Abstract
Background:
TP53 mutation (TP53mut) confers adverse prognosis in acute myeloid leukemia (AML). Venetoclax with hypomethylating agents is a current standard for older patients, however recent reports suggest that TP53mut confers resistance to venetoclax. We investigated outcomes of patients with TP53mut AML treated with 10-day decitabine and venetoclax (DEC10-VEN, NCT03404193).
Methods:
Patients with newly diagnosed AML received decitabine 20mg/m2 for 10-days every 4–6 weeks for induction, followed by decitabine 5-days after response. Venetoclax dose was 400 mg daily. TP53mut was identified in bone marrow samples using NGS with sensitivity of 5%. Outcomes were analyzed per ELN 2017 guidelines.
Results:
Among 118 patients (median age: 72 years, range 49–89), 53% (n=63) had secondary AML, 33% (n=39) had AML with complex karyotype and 30% (n=35) had TP53mut AML. The median TP53 variant allele frequency was 32% (interquartile range 16%−65%), 23% (n=8) patients had only a single TP53 mutation, 43% (n=15) had multiple mutations, and 34% (n=12) had mutation+deletion. Outcomes were significantly worse in TP53mut compared to TP53WT AML with overall response rate of 66% vs 89% (p=.002), CR/CRi of 57% vs 77% (p=.029) and 60-day mortality of 26% vs 4% (p<.001), respectively. Patients with TP53mut vs TP53WT had shorter overall survival at 5.2 vs. 19.4 months (hazard ratio [HR] 4.67, 95%CI 2.44–8.93, p<.0001), and shorter relapse-free survival at 3.4 vs 18.9 months (HR 4.80, 95%CI 1.97–11.69, p<.0001), respectively. Outcomes with DEC10-VEN in TP53mut AML were comparable to historical results with 10-day decitabine alone.
Conclusion:
Patients with TP53mut AML have lower response rates and shorter survival with DEC10-VEN.
Keywords: TP53, AML, acute myeloid leukemia, decitabine, venetoclax, outcome
INTRODUCTION
TP53 is the most frequently mutated gene in human cancer. TP53 functions as a tumor suppressor protecting against cellular stress and serves as the “guardian of the genome” preserving genomic integrity.1 TP53 mutations (TP53mut) occur in 5–10% cases of de-novo AML, with higher frequency in older patients, and in 20–35% cases of therapy-related AML.2–4 AML with TP53mut is associated with complex karyotype, poor response to intensive chemotherapy and has dismal outcomes with short median overall survival (OS) of 5 to 9 months.5,6
Older patients are frequently unfit for intensive chemotherapy and epigenetic therapy with hypomethylating agents (HMA) offer modest advantage over chemotherapy in TP53mut AML.7 The 10-day regimen of decitabine (DEC10) has been noted to be active in adverse-risk AML and those with relapsed/refractory disease.8–11 One study showed 100% response rate in TP53mut AML and MDS and high mutation clearance with the 10-day regimen of decitabine.11 Venetoclax in combination with low-intensity regimens is now a standard therapy for older or unfit patients with AML.12,13 Combining a 10-day regimen of decitabine with venetoclax (DEC10-VEN) showed high activity in adverse-risk AML.14 However, recent preclinical studies have suggested that TP53mut may confer resistance to venetoclax.15–18
Hence, we investigated outcomes of patients with TP53mut AML treated on a prospective clinical trial of DEC10-VEN and compared outcomes to TP53WT AML. Additionally, we evaluated the benefit of adding venetoclax to 10-day decitabine by comparing the results to a historical cohort of patients treated with 10-day decitabine alone from another prospective trial.
METHODOLOGY
Study design and participants
We conducted a post-hoc analysis of a phase 2 trial of 10-day decitabine and venetoclax (DEC10-VEN, NCT03404193). This trial enrolled patients who were 60 years of age and older with newly diagnosed AML who were unfit for intensive chemotherapy, or had secondary AML; or relapsed or refractory (R/R) AML. Patients were eligible if they had an Eastern Cooperative Oncology Group (ECOG) performance status of 3 or less, white blood cell count less than 10 ×109/L, and adequate end-organ function. Patients with European Leukemia Net (ELN) favorable risk cytogenetics and prior exposure to BCL2 inhibitor were excluded.
Patients received decitabine 20 mg/m2 for 10 days every 4 to 6 weeks for induction followed by decitabine for 5 days after CR/CRi. Venetoclax dose was 400 mg daily or equivalent with concomitant azole antifungal. Reduction of venetoclax duration was allowed in cases of prolonged myelosuppression. The full protocol of the study has been published previously.14 In this analysis, we included patients receiving frontline therapy for AML. Additionally, we compared outcomes of these patients treated with DEC10-VEN with individual patient level data of older patients with newly diagnosed TP53mut AML treated with 10-day decitabine alone from another prospective trial at our institution (NCT01786343).8
TP53 sequencing was performed on DNA obtained from bone marrow aspirate using a next generation sequencing (NGS) panel targeting the entire coding or hot spot regions of 81 genes, or TP53 alone, as described previously.19 The covered regions of TP53 included the following exons and (codons): 2 (1–25), 4–11 (80–394). Bidirectional paired-end sequencing was performed using an Illumina MiSeq NGS platform (Illumina, San Diego, CA, USA) to screen for single nucleotide variants, and insertions/deletions (up to 52 base-pairs). The analytical sensitivity of the platform is variable for different genes but is generally 1–3% mutant reads in a background of wild type reads (supplemental methods). Measurable residual disease (MRD) was assessed using bone marrow aspirate samples using multiparametric flow cytometry (FCM) validated to a sensitivity level of 0.01–0.1%.20 All cytogenetic and molecular analyses were conducted in a CLIA-certified laboratory.
Outcomes
The studied outcomes included response, relapse free survival (RFS) and overall survival (OS) defined per the ELN 2017 criteria.21 Overall response rate (ORR) included complete response (CR), CR with incomplete hematologic recovery (CRi), and morphologic leukemia-free state (MLFS). OS was defined from time from treatment initiation until death or censored at the last follow-up. RFS was defined from the time from achievement of CR or CRi until relapse or death or censored at last follow-up. In patients with TP53mut AML outcomes were compared between responding patients without relapse until last follow-up; relapse after response, defined as morphologic relapse in bone marrow or peripheral blood after achieving a response; and primary refractory disease, defined as no response by four cycles of therapy.
Statistical Analysis
Chi-square test or Fisher’s exact test was used to compare distribution of categorical variables between groups. Wilcoxon-Rank sum test was used for continuous variables between groups as appropriate. The distributions of time-to-event endpoints including RFS and OS were estimated using the Kaplan-Meier method and compared using the log-rank test. Cox proportional hazard model was used to determine the hazard ratio for outcome related to TP53 mutation status. Univariate and multivariate logistic and Cox-regression models were used to evaluate the association between patient characteristics and outcomes. For multivariate regression, variables were selected using a backward selection with a p-value cut-off at 0.05. TP53 variant allelic frequencies (VAF) at screening and after cycle 1 were compared using paired t-test. All analyses were conducted using STATA version 13.0 (StataCorp, College Station, TX), Prism v 8.4 (GraphPad software, San Diego, California), and R v3.4.3 (R Core Team, Vienna, Austria).
RESULTS
Between January 20, 2018 and April 15, 2020, 118 patients received frontline therapy with DEC10-VEN and 35 (30%) patients had TP53mut AML. The median age was 72 years (range 49–89). Eighty (68%) patients were older than 70 years, 32 (27%) patients had ECOG performance status of 2 or higher, 63 (53%) patients had secondary AML including 25 (21%) patients with therapy-related AML. Seventy-eight (66%) patients had ELN adverse risk AML and 39 (33%) patients had AML with complex karyotype (Table 1). Patients with TP53mut AML were more likely to have therapy-related AML (t-AML) in 46% (n=16/35) patients compared to 11% (n=9/83) in patients with TP53WT AML (p<.001). Patients with TP53mut AML were less likely to have co-mutations compared to patients with TP53WT AML including NPM1 (3% vs 33%, p=.001), RUNX1 (2% vs 22%, p=.035), ASXL1 (3% vs 23%, p=.008) and K/NRAS (11% vs 28%, p=.005) and were more likely to have AML with complex karyotype compared to patients with TP53WT AML (89% vs 10%, p< .001). The proportion of prior therapies for antecedent hematological disorder including hypomethylating agents, intensive chemotherapy, and stem-cell transplantation were comparable among patients with TP53mut and TP53WT AML (Table 1).
Table 1.
Baseline characteristics of patients with acute myeloid leukemia with and without TP53 mutation treated with 10-day decitabine and venetoclax.
| Patient characteristics | TP53 mutated AML (N= 35) | TP53 wild type AML (N= 83) | P |
|---|---|---|---|
|
| |||
| Age, years | 74 [69–78] | 71 [68–77] | |
| ≥ 70 years | 25 (71) | 55 (66) | .583 |
| Male sex | 18 (51) | 46 (55) | .691 |
| ECOG Performance Status 0–1 |
25 (71) | 61 (73) | .818 |
| ≥2 | 10 (29) | 22 (27) | |
| Peripheral blood blasts, % | 5 [0–17] | 10 [0–37] | .247 |
| Bone marrow blasts, % | 28 [14–54] | 40 [23–62] | .056 |
| Diagnosis De novo AML |
11 (31) | 44 (53) | .032 |
| sAML with AHD | 12 (34) | 32 (39) | .661 |
| Therapy-related AML | 16 (46) | 9(11) | <.001 |
| ELN 2017 cytogenetic risk Favorable |
0 (0) | 0 (0) | |
| Intermediate | 2 (6) | 58 (70) | |
| Adverse | 33 (94) | 25 (30)1 | <.001 |
| Complex cytogenetics | 31 (89) | 8 (10) | <.001 |
| Co-mutations NPM1 |
1 (3) | 27 (33) | .001 |
| FLT3-ITD/TKD | 0 (0) | 18 (22) | .001 |
| IDH1/2 | 4 (11) | 21 (25) | .092 |
| RUNX1 | 2 (6) | 18 (22) | .035 |
| ASXL1 | 1 (3) | 19 (23) | .008 |
| K/NRAS | 4 (11) | 23 (28) | .005 |
| ELN 2017 risk group Favorable |
0 (0) | 26 (31) | |
| Intermediate | 0 (0) | 14 (17) | <.001 |
| Adverse | 35 (100) | 43 (52) | |
| Prior therapy for AHD | 7 (20) | 21 (25) | .536 |
| Hypomethylating agent (HMA) | 7 (20) | 18 (22) | .838 |
| Intensive chemotherapy (IC) | 0 (0) | 4 (5) | .186 |
| HMA and IC | 0 (0) | 2 (2) | .354 |
| Stem-cell transplantation | 3 (9) | 5 (6) | .615 |
All results expressed as no. (%) or median [interquartile range], unless specified. ECOG: Eastern Cooperative Oncology Group; sAML: secondary acute myeloid leukemia; AHD: antecedent hematological disorder; ELN: European Leukemia Net.
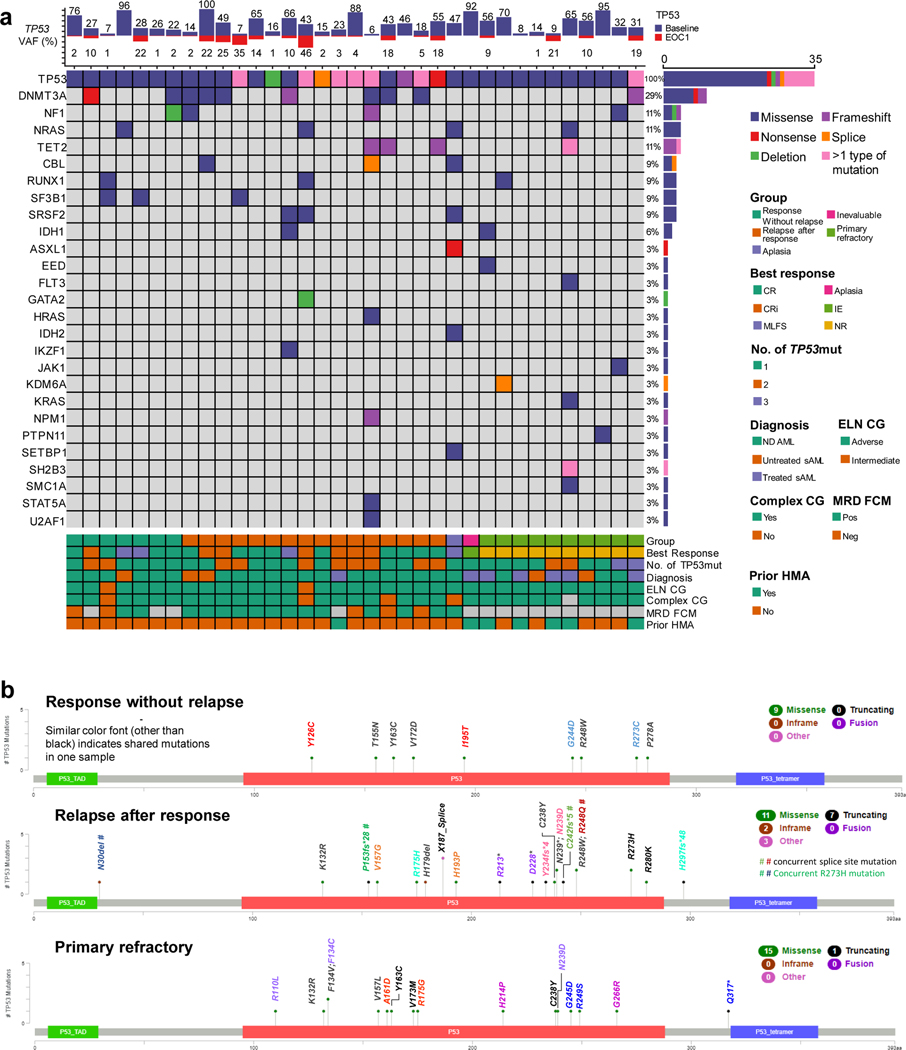
The most frequent co-mutations in TP53mut AML included DNMT3A in 29% patients (Fig. 1a), NF1, NRAS, TET2 (11% each) and CBL, RUNX1, SF3B1 or SRSF2 (9%, each). The median variant allele frequency (VAF) of TP53mut was 32% (IQR 16–65%). At least one mutation per case involved the DNA binding domain of TP53 in all TP53mut AML. TP53-altered subgroups included single mutation only without deletion of TP53 (n = 8/35, 23%) and multi-hit alterations (n=27/35, 77%) including multiple mutations without chromosomal deletion involving the TP53 locus (n =12/35, 34%), or TP53 mutation(s) with concomitant deletion noted on karyotype, array CGH or FISH (n = 15/35, 43%). Copy neutral loss of heterozygosity data was not available in this study.
Fig 1.
a. TP53 mutations mapped according to response b. mutational landscape of TP53 mutated acute myeloid leukemia treated with 10-day decitabine and venetoclax.
>1 type of mutation refers to patients with a missense and frameshift mutation or missense and nonsense mutation, etc.
Mutations in patients with response without relapse (n=7, 20%) vs. relapse (n=16, 48%) vs. primary refractory disease (n=10, 30%) are shown in Fig. 1b. Proportion of multi-hit TP53 alterations were noted in 4/7 (57%) of responding patients without relapse vs. 13/16 (81%) in patients with relapse after response vs. 10/10 (100%) in refractory AML with statistical significance (p= .049). Median baseline VAF was similar in these populations with median VAF of 27% in patients with response without relapse vs. 36% in patients with relapse after response vs. 44% in refractory AML, (p=.918). Twenty-five (76%) patients with TP53mut AML had follow-up NGS testing at the end of cycle 1 (EOC1) including 20 responders (with or without relapse) and 5 refractory patients. Responding patients had significant reduction of mutant TP53 VAF (mean change −28.5%, 95% CI −15.4, −41.6%, p<.001). Among 5 patients who had refractory disease, the VAF change was not significant (mean change −21.4%, 95% CI −9.5, 52.2%, p=.126), and none obtained a TP53 VAF <5% at EOC1. Thirteen patients with relapsed disease after response had NGS at time of progression; there was significant increase in TP53mut VAF compared to VAF after the end of cycle 1 (mean change +22.6%, 95% CI 4.8, 40.5%, p=.018).
The number of co-mutations were comparable among 3 groups with a median of one co-mutation in each of the 3 aforementioned groups. There were no identifiable differences in characteristics of TP53 mutation between patients without relapse vs. those experiencing relapse including multi-hit alterations vs single mutation only (p=.312), VAF (p=.806), co-mutations (p=.830) or complex karyotypes (p= .791). Nine out of 10 (90%) patients with refractory disease had secondary AML, compared to 60% of responding patients (with or without relapse, p=.084). History of antecedent hematological disorder was present in 7 out of 10 patients (70%) with refractory disease compared to 4 out of 23 of patients (17%) with responsive disease (p = .006). Five out of 10 (50%) patients with refractory disease had prior HMA exposure, compared to 1 out of 23 (4%) patients with responsive disease (p=.005). Patients with response without relapse had longer OS compared to patients with primary refractory disease (9.6 vs 1.9 months, HR 4.81, 95% CI 1.45–15.96, p= .010) but their OS was not significantly different compared to patients with relapsed disease (9.6 vs 6.9 months, HR 0.95, 95% CI 0.30–2.96, p=.928; Fig. S1).
Patients with TP53mut AML had significantly lower response rates compared to patients with TP53WT AML (Table 2). The ORR in patients with TP53mut compared to TP53WT AML was 66% vs 89% (p=.002), with CR/CRi in 57% vs 77% (p=.029) with lower rates of MRD negativity by FCM at 29% compared to 59% (p=.012). Incidence of primary refractory disease was 34% in patients with TP53mut AML versus 11% in patients with TP53WT AML (p=.002). On univariate and multivariate analysis, TP53mut AML conferred significantly lower odds ratio of achieving CR (odds ratio [OR] 0.17, p<.001) and CR/CRi (OR 0.22, p=.003; Table 3, S1, S2, S3). Compared to patients with TP53WT AML, those with TP53mut AML had a higher 30-day mortality (1% vs 3%, p= .525) and higher 60-day mortality (4% vs 26%, p<.001). All early deaths (n=9) in patients with TP53mut AML within 60 days occurred in those who had refractory disease. Mortality due to uncontrolled infection or sepsis was not statistically significant in patients with TP53mut AML compared to TP53WT AML patients (21% vs. 17%, p=.586). Patients with response MLFS or better had lower infection-related mortality (6% vs. 4%, p=.230). Six (67%) patients had sepsis and 3 (33%) patients transitioned to hospice. Sixteen patients (19%) with TP53WT AML and only 1 patient (3%) with TP53mut AML underwent stem-cell transplantation (SCT) after response.
Table 2.
Outcomes of patients with acute myeloid leukemia with and without TP53 mutation treated with 10-day decitabine and venetoclax.
| Outcome | TP53 mutated AML (N= 35) | TP53 wild type AML (N= 83) | p |
|---|---|---|---|
|
| |||
| Overall response rate | 23 (66) | 74 (89) | .002 |
| CR | 13 (37) | 48 (58) | .040 |
| CRi | 7 (20) | 16 (19) | .928 |
| CR/CRi | 20 (57) | 64 (77) | .029 |
| Morphologic leukemia-free state | 3 (9) | 10 (12) | .582 |
| MRD negative by FCM | 6 (29) | 44 (59) | .012 |
| No response | 10 (29) | 9 (11) | .017 |
| Inevaluable/Aplasia1 | 2 (6) | 0 (0) | .028 |
| 30-day mortality | 1 (3) | 1 (1) | .525 |
| 60-day mortality | 9 (26) | 3 (4) | < .001 |
All results expressed as no. (%). CR=complete remission, CRi = CR with incomplete hematologic recovery; MRD = minimal residual disease FCM = flow cytometry.
One patient had early death before first evaluation and one patient with aplasia on initial evaluation passed away prior to repeat bone marrow evaluation.
Table 3.
Multivariate analysis for achievement of CR and overall survival
| Parameter | OR (95% CI) | p |
|---|---|---|
| Achievement of CR | ||
| TP53 mutated vs wild type | 0.17 (0.06–0.47) | <.001 |
| ECOG PS ≥2 vs 0–1 | 0.24 (0.08–0.71) | .010 |
| Prior HMA for AHD vs none | 0.15 (0.01–0.24) | .002 |
| RUNX1 mutated vs wild type | 0.23 (0.06–0.88) | .031 |
| ASXL1 mutated vs wild type | 0.05 (0.12–0.24) | <.001 |
| Overall survival | HR (95% CI) | |
| TP53 mutated vs wild type | 6.96 (3.76–12.88) | <0.001 |
| sAML with AHD vs de novo AML | 2.97 (1.78–4.94) | <0.001 |
| DNMT3A mutated vs wild type | 0.44 (0.24–0.81) | 0.009 |
| K/NRAS mutated vs wild type | 2.82 (1.58–5.02) | <0.001 |
PS = performance status, HMA = hypomethylating agent, AHD = antecedent hematological disorder, sAML = secondary AML
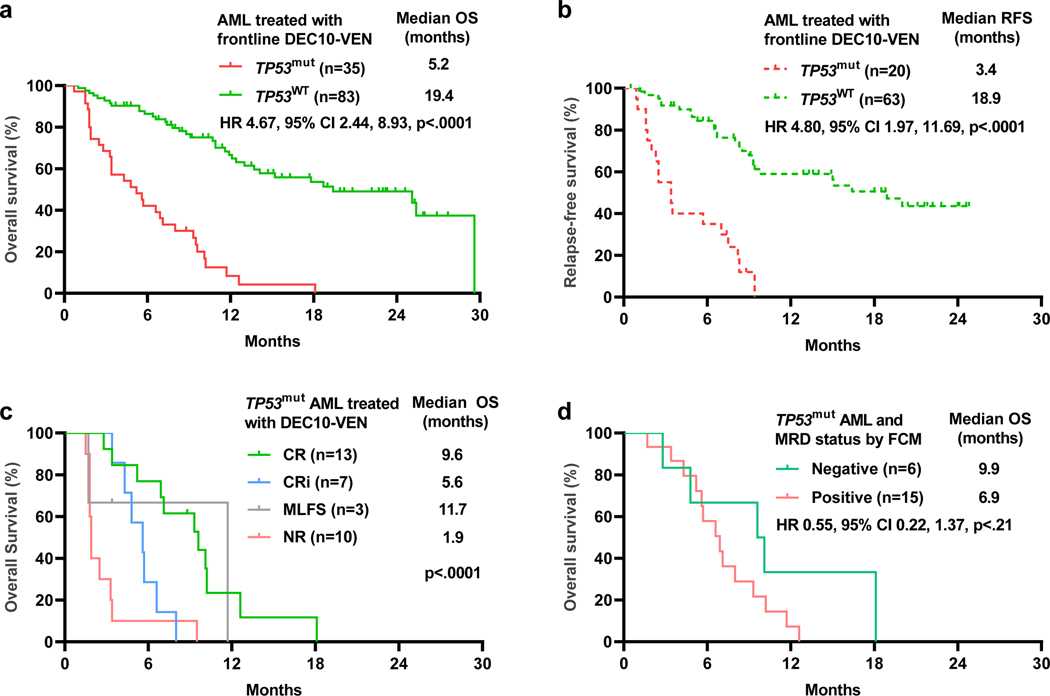
After a median follow-up of 20.2 months (95% CI 15.6, 22.9), the median OS in patients with TP53mut AML was inferior compared to patients with TP53WT AML (5.2 vs 19.4 months, HR 4.67, 95% CI 2.44–8.93, p <.0001, Fig. 2a). Similarly, median RFS in patients with TP53mut AML was significantly shorter compared to patients with TP53WT AML (3.4 vs 18.9 months, HR 4.80, 95% CI 3.26–14.99, p<0.001, Fig. 2b). Patients with TP53mut AML patients achieving CR had a median OS of 9.6 months compared to patients with CRi who had a median OS of 5.6 months vs non-responding patients who had a median OS of 1.9 months (Fig. 2c). Patients with TP53mut AML who achieved negative MRD status by FCM had numerically higher median OS at 9.9 months, compared to patients with persistent MRD who had a median OS at 6.9 months, however this analysis was limited by the small sample size (HR 0.55, 95% CI 0.22–1.37, p=.21, Fig. 2d). Patients with TP53mut AML had significantly shorter duration of response compared to TP53WT AML (3.5 months vs NR, HR 7.21, 95% CI 3.34–15.56, p<0.001, Fig. S2).
Fig 2.
a. Overall survival (OS) by TP53 mutation status. b. Relapse Free Survival (RFS) by TP53 mutation status. c. OS by response in TP53 mutated acute myeloid leukemia (AML). d. OS in TP53 mutated AML patients by measurable residual disease (MRD) status.
TP53 VAF cut-offs ranging from 20% to 40% did not have prognostic value for OS or RFS with DEC10-VEN. Patients with multi-hit alterations did not have significant survival difference compared to patients with single mutation only in OS (HR 1.24, 95% CI 0.50–3.05, p= .643; Fig. S3) or RFS (HR 1.65, 95% CI 0.37–7.34, p= .512). Fifteen patients with TP53 mutation with noted deletion on karyotype, array CGH or FISH did not show difference in OS compared to TP53mut patients without noted deletion (HR 0.95, 95% CI 0.46–1.93, p= .876).
Other independent adverse prognostic factors for achievement of CR included ECOG PS ≥2, RUNX1mut, ASXL1mut, and prior therapy for antecedent hematological disorder (Table 3). Univariate and multivariate analyses for OS confirmed that TP53mut was associated with significantly higher risk of death (HR 6.96, 95% 3.76–12.88), along with secondary AML with antecedent hematologic disorder, K/NRAS and DNMT3A mutations (Table S5, 3). Similarly, univariate and multivariate analyses of RFS confirmed that TP53mut was independently associated with high risk of relapse for patients with CR/CRi (HR 5.52, 95% CI 2.70–11.28, p<0.001, Table S6, S7).
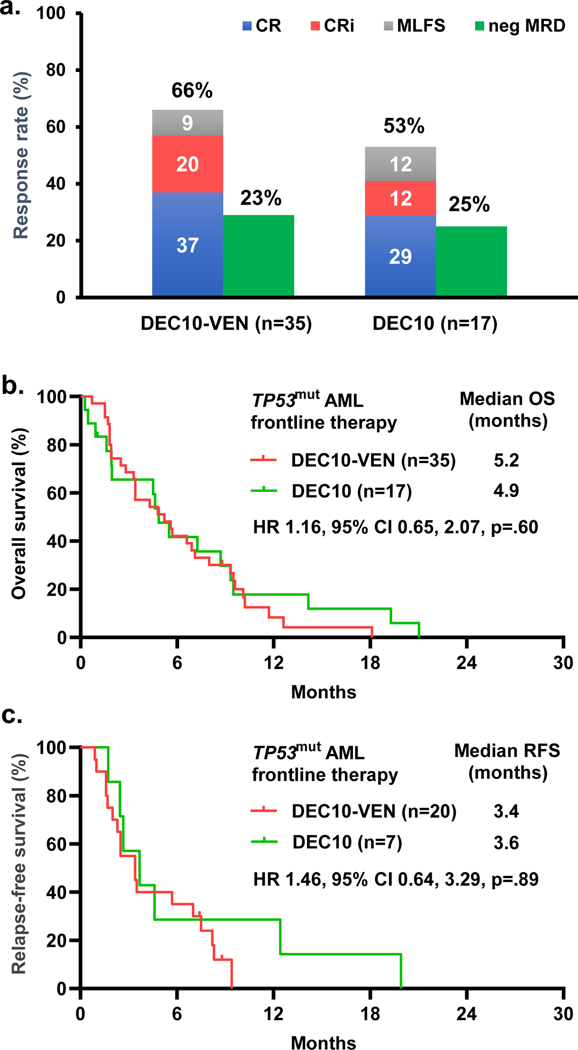
Fig 3.
Outcomes in patients with newly diagnosed TP53mut AML with 10-day decitabine with venetoclax vs 10-day decitabine alone, a. morphologic response and measurable residual disease (MRD) status, b. overall survival, and c. relapse-free survival. CR = complete response, CRi = CR with incomplete hematologic response, MLFS = morphologic leukemia-free state
Finally, we compared outcomes of patients with newly diagnosed TP53mut AML treated with DEC10-VEN (n=35) versus 10-day decitabine alone (n=17) treated on a separate prospective clinical trial (NCT01786343). The baseline characteristics of these patients were comparable (Table S4). Overall response rate was numerically higher at 66% with DEC10-VEN compared to 53% with 10-day decitabine. Negative MRD status was achieved in 29% (n=6/20) with DEC10-VEN compared to 25% (n=2/8) patients with 10-day decitabine. Time to morphologic response was comparable with DEC10-VEN vs 10-day decitabine at 1.2 months (IQR 1.1–1.4) vs. 1.3 months (IQR 1.2–2.4), respectively (p=.197). There was no significant difference in OS or RFS (Fig. 3). None of the patients who received 10-day decitabine underwent SCT. 7 patients with TP53mut AML treated with 5-day decitabine alone in same trial had lower response rate at 43% including 1 CR, 1 CRi and 1 MLFS. 2 patients achieved MRD negative status (66%). While the sample size is very limited, there was no significant difference in OS or RFS compared to DEC10-VEN trial (Fig. S4).
DISCUSSION
Development of venetoclax has been an important breakthrough for the field of AML therapy, however primary and acquired resistance to venetoclax-based regimens continues to be a major problem. To our knowledge, this report represents the largest analysis to date which validates pre-clinical findings and smaller prior reports on the adverse impact of TP53mut with venetoclax and HMA. Our study showed that patients with TP53mut AML experienced significantly lower response rates and survival with DEC10-VEN compared to patients with TP53WT AML despite reasonable response rates. Patients with TP53mut AML with prior HMA exposure were significantly less likely to respond to DEC10-VEN. This is in contrast to our previous findings where patients failing frontline HMA for AML are still likely respond to DEC10-VEN compared to salvage intensive chemotherapy.22
Patients who achieved a CR or CRi had modestly better survival compared to patients who had refractory disease, however the number of patients in these sub-analyses were small. Achieving negative MRD status did not show significant benefit in our study. Overall, these results were comparable to prior reports of HMA with venetoclax showing ORR of 14 to 62% in TP53mut AML and short overall survival.16–18 Previous reports have shown median OS of 2.1 to 10.1 months in TP53mut AML with HMA or low-intensity therapy.5,11,23,24 No TP53 VAF cut-off showed prognostic value for OS with DEC10-VEN in our study, consistent with prior studies investigating HMA or HMA with venetoclax in AML.8,17,25 Interestingly, there was no direct therapy-related mortality in patients with TP53mut, yet these patients had significantly higher early mortality due to refractory disease and infections (66%).
In our study, only 1 (3%) patient with TP53mut underwent SCT; the majority of patients were ineligible for SCT due to co-morbidities, development of complications including infections, or refractory disease. It is debatable if outcomes would have been different if more patients could have received SCT as patients with TP53mut AML are at significant risk of relapse following SCT with long-term survival of less than 10%.6,11 Pre-clinical studies have suggested that TP53mut confers intrinsic resistance to venetoclax through perturbation of mitochondrial homeostasis and cellular metabolism including increased oxidative phosphorylation.15 This study along with prior reports provide clinical validation of these pre-clinical findings and highlight the urgent need for novel therapies for TP53mut AML. TP53mut AML remains a therapeutic challenge and optimal backbone for combination with novel therapies remains to be evaluated in prospective trials. Potential approaches to overcome such mutant p53 mediated resistance include tropomyosin receptor kinase (TRK) inhibition, targeting oxidative phosphorylation or glutamine metabolism, p53 reactivators and harnessing other p53 independent mechanisms.6,15,26 Novel immunotherapeutic approaches including magrolimab, flotetuzumab, cusatuzumab are currently advancing in clinical trials and offer hope for patients with TP53mut AML.
This was a post-hoc analysis which has inherent limitations. Detailed comparisons within subgroups of TP53mut AML were limited by the small number of patients. TP53 mutation analysis was conducted in a clinical laboratory and detailed information beyond standard clinical testing were not available, e.g., allelic status, copy neutral loss of heterozygosity, single-cell level data, etc.27 We only had three patients with responses lasting beyond six months. Consequently, we could not evaluate patients with TP53mut AML who may have truly durable responses to venetoclax, and some patients without relapse had short follow-up and may relapse with longer follow-up. There was no preferential TP53 mutations that we identified specific to responders or non-responders in the study. Further larger cohort study with TP53mut AML patients with venetoclax-based therapy would provide insight in the role of specific TP53 mutations in treatment response. Our exploratory comparison of DEC10-VEN vs 10-day decitabine alone should be interpreted with caution due to the small number of patients and ineluctable differences between the two trial populations. The backbone of 10-day decitabine offers a different risk-benefit ratio compared to the more widely adopted 5-day regimen, or the 7-day regimen of azacitidine used with venetoclax, thus limiting cross-trial comparisons.
In summary, we report the largest series of patients with TP53mut treated on a prospective trial of DEC10-VEN and show that outcomes in these patients are significantly worse compared to patients with TP53WT AML. These results highlight the urgent need for novel therapies for TP53mut AML.
Supplementary Material
Acknowledgements:
This study was supported in part by the MD Anderson Cancer Center Support Grant CA016672 from the National Cancer Institute and the Research Project Grant Program (R01CA235622) from the National Institutes of Health. A. Maiti was supported by the American Society of Clinical Oncology Young Investigator Award.
Conflict of Interests
KK : None
AM : Celgene: Research Funding
SL : None
RP : None
TMK : Abbvie: Honoraria, Research Funding; Pulmotec: Research Funding; Celgene: Research Funding; Amgen: Research Funding; JAZZ: Honoraria, Research Funding; Cyclacel: Research Funding; Genentech: Honoraria, Research Funding; Ascentage: Research Funding; Incyte: Research Funding; Astra Zeneca: Research Funding; Astellas: Research Funding; Novartis: Honoraria; Pfizer: Honoraria, Research Funding; BMS: Honoraria, Research Funding; Cellenkos: Research Funding.
CRR : None
KF : None
ND : Gilead: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Amgen: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Daiichi Sankyo: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; ImmunoGen: Research Funding; Novartis: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Celgene: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Jazz: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Trillium: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Syndax: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Amgen: Consultancy, Membership on an entity’s Board of Directors or advisory committees; KITE: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Agios: Consultancy, Membership on an entity’s Board of Directors or advisory committees; Trovagene: Research Funding; Fate Therapeutics: Research Funding; Bristol-Myers Squibb: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Pfizer: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Karyopharm: Research Funding; Servier: Research Funding; Genentech: Research Funding; AbbVie: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Astellas: Consultancy, Membership on an entity’s Board of Directors or advisory committees, Research Funding; Novimmune: Research Funding.
YA : None
MO : None
KS : None
NJS : Amgen: Honoraria; AstraZeneca: Consultancy; Astellas: Research Funding; Takeda Oncology: Consultancy, Honoraria, Research Funding.
KT : Symbio Pharmaceuticals: Advisory board, Consultancy; GSK: Consultancy; Celgene: Consultancy
MY : Pint Pharma: Honoraria; Pfizer: Research Funding; Daicho Sankyo: Research Funding. Alvarado: Daiichi-Sankyo: Research Funding; BerGenBio ASA: Research Funding; MEI Pharma: Research Funding; Jazz Pharmaceuticals: Research Funding; Astex Pharmaceuticals: Research Funding; Sun Pharma: Research Funding; Tolero Pharmaceuticals: Research Funding; FibroGen: Research Funding.
FR : Amgen: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria; BMS: Consultancy, Honoraria, Research Funding; Xencor: Consultancy, Honoraria, Research Funding; AstraZeneca: Consultancy, Honoraria; Macrogenics: Research Funding; Abbvie: Consultancy, Honoraria, Research Funding; Jazz Pharmaceuticals: Consultancy, Honoraria, Research Funding; Astellas: Consultancy, Honoraria, Research Funding; Orsenix: Consultancy, Honoraria, Research Funding.
HMK : Daiichi-Sankyo: Honoraria, Research Funding; Ascentage: Research Funding; Amgen: Honoraria, Research Funding; BMS: Research Funding; Abbvie: Honoraria, Research Funding; Jazz: Research Funding; Novartis: Honoraria, Research Funding; Pfizer: Honoraria, Research Funding; Sanofi: Research Funding; Actinium: Honoraria, Membership on an entity’s Board of Directors or advisory committees; Adaptive biotechnologies: Honoraria; Aptitute Health: Honoraria; BioAscend: Honoraria; Delta Fly: Honoraria; Janssen: Honoraria; Oxford Biomedical: Honoraria; Immunogen: Research Funding.
CDD : Takeda: Honoraria; Daiichi Sankyo: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Calithera: Research Funding; MedImmune: Honoraria; Notable Labs: Membership on an entity’s Board of Directors or advisory committees; Jazz: Honoraria; Agios: Consultancy, Honoraria, Research Funding; ImmuneOnc: Honoraria, Research Funding; AbbVie: Consultancy, Honoraria, Research Funding; Novartis: Consultancy.
MYK : Amgen: Consultancy; Stemline Therapeutics: Consultancy, Research Funding; Agios: Research Funding; Calithera: Research Funding; Cellectis: Research Funding; Reata Pharmaceutical Inc.;: Patents & Royalties: patents and royalties with patent US 7,795,305 B2 on CDDO-compounds and combination therapies, licensed to Reata Pharmaceutical; AstraZeneca: Research Funding; Eli Lilly: Research Funding; Ablynx: Research Funding; Ascentage: Research Funding; Rafael Pharmaceutical: Research Funding; Kisoji: Consultancy; Sanofi: Research Funding; Forty-Seven: Consultancy, Research Funding; AbbVie: Consultancy, Research Funding; Genentech: Consultancy, Research Funding; F. Hoffmann La-Roche: Consultancy, Research Funding.
Footnotes
Prior Presentation:
These results were presented in an abstract form at the Annual Meeting of the American Society of Hematology, December 17th 2020.
Data sharing:
At this time, we will not be able to share individual patient level data outside of our institution.
REFERENCES
- 1.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2(1):a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen D, Groves MJ, Burnett AK, et al. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia 2009;23(1):203–6. [DOI] [PubMed] [Google Scholar]
- 3.Ok CY, Patel KP, Garcia-Manero G, et al. Mutational profiling of therapy-related myelodysplastic syndromes and acute myeloid leukemia by next generation sequencing, a comparison with de novo diseases. Leukemia Research 2015;39(3):348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New England Journal of Medicine 2016;374(23):2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadia TM, Jain P, Ravandi F, et al. TP53 mutations in newly diagnosed acute myeloid leukemia: Clinicomolecular characteristics, response to therapy, and outcomes. Cancer 2016;122(22):3484–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter AM, Sallman DA. Current status and new treatment approaches in TP53 mutated AML. Best Practice & Research Clinical Haematology 2019;32(2):134–44. [DOI] [PubMed] [Google Scholar]
- 7.Montalban-Bravo G, Takahashi K, Garcia-Manero G. Correspondence from The New England Journal of Medicine — Decitabine in TP53-Mutated AML. New Engl J Med 2017;376(8):796–7. [DOI] [PubMed] [Google Scholar]
- 8.Short NJ, Kantarjian HM, Loghavi S, et al. Treatment with a 5-day versus a 10-day schedule of decitabine in older patients with newly diagnosed acute myeloid leukaemia: a randomised phase 2 trial. The Lancet Haematology 2019;6(1):e29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum W, Garzon R, Klisovic RB, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. PNAS 2010;107(16):7473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatnagar B, Duong VH, Gourdin TS, et al. Ten-day decitabine as initial therapy for newly diagnosed patients with acute myeloid leukemia unfit for intensive chemotherapy. Leukemia & Lymphoma 2014;55(7):1533–7. [DOI] [PubMed] [Google Scholar]
- 11.Welch JS, Petti AA, Miller CA, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med 2016;375(21):2023–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiNardo CD, Pratz KW, Letai A, et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol 2018;19(2):216–28. [DOI] [PubMed] [Google Scholar]
- 13.DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. New England Journal of Medicine 2020;383(7):617–29. [DOI] [PubMed] [Google Scholar]
- 14.DiNardo CD, Maiti A, Rausch CR, et al. 10-day decitabine with venetoclax for newly diagnosed intensive chemotherapy ineligible, and relapsed or refractory acute myeloid leukaemia: a single-centre, phase 2 trial. Lancet Haematol 2020;7:e724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nechiporuk T, Kurtz SE, Nikolova O, et al. The TP53 Apoptotic Network is a Primary Mediator of Resistance to BCL2 inhibition in AML Cells. Cancer Discov 2019;CD-19–0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNardo CD, Tiong IS, Quaglieri A, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020;135(11):791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aldoss I, Zhang J, Pillai R, et al. Venetoclax and hypomethylating agents in TP53-mutated acute myeloid leukaemia. Br J Haematol 2019;187(2):e45–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y-W, Tsai C-H, Lin C-C, et al. Cytogenetics and mutations could predict outcome in relapsed and refractory acute myeloid leukemia patients receiving BCL-2 inhibitor venetoclax. Ann Hematol 2020;99(3):501–11. [DOI] [PubMed] [Google Scholar]
- 19.Quesada AE, Routbort MJ, DiNardo CD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol 2019;94(7):757–66. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Jorgensen JL, Wang SA. How Do We Use Multicolor Flow Cytometry to Detect Minimal Residual Disease in Acute Myeloid Leukemia? Clin Lab Med 2017;37(4):787–802. [DOI] [PubMed] [Google Scholar]
- 21.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129(4):424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiti A, DiNardo CD, Kadia TM, et al. Ten-Day Decitabine with Venetoclax Versus Intensive Chemotherapy in Relapsed or Refractory Acute Myeloid Leukemia: A Propensity Score Matched Analysis. Blood 2020;136(Supplement 1):30–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boddu P, Kantarjian H, Ravandi F, et al. Outcomes with lower intensity therapy in TP53 -mutated acute myeloid leukemia. Leukemia & Lymphoma 2018;59(9):2238–41. [DOI] [PubMed] [Google Scholar]
- 24.Bewersdorf JP, Shallis RM, Gowda L, et al. Clinical Outcomes of Patients (pts) with TP53-Mutated Acute Myeloid Leukemia (AML) or Myelodysplastic Syndromes (MDS): A Single Center Experience. Blood 2019;134(Supplement_1):5173–5173. [Google Scholar]
- 25.Short NJ, Montalban-Bravo G, Hwang H, et al. Prognostic and therapeutic impacts of mutant TP53 variant allelic frequency in newly diagnosed acute myeloid leukemia. Blood Adv 2020;4(22):5681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan R, Ruvolo V, Mu H, et al. Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell 2017;32(6):748–760.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard E, Nannya Y, Hasserjian RP, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nature Medicine 2020;26(10):1549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
At this time, we will not be able to share individual patient level data outside of our institution.