Abstract
Protein phosphorylation is a universal mechanism regulating a wide range of cellular responses across all domains of life. The antagonistic activities of kinases and phosphatases can orchestrate the life cycle of an organism. The availability of bacterial genome sequences, particularly Bacillus species, followed by proteomics and functional studies have aided in the identification of putative protein kinases and protein phosphatases, and their downstream substrates. Several studies have established the role of phosphorylation in different physiological states of Bacillus species as they pass through various life stages such as sporulation, germination, and biofilm formation. The most common phosphorylation sites in Bacillus proteins are histidine, aspartate, tyrosine, serine, threonine, and arginine residues. Protein phosphorylation can alter protein activity, structural conformation, and protein–protein interactions, ultimately affecting the downstream pathways. In this review, we summarize the knowledge available in the field of Bacillus signaling, with a focus on the role of protein phosphorylation in its physiological processes.
Keywords: Bacillus, phosphorylation, protein kinases, protein phosphatases, sporulation, germination, biofilm, virulence
This article reviews the literature on protein phosphorylation events in Bacillus species during different stages of their life cycle and pathogenesis.
Introduction
The Bacillus genus comprises ~300 annotated species, which are divided into different phylogenetic clusters based on 16S rRNA and evolutionary relationships (Ash et al. 1991, Parte 2018). Members of the Bacillus genus are rod-shaped, mostly aerobic, or facultative anaerobes, Gram-positive spore-forming bacteria. Two of the most studied clades in these clusters are the Bacillus subtilis and Bacillus cereus clades. The subtilis clade comprises ten species including B. subtilis, B. amyloliquefaciens, B. aerophilus, B. pumilus, B. atrophaeus, B. licheniformis, Bacillus sp. 586, Bacillus sp. 916, Bacillus sp. JS, and Bacillus sp. BT1B_CT2, and the pathogenic species such as B. anthracis, B. cereus, and B. thuringiensis are classified as part of the cereus clade (Rooney et al. 2009, Bhandari et al. 2013). Among these, B. subtilis, commonly found in soil environments and plant roots, is the most characterized species. It is used as a model organism to decipher physiological and regulatory mechanisms in all the Bacillus species (Vlamakis et al. 2013, Harwood et al. 2018, Ravikumar et al. 2018, Richts et al. 2019, Errington and Aart 2020). Subtilis clade members have a wide range of industrial and medical applications owing to their phenotypic and genotypic variations (Cui et al. 2018, Harwood et al. 2018, Jezewska-Frackowiak et al. 2018, Su et al. 2020).
The Bacillus life cycle is composed of three distinct phases: spore formation (the dormant stage), spore germination, and a replicating vegetative phase (Soule 1932, Sella et al. 2014). Bacillus spores, formed under nutrition-deprived conditions, are metabolically inactive entities that can remain viable for extended periods (Graham-Smith 1930, Khanna et al. 2020). These endospores are highly resistant to stress conditions encountered in nature and, therefore, can be found in a wide range of environments including water, air, and soil. Under favorable conditions, these spores germinate into the metabolically active vegetative form (Watabe 2013, Bressuire-Isoard et al. 2018, Christie and Setlow 2020). Pathogenic Bacillus species proliferate inside their vertebrate hosts (Nicholson et al. 2000, Nicholson 2002, Arora et al. 2017a). When pathogenic Bacilli infect their hosts, the spores germinate and secrete toxins such as enterotoxin and emetic toxin, as well as phospholipases and proteases that help the bacteria to survive in the host environment (Moayeri et al. 2015, Sharma et al. 2017, Ehling-Schulz et al. 2019, Enosi Tuipulotu et al. 2021). Another survival strategy adopted by the members of this genus is biofilm formation. A biofilm is a matrix of exopolysaccharides, proteins, and nucleic acids that acts as a shield and protects the bacterium from various antibiotics and the host’s immune response (Hobley et al. 2015, Hall and Mah 2017, Huang et al. 2020, Shemesh and Ostrov 2020). The transition of Bacillus from one phase to another depends on environmental and nutritional conditions, which are sensed by the bacterial regulatory proteins. The Bacillus regulatory network involves various sensory proteins present on the surface and their post-translationally modified forms, particularly phosphorylation. This review article focuses on the role of protein phosphorylation events during transitioning of Bacillus species between different stages as well as during infection in the human host. We tried to include the most relevant articles but may have missed some studies due to limitations of search engine criteria and article length.
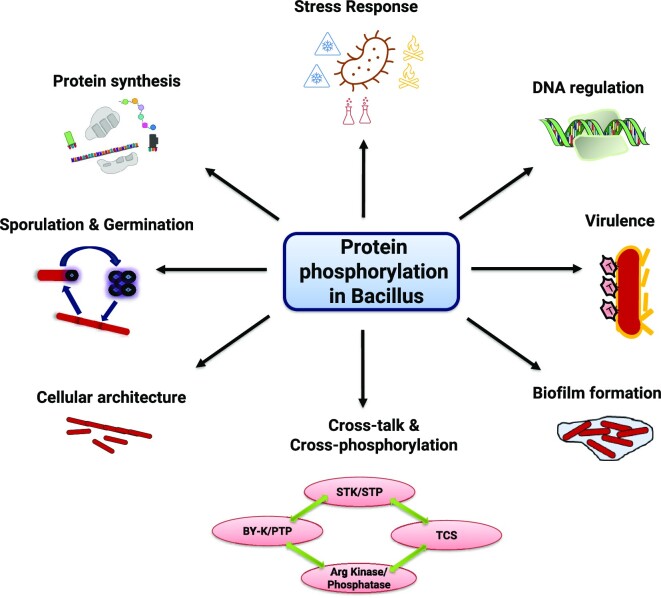
Post-translational modifications (PTMs) play important roles in providing the required proteomic diversity in bacteria, despite smaller genome sizes. PTMs such as phosphorylation, glycosylation, acetylation, and methylation influence almost all aspects of cellular physiology (Arora et al. 2010, 2021, Higgins and Dworkin 2012, Sajid et al. 2015, Manuse et al. 2016, Mijakovic et al. 2016, Khan et al. 2018, Macek et al. 2019, Bonne Kohler et al. 2020, Singhal et al. 2020, Wang and Cole 2020). Among these PTMs, protein phosphorylation is one of the most widely documented and investigated modifications across the bacterial system (Fig. 1). There are dedicated protein kinases and phosphatases that are categorized based on the phosphate group transfer from ATP to specific amino acids, such as serine (Ser)/threonine (Thr) (Ser/Thr protein kinase-STK and Ser/Thr protein phosphatase-STP), tyrosine (Tyr) (bacterial protein Tyr kinase-BY kinase and protein Tyr phosphatase-PTP), histidine (His)/aspartate (Asp) (two-component system-TCS/His sensor kinase-phosphorelay), srginine (Arg) (Arg kinase and Arg phosphatase), cysteine (Cys) [phosphotransferase system (PTS) regulated protein (such as EIIA) phosphorylation of Cys residues using PEP as energy source], and atypical protein kinases that are unlike characterized kinases but still possess kinase activity. The major phosphorylation systems are shown in the Featured image (Hoch 2000, Stock et al. 2000, Pereira et al. 2011, Capra and Laub 2012, Grangeasse et al. 2012, Fuhrmann et al. 2013, Chao et al. 2014, Deutscher et al. 2014, Mijakovic et al. 2016, Elhawy et al. 2021, Huang et al. 2021, Zhang et al. 2021). Interestingly, many proteins are phosphorylated on multiple sites, as also identified in most phosphoproteome-based studies (Macek et al. 2007, Elsholz et al. 2012, Ravikumar et al. 2014, Birk et al. 2021). For example, 214 Ser/Thr/Tyr phosphosites were identified in 153 proteins (Birk et al. 2021) and 121 Arg phosphosites were identified in 87 proteins of Bacillus subtilis (Elsholz et al. 2012). It also indicates promiscuous activities of several kinases, with overlapping specificity, and the primary reason for the fallible connection between different studies.
Figure 1.
Network of protein phosphorylation in Bacillus. Protein phosphorylation in Bacillus regulates several aspects of their life cycle, including sporulation, germination, biofilm formation, protein synthesis, DNA regulation, cell architecture, stress responses, and virulence. The detailed regulatory processes are discussed in the text along with the unique aspects of cross-talk and cross-phosphorylation within the kinases of different classes.
TCSs are ubiquitous among bacterial systems, consisting of two proteins, a sensor His kinase (HK) and a response regulator (RR). When an external stimulus is sensed, the sensor HK is activated by autophosphorylation of a conserved His residue. This signal is then subsequently transferred to its cognate RR, which is usually a transcription factor, resulting in differential altered gene expression (Fabret et al. 1999, Bijlsma and Groisman 2003, Capra and Laub 2012, Groisman 2016). TCSs are known to be involved in various cellular pathways in Bacillus sp, such as stress response (Darmon et al. 2002, Dhiman et al. 2014, Diomande et al. 2014, Mike et al. 2014, Groisman 2016, Gupta et al. 2018), protease regulation (Perchat et al. 2011, Gupta et al. 2017), sporulation (Stephenson and Hoch 2002, Fujita and Losick 2003, White et al. 2006, Gopalani et al. 2016, Peng et al. 2017), metabolic pathways (Birkey et al. 1998, Tanaka et al. 2003, Repizo et al. 2006, Geng et al. 2007, Myers et al. 2016, Aggarwal et al. 2017, van den Esker et al. 2017), cell wall regulation (Howell et al. 2003, Dobihal et al. 2019, Wu et al. 2019), biofilm formation (Verhamme et al. 2007, Stubbendieck and Straight 2017, Zhou et al. 2018), antibiotic tolerance (Kesel et al. 2013, Dintner et al. 2014, Zhang et al. 2015, Koh et al. 2021), and pathogenesis (Duport et al. 2006, Vetter and Schlievert 2007, Brillard et al. 2008, Gohar et al. 2008, Stauff and Skaar 2009, van Schaik et al. 2009).
Our understanding of phosphorylation events at different residues such as Ser, Thr, Tyr, Arg, or Cys has evolved over the last three decades. For example, the kinases responsible for Tyr phosphorylation were named bacterial tyrosine (BY) kinases and were identified based on sequence homology with known Tyr kinases, such as the YwqD/YwqE Tyr kinase and phosphatase pair in B. subtilis (Mijakovic et al. 2003, 2005). Subsequently, more such pairs were characterized in B. subtilis with roles predicted in various cellular pathways based on substrate identification in phosphoproteome studies (Mijakovic and Deutscher 2015).
The importance of Ser/Thr phosphorylation was soon realized and with the advancement in genome sequencing and -omics technologies, various Ser/Thr kinases (STKs) and Ser/Thr phosphatases (STPs) were identified and characterized in B. subtilis and other microbes (Fischer et al. 1996, Moszer 1998, Obuchowski et al. 2000, Vijay et al. 2000, Iwanicki et al. 2002, Madec et al. 2002, Shakir et al. 2010, Lima et al. 2011, Pereira et al. 2011, Arora et al. 2012, Bidnenko et al. 2013, Borriss et al. 2018, Rajagopalan et al. 2018, Baros et al. 2020). Global phosphoproteome and in silico analysis further helped in the identification of substrates regulated by Ser/Thr phosphorylation (Levine et al. 2006, Macek et al. 2007, Miller et al. 2009, Ravikumar et al. 2014, Pan et al. 2015, Rosenberg et al. 2015, Arora et al. 2017b, Zhang et al. 2018, Shi et al. 2020, Birk et al. 2021). The Bacillus genus encodes two classes of STKs that are categorized based on sequence homology and conserved sequence motifs. These are eukaryotic-like Ser/Thr kinases (eSTKs) or Hanks-type kinases and atypical STKs (non-eSTKs), and two-component kinases such as Ser/Thr kinases with specific single substrates (Pereira et al. 2011, Arora et al. 2012, Shi et al. 2014a, Mijakovic et al. 2016, Nguyen et al. 2016, Stancik et al. 2018, Zhang et al. 2021).
The global phosphoproteome of Arg phosphorylation in B. subtilis identified >100 proteins with phosphorylated Arg sites across diverse classes of proteins involved in metabolism, cellular architecture, sporulation, and various stress-induced responses (Elsholz et al. 2012, Schmidt et al. 2014, Trentini et al. 2014, Singh et al. 2015, Fuhrmann et al. 2016, Mijakovic et al. 2016, Zhou et al. 2019). In the phosphoproteome study by Elsholz et al. (2012), of the 87 proteins identified to be phosphorylated on Arg residues, 17 were phosphorylated on multiple sites. These proteins were ClpC, RpoB, AroA, MtnK, OdhA, BdhA, ComfA, ComGA, ComK, RpoC, RpsM, GltA, Gmk, GroEL, GudB, MenB, and MtnA (Elsholz et al. 2012). Interestingly, genetic deletion of Arg kinase McsB in B. anthracis caused alteration in vegetative cells and spore morphology, defective growth at elevated temperatures, and reduced sporulation and germination efficiencies (Singh et al. 2015). These studies signify the physiological relevance of Arg phosphorylation in the Bacillus species.
In the following sections, we summarize the role of protein phosphorylation during different stages and pathways in the Bacillus (Table 1).
Table 1.
Phosphorylation targets in Bacillus sp. in various cellular pathways.
| Kinase/phosphatase | Substrate/target | Experimental validation | References |
|---|---|---|---|
| Cell morphogenesis | |||
| Bsu PrkC | YvcK, YmfM (RodZ), WalR, CpgA, LtaS, YfnI, YvgJ, YqgS, FtsZ | I, II, IIIa, IIIb | Foulquier et al. (2014), Ravikumar et al. (2014), Libby et al. (2015), Absalon et al. (2008), Pompeo et al. (2018a) |
| Bsu PrpC | YvcK, YmfM (RodZ), WalR, CpgA, YfnI, FtsZ | I, IIIa, IIIb | Foulquier et al. (2014), Ravikumar et al. (2014), Libby et al. (2015), Absalon et al. (2008), Pompeo et al. (2018a) |
| Bsu WalRK | cwlO, lytE, iseA | I, IIIb | Salzberg et al. (2013), Dobihal et al. (2019) |
| Bas WalRK | ftsE, eag, kinB3 | I | Dhiman et al. (2014) |
| DNA metabolism/regulation | |||
| Bsu PtkA | SSBs, YorK, SalA, MinD, RecA, DivIVA | I, II, IIIa, IIIb | Mijakovic et al. (2006), Macek et al. (2007), Jers et al. (2010), Shi et al. (2014b) |
| Bsu PtkB | MutL, RpoB, MinD, PolA | II | Shi et al. (2014b) |
| Bsu PtpZ | SSBs, MinD, PolA, RpoB, MutL | I, II, IIIb | Mijakovic et al. (2006), Shi et al. (2014b) |
| Bsu YabT | MinD, MutL, AbrB | I, II | Shi et al. (2014b), Kobir et al. (2014) |
| Bsu PrkD | SalA, AbrB | I, II | Shi et al. (2014b), Kobir et al. (2014) |
| Bsu PrkC | AbrB | I | Kobir et al. (2014) |
| Bsu STK | CodY, AbrB, RecA | IIIa | Macek et al. (2007), Soufi et al. (2010) |
| Bsu McsB | RecA, RpoB, Ssb, DivIVA | IIIa | Elsholz et al. (2012), Schmidt et al. (2014) |
| Bsu YwlE | RecA, RpoB, Ssb, DivIVA | IIIa | Elsholz et al. (2012), Schmidt et al. (2014) |
| Bsu DegSU | ComK | I | Dahl et al. (1992), Hamoen et al. (2003) |
| Metabolsim | |||
| Bsu HprK/P | Crh | I, IIIb | Landmann et al. (2012) |
| Bsu PrkC | YwjH, GlnA, Icd, AlsD | I | Pietack et al. (2010) |
| Bsu PtkA | Ugd, Asd, Ldh, Eno, FatR, SalA | I, II | Jers et al. (2010), Mijakovic et al. (2003), Petranovic et al. (2009), Derouiche et al. (2013), Shi et al. (2014b), Derouiche et al. (2015) |
| Bsu PtpZ | Ugd | I | Mijakovic et al. (2003) |
| Bsu STK | Pyk, Asd, Eno, GlnA, Icd, YwjH | IIIa | Eymann et al. (2007), Macek et al. (2007) |
| Bsu McsB | Eno, GlnA | IIIa | Schmidt et al. (2014) |
| Bsu YwlE | Eno, GlnA | IIIa | Schmidt et al. (2014) |
| Bsu CitST | citM | I | Repizo et al. (2006) |
| Bsu GlnKL | glsA, glnT | I | Satomura et al. (2005) |
| Bsu YufLM | maeN, yflS | I | Tanaka et al. (2003) |
| Bsu PhoPR | ResDE | I | Birkey et al. (1998) |
| Bsu LytST | ysbA | I | van den Esker et al. (2017) |
| Bas PrkD | Pyk | I | Arora et al. (2012) |
| Bas PrkC | Eno | I | Virmani et al. (2019) |
| Bas PhoPR | phoA, pst | I | Aggarwal et al. (2017) |
| Protein synthesis | |||
| Bsu PrkC | Ef-Tu, Ef-G, CpgA | I, IIIa, IIIb | Absalon et al. (2009), Ravikumar et al. (2014), Gaidenko et al. (2002), Shah et al. (2008), Dworkin and Shah (2010), Ravikumar et al. (2014), Absalon et al. (2009), Pompeo (2012) |
| Bsu PrpC | Ef-Tu, Ef-G, CpgA | I, IIIa, IIIb | Gaidenko et al. (2002), Ravikumar et al. (2014), Absalon et al. (2009), Ravikumar et al. (2014) |
| Bsu STK | Ef-Tu, Ef-G | IIIa | Levine et al. (2006), Eymann et al. (2007) |
| Bsu McsB | Ef-Tu, Ef-G | IIIa | Schmidt et al. (2014) |
| Bsu YwlE | Ef-Tu, Ef-G | IIIa | Schmidt et al. (2014) |
| Bsu WalRK | sasA | I | Libby et al. (2019) |
| Bas PrkC | Ef-Tu, Ef-G | I | Arora et al. (2017b), Arora (2013) |
| Bas PrpC | Ef-Tu | I | Arora et al. (2013) |
| Sporulation | |||
| Bsu YabT | RecA, RacA, SsbA, AbrB, YabA, Ef-Tu | I, II, IIIb | Bidnenko et al. (2013), Shi et al. (2014b), Derouiche et al. (2016), Kobir et al. (2014), Garcia Garcia et al. (2018), Pereira et al. (2015) |
| Bsu SpoIIE | RecA, RacA | I, II | Shi et al. (2014b) |
| Bsu PrkA | σK | I | Yan et al. (2015) |
| Bsu CotH | CotB, CotG | I | Nguyen et al. (2016) |
| Bsu SpoIIAB | SpoIIAA, σF | I | Clarkson et al. (2004), Duncan and Losick (1993) |
| Bsu SpoIIE | SpoIIAA | I | Duncan et al. (1995) |
| Bsu KinA-E | Spo0F, Spo0B, Spo0A | I | Burbulys et al. (1991), Jiang et al. (2000) |
| Bsu RapA | Spo0F | I | Perego et al. (1994) |
| Bsu Spo0E | Spo0A | I | Ohlsen et al. (1994) |
| Bsu CotH | CotB, CotG | I | Nguyen et al. (2016) |
| Bas BAS1213–1214 | KinD | I | Gupta et al. (2018) |
| Bce LytSR | lrgAB, clhAB2 | I | Chandramohan et al. (2009) |
| Bth LytSR | SpoIIP | I | Peng et al. (2017) |
| Germination | |||
| Bsu PrkC | Ef-G | I, IIIa, IIIb | Gaidenko et al. (2002), Shah et al. (2008), Dworkin and Shah (2010), Ravikumar et al. (2014) |
| Bsu PrpC | Ef-G | I, IIIa, IIIb | Gaidenko et al. (2002), Ravikumar et al. (2014) |
| Bsu McsB | Tig, SigA | I, IIIa | Elsholz et al. (2012), Zhou et al. (2019) |
| Bsu YwlE | Tig, SigA | I, IIIa | Elsholz et al. (2012), Zhou et al. (2019) |
| Bsu CotH | CotB, CotG | I | Nguyen et al. (2016) |
| Bas PrkC | Eno | I | Virmani et al. (2019) |
| Biofilm | |||
| Bsu KinC | Spo0A | I | Lopez et al. (2009), Shemesh et al. (2010) |
| Bsu KinD | Spo0A | I | Aguilar et al. (2010) |
| Bsu CssRS | TasA | I | Steinberg et al. (2020) |
| Bsu DegSU | YiT | – | Kobayashi and Ikemoto (2019) |
| Bsu PrkC | AhpA via AbrB, GroEL | IIIa | Zwick et al. (2017), Ravikumar et al. (2014) |
| Bsu PrpC | GroEL | III | Ravikumar et al. (2014) |
| Bsu YabT | YabA | I | Garcia Garcia et al. (2018) |
| Bas PrkC | GroEL | I, IIIb | Arora et al. (2017b) |
| Stress response | |||
| Bsu McsB | CtsR, GroEL, ClpC, ClpP | I, IIIa | Fuhrmann et al. (2009), Elsholz et al. (2012), Schmidt et al. (2014) |
| Bsu YwlE | CtsR, GroEL, ClpC, ClpP | IIIa | Elsholz et al. (2012), Schmidt et al. (2014) |
| Bsu PtkA | DnaK | I, IIIa | Shi et al. (2016) |
| Bsu PtkB | DnaK | I, IIIa | Shi et al. (2016) |
| Bsu DesKR | desA | I | Cybulski et al. (2004) |
| Bsu LiaSR | liaI | I | Jordan et al. (2007), Kesel et al. (2013) |
| Bsu BceRS | bceA | I, IIIb | Dintner et al. (2014) |
| Bsu WalRK | cwlO, lytE | I | Takada et al. (2018) |
| Bsu CssRS | HtrA,HtrB | – | Westers et al. (2006), Noone et al. (2012) |
| Bsu RsbW | RsbV | I | Alper et al. (1996), Yang et al. (1996) |
| Bsu RsbP, RsbU | RsbV | I | Yang et al. (1996), Vijay et al. (2000) |
| Bsu RsbT | RsbS, RsbRA | I, IIIb | Yang et al. (1996), Chen et al. (2004), Kim et al. (2004b) |
| Bsu RsbX | RsbS, RsbRA | I | Yang et al. (1996), Chen et al. (2004) |
| Bas HssRS | – | – | Mike et al. (2014) |
| Bas HitRS | – | – | Mike et al. (2014), Pi et al. (2020) |
| Bas EdsRS | clsT | I | Laut et al. (2020) |
| Bas WalRK | – | – | Dhiman et al. (2015) |
| Bce CasKR | desA | I | Diomande et al. (2016) |
| Bce RsbK | RsbY | I | van Schaik et al. (2005), de Been et al. (2010) |
| Bth YvqEC | – | – | Zhang et al. (2015) |
| Bth YvcPQ | yvcS1S2, yvcR, kapD, BMB171_C4835 | I | Zhang et al. (2016) |
| Virulence | |||
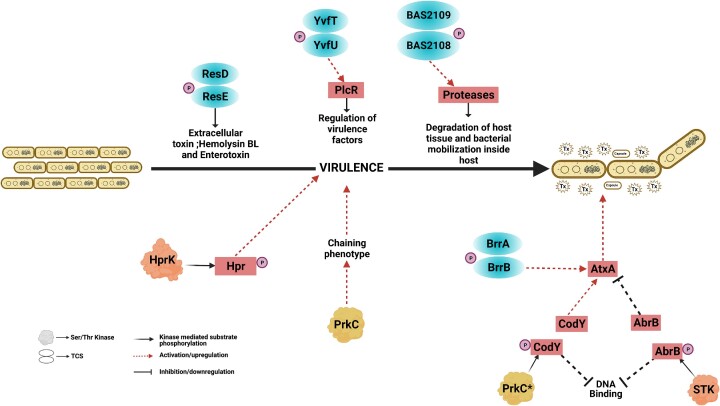
| Bas PTS | AtxA | IIIb | Tsvetanova et al. (2007), Raynor et al. (2018), Bier et al. (2020) |
| Bas STK | AtxA via CodY and AbrB | I | Strauch et al. (2005), van Schaik et al. (2009), Chateau et al. (2013) |
| Bas PrpN | CodY | I, IIIb | Gangwal et al. (2022) |
| Bas PrkC | CodY | I | Gangwal et al. (2022) |
| Bas HprK/P | HpR | I | Poncet et al. (2004), Choi et al. (2006) |
| Bas BrrAB | lef, pag, cya, atxA | I | Vetter and Schlievert (2007) |
| Bas BAS2109–2108 | Proteases | I, IIIb | Gupta et al. (2017) |
| Bce ResDE | Enterotoxins (without PlcR) | I | Duport et al. (2006) |
| Bce YvfTU | Via PlcR | I | Brillard et al. (2008) |
Protein kinases and phosphatases are represented by different colors. Pink: STK (Ser/Thr kinases); light blue: STP (Ser/Thr phosphatases); dark blue: BY-K (Bacillus Tyr kinases); orange: PTP (Bacillus Tyr phosphatases); maroon: Arg kinases; gold: Arg phosphatases; green: histidine (His)/aspartate (Asp) (two-component system-TCS/His sensor kinase-phosphorelay); red: atypical STKs and two-component like Ser/Thr kinases and phosphatases. Bacillus species are abbreviated as B. subtilis (Bsu), B. anthracis (Bas), B. cereus (Bce), and B. thuringiensis (Bth).
The experimental validation of proteins/targets is shown as:
I: in-vitro; II: interactomics-based study; IIIa: phosphoproteome; and IIIb: in-vivo.
Cellular architecture and growth
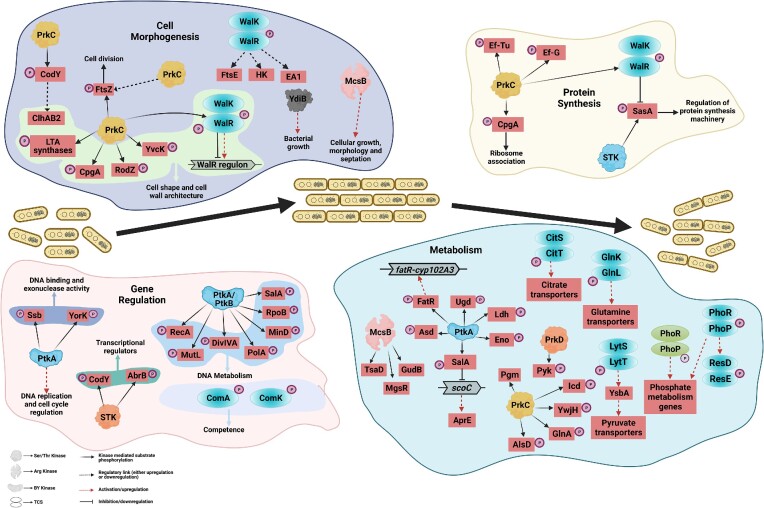
The Gram-positive bacterial cell wall is comprised mostly of multilayered peptidoglycan sheets and teichoic acids that serve as a protective barrier and help to maintain the bacterial shape and integrity. Bacterial growth and cellular integrity is a dynamic process that requires continuous hydrolysis of the cell wall to introduce breaks for the accumulation of newly formed peptidoglycan units into the cell wall (Vermassen et al. 2019). In B. subtilis, this controlled autolysis is carried out by endopeptidases such as CwlO and LytE, which are further regulated by the WalRK TCS (Yamaguchi et al. 2004, Salzberg et al. 2013, Dobihal et al. 2019, 2022). The WalRK regulon is mostly composed of genes encoding autolysins, autolysin inhibitors, ABC transporters, and genes involved in teichoic acid synthesis (Salzberg et al. 2013). Phosphorylation of the WalR RR at Asp53 by its cognate HK, WalK, blocks the promoter of autolysin inhibitor iseA, thereby causing an increase in the expression of these endopeptidases (Libby et al. 2015, Dobihal et al. 2019). Moreover, mutant walR bacilli have an L-shaped morphology and a complete loss of the bacterial cell wall, implicating the role of WalR in cell wall morphogenesis (Dominguez-Cuevas et al. 2012). In B. anthracis, though the functional relevance of the WalRK TCS has not been well-explored, its expression is induced in the presence of an inhibitor of cell wall synthesis, fosfomycin (Dhiman et al. 2015). Furthermore, WalR has been shown to bind with the promoter regions of eag (S-layer protein EA1), ftsE (ABC transporter), and kinB3 (HK) (Dhiman et al. 2014). Among these, EA1 is an important component of the S-layer surrounding the cellular envelope that imparts structural stability to the cell wall (Chateau et al. 2020, Fioravanti et al. 2022). Phosphorylation of WalR at Thr101 by PrkC, a membrane-localized eSTK with surface-exposed PASTA (peptidoglycan and Ser/Thr kinase-associated) domain, is important for the regulation of cell wall homeostasis during the stationary phase in B. subtilis (Libby et al. 2015). The two-site phosphorylation of WalR at Asp53 and Thr101 by WalK and PrkC, respectively, enhances WalR activity and blocks the expression of SasA, a protein involved in antibiotic tolerance (Libby et al. 2019) (Fig. 2).
Figure 2.
Role of protein kinases in Bacillus cell development and growth. The schematic diagram shows multiple proteins being regulated by phosphorylation during general cell development and growth with four specific processes illustrated: cell morphogenesis, protein synthesis, DNA regulation, and metabolism. Different classes of kinases are involved in each process that targets the substrates, which are involved in multiple pathways. Black solid arrows represent kinase-mediated substrate phosphorylation, red dotted arrows represent activation/upregulation, black lines with terminal bars represent inhibition or downregulation, and black dotted arrows represent regulatory links (either upregulation or downregulation).
Lipoteichoic acid (LTA) is another important component of the cell wall, i.e. threaded across peptidoglycan layers and is required for optimal bacterial growth and physiology. Synthesis of LTA in B. subtilis involves four enzymes: LtaS, YfnI, YvgJ, and YqgS, which are phosphorylated by PrkC (Pompeo et al. 2018a). CpgA (YloQ), a multidomain GTPase, is another target of the PrkC/PrpC pair that plays an important role in the maintenance of bacterial shape and peptidoglycan deposition in B. subtilis (Cladiere et al. 2006, Absalon et al. 2008, 2009). Though the studies on CpgA phosphorylation lack functional and genetic evidence, they suggest a possible link between peptidoglycan biogenesis and Ser/Thr phosphorylation (Fig. 2).
In the vegetative stage, division at the septal region results in the generation of two daughter cells termed vegetative cells. These vegetative cells display a specific rod-shaped morphology, which is mainly determined by various proteins associated with the cell wall and cytoskeleton (Jones et al. 2001, Errington and Wu 2017, Angeles and Scheffers 2021). MreB, an actin-like cytoskeletal protein is a key factor that determines the characteristic rod-shape of Bacilli, and the absence of mreB in B. subtilis alters the localization of penicillin-binding protein (PBP1), resulting in bulging cell phenotype (Formstone and Errington 2005, Carballido-Lopez 2006, Kawai et al. 2009). Interestingly, overexpression of YvcK, a protein with a similar localization pattern as MreB, was able to rescue the mreB mutant phenotype with proper PBP1 localization (Gorke et al. 2005, Foulquier et al. 2011). Ser/Thr phosphorylation-mediated regulation of YvcK (Thr304) by the PrkC/PrpC pair is shown to be critical for the rod-shaped morphology in mreB mutant cells of B. subtilis. A null mutant strain of mreB overexpressing the phosphoablative mutant YvcK (Thr304Ala) exhibits a bulging-type phenotype with mis-localized PBP1. However, mreB mutant bacteria overexpressing the phosphomimetic mutant YvcK (Thr304Glu) were able to restore rod-shaped morphology (Foulquier et al. 2014). Another cell shape-determining protein, RodZ (YmfM), is a target of the PrkC/PrpC pair in B. subtilis, although its role and functional implication is still unknown (Alyahya et al. 2009, Ravikumar et al. 2014, van Beilen et al. 2016) (Fig. 2).
Ser/Thr phosphorylation of the cell division protein FtsZ is important for efficient septum formation in prokaryotes (Garcia et al. 2016, Manuse et al. 2016, Hardt et al. 2017, Maurya et al. 2018, Rajpurohit et al. 2022). Interestingly, phosphoproteome analysis by Ravikumar et al. (2014) also identified FtsZ as a substrate of PrkC/PrpC in B. subtilis, and a study by our group reported septation defects and upregulated ftsZ levels in a prkC deletion mutant strain in B. anthracis (Dhasmana et al. 2021). These reports may provide a functional connection between PrkC and the regulation of cell division machinery in Bacillus genus via phosphorylation of FtsZ. In addition, a study on McsB Arg kinase (previously characterized as a Tyr kinase) in B. anthracis showed attenuated growth, elongated cell morphology (elongated cells), and defective septum formation in an mcsB null mutant strain, signifying the importance of Arg phosphorylation in bacterial growth and development (Mattoo et al. 2008, Singh et al. 2015). Furthermore, a new family of protein kinases belonging to the ubiquitous bacterial kinase family YdiB was shown to be important for normal bacterial growth (Karst et al. 2009, Nguyen et al. 2017).
Altogether, these studies highlight the widespread role of protein phosphorylation in the regulation of cell morphogenesis and growth in the Bacillus genus (Fig. 2).
Gene regulation
Protein phosphorylation is important in the regulation of various genetic processes including replication, transcription initiation, DNA condensation, and repair, via the phosphorylation of DNA/RNA binding proteins (Garcia-Garcia et al. 2016). Phosphoproteomics and interactomics studies have identified several proteins in B. subtilis that are phosphorylated at Ser/Thr/Tyr residues and are part of the gene regulatory network (Ravikumar et al. 2014, Shi et al. 2014b, Rosenberg et al. 2015). For instance, phosphorylation of a DNA single-stranded binding protein (Ssb) by the BY-kinase PtkA (formerly known as YwqD) and the YorK exonuclease specific for single-stranded DNA is important for DNA binding and exonuclease activities, respectively (Mijakovic et al. 2006, Jers et al. 2010) (Fig. 2). Inactivation of ptkA in B. subtilis causes the formation of multiple nucleoids the and accumulation of additional genetic material in the cells, indicating the role of Tyr phosphorylation in DNA replication and cell cycle regulatory pathways (Petranovic et al. 2007). Several other proteins involved in DNA and RNA metabolism that were identified as the target of BY-kinases (PtkA or PtkB) based on interactome analysis include RecA (recombinase), DivIVA (cell division protein), MinD (cell-division regulator), SalA (negative regulator of scoC expression, activator of PtkA kinase activity), PolA (DNA polymerase I), RpoB (DNA-directed RNA polymerase beta subunit), MutL (DNA mismatch repair protein), and TkmA/TkmB (protein tyrosine kinase activator) (Shi et al. 2014b). DivIVA is known to be regulated by protein phosphorylation in other bacterial species indicating that kinase-mediated regulation of cell division is conserved in diverse bacteria (Arora et al. 2014, Fleurie et al. 2014, Lee et al. 2014, Chaudhary et al. 2023).
The most studied Ser/Thr kinase and phosphatase pair involved in DNA regulation is the YabT kinase (Bidnenko et al. 2013) and the SpoIIE phosphatase (Duncan et al. 1995). These two enzymes are sporulation stage-specific and are critical to the development and maintenance of the spore genome (discussed in the sporulation section). Transcriptional regulators are another class of DNA-binding proteins that are targets of phosphorylation networks in the bacterial system (Dworkin 2015, Kalantari et al. 2015). The functional impact of Ser/Thr phosphorylation on the global transcriptional regulators CodY and AbrB has been studied using phosphorylation site mutants and DNA-binding experiments. Phosphorylation of AbrB at Ser86 hinders its DNA binding ability, leading to deregulation of target genes involved in the production of exoprotease, sporulation, and competence development in B. subtilis (Soufi et al. 2010, Kobir et al. 2014), while in both B. subtilis and B. anthracis, CodY phosphorylation was detected at a residue (Ser215) critical for its DNA binding activity (Joseph et al. 2005, Macek et al. 2007, Joon et al. 2017). Furthermore, our recent study in B. anthracis shows that CodY phosphorylation at Ser215 completely abolishes DNA binding with the promoter region of one of its target genes, atxA, leading to defects in toxin synthesis (Gangwal et al. 2022) (Fig. 2). Apart from these, protein phosphorylation also regulates various pathways involved in bacterial competence, the ability to uptake DNA from the environment, and its subsequent incorporation in the genome by specific protein channels and import machinery, leading to variations in the bacterial genotype and phenotype (O’Connell et al. 2022). In B. subtilis, the TCS RRs ComA and ComK are the most widely studied positive regulators of various early and late competence genes, respectively (Weinrauch et al. 1990, Dubnau et al. 1994, Maier 2020). ComK is shown to be phosphorylated by Arg kinase McsB and dephosphorylated by cognate phosphatase ywlE. Further, in the ywlE mutant of B. subtilis, ComK was found to be phosphorylated at six Arginine sites (R65, R157, R161, R165, R186, and R19). In the ywlE mutant, ComK-dependent gene expression was upregulated which suggests ComK activity is positively regulated by Arg phosphorylation (Elsholz et al. 2012). The phosphorylation status of another TCS RR, DegU, plays a critical role in the activation of competence genes and assists in the binding of ComK during competence (Dahl et al. 1992, Hamoen et al. 2003).
Cellular metabolism
Cellular metabolism plays a key role in bacterial growth and development via the modulation of important processes depending on the nutritional status and metabolic activity of the bacteria. In Bacillus, metabolic changes help to drive cellular machinery down to one of two paths, to produce either replicating vegetative cells or dormant spores. Glucose serves as a common energy source for heterotrophic bacteria and its absence triggers the uptake of other carbon sources, such as citrate, glutamine, pyruvate, or malate (Schilling et al. 2007). In B. subtilis, dedicated TCSs sense the presence of these additional carbon sources and thus increase the expression of membrane transporters facilitating their cellular intake. For example, CitST and GlnKL TCSs regulate the expression of Mg2+–citrate transporters and glutamine transporters, respectively (Satomura et al. 2005, Repizo et al. 2006). The presence of citrate or glutamine causes autophosphorylation of the respective HKs, CitS, or GlnK, followed by the activation of their respective RRs, which in turn upregulate the expression of transporters involved in the uptake of citrate or glutamine (Satomura et al. 2005, Repizo et al. 2006).
In B. subtilis, in the absence of glucose as a major carbon source, LytST TCS is required for the uptake of pyruvate via inducing the transcription of YsbA, a protein involved in the upregulation of pyruvate transporters (van den Esker et al. 2017). Similarly, in a minimal medium, the presence of malate as the sole carbon source activates the YufLM TCS. Phosphorylated YufM binds to the promoter region of maeN and increases the surface expression of malate transporters to facilitate cellular growth on malate (Tanaka et al. 2003). In B. anthracis, PhoPR is another functional TCS, which is activated upon phosphate starvation. PhoP regulates the expression of phosphate metabolism-associated genes, e.g. phoA and pst, which help to overcome the limiting phosphate availability (Aggarwal et al. 2017). A similar PhoPR TCS exists in B. subtilis, which is activated under phosphate starvation and stimulates the degradation of teichoic acid in the cell wall, releasing phosphate ions, and hence fulfilling the metabolic need for phosphate (Pragai et al. 2004, Myers et al. 2016). Furthermore, phosphorylated PhoP directly increases the expression of another important TCS called ResDE, which is important for aerobic as well as anaerobic respiration during phosphate starvation conditions in B. subtilis (Birkey et al. 1998) (Fig. 2).
In B. subtilis, HPrK/P-mediated phosphorylation of the carbon-flux-regulating histidine protein Crh (a paralog of HPr) acts as a regulatory switch in carbon metabolism (Landmann et al. 2012). Additionally, PrkC phosphorylates four major metabolic enzymes, namely Transaldolase (YwjH), Glutamine Synthetase (GlnA), Isocitrate Dehydrogenase (Icd), and α-Acetolactate Decarboxylase (AlsD) (Pietack et al. 2010). Bacillus anthracis dual-specificity protein kinase (DSPK) PrkD phosphorylates pyruvate kinase (Pyk), an enzyme that catalyzes the concluding step of glycolysis, resulting in the inhibition of its specific activity (Arora et al. 2012). Pyk was also detected in the phosphoproteome study of B. subtilis (Macek et al. 2007) (Fig. 2). Since pyk mutant in B. subtilis produce more carbon dioxide and have a reduced growth rate (Fry et al. 2000), the role of Pyk phosphorylation in the growth, metabolism, and pathogenesis of different Bacillus species needs to be studied further.
Several other reports on B. subtilis have identified the metabolic enzymes that are phosphorylated at Arg residues, indicating a possible role of the McsB/YwlE system in their regulation (Elsholz et al. 2012, Schmidt et al. 2014, Trentini et al. 2016, Zhou et al. 2019, Ogura 2020). A study by Ogura et al. (2004) showed glucose-mediated regulation of mcsB and ywlE expression in B. subtilis, suggesting that Arg phosphorylation plays a role in cellular growth in glucose-containing media. McsB and YwlE mediate reversible Arg phosphorylation of ClpCP protease and TsaD, a tRNA modification enzyme that regulates ylxR (a nucleoid-associated protein) expression through PylxS promoter. YlxR is known to regulate >400 genes, indicating the importance of glucose mediated induction of gene expression through McsB and YwlE (Ogura 2020). Interestingly, McsB has been shown to be involved in regulating protein turnover of its substrates, causing degradation of anomalous proteins through the ClpCP protease system, as discussed later in stress response section. B. subtilis McsB and its cognate Arg phosphatase YwlE were shown to reversibly phosphorylate and regulate Glutamate dehydrogenase GudB degradation in the cell (Stannek et al. 2014). The redox-sensitive modulator MgsR of SigB regulon, controls the expression of genes involved in oxidative or thiol-specific stress in B. subtilis. MgsR is phosphorylated by McsB, which mediates its degradation through Clp proteases (Lilge et al. 2020) (Fig. 2). Hajdusits et al. (2021) delineated that under stress conditions McsB forms a closed octamer-like compartment, which interconverts with monomers and other oligomers in a phosphorylation dependent manner. Interestingly, the active sites in the octamer are sequestered and only phosphorylate unfolded proteins that can enter the compartment, thus mediating their degradation. Contrarily, dimerized McsB can phosphorylate its substrates and regulate their activity (Hajdusits et al. 2021). Additional metabolic studies showed that PtkA-mediated phosphorylation of a Ugd family protein—UDP glucose dehydrogenase, is important for its catalytic activation in B. subtilis (Mijakovic et al. 2003, Petranovic et al. 2009) (Fig. 2). A similar PtkA-dependent phosphorylation mechanism activates aspartate semialdehyde dehydrogenase (Asd), converting aspartyl phosphate to aspartyl semialdehyde and inorganic phosphate (Jers et al. 2010). In different species of Bacillus, the role of UDP glucose dehydrogenase is implicated in the production of exopolysaccharides and cell wall organization, while Asd is involved in amino acid biosynthesis (Mijakovic et al. 2004, Jakobsen et al. 2009, Naerdal et al. 2011). In B. subtilis, PtkA has been shown to regulate the cellular localization of enzymes required for carbon metabolism, namely Ldh (Lactate dehydrogenase) and Eno (Enolase) (Jers et al. 2010). Phosphorylation of Eno by PrkC also affects its cellular localization and expression in B. anthracis (Virmani et al. 2019). Additionally, phosphorylation of Pgm by PrkC regulates its activity, indicating that PrkC regulates glycolysis at multiple steps (Virmani et al. 2023) (Fig. 2).
Protein phosphorylation can indirectly regulate metabolic enzymes through phosphorylation-mediated activation or inhibition of various transcriptional regulators. In B. subtilis, PtkA-mediated phosphorylation of a transcriptional regulator FatR, involved in the metabolism of polyunsaturated fatty acids, abrogates its DNA binding ability consequently leading to the derepression of the fatR-cyp102A3 operon (Derouiche et al. 2013). PtkA-mediated phosphorylation of another transcriptional regulator, SalA, represses scoC (an aprE repressor) and activates the expression of a B. subtilis exoprotease, AprE (Derouiche et al. 2015). AprE is an important metabolic enzyme, i.e. required by growing bacteria for the supply of nutrients via extracellular protein degradation. In different Bacillus species, AprE synthesis is regulated by various transcriptional regulators including CodY, AbrB, DegU, ScoC, Hpr, SinR, and SalA that are in turn regulated by protein phosphorylation events (Ogura et al. 2004, Derouiche et al. 2015, Barbieri et al. 2016, Liu et al. 2020, Zhou et al. 2021, Zolfaghari Emameh et al. 2022). These studies, therefore, suggest a functional correlation between metabolism and protein phosphorylation.
Protein synthesis
The optimal functioning of protein synthesis machinery is vital for cell survival and growth. Protein synthesis comprises of three basic steps: initiation, elongation, and termination. After initiation, there is a multistep elongation cycle involving three major elongation factors, EF-Tu, EF-Ts, and EF-G (Xu et al. 2021). These are highly conserved proteins that are essential for the survival of bacteria. EF-Tu is a GTPase that has diverse functional roles ranging from translation to pathogenesis and can interact with a variety of macromolecules including RNA, nucleotides, and other proteins (Krab and Parmeggiani 2002, Maracci and Rodnina 2016, Harvey et al. 2019). It alternates between active (GTP-bound) and inactive (GDP-bound) states, which is responsible for accurate selection of aminoacyl-tRNA and its binding to the ribosome (Bourne et al. 1991, Schmeing et al. 2009, Sajid et al. 2011a, Talavera et al. 2018). These states of EF-Tu are controlled by EF-Ts, the guanine nucleotide exchange factor. EF-G is also an essential GTPase that translocates the ribosomes along the translating mRNA (Rodnina et al. 1997, Agirrezabala and Frank 2009).
PrkC/PrpC-dependent reversible phosphorylation of the elongation factors EF-G and EF-Tu has been widely reported in the Bacillus genus (Gaidenko et al. 2002, Levine et al. 2006, Shah et al. 2008, Absalon et al. 2009, Shah and Dworkin 2010, Arora et al. 2013, 2017). Ribosome-associated proteins have also been shown to be the targets of Ser/Thr phosphorylation in B. subtilis (Absalon et al. 2009, Pompeo et al. 2012). For example, the ribosome associated GTPase CpgA is phosphorylated by PrkC in B. subtilis. Mutation of the phosphorylated residue Thr166 decreases the GTPase activity of CpgA as well as its affinity to 30S ribosomal subunits. The Bacillus strains expressing the CpgA-Thr166Ala variant show growth defects and exhibit a curly morphology, indicating the importance of ribosome-associated protein phosphorylation in maintaining B. subtilis growth and morphology (Pompeo et al. 2012) (Fig. 2).
Bacillus sp. also utilize secondary messengers known as alarmones or hyperphosphorylated (p)ppGpp nucleosides for protein synthesis during nutrient deprivation (Bange and Bedrunka 2020). These alarmones act as a cue for starving bacteria to shut down various essential cellular pathways, such as transcription and DNA replication, thereby helping them to conserve their energy resources (Potrykus and Cashel 2008, Steinchen and Bange 2016, Gourse et al. 2018). In B. subtilis, the cellular level of these alarmones is determined by dedicated (p)ppGpp synthetases such as SasA (small alarmone synthetase A) (Nanamiya et al. 2008), which are known to be regulated by Ser/Thr phosphorylation (Libby et al. 2019). The expression of sasA is regulated by the WalR transcription factor, a component of the WalRK TCS. Interestingly, WalR itself is regulated by PrkC/PrpC-mediated reversible phosphorylation, resulting in further activation of WalR activity and repression of sasA expression (Libby et al. 2019). Thus, the protein synthesis machinery in B. subtilis is subjected to regulation by phosphorylation, either directly by phosphorylation of translational factors or by (p)ppGpp-mediated inhibition (Fig. 2).
Sporulation
The process of spore formation and revival provides an excellent model for understanding the developmental processes in the bacterial system. During sporulation, the entire Bacillus metabolic pathways are reset with the help of various regulatory modifications such as protein phosphorylation to form a dormant spore (Errington 2003, Hoch 2017, Khanna et al. 2020). In B. subtilis, initiation of sporulation is triggered by adverse environmental conditions, such as, nutritional stress, oxygen tension, or redox changes. These signals are perceived by KinA-E HKs, which phosphorylate Spo0A, the master regulator of sporulation (Fig. 3) (LeDeaux and Grossman 1995, LeDeaux et al. 1995, Jiang et al. 1999, Fujita and Losick 2005, Aguilar et al. 2010). The activation of Spo0A is managed by a phosphorelay cascade emanating from Kin HKs (Jiang et al. 2000). For example, KinA-mediated phosphorelay involves Spo0F, Spo0B, and Spo0A (Burbulys et al. 1991). Activated Spo0A, in turn, represses the expression of AbrB, a transcription repressor of sporulation (Strauch et al. 1990). On the other hand, phosphatases RapA and Spo0E reset Spo0F and Spo0A to an unphosphorylated state (Ohlsen et al. 1994, Perego et al. 1994, Perego 2001). In B. anthracis, out of nine sensor HKs, five have been characterized and shown to initiate sporulation via this classic phosphorelay (Brunsing et al. 2005). Apart from these Kin HKs, two functional TCSs, BAS1213–1214 and BAS0540–0541, are characterized in B. anthracis with a possible role in sporulation (Gopalani et al. 2016, Gupta et al. 2018). The BAS1214 HK senses oxidative stress and phosphorylates its cognate RR, BAS1213. The phosphorylated RR thus increases its expression and that of sporulation kinase D, which causes a reduction in sporulation efficiency (Gupta et al. 2018). In the BAS0540–0541 TCS, the overexpression of the BAS0540 RR causes a reduction in spore counts, although the molecular mechanism is still unknown (Gopalani et al. 2016).
Figure 3.
Role of protein kinases during sporulation in Bacillus. The schematic diagram shows several pathways being regulated by phosphorylation during sporulation. Every pathway involves multiple proteins’ activity regulated through phosphorylation. Black solid arrows represent kinase-mediated substrate phosphorylation, red dotted arrows represent activation or upregulation, and black lines with terminal bars represent inhibition or downregulation.
The significance of TCSs is not limited to the initiation of sporulation, they are also involved in later stages of sporulation including forespore engulfment and mother cell lysis. In B. anthracis, LytSR TCS functions by dissipating proton motive force, acting as a signal for the induction of lrgAB and clhAB2 transcription, which can affect sporulation efficiency and cell viability during the stationary growth phase (Chandramohan et al. 2009). In B. thuringiensis, the LytSR TCS is induced by the mother cell compartment sigma factor, SigE. Deletion of lytS-lytR causes defects in forespore engulfment by the mother cell. LytSR can induce the expression of a cell wall hydrolase, SpoIIP, however, the exact mechanism of this regulation is not known (Peng et al. 2017). Thus, multiple TCSs are important for controlling sporulation (Fig. 3).
The importance of Ser/Thr phosphorylation in context of sporulation in B. subtilis has been studied and reviewed extensively (Pompeo et al. 2016). Activation of prespore specific RNA polymerase (SigF) gene by Ser phosphorylation was one of the first discovered examples of regulatory Ser phosphorylation in B. subtilis (Min et al. 1993). This is achieved by the proteins present in the same operon: SpoIIAB (anti-SigF), SpoIIAA (anti-anti-SigF), and SpoIIE (serine phosphatase). SpoIIAB binds and inactivates SigF, while phosphorylation of SpoIIAA by SpoIIAB at a serine residue results in the release of SigF from the complex (Duncan and Losick 1993, Clarkson et al. 2004).
As cell-type-specific transcription factors are activated during different sporulating stages, the expression of specific kinases and phosphatases is also triggered during sporulation, indicating their requirement during this process (Kroos et al. 1999, Piggot and Hilbert 2004). For example, among the four characterized Hanks family Ser/Thr kinases in B. subtilis, the expression of YabT and PrkA peaks during sporulation initiation (Bidnenko et al. 2013, Yan et al. 2015). In B. subtilis, PrkA was shown to phosphorylate a 60-kDa protein in the crude extract at a Ser residue, but its functional relevance remained inconclusive due to lack of in-vivo data in the absence of this protein (Fischer et al. 1996). Later, the role of PrkA was linked to the sporulation process due to defective sporulation in prkA null mutant strain, possibly due to the downregulation of spore-specific transcription factor σK (Yan et al. 2015) (Fig. 3). Interestingly, another independent study in B. subtilis revealed sequence similarity of PrkA with ATP-dependent protease family proteins and PrkC-mediated regulation of PrkA protease activity was shown to be critical for the initiation of the sporulation process (Zhang et al. 2022). The expression of YabT kinase during sporulation initiation is required for the development and maturation of bacterial endospores in B. subtilis (Bidnenko et al. 2013). DNA binding proteins such as RecA, RacA, YabA, and single-stranded DNA binding proteins (Ssb) are the targets of YabT kinase (Bidnenko et al. 2013, Shi et al. 2014b, Derouiche et al. 2016, Garcia-Garcia et al. 2016, 2018) (Fig. 3). Among these, RecA (a DNA recombinase), and RacA (a chromosome-anchoring protein), are involved in maintaining the chromosomal integrity of the developing spore (Bidnenko et al. 2013, Shi et al. 2014b, Ramirez-Guadiana et al. 2016), while YabA is crucial for replication initiation during sporulation (Noirot-Gros et al. 2006, Garcia Garcia et al. 2018). Bacillus subtilis has two Ssb proteins, SsbA and SsbB, that are implicated in various pathways related to genome maintenance and natural competence (Lindner et al. 2004, Yadav et al. 2013, 2014, Paschalis et al. 2017). Mass spectrometry (MS) analysis revealed a novel phosphorylation site (Thr38) on the SsbA in B. subtilis (Derouiche et al. 2016), which was previously found to be phosphorylated on Tyr82 (Mijakovic et al. 2006) and Arg76 residues (Elsholz et al. 2012). This phosphorylation was primarily mediated by YabT, which enhanced the cooperative binding to single-stranded DNA (Derouiche et al. 2016). RecA is an SOS repair protein that plays an important role in bacterial DNA damage repair pathways (Nahrstedt et al. 2005, Million-Weaver et al. 2015, Torres et al. 2019). The deletion mutant of yabT and nonphosphorylatable mutant of RecA in B. subtilis exhibits same phenotype of increased sensitivity to DNA damage, indicating its role in DNA integrity maintenance (Bidnenko et al. 2013).
YabA negatively regulates replication initiation by decreasing cooperative binding of replication initiator DnaA to DNA (Noirot-Gros et al. 2006, Schenk et al. 2017). Phosphorylation of YabA by YabT kinase causes enhanced sporulation in B. subtilis, possibly correlating with decreased DNA replication and cell division (Garcia Garcia et al. 2018) (Fig. 3). YabT also phosphorylates the transition-phase transcriptional regulator AbrB, which is important for sporulation (Kobir et al. 2014). AbrB phosphorylation abrogates its DNA binding to the promoter of Spo0E, which is essential for maintaining the level of the active form of Spo0A-P (Molle et al. 2003, Shafikhani and Leighton 2004) (Fig. 3). YabT also phosphorylates EF-Tu, leading to decreased GTP hydrolysis and stabilization of interaction with the ribosome. This results in downregulation of protein synthesis by causing a halt in the protein elongation cycle in the cells undergoing a dormant state (Pereira et al. 2015). Furthermore, the functional implication of Ser/Thr phosphorylation in the context of sporulation has not been well-explored in the pathogenic Bacillus species. A recent report on B. anthracis demonstrated complete inhibition of the sporulation process in the absence of the STP PrpN (Gangwal et al. 2022). In a nutshell, these studies indicate the critical role of Ser/Thr phosphorylation in the sporulation pathway.
Bacillus spores have a multilayered structure consisting of exosporium (present in only a few species), a spore coat, cortex, and the core wall (Khanna et al. 2020). The spore coat is a thick sieve-like protein layer that protects the endospore. In B. subtilis, CotH, an atypical kinase present in the inner layer of the spore coat regulates the assembly of spore coat proteins (Naclerio et al. 1996, Nguyen et al. 2016, Scott and Newton 2016). CotH in B. subtilis and B. cereus acts as a kinase, phosphorylating two other spore coat proteins (CotB and CotG) on Ser residues, which is essential for efficient spore coat assembly (Nguyen et al. 2016, Freitas et al. 2020, Di Gregorio Barletta et al. 2022). Moreover, phosphorylation levels of CotB and CotG by CotH were shown to be sensitive to thermal variations, with higher efficiency at lower temperature (25°C) than at higher temperature (42°C) (Isticato et al. 2020, Di Gregorio Barletta et al. 2022). This makes CotH-mediated phosphorylation of CotB and CotG essential for proper spore coat morphogenesis (Fig. 3). Exosporium, an irregular-shaped layer consisting of hair-like projections is present in the outer layer of spore coat and is required for interaction with host cells and the surrounding environment (Bozue et al. 2007, Stewart 2015, Wang et al. 2016). In B. anthracis, ExsB, a CotG homolog present at the basal layer, is essential for stable attachment of exosporium to the spore coat. ExsB was found to be the highly phosphorylated exosporium protein, with at least 14 of its 19 Thr residues modified in its central region. This phosphorylation event is speculated to be controlled by a homolog of CotH, as occurs in the case of B. subtilis (McPherson et al. 2010, Freitas et al. 2020). Besides this, the deletion strain of McsB Arg kinase in B. anthracis showed defects in sporulation, in addition to cell growth and germination, confirming the role of Arg phosphorylation during the sporulation process (Singh et al. 2015) (Fig. 3).
Germination
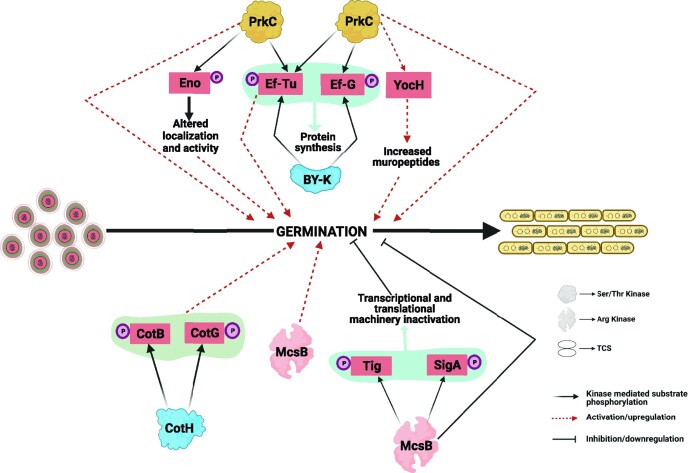
The process of germination involves the progressive metabolic awakening of the dormant spore by reactivation of major biological processes including cell growth and protein synthesis machinery. This phenomenon is triggered by stimuli such as nutrient germinants that are sensed by surface receptors on the spore membrane and involve the temporal expression of about 30% of the bacterial genome as studied in the model organism B. subtilis (Keijser et al. 2007). Bacterial kinases are reported to be important in spore germination. The Ser/Thr kinase PrkC with surface-exposed PASTA domain, is a crucial germinant receptor, which mediates the muropeptide (small peptide fragments released from growing bacterial membranes)-dependent germination process in B. subtilis (Shah et al. 2008, Shah and Dworkin 2010, Squeglia et al. 2011) (Fig. 4). Furthermore, PrkC-mediated induction of the secretory peptidoglycan hydrolase YocH ensures the availability of muropeptides during the germination process by digestion of the surrounding bacterial peptidoglycan (Shah and Dworkin 2010). Mutant spores lacking prkC showed a muropeptide-dependent defective germination profile in B. subtilis and B. anthracis (Shah et al. 2008).
Figure 4.
Role of protein kinases during spore germination in Bacillus. The schematic diagram shows multiple proteins regulated by phosphorylation during the spore germination process. The phosphorylation of these proteins is shown by black solid arrows. Red dotted arrows represent activation/upregulation, and black lines with terminal bars represent inhibition/downregulation.
Following signal acquisition to exit dormancy, the protein machinery is activated to stimulate the growth of metabolically active vegetative cells (Sinai et al. 2015, Xing and Harper 2020). The translation factor EF-G is identified as a common substrate of PrkC/PrpC (Fig. 4), and its phosphorylation is important in the regulation of protein synthesis during germination in B. subtilis (Gaidenko et al. 2002, Shah et al. 2008, Shah and Dworkin 2010). As mentioned, the surface-exposed PASTA domain of PrkC can sense muropeptides, which activate PrkC and help in spore germination. Activated PrkC, in turn, transmits the signal inside the germinating spore and phosphorylates EF-G. This signaling module suggests the role of PrkC in exiting dormancy and facilitating spore germination through protein synthesis (Shah et al. 2008). Another translation factor, EF-Tu, was also identified as a common substrate of the PrkC/PrpC pair in B. subtilis and B. anthracis, though the functional relevance of this phosphorylation is still unknown (Absalon et al. 2009, Arora et al. 2013) (Fig. 4).
Arg phosphorylation also plays an important role during spore germination in B. subtilis (Zhou et al. 2019). A transposon-based genetic screen in B. subtilis showed that genetic disruption of the Arg phosphatase gene ywlE results in severe germination defects (Zhou et al. 2019). In this study, the significance of Arg phosphorylation in spore germination was also corroborated by an accelerated germination process in the absence of Arg kinase-McsB. Furthermore, Arg phosphoproteome of spores identified 18 proteins, including the translation factor Tig and housekeeping sigma factor SigA. The impact of Arg phosphorylation on these two proteins in the context of germination was assessed by using phosphomimetic and phosphoablative mutants and germination defects were observed in Tig (Arg45Asp) and SigA (Arg365Asp) mutants. The study showed arginine dephosphorylation of Tig and SigA as an important regulatory step for the re-establishment of bacterial transcriptional and translational machinery by enabling Tig association with ribosomes and SigA activation during the germination process (Zhou et al. 2019). Interestingly, the deletion of mcsB in B. anthracis leads to reduced germination efficiency, thus highlighting a strain-specific role of McsB in spore germination (Singh et al. 2015).
Apart from the protein synthesis pathway, the germination of bacterial spores involves the reprogramming of cellular metabolism. In fact, various studies have shown glycolytic enzymes as a target of Ser/Thr phosphorylation in B. subtilis and B. anthracis (Arora et al. 2012, Rosenberg et al. 2015, Virmani et al. 2019). In B. anthracis, phosphorylation of a glycolytic enzyme, enolase, by PrkC plays an important role in spore germination by modulation of enolase activity, expression, and cellular localization (Virmani et al. 2019) (Fig. 4).
In another class of atypical STKs, the Bacillus spore coat protein CotH was shown to be important during germination in B. subtilis (Naclerio et al. 1996). CotH-mediated Ser phosphorylation of two other spore coat proteins, CotB and CotG, is an essential requirement for spore germination (Nguyen et al. 2016). Although the mechanistic aspects of the observed germination defect have not been explored yet, it would be interesting to investigate the functional relevance of CotH in the Bacillus genus owing to the presence of its orthologs across different genera including spore-forming bacteria and even eukaryotic species (Nguyen et al. 2016) (Fig. 4).
Biofilm
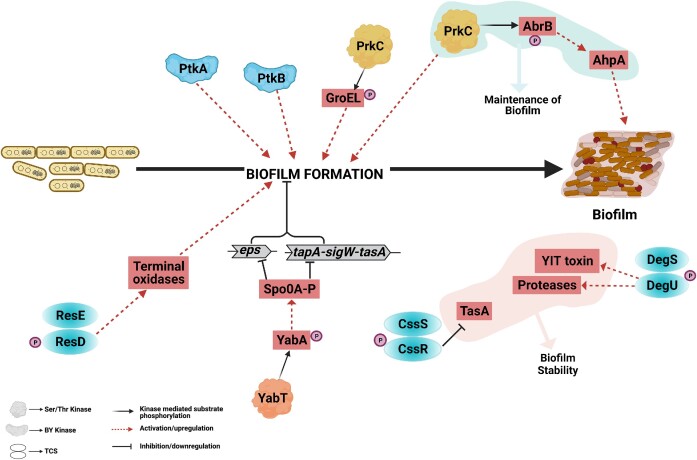
Biofilm is a multicellular structure with complex cellular differentiation that helps bacteria to deal with external stresses such as nutrient deprivation, antibiotics, and low oxygen tension, allowing them to survive in adverse conditions (Hoiby et al. 2010, Kostakioti et al. 2013, Yin et al. 2019, Arnaouteli et al. 2021). As mentioned before, B. subtilis has five HKs (KinA–E) that autophosphorylate upon sensing environmental signals and regulate Spo0A phosphorylation (Burbulys et al. 1991). Apart from inducing sporulation, Spo0A is known to be vital in the formation of biofilm under low oxygen conditions, and deletion of spo0A restricts the cells to a monolayer pattern rather than a three-dimensional structure (Hamon and Lazazzera 2001, Mielich-Suss and Lopez 2015). Also, while KinA primarily controls sporulation, KinC-mediated activation of Spo0A is mainly linked to biofilm formation (Shemesh et al. 2010, Devi et al. 2015). With the aid of mathematical modeling, the activation of Spo0A by KinC was shown to be dependent on the bacterial growth phase and concentration of KinA (Chen et al. 2022). Another HK, KinD is reported to possess both kinase and phosphatase activities and is activated by lipoprotein Med (Aguilar et al. 2010, Banse et al. 2011). KinD-mediated phosphorelay fine-tunes Spo0A phosphorylation levels, thus becoming a switch to trigger either sporulation or biofilm formation (Aguilar et al. 2010). Small molecule inducers including l-malic acid released by tomato roots activate KinD, hence triggering the transcription of matrix-producing genes leading to bacterial biofilm formation on tomato roots (Chen et al. 2012). A combination of glycerol and manganese is also reported to initiate biofilm formation via specifically activating KinD-mediated signaling (Shemesh and Chai 2013). Also, in B. subtilis biofilm defects in spo0A mutants were rescued by mutations in abrB transcription factor, suggesting that Spo0A-mediated repression of abrB is essential for biofilm formation (Hamon and Lazazzera 2001).
In B. subtilis, the CssRS TCS stabilizes the biofilm by limiting the expression of the repressor tasA, thereby increasing the production of the extracellular matrix, which is crucial for successful biofilm formation (Steinberg et al. 2020). Also, the deletion of cssRS causes a sharp increase in the population of motile bacteria (Steinberg et al. 2020). Even after the biofilm is formed, another TCS, DegS-DegU induces the secretion of YIT toxin within the biofilm to destroy sensitive cells and attack any incoming competitor cells (Kobayashi and Ikemoto 2019). DegSU also controls the expression of extracellular proteases and enzymes that are crucial for biofilms (Kobayashi 2007). In rhizobacterium, B. amyloliquefaciens, the ResDE TCS is reported to sense oxygen deprivation. It triggers biofilm formation by increasing the expression of terminal oxidases (Zhou et al. 2018). Apart from histidine kinases, Ser/Thr kinases also regulate biofilm formation and maintenance (Fig. 5).
Figure 5.
Role of protein kinases in Bacillus biofilm formation. Biofilm formation in bacteria is a complex process and requires regulation at several steps. The figure is a schematic representation of phosphorylation-mediated regulation of biofilm formation in bacteria. As shown, different classes of kinases and their substrates are required for biofilm formation. Black solid arrows represent kinase-mediated substrate phosphorylation, red dotted arrows represent activation/upregulation, and black lines with terminal bars represent inhibition/downregulation.
The formation of biofilm, as already mentioned, depends on environmental cues to which the bacteria are exposed. These cues are sensed by surface proteins that mediate proper colonization. The Ser/Thr kinase PrkC, which possesses a surface-exposed sensor domain is crucial for biofilm formation. Interestingly, prkC deletion in B. anthracis also leads to the abrogation of biofilm formation. Mechanistic insights on the signaling pathway mediated by Ser/Thr phosphorylation driving biofilm formation in B. anthracis have emerged from the linkage of PrkC and one of its substrates, the GroEL chaperone (Arora et al. 2017b). GroEL has also been shown to be involved in biofilm formation in various other organisms like Mycobacteria and Streptococci (Ojha et al. 2005, Yin et al. 2019). MS studies revealed GroEL as one of the substrates of PrkC, and overexpression of native GroEL resulted in partial resumption of biofilm formation in a biofilm-defective prkC deletion strain (Fig. 5) (Arora et al. 2017b). Furthermore, phosphorylation of YabA by Ser/Thr kinase YabT negatively regulates biofilm formation by increasing the cellular level of Spo0A-P, the key regulator of genes involved in biofilm formation and sporulation (Garcia Garcia et al. 2018). Although the mechanism of tyrosine phosphorylation in biofilm formation is still unknown, null mutant strains of ptkA and ptpZ showed an altered biofilm phenotype in B. subtilis (Kiley and Stanley-Wall 2010). The biofilm formed in these conditions showed loss of “fruiting bodies” for sporulation and the absence of typical biofilm complex radial structures. The reason for this phenotype was attributed to the defective sporulation efficiency of the bacterial cells that were growing in the biofilm colony. Complete loss of biofilm formation was observed in the strains lacking both the BY kinases, ptkA and ptkB (Gerwig et al. 2014). Apart from this, flagellar motility of several Bacillus species that helps the bacteria to swim and slide across surfaces is often linked to biofilm initiation and development (Houry et al. 2010, Guttenplan and Kearns 2013, Liaqat et al. 2018, Li et al. 2022). Inhibition of motility promotes biofilm formation and in B. subtilis, flagellar motility and swarming properties of the bacteria are regulated by the phosphorylation status of the transcriptional regulator DegU by its cognate HK, DegS (Verhamme et al. 2007, Murray et al. 2009). Altogether, these studies indicate the central role of protein phosphorylation during biofilm formation in Bacillus (Fig. 5).
Stress response
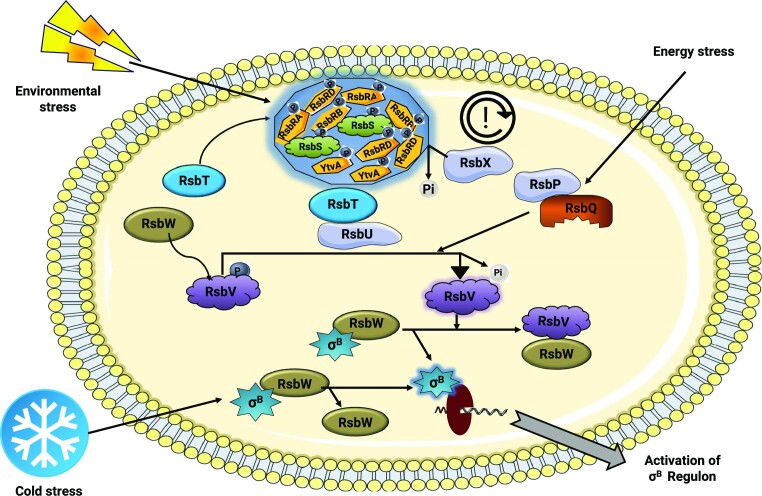
Bacterial populations encounter various stressful conditions throughout their life cycle ranging from alterations in temperature, pH, osmolytes, nutrient deficiency, and exposure to antibiotics. To combat this, bacteria have evolved sophisticated stress responses that involve a wide range of cellular and morphological changes (Marles-Wright and Lewis 2007, Ultee et al. 2019, Cheng-Guang and Gualerzi 2020). This section presents current knowledge on the role of protein phosphorylation during stress conditions in Bacillus sp. including activation of SigB, the master regulator of general stress response (GSR) via a protein phosphorylation cascade involving Rsb (Regulator of sigma B) family proteins (Fig. 6).
Figure 6.
Ser/Thr phosphorylation in the activation of the SigB (σB) regulon. In unstressed conditions, the RsbT kinase forms a complex with other Rsb family proteins (RsbS antagonist and RsbR coantagonist with its paralogues). Upon environmental stress, the RsbT kinase phosphorylates RsbS and RsbR with its paralogues at Ser/Thr residues and is released from the complex. RsbT then interacts with the RsbU phosphatase resulting in its activation and dephosphorylation of a downstream antagonist, RsbV, which is further regulated by RsbW-mediated phosphorylation. Unphosphorylated RsbV dissociates the σB–RsbW kinase complex by binding to RsbW, thereby releasing σB, which in turn associates with RNA Polymerase and activates σB regulon gene expression. The RsbX phosphatase resets the entire stressosome machinery by dephosphorylation of RsbS and RsbR. Under energy-deficient conditions, a separate pathway is activated involving dissociation of another complex comprised of the RsbP phosphatase and a stress RR, RsbQ. The released RsbP results in the dephosphorylation of RsbV, which triggers downstream signaling pathways. During cold stress, bacteria respond by triggering another independent pathway involving RsbW. The complex of RsbW and σB modulates the expression of genes regulated by σB.
Protein Arg phosphorylation in the Bacillus is primarily linked to the maintenance of overall protein quality by regulated proteolysis of misfolded or aggregated proteins during stress conditions (Mijakovic et al. 2016). This is achieved by a regulatory system that involves McsB (Arg kinase)-mediated inhibition of CtsR (stress response transcriptional regulator) repressor activity, activation of the ClpCP proteolytic machinery, and phospho-Arg tagging of the misfolded proteins (Fuhrmann et al. 2009, Elsholz et al. 2011, Tao et al. 2012, Trentini et al. 2016). The kinase activity of McsB is further modulated by its repressor CtsR, binding of phospho-Arg polypeptides at its catalytic site, ClpC (stress response-related ATPase, AAA+ superfamily), McsA (Arg kinase activator protein), and finally by the activity of its cognate Arg phosphatase (YwlE) (Kirstein et al. 2005, 2007, Elsholz et al. 2011, 2012, Fuhrmann et al. 2013, Suskiewicz et al. 2019). Strains lacking mcsB and clpC in B. anthracis show growth defects at elevated temperature (43°C) (Singh et al. 2015). The importance of Arg phosphorylation in stress response pathways was demonstrated by the global phosphoproteome of the B. subtilis Arg phosphatase (ywlE) mutant strain. Phospho-Arg sites were detected in proteins involved in GSR (controlled by SigB), such as stress on the cell envelope by antibiotics and osmolarity changes, heat shock, cold shock, and oxidative stress (Elsholz et al. 2012). Given the widespread role of Arg phosphorylation in bacterial stress response, another Arg phosphoproteome study, focusing specifically on heat and oxidative stress, identified key bacterial stress RRs such as CtsR, GroEL, ClpC, and ClpP as targets of Arg phosphorylation in B. subtilis (Schmidt et al. 2014). Furthermore, the YwlE phosphatase is inactivated during oxidative stress conditions as a regulatory mechanism for the induction of stress-related genes and McsB-mediated Arg phosphorylation (Fuhrmann et al. 2016). These studies highlight the importance of protein Arg phosphorylation during stress conditions in Bacillus.
The documented role of Tyr phosphorylation in Bacillus stress pathways, on the other hand, is limited to only a few studies. The chaperone protein DnaK (Hsp70 family protein) is a widely studied heat shock protein, i.e. a member of chaperone machinery activated during stress conditions and is also reported to be a target of protein phosphorylation in other bacteria (Sherman and Goldberg 1993, Seeger et al. 1996, Peake et al. 1998, Mayer and Bukau 2005, Roncarati and Scarlato 2017, Rigo et al. 2020). DnaK was identified as a substrate in Ser/Thr/Tyr phosphoproteomes (Eymann et al. 2007). Later, it was identified that PtkA/PtpZ-mediated regulation of DnaK by phosphorylation at a Tyr residue was responsible for controlling its chaperone activity, which affects survival of B. subtilis under heat shock conditions. (Shi et al. 2016). Deletion mutants of two low molecular weight Tyr phosphatases, YwlE (also an Arg phosphatase) and YkfJ showed reduced bacterial resistance to ethanol stress in B. subtilis, suggesting their possible roles in generating resistance to stress (Musumeci et al. 2005).
Various environmental stresses encountered by bacteria activate specific TCSs. Two different TCS can activate the same set of downstream target genes. One such example is the Heme sensing TCS (HssRS) and the HssRS-interfacing TCS (HitRS) in B. anthracis (Mike et al. 2014, Pi et al. 2020). Alterations in cell envelope integrity activates HitRS, that in turn interacts with HssRS and coordinate heme and cell envelope stress response (Mike et al. 2014). Similarly, the HssRS and HitRS TCSs exist in B. thuringiensis, which indirectly control the growth in the presence of heme through the uncharacterized operon hrmXY (Schmidt et al. 2016).
In B. subtilis, adaptation to cold temperature is achieved by the DesKR TCS. DesK (HK) senses the ordered lipid pattern in the membrane and autophosphorylates, followed by DesR (RR) phosphorylation (Albanesi et al. 2004, Abriata et al. 2017). Active phospho-DesR then binds to the promoters of genes involved in the synthesis of the fatty acid desaturase DesA (Cybulski et al. 2004). DesK also responds to changes in pH; acidic pH breaks hydrogen bonds in the helix connecting the DesK transmembrane domain to the cytosolic domain, and thus abolishes the DesK-dependent synthesis of unsaturated acids, providing rigidity to the bacilli (Bortolotti et al. 2020). Apart from temperature and pH sensing, DesKR is highly expressed in low-pressure conditions in B. subtilis strain WN1106 (Fajardo-Cavazos et al. 2012). In B. cereus, adaptation to a cold environment occurs by higher expression of unsaturated fatty acid to maintain membrane fluidity at a lower temperature (Diomande et al. 2014). The CasKR TCS activates the expression of fatty acid desaturase DesA under cold conditions by ceasing repression of the desA promoter (Diomande et al. 2015, 2016).
In B. anthracis, a novel TCS named the Envelope Disruption System EdsRS is critical for managing the stress induced by targocil, an inhibitor that targets the cell envelope (Laut CL 2020). Compromised membrane integrity is detected by EdsS (HK), which results in its autophosphorylation and subsequent phosphorylation of EdsR (RR). Phosphorylated EdsR binds and activates the promoter of the BAS1661–BAS1663 operon encoding the cardiolipin synthase ClsT for the repair of the cell envelope in targocil-exposed vegetative cells. Exposure of Bacillus spores to targocil is reported to be highly toxic, as spore outgrowth requires rapid membrane generation (Laut CL 2020). In B. thuringiensis, the YvqEC and YvcPQ TCSs sense disturbances in the cell envelope and provide resistance against the cell wall targeting compounds vancomycin and bacitracin, respectively (Zhang et al. 2015, 2016). In B. subtilis, the LiaSR TCS manages the exogenous stress induced by the presence of peptide antibiotics targeting cell wall synthesis (bacitracin, nisin, vancomycin, and ramoplanin) (Mascher et al. 2004, Kesel et al. 2013). The LiaR-dependent promoter liaI is expressed during the transition from the exponential to stationary phase, suggesting an additional sensor signal for LiaS, other than antibiotics (Jordan et al. 2006, 2007). In B. subtilis, BceRS is a dedicated TCS that responds to the presence of bacitracin via its specific binding to BceB (permease) (Ohki et al. 2003, Dintner et al. 2014). BceS (HK) works in coordination with BceB (permease) and the level of BceS (HK) autophosphorylation is dependent on the BceA-bacitracin complex (Dintner et al. 2014).
WalRK plays an important role in cell wall homeostasis under physiological conditions in B. subtilis. Apart from this, WalRK is equally important for growth and cell proliferation under heat stress (Takada et al. 2018). WalK (HK) is activated upon heat stress and its activity is negatively modulated by two membrane proteins WalH and WalI (Takada et al. 2018). The presence of WalH and WalI is crucial to avoid overexpression of downstream endopeptidase genes, thus avoiding cell lysis. Even in B. anthracis, the expression of WalRK is induced either by temperature or in the presence of the bactericidal drug fosfomycin, an inhibitor of cell wall synthesis (Dhiman et al. 2015). In B. subtilis, the CssRS TCS is important in managing secretion stress, which happens when misfolded proteins accumulate outside the membrane and interfere with protein secretion. This inefficient protein secretion is detected by CssRS, which then increases the expression of the chaperonic proteases HtrA and HtrB (Westers et al. 2006, Noone et al. 2012).
Another bacterial GSR involves the activation of sigma factor, SigB (master regulator of GSR), which regulates the expression of over 100 genes in B. subtilis during stress conditions (Haldenwang and Losick 1979, Bernhardt et al. 1997, Hecker and Völker 1998). Activation of the SigB regulon is achieved by a signaling cascade that involves Ser/Thr phosphorylation of Rsb (regulator of sigma factor) family proteins comprising TCS such as Ser/Thr kinases (RsbT and RsbW); PP2C-type phosphatases (RsbP, RsbX, and RsbU); antagonists (RsbS and RsbV), and coantagonists (RsbRA, RsbRB, RsbRC, and RsbRD) (Price et al. 2001, Hecker et al. 2007, Rodriguez Ayala et al. 2020).
In normal conditions, Ser/Thr kinase RsbW phosphorylates the antagonist RsbV, causing its inactivation and also acts as a negative regulator of SigB that blocks its binding to the RNA polymerase core enzyme (Alper et al. 1996, Yang et al. 1996, Rodriguez Ayala et al. 2020). Environmental stress conditions (acid/ethanol/salt) trigger dephosphorylation of RsbV by RsbU. RsbU is activated by interacting with RsbT Ser/Thr kinase that acts as a mediator and conveys environmental stress signals to initiate the downstream signaling. In the absence of stress, RsbT is entrapped in a 25S multiprotein oligomeric complex consisting of RsbS (antagonist) and RsbRAs (coantagonist) and its paralogues such as YkoB (RsbRB), YojH (RsbRC), YqhA (RsbRD), and YtvA (Akbar et al. 2001, Chen et al. 2003, Kim et al. 2004a). Under stress conditions, RsbT phosphorylates RsbRA, which facilitates phosphorylation of RsbS. Then RsbT is released from the complex, thus activating RsbU (Chen et al. 2004, Kim et al. 2004b). Activated RsbU dephosphorylates RsbV, which then binds to the RsbW kinase (RsbV–RsbW), thereby dissociating the RsbW–SigB complex. The SigB released from the complex binds to RNA polymerase, thus activating the SigB regulon (Dufour and Haldenwang 1994). This signaling pathway is reset by RsbX STP that dephosphorylates RsbS and RsbRA, and serves as a fine-tuning mechanism resulting in the sequestration of RsbT in the stressosome complex (Yang et al. 1996, Price et al. 2001, Chen et al. 2004). Furthermore, the absence of the RsbX protein in the B. subtilis system results in uncontrollable activation of the SigB regulon (Voelker et al. 1997) (Fig. 6). Under energy stress conditions (glucose, oxygen, and phosphate starvation), RsbV dephosphorylation is mediated by a complex of RsbP phosphatase and RsbQ hydrolase, thus activating the SigB regulon (Vijay et al. 2000, Brody et al. 2001). In the case of cold shock, the signaling pathway works independently of RsbP, RsbU, and RsbV. The degree of stability of the RsbW–SigB complex regulates the silencing or transcription of stress-related genes (Brigulla et al. 2003) (Fig. 6).
In B. cereus, SigB activation is achieved by a protein complex comprising RsbK (sensor kinase), RsbW (kinase), RsbV (antagonist), and RsbY (phosphatase). Following stress conditions, the sensor kinase RsbK phosphorylates and activates RsbY, which in turn dephosphorylates the RsbV antagonist. Unphosphorylated RsbV sequesters RsbW (a negative regulator of SigB), thus activating the SigB regulon (van Schaik et al. 2005, de Been et al. 2010).
Virulence
While the majority of Bacillus species are nonpathogenic, a few of them belonging to the B. cereus group are known to cause disease in animals and humans (Ehling-Schulz et al. 2019). The pathogenicity of this group is attributed to two major factors-secreted toxins or capsule. These factors are paramount for the survival of the bacteria inside the host, and thus it is important to study the network that regulates their synthesis for the development of novel prophylactics (Moayeri et al. 2015). In B. anthracis, the most studied protein, which is involved in the synthesis of toxins and capsule is the master virulence regulator, AtxA (Anthrax toxin activator) (Fouet 2010, McCall et al. 2019). The B. anthracis null mutant of atxA is avirulent in the mouse model of infection (Dai et al. 1995). Furthermore, the cellular level of AtxA decides the fate of associated pathways and thereby modulates the pathogenic cycle. High levels of AtxA in the cell are conducive to toxin gene expression while inhibiting sporulation in B. anthracis (Dale et al. 2018). The global transcriptional regulators CodY and AbrB regulate the synthesis of AtxA (Strauch et al. 2005, van Schaik et al. 2009, Gangwal et al. 2022), and are themselves targeted by Ser/Thr phosphorylation machinery (Macek et al. 2007, Kobir et al. 2014). This regulation is achieved either by direct binding of the transcriptional regulator to the promoter region of the toxin-producing genes (Dale et al. 2012, Chateau et al. 2013) or by an unknown post-translational mechanism (van Schaik et al. 2009).
In the nonpathogenic B. subtilis, phosphorylation of AbrB at a Ser residue inhibits its DNA binding property (Kobir et al. 2014), whereas in B. anthracis, CodY phosphorylation at a Ser residue abrogates its DNA binding to the atxA promoter, leading to depletion of toxin proteins (Gangwal et al. 2022). In B. anthracis, PTS-mediated phosphorylation of AtxA at conserved His residues in the PRD domain (PTS-regulated domain) regulates its activity and affects toxin synthesis (Tsvetanova et al. 2007, Raynor et al. 2018). Interestingly, PTS proteins HPr and Enzyme I were also shown to be required for atxA expression and their deletion mutant B. anthracis strain showed defective toxin production and attenuated virulence in an animal model of anthrax (Bier et al. 2020). More recently, another STP—PrpN was shown to regulate AtxA synthesis via phosphorylation-mediated regulation of CodY DNA binding activity. CodY was confirmed as a target of the PrkC and PrpN and the null mutant strain of prpN demonstrated a significant reduction in anthrax toxins and AtxA. Functional implication of PrpN-mediated dephosphorylation on CodY activity was examined using phosphomimetic and phosphoablative CodY mutants. While phosphorylation of CodY at a Ser residue resulted in complete abolishment of its DNA-binding ability at the atxA promoter region, unphosphorylated CodY was reported as a positive regulator of anthrax toxins (Gangwal et al. 2022). This highlights the importance of protein Ser/Thr and His phosphorylation in maintaining the optimum AtxA level for toxin production (Fig. 7).
Figure 7.
Role of protein kinases in Bacillus virulence. The pathways associated with virulence are activated in pathogenic species such as B. anthracis. These pathways follow specific signals during the infection stage within the host, which activate a unique set of signaling events emanating usually from surface-bound kinases and phosphatases. The schematic shows these specific proteins and their interacting partners, which carry out the indicated functions. Black solid arrows represent kinase-mediated substrate phosphorylation, red dotted arrows represent activation/upregulation, and black lines with terminal bars represent inhibition/downregulation.
Toxin biosynthesis is often correlated with carbon sources in pathogenic bacteria (Poncet et al. 2009, Chiang et al. 2011). In Bacillus sp., this is mediated by a carbon catabolite protein (CcpA) and phosphotransferase system (PTS) component, HPr, via the classical carbon catabolite repression (CCR) mechanism (Khan and Banerjee-Bhatnagar 2002, Lorca et al. 2005, Singh et al. 2008). Hpr is phosphorylated by Hpr Kinase (Poncet et al. 2004, Choi et al. 2006) and PrkC (Macek et al. 2007, Pietack et al. 2010). While the functional relevance of phosphorylation by PrkC is still unknown, the phosphorylation by Hpr kinase plays an important role in modulating Hpr activity and binding to CcpA, resulting in differential expression of multiple genes in B. subtilis (Blencke et al. 2003, Deutscher et al. 2014) (Fig. 7).
Chain formation is another factor that contributes to the virulence of bacteria including B. anthracis by helping the pathogen to evade the host immune response (Moller et al. 2012, Rodriguez et al. 2012, Prashar et al. 2013, Jouvion et al. 2016). In B. anthracis, prkC deletion results in severe defects in chain formation as compared to wild type strain, indicating the role of PrkC in the regulation of chain length in B. anthracis (Dhasmana et al. 2021). Earlier studies have also shown that B. anthracis lacking the prkC/prpC pair showed impaired survival within macrophages and attenuated virulence in an animal model (Shakir et al. 2010). These studies provide a possible regulatory link between Ser/Thr phosphorylation and virulence via chain formation in B. anthracis.
When B. anthracis infects its host, it secretes lethal and edema toxins, which target smooth muscle cells and epithelial cells, exacerbating the infection. The BrrAB TCS regulates the expression of these toxins during infection. Deletion of any one of the components of BrrAB causes downregulation of atxA and a significant reduction in the toxin-producing ability of the pathogen, thus decreasing the virulence (Vetter and Schlievert 2007). In addition to exotoxins, secreted proteases facilitate the infection via degrading the host tissue and mobilizing the bacteria inside the host (Chung et al. 2006). A novel TCS encoded by BAS2109–2108 regulates the expression of eight intracellular and extracellular proteases (Gupta et al. 2017). Activated BAS2108 (HK) transfers the phosphate group to its cognate RR, BAS2109, which binds to the promoters of many proteases, such as metalloendopeptidase, bacillolysin, and several Ser proteases; few of these proteases are involved in pathogenesis. Additionally, the expression of BAS2109 (RR) increases in the presence of carbon dioxide or nutritional stress (Gupta et al. 2017) (Fig. 7).
The food-borne pathogen B. cereus has a TCS, ResDE, which controls the production of hemolysin BL and nonhemolytic enterotoxins under anaerobic conditions, thus controlling levels of virulence (Duport et al. 2006). ResE responds to the acidic environment and oxygen limitation in the gut and helps bacteria to secrete extracellular toxins to establish infection (Duport et al. 2006). YvfTU TCS controls the expression of a major pleiotropic virulence gene regulator, PlcR in B. cereus (Gohar et al. 2008, Brillard et al. 2008). Although plcR expression is decreased in yvfTU mutant bacilli, there are only minor differences reported in the expression of the plcR regulon (Brillard et al. 2008).
Cross-talk and cross-phosphorylation
A recent surge in the number of protein–protein interactome and phosphoproteome studies of bacterial systems including that of Bacillus have revealed multisite phosphorylation of several proteins by different kinases. These kinases exhibit promiscuous activity and can phosphorylate each other as well as have common substrates, thereby contributing to the biological complexity, adding another regulatory layer to the protein phosphorylation network in the bacterial system (Lin et al. 2009, Chao et al. 2010, Kobir et al. 2011, Baer et al. 2014, Shi et al. 2014a, b, Willett and Crosson 2017, Pinas et al. 2018, Shen et al. 2020). To differentiate these two regulatory networks, we have used the term “cross-talk” to define phosphorylation and dephosphorylation of a substrate at multiple amino acid residues by different kinases/phosphatases and “cross-phosphorylation” to define phosphorylation/dephosphorylation between different kinases/phosphatases (Fig. 8).
Figure 8.
Cross-talk and cross-phosphorylation in Bacillus. (A) Cross-talk network representing multisite phosphorylation of proteins by different classes of kinases and phosphatases in Bacillus. A protein–protein interaction network was created using Cytoscape 3.8.2 software. Interaction among different kinases/phosphatases and their substrates are represented with colored nodes and colored lines. STK/STP: Ser/Thr kinase and phosphatase, BY-K/PTP: Bacillus Tyr kinase and phosphatase, and Hpr K: Hpr kinase. Among the kinases depicted in the figure, only PrkC and PrpC have validated substrates in both B. anthracis and B. subtilis (labeled BA in parentheses). (B) The cross-phosphorylation network between kinases is shown by a protein–protein interaction network. Different classes of kinases and phosphatases are represented by colored nodes, and interaction among different kinases/phosphatases are determined by colored arrows and lines. Blue arrows represent validated in-vitro assays, pink lines indicate phosphoproteome validations, and orange lines denote yeast two-hybrid interactions between bait and prey. Arrows point to the target proteins that are phosphorylated.
In B. subtilis, an interaction network linking different classes of protein kinases/phosphatases was generated using yeast two-hybrid interactome studies and in-vitro kinase assays (Shi et al. 2014a, b). A direct interaction was predicted between Ser/Thr kinase-YabT and BY-kinase/PTP PtkB and cognate Phosphatase PtpZ. An indirect interaction was also observed via TkmA (the specific modulator for BY-kinase PtkA) between YabT and another BY-kinase, PtkB. The two BY-kinases, PtkA and PtkB show possible cross-interaction through the PtkA-modulator TkmA. Also, the Tyr phosphatase PtpZ interacts with TkmA and PtkA. RsbT, a two-component-like Ser/Thr kinase regulating the stressosome complex of B. subtilis interacts with BY-kinase/phosphatase PtkA/PtpZ and Ser/Thr kinase PrkD. DegS, an HK, was shown to be phosphorylated by Ser/Thr kinases including interacts with the PrkD and YabT/SpoIIE pair (Jers et al. 2011, Shi et al. 2014a, b).
Furthermore, cross phosphorylation among the kinases has been defined by categorizing the kinases into two broad classes. The first class consists of PrkD, PrkC, YabT, and PtkA kinases that can autophosphorylate and phosphorylate other kinases. Among these, PrkC, PrkD, and YabT show intermolecular phosphorylation, while YabT and PtkA can cross-phosphorylate each other. Also, PrkC can phosphorylate PtkA near its autophosphorylating site. The other class includes PtkA and two-component Ser/Thr kinases such as RsbW and SpoIIAB, which are phosphorylated by other Ser/Thr kinases. Additionally, HPr kinase/phosphorylase (PTS component) is phosphorylated by PrkD, PrkC, and PtkA, while RsbT interacts with PrkD, and YabT (Shi et al. 2014a). Another example of cross-phosphorylation involves phosphorylation of TCS proteins DegS (HK) and WalR (RR) by STKs. Phosphorylation of DegS by STKs (PrkD and YabT) at the Ser76 residue enhances the phosphotransfer to DegU (RR) and was found to be important in the regulation of competence development, complex colony formation, and swarming (Macek et al. 2007, Jers et al. 2011). Furthermore, PrkC-mediated phosphorylation of WalR (RR) at Thr101 regulates the expression of genes involved in cell wall metabolism (Libby et al. 2015). These studies suggest different kinase classes work in tandem in a network and illustrate the biological complexity of cross-phosphorylation in Bacillus genus (Fig. 8).
Interestingly, in a high-throughput experiment, multisite phosphorylation of substrates was predicted with different classes of kinases/phosphatases. For example, RecA is phosphorylated by YabT, PtkA and Arg kinase (Soufi et al. 2010, Elsholz et al. 2012, Bidnenko et al. 2013, Shi et al. 2014b). Phosphorylation of SalA by PtkA was also validated by an in-vitro kinase assay (Derouiche et al. 2015). Another phosphoproteome study reported phosphorylation of known substrates of Tyr kinase PtkA such as aspartate semialdehyde dehydrogenase (Asd), Enolase (Eno), and YjoA at Ser/Thr residues (Jers et al. 2010, Ravikumar et al. 2014). Out of these, Eno was identified as a substrate of the McsB/YwlE pair in an Arg phosphoproteome analysis (Schmidt et al. 2014). In B. anthracis Eno was also shown to be phosphorylated at Ser/Thr residues by PrkC (Virmani et al. 2019). This PrkC-mediated multisite phosphorylation of Eno imprints phenotypic memory in B. anthracis spores and is important for spore germination. The global transcriptional regulator AbrB is also phosphorylated at Ser and Tyr residues. Phosphorylation at the Ser residue is mediated by STKs (PrkC/PrkD/YabT), while Tyr phosphorylation was detected in the B. subtilis phosphoproteome (Kobir et al. 2014, Ravikumar et al. 2014).
Cross-talk among B. subtilis phosphorylation pathways involve Ssb (SsbA and SsbB) required for DNA replication that are phosphorylated by PtkA and McsB (Mijakovic et al. 2006, Elsholz et al. 2012). Interestingly, SsbA is also phosphorylated by Ser/Thr kinases (PrkD, PrkC, and most efficiently by YabT), which aids in the cooperative binding of SsbA tetramers to DNA (Derouiche et al. 2016). Another example of cross-talk involves phosphorylation of a PTS protein, Hpr, at multiple residues. Phosphorylation at Ser residues is mediated by either a bifunctional ATP dependent Hpr kinase/phosphorylase (Reizer 1989) or Ser/Thr kinase/phosphatase pair PrkC/PrpC (Singh et al. 2007, Pietack et al. 2010), while phosphorylation at His is catalyzed by another PTS protein, Enzyme I (Reizer 1989). Phosphorylation of Hpr at the Ser/Thr/Tyr residues was also observed in phosphoproteome studies of B. subtilis (Ravikumar et al. 2014, 2018) (Fig. 8).
Arg phosphoproteome analysis identified several substrates such as GroEL, DegU, and the elongation factors EF-Tu and EF-G in B. subtilis that were previously shown to be phosphorylated at other residues (described in above sections), indicating multisite phosphorylation and possible cross-talk among the different families of kinases (Elsholz et al. 2012, Schmidt et al. 2014, Trentini et al. 2014). EF-Tu and EF-G were also identified as targets of Tyr phosphorylation in the B. subtilis germinating spore phosphoproteome (Rosenberg et al. 2015).
Compared to B. subtilis, B. anthracis has lost some Tyr kinases during evolution. Consequently, two unique DSPKs, PrkD, and PrkG, were identified in B. anthracis, belonging to different classes defined on the basis of their target residues. PrkD belongs to “dual-specificity tyrosine phosphorylation-regulated kinase” (DYRK), which autophosphorylates on Ser, Thr, and Tyr but phosphorylates its substrates only on Ser and Thr. PrkG was identified as a bonafide DSPK that can autophosphorylate and mediate substrate phosphorylation on Ser/Thr/Tyr residues. Interestingly, the STP PrpC also exhibits dual specificity (Arora et al. 2012). It can dephosphorylate both PrkD and PrkG, in addition to PrkC (Obuchowski et al. 2000, Shakir et al. 2010).
Conclusions and future directions
In this review, we assessed the role of protein phosphorylation in different life stages of Bacillus species (Table 1). Bacillus sp. have the ability to metamorphosize into different forms such as spores, vegetative cells, or biofilms, which is vital for their survival in the environment or in the host. The shift in metabolic needs between these stages is a complex process (Fouet and Mock 2006, Tan and Ramamurthi 2014, Christie and Setlow 2020, Setlow and Christie 2020). Besides basic control of gene expression, PTMs such as phosphorylation, provide an additional layer of regulation. Over the years, the importance of His/Asp and Ser/Thr/Tyr protein phosphorylation has been unraveled in different cellular pathways. More recently, phosphorylation of other residues including Lys, Arg, and Cys have also been discovered. Genetic regulation extends beyond the identification of direct signaling pathways, as many of these phosphorylation events are controlled by more than one phosphorylation scheme, as discussed in the cross-talk section.
Most of the early research on the role of protein phosphorylation was performed using the model organism B. subtilis (Cozzone 1988, Perego et al. 1989, Mukai et al. 1990). Over the years, it has been established that protein phosphorylation schemes are ubiquitous throughout the Bacillus species, though some members of the Cereus clade have lost some Tyr kinases and gained dual-specificity protein kinases that can perform the function of both Ser/Thr and Tyr protein phosphorylation (Arora et al. 2012).
Of the different phosphorylation systems, early discoveries emphasized the role of His/Asp-based TCSs in sensing environmental stress and regulating cellular behavior, while recent advances in this field indicate that Ser/Thr/Tyr/Arg protein phosphorylation may play an even larger role. Though many phosphorylated proteins have been recognized in recent literature, our understanding of upstream sensing stimuli is still limited. Among the Ser/Thr protein kinases, the role of muropeptides as sensory signals to PrkC is well-established (Dworkin and Shah 2010, Squeglia et al. 2011, Pompeo et al. 2018b). Recent studies on the PrkC homolog PknB in Mycobacterium tuberculosis indicate that lipid moieties can also be sensory signals (Kaur et al. 2019). Further work is needed to understand if in addition to muropeptides, lipid II, can also act as sensing signals to PrkC. These studies show that protein kinase signaling in Bacillus species is highly evolved and regulated by specific sensory signals.
Recent advancement in the MS techniques have facilitated increase in phosphoproteome studies. They range from nonspecific simple gel-based studies to more specific and complex MS studies. Improved phosphopeptide enrichment methods coupled with novel MS-based systemic approaches provides a better blueprint of the global phosphoproteome (Liu et al. 2021, Paulo and Schweppe 2021, Pina et al. 2021, Xiao et al. 2023). An in-depth description of these techniques is beyond the scope of this review but has been reported elsewhere (Engholm-Keller and Larsen 2013, Ed Dudley 2014, Yague et al. 2020). Importantly, phosphorylation at specific residues in a protein differs in lability, stoichiometry, and abundance. For instance, compared to phosphorylation at other residues, phospho-Asp/His and Arg are relatively unstable (Gonzalez-Sanchez et al. 2013, Huang et al. 2021). For this reason, enrichment and phosphoproteome studies on Asp/His/Arg phosphorylation are challenging and extensive research is needed in this field.
Unlike protein kinases, there is limited literature on the role of cognate protein phosphatases in cellular processes (Kennelly 2002, Sajid et al. 2015). Also, many regulatory mechanisms of phosphatases have been reported. For example, in M. tuberculosis, in a feedback loop system, PstP (homolog of B. anthracis PrpC) is phosphorylated by its cognate kinases PknA and PknB, causing its activation, though a similar mode of regulation has not been reported in Bacillus (Sajid et al. 2011b). The phosphatases of stressosome complexes are also shown to be regulated by a similar mechanism (Misra et al. 2019). The existing literature also suggests that similar to eukaryotes, a cross-talk and cross-phosphorylation network is also present in the bacterial systems and might provide another layer of regulation for phosphorylation pathways, thus enabling the efficient regulation of principal processes such as growth, DNA repair, cellular metabolism, and stress response.
In conclusion, phosphorylation is the key regulator of every aspect of Bacillus life, yet information about what triggers these events and how they maintain control over different pathways and cross-talk with each other is somewhat limited. Having several conserved processes, the Bacillus species are one of the best prokaryotic model systems utilized to study various aspects of bacterial physiology (Getz et al. 2019). Continued studies in this field can help to unravel more bacterial signaling systems and networks.
Acknowledgments
We thank George Leiman (National Institutes of Health) for reading and editing the manuscript. Support of SERB grant number CRG/2018/000847/HS; J.C. Bose fellowship and grant from Institutes of Eminence (IoE, UGC) to Y.S. are acknowledged. Figures were prepared using Biorender and Cytoscape.
Contributor Information
Aakriti Gangwal, Department of Zoology, University of Delhi, Faculty of Science, Delhi- 110007, India.
Nishant Kumar, Department of Zoology, University of Delhi, Faculty of Science, Delhi- 110007, India.
Nitika Sangwan, Department of Zoology, University of Delhi, Faculty of Science, Delhi- 110007, India; Department of Biomedical Science, Bhaskaracharya College of Applied Sciences, University of Delhi, New Delhi-110075, India.
Neha Dhasmana, School of Medicine, New York University, 550 First Avenue New York-10016, New York, United States.
Uma Dhawan, Department of Biomedical Science, Bhaskaracharya College of Applied Sciences, University of Delhi, New Delhi-110075, India.
Andaleeb Sajid, 300 Cedar St, Yale School of Medicine, Yale University, New Haven, Connecticut 06520, New Haven CT, United States.
Gunjan Arora, 300 Cedar St, Yale School of Medicine, Yale University, New Haven, Connecticut 06520, New Haven CT, United States.
Yogendra Singh, Department of Zoology, University of Delhi, Faculty of Science, Delhi- 110007, India; Delhi School of Public Health, Institution of Eminence, University of Delhi, Delhi-110007, India.
Conflict of interest
None declared
References
- Abriata LA, Albanesi D, Dal Peraro Met al. Signal sensing and transduction by histidine kinases as unveiled through studies on a temperature sensor. Acc Chem Res. 2017;50:1359–66. [DOI] [PubMed] [Google Scholar]
- Absalon C, Hamze K, Blanot Det al. The GTPase CpgA is implicated in the deposition of the peptidoglycan sacculus in Bacillus subtilis. J Bacteriol. 2008;190:3786–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absalon C, Obuchowski M, Madec Eet al. CpgA, EF-Tu and the stressosome protein YezB are substrates of the ser/thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology. 2009;155:932–43. [DOI] [PubMed] [Google Scholar]
- Aggarwal S, Somani VK, Gupta Vet al. Functional characterization of PhoPR two component system and its implication in regulating phosphate homeostasis in Bacillus anthracis. Biochim Biophys Gen Sub. 2017;1861:2956–70. [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Quart Rev Biophys. 2009;42:159–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar C, Vlamakis H, Guzman Aet al. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio. 2010;1. 10.1128/mbio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar S, Gaidenko TA, Kang CMet al. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J Bacteriol. 2001;183:1329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi D, Mansilla María, De Mendoza D. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol. 2004;186:2655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper S, Dufour A, Garsin DAet al. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–77. [DOI] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa Tet al. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA. 2009;106:1239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles DM, Scheffers D-J. The cell wall of Bacillus subtilis. Curr Iss Mol Biol. 2021;41:539–96. [DOI] [PubMed] [Google Scholar]
- Arnaouteli S, Bamford NC, Stanley-Wall NRet al. Bacillus subtilis biofilm formation and social interactions. Nat Rev Micro. 2021;19:600–14. [DOI] [PubMed] [Google Scholar]
- Arora G, Bothra A, Prosser Get al. Role of post-translational modifications in the acquisition of drug resistance in Mycobacterium tuberculosis. FEBS J. 2021;288:3375–93. [DOI] [PubMed] [Google Scholar]
- Arora G, Misra R, Sajid A. Model systems for pulmonary infectious diseases: paradigms of anthrax and tuberculosis. CTMC. 2017a;17:2077–99. [DOI] [PubMed] [Google Scholar]
- Arora G, Sajid A, Arulanandh MDet al. Unveiling the novel dual specificity protein kinases in Bacillus anthracis: identification of the first prokaryotic dual specificity tyrosine phosphorylation-regulated kinase (DYRK)-like kinase. J Biol Chem. 2012;287:26749–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G, Sajid A, Arulanandh MDet al. Zinc regulates the activity of kinase-phosphatase pair (BasPrkC/BasPrpC) in Bacillus anthracis. Biometals. 2013;26:715–30. [DOI] [PubMed] [Google Scholar]
- Arora G, Sajid A, Gupta Met al. Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS ONE. 2010;5:e10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G, Sajid A, Singhal Aet al. Identification of ser/thr kinase and forkhead associated domains in Mycobacterium ulcerans: characterization of novel association between protein kinase Q and MupFHA. PLoS Negl Trop Dis. 2014;8:e3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora G, Sajid A, Virmani Ret al. Ser/thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. npj Biofilms Microbiomes. 2017b;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash C, Farrow JAE, Wallbanks Set al. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative-analysis of small-subunit-ribosomal RNA sequences. Lett Appl Microbiol. 1991;13:202–6. [Google Scholar]
- Baer CE, Iavarone AT, Alber Tet al. Biochemical and spatial coincidence in the provisional ser/thr protein kinase interaction network of Mycobacterium tuberculosis. J Biol Chem. 2014;289:20422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Bedrunka P. Physiology of guanosine-based second messenger signaling in Bacillus subtilis. Biol Chem. 2020;401:1307–22. [DOI] [PubMed] [Google Scholar]
- Banse AV, Hobbs EC, Losick R. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein med in Bacillus subtilis. J Bacteriol. 2011;193:3949–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri G, Albertini AM, Ferrari Eet al. Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J Bacteriol. 2016;198:907–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baros SS, Blackburn JM, Soares NC. Phosphoproteomic approaches to discover novel substrates of mycobacterial Ser/Thr protein kinases. Mol Cell Proteomics. 2020;19:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt JR, Lker UV, Lker AVet al. Specific and general stress proteins in Bacillus subtilis–a two-deimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. [DOI] [PubMed] [Google Scholar]
- Bhandari V, Ahmod NZ, Shah HNet al. Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int J Syst Evol Microbiol. 2013;63:2712–26. [DOI] [PubMed] [Google Scholar]
- Bidnenko V, Shi L, Kobir Aet al. Bacillus subtilis serine/threonine protein kinase YabT is involved in spore development via phosphorylation of a bacterial recombinase. Mol Microbiol. 2013;88:921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier N, Hammerstrom TG, Koehler TM. Influence of the phosphoenolpyruvate:carbohydrate phosphotransferase system on toxin gene expression and virulence in Bacillus anthracis. Mol Microbiol. 2020;113:237–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11:359–66. [DOI] [PubMed] [Google Scholar]
- Birk MS, Charpentier E, Frese CK. Automated phosphopeptide enrichment for Gram-positive bacteria. J Proteome Res. 2021;20:4886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkey S, Zhang X, Duggan MFet al. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol Microbiol. 1998;30:943–53. [DOI] [PubMed] [Google Scholar]
- Blencke HM, Homuth G, Ludwig Het al. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 2003;5:133–49. [DOI] [PubMed] [Google Scholar]
- Bonne Køhler J, Jers C, Senissar Met al. Importance of protein Ser/Thr/Tyr phosphorylation for bacterial pathogenesis. FEBS Lett. 2020;594:2339–69. [DOI] [PubMed] [Google Scholar]
- Borriss R, Danchin A, Harwood CRet al. Bacillus subtilis, the model Gram-positive bacterium: 20 years of annotation refinement. Microb Biotechnol. 2018;11:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti A, Vazquez DB, Almada JCet al. A transmembrane histidine kinase functions as a pH sensor. Biomolecules. 2020;10:1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, Mccormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–27. [DOI] [PubMed] [Google Scholar]
- Bozue J, Moody KL, Cote CKet al. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect Immun. 2007;75:4498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressuire-Isoard C, Broussolle V, Carlin F. Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol Rev. 2018;42:614–26. [DOI] [PubMed] [Google Scholar]
- Brigulla M, Hoffmann T, Krisp Aet al. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J Bacteriol. 2003;185:4305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillard J, Susanna K, Michaud Cet al. The YvfTU two-component system is involved in plcR expression in Bacillus cereus. BMC Microbiol. 2008;8:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody MS, Vijay K, W. Price C. Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis. J Bacteriol. 2001;183:6422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsing RL, La Clair C, Tang Set al. Characterization of sporulation histidine kinases of Bacillus anthracis. J Bacteriol. 2005;187:6972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–52. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R. The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev. 2006;70:888–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan L, Ahn J-S, Weaver KEet al. An overlap between the control of programmed cell death in Bacillus anthracis and sporulation. J Bacteriol. 2009;191:4103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JD, Papavinasasundaram KG, Zheng Xet al. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J Biol Chem. 2010;285:29239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JD, Wong D, Av-Gay Y. Microbial protein-tyrosine kinases. J Biol Chem. 2014;289:9463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateau A, Van der Verren SE, Remaut Het al. The Bacillus anthracis cell envelope: composition, physiological role, and clinical relevance. Microorganisms. 2020;8:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateau A, van Schaik W, Joseph Pet al. Identification of CodY targets in Bacillus anthracis by genome-wide in vitro binding analysis. J Bacteriol. 2013;195:1204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Kota S, Misra HS. DivIVA phosphorylation affects its dynamics and cell cycle in radioresistant Deinococcus radiodurans. Microbiol Spectr. 2023;11:e0314122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Lewis RJ, Harris Ret al. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol Microbiol. 2003;49:1657–69. [DOI] [PubMed] [Google Scholar]
- Chen CC, Yudkin MD, Delumeau O. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J Bacteriol. 2004;186:6830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao S, Chai Yet al. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol. 2012;85:418–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Srivastava P, Zarazua-Osorio Bet al. Bacillus subtilis histidine kinase KinC activates biofilm formation by controlling heterogeneity of single-cell responses. mBio. 2022;13:e0169421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng-Guang H, Gualerzi CO. The ribosome as a switchboard for bacterial stress response. Front Microbiol. 2020;11:619038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Bongiorni C, Perego M. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J Bacteriol. 2011;193:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ID, Kim KN, Yun CWet al. Cloning and characterization of the hpr kinase/phosphorylase gene from Bacillus stearothermophilus No. 236. Biosci Biotechnol Biochem. 2006;70:1089–101. [DOI] [PubMed] [Google Scholar]
- Christie G, Setlow P. Bacillus spore germination: knowns, unknowns and what we need to learn. Cell Signal. 2020;74:109729. [DOI] [PubMed] [Google Scholar]
- Chung MC, Popova TG, Millis BAet al. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J Biol Chem. 2006;281:31408–18. [DOI] [PubMed] [Google Scholar]
- Cladiere L, Hamze K, Madec Eet al. The GTPase, CpgA(YloQ), a putative translation factor, is implicated in morphogenesis in Bacillus subtilis. Mol Genet Genomics. 2006;275:409–20. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Campbell ID, Yudkin MD. Efficient regulation of sigmaF, the first sporulation-specific sigma factor in B. subtilis. J Mol Biol. 2004;342:1187–95. [DOI] [PubMed] [Google Scholar]
- Cozzone AJ. Protein phosphorylation in prokaryotes. Annu Rev Microbiol. 1988;42:97–125. [DOI] [PubMed] [Google Scholar]
- Cui W, Han L, Suo Fet al. Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J Microbiol Biotechnol. 2018;34:145. [DOI] [PubMed] [Google Scholar]
- Cybulski LE, del Solar G, Craig POet al. Bacillus subtilis DesR functions as a phosphorylation-activated switch to control membrane lipid fluidity. J Biol Chem. 2004;279:39340–7. [DOI] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst Fet al. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14. [PubMed] [Google Scholar]
- Dai Z, Sirard JC, Mock Met al. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol. 1995;16:1171–81. [DOI] [PubMed] [Google Scholar]
- Dale JL, Raynor MJ, Dwivedi Pet al. cis-acting elements that control expression of the master virulence regulatory gene atxA in Bacillus anthracis. J Bacteriol. 2012;194:4069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JL, Raynor MJ, Ty MCet al. A dual role for the Bacillus anthracis master virulence regulator AtxA: control of sporulation and anthrax toxin production. Front Microbiol. 2018;9:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E, Noone D, Masson Aet al. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J Bacteriol. 2002;184:5661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been M, Tempelaars MH, van Schaik Wet al. A novel hybrid kinase is essential for regulating the sigma(B)-mediated stress response of Bacillus cereus. Environ Microbiol. 2010;12:730–45. [DOI] [PubMed] [Google Scholar]
- Derouiche A, Bidnenko V, Grenha Ret al. Interaction of bacterial fatty-acid-displaced regulators with DNA is interrupted by tyrosine phosphorylation in the helix-turn-helix domain. Nucleic Acids Res. 2013;41:9371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A, Petranovic D, Macek Bet al. Bacillus subtilis single-stranded DNA-binding protein SsbA is phosphorylated at threonine 38 by the serine/threonine kinase YabT. Periodicum Biologorum. 2016;118:399–404. [Google Scholar]
- Derouiche A, Shi L, Bidnenko Vet al. Bacillus subtilis SalA is a phosphorylation-dependent transcription regulator that represses scoC and activates the production of the exoprotease AprE. Mol Microbiol. 2015;97:1195–208. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Ake FM, Derkaoui Met al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SN, Vishnoi M, Kiehler Bet al. In vivo functional characterization of the transmembrane histidine kinase KinC in Bacillus subtilis. Microbiology. 2015;161:1092–104. [DOI] [PubMed] [Google Scholar]
- Dhasmana N, Kumar N, Gangwal Aet al. Bacillus anthracis chain length, a virulence determinant, is regulated by membrane localized serine/threonine protein kinase PrkC. J Bacteriol. 2021;203:e00582–00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman A, Bhatnagar S, Kulshreshtha Pet al. Functional characterization of WalRK: a two-component signal transduction system from Bacillus anthracis. FEBS Open Bio. 2014;4:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman A, Gopalani M, Bhatnagar R. WalRK two component system of Bacillus anthracis responds to temperature and antibiotic stress. Biochem Biophys Res Commun. 2015;459:623–8. [DOI] [PubMed] [Google Scholar]
- Di Gregorio Barletta G, Vittoria M, Lanzilli Met al. CotG controls spore surface formation in response to the temperature of growth in Bacillus subtilis. Environ Microbiol. 2022;24:2078–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintner S, Heermann R, Fang Cet al. A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J Biol Chem. 2014;289:27899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diomande SE, Chamot S, Antolinos Vet al. The CasKR two-component system is required for the growth of mesophilic and psychrotolerant Bacillus cereus strains at low temperatures. Appl Environ Microb. 2014;80:2493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diomande SE, Doublet B, Vasai Fet al. Expression of the genes encoding the CasK/R two-component system and the DesA desaturase during Bacillus cereus cold adaptation. FEMS Microbiol Lett. 2016;363:fnw174. [DOI] [PubMed] [Google Scholar]
- Diomande SE, Nguyen-the C, Abee Tet al. Involvement of the CasK/R two-component system in optimal unsaturation of the Bacillus cereus fatty acids during low-temperature growth. Int J Food Microbiol. 2015;213:110–7. [DOI] [PubMed] [Google Scholar]
- Dobihal GS, Brunet YR, Flores-Kim Jet al. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. eLife. 2019;8:e52088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobihal GS, Flores-Kim J, Roney IJet al. The WalR-WalK signaling pathway modulates the activities of both CwlO and LytE through control of the peptidoglycan deacetylase PdaC in Bacillus subtilis. J Bacteriol. 2022;204:e0053321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Cuevas P, Mercier R, Leaver Met al. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol Microbiol. 2012;83:52–66. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Hahn J, Roggiani Met al. Two-component regulators and genetic competence in Bacillus subtilis. Res Microbiol. 1994;145:403–11. [DOI] [PubMed] [Google Scholar]
- Dufour A, Haldenwang WG. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J Bacteriol. 1994;176:1813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Alper S, Arigoni Fet al. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–4. [DOI] [PubMed] [Google Scholar]
- Duncan L, Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc Natl Acad Sci USA. 1993;90:2325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duport C, Zigha A, Rosenfeld Eet al. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J Bacteriol. 2006;188:6640–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Micro. 2010;8:890–6. [DOI] [PubMed] [Google Scholar]
- Dworkin J. Ser/thr phosphorylation as a regulatory mechanism in bacteria. Curr Opin Microbiol. 2015;24:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ed Dudley AEB. Advances in Protein Chemistry and Structural Biology. Cambridge: Academic Press, 2014. [Google Scholar]
- Ehling-Schulz M, Lereclus D, Koehler TM. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr. 2019;7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhawy MI, Huc-Brandt S, Patzold Let al. The phosphoarginine phosphatase PtpB from Staphylococcus aureus is involved in bacterial stress adaptation during infection. Cells. 2021;10:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz AK, Hempel K, Michalik Set al. Activity control of the ClpC adaptor McsB in Bacillus subtilis. J Bacteriol. 2011;193:3887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz AK, Turgay K, Michalik Set al. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci USA. 2012;109:7451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engholm-Keller K, Larsen MR. Technologies and challenges in large-scale phosphoproteomics. Proteomics. 2013;13:910–31. [DOI] [PubMed] [Google Scholar]
- Enosi Tuipulotu D, Mathur A, Ngo Cet al. Bacillus cereus: epidemiology, virulence factors, and host-pathogen interactions. Trends Microbiol. 2021;29:458–71. [DOI] [PubMed] [Google Scholar]
- Errington J, Aart LTV. Microbe profile: Bacillus subtilis: model organism for cellular development, and industrial workhorse. Microbiology. 2020;166:425–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Wu LJ. Cell cycle machinery in Bacillus subtilis. Subcell Biochem. 2017;84:67–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Micro. 2003;1:117–26. [DOI] [PubMed] [Google Scholar]
- Eymann C, Becher D, Bernhardt Jet al. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics. 2007;7:3509–26. [DOI] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo-Cavazos P, Waters SM, Schuerger ACet al. Evolution of Bacillus subtilis to enhanced growth at low pressure: up-regulated transcription of des-desKR, encoding the fatty acid desaturase system. Astrobiology. 2012;12:258–70. [DOI] [PubMed] [Google Scholar]
- Fioravanti A, Mathelie-Guinlet M, Dufrene YFet al. The Bacillus anthracis S-layer is an exoskeleton-like structure that imparts mechanical and osmotic stabilization to the cell wall. PNAS Nexus. 2022;1:pgac121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C, Geourjon C, Bourson Cet al. Cloning and characterization of the Bacillus subtilis prkA gene encoding a novel serine protein kinase. Gene. 1996;168:55–60. [DOI] [PubMed] [Google Scholar]
- Fleurie A, Manuse S, Zhao Cet al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10:e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–57. [DOI] [PubMed] [Google Scholar]
- Fouet A, Mock M. Regulatory networks for virulence and persistence of Bacillus anthracis. Curr Opin Microbiol. 2006;9:160–6. [DOI] [PubMed] [Google Scholar]
- Fouet A. AtxA, a Bacillus anthracis global virulence regulator. Res Microbiol. 2010;161:735–42. [DOI] [PubMed] [Google Scholar]
- Foulquier E, Pompeo F, Bernadac Aet al. The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol Microbiol. 2011;80:309–18. [DOI] [PubMed] [Google Scholar]
- Foulquier E, Pompeo F, Freton Cet al. PrkC-mediated phosphorylation of overexpressed YvcK protein regulates PBP1 protein localization in Bacillus subtilis mreB mutant cells. J Biol Chem. 2014;289:23662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas C, Plannic J, Isticato Ret al. A protein phosphorylation module patterns the Bacillus subtilis spore outer coat. Mol Microbiol. 2020;114:934–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B, Zhu T, Domach MMet al. Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant. Appl Environ Microb. 2000;66:4045–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann J, Mierzwa B, Trentini DBet al. Structural basis for recognizing phosphoarginine and evolving residue-specific protein phosphatases in Gram-positive bacteria. Cell Rep. 2013;3:1832–9. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J, Schmidt A, Spiess Set al. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science. 2009;324:1323–7. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J, Subramanian V, Kojetin DJet al. Activity-based profiling reveals a regulatory link between oxidative stress and protein arginine phosphorylation. Cell Chem Biol. 2016;23:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 2003;17:1166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidenko TA, Kim TJ, Price CW. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J Bacteriol. 2002;184:6109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwal A, Sangwan N, Dhasmana Net al. Role of serine/threonine protein phosphatase PrpN in the life cycle of Bacillus anthracis. PLoS Pathog. 2022;18:e1010729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Garcia T, Ventroux M, Derouiche Aet al. Phosphorylation of the Bacillus subtilis replication controller YabA plays a role in regulation of sporulation and biofilm formation. Front Microbiol. 2018;9:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PS, Simorre JP, Brochier-Armanet Cet al. Cell division of Streptococcus pneumoniae: think positive!. Curr Opin Microbiol. 2016;34:18–23. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia T, Poncet S, Derouiche Aet al. Role of protein phosphorylation in the regulation of cell cycle and DNA-related processes in bacteria. Front Microbiol. 2016;7:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Zhu Y, Mullen Ket al. Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. J Bacteriol. 2007;189:1745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwig J, Kiley TB, Gunka Ket al. The protein tyrosine kinases EpsB and PtkA differentially affect biofilm formation in Bacillus subtilis. Microbiology. 2014;160:682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz LJ, Runte CS, Rainey JK&et al. Tyrosine phosphorylation as a widespread regulatory mechanism in prokaryotes. J Bacteriol. 2019;201:e00205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohar M, Faegri K, Perchat Set al. The PlcR virulence regulon of Bacillus cereus. PLoS ONE. 2008;3:e2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sanchez MB, Lanucara F, Helm Met al. Attempting to rewrite history: challenges with the analysis of histidine-phosphorylated peptides. Biochem Soc Trans. 2013;41:1089–95. [DOI] [PubMed] [Google Scholar]
- Gopalani M, Dhiman A, Rahi Aet al. Identification, functional characterization and regulon prediction of a novel two component system comprising BAS0540-BAS0541 of Bacillus anthracis. PLoS ONE. 2016;11:e0158895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B, Foulquier E, Galinier A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology. 2005;151:3777–91. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Chen AY, Gopalkrishnan Set al. Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol. 2018;72:163–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham-Smith GS. The longevity of dry spores of B. anthracis. J Hyg. 1930;30:213–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C, Nessler S, Mijakovic I. Bacterial tyrosine kinases: evolution, biological function and structural insights. Phil Trans R Soc B. 2012;367:2640–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA. Feedback control of two-component regulatory systems. Annu Rev Microbiol. 2016;70:103–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Chaudhary N, Aggarwal Set al. Functional analysis of BAS2108-2109 two component system: evidence for protease regulation in Bacillus anthracis. Int J Biochem Cell Biol. 2017;89:71–84. [DOI] [PubMed] [Google Scholar]
- Gupta V, Jain K, Garg Ret al. Characterization of a two component system, Bas1213-1214, important for oxidative stress in Bacillus anthracis. J Cell Biochem. 2018;119:5761–74. [DOI] [PubMed] [Google Scholar]
- Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37:849–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdusits B, Suskiewicz MJ, Hundt Net al. McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation. eLife. 2021;10:e63505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang WG, Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979;282:256–60. [DOI] [PubMed] [Google Scholar]
- Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–209. [DOI] [PubMed] [Google Scholar]
- Hardt P, Engels I, Rausch Met al. The cell wall precursor lipid II acts as a molecular signal for the ser/thr kinase PknB of Staphylococcus aureus. Int J Med Microbiol. 2017;307:1–10. [DOI] [PubMed] [Google Scholar]
- Harvey KL, Jarocki VM, Charles IGet al. The diverse functional roles of elongation factor tu (EF-Tu) in microbial pathogenesis. Front Microbiol. 2019;10:2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Mouillon JM, Pohl Set al. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol Rev. 2018;42:721–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Pane-Farre J, Völker U. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–36. [DOI] [PubMed] [Google Scholar]
- Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–36. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36:131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobley L, Harkins C, MacPhee CE&et al. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39:649–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA. A life in Bacillus subtilis signal transduction. Annu Rev Microbiol. 2017;71:1–19. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–70. [DOI] [PubMed] [Google Scholar]
- Hoiby N, Bjarnsholt T, Givskov Met al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–32. [DOI] [PubMed] [Google Scholar]
- Houry A, Briandet R, Aymerich Set al. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology. 2010;156:1009–18. [DOI] [PubMed] [Google Scholar]
- Howell A, Dubrac S, Andersen KKet al. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol. 2003;49:1639–55. [DOI] [PubMed] [Google Scholar]
- Huang B, Zhao Z, Zhao Yet al. Protein arginine phosphorylation in organisms. Int J Biol Macromol. 2021;171:414–22. [DOI] [PubMed] [Google Scholar]
- Huang Y, Flint SH&, Palmer JS. Bacillus cereus spores and toxins – the potential role of biofilms. Food Microbiol. 2020;90:103493. [DOI] [PubMed] [Google Scholar]
- Isticato R, Lanzilli M, Petrillo Cet al. Bacillus subtilis builds structurally and functionally different spores in response to the temperature of growth. Environ Microbiol. 2020;22:170–82. [DOI] [PubMed] [Google Scholar]
- Iwanicki A, Herman-Antosiewicz A, Pierechod Met al. PrpE, a PPP protein phosphatase from Bacillus subtilis with unusual substrate specificity. Biochem J. 2002;366:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen OM, Brautaset T, Degnes KFet al. Overexpression of wild-type aspartokinase increases L-lysine production in the thermotolerant methylotrophic bacterium Bacillus methanolicus. Appl Environ Microb. 2009;75:652–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jers C, Kobir A, Sondergaard EOet al. Bacillus subtilis two-component system sensory kinase DegS is regulated by serine phosphorylation in its input domain. PLoS ONE. 2011;6:e14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jers C, Pedersen MM, Paspaliari DKet al. Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol Microbiol. 2010;77:287–99. [DOI] [PubMed] [Google Scholar]
- Jezewska-Frackowiak J, Seroczynska K, Banaszczyk Jet al. The promises and risks of probiotic Bacillus species. Acta Biochim Pol. 2018;65:509–19. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego Met al. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–42. [DOI] [PubMed] [Google Scholar]
- Jiang M, Tzeng YL, Feher VAet al. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–95. [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. [DOI] [PubMed] [Google Scholar]
- Joon S, Gopalani M, Rahi Aet al. Biochemical characterization of the GTP-sensing protein, CodY of Bacillus anthracis. Pathog Dis. 2017;75:28472295. [DOI] [PubMed] [Google Scholar]
- Jordan S, Junker A, Helmann JDet al. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol. 2006;188:5153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Rietkotter E, Strauch MAet al. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology. 2007;153:2530–40. [DOI] [PubMed] [Google Scholar]
- Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL. A region of Bacillus subtilis CodY protein required for interaction with DNA. J Bacteriol. 2005;187:4127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvion G, Corre JP, Khun Het al. Physical sequestration of Bacillus anthracis in the pulmonary capillaries in terminal infection. J Infect Dis. 2016;214:281–7. [DOI] [PubMed] [Google Scholar]
- Kalantari A, Derouiche A, Shi Let al. Serine/threonine/tyrosine phosphorylation regulates DNA binding of bacterial transcriptional regulators. Microbiology. 2015;161:1720–9. [DOI] [PubMed] [Google Scholar]
- Karst JC, Foucher AE, Campbell TLet al. The atpase activity of an ‘essential’ Bacillus subtilis enzyme, YdiB, is required for its cellular function and is modulated by oligomerization. Microbiology. 2009;155:944–56. [DOI] [PubMed] [Google Scholar]
- Kaur P, Rausch M, Malakar Bet al. LipidII interaction with specific residues of Mycobacterium tuberculosis PknB extracytoplasmic domain governs its optimal activation. Nat Commun. 2019;10:1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–44. [DOI] [PubMed] [Google Scholar]
- Keijser BJ, Ter Beek A, Rauwerda Het al. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J Bacteriol. 2007;189:3624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett. 2002;206:1–8. [DOI] [PubMed] [Google Scholar]
- Kesel S, Mader A, Hofler Cet al. Immediate and heterogeneous response of the LiaFSR two-component system of Bacillus subtilis to the peptide antibiotic bacitracin. PLoS ONE. 2013;8:e53457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MZ, Kaur P, Nandicoori VK. Targeting the messengers: serine/threonine protein kinases as potential targets for antimycobacterial drug development. IUBMB Life. 2018;70:889–904. [DOI] [PubMed] [Google Scholar]
- Khan SR, Banerjee-Bhatnagar N. Loss of catabolite repression function of hpr, the phosphocarrier protein of the bacterial phosphotransferase system, affects expression of the cry4A toxin gene in Bacillus thuringiensis subsp. Israelensis. J Bacteriol. 2002;184:5410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K, Lopez-Garrido J, Pogliano K. Shaping an endospore: architectural transformations during Bacillus subtilis sporulation. Annu Rev Microbiol. 2020;74:361–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley TB, Stanley-Wall NR. Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation. Mol Microbiol. 2010;78:947–63. [DOI] [PubMed] [Google Scholar]
- Kim TJ, Gaidenko TA, Price CW. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J Mol Biol. 2004a;341:135–50. [DOI] [PubMed] [Google Scholar]
- Kim TJ, Gaidenko TA, Price CW. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J Bacteriol. 2004b;186:6124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Dougan DA, Gerth Uet al. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 2007;26:2061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Zuhlke D, Gerth Uet al. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 2005;24:3435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ikemoto Y. Biofilm-associated toxin and extracellular protease cooperatively suppress competitors in Bacillus subtilis biofilms. PLoS Genet. 2019;15:e1008232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. [DOI] [PubMed] [Google Scholar]
- Kobir A, Poncet S, Bidnenko Vet al. Phosphorylation of Bacillus subtilis gene regulator AbrB modulates its DNA-binding properties. Mol Microbiol. 2014;92:1129–41. [DOI] [PubMed] [Google Scholar]
- Kobir A, Shi L, Boskovic Aet al. Protein phosphorylation in bacterial signal transduction. Biochim Biophys Gen Sub. 2011;1810:989–94. [DOI] [PubMed] [Google Scholar]
- Koh A, Gibbon MJ, Van der Kamp MWet al. Conformation control of the histidine kinase BceS of Bacillus subtilis by its cognate ABC-transporter facilitates need-based activation of antibiotic resistance. Mol Microbiol. 2021;115:157–74. [DOI] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3:a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krab IM, Parmeggiani A. Mechanisms of EF-Tu, a pioneer GTPase. Prog Nucleic Acid Res Mol Biol. 2002;71:513–51. [DOI] [PubMed] [Google Scholar]
- Kroos L, Zhang B, Ichikawa Het al. Control of sigma factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–94. [DOI] [PubMed] [Google Scholar]
- Landmann JJ, Werner S, Hillen Wet al. Carbon source control of the phosphorylation state of the Bacillus subtilis carbon-flux regulator Crh in vivo. FEMS Microbiol Lett. 2012;327:47–53. [DOI] [PubMed] [Google Scholar]
- Laut CLPW, Metzger AL, Weiss Aet al. Bacillus anthracis responds to Targocil-induced envelope damage through EdsRS activation of Cardiolipin synthesis. mBio. 2020;11:e03375–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux JR, Grossman AD. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Kan CM, Lee JHet al. Phosphorylation-dependent interaction between a serine/threonine kinase PknA and a putative cell division protein Wag31 in Mycobacterium tuberculosis. New Microbiol. 2014;37:525–33. [PubMed] [Google Scholar]
- Levine A, Vannier F, Absalon Cet al. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics. 2006;6:2157–73. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen N, Wu Qet al. A flagella hook coding gene flgE positively affects biofilm formation and cereulide production in emetic Bacillus cereus. Front Microbiol. 2022;13:897836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaqat I, Mirza SA, Iqbal Ret al. Flagellar motility plays important role in biofilm formation of Bacillus cereus and Yersinia enterocolitica. Pak J Pharm Sci. 2018;31:2047–52. [PubMed] [Google Scholar]
- Libby EA, Goss LA, Dworkin J. The eukaryotic-like ser/thr kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis. PLoS Genet. 2015;11:e1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Reuveni S, Dworkin J. Multisite phosphorylation drives phenotypic variation in (p)ppGpp synthetase-dependent antibiotic tolerance. Nat Commun. 2019;10:5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilge L, Reder A, Tippmann Fet al. The involvement of the McsB arginine kinase in clp-dependent degradation of the MgsR regulator in Bacillus subtilis. Front Microbiol. 2020;11:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima A, Duran R, Schujman GEet al. Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J Proteomics. 2011;74:1720–34. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Walthers D, Connelly JEet al. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol. 2009;71:1477–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner C, Nijland R, van Hartskamp Met al. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J Bacteriol. 2004;186:1097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fields R, Schweppe DKet al. Strategies for mass spectrometry-based phosphoproteomics using isobaric tagging. Expert Rev Proteomics. 2021;18:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Han LL, Chen TYet al. Characterization of a protease hyper-productive mutant of Bacillus pumilus by comparative genomic and transcriptomic analysis. Curr Microbiol. 2020;77:3612–22. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu Fet al. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106:280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca GL, Chung YJ, Barabote RDet al. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, hpr, and HprK. J Bacteriol. 2005;187:7826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall RM, Sievers ME, Fattah Ret al. Bacillus anthracis virulence regulator AtxA binds specifically to the pagA promoter region. J Bacteriol. 2019;201:e00569–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson SA, Li M, Kearney JFet al. ExsB, an unusually highly phosphorylated protein required for the stable attachment of the exosporium of Bacillus anthracis. Mol Microbiol. 2010;76:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B, Forchhammer K, Hardouin Jet al. Protein post-translational modifications in bacteria. Nat Rev Micro. 2019;17:651–64. [DOI] [PubMed] [Google Scholar]
- Macek B, Mijakovic I, Olsen JVet al. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6:697–707. [DOI] [PubMed] [Google Scholar]
- Madec E, Laszkiewicz A, Iwanicki Aet al. Characterization of a membrane-linked ser/thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol Microbiol. 2002;46:571–86. [DOI] [PubMed] [Google Scholar]
- Maier B. Competence and transformation in Bacillus subtilis. Curr Iss Mol Biol. 2020;37:57–76. [DOI] [PubMed] [Google Scholar]
- Manuse S, Fleurie A, Zucchini Let al. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev. 2016;40:41–56. [DOI] [PubMed] [Google Scholar]
- Maracci C, Rodnina MV. Review: translational GTPases. Biopolymers. 2016;105:463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles-Wright J&, Lewis RJ. Stress responses of bacteria. Curr Opin Struct Biol. 2007;17:755–60. [DOI] [PubMed] [Google Scholar]
- Mascher T, Zimmer SL, Smith TAet al. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother. 2004;48:2888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AR, Arora A, Maiti Set al. Identification, characterization and activation mechanism of a tyrosine kinase of Bacillus anthracis. FEBS J. 2008;275:6237–47. [DOI] [PubMed] [Google Scholar]
- Maurya GK, Modi K, Banerjee Met al. Phosphorylation of FtsZ and FtsA by a DNA damage-responsive Ser/Thr protein kinase affects their functional interactions in Deinococcus radiodurans. mSphere. 2018;3:e00325–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielich-Suss B, Lopez D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol. 2015;17:555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Deutscher J. Protein-tyrosine phosphorylation in Bacillus subtilis: a 10-year retrospective. Front Microbiol. 2015;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Grangeasse C, Turgay K. Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol Rev. 2016;40:398–417. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, Musumeci L, Tautz Let al. In vitro characterization of the Bacillus subtilis protein tyrosine phosphatase YwqE. J Bacteriol. 2005;187:3384–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Petranovic D, Deutscher J. How tyrosine phosphorylation affects the UDP-glucose dehydrogenase activity of Bacillus subtilis YwqF. J Mol Microbiol Biotechnol. 2004;8:19–25. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, Petranovic D, Macek Bet al. Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res. 2006;34:1588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Poncet S, Boel Get al. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mike LA, Choby JE, Brinkman PRet al. Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathog. 2014;10:e1004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ML, Soufi B, Jers Cet al. NetPhosBac – a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics. 2009;9:116–25. [DOI] [PubMed] [Google Scholar]
- Million-Weaver S, Samadpour AN, Merrikh H. Replication restart after replication-transcription conflicts requires RecA in Bacillus subtilis. J Bacteriol. 2015;197:2374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KT, Hilditch CM, Diederich Bet al. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–42. [DOI] [PubMed] [Google Scholar]
- Misra R, Menon D, Arora Get al. Tuning the Mycobacterium tuberculosis alternative sigma factor SigF through the multidomain regulator Rv1364c and osmosensory kinase protein kinase D. J Bacteriol. 2019;201:e00725–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Leppla SH, Vrentas Cet al. Anthrax pathogenesis. Annu Rev Microbiol. 2015;69:185–208. [DOI] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen STet al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–701. [DOI] [PubMed] [Google Scholar]
- Moller J, Luehmann T, Hall H&et al. The race to the pole: how high-aspect ratio shape and heterogeneous environments limit phagocytosis of filamentous Escherichia coli bacteria by macrophages. Nano Lett. 2012;12:2901–5. [DOI] [PubMed] [Google Scholar]
- Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. [DOI] [PubMed] [Google Scholar]
- Mukai K, Kawata M, Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990;265:20000–6. [PubMed] [Google Scholar]
- Murray EJ, Kiley TB, Stanley-Wall NR. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology. 2009;155:1–8. [DOI] [PubMed] [Google Scholar]
- Musumeci L, Bongiorni C, Tautz Let al. Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis. J Bacteriol. 2005;187:4945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CL, Li FK, Koo BMet al. Identification of two phosphate starvation-induced wall teichoic acid hydrolases provides first insights into the degradative pathway of a key bacterial cell wall component. J Biol Chem. 2016;291:26066–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naclerio G, Baccigalupi L, Zilhao Ret al. Bacillus subtilis spore coat assembly requires cotH gene expression. J Bacteriol. 1996;178:4375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naerdal I, Netzer R, Ellingsen TEet al. Analysis and manipulation of aspartate pathway genes for L-lysine overproduction from methanol by Bacillus methanolicus. Appl Environ Microb. 2011;77:6020–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrstedt H, Schroder C, Meinhardt F. Evidence for two recA genes mediating DNA repair in Bacillus megaterium. Microbiology. 2005;151:775–87. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa Aet al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. [DOI] [PubMed] [Google Scholar]
- Nguyen HA, El Khoury T, Guiral Set al. Expanding the kinome world: a new protein kinase Family widely conserved in bacteria. J Mol Biol. 2017;429:3056–74. [DOI] [PubMed] [Google Scholar]
- Nguyen KB, Sreelatha A, Durrant ESet al. Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc Natl Acad Sci USA. 2016;113:E3482–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Munakata N, Horneck Get al. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL. Roles of Bacillus endospores in the environment. Cell Mol Life Sci. 2002;59:410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot-Gros MF, Velten M, Yoshimura Met al. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103:2368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noone D, Botella E, Butler Cet al. Signal perception by the secretion stress-responsive CssRS two-component system in Bacillus subtilis. J Bacteriol. 2012;194:1800–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell LM, Kelleher P, van Rijswijck IMHet al. Natural transformation in Gram-positive bacteria and its biotechnological relevance to lactic acid bacteria. Annu Rev Food Sci Technol. 2022;13:409–31. [DOI] [PubMed] [Google Scholar]
- Obuchowski M, Madec E, Delattre Det al. Characterization of PrpC from Bacillus subtilis, a member of the PPM phosphatase family. J Bacteriol. 2000;182:5634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Matsuzawa A, Yoshikawa Het al. Bacillus subtilis SalA (YbaL) negatively regulates expression of scoC, which encodes the repressor for the alkaline exoprotease gene, aprE. J Bacteriol. 2004;186:3056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M. Glucose-mediated protein arginine phosphorylation/dephosphorylation regulates ylxR encoding nucleoid-associated protein and cell growth in Bacillus subtilis. Front Microbiol. 2020;11:590828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Giyanto TK, Masuyama Wet al. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol. 2003;49:1135–44. [DOI] [PubMed] [Google Scholar]
- Ohlsen KL, Grimsley JK, Hoch JA. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha A, Anand M, Bhatt Aet al. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–73. [DOI] [PubMed] [Google Scholar]
- Pan Z, Wang B, Zhang Yet al. dbPSP: a curated database for protein phosphorylation sites in prokaryotes. Database. 2015;2015:bav031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC. LPSN – list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–9. [DOI] [PubMed] [Google Scholar]
- Paschalis V, Le Chatelier E, Green Met al. Interactions of the Bacillus subtilis DnaE polymerase with replisomal proteins modulate its activity and fidelity. Open Biol. 2017;7:170146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, Schweppe DK. Advances in quantitative high-throughput phosphoproteomics with sample multiplexing. Proteomics. 2021;21:2000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peake P, Winter N, Britton W. Phosphorylation of Mycobacterium leprae heat-shock 70 protein at threonine 175 alters its substrate binding characteristics. Biochim Biophy Acta Prot Struct Mol Enzymol. 1998;1387:387–94. [DOI] [PubMed] [Google Scholar]
- Peng Q, Wu J, Chen Xet al. Disruption of two-component system LytSR affects forespore engulfment in Bacillus thuringiensis. Front Cell Infect Microbiol. 2017;7:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchat S, Dubois T, Zouhir Set al. A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol Microbiol. 2011;82:619–33. [DOI] [PubMed] [Google Scholar]
- Perego M, Cole SP, Burbulys Det al. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Hanstein C, Welsh KMet al. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–55. [DOI] [PubMed] [Google Scholar]
- Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol. 2001;42:133–43. [DOI] [PubMed] [Google Scholar]
- Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SFF, Gonzalez RL, Dworkin Jet al. Protein synthesis during cellular quiescence is inhibited by phosphorylation of a translational elongation factor. Proc Natl Acad Sci USA. 2015;112:E3274–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D, Grangeasse C, Macek Bet al. Activation of Bacillus subtilis ugd by the BY-kinase PtkA proceeds via phosphorylation of its residue tyrosine 70. J Mol Microbiol Biotechnol. 2009;17:83–89. [DOI] [PubMed] [Google Scholar]
- Petranovic D, Michelsen O, Zahradka Ket al. Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol Microbiol. 2007;63:1797–805. [DOI] [PubMed] [Google Scholar]
- Pi H, Chu ML, Ivan SJet al. Directed evolution reveals the mechanism of HitRS signaling transduction in Bacillus anthracis. PLoS Pathog. 2020;16:e1009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietack N, Becher D, Schmidl SRet al. In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: prkC phosphorylates enzymes from different branches of basic metabolism. J Mol Microbiol Biotechnol. 2010;18:129–40. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–86. [DOI] [PubMed] [Google Scholar]
- Pina AS, Batalha IL, Dias Aet al. Affinity tags in protein purification and peptide enrichment: an overview. Methods Mol Biol. 2021;2178:107–32. [DOI] [PubMed] [Google Scholar]
- Pinas GE, Reinoso-Vizcaino NM, Yandar Barahona NYet al. Crosstalk between the serine/threonine kinase StkP and the response regulator ComE controls the stress response and intracellular survival of Streptococcus pneumoniae. PLoS Pathog. 2018;14:e1007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F, Byrne D, Mengin-Lecreulx Det al. Dual regulation of activity and intracellular localization of the PASTA kinase PrkC during Bacillus subtilis growth. Sci Rep. 2018b;8:1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F, Foulquier E, Galinier A. Impact of serine/threonine protein kinases on the regulation of sporulation in Bacillus subtilis. Front Microbiol. 2016;7:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F, Freton C, Wicker-Planquart Cet al. Phosphorylation of CpgA protein enhances both its GTPase activity and its affinity for ribosome and is crucial for Bacillus subtilis growth and morphology. J Biol Chem. 2012;287:20830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F, Rismondo J, Grundling Aet al. Investigation of the phosphorylation of Bacillus subtilis LTA synthases by the serine/threonine kinase PrkC. Sci Rep. 2018a;8:17344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet S, Mijakovic I, Nessler Set al. HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria. Biochim Biophys Acta Proteins Proteomics. 2004;1697:123–35. [DOI] [PubMed] [Google Scholar]
- Poncet S, Milohanic E, Maze Aet al. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol. 2009;16:88–102. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical?. Annu Rev Microbiol. 2008;62:35–51. [DOI] [PubMed] [Google Scholar]
- Pragai Z, Allenby NE, O'Connor Net al. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J Bacteriol. 2004;186:1182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar A, Bhatia S, Gigliozzi Det al. Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J Cell Biol. 2013;203:1081–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CW, Fawcett P, Ceremonie Het al. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol Microbiol. 2001;41:757–74. [DOI] [PubMed] [Google Scholar]
- Rajagopalan K, Dworkin J, Nagle E. Identification and biochemical characterization of a novel protein phosphatase 2C-like Ser/Thr phosphatase in Escherichia coli. J Bacteriol. 2018;200:e00225–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit YS, Sharma DK, Misra HS. Involvement of serine /threonine protein kinases in DNA damage response and cell division in bacteria. Res Microbiol. 2022;173:103883. [DOI] [PubMed] [Google Scholar]
- Ramirez-Guadiana FH, Barajas-Ornelas Rdel C, Corona-Bautista SUet al. The RecA-dependent SOS response is active and required for processing of DNA damage during Bacillus subtilis sporulation. PLoS ONE. 2016;11:e0150348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar V, Nalpas NC, Anselm Vet al. In-depth analysis of Bacillus subtilis proteome identifies new ORFs and traces the evolutionary history of modified proteins. Sci Rep. 2018;8:17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar V, Shi L, Krug Ket al. Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol Cell Proteomics. 2014;13:1965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynor MJ, Roh JH, Widen SGet al. Regulons and protein-protein interactions of PRD-containing Bacillus anthracis virulence regulators reveal overlapping but distinct functions. Mol Microbiol. 2018;109:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J. Regulation of sugar uptake and efflux in Gram-positive bacteria. FEMS Microbiol Rev. 1989;5:149–56. [DOI] [PubMed] [Google Scholar]
- Repizo GD, Blancato VS, Sender PDet al. Catabolite repression of the citST two-component system in Bacillus subtilis. FEMS Microbiol Lett. 2006;260:224–31. [DOI] [PubMed] [Google Scholar]
- Richts B, Rosenberg J, Commichau FM. A survey of pyridoxal 5'-phosphate-dependent proteins in the Gram-positive model bacterium Bacillus subtilis. Front Mol Biosci. 2019;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo MM, Borges TJ, Lang BJet al. Host expression system modulates recombinant Hsp70 activity through post-translational modifications. FEBS J. 2020;287:4902–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VIet al. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. [DOI] [PubMed] [Google Scholar]
- Rodriguez Ayala F, Bartolini M, Grau R. The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: an old friend with new functions. Front Microbiol. 2020;11:1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JL, Dalia AB, Weiser JN. Increased chain length promotes pneumococcal adherence and colonization. Infect Immun. 2012;80:3454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati D, Scarlato V. Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol Rev. 2017;41:549–74. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Price NP, Ehrhardt Cet al. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. Inaquosorum subsp. nov. Int J Syst Evol Microbiol. 2009;59:2429–36. [DOI] [PubMed] [Google Scholar]
- Rosenberg A, Soufi B, Ravikumar Vet al. Phosphoproteome dynamics mediate revival of bacterial spores. BMC Biol. 2015;13:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A, Arora G, Gupta Met al. Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J Bacteriol. 2011a;193:5347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A, Arora G, Gupta Met al. Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS ONE. 2011b;6:e17871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A, Arora G, Singhal Aet al. Protein phosphatases of pathogenic bacteria: role in physiology and virulence. Annu Rev Microbiol. 2015;69:527–47. [DOI] [PubMed] [Google Scholar]
- Salzberg LI, Powell L, Hokamp Ket al. The WalRK (YycFG) and sigma(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol Microbiol. 2013;87:180–95. [DOI] [PubMed] [Google Scholar]
- Satomura T, Shimura D, Asai Ket al. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J Bacteriol. 2005;187:4813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk K, Hervas AB, Rosch TCet al. Rapid turnover of DnaA at replication origin regions contributes to initiation control of DNA replication. PLoS Genet. 2017;13:e1006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling O, Frick O, Herzberg Cet al. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl Environ Microb. 2007;73:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley ACet al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Trentini DB, Spiess Set al. Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response. Mol Cell Proteomics. 2014;13:537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RM, Carter MM, Chu MLet al. Heme sensing in Bacillus thuringiensis: a supplementary HssRS-regulated heme resistance system. FEMS Microbiol Lett. 2016;363:fnw076. [DOI] [PubMed] [Google Scholar]
- Scott JD, Newton AC. Bacterial spore coat protein kinases: a new twist to an old story. Proc Natl Acad Sci USA. 2016;113:6811–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Osorio G, Jerez CA. Phosphorylation of GroEL, DnaK and other proteins from Thiobacillus ferrooxidans grown under different conditions. FEMS Microbiol Lett. 1996;138:129–34. [DOI] [PubMed] [Google Scholar]
- Sella SR, Vandenberghe LP, Soccol CR. Life cycle and spore resistance of spore-forming Bacillus atrophaeus. Microbiol Res. 2014;169:931–9. [DOI] [PubMed] [Google Scholar]
- Setlow P, Christie G. Bacterial spore mRNA – what’s up with that?. Front Microbiol. 2020;11:596092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafikhani SH, Leighton T. AbrB and Spo0E control the proper timing of sporulation in Bacillus subtilis. Curr Microbiol. 2004;48:262–9. [DOI] [PubMed] [Google Scholar]
- Shah IM, Dworkin J. Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides. Mol Microbiol. 2010;75:1232–43. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DLet al. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir SM, Bryant KM, Larabee JLet al. Regulatory interactions of a virulence-associated serine/threonine phosphatase-kinase pair in Bacillus anthracis. J Bacteriol. 2010;192:400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Dhasmana N, Dubey Net al. Bacterial virulence factors: secreted for survival. Ind J Microbiol. 2017;57:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Chai Y. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol. 2013;195:2747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Kolter R, Losick R. The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J Bacteriol. 2010;192:6352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Ostrov I. Role of Bacillus species in biofilm persistence and emerging antibiofilm strategies in the dairy industry. J Sci Food Agric. 2020;100:2327–36. [DOI] [PubMed] [Google Scholar]
- Shen D, Perpich JD, Stocke KSet al. Role of the RprY response regulator in P. gingivalis community development and virulence. Mol Oral Microbiol. 2020;35:231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Heat shock of Escherichia coli increases binding of dnaK (the hsp70 homolog) to polypeptides by promoting its phosphorylation. Proc Natl Acad Sci USA. 1993;90:8648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Pigeonneau N, Ravikumar Vet al. Cross-phosphorylation of bacterial serine/threonine and tyrosine protein kinases on key regulatory residues. Front Microbiol. 2014a;5:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Pigeonneau N, Ventroux Met al. Protein-tyrosine phosphorylation interaction network in Bacillus subtilis reveals new substrates, kinase activators and kinase cross-talk. Front Microbiol. 2014b;5:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ravikumar V, Derouiche Aet al. Tyrosine 601 of Bacillus subtilis DnaK undergoes phosphorylation and is crucial for chaperone activity and heat shock survival. Front Microbiol. 2016;7:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang Y, Lin Set al. dbPSP 2.0, an updated database of protein phosphorylation sites in prokaryotes. Sci Data. 2020;7:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L, Rosenberg A, Smith Yet al. The molecular timeline of a reviving bacterial spore. Mol Cell. 2015;57:695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KD, Halbedel S, Gorke Bet al. Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC. J Mol Microbiol Biotechnol. 2007;13:165–71. [DOI] [PubMed] [Google Scholar]
- Singh KD, Schmalisch MH, Stülke Jet al. Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources. J Bacteriol. 2008;190:7275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LK, Dhasmana N, Sajid Aet al. clpC operon regulates cell architecture and sporulation in Bacillus anthracis. Environ Microbiol. 2015;17:855–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, Virmani R, Naz Set al. Methylation of two-component response regulator MtrA in mycobacteria negatively modulates its DNA binding and transcriptional activation. Biochem J. 2020;477:4473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Kumar C, Gnad Fet al. Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis. J Proteome Res. 2010;9:3638–46. [DOI] [PubMed] [Google Scholar]
- Soule MH. Identity of Bacillus subtilis, Cohn 1872. J Infect Dis. 1932;51:191–215. [Google Scholar]
- Squeglia F, Marchetti R, Ruggiero Aet al. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J Am Chem Soc. 2011;133:20676–9. [DOI] [PubMed] [Google Scholar]
- Stancik IA, Sestak MS, Ji Bet al. Serine/threonine protein kinases from bacteria, Archaea and Eukarya share a common evolutionary origin deeply rooted in the tree of life. J Mol Biol. 2018;430:27–32. [DOI] [PubMed] [Google Scholar]
- Stannek L, Gunka K, Care RAet al. Factors that mediate and prevent degradation of the inactive and unstable GudB protein in Bacillus subtilis. Front Microbiol. 2014;5:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff DL, Skaar EP. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol Microbiol. 2009;72:763–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg N, Keren-Paz A, Hou Qet al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci Signal. 2020;13:eaaw8905. [DOI] [PubMed] [Google Scholar]
- Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol. 2016;101:531–44. [DOI] [PubMed] [Google Scholar]
- Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol. 2002;46:297–304. [DOI] [PubMed] [Google Scholar]
- Stewart GC. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev. 2015;79:437–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. [DOI] [PubMed] [Google Scholar]
- Strauch M, Webb V, Spiegelman Get al. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA, Ballar P, Rowshan AJet al. The DNA-binding specificity of the Bacillus anthracis AbrB protein. Microbiology. 2005;151:1751–9. [DOI] [PubMed] [Google Scholar]
- Stubbendieck RM, Straight PD. Linearmycins activate a two-component signaling system involved in bacterial competition and biofilm morphology. J Bacteriol. 2017;199:e00186–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Liu C, Fang Het al. Bacillus subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microb Cell Fact. 2020;19:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suskiewicz MJ, Hajdusits B, Beveridge Ret al. Structure of McsB, a protein kinase for regulated arginine phosphorylation. Nat Chem Biol. 2019;15:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada H, Shiwa Y, Takino Yet al. Essentiality of WalRK for growth in Bacillus subtilis and its role during heat stress. Microbiology. 2018;164:670–84. [DOI] [PubMed] [Google Scholar]
- Talavera A, Hendrix J, Versees Wet al. Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors. Sci Adv. 2018;4:eaap9714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kobayashi K, Ogasawara N. The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology. 2003;149:2317–29. [DOI] [PubMed] [Google Scholar]
- Tao L, Chattoraj P, Biswas I. CtsR regulation in mcsAB-deficient Gram-positive bacteria. J Bacteriol. 2012;194:1361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Serrano E&, Alonso JC. Bacillus subtilis RecA interacts with and loads RadA/Sms to unwind recombination intermediates during natural chromosomal transformation. Nucleic Acids Res. 2019;47:9198–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentini DB, Fuhrmann J, Mechtler Ket al. Chasing phosphoarginine proteins: development of a selective enrichment method using a phosphatase trap. Mol Cell Proteomics. 2014;13:1953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentini DB, Suskiewicz MJ, Heuck Aet al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova B, Wilson AC, Bongiorni Cet al. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol Microbiol. 2007;63:644–55. [DOI] [PubMed] [Google Scholar]
- Ultee E, Ramijan K, Dame RTet al. Stress-induced adaptive morphogenesis in bacteria. Adv Microb Physiol. 2019;74:97–141. [DOI] [PubMed] [Google Scholar]
- van Beilen J, Blohmke CJ, Folkerts Het al. RodZ and PgsA play intertwined roles in membrane homeostasis of Bacillus subtilis and resistance to weak organic acid stress. Front Microbiol. 2016;7:1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Esker MH, Kovacs AT, Kuipers OP. YsbA and LytST are essential for pyruvate utilization in Bacillus subtilis. Environ Microbiol. 2017;19:83–94. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Chateau A, Dillies MAet al. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect Immun. 2009;77:4437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, Tempelaars MH, Zwietering MHet al. Analysis of the role of RsbV, RsbW, and RsbY in regulating sigmaB activity in Bacillus cereus. J Bacteriol. 2005;187:5846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–68. [DOI] [PubMed] [Google Scholar]
- Vermassen A, Leroy S, Talon Ret al. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol. 2019;10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter SM, Schlievert PM. The two-component system Bacillus respiratory response A and B (BrrA-BrrB) is a virulence factor regulator in Bacillus anthracis. Biochemistry. 2007;46:7343–52. [DOI] [PubMed] [Google Scholar]
- Vijay K, Brody MS, Fredlund Eet al. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the SigmaB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–8. [DOI] [PubMed] [Google Scholar]
- Virmani R, Pradhan P, Joshi Jet al. Phosphorylation-mediated regulation of the Bacillus anthracis phosphoglycerate mutase by the Ser/Thr protein kinase PrkC. Biochem Biophys Res Commun. 2023;665:88–97. [DOI] [PubMed] [Google Scholar]
- Virmani R, Sajid A, Singhal Aet al. The Ser/Thr protein kinase PrkC imprints phenotypic memory in Bacillus anthracis spores by phosphorylating the glycolytic enzyme enolase. J Biol Chem. 2019;294:8930–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard Pet al. Sticking together: building A biofilm the Bacillus subtilis way. Nat Rev Micro. 2013;11:157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker U, Luo T, Smirnova Net al. Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J Bacteriol. 1997;179:1980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jenkins SA, Gu Cet al. Bacillus anthracis spore surface protein BclA mediates complement factor H binding to spores and promotes spore persistence. PLoS Pathog. 2016;12:e1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Cole PA. The chemical biology of reversible lysine post-translational modifications. Cell Chem Biol. 2020;27:953–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe K. [Overview of study on Bacillus subtilis spores]. Yakugaku Zasshi. 2013;133:783–97. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y, Penchev R, Dubnau Eet al. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–72. [DOI] [PubMed] [Google Scholar]
- Westers H, Westers L, Darmon Eet al. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 2006;273:3816–27. [DOI] [PubMed] [Google Scholar]
- White AK, Hoch JA, Grynberg Met al. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J Bacteriol. 2006;188:6354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett JW, Crosson S. Atypical modes of bacterial histidine kinase signaling. Mol Microbiol. 2017;103:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Song Q, Han A. Interacting proteins of the essential two-component system YycFG in Bacillus subtilis. J Basic Microbiol. 2019;59:950–9. [DOI] [PubMed] [Google Scholar]
- Xiao D, Chen C, Yang P. Computational systems approach towards phosphoproteomics and their downstream regulation. Proteomics. 2023;23:2200068. [DOI] [PubMed] [Google Scholar]
- Xing Y, Harper WF. Bacillus spore awakening: recent discoveries and technological developments. Curr Opin Biotechnol. 2020;64:110–5. [DOI] [PubMed] [Google Scholar]
- Xu B, Liu L, Song G. Functions and regulation of translation elongation factors. Front Mol Biosci. 2021;8:816398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T, Carrasco B, Hejna Jet al. Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J Biol Chem. 2013;288:22437–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T, Carrasco B, Serrano Eet al. Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination. J Biol Chem. 2014;289:27640–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yague P, Gonzalez-Quinonez N, Fernandez-Garcia Get al. Correction: Yague, P., et al. Goals and challenges in bacterial phosphoproteomics. Int. J. Mol. Sci. 2019, 20, 5678. Int J Mol Sci. 2020;21:9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Furuhata K, Fukushima Tet al. Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J Biosci Bioeng. 2004;98:174–81. [DOI] [PubMed] [Google Scholar]
- Yan J, Zou W, Fang Jet al. Eukaryote-like Ser/Thr protein kinase PrkA modulates sporulation via regulating the transcriptional factor sigma(K) in Bacillus subtilis. Front Microbiol. 2015;06:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kang CM, Brody MSet al. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–75. [DOI] [PubMed] [Google Scholar]
- Yin W, Wang Y, Liu Let al. Biofilms: the microbial “protective clothing” in extreme environments. Int J Mol Sci. 2019;20:3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Lebrun R, Espinosa Let al. PrkA is an ATP-dependent protease that regulates sporulation in Bacillus subtilis. J Biol Chem. 2022;298:102436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Pompeo F, Galinier A. Overview of protein phosphorylation in bacteria with a main focus on unusual protein kinases in Bacillus subtilis. Res Microbiol. 2021;172:103871. [DOI] [PubMed] [Google Scholar]
- Zhang QB, Yu K, Liu Zet al. Prediction of prkC-mediated protein serine/threonine phosphorylation sites for bacteria. PLoS ONE. 2018;13:e0203840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hu Y, Fan Qet al. Two-component system YvqEC-dependent bacterial resistance against vancomycin in Bacillus thuringiensis. Antonie Van Leeuwenhoek. 2015;108:365–76. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li X, Wang Xet al. The two-component signal transduction system YvcPQ regulates the bacterial resistance to bacitracin in Bacillus thuringiensis. Arch Microbiol. 2016;198:773–84. [DOI] [PubMed] [Google Scholar]
- Zhou B, Semanjski M, Orlovetskie Net al. Arginine dephosphorylation propels spore germination in bacteria. Proc Natl Acad Sci USA. 2019;116:14228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhang H, Fang Het al. Transcriptome based functional identification and application of regulator AbrB on alkaline protease synthesis in Bacillus licheniformis 2709. Int J Biol Macromol. 2021;166:1491–8. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhang N, Xia Let al. ResDE two-component regulatory system mediates oxygen limitation-induced biofilm formation by Bacillus amyloliquefaciens SQR9. Appl Environ Microb. 2018;84:e02744–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari Emameh R, Kazokaite J, Yakhchali B. Bioinformatics analysis of extracellular subtilisin E from Bacillus subtilis. J Biomol Struct Dyn. 2022;40:7183–90. [DOI] [PubMed] [Google Scholar]
- Zwick JV, Noble S, Ellaicy YKet al. AhpA is a peroxidase expressed during biofilm formation in Bacillus subtilis. Microbiologyopen. 2017;6:e00403. [DOI] [PMC free article] [PubMed] [Google Scholar]