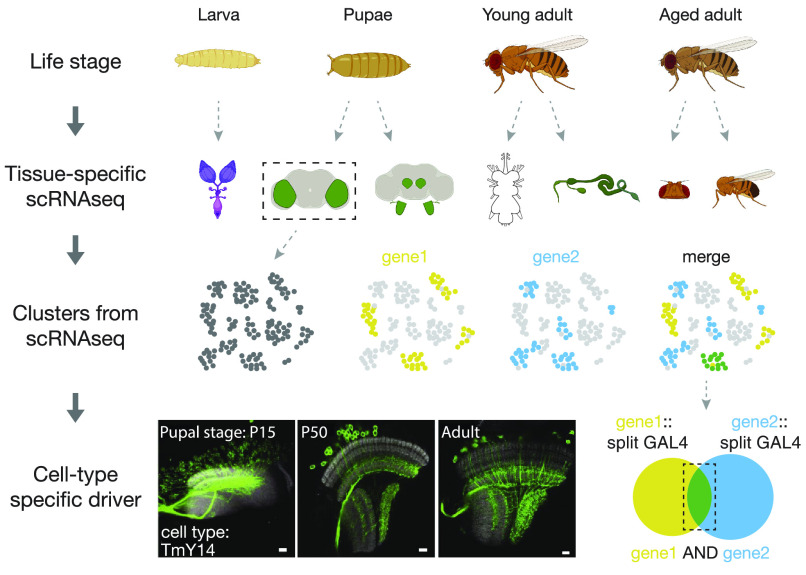
Most multicellular organisms consist of tremendous diversity of cells. Genetic drivers that target specific groups of cells have played a key role in dissecting complex biological processes. Tens of thousands of driver lines for the popular GAL4/UAS-based binary expression in a specific group of cells have been produced in the fruit fly Drosophila (1–3), which enable genetic manipulation of these cells to study their function. Researchers can search from these drivers for their cell types of interests (4). More recently, the emergence of a large collection of split-GAL4 drivers (see below), which in principle generate the intersection of gene expression patterns from two different drivers (3, 5), is gradually making single-cell-type labeling a routine. However, since expression patterns of most drivers have been characterized only in adults, they cannot be directly utilized in studying developmental processes. Writing in PNAS, Chen et al. (6) start to fill this gap by proposing a working pipeline of generating cell type–specific drivers for developmental study (Fig. 1).
Fig. 1.
An approach to generate cell type–specific drivers throughout different life stages of the fly. Top row: the life cycle of a fly begins with an egg that develops into the stage of larvae, pupae, young adult, and aged adult. Second row: scRNAseq datasets from various tissues or brain regions could provide the related gene expression profiles across life stages. Left to right, fly ovary, optic lobes, antennae and antennal lobes, ventral nerve cord, midgut, head, and body. Third and bottom rows: if two genes are both expressed only in one or a few transcriptomic clusters (corresponding to cell types) across multiple life stages, the genetic intersection of this gene pair could lead to cell type–specific drivers across life stages. Hypothetical transcriptomic clusters from only one life stage are shown. As an example, Chen et al. show consistent labeling of a cell type–specific driver in the developing and adult Drosophila optic lobe. (Scale bar: 10 µm.) P15, P50: time when pupal development reaches 15% and 50%, respectively. Images are adapted from Chen et al. (6).
The first obstacle that Chen et al. overcame is to generate drivers whose expression patterns can be accurately predicted throughout development. In general, genetic drivers are prone to changing expression patterns across development, a reflection of the dynamic nature of gene expressions. For the GAL4 drivers from the large existing collections (1, 2, 7), their expression patterns are characterized mainly in adult flies with little expression information during development. Moreover, these patterns often do not match the expression patterns of the genes whose regulatory elements were used in generating these drivers. To tackle these problems, Chen et al. applied two strategies to generating drivers that could recapitulate the expression of the corresponding endogenous genes: One is based on CRISPR knock-in (8) and the other on existing coding intron lines (9, 10). Both strategies support inserting self-cleavable T2A-split GAL4 (half of a GAL4, see below) into the endogenous coding sequence of a gene, which largely recapitulates the original expression pattern of this gene. To make this process more convenient to researchers, the authors also generated ready-to-use donor flies which can assist the generation of gene-specific drivers for over 3,000 genes. The availability of these gene-specific drivers will greatly expand the toolkits of fly genetics for many applications.
The second obstacle that Chen et al. overcame is to reliably predict the outcome cell types from genetic intersections. To more specifically target cell types than GAL4 drivers do, the fly field has been using the split-GAL4 system to generate drivers that express either the N- or C-terminal half of the GAL4 proteins containing the DNA-binding domain and transcriptional activation domain, respectively. Only cells that express both halves of the split GAL4 can have a functioning GAL4 (11). But the question is which two split-GAL4 drivers should one choose when one aims to specifically label a single cell type during development? Using the Drosophila optic lobe as an example, Chen et al. demonstrated that this can be done in a highly predictable way. Based on single-cell RNA sequencing (scRNAseq) data across multiple developmental stages (12, 13), the authors identified candidate gene pairs that are coexpressed in the cell type(s) of interest. They also developed a user-friendly software called scMarco to assist the search for candidate genes (https://apps.ycdavidchen.com/scMarco). From the gene pairs identified, around a hundred gene-specific split-GAL4 drivers were generated in this study (https://splitgal4.org). The authors validated the expression patterns of the drivers throughout development and demonstrated a high success rate for labeling the cell type(s) of interest. Based on the authors’ calculation, this approach could in theory grant access to >90% cell types in the Drosophila optic lobe during development.
While scRNAseq data were used to instruct the generation of the cell type–specific drivers, can cell type–specific drivers in turn assist the analysis of scRNAseq datatsets? One challenging step in analyzing large scRNAseq datatsets is to assign all clusters to specific cell types. Leveraging on the newly built cell type–specific drivers, Chen et al. successfully revealed the identity of multiple previously unannotated clusters in the developing Drosophila optic lobe scRNAseq dataset. They even discovered new cell types that have not been reported before. This reiterative approach could potentially lead to a continual refinement of cell type identity in the entire fly brain, still a major challenge, especially in combination with the recently released serial electron microscopy–based connectome datasets (14–16).
Leveraging on the newly built cell-type specific drivers, Chen et al. successfully revealed the identity of multiple previously unannotated clusters in the developing Drosophila optic lobe scRNAseq dataset.
Moving forward, with the scRNAseq data from more and more tissues or brain regions at various life stages becoming available (17, 18), genetic drivers based on the native genetic regulatory elements can be directly extended to the sequenced tissues and brain regions; an example is the antennal lobe where cell type–decoded scRNAseq datasets are available (19, 20). The pipeline described by Chen et al. could also be expanded to regulators other than GAL4, such as split QF (21), split LexA (22), GAL80 (23), and GAL80-repressible split GAL4 (24). Imagine one day you can order whatever components for the cell type–specific drivers of your interest at will at any developmental stage. Your life as a researcher to figure out the function of that cell type will be made much easier. Hopefully, this day won’t be too far away, with Chen et al. as a start, continued with the joint efforts from the entire fly community. Altogether, these efforts could help make fruit flies even more attractive in investigating the fundamental biology of multicellular organisms.
Acknowledgments
C.L. was supported by the Stanford Science Fellows Program.
Author contributions
C.L., Z.L. and L.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
See companion article, “Using single-cell RNA sequencing to generate predictive cell-type-specific split-GAL4 reagents throughout development,” 10.1073/pnas.2307451120.
References
- 1.Jenett A., et al. , A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kvon E. Z., et al. , Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512, 91–95 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Tirian L., Dickson B. J., The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. bioRxiv [Preprint] (2017). 10.1101/198648 (Accessed 20 July 2023). [DOI]
- 4.Meissner G. W., et al. , A searchable image resource of Drosophila GAL4 driver expression patterns with single neuron resolution. Elife 12, 1–20 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dionne H., Hibbard K. L., Cavallaro A., Kao J. C., Rubin G. M., Genetic reagents for making split-GAL4 lines in Drosophila. Genetics 209, 31–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y.-C.D., et al. , Using single-cell RNA sequencing to generate predictive cell-type-specific split-GAL4 reagents throughout development. Proc. Natl. Acad. Sci. U.S.A., this issue. [DOI] [PMC free article] [PubMed]
- 7.Hayashi S., et al. , GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 34, 58–61 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Gratz S. J., et al. , Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venken K. J. T., et al. , MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee P. T., et al. , A gene-specific T2A-GAL4 library for Drosophila. Elife 7, 1–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan H., Peabody N. C., Vinson C. R. R., White B. H., Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurmangaliyev Y. Z., Yoo J., Valdes-Aleman J., Sanfilippo P., Zipursky S. L., Transcriptional programs of circuit assembly in the Drosophila visual system. Neuron 108, 1045–1057.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Özel M. N., et al. , Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheffer L. K., et al. , A connectome and analysis of the adult Drosophila central brain. Elife 9, e57443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorkenwald S., et al. , Neuronal wiring diagram of an adult brain. bioRxiv [Preprint] (2023). 10.1101/2023.06.27.546656 (Accessed 20 July 2023). [DOI] [PMC free article] [PubMed]
- 16.Schlegel P., et al. , A consensus cell type atlas from multiple connectomes reveals principles of circuit stereotypy and variation. bioRxiv [Preprint] (2023). 10.1101/2023.06.27.546055 (Accessed 20 July 2023). [DOI]
- 17.Li H., Single-cell RNA sequencing in Drosophila: Technologies and applications. Wiley Interdiscip. Rev. Dev. Biol. 10, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., et al. , Fly cell atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaughlin C. N., et al. , Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. Elife 10, e63856 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Q., et al. , Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. Elife 10, e63450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riabinina O., Vernon S. W., Dickson B. J., Baines R. A., Split-QF system for fine-tuned transgene expression in Drosophila. Genetics 212, 53–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting C. Y., et al. , Focusing transgene expression in Drosophila by Coupling Gal4 with a novel split-Lex A expression system. Genetics 188, 229–233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suster M. L., Seugnet L., Bate M., Sokolowski M. B., Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 39, 240–245 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Ewen-Campen B., et al. , split-intein Gal4 provides intersectional genetic labeling that is repressible by Gal80. Proc. Natl. Acad. Sci. U.S.A. 120, e2304730120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]