Abstract
Background
The number of exposures to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to vaccine antigens affect the magnitude and avidity of the polyclonal response.
Methods
We studied binding and avidity of different antibody isotypes to the spike, the receptor-binding domain (RBD), and the nucleoprotein (NP) of wild-type (WT) and BA.1 SARS-CoV-2 in convalescent, mRNA vaccinated and/or boosted, hybrid immune individuals and in individuals with breakthrough cases during the peak of the BA.1 wave.
Results
We found an increase in spike-binding antibodies and antibody avidity with increasing number of exposures to infection and/or vaccination. NP antibodies were detectible in convalescent individuals and a proportion of breakthrough cases, but they displayed low avidity. Omicron breakthrough infections elicited high levels of cross-reactive antibodies between WT and BA.1 antigens in vaccinated individuals without prior infection directed against the spike and RBD. The magnitude of the antibody response and avidity correlated with neutralizing activity against WT virus.
Conclusions
The magnitude and quality of the antibody response increased with the number of antigenic exposures, including breakthrough infections. However, cross-reactivity of the antibody response after BA.1 breakthroughs, was affected by the number of prior exposures.
Keywords: SARS-CoV-2, antibodies, avidity, neutralization, omicron
Antibody binding, neutralization, and avidity increase with more antigenic exposures. Nucleoprotein-directed antibodies increase after Omicron breakthrough infection but display low avidity, and antibodies induced by Omicron breakthrough infection in double-vaccinated individuals broadly react against ancestral and Omicron receptor-binding domain/spike antigens.
Infections with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019, lead to a rapid induction of long-lasting antibodies, albeit at variable levels [1]. A large proportion of the antibody response elicited during SARS-CoV-2 infections is directed to the spike surface glycoprotein, which mediates viral entry into the target cells and is expressed on the surface of infected cells [2]. Among the 3 major domains of the spike protein, which include the N-terminal domain, the S2 domain and the receptor-binding domain (RBD), the RBD represents the main target of the neutralizing antibody response because of the domain’s direct interaction with the cellular receptor angiotensin-converting enzyme 2 [3]. Importantly, levels of binding antibodies correlate well with virus neutralization titers [1, 4], and induction of neutralizing antibodies was initially reported to correlate with the severity of infection [4].
The introduction of vaccination (including booster doses), the emergence of diverse variants of interest and/or concern in the pre-Omicron era [5], and a large number of breakthrough infections during the Omicron era have increased the complexity of individual exposure histories. It is expected that the magnitude as well as the affinity of antibodies will increase with the number of exposures. Affinity maturation of neutralizing antibodies can alter their capacity to control SARS-CoV-2 variants and expand the breadth of neutralization to other sarbecoviruses [6]. For polyclonal antibodies, assessment of antibody avidity provides a measure of the overall strength of bivalent or multivalent interactions between antibodies and their epitopes [7]. Antibody avidity is influenced by the affinity of the individual antibody clones and by the valency of the specific antibody isotype [8]. Avidity can be assessed by an enzyme-linked immunosorbent assay (ELISA) incorporating a chaotropic agent after incubation of polyclonal serum samples with the antigen [9, 10]. Alternatively, the dissociation rate (constant of dissociation [Kdis]) of polyclonal serum measured using biolayer interferometry (BLI) or surface plasmon resonance can be used as a proxy for polyclonal avidity [11, 12].
To investigate how different exposures to SARS-CoV-2 infection or vaccination influence the polyclonal immune response, we characterized the antibody responses and avidity of different antibody isotypes—immunoglobulin (Ig) G, IgM, and IgA—against the recombinant spike, RBD, and nucleoprotein (NP) of SARS-CoV-2. Given that avidity often correlates well with viral neutralization [13], we assessed the correlation between binding antibodies, neutralizing antibodies, and polyclonal antibody avidity. The use of samples from 3 distinct groups of study participants (eg, convalescent individuals, those who received 2 vaccine doses and a booster vaccine dose, and those with breakthrough infection) allowed us to identify specific signatures associated with the type of initial exposure or preexisting immunity at different time points after infection or vaccination. Moreover, we identified specific antibody signatures in individuals who had experienced breakthrough infections with the antigenically distinct Omicron variant.
METHODS
Study Cohort and Serum Samples
Forty convalescents’ serum samples were obtained as residual, deidentified samples from the chemistry laboratories at the Department of Pathology at the Icahn School of Medicine at Mount Sinai in the metropolitan area of New York during the first wave of SARS-CoV-2 infections. Individuals were tested for SARS-CoV-2 antibodies using a Mount Sinai Health System clinical pathology laboratory assay certified by the Clinical Laboratory Improvement Amendments. Samples with variable levels (1:80, 1:160, 1:320, 1:960, or ≥1:2880) were selected for this study. The “vaccination” group comprised 40 serum samples, including 20 from participants who received 2 doses of the BNT162b2 vaccine and 20 from those receiving 3 doses. “Breakthrough infection” samples were collected from 25 individuals with Omicron (BA.1) breakthrough infections after vaccination. No comorbidities were reported by the participants.
Vaccination and breakthrough infection serum samples were obtained from the observational PARIS (Protection Associated with Rapid Immunity to SARS-CoV-2) study (approved by the Mount Sinai Hospital Institutional Review Board; IRB20-03374). All PARIS study participants signed written consent forms before sample and data collection. Detailed demographic characteristics and vaccination information for the individuals from the different groups are shown in Supplementary Tables 1 and 2.
Cells and Viruses
African green monkey Vero. E6 cells expressing transmembrane protease serine 2 were cultured at 37°C with 5% carbon dioxide in Dulbecco's modified Eagle medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, 1× non-essential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin, 3 μg/mL puromycin (InvivoGen) and 100 µg/mL Normocin (InvivoGen). The SARS-CoV-2 isolate USA-WA1/2020 was used as the wild-type reference (BEI Resources; NR-52281).
Recombinant Proteins
Recombinant soluble SARS-CoV-2 proteins were expressed using a mammalian cell protein expression system, as described elsewhere [14] (details in Supplementary Methods).
ELISAs and Other Assays
SARS-CoV-2 antibody titers were measured with a research-grade in-house ELISA using recombinant RBD, spike, and NP antigens from the original Wuhan-Hu-1 SARS-CoV-2 isolate (wild type) and Omicron (BA.1) SARS-CoV-2 strains, according to manufacturer’s instructions and as described elsewhere [15, 16]. The binding of IgG, IgM, and IgA antibody isotypes was assessed. Antibody titers in serum samples from convalescent individuals were measured using another ELISA, the commercial COVID-SeroKlir Kantaro Semi-Quantitative SARS-CoV-2 IgG Antibody Kit (Kantaro Biosciences), according to the manufacturer’s instructions and as described elsewhere [17]
Serum samples were screened for neutralizing antibodies against ancestral SARS-CoV-2 (USA-WA1/2020), using our standard microneutralization assay, as described elsewhere [15, 18]. Finally, the binding avidity of polyclonal serum samples was measured using BLI performed with an Octet Red96 instrument (ForteBio), as described elsewhere [12]. For all assays, see the Supplementary Methods for details.
Statistical Analysis
A Pearson correlation coefficient was used to assess correlations between binding area under the curve (AUC), avidity index (AI), neutralization titers, and Kdis. We used a Mann-Whitney U test to compare differences in binding AUC and AI between different subgroups of the “convalescent” and vaccination groups,. For the breakthrough infection group, we performed statistical analyses using a parametric paired t test and assuming a normal distribution. In this group, normality could not be assessed uniformly owing to the small sample size in some groups. Differences were considered statistically significant at P < .03 with a 95% confidence interval. Statistical analyses were performed using Prism 9 software (GraphPad).
RESULTS
Binding, Avidity and Neutralization of Serum Antibodies from Convalescent Individuals
Initially, we characterized serum samples from convalescent individuals (Figure 1). Antibodies induced after SARS-CoV-2 infection typically display low avidity against the spike and against internal proteins of the virus [19, 20]. To analyze in detail the antibody levels and avidity induced after infection using serum samples from the convalescent group, we stratified samples based on their initial IgG anti-spike antibody response measured before the current study in a Mount Sinai's Clinical Laboratory Improvement Amendments–certified laboratory, as described elsewhere [1]. Titers of 1:80 and 1:160 were categorized as low (+), 1:320 as moderate (++), and 1:960 and ≥1:2880 as high (+++).
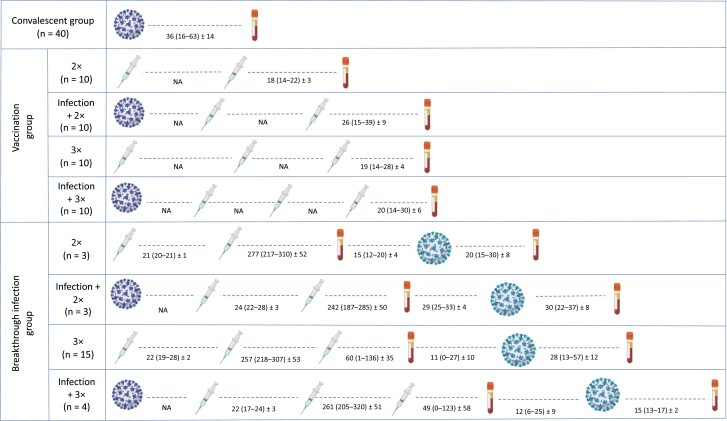
Figure 1.
Schematic representation of the groups with different exposure histories used in the study. Viral particles depicted in the left side indicate prior infection (by ancestral severe acute respiratory syndrome coronavirus 2 strains); those depicted in the right side, breakthrough infection (by Omicron BA.1). The number and sequence of vaccinations are indicated by syringes; the serum collection time points, by blood tubes. The intervals (days) between vaccination or breakthrough infection and sample collection are given as mean (range) ± standard deviation. Further details are provided in Supplementary Table 1. Abbreviations: 2× and 3×, 2 or 3 vaccine doses; NA, not available.
Importantly, IgM and IgA antibody isotypes were correlated well with IgG titers (Figure 2A) and this was consistent with binding assessed using the SeroKlir commercial RBD-Spike ELISA from Kantaro Biosciences, as reported elsewhere [17] (Figure 2B). However, neutralization titers against USA-WA1/2020 SARS-CoV-2, indicated by the inhibitory dilution 50% (ID50), were low in most serum samples (ID50, <100) and moderate (ID50, 100–200) in those with the highest binding titers (Figure 2C). Of note, the IgG antibody levels measured with the SeroKlir assay and neutralization titers showed a good correlation with IgG measured using our SARS-CoV-2 spike-binding IgG in-house ELISA (Figure 2A–2C) and were moderately correlated with IgM and IgA (Supplementary Figure 1).
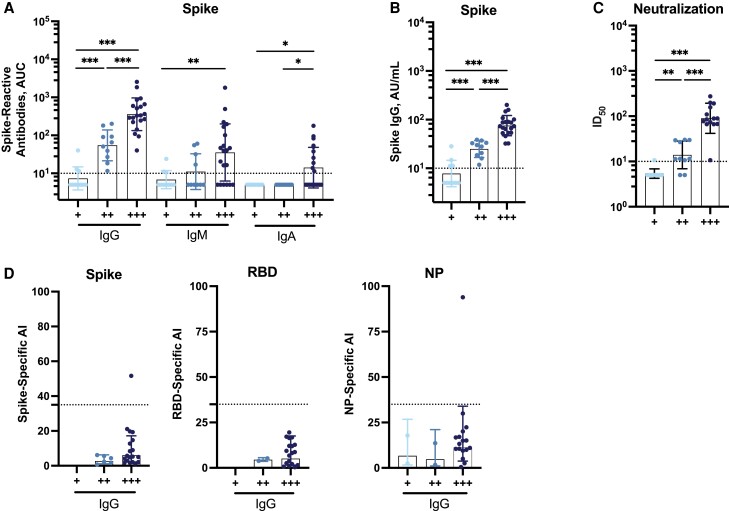
Figure 2.
Binding, neutralization, and avidity profiles of antibodies from individuals in the “convalescent” group. A, Analysis of different antibody isotypes (immunoglobulin [Ig] G, IgM, and IgA) against wild-type (WT) spike measured using our in-house enzyme-linked immunosorbent assay (ELISA; n = 40). Abbreviation: AUC, area under the curve. B, IgG levels measured using the SeroKlir commercial ELISA from Kantaro Biosciences (n = 40). Abbreviation: AU, arbitrary units. C, Neutralization titers against USA-WA1/2020 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), expressed as the inhibitory dilution 50% (ID50; n = 40). D, Antibody avidity, expressed as avidity index (AI), against the spike, the receptor-binding domain (RBD), and the nucleoprotein (NP) (n = 40). In D, samples with AIs <35, indicated by the dotted lines, are considered to have low avidity. WT refers to the sequence of the original Wuhan-Hu-1 SARS-CoV-2 isolate. Samples were stratified based on their initial IgG anti-spike antibody response measured in a Mount Sinai Clinical Laboratory Improvement Amendments–certified laboratory and categorized as low (+; titer, 1:80 or 1:160), moderate (++; titer, 1:320), or high (+++; titer, 1:960 or ≥1:2880). Each symbol represents a single participant. A Mann-Whitney U test for comparisons among different groups was performed. In A–C, the limit of detection is indicated by the horizontal dotted line. *P < .03; **P < .002; ***P < .001. Bars represent geometric means; error bars, geometric standard deviations.
To study the avidity of antibodies contained in polyclonal serum samples from our study participants, we first used an established ELISA that uses urea as chaotropic agent. Urea-treated versus nontreated samples were analyzed, and an AI was calculated as reported elsewhere [21]. IgG, IgM, and IgA avidity was evaluated against the spike, the RBD, and the NP from Wuhan-1 SARS-CoV-2. The AI (calculated as urea-treated sample AUC/nontreated sample AUC × 100) used here ranges from 0–100, where 0–30 indicates low avidity, 30–50, intermediate avidity, and 50–100, high avidity. Only the AIs of serum samples with an AUC >50 (under no urea treatment) were included in the analysis owing to a limitation in calculating with high accuracy the AIs in samples with low antibody levels, but all antibody titers are shown in Supplementary Figures 2, 5, and 6. Overall, the avidity of the different antibody isotypes—IgG, IgM, and IgA—was low against the Wuhan-1 spike, RBD, and NP (Figure 2D and Supplementary Figure 2).
Correlation of Neutralizing Activity of Infection-Derived Antibodies With Binding and Avidity
To assess the association between binding antibodies, neutralizing antibodies, and antibody avidity, we assessed the correlation between virus neutralization, measured by the ID50, and spike-specific IgG, measured using our in-house ELISA (Figure 3A) or the SeroKlir commercial RBD-spike ELISA (Figure 3B). In both cases, we detected a strong correlation between binding and neutralizing antibodies (r = 0.8845 and r = 0.9486, respectively; both P < .001). Given the limitation in calculating with high accuracy the AI in samples with low antibody levels, we assessed the correlation of IgG avidity—given by the AI—with virus neutralization in samples with high antibody titers (+++; 1:960 or ≥1:2880 titer) (Figure 3C). We found that these individuals had low IgG avidity, regardless of the neutralization titer. These results are in line with previous reports indicating that people displayed low antibody avidity after initial SARS-CoV-2 infection [21].
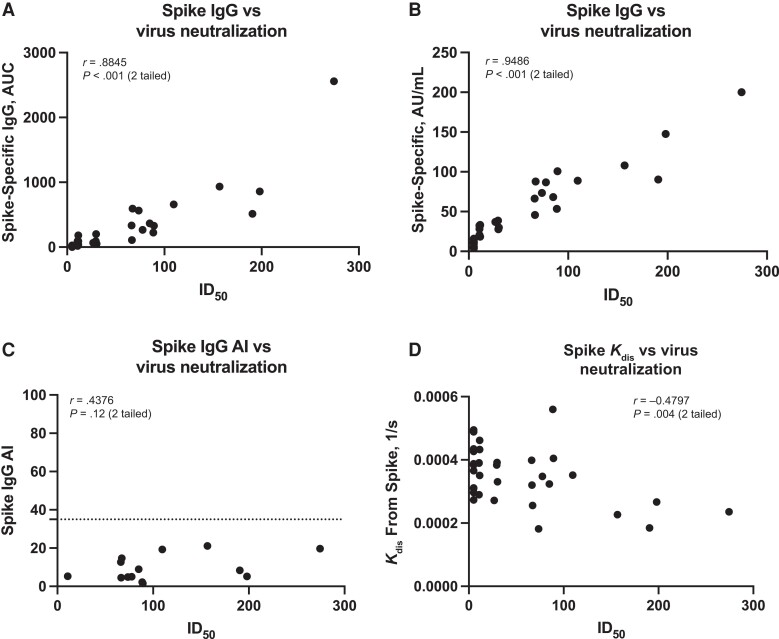
Figure 3.
Correlation of neutralizing activity of infection-derived antibodies with binding and avidity. A, B, Correlation of neutralization titers, expressed as inhibitory dilution 50% (ID50), with wild-type (WT) spike-specific immunoglobulin (Ig) G measured with the in-house enzyme-linked immunosorbent assay (ELISA) (A) or with the SeroKlir commercial ELISA from Kantaro Biosciences (B). Abbreviations: AU, arbitrary units; AUC, area under the curve. C, D, WT spike IgG avidity index (AI) (C) and avidity measured using biolayer interferometry (D). Abbreviation, Kdis, constant of dissociation. In C, samples with an AI <35, indicated by the dotted line, are considered to have low avidity. WT refers to the sequence of the original Wuhan-Hu-1 severe acute respiratory syndrome coronavirus 2 isolate. Each symbol represents a single participant. Pearson correlation was used; significance and correlation coefficients are shown. A, B, D, n = 34; C, n = 20.
We then analyzed whether avidity would correlate with neutralization by means of other, perhaps more sensitive methods. Biolayer interferometry (BLI) is typically used to assess the affinity of monoclonal antibodies, and the dissociation rate has been used as surrogate for the avidity of polyclonal serum samples using BLI [22]. We standardized the conditions for measuring this response in polyclonal serum samples using Niquel2+-nitriloacetic acid sensors that capture the recombinant RBD or spike by binding to their hexa-histidine tag. We assessed the correlation between antibody dissociation from the Wuhan-1 spike—as measured by the constant of dissociation or Kdis (lower values indicate higher avidity)—and neutralization of USA-WA1/2020 SARS-CoV-2 (Figure 3D).
A modest negative correlation was detected, indicating that antibodies with the highest neutralization titers display higher avidity. Likewise, we detected negative correlations between dissociation from spike/RBD and neutralization levels and between dissociation from spike/RBD and binding IgG to the spike and RBD (SupplementaryFigure 3A and 3B). A similar pattern was observed with BLI sensors preloaded with an anti-IgG, in which antibodies are first captured from the serum samples and the recombinant spike is added afterward (Supplementary Figure 3C). Overall, these data indicate that, although serum samples from convalescent individuals have variable levels of antibodies that correlate well with neutralization, the majority of individuals display low antibody avidity, which can be evidenced by more sensitive methods, such as BLI.
Diversification of the Antibody Responses to SARS-CoV-2 Is Highly Dependent on Infection and Vaccination Histories
Next, we assessed the binding and avidity of antibodies in the vaccination and breakthrough infection groups. Overall, serum samples from study participants with different immune histories displayed responses with variable magnitudes but with specific signatures. We found low levels of IgM against most antigens and low levels of IgA against the spike and RBD antigens (Figure 4A [vaccination group] and 4B [breakthrough infection group]), while NP levels were undetectable for these isotypes (data not shown). Moreover, we found that most individuals with hybrid immunity showed the highest levels of IgA (Figure 4Aand 4B ). Interestingly, IgA levels were boosted by breakthrough infections in individuals with only 2 vaccine doses, and several individuals in this group had high IgM induction following breakthrough infection, suggestive of potential de novo responses to the variant spike antigen (Figure 4B).
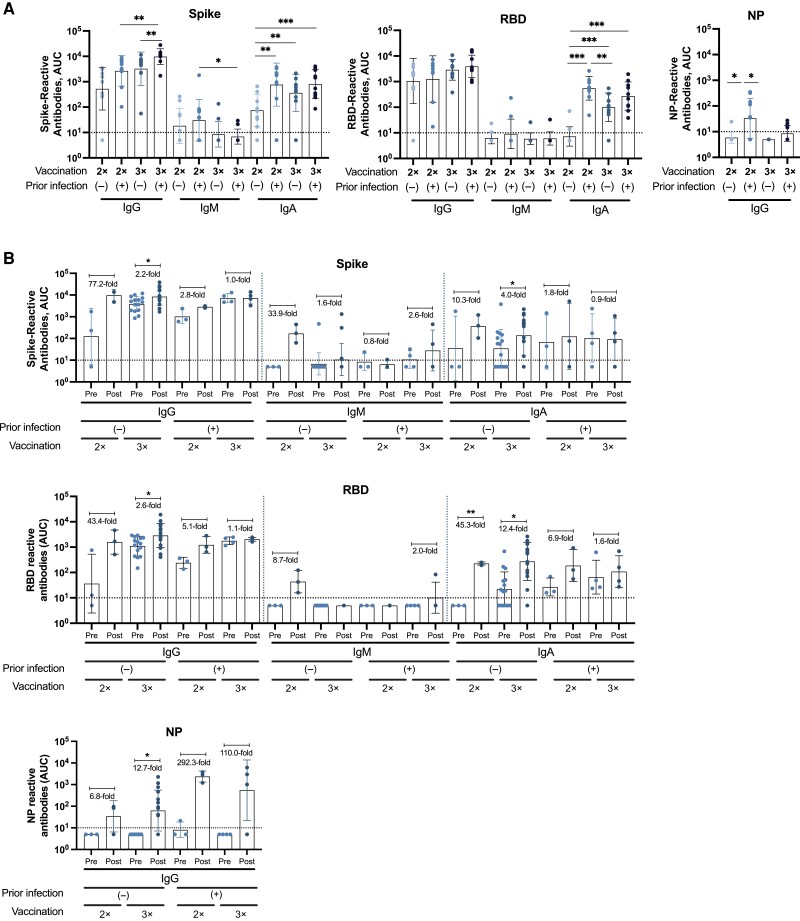
Figure 4.
Binding profile of antibodies against Wuhan-1 antigens from individuals in the “vaccination” and “breakthrough infection” groups. Immunoglobulin (Ig) G, IgM, and IgA antibody levels against wild-type (WT) spike, WT receptor-binding domain (RBD), and WT nucleoprotein (NP) antigens, expressed as the area under the curve (AUC), are shown for the vaccination (n = 40) (A) and breakthrough infection (n = 25) (B) groups. The limit of detection is indicated by the horizontal dotted lines. WT refers to the sequence of the original Wuhan-Hu-1 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate. Analyses were performed using a Mann-Whitney U test for comparisons among different groups (A), and a parametric paired t test for comparing prebreakthrough (Pre) and postbreakthrough (Post) infection responses (B). The average fold change occurring with breakthrough infection is indicated for each Pre-Post pair, represented by dots. *P < .03; **P < .002; ***P < .001. Each symbol represents a single participant; bars, geometric means; and error bars, geometric standard deviations.
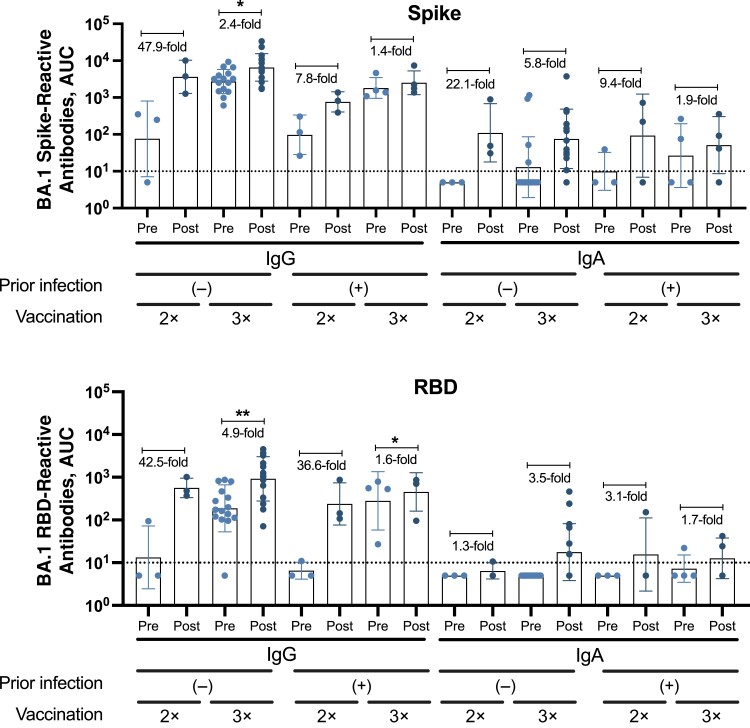
Most individuals in the vaccination and breakthrough infection groups displayed medium to high IgG levels against the spike and RBD of Wuhan-1 SARS-CoV-2 and Omicron BA.1 (assessed only in the breakthrough infection group). In the vaccination group, individuals who had received 3 doses of the BNT162b2 messenger RNA (mRNA), vaccine with or without prior SARS-CoV-2 infection, showed the highest IgG levels (Figure 4A). Interestingly, in the breakthrough infection group, double-vaccinated individuals without prior infection presented the highest antibody induction against all of the Wuhan-1 and Omicron BA.1 antigens (spike and RBD) (Figure 4B and Figure 5) after breakthrough infection, while double-vaccinated individuals with prior infection had higher antibody induction against Omicron BA.1 spike/RBD compared with Wuhan-1 antigens (Figure 5).
Figure 5.
Binding profile of antibodies from individuals of the “breakthrough infection” group against Omicron BA.1 antigens. Immunoglobulin (Ig) G, IgM, and IgA antibody levels against BA.1 spike and BA.1 receptor-binding domain (RBD) antigens of Omicron BA.1, expressed as area under the curve (AUC) (n = 24). BA.1 refers to the sequence of the first isolate of the Omicron lineage. The limit of detection is indicated by the horizontal dotted line. A parametric paired t test was performed to compare prebreakthrough (Pre) and postbreakthrough (Post) infection responses. The average fold change occurring with breakthrough infection is indicated for each Pre-Post pair, represented by dots. *P < .03; **P < .002. Each symbol represents a single participant; bars, geometric mean; and error bars, geometric standard deviations.
We next measured antibodies against the NP from Wuhan-1 SARS-CoV-2 in serum samples from study participants with or without a documented SARS-CoV-2 infection. Overall, and as expected, low to undetectable levels of IgM and IgA NP antibodies were seen in individuals who had not been infected previously with SARS-CoV-2 (Figure 4A and 4B). In the vaccination group, anti-NP levels were detected in several individuals who had had a prior infection (Figure 4A). In those who had experienced infection followed by 3 vaccine doses, the IgG NP titers were lower than in double-vaccinated individuals, likely a function of waning of NP antibodies, since the time of infection would have been longer for this group. In the breakthrough infection group, the highest IgG reactivity against NP was detected after the breakthrough infection in individuals with prior SARS-CoV-2 infection (Figure 4B), likely owing to a recall response since these individuals had already seen the NP antigen once.
Diversity of Immunity Acquired Through SARS-CoV-2 Infection or Vaccination Affects the Avidity of the Polyclonal Antibody Response
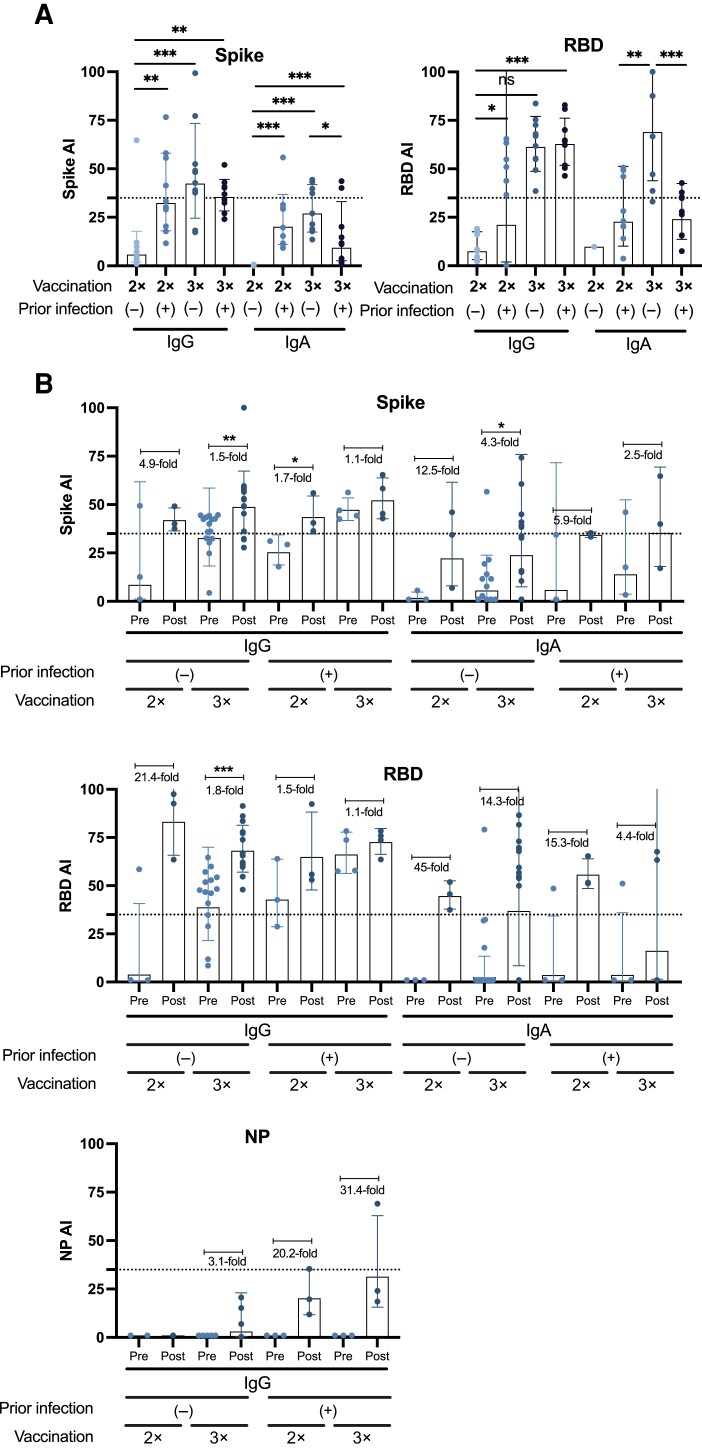
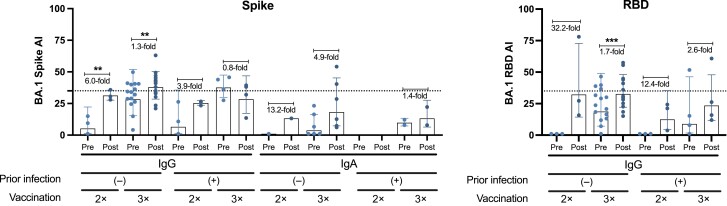
We found that individuals in the vaccination group who had received 3 doses of the BNT162b2 mRNA vaccine without prior SARS-CoV-2 infection developed the highest IgG and IgA avidity against the RBD and spike (Figure 6A). Similarly, in the breakthrough infection group, serum samples from individuals with 3 vaccine doses without prior infection had the highest avidity against the RBD/spike proteins of Wuhan-1 (Figure 6B) or Omicron BA.1 (Figure 7) after a breakthrough infection. In general, most individuals from the breakthrough infection group had increased antibody avidity after infection. Some individuals from both groups displayed detectable IgG levels against NP; however, the avidity of these antibodies was very low (Figure 6B). Supplementary Figures 4–6 show AUC values for all antigens in the presence or absence of urea, in both vaccination and breakthrough infection groups.
Figure 6.
Avidity profile of antibodies from individuals of the “vaccination” and “breakthrough infection” groups against Wuhan-1 antigens. Immunoglobulin (Ig) G, IgM, and IgA avidity against wild-type (WT) spike or WT receptor-binding domain (RBD) antigens is shown for the vaccination (n = 40) (A) and breakthrough infection (n = 25) (B) groups, expressed as the avidity index (AI). WT refers to the sequence of the original Wuhan-Hu-1 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate. Analyses were performed using Mann-Whitney U test for comparisons among different groups (A) and a parametric paired t test for comparing prebreakthrough and postbreakthrough infection responses (B). *P < .03; **P < .002; ***P < .001. The average fold change in AI occurring with breakthrough infection is indicated for each Pre-Post pair, represented by dots. Each symbol represents a single participant; bars, geometric mean; and error bars, geometric standard deviations. Samples with an AI <35, indicated by the dotted line, are considered to have low avidity.
Figure 7.
Avidity profile of antibodies from individuals of the “breakthrough infection” group against Omicron BA.1 antigens, with immunoglobulin (Ig) G, IgM, and IgA avidity, expressed as avidity index (AI), against recombinant spike and receptor-binding domain (RBD) antigens of Omicron BA.1 (n = 24). BA.1 refers to the sequence of the first isolate of the Omicron lineage. A parametric paired t test was performed to compare prebreakthrough (Pre) and postbreakthrough (Post) infection responses. ***P < .002; ***P < .001. Each symbol represents a single participant; bars, geometric means; and error bars, geometric standard deviations. Samples with AI <35, indicated by horizontal dotted lines, are considered to have low avidity.
Finally, we evaluated the correlation between the AI and viral neutralization using serum samples from the breakthrough infection group. We detected a moderate correlation between neutralization as measured by inhibitory dilution 50% (ID50) and IgG AI against the ancestral spike; however, the correlation between virus neutralization and IgA AI against the ancestral RBD or the spike was weaker (Supplementary Figure 7). Three samples displayed low neutralization titers (ID50, <100) but high RBD IgG antibody avidity (AI, >50), which suggests the presence—in some individuals—of high-affinity nonneutralizing antibodies. Importantly, most of the strongly neutralizing samples displayed IgG AIs of >50 against RBD and the spike, indicating that these individuals have spike-reactive neutralizing antibodies with high avidity. Overall, our results indicate that the magnitude and antigen specificity of the IgG responses against the spike and the RBD induced after SARS-CoV-2 infection—with exception of NP antibodies—correlates well with antibody avidity and that vaccination increases neutralizing antibodies that display high avidity.
DISCUSSION
In the current work, we evaluated the binding and avidity profiles of different antibody isotypes in individuals with variable histories of preexisting immunity, including some who had an Omicron BA.1 infection despite having received 3 mRNA vaccine doses. We characterized the antibody responses against the RBD, spike, and NP from the ancestral SARS-CoV-2 strain and against the RBD and spike from the BA.1 Omicron sublineage. Overall, our data agree with previous reports indicating that levels of IgM and IgA antibodies are lower than those for IgG after infection or vaccination [23, 24].
Notwithstanding, we identified distinct antibody signatures. First, individuals in the convalescent group and those with lower levels of preexisting immunity, displayed the highest levels of IgM, likely owing to a de novo response given by the first encounter to SARS-CoV-2 antigens [25]. Second, in both the vaccination and breakthrough infection groups, levels of spike- and RBD-reactive IgA, mostly against the ancestral virus, were remarkably higher in individuals exposed to the virus, by a either primary or a breakthrough infection. Although data related to the boosting effect of IgA antibodies are scarce, a few reports suggest that there is antibody boost during SARS-CoV-2 vaccination [26, 27], as has been described for other vaccine antigens [28]. Importantly, serum IgA has been shown to appear early during a primary infection and displays potent neutralizing activity [24]. Hence, the serum IgA detected after breakthrough infections might represent a significant arm of immunity to tackle the virus and prevent severe disease.
Interestingly, individuals from the breakthrough infection group who received 2 doses of an mRNA vaccine without prior SARS-CoV-2 infections presented the highest induction of IgG against the Wuhan-1 and Omicron BA.1 spikes and RBDs after breakthrough infection. We hypothesize that a less antigen-experienced repertoire of B cells engages conserved epitopes between ancestral SARS-CoV-2 strains and Omicron variants and is responsible for the majority of the response observed. In addition, a very small fraction of naive B cells would be able to engage unique epitopes on the Omicron BA.1 spike. However, elucidation of these mechanisms would require the study of B cells at the monoclonal level, and the analysis of the evolution of these responses by the sequences of the rearranged variable-domain genes [29, 30].
In general, antibody avidity was correlated with the number of exposures to SARS-CoV-2 infection or vaccination. In particular, avidity increased after breakthrough infection for the majority of the antigens tested. Interestingly, similar to the magnitude of the antibody response, antibody avidity was higher after breakthrough infection in individuals without prior infection, and this increased avidity was detected against the spike and RBD proteins from both Wuhan-1 and Omicron BA.1. Again, this suggests that the affinity maturation of antibodies against broadly conserved epitopes can be shaped by the number and nature of the exposures over time.
Similar to other groups [31, 32], we detected a good correlation between binding of different antibody isotypes to the spike or the RBD and virus neutralization, with the highest correlation detected for IgG. We found that individuals with high virus neutralization titers display high IgG avidity; however, few individuals with these characteristics were present in the convalescent group, comprising SARS-CoV-2 infected individuals without prior vaccination. This is in line with incomplete avidity maturation after a primary SARS-CoV-2 infection versus vaccination [33]. We detected 3 individuals with high IgG avidity and low IgG neutralization titers (ID50, <100), suggesting the presence of nonneutralizing antibodies with high-affinity maturation that result in increased avidity at the polyclonal level.
In contrast to the antibody responses elicited against the spike protein during SARS-CoV-2 infection, which are of high magnitude and long-lasting, the levels of antibodies against NP are typically lower in magnitude, and the rate of antibody decay over time seems to be significantly lower [17]. In the current study, we looked at NP antibody levels in convalescent individuals from a few weeks up to 3 months after a primary infection and after breakthrough infections. As expected, NP reactivity was detectable and relatively high after a primary infection; however, the avidity of the antibodies induced was very low. Importantly, we found that the magnitude of the anti-NP antibody response increased after breakthrough infection, but the boosted anti-NP antibodies displayed low avidity.
In summary, we tested the binding and avidity profiles of serum antibodies of different isotypes against distinct antigens of SARS-CoV-2, including the spike, RBD, and NP. Our analyses allowed us to identify distinct antibody signatures based on the infection and/or vaccination exposure histories.
Our study had some limitations. First, the timing from infection to the first vaccine dose is not available for all participants included in the study. Second, we lack samples collected before the first SARS-CoV-2 infection to assess the serostatus of the participants. Third, the size is limited for some groups, especially in the breakthrough infection group. Although statistical analyses were performed assuming a normal distribution—given by the larger groups—normality could not be assessed in the smaller groups (ie, n = 3). Fourth, the timing of sample collection was not identical among the different groups, particularly among the breakthrough infection groups, in which the average timing of sample collection after breakthrough infection ranged from 15 to 30 days. Owing to the relatively low sample size in each of the stratified groups, we did not adjust for age or days since last exposure. Unfortunately, these factors are not in our control, given that we worked with samples available in our cohorts. Studies with larger sample sizes focused on selected populations would be ideal to explore the outcomes of this work.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Gagandeep Singh, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Anass Abbad, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Johnstone Tcheou, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Demodara Rao Mendu, Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Adolfo Firpo-Betancourt, Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Charles Gleason, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Komal Srivastava, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Carlos Cordon-Cardo, Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Viviana Simon, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Division of Infectious Diseases, Department of Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA; The Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Juan Manuel Carreño, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA; Center for Vaccine Research and Pandemic Preparedness, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Notes
Acknowledgments. The authors thank all participants in the longitudinal PARIS (Protection Associated with Rapid Immunity to SARS-CoV-2) study for their generous and continued support of research. They also thank D. Noah Sather, PhD from the University of Washington for providing the BA.1 spike protein used in this study and Shu Horiuchi from the NYU Grossman School of Medicine for thoroughly revising this work.
The authors also thank the members of the PARIS study group: Ariel Raskin, Dominika Bielak, Temima Yellin, Angela A. Amoako, Dalles Andre, Maria C. Bermúdez-González, Gianna Cai, Christian Cognigni, Giulio Kleiner, Neko Lyttle, Jacob Mauldin, Brian Monahan, Annika Oostenink, Aria Rooker, Ashley Salimbangon, and Morgan Van Kesteren.
Author contributions. F. K. and J. M. C. conceptualized the study; D. R. M., A. F. B., C. G., K. S., and the PARIS study group enrolled participants, collected data, evaluated surveys and provided biospecimen and metadata; G. S., A. A., and J. M. C performed experiments; G. S., A. A., J. T., and J. M. C. analyzed data; C. C. C., V. S., F. K., and J. M. C. administered the project; V. S. and F. K. provided resources; G. S., A. A., V. S., F. K., and J. M. C. wrote original draft. All authors reviewed, edited, and approved the final version of the manuscript and have had access to the raw data. Members of the PARIS study group collected, processed, stored biospecimen, curated metadata, and assisted with serological antibody measurements.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. Open Access publication charges for this article were paid from institutional funds.
Financial support. This work was supported by the Serological Sciences Network (SeroNet), in part with federal funds from the National Cancer Institute, National Institutes of Health (contract 75N91019D00024; task order 75N91021F00001; by the Centers of Excellence for Influenza Research and Surveillance (contract HHSN272201400008C), the Centers of Excellence for Influenza Research and Response (contract 75N93021C00014), and the Collaborative Influenza Vaccine Innovation Centers (contract 75N93019C00051); and by institutional funds.
References
- 1. Wajnberg A, Amanat F, Firpo A, et al. . Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020; 370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181: 281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li W, Moore MJ, Vasilieva N, et al. . Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Legros V, Denolly S, Vogrig M, et al. . A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol 2021; 18:318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carreno JM, Alshammary H, Singh G, et al. . Evidence for retained spike-binding and neutralizing activity against emerging SARS-CoV-2 variants in serum of COVID-19 mRNA vaccine recipients. EBioMedicine 2021; 73:103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muecksch F, Weisblum Y, Barnes CO, et al. . Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity 2021; 54:1853–68.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eisen HN. Affinity enhancement of antibodies: how low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol Res 2014; 2:381–92. [DOI] [PubMed] [Google Scholar]
- 8. Klasse PJ. How to assess the binding strength of antibodies elicited by vaccination against HIV and other viruses. Expert Rev Vaccines 2016; 15:295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaspar EB, De Gaspari E. Avidity assay to test functionality of anti-SARS-Cov-2 antibodies. Vaccine 2021; 39:1473–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim W, Zhou JQ, Horvath SC, et al. . Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature 2022; 604:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khurana S, King LR, Manischewitz J, Coyle EM, Golding H. Novel antibody-independent receptor-binding SPR-based assay for rapid measurement of influenza vaccine potency. Vaccine 2014; 32:2188–97. [DOI] [PubMed] [Google Scholar]
- 12. Nachbagauer R, Feser J, Naficy A, et al. . A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med 2021; 27:106–14. [DOI] [PubMed] [Google Scholar]
- 13. Benner SE, Patel EU, Laeyendecker O, et al. . SARS-CoV-2 antibody avidity responses in COVID-19 patients and convalescent plasma donors. J Infect Dis 2020; 222:1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amanat F, Stadlbauer D, Strohmeier S, et al. . A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carreno JM, Alshammary H, Tcheou J, et al. . Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2022; 602:682–8. [DOI] [PubMed] [Google Scholar]
- 16. Stadlbauer D, Amanat F, Chromikova V, et al. . SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 2020; 57:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carreno JM, Mendu DR, Simon V, et al. . Longitudinal analysis of severe acute respiratory syndrome coronavirus 2 seroprevalence using multiple serology platforms. iScience 2021; 24:102937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amanat F, White KM, Miorin L, et al. . An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol 2020; 58:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klein SL, Pekosz A, Park HS, et al. . Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest 2020; 130:6141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo YR, Chakraborty I, Yun C, Wu AHB, Lynch KL. Kinetics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody avidity maturation and association with disease severity. Clin Infect Dis 2021; 73:e3095–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moura AD, da Costa HHM, Correa VA, et al. . Assessment of avidity related to IgG subclasses in SARS-CoV-2 Brazilian infected patients. Sci Rep 2021; 11:17642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaw AE, Burman A, Asfor A, et al. . Avidity of polyclonal antibodies to foot-and-mouth disease virus in bovine serum measured using bio-layer interferometry. Viruses 2022; 14:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruggiero A, Piubelli C, Calciano L, et al. . SARS-CoV-2 vaccination elicits unconventional IgM specific responses in naive and previously COVID-19-infected individuals. EBioMedicine 2022; 77:103888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sterlin D, Mathian A, Miyara M, et al. . IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med 2021; 13:eabd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones K, Savulescu AF, Brombacher F, Hadebe S. Immunoglobulin M in health and diseases: how far have we come and what next? Front Immunol 2020; 11:595535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zurac S, Nichita L, Mateescu B, et al. . COVID19 vaccination and IgG and IgA antibody dynamics in healthcare workers. Mol Med Rep 2021; 24:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mao T, Israelow B, Suberi A, et al. . Unadjuvanted intranasal spike vaccine booster elicits robust protective mucosal immunity against sarbecoviruses. Science. 2022;378:eabo2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mascart-Lemone F, Duchateau J, Conley ME, Delacroix DL. A polymeric IgA response in serum can be produced by parenteral immunization. Immunology 1987; 61:409–13. [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt AG, Do KT, McCarthy KR, et al. . Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep 2015; 13:2842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moody MA, Zhang R, Walter EB, et al. . H3n2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 2011; 6:e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klingler J, Weiss S, Itri V, et al. . Role of immunoglobulin M and A antibodies in the neutralization of severe acute respiratory syndrome coronavirus 2. J Infect Dis 2021; 223:957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dispinseri S, Secchi M, Pirillo MF, et al. . Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun 2021; 12:2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Struck F, Schreiner P, Staschik E, et al. . Vaccination versus infection with SARS-CoV-2: establishment of a high avidity IgG response versus incomplete avidity maturation. J Med Virol 2021; 93:6765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.