Dataset S03 (PDF)
Acknowledgments
We thank Francis James for constructing the scRNA-seq, ATAC-seq, and CUT&RUN data analysis pipelines. We thank Dr. Andrew Groves for providing insightful feedback and editing the manuscript. We also thank University of Southern California Stem Cell Flow Cytometry Facility for cell sorting assistance, Children’s Hospital Los Angeles Molecular Pathology Genomics Core for Next-generation sequencing, Dr. Richard Sandberge for the Tn5 plasmid, Dr. Steven Henikoff for CUT&RUN reagents. We thank Dr. Elisabeth Sock for Sox4 and Sox11 antibody, Dr. Jennifer Stone for Pou4f3DTR/+ mice, Dr. Veronique Lefebvre for Sox4fl/fl Sox11fl/fl mice, and Dr. Andrew Groves for LfngCreERt2 and Pax2Cre mice. This work was supported by grants to K.G. from the National Institute on Deafness and Other Communication Disorders (R21 DC016984) and to N.S. from the Hearing Restoration Program of the Hearing Health Foundation and from the National Institute on Deafness and Other Communication Disorders (R01DC015829). X.W. was supported through NIDCD training grant (T32DC009975).
Author contributions
X.W., N.S., and K.G. designed research; X.W., J.L., T.T., T.S., and K.G. performed research; W.M. and J.G.C. contributed new reagents/analytic tools; X.W., T.T., L.T., and K.G. analyzed data; J.L. and W.M. provided assistance with experimental design, mouse colony maintenance, and dissection expertise; J.G.C., N.S., and K.G. supervised the research; and X.W. and K.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All sequencing datasets for scRNA-seq, ATAC-seq, and CUT&RUN ChIP-seq are deposited to NCBI’s Gene Expression Omnibus (GEO: GSE215171) database (93).
Supporting Information
References
- 1.Gage F. H., Mammalian neural stem cells. Science (80-) 287, 1433–1438 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Huttner W. B., [S17]: The cell biology of neurogenesis. Int. J. Dev. Neurosci. 24, 478–478 (2006). [Google Scholar]
- 3.Marquardt T., Gruss P., Generating neuronal diversity in the retina: One for nearly all. Trends Neurosci. 25, 32–38 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Ruben R. J., Development of the inner ear of the mouse: A radioautographic study of terminal mitoses. Acta Otolaryngol. (suppl. 220), 1–44 (1967). [PubMed]
- 5.Lowenheim H., et al. , Gene disruption of p27Kip1 allows cell proliferation in the postnatal and adult organ of Corti. Proc. Natl. Acad. Sci. U.S.A. 96, 4084–4088 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P., Segil N., p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Chen P., Johnson J. E., Zoghbi H. Y., Segil N., The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Kiernan A. E., et al. , Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–5 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Matei V., et al. , Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 234, 633–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y.-S., A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133, 2817–2826 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Groves A. K., Zhang K. D., Fekete D. M., The genetics of hair cell development and regeneration. Annu. Rev. Neurosci. 36, 361–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fekete D. M., Muthukumar S., Karagogeos D., Hair cells and supporting cells share a common progenitor in the avian inner ear. J. Neurosci. 18, 7811–7821 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oesterle E. C., Chien W.-M., Campbell S., Nellimarla P., Fero M. L., p27 Kip1 is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell Cycle 10, 1237–1248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adler H. J., Raphael Y., New hair cells arise from supporting cell conversion in the acoustically damaged chick inner ear. Neurosci. Lett. 205, 17–20 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Duncan L. J., et al. , Differential expression of unconventional myosins in apoptotic and regenerating chick hair cells confirms two regeneration mechanisms. J. Comp. Neurol. 499, 691–701 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberson D. W., Alosi J. A., Cotanche D. A., Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J. Neurosci. Res. 78, 461–471 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Bermingham N. A., et al. , Math1: An essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Gao Z., et al. , Spatial and age-dependent hair cell generation in the postnatal mammalian utricle. Mol. Neurobiol. 53, 1601–1612 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Izumikawa M., et al. , Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 11, 271–276 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Kelly M. C., Chang Q., Pan A., Lin X., Chen P., Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 32, 6699–6710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., et al. , Age-dependent in vivo conversion of mouse cochlear pillar and deiters’ cells to immature hair cells by atoh1 ectopic expression. J. Neurosci. 32, 6600–6610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shou J., Zheng J. L., Gao W.-Q., Robust generation of new hair cells in the mature mammalian inner ear by adenoviral expression of Hath1. Mol. Cell. Neurosci. 23, 169–179 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Doetzlhofer A., et al. , Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bramhall N. F., Shi F., Arnold K., Hochedlinger K., Edge A. S. B., Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2, 311–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox B. C., et al. , Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L., et al. , Diphtheria toxin-induced cell death triggers wnt-dependent hair cell regeneration in neonatal mice. J. Neurosci. 36, 9479–9489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maass J. C., et al. , Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front. Cell. Neurosci. 9, 110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGovern M. M., Zhou L., Randle M. R., Cox B. C., Spontaneous hair cell regeneration is prevented by increased notch signaling in supporting cells. Front. Cell. Neurosci. 12, 120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao L., et al. , Enhancer decommissioning imposes an epigenetic barrier to sensory hair cell regeneration. Dev. Cell 56, 2471–2485.e5 (2021), 10.1016/j.devcel.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H. V., et al. , POU4F3 pioneer activity enables ATOH1 to drive diverse mechanoreceptor differentiation through a feed-forward epigenetic mechanism. Proc. Natl. Acad. Sci. U.S.A. 118, e2105137118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley M., Talreja D., Corwin J., Replacement of hair cells after laser microbeam irradiation in cultured organs of corti from embryonic and neonatal mice. J. Neurosci. 15, 3013–3026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montcouquiol M., Kelley M. W., Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J. Neurosci. 23, 9469–9478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haque K. D., Pandey A. K., Kelley M. W., Puligilla C., Culture of embryonic mouse cochlear explants and gene transfer by electroporation. J. Vis. Exp. 52260 (2015), 10.3791/52260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gnedeva K., Hudspeth A. J., Segil N., Three-dimensional organotypic cultures of vestibular and auditory sensory organs. J. Vis. Exp. 57527 (2018), 10.3791/57527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helms A. W., Abney A. L., Ben-Arie N., Zoghbi H. Y., Johnson J. E., Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185–1196 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Ikeda R., Pak K., Chavez E., Ryan A. F., Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol. Neurobiol. 51, 672–684 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda M., Pak K., Chavez E., Ryan A. F., TFE2 and GATA3 enhance induction of POU4F3 and myosin VIIa positive cells in nonsensory cochlear epithelium by ATOH1. Dev. Biol. 372, 68–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doetzlhofer A., White P. M., Johnson J. E., Segil N., Groves A. K., In vitro growth and differentiation of mammalian sensory hair cell progenitors: A requirement for EGF and periotic mesenchyme. Dev. Biol. 272, 432–447 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Gnedeva K., et al. , Organ of Corti size is governed by Yap/Tead-mediated progenitor self-renewal. Proc. Natl. Acad. Sci. U.S.A. 117, 13552–13561 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean C. Y., et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed M., et al. , Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Appler J. M., Goodrich L. V., Connecting the ear to the brain: Molecular mechanisms of auditory circuit assembly. Prog. Neurobiol. 93, 488–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karis A., et al. , Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol. 429, 615–630 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Milo M., et al. , Genomic analysis of the function of the transcription factor gata3 during development of the mammalian inner ear. PLoS One 4, e7144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng W., et al. , The role of Six1 in mammalian auditory system development. Development 130, 3989–4000 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou D., Silvius D., Fritzsch B., Xu P.-X., Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 131, 5561–5572 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowles J., Schepers G., Koopman P., Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227, 239–255 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Bhattaram P., et al. , Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 1, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dy P., et al. , The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36, 3101–3117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mu L., et al. , Soxc transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 32, 3067–3080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gnedeva K., Hudspeth A. J., SoxC transcription factors are essential for the development of the inner ear. Proc. Natl. Acad. Sci. U.S.A. 112, 14066–14071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohyama T., Groves A. K., Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis 38, 195–199 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Lefebvre V., Dumitriu B., Penzo-Méndez A., Han Y., Pallavi B., Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 39, 2195–2214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S., et al. , Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev. Biol. 8, 20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morsli H., et al. , Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development 126, 2335–2343 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Morsli H., Choo D., Ryan A., Johnson R., Wu D. K., Development of the mouse inner ear and origin of its sensory organs. J. Neurosci. 18, 3327–3335 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dabdoub A., et al. , Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc. Natl. Acad. Sci. U.S.A. 105, 18396–18401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zine A., Van De Water T. R., de Ribaupierre F., Notch signaling regulates the pattern of auditory hair cell differentiation in mammals. Development 127, 3373–3383 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Radde-Gallwitz K., et al. , Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J. Comp. Neurol. 477, 412–421 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou D., et al. , Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum. Mol. Genet. 17, 3340–3356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrionuevo F., et al. , Sox9 is required for invagination of the otic placode in mice. Dev. Biol. 317, 213–224 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Saint-Germain N., Lee Y.-H., Zhang Y., Sargent T. D., Saint-Jeannet J.-P., Specification of the otic placode depends on Sox9 function in Xenopus. Development 131, 1755–1763 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Yan Y.-L., et al. , A pair of Sox: Distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069–1083 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Hayashi T., Ray C. A., Bermingham-McDonogh O., Fgf20 is required for sensory epithelial specification in the developing cochlea. J. Neurosci. 28, 5991–5999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garel S., Marín F., Grosschedl R., Charnay P., Ebf1 controls early cell differentiation in the embryonic striatum. Development 126, 5285–5294 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Kuroda T., et al. , SALL3 expression balance underlies lineage biases in human induced pluripotent stem cell differentiation. Nat. Commun. 10, 2175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hawkins R. D., et al. , Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS One 2, e525 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semerci F., et al. , Lunatic fringe-mediated Notch signaling regulates adult hippocampal neural stem cell maintenance. Elife 6, e24660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golub J. S., et al. , Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J. Neurosci. 32, 15093–15105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landegger L. D., et al. , A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 35, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haslinger A., Schwarz T. J., Covic M., Chichung Lie D., Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur. J. Neurosci. 29, 2103–2114 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Kuwajima T., Soares C. A., Sitko A. A., Lefebvre V., Mason C., Soxc transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron 93, 1110–1125.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergsland M., Werme M., Malewicz M., Perlmann T., Muhr J., The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 20, 3475–3486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanaka S., et al. , Interplay of SOX and POU factors in regulation of the nestin gene in neural primordial cells. Mol. Cell. Biol. 24, 8834–8846 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buttitta L. A., Edgar B. A., Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697–704 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Götz M., Huttner W. B., The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Ruijtenberg S., van den Heuvel S., Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kavyanifar A., Turan S., Lie D. C., SoxC transcription factors: Multifunctional regulators of neurodevelopment. Cell Tissue Res. 371, 91–103 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Smith D. K., Yang J., Liu M.-L., Zhang C.-L., Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 7, 955–969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tiwari N., et al. , Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Kempfle J. S., Turban J. L., Edge A. S. B., Sox2 in the differentiation of cochlear progenitor cells. Sci. Rep. 6, 23293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neves J., Uchikawa M., Bigas A., Giraldez F., The prosensory function of Sox2 in the chicken inner ear relies on the direct regulation of Atoh1. PLoS One 7, e30871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steevens A. R., et al. , SOX2 is required for inner ear growth and cochlear nonsensory formation prior to sensory development. Development 146, dev170522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dvorakova M., et al. , Incomplete and delayed Sox2 deletion defines residual ear neurosensory development and maintenance. Sci. Rep. 6, 38253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basch M. L. M. M. L., et al. , Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates. Elife 5, 841–850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu R., et al. , Lineage tracing of Sox2-expressing progenitor cells in the mouse inner ear reveals a broad contribution to non-sensory tissues and insights into the origin of the organ of Corti. Dev. Biol. 414, 72–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lumpkin E. A., et al. , Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–95 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Menendez L., et al. , Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. Elife 9, e5524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baek S., et al. , Single-cell transcriptome analysis reveals three sequential phases of gene expression during zebrafish sensory hair cell regeneration. Dev. Cell 57, 799–819.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arnold K., et al. , Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X., et al. , SoxC transcription factors shape the epigenetic landscape to establish competence for sensory differentiation in the mammalian organ of Corti. The National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE215171. Deposited 10 October 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (PDF)
Dataset S03 (PDF)
Data Availability Statement
All sequencing datasets for scRNA-seq, ATAC-seq, and CUT&RUN ChIP-seq are deposited to NCBI’s Gene Expression Omnibus (GEO: GSE215171) database (93).