Abstract
The ubiquitin-proteasome system (UPS) plays crucial roles in numerous cellular functions. Dysfunction of the UPS shows certain correlations with the pathological changes in Alzheimer’s disease (AD). This study aimed to explore the different impairments of the UPS in multiple brain regions and identify hub ubiquitin ligase (E3) genes in AD. The brain transcriptome, blood transcriptome and proteome data of AD were downloaded from a public database. The UPS genes were collected from the Ubiquitin and Ubiquitin-like Conjugation Database. The hub E3 genes were defined as the differentially expressed E3 genes shared by more than three brain regions. E3Miner and UbiBrowser were used to predict the substrate of hub E3. This study shows varied impairment of the UPS in different brain regions in AD. Furthermore, we identify seven hub E3 genes (CUL1, CUL3, EIF3I, NSMCE1, PAFAH1B1, RNF175, and UCHL1) that are downregulated in more than three brain regions. Three of these genes (CUL1, EIF3I, and NSMCE1) showed consistent low expression in blood. Most of these genes have been reported to promote AD, whereas the impact of RNF175 on AD is not yet reported. Further analysis revealed a potential regulatory mechanism by which hub E3 and its substrate genes may affect transcription functions and then exacerbate AD. This study identified seven hub E3 genes and their substrate genes affect transcription functions and then exacerbate AD. These findings may be helpful for the development of diagnostic biomarkers and therapeutic targets for AD.
Keywords: Alzheimer’s disease, brain region, gene expression, ubiquitin-proteasome system, ubiquitin ligases
Introduction
The ubiquitin-proteasome system (UPS) includes the following enzymes: ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2), ubiquitin ligase enzymes (E3), deubiquitinating enzymes (DUB), and the 26S proteasome.1,2 The UPS is a multicomponent system for protein degradation that is necessary for eukaryotic organisms to degrade approximately 80% of intracellular proteins. 3 Ubiquitin is an evolutionarily conserved peptide containing 76 amino acids; ubiquitin-related enzymes bind ubiquitin molecules to substrates through a multistep catalytic process, and the substrate protein is then degraded by the proteasome. 4 The UPS also plays crucial roles in posttranslational modification, regulation of cellular signal transduction, cell cycle processes, and repair of DNA damage.4,5 Numerous metabolic pathways and cellular regulatory networks require precise spatial and temporal control of effector protein levels by the UPS. 6
The UPS is involved in protein quality control and the removal of misfolded or aggregated proteins, and UPS dysfunction is correlated with neurodegenerative diseases. Damage to the UPS will affect the degradation of amyloid β (Aβ) and cause abnormal aggregation of Aβ in Alzheimer’s disease (AD). Aggregated Aβ inhibits proteasome activity and subsequently causes damage to the multivesicular body sorting pathway. 7 The impaired UPS also causes increased β-secretase and γ-secretase, which accelerates the hydrolysis of amyloid precursor protein (APP) to produce Aβ and promotes Aβ aggregation. 7 Ubiquitinated APP cannot be degraded by the proteasome when the activity of the proteasome is inhibited, which results in the accumulation of ubiquitinated proteins in the cell. 8 Dysfunction of the UPS may also promote the aggregation of tau protein in AD, and the higher-order oligomers and aggregates of tau cannot enter the narrow proteasome open channels. 9 Recent studies have suggested that UPS-dependent protein degradation is associated with synaptic plasticity and learning or memory functions, that the UPS plays a vital role in hippocampal long-term memory consolidation and that substrate-specific E3 may be the key factor in memory regulation. 10
The above studies suggested that the impaired UPS correlated to the pathogenesis of AD, and the decreased UPS activity in AD may lead to further deterioration of the disease. It is worth noting that increasing the UPS activity will enhance the organism’s ability to resist oxidative stress and prolong life in various animal models and human cell lines. 11 Our previous work revealed the differences in the expression of learning- and memory-related genes in different brain regions in AD. 12 In this study, we aimed to explore the different impairments of the UPS in multiple brain regions of AD through transcriptome data analysis and to identify hub E3 genes and their potential substrate genes.
Methods
Data collection
The microarray data of Alzheimer’s disease were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). Data screening was based on the following criteria: (1) the data are genome-wide; (2) the data should include AD and control samples; (3) the data should include multiple brain regions; (4) no age difference exists between AD and controls in each brain region; and (5) raw microarray data are available. With these criteria, we finally chose the dataset of GSE5281 13 for our reanalysis. The dataset contains 87 AD samples and 74 control samples. The brain regions in this dataset included the entorhinal cortex (EC), hippocampus (HIP), medial temporal gyrus (MTG), posterior cingulate (PC), superior frontal gyrus (SFG), and primary visual cortex (PVC). There was no age difference between AD and controls in each brain region (Table 1). The datasets of GSE36980 14 and GSE48350 15 also contain AD and control transcriptome data for multiple brain regions and were used as validation sets. In order to explore the expression differences in the blood transcriptome and proteome in patients with AD, we chose the GSE63060 dataset 16 to analyze the gene expression profiles in blood, which contains 145 patients with AD and 104 age- and sex-matched controls. The GSE29676 dataset 17 contains serum proteome data from 50 patients with AD and 40 controls and was used to explore the changes in protein levels. For details on data preprocessing, see our previous reports.12,18
Table 1.
Age information between patients with Alzheimer’s disease and controls.
| Brain regions | Alzheimer’s disease |
Control |
p b | ||
|---|---|---|---|---|---|
| N a | Age | N a | Age | ||
| Entorhinal cortex | 10 | 85.6 ± 6.3 | 13 | 80.3 ± 9.2 | 0.118 |
| Hippocampus | 10 | 77.8 ± 5.7 | 13 | 79.6 ± 9.4 | 0.574 |
| Medial temporal gyrus | 16 | 79.1 ± 6.4 | 12 | 80.1 ± 9.8 | 0.771 |
| Posterior cingulate | 9 | 77.6 ± 6.5 | 13 | 79.8 ± 9.4 | 0.522 |
| Superior frontal gyrus | 23 | 79.2 ± 7.5 | 11 | 79.3 ± 10.2 | 0.978 |
| Primary visual cortex | 19 | 80.2 ± 6.7 | 12 | 77.9 ± 6.9 | 0.385 |
Number of samples.
The p value was calculated by student’s t-test.
Differential expression gene analysis
Bioinformatics analysis of the microarray data was carried out by R statistical software v3.6.1 (https://www.r-project.org/) and Bioconductor Library (https://www.bioconductor.org/). Differential gene expression analysis was performed using the empirical Bayesian algorithm in the limma package in R. 19 Up- and downregulated genes were defined as a log2 transformed fold-change (logFC) ≥ 1 or ≤−1 for AD samples compared with controls. A false discovery rate (FDR)-corrected p value ≤0.05 was considered significant. Differentially expressed genes shared by more than three brain regions are considered to be hub genes. The heatmap package in R was used to show the gene expression profiles, and the clustering method was chosen as “ward.D2.”
GO and KEGG enrichment analysis
Information on human genes and related GO terms (including biological process, cellular component and molecular function) was downloaded from the QuickGO database (http://www.ebi.ac.uk/QuickGO-Beta/). The reference human genes and pathways were downloaded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/). GO terms and KEGG pathways with less than 10 genes were removed. GO and KEGG enrichment analysis was performed using the hypergeometric test and the formula shown in a previous report. 20 The reference gene set is all detected genes in the GSE5281 dataset. An FDR-corrected p value ≤0.05 was considered significantly enriched.
UPS gene collection
The UPS genes were downloaded from the Ubiquitin and Ubiquitin-like Conjugation Database (http://uucd.biocuckoo.org). 21 This database collected 129,416 potential proteins from 148 different species and protein types, including E1, E2, E3, DUB, ubiquitin-binding domain-containing protein (UBD) and ubiquitin-like domain-containing protein (ULD). A total of 878 human UPS genes (including 10 E1 genes, 42 E2 genes, 700 E3 genes, and 126 DUB genes) were downloaded in this study. Proteasome-related genes were extracted from the proteasome pathway in the KEGG database. This study mainly focused on the E3 genes, and the hub E3 genes were defined as the differentially expressed E3 genes shared by more than three brain regions.
Construction of AD prediction models
The prediction models were built using the blood transcriptome and proteome data of patients with AD and controls. The biomarkers were chosen as the hub E3 genes and proteins. The receiver operating characteristic (ROC) curve and the area under the curve (AUC) of the single marker were calculated using the pROC package in R. A stepwise modeling strategy was used to screen the optimal multigene and multiprotein models. The model with the largest AUC was defined as the optimal model. For example, in the multigene prediction model, the gene with the largest AUC was first selected. Then, we used a multivariate logistic regression model to generate the combined effect of the selected gene and each of the remaining genes. Next, we selected the best two-gene model with the highest AUC and repeated the previous steps. Finally, we selected the optimal model with the highest AUC in each multigene combination model.
E3 substrate prediction and correlation analysis
Two web server tools, including E3Miner (http://e3miner.biopathway.org) 22 and UbiBrowser (http://ubibrowser.ncpsb.org), 23 were used to predict the substrate of hub E3. The hub E3 genes and the top 10 predicted substrate genes based on confidence were used to construct the E3-substrate interactive network. Network visualization was performed using Cytoscape v3.4.0 (https://cytoscape.org/). Pearson correlation analysis was used to analyze the correlation between the E3 genes and the predicted substrate genes. A p value ≤0.05 was considered significant. The mouse dataset GSE113436 was used to validate the expression of NSMCE1 and the predicted target genes. The data is the transcriptome profile of Nsmce1 overexpression in mouse hippocampal neuronal cells. 24
Results
Commonly differentially expressed genes correlate to AD pathology
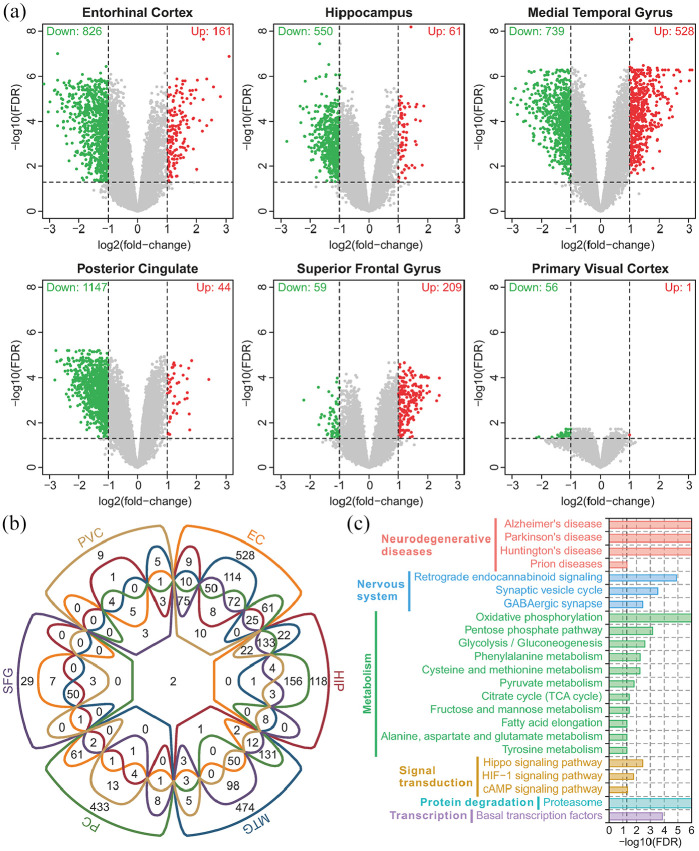
Differential expression analysis showed that there were a large number of downregulated genes in EC, HIP, MTG, and PC, and relatively few genes were affected in SFG and PVC (Figure 1(a)). Furthermore, there were also a considerable number of upregulated genes in the MTG and SFG. Most of these genes were differentially expressed in multiple brain regions, and few genes were differentially expressed in specific brain regions (Figure 1(b)). There were two differentially expressed genes shared by all six brain regions (APOO and PCYOX1L). A Venn diagram showed that a total of 462 genes were differentially expressed in more than three brain regions (Figure 1(b)). The KEGG pathway enrichment results suggested that these genes were mainly involved in the pathways of neurodegenerative diseases (such as Alzheimer’s disease), nervous system (such as synaptic vesicle cycle and GABAergic synapse), metabolism (such as oxidative phosphorylation, glycolysis and the TCA cycle), signal transduction (such as the Hippo signaling pathway and cAMP signaling pathway), protein degradation (such as the proteasome) and transcription (such as basal transcription factors) (Figure 1(c)). Numerous studies have reported that these pathways are correlated with the Aβ and tau protein processes, synaptic dysfunction and neurodegeneration in AD.7,25–28 This evidence suggests that the damaged genes and pathways shared by multiple brain regions can accurately reflect the pathological changes of AD.
Figure 1.
Overview of the differentially expressed genes and their functions: (a) volcano plot of all genes in six brain regions. Red points indicate upregulated genes, and green points indicate down regulated genes, (b) Venn diagram of differentially expressed genes in six brain regions, and (c) enriched KEGG pathways of differentially expressed genes shared by more than three brain regions.
Each color represents a pathway class. The dashed line indicates statistical significance, and the bar width indicates the enrichment percentage.
UPS-related functions were damaged in multiple brain regions in AD
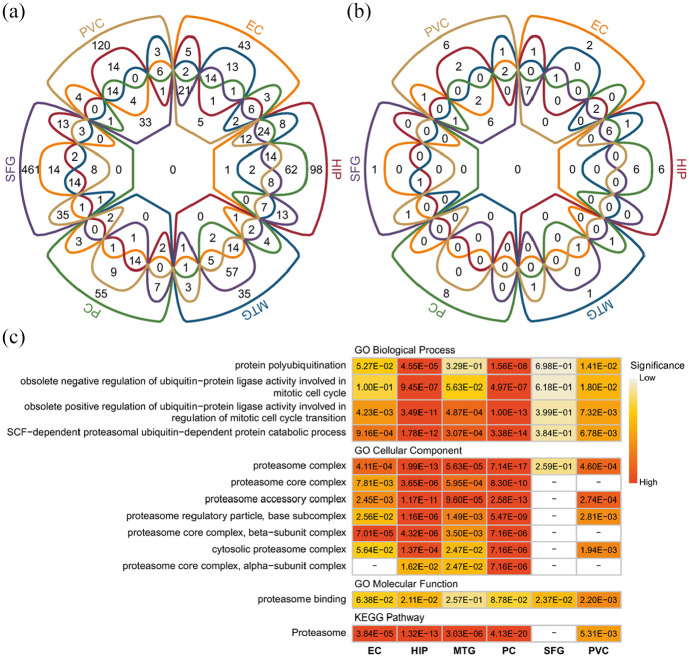
GO and KEGG enrichment analyses were performed using the differentially expressed genes in each brain region. The results showed that there were 193, 357, 272, 283, 666, and 265 enriched GO items (Figure 2(a)) and 23, 40, 29, 38, 3, and 22 enriched KEGG pathways (Figure 2(b)) in the EC, HIP, MTG, PC, SFG, and PVC, respectively. There were 195 enriched GO items and 27 enriched KEGG pathways shared by more than three brain regions. These enriched GO items and KEGG pathways included multiple UPS-related functions. By analyzing the enriched UPS-related functions in different brain regions, we found that HIP and PC showed the highest significance, EC and MTG showed moderate significance, and SFG and PVC showed relatively low significance (Figure 2(c)).
Figure 2.
Enriched GO functions and KEGG pathways in six brain regions: (a) Venn diagram of enriched GO functions (including biological process, cellular component and molecular function) in six brain regions, (b) Venn diagram of enriched KEGG pathways in six brain regions, and (c) significantly enriched ubiquitin-proteasome system-related functions and pathways in more than three brain regions.
The color bar indicates the FDR-corrected p value. The blank indicates that there are no differentially expressed genes in the pathway corresponding to the brain region.
E3 genes were downregulated in multiple brain regions
There were multiple differentially expressed UPS genes in the EC, HIP, MTG, and PC, whereas only a few genes were affected in the SFG and PVC (Figure 3(a)). Because E3 has substrate specificity, this study mainly focused on the E3 genes. These differential E3 genes were almost all downregulated in the EC, HIP, and PC, and only MTG showed more than a dozen upregulated genes. The clustering results of the 73 combined differentially expressed E3 genes in six brain regions confirmed this trend (Figure 3(b)). The Venn diagram analysis showed that there were seven differentially expressed E3 genes shared by more than three brain regions (Figure 3(c)), and these genes were identified as hub E3 genes. Among these genes, UCHL1 was downregulated in the EC, HIP, MTG, and PC; CUL3, EIF3I, and NSMCE1 were downregulated in the EC, HIP, and PC; and CUL1, PAFAH1B1, and RNF175 were downregulated in the HIP, MTG, and PC (Figure 3(d)). The clustering results suggested that the HIP and PC showed the most serious downregulation of the hub E3 genes, followed by the MTG and EC, whereas the SFG and PVC showed relatively slight damage. The clustering results of the proteasome genes also showed a consistent trend (Figure 3(e)). Furthermore, most of the combined differentially expressed E3 genes showed a low expression trend in different brain regions, and the hub E3 genes also showed a consistent downregulation trend in almost all brain regions in the two validation datasets (Figure 3(f)–(i)). Combining the above analysis results, we infer that these hub E3 genes may play crucial roles in the pathogenesis of AD. The role of these hub E3 genes in AD pathology showed in Supplemental Table 1, and the brief signaling cascade of these hub E3 genes showed in Supplemental Figure 1. To explore the possible E2 conjugating enzyme in connection with identified E3 ligases, we used Pearson correlation analysis to explore the combination possibility between hub E3 and E2 genes. Among these hub E3 genes, CUL1, CUL3, PAFAH1B1, and RNF175 showed a strong correlation with multiple E2 genes (Supplemental Figure 2).
Figure 3.
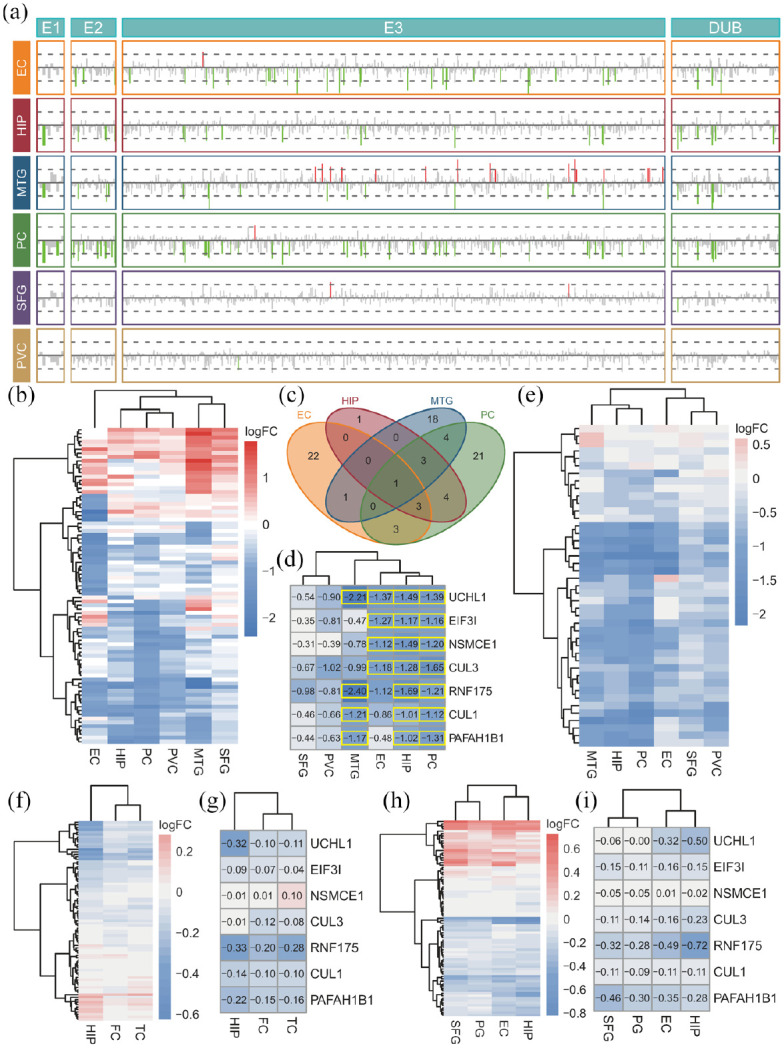
Expression profiles of ubiquitin-proteasome system-related genes and hub E3 genes: (a) gene expression of ubiquitin-related genes in different brain regions. The line length indicates the log2 (fold-change). The red, green and gray colors indicate that the gene is upregulated, downregulated and not statistically significant, (b) heatmap of the combined differentially expressed E3 genes in six brain regions, (c) Venn diagram of the differentially expressed E3 genes in the EC, HIP, MTG and PC, (d) heatmap of the hub E3 genes in six brain regions, (e) heatmap of the proteasome-related genes in six brain regions, (f) heatmap of the combined differentially expressed E3 genes in the validation set of GSE36980, (g) heatmap of the hub E3 genes in the validation set of GSE36980, (h) heatmap of the combined differentially expressed E3 genes in the validation set of GSE48350, and (i) heatmap of the hub E3 genes in the validation set of GSE48350.
The color bar and the number in the box of the heatmap indicate the log2 (fold-change). The yellow box indicates statistical significance.
Expression of hub E3 genes and proteins in the blood of AD
To explore the expression difference in E3 genes in blood, we mapped the AD brain transcriptome data with blood transcriptome and proteome data. There were 73 combined differentially expressed E3 genes in the GSE63060 dataset and 28 combined differentially expressed E3 proteins in the GSE29676 dataset. The clustering results showed that the total expression profiles of these genes or proteins may not distinguish patients with AD and controls (Figure 4(a) and (b)). Three hub E3 genes (CUL1, EIF3I, and NSMCE1) were significantly expressed at low levels in patients with AD compared to controls in blood, which is consistent with the expression trend in the brain (Figure 4(c)). However, the protein expression of CUL1 and RNF175 showed the opposite trend between AD and controls in blood (Figure 4(d)). We constructed prediction models to explore the diagnostic accuracy of these hub E3 genes and proteins in AD. The optimal multigene model is the combination of five hub E3 genes, including NSMCE1, PAFAH1B1, CUL1, RNF175, and EIF3I (Figure 4(e)), which had the highest AUC of 0.726 (Figure 4(f)). The optimal multiprotein model is the combination of five hub E3 proteins, including RNF175, PAFAH1B1, CUL3, CUL1, and UCHL1 (Figure 4(g)), which had the highest AUC of 0.789 (Figure 4(h)). Unfortunately, neither the multigene nor the multiprotein model achieved high prediction accuracy.
Figure 4.
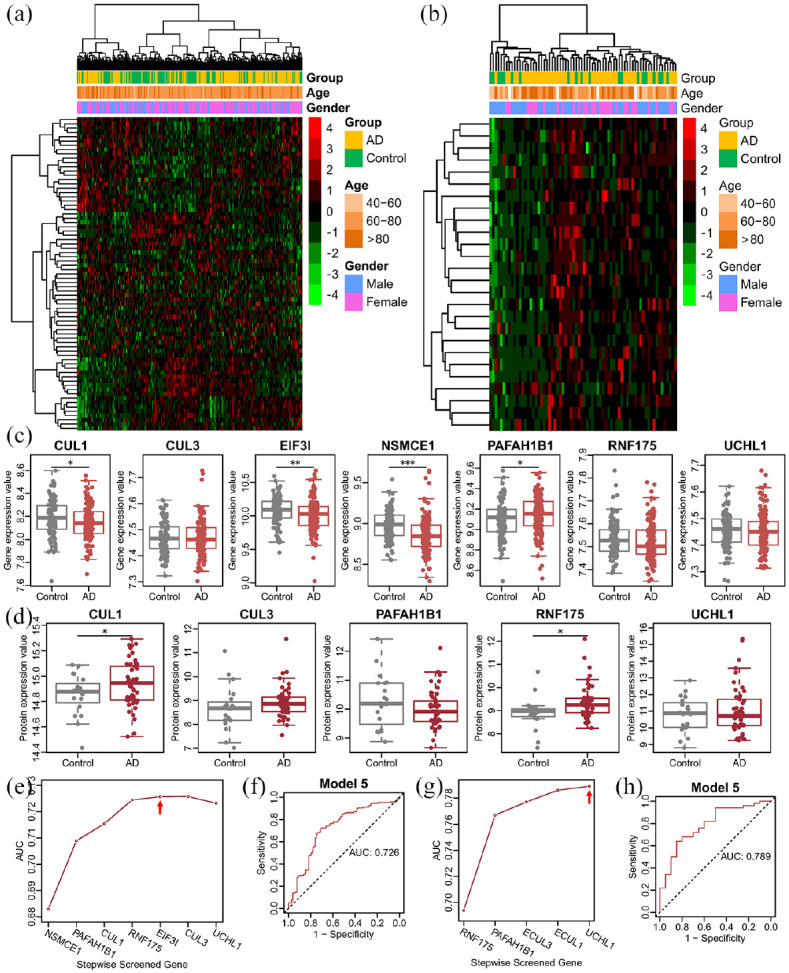
Expression of hub E3 genes and proteins in the blood of AD and diagnostic model construction: (a) heatmap of the combined differentially expressed E3 genes in the GSE63060 dataset, (b) heatmap of the combined differentially expressed E3 proteins in the GSE29676 dataset. The gene and protein expression values were z-score converted, and the color bar indicates the z-score, (c) gene expression of the hub E3 genes in the blood between patients with AD and controls, (d) protein expression of the hub E3 genes in the blood between patients with AD and controls (no protein expression data of EIF3I or NSMCE1). Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, (e) stepwise screened prediction models using hub E3 gene expression values in blood transcriptome data. From left to right on the x-axis (stepwise screened genes), each additional gene corresponds to a model (for example, the gene of NSMCE1 represents model 1, which contains one gene of NSMCE1, PAFAH1B1 represents model 2, which contains two genes including NSMCE1 and PAFAH1B1). The red arrow shows the optimal model, (f) ROC curve of the screened optimal multigene model, (g) stepwise screened prediction models using hub E3 protein expression values in blood proteome data, and (h) ROC curve of the screened optimal multiprotein model.
Hub E3 and its substrates aggravate AD by affecting transcription functions
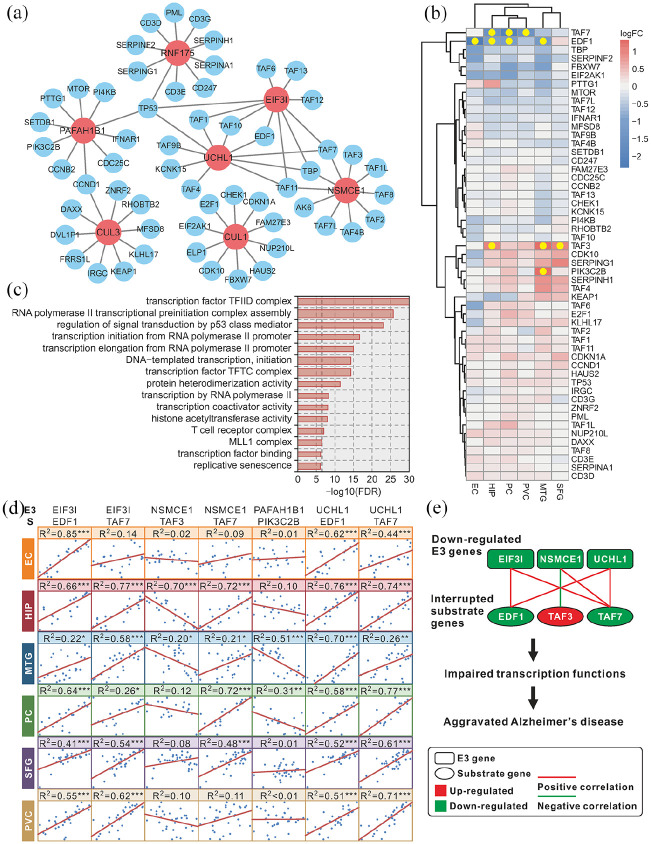
The interactive network of the hub E3 genes and the predicted substrate genes showed that there were eight substrates that were regulated by multiple E3 (Figure 5(a)). The heatmap showed that there were four differentially expressed substrate genes: EDF1 was downregulated in the EC, HIP, MTG, and PC; TAF7 was downregulated in the HIP, PC, and PVC; TAF3 was upregulated in the HIP, MTG, and SFG; and PIK3C2B was upregulated in the MTG (Figure 5(b)). The KEGG enrichment results showed that these substrate genes are involved in transcription functions (Figure 5(c)). Pearson correlation analysis of the hub E3 genes and the differentially expressed substrate genes showed that EIF3I and UCHL1 positively correlated with EDF1 in all brain regions; EIF3I, NSMCE1, and UCHL1 positively correlated with TAF7 in most brain regions; NSMCE1 negatively correlated with TAF3 in the HIP and MTG; and PAFAH1B1 negatively correlated with PIK3C2B in the MTG and PC (Figure 5(d)). Based on the above results, we concluded that the downregulated E3 genes may affect the substrate genes, damage the transcription functions and aggravate AD (Figure 5(e)). In the validation dataset, there was no difference in the expression of Taf3 or Taf7 after Nsmce1 was overexpressed (Supplemental Figure 3).
Figure 5.
Network and function of hub E3 and their substrate genes: (a) hub E3 and their substrate gene interactive network. The red color indicates E3 genes, and the blue color indicates substrate genes, (b) heatmap of the hub E3 substrate genes in six brain regions. The color bar indicates the log2 (fold-change), and the yellow point indicates statistical significance, (c) top 15 enriched GO biological processes of the hub E3 substrate genes. The dashed line indicates statistical significance, and the bar width indicates the enrichment percentage, (d) Pearson correlation of hub E3 genes and differentially expressed substrate genes (represented by S). The R2 indicates the square of the Pearson correlation coefficient. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, and (e) the potential mechanism of down regulated hub E3 genes aggravating AD.
Discussion
This study showed varied impairment of the UPS in different brain regions in AD. The UPS was seriously damaged in the HIP and PC, moderately damaged in the EC and MTG, and slightly damaged in the SFG and PCV. The seven identified hub E3 genes were downregulated in most brain regions, and three of these genes were also expressed at low levels in blood. These hub E3 genes and their substrate genes may affect transcription functions and then exacerbate AD.
Different brain regions have specific functions, and there are strong connections among one another. The EC is the main channel between the HIP and neocortex and is involved in the formation of long-term memory. 29 EC receives information from the HIP and transmits it to the neocortex through neurotransmitters such as glutamate and is one of the brain regions affected in the early stages of AD. 30 The HIP is a part of the temporal lobe, which is essential for the formation of new memories. Usually, HIP function is impaired earlier than other cortices in patients with AD, suggesting that the first process in AD is memory impairment. 31 The MTG involves many cognitive processes, such as semantic memory and language processing, and integrates information from different senses. Studies have shown that there is active neuronal loss in the MTG in AD.32,33 The PC is a part of the cingulate cortex, which is a highly connected and metabolically active brain region and is functionally involved in learning and spatial memory. It has been identified that there was amyloid deposition and decreased metabolism in PC during AD progression, and the volume of the PC in patients with AD is smaller than that of healthy controls.34,35 The SFG is located in the upper part of the prefrontal cortex, accounting for approximately one-third of the frontal lobe. Impaired SFG will lead to disorders of working memory and self-awareness.36,37 Several studies have shown that reduced metabolic functions in FC are associated with AD. 38 The PVC occupies the entire surface of the occipital lobe and acts as a receiver of visual data. Some evidence suggests that the changes in PVC are associated with normal aging, and Aβ plaques and neurofibrillary tangles usually occur in the late stages of AD.39,40 Our previous study found that multiple learning and memory-related genes were affected in the HIP and temporal lobe in AD. 12 Combined with the findings in this study that UPS showed the most serious damage in the HIP and PC, we speculate that there may be a causal relationship between UPS impairment and reduced learning or memory functions.
The number of E1, E2, and E3 has a pyramid structure, and a previous report showed that there were approximately 2 E1 genes, 30-40 E2 genes and more than 600 E3 genes in humans. 11 Recent studies have revealed more genes in the human ubiquitination pathway (including E1, E2, E3, and DUB). 21 E3 has substrate specificity and is the most critical enzyme in the UPS. This study identified seven hub E3 genes, including CUL1, CUL3, EIF3I, NSMCE1, PAFAH1B1, RNF175, and UCHL1. The proteins encoded by CUL1 and CUL3 belong to the cullin protein family. Multisubunit Cullin-RING structure ligase is the most diverse E3 and plays important roles in AD pathology. The CUL1-dependent ubiquitination process is activated by the Neddylation process, which regulates the degradation process of APP and causes APP to be degraded by endocytosis. Studies have shown that downregulated CUL1 leads to APP aggregation and promotes Aβ production. 41 Under normal conditions, the KEAP1-CUL3 complex polyubiquitylates NRF2 and is subsequently degraded by the proteasome. However, the interaction between the KEAP1-CUL3 complex and NRF2 is unstable under oxidative stress conditions, 42 which may cause NRF2 aggregation and inhibit its downstream gene expression in AD. 43 EIF3I encodes eukaryotic translation initiation factor 3 subunit I and is involved in the transcription process of Aβ23. 44 NSMCE1 encodes a subunit of the SMC5-SMC6 complex and plays an important role in DNA repair, and overexpression of NSMCE1 inhibits APP and other AD marker genes in mice. 24 A previous study showed that heterozygous mutations in PAFAH1B1 cause learning and behavior disorders in mice. 45 Compared with cerebral malaria-resistant mice, PAFAH1B1 was reduced in cerebral malaria-susceptible mice and was associated with AD. 46 Several studies reported that UCHL1 mRNA and protein levels were reduced in AD.47–50 A previous study showed identified UCHL1 was deregulated in four out of six brain regions, suggested that there were region-specific changes in UPS components. 50 Overexpression of UCHL1 in APP23 transgenic mice delayed AD progression. 51 However, a correlation between RNF175 and AD has not yet been reported. Furthermore, studies have reported that the substrate genes of hub E3 have certain correlations with AD, such as EDF1, 52 TAF7, 53 and TAF3. 54 These substrate genes are correlated to transcription functions.
There were several limitations to this study. Firstly, the potential substrate genes of hub E3 genes were obtained through database mining and literature search, the interactions between hub E3 genes and the substrate genes still need experimental verification. Secondly, the combination of multiple E1-E2-E3 axes is still unknown, and more in-depth research is needed. Lastly, this study performed only transcriptome level analysis. Whether the protein expression of these E3 or substrates are consistent with gene expression remains to be confirmed.
Conclusion
In summary, the damaged UPS extensively and persistently exacerbates the AD process. This study identified seven hub E3 genes (CUL1, CUL3, EIF3I, NSMCE1, PAFAH1B1, RNF175, and UCHL1) and their substrate genes (EDF1, TAF3, TAF7, etc.) affect transcription functions and then exacerbate AD. Among these E3 genes, CUL1, EIF3I, and NSMCE1 were both downregulated in blood and brain and may be used as potential markers for AD diagnosis. However, the underlying mechanism of the interactions between E3 and substrates affecting AD remains unclear. Therefore, revealing the E3-substrate regulatory mechanisms in future works will help develop diagnostic biomarkers and therapeutic targets for AD.
Supplemental Material
Supplemental material, sj-docx-1-sci-10.1177_00368504211001146 for Identification of hub ubiquitin ligase genes affecting Alzheimer’s disease by analyzing transcriptome data from multiple brain regions by Dahai Liu, Shao-Xing Dai, Kan He, Gong-Hua Li, Justin Liu, Leyna G Liu, Jing-Fei Huang, Lin Xu and Wen-Xing Li in Science Progress
Author biographies
Dahai Liu earned both a Master’s degree and a Doctoral degree in Biochemistry and Molecular Biology from University of California, Los Angeles (UCLA) after he left a faculty position at Fudan University School of Medicine in China. He became Professor and Dean of School of Life Sciences at Anhui University in 2012 and a present member of University Academic Committee at Foshan University. He founded the first Center for Stem Cell and Translational Medicine in Anhui Province of China. Much of his current interests concentrate on various aspects of biomedicine ranging from stem cell research, bioinformatics to translational research. He is Principal Investigator of dozens of grants including the National Natural Science Foundation of China as well as the author/co-author of 5 books, around 70 scientific papers and more than 10 patents. He serves on the editorial board of the Chinese Journal of Cell Biology and other scientific journals.
Shao-Xing Dai earned a Doctoral degree in Kunming Institute of Zoology, Chinese Academy of Sciences. He worked as an assistant researcher at the Kunming Institute of Zoology, Chinese Academy of Sciences from 2013 to 2018. He currently is working as an associate professor at the Institute of Primate Translational Medicine, Kunming University of Science and Technology. He mainly focuses on using bioinformatics or machine learning tools to integrate multiple omics data of human diseases for target identification and drug discovery.
Kan He received his Doctoral degree in School of Agriculture and Biology, Shanghai Jiao Tong University. He has conducted post-doctoral research in the Department of Biology, Hong Kong Baptist University. He is now a lecturer in the School of Life Sciences, Anhui University. He mainly focuses on the research of statistical genomics and bioinformatics.
Gong-Hua Li earned a Doctoral degree in Kunming Institute of Zoology, Chinese Academy of Sciences. He successively served as assistant researcher and associate researcher at Kunming Institute of Zoology, Chinese Academy of Sciences. He served as a visiting scholar at Massachusetts General Hospital of Harvard Medical School in 2014-2015. His research directions are bioinformatics, systems biology and metabolic network modeling. He published more than 10 academic papers in SCI journals such as Bioinformatics, BMC bioinformatics, PeerJ, etc.
Justin Liu received his Bachelor’s degree in microbiology at University of California, San Diego (UCSD) which is one of the world’s leading public research universities. Shortly after he graduated from UCSD, he worked as a research coordinator at Edwards Lifesciences, a public company in California. Currently, he entered a graduate program as Master student in statistics at University of California, Riverside (UCR). He had published several research papers in the field of life sciences.
Leyna G Liu is currently a senior student in Portola High School located in Irvine, California. She is a great dancer, key school cheerleader and President of Animals Need Love Club. She won the Varsity Scholar Athlete Award at school. She is especially interested in biological research and published a few papers on translational medical research using data analysis and statistical tools.
Jing-Fei Huang received his Bachelor’s degree in Biology Department of Yunnan University. He was engaged in computational biology research at the Kunming Institute of Zoology, Chinese Academy of Sciences and successively served as a research intern, assistant researcher, and associate researcher from 1982 to 1996. In 1997, he was awarded the “Royal Society K.C. Wong Fellowships” in the United Kingdom. As a researcher of the Royal Society, he was engaged in the evolution of genes and protein families in the Department of Biochemistry, University of Cambridge. He became Professor and Principal Investigator of Bioinformatics and Systems Biology Laboratory in 1998. He has undertaken a number of scientific research projects such as the National Fund, the “Tenth Five-Year Plan” major project of the Chinese Academy of Sciences, the key and general projects of the Yunnan Provincial Applied Basic Research Fund. He has published more than 100 papers in SCI journals.
Lin Xu received his Doctoral degree in Department of Pharmacology, Trinity College, Dublin, Ireland. He became Professor and Principal Investigator of Learning and Memory Laboratory in 1998. In 1999, he was awarded the Outstanding Youth Fund of the National Foundation of China. He is currently a doctoral supervisor in neurobehaviour of Yunnan University and University of Science and Technology of China. He is deputy chairman of the “Basic and Clinical Professional Committee of Psychiatry” of the Chinese Society of Neuroscience, and the “Physiological Psychology Professional Committee” of the Chinese Society of Physiology. He published more than 100 SCI papers in academic journals such as Nature, PNAS, Cell, Neuron, etc., and participated in the editing of 2 monographs. The Nature paper published in 1998 was included in one of the 28 papers in the modern memory research catalog (Bibliography on Memory since 1950) by Thomas Donaldson.
Wen-Xing Li earned a Doctoral degree in Kunming Institute of Zoology, Chinese Academy of Sciences. Now he engages in post-doctoral research at the School of Basic Medicine, Southern Medical University. He mainly focuses on bioinformatics and drug research and development. He published more than 40 SCI papers in academic journals such as Aging, J Alzheimers Dis, Oncotarget, Sci Rep, PeerJ, etc., and applied for a number of patents and obtained 4 authorizations.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Basic Research Program of China (No. 2013CB835100), the Strategic Priority Research Program of Chinese Academy of Sciences (No. XDBS01020100), the National Natural Science Foundation of China (No. 81570376, No. 81870307), the University Special Innovative Research Program of Department of Education of Guangdong Province (No. 2017KTSCX189), the Start-up Fund of Kunming University of Science and Technology (No. KKZ3201927005), and Yunnan Fundamental Research Projects (No. 2019FB050).
ORCID iD: Wen-Xing Li  https://orcid.org/0000-0001-9984-8439
https://orcid.org/0000-0001-9984-8439
Supplemental material: Supplemental material for this article is available online.
References
- 1.Gilon T, Chomsky O, Kulka RG.Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J 1998; 17(10): 2759–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A.The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM.Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70: 503–533. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Q, Huang T, Zhang L, et al. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front Aging Neurosci 2016; 8: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iconomou M, Saunders DN.Systematic approaches to identify E3 ligase substrates. Biochem J 2016; 473(22): 4083–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guharoy M, Bhowmick P, Tompa P.Design principles involving protein disorder facilitate specific substrate selection and degradation by the ubiquitin-proteasome system. J Biol Chem 2016; 291(13): 6723–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong L, Huang HC, Jiang ZF.Relationship between amyloid-beta and the ubiquitin-proteasome system in Alzheimer’s disease. Neurol Res 2014; 36(3): 276–282. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Saunders AJ.The role of ubiquitin-proteasome in the metabolism of amyloid precursor protein (APP): implications for novel therapeutic strategies for Alzheimer’s disease. Discov Med 2014; 18(97): 41–50. [PubMed] [Google Scholar]
- 9.Lee MJ, Lee JH, Rubinsztein DC.Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol 2013; 105: 49–59. [DOI] [PubMed] [Google Scholar]
- 10.Lip PZ, Demasi M, Bonatto D.The role of the ubiquitin proteasome system in the memory process. Neurochem Int 2017; 102: 57–65. [DOI] [PubMed] [Google Scholar]
- 11.Papaevgeniou N, Chondrogianni N.UPS activation in the battle against aging and aggregation-related diseases: an extended review. Methods Mol Biol 2016; 1449: 1–70. [DOI] [PubMed] [Google Scholar]
- 12.Li WX, Dai SX, Liu JQ, et al. Integrated analysis of Alzheimer’s disease and schizophrenia dataset revealed different expression pattern in learning and memory. J Alzheimers Dis 2016; 51(2): 417–425. [DOI] [PubMed] [Google Scholar]
- 13.Liang WS, Reiman EM, Valla J, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A 2008; 105(11): 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hokama M, Oka S, Leon J, et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cereb Cortex 2014; 24(9): 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berchtold NC, Cribbs DH, Coleman PD, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A 2008; 105(40): 15605–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sood S, Gallagher IJ, Lunnon K, et al. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol 2015; 16: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagele E, Han M, DeMarshall C, et al. Diagnosis of Alzheimer’s disease based on disease-specific autoantibody profiles in human sera. PLoS One 2011; 6(8): e23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WX, Dai SX, Wang Q, et al. Integrated analysis of ischemic stroke datasets revealed sex and age difference in anti-stroke targets. PeerJ 2016; 4: e2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu DI, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43(7): e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HY, Zhao H, Li WX.Integrated analysis of transcriptome and prognosis data identifies FGF22 as a prognostic marker of lung adenocarcinoma. Technol Cancer Res Treat 2019; 18: 1533033819827317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao T, Liu Z, Wang Y, et al. UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res 2013; 41(D1): D445–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H, Yi GS, Park JC.E3Miner: a text mining tool for ubiquitin-protein ligases. Nucleic Acids Res 2008; 36(Web Server issue): W416–W422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Xie P, Lu L, et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat Commun 2017; 8(1): 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong M, Wang Z, Liu Y, et al. A transcriptomic analysis of Nsmce1 overexpression in mouse hippocampal neuronal cell by RNA sequencing. Funct Integr Genomics 2020; 20(3): 459–470. [DOI] [PubMed] [Google Scholar]
- 25.Querfurth HW, LaFerla FM.Alzheimer’s disease. N Engl J Med 2010; 362(4): 329–344. [DOI] [PubMed] [Google Scholar]
- 26.Selkoe DJ.Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev 2001; 81(2): 741–766. [DOI] [PubMed] [Google Scholar]
- 27.Zlokovic BV.Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat Rev Neurosci 2011; 12(12): 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathys H, Davila-Velderrain J, Peng Z, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019; 570(7761): 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fransen E.Functional role of entorhinal cortex in working memory processing. Neural Netw 2005; 18(9): 1141–1149. [DOI] [PubMed] [Google Scholar]
- 30.Khan UA, Liu L, Provenzano FA, et al. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer’s disease. Nat Neurosci 2014; 17(2): 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mu Y, Gage FH.Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol Neurodegener 2011; 6: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Convit A, De Asis J, De Leon MJ, et al. Atrophy of the medial occipitotemporal, inferior, and middle temporal gyri in non-demented elderly predict decline to Alzheimer’s disease. Neurobiol Aging 2000; 21(1): 19–26. [DOI] [PubMed] [Google Scholar]
- 33.Grady CL, McIntosh AR, Beig S, et al. Evidence from functional neuroimaging of a compensatory prefrontal network in Alzheimer’s disease. J Neurosci 2003; 23(3): 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leech R, Sharp DJ.The role of the posterior cingulate cortex in cognition and disease. Brain 2014; 137(Pt 1): 12–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leech R, Braga R, Sharp DJ.Echoes of the brain within the posterior cingulate cortex. J Neurosci 2012; 32(1): 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.du Boisgueheneuc F, Levy R, Volle E, et al. Functions of the left superior frontal gyrus in humans: a lesion study. Brain 2006; 129(Pt 12): 3315–3328. [DOI] [PubMed] [Google Scholar]
- 37.Stuss DT, Gow CA, Hetherington CR.“No longer Gage”: frontal lobe dysfunction and emotional changes. J Consult Clin Psychol 1992; 60(3): 349–359. [DOI] [PubMed] [Google Scholar]
- 38.Vasconcelos LG, Jackowski AP, Oliveira MO, et al. The thickness of posterior cortical areas is related to executive dysfunction in Alzheimer’s disease. Clinics 2014; 69(1): 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brewer AA, Barton B.Visual cortex in aging and Alzheimer’s disease: changes in visual field maps and population receptive fields. Front Psychol 2014; 5: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusne Y, Wolf AB, Townley K, et al. Visual system manifestations of Alzheimer’s disease. Acta Ophthalmol 2017; 95(8): e668–e676. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Neve RL, Liu H.Neddylation dysfunction in Alzheimer’s disease. J Cell Mol Med 2012; 16(11): 2583–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canning P, Bullock AN.New strategies to inhibit KEAP1 and the Cul3-based E3 ubiquitin ligases. Biochem Soc Trans 2014; 42(1): 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Li WX, Dai SX, et al. Meta-analysis of Parkinson’s disease and Alzheimer’s disease revealed commonly impaired pathways and dysregulation of NRF2-dependent genes. J Alzheimers Dis 2017; 56(4): 1525–1539. [DOI] [PubMed] [Google Scholar]
- 44.Olzscha H, Schermann SM, Woerner AC, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 2011; 144(1): 67–78. [DOI] [PubMed] [Google Scholar]
- 45.Paylor R, Hirotsune S, Gambello MJ, et al. Impaired learning and motor behavior in heterozygous Pafah1b1 (Lis1) mutant mice. Learn Mem 1999; 6(5): 521–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delahaye NF, Coltel N, Puthier D, et al. Gene expression analysis reveals early changes in several molecular pathways in cerebral malaria-susceptible mice versus cerebral malaria-resistant mice. BMC Genomics 2007; 8: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M, Deng Y, Luo Y, et al. Control of BACE1 degradation and APP processing by ubiquitin carboxyl-terminal hydrolase L1. J Neurochem 2012; 120(6): 1129–1138. [DOI] [PubMed] [Google Scholar]
- 48.Esteve-Rudd J, Campello L, Herrero MT, et al. Expression in the mammalian retina of parkin and UCH-L1, two components of the ubiquitin-proteasome system. Brain Res 2010; 1352: 70–82. [DOI] [PubMed] [Google Scholar]
- 49.Tramutola A, Di Domenico F, Barone E, et al. It is all about (U)biquitin: role of altered ubiquitin-proteasome system and UCHL1 in Alzheimer disease. Oxid Med Cell Longevity 2016; 2016: 2756068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang WS, Dunckley T, Beach TG, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics 2008; 33(2): 240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Cai F, Zhang S, et al. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci Rep 2014; 4: 7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai MK, Esiri MM, Tan MG.Genome-wide profiling of alternative splicing in Alzheimer’s disease. Genomics Data 2014; 2: 290–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amber S, Zahid S.Data integration for functional annotation of regulatory single nucleotide polymorphisms associated with Alzheimer’s disease susceptibility. Gene 2018; 672: 115–125. [DOI] [PubMed] [Google Scholar]
- 54.Kreuz S, Fischle W.Oxidative stress signaling to chromatin in health and disease. Epigenomics 2016; 8(6): 843–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-sci-10.1177_00368504211001146 for Identification of hub ubiquitin ligase genes affecting Alzheimer’s disease by analyzing transcriptome data from multiple brain regions by Dahai Liu, Shao-Xing Dai, Kan He, Gong-Hua Li, Justin Liu, Leyna G Liu, Jing-Fei Huang, Lin Xu and Wen-Xing Li in Science Progress