Abstract
Current classifications (World Health Organization-HAEM5/ICC) define up to 26 molecular B-cell precursor acute lymphoblastic leukemia (BCP-ALL) disease subtypes by genomic driver aberrations and corresponding gene expression signatures. Identification of driver aberrations by transcriptome sequencing (RNA-Seq) is well established, while systematic approaches for gene expression analysis are less advanced. Therefore, we developed ALLCatchR, a machine learning-based classifier using RNA-Seq gene expression data to allocate BCP-ALL samples to all 21 gene expression-defined molecular subtypes. Trained on n = 1869 transcriptome profiles with established subtype definitions (4 cohorts; 55% pediatric / 45% adult), ALLCatchR allowed subtype allocation in 3 independent hold-out cohorts (n = 1018; 75% pediatric / 25% adult) with 95.7% accuracy (averaged sensitivity across subtypes: 91.1% / specificity: 99.8%). High-confidence predictions were achieved in 83.7% of samples with 98.9% accuracy. Only 1.2% of samples remained unclassified. ALLCatchR outperformed existing tools and identified novel driver candidates in previously unassigned samples. Additional modules provided predictions of samples blast counts, patient’s sex, and immunophenotype, allowing the imputation in cases where these information are missing. We established a novel RNA-Seq reference of human B-lymphopoiesis using 7 FACS-sorted progenitor stages from healthy bone marrow donors. Implementation in ALLCatchR enabled projection of BCP-ALL samples to this trajectory. This identified shared proximity patterns of BCP-ALL subtypes to normal lymphopoiesis stages, extending immunophenotypic classifications with a novel framework for developmental comparisons of BCP-ALL. ALLCatchR enables RNA-Seq routine application for BCP-ALL diagnostics with systematic gene expression analysis for accurate subtype allocation and novel insights into underlying developmental trajectories.
INTRODUCTION
Improved outcomes in B-cell precursor acute lymphoblastic leukemia (BCP-ALL)—both, in pediatric and adult patients—have been achieved by precise risk stratification and target-specific treatments. Molecular BCP-ALL subtypes and immunophenotype are the most important baseline prognosticators for BCP-ALL besides white blood cell counts and age. They inform risk-adapted treatments and targeted therapies. Currently, the revised World Health Organization (WHO) classification of lymphoid neoplasms (WHO-HAEM5)1 and the International Consensus Classification (ICC) of Myeloid Neoplasms and Acute Leukemia2 have acknowledged 11 and 26 molecular-defined BCP-ALL subtypes as distinct diagnostic entities, respectively, including 5 provisional entities (ICC classification). A total of 21 of these subtypes have been characterized by distinct gene expression profiles,3–8 while the remaining subtypes2,5 are rare (IGH::IL3) or were defined by specific sets of underlying genomic drivers (Ph-like: ABL class / JAK-STAT / NOS) or their absence (KMT2A- / ZNF384-like). This heterogeneity of diagnostic subtypes exceeds the capabilities of cytogenetic (chromosome banding analysis and FISH) and molecular genetic methods (breakpoint specific PCR, multiplex ligation-dependent probe amplification, and SNP-array/array-CGH), which so far have been combined for identification of BCP-ALL subtypes. Transcriptome sequencing (RNA-Seq) enables identification of all BCP-ALL subtypes with a single method, establishing a new diagnostic standard. Further implementation as routine clinical diagnostic requires unified analysis methods. Calling of driver gene fusions9,10 is well established and novel approaches for the identification of hotspot single-nucleotide10 variants and virtual karyoytpes11 exist. Yet only few approaches for systematic gene expression analysis are currently available.12–14
Gene expression signatures represent the signaling equivalent of heterogeneous genomic driver alterations, and have been used to define BCP-ALL subtypes. Initially, unsupervised clustering or prediction analysis for microarrays were used to define subtype-specific gene sets resulting in considerable heterogeneity regarding gene set definitions and subtype allocation of individual samples.15 More recent systematic approaches for BCP-ALL subtype allocations have employed machine learning methods to train classifiers for BCP-ALL subtype allocation mainly on pediatric ALL datasets.12,13 Yet the optimal method still needs to be defined—especially for rare and difficult to classify subtypes and subtypes with predominance in adults. Additionally, correct assignment of samples, which do not fall into established subtype categories either due to interfering biological conditions (e.g., low blast count or poor RNA quality) or because these samples represent novel candidate subtypes, remains a challenge. In addition to molecular subtype definitions, gene expression profiles might be informative for clinical baseline parameters such as leukemic blast proportion, immunophenotype, or more detailed analysis of lymphopoiesis trajectories underlying BCP-ALL development. However, systematic approaches and especially RNA-Seq data that link BCP-ALL subtypes to human B-lymphopoiesis differentiation stages are lacking.
Here, we describe ALLCatchR, a machine learning-based classifier pretrained for allocation of BCP-ALL gene expression profiles to all 21 gene expression-defined molecular subtypes of the WHO-HAEM5 and ICC classifications. High accuracies in independent validation cohorts are achieved by integrating machine learning and gene set-based nearest-neighbor models into a compound classifier. ALLCatchR infers clinical baseline variables such as blast proportion and patient’s sex from RNA-Seq data and provides insights into underlying developmental trajectories of BCP-ALL based on our newly established reference of human B-lymphopoiesis. ALLCatchR sustains routine diagnostic application of RNA-Seq with systematic gene expression analysis providing subtype allocations and insights into underlying biology for further exploratory analysis.
MATERIALS AND METHODS
The 3532 sample BCP-ALL transcriptome reference data set
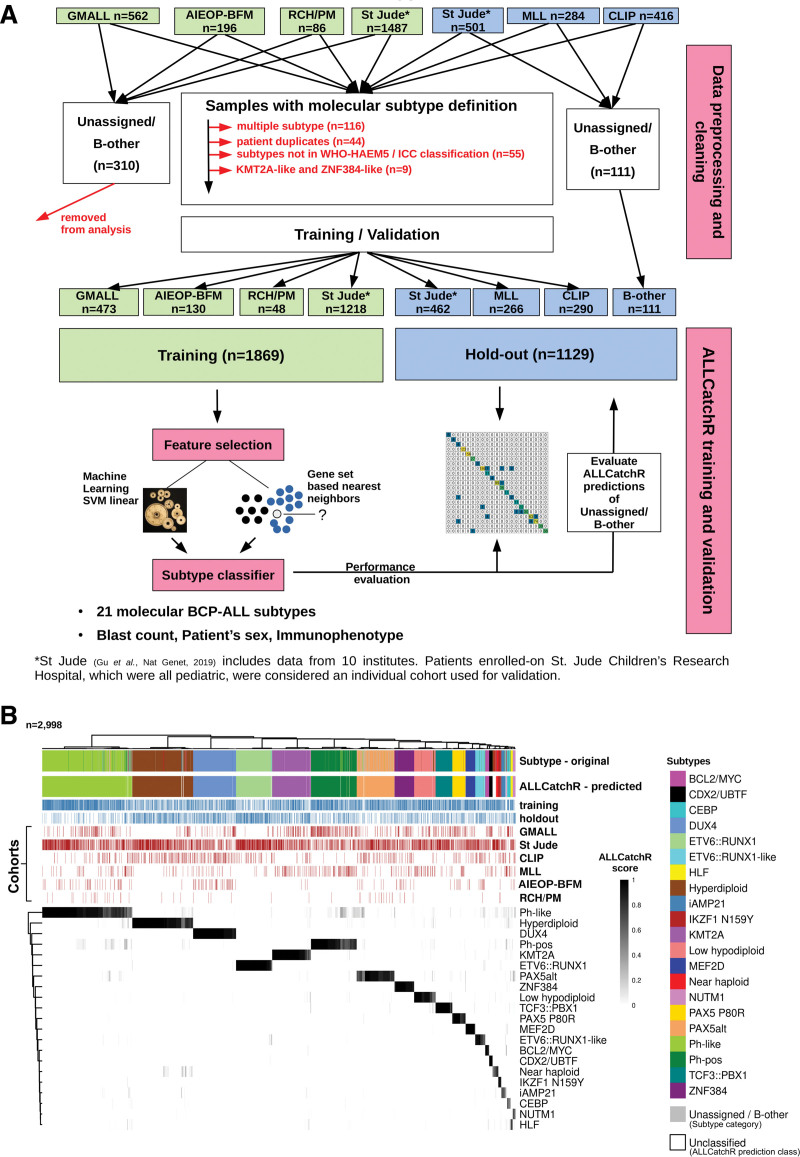
We aggregated RNA-Seq count data from n = 3532 BCP-ALL samples including 64.5% pediatric5–7,12 and 35.5% adult3–5,8,12 cases combined from 6 independent datasets (Figure 1A; Suppl. Table S1). Excluded were samples with multiple subtype assignments (n = 116), multiple representations of the same patient (n = 44), subtypes that are not part of WHO-HAEM5/ICC classification (low hyperdiploid, n = 51; IDH1/2, n = 4) or that are mainly defined by absence of a genomic driver (KMT2A-like, n = 4; ZNF384-like, n = 5). A total of n = 421 samples were defined ‘unassigned’ or B-other in the original studies. Subtype-defining genomic events were identified in >90% of cases either by RNA-Seq (gene fusions, hotspot single-nucleotide variants, and virtual karyotypes) or by genomic profiling (whole genome- / whole exome- / gene panel-sequencing, SNP-arrays, and array-CGH). The data set was split into a data set used for training of the classifier (n = 1869) and 3 hold-out studies (n = 1129) (Figure 1A). Complete hold-out-cohorts were used to challenge the classifier with new independent data structures mimicking real-world application. Selection of hold-out-data sets was based on best representation of all subtypes and age groups. Out of n = 421 samples defined ‘unassigned’ or B-other in the original studies, n = 111 belonged to the 3 hold-out studies and were kept for evaluating ALLCatchR predictions on these cohorts (Figure 1A). All WHO-HAEM5/ICC-defined BCP-ALL molecular subtypes, which were characterized by distinct gene expression signatures in their original description (n = 21), were represented in the data set. Ph-like was considered one subtype without subdivision. CEBP/ZEB2 subtype lacks final definitions so far and was defined here as CEBP by the presence of IGH::CEBPA/CEBPE/CEBPD fusions and the absence of other drivers (Suppl. Table S2). Raw read counts for 15,728 protein-coding genes represented in all cohorts were used including heterogenous sequencing approaches (poly-A selection/depletion of ribosomal RNAs), different sequencing depths, and different read count quantification methods. Counts were normalized by log10(count + 1), followed by z-transformation and scaling between 0 and 1.
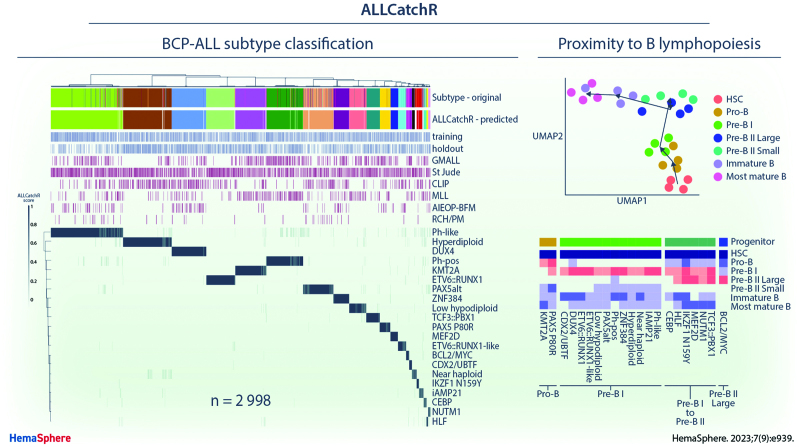
Figure 1.
ALLCatchR predicts molecular BCP-ALL subtypes based on the gene expression count data with high accuracy. (A) ALLCatchR workflow for BCP-ALL molecular subtype classification. Gene wise count data of a total of n = 3532 BCP-ALL patients from RNA-Seq experiments comprising 6 datasets were included in this study. Four data sets (n = 1869) were used for training and validation was performed on 3 hold-out data sets (n = 1129). The largest data set was St Jude (n = 1988) and included patients from St Jude Children’s Research Hospital, Children’s Oncology Group, ECOG-ACRIN Cancer Research Group, the Alliance for Clinical Trials in Oncology, M.D. Anderson Cancer Center, University of Toronto, Northern Italian Leukemia Group, Southwestern Oncology Group, Medical Research Council UK, and City of Hope. Samples marked with institute as St Jude (n = 501), which were all pediatric, were considered an individual cohort used for validation. ALLCatchR is a compound classifier based on the deterministic linear SVM predictions and a classifier based on the sample-to-sample distances to subtype-specific gene sets. In addition to the n = 1018 hold-out samples with an assigned subtype, n = 111 were defined ‘unassigned’ or B-other in the original studies and kept for evaluating ALLCatchR predictions. (B) Heatmap showing the ALLCatchR prediction scores for 21 gene expression-defined BCP-molecular subtypes (WHO-HAEM5 / ICC) in n = 2998 samples of the entire BCP-ALL cohort (after removal of duplicate samples, samples with 2 primary subtype allocations and ‘unassigned’/B-other samples in the training data; n = 534) samples. Molecular subtypes had been defined in the 6 original studies (GMALL, St Jude, CLIP, MLL, AIEOP-BFM, and RCH/PM) based on the genomic driver aberrations and corresponding gene expression signatures in n = 2887 cases (ground truth). Remaining cases were deemed ‘unassigned’ or B-other. ALLCatchR scores are shown for the combined data set of training and hold-out cohorts. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; CLIP = Childhood Leukaemia Investigation Prague; GMALL = German Multicenter Study Group on Adult Acute Lymphoblastic Leukemia; MLL = Munich Leukemia Laboratory; RCH/PM = The Royal Children’s Hospital / Peter MacCallum Cancer Centre.
Integration of machine learning and gene set-based nearest-neighbor models for BCP-ALL subtype allocation
To perform molecular subtype allocation based exclusively on gene expression data, we developed ALLCatchR, a classifier that integrates linear support vector machine (SVM) and nearest-neighbor association models for BCP-ALL subtypes derived from the training data (Figure 1A). Training was performed in a 10-fold randomized stratified cross-validation scheme. For feature selection, we applied least absolute shrinkage and selection operator (LASSO) regression with 4 different alpha parameters (0.1, 0.3, 0.5, and 1), where higher values result in a more stringent selection of features. LASSO16 was run in an internal 10-fold cross-validation with type.measure = deviance and family = multinomial logistic regression using the cv.glmnet function of the glmnet R package.17 We used also Boruta18—a Random Forest-based feature selection method—allowing for nonlinear feature to class associations. Each feature selection method was used for training 5 machine learning algorithms of which linear SVM19 performed best (Suppl. Figure S1). The best feature selection method was LASSO (alpha = 0.1) resulting in 2802 genes with high discriminative power for 21 molecular subtypes (Suppl. Figure S2; Suppl. Table S3). Linear SVM achieved a remarkable accuracy of subtype prediction in the training data (0.963), thus outperforming all other machine learning methods. However, linear SVM is restricted to predefined classes and does not compute probabilities for individual subtype predictions, which prevents it from correctly handling cases that are unassigned or ambiguous due to multiple drivers or cases that represent novel candidates. To achieve a probabilistic compound model, we incorporated single-sample gene set enrichment analyses (ssGSEA) using singscore20 of the same subtype-defining LASSO gene sets. By this approach, batch effects between cohorts were removed (Suppl. Figure S3). Euclidean distance of each test sample to each training sample was computed and the 10 nearest-neighbors were considered for subtype allocations of each test sample (accuracy for subtype prediction based on highest enrichment for each sample: 0.912). Both models—SVM linear predictions and sample-to-samples-distances in subtype-defining gene sets—were integrated into our newly established compound classifier, ALLCatchR, which provides dynamic ranges of subtype-specific scores. To achieve a better separation between highly similar high hyperdiploid and near haploid ALL, both subtypes where first represented as 1 class in the overall classifier and then separated by a second 2-class compound classifier with the same design as the overall classifier.
RNA-Seq reference of human B-lymphopoiesis
Bone marrow samples from healthy adult donors (n = 4; M:F = 1:3; age, 27–39 years; study registration DRKS00023583; ethical approval of ethics committee, Kiel University: D 583/20) were subjected to immunodensity cell separation (RosetteSep, STEMCELL Technologies; Inc., Vancouver, BC, Canada; purging: CD16, CD36, CD66b, CD235a, CD3). Nondepleted cells were stained with a 9-color antibody panel (Suppl. Table S4; Suppl. Figure S4) and used for fluorescence activated cell sorting (FACS; FACSAria fusion; BD Biosciences, Franklin Lakes, NJ) to 7 lymphoid differentiation stages. RNA was extracted from 5000 to 320,000 cells per differentiation stage (AllPrep DNA/RNA Micro Kit, Qiagen, Venlo, Netherlands) and subjected to ultra-low-input RNA sequencing after generation of stranded sequencing libraries (SMART-Seq Stranded Kit, Takara Bio Inc., Kusatsu, Shiga, Japan; NovaSeq 6000, Illumina, San Diego, CA).
Data availability
ALLCatchR is freely available as an R-package through https://github.com/ThomasBeder/ALLCatchR. Transcriptome sequencing data of bone marrow samples from healthy donors were deposited at the European Genome Phenome archive (EGAS00001007305). BCP-ALL transcriptome profiles have been deposited in open or controlled access archives (EGAS00001006107; https://viz.stjude.cloud/st-jude-childrens-research-hospital/visualization/pax5-driven-subtypes-of-b-progenitor-acute-lymphoblastic-leukemia-genomepaint~16; https://github.com/Oshlack/ALLSorts) or can be obtained by the authors of the original publications upon reasonable request.4–8
RESULTS
ALLCatchR predicts BCP-ALL molecular subtypes with high accuracy
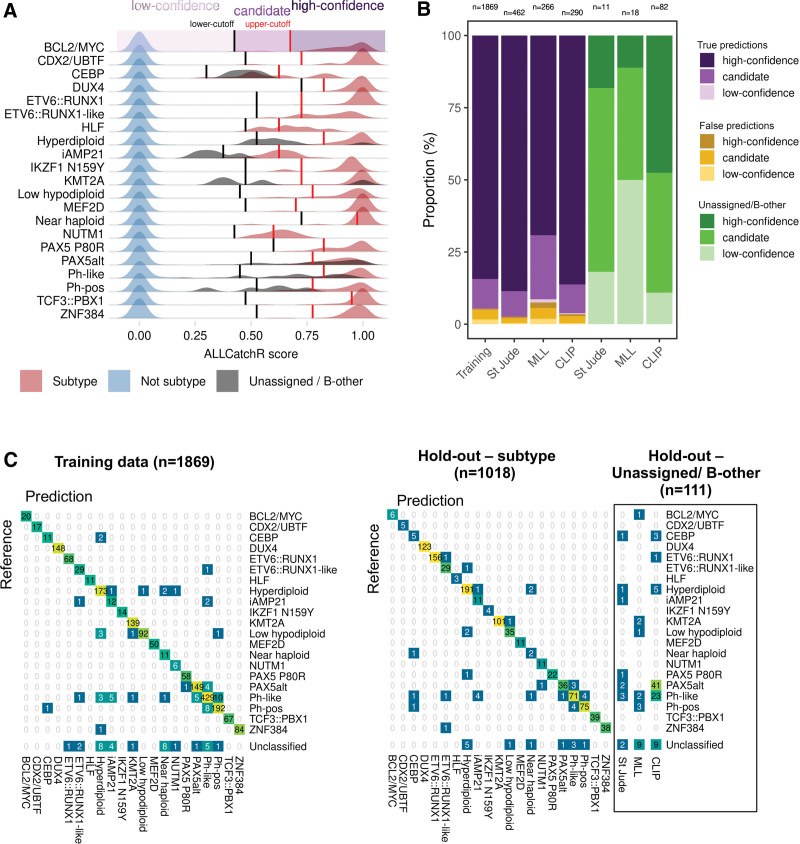
We used aggregated BCP-ALL gene expression profiles (n = 2998 samples; n = 6 cohorts) to develop and validate ALLCatchR, a pretrained machine learning classifier, which performs BCP-ALL molecular subtype allocation based on gene expression alone (detailed in Methods; Figure 1A). ALLCatchR provides scores for each sample and all gene expression-defined BCP-ALL subtypes. Using these scores, samples were grouped according to their subtype independent of cohort and age group (Figure 1B). Final prediction scores varied in their range for individual subtypes depending on number of samples and strength of subtype-specific gene expression signature. Rare subtypes (e.g., HLF or NUTM1) or subtypes with less well distinguishable gene expression signatures (e.g., iAMP21) achieved overall lower prediction scores compared with more frequent subtypes (e.g., KMT2A or DUX4) or subtypes with very specific gene expression profiles (e.g., CDX2/UBTF; Figure 1B). Therefore, we defined subtype-specific cutoffs (Suppl. Table S5) based on the comparison of scores from samples belonging to the corresponding subtype and all remaining samples of the cohort (Figure 2A). This resulted in the following: (1) high-confidence predictions; (2) candidate predictions; and (3) low-confidence predictions that is unclassified samples. Cutoffs for high-confidence predictions were defined to include >90% of correct predictions. Cutoffs for candidate predictions were defined to exclude all samples from other subtypes but allowed ‘unassigned’/B-other samples to be classified (n = 111; Figure 2A). Low-confidence prediction scores represented overlaps between different subtypes and were therefore considered unclassified.
Figure 2.
Confidence categories and predictions of samples previously unassigned/B-other. (A) Cutoffs were defined for each BCP-ALL subtype based on distribution of all ALLCatchR scores in every subtype. (B) The proportions of confidence categories for true and false predictions in the training and hold-out data sets are shown. A prediction was considered true if the sample received the same subtype allocation as in the original study. False predictions represent allocations to other subtypes than the subtype assigned in the original study. For comparison, ‘unassigned’/B-other samples from the hold-out data sets are shown. (C) Confusion matrices relate ALLCatchR predictions to the ground truth in training samples (left) and hold-out cohorts (right). The training cohort did not contain ‘unassigned’/B-other samples, because these do not represent a homogenous group a classifier can be trained for. In the hold-out data, n = 111 samples had been defined as ‘unassigned’/B-other and predictions for these are also shown. Suppl. Figure S5A and Suppl. Table S6 indicate how ALLCatchR predictions in ‘unassigned’/B-other samples are supported by corresponding genomic drivers in 72.1% of high confidence and 27.1% of candidate predictions. BCP-ALL = B-cell precursor acute lymphoblastic leukemia.
In the training data, 84.6% of samples achieved high-confidence predictions with an accuracy of 0.997, while 13.7% achieved candidate predictions with an accuracy of 0.797 to guide further validation based on genomic drivers in well prespecified directions (Figure 2B). Only 1.7% of samples achieved low-confidence predictions and were considered unclassified. To validate ALLCatchR performance, we used independent validation data from 3 hold-out cohorts (n = 1018 with assigned subtype; Figure 1A), not previously seen by the classifier. A total of n = 1006 of 1018 (98.8%) samples was allocated to 1 of 21 subtypes (high-confidence and candidate predictions) with an accuracy of 0.957, demonstrating the feasibility of highly accurate subtype allocations based on gene expression alone. High-confidence and candidate predictions were achieved in 83.7% and 15.1% of samples with accuracies of 0.989 and 0.851, respectively. A total of n = 44 samples were assigned to the wrong subtype (n = 32; 3.1%) or received no subtype allocation (Figure 2C; n = 12; 1.2%). Most frequently Ph-like samples were misclassified to Ph-pos (n = 4) or PAX5alt (n = 3), followed by Ph-pos samples being misclassified to Ph-like (n = 4) and iAMP21 cases being misclassified to Ph-like (n = 4) or hyperdiploid (n = 1). Highly similar signaling patterns and even co-occurance of drivers21,22 have been described between Ph-pos−/−like and iAMP21 ALL, which might be the underlying reason for these misclassifications. Next frequently, hyperdiploid cases (n = 4) were misclassified to different subtypes (low hypodiploid: n = 2; PAX5 P80R: n = 1; ETV6::RUNX1-like: n = 1). Different subtypes were involved in the remaining n = 12 misclassified cases (Figure 2C). Importantly, most misclassified samples (n = 23/32; 71.9%) had received candidate (not high-confidence) predictions, indicating the need to validate these predictions based on genomic drivers.
ALLCatchR provides subtype allocations for previously unassigned/B-other samples
In addition to the n = 1018 hold-out samples with assigned subtype, n = 111 samples (Figure 2B and 2C) had been defined as unassigned/B-other in the original studies. ALLCatchR concordantly identified n = 20 (18.0%) of these as unclassified (Figure 2C; Suppl. Figure S5). However, n = 43 (38.7%) and n = 48 (43.2%) cases received high-confidence or candidate predictions, respectively (Figure 2C). Analysis of available RNA-Seq gene fusion calls or cytogenetic profiles and/or virtual karyotyping (whole genome sequencing [WGS]/SNP-arrays) identified driver candidates supporting the corresponding subtype allocations in n = 31 (72.1%) of high-confidence and n = 13 (27.1%) of candidate predictions (Suppl. Table S6; Suppl. Figure S5). These newly suggested subtype allocations consisted of PAX5alt predictions (n = 25), which had not shown a clear PAX5alt gene expression profile in the original cohort (n = 1), or which were contributed from the CLIP cohort where this subtype had not been annotated previously. Next frequently, n = 11 CRLF2-rearranged cases from CLIP and St Jude cohorts without Ph-like gene expression profile in the original cohorts received ALLCatchR Ph-like predictions. Of the remaining n = 7 samples, n = 4 were predicted to be KMT2A of which 2 cases had KMT2A amplifications and 1 case with an ALLCatchR high-confidence KMT2A prediction was found to harbor a KMT2A partial tandem duplication by WGS (Suppl. Table S6; Suppl. Figure S5). To the best of our knowledge, this is the first identification in BCP-ALL of this aberration, which is recurrently observed in acute myeloid leukemia. In a second of these n = 7 cases, an IGH::MYC gene fusion was identified in support of a BCL2/MYC ALLCatchR prediction. Further ALLCatchR high-confidence predictions for unassigned/B-other samples without corresponding drivers included PAX5alt (n = 9) and Ph-like (n = 3) predictions, which generally are defined in a proportion of samples by gene expression alone. Thus, ALLCatchR suggested molecular subtype allocations in previously unassigned cases with atypical and less well-defined gene expression signatures and supported the identification of novel driver candidates.
High accuracy of ALLCatchR predictions is observed across cohorts and molecular subtypes
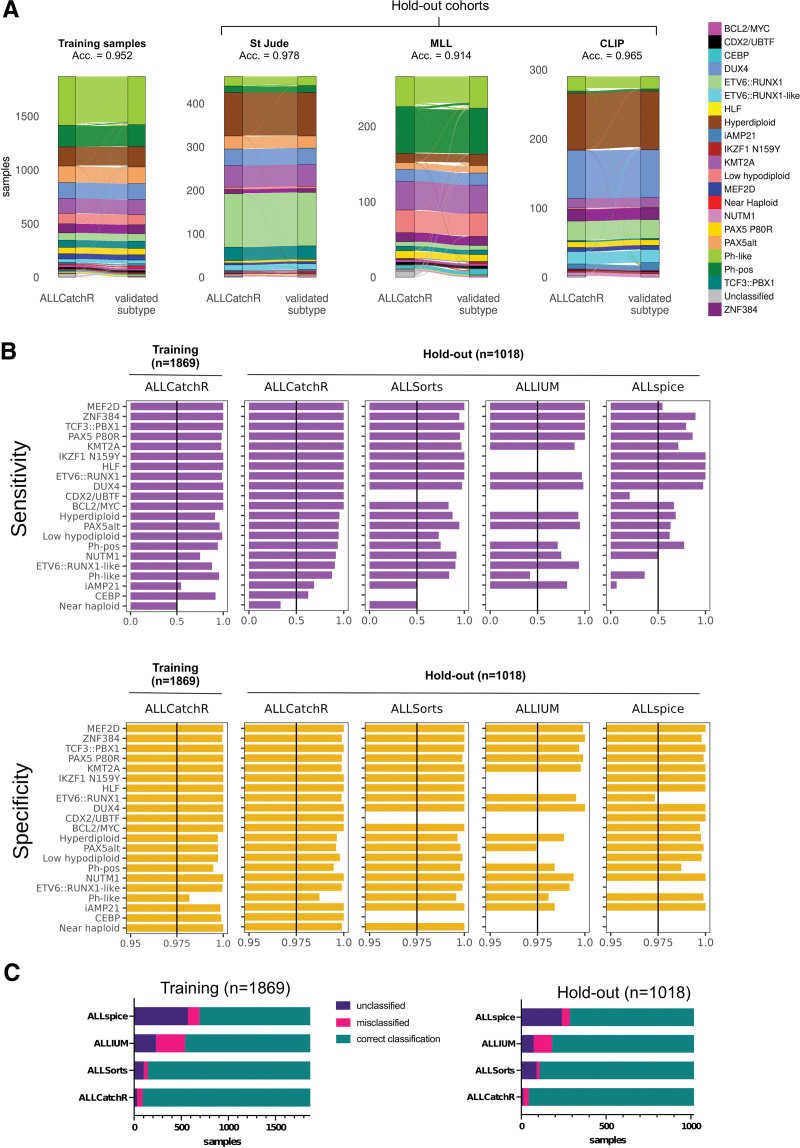
The accuracy of predictions was consistently high in the training and hold-out data with 0.952 and 0.957, respectively. Almost congruent predictions were achieved in St Jude and CLIP cohorts with accuracies of 0.978 and 0.965, respectively. St Jude and CLIP represent pediatric data from clinical trials. In the MLL hold-out set, the accuracy was slightly lower with 0.914 (Figure 3A), possibly due to less stringent preselection criteria (blast count and selection of subtypes) in a real-world diagnostic laboratory, indicating that ALLCatchR achieves reliable predictions also in less preselected samples outside from clinical trials. Despite the overall high accuracies, classification performance varied between molecular subtypes (Figure 3B). ALLCatchR achieved specificities >0.99 for all 21 subtypes, both in training and hold-out data sets. The average sensitivity across subtypes was 0.919 ± 0.145 and 0.911 ± 0.167 in the training and hold-out data, respectively. For n = 17/21 subtypes, sensitivities were ≥0.85 both on training and hold-out data (Figure 3B). Only 4 remaining subtypes (n = 106 samples; 3.7% of entire cohort) achieved sensitivities below 0.85 (NUTM1, CEBP, iAMP21, and near haploid), which was mainly related to the small number of samples representing these subtypes. Importantly, age and different subtype prevalence did not affect the results as accuracies were high in pediatric and adult samples, and sensitivity and specificity did not differ across age groups in the training and testing sets (Suppl. Table S7).
Figure 3.
ALLCatchR accuracy for subtype allocation is consistently high across cohorts and BCP-ALL subtypes. (A) Sankey diagrams indicate ALLCatchR subtype allocations and corresponding subtype ground truth in the training cohort and the individual hold-out data sets. Acc. indicates accuracy in the corresponding data set. (B) Bar charts indicated sensitivity and specificity of ALLCatchR predictions in the training data and ALLCatchR, ALLSorts, ALLIUM (GEX) and ALLspice predictions in the hold-out data. (C) Bar charts with correct classified, misclassified, and unclassified samples in the training and hold-out data, for different tools using gene expression for molecular BCP-ALL subtype allocation. Subtypes with missing values cannot be predicted by the individual tools. Validated ground truth was used to define true positive cases, that is, belonging to this subtype and true negative cases, that is, not belonging to this subtype. Values were obtained as fraction of true positive cases from all cases defined by ALLCatchR as belonging to this subtype (sensitivity) and as fraction of true negative cases from all cases defined by ALLCatchR as not belonging to this subtype (specificity). BCP-ALL = B-cell precursor acute lymphoblastic leukemia.
ALLCatchR subtype allocation outperforms current tools
Recently, 3 other tools—ALLSorts,12 Allspice,13 and ALLIUM GEX14—were independently developed for BCP-ALL subtype allocation based on gene expression profiles. For performance comparison, subtype allocation was performed on our hold-out data set (n = 1018 with assigned subtype). All tools achieved correct subtype allocations in the majority of cases (Figure 3B), but highest accuracy was achieved by ALLCatchR (0.957), leaving only n = 12/1018 samples unclassified and n = 32/1018 samples with an incorrect subtype allocation (Figure 3B; Suppl. Figure S6). ALLSorts performed well with an accuracy of 0.913 (n = 14/1018 samples misclassified) but left more samples unclassified (n = 94/1018). The number of unclassified samples was also higher with ALLIUM GEX (n = 73/1018) and ALLSpice (n = 239/1018), partially because all these tools were trained on less subtypes (ALLSorts: n = 19/21 subtypes; Allspice: n = 18/21; ALLIUM GEX: n = 14/21), precluding classification in part to some of the rarer subtypes such as IKZF1 N159Y, HLF, CDX2/UBTF, BCL2/MYC, low hypodipolid, CEBP, and near haploid ALL (Figure 3C; Suppl. Figure S6). Compared with ALLCatchR, ALLIUM GEX achieved higher sensitivity for classification of iAMP21 (0.81 versus 0.69 with ALLCatchR) and ALLSorts was more sensitive in detecting near haploid cases (n = 6; sensitivity: 0.5 versus 0.33 with ALLCatchR). For all other subtypes, sensitivity and specificity in all cases was higher with ALLCatchR (Figure 3B and 3C).
Gene expression-based modeling predicts clinical baseline variables
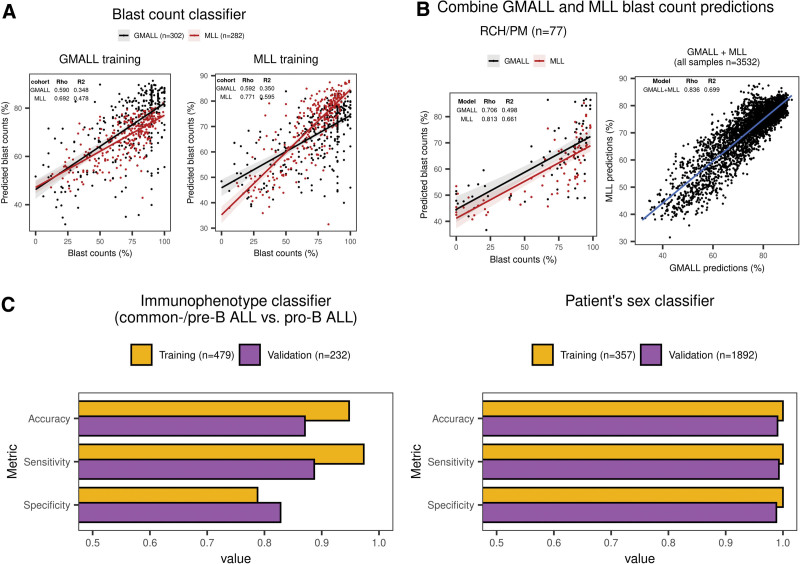
Blast count proportions impact accuracy of gene expression-based molecular subtype allocation, as sequencing reads from nonleukemic compartments contribute to bulk transcriptome profiles. To infer sample blast proportions, we trained 2 machine learning regression models on data sets of our combined cohort with available blast counts obtained by manual counting or flow cytometry (GMALL and MLL) and used these and the RCH/PM cohort for validation. Blast count predictions from single cohorts achieved good accuracies when applied to each other (Figure 4A and 4B) with a high concordance between GMALL and MLL training sets (Figure 4B), which were therefore combined for the final classifier. Only 1.85% of samples with high-confidence subtype predictions had blast count predictions <50% while these were observed in 9.83% of candidate predictions and in 17.95% of unclassified samples of the entire cohort (Suppl. Figure S7). Thus, ALLCatchR can identify a subset of samples with worse performance for subtype allocation due to lower blast infiltration. Gene expression profiles were also informative for patient’s sex and disease immunophenotype. To enable gene expression-based cross-validation of these important clinical baseline characteristics, we implemented subclassifiers to the samples immunophenotype (pro-B versus common-/pre-B ALL; accuracy of 0.871 in the validation data) and patient’s sex (accuracy: 0.991 in validation data set; Figure 4C). ALLCatchR thus provides a cross-validation of clinical baseline variables and allows imputation of missing values.
Figure 4.
ALLCatchR predicts sample blast counts, patient’s sex, and immunophenotype based on the gene expression data. (A) For GMALL (n = 302), MLL (n = 282), and RCH/PM (n = 77), sample blast counts obtained by cytology or flow cytometry were available. GMALL and MLL cohorts were separately used for training 2 classifiers in a 10-fold cross-validation scheme with the same machine learning algorithms used for subtype prediction. GMALL and MLL classifiers were validated on each other, and both were validated on the RCH/PM data. Best performing methods in terms of the RSME on the training data are shown. Training 2 classifiers on independent data sets allowed for the validation on each other and both were combined for final predictions. Blast count predictions had a good correlation to measured counts, that is, rho = 0.590 in GMALL and rho = 0.771 in MLL. Moreover, predicting MLL samples with the classifier trained on GMALL achieved a similar performance as the classifier trained on MLL samples and vice versa. (B) Because both GMALL and MLL classifiers had a good performance and were generalizable, predictions from both are combined in ALLCatchR. (C) Subclassifiers for immunophenotype and patient’s sex were developed using SVM linear and ranger machine learning models, respectively. An immunophenotype classifier was trained on GMALL samples (n = 413 common-B/pre-B and n = 66 pro-B) and validated on MLL data (n = 168 common-B/pre-B and n = 64 pro-B) with available EGIL immunophenotypes. A patient sex classifier was trained on n = 357 GMALL samples (female = 165; male = 192) analogous to the subtype classifier. For validation n = 1892 St Jude samples with known sex (female = 850; male = 1042) were used. Corresponding accuracies, sensitivities, and specificities are shown for these subclassifiers. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; RSME = root mean squared error.
Shared gene expression patterns suggest distinct developmental trajectories for BCP-ALL subtypes
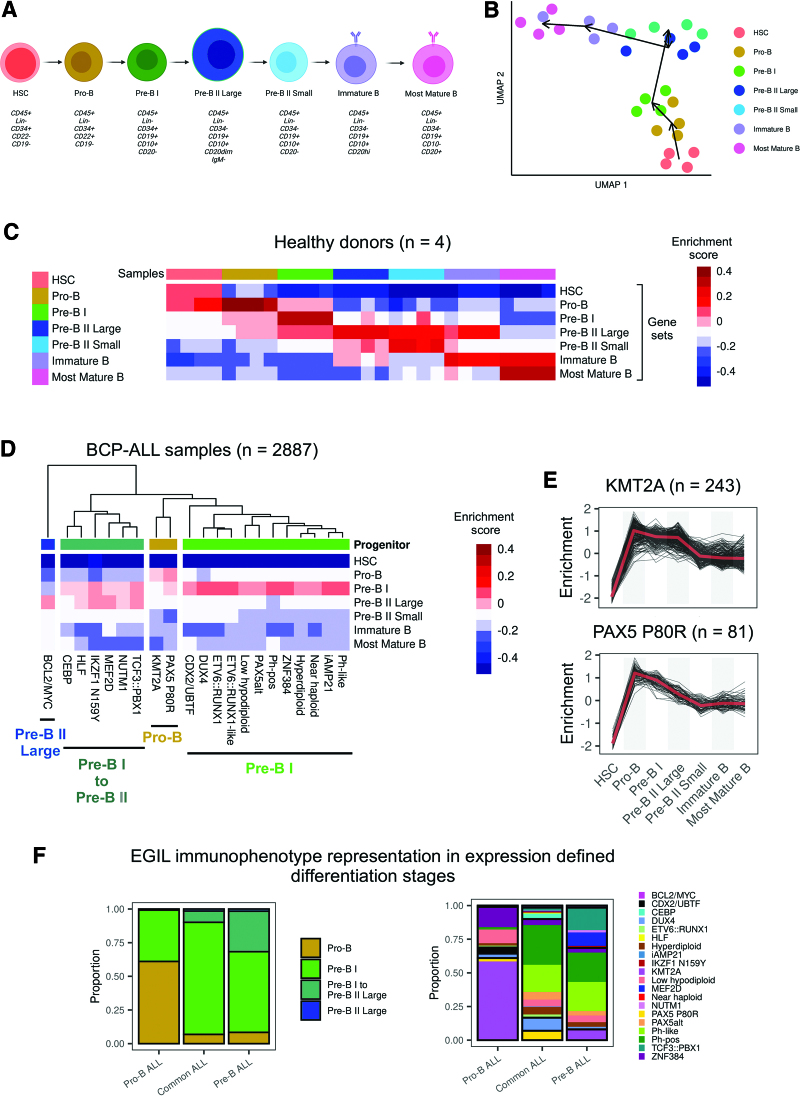
The cell of origin for BCP-ALL cases remains to be defined, with immunophenotyping according to European Group for Immunological Classification of Leukemias (EGIL) criteria23 representing a framework for orientation. An improved understanding of underlying lymphopoiesis trajectories is especially warranted regarding current immunotherapies, which rely on differentiation stage- and lineage-specific markers as therapeutic targets. To map BCP-ALL subtypes to underlying B-lymphopoiesis trajectories, we established a reference of normal human B-lymphopoiesis for 7 differentiation stages from hematopoietic stem cells to mature bone marrow B-cell subsets (Figure 5A), based on established definitions.24 Expression profiles were obtained from ultra-low input RNA-Seq of FACS-sorted bone marrow samples of healthy adult donors (n = 4). A high sequencing depth was achieved despite limited input cell numbers (5000–320,000), enabling quantification of 31,787 ± 4008 genes (89.7% of all human genes). Marker gene expression confirmed on the transcript level surface protein profiles used for FACS_sorting (Suppl. Figure S8). Unsupervised analysis of variable expressed genes grouped samples according to the developmental course with high concordance between donors (Figure 5B). Stage-specific gene sets were obtained by multicomparison ANOVA on normalized counts (vst), yielding well discriminative definitions (Figure 5C; Suppl. Table S8). Analysis of immunoglobulin rearrangements using droplet PCR indicated a germline configuration in hematopoietic stem cells, initiation of DH-JH rearrangements in sorted pro-B cells, while VH-(D)JH rearrangements were first observed in pre-B II large cells and class switch recombination occurred exclusively in the most mature B cells, providing an immunogenomic differentiation trajectory25 which independently confirms our sorting strategy (Suppl. Figures S4 and S9). We implemented this newly established model of human B-lymphopoiesis in ALLCatchR using ssGSEA to define the proximity of each BCP-ALL sample to all 7 lymphopoiesis stages (Figure 5D; Suppl. Figure S10). Medians of these enrichment scores across samples revealed distinct patterns of enrichments for BCP-ALL subtypes (pro-B /pre-B I /pre-B I to pre-B II large transition / pre-B II large; Suppl. Figure S10) with similar patterns in pediatric and adult data sets (Suppl. Figure S11). Most BCP-ALL subtypes and the majority of all cases showed highest similarity to the pre-B I stage (Figure 5D). However, KMT2A-rearranged and PAX5 P80R ALL showed a clearly distinct enrichment pattern similar to an earlier pro-B differentiation stage (Figure 5E). In contrast, CEBP, HLF, IKZF1 N159Y, MEF2D, NUTM1, and TCF3::PBX1 were grouped in a cluster with highest enrichment in transition of pre-B-I to pre-B-II large stage and BCL2/MYC showed the highest degree of similarity exclusively to pre-B II large differentiation stage (Figure 5D). These observations confirm expectations for the extremes of this trajectory (KMT2A and BCL2/MYC).26,27 A recently reported mouse model of PAX5 P80R ALL28 established a pro-B differentiation arrest as initial event in PAX5 P80R homozygous models, supporting a pro-B origin of this leukemia subtype or at least an altered PAX5 function inducing a pro-B like phenotype in PAX5 P80R mutated cases. Thus, specific enrichment patterns of normal lymphopoiesis are shared between molecular subtypes, suggesting distinct stages of transition from normal to leukemic lymphopoiesis. We have included this model in ALLCatchR. Comparison of EGIL immunophenotypes to gene-expression-defined stages indicated expected enrichments (pro-B stage in pro-B immunophenotype / pre-B II large in pre-B immunophenotypes; Figure 5F) but nearly all gene-expression-based differentiation stages were represented in each immunophenotype. BCP-ALL subtypes were more closely related to gene-expression-based differentiation stages as to EGIL immunophenotypes, suggesting that ALLCatchR identifies developmental underpinnings of BCP-ALL drivers at higher resolution.
Figure 5.
ALLCatchR identifies B-cell developmental trajectories underlying BCP-ALL subtypes. (A) To establish a reference map of human B-lymphopoiesis, we obtained bone marrow samples from healthy adult donors (n = 4) and used a 9-color antibody panel for FACS sorting of 7 B-lymphopoiesis stages following described definitions24 after pre-enrichment of wanted populations. Lin-selection included CD3, CD33, CD56, CD14, CD66c, and CD138. Antibodies used are shown in Suppl. Table S4. Suppl. Figure S9 shows immunogenomic profiling of immune gene rearrangements in support of the applied sorting strategy. (B) Ultra-low input RNA-Seq was performed for total RNA to obtain stage-specific gene expression. UMAP shows clustering of human B-lymphopoiesis stages based on 400 most variable expressed genes. (C) Multi comparison ANOVA on normalized (vst) count data was performed to obtain differentiation stage-specific gene sets. Heatmap depicts single-sample gene set enrichment analyses (singscore)20 of B-lymphopoiesis subsets (columns) to stage defining gene sets (rows). (D) BCP-ALL samples with known subtype allocation (n = 2887) were used for single-sample gene set enrichment analysis with B-lymphopoiesis-specific gene sets obtained from (C). Suppl. Figure S10 shows enrichment patterns of individual samples from all BCP-ALL subtypes for all differentiation stages. Heatmap depicts averaged enrichment scores for all BCP-ALL subtypes and all B-lymphopoiesis stages grouped by unsupervised clustering. Normal progenitors with proximity to BCP-ALL subtypes are annotated on top. Suppl. Figure S11 provides separate analyses for pediatric and adult patients indicating a high degree of similarity. (E) KMT2A rearranged and PAX5 P80R ALL had both the highest enrichment toward pro-B supporting a shared developmental origin (also depicted in Suppl. Figure S10). (F) Comparison of gene expression-defined differentiation stages and EGIL immunophenotypes are shown for n = 711 samples with available gene expression data. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; UMAP = uniform manifold approximation plot.
BCP-ALL subtype-defining gene sets indicate shared signaling trajectories
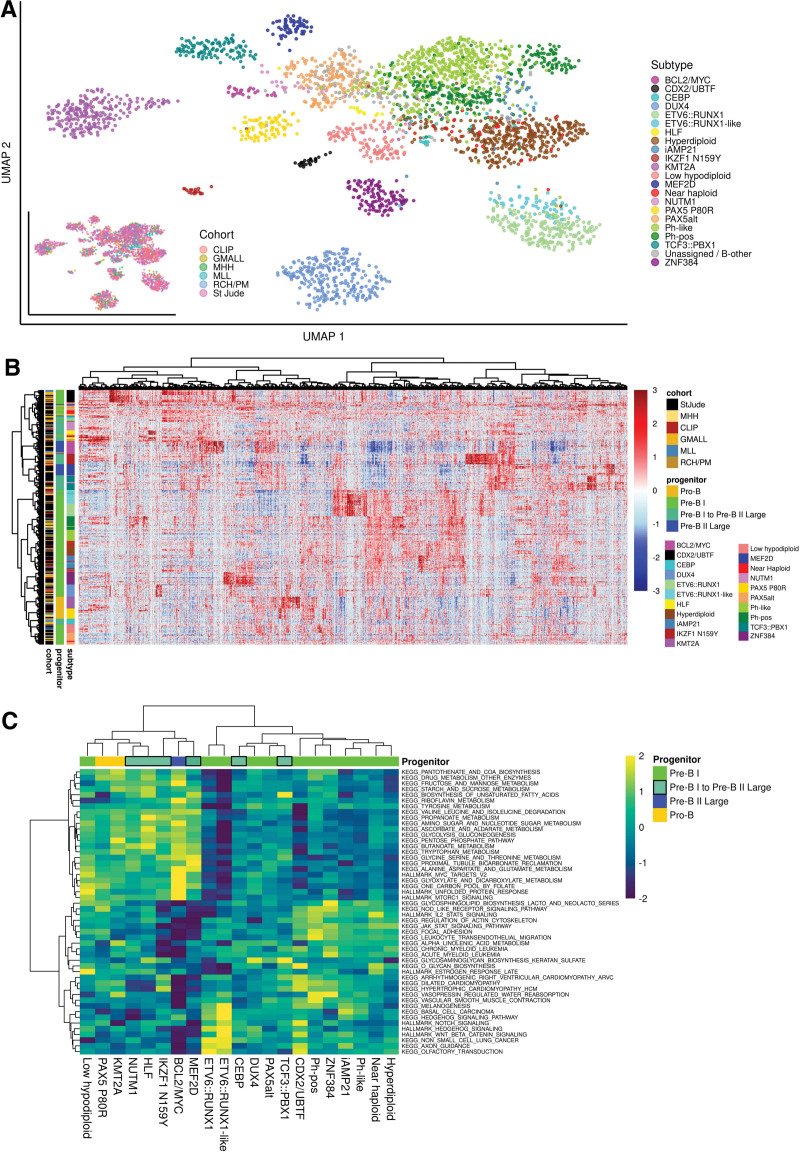
Definitions of BCP-ALL subtype-specific gene expression signatures depend on the size and composition of the remaining cohort used as comparator. We made use of the aggregated transcriptome profiles of 21 BCP-ALL subtypes to define subtype-specific gene expression profiles based on the largest data set (n = 2998) available till date, representing different age groups, cohorts, and sequencing methods. Uniform manifold approximation plot clustering of all batch corrected samples according to the LASSO-selected subtype-specific gene sets indicated a clear separation of molecular subtypes independently of the contributing cohorts (Figure 6A). To characterize subtype-specific gene expression profiles beyond top discriminative features, we performed differential gene expression analysis for each subtype compared with the remaining cohort. A median of 673 differentially expressed genes per subtype were identified (range: 144–1465; fold change: <1.5-log2-fold; FDR: <0.001; Figure 6B). Overlap between these gene sets was very low (Suppl. Figure S12) indicating that subtype-specific differences are represented in broad gene regulatory programs. Subtype-specific gene expression profiles were provided as a resource in Suppl. Tables S9-S16, S17-S22, and S23-S29. To explore the potential of this dataset to reveal underlying biological functions, we performed ssGSEA for canonical signaling pathways (MSigDB Hallmark/KEGG gene sets). Analysis of pathways top differentially enriched in BCP-ALL subtypes (1-way ANOVA) indicated previously unrecognized clusters of subtypes with enrichment in cytokine receptor/JAK-STAT signaling (Ph-pos, Ph-like, ZNF384, Hyperdiploid, iAMP21) or WNT-/beta catenin/hedgehog signaling (ETV6::RUNX1 and -like, CDX2/UBTF), which together represented the majority of subtypes with proximity to normal pre-B-I cells (Figure 6C). For the remaining subtypes, an enrichment in MYC-/MTOR signaling was observed in subtypes similar to either a more or less advanced differentiation stage (pro-B: KMT2A, PAX5 P80R / pre-B I to pre-B II large: BCL2/MYC, IKZF1 N159Y, MEF2D; Figure 6C). Interestingly, Ph-pos, Ph-like, iAMP21 and ZNF384 subtypes were grouped together here by unsupervised clustering. Shared enrichment of JAK/STAT signaling pathways supports previously suggested31,32 shared signaling trajectories in these otherwise independent subtypes. Thus, enrichment analysis for canonical signaling pathways independently grouped together BCP-ALL subtypes form similar underlying B-lymphopoiesis differentiation stages. ALLCatchR not only provides a systematic gene expression analysis for accurate identification of molecular BCP-ALL subtypes but also enables insights into underlying disease biology, which is closely interconnected with subtype nosology.
Figure 6.
The gene expression landscape in BCP-ALL. (A) UMAP plot showing all n = 2998 samples used in this study. Count data from the 6 data sets was batch corrected using the sva package29 and TPM values were calculated. The plot is based on 2802 genes selected by LASSO for training of ALLCatchR. Cohorts are highlighted on the bottom left plot. The expression data before batch correction is shown in Suppl. Figure S3A. (B) ALLCatchR predictions were used to define samples that best represented their respective molecular subtype. A total of n = 20 top ranking samples per subtype (exceptions with lesser samples available: HLF n = 14, CEBP n = 16, NUTM1 n = 17, IKZF1 N159Y n = 18) were used to obtain a homogenous data set representing all 21 BCP-ALL subtypes (n = 405). Differential gene expression analyses for each subtype versus the remaining cohort using DESeq230 revealed 5110 differentially expressed genes (cutoff: 1.5-log2-fold change, FDR: 0.001) used for unsupervised clustering. Suppl. Figure S12 and Suppl. Tables S9-S16, S17-S22, and S23-S29 provide detailed information on the derived gene sets. (C) Canonical signaling pathways (KEGG, HALLMARK gene sets; MSigDB) were used for single-sample gene set enrichment analysis using the BCP-ALL subcohort from (B) for balanced representation of all subtypes. Enrichment scores for top variable enriched pathways are shown. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; UMAP = uniform manifold approximation plot.
DISCUSSION
Risk stratification based on the molecular disease subtypes has contributed to the remarkable improvement in outcomes of patients with BCP-ALL in the last decades and has provided guidance for target-specific treatments. Current nosology of BCP-ALL includes up to 26 specific subtypes (WHO-HAEM5 / ICC),1,2 exceeding the capability of cytogenetic and molecular genetic techniques, which have so far been combined for molecular subtype allocation. Transcriptome sequencing provides informative gene expression profiles and allows identification of underlying driver gene fusions and more recently also driver single-nucleotide variants and karyotypes. Analysis of gene expression profiles for molecular subtype allocation is still not standardized, despite its potential for validating genomic driver calls and for subtype allocation of samples with missed genomic drivers.4
We have developed ALLCatchR, a pretrained machine learning classifier, which allows molecular subtype allocation in independent hold-out data with >95% accuracy. ALLCatchR is the only tool, which systematically provides allocation to all gene expression-defined subtypes of the ICC classification, including novel CDX2/UBTF ALL4,33–35 and CEBP/ZEB2.36–38 Comparable published approaches (ALLSorts, ALLIUM, and ALLspice) also achieved accurate predictions. However, ALLCatchR achieved superior performance through enabling more correct subtype allocations especially for MLL cohort.8 Immunophenotyping is a routine diagnostic in BCP-ALL and provides putative differentiation stages of origin with pro-B immunophenotype used as high-risk marker in some treatment stratification systems. EGIL definitions23 were derived from murine B-lymphopoiesis. Projecting BCP-ALL samples to our newly established reference of normal lymphopoiesis yielded novel insights into similarities between differentiation stages and BCP-ALL subtypes. Interestingly, KMT2A and PAX5 P80R ALL showed a strong proximity to normal pro-B cells, the most immature B lymphoid stage analyzed. These observations are in line with very recent single-cell analyses suggesting a pro-B or even pre-pro-B origin of KMT2A ALL27,39 and murine models of PAX5 P80R ALL showing that homozygous PAX5 P80R induces a pro-B differentiation arrest in lymphopoiesis before full transformation through acquisition of additional driver events.28 Here, ALLCatchR analysis based on our large aggregated reference cohort confirmed these observations of smaller cohorts,27,39 preclinical models,28 and previous assumptions on redirected PAX5 functionality in PAX5 P80R ALL.3,5 Gene-expression-based definitions of developmental stages in BCP-ALL were more closely related to BCP-ALL subtypes than immunophenotypes, suggesting that selection for leukemogenic drivers occurs in a differentiation stage-specific manner.
Diagnostic definitions of molecular BCP-ALL subtypes1,2 rely primarily on genomic drivers. Gene fusion calling9,40 and identification of driver hotspot variants5,10 from RNA-Seq data is well established. Recently, it has been shown that virtual karyotypes can also be imputed from RNA-Seq data.11 Gene expression profiles, however, represent the downstream signaling equivalent of these genomic events and by that inform biological insights. Shared gene expression patterns serve as validation of the functional relevance of the observed drivers—also in cases with multiple drivers. They establish subtype allocations for samples with missed driver calls due to difficult to identify targets (e.g., IGH::DUX4 or other IGH-fusions) and provide unifying definitions for subtypes with heterogeneous drivers (e.g., Ph-like ALL). We see ALLCatchR as the central component of an integrated workflow for RNA-Seq in BCP-ALL, which incorporates gene fusion calling, identification of hotspot variants, and virtual karyotypes together with gene expression profiling for a subtype allocation with highest diagnostic precision.
ALLCatchR is based on the largest cohort of BCP-ALL gene expression profiles across age groups and molecular subtypes available till date. We make use of this aggregated data to provide subtype-defining gene sets for normal and leukemic B-lymphopoiesis as an independent research resource. Although only a small minority of samples remain unassigned, novel subtype candidates are being discussed (e.g., IDH1/2 mutated ALL and low hyperdiploid ALL).5,33 ALLCatchR is a freely available open-source tool providing a conceptual and technical framework, which can easily be extended for incorporation of novel subtypes and additional predictive models. When combined with already established approaches for calling of genomic drivers (e.g., gene fusions), ALLCatchR will complement the essential prerequisites for the transition of RNA-Seq from research to routine application in clinical diagnostics.
ACKNOWLEDGMENTS
We gratefully appreciate critical contributions from Saskia Kohlscheen and Matthias Ritgen for the development of the healthy donor FACS sort panel and Monika Szczepanowski for contributing to sample collection and critical discussion of the manuscript. We are indebted to Christian Peters and Esther Schiminsky for performing the FACS sorts.
AUTHOR CONTRIBUTIONS
TB, MBr, CDB, and LBas designed the study. TB and LBas established models for molecular subtype allocation and B cell developmental stages and developed the classifier. BTH, LBas, and CDB conceived the clinical trial to obtain healthy bone marrow samples. BTH, EA, and LBas established the normal donor FACS panel. BTH and MBu performed FACS sorting. TB, BTH, AMH, NK, LBar, SB, JK, and MJB established bioinformatic workflows and performed analyses of BCP-ALL and healthy donor gene expression profiles. JZ and CK developed and tested the CRAN package for ALLCatchR distribution. WW, MZ, ZA, PC, GC, MS, MN, NG, AKB, JT, and CH contributed BCP-ALL sequencing data and validated ground truth and/or contributed to the classifier concept. LBas and CDB supervised the project. TB, CDB, and LBas drafted the first version of the article. All authors revised and approved the final version of the article.
DISCLOSURES
The authors have no conflicts of interests to disclose.
SOURCES OF FUNDING
This study was in part funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—project number 444949889 (KFO 5010/1 Clinical Research Unit “CATCH ALL” to LB, AH, MPH, MN, MB, and CDB), and project number 413490537 (Clinician Scientist Program in Evolutionary Medicine to BTH) and Deutsche Jose Carreras Leukämie Stiftung (DJCLS 01R/2016 to LB and CDB, DJCLS R 15/11 and DJCLS 06R/2019 to MBr) and the Czech Health Research Council (NU20-07-00322 to MZ and JT).
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
REFERENCES
- 1.Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemia: integrating morphological, clinical, and genomic data. Blood. 2022;140:1200–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastian L, Schroeder MP, Eckert C, et al. PAX5 biallelic genomic alterations define a novel subgroup of B-cell precursor acute lymphoblastic leukemia. Leukemia. 2019;33:1895–1909. [DOI] [PubMed] [Google Scholar]
- 4.Bastian L, Hartmann AM, Beder T, et al. UBTF::ATXN7L3 gene fusion defines novel B cell precursor ALL subtype with CDX2 expression and need for intensified treatment. Leukemia. 2022;36:1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaliova M, Stuchly J, Winkowska L, et al. Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematologica. 2019;104:1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chouvarine P, Antić Z, Lentes J, et al. Transcriptional and mutational profiling of B-other acute lymphoblastic leukemia for improved diagnostics. Cancers. 2021;13:5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter W, Shahswar R, Stengel A, et al. Clinical application of whole transcriptome sequencing for the classification of patients with acute lymphoblastic leukemia. BMC Cancer. 2021;21:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014:011650. [Google Scholar]
- 10.Rehn J, Mayoh C, Heatley SL, et al. RaScALL: Rapid (Ra) screening (Sc) of RNA-seq data for prognostically significant genomic alterations in acute lymphoblastic leukaemia (ALL). PLoS Genet. 2022;18:e1010300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bařinka J, Hu Z, Wang L, et al. RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data. Leukemia. 2022;36:1492–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt B, Brown LM, Ryland GL, et al. ALLSorts: an RNA-Seq subtype classifier for B-cell acute lymphoblastic leukemia. Blood Adv. 2022;6:4093–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mäkinen V-P, Rehn J, Breen J, et al. Multi-cohort transcriptomic subtyping of B-cell acute lymphoblastic leukemia. Int J Mol Sci. 2022;23:4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krali O, Marincevic-Zuniga Y, Arvidsson G, et al. Multimodal classification of molecular subtypes in pediatric acute lymphoblastic leukemia. Hematology. 2023:2023.03.24.23287613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boer JM, Marchante JRM, Evans WE, et al. BCR-ABL1-like cases in pediatric acute lymphoblastic leukemia: a comparison between DCOG/Erasmus MC and COG/St. Jude signatures. Haematologica. 2015;100:e354–e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibshirani R. The lasso method for variable selection in the cox model. Stat Med. 1997;16:385–395. [DOI] [PubMed] [Google Scholar]
- 17.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Kursa MB, Rudnicki WR. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 19.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26.27774042 [Google Scholar]
- 20.Foroutan M, Bhuva DD, Lyu R, et al. Single sample scoring of molecular phenotypes. BMC Bioinf. 2018;19:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison CJ, Moorman AV, Schwab C, et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia. 2014;28:1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwab CJ, Chilton L, Morrison H, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98:1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995;9:1783–1786. [PubMed] [Google Scholar]
- 24.van Zelm MC, van der Burg M, de Ridder D, et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol Baltim Md 1950. 2005;175:5912–5922. [DOI] [PubMed] [Google Scholar]
- 25.Zoutman WH, Nell RJ, Versluis M, et al. A novel digital PCR-based method to quantify (switched) B cells reveals the extent of allelic involvement in different recombination processes in the IGH locus. Mol Immunol. 2022;145:109–123. [DOI] [PubMed] [Google Scholar]
- 26.Wagener R, López C, Kleinheinz K, et al. IG-MYC+ neoplasms with precursor B-cell phenotype are molecularly distinct from Burkitt lymphomas. Blood. 2018;132:2280–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Yu W, Alikarami F, et al. Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood. 2022;139:2198–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Z, Hu Z, Damirchi B, et al. Characterization of PAX5 Mutations in B Progenitor Acute Lymphoblastic Leukemia. Blood. 2022;140:1001–1002. [Google Scholar]
- 29.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinclair PB, Ryan S, Bashton M, et al. SH2B3 inactivation through CN-LOH 12q is uniquely associated with B-cell precursor ALL with iAMP21 or other chromosome 21 gain. Leukemia. 2019;33:1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClure BJ, Heatley SL, Kok CH, et al. Pre-B acute lymphoblastic leukaemia recurrent fusion, EP300-ZNF384, is associated with a distinct gene expression. Br J Cancer. 2018;118:1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda T, Sanada M, Kawazu M, et al. Two novel high-risk adult B-cell acute lymphoblastic leukemia subtypes with high expression of CDX2 and IDH1/2 mutations. Blood. 2022;139:1850–1862. [DOI] [PubMed] [Google Scholar]
- 34.Passet M, Kim R, Gachet S, et al. Concurrent CDX2 cis-deregulation and UBTF-ATXN7L3 fusion define a novel high-risk subtype of B-cell ALL. Blood. 2022;139:3505–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura S, Montefiori L, Iacobucci I, et al. Enhancer retargeting of CDX2 and UBTF::ATXN7L3 define a subtype of high-risk B-progenitor acute lymphoblastic leukemia. Blood. 2022;139:3519–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J-F, Dai Y-T, Lilljebjörn H, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115:E11711–E11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109:3451–3461. [DOI] [PubMed] [Google Scholar]
- 38.Zaliova M, Potuckova E, Lukes J, et al. Frequency and prognostic impact of ZEB2 H1038 and 1072 mutations in childhood B-other acute lymphoblastic leukemia. Haematologica. 2020;106:886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khabirova E, Jardine L, Coorens THH, et al. Single-cell transcriptomics reveals a distinct developmental state of KMT2A-rearranged infant B-cell acute lymphoblastic leukemia. Nat Med. 2022;28:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhrig S, Ellermann J, Walther T, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31:448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ALLCatchR is freely available as an R-package through https://github.com/ThomasBeder/ALLCatchR. Transcriptome sequencing data of bone marrow samples from healthy donors were deposited at the European Genome Phenome archive (EGAS00001007305). BCP-ALL transcriptome profiles have been deposited in open or controlled access archives (EGAS00001006107; https://viz.stjude.cloud/st-jude-childrens-research-hospital/visualization/pax5-driven-subtypes-of-b-progenitor-acute-lymphoblastic-leukemia-genomepaint~16; https://github.com/Oshlack/ALLSorts) or can be obtained by the authors of the original publications upon reasonable request.4–8