Abstract
Traumatic brain injury (TBI) is characterized by heterogeneity in terms of injury severity, mechanism, outcome, and pathophysiology. A single biomarker alone is unlikely to capture the heterogeneity of even one injury subtype, necessitating the use of panels of biomarkers. Herein, we focus on traumatic cerebrovascular injury and investigate associations of a panel of 16 vascular injury-related biomarkers with indices of TBI severity and outcomes using data from 159 participants in the Transforming Research and Clinical Knowledge in TBI (TRACK-TBI) Pilot Study. Associations of individual biomarkers and clusters of biomarkers identified using non-linear principal components analysis with TBI severity and outcomes were assessed using logistic regression models and Spearman's correlations. As individual biomarkers, higher levels of thrombomodulin, angiopoietin (Ang)-2, von Willebrand factor, and P-selectin were associated with more severe injury; higher levels of Ang-1, Tie2, vascular endothelial growth factor (VEGF)-C, and basic fibroblast growth factor (bFGF) were associated with less severe injury (all p < 0.05 in age-adjusted models). After false discovery rate correction for multiple comparisons, higher levels of Ang-2 remained associated with more severe injury and higher levels of Ang-1, Tie2, and bFGF remained associated with less severe injury at a p < 0.05 level. In principal components analysis, principal component (PC)1, comprised of Ang1, bFGF, P-selectin, VEGF-C, VEGF-A, and Tie2, was associated with less severe injury (age-adjusted odds ratio [OR]: 0.63, 95% confidence interval [CI]: 0.44-0.88 for head computer tomography [CT] positive vs. negative) and PC2 (Ang-2, E-selectin, Flt-1, placental growth factor, thrombomodulin, and vascular cell adhesion protein 1) was associated with greater injury severity (age-adjusted OR: 2.29, 95% CI: 1.49-3.69 for Glasgow Coma Scale [GCS] 3-12 vs. 13-15 and age-adjusted OR 1.59, 95% CI: 1.11-2.32 for head CT positive vs. negative). Neither individual biomarkers nor PCs were associated with outcomes in adjusted models (all p > 0.05). In conclusion, in this trauma-center based population of acute TBI patients, biomarkers of microvascular injury were associated with TBI severity.
Keywords: biomarkers, injury severity, microvascular injury, outcome, traumatic brain injury
Introduction
Traumatic brain injury (TBI) is a heterogenous disease that is associated with significant morbidity and mortality.1 Although existing classification systems, which incorporate information about loss of consciousness, alteration of consciousness, post-traumatic amnesia, neuroimaging, and Glasgow Coma Scale (GCS), distinguish broadly among injury severity categories,2,3 there is growing appreciation that markers of specific injury subtypes (or endophenotypes) are needed to further characterize the pathogenesis of TBI and inform the pursuit of mechanism-based therapies. Indeed, TBI is defined by neuronal injury, glial injury, axonal injury/shearing, vascular and microvascular injury, cerebral edema, and inflammation.4-6 The heterogeneity in the type and severity of the underlying neuropathology presents a formidable challenge to clinical management.
Traumatic cerebral microvascular injury is one common endophenotype in TBI6,7 that is associated with long-term impaired microvascular and blood-brain barrier function.6,8 Several biomarkers have previously been studied for diagnostic and prognostic purposes in relation to traumatic cerebral microvascular injury and microvascular repair in humans and in animal models, including von Willebrand factor (vWF),9 cellular cibronectin (c-Fibronectin),9 thrombomodulin,10 endothelium-specific receptor tyrosine kinase receptor (Tie2),11 and angiopoietin 1 (Ang1)12 and angiopoietin 2 (Ang2).12 However, these preliminary studies had small sample sizes, and there is a need to investigate how microvascular-related biomarkers may be related to TBI severity and to TBI outcomes. Further, it is important to evaluate associations of these biomarkers with TBI severity and outcomes not just in isolation (as one biomarker may not fully capture the heterogeneity of TBI),13 but also in combination using analytic dimensional reduction approaches to help inform which clusters of biomarkers may be most relevant in the setting of complex brain injuries.
The Transforming Research and Clinical Knowledge in TBI (TRACK-TBI) Pilot Study is a comprehensively phenotyped cohort of acute TBI patients followed for 6 months, containing data from a panel of 16 biomarkers related to microvascular injury. In the present analyses, we investigated associations of a priori hypothesized and more novel, exploratory biomarkers of microvascular injury with measures of TBI severity and with outcomes over 6 months of follow-up.
Methods
Study design and study population
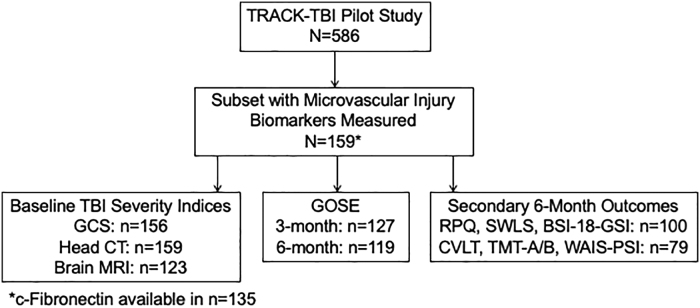
The multi-center, prospective observational TRACK-TBI Pilot Study recruited a total of 650 participants via convenience sampling at three level 1 trauma centers (San Francisco General Hospital, University of Pittsburgh Medical Center, and University Medical Center Brackenridge in Austin, Texas) and one rehabilitation center (Mount Sinai Rehabilitation Center in New York City) between April 2010 and January 2011.14 Participants were followed for 6 months. For the three level 1 trauma centers, eligible participants included all traumatic brain injury patients presenting to the emergency department who underwent a clinically indicated head computed tomography (CT) scan within 24 h of injury. Study exclusion criteria consisted of pregnancy, being in law enforcement custody, being on psychiatric hold, non-English speaking, and having contraindications to magnetic resonance imaging (MRI).14 The TRACK-TBI Pilot Study was approved by the Institutional Review Boards of all participating institutions and written informed consent was obtained at each study visit from participants or proxies. Of the 650 TRACK-TBI Pilot Study participants, 13 were age <16 years and 51 were recruited from the rehabilitation center site. Of the 586 participants age 16+ years with acute TBI, a subset of 159 participants with data on blood-based biomarkers of microvascular injury were included in the present analysis (Fig. 1).
FIG. 1.
Study design. BSI-18-GSI, 18-Item Brief Symptom Inventory Global Severity Index; CT, computed tomography; CVLT, California Verbal Learning Test; GCS, Glasgow Coma Scale; GOSE, Glasgow Outcome Scale-Extended; RPQ, Rivermead Post-Concussion Symptoms Questionnaire; SWLS, Satisfaction with Life Scale; TBI, traumatic brain injury; TMT-A/B, Trail Making Test Parts A and B; WAIS-PSI, Processing Speed Index from the Wechsler Adult Intelligence Scale, 4th Edition.
Blood sample collection and biomarker measurement
Blood samples were collected within 24 h of injury in accordance with the TBI Common Data Elements Biospecimens and Biomarkers Working Group Guidelines.15 Among the subset of 33 participants with data on hours from injury to blood collection, the median (25th-75th percentile) time was 8.3 (3.8-14.5) h. Blood samples were centrifuged, aliquoted, and frozen at -80°C within 1 h of collection. In 2021, available stored frozen samples were provided to MesoScale Diagnostics, LLC. (Rockville, Maryland) for measurement of plasma levels of 16 biomarkers related to microvascular injury/function (biomarkers a priori hypothesized to be related to TBI severity and outcomes based on literature review: vWF, thrombomodulin, Ang-1, Ang-2, c-Fibronectin, Tie-2 and exploratory biomarkers: vascular endothelial growth factor receptor 1 [Flt-1], placental growth factor [PIGF], vascular endothelial growth factor A [VEGF-A], VEGF-C, VEGF-D, E-selectin, platelet-derived growth factor receptor ß [PDGFR-ß], P-selectin, vascular cell adhesion protein 1 [VCAM-1], and basic fibroblast growth factor [bFGF]).
Biomarkers were measured using an electrochemiluminescence (ECL) immunoassay and plasma levels were quantified using ECL detection in an array-based multiplex format.16 The 15 biomarkers of vWF, thrombomodulin, Ang-1, Ang-2, Tie-2, Flt-1, PIGF, VEGF-A, VEGF-C, VEGF-D, E-selectin, PDGFR-ß, P-selectin, VCAM-1, and bFGF were measured together while c-Fibronectin was measured in a separate sub-cohort (n = 160) of TRACK-TBI participants as part of the MesoScale Diagnostics, LLC. (Rockville, MD) V-Plex Panel (135 participants with available c-fibronectin were also included in the present sub-cohort of 159 participants). A capture antibody for each biomarker was printed on an electrode array spot within the wells of 96-well MULTI-ARRAY® plates. Detection antibody was conjugated with electrochemiluminescent SULFO-TAG™. To reduce background and reduce heterophilic antibody interference, diluents contained various blockers including purified mouse immunoglobulin. Plates were read on a SECTOR® Imager 6000 reader. Calibrator dilutions were assayed in duplicate, and samples were measured in singlicate in order to maximize the number of assays run with limited sample volume.17 Each plate also contained a control sample made from a plasma pool and eight dilutions of a recombinant calibrator for each biomarker. The calibrator data was fitted with a four-parameter logistic curve fit and used to quantitate control and sample biomarker concentrations. The calibration curve also was used to estimate the upper limit of each immunoassay's linear range.
TBI severity indices and outcome measures
TBI severity was assessed by the initial Glasgow Coma Scale2 (GCS; classified as mild [score 13-15] vs. moderate/severe injury [score 3-12]) and the initial head CT (classified as positive vs. negative for acute intracranial injury defined according to the TBI Common Data Elements [CDE] Neuroimaging Working Group expert consensus recommendations).18,19 In secondary analyses, we also considered the CT findings of the presence versus absence of contusions, subdural hemorrhages (SDHs), subarachnoid hemorrhages (SAHs), intraventricular hemorrhages (IVHs), and diffuse axonal injury, as well as MRI findings in a subsample of 123 participants (presence vs. absence of any injury-related findings and the presence vs. absence of microhemorrhages; all defined according to the TBI CDE Neuroimaging Working Group expert consensus recommendations).18,19
Global functional outcome was assessed at 3 and 6 months by the Glasgow Outcome Scale, Extended-TBI Version (GOSE-TBI), which is a self- or proxy-reported global measurement of functional impairment due only to the TBI (i.e., not due to co-occurring polytrauma).20 The GOSE-TBI was categorized as 1-4 (dead; vegetative state; lower/upper severe disability) versus 5-8 (lower/upper moderate disability; lower/upper good recovery). Secondary symptom-related and cognitive outcomes were assessed at 6 months, including the Rivermead Post-Concussion Symptoms Questionnaire21 (RPQ; measure of self-reported post-TBI symptoms, higher scores indicate more severe symptoms), the Satisfaction with Life Scale22 (SWLS; measure of general life satisfaction, higher scores indicate greater life satisfaction), the 18-Item Brief Symptom Inventory Global Severity Index23 (BSI-18-GSI; measure of psychological distress, higher scores indicate more severe psychological symptoms), the California Verbal Learning Test24 (CVLT; test of verbal learning and memory, sum of trials 1-5, higher scores indicate better performance), the Trail Making Test Parts A and B25 (TMT-A and TMT-B; test of executive function and processing speed, time to completion, higher scores indicate worse performance), and the Processing Speed Index from the Wechsler Adult Intelligence Scale, 4th Edition26 (WAIS-PSI; test of processing speed, higher scores indicate better performance).
Statistical analysis
Baseline characteristics of TRACK-TBI Pilot Study participants with (n = 159) and without (n = 427) available biomarker data are shown using means and standard deviations (SDs) for continuous variables, and numbers and proportions for categorical variables. Characteristics were compared between groups using Wilcoxon Rank Sum tests for continuous variables and Fisher's exact tests for categorical variables. Baseline characteristics were also compared between included participants with and without 3- and 6-month GOSE-TBI outcomes.
Spearman's correlations were calculated between each microvascular injury biomarker pair. Unadjusted and adjusted logistic regression models were used to estimate odds ratios (ORs) and 95% confidence intervals (95% CIs) for associations of each biomarker with measures of injury severity (age-adjusted) and GOSE (age- and GCS-adjusted) outcomes. We additionally used false discovery rate (FDR) correction to control for multiple comparisons. Spearman's correlations were used to assess the associations of each individual biomarker with the secondary outcomes of RPQ, SWLS, BSI-18-GSI, CVLT, TMT-A, TMT-B, and WAIS-PSI scores. Due to the skewness of the biomarker distributions, log-transformed values were used for all the modeling.
In addition to examining associations of injury severity and outcomes with each biomarker separately, we performed a non-linear principal components analysis on 15 biomarkers (excluding c-Fibronectin as it was not measured in all included participants) for dimension reduction. A scree plot was generated to determine the percent of variance accounted for by each principal component (PC). A 1000 times permutation test was used to test whether the variance accounted for was greater than chance, and results were used to identify components for subsequent analyses. Individual scores for each participant on the PCs were calculated based on the weight of the loadings for all biomarkers. Logistic regression and Spearman's correlations were used to estimate the associations of the first and second principal component (PC1 and PC2) scores with injury severity and GOSE-TBI and secondary outcomes. The PC analysis was conducted using the syndRomics package27 in statistical software R (version 4.1.2).28 All other analyses were performed using statistical software R (version 3.6.1)28 and a two-sided p value <0.05 was considered statistically significant.
Results
The included 159 participants had a mean age of 44 years (SD 18 years), 35% were women, and 18% were of non-white race. Seventy-nine percent of included participants were classified with TBI of mild severity (GCS 13-15), 64% had associated loss of consciousness, 54% had associated post-traumatic amnesia, and 50% had acute TBI-related intracranial abnormalities on CT scan. Compared with TRACK-TBI Pilot Study participants without available biomarker data, participants included in the present analysis were more likely to be women (35% vs. 26%, p = 0.031), but were similar in terms of other demographics and injury characteristics (Table 1). Compared with individuals with missing GOSE-TBI data (primary outcome) at 3 and 6 months, participants with non-missing GOSE-TBI were more likely to have high school or greater education (p < 0.05) but were similar in terms of other demographics and injury characteristics (Table 2).
Table 1.
Baseline Characteristics Comparing Participants With and without Microvascular Injury-Related Biomarker Data, TRACK-TBI Pilot Study
| Biomarker data available* (n = 159) | Biomarker data not available (n = 427) | p Value | |
|---|---|---|---|
| Age (years), mean (SD) | 44.3 (18.0) | 43.0 (18.7) | 0.338 |
| Female, n (%) | 56 (35.2) | 111 (26.0) | 0.031 |
| Race, n (%) | 0.895 | ||
| White | 130 (81.8) | 341 (79.9) | |
| Black | 12 (7.6) | 38 (8.9) | |
| Other | 17 (10.7) | 48 (11.2) | |
| Hispanic ethnicity, n (%) | 25 (16.0) | 59 (14.0) | 0.510 |
| Study site, n (%) | <0.001 | ||
| University of California San Francisco | 104 (65.4) | 222 (52.0) | |
| University of Pittsburg Medical Center | 39 (30.8) | 131 (30.7) | |
| University Medical Center Brackenridge | 6 (3.8) | 74 (17.3) | |
| Education, n (%) | 0.653 | ||
| <High school | 18 (11.9) | 50 (12.4) | |
| High school, GED, or equivalent | 92 (60.9) | 228 (56.4) | |
| Some college or above | 41 (27.2) | 126 (31.2) | |
| Pre-injury employment, n (%) | 0.332 | ||
| Full-time | 59 (38.8) | 160 (40.0) | |
| Part-time | 19 (12.5) | 73 (18.3) | |
| Unemployed | 38 (25.0) | 82 (20.5) | |
| Retired/disabled/student | 36 (23.7) | 85 (21.3) | |
| Current smoking, n (%) | 59 (37.1) | 132 (30.9) | 0.166 |
| Current alcohol consumption, n (%) | 91 (57.2) | 249 (58.3) | 0.851 |
| Current illicit drug use, n (%) | 51 (32.1) | 111 (26.0) | 0.147 |
| Hypertension, n (%) | 43 (27.0) | 105 (24.6) | 0.593 |
| Diabetes, n (%) | 16 (10.1) | 36 (8.4) | 0.518 |
| Depression, n (%) | 42 (26.4) | 87 (20.4) | 0.118 |
| Prior TBI, n (%) | 71 (47.3) | 190 (47.1) | 0.544 |
| TBI Severity, n (%) | 0.174 | ||
| Mild (GCS 13-15) | 123 (78.9) | 357 (84.0) | |
| Moderate/Severe (GCS 3-12) | 33 (21.2) | 68 (16.0) | |
| Injury cause, n (%) | 0.705 | ||
| Motor vehicle crash (occupant) | 38 (24.1) | 99 (23.3) | |
| Motor vehicle crash (bicyclist/pedestrian) | 31 (19.6) | 68 (16.0) | |
| Motorcycle crash | 6 (3.8) | 24 (5.7) | |
| Fall | 57 (36.1) | 145 (34.1) | |
| Assault | 22 (13.9) | 72 16.9) | |
| Other | 4 (2.5) | 17 (4.0) | |
| Loss of consciousness, n (%) | 101 (64.3) | 302 (71.6) | 0.237 |
| Post-traumatic amnesia, n (%) | 85 (54.1) | 249 (59.0) | 0.540 |
| Head CT positive for acute intracranial injury, n (%) | 79 (49.7) | 211 (49.4) | >0.99 |
| Injury severity score, mean (SD) | 12.3 (11.2) | 11.6 (11.5) | 0.357 |
The following variables contained missing data among the 159 participants with biomarker data included in the present analyses: Hispanic ethnicity (n = 3), education (n = 8), employment (n = 7), APOE ɛ4 genotype (n = 18), TBI severity (n = 3), injury cause (n = 1), loss of consciousness (n = 2), post-traumatic amnesia (n = 2).
TRACK-TBI, Transforming Research and Clinical Knowledge in Traumatic Brain Injury; SD, standard deviation; GED, general educational development; TBI, traumatic brain injury; GCS, Glasgow Coma Scale; CT, computed tomography; APOE, apolipoprotein.
Table 2.
Baseline Characteristics Comparing Participants With and Without 3- and 6-Month GOSE Outcomes
| 3-Month GOSE Available (n = 127) | 3-Month GOSE Not Available (n = 32) | p Value | 6-Month GOSE Available (n = 119) | 6-Month GOSE Not Available (n = 40) | p Value | |
|---|---|---|---|---|---|---|
| Age (years), mean (SD) | 45.8 (18.2) | 38.6 (15.7) | 0.046 | 45.8 (19.0) | 40.0 (13.8) | 0.119 |
| Female, n (%) | 47 (37.0) | 9 (28.1) | 0.411 | 45 (37.8) | 11 (27.5) | 0.258 |
| Race, n (%) | 0.795 | 0.737 | ||||
| White | 105 (82.7) | 25 (78.1) | 98 (82.4) | 32 (80.0) | ||
| Black | 9 (7.1) | 3 (9.4) | 8 (6.7) | 4 (10.0) | ||
| Other | 13 (10.2) | 4 (12.5) | 13 (10.9) | 4 (10.0) | ||
| Hispanic ethnicity, n (%) | 19 (15.3) | 6 (18.8) | 0.599 | 18 (15.5) | 7 (17.5) | 0.804 |
| Study Site, n (%) | 0.526 | 0.232 | ||||
| University of California San Francisco | 85 (66.9) | 19 (59.4) | 77 (64.7) | 27 (67.5) | ||
| University of Pittsburg Medical Center | 38 (29.9) | 11 (34.4) | 39 (32.8) | 10 (25.0) | ||
| University Medical Center Brackenridge | 4 (3.2) | 2 (6.3) | 3 (2.5) | 3 (7.5) | ||
| Education, n (%) | 0.006 | 0.037 | ||||
| < High school | 9 (7.4) | 9 (31.0) | 9 (8.0) | 9 (23.7) | ||
| High school, GED, or equivalent | 77 (63.1) | 15 (51.7) | 70 (62.0) | 22 (57.9) | ||
| Some college or above | 36 (29.5) | 5 (17.24) | 34 (30.1) | 7 (18.4) | ||
| Pre-injury employment, n (%) | 0.075 | 0.743 | ||||
| Full-time | 51 (41.5) | 8 (27.6) | 46 (40.4) | 13 (34.2) | ||
| Part-time | 18 (14.6) | 1 (3.5) | 15 (13.2) | 4 (10.5) | ||
| Unemployed | 27 (22.0) | 11 (37.9) | 26 (22.8) | 12 (31.6) | ||
| Retired/disabled/student | 27 (22.0) | 9 (31.0) | 27 (23.7) | 9 (23.7) | ||
| Current smoking, n (%) | 44 (34.7) | 15 (46.9) | 0.223 | 40 (33.6) | 19 (47.5) | 0.132 |
| Current alcohol consumption, n (%) | 72 (56.7) | 19 (59.4) | 0.843 | 67 (56.3) | 24 (60.0) | 0.716 |
| Current illicit drug use, n (%) | 37 (29.1) | 14 (43.8) | 0.139 | 40 (33.6) | 11 (27.5) | 0.559 |
| Hypertension, n (%) | 34 (26.8) | 9 (28.1) | >0.999 | 33 (27.7) | 10 (25.0) | 0.838 |
| Diabetes, n (%) | 13 (10.2) | 3 (9.4) | >0.999 | 12 (10.1) | 4 (10.0) | >0.999 |
| Depression, n (%) | 34 (26.8) | 8 (25.0) | >0.999 | 33 (27.7) | 9 (22.5) | 0.679 |
| Prior TBI, n (%) | 57 (46.3) | 14 (51.9) | 0.667 | 54 (46.6) | 17 (50.0) | 0.599 |
| APOE ɛ4 genotype, n (%) | 0.653 | 0.273 | ||||
| 0 APOE ɛ4 alleles | 83 (72.8) | 18 (66.7) | 80 (74.1) | 21 (63.6) | ||
| 1 or 2 APOE ɛ4 alleles | 31 (27.2) | 9 (33.3) | 28 (25.9) | 12 (36.4) | ||
| TBI Severity, n (%) | 0.541 | 0.177 | ||||
| Mild (GCS 13-15) | 99 (79.2) | 24 (77.4) | 89 (76.1) | 34 (87.2) | ||
| Moderate/severe (GCS 3-12) | 26 (20.8) | 7 (22.6) | 28 (23.9) | 5 (12.8) | ||
| Injury cause, n (%) | 0.045 | 0.313 | ||||
| Car accident (occupant) | 31 (24.4) | 7 (22.6) | 28 (23.5) | 10 (25.6) | ||
| Car accident (bicyclist/pedestrian) | 27 (21.3) | 4 (12.9) | 23 (19.3) | 8 (20.5) | ||
| Motorcycle accident | 6 (4.7) | 0 (0.0) | 6 (5.0) | 0 (0.0) | ||
| Fall | 48 (37.8) | 9 (29.0) | 46 (38.7) | 11 (28.2) | ||
| Assault | 12 (9.5) | 10 (32.3) | 13 (10.9) | 9 (23.1) | ||
| Other | 3 (2.4) | 1 (3.2) | 3 (2.5) | 1 (2.6) | ||
| Loss of consciousness, n (%) | 83 (66.4) | 18 (56.3) | 0.280 | 77 (65.8) | 24 (60.0) | 0.788 |
| Post-traumatic amnesia, n (%) | 68 (54.4) | 17 (53.1) | 0.541 | 60 (51.3) | 25 (62.5) | 0.307 |
| Head CT positive for acute intracranial injury, n (%) | 65 (51.2) | 14 (43.8) | 0.554 | 62 (52.1) | 17 (42.5) | 0.361 |
| Injury severity score, mean (SD) | 13.0 (11.5) | 9.6 (9.7) | 0.118 | 13.2 (11.5) | 9.7 (9.9) | 0.082 |
The following variables contained missing data: Hispanic ethnicity (n = 3), education (n = 8), employment (n = 7), APOE ɛ4 genotype (n = 18), TBI severity (n = 3), injury cause (n = 1), loss of consciousness (n = 2), post-traumatic amnesia (n = 2).
GOSE, Glasgow Outcome Scale-Extended; SD, standard deviation; GED, general educational development; TBI, traumatic brain injury; APOE, apolipoprotein; GCS, Glasgow Coma Scale; CT, computed tomography.
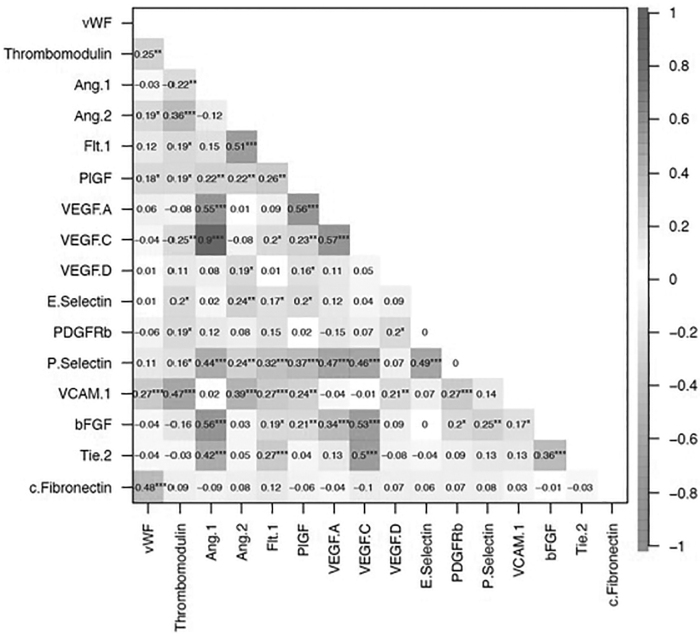
Median (25th-75th percentile) biomarker concentrations are shown overall and stratified by injury severity (GCS 13-15 vs. 3-12) in Supplementary Table S1. Spearman's correlations between microvascular injury biomarkers are shown in Figure 2. The strongest positive Spearman's correlations were seen between VEGF-C and Ang-1 (ρ = 0.90, p < 0.001), VEGF-C and VEGF-A (ρ = 0.57, p < 0.001), VEGF-A and PIGF (ρ = 0.56, p < 0.001), bFGF and Ang-1 (ρ = 0.56, p < 0.001), and VEGF-A and Ang-1 (ρ = 0.55, p < 0.001). The strongest negative Spearman's correlation was between c-Fibronectin and vWF (ρ = -0.48, p < 0.001).
FIG. 2.
Spearman's correlations between microvascular injury–related biomarkers. Note: Sample size for c-Fibronectin is 135; sample size for all other biomarkers is 159. *p < 0.05; **p < 0.01; ***p < 0.001.
Table 3 shows unadjusted and age-adjusted associations of biomarkers with TBI severity. Of the six a priori hypothesized biomarkers, higher levels of vWF were significantly associated with head CT positivity (age-adjusted odds ratio [OR]: 1.62, 95% confidence interval [CI]: 1.04-2.54 per 1 log unit increase); and higher levels of thrombomodulin and Ang-2 were significantly associated with greater injury severity GCS 3-12 versus 13-15 (age-adjusted OR: 4.14, 95% CI: 1.12-15.63 per 1 log unit increase and age-adjusted OR = 3.37, 95% CI = 1.75-6.49 per 1 log unit increase, respectively). Higher levels of Ang-1 and Tie-2 were inversely associated with head CT positivity (age-adjusted OR: 0.63, 95% CI: 0.46-0.85 per 1 log unit increase and age-adjusted OR: 0.26, 95% CI: 0.10-0.72, per 1 log unit increase, respectively). Of the 11 exploratory biomarkers, higher levels of P-selectin were associated with greater injury severity (GCS 3-12) and higher levels of VEGF-C and bFGF were inversely associated with CT positivity (all p < 0.05). After false discovery rate correction for multiple comparisons, higher levels of Ang-2 remained associated with more severe injury and higher levels of Ang-1, Tie2, and bFGF remained associated with less severe injury at a p < 0.05 level. In secondary analyses of the a priori hypothesized biomarkers, higher levels of Ang-1 were inversely associated with contusion, SDH, and SAH presence, higher levels of vWF were associated with SDH and IVH, higher levels of thrombomodulin were associated with SAH and IVH, and higher levels of Ang-2 were associated with SDH and SAH; however, only the association of thrombomodulin with IVH remained significant at p < 0.05 after FDR correction (Table 4). No individual biomarkers were associated with MRI findings in the subset of the population with MRI data.
Table 3.
Associations of Microvascular Injury-Related Biomarkers With Indices of TBI Severity
| GCS 3-12 vs. 13-15 |
Head CT positive vs. negative |
|||||
|---|---|---|---|---|---|---|
| Unadjusted OR (95% CI) | Age Adjusted OR (95% CI) | Unadjusted AUC (95% CI) | Unadjusted OR (95% CI) | Age Adjusted OR (95% CI) | Unadjusted AUC (95% CI) | |
| A priori hypothesized | ||||||
| vWF | 1.31 (0.77, 2.22) | 1.27 (0.75, 2.16) | 0.62 (0.53, 0.70) | 1.75 (1.14, 2.70)* | 1.62 (1.04, 2.54) | 0.60 (0.46, 0.75) |
| Thrombomodulin | 4.16 (1.25, 13.86) | 4.14 (1.12, 15.63) | 0.56 (0.47, 0.65) | 1.80 (0.72, 4.54) | 1.17 (0.44, 3.12) | 0.59 (0.44, 0.74) |
| Ang-1 | 0.84 (0.60, 1.19) | 0.85 (0.60, 1.21) | 0.66 (0.57, 0.75) | 0.61 (0.45, 0.83)* | 0.63 (0.46, 0.85)* | 0.59 (0.44, 0.73) |
| Ang-2 | 3.37 (1.75, 6.49)* | 3.39 (1.73, 6.66)* | 0.59 (0.50, 0.68) | 1.44 (0.88, 2.35) | 1.22 (0.73, 2.03) | 0.68 (0.55, 0.82) |
| c-Fibronectin | 1.91 (0.80, 4.57) | 1.91 (0.79, 4.59) | 0.53 (0.43, 0.63) | 1.26 (0.63, 2.52) | 1.25 (0.61, 2.57) | 0.59 (0.42, 0.77) |
| Tie-2 | 0.44 (0.17, 1.14) | 0.47 (0.18, 1.27) | 0.62 (0.50, 0.73) | 0.21 (0.08, 0.55)* | 0.26 (0.10, 0.72)* | 0.55 (0.40, 0.69) |
| Exploratory | ||||||
| Flt-1 | 1.93 (1.01, 3.72) | 1.88 (0.97, 3.61) | 0.54 (0.45, 0.63) | 1.15 (0.68, 1.96) | 1.08 (0.62, 1.87) | 0.50 (0.35, 0.64) |
| PIGF | 1.40 (0.64, 3.30) | 1.33 (0.60, 2.92) | 0.58 (0.49, 0.67) | 1.41 (0.74, 2.68) | 1.21 (0.63, 2.33) | 0.59 (0.45, 0.73) |
| VEGF-A | 1.23 (0.86, 1.75) | 1.22 (0.86, 1.74) | 0.55 (0.46, 0.67) | 1.18 (0.89, 1.57) | 1.16 (0.86, 1.56) | 0.58 (0.43, 0.73) |
| VEGF-C | 0.88 (0.56, 1.40) | 0.91 (0.57, 1.46) | 0.55 (0.43, 0.66) | 0.62 (0.42, 0.91)* | 0.66 (0.44, 0.98) | 0.58 (0.44, 0.73) |
| VEGF-D | 0.90 (0.43, 1.88) | 0.83 (0.40, 1.76) | 0.53 (0.41, 0.65) | 1.17 (0.65, 2.12) | 0.95 (0.50, 1.77) | 0.65 (0.51, 0.79) |
| E-Selectin | 1.91 (0.95, 3.86) | 1.97 (0.97, 4.02) | 0.59 (0.48, 0.70) | 1.05 (0.61, 1.80) | 1.09 (0.62, 1.91) | 0.52 (0.37, 0.67) |
| PDGFR-ß | 0.92 (0.50, 1.69) | 0.89 (0.48, 1.64) | 0.54 (0.44, 0.64) | 0.97 (0.59, 1.59) | 0.89 (0.53, 1.48) | 0.54 (0.40, 0.69) |
| P-Selectin | 2.33 (1.11, 4.89) | 2.38 (1.12, 5.04) | 0.65 (0.55, 0.75) | 1.42 (0.78, 2.58) | 1.47 (0.79, 2.71) | 0.47 (0.32, 0.61) |
| VCAM-1 | 1.14 (0.42, 3.12) | 1.03 (0.38, 2.82) | 0.53 (0.42, 0.63) | 0.59 (0.25, 1.39) | 0.36 (0.13, 0.98) | 0.56 (0.42, 0.71) |
| bFGF | 0.84 (0.61, 1.15) | 0.83 (0.61, 1.15) | 0.58 (0.47, 0.69) | 0.62 (0.47, 0.82)* | 0.61 (0.46, 0.81)* | 0.62 (0.47, 0.76) |
| Principal Components Analysis | ||||||
| PC1 | 0.85 (0.54, 1.27) | 0.83 (0.53, 1.25) | 0.77 (0.66, 0.88) | 0.60 (0.42, 0.84) | 0.63 (0.44, 0.88) | 0.73 (0.59, 0.86) |
| PC2 | 2.19 (1.47, 3.42) | 2.29 (1.49, 3.69) | 1.75 (1.25, 2.51) | 1.59 (1.11, 2.32) | ||
Denotes associations with individual biomarkers where p < 0.05 after false discovery rate (FDR) correction for multiple comparisons.
Bolded data represents p < 0.05.
Sample size for c-Fibronectin is 135; sample size for all other biomarkers and principal components analysis is 159. OR is expressed per 1 log unit increase in each biomarker or 1 unit PC score. Individual models were run for each biomarker separately. PC1 and PC2 were included in the same model.
TBI, traumatic brain injury; GCS, Glasgow Coma Scale; CT, computed tomography; OR, odds ratio; CI, confidence interval; AUC, area under the curve; vWF, von Willebrand factor; Ang, angiopoietin; PIGF, placental growth factor; VEGF, vascular endothelial growth factor; PDGFR, platelet-derived growth factor receptor; VCAM-1, vascular cell adhesion protein 1; bFGF, basic fibroblast growth factor; PC, principal component.
Table 4.
Unadjusted Associations (OR [95% CIs]) of Microvascular Injury-Related Biomarkers With Vascular-Related CT and MRI Findings
| CT contusion (yes vs. no) | CT subdural hemorrhage (yes vs. no) | CT subarachnoid hemorrhage (yes vs. no) | CT intraventricular hemorrhage (yes vs. no) | CT diffuse axonal injury (yes vs. no) | MRI positive vs. negative | MRI microhemorrhage (yes vs. no) | |
|---|---|---|---|---|---|---|---|
| A priori hypothesized | |||||||
| vWF | 1.21 (0.72, 2.02) | 2.20 (1.29, 3.75) | 1.44 (0.92, 2.26) | 2.89 (1.05, 7.95) | 1.13 (0.54, 2.34) | 1.55 (0.84, 2.84) | 1.16 (0.65, 2.08) |
| Thrombomodulin | 0.98 (0.33, 2.95) | 2.09 (0.72, 6.06) | 3.38 (1.18, 9.65) | 42.72 (4.61, 396.20)* | 0.86 (0.18, 4.08) | 1.75 (0.47, 6.57) | 1.36 (0.37, 5.04) |
| Ang-1 | 0.67 (0.47, 0.96) | 0.72 (0.52, 1.00) | 0.73 (0.54, 1.00) | 0.70 (0.39, 1.25) | 1.04 (0.63, 1.71) | 0.78 (0.50, 1.22) | 0.70 (0.44, 1.10) |
| Ang-2 | 1.57 (0.87, 2.84) | 1.76 (1.01, 3.09) | 1.89 (1.11, 3.23) | 1.96 (0.76, 5.08) | 0.32 (0.13, 0.82) | 2.20 (0.94, 5.16) | 1.39 (0.63, 3.06) |
| c-Fibronectin | 1.11 (0.49, 2.55) | 0.68 (0.32, 1.46) | 1.59 (0.76, 3.33) | 0.79 (0.20, 3.10) | 1.25 (0.38, 4.12) | 1.88 (0.65, 5.44) | 2.13 (0.72, 6.27) |
| Tie-2 | 0.43 (0.17, 1.10) | 0.55 (0.23, 1.32) | 0.63 (0.27, 1.43) | 0.39 (0.11, 1.42) | 0.46 (0.14, 1.48) | 0.57 (0.15, 2.10) | 0.46 (0.13, 1.68) |
| Exploratory | |||||||
| Flt-1 | 1.39 (0.73, 2.65) | 1.32 (0.72, 2.42) | 1.84 (1.02, 3.23) | 0.81 (0.27, 2.46) | 0.45 (0.17, 1.19) | 0.86 (0.37, 2.01) | 0.73 (0.31, 1.71) |
| PIGF | 1.07 (0.49, 2.33) | 1.53 (0.74, 3.18) | 1.76 (0.88, 3.52) | 2.14 (0.62, 7.32) | 0.88 (0.29, 2.69) | 1.73 (0.67, 4.47) | 1.69 (0.67, 4.26) |
| VEGF-A | 1.32 (0.93, 1.89) | 1.07 (0.77, 1.48) | 1.20 (0.89, 1.63) | 1.21 (0.67, 2.17) | 1.25 (0.75, 2.06) | 1.26 (0.81, 1.97) | 1.04 (0.68, 1.60) |
| VEGF-C | 0.76 (0.47, 1.22) | 0.74 (0.47, 1.14) | 0.77 (0.51, 1.16) | 0.33 (0.12, 0.92) | 1.07 (0.56, 2.05) | 0.78 (0.45, 1.37) | 0.55 (0.29, 1.01) |
| VEGF-D | 0.73 (0.34, 1.56) | 0.80 (0.40, 1.60) | 1.44 (0.77, 2.69) | 3.02 (1.08, 8.39) | 1.16 (0.42, 3.19) | 2.75 (0.87, 8.74) | 2.90 (0.91, 9.22) |
| E-Selectin | 1.60 (0.80, 3.19) | 1.12 (0.60, 2.09) | 1.70 (0.93, 3.11) | 3.03 (0.91, 10.16) | 0.84 (0.32, 2.16) | 1.04 (0.45, 2.36) | 0.85 (0.38, 1.93) |
| PDGFR-ß | 0.86 (0.47, 1.57) | 1.21 (0.68, 2.14) | 1.13 (0.66, 1.91) | 1.17 (0.42, 3.30) | 0.82 (0.35, 1.94) | 0.80 (0.36, 1.79) | 0.66 (0.30, 1.46) |
| P-Selectin | 1.71 (0.84, 3.49) | 1.20 (0.62, 2.33) | 2.24 (1.15, 4.36) | 2.23 (0.76, 6.58) | 0.70 (0.25, 1.99) | 1.37 (0.59, 3.18) | 0.80 (0.35, 1.81) |
| VCAM-1 | 0.53 (0.19, 1.50) | 0.97 (0.38, 2.48) | 1.45 (0.60, 3.52) | 0.55 (0.30, 0.99) | 0.49 (0.35, 0.94) | 1.03 (0.28, 3.79) | 0.69 (0.19, 2.54) |
| bFGF | 0.73 (0.53, 1.01) | 0.75 (0.55, 1.00) | 0.82 (0.63, 1.07) | 0.39 (0.11, 1.42) | 0.46 (0.14, 1.48) | 0.74 (0.49, 1.11) | 0.65 (0.13, 1.68) |
| Principal Components Analysis | |||||||
| PC1 | 0.75 (0.48, 1.13) | 0.75 (0.50, 1.08) | 0.80 (0.56, 1.14) | 0.36 (0.10, 0.91) | 0.75 (0.40, 1.32) | 0.67 (0.39, 1.11) | 0.49 (0.25, 0.85) |
| PC2 | 1.43 (1.98, 2.10) | 1.55 (1.08, 2.25) | 2.00 (1.40, 2.95) | 2.40 (1.26, 4.96) | 0.75 (0.41, 1.29) | 1.95 (1.12, 3.72) | 1.74 (1.03, 3.20) |
Denotes associations with individual biomarkers where p < 0.05 after false discovery rate correction for multiple comparisons.
Bolded data represents p < 0.05.
Sample size for c-Fibronectin is 135 and sample size for all other biomarkers and principal components analysis is 159 for CT Findings. Sample size for c-Fibronectin is 53 and sample size for all other biomarkers and principal components analysis is 123 for MRI Findings. OR is expressed per 1 log unit increase in each biomarker or 1 unit PC score. Individual models were run for each biomarker separately. PC1 and PC2 were included in the same model.
OR, odds ratio; CI, confidence interval; MRI, magnetic resonance imaging; vWF, von Willebrand factor; Ang, angiopoietin; PIGF, placental growth factor; VEGF, vascular endothelial growth factor; PDGFR, platelet-derived growth factor receptor; VCAM-1, vascular cell adhesion protein 1; bFGF, basic fibroblast growth factor; PC, principal component.
In unadjusted models, of the a priori hypothesized biomarkers, higher levels of vWF, thrombomodulin, and Ang-2 were associated with worse global functional outcome (GOSE 1-4 vs, 5-8) at 3-months (all p < 0.05). Associations were attenuated and no longer significant after adjustment for age and injury severity (GCS; Table 5). Of the exploratory biomarkers, higher levels of VEGF-D and VCAM-1 were associated with worse 3-month GOSE in unadjusted models, but these associations were no longer significant after adjusting for age and GCS. There were no significant associations of any biomarkers with 6-month GOSE. Individual biomarkers were largely not significantly correlated with 6-month symptom outcomes or cognitive outcomes (Table 6).
Table 5.
Associations of Microvascular Injury-Related Biomarkers With 3- and 6-Month GOSE
| 3-Month GOSE 1-4 vs. 5-8 |
6-Month GOSE 1-4 vs. 5-8 |
|||||
|---|---|---|---|---|---|---|
| Unadjusted OR (95% CI) | Age and GCS Adjusted OR (95% CI) | Unadjusted AUC (95% CI) | Unadjusted OR (95% CI) | Age and GCS Adjusted OR (95% CI) | Unadjusted AUC (95% CI) | |
| A priori hypothesized | ||||||
| vWF | 2.27 (1.17, 4.39) | 1.80 (0.83, 3.90) | 0.69 (0.57, 0.81) | 1.82 (0.90, 3.69) | 1.37 (0.60, 3.10) | 0.65 (0.50, 0.80) |
| Thrombomodulin | 6.84 (1.66, 28.25) | 2.24 (0.46, 10.87) | 0.70 (0.58, 0.82) | 2.01 (0.48, 8.41) | 0.49 (0.10, 2.39) | 0.59 (0.46, 0.72) |
| Ang-1 | 0.70 (0.46, 1.05) | 0.89 (0.54, 1.49) | 0.64 (0.52, 0.75) | 0.74 (0.47, 1.15) | 0.94 (0.54, 1.65) | 0.61 (0.47, 0.75) |
| Ang-2 | 2.79 (1.30, 6.00) | 0.79 (0.33, 1.90) | 0.67 (0.54, 0.80) | 2.12 (0.97, 4.61) | 0.57 (0.22, 1.47) | 0.66 (0.50, 0.82) |
| c-Fibronectin | 0.87 (0.32, 2.31) | 0.53 (0.13, 2.18) | 0.53 (0.39, 0.67) | 1.02 (0.32, 3.27) | 0.82 (0.15, 4.50) | 0.50 (0.34, 0.66) |
| Tie-2 | 0.38 (0.14, 1.08) | 1.62 (0.41, 6.35) | 0.65 (0.52, 0.79) | 0.38 (0.13, 1.17) | 1.85 (0.31, 11.07) | 0.68 (0.52, 0.83) |
| Exploratory | ||||||
| Flt-1 | 1.63 (0.76, 3.48) | 0.99 (0.44, 2.22) | 0.60 (0.45, 0.75) | 1.57 (0.72, 3.43) | 0.98 (0.40, 2.44) | 0.60 (0.44, 0.77) |
| PIGF | 2.59 (0.98, 6.86) | 2.17 (0.63, 7.51) | 0.64 (0.51, 0.77) | 1.94 (0.72, 5.26) | 1.21 (0.33, 4.41) | 0.60 (0.45, 0.75) |
| VEGF-A | 1.39 (0.91, 2.14) | 1.56 (0.90, 2.71) | 0.58 (0.46, 0.70) | 1.10 (0.69, 1.76) | 1.00 (0.56, 1.81) | 0.50 (0.36, 0.64) |
| VEGF-C | 0.61 (0.34, 1.11) | 0.93 (0.46, 1.89) | 0.61 (0.48, 0.73) | 0.65 (0.35, 1.24) | 0.98 (0.47, 2.06) | 0.59 (0.45, 0.74) |
| VEGF-D | 3.12 (1.26, 7.70) | 2.20 (0.82, 5.91) | 0.62 (0.48, 0.76) | 2.24 (0.88, 5.76) | 1.78 (0.63, 5.09) | 0.58 (0.42, 0.74) |
| E-Selectin | 2.21 (0.95, 5.15) | 3.53 (1.15, 10.82) | 0.62 (0.50, 0.75) | 1.38 (0.58, 3.32) | 1.34 (0.44, 4.07) | 0.55 (0.40, 0.69) |
| PDGFR-ß | 2.09 (0.92, 4.79) | 1.79 (0.63, 5.12) | 0.60 (0.47, 0.73) | 1.08 (0.46, 2.50) | 0.70 (0.23, 2.15) | 0.51 (0.38, 0.64) |
| P-Selectin | 1.56 (0.68, 3.57) | 1.48 (0.51, 4.31) | 0.59 (0.46, 0.71) | 0.74 (0.30, 1.86) | 0.47 (0.15, 1.44) | 0.52 (0.38, 0.65) |
| VCAM-1 | 4.33 (1.26, 14.83) | 3.19 (0.79, 12.95) | 0.66 (0.53, 0.78) | 2.75 (0.64, 11.81) | 1.82 (0.27, 12.40) | 0.61 (0.47, 0.74) |
| bFGF | 0.99 (0.69, 1.42) | 1.24 (0.75, 2.05) | 0.51 (0.39, 0.63) | 0.83 (0.55, 1.25) | 1.02 (0.60, 1.72) | 0.56 (0.41, 0.71) |
| Principal Components Analysis | ||||||
| PC1 | 0.73 (0.40, 1.22) | 0.99 (0.51, 1.81) | 0.77 (0.66, 0.88) | 0.66 (0.34, 1.45) | 0.97 (0.48, 1.81) | 0.73 (0.59, 0.86) |
| PC2 | 2.53 (1.55, 4.38) | 1.56 (0.85, 2.98) | 1.90 (1.16, 3.25) | 0.97 (0.49, 1.91) | ||
Sample size for c-Fibronectin is 135; sample size for all other biomarkers and principal components analysis is 159. OR is expressed per 1 log unit increase in each biomarker or 1 unit increase in PC score. Individual models were run for each biomarker separately. PC1 and PC2 were included in the same model. Bolded data represents p < 0.05.
No associations with individual biomarkers had p < 0.05 after false discovery rate correction for multiple comparisons.
GOSE, Glasgow Outcome Scale-Extended; OR, odds ratio; CI, confidence interval; GCS, Glasgow Coma Scale; AUC, area under the curve; vWF, von Willebrand factor; Ang, angiopoietin; PIGF, placental growth factor; VEGF, vascular endothelial growth factor; PDGFR, platelet-derived growth factor receptor; VCAM-1, vascular cell adhesion protein 1; bFGF, basic fibroblast growth factor; PC, principal component.
Table 6.
Spearman's Correlations of Microvascular Injury-Related Biomarkers With Secondary 6-Month Outcomes
| RPQ | SWLS | BSI-18 | CVLT | TMT-A | TMT-B | WAIS-PSI | |
|---|---|---|---|---|---|---|---|
| A priori hypothesized | |||||||
| vWF | -0.08 | 0.00 | 0.00 | 0.10 | -0.05 | -0.09 | 0.12 |
| Thrombomodulin | 0.08 | 0.06 | -0.03 | -0.03 | -0.01 | 0.14 | 0.00 |
| Ang-1 | -0.06 | 0.04 | -0.06 | 0.00 | 0.13 | 0.08 | 0.03 |
| Ang-2 | -0.14 | 0.07 | -0.21 | -0.08 | 0.15 | 0.18 | -0.06 |
| c-Fibronectin | 0.05 | 0.09 | -0.07 | -0.25* | -0.05 | -0.01 | -0.05 |
| Tie-2 | -0.04 | 0.06 | 0.04 | 0.05 | 0.12 | 0.11 | 0.00 |
| Exploratory | |||||||
| Flt-1 | -0.02 | 0.04 | -0.03 | -0.04 | 0.08 | 0.11 | 0.01 |
| PIGF | -0.03 | 0.04 | -0.13 | 0.09 | 0.01 | 0.07 | 0.18 |
| VEGF-A | -0.05 | 0.02 | -0.04 | 0.03 | 0.08 | 0.05 | 0.17 |
| VEGF-C | -0.02 | 0.00 | 0.03 | -0.02 | 0.16 | 0.09 | 0.05 |
| VEGF-D | 0.08 | -0.13 | 0.06 | 0.04 | 0.06 | 0.02 | 0.07 |
| E-Selectin | 0.16 | -0.07 | 0.12 | -0.19 | -0.06 | 0.14 | -0.11 |
| PDGFR-ß | 0.03 | -0.04 | -0.08 | 0.03 | -0.11 | -0.06 | 0.08 |
| P-Selectin | 0.00 | 0.11 | -0.16 | -0.21 | 0.04 | 0.09 | 0.04 |
| VCAM-1 | 0.11 | -0.14 | -0.01 | 0.03 | 0.13 | 0.13 | 0.03 |
| bFGF | -0.04 | 0.16 | -0.09 | 0.04 | 0.14 | 0.08 | 0.16 |
| Principal Components Analysis | |||||||
| PC1 | -0.04 | 0.04 | 0.00 | -0.01 | 0.12 | 0.05 | 0.10 |
| PC2 | 0.01 | 0.00 | -0.01 | -0.14 | 0.08 | 0.20 | -0.06 |
p < 0.05
Sample size for c-Fibronectin is 135; sample size for all other biomarkers and principal components analysis is 159.
RPQ, Rivermead Post-Concussion Symptoms Questionnaire; SWLS, Satisfaction with Life Scale; BSI-18, 18-Item Brief Symptom Inventory; CVLT, California Verbal Learning Test; TMT, Trail Making Test; WAIS-PSI, Processing Speed Index from the Wechsler Adult Intelligence Scale, 4th Edition; vWF, von Willebrand factor; Ang, angiopoietin; PIGF, placental growth factor; VEGF, vascular endothelial growth factor; PDGFR, platelet-derived growth factor receptor; VCAM-1, vascular cell adhesion protein 1; bFGF, basic fibroblast growth factor; PC, principal component.
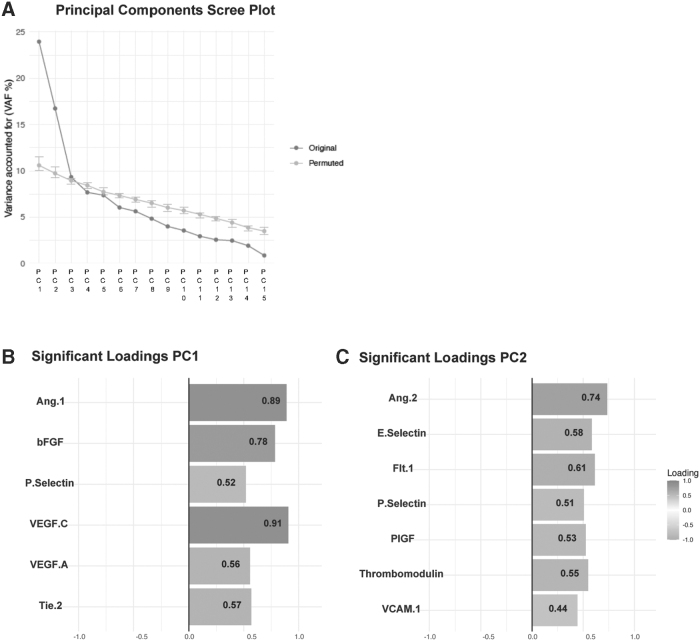
Using non-linear principal components analyses, the variance was partitioned into orthogonal components, with PC1 accounting for 23.9% of the variance and PC2 accounting for 16.7% of the variance (Fig. 3). Higher PC1 score was associated with less severe injury (age-adjusted OR per 1 unit increase in PC score: 0.63, 95% CI: 0.44-0.88 for CT positivity). In contrast, higher PC2 score was associated with greater injury severity (age-adjusted OR per 1 unit increase in PC score: 1.59, 95% CI: 1.11-2.32 for CT positivity and age-adjusted OR per 1 unit increase in PC score: 2.29, 95% CI: 1.49-3.69 for GCS 3-12 vs. 13-15; Table 3). Higher PC2 score was associated with the presence of SDH, SAH, and IVH on CT and with MRI positivity (unadjusted OR per 1 unit increase in PC score: 1.95, 95% CI 1.12-3.72), and higher PC1 score was inversely associated with the presence of microhemorrhages on brain MRI (unadjusted OR per 1 unit increase in PC score: 0.49, 95% CI: 0.25-0.85; Table 4). Higher PC2 score was also associated with worse functional outcome (GOSE 1-4 vs. 5-8) at 3 months in unadjusted models (OR per 1 unit increase in PC score: 2.53, 95% CI: 1.55-4.38) and at 6 months (OR per 1 unit increase in PC score: 1.90, 95% CI: 1.16-3.25) but these associations were no longer significant after adjusting for age and injury severity. PC1 was not associated with 3- or 6-month GOSE-TBI (Table 5) and PC1 and PC2 were not associated with 6-month symptom outcomes or cognitive outcomes (Table 6).
FIG. 3.
Principal components analysis. (A) Scree plot showing variance accounted for by each orthogonal component. The first principal component (PC1) accounted for 23.9% of the variance and the second principal component (PC2) accounted for 16.7% of the variance. (B) The subset of vascular biomarkers included in PC1 that were significant (p < 0.05) after permutation testing. (C) The subset of vascular biomarkers included in PC2 that were significant (p < 0.05) after permutation testing.
Discussion
In this level 1 trauma-center-based population of acute TBI patients enrolled in the TRACK-TBI Pilot Study, we found associations between biomarkers of microvascular injury with TBI severity. Further work investigating vascular function-related biomarkers in larger sample sizes of TBI patients is warranted to better characterize the endophenotype of traumatic cerebral microvascular injury and to examine potential links between injury endophenotype and injury severity and outcomes.
Biomarkers have the potential to contribute to multiple facets of clinical care in TBI, including diagnostic, prognostic, mechanistic, and monitoring response to therapeutics, among others.29,30 The microvascular injury-related biomarkers evaluated in the present study best represent mechanistic biomarkers.31 Consistent with categorization, we found more robust associations between microvascular injury-related biomarkers with indices of TBI severity (GCS and head CT) compared with indices of TBI outcomes. We also found that our principal components analysis provided more robust results than the results of associations of individual a priori hypothesized and exploratory biomarkers with severity and outcomes.
Although the number of individual biomarkers entered into the principal components analysis was relatively small, two subsets of biomarkers loaded highly together in PC1 and PC2 and reflect distinct domains, both with biologic plausibility. After a TBI event, there is a complex interplay between various forms of primary and secondary vascular injury (such as hemorrhage, edema, altered cerebral blood flow, blood brain barrier disruption, coagulopathy, and chronic inflammation) and subsequent vascular repair (angiogenesis [formation of new blood vessels from existing vasculature], vasculogenesis [de novo formation of new blood vessels], and repair of the blood–brain barrier).32 Indeed, in our principal components analysis, two clusters of biomarkers emerged. The first cluster (PC1) contained the biomarkers Ang-1, bFGF, P-Selectin, VEGF-C, VEGF-A, and Tie2. Several of the biomarkers in this cluster have previously been studied in the context of human and animal studies of TBI and have been associated with various mechanisms of vascular repair and neuroprotection after injury.33,34 Specifically, increased expression of Ang-1 and Tie-2 in the acute post-injury period have been shown to prevent vascular leakage and promote blood brain barrier integrity11,12 and increased VEGF-C has been shown to induce alternative activation of microglia, promoting TBI recovery.35
The second cluster (PC2) was comprised of the biomarkers of Ang-2, E-Selectin, Flt-1, P-Selectin, PIGF, Thrombomodulin, and VCAM-1. Higher levels of Ang-2 have been associated with greater injury severity and with blood-brain barrier breakdown and endothelial apoptosis after TBI.12 Several other biomarkers of endothelial injury in this cluster have been found to elevated in acute TBI (E-selectin, VCAM-1, Flt-1, Thrombomodulin).36-38 Taken together, the biomarkers loading on PC1 are generally considered to be vaso-protective and pro-angiogenic, while biomarkers loading on PC2 are generally considered to reflect vaso-toxic and inflammatory responses that contribute to breakdown of the blood–brain barrier.
Three of the biomarkers were not included in the clusters identified in our principal components analysis (vWF, VEGF-D, and PDGFR-ß). Similar to prior work, we found that vWF was associated with greater head CT severity.9,39 Several studies have investigated PDGFR-ß measured in the cerebral spinal fluid as a marker of pericyte injury and blood–brain barrier breakdown in populations with dementia,40,41 but this biomarker is less well understood in relation to TBI and in blood-based assays. Similarly, the potential role of VEGF-D is less well characterized in TBI and other neurologic diseases compared with diseases of other systems.42 It is important to note that all biomarkers investigated in the present analyses may not be solely biomarkers of microvascular injury; indeed, many have also been shown to be associated with inflammation or other biological processes.
The present analysis within the TRACK-TBI Pilot Study population showed the feasibility of measuring and analyzing a panel of microvascular function-related biomarkers using both hypothesis-driven and data-driven methods, but we recognize several limitations. The results from this pilot study are hypothesis-generating and require replication in larger samples of TBI patients that are diverse in terms of sex and race/ethnicity. Our sample is small relative to the number of associations tested, and although some associations with injury severity remained statistically significant after applying FDR correction for multiple comparisons, no associations with outcomes remained statistically significant. In addition, given limited sample volume, samples were measured in singlicate in order to maximize the number of assays run; however, this method is increasingly used and accepted in the field.17
Our population consisted of relatively milder injuries. Eighty percent had GCS score between 13 to 15, but it is important to note that our “mild” population is likely comprised of more severe injuries than other “mild” TBI populations, given that they presented to a level 1 trauma center emergency department for evaluation and 50% had acute TBI-related intracranial abnormalities on CT scan. Additionally, the microvascular-related biomarkers were only measured in a subset of the TRACK-TBI Pilot Study population; however, participants with and without biomarker and with and without outcome data were similar in terms of key demographic and injury-related variables. Further, only a subset (n = 33) of included participants had data on hours from injury to blood collection, so we were unable to investigate associations of time between injury and blood collection with biomarker levels, but all participants had blood collected within 24 h of injury.
In conclusion, in this population of acute TBI patients presenting to a level 1 trauma center, biomarkers of microvascular injury were associated with TBI severity. The results of this study require further replication in larger studies but indicate that biomarkers associated with traumatic cerebral microvascular injury and vascular repair after TBI can be measured peripherally and are associated with injury severity.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the TRACK-TBI Pilot Study for their important contributions.
Contributor Information
Collaborators: the TRACK-TBI Study Investigators
TRACK-TBI Investigators
Neeraj Badjatia, MD, University of Maryland, College Park, MD; Ann-Christine Duhaime, MD, MassGeneral Hospital for Children, Boston, MA; Adam R. Ferguson, PhD, University of California, San Francisco, San Francisco, CA; Brandon Foreman, MD, University of Cincinnati, Cincinnati, OH; Shankar Gopinath, MD, Baylor College of Medicine, Houston, TX; Ramesh Grandhi, MD MS, University of Utah, Salt Lake City, Utah; Ruchira Jha, MD, MSc, Barrow Neurological Institute, Phoenix, AZ; Frederick K. Korley, MD, PhD, University of Michigan, Ann Arbor, MI; Christopher Madden, MD, UT Southwestern, Dallas, TX; Michael McCrea, PhD, Medical College of Wisconsin Milwaukee, WI; Randall Merchant, PhD, Virginia Commonwealth University, Richmond, VA; Pratik Mukherjee, MD PhD, University of California, San Francisco, San Francisco, CA; Laura B. Ngwenya, MD, PhD, University of Cincinnati, Cincinnati, OH; David Okonkwo, MD PhD, University of Pittsburgh, Pittsburgh, PA; Ava Puccio, PhD, University of Pittsburgh, Pittsburgh, PA; Claudia Robertson, MD, Baylor College of Medicine, Houston, TX; Richard B Rodgers, MD, Goodman Campbell Brain and Spine, Carmel, IN; David Schnyer, PhD, UT Austin, Austin, Tx; Sabrina R. Taylor, PhD, University of California, San Francisco, San Francisco, CA; Mary Vassar, RN MS, University of California, San Francisco, San Francisco, CA; Kevin Wang, PhD, University of Florida, Gainesville, FL; Ross Zafonte, DO, Harvard Medical School, Boston, MA.
Authors' Contributions
Andrea L.C. Schneider, MD, PhD: Conceptualization; writing—original draft.
J. Russell Huie, PhD: Conceptualization; Formal analysis; Writing—Review and Editing.
Sonia Jain, PhD: Conceptualization; formal analysis; writing—review and editing.
Xiaoying Sun, MS: Conceptualization; formal analysis; writing—review and editing.
Adam R. Ferguson, PhD: Conceptualization; writing—review and editing.
Cillian Lynch, PhD: Conceptualization; writing—review and editing.
John K. Yue, MD: Conceptualization; writing—review and editing.
Geoffrey T. Manley, MD, PhD: Conceptualization; writing—review and editing; resources; supervision; funding acquisition.
Kevin K.W. Wang, PhD: Conceptualization; writing—review and editing.
Danielle K. Sandsmark, MD, PhD: Conceptualization; writing—review and editing.
Christopher Campbell, MD, PhD: Resources; writing—review and editing.
Ramon Diaz-Arrastia, MD, PhD: Conceptualization; writing—review and editing; supervision; funding acquisition.
Funding Information
The TRACK-TBI Study was funded by National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) grants RC2NS069409 and U01NS086090 and by the Department of Defense grant W81XWH-14-0176. Biomarker testing was supported by the Office of Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Alzheimer's Research Program, Convergence Science Research Award under Award No. W81XWH-17-1-0648. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. Dr. Diaz-Arrastia is supported by NIH/NINDS grant U01NS114140. Dr. Schneider is supported by NIH/NINDS grant K23NS123340.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Schneider ALC, Wang D, Ling G, et al. Prevalence of self-reported head injury in the United States. N Engl J Med 2018;379(12):1176–1178; doi: 10.1056/NEJMc1808550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teasdale G, Jennett B. Assessment of coma and impaired consciousness. A practical scale. Lancet 1974;2(7872):81–84; doi: 10.1016/s0140-6736(74)91639-0 [DOI] [PubMed] [Google Scholar]
- 3. Management of Concussion/mTBI Working Group. VA/DoD Clinical Practice Guideline for Management of Concussion/Mild Traumatic Brain Injury. J Rehabil Res Dev 2009;46(6):CP1–CP68. [PubMed] [Google Scholar]
- 4. Ojo JO, Mouzon B, Algamal M, et al. Chronic repetitive mild traumatic brain injury results in reduced cerebral blood flow, axonal injury, gliosis, and increased T-tau and tau oligomers. J Neuropathol Exp Neurol 2016;75(7):636–655; doi: 10.1093/jnen/nlw035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jassam YN, Izzy S, Whalen M, et al. Neuroimmunology of traumatic brain injury: time for a paradigm shift. Neuron 2017;95(6):1246–1265; doi: 10.1016/j.neuron.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sandsmark DK, Bashir A, Wellington CL, et al. Cerebral microvascular injury: a potentially treatable endophenotype of traumatic brain injury-induced neurodegeneration. Neuron 2019;103(3):367–379; doi: 10.1016/j.neuron.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Logsdon AF, Lucke-Wold BP, Turner RC, et al. Role of microvascular disruption in brain damage from traumatic brain injury. Compr Physiol 2015;5(3):1147–1160; doi: 10.1002/cphy.c140057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hay JR, Johnson VE, Young AM, et al. Blood-brain barrier disruption is an early event that may persist for many tears after traumatic brain injury in humans. J Neuropathol Exp Neurol 2015;74(12):1147–1157; doi: 10.1097/NEN.0000000000000261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandsmark DK, Bogoslovsky T, Qu BX, et al. Changes in plasma von Willebrand factor and cellular fibronectin in MRI-defined traumatic microvascular injury. Front Neurol 2019;10:246; doi: 10.3389/fneur.2019.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Albert V, Subramanian A, Agrawal D, et al. Acute traumatic endotheliopathy in isolated severe brain injury and its impact on clinical outcome. Med Sci (Basel) 2018;6(1); doi: 10.3390/medsci6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brickler TR, Hazy A, Guilhaume Correa F, et al. Angiopoietin/tie2 axis regulates the age-at-injury cerebrovascular response to traumatic brain injury. J Neurosci 2018;38(45):9618–9634; doi: 10.1523/JNEUROSCI.0914-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chittiboina P, Ganta V, Monceaux CP, et al. Angiopoietins as promising biomarkers and potential therapeutic targets in brain injury. Pathophysiology 2013;20(1):15–21; doi: 10.1016/j.pathophys.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 13. Huie JR, Diaz-Arrastia R, Yue JK, et al. Testing a multivariate proteomic panel for traumatic brain injury biomarker discovery: A TRACK-TBI Pilot Study. J Neurotrauma 2019;36(1):100–110; doi: 10.1089/neu.2017.5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yue JK, Vassar MJ, Lingsma HF, et al. Transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the common data elements for traumatic brain injury. J Neurotrauma 2013;30(22):1831–1844; doi: 10.1089/neu.2013.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manley GT, Diaz-Arrastia R, Brophy M, et al. Common data elements for traumatic brain injury: recommendations from the biospecimens and biomarkers working group. Arch Phys Med Rehabil 2010;91(11):1667–1672; doi: 10.1016/j.apmr.2010.05.018 [DOI] [PubMed] [Google Scholar]
- 16. Debad JD, Wohlstadter J, Sigal GB. Clinical and biological applications of electrogenerated chemiluminescence. In: Electrogenerated Chemiluminescence. (Bard AJ. ed.) CRC Press: New York, N.Y.; 2004; pp. 359–396. [Google Scholar]
- 17. Ye Z, Tu J, Midde K, et al. Singlicate analysis: should this be the default for biomarker measurements using ligand-binding assays? Bioanalysis 2018;10(12):909–912; doi: 10.4155/bio-2018-0067 [DOI] [PubMed] [Google Scholar]
- 18. Duhaime AC, Gean AD, Haacke EM, et al. Common data elements in radiologic imaging of traumatic brain injury. Arch Phys Med Rehabil 2010;91(11):1661–1666; doi: 10.1016/j.apmr.2010.07.238 [DOI] [PubMed] [Google Scholar]
- 19. Haacke EM, Duhaime AC, Gean AD, et al. Common data elements in radiologic imaging of traumatic brain injury. J Magn Reson Imaging 2010;32(3):516–543; doi: 10.1002/jmri.22259 [DOI] [PubMed] [Google Scholar]
- 20. Jennett B, Snoek J, Bond MR, et al. Disability after severe head injury: observations on the use of the Glasgow Outcome Scale. J Neurol Neurosurg Psychiatry 1981;44(4):285–293; doi: 10.1136/jnnp.44.4.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King NS, Crawford S, Wenden FJ, et al. The Rivermead Post Concussion Symptoms Questionnaire: a measure of symptoms commonly experienced after head injury and its reliability. J Neurol 1995;242(9):587–592; doi: 10.1007/BF00868811 [DOI] [PubMed] [Google Scholar]
- 22. Diener E, Emmons RA, Larsen RJ, et al. The Satisfaction With Life Scale. J Pers Assess 1985;49(1):71–75; doi: 10.1207/s15327752jpa4901_13 [DOI] [PubMed] [Google Scholar]
- 23. Derogatis L. Brief Symptom Inventory 18 (BSI-18): Administration, Scoring, and Procedures Manual. Pearson: Bloomington, MN; 2001. [Google Scholar]
- 24. Delis DC., Kramer JH., Kaplan E, et al. California Verbal Learning Test: 2nd Edition. Psychological Corporation: San Antonio, TX; 2000. [Google Scholar]
- 25. Reitan RM. Validity of the Trail Making Test as an indicator of organic brain damage. Percept Mot Skills 1958;8:271–276. [Google Scholar]
- 26. Wechsler D. Wechsler Adult Intelligence Scale: 4th Edition. Psychological Corporation: San Antonia, TX; 2008. [Google Scholar]
- 27. Torres-Espin A, Chou A, Huie JR, et al. Reproducible analysis of disease space via principal components using the novel R package syndRomics. Elife 2021;10; doi: 10.7554/eLife.61812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R Core Team. R: A language and environment for statistical computing. Vienna, Austria; 2020. Available from: https://www.R-project.org [Last accessed April 24, 2023].
- 29. Gan ZS, Stein SC, Swanson R, et al. Blood biomarkers for traumatic brain injury: a quantitative assessment of diagnostic and prognostic accuracy. Front Neurol 2019;10:446; doi: 10.3389/fneur.2019.00446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang KK, Yang Z, Zhu T, et al. An update on diagnostic and prognostic biomarkers for traumatic brain injury. Expert Rev Mol Diagn 2018;18(2):165–180; doi: 10.1080/14737159.2018.1428089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson WH, Lindstrom TM, Cheung RK, et al. Mechanistic biomarkers for clinical decision making in rheumatic diseases. Nat Rev Rheumatol 2013;9(5):267–276; doi: 10.1038/nrrheum.2013.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salehi A, Zhang JH, Obenaus A. Response of the cerebral vasculature following traumatic brain injury. J Cereb Blood Flow Metab 2017;37(7):2320–2339; doi: 10.1177/0271678X17701460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattson MP, Scheff SW. Endogenous neuroprotection factors and traumatic brain injury: mechanisms of action and implications for therapy. J Neurotrauma 1994;11(1):3–33; doi: 10.1089/neu.1994.11.3 [DOI] [PubMed] [Google Scholar]
- 34. Siddiq I, Park E, Liu E, et al. Treatment of traumatic brain injury using zinc-finger protein gene therapy targeting VEGF-A. J Neurotrauma 2012;29(17):2647–2659; doi: 10.1089/neu.2012.2444 [DOI] [PubMed] [Google Scholar]
- 35. Ju S, Xu C, Wang G, et al. VEGF-C induces alternative activation of microglia to promote recovery from traumatic brain injury. J Alzheimers Dis 2019;68(4):1687–1697; doi: 10.3233/JAD-190063 [DOI] [PubMed] [Google Scholar]
- 36. Balabanov R, Goldman H, Murphy S, et al. Endothelial cell activation following moderate traumatic brain injury. Neurol Res 2001;23(2-3):175–182; doi: 10.1179/016164101101198514 [DOI] [PubMed] [Google Scholar]
- 37. Skold MK, von Gertten C, Sandberg-Nordqvist AC, et al. VEGF and VEGF receptor expression after experimental brain contusion in rat. J Neurotrauma 2005;22(3):353–367; doi: 10.1089/neu.2005.22.353 [DOI] [PubMed] [Google Scholar]
- 38. Yokota H, Naoe Y, Nakabayashi M, et al. Cerebral endothelial injury in severe head injury: the significance of measurements of serum thrombomodulin and the von Willebrand factor. J Neurotrauma 2002;19(9):1007–1015; doi: 10.1089/089771502760341929 [DOI] [PubMed] [Google Scholar]
- 39. De Oliveira CO, Reimer AG, Da Rocha AB, et al. Plasma von Willebrand factor levels correlate with clinical outcome of severe traumatic brain injury. J Neurotrauma 2007;24(8):1331–1338; doi: 10.1089/neu.2006.0159 [DOI] [PubMed] [Google Scholar]
- 40. Miners JS, Kehoe PG, Love S, et al. CSF evidence of pericyte damage in Alzheimer's disease is associated with markers of blood-brain barrier dysfunction and disease pathology. Alzheimers Res Ther 2019;11(1):81; doi: 10.1186/s13195-019-0534-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, Fan DY, Li HY, et al. Dynamic changes of CSF sPDGFRbeta during ageing and AD progression and associations with CSF ATN biomarkers. Mol Neurodegener 2022;17(1):9; doi: 10.1186/s13024-021-00512-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stacker SA, Achen MG. Emerging roles for VEGF-D in human disease. Biomolecules 2018;8(1); doi: 10.3390/biom8010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.