Abstract
The cell cycle regulator cyclin D3 (CCND3) is highly expressed in multiple myeloma (MM) and it promotes MM cell proliferation. After a certain phase of cell cycle, CCND3 is rapidly degraded, which is essential for the strict control of MM cell cycle progress and proliferation. In the present study, we investigated the molecular mechanisms regulating CCND3 degradation in MM cells. By utilizing affinity purification-coupled tandem mass spectrometry, we identified the deubiquitinase USP10 interacting with CCND3 in human MM OPM2 and KMS11 cell lines. Furthermore, USP10 specifically prevented CCND3 from K48-linked polyubiquitination and proteasomal degradation, therefore enhancing its activity. We demonstrated that the N-terminal domain (aa. 1–205) of USP10 was dispensable for binding to and deubiquitinating CCND3. Although Thr283 was important for CCND3 activity, it was dispensable for CCND3 ubiquitination and stability modulated by USP10. By stabilizing CCND3, USP10 activated the CCND3/CDK4/6 signaling pathway, phosphorylated Rb, and upregulated CDK4, CDK6 and E2F-1 in OPM2 and KMS11 cells. Consistent with these findings, inhibition of USP10 by Spautin-1 resulted in accumulation of CCND3 with K48-linked polyubiquitination and degradation that synergized with Palbociclib, a CDK4/6 inhibitor, to induce MM cell apoptosis. In nude mice bearing myeloma xenografts with OPM2 and KMS11 cells, combined administration of Spautin-l and Palbociclib almost suppressed tumor growth within 30 days. This study thus identifies USP10 as the first deubiquitinase of CCND3 and also finds that targeting the USP10/CCND3/CDK4/6 axis may be a novel modality for the treatment of myeloma.
Keywords: cyclin D3, multiple myeloma, ubiquitin specific peptidase 10, cyclin-dependent kinase, mass spectrometry
Introduction
The D-type cyclin family members including CCND1, CCND2, and CCND3 are critical regulators of the cell cycle G1-S transition in response to growth factor stimulation in somatic cells. Upon stimulation of cell proliferation signaling, D-cyclins interact with cyclin-dependent kinases (CDKs), specifically CDK4/6, as regulatory subunits and trigger autophosphorylation of CDK4/6. The CDK4/6 kinases further phosphorylate and inactivate the retinoblastoma (Rb) protein and promote the transcription of key genes for DNA synthesis [1]. D-cyclins therefore display an oncogenic role in promoting cancer cell proliferation, including multiple myeloma (MM), an incurable hematological malignancy derived from clonal plasma cells [2]. Of the D-cyclin family, CCND3 is highly dysregulated in MM cells in association with the t(6;14) chromosomal translocation and other molecular alterations [3]. The specific expression pattern of CCND1 and CCND3 has been considered as a molecular subtype of MM [4, 5].
CCND3 is cell-cycle dependent and it should be rapidly degraded after a certain phase of the cell cycle, which is essential for the strict modulation of cell cycle progress and cell proliferation. CCND3 contains a PEST domain (rich in proline (P), glutamic acid (E), serine (S), and threonine (T)), a signal peptide for protein degradation. It has been demonstrated that CCND3 undergoes rapid degradation via the ubiquitin proteasomal pathway mediated by the ubiquitin ligases FBXL2 [6] and FBXL8 [7]. Cul-1, a component of the SCF ubiquitin ligase complex, is also associated with CCND3 ubiquitination and degradation [8]. However, protein ubiquitination is a dynamic and reversible process. The covalently attached ubiquitin molecules could be hydrolyzed by specific enzymes called deubiquitinases (Dubs) or ubiquitin-specific proteases (USPs). It is known that CCND1 could be deubiquitinated by several Dubs including USP2 [9], USP2a [10], USP10 [11], and USP22 [12]. However, no Dubs of CCND3 have been reported. In the present study, we identified ubiquitin-specific protease 10 (USP10) by tandem mass spectrometry from the CCND3-interactome. USP10 binds to CCND3 and prevents its K48-linked polyubiquitination, therefore, increasing its stability and promoting MM cell proliferation. Moreover, we found that inhibition of USP10 synergizes with CDK4/6 inhibition in MM cell apoptosis.
Materials and methods
Cell culture
Human embryonic kidney cells (HEK293T) were kindly provided by Dr. Michael F. Moran, University of Toronto, Canada. MM cell lines OPM2 and KMS11 were kindly provided by Dr. A. Keith Stewart, University Health Network, Toronto, Canada. HK293T was cultured in DMEM (Hyclone®, Logan, UT, USA) and MM cell lines were cultured in IMDM (Hyclone®), supplemented with 10% fetal bovine serum (ExCell Bio, Inc., Shanghai, China) and 100 μg/mL penicillin, and 100 units/mL streptomycin (Beyotime Biotech. Inc., Shanghai, China). Cells were cultured at 37 °C with steady 5% CO2.
Plasmids and gene transfection
The complete cDNA fragments for USP10 and CCND3 were cloned into pcDNA3.1 vectors. The siUSP10 (5′-CCAUAAAGAUUGCAGAGUUTT-3′) and siCCND3 (5′-UGCGGAAGAUGCUGGCUUA-3′) were provided by Guangzhou RiboBio Co., Ltd (Guangzhou, China). The sgRNA (single-guide RNA) sequence targeting USP10 was 5′-GACTCCTCGATCTTCAGTTGAGG(PAM)-3′ that was packed into a lentivirus as described previously [13]. Genes were delivered into MM cells by using Lipofectamine 2000 (Thermo Fisher®, Waltham, MA, USA) according to the manufacturer’s instructions. Polyethyleneimine (PEI, Sigma-Aldrich, Saint Louis, MO, USA) was used as the carrier for gene transfection into HEK293T cells as described previously [14].
Lentiviral USP10
The USP10 cDNA generated by using the following primers: 5′-CCGCTCGAGATGGCCCTCCACAGCCC-3′ (Forward) and 5′-TGCTTCGAATTACAGCAGGTCCACTCGG-3′ (Reverse) was inserted into a pLVX-AcGFP lentiviral vector (Clontech Laboratories). To generate lentiviral particles, HEK293T cells at 80% confluence were transfected with pLVX-AcGFP-USP10 (10 µg) and plasmids for VSV-G envelope glycoprotein (3.5 µg), Rev (2.5 µg), and ΔR8.74 packaging proteins (6.5 µg) by PEI (Sigma-Aldrich). Forty hours later, the lentiviral particle-enriched supernatants were harvested, filtered, and stored frozen at −80 °C for further use.
Constructs of truncated USP10
The USP10 truncates were cloned by using the wild-type USP10 as the template. Primers for the specific domains to generate USP10 truncates were designed as shown in Table 1.
Table 1.
Primers for USP10 truncates
| aa 1-205 | Forward Primer | 5′-GCTCTAGAATGGCCCTCCACAGCCC-3' |
| Reverse Primer | 5′-CCGCTCGAGCGTAAGTGGCGGAGGCATG-3' | |
| aa 206-798 | Forward Primer | 5′-GCTCTAGACCCAGGACTTGTAACAGCCC-3' |
| Reverse Primer | 5′-CCGCTCGAGTTACAGCAGGTCCACTCGG-3' | |
| aa 1-399 | Forward Primer | 5′-GCTCTAGAATGGCCCTCCACAGCCC-3' |
| Reverse Primer | 5′-CCGCTCGAGCAACTCTGCAATCTTTATGGCTACAGG-3' | |
| aa 400-798 | Forward Primer | 5′-GCTCTAGACTGGAGAATGTAACCCTAATCCATAAACCA-3' |
| Reverse Primer | 5′-CCGCTCGAGTTACAGCAGGTCCACTCGG-3' |
Chemicals and antibodies
Spautin-1 (Cat# S7888) was purchased from Selleck Chemicals (Houston, TX, USA). MG132 (Cat# HY-13259) and cycloheximide (CHX, Cat# HY-12320) were provided by MedChemExpress LLC (Shanghai, China). Anti-HA (Cat# M180-3), anti-Myc (Cat# M192-3), and anti-Flag (Cat# M185-3L) were from MBL Biotech Co., Ltd. (Nagoya, Japan). Anti-β-tubulin (Cat# AC008) was obtained from ABclonal Biotechnology Co., Ltd (Woburn, MA, USA). Anti-CCND3 (Cat# 26755-1-AP), anti-E2F (Cat# 66515-1-Ig), and anti-GAPDH (Cat# 60004-1-Ig) were from Proteintech Group, Inc (Wuhan, China). Anti-USP10 (Cat# 8501), anti-CCND1 (Cat# 55506), anti-CCND2 (Cat# 3741), anti-CDK4 (Cat# 12790), anti-K48Ub (Cat# 4289), anti-K63Ub (Cat# 12930), anti-CDK6 (Cat# 3136), and anti-p-Rb (Cat# 8516) were obtained from Cell Signaling Technology, Danvers, MA, USA.
Immunoblotting (IB) assays
After cell lysates were prepared, protein concentrations were determined by a BCA assay (Beyotime). Equal amounts of protein (10–30 µg) were separated by sodium dodecyl sulfate-poly-acrylamide gel electrophoresis (SDS-PAGE). The isolated proteins were then transferred to polyvinylidene difluoride membranes (Millipore). After normalization, the blots were probed with appropriate antibodies, as described previously [14].
Immunoprecipitation (IP)
Cell lysates of interest were prepared in a PCL lysis buffer [14]. After being clarified at high speed, the supernatants were first pre-incubated with Protein A + G beads (Sigma-Aldrich). The pre-treated proteins were then incubated with specific primary antibodies as needed followed by incubation with 40 μL of a 50% slurry of protein A + G agarose beads with gentle rotation at 4 °C for 2 h. The agarose beads were then collected and washed 5 times with the lysis buffer, followed by re-suspension in 20 μL of 2× SDS loading buffer. Samples were then boiled before being subjected to fractionation on SDS-PAGE and immunoblotting (IB) analysis.
Affinity purification-coupled mass spectrometry (AP/MS)
HEK293T cells were transfected with a Flag-CCND3 plasmid or empty vector for 36 h before being treated with MG132 (10 μM) for another 4 h. The cells were then collected for protein extraction by using a lysis buffer as described previously [14]. After clarification, 10 mg of each cell lysate was subjected to co-IP by using anti-FLAG M2 Affinity Gel (Sigma-Aldrich) overnight at 4 °C. After gentle washing, proteins were fractionated by SDS-PAGE. After silver staining (Beyotime), the protein bands from the gel were excised and processed for high performance liquid chromatography (HPLC) and tandem mass spectrometry (MS/MS) analysis as described previously [14].
MS data analysis
MS/MS raw files were analyzed by using Proteome Discoverer (version 1.4) as shown previously [15]. Specifically, the Andromeda probabilistic search engine was used to search peak lists against the UniProt database (Releases in July 2017, containing 182,230 entries). Cysteine carbamidomethylation was set as a fixed modification and methionine oxidation was set as variable modifications. The digestion enzyme was set as trypsin and a maximum of two missed-cleavage sites was allowed. A minimum of seven amino acids per identified peptides were required. The mass tolerance for precursor ions was set to 20 ppm for the first search and 4.5 ppm for the main search. The mass tolerance for fragment ions was set to 0.5 Da. The false discovery rate was determined by searching a reverse database and was set as 1% for proteins and peptides.
Cell viability assay and flow cytometry
MM cells were subjected to USP10 knockdown by siRNA. Seventy-two hours later, cells were replated in 96-well plates. Seventy-two hours later, relative cell proliferation was measured by MTT assay. Apoptosis was measured by flow cytometry to determine Annexin V and propidium iodide (MultiSciences Biotech Co., Ltd, Hangzhou, China) staining as described previously [14].
Cell cycle analysis
After being transfected with siCCND3 for 72 h, MM cells were collected and treated with cold 70% ethanol and stained with propidium iodide (PI). Cell cycle was analyzed on a cytometer (FACSCalibur, BD Biosciences, San Jose, CA, USA).
In-tube ubiquitination assay
The polyubiquitinated Flag-CCND3 and the Myc-USP10 protein were purified by IP from HEK293T cells in the presence of MG132 as described previously [15, 16]. To exclude nonspecific ubiquitin-modified species from the purified protein, the immunoprecipitates were washed extensively using the ubiquitination wash buffer (50 mM Tris-HCl pH 6.8, 150 mM NaCl, 0.5% Triton X-100, 0.5% NP-40, 0.5% DCA, 1 M Urea, 1 mM NEM, and protease inhibitors). Both purified proteins were then incubated in the deubiquitination buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, and 5% glycerol) at 37 °C for 2 h according to the previous report [16]. The resulting reactions were subjected to IB analysis, and CCND3 ubiquitination was detected with an anti-Ub antibody. After the reaction was terminated, the mixture was subjected to IB assay.
Human myeloma xenografts in immunodefficient nude mice
Female BALB/c nude mice (5 weeks old) were purchased from Guangdong Vital River Laboratory Animal Technology Co. Ltd (Foshan, China). KMS11 and OPM2 cells (2 × 107) were resuspended in 200 μL PBS and injected subcutaneously into the left flank of each mouse to establish tumor xenografts model. When tumors were palpable, the mice were randomly divided into 4 groups (5 mice/group) and treated with either vehicle, Spautin-1(20 mg/kg, intraperitoneal injection), Palbociclib (80 mg/kg, oral gavage) or their combination (Spautin-1/Palbociclib) every other day for a month. The body weight and the tumor volume (L × W2/2) of the mice were measured every 3 days. At the end of the experiment, mice were sacrificed. Dissected tumors were stored at −80°C for further study. All experiments were performed in accordance with the relevant guidelines and regulations abided by animal welfare. The animal study was conducted with the approval of the Institutional Animal Care and Use Committee of Guangzhou Medical University.
Statistical analysis
Student’s t-test was used to calculate P values for differences. Differences were considered significant at P < 0.05. All experiments were repeated independently three times (except for the MS/MS assays that were repeated twice).
Results
Knockdown of CCND3 results in MM cell apoptosis
The D-type cyclin family includes three members, CCND1, CCND2, and CCND3, of which all are overexpressed in MM, and CCND1 and D2 have been well demonstrated to promote MM cell cycle progress and their inhibition leads to MM cell apoptosis [17]. To find out the effects of CCND3 in MM cell proliferation and survival, we knocked down CCND3 in two typical MM cell lines that harbor t(6;14) chromosomal translocation and highly express CCND3 (Supplementary Fig. S1a), followed by cell cycle analysis and apoptosis. As shown in Supplementary Fig. S1b, compared to the scrambled transfection, knockdown of CCND3 markedly increased the fraction of cells at the G1 phase in both cell lines. Moreover, CCND3 knockdown strikingly increased the fractions with positive Annexin V staining (Supplementary Fig. S1c), indicating that CCND3 promotes MM cell survival. These findings were consistent with the results from the CCND1 and D2 knockdown [17].
USP10 interacts with CCND3 in MM cells
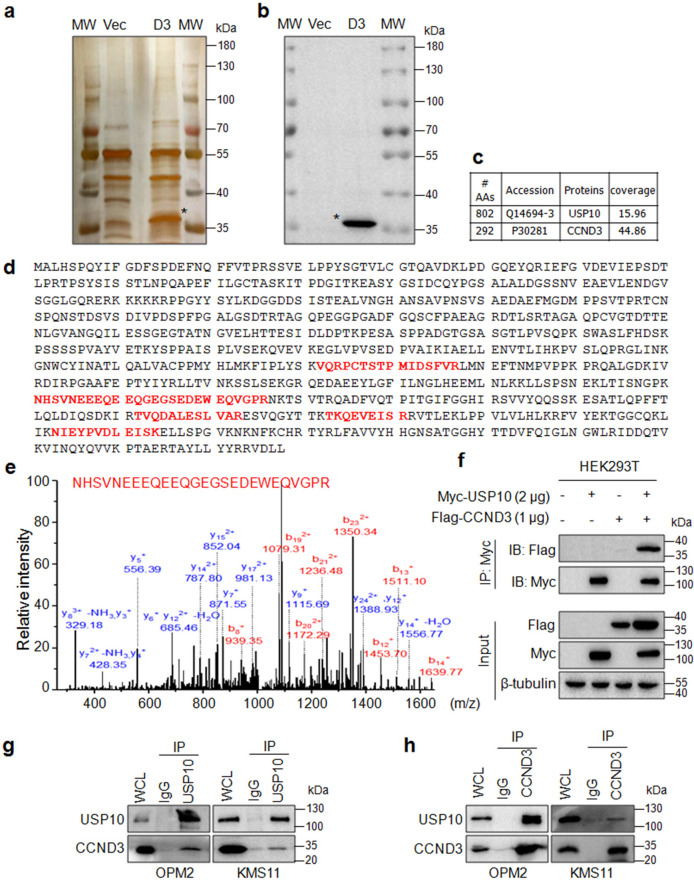
Given that CCND3 undergoes rapid degradation via the ubiquitin-proteasomal pathway in the direction of the ubiquitin ligases FBXL2 [6] and FBXL8 [7], we wondered which Dub might be responsible for CCND3 deubiquitination. To this end, we made a Flag-CCND3 construct and transfected it into HEK293T cells followed by affinity-purification using an anti-Flag antibody. Before being subjected to the tandem MS, the IP samples were analyzed by both silver staining and IB for the expression of CCND3 (Fig. 1a). The IB assay clearly demonstrated that CCND3 was found in the experimental sample but not in the control one (Fig. 1b). The subsequent tandem MS analysis revealed that USP10 was the only Dub that was found in the CCND3-immunoprecipitates with more than 2 unique peptides but not found in the control (Fig. 1c). USP10 was identified with 5 unique peptides by the tandem MS (Fig. 1d). One of mass spectra of the unique peptides was shown in Fig. 1e. To verify the interaction between USP10 and CCND3, the Flag-CCND3 and the Myc-USP10 plasmids were co-transfected into HEK293T cells, followed by the IP/IB assay. The IB assay revealed that CCND3 was present in the USP10 IP complex (Fig. 1f). Moreover, the reciprocal IP/IB assay by using anti-USP10 and anti-CCND3, respectively, also showed that USP10 bound to CCND3 in intact MM cells (Fig. 1g, h). Therefore, these findings suggest that USP10 interacted with CCND3 in MM cells.
Fig. 1. USP10 interacts with CCND3.
a a Flag-CCND3 plasmid (D3) or empty vector (Vec) was transfected into HEK293T cells for 48 h. The cell lysates were then subjected to IP with an anti-Flag antibody. The eluted proteins from agarose beads were subjected to SDS-PAGE and silver staining. MW: protein molecular weight. b The cell lysates from (a) were subjected to IB against CCND3. c The statistical analysis of CCND3 and USP10 identified by MS. d The unique peptides identified by MS were highlighted in red. e The MS spectrum of a representative polypeptide sequence. f HEK239T cells were co-transfected with USP10 and CCND3 plasmids, followed by IP/IB assays as indicated. g, h The OPM2 and KMS11 cell lysates were subjected to reciprocal IP/IB assays with specific antibodies as indicated.
USP10 deubiquitinates CCND3 in cells and in vitro
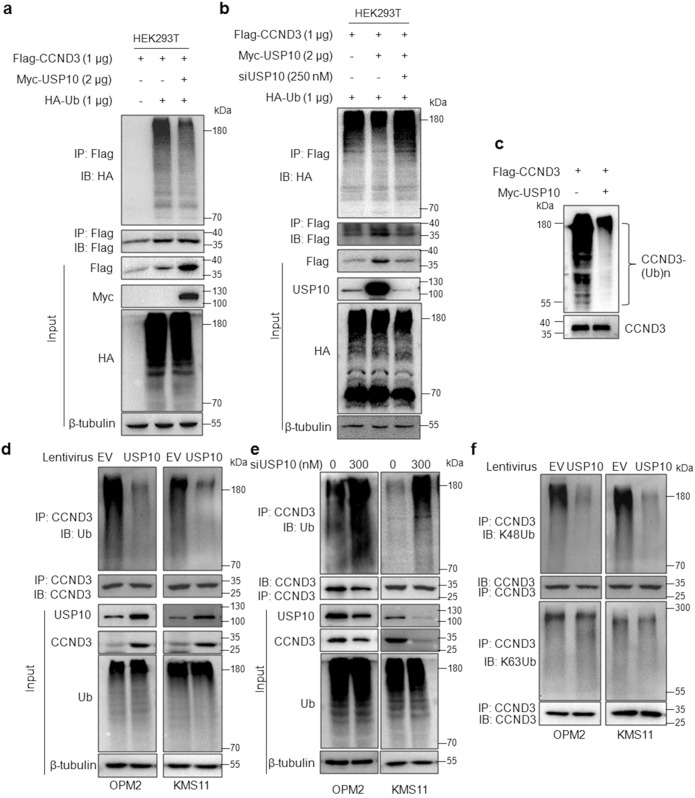
Given USP10 is a ubiquitin-specific peptidase and it interacts with CCND3, we wondered whether USP10 could decrease CCND3 from ubiquitination as a Dub. To this end, we first evaluated the effects of USP10 on CCND3 ubiquitination levels by co-transfecting Myc-USP10, Flag-CCND3 and HA-ubiquitin (Ub) plasmids into HEK293T cells, followed by IP/IB assays. The results showed that the polyubiquitination level of CCND3 was strikingly decreased in cells co-transfected with USP10 (Fig. 2a). In agreement with this finding, the specific siRNA of USP10 partly recovered the ubiquitination level of CCND3 reduced by USP10 (Fig. 2b), further suggesting USP10 indeed deubiquitinated CCND3. Next, we evaluated the deubiquitination activity of USP10 toward CCND3 in a cell-free system. As shown in Fig. 2c, the heavy ubiquitination was drastically decreased from CCND3 in the presence of USP10, a direct evidence that USP10 was a Dub of CCND3. To further evaluate the effect of USP10 on CCND3 ubiquitination in MM cells, USP10 was carried into MM cell lines by lentivirus followed by IP/IB assays. Consistent with the in-vitro assay, USP10 markedly reduced the polyubiquitination level from CCND3 (Fig. 2d). In contrast, when USP10 was knocked down, the ubiquitination level of CCND3 was increased (Fig. 2e). More specifically, we found that USP10 markedly decreased the K48-linked but not the K63-linked polyubiquitination from CCND3 (Fig. 2f). Therefore, we could conclude that USP10 was a Dub of CCND3 and it specifically prevented CCND3 from K48-linked polyubiquitination.
Fig. 2. USP10 prevents CCND3 from K48-linked polyubiquitination.
a USP10 and CCND3 plasmids were co-transfected into HEK293T cells for 48 h, followed by IP/IB assays as indicated. b USP10 and CCND3 plasmids were co-transfected with or without siUSP10 into HEK293T cells for 48 h, followed by IP/IB assays as indicated. c Purified CCND3 proteins were co-incubated with or without purified USP10 protein in a tube for 2 h, followed by IB assay, as stated in the Methods section (In-tube ubiquitination assay). d OPM2 and KMS11 cells were infected with lentiviral USP10 or empty virus (EV) for 72 h, followed by IP/IB assays as indicated. e USP10 was knocked down from OPM2 and KMS11 cells for 72 h, followed by IP/IB assays as indicated. f OPM2 and KMS11 cells were infected with lentiviral USP10 or empty virus (EV) for 72 h, followed by IP/IB assays with specific antibodies as indicated. All the experiments were performed three times independently.
USP10 increases CCND3 stability in MM cells
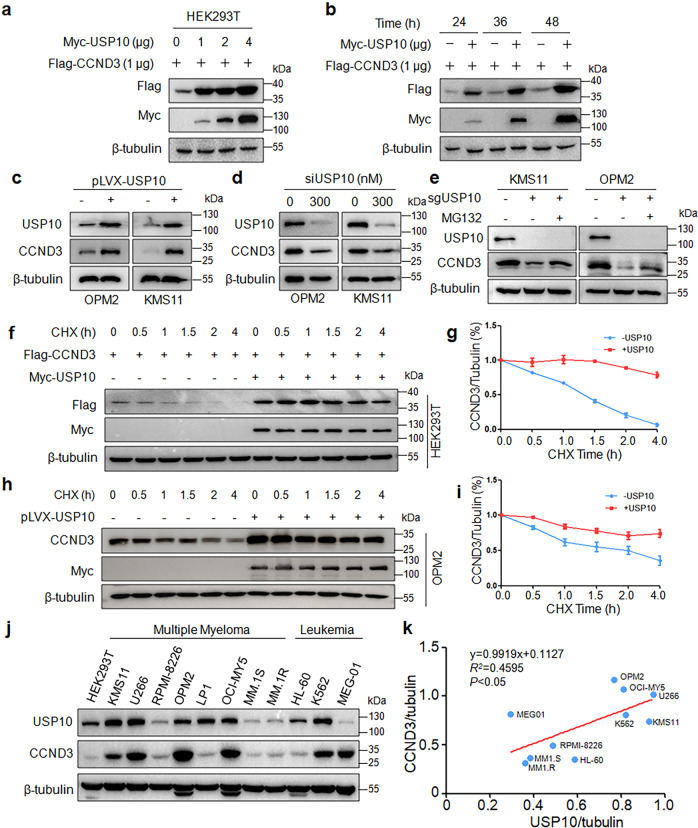
The above study showed that USP10 hydrolyzes the K48-linked ubiquitin chain from CCND3 and it is well known that the K48-linked polyubiquitination leads to protein degradation in proteasomes. To find out whether USP10 could prevent CCND3 from degradation, we evaluated the effects of USP10 on CCND3 stability in HEK293T cells. As shown in Fig. 3a, b, CCND3 was strikingly increased following USP10 treatment in a concentration- and time-dependent manner. Moreover, CCND3 was increased in MM cells after infection of lentiviral USP10 but it was decreased when USP10 was knocked down (Fig. 3c–e). This effect was confirmed by CHX chase assay. As shown in Fig. 3f–i, when protein synthesis was inhibited by CHX, overexpression of USP10 significantly prolonged the half-life of CCND3 in both HEK293T and MM cells. Lastly, we measured the expression levels of CCND3 and USP10 in a panel of MM and other cancer cell lines by IB. The subsequent association assay revealed that the expression level of USP10 was positively correlated with CCND3 (Fig. 3j, k). Therefore, USP10 stabilized CCND3 in accordance with its deubiquitination activity.
Fig. 3. USP10 stabilizes CCND3 in MM cells.
a A CCND3 plasmid was co-transfected with increased USP10 into HEK293T cells for 48 h, followed by IB assay. b CCND3 and USP10 were co-transfected into HEK293T cells for 24 to 48 h, followed by cell lysate preparation and IB assays. c OPM2 and KMS11 cells were infected with lentiviral USP10 for 72 h, followed by IB assays as indicated. d USP10 was knocked down from OPM2 and KMS11 cells by specific siRNA for 48 h, followed by IB assays. e USP10 was knocked out by its sgRNA, followed by MG132 treatment. The cell lysates were then subjected to IB assays. f USP10 and CCND3 were co-transfected into HEK293T cells for 36 h, followed by CHX treatment at indicated periods before collecting for IB assay. g The blots in f was subjected to desitometry for CCND3 expression against GAPDH. h The USP10 lentivirus was infected into OPM2 cells for 48 h, followed by CHX treatment at indicated periods before collecting for IB assay. i The blots in h was subjected to densitometry for CCND3 expression against GAPDH. j Cell lysates from cell lines were subjected to IB assays against CCND3 and USP10. k The relationship analysis between the CCND3 and USP10 expression levels from i.
Given CCND1, D2 and D3 belong to the D-cyclin family and share high similarity in amino acid composition, we therefore wondered whether USP10 also displayed activity on D1 and D2. Similar to D3, USP10 also decreased the ubiquitination levels on CCND1 and D2 in both HEK293T and MM cells, therefore USP10 also upregulated the protein levels of both CCND1 and D2, in a similar manner to CCND3 (Supplementary Fig. S2), which is consistent with previous findings in glioblastoma cells [11]. Therefore, USP10 might deubiquitinate and stabilize all three members of D-cyclin family.
USP10 deubiquitinates CCND3 independent of its N-terminus
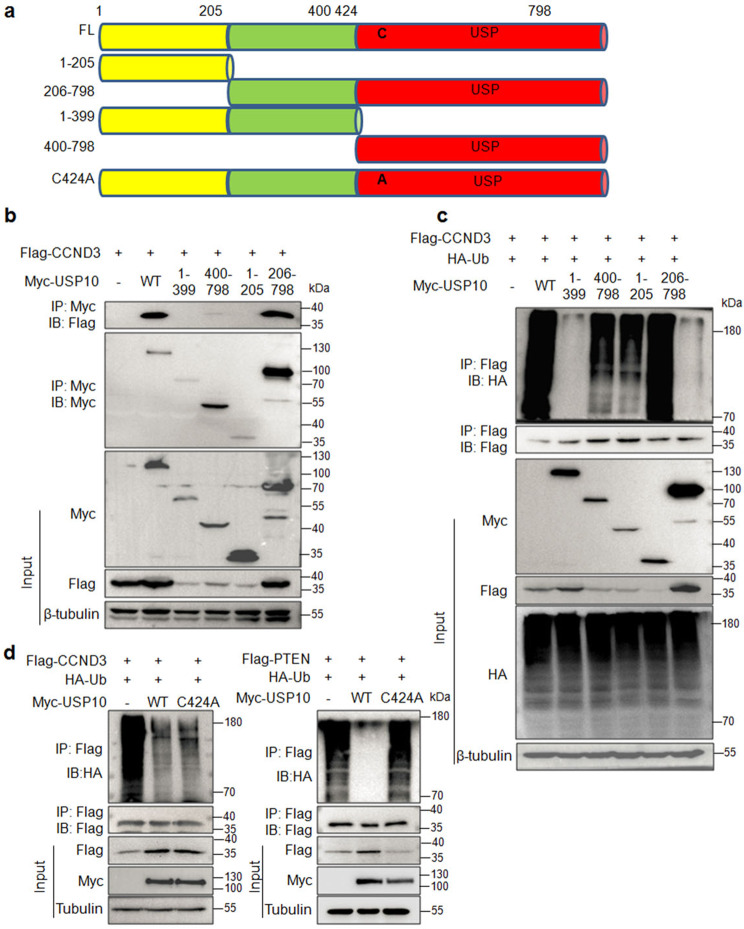
USP10 is a cysteine protease containing an N-terminal unknown domain and a C-terminal catalytic domain (USP domain) in which Cyc424 (C424) may be essential to maintain its catalytic activity [14, 18]. To find out the detailed interaction and the deubiquitinase activity, we constructed several truncates of USP10, including the fragments of aa. 1–205, aa. 206–798, aa. 1–399, and 400–798 (Fig. 4a) according to previous reports [19, 20]. These truncates were then co-transfected into HEK293T cells with Flag-CCND3. Subsequent IP/IB assays found that only the aa. 206–798 fragment bound to CCND3 similar to the wild-type (Fig. 4b). Moreover, the aa. 206–798 fragment deubiquitinated CCND3 but the aa. 1–399 and aa. 400–798 fragments lost this activity (Fig. 4c). These results suggested that the first 205 amino acids at the N-terminus was dispensable for USP10 to exert its cysteine protease on CCND3 ubiquitination. Moreover, we also found that USP10 C424A mutant failed to deubiquitinate PTEN as reported previously [18], however, it remained activity to deubiquitinate CCND3 (Fig. 4d). This finding suggests that C424A might be not important for USP10 to deubiquitinate CCND3.
Fig. 4. The N-terminal aa. 1–205 fragment is dispensable for USP10 to bind to and deubiquitinate CCND3.
a The schematic illustration of the USP10 protein. b Each truncate of USP10 was co-transfected with a CCND3 plasmid in HEK293T cells for 24 h, followed by cell lysate preparation and IP/IB assays with specific antibodies as indicated. c The truncates of USP10 were co-transfected with a CCND3 plasmid and a Ub plasmid for 24 h, followed by the measurement of CCND3 polyubiquitination with the IP/IB assays. d HEK293T cells were co-transfected with HA-Ub, Flag-CCND3 or Flag-PTEN with WT or C424 USP10 plasmids for 24 h, followed by IP/IB assays as indicated.
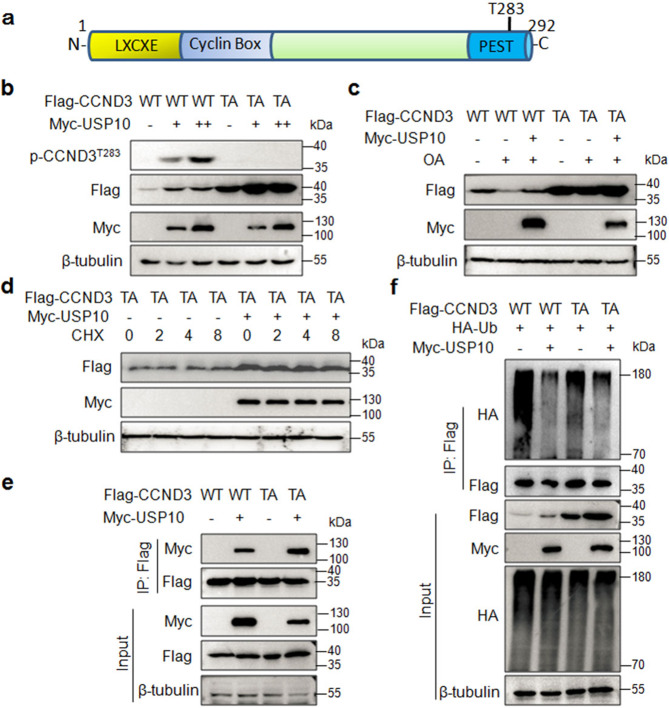
USP10 stabilizes both wild-type and T283A mutant CCND3
CCND3 degradation following phosphorylation has been well demonstrated [21] and a recent study reported that the ubiquitin ligase SCF-FBXL8 mediated CCND3 upon its Thr283 (T283) phosphorylation [7]. Moreover, basal and cAMP-induced degradation of CCND3 in Reh cells is dependent on T283 phosphorylation by glycogen synthase kinase-3β (GSK-3β) [21]. However, another study showed that CCND3 degradation is independent of T283 phosphorylation [22]. To find out whether T283 is required for CCND3 ubiquitination and stability modulated by USP10, we mutated T283 to A (TA) CCND3 (Fig. 5a), followed by a serial studies on its stability and ubiquitination in the presence of USP10. Firstly, we found that T283 mutation indeed lost its phosphorylation at T283 and increased CCND3 stability (Fig. 5b), interestingly, USP10 also promoted CCND3 phosphorylation at T283 in association with the increase of CCND3 stability (Fig. 5b). Moreover, we found that USP10 also raised the stability of T283A CCND3 although no changes were observed on its phosphorylation on T283 (Fig. 5b), suggesting that T283 phosphorylation indeed triggers CCND3 degradation, but USP10 stabilizes CCND3 independent of T283 phosphorylation. To confirm this hypothesis, cells expressing WT or T283A CCND3 and USP10 were treated with okadaic acid (OA), an inhibitor of protein phosphatases 1 and 2 A (PP1 and PP2A) thereby accumulating CCND3 phosphorylation [22]. As expected, OA markedly induced the degradation of wild-type CCND3 and it was ablated by USP10 (Fig. 5c). However, T283A CCND3 had no response to OA treatment, and in the presence of OA, USP10 only slightly increased the protein level of T283A CCND3, suggesting that USP10 might stabilize CCND3 independent of T283 phosphorylation. Subsequently, we performed a CHX chase assay. Compared with the wild-type CCND3 that was almost degraded within 1.5 h (Fig. 5d), the T283A mutant was not degraded within 8 h. Moreover, this mutant could be markedly increased by USP10 (Fig. 5d), further suggesting USP10 might deubiquitinate T283A CCND3. Therefore, we next evaluated the interaction between USP10 and T283A CCND3 and found that USP10 bound to mutant CCND3 at a similar level to the wtCCND3 (Fig. 5e). Furthermore, the T283A CCND3 could be polyubiquitinated and this ubiquitination could be reduced by USP10 (Fig. 5f). The above study thus collectively demonstrated that CCND3 could be deubiquitinated and stabilized by USP10 independent of its T283 residue.
Fig. 5. USP10 stabilizes both wild-type and T283A mutant CCND3.
a The schematic illustration of the CCND3 protein. b The wild-type (WT) or T283A (TA) CCND3 plasmid was co-transfected with USP10 into HEK293T cells for 24 h, the cell lysates were then subjected to IB assays with specific antibodies as indicated. c The WT or TA CCND3 plasmid was co-transfected with USP10 into HEK293T cells for 36 h, followed by okadaic acid (OA) treatment for 24 h. The cell lysates were then subjected to IB assays with specific antibodies as indicated. d The WT or TA CCND3 plasmid was co-transfected with USP10 into HEK293T cells for 36 h, followed by CHX treatment. Cell lysates were prepared at indicated periods after CHX treatment for IB assays. e Wild-type (WT) or TA CCND3 was co-transfected with USP10 into HEK293T cells for 36 h, followed by IP/IB assays as indicated. f WT or TA CCND3 was co-transfected with Ub and USP10 into HEK293T cells for 36 h, followed by IP/IB assays as indicated.
USP10 activates the CCND3/CDK4/6 signaling pathway and promotes MM cell proliferation
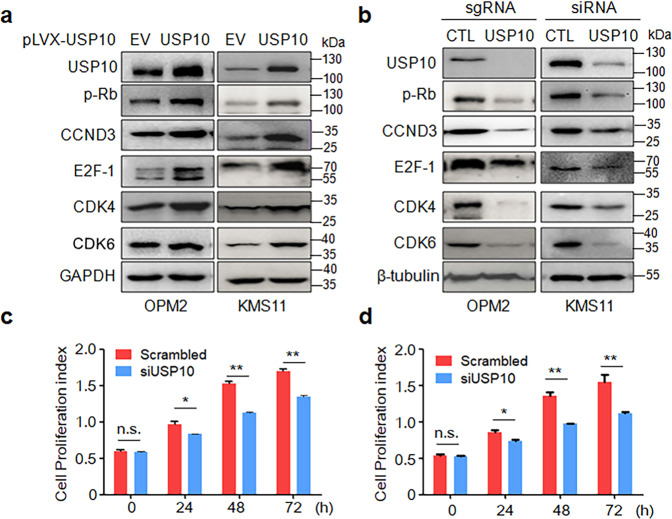
It is well known that CCND3 is a regulatory subunit of CDK4/6 in phosphorylating Rb protein, therefore, triggering cell cycle progress from G1 to S phase [1]. The above experiments have demonstrated that USP10 stabilized CCND3, we thus wondered whether USP10 could activate the CCND3/CDK4/6 signaling pathway. To this end, KMS11 and OPM2 cells were infected with lentiviral USP10. The subsequent IB assays revealed that USP10 increased CCND3, along with phosphorylated Rb (p-Rb) and upregulated expression of CDK4, CDK6, E2F-1 (Fig. 6a). Consistent with this finding, knockdown of USP10 resulted in decreased CCND3, CDK4, CDK6, E2F-1, and p-Rb (Fig. 6b). Therefore, USP10 indeed activated the CCND3-associated cell cycle progress signaling in MM cells. Given cell cycle progress promotes MM cell proliferation, we next knocked down USP10 from MM cell lines and measured its effects on cell proliferation as assayed by MTT. The results showed that the knockdown of USP10 markedly inhibited MM cell proliferation (Fig. 6c, d), consistent with its effects on the cell cycle regulators.
Fig. 6. USP10 activates the CCND3 signaling pathway and promotes MM cell proliferation.
a Lentiviral USP10 or empty virus (EV) was infected into OPM2 and KMS11 cells for 72 h, followed by cell lysate preparations and IB assays as indicated. b USP10 was knocked out or knocked down by using sgRNA or siRNA, respectively, for 72 h. Cells were then subjected to protein extraction and IB assays as indicated. CTL: control. OPM2 (c) and KMS11 (d) cells were subjected to USP10 knockdown by siRNA, 72 h later, cells were replated in 96-well plates. Cell proliferation was further measured by MTT assay at indicated time points. n.s., not significant; *P < 0.05, **P < 0.01.
Inhibition of USP10 synergizes with Palbociclib to induce MM cell apoptosis
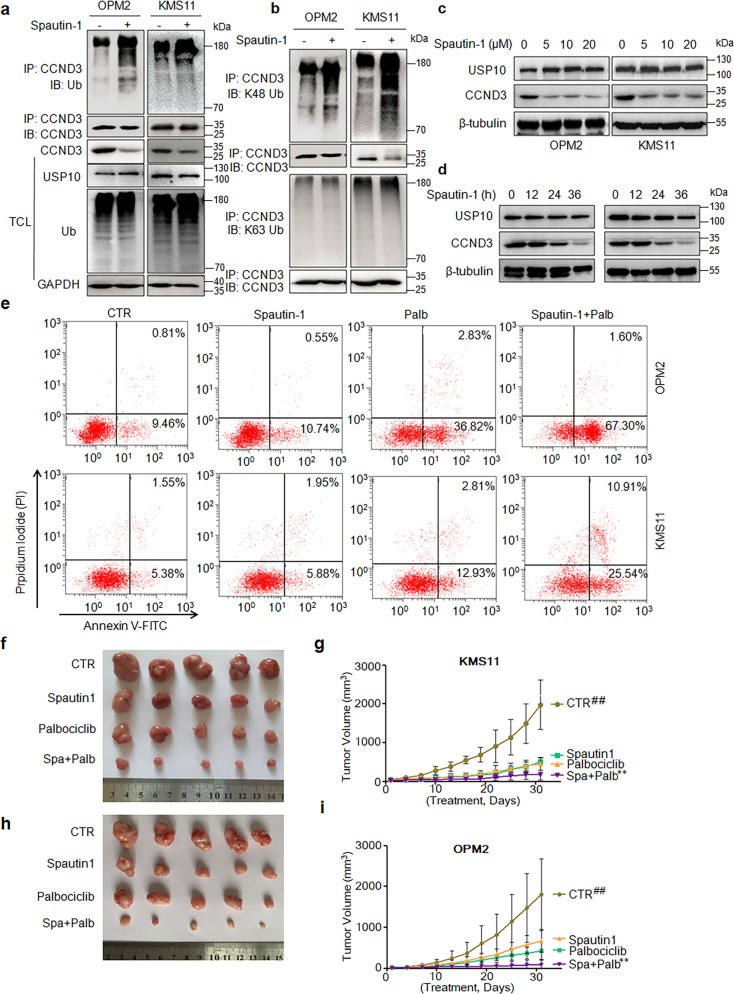
The above studies have demonstrated that USP10 stabilized CCND3 by downregulating its polyubiquitination, while knockdown of CCND3 leads to MM cell apoptosis, suggesting the USP10/CCND3 axis could be a therapeutic target for the treatment of MM. To further examine our hypothesis, we first evaluated the ubiquitination level of CCND3 when Spautin-1, a small molecule inhibitor of USP10 [23], was applied. The IB assays showed that Spautin-1 drastically increased the polyubiquitination levels of CCND3 in MM cells (Fig. 7a). Moreover, this ubiquitination on CCND3 was specifically seen as the K48- but not K63-linked form (Fig. 7b), which was consistent with the effects of USP10 on CCND3 polyubiquitination (Fig. 2). Furthermore, Spautin-1 downregulated CCND3 protein in a concentration- and a time-dependent manner (Fig. 7c, d). We next measured MM cell apoptosis by flow cytometry after Spautin-1 treatment. It revealed that Spautin-1 only induced a minimal apoptosis in both KMS11 and OPM2 cells in terms of Annexin V staining, a hallmark of cell apoptosis (Fig. 7e). Given CCND3 acts with CDK4/6 in control of cell cycle progress, we wondered whether inhibition of USP10 could synergize with CDK4/6 inhibition. To this end, Palbociclib, a CDK4/6 inhibitor and a potential anti-MM drug [24], was used to treat MM cells alone or with Spautin-1. The subsequent apoptosis analysis showed that Spautin-1 at a low concentration strikingly enhanced MM cell apoptosis induced by Palbociclib in both OPM2 and KMS11 cells (Fig. 7e). To further evaluate this therapeutic potential, we established myeloma xenografts with OPM2 and KMS11 cells in immunodefficient nude mice followed by administration of Spautin-l, Palbociclib alone or together. The results showed that, within the one-month treatment, both Spautin-1 and Palbociclib could markedly slowed down the myeloma tumor growth, but only the combined treatment almost suppressed tumor growth within 30 days (Fig. 7f–i). Taken together, the cellular and animal studies demonstrated that the USP10/CCND3 might be a potential target for MM treatment.
Fig. 7. Inhibition of USP10 synergizes with CDK4/6 inhibitor to induce MM cell apoptosis.
a, b OPM2 and KMS11 cells were treated with Spautin-1 for 24 h, followed by cell lysate preparation and IP/IB assays as indicated. c, d OPM2 and KMS11 cells were treated with Spaulin-1 at increased concentrations for 24 h (c) or 10 µM for indicated periods (d), followed by IB assays. e Cells were treated with Spaulin-1 and/or Palbociclib (Palb) for 24 h, followed by flow cytometric analysis after Annexin V-FITC and PI staining. f–i Balb/c immunodefficient mice were injected subcutaneously with KMS11 or OPM2 cells. When tumors were palpable, mice were randomly divided into 4 groups in each model, followed by administration of vehicle, Spautin-1, Palbociclib or Spau + Palb for 30 days. Tumor size was measured every three days (g, KMS11; i, OPM2). Tumors were excised at the end of the study (f, KMS11; h, OPM2). ##P < 0.01 when controls were analyzed against the treatments. **P < 0.01, the Spau+Pal combined treatment against the Spautin-1 or Palbociclib treatment.
Discussion
The above study identified that USP10 is a putative Dub of CCND3 and increases its stability by decreasing its K48-linked polyubiquitination therefore stabilizing CCND3 and activating the CCND3/CDK/4/6 signaling pathway. We also found that inhibition of USP10 leads to CCND3 degradation and synergizes with anti-MM activity of the CDK4/6 inhibitor.
MM is a hematological malignancy featured with several chromosomal translocations, including t(6;14) and t(11;14), that lead to the high expression of cyclin D3 (CCND3) and cyclin D1 (CCND1), respectively [2]. Given their roles in control of cell cycle progress, these D-cyclins are believed to participate in promoting MM cell proliferation and survival. Actually, earlier studies have proposed that dysregulation of D-cyclins is an early and unifying event in MM pathophysiological process [5] and could be used for the classification of MM [25]. Overexpression of CCND1 and CCND2 has been demonstrated to promote the transition of cell cycle from G1 to S in MM cells, in contrast, silence of CCND1 and CCND2 results in MM cell cycle arrest and cell apoptosis [17]. In the present study, we fill this gap and find that CCND3 is important for MM cell proliferation and survival because knockdown of CCND3 results in cell cycle arrest and MM cell apoptosis as that seen in CCND1 and CCND2. These findings suggest that D-cyclins are critical for MM cell proliferation. More importantly, as cell-cycle dependent protein, the expression of D-cyclins is strictly modulated in both RNA and protein levels during cell cycle progress. It has clearly documented that D-cyclins contain the PEST domain for rapid degradation via the ubiquitin-proteasomal pathway [26]. The modulation of CCND1 and D2 has been elucidated. For example, the ubiquitination of CCND1 is fine-tuned by ubiquitin ligases Fbxo4, Fbxo31, β-TrCP [27], and deubiquitinase USP2 [28], USP10 [11], and USP22 [12]. Although the ubiquitin ligases FBXL2 [6] and FBXL8 [7] have been identified to induce CCND3 ubiquitination and degradation, its Dub(s) was not reported yet. The present study finds USP10 binds to and reduces the K48-linked polyubiquitination on CCND3, therefore upregulating its stability in MM cells. Regrettably, the present study could not verify this finding in non-malignant and malignant primary plasma cells, given the low percentage and availability of plasma cells in healthy bone marrow and peripheral blood.
USP10 belongs to the ubiquitin-specific peptidase (USP) subfamily of deubiquitinates and can remove conjugated ubiquitin from a series of important target proteins such as PTEN [18], AMPK [29], and p53 [30]. Our current study adds CCND3 as a novel member to the substrate list of USP10, however, we found that the specific mechanism in which USP10 deubiquitinates CCND3 might differ from its other known proteins. One of those is that USP10 deubiquitinates CCND3 independent of its C424, one of the key residues for the deubiquitinase activity of USP10 [31]. We found that the C424A USP10 fails to deubiquitinate PTEN which is consistent with the previous report [18], however, this mutant is still able to deubiquitinate CCND3 at a similar level comparable to the wild-type one. This finding suggests that C424 is probably dispensable for USP10 deubiquitinating activity against CCND3. Interestingly, the C424A USP10 also remains certain deubiquitinating activity against MSH2 [19]. Moreover, although USP10 binds to TP53 via N-terminal aa. 1–100 fragment [20], it can interact with p62 via the aa. 275–798 [32]. In the present study, we found that USP10 binds to CCND3 via the aa. 206–798 fragment but not the N-terminal aa. 1–205 fragment. These findings suggest USP10 might display specific activity toward individual target substrates.
CCND3 degradation is quickly processed, but previous studies reported controversial findings in terms of its phosphorylation status. A group from Norway finds that T283 is critical for CCND3 stability, because T283A CCND3 fails to be degraded [21]. The same group further finds that okadaic acid, an inhibitor of protein phosphatase 1 and 2 A, induces CCND3 phosphorylation at T283 and subsequent degradation via proteasomes [22]. However, they also found that T283 mutation fails to prevent OA-mediated CCND3 degradation, suggesting CCND3 degradation is a complicated process. In our study, we also found that T283A mutation markedly increases CCND3 stability, however, it does not prevent the interaction between CCND3 and USP10. Moreover, this T283A mutant could be further stabilized by USP10 via deubiquitination. All these findings confirm that T283 is important for CCND3 phosphorylation and degradation, however, USP10-mediated CCND3 stability and deubiquitination is not affected by the status of CCND3 phosphorylation at T283. USP10 might protect CCND3 from proteasomal degradation independent of T283.
CDK4/6 is the catalytic subunit of the CCND3/CDK4/6 complex. We found that USP10 upregulates the expression of all the downstream components in the CCND3/CDK4/6 axis, typically including increased phosphorylation level of Rb and the expression of E2F-1, suggesting USP10 promotes MM cell proliferation via the CCND3/CDK4/6/Rb/E2F-1 pathway. Notably both chemical and genetic inhibition of USP10 leads to suppressed signaling in cell cycle progress and induces MM cell proliferation, which is consistent with the previous report [11, 33]. Moreover, given the nature of the CCND3/CDK4/6 complex, we found Spautin-1, the inhibitor of USP10, synergizes with the anti-MM activity of Palbociclib, a CKD4/6 specific inhibitor [34]. Co-treatment of Spautin-1 and Palbociclib markedly induces MM cell apoptosis and strikingly inhibits the growth of myeloma xenografts in immunodefficient mice. Our finding is consistent with a previous report in that the CCND3-CDK6 inhibition leads to tumor cell apoptosis [35]. Therefore, USP10 could be a novel therapeutic target for MM and its inhibitors could be used in combination with CDK4/6 inhibitors as a novel modality for MM treatment.
In summary, the present study identifies USP10 as the first Dub of CCND3. By stabilizing CCND3, USP10 activates CDK4/6 and its downstream E2F signaling thereby promoting MM cell survival and proliferation. Moreover, inhibition of USP10 could synergize with CDK4/6 inhibitors for the treatment of MM. The USP10/CCND3/CDK4/6 axis could be a novel therapeutic target of myeloma.
Supplementary information
Acknowledgements
This study was supported in part by National Natural Science Foundation of China (#81970194 and #82170176 to XLM), the National Key Research and Development Program of China (#2022YFC2705003 to XLM), Guangzhou Medical University Discipline Construction Funds (Basic Medicine) (Grant No. JCXKJS2022A05 to XLM), and Department of Education of Guangdong Province of China (Grant No. 2021ZDZX2009 to XLM).
Author contributions
XLM designed the study. KZ, YJX, and XLM developed methods. KZ, YJX, YR, ZQH, CYM, YHY, XTZ, and ZYS performed experiments. XLM, ZQH, YJX, and GQX analyzed and interpreted data. XLM and YJX wrote and composed the manuscript. XLM, ZQH, BYC, and ZBZ administrated the projected.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yu-jia Xu, Kun Zeng.
Contributor Information
Zhen-qian Huang, Email: huangzq2013@163.com.
Xin-liang Mao, Email: xinliangmao@gzhmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-023-01083-w.
References
- 1.Zhang Q, Sakamoto K, Wagner KU. D-type Cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol Cell Endocrinol. 2014;382:583–92. doi: 10.1016/j.mce.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabris S, Agnelli L, Mattioli M, Baldini L, Ronchetti D, Morabito F, et al. Characterization of oncogene dysregulation in multiple myeloma by combined FISH and DNA microarray analyses. Genes Chromosomes Cancer. 2005;42:117–27. doi: 10.1002/gcc.20123. [DOI] [PubMed] [Google Scholar]
- 3.Agnelli L, Bicciato S, Mattioli M, Fabris S, Intini D, Verdelli D, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23:7296–306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 4.Kuehl WM, Bergsagel PL. Early genetic events provide the basis for a clinical classification of multiple myeloma. Hematology Am Soc Hematol Educ Program. 2005;1:346–52. doi: 10.1182/asheducation-2005.1.346. [DOI] [PubMed] [Google Scholar]
- 5.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012;31:2566–79. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida A, Choi J, Jin HR, Li Y, Bajpai S, Qie S, et al. Fbxl8 suppresses lymphoma growth and hematopoietic transformation through degradation of cyclin D3. Oncogene. 2021;40:292–306. doi: 10.1038/s41388-020-01532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell A, Thompson MA, Hendley J, Trute L, Armes J, Germain D. Cyclin D1 and D3 associate with the SCF complex and are coordinately elevated in breast cancer. Oncogene. 1999;18:1983–91. doi: 10.1038/sj.onc.1202511. [DOI] [PubMed] [Google Scholar]
- 9.Davis MI, Pragani R, Fox JT, Shen M, Parmar K, Gaudiano EF, et al. Small molecule inhibition of the ibiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem. 2016;291:24628–40. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magiera K, Tomala M, Kubica K, De Cesare V, Trost M, Zieba BJ, et al. Lithocholic acid hydroxyamide destabilizes cyclin D1 and induces G0/G1 arrest by inhibiting deubiquitinase USP2a. Cell Chem Biol. 2017;24:458–70. doi: 10.1016/j.chembiol.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun T, Xu YJ, Jiang SY, Xu Z, Cao BY, Sethi G, et al. Suppression of the USP10/CCND1 axis induces glioblastoma cell apoptosis. Acta Pharmacol Sin. 2021;42:1338–46. doi: 10.1038/s41401-020-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gennaro VJ, Stanek TJ, Peck AR, Sun Y, Wang F, Qie S, et al. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc Natl Acad Sci USA. 2018;115:E9298–307. doi: 10.1073/pnas.1807704115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Huang X, Xin HB, Fu M, Xue A, Wu ZH. TRAF family member-associated NF-κB activator (TANK) inhibits genotoxic nuclear factor κB activation by facilitating deubiquitinase USP10-dependent deubiquitination of TRAF6 ligase. J Biol Chem. 2015;290:13372–85. doi: 10.1074/jbc.M115.643767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Tong J, Tang X, Juan J, Cao B, Hurren R, et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood. 2016;127:1676–86. doi: 10.1182/blood-2015-07-658203. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang H, Ren Y, Mao C, Zhong Y, Zhang Z, Cao B, et al. Induction of zinc finger protein RNF6 auto-ubiquitination for the treatment of myeloma and chronic myeloid leukemia. J Biol Chem. 2022;298:102314. doi: 10.1016/j.jbc.2022.102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Zhang Q, Lou Y, Wang J, Zhao X, Wang L, et al. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res. 2019;7:1580–90. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 17.Tiedemann RE, Mao X, Shi CX, Zhu YX, Palmer SE, Sebag M, et al. Identification of kinetin riboside as a repressor of CCND1 and CCND2 with preclinical antimyeloma activity. J Clin Invest. 2008;118:1750–64. doi: 10.1172/JCI34149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Jiang S, Mao C, Zheng H, Cao B, Zhang Z, et al. The deubiquitinase USP10 restores PTEN activity and inhibits non-small cell lung cancer cell proliferation. J Biol Chem. 2021;297:101088. doi: 10.1016/j.jbc.2021.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Hu C, Tong D, Xiang S, Williams K, Bai W, et al. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage. J Biol Chem. 2016;291:10783–91. doi: 10.1074/jbc.M115.700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–96. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naderi S, Gutzkow KB, Låhne HU, Lefdal S, Ryves WJ, Harwood AJ, et al. cAMP-induced degradation of cyclin D3 through association with GSK-3beta. J Cell Sci. 2004;117:3769–83. doi: 10.1242/jcs.01210. [DOI] [PubMed] [Google Scholar]
- 22.Låhne HU, Kloster MM, Lefdal S, Blomhoff HK, Naderi S. Degradation of cyclin D3 independent of Thr-283 phosphorylation. Oncogene. 2006;25:2468–76. doi: 10.1038/sj.onc.1209278. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niesvizky R, Badros AZ, Costa LJ, Ely SA, Singhal SB, Stadtmauer EA, et al. Phase 1/2 study of cyclin-dependent kinase (CDK)4/6 inhibitor palbociclib (PD-0332991) with bortezomib and dexamethasone in relapsed/refractory multiple myeloma. Leuk Lymphoma. 2015;56:3320–8. doi: 10.3109/10428194.2015.1030641. [DOI] [PubMed] [Google Scholar]
- 25.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–72. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 27.Qie S, Diehl JA. Cyclin D degradation by E3 ligases in cancer progression and treatment. Semin Cancer Biol. 2020;67:159–70. doi: 10.1016/j.semcancer.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36:469–76. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng M, Yang X, Qin B, Liu T, Zhang H, Guo W, et al. Deubiquitination and activation of AMPK by USP10. Mol Cell. 2016;61:614–24. doi: 10.1016/j.molcel.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharya U, Neizer-Ashun F, Mukherjee P, Bhattacharya R. When the chains do not break: The role of USP10 in physiology and pathology. Cell Death Dis. 2020;11:1033. doi: 10.1038/s41419-020-03246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284:18778–89. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi M, Kitaura H, Kakita A, Kakihana T, Katsuragi Y, Nameta M, et al. USP10 is a driver of ubiquitinated protein aggregation and aggresome formation to inhibit apoptosis. iScience. 2018;9:433–50. doi: 10.1016/j.isci.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Fang ZX, Wang WW, Zhang Y, Bu ZL, Liu M, et al. Wu-5, a novel USP10 inhibitor, enhances crenolanib-induced FLT3-ITD-positive AML cell death via inhibiting FLT3 and AMPK pathways. Acta Pharmacol Sin. 2021;42:604–12. doi: 10.1038/s41401-020-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, Zhou H, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem. 2005;48:2388–406. doi: 10.1021/jm049354h. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Nicolay BN, Chick JM, Gao X, Geng Y, Ren H, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546:426–30. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.