Determining the molecular pathways by which supraphysiological androgens promote growth arrest and treatment responses in prostate cancer provides opportunities for biomarker-selected clinical trials and the development of strategies to augment responses.
Abstract
The androgen receptor (AR) pathway regulates key cell survival programs in prostate epithelium. The AR represents a near-universal driver and therapeutic vulnerability in metastatic prostate cancer, and targeting AR has a remarkable therapeutic index. Though most approaches directed toward AR focus on inhibiting AR signaling, laboratory and now clinical data have shown that high dose, supraphysiological androgen treatment (SPA) results in growth repression and improved outcomes in subsets of patients with prostate cancer. A better understanding of the mechanisms contributing to SPA response and resistance could help guide patient selection and combination therapies to improve efficacy. To characterize SPA signaling, we integrated metrics of gene expression changes induced by SPA together with cistrome data and protein-interactomes. These analyses indicated that the dimerization partner, RB-like, E2F, and multivulval class B (DREAM) complex mediates growth repression and downregulation of E2F targets in response to SPA. Notably, prostate cancers with complete genomic loss of RB1 responded to SPA treatment, whereas loss of DREAM complex components such as RBL1/2 promoted resistance. Overexpression of MYC resulted in complete resistance to SPA and attenuated the SPA/AR-mediated repression of E2F target genes. These findings support a model of SPA-mediated growth repression that relies on the negative regulation of MYC by AR leading to repression of E2F1 signaling via the DREAM complex. The integrity of MYC signaling and DREAM complex assembly may consequently serve as determinants of SPA responses and as pathways mediating SPA resistance.
Significance:
Determining the molecular pathways by which supraphysiological androgens promote growth arrest and treatment responses in prostate cancer provides opportunities for biomarker-selected clinical trials and the development of strategies to augment responses.
Introduction
The signaling program regulated by the androgen receptor (AR) is an important therapeutic target for the treatment of metastatic prostate cancer (mPC) as it mediates critical cell survival, proliferation, and metabolic functions (1, 2). Treatment strategies center on suppressing AR signaling, either by androgen deprivation therapy (ADT), and/or administering AR signaling inhibitors (ARSI) including competitive antagonists (3–6). Initially these approaches are highly effective, resulting in multiyear responses and disease control. However, resistance to ADT and ARSI treatment is nearly universal, leading to a clinical state termed castration resistant prostate cancer (CRPC). In this setting, molecular assessments of resistant prostate cancers have determined that the vast majority of these cancers retain or regain AR signaling, a situation usually accompanied by activating AR mutations or structural alterations involving AR enhancer and/or gene body amplifications that serve to maintain AR activity in low androgen environments (7–10). These findings suggest that AR signaling remains a therapeutic target in the context of ADT/ARSI-resistant disease progression.
Paradoxically, an approach demonstrating clinical responses in patients resistant to ADT/ARSI therapy involves administering high concentrations of AR-agonists. Although treating prostate cancer cells or patients with CRPC with testosterone concentrations that approximate physiologic levels found in adult men results in cell proliferation and tumor progression (11), exposing CRPCs to supraphysiological levels of steroidal androgens, or selective AR modulators (SARM), can potently suppress growth and maintain differentiation (11–15). Clinically, this approach has been explored using a strategy termed bipolar androgen therapy (BAT; ref. 16), which has shown promise in clinical trials (17–19). In BAT, testosterone is administered in monthly cycles with low ligand levels alternating with high supraphysiological androgen (SPA) concentrations.
The molecular mechanisms that govern responses to SPA are not well understood (20). Notably, the normal AR cistrome includes both transcriptionally activated and repressed target genes. Prior studies have characterized cellular responses to SPA, including G1 cell-cycle arrest and reduced transcription of genes regulated by RB1/E2F and MYC (12, 13, 21). The AR directly binds to a subset of E2F1 target genes, including those involved in cell-cycle progression (22, 23). In addition, RB1 has been reported to mediate repressive transcriptional functions of AR with RB1 knockdown, attenuating the growth inhibitory effects of SPA (22, 23). The relationship between AR and MYC signaling is complex. AR activation can repress MYC expression and MYC upregulation can repress components of the AR program (24). As MYC also functions to promote cell-cycle progression, the growth arrest phenotype resulting from SPA may occur in part via MYC (21). Given this, AR-mediated gene repression is likely a key determinant of response to SPA, though key relationships between the AR, RB1/E2F, and MYC signaling remain poorly defined. Understanding these interactions may have clinical relevance in the identification of patients with CRPC likely to respond or resist SPA treatment.
In this study, we sought to identify the mechanisms by which SPA represses prostate cancer growth. Using integrative analyses of RNA-seq, protein-interactome, and cistrome data, we determined that the dimerization partner, RB-like, E2F, and multivulval class B (DREAM) complex mediates growth repression and downregulation of E2F targets in response to SPA. Notably, mPCs with complete genomic loss of RB1 responded to SPA treatment and AR-mediated suppression of MYC was not affected by loss of RB-family members. Overexpression of MYC promoted complete resistance to SPA and AR-mediated repression of E2F target genes. Collectively, these data indicate that AR regulates cell-cycle and E2F gene expression through MYC signaling and the DREAM complex.
Materials and Methods
In vitro models and cell culture
LNCaP cells were obtained from ATCC (CRL-1740) and cultured in RPMI1640 (Thermo Fisher Scientific, no. 11835030) with 10% FBS (Thermo Fisher Scientific, no. 10437–02). TP53 and RB1 knockout cells were generated using CRISPR/CAS9 engineering in a previous study (25). Overexpression of MYC and RBL2 was achieved using a custom vector made by replacing the promoter of pLenti6.3 (Thermo Fisher Scientific, no. V53306) with EF1α and cloning in the MYC or RBL2 ORF from a custom entry vector. RB1 overexpression was achieved by direct transduction into cells using an RB1-overexpressing lentivirus (Genecopoeia, no. LPP-B0065-Lv105–200-S). siRNA knockdowns were performed using the following siRNAs supplied by Qiagen: RBL2, no. SI02664473; RBL1, no. SI02629921; and AllStars Negative Control, no. 1027281. siRNAs were delivered into cells using HiPerFect Transfection Reagent (Qiagen, no. 301705), according to the manufacturer's protocol.
Isogenic knockouts were generated by transducing DKO-B72 cells with the plentiCRISPRv2 lentivector (Addgene, no. 52961) with the protospacer sequences: RBL2:CATGAGCGAAAGCTACACGC or RBL1:AACAGATCCTTAACACTGCA. 4xKO lines were generated by knocking out RBL2 in the TKO.L1–32 line. Engineered cell pools were single cell cloned and screened by Western blot. Cultures were maintained in a 5% CO2 incubator at 37°C. Cells were tested for Mycoplasma and validated by the DNA Diagnostic Center and were used within 20 passages of receipt.
In vitro cell count assay
For cell counting assays, 150k cells were plated into 12-well dishes in 1 mL of growth media. An additional 1 mL of media was added with 2× drug concentration for 1× final drug concentration. After 5 days in culture, cells were harvested with trypsin and counted on the Vi-CELL cell counter (Beckman-Coulter). Each sample was represented in triplicate.
Dose-response assay
A total of 5K cells were plated in 96-well black-walled tissue culture–coated plates (Corning, catalog no. 3764BC) in 50 μL of normal growth media. A 2× concentration drug dilution series was added to the wells in another 50 μL media. Each sample and dose-point was represented in quadruplicate. Growth curves were normalized to the lowest dose point. After four days Celltiter-Glo (Promega, no. G7572) was measured on the Synergy H1 microplate reader (BioTek).
RNA sequencing
RNA was harvested using the RNeasy Mini Kit with incorporation of DNAse treatment (Qiagen). Each genotype/knockout was represented by two separate clones. Each sample group was represented by two biological replicates. Library preparation was performed as described previously (13). Sequence reads were aligned to the h38 human genome using STAR.v2.7.3a. All subsequent analyses were performed in R. Gene-level abundance was quantitated using GenomicAlignments. Differential expression was assessed using limma and used the Benjamin–Hochberg FDR adjustment. Gene expression results were ranked by their limma statistics and used to conduct Gene Set Enrichment Analysis (GSEA), utilizing the pathways from within the MSigDBv7.5.1. Single-sample enrichment scores were calculated using GSVA with default parameters using genome-wide log2 fragments per kilobase of transcript per million mapped read (FPKM) values as input and the 10 signatures listed in Supplementary Table S1.
Cleavage under targets and release using nuclease assays (CUT&RUN)
For each CUT&RUN sample, 5 × 106 cells were cultured for 48 hours with drug or vehicle control in growth media, 10% FBS in RPMI1640 (Thermo Fisher Scientific, catalog no. SH3006803). Each sample was represented by biological triplicates. Samples were processed using the low-salt protocol (26) and pAG-MNase (EpiCypher, SKU:15–1016). Antibodies used for CUT&RUN were: RB1 (Thermo Fisher Scientific, no. PA5–27215), RBL1 (Invitrogen, no. PA5–84228), RBL2 (Atlas Antibodies, no. HPA019703), and IgG (CST, no. 3900S). DNA libraries were prepared using MicroPlex Library Preparation Kit v2 (Diagenode, catalog no. C05010014) then sequenced on the Illumina HiSeq 2500, 50bp paired-end reads. Sequence reads were aligned to h38 using TopHat.v2 (27). Peaking calling was performed using MACS2. Differentially bound peaks were determined using DiffBind. Peak annotation was performed using ChIPseeker. Transcription factor similarity scoring was performed using GIGGLE (28). Peaks were visualized using IGV.
Chromatin immunoprecipitation sequencing
For chromatin immunoprecipitation sequencing (ChIP-seq) sample preparation, 1 × 107 cells were fixed, and their nuclei isolated using the iDeal ChIP-seq Kit for Transcription Factors (Diagenode, no. C01010170), following the manufacturer's protocol. Libraries were prepared for sequencing using MicroPlex Library Preparation Kit v3 (Diagenode, no. C05010014) and 24 Dual indexes for MicroPlex Kit v3 (Diagenode, no. C05010003). The antibodies used were E2F1 (BD Biosciences, no. 554213), LIN9 (Bethyl Laboratories, A300-BL2981), LIN37 (Bethyl Laboratories, A300-BL2983), and IgG (Cell Signaling Technology, no. 3900S). Chromatin was sonicated using the Covaris M220. Sequencing and analysis was performed using the same methods for CUT&RUN except BWA was used to align sequence reads. Pathway and gene ontology (GO) analysis was performed using PantherGO.
Immunoblotting
Western blots were run using NuPAGE 4%–12% Bis-Tris gels (Thermo Fisher Scientific, no. NP0321) with MOPS buffer (Thermo Fisher Scientific, no. NP0001) and transferred to nitrocellulose membranes (Thermo Fisher Scientific, no. LC2000) using NuPAGE transfer buffer (Thermo Fisher Scientific, no. NP0006) according to the manufacturer's instructions. Chemiluminescence was achieved using Supersignal Pico (Thermo Fisher Scientific, no. PI34080). The following antibodies were used for Western blots: GAPDH (Cell Signaling Technology, no. 2118L), RB1 (Cell Signaling Technology, no. 9309S), RBL1 (Cell Signaling Technology, no. 89798S), RBL2 (Cell Signaling Technology, no. 13610S), TP53 (Santa Cruz Biotechnology, no. sc-126), AR (Cell Signaling Technology, no. 5153S), and p27 (Abcam, ab32034).
Cell-cycle analysis
For cell-cycle analysis, 1 × 106 cells were cultured for 48 hours with drug or vehicle control. Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific, no. C10420) was used as the S-phase marker and FxCycle Violet Stain (Thermo Fisher Scientific, no. F10347) was used to measure DNA-content. Stained cells were measured by flow cytometry using the Sony SH800 cytometer.
RIME
For RIME analysis, 107 LNCaP cells were cultured for 48 hours in normal growth media with 10 nmol/L R1881 or EtOH control. Both treatment conditions are represented by three biological replicates. Cells were fixed, permeabilized, and immunoprecipitated as published previously (29). Chromatin was sonicated using the Covaris M220 then immunoprecipitated using antibodies to LIN37 (Bethyl Laboratories, A300-BL2983) or IgG (Cell Signaling Technology, no. 3900S). Mass-spec data were analyzed using Proteome Discoverer (Thermo Fisher Scientific) and filtered using the CRAPome repository to remove likely contaminants (30).
Statistical analysis
Statistical analyses pertaining to each figure are included within the figure legends. For categorical comparisons, we performed Fisher exact test or for pairwise comparisons using the fisher.multcomp function in R with Benjamini–Hochberg multiple testing correction. Gene expression (log2 FPKM) or GSVA signature scores were compared using Wilcoxon-rank tests with Benjamini–Hochberg multiple testing correction. Pearson correlation coefficient was used to study the relationships between variables shown in scatterplots using the cor.test function in R.
Data availability
The RNAseq data used in this study are available in the Gene Expression Omnibus repository (GEO) under accession number GSE225481. The CUT&RUN and ChIP-seq data generated by the authors are publicly available and were deposited in the Gene Expression Omnibus under study numbers: GSE228897, GSE228895, and GSE228896. Publicly available data generated by others were used by the authors. The public data analyzed in this study were obtained from the GEO: GSE147250, GSE135879, GSE225481, GSE124704, and GSE157107.
Results
SPAs repress the proliferation of prostate cancers with loss of RB1 and TP53
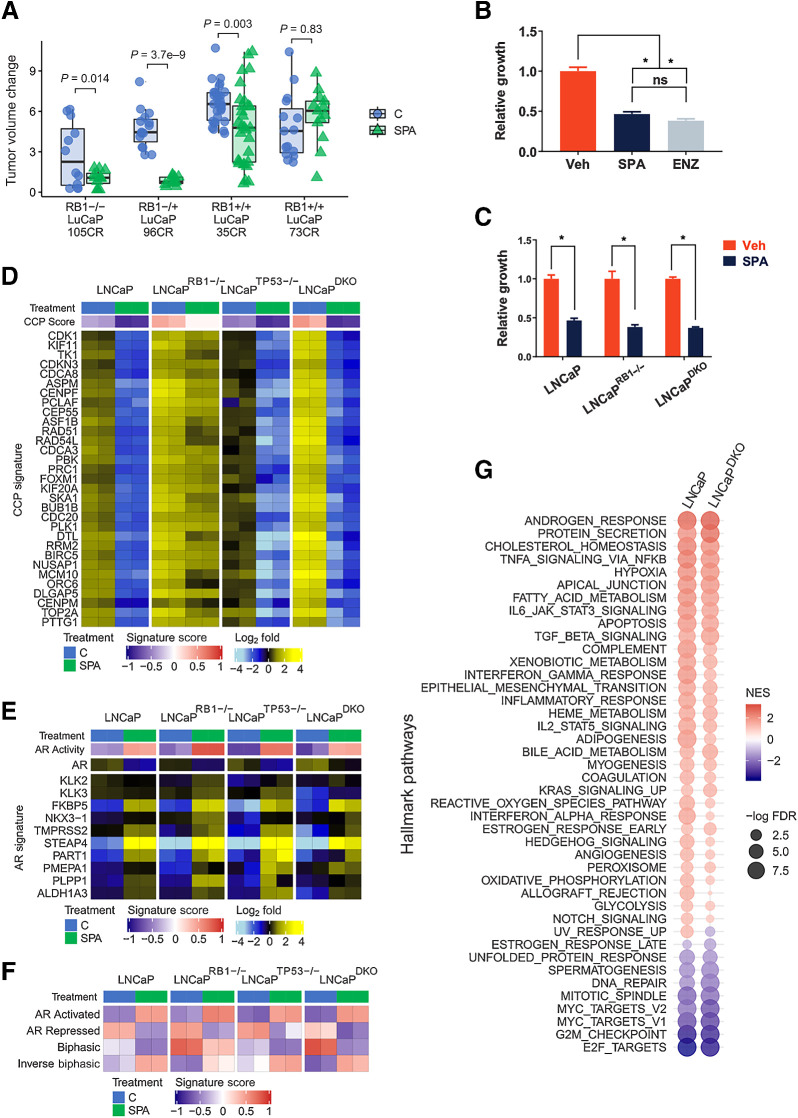
SPA exhibits variable antitumor responses across prostate cancer cell lines, patient-derived xenografts (PDX) and patients with marked growth repression in some, and de novo resistance in others (12, 16–18, 31, 32). Other than a requirement for AR expression, strong determinants and biomarkers of response have not been identified. Prior studies determined that AR signaling, particularly those functions involving gene repression, are influenced by loss of the RB1 tumor suppressor (22, 33). SPA has previously been shown to repress E2F target genes, and RB1 loss in prostate cancer models can attenuate the growth repressive effects of SPA (12). We reanalyzed in vivo prostate cancer PDX responses to SPA based on tumor RB1 status and found that SPA inhibited the growth of tumors with intact RB1 as well CRPCs with biallelic RB1 loss (Fig. 1A; Supplementary Fig. S1A). To confirm that RB1 loss alone does not substantially influence the growth repressive effects of SPA, we generated isogenic LNCaP sublines with CRISPR/Cas9 deletion of RB1 (LNCaPRB1−/−). Confirming prior studies, treatment with 10 nmol/L R1881, a potent steroidal androgen (hereafter SPA) repressed the growth of RB1-intact LNCaP cells by 53% relative to control (P = 8 × 10−5), which approximated the growth inhibitory effects of the AR antagonist enzalutamide (ENZ; Fig. 1B). SPA also inhibited the growth of LNCaPRB1−/− cells by 62%, (P = 4.6 × 10−4; Fig. 1C). Because homozygous RB1 loss predominantly occurs with TP53 loss in a clinical context (25), we tested isogenic lines with double knockout of RB1 and TP53 (LNCaP-RB1−/−/TP53−/−; hereafter LNCaPDKO). SPA also inhibited the proliferation of cells harboring loss of both tumor suppressors with a 63% reduction in cell growth compared with control (P = 2 × 10−6; Fig. 1C).
Figure 1.
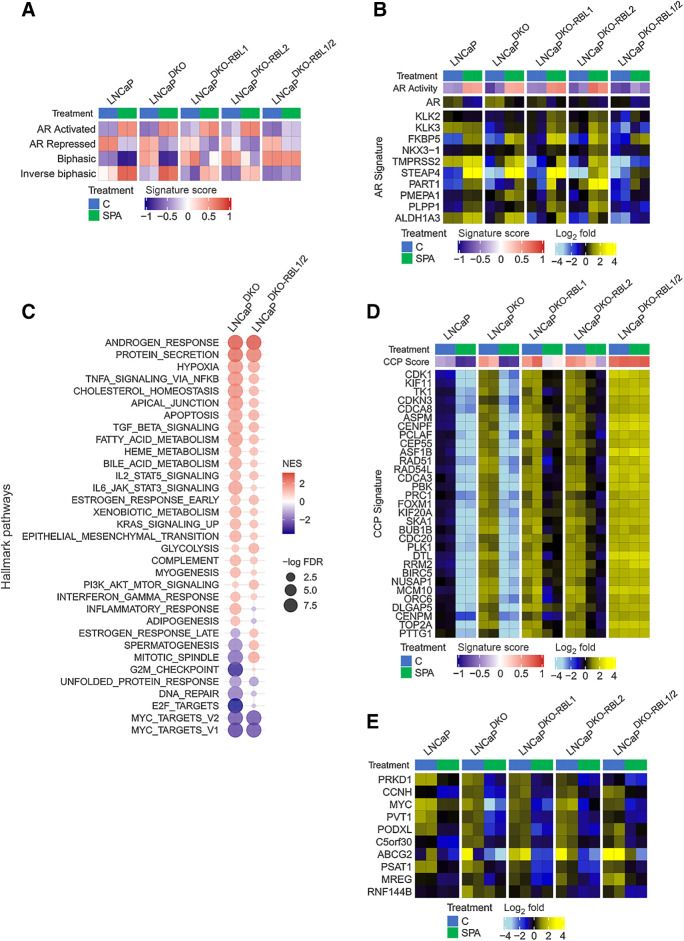
RB1 loss is not sufficient to mediate resistance to SPA. A, Growth responses of prostate cancer PDX to a 28-day course of supraphysiological testosterone. PDX lines were categorized by intact, monoallelic, or biallelic loss of RB1. Changes in tumor volume during the last 2 weeks of the study were compared by one-sided t tests with Benjamini–Hochberg-adjusted P values. B, Five-day growth assay comparing 10 nmol/L R1881 to EtOH vehicle control in LNCaP cells. C, Five-day growth assay comparing 10 nmol/L R1881 to vehicle control for LNCaP, LNCaP-RB1 knockout, and LNCaP RB1/TP53 double knockout (LNCaPDKO). D, The expression of cell-cycle progression (CCP) genes measured by RNA-seq in prostate cancer cells treated for 48 hours with vehicle control or 10 nmol/L R1881 (SPA). Data are shown as heatmaps of RNA-seq mean-centered log2 (FPKM) gene expression values. E, The expression of canonical AR-regulated genes measured by RNA-seq in prostate cancer cells treated for 48 hours with vehicle control or 10 nmol/L R1881 (SPA). F, Molecular signature (GSVA) scores determined by RNA-seq data. G, Plot of GSEA normalized enrichment scores (NES) of MSigDB Hallmark gene sets comparing 10 nmol/L R1881 (SPA)-induced gene expression changes in LNCaP and DKO cells. *, P < 0.05, by one-way ANOVA with Dunnett multiple-comparison test. Data represent the mean ± SD.
We performed RNA-seq to compare transcriptional changes associated with SPA on parental LNCaP cells and two independent LNCaPRB1−/− and LNCaPDKO clonal lines. Congruent with the growth repressive effects of SPA on each LNCaP genotype, transcripts comprising genes involved in cell-cycle progression were significantly repressed regardless of RB1 status, although RB1 loss attenuated SPA-mediated CCP score reduction (Fig. 1D). The AR program in LNCaP cells was induced by SPA in both LNCaPRB1−/− and LNCaPDKO lines (Fig. 1E).
A subset of AR-regulated genes exhibit a biphasic expression pattern: repression when AR activity is inhibited, upregulation at physiological androgen levels, and repression with high AR ligand levels (12). These patterns were also maintained in LNCaPRB1−/− and LNCaPDKO lines treated with SPA (Fig. 1F). In agreement, a comparison of GSEA scores for LNCaPDKO and parental LNCaP showed largely overlapping responses to SPA with enrichments of luminal prostate epithelial cell differentiation pathways including androgen response and protein secretion; and a reduction of mitotic signaling signatures including E2F targets, G2M checkpoint, DNA repair, mitotic spindle, and MYC targets (Fig. 1G).
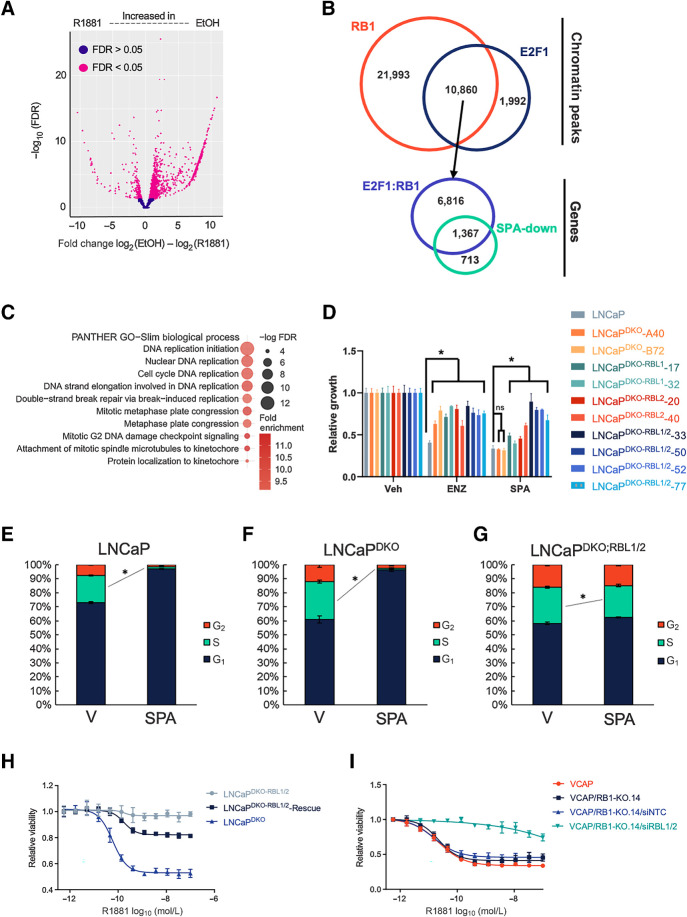
To explain the SPA-mediated downregulation of cell cycle and E2F-regulated gene expression in an RB1-null context, we performed ChIP-seq to determine SPA-induced changes to the E2F1 cistrome in LNCaPDKO cells. Differential binding analysis indicated that E2F1 localization to chromatin sites was broadly reduced in LNCaPDKO cells treated with SPA (Fig. 2A) compared with cells grown in typical LNCaP culture medium comprising 10% FBS with free T concentrations of ∼5 to 10 pmol/L, approximating the serum levels of men with CRPC (11). Given the relationship of AR signaling with PC proliferation, we sought to determine the prevalence of E2F targets among the genes that are repressed by SPA. To achieve this, we mapped the 12,819 E2F1 binding sites identified in LNCaPDKO cells by ChIP-seq under normal growth conditions and determined that E2F1 localized to promoters of 10,493 different genes. Of the 2,081 genes significantly repressed by SPA (FDR < 0.05), 1,477 (71%) of the genes were bound by E2F1 in normal growth conditions (Fig. 2B). Consistent with canonical E2F functions, the genes with regulatory sites occupied by E2F in normal conditions, but with diminished expression following SPA treatment, were enriched for genes involved in DNA replication, mitosis, and DNA-metabolism (Fig. 2C).
Figure 2.
SPA repression of E2F target genes and prostate cancer proliferation depends on RBL1 and RBL2. A, E2F1 ChIP-seq volcano plot showing differentially bound E2F1 binding sites between EtOH- and SPA-treated LNCaPDKORB1−/−;TP53−/− cells. B, Overlap between RB1-bound sites in LNCaP and E2F1-bound sites in LNCaPDKO cells. These E2F1:RB1 bound sites were mapped to promoters of genes, which were overlapped with genes transcriptionally repressed by SPA. C, Graph of over-represented GO categories in overlapping RB1/E2F-bound and SPA-repressed genes. D, Growth assay results for knockout and parental cells treated with 10 μmol/L ENZ or SPA for 5 days. E–G, Cell-cycle profile changes induced by SPA on LNCaP (E), LNCaPDKO (F), or LNCaPDKO;RBL1/2 (G) cells using flow cytometry to measure EdU incorporation (S-phase) and FxCycle violet staining (DNA-content). H, R1881 dose–response assay comparing LNCaPDKO cells with LNCaPDKO;RBL1/2 cells with and without ectopic expression of RBL2. I, R1881 dose–response assay on VCAP and the VCAPRB1–14 RB1-knockout clonal cell line transfected with siRNA to RBL1/2 or a nontargeting control (NTC). *, P < 0.05, by one-way ANOVA with Dunnett multiple-comparison test. Data represent the mean ± SD.
SPA-induced growth repression depends on DREAM complex members p107/RBL1 and p130/RBL2
Because many E2F1 targets were repressed by SPA in LNCaPDKO cells, despite the loss of RB1, we hypothesized that other RB-family members such as p107/RBL1 and p130/RBL2 may negatively regulate E2F1 target genes such as those involved in cell cycle in the context of SPA treatment. Instead of binding E2F1/2 directly, RBL1/2 bind to E2F4/5, dimerization partner (DP), and the MuvB complex—comprised of LIN9, LIN37, LIN52, LIN54, and RBBP4—to form the DREAM complex (34). The DREAM complex negatively regulates G1–S genes during quiescence by binding to E2F sites in promoters and preventing their activation (34, 35).
To evaluate the role of the DREAM complex in mediating the growth suppressive effects of SPA, we sought to disrupt the complex by deleting RBL1 and RBL2. Prior studies of DREAM activation implicate the TP53/p21 signaling pathway in supporting DREAM formation (36–38). As we found that SPA can repress growth in the absence of TP53 (Fig. 1C), we used LNCaPDKO cells to determine if AR mediates DREAM complex activation through a novel, TP53/RB1-independent, mechanism. Because RBL1 and RBL2 regulate largely overlapping gene sets, we used CRISPR/Cas9 to engineer isogenic triple knockout clones of RB1, TP53, and RBL1 (LNCaPDKO;RBL1) or RBL2 (LNCaPDKO;RBL2) and quadruple knockouts of RB1, TP53, RBL1, and RBL2 (LNCaPDKO;RBL1/2) to study the contribution of partial and complete DREAM complex disruption to SPA growth repression (Supplementary Fig. S1B).
To determine if RBL1 and RBL2 influenced differential sensitivities to SPA, we performed growth assays on the isogenic lines. We first evaluated cellular responses to ENZ, where we previously observed that loss of RB1 and TP53 attenuated ENZ growth repression (25). Consistent with our previous results, LNCaPDKO cells with RB1 and TP53 loss were more resistant to 10 μmol/L ENZ than parental LNCaP cells (Fig. 2D; Supplementary Fig. S1C). The loss of RBL1, RBL2, or both did not further increase ENZ resistance (Fig. 2D). As noted above, LNCaPDKO cells retained sensitivity to SPA with a 68% growth reduction over a 96-hour hour time span compared with vehicle treatment (P = 3.8 × 10−12), which was not different than LNCaP parental cells. However, RBL1 or RBL2 loss promoted resistance to SPA and the growth repressive effect was enhanced with combined RBL1 and RBL2 loss. Compared with the 68% growth repression with SPA in LNCaPDKO cells, SPA reduced the growth of LNCaPDKO;RBL1 by 56%, LNCaPDKO;RBL2 by 46%, and LNCaPDKO;RBL1/2 by only 21% (P = 1.4 × 10−6; Fig. 2D).
Cell-cycle analysis of parental LNCaP cells treated with SPA showed cycle arrest in the G1 phase (Fig. 2E). SPA also induced LNCaPDKO cells to arrest in the G1 phase (Fig. 2F). In agreement with the in vitro growth data, LNCaPDKO;RBL1/2 cells showed minimal, albeit significant, changes to the S-phase compartment going from 26% of cells in S-phase with vehicle to 23% of cells in S-phase with SPA (Fig. 2G). As with wild-type LNCaP cells, R1881 repressed the viability of LNCaPDKO cells at concentrations as low as 0.1 nmol/L while having little effect on LNCaPDKO;RBL1/2 cells (Fig. 2H). The resistance to SPA-mediated repressive effects observed in LNCaPDKO;RBL1/2 cells was rescued by the ectopic expression of RBL2 (Fig. 2H). The ectopic expression of RB1 did not rescue the growth repressive effects of SPA (Supplementary Figs. S1D and S1E). We also used CRISPR/Cas9 to delete RB1 in the VCaP prostate cancer cell model and generated several independent VCaPRB1−/− lines. SPA repressed the growth of parental VCaP cells as well as VCaP cells with RB1 loss, although the extent of repression varied by subline (Supplementary Figs. S1F and S1G). Of note, VCaP cells harbor a native frameshift insertion mutation (E607fs) in RBL2 (depmap.org). Consistent with previous studies (12, 22), knockout of RB1 appears to contribute to partial resistance to SPA in VCaP in some subclones. Nonetheless, siRNA-mediated knockdown of RBL1 and RBL2 induced strong resistance to SPA in all VCaPRB1−/− clones (Fig. 2I; Supplementary Fig. S1H).
Regulatory regions of cell-cycle genes repressed by SPA are bound by the DREAM complex
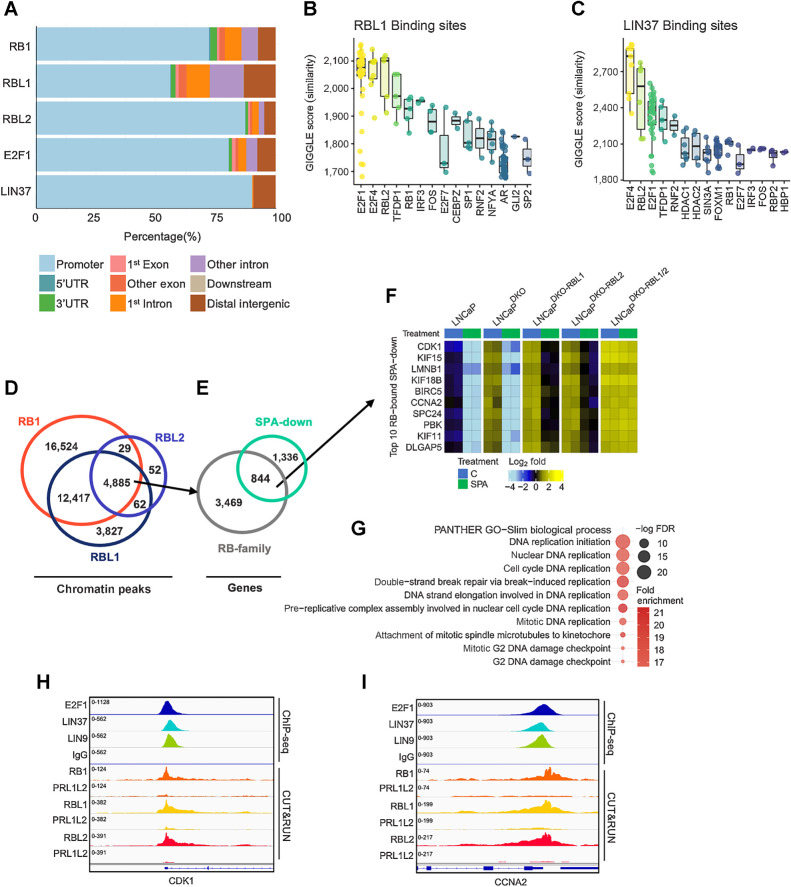
GO signatures of LNCaPDKO cells treated with SPA reflected a shift in differentiation to a quiescent, secretory luminal-like phenotype, and reduced expression of genes involved in nucleic acid and ribosome biosynthesis (Supplementary Fig. S2A). We next sought to confirm the direct contribution of DREAM complex members in SPA-mediated growth repression, specifically in the context of RB1 loss. We identified RBL1-bound and RBL2-bound sites by performing genome-wide CUT&RUN for RBL1 on LNCaPDKO;RBL2 and for RBL2 on LNCaPDKO;RBL1 cells treated for 48 hours with SPA. Because of the limited availability of suitable antibodies to human LIN37, we performed ChIP-seq for LIN37-bound chromatin sites using an antibody to the orthologous gene in Drosophila, mip40. To determine the contribution of nonspecific reads, we also performed CUT&RUN on LNCaPDKO;RBL1/2 cells as negative controls. Analyses to identify differential RB1 binding in parental LNCaP versus LNCaPDKO;RBL1/2 cells demonstrated that the vast majority of CUT&RUN peaks were on-target signal (Supplementary Fig. S2B). Peaks bound by RB1, RBL1, and RBL2 predominately mapped to gene promoters, which mirrored binding profiles of E2F1 and LIN37/mip40 (Fig. 3A). We used the GIGGLE platform to perform similarity analysis on the RBL1 and RBL2 cistromes to those from other published studies. The transcription factors exhibiting profiles most similar to both RBL1 and RBL2 peak profiles were E2F1, TFDP1, E2F4, and RBL2 (Fig. 3B; Supplementary Fig. S2C). The RBL1 and RBL2 cistromes were also highly similar to LIN9 and LIN37/mip40 cistromes determined by ChIP-seq performed on LNCaP cells (Fig. 3C; Supplementary Fig. S2D).
Figure 3.
Androgen-repressed cell-cycle genes bound by the DREAM complex. A, RB-family, E2F1, and LIN37 genomic binding sites determined by ChIP-seq from LNCaP cells and mapped to annotated regions. B, Boxplots of GIGGLE similarity scores for RBL1 ChIP-seq from LNCaP cells. C, Boxplots of GIGGLE similarity scores for LIN37 ChIP-seq from LNCaP cells. D, Overlapping CUT&RUN peaks bound by RB1 LNCaP cells, RBL1 in LNCaPDKO-RBL2 cells, and RBL2 LNCaPDKO-RBL1 cells. E, RB-family co-bound sites mapped to gene promoters and overlapped with SPA-repressed genes in LNCaPDKO cells (FDR < 0.05). F, Heatmap of mean-centered log2(FPKM) RNA-seq values for the top 10 androgen-repressed genes bound by all three RB-family members in LNCaP and isogenic gene knockout sublines. G, GO enrichment analysis on androgen repressed genes co-bound by RB-family members. H and I, Chromatin peaks for ChIP-seq datasets for E2F1 (LNCaPDKO), LIN37 (LNCaP), LIN9 (LNCaP), and IgG (LNCaPDKO); and for CUT&RUN peaks for RB1 (LNCaP), RBL1 (LNCaPDKO-RBL2), RBL2 (LNCaPDKO-RBL1), and LNCaP DKO-RBL1/2 (PRL1L2) as a negative control for each antibody on the promoters for SPA-repressed genes CDK1 (H) and CCNA2 (I).
We overlapped the RBL1, RBL2, and RB1 cistromes to determine high-confidence consensus sites (Fig. 3D and E). Although differing in their total number of called peaks, the sites of chromatin localization for all 3 RB-family members overlapped significantly: RBL1 peaks were concordant with 17,302 of 21,191 (82%) RB1 sties (FDR q < 0.05). RBL2 peaks were concordant with 4,885 of 5,028 (97%) sites occupied by RB1 and RBL2 (FDR q < 0.05; Fig. 3D). Overall, the 4,885 consensus RB1, RBL1, and RBL2 sites mapped to 4,313 gene promoters and of these, 844 corresponded to genes that were repressed by SPA in LNCaPDKO cells (FDR q < 0.05; Fig. 3E). The top 10 SPA-repressed DREAM targets include known cell-cycle-associated factors CDK1, survivin (BIRC5), cyclin A (CCNA2), and KIF11 (Fig. 3F). In agreement, we found GO categories related to mitosis and DNA metabolism to be highly enriched (Fig. 3G). Further, a comparison of LIN9, LIN37, and E2F1 binding with RB-family members showed both E2F/RB and DREAM complexes bind to promoters of key cell-cycle regulators repressed with SPA such as CDK1 (Fig. 3H) and CCNA2 (Fig. 3I). Collectively, these results nominate a cohort of genes regulated by the DREAM complex that arrest the cell cycle in response to SPA.
DREAM complex assembles on chromatin with the SIN3A–HDAC complex
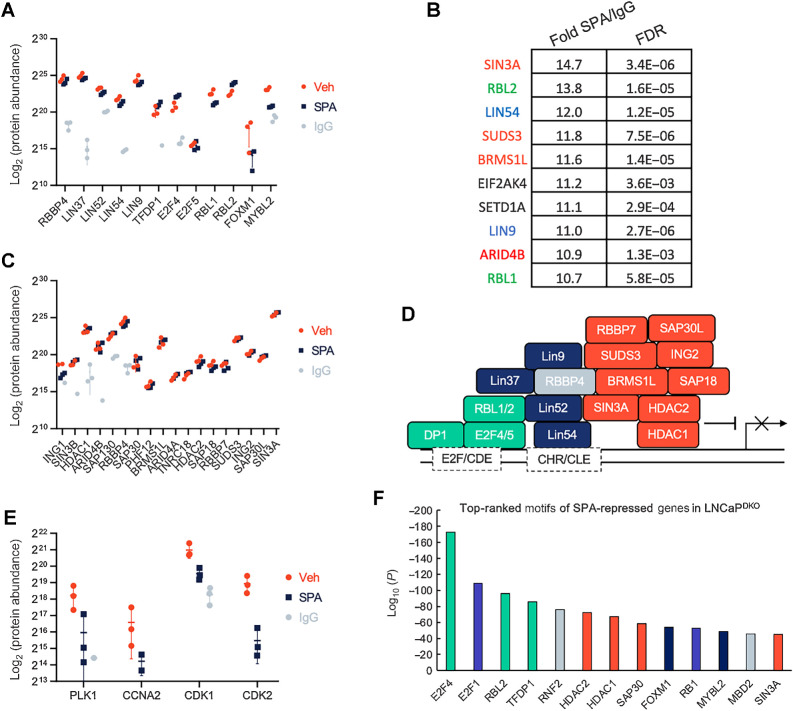
The analyses of chromatin binding of individual DREAM members demonstrating colocalization to common chromosomal regions suggested that the DREAM complex is assembled in the context of SPA exposure. To further evaluate this hypothesis and identify interacting factors that may be responsible for repression of DREAM targets, we performed rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) analysis (29) for LIN37, a DREAM/MuvB core complex member with suitable antibody reagents. All known members of the MuVB and DREAM complexes were detected by RIME and were significantly enriched over the IgG control (Fig. 4A). The 10 most enriched proteins by LIN37 RIME included members of the DREAM complex LIN9, LIN54, RBL1, and RBL2 (Fig. 4B). In contrast, MYBL2, a necessary factor for G2–M gene activation and part of the MuVB–MYLB2 complex, was depleted 1.8-fold in the SPA group relative to control (FDR = 0.06; P = 0.001).
Figure 4.
DREAM complex interacting proteins identified by LIN37 RIME. A, Log2-protein abundance scores of proteins enriched with LIN37-bound chromatin in LNCaPDKO cells treated with 10 nmol/L R1881 (SPA) or EtOH (Veh) control for the DREAM and MuvB-FOXM1/MYBL2 complexes. B, Top 10 enriched proteins by LIN37-bound chromatin precipitation of LNCaPDKO cells. Green, DREAM complex members; blue, MuvB core; red, SIN3A complex members. C, Log2 protein abundance scores of proteins enriched with LIN37-bound chromatin in LNCaPDKO cells treated with 10 nmol/L R1881 or EtOH control for the SIN3A complex. All proteins shown were significantly enriched (FDR < 0.05) over the IgG pulldown control. D, Diagram of DREAM/SIN3A repression complex members precipitated with LIN37-bound chromatin. E, Log2-protein abundance scores of proteins enriched with LIN37-bound chromatin in LNCaPDKO cells treated with 10 nmol/L R1881 or EtOH control for DREAM destabilizing factors. All proteins shown were significantly enriched (FDR < 0.05) over the IgG pulldown control. F, Top motifs of SPA-repressed genes in LNCaPDKO RNA-seq datasets as predicted by LISA analysis. MuvB core, dark blue; repressive DREAM components, green; SIN3A complex members, orange; E2F1/RB1, light blue; other, gray.
In addition to established DREAM complex members, LIN37 RIME also identified interactions with components of the SIN3A repressive complex (Fig. 4B and C). The entire SIN3A complex was significantly enriched over the IgG control for both treatment groups (Fig. 4C). Both the DREAM and SIN3A complexes contain RBBP4, which could facilitate the interaction between DREAM and the SIN3A histone deacetylase complex to effect chromatin silencing (Fig. 4D). Several cell-cycle progression proteins were reduced in SPA-treated cells versus controls, with CDK2 reaching statistical significance (P = 0.014; Fig. 4E). With some notable differences, the LIN37-interactomes are largely similar between the R1881 and vehicle groups—this may be explained by the observation that G1 is the dominant cell-cycle phase in both treatment conditions and the competing complex for LIN37, MuVB, is formed in G2, which only comprises a small percentage of cells in an unsynchronized population (Fig. 2F).
Other chromatin modifying proteins that regulate promoter and enhancer activation were also significantly enriched in LIN37 complexes including the RBBP5, SETD1A, and ASHL2 histone methyltransferase complex as well as SWI/SNF members SMARCA4, BRD2 (Supplementary Fig. S3A).
We next used Lisa (epigenetic Landscape In Silico deletion Analysis and the second descendent of MARGE) analysis as an orthogonal approach to predict transcription factors that regulate SPA-repressed genes identified by RNA-seq in the LNCaPDKO cells (39). The set of transcription factors predicted to regulate genes repressed by SPA largely mirrored the RIME results, with DREAM/MuVB, SIN3A, and E2F/RB1 members as the most significant contributors (Fig. 4F). In contrast, SPA-activated genes were predicted to be regulated by AR and AR-cofactors like FOXA1, HOXB13, and NKX3–1 (Supplementary Fig. S3B). Taken together, these data are consistent with the hypothesis that cell-cycle gene downregulation occurs via DREAM-mediated repression in RB1-null prostate cancer cells, potentially through recruitment of the SIN3A–HDAC complex.
SPA-mediated repression of MYC is insufficient to arrest prostate cancer growth in the context of RBL1 and RBL2 loss
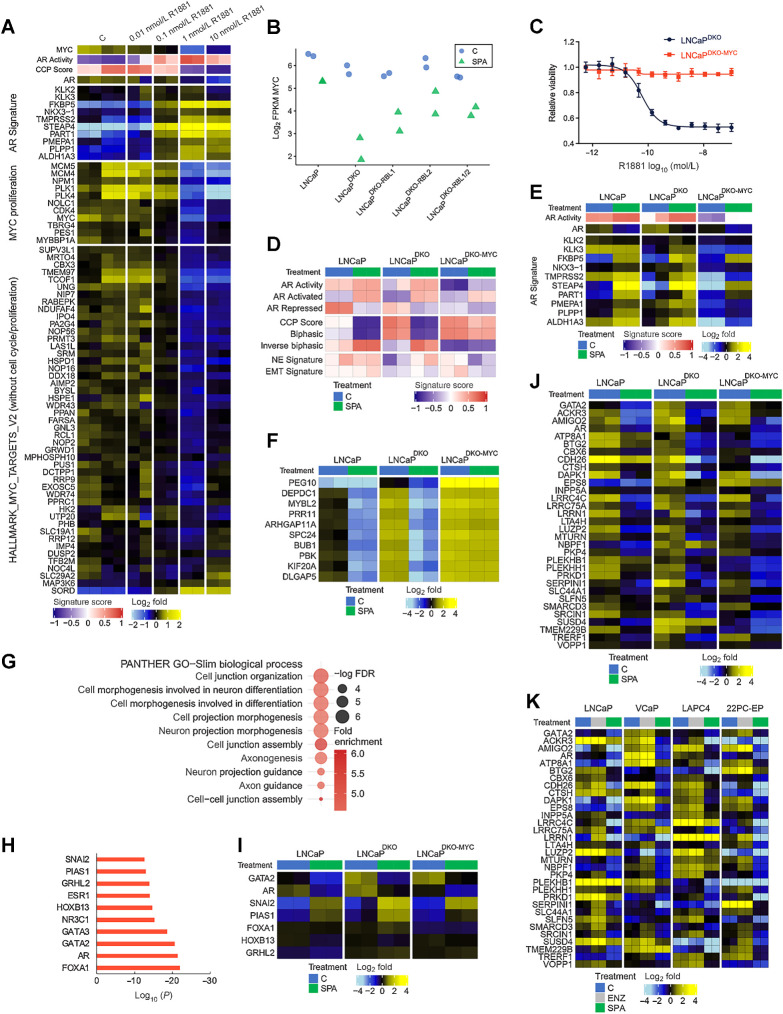
Although the majority of genes repressed by SPA were E2F/RB1 targets as determined by chromatin localization assays, roughly a third of genes repressed by SPA were not associated with E2F1/RB1 binding (Fig. 2B). To further study SPA-repressed genes that are E2F-independent, we performed RNA-seq on the LNCaP and LNCaP RB-family knockout models treated with SPA or vehicle control. GSEA of the RNA-seq transcript abundance measurements demonstrated consistent SPA-mediated activation and repression of AR-regulated gene programs in the LNCaPDKO, LNCaPDKO;RBL1, and LNCaPDKO;RBL2 models with moderately reduced activity in the LNCaPDKO;RBL1/2 lines (Fig. 5A). As expected, genes with biphasic regulation, which closely associate with cell-cycle gene sets (12), were constitutively upregulated in the LNCaPDKO;RBL1/2 lines (Fig. 5A). Expression levels of individual genes comprising AR-regulated gene signatures showed heterogenous effects of RBL1/2 loss on specific AR-activated genes (Fig. 5B): KLK3, ALDH1A3, and TMPRSS2 had lower levels of induction by SPA in the LNCaPDKO;RBL1/2 whereas FKBP5 was less affected by the additional loss of RBL1/2 (Fig. 5B). Because AR protein levels were maintained in LNCaPDKO;RBL1/2 cells (Supplementary Fig. S4A), the lower inducibility of KLK3 and other AR-regulated markers of prostate epithelial cells may be due to cell-cycle induced perturbations to the AR program (40, 41).
Figure 5.
Androgen-mediated repression of cell-cycle genes is dependent on RBL1 and RBL2. A, GSVA signature score heatmaps of gene expression signatures on RNA-seq data generated from LNCaP models treated with SPA (10 nmol/L R1881) for 48 hours. B, Heatmap of mean-centered log2 (FPKM) RNA-seq values for the androgen-upregulated AR Activity signature genes. LNCaP and LNCaP-DKO data are replotted from Fig. 1. C, Plot of GSEA enrichment scores of MSigDB Hallmark gene sets representing SPA-induced gene expression changes in LNCaPDKO and LNCaPDKO-RBL1/2 cells. D, Heatmap of mean-centered log2(FPKM) RNA-seq values for the cell-cycle progression (CCP) signature genes. LNCaP and LNCaP-DKO data are replotted from Fig. 1. E, Heatmap of mean-centered log2(FPKM) RNA-seq values for the top 10 SPA-repressed genes in the LNCaPDKO-RBL1/2 cells. LNCaP and LNCaP-DKO data are replotted from Fig. 1.
To broadly characterize signaling changes in LNCaPDKO;RBL1/2 cells treated with SPA, we performed GSEA to compare alterations in cellular pathways and regulatory programs following SPA treatment. As with parental LNCaP and LNCaPDKO cells, pathways of androgen response and protein secretion were upregulated in LNCaPDKO;RBL1/2 cells by SPA treatment (Fig. 5C). The major distinguishing pathway between cell genotypes was a lack of E2F pathway repression in LNCaPDKO;RBL1/2 compared with cells with intact RBL1/2 (Fig. 5C). Cell-cycle progression (CCP.31) signature genes, 15 of which overlap with the Hallmark E2F target gene set (42), showed diminishing SPA-mediated repression comparing LNCaPDKO with LNCaPDKO;RBL1 or LNCaPDKO;RBL2 and LNCaPDKO;RBL1 or LNCaPDKO;RBL2 with LNCaPDKO;RBL1/2 (Fig. 5D).
Of interest, the group of genes most repressed by SPA in LNCaPDKO;RBL1/2 cells included cMYC and cMYC-regulated genes such as the lncRNA PVT1 (Fig. 5E). Overall, gene sets comprising MYC targets were consistently reduced in LNCaP, LNCaPDKO, and LNCaPDKO;RBL1/2 cells following SPA, though the concordant repression of both MYC and proliferation were uncoupled in the context of RBL1/2 loss where MYC targets were repressed but cell-cycle-associated genes were not (Fig. 5C).
To further explore the effects of SPA on the repression of MYC activity and DREAM targets, we analyzed previously published datasets of SPA-treated PDX models (43) and cell lines (12) for changes in expression of MSigDB cell-cycle DREAM (35) and MYC (44) gene signatures in response to SPA. SPA treatment resulted in a repression of DREAM-target genes in responsive models as opposed to nonresponder models (Supplementary Figs. S4B and S4C).
Downregulation of MYC has been shown to attenuate cell proliferation in multiple prostate cancer models (45, 46) and prior studies have suggested that SPA-mediated growth arrest is due to SPA repression of MYC (21, 47–49). The maintenance of cell proliferation in the setting of RBL1/2 loss indicates that SPA-mediated repression of MYC and reduced MYC-signaling cannot overcome loss of DREAM complex activity. Collectively, these results indicate that AR-mediated repression of MYC transcription is independent of cell cycle and E2F signaling, and SPA-induced MYC repression requires an intact DREAM complex to arrest the cell cycle.
MYC expression is a modulator of AR signaling and a key determinant of response to SPA
Prior studies have determined that enforced MYC expression can promote the growth of prostate cancer cells in the setting of castrate androgen levels and override SPA-mediated proliferation arrest (48, 50). MYC was strongly repressed by SPA in LNCaP, LNCaPDKO, and LNCaPDKO;RBL1/2 cells (Fig. 6A and B). MYC downregulation was accompanied by loss of E2F activity and growth arrest in LNCaP and LNCaPDKO cells, but not in LNCaPDKO;RBL1/2 cells (Figs. 1C and 2D–H). Canonical MYC target genes include the cyclins and cyclin-dependent kinases (CDK) cyclinD-CDK4/6 and cyclinE-CDK2 that serve to drive G1–S cell-cycle progression (51). Accordingly, ectopic expression of MYC from a constitutive promoter in LNCaPDKO cells (LNCaPDKO-MYC) resulted in complete resistance to SPA (Fig. 6; Supplementary Fig. S4D). Downregulation of MYC by SPA treatment would be expected to arrest cell growth in cells capable of assembling the DREAM complex. To explore this, we analyzed previously published RNA-seq datasets of siRNA-mediated MYC knockdown in VCaP cells (21) for changes to DREAM target gene signature expression. As expected, knockdown of MYC resulted in the repression of DREAM target genes (Supplementary Fig. S4E).
Figure 6.
MYC expression is a key determinant of response to high-dose androgen. A, Heatmaps of RNA-seq mean-centered log2(FPKM) gene expression values and AR and MYC-regulated genes in LNCaP cells treated with a dose-range of R1881. C, vehicle control. B, RNA-seq log2(FPKM) expression of MYC is plotted for LNCaP and isogenic knockout lines treated with SPA for 48 hours or vehicle control. C, Dose–response curve to R1881 for LNCaPDKO cells with or without ectopic MYC overexpression. D, Heatmap of molecular signature (GSVA) scores derived from RNA-seq data from LNCaPDKO cells with or without ectopic expression of MYC treated with SPA for 48 hours. CCP, cell-cycle proliferation; EMT, epithelial-to-mesenchymal transition; NE, neuroendocrine. E, Heatmap of mean-centered log2(FPKM) RNA-seq values for the AR activity signature genes determined from RNA-seq data from SPA or vehicle control in LNCaP, LNCaPDKO, and LNCaPDKO-MYC cells. GSVA scores and treatment groups are shown at the top of plot and colored according to legends at the bottom. LNCaP and LNCaP-DKO data are replotted from Fig. 1. F, Heatmap of mean-centered log2(FPKM) RNA-seq values for the top 10 genes differentially regulated by MYC overexpression in LNCaPDKO cells treated with SPA. G, GO enrichment analysis of MYC/E2F-independent AR-repressed genes in LNCaPDKO cells. H, Top motifs predicted to regulate MYC/E2F-independent AR-repressed genes by LISA. I, Heatmap of mean-centered log2(FPKM) RNA-seq values for predicted regulators of MYC/E2F-independent AR-repressed genes. J, Heatmap of mean-centered log2(FPKM) RNA-seq values for consensus MYC/E2F-independent AR-repressed genes (AR-repressed signature genes repressed by SPA in LNCaPDKO-MYC cells). LNCaP and LNCaP-DKO data are replotted from Fig. 1. K, Heatmap of mean-centered log2(FPKM) RNA-seq values for consensus MYC/E2F-independent AR-repressed genes comparing SPA versus vehicle control in LNCaP, VCaP, LAPC4, and 22PC-EP cell lines.
To further explore the role of MYC in transcriptional responses induced by SPA, we performed RNA-seq on LNCaPDKO-MYC cells treated with SPA or vehicle control. MYC overexpression diminished AR activity and the expression of canonical AR target genes such as PSA/KLK3 and TMPRSS2 (Fig. 6D and E), and induced cell-cycle-related genes (Fig. 6D). Further, the top 10 genes induced by MYC expression included cell-cycle regulators that are repressed by both ENZ and SPA in LNCaP and LNCaPDKO cells. One of these genes is MYBL2, a DREAM-repressed oncogene that binds to the MuVB core complex in a mutually exclusive fashion with E2F4/5 to activate DREAM target genes during cell cycle (Fig. 6F). Interestingly, PEG10, an established MYC target involved in the transdifferentiation of prostate cancer to a neuroendocrine subtype (52), was the top ranked result by edgeR analysis (Fig. 6F). However, the induction of a neuroendocrine gene expression program was not observed in LNCaPDKO-MYC cells (Fig. 6D).
MYC/E2F-independent SPA-repressed genes regulate distinct cellular processes
Although most genes repressed by SPA were MYC/E2F targets in LNCaP and LNCaPDKO cells, 360 genes were still repressed by SPA in the LNCaPDKO-MYC cells, indicating these genes were regulated independently of MYC and cell cycle. GO analysis identified enrichment for processes related to cell migration, morphogenesis, and extracellular contact (Fig. 6G). LISA analysis predicted their transcriptional regulation by the AR, and known AR-interacting factors including FOXA1, GATA2, GRHL2, and HOXB13 (Fig. 6H). Of these regulators, AR and GATA2 were themselves repressed by SPA (Fig. 6I). Of note, the expression of transcriptional repressors SNAI2/SLUG and PIAS1 were increased by SPA (Fig. 6I; refs. 53, 54). To extend these results, we determined the overlap of the 360 MYC-independent SPA-repressed genes with a consensus set of 260 AR-repressed genes from a prior study of LNCaP, VCaP, 22PC, and LAPC4 cells (12); 32 genes, including AR and GATA2, overlapped between these two gene sets and so represent a MYC/E2F-independent, AR-repressed program (Fig. 6J and K). Taken together, these data indicate that many SPA-repressed genes are regulated indirectly through the repression of transcriptional activators such as MYC and GATA2 or the activation of transcriptional repressors such as SNAI2 and PIAS1.
Discussion
The AR is a central regulator of prostate cancer cell survival and proliferation throughout the entire course of disease and inhibiting AR signaling remains the foundational therapeutic intervention for the treatment of metastatic disease. Despite initial responses, resistance to AR pathway repression is nearly universal and is accompanied by structural genomic alterations to AR that maintain or enhance AR expression. The re-activation of AR signaling is increasingly difficult to suppress using conventional approaches that reduce androgens or block ligand–receptor interactions. However, the AR-enhancer and gene body amplifications that contribute to high receptor levels also confer a vulnerability to high concentrations of AR ligands that can repress growth. Although several mechanisms have been proposed to explain the paradoxical effects of SPA, most ultimately involve processes that contribute to cell-cycle arrest (11, 20, 55). The central role of RB1 and E2F in regulating cell cycle has consequently been a focus of AR-mediated growth regulation. When RB1 is lost, AR/E2F co-bound genes increase in expression and their associations on chromatin are enhanced (22, 56). Because of AR/E2F/RB1 associations, RB1 has been implicated as a mediator of SPA growth suppression via the co-regulation of chromatin sites that are also bound by AR (22, 23, 57). However, the findings of prior studies are complicated by proliferation as a confounding variable, leading to difficulties in establishing cause–effect relationships.
In this study, we used isogenic knockout models and functional genomic approaches to establish the relationships between RB1/E2F, MYC, and AR signaling in prostate cancers. RB1 loss had only minor effects on SPA-mediated growth suppression and the repression of cell-cycle-related genes. These findings were unexpected because earlier preclinical studies, cited above, linked RB1 to AR-mediated gene repression, and we previously demonstrated that indirect modulation of E2F/RB signaling through oveexpression of CDK4/6 and cyclin-D or the knockout of negative cell-cycle regulators including p21, p57, and p27, mediated partial resistance to SPA (12). To test our hypothesis that the DREAM complex was mediating repression of E2F1-target genes, we deleted RBL1 and RBL2 on an RB1/TP53-null background. These RB/DREAM deficient lines demonstrated strong resistance to SPA with respect to proliferation and the suppression of E2F genes, confirming prior studies implicating E2F1 as critical mediator of SPA-mediated growth repression (12, 22).
Given that MYC overexpression resulted in resistance to SPA and constitutively high E2F target gene expression, the most parsimonious interpretation of our data is that AR regulates both cell-cycle progression and expression of E2F gene targets through the regulation of MYC. Although DREAM-mediated suppression of E2F1 signaling and cell-cycle arrest in the absence of RB1 is well described, the most established mechanism that activates DREAM complex in response to growth arresting signals requires p53 (58). This study presents a TP53/RB1 independent mechanism of DREAM complex formation in prostate cancer though AR repression of MYC and downstream MYC targets such as cyclin/CDK complexes (Supplementary Fig. S4F). Importantly, MYC expression was repressed by SPA even in RB/DREAM-deficient cells, indicating an AR regulatory mechanism independent of RB1/E2F and cell cycle.
MYC and E2F targets constitute the majority of SPA-repressed genes. However, key genes, including AR itself, are also consistently repressed by AR signaling. These include the downregulation of transcriptional activators like MYC, GATA2, and AR itself, as well as upregulation of transcriptional repressors like SNAI2 and PIAS1. It is notable that several of the biological processes regulated by AR-repressed genes include migration and cell morphology, processes regulated by SNAI2. The significance of these observations toward prostate cancer and its progression remains unclear and warrant further study.
Our findings support a model of SPA-mediated growth repression that relies on the negative regulation of E2F1 signaling via the DREAM–SIN3A–HDAC complex. Since DREAM complex stability is regulated by MYC, a key determinant of tumor sensitivity to SPA treatment may be the ability to effectively repress MYC activity. Further study of MYC regulation by AR may yield co-therapeutic approaches that synergize with SPA to enhance growth suppression or prolong sensitivity to subsequent ADT.
Supplementary Material
Table of genes comprising signatures.
Supplemental Figures 1-4
Acknowledgments
The authors are grateful to colleagues including Steve Balk, Myles Brown, Joshua Russo, and Henry Long for helpful comments and productive discussions. They thank members and colleagues in the Nelson laboratory and Prostate Cancer Research Program for advice and input. The authors gratefully acknowledge research support from awards from the NCI P30CA015704, P50CA97186, P01CA163227, and CDMRP W81XWH-18-1-0347 and W81XWH-18-1-0354. This work was also supported by the Prostate Cancer Foundation and the IPCR. Scientific Computing Infrastructure at FHCC is funded by ORIP grant no. S10OD028685.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
M.D. Nyquist is currently employed by Gilead Sciences. E. Corey reports other support from AbbVie, Gilead, Zenith Epigenetics, Bayer Pharmaceuticals, Gennetech, Janssen Research, Kronos Bio, Forma Therapeutics, Foghorn, and MacroGenics outside the submitted work. P.S. Nelson reports personal fees from Janssen, Bristol Myers Squibb, Pfizer, and Merck and grants from Janssen outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
M.D. Nyquist: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. I.M. Coleman: Data curation, formal analysis, visualization. J.M. Lucas: Investigation, methodology, writing–review and editing. D. Li: Investigation. B. Hanratty: Formal analysis. H. Meade: Formal analysis. E.A. Mostaghel: Conceptualization, writing–review and editing. S.R. Plymate: Conceptualization, writing–review and editing. E. Corey: Resources, methodology, writing–review and editing. M.C. Haffner: Conceptualization, formal analysis, writing–review and editing. P.S. Nelson: Conceptualization, formal analysis, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Nelson PS. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J Clin Oncol 2012;30:644–6. [DOI] [PubMed] [Google Scholar]
- 2. Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002;99:11890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ryan CJ, Tindall DJ. Androgen receptor rediscovered: the new biology and targeting the androgen receptor therapeutically. J Clin Oncol 2011;29:3651–8. [DOI] [PubMed] [Google Scholar]
- 4. Huggins C, Hodges CV. Studies on prostate cancer 1: the effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res 1941;1:293–7. [Google Scholar]
- 5. Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324:787–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ware KE, Garcia-Blanco MA, Armstrong AJ, Dehm SM. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer 2014;21:T87–T103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murillo H, Schmidt LJ, Karter M, Hafner KA, Kondo Y, Ballman KV, et al. Prostate cancer cells use genetic and epigenetic mechanisms for progression to androgen independence. Genes Chromosomes Cancer 2006;45:702–16. [DOI] [PubMed] [Google Scholar]
- 9. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viswanathan SR, Ha G, Hoff AM, Wala JA, Carrot-Zhang J, Whelan CW, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 2018;174:433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Isaacs JT, D'Antonio JM, Chen S, Antony L, Dalrymple SP, Ndikuyeze GH, et al. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate 2012;72:1491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nyquist MD, Corella A, Mohamad O, Coleman I, Kaipainen A, Kuppers DA, et al. Molecular determinants of response to high-dose androgen therapy in prostate cancer. JCI insight 2019;4:e129715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nyquist MD, Ang LS, Corella A, Coleman IM, Meers MP, Christiani AJ, et al. Selective androgen receptor modulators activate the canonical prostate cancer androgen receptor program and repress cancer growth. J Clin Invest 2021;131:e146777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Umekita Y, Hiipakka RA, Kokontis JM, Liao S. Human prostate tumor growth in athymic mice: inhibition by androgens and stimulation by finasteride. Proc Natl Acad Sci U S A. 1996;93:11802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kokontis JM, Hay N, Liao S. Progression of LNCaP prostate tumor cells during androgen deprivation: hormone-independent growth, repression of proliferation by androgen, and role for p27Kip1 in androgen-induced cell cycle arrest. Mol Endocrinol 1998;12:941–53. [DOI] [PubMed] [Google Scholar]
- 16. Schweizer MT, Antonarakis ES, Wang H, Ajiboye AS, Spitz A, Cao H, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med 2015;7:269ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Markowski MC, Kachhap S, De Marzo AM, Sena LA, Luo J, Denmeade SR, et al. Molecular and clinical characterization of patients with metastatic castration resistant prostate cancer achieving deep responses to bipolar androgen therapy. Clin Genitourin Cancer 2022;20:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denmeade SR, Wang H, Agarwal N, Smith DC, Schweizer MT, Stein MN, et al. TRANSFORMER: a randomized phase II study comparing bipolar androgen therapy versus enzalutamide in asymptomatic men with castration-resistant metastatic prostate cancer. J Clin Oncol 2021;39:1371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teply BA, Wang H, Luber B, Sullivan R, Rifkind I, Bruns A, et al. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol 2018;19:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohammad OS, Nyquist MD, Schweizer MT, Balk SP, Corey E, Plymate S, et al. Supraphysiologic testosterone therapy in the treatment of prostate cancer: models, mechanisms and questions. Cancers (Basel) 2017;9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo H, Wu Y, Nouri M, Spisak S, Russo JW, Sowalsky AG, et al. Androgen receptor and MYC equilibration centralizes on developmental super-enhancer. Nat Commun 2021;12:7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao S, Gao Y, He HH, Han D, Han W, Avery A, et al. Androgen receptor tumor suppressor function is mediated by recruitment of retinoblastoma protein. Cell Rep 2016;17:966–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han W, Liu M, Han D, Toure AA, Li M, Besschetnova A, et al. Exploiting the tumor-suppressive activity of the androgen receptor by CDK4/6 inhibition in castration-resistant prostate cancer. Mol Ther 2022;30:1628–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barfeld SJ, Urbanucci A, Itkonen HM, Fazli L, Hicks JL, Thiede B, et al. c-Myc antagonises the transcriptional activity of the androgen receptor in prostate cancer affecting key gene networks. EBioMedicine 2017;18:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep 2020;31:107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meers MP, Bryson TD, Henikoff JG, Henikoff S. Improved CUT&RUN chromatin profiling tools. Elife 2019;8:e46314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods 2018;15:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D'Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc 2016;11:316–26. [DOI] [PubMed] [Google Scholar]
- 30. Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 2013;10:730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chatterjee P, Schweizer MT, Lucas JM, Coleman I, Nyquist MD, Frank SB, et al. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J Clin Invest 2019;130:4245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu X, Brown LG, Conner JL, Nguyen HM, Boufaied N, Abou Alaiwi S, et al. Response to supraphysiological testosterone is predicted by a distinct androgen receptor cistrome. JCI insight 2022;7:e157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 2010;120:4478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 2013;13:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 2016;44:6070–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uxa S, Bernhart SH, Mages CFS, Fischer M, Kohler R, Hoffmann S, et al. DREAM and RB cooperate to induce gene repression and cell-cycle arrest in response to p53 activation. Nucleic Acids Res 2019;47:9087–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan L, Perez RE, Calhoun S, Maki CG. RBL2/DREAM-mediated repression of the Aurora kinase A/B pathway determines therapy responsiveness and outcome in p53 WT NSCLC. Sci Rep. 2022;12:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ 2022;29:946–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin Q, Fan J, Zheng R, Wan C, Mei S, Wu Q, et al. Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol 2020;21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McNair C, Urbanucci A, Comstock CE, Augello MA, Goodwin JF, Launchbury R, et al. Cell cycle-coupled expansion of AR activity promotes cancer progression. Oncogene 2017;36:1655–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schiewer MJ, Augello MA, Knudsen KE. The AR dependent cell cycle: mechanisms and cancer relevance. Mol Cell Endocrinol 2012;352:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lam HM, Nguyen HM, Labrecque MP, Brown LG, Coleman IM, Gulati R, et al. Durable response of enzalutamide-resistant prostate cancer to supraphysiological testosterone is associated with a multifaceted growth suppression and impaired DNA damage response transcriptomic program in patient-derived xenografts. Eur Urol 2020;77:144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 2001;29:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Civenni G, Malek A, Albino D, Garcia-Escudero R, Napoli S, Di Marco S, et al. RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer. Cancer Res 2013;73:6816–27. [DOI] [PubMed] [Google Scholar]
- 46. Antony L, van der Schoor F, Dalrymple SL, Isaacs JT. Androgen receptor (AR) suppresses normal human prostate epithelial cell proliferation via AR/beta-catenin/TCF-4 complex inhibition of c-MYC transcription. Prostate 2014;74:1118–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kokontis J, Takakura K, Hay N, Liao S. Increased androgen receptor activity and altered c-myc expression in prostate cancer cells after long-term androgen deprivation. Cancer Res 1994;54:1566–73. [PubMed] [Google Scholar]
- 48. Chuu CP, Kokontis JM, Hiipakka RA, Fukuchi J, Lin HP, Lin CY, et al. Androgen suppresses proliferation of castration-resistant LNCaP 104-R2 prostate cancer cells through androgen receptor, Skp2, and c-Myc. Cancer Sci 2011;102:2022–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sena LA, Kumar R, Sanin DE, Thompson EA, Rosen DM, Dalrymple SL, et al. Androgen receptor activity in prostate cancer dictates efficacy of bipolar androgen therapy through MYC. J Clin Invest 2022;132:e162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vander Griend DJ, Litvinov IV, Isaacs JT. Conversion of androgen receptor signaling from a growth suppressor in normal prostate epithelial cells to an oncogene in prostate cancer cells involves a gain of function in c-Myc regulation. Int J Biol Sci 2014;10:627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Involvement of Myc activity in a G(1)/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol 2000;20:3497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep 2015;12:922–36. [DOI] [PubMed] [Google Scholar]
- 53. Ledsaak M, Bengtsen M, Molvaersmyr AK, Fuglerud BM, Matre V, Eskeland R, et al. PIAS1 binds p300 and behaves as a coactivator or corepressor of the transcription factor c-Myb dependent on SUMO-status. Biochim Biophys Acta 2016;1859:705–18. [DOI] [PubMed] [Google Scholar]
- 54. Toropainen S, Malinen M, Kaikkonen S, Rytinki M, Jaaskelainen T, Sahu B, et al. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res 2015;43:848–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hedayati M, Haffner MC, Coulter JB, Raval RR, Zhang Y, Zhou H, et al. Androgen deprivation followed by acute androgen stimulation selectively sensitizes AR-positive prostate cancer cells to ionizing radiation. Clin Cancer Res 2016;22:3310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mandigo AC, Shafi AA, McCann JJ, Yuan W, Laufer TS, Bogdan D, et al. Novel oncogenic transcription factor cooperation in RB-deficient cancer. Cancer Res 2022;82:221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han D, Chen S, Han W, Gao S, Owiredu JN, Li M, et al. ZBTB7A mediates the transcriptional repression activity of the androgen receptor in prostate cancer. Cancer Res 2019;79:5260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peuget S, Selivanova G. p53-dependent repression: DREAM or reality? Cancers (Basel) 2021;13:4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of genes comprising signatures.
Supplemental Figures 1-4
Data Availability Statement
The RNAseq data used in this study are available in the Gene Expression Omnibus repository (GEO) under accession number GSE225481. The CUT&RUN and ChIP-seq data generated by the authors are publicly available and were deposited in the Gene Expression Omnibus under study numbers: GSE228897, GSE228895, and GSE228896. Publicly available data generated by others were used by the authors. The public data analyzed in this study were obtained from the GEO: GSE147250, GSE135879, GSE225481, GSE124704, and GSE157107.