Abstract
Traumatic spinal cord injuries (SCI) are a group of highly debilitating pathologies affecting thousands annually, and adversely affecting quality of life. Currently, no fully restorative therapies exist, and SCI still results in significant personal, societal and financial burdens. Inflammation plays a major role in the evolution of SCI, with myeloid cells, including bone marrow derived macrophages (BMDMs) and microglia (MG) being primary drivers of both early secondary pathogenesis and delayed wound healing events.
The precise role of myeloid cell subsets is unclear as upon crossing the blood-spinal cord barrier, infiltrating bone marrow derived macrophages (BMDMs) may take on the morphology of resident microglia, and upregulate canonical microglia markers, thus making the two populations difficult to distinguish.
Here, we used time-resolved scRNAseq and transgenic fate-mapping to chart the transcriptional profiles of tissue-resident and -infiltrating myeloid cells in a mouse model of thoracic contusion SCI.
Our work identifies a novel subpopulation of foam cell-like inflammatory myeloid cells with increased expression of Fatty Acid Binding Protein 5 (Fabp5) and comprise both tissue-resident and -infiltrating cells. Fabp5+ inflammatory myeloid cells display a delayed cytotoxic profile that is predominant at the lesion epicentre and extends into the chronic phase of SCI.
Keywords: Single cell RNA sequencing, Neuroinflammation, Fatty acid binding protein 5, Spinal cord injury, Fate-mapping myeloid cells, Myeloid cells, Microglia, Lipid metabolism, Neurodegeneration
1. Introduction
Acute traumatic spinal cord injuries (SCI) typically result in permanent disability due to the inefficiency of spontaneous regenerative processes in the central nervous system (CNS). In comparison to normal wound healing, the mechanisms of tissue recovery after SCI are dysregulated, leading to a chronic wound state rather than a return to homeostasis [1]. One of the major hallmarks of this chronic state is a persistent state of local inflammation, which is driven by CNS-resident microglia (MG) and -infiltrating myeloid cells [2]. However, the precise roles of these myeloid cell subsets in chronic SCI remains unclear, as CNS infiltrating monocyte-derived macrophages become difficult to distinguish from MG in the injured CNS using canonical morphology and lineage markers [3].
Fatty acid binding proteins (FABPs) are cytosolic lipid chaperones that reversibly bind to hydrophobic molecules, including free fatty acids (FAs). FABPs transport cargo to specific nuclear compartments for lipoprotein, triglyceride, and cholesterol synthesis [[4], [5], [6]], or to nuclear receptors like PPARγ [7]. FABP5 loss of function leads to a build-up of intracellular FAs and a decrease in cholesterol [8]. In bone marrow-derived macrophages (BMDM) with FABP5 loss of function, both pro- and anti-inflammatory stimulation results in significantly higher expression of anti-inflammatory factors [8,9]. These findings demonstrate that loss of FABP5 promotes anti-inflammatory responses in macrophages. While little is known about the role of FABP5 in microglia, it represents an interesting target that could be manipulated to alter lipid metabolism and reduce chronic neuroinflammation.
Here, we utilized in vivo fate-mapping, ex vivo FACS isolation (S. Fig. 1), and single-cell RNA sequencing (scRNAseq) to investigate the ontology and temporal dynamics of myeloid cells in a mouse model of low thoracic contusion SCI (Fig. 1A).
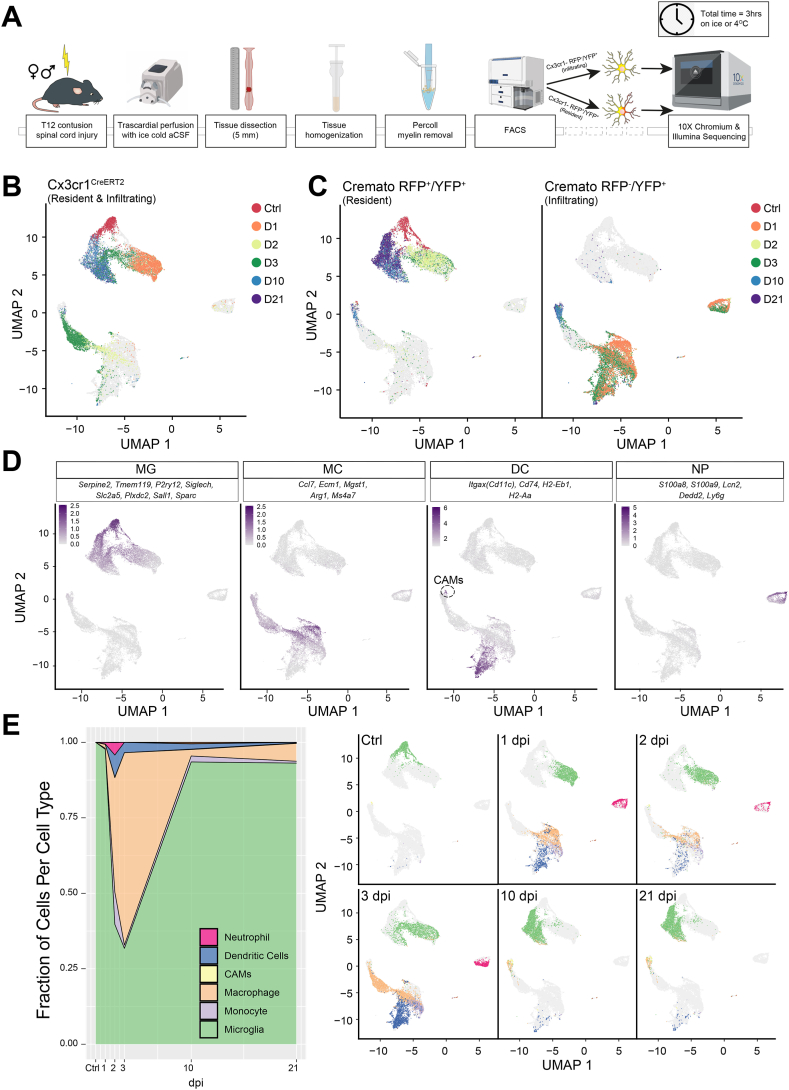
Fig. 1.
Characterization of CNS infiltrating and resident myeloid cells upon SCI. A, Myeloid lineage-specific cell isolation protocol used in the study. The illustration depicts the Cremato mouse line FACS only. B–C, UMAPs of the batch corrected dataset separated by strain and fate mapping in the Cx3cr1CreERT2 and Cremato mouse lines. Each dot represents a cell and is coloured by the time of collection. D, UMAPs with cells (dots) coloured by the gene expression of the listed cell-type markers in the Cremato mouse line. Scale bars are the averaged log2-transformed counts from the listed marker genes. E, Area plot of the myeloid cell types comprising the Cx3cr1 dataset (left) and UMAP of the entire dataset (right) separated by time point and coloured by cell type.
We first employed a G-protein coupled chemokine (C-X3-C motif) receptor 1 (Cx3cr1)CreERT2 mouse line, which expresses yellow fluorescent protein (YFP) under the Cx3cr1 promoter, to isolate all YFP + myeloid cells from the spinal cord of mice in the acute [1, 2, 3 days post-injury (dpi)], subacute (10 dpi), and chronic (21 dpi) phases of SCI. Mice subjected to laminectomy only were used as controls (Ctrl) (Fig. 1B).
Through scRNAseq, we identified transcriptionally heterogeneous cell populations, which we hypothesized to include both CNS-resident and -infiltrating myeloid cells. To unambiguously evaluate the relative contribution of these two different myeloid cell subpopulations, we next adopted a Cx3cr1CreERT2:R26tdTomato fate mapping mouse line (hereafter called Cremato), which exploits the differing lifespans of CNS-resident vs. -infiltrating myeloid cells [10,11]. This approach allowed us to enrich for double-positive (RFP+/YFP+) CNS-resident myeloid cells vs. single-positive (RFP−/YFP+) -infiltrating cells (Fig. 1C), and to quantify their respective spatial distribution over time (S. Fig. 2A), ultimately bringing a unique myeloid-centred perspective to SCI, compared to previously published scRNAseq data [12,13].
2. Results
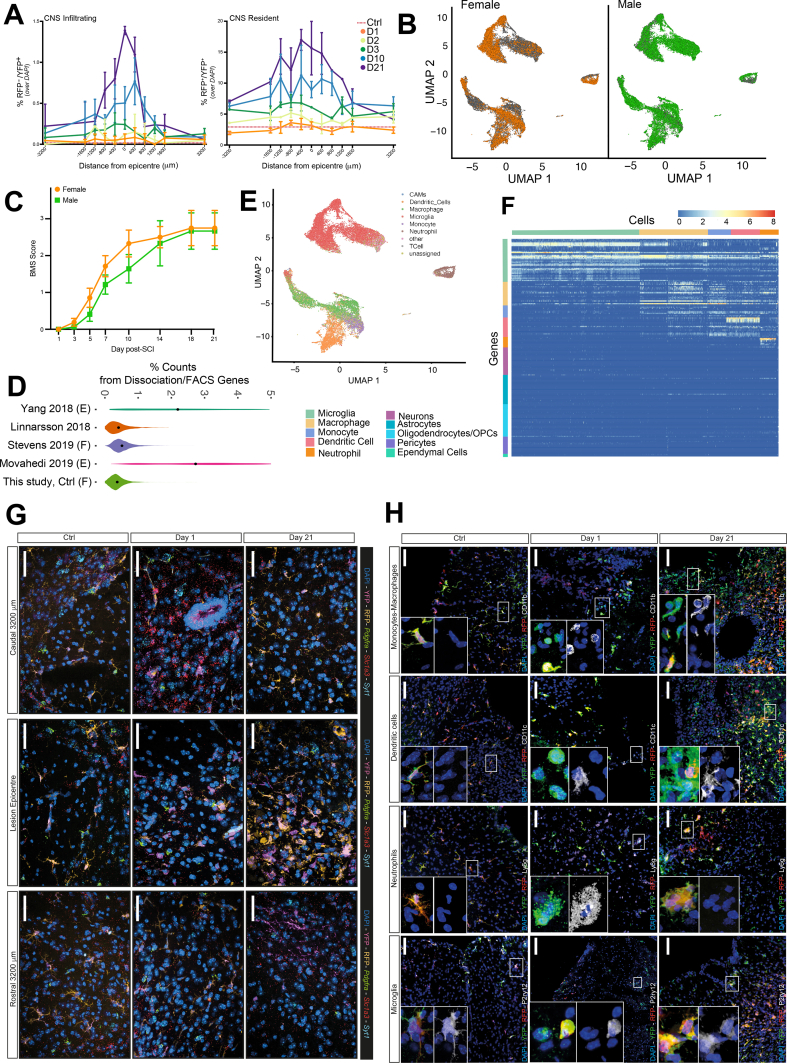
In total, we analysed 31,423 cells from Cx3cr1CreERT2 and Cremato mice (S. Table S1). We observed neither behavioural nor transcriptional differences between males and females after contusion SCI, allowing us to pool these datasets (S. Fig. 2B and C) [3,14,15]. Compared to previous studies [[15], [16], [17]], we observed lower-levels of dissociation- and FACS-induced gene expression [18] in Ctrls, which suggests that our protocol minimises cellular activation and is optimal for ex vivo cell isolation and gene expression analyses (S. Fig. 2D). After batch correction, we annotated the dataset using a combination of CellAssign and manual annotations based on fate mapping labels, Gene Ontology (GO), and core gene signatures from previous CNS scRNAseq studies (S. Figs. 2E and 1D) [3,10,17,19]. Finally, we confirmed the myeloid-lineage specificity of the Cremato mouse line via scRNAseq (S. Fig. 2F), single-molecule fluorescent in situ hybridization (smFISH) (S. Fig. 2G), and confocal immunofluorescence microscopy (S. Fig. 2H).
We isolated only CNS-resident MG cells from Ctrls (Fig. 1E and S. Fig. 2G and H). In the acute phase following SCI, CNS-infiltrating myeloid cells (including monocytes, neutrophils [NPs], and myeloid dendritic cells [DCs]) [20] increased considerably (Fig. 1E and S. Fig. 2A). During the subacute and chronic phases, most of the isolated myeloid cells were either MG or, to a lower extent, monocyte-derived macrophages (Fig. 1E and S. Fig. 2A).
Recent scRNAseq data has demonstrated that MG downregulate canonical markers upon activation [3,10,14,21]. Indeed, we found that the MG-specific markers Serpine2, P2ry12, Plxdc2, Tmem119, Siglech, and Sparc were expressed in Ctrl MG, but were downregulated in MG at 1 dpi (Fig. 2A). MG markers, such as Olfml3 (Fig. 2A and B), Grp34 (Fig. 2A), and Fcrls (S. Table S1) were also downregulated in MG at 1 dpi but were upregulated in CNS-associated macrophages (CAMs) at 10 dpi. Other MG markers, like Hexb (Fig. 2A–B), C1qa (Fig. 2A), Trem2 (Fig. 2A), and Aif1/Iba-1 (S. Table S1), were stably expressed in MG, but showed increased expression in CNS-infiltrating myeloid cells, thus confirming that subsets of monocyte-derived macrophages adopt a MG-like phenotype upon entering the CNS [3].
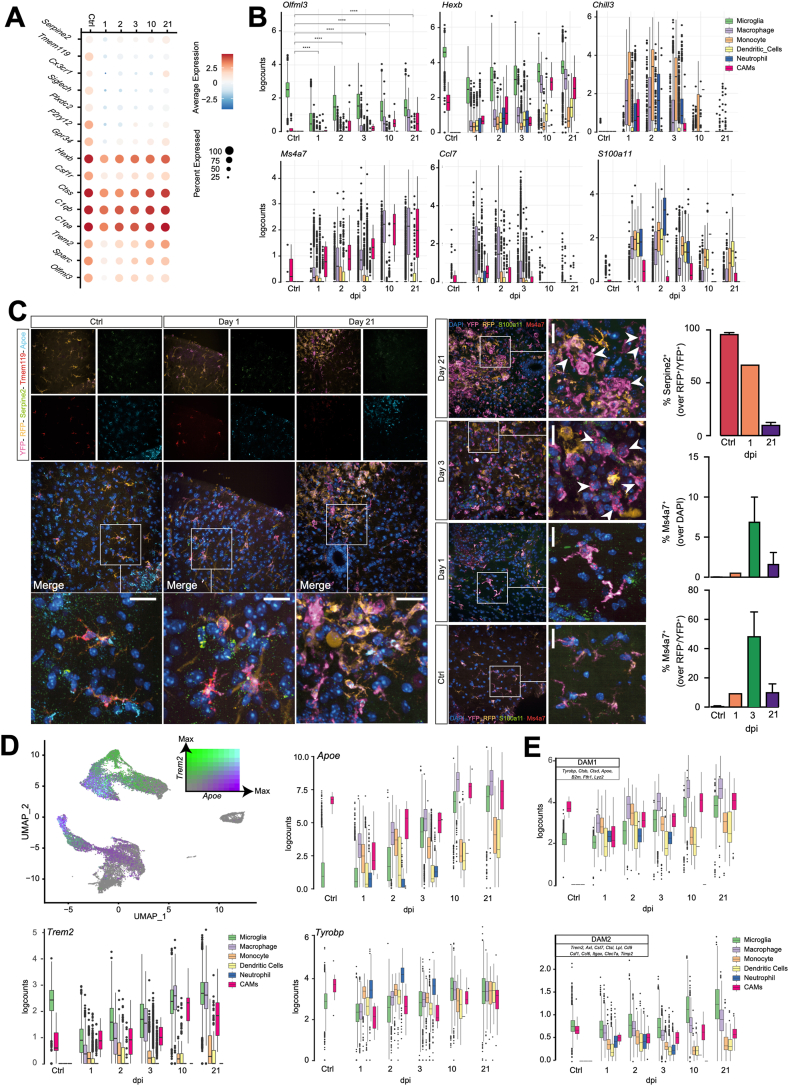
Fig. 2.
Validation against published myeloid cell studies via both scRNAseq and smFISH. A, Seurat dot plot showing canonical MG marker expression by dpi. Scale bar is the natural log of the average log2-transformed counts. B, Boxplots by cell type and time point of the log2-transformed expression of selected genes. For Olfml3: Welch t-test with Benjamini-Hochberg post-hoc correction.ns p ≥ 0.05; *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). Boxplots for every gene in this dataset can be found on the SCI Myeloid Cell Atlas shiny app (https://marionilab.cruk.cam.ac.uk/SCI_Myeloid_Cell_Atlas/). C, smFISH data depicting the expression of Tmem119, Serpine2 and Apoe from Ctrl, 1 dpi, and 21 dpi (left) and Ms4a7 and S100a11 from Ctrl, 1, 3, and 21 dpi (right). The bar graph shows the quantification of Ms4a7+ and Serpine2+ cells. Arrowheads indicate Ms4a7+/RFP−/YFP+ cells. Data are mean % (±SD) from n = 2 mice per time point. Nuclei were stained with DAPI. Scale bars: 20 μm. D, UMAP coloured by the scaled log2-transformed counts of Apoe and Trem2. Boxplots by time and cell type demonstrating the upregulation of TREM2-induced APOE pathway-associated genes Apoe, Trem2, and Tyrobp upon SCI. E, Boxplots by time point coloured by cell type, quantifying the average log2-transformed counts of DAM1-vs. DAM2-associated genes expressed per cell.
We also observed several genes that appeared to be strictly lineage specific, even after SCI. Among these, we confirmed that Chil3/Ym1 was expressed by monocyte-derived macrophages and NPs only [1] (Fig. 2B). Ms4a7 [3] (Fig. 2B), Ccl7 [22] (Fig. 2B), Arg1 [10], and Mgst1 [19] (S. Table S1) were restricted specifically to CAMs and monocyte-derived cells. We further validated the dynamic expression of a selection of resident-specific (i.e., Serpine2, Tmem119) and infiltrating-specific (i.e., Ms4a7 and S100a11) markers in MG and CNS-infiltrating myeloid cells with smFISH (Fig. 2C). We found that S100a11 was expressed by infiltrating myeloid cells only, but not by resident MGs (Fig. 2B and C), which anticipates a putative marker to distinguish infiltrating vs. resident myeloid cells in the injured CNS. Altogether, these findings indicate that our myeloid cell scRNAseq dataset is consistent with, but importantly also expands upon, previous scRNAseq studies in CNS disease models [12,13].
To gain further insight into the cellular and molecular mechanisms driving these transcriptional changes, we focused on the TREM2-induced APOE pathway, which has been shown to mediate the activation of myeloid cells in disease [23]. Upon SCI, both MG and infiltrating myeloid cells upregulated apolipoprotein E (Apoe) (Fig. 2C and D) and the TREM2-APOE pathway component, transmembrane immune signalling adaptor (Tyrobp) (Fig. 2D). The upregulation of Apoe and Tyrobp, along with the downregulation of several homeostatic MG markers, has recently been described as the first step towards the acquisition of a neuroprotective, phagocytic, ‘Disease Associated MG’ (DAM) phenotype in murine models of Alzheimer's disease and amyotrophic lateral sclerosis [21]. As such, DAMs are described to comprise two sequential stages (DAM1 and DAM2), which are conserved between mice and humans [21]. The transcriptional profile of DAMs is yet to be described in SCI.
We found that the expression of DAM1-associated genes increases in both MG and monocyte-derived macrophages in the acute phases post-SCI (Fig. 2E). Conversely, the expression of DAM2-associated genes increases over time in MG only (Fig. 2E), with all (but two) DAM2-associated genes being upregulated in MG by the subacute and chronic phases of SCI (S. Table S1).
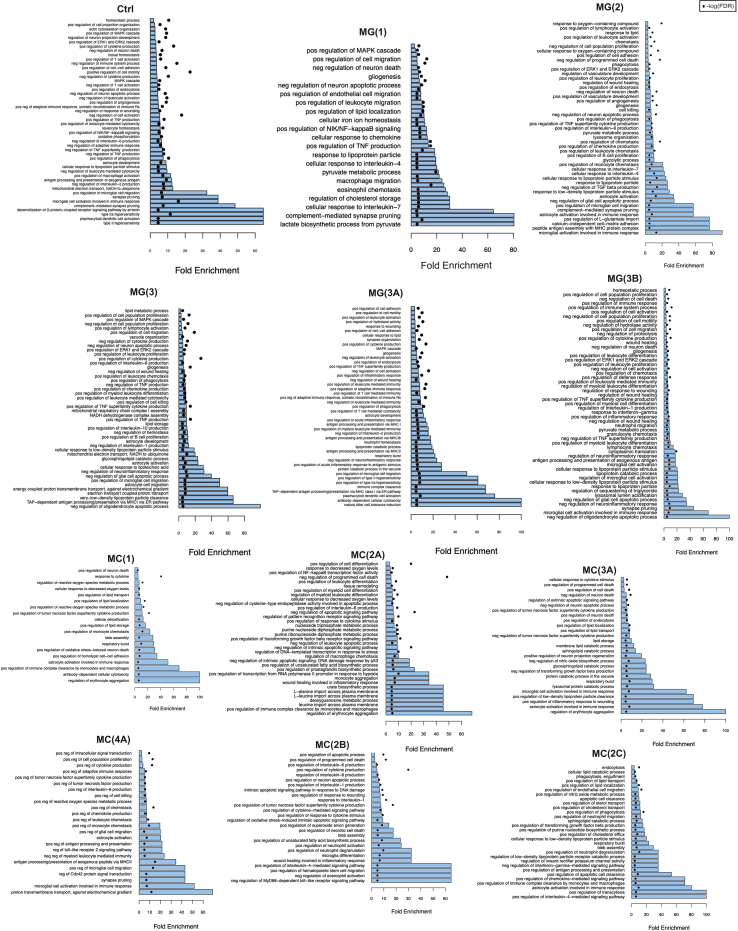
To identify novel molecular processes involved in the immune response to SCI, we used unsupervised clustering to study resident MG vs. infiltrating myeloid cell and monocyte-derived cell (MC) subsets (Fig. 3A), and generated cluster-specific differentially expressed genes (DEG) followed by GO enrichment analysis (S. Fig. 3).
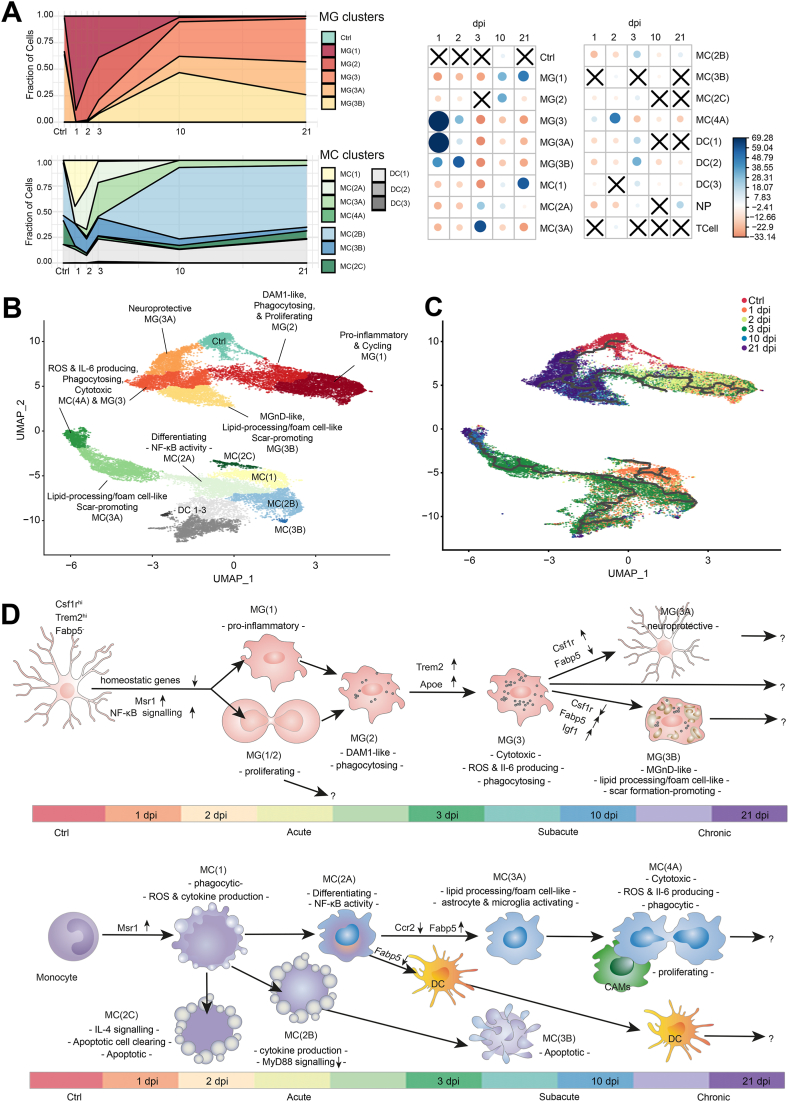
Fig. 3.
Dynamic trajectories of myeloid cells post-SCI. A, Breakdown of the MG and MC clusters by the fraction of cells from each time point (left), and correlation plot of the Pearson's chi-squared residuals (right). The size of each circle is proportional to the absolute value of the standardized chi-square residual. Blue signifies a positive standardized chi-square residual, while red signifies the opposite. “X” represents a non-significant contribution. B, UMAP coloured by cluster, as determined by Leiden community detection using Monocle 3. Clusters are named based on their majority cell-type and their position in the trajectory. The UMAP is superimposed with the unsupervised reversed graph embedding-derived trajectory inferred via Monocle 3. C, UMAP coloured by time post-SCI, giving directionality to the trajectory. Trajectories of B are represented by the black lines. D, Proposed map of MG (top) or MC (bottom) states over time.
We first noted that the fraction of myeloid cells per cluster changed over time (Fig. 3A) and the correlation between the time of collection and the assigned cluster was significant (S. Table S1). Therefore, we performed unsupervised trajectory analysis to infer the progression of myeloid cell states across the different phases of SCI (Fig. 3B and C). This allowed us to generate a map of MG and MC subset transcriptional profiles across the acute, subacute and chronic phases of SCI injury (Fig. 3D).
We found that at 1 dpi, the majority of MG were in a highly activated MG(1) state (Fig. 3B) characterized by GO terms for cell cycling and proliferation (S. Fig. 3) [11,12]. These findings were then confirmed by cell cycle analysis performed using the cyclone function from the scran package (Fig. 4A). As expected, MG(1) were also highly enriched for GO terms related to the production and response to cytokines, recruitment of blood-borne leukocytes, aerobic glycolysis, as well as astrocyte activation and leukocyte migration (S. Fig. 3) [2,12]. We also found that MG(1) lacked Csf1r, a gene required for MG proliferation and survival during homeostasis [24], and upregulated fatty acid-binding protein 5 (Fabp5) (Fig. 4B–D), a target of both the NF-κB and PPAR-γ pathways that is involved in lipid transport/localization [25,26]. Notably, Fabp5 is also upregulated in glial and neuronal cells, including in BMDMs post-SCI [27], in response to various animal models of neurotrauma [28]. Upregulation of Fabp5 appeared to occur at the root of the MG(1) trajectory (Fig. 3C). From this point, the trajectory analysis predicts that cells pass through the highly proliferative acute-phase MG(1) and MG(2).
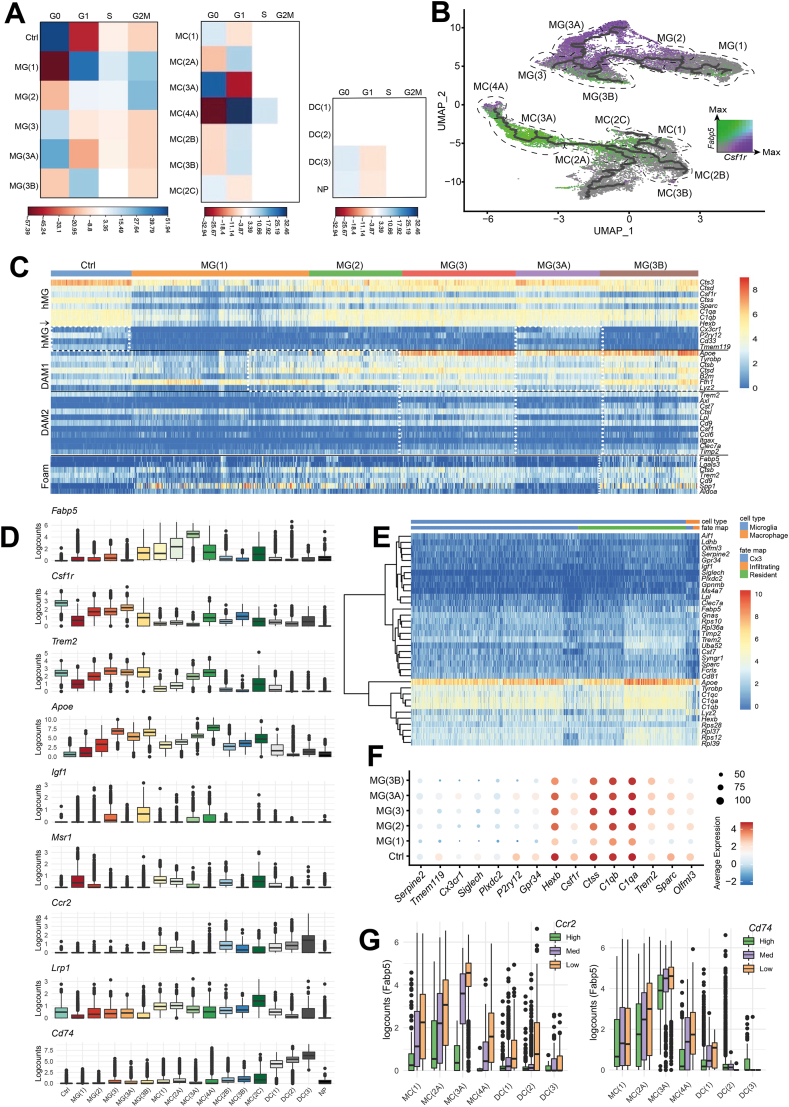
Fig. 4.
Clusters represent the cell states of myeloid cell subpopulations post-SCI. A, Correlation plot of the Pearson's chi-squared residuals between cell cycle phase (columns) and MG/MC cluster (rows) membership: MG Pearson's Chi-squared test X-squared = 7417.2, df = 18, p-value <2.2e-16 or MC Pearson's Chi-squared test X-squared = 2414, df = 33, p-value <2.2e-16. B, UMAP coloured by the scaled log2-transformed counts of Fabp5 (green) or Csf1r (purple). Dotted lines depict relevant clusters. C, Heatmap of homeostatic, DAM-associated, and foam cell genes in MG across the different clusters. Profiles are separated by solid black line. Each row is a gene, and each column is a cell. Dotted white boxes highlight MG that fit the designated profile. hMG = homeostatic MG, hMG↓ = homeostatic MG genes that are downregulated in the DAM1 signature. D, Boxplots of the log2-transformed counts of selected genes by cluster. E, Heatmap log2-transformed counts for the top 20 differentially expressed genes for MG(A2) plus Serpine2 and Gpnmb. Cells (columns) are labelled by their fate mapping status. F, Seurat dot plot showing canonical MG marker expression by cluster. Scale bar is the natural log of the average log2-transformed counts. G, Boxplot of the Fabp5 expression across the MC(1), MC(A), and DC clusters. Left: Cells are binned by Ccr2 expression levels; High ≥ 10 counts; 1 < Med <10; Low. Right: Cells are binned by Cd74 expression levels; High ≥ 20 counts; 1 < Med <20; Low =< 1.
By 2 dpi, half of MG remained in the MG(1) stage, while the other half adopted a MG(2) phenotype (Fig. 3A–D), which closely resembled DAM1 (Fig. 4C) [21]. MG(2) were more likely to be in G2-M phase (Fig. 4A), downregulated Fabp5, but upregulated Csf1r (Fig. 4B–D). According to GO analysis, MG(2) continued to produce cytokines, perform aerobic glycolysis, migrate, activate astrocytes, promote angiogenesis, recruit blood borne leukocytes, and metabolise lipids (S. Fig. 3). Unlike MG(1), MG(2) showed GO terms for phagocytosis (S. Fig. 3) and upregulated Apoe and Trem2 (Fig. 4C and D), which are both further predictive of a phagocytic phenotype [29].
By 3 dpi, MG remained largely split between the proliferating, proinflammatory clusters, MG(1) and MG(2), with ∼11% clustering in MG(3) (Fig. 3A). MG(3) adopted a more DAM2-like phenotype (Fig. 4C), but still showed GO terms for blood-borne leukocyte recruitment, phagocytosis and lipid processing (S. Fig. 3), further supported by high expression levels of lipid processing genes Trem2, Apoe, and Fabp5 (Fig. 4B and D). Unlike MG(1/2), MG(3) was no longer proliferative (Fig. 4A), and had GO terms for cytotoxicity, reactive oxygen species (ROS)-producing reverse electron transport [5], and the production of IL-6 (S. Fig. 3). IL-6 is a cytokine known to sustain chronic inflammation in SCI by promoting monocyte differentiation from a DC to macrophage fate [30,31], but also promote corralling of pro-inflammatory infiltrating cells by targeting astrocytes [11,32]. MG(3) showed GO terms for activation of astrocyte migration and began to upregulate Igf1 (Fig. 4D), which has been shown to promote astrogliosis and ultimately scar formation in SCI [11]. MG(3) also presented several additional GO terms for both the positive and negative regulation of cytokine production (S. Fig. 3), which might be explained by the heterogeneity within this cluster, as depicted by the branching trajectory that passes through this cluster and the differential expression of Fabp5 and Csf1r within the cluster (Fig. 4B).
At 10 and 21 dpi, MG were split between MG(3), MG(3A), and MG(3B) (Fig. 3A). MG(3A) closely resembled Ctrls regarding its homeostatic gene expression (Fig. 4C), cell cycle phase (Fig. 4A), and GO terms, which included antigen presentation, leukocyte homeostasis, phagocytosis, synapse pruning, cell adhesion, and motility (S. Fig. 3). The major difference between Ctrls and MG(3A), was instead the upregulation of Apoe (Fig. 4D) and other DAM1-genes such as Tyrobp and Lyz2 (Fig. 4C) in MG(3A). Compared to MG(3) and MG(3B), MG(3A) lacked a DAM2-like expression pattern (Fig. 4C). MG(3B) comprised of ∼8% 3 dpi cells but was predominantly populated by cells from 10 to 21 dpi. Of all these MG(3B), approximately 5% were monocyte-derived macrophages, which we distinguished by their fate mapping label and differential expression of MG markers (particularly Siglech, Serpine2, and Sparc, and the upregulation of Ms4a7) (Fig. 4E), as described [10,33]. MG(3B) adopted a DAM2-like phenotype (Fig. 4C) and downregulated homeostatic MG genes (Fig. 4D–F). MG(3B) also had the highest expression of scar-formation promoting, Igf1 (Fig. 4D) [11]. In terms of GO analysis, cluster MG(3B), like MG(3) and MG(3A), showed signs of migration, but lacked antigen presentation, metabolism-related, and phagocytosis (S. Fig. 3). GO terms for processing phagocytic cargo, such as lysosomal acidification and triglyceride metabolism, were also enriched [34,35] (S. Fig. 3), and genes for lipid processing (including Fabp5 and Apoe) were upregulated (Fig. 4C and D). Altogether this data led us to (i) consider that the lipid-processing GO terms of MG(3B) reflected a foam-cell like phenotype, and (ii) investigate the expression of peripheral macrophage foam cell markers [22,36].
Monocyte-derived macrophage Msr1 expression was recently demonstrated to promote foamy macrophage formation and neuronal apoptosis through myelin-mediated NF-κB signalling in rodents with experimental contusion SCI [37]. Indeed, MG(3B) cells upregulated many foam-cell markers, including the Msr1 (Fig. 4C and D). Also, we found that Msr1 was upregulated in MG(1) and (2) at 1 dpi, suggesting that the foam cell-like state of MG(3B) is initiated very early on in the MG trajectory (Fig. 4D). To the best of our knowledge, while foamy macrophages have been well described in hematopoietic macrophages, this is the first report of foam cell-like MG in the context of SCI.
Infiltrating cell subsets were instead dominated by cells from the acute phases (1–3 dpi) (Fig. 3A). At 1 dpi, two infiltrating myeloid cell types were present: NPs and MC, in line with previous findings [2,12,38]. NPs were present only in the acute phase of SCI (Fig. 1D and E), showed little evidence of cell cycle (Fig. 4A), and presented the expected GO terms of phagocytosis, NP aggregation, and intrinsic apoptosis (S. Fig. 3) [39].
MC clusters were more populated and dynamic than NPs. MC(1) comprised mainly infiltrating cells from 1 dpi. Based on IF-based quantification (S. Fig. 2) and the known expression [40] of Ccr2 (Fig. 4D), MC(1) had yet to fully infiltrate the spinal cord parenchyma and differentiate into monocyte-derived macrophages or DCs [2,12,38]. This cluster was characterised by GO terms for the response to low oxygen levels and angiogenesis, and the initiation of fibrosis (S. Fig. 3), both of which have been reported in monocytes infiltrating the spinal cord parenchyma after 3 dpi [41,42]. MC(1) was also enriched with GO terms for the production and response to cytokines and ROS, oxidative stress-induced neuronal death, respiratory bursts, phagocytosis, cellular detoxification, blebbing, and lipid processing (S. Fig. 3). MC(1) also expressed Fabp5, which supports a lipid processing function (Fig. 4B and D) [22].
From MC(1), MC clusters projected down three main trajectories, arbitrarily labelled from A-to-C (Fig. 3D). The shortest trajectory, MC(2C) comprised cells from 1 to 3 dpi (Fig. 3A–C), upregulated Fabp5 (Fig. 4B and D), and expressed GO terms for IL-4 signalling (S. Fig. 3), which has been shown to be neuroprotective in the context of SCI [43]. This cluster was uniquely defined by an increase in the expression of Lrp1 (Fig. 4D), an endocytic receptor involved in the phagocytosis of apoptotic cells [44]. Thus, MC(2C) might represent a small subset of infiltrating myeloid cells that are activated by IL-4 signalling and clear apoptotic cells after SCI, before undergoing apoptosis themselves.
MC(2B) and its derivate cluster, MC(3B), were Ccr2-expressing (Fig. 4D), acute phase clusters with minimal (<2.5%) contributions from 10 to 21 dpi (Fig. 3A), and no strong evidence of cell cycle (Fig. 4A). These clusters were all characterised by GO terms for blebbing (S. Fig. 3) and were fated towards cytokine production and apoptosis [45]. However, these clusters also contained GO terms for the negative regulation of MyD88 signalling (a pro-inflammatory, NF-κB-activating, MC-recruiting signalling pathway that has been demonstrated to have degenerative effects in rat models of compression SCI) [[46], [47], [48]], and showed a positive regulation of neuroprotective IL-4 signalling (S. Fig. 3). This data identifies MC(2B) as a potentially neuroprotective cluster of infiltrating monocytes/monocyte-derived macrophages. Of note, MC(2B) expressed very low levels of Fabp5, compared to other infiltrating clusters (Fig. 4B and D), which supports the hypothesis that Fabp5 expression, like fatty acid binding protein 4 [49], may have detrimental effects on the SCI pathology.
Finally, MC(2A) was contributed to by infiltrating cells from 1 to 2 dpi but dominated by cells from 3 dpi (Fig. 3A). This cluster was characterised by GO terms for the negative regulation of intrinsic apoptosis, response to hypoxia, and NF-κB transcription factor activity (S. Fig. 3). Notably, NF-κB is upstream of Fabp5, which was drastically upregulated in MC(3A) (Fig. 4D) [26], the next stage in the Monocle3 predicted trajectory (Fig. 3B and C). In fact, the fate of MC(2A) correlated with its Fabp5 expression; Fabp5lo cells differentiated into pro-regenerative DCs [50], whereas Fabp5hi cells became pro-inflammatory monocyte-derived macrophage clusters, MC(3A) and MC(4A) (Fig. 3D). Indeed, when we binned cells based on Cd74 expression (which is highly upregulated in DCs), or Ccr2 (which is downregulated upon macrophage differentiation) [40], we found that both high Cd74 and high Ccr2 expression correlated with low Fabp5 expression, even as early as in MC(1) (Fig. 4G).
The Cd74+ monocyte-derived DC clusters were predominant in the acute phase, while a portion remained present at 10 and 21 dpi (Fig. 3A–D). This possibly occurred through local proliferation [51], as a small portion of DC(2) and DC(3) cells were assigned to the G1 phase (Fig. 4A and S. Fig. 3). These DC clusters were also dominated by the expression of MHC-II genes and GO terms for antigen presentation, phagocytosis, and T-cell chemotaxis (S. Fig. 3) [52].
The Ccr2- MC(3A) and MC(4A) comprised 3 dpi and 10–21 dpi cells, respectively (Fig. 3A), in line with previous findings [2,12,38,45]. MC(3A) was highly correlated with the G0 phase (Fig. 4A) and was characterised by a uniform upregulation of Fabp5 (Fig. 4B). GO analysis showed highly enriched terms for lipid metabolism and processing phagocytic cargo, but terms for phagocytosis were absent (S. Fig. 3). These features closely resembled those we observed in the foam cell-like MG(3B). Thus, we investigated whether MC(3A) also acquired a foam cell-like profile and found that it did, including the upregulation of Msr1 in the preceding clusters (Fig. 4D) [37].
Trajectory analysis showed that MC(3A) transitioned from a predominantly lipid processing - Fabp5hi - state at 3 dpi, to a Trem2hi - MG(4A)-like - state at 10–21 dpi (Fig. 3, Fig. 4D). Unlike MC(3A), the 10–21 dpi MC(4A) was highly correlated with the G1 cell cycle phase (Fig. 4A), suggesting that infiltrating macrophages may have undergone local proliferation in the subacute and chronic phases of SCI, as described [45,51]. Surprisingly, GO analysis of MG(4A) returned many of the same terms as MG(3) (S. Fig. 3), which included glia activation, antigen presentation, cytokine production and monocyte chemotaxis, ROS-producing reverse electron transport [5], cell killing, and inflammation-perpetuating IL-6 production [30]. Furthermore, MC(4A) comprised not only infiltrating macrophages, but also a population of CAMs, distinguished by a unique expression pattern and the resident myeloid cell fate mapping label (Fig. 1C–E). Altogether the data anticipate the key role of Fabp5 in guiding the temporal dynamics of different myeloid states in SCI, both in resident and infiltrating myeloid cells.
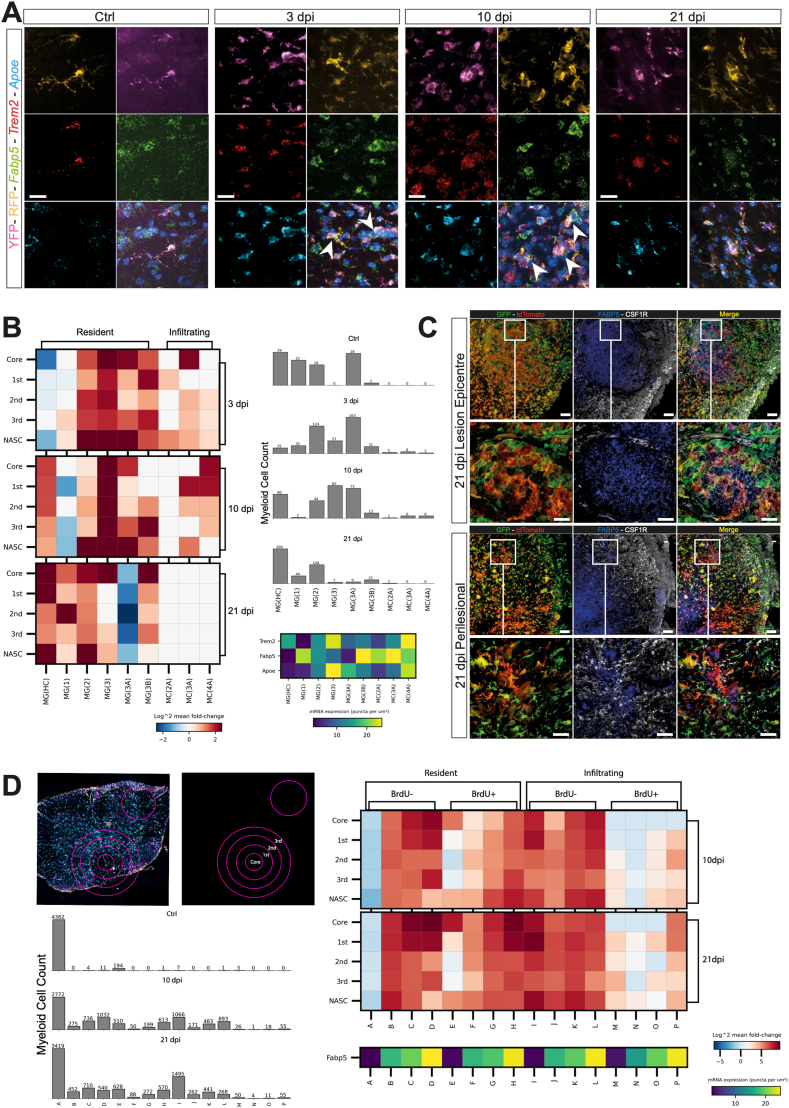
To validate some of these findings, we first used smFISH on Cremato tissue at 3, 10, and 21 dpi (Fig. 5A). We confirmed that Fabp5hi/med/Trem2med/lo/Apoemed/lo MG and monocyte-derived macrophages, likely corresponding to foam cell-like resident MG(3B), dominated the lesion core, while the majority of Fabp5lo/-/Trem2+/Apoe+ MG, possibly MG(3A), remained perilesional (Fig. 5A and B). We next verified this finding at the protein level, via quadruple immunofluorescence, and confirmed that FABP5+ cells were clearly seen in the lesion epicentre at 21 dpi, and that the expression of FABP5 and CSF1R was mutually exclusive (Fig. 5C).
Fig. 5.
smFISH and confocal immunofluorescence microscopy validation and spatial context. A, Expression of Fabp5, Trem2 and Apoe. Nuclei were stained with DAPI. Arrowheads indicate Fabp5+/RFP+/YFP+ cells. Scale bars: 20 μm. B, Heatmap of smFISH data depicting the spatial distribution of the scRNAseq clusters at the lesion epicentre and in the perilesional cord, as in the representative images. Data are presented as mean fold change over Ctrl. Quantification of cells per bin is shown in the bar graphs on the right. Bottom right is the expression level of the three smFISH probes, Apoe, Fabp5, and Trem2 defining each cluster. C, Confocal microscopy images of the expression of FABP5 and CSF1R at the lesion epicentre (top) and in the perilesional cord (bottom) at 21 dpi. Nuclei were stained with DAPI. Scale bars: main panels: 60 μm; insert panels: 20 μm. D, Heatmap of smFISH data from the BrdU, acute proliferation experiment at the lesion epicentre and in the perilesional cord, as in B. Data are presented as mean fold change over Ctrl. Bottom left heatmap of smFISH data depicts the spatial distribution of binned cells and the expression level of Fabp5 defining each bin. Data are binned by BrdU expression (±) and Fabp5 expression (-/lo/med/hi). Quantification of cells per bin is shown in the bar graphs on the right. Data are mean % (±SD) from n = 2 mice per time point.
Finally, to understand the spatio-temporal dynamics of Fabp5 in proliferating myeloid cells, we used BrdU to label S-phase cells during the acute SCI in CreMato mice, and employed smFISH at 10 and 21 dpi to probe Fabp5 expression in BrdU− vs. BrdU + resident and infiltrating myeloid cells. We found that BrdU−/Fabp5- resident MG decreased after the initial injury across all the tissue regions sampled (Fig. 5D). Conversely, BrdU−/Fabp5lo MG increased across all regions. BrdU−/Fabp5med/high MG were enriched across all regions, but particularly in the core and first concentric ring (Fig. 5D). BrdU+ resident MG followed the same trends with the exception of BrdU+/Fabp5- MG, which decreased in the perilesional areas, but increased in the core and normal appearing spinal cord (NASC) (Fig. 5D). These findings suggest that acutely proliferating Fabp5- cells localise to the core and NASC. Most importantly, we found that Fabp5med/hi foam cell-like MG are most prominent in the lesion core, but are also present throughout the lesion epicentre, including in the NASC. When we focused on infiltrating myeloid cells, we found that by 10 and 21 dpi these cells distributed across the tissue irrespective of their Fabp5 expression. As expected, very few BrdU+ infiltrating myeloid cells were observed at these time points, and they were predominantly absent from the lesion core.
3. Discussion
Our data suggests that the myeloid cell response to SCI is highly dynamic with both beneficial and detrimental effects. Thanks to our unique approach, we were able to (i) identify S100a11 as novel specific marker for infiltrating myeloid cells in SCI; (ii) characterise Fabp5 expression as a predictor of the spatio-temporal dynamics of different myeloid states in SCI, and (iii) describe for the first time the acquisition of a foam cell-like MG(3B) transcriptional profile in resident microglia.
This study is not without limitations. The single cell isolation protocol from neural tissue inevitably damages the cells and, in the case of microglia, activates them [12,14]. The cell isolation protocol can result in a sampling bias even within the same cell type where certain cell states – based on morphology, surrounding tissue composition, or other unknown factors – have different probabilities of surviving the isolation protocol and giving rise to high quality cell libraries [53,54]. In this mouse myeloid cell atlas, this sampling bias might account for the diminished 21 dpi samples, or the inconsistent proportions of foam cell-like MG(3B) cells observed in the scRNAseq vs smFISH data. In lieu of equivalently high-throughput protein measurements we have characterised cells on an mRNA-level to elucidate protein-level functions, which do not correlate perfectly [55]. Another assumption was that mRNA within a cell reflects the cell's own transcriptional activity. Pertinently for the characterisation of phagocytes, this is not necessarily true. A comparison of the transcriptome vs RiboTag-acquired translatome of both homeostatic and activated microglia found significant phagocytic cargo contamination in the transcriptome [53].
Cumulatively, our data suggests that the myeloid cell response to SCI is highly dynamic with both beneficial and detrimental effects. Thanks to our unique approach, we were able to (i) identify S100a11 as novel specific marker for infiltrating myeloid cells in SCI; (ii) characterise Fabp5 expression as a predictor of the spatio-temporal dynamics of different myeloid states in SCI, and (iii) describe for the first time the acquisition of a foam cell-like MG(3B) transcriptional profile in resident microglia.
Whether or not this profile is transient, or can be targeted with specific therapies, requires further investigation. Nonetheless, we are confident that our novel, publicly available (https://marionilab.cruk.cam.ac.uk/SCI_Myeloid_Cell_Atlas/) scRNAseq dataset will serve as a valuable resource for future investigations into the dynamic changes and role of the innate immune response after SCI.
Author contributions
Regan Hamel: Conceived and designed the experiments; Performed the experiments; Analysed and interpreted the data; Wrote the paper. Luca Peruzzotti-Jametti: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper. Katherine Ridley: Performed the experiments; Analysed and interpreted the data. Veronica Testa: Performed the experiments; Analysed and interpreted the data. Bryan Yu: Performed the experiments. David Rowitch: Contributed reagents, materials, analysis tools or data; Analysed and interpreted the data. John Marioni: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Analysed and interpreted the data; Wrote the paper. Stefano Pluchino: Conceived and designed the experiments; Analysed and interpreted the data; Wrote the paper.
Data availability
Raw and processed data files for mouse scRNA-seq data are available through the Gene Expression Omnibus (GEO) under the accession number GSE159638. Processed data can be explored and downloaded via a shiny app at https://marionilab.cruk.cam.ac.uk/SCI_Myeloid_Cell_Atlas/. The code used to process this data and generate the figures can be found on GitLab at https://github.com/regan-hamel/SCI_2020.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge Aviva Tolkovsky, Alice Braga, Jayden A. Smith, Yutong Chen, Tommaso Leonardi, Christian Frezza, Michele De Palma, and Giovanni Pluchino for their technical contributions and critical insights throughout the execution of the study. The authors acknowledge the Cambridge NIHR BRC Cell Phenotyping Hub for their cell sorting support and the CRUK Genomics Core Facility for processing the scRNAseq samples and for their technical support. This work was supported by Wings for Life (RG 82921 to S.P.), the Bascule Charitable Trust (RG 75149 and RG98181 to S.P.), the Rosetrees Trust (A1850 to R.H. and S.P.), and by core funding from EMBL (to J.M.) and Cancer Research UK (C9545/A29580) (to J.M), and the Wellcome Trust (D.H.R.). R.H was supported by the Cambridge Trust (10468562) and is the recipient of a Canadian Scholarship Trust Foundation, an MNI-Cambridge Douglas Avrith Graduate Studentship, and a Rosetrees Trust Studentship (A1850). L.P.-J. was supported by a senior research fellowship FISM - Fondazione Italiana Sclerosi Multipla - cod. 2017/B/5 and financed or co-financed with the ‘5‰’ public funding, a Wellcome Trust CRCD Fellowship (RG G105713), and the Addenbrooke's Charitable Trust (RG 97519).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e18339.
Contributor Information
Regan Hamel, Email: regan.hamel@mail.mcgill.ca.
John C. Marioni, Email: marioni@ebi.ac.uk.
Stefano Pluchino, Email: spp24@cam.ac.uk.
Appendix.
Methods, methods
Contusion models of spinal cord injury
Male and female mice aged 8–10 weeks were bred and housed in pathogen-free conditions and provided chow and water ad libitum. The following strains were used in this experiment: Cx3Cr1CreERT2 (purchased in 2018 from Jax, strain name: B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ) to isolate Cx3cr1-YFP+ myeloid-lineage cells; Cx3cr1CreERT2 crossed with TdTomato flox (Cremato) (purchased in 2018 from Jax, strain name: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) to distinguish between YFP+ infiltrating myeloid cells and double-positive RFP/YFP CNS resident microglia (MG), as recently describe3 dpi; C57BL/6 (purchased as needed from Charles River), hereon referred to as WT, for tissue pathology. We administered Cremato mice with daily intraperitoneal injections of 125 mg/kg of body weight of Tamoxifen (Sigma Life Science) dissolved in corn oil for 5 consecutive days to activate the Cre-recombinase, labelling all CX3CR1+ cells as double-positive for RFP and YFP. We induced SCI after a 28-day wash-out period, during which peripheral monocytes were replaced by single positive YFP bone marrow-derived monocytes.
After deeply anaesthetizing the animals with isoflurane (4% induction, 2% maintenance) in oxygen (1.5 l/min) we provided buprenorphine (Temgesic, RB Pharmaceuticals) (pre- and post-operatively), applied ocular ointment to prevent the eyes from drying, shaved the hair on the back of the mice, and swabbed it with a germicide prior to surgery. We placed the animals in prone position and under a surgical microscope, created a dorsal midline incision over the thoracic vertebrae with a sterile scalpel and separated the paravertebral muscles from using a spring scissor (Fine Science Tools). We identified T12 as the apex of the dorsal aspect and performed the laminectomy at T12 using Dumont #2 laminectomy forceps (Fine Science Tools) and spring forceps (Fine Science Tools) leaving the dura intact. In order to maintain vertebral column stability, we did not remove the lateral part of the vertebra at the site of laminectomy. The extension of the laminectomy was consistent between animals at approximately 3 mm in width and 5 mm in length, sufficient to allow room for the 1.3 mm diameter impactor tip.
For the control group (Ctrl), we performed a laminectomy at T12 and omitted the contusion injury step. For the spinal cord injury (SCI) group, we induced a bilateral contusion injury on the exposed spinal cord at T12 using the Infinite Horizon (IH) impactor device (Precision Systems and Instrumentation, Lexington, KY) as previously described [56]. First, we aligned the mouse-impacting tip over the exposed cord and centred it over the central vein, avoiding any overlap with the transverse processes. Then a moderate contusion injury (70 kdyne force) was induced using the IH impactor. After the injury, we closed the incision with 7-mm AutoClips (Fine Science Tool), which were removed after 7 dpi. We expelled the bladders of mice subjected to SCI by applying manual abdominal pressure twice a day until they regained urinary function or reached their endpoint.
We performed all experimental animal procedures in accordance with the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). Animal work was covered by the PPL 7008840 (to Stefano Pluchino).
Bromodeoxyuridine (BrdU) injections
To label acutely proliferating cells, we injected CreMato mice intraperitoneally every 12 h with BrdU (100 mg/kg of body weight in 0.9% saline; ab142567) for 5 consecutive days, starting on the day of SCI. Tissue was collected at 10 and 21 dpi.
Motor behavioural assessment
We assessed hindlimb motor performance using the open-field Basso Mouse Scale (BMS) [57], with scores ranging from 0 (complete hindlimb paralysis) to 9 (healthy). We scored the animals for 4 min in an open field prior to the SCI induction and 1, 3, 5, 7, 10, 14, 18, and 21-days post to ensure that the mice displayed the level of hind-limb locomotor impairment expected from a moderate contusion SCI. Only animals with left and right hindlimb BMS scores within 2 points of each other and a BMS score of 0 at day 1 post-SCI were used in this study. For statistical analysis of the BMS scores, we took the average scores of the left and right hind limbs resulting in a single BMS score for each animal.
Spinal cord extraction for tissue pathology
Mice were deeply anaesthetized with intraperitoneal injections of 100 μl of pentobarbital sodium and transcardially perfused with ice-cold artificial cerebral spinal fluid (aCSF) for 7 min followed by ice-cold 4% paraformaldehyde (PFA) in PBS with a pH 7.4. We post-fixed the spinal cords in 4% PFA for 12 h at 4 °C and then washed with 1X PBS. After dissecting out the spinal cords, we cryoprotected them for at least 24 h in 30% sucrose (Sigma) in 1X PBS at 4 °C before embedding them in Optimal Cutting Temperature (OCT) Compound (VWR Chemicals) and snap freezing them with isopentane on dry ice. We stored frozen cord blocks at −80 °C until a total of 15 mm of each spinal cord segment, centred on the lesion, was sectioned coronally at 20 μm thickness (cryostat CM1850; Leica) and collected onto SuperfrostPlus slides (ThermoScientific). We stored sections at −80 °C until needed.
Immunofluorescence
In preparation for immunohistochemistry, we baked spinal cord sections mounted on slides at 37 °C for 30 min then washed with MilliQ water and PBS, for a total of 15 min. For BrdU, tissue sections were immersed in HCl (2.0 N) at 37 °C for 30 min then 0.1 M sodium borate (pH 8.5) at room temperature for 10 min. Then, we incubated the sections in blocking solution (1X PBS with 0.1% Triton X-100 (Sigma) and 10% normal goat serum (NGS; Sigma)) for 1 h at room temperature (RT). We used the following primary antibodies: chicken anti-GFP (Abcam, ab13970, 1:500), rabbit anti-FABP5 (invitrogen, PA5-92929, 1:100), mouse anti-CD11c (Abcam, ab11029, 1:50), rat anti-CD11b (Abcam, A86819, 1:100), rat anti-Ly6g (Abcam, ab25377, 1:100), rabbit anti-P2Ry12 (Alamone, APR-012, 1:100), rat anti-BrdU (1:750, Abcam, ab6326). We then incubated the sections overnight with the primary antibody in a solution of PBS 1X + 0.1% Triton X100 (Sigma) + 1% NGS at 4 °C. Then, after 3 washes with PBS 1X + 0.1% Triton X100 for a total of 15 min, we incubated the slides for 1 h in a humid chamber with the species-appropriate fluorochrome-conjugated secondary antibodies in a solution of PBS 1X + 0.1% Triton X100 + 1% NGS. We used the following secondary antibodies: goat anti-chicken AF488 (Thermo-Fisher, A11039, 1:1000), goat anti-rabbit AF647 (Thermo-Fisher, A21244, 1:1000), goat anti-mouse AF647 (Thermo-Fisher, A21238, 1:1000), goat anti-rat AF647 (Abcam, ab150159, 1:1000), goat anti-rabbit AF405 (Abcam, ab175671, 1:1000). We blocked the staining by washing the slides first 3 times with PBS 1X + 0.1% Triton X100 and then with PBS for a total of 20 min. We counterstained the sections with DAPI (Roche) diluted 1:10000 in PBS for 10 min at RT. Finally, we washed the sections with PBS and mounted them on glass coverslips with fluorescence mounting medium (Dako). We acquired images of the stained spinal cord sections using a vertical epifluorescence microscope (Leica DM6000) or a confocal microscope (Leica TCS SP5).
Quantification
We acquired images of the stained spinal cord sections using a vertical epifluorescence microscope (Leica DM6000) at a magnification of 20X and spaced approximately 400 μm apart. We converted single 20X images to mosaics using ImageJ 1.52p and quantified cells using 2 ROIs of 700 μm2 per tissue section. We then quantified RFP and YFP positive cells by manual counting, while DAPI positive cells were quantified using CellProfiler. Quantification results were presented as a percentage of positive cells out of all DAPI positive cells. We used a total of n = 2 slices for each distance from the lesion epicentre.
smFISH multiplex automated staining
Spinal cord sections were baked at 65°c for 1 h and fixed in 4% paraformaldehyde (PFA) solution at 4°c for 15mins. Slides were then washed and dehydrated in PBS (1x) and ethanol gradients from 50% to 100% for a total of 30 min. Slides were air dried before automated single molecule fluorescent in situ hybridization (smFISH) protocol.
Multiplex smFISH was performed on a Leica BondRX fully automated stainer, using RNAScope© Multiplex Fluorescent V2 technology (Advanced Cell Diagnostics, 322000). Slides underwent heat-induced epitope retrieval with Epitope Retrieval Solution 2 (pH 9.0, Leica AR9640) at 95 °C for 5 min. Slides were then incubated in RNAScope© Protease III reagent (ACD 322340) at 42 °C for 15 min, before being treated with RNAScope© Hydrogen Peroxide (ACD 322330) for 10 min at RT to inactivate endogenous peroxidases.
All double-Z mRNA probes were designed against mouse genes by ACD for RNAScope© on Leica Automated Systems. Slides were incubated in RNAScope 2.5 LS probes for 2 h at RT DNA amplification trees were built through consecutive incubations in AMP1 (preamplifier, ACD 323101), AMP2 (background reduction, ACD 323102) and AMP3 (amplifier, ACD 323103) reagents for 15 to 30 min each at 42 °C. Slides were washed in LS Rinse buffer (ACD 320058) between incubations.
After amplification, probe channels were detected sequentially via HRP–TSA labelling. To develop the C1–C3 probe signals, samples were incubated in channel-specific horseradish peroxidase (HRP) reagents for 30 min, TSA fluorophores for 30 min and HRP-blocking reagent for 15 min at 42 °C. The probes in C1, C2 and C3 channels were labelled using Opal 520 (Akoya FP1487001KT), and Opal 650 (Akoya FP1496001KT) fluorophores (diluted 1:500). The C4 probe complexes were first incubated with TSA–Biotin (Akoya NEL700A001KT, 1:250) for 30 min at RT, followed by streptavidin-conjugated Atto425 (Sigma 56759, 1:400) for 30 min at RT. Directly following the smFISH assay, tissue was incubated with anti-RFP (ThermoFisher, MA515257, 1:200) and anti-GFP (Abcam, ab13970, 1:400) antibodies in blocking solution for 1 h. Samples treated with BrdU during acute injury were also incubated with anti-BrdU (Abcam, ab6326, 1:750). To develop the GFP antibody signal, samples were incubated in donkey anti-goat HRP (ThermoFisher - A15999, 1:200) for 1 h, TSA biotin (PerkinElmer - NEL700A001KT, 1:200) for 10 min and streptavidin-conjugated Alexa 700-streptavidin (Sigma - S21383, 1:200) for 30 min. To develop the RFP antibody signal, samples were then incubated in donkey anti-mouse Alexa 594 (ThermoFisher, A21203, 1:500). BrdU was developed using goat anti-rat Alexa 488 (Abcam, A-11006, 1:500). Samples were then incubated in DAPI (Sigma, 0.25 μg/ml) for 20 min at RT, to mark cell nuclei. Slides were briefly air-dried and manually mounted using ∼90 μl of Prolong Diamond Antifade (Fisher Scientific) and standard coverslips (24 × 50 mm2; Fisher Scientific). Slides were dried at RT for 24 h before storage at 4 °C for >24 h before imaging.
smFISH confocal imaging
smFISH stained mouse spinal cord slides were imaged on an Operetta CLS high-content screening microscope (PerkinElmer). Image acquisition was controlled using PerkinElmer's Harmony software. High resolution smFISH images were acquired in confocal mode using an sCMOS camera and x40 NA 1.1 automated water-dispensing objective. Each field and channel were imaged with a z-stack of 20 planes with a 1 μm step size between planes. All appropriate fields of the tissue section were manually selected and imaged with an 8% overlap.
smFISH quantification of cell types and gene expression in situ
To segment single cells and quantify RNA spots from high-resolution images, analysis scripts were created on Harmony software (PerkinElmer) using customizable, predefined, function blocks. Maximum intensity z-projection images were generated from confocal z-stacks per field. Single nuclei were selected from DAPI signal, and doublet and partial nuclei discarded via a DAPI morphology, intensity and texture driven linear classification model. The cytoplasm of single macrophages was identified by intensity thresholding of RFP/YFP immunofluorescence signals in the surrounding perinuclear region. Cells were segmented and classified based on RFP/YFP/BrdU expression characteristics. mRNA puncta were then automatically identified by the detection of local intensity maxima across each smFISH channel within the 2D area of the segmented cell populations. All cells with an mRNA puncta count <5 were considered positive for that gene.
smFISH data processing and annotation
The analysis was performed on a desktop workstation with two 6-core Intel i7–4930K 3.4 GHz CPUs and 64 GB of RAM. The data were exported from Harmony as a single-cell matrix showing cell coordinates, morphologic and intensity measurements, and RNA spot counts per cell. Data visualisation and spinal cord region segmentation was performed in OMERO (Open Microscopy Environment). To annotate tissue regions for each section, the lesion core was identified using DAPI landmarks in the dorsal spinal cord and annotated with a circle of 30,000 μm2 area. The lesion border regions were annotated with three concentric rings leading out from the lesion core, each with 100 μm radius. Furthermore, normal appearing spinal cord (NASC) was defined as a 150,000 μm2 area of the ventral horn per section. Annotation area and shape were identical per section to control for lesion morphological diversity across the samples (see Fig. 4E, region annotation).
Celltype clustering and plotting
We then aimed to identify distinct macrophage subtype clusters, pulled from scRNASeq analysis (see ‘Quality Control, Batch Correction & Clustering’), in the smFISH data based on expression of Fabp5, Trem2 and Apoe (see Fig. 4B). For downstream analysis, cells were identified as ‘Resident’ or ‘Infiltrating’, ‘Proliferating ‘or ‘Non-Proliferating’ based on the YFP/RFP intensity characteristics in the Cremato mouse, and BrdU intensity characteristics in BrdU-treated tissue (see Fig. 4D). Cells were filtered with a minimum cumulative spot count threshold of 20. Outlier cells were removed by filtering positive cells based on spot counts between the 1st and 99th percentile. Cells were then clustered into 5 spot count bins per gene – negative (<5 spots & <1st percentile), low (1st-33rd), med (33rd-66th), high (66th-99th percentile), and positive (low, med, high).
From the scRNASeq analysis, nine distinct clusters were identified based on Resident or Infiltrating status and Fabp5, Trem2 and Apoe bin classification. E.g. Resting microglia (MG(HC)) were identified as Resident/Fabp5 negative/Apoe negative/Trem2 positive. The clusters were quantified, and log transformed (log2(spot counts +1)) using Python NumPy functions. The cluster quantifications were then plotted spatially, and across timepoints (post-SCI) as a function of mean fold change (log2FC) from the control, using Seaborn and ScanPy matrixplot functions (see Fig. 4B and D).
Spinal cord extraction and cell isolation for scRNAseq
Mice were deeply anaesthetized with intraperitoneal injections of 100 μl of pentobarbital sodium and transcardially perfused with ice-cold artificial cerebral spinal fluid (aCSF) for 7 min. After perfusion, we voided the spinal columns using a 5 ml syringe filled with ice-cold aCSF. We placed the spinal cords on a Petri dish above millimetre graph paper and cut a 5 mm section centred on the lesion using a sterile blade. For the first 2 batches, mice were not pooled before sequencing. After establishing that sex- and condition-matched littermates showed no transcriptional differences, up to 3 mice were pooled in order to maximize cell yields for sequencing. We mechanically dissociated the tissue in a glass homogenizer with 6 ml of aCSF-based Homogenization buffer [aCSF plus 10 mM HEPES (Sigma) to maintain stable pH despite CO2 released from the cells, 1% BSA (Sigma) to reduce cell clumping, 1 mM EDTA (Thermo Scientific) to chelate Ca++ and Mg++, reducing cell adhesion, 10 mg/ml of DNAse (Roche) 3000U to degrade any free-floating DNA and reduce clumping of cells, and 40 units/μl of RNAse inhibitor (Invitrogen) to minimize the degradation of the cells mRNA transcripts]. We chose mechanical dissociation over enzymatic methods because it is more rapid, can be performed at ice-cold temperatures, and is less likely to cause conformational changes of surface receptors, which could alter cell function.
After tissue dissociation, we filtered the suspension with a pre-wet 40 μm strainer and rinsed the homogenizer with 2 ml of Homogenization buffer to increase cell yield. To remove myelin and debris from the cell suspension, we added 2.7 ml of isotonic 9:1 Percoll (Sigma) to 10X PBS to each sample. We gently mixed the samples and centrifuged at 800 g for 20 min at 4 °C with a brake speed of 0. Myelin debris visibly layered at the surface and was carefully removed with a pipette. To remove the remaining Percoll, we added an ice-cold buffer of 5% autoMACS Rinsing Solution (Miltenyi Biotec) in 1X MACS BSA solution (Miltenyi Biotec) and centrifuged at 800 g for 5 min at 4 °C. Then we resuspended the pelleted cells in 200 μl Fluorescent Activated Cell Sorter (FACS) buffer [see Homogenization Buffer ingredients but dissolved in Cell Staining Buffer (Biolegend) not aCSF, and with 7-AAD live/dead stain (Invitrogen) at a concentration of 1:50].
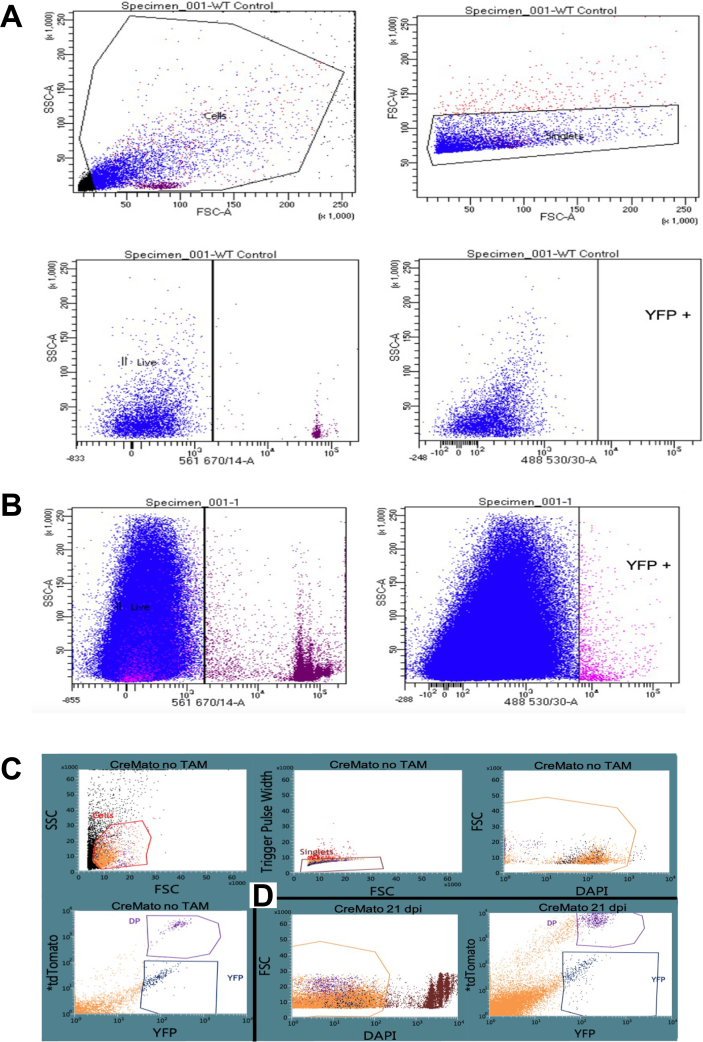
The ex vivo FACS samples were sorted using a BD FACS Aria III cell sorter set to 3-way purity with a 100 μm nozzle at 20 psi. As our panel consisted of RFP, YFP and 7-AAD, fluorochrome compensation was not required. All gates were set based on the following: unstained WT sample, 7-AAD stained WT sample treated with DMSO to kill the cells, and for the Cremato samples, non-tamoxifen treated Cremato mice (RFP−, YFP+) to control for bleed-through of the non-tamoxifen activated RFP.
Single-cell isolation and sequencing
FACs-isolated cells were sequenced on a single-cell level using the microdroplet based protocol, 10X Genomics Chromium Single Cell 3’ Solution (v2 chemistry for 2 dpi Cx3cr1 samples, v3 for all others). As per the manufacturer instructions, approximately 18,000 cells per sample were loaded into each well. Single cells were isolated with a gel bead in emulsion, and mRNA transcripts were barcoded by cell and transcript (via unique molecular identifiers) before the generation of barcoded libraries, which were sequenced with the NovaSeq 6000 (Illumina) to an average depth of at least 40,000 reads per cell and >85% sequencing saturation, as calculated by Cell Ranger.
Single cell RNA-seq data pre-processing
Sequenced samples were demultiplexed and aligned to the GRCm38 (mm10) using the Cell Ranger 3.1.0 pipeline (10X Genomics). Notably, the gene Clec7a was not present on the mm10 dataset supplied by Cell Ranger, so we generated a custom Cell Ranger mm10 dataset to include this gene. We performed all scRNAseq data processing (post-Cell Ranger) in R version 3.6.3. All packages used are free of charge and publicly available at the Comprehensive R Archive Network (https://cran.r-project.org), the Bioconductor project (http://bioconductor.org), or their respective GitHub pages. Our complete R analysis workflow, including scripts for generating the figures, is available on GitHub (regan-hamel/SCI_2020).
Quality control, Batch Correction & Clustering
We first corrected each batch of unfiltered gene/cell matrices for the effects of barcode swapping (DropletUtils package) and then filtered out empty droplets (DropletUtils). We removed low-quality cells using fixed thresholds. Cells with more than 5% [58] mitochondrial transcripts. For 10x chemistry version 2 samples, we removed cells with <450 UMIs and <450 unique genes per cell. For 10x chemistry version 3 samples, we removed cells with <1000 UMIs and <600 unique genes per cell. Samples in which the majority of cells did not meet these thresholds were considered failed runs and were not included for downstream analysis.
We predicted and removed likley doublets using the cxds_bcds_hybrid(.) function from the scds package (version 1.2.0) with the parameter estNdbl = TRUE [59]. Briefly, this approach predicts doublets by combining the outputs of two scoring methods: cxds, which evaluates the co-expression of genes across cells, looking for instances of unlikely combinations; and bcds, which generates artificial doubles based on the dataset, then looks for cell libraries that are similar to the artificial doublets.
We annotated cell types by calling the function, cellassign, from the cellassign package (version 0.99.21) using the default parameters and a custom marker gene list (Microglia: Sparc, C1qa, Plxdc2, Serpine2, P2ry12, Tmem119, Siglech, Ctss, Cst3, Slc2a5, Sall1; Macrophages: Ms4a7, Ecm1, Arg1, Ccl7, Mgst1; Monocytes: Chil3, Plac8, Ccr2, Rgs1, Fn1; Dendritic Cells: Cd74, H2-Eb1, H2-Aa, H2-Ab1, Plac8, H2-DMb1, H2-DMa, Klrd1, Flt3, Zbtb46; Neutrophils: S100a8, S100a9, Lcn2, Dedd2, Retnlg, Wfdc21, Mmp9; CAMs: Ms4a7, Cd74, Cd163, Cbr2, Lyve1, Mrc1; T-Cells: Cd2, Cd3d, Cd3e, Cd3g, Ptprc).
Before batch correction, we performed multi-batch normalization (batchelor), modelled the variance (scran) and selected the top 20% highly variable genes to be used in the batch correction (batchelor). We batch corrected based using the mutual nearest neighbours (MNN) approach [60] by calling fastMNN(.) from the batchelor package with following parameters: batch = the strain and 10x chemistry version, subset.row = HVGs, merge.order = the two version 3 datasets, and then the version 2 batch. We used the batch corrected data to visualize the entire dataset using Uniform Manifold Approximation and Projection (UMAP) (scater) with the default parameters. We clustered the dataset by building a shared nearest neighbour graph (scater) and used the walktrap algorithm (igraph) to identify clusters. Note these clusters were used for quality control purpose and were not used downstream. We removed clusters that expressed canonical marker genes of more than one cell type and had a higher than average UMI counts, as they likely represented doublets not detected by the scds package. Three clusters with <55 cells and a cluster originating from a single sample were not considered for further analysis. After removing low-quality clusters of cells, we re-performed the batch correction and clustering and used this post-quality control dataset for all downstream analysis.
Trajectory analysis
We performed trajectory analysis using the beta version of Monocle3. To prepare for the trajectory analysis, we processed the post-quality control dataset following the Monocle3 workflow. First, we converted the non-log transformed counts matrix to a cell dataset object (cds). Then we called preprocess_cds with the default parameters to normalize the dataset and perform principal component analysis. We re-performed the mutual nearest neighbour batch correction (batchelor) on this cds by calling align_cds and correcting, as before, only between samples collected on different dates. Next we computed the UMAP with the following parameters: minimum distance = 0.1; metric = cosine; number of neighbours = 15. We performed unsupervised clustering via Leiden community detection by calling cluster_cells on the batch corrected cells with the parameters reduction_method = UMAP and the number of nearest neighbours (k) = 16. Finally, we built the trajectory by calling the function learn_graph with the parameters use_partitions = TRUE and close_loop = FALSE. Note that for the downstream analysis we used only the cluster membership, trajectory, and UMAP, but not the normalized gene expression matrix from this Monocle3-processed dataset.
Differential gene expression & Gene Ontology enrichment analysis
We generated differentially expressed genes (DEGs) for each cluster by calling the scran function, findMarkerson the multi-batch normalized log2-transformed counts (batchelor; as described in the Quality Control, Batch Correction & Clustering section), with the parameters group = cluster membership, direction = upregulated, pval.type = some. The findMarkers function identifies DEGs by pairwise Welch t-Tests comparisons corrected for multiple testing via the Benjamini-Hochberg method and combines the comparisons into a ranked list of markers for each cluster. We used these DEGs for Gene Ontology (GO) enrichment analysis. To this end, we input the significant DEGs (FDR< 0.0001, FC > 1.25, ranging from 455 to 848 genes) for each cluster into the PANTHER Classification System overrepresentation test (release 2021-07-02) following the instructions on the Gene Enrichment analysis page (http://geneontology.org/docs/go-enrichment-analysis/), including the optional use of a custom reference list. P-values computed using the default setting of Fisher's Exact Test were corrected for multiple testing using the Benjamini-Hochberg method. We examined only biological processes that were over-represented.
Cell type annotation
We assigned a cell type identity to each cluster using its top DEGs, GO terms, and the expression of canonical marker genes. We then refined these annotations, for example in the case of MG(3B), which contained both MG and monocyte-derived macrophages, using the CNS resident vs. CNS infiltrating labels from our fate-mapping Cremato mouse line.
Dissociation and FACS-associated expression analysis
For S. Fig. 2C, we analysed the expression of dissociation and FACS-associated gene expression across several studies. The genes considered were those identified by van den Birk et al., 2017 [18] that intersected with the genes of all 5 datasets: Mt1, Ier2, Dnaja1, Socs3, Atf3, Jund, Ppp1r15a, Hspe1, Dusp1, Nfkbia, Hsph1, Jun, Junb, Egr1, Ubc, Zfp36, Hsp90aa1, Mt2, Dnajb1, Btg2, Nr4a1. We downloaded gene-matrix text files from Genome Expression Omnibus, read them into R and converted them to Single Cell Experiment objects (SingleCellExperiment). We removed cells which did not meet the following quality control (including in our data): greater than 600 UMI counts, greater than 600 unique genes per cell, and less than 10% counts from mitochondrial genes. Notably, samples differed in their platforms and disease states, but all cells analysed were MG. The following samples were used: Yang 2018 [61] (Drop-seq, whole-brain enzymatic dissociation, C1q+ Sham control MG), Linnarsson 2018 [15] (10X Chromium, whole-brain mechanical homogenization, homeostatic MG), Stevens 2019 [14] (10X Chromium, mechanical homogenization, FACS of homeostatic MG – note this dataset consisted only of highly variable genes and all mitochondrial genes had been removed), Movahedi 2019 [17] (10X Chromium, whole-brain enzymatic dissociation, homeostatic MG), and Ctrl RFP+/YFP+ MG from the current study.
Statistics
We analysed the statistical significance of the BMS scores between males and females using an unpaired two-tailed t-Test. We performed statistical analyses on the scRNAseq data in R. We analysed the significance of the downregulation of MG-specific genes (Fig. 2B) using a Welch t-Test corrected for multiple testing via the Benjamini-Hochberg method based on the total number of genes in the dataset (scran). We considered differences to be significant with the P values were <0.05. To test for independence between the categorical variables, cluster membership and time point, we used Pearson's chi-squared test for independence. First, we generated a contingency table of cluster vs. time point, then tested for independence using Pearson's chi-squared test by calling chisq.test (stats) on the table. To understand how each comparison contributed to the significant result, we investigated the standardized residuals. To determine if a standardized residual was significant, we squared them to obtain chi-squared values then called pchisq (stats) with df = 1 and lower.tail = FALSE to obtain a P-value, which we compared with the Bonferroni corrected P values as described [62].
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Per cell and per sample metrics of the entire dataset after quality control. Cluster-Based Differentially Expressed Genes and Gene Ontology Enrichment Analysis Results are shown.
S. Fig. 1.
Representative FACS gating strategy. The gates were set on A, 7-AAD stained WT sample for B, 21 dpi Cx3cr1 sample; C, unstained CreMato sample with no tamoxifen treatment. Even without tamoxifen treatment, some cells are double positive or YFP-/tdTomato+. D, Gates from (C) to a 21 dpi CreMato sample stained for DAPI. SSC: side scatter; FSC: forward scatter.
S. Fig. 2.
Quality controls. A, Quantification of RFP-/YFP+ infiltrating myeloid cells (left) or RFP+/YFP+ CNS resident myeloid cells (right) as a proportion of DAPI+ at set distances rostral (+) or caudal (-) of the lesion epicentre. Data are mean % (± SD) and have been collected from n= 3 mice per time point. B, UMAPs of cells from male vs. female mice. C, Basso Mouse Score (BMS) from male and female mice. Data are mean numbers (± SEM) and have been collected from n ≥ 4 mice per time point. D, Violin plots showing the percentage of all UMI counts originating from dissociation and FACS associated genes, per Ctrl MG cell, across several droplet-based scRNAseq studies after applying the same quality control metrics. Black dots are the mean. F= FACS, E= enzymatic (vs. mechanical) dissociation. E, UMAP of the entire dataset, coloured by CellAssign annotation, before manual assignments. F, Heatmap of the log2-transformed counts of canonical marker genes for CNS-associated cell types. G, smFISH analysis of the expression of cell-type markers Pdgfra (oligodendrocyte progenitor cells), Slc1a3 (astrocytes), and Syt1 (neurons) from Ctrl, 1 dpi, and 21 dpi. H, Confocal microscopy images of the expression of P2ry12, Ly6g, CD11c and CD11b by MG, NPss, DCs and monocytes-macrophages, respectively, at the lesion epicentre. Nuclei were stained with DAPI. Scale bars: 60 μm.
S. Fig. 3.
Select GO terms for the MG and MC clusters. The fold enrichment and FDR of select GO terms for each cluster. Pos = positive; Neg=negative, reg = regulation.
References
- 1.Gensel J.C., Zhang B. Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res. 2015;1619:1–11. doi: 10.1016/j.brainres.2014.12.045. [DOI] [PubMed] [Google Scholar]
- 2.Donnelly D.J., Popovich P.G. Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp. Neurol. 2008;209:378–388. doi: 10.1016/j.expneurol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett F.C., et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron. 2018;98 doi: 10.1016/j.neuron.2018.05.014. 1170–1183.e1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotamisligil G.S., Bernlohr D.A. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015;11:592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peruzzotti-Jametti L., Willis C.M., Hamel R., Krzak G., Pluchino S. Metabolic control of smoldering neuroinflammation. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.705920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu T., et al. Knockdown of FABP5 mRNA decreases cellular cholesterol levels and results in decreased apoB100 secretion and triglyceride accumulation in ARPE-19 cells. Lab. Invest. 2010;90:906–914. doi: 10.1038/labinvest.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levi L., et al. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res. 2013;73:4770–4780. doi: 10.1158/0008-5472.CAN-13-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., et al. Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-β responses in tumor-associated macrophages. Cancer Res. 2014;74:2986–2998. doi: 10.1158/0008-5472.CAN-13-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore S.M., Holt V.V., Malpass L.R., Hines I.N., Wheeler M.D. Fatty acid-binding protein 5 limits the anti-inflammatory response in murine macrophages. Mol. Immunol. 2015;67:265–275. doi: 10.1016/j.molimm.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordão M.J.C., et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363 doi: 10.1126/science.aat7554. eaat7554. [DOI] [PubMed] [Google Scholar]
- 11.Bellver-Landete V., et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019;10:518. doi: 10.1038/s41467-019-08446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milich L.M., et al. Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matson K.J.E., et al. A single cell atlas of spared tissue below a spinal cord injury reveals cellular mechanisms of repair. bioRxiv. 2021;2021 [Google Scholar]
- 14.Hammond T.R., et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271.e256. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisel A., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174 doi: 10.1016/j.cell.2018.06.021. 999-1014.e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q., et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223.e210. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Hove H., et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 2019;22:1021–1035. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 18.van den Brink S.C., et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 19.Haage V., et al. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropath. Commun. 2019;7:20. doi: 10.1186/s40478-019-0665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling D.P., Yong V.W. Dynamics of the inflammatory response after murine spinal cord injury revealed by flow cytometry. J. Neurosci. Res. 2008;86:1944–1958. doi: 10.1002/jnr.21659. [DOI] [PubMed] [Google Scholar]
- 21.Keren-Shaul H., et al. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169 doi: 10.1016/j.cell.2017.05.018. 1276–1290.e1217. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y., et al. Macrophage transcriptional profile identifies lipid catabolic pathways that can Be therapeutically targeted after spinal cord injury. J. Neurosci. 2017;37:2362–2376. doi: 10.1523/JNEUROSCI.2751-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krasemann S., et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–581.e569. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elmore M.R.P., et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senga S., Kobayashi N., Kawaguchi K., Ando A., Fujii H. Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-κB) signaling in cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:1057–1067. doi: 10.1016/j.bbalip.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Kaczocha M., et al. Inhibition of fatty acid binding proteins elevates brain anandamide levels and produces analgesia. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan F.H., et al. Microglia coordinate cellular interactions during spinal cord repair in mice. Nat. Commun. 2022;13:4096. doi: 10.1038/s41467-022-31797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa J.D., et al. Fatty acid binding protein 5 modulates docosahexaenoic acid-induced recovery in rats undergoing spinal cord injury. J. Neurotrauma. 2016;33:1436–1449. doi: 10.1089/neu.2015.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atagi Y., et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2) J. Biol. Chem. 2015;290:26043–26050. doi: 10.1074/jbc.M115.679043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 31.Chomarat P., Banchereau J., Davoust J., Karolina Palucka A. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat. Immunol. 2000;1:510–514. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 32.Penkowa M., et al. Astrocyte-targeted expression of IL-6 protects the CNS against a focal brain injury. Exp. Neurol. 2003;181:130–148. doi: 10.1016/s0014-4886(02)00051-1. [DOI] [PubMed] [Google Scholar]
- 33.Konishi H., et al. Siglec-H is a microglia-specific marker that discriminates microglia from CNS-associated macrophages and CNS-infiltrating monocytes. Glia. 2017;65:1927–1943. doi: 10.1002/glia.23204. [DOI] [PubMed] [Google Scholar]
- 34.Chandak P.G., et al. Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J. Biol. Chem. 2010;285:20192–20201. doi: 10.1074/jbc.M110.107854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marschallinger J., Iram T., Zardeneta M., et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willemsen L., de Winther M.P. Macrophage subsets in atherosclerosis as defined by single-cell technologies. J. Pathol. 2020;250:705–714. doi: 10.1002/path.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong F.-Q., et al. Macrophage MSR1 promotes the formation of foamy macrophage and neuronal apoptosis after spinal cord injury. J. Neuroinflammation. 2020;17:62. doi: 10.1186/s12974-020-01735-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alizadeh A., Dyck S.M., Karimi-Abdolrezaee S. Traumatic spinal cord injury: an overview of pathophysiology, models and acute injury mechanisms. Front. Neurol. 2019;10 doi: 10.3389/fneur.2019.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zivkovic S., Ayazi M., Hammel G., Ren Y. For better or for worse: a look into neutrophils in traumatic spinal cord injury. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.648076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips R.J., Lutz M., Premack B. Differential signaling mechanisms regulate expression of CC chemokine receptor-2 during monocyte maturation. J. Inflamm. 2005;2:14. doi: 10.1186/1476-9255-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kigerl K.A., et al. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J. Neurosci. 2009;29:13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whetstone W.D., Hsu J.-Y.C., Eisenberg M., Werb Z., Noble-Haeusslein L.J. Blood-spinal cord barrier after spinal cord injury: relation to revascularization and wound healing. J. Neurosci. Res. 2003;74:227–239. doi: 10.1002/jnr.10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fenn A.M., Hall J.C.E., Gensel J.C., Popovich P.G., Godbout J.P. IL-4 signaling drives a unique arginase+/IL-1β+ microglia phenotype and recruits macrophages to the inflammatory CNS: consequences of age-related deficits in IL-4Rα after traumatic spinal cord injury. J. Neurosci. 2014;34:8904–8917. doi: 10.1523/JNEUROSCI.1146-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May P., Reddy Y.K., Herz J. Proteolytic processing of low density lipoprotein receptor-related protein mediates regulated release of its intracellular domain. J. Biol. Chem. 2002;277:18736–18743. doi: 10.1074/jbc.M201979200. [DOI] [PubMed] [Google Scholar]
- 45.Greenhalgh A.D., David S. Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J. Neurosci. 2014;34:6316–6322. doi: 10.1523/JNEUROSCI.4912-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao A.-H., et al. Early blockade of TLRs MyD88-dependent pathway may reduce secondary spinal cord injury in the rats. Evid. Bas. Comp. Alter. Med. 2012;2012:591298. doi: 10.1155/2012/591298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boivin A., et al. Toll-like receptor signaling is critical for wallerian degeneration and functional recovery after peripheral nerve injury. J. Neurosci. 2007;27 doi: 10.1523/JNEUROSCI.3027-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu L., Botchway B.O.A., Zhang S., Zhou J., Liu X. Inhibition of NF-κB signaling pathway by resveratrol improves spinal cord injury. Front. Neurosci. 2018;12:690. doi: 10.3389/fnins.2018.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licero Campbell J., et al. Fatty acid-binding protein 4 inhibition promotes locomotor and autonomic recovery in rats following spinal cord injury. J. Neurotrauma. 2022;39:1099–1112. doi: 10.1089/neu.2021.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaguchi M., et al. Transplantation of dendritic cells promotes functional recovery from spinal cord injury in common marmoset. Neurosci. Res. 2009;65:384–392. doi: 10.1016/j.neures.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Swirski F.K., Hilgendorf I., Robbins C.S. From proliferation to proliferation: monocyte lineage comes full circle. Semin. Immunopathol. 2014;36:137–148. doi: 10.1007/s00281-013-0409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guilliams M., et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haimon Z., et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018;19:636–644. doi: 10.1038/s41590-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denisenko E., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21:130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reimegård J., et al. A combined approach for single-cell mRNA and intracellular protein expression analysis. Commun. Biol. 2021;4:624. doi: 10.1038/s42003-021-02142-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cusimano M., et al. Transplanted neural stem/precursor cells instruct phagocytes and reduce secondary tissue damage in the injured spinal cord. Brain. 2012;135:447–460. doi: 10.1093/brain/awr339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basso M.D., et al. Basso mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma. 2006;23:635–659. doi: 10.1089/neu.2006.23.635. [DOI] [PubMed] [Google Scholar]
- 58.Osorio D., Cai J.J. Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control. Bioinformatics. 2021;37:963–967. doi: 10.1093/bioinformatics/btaa751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bais A.S., Kostka D. scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2020;36:1150–1158. doi: 10.1093/bioinformatics/btz698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arneson D., et al. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat. Commun. 2018;9:3894. doi: 10.1038/s41467-018-06222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beasley T.M., Schumacker R.E. Multiple regression approach to analyzing contingency tables: post hoc and planned comparison procedures. J. Exp. Educ. 1995;64:79–93. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Per cell and per sample metrics of the entire dataset after quality control. Cluster-Based Differentially Expressed Genes and Gene Ontology Enrichment Analysis Results are shown.
Data Availability Statement
Raw and processed data files for mouse scRNA-seq data are available through the Gene Expression Omnibus (GEO) under the accession number GSE159638. Processed data can be explored and downloaded via a shiny app at https://marionilab.cruk.cam.ac.uk/SCI_Myeloid_Cell_Atlas/. The code used to process this data and generate the figures can be found on GitLab at https://github.com/regan-hamel/SCI_2020.