Abstract
The centrosome, as the main microtubule organizing centre, plays key roles in cell polarity, genome stability and ciliogenesis. The recent identification of ribosomes, RNA-binding proteins and transcripts at the centrosome suggests local protein synthesis. In this context, we hypothesized that TDP-43, a highly conserved RNA binding protein involved in the pathophysiology of amyotrophic lateral sclerosis and frontotemporal lobar degeneration, could be enriched at this organelle. Using dedicated high magnification sub-diffraction microscopy on human cells, we discovered a novel localization of TDP-43 at the centrosome during all phases of the cell cycle.
These results were confirmed on purified centrosomes by western blot and immunofluorescence microscopy. In addition, the co-localization of TDP-43 and pericentrin suggested a pericentriolar enrichment of the protein, leading us to hypothesize that TDP-43 might interact with local mRNAs and proteins. Supporting this hypothesis, we found four conserved centrosomal mRNAs and 16 centrosomal proteins identified as direct TDP-43 interactors. More strikingly, all the 16 proteins are implicated in the pathophysiology of TDP-43 proteinopathies, suggesting that TDP-43 dysfunction in this organelle contributes to neurodegeneration.
This first description of TDP-43 centrosomal enrichment paves the way for a more comprehensive understanding of TDP-43 physiology and pathology.
Keywords: TDP-43, centrosome, pericentriolar matrix, ALS, FTLD
Using sub-diffraction imaging, Bodin et al. reveal localization of TDP-43 at the centrosome throughout all phases of the cell cycle. This first description of TDP-43 centrosomal enrichment in human cells paves the way to a more comprehensive understanding of TDP-43 physiology and pathology.
See Megan Dykstra and Sami J. Barmada (https://doi.org/10.1093/brain/awad268) for a scientific commentary on this article.
Introduction
Transactive response DNA-binding protein 43 (TDP-43), encoded by the TARDBP gene, is a highly conserved and ubiquitously expressed RNA-binding protein (RBP) predominantly localized in the nucleus of cells under physiological conditions. The protein modulates RNA metabolism through its N-terminal RNA recognition motifs and interacts with multiple protein partners through its C-terminal end. TDP-43 has received increasing attention since its identification as one of the main actors in the pathophysiology of amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) and limbic-predominant age-related TDP-43 encephalopathy (LATE). Indeed, cytosolic translocation and aggregation of phosphorylated and ubiquitinated TDP-43 in degenerating neurons is a pathologicalal hallmark of these diseases,1–3 and TARDBP mutations are responsible for familial forms of ALS and FTLD.4,5 However, the precise mechanisms by which TDP-43 pathology contributes to neurodegeneration remain unclear.
In recent years, interest has been growing in the presence of RBPs at the centrosome of cells since mRNAs and ribosomes have been identified within this organelle.6–8 The centrosome is known to be the main microtubule organizing centre in human cells, controlling cell shape, polarity, trafficking, motility, ciliogenesis and cell cycle. RBPs transport RNAs to the pericentriolar matrix (PCM) and mediate their local translation.6 Since TDP-43 is an RBP that belongs to the heterogeneous nuclear ribonucleoproteins (hnRNP) family, we hypothesized that it might be localized at the centrosome. To this end, we performed dedicated sub-diffraction imaging and centrosome purification techniques in cultured cells to detect TDP-43 in this organelle.
Materials and methods
Cell culture
Primary skin fibroblasts were obtained from healthy subjects and patients with sporadic9,10 and familial ALS (Supplementary Table 1). Fibroblasts carrying the TARDBPG348C mutation were obtained from the Erasmus Hospital (Brussels, Belgium) with informed consent. Primary skin fibroblasts were cultured in Dulbecco’s modified Eagle medium (DMEM): nutrient mixture F-12 in the presence of 10% fetal calf serum (FCS), 1% uridine and 1% pyruvate. HeLa (human cervical carcinoma), U-87 MG (human glioblastoma cancer) and SH-SY5Y (human neuroblastoma cancer) cell lines were grown in high glucose Dulbecco’s minimum essential medium, supplemented with 10% FCS and 1% glutamine. Human lymphoblastic KE-37 cells were grown in RPMI and DMEM/F12 medium supplemented with 10% FCS in the presence of penicillin-streptomycin (1000 units/ml and 1000 µg/ml, respectively HyClone). All cell lines were cultured in T25 flasks and maintained in a humidified atmosphere (95% air, 5% CO2) at 37°C. Experiments were conducted on cells with similar passage numbers, ranging from 6 to 20, to avoid artefacts due to senescence. If needed, cells were treated with nocodazole (5 mg/ml) prior to fixation.
Immunofixation
Approximately 30 000 cells were seeded into 22 mm borosilicate glass coverslips (Dutscher D.) in a humidified atmosphere (95% air, 5% CO2) at 37°C for at least 24 h. Cells were then fixed with cold methanol at −20°C for 6 min. After three washes in PBS, cells were blocked in 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Cells were then washed and incubated overnight at 4°C with primary antibodies: rabbit anti-TDP-43 polyclonal antibody (10782-2-AP, 1:1000), rabbit recombinant anti-TDP-43 monoclonal antibody (ab109535, 1:750), rabbit anti-TDP-43 (C-terminal) polyclonal antibody (12892-1-AP, 1:1500), mouse anti-TDP-43 (human specific) monoclonal antibody (60019-2-Ig, 1:50 000), mouse anti-TDP-43 monoclonal antibody (ab57105, 1:6000), mouse recombinant anti-pericentrin monoclonal antibody (ab28144, 1:5000), mouse anti-centrin monoclonal antibody (ZMS1054, 1:5000), mouse anti-γ-tubulin monoclonal antibody (T5326, 1:5000), rabbit anti-γ-tubulin polyclonal antibody,11 mouse anti-polyglutamylated tubulin (GT335) monoclonal antibody (C. Janke, gift),12 CoraLite®488 rabbit anti-CEP164 polyclonal antibody (CL488-22227, 1:1000), mouse anti-alpha-tubulin monoclonal antibody (T9026, 1:500), mouse anti-ataxin 2 monoclonal antibody (68316-1-Ig, 1:100), mouse anti-p150 (Dynactin) monoclonal antibody (610473, 1:1000), mouse anti-VCP monoclonal antibody (60316-1-Ig, 1:100), mouse anti-cyclophilin A monoclonal antibody (PPIA) (67880-1-Ig, 1:1000), rabbit anti-CHMP2B polyclonal antibody (12527-1-AP, 1:50), rabbit anti-Cyclin F polyclonal antibody (ab203117, 1:100) and rabbit anti-PFN1 polyclonal antibody (P7749, 1:100). Finally, cells were washed in PBS and incubated with Alexa Fluor conjugated secondary antibodies (Invitrogen, 1:2000) for 90 min at room temperature. Nuclei were counterstained with Hoechst 33 342 (Thermo Fisher Scientific) 1:10 000 in PBS. References of reagents and antibodies are listed in Supplementary Table 2.
Total internal reflection fluorescence and stochastic optical reconstruction microscopy
To perform sub-diffraction imaging, the cavity of a clean single depression slide (Paul Marienfeld) was filled with 50 μl of PBS for total internal reflection fluorescence (TIRF) and switching buffer (Abbelight) for stochastic optical reconstruction microscopy (STORM), and covered the cells with the coverslip facing downward. Excess buffer was carefully wiped away, and the coverslip was sealed with a two-component silicone-glue (Twinsil®, Picodent). After a 10 min drying, the device was placed on the stage of an inverted motorized microscope NIKON ECLIPSE Ti-E (Nikon Instruments Europe) equipped with a CFI SR APO TIRF 100× ON1.49 objective, a Perfect Focus System, a TIRF ILas2 module (Roper Scientific) and a single-photon sensitive camera Evolve 128TM EMCCD 512 × 512 imaging array, 16 × 16 μm pixels (Photometrics). Acquisitions were performed at 25°C in a dark heating chamber (Okolab NA). Multichannel 2D- and 3D-TIRF microscopy was performed using Metamorph 7.7 software (Molecular Devices, CA, USA). For complementary STORM imaging, the excitation power of either the 647 nm or 568 laser line was then increased (∼50 to 100 mW before the objective lens) to induce fluorophore blinking. The em-gain of the camera was set to high amplification to optimize the signal-to-noise ratio. Images were acquired with an integration time of 30 ms per frame. The total acquisition time points for each sequence were adapted to the labelling density (5000 to 10 000 frames). The centre position of the fluorophores was determined by fitting of 2D Gaussian function with the parameters of microscope’s point spread function (PSF) using WaveTracer software (Roper Scientific). Imaris 8.0 (Bitplane, Zurich, Switzerland) was used for image processing and analysis (length and volume measurements, Pearson coefficient).13
Cellular fractionation
Triton X-100 soluble and insoluble protein fractions were prepared as follows: KE37cells were washed in PBS and lysed in a PHEM buffer (45 mM PIPES, 45 mM HEPES, 10 mM EGTA, 5 mM MgCl2, 1 mM phenylmethanesulphonyl fluoride adjusted at pH 6.8) containing 1% Triton X-100 and protease inhibitors. Insoluble proteins were pelleted at 300g, solubilized in SDS-PAGE sample buffer (Laemmli, 1970), and then boiled for 5 min. Soluble proteins were precipitated with 9 vol of methanol at 4°C for 1 h and pelleted. The pellet was resuspended in the same volume of sample buffer that was used for the insoluble proteins.
Centrosome purification
Centrosomes were isolated from KE-37 cells as described previously.14 Briefly, cells were pretreated for 1 h with nocodazole (2 × 10−7 M) and cytochalasin D (2 × 10−6 M). Cells were then washed in PBS and resuspended in 8% sucrose in 10× diluted PBS before cell lysis in lysis buffer (1 mM HEPES, 0.5 mM MgCl2, 0.5% NP40, 1 mM phenylmethanesulphonyl fluoride and antiproteases). After centrifugation at 4000 rpm, the supernatant was filtered through a nylon mesh, readjusted to 10 mM HEPES and treated with DNAse and benzonase. Concentrated centrosomes were overlaid on a discontinuous sucrose gradient (70%, 50% and 40%) in a SW32Ti tube and centrifuged at 25 000 rpm for 1 h 15 min. Fractions were then collected and analysed for their purity using an anti-γ-tubulin rabbit polyclonal antibody,11 a mouse monoclonal antibody CTR453 (a gift from M. Bornens15) and DAPI (4′,6-diamidino-2-phenylindole, D8417) to stain DNA contamination.
Western blotting of purified centrosomes
Triton X-100-soluble and insoluble fractions as well as centrosomal fraction were run on an SDS-PAGE gel of 10% acrylamide/bis-acrylamide. Gels were transferred on nitrocellulose membranes with a semi-dry transfer device (Bio-Rad). Membranes were blotted with primary antibodies: rabbit anti-TDP-43 polyclonal antibody (10782-2-AP, 1:5000), rabbit anti-TDP-43 (C-terminal) polyclonal antibody (12892-1-AP, 1:1000), mouse anti-TDP-43 (human specific) monoclonal antibody (60019-2-Ig, 1:1000), mouse anti-γ-tubulin monoclonal antibody (T5326, 1:5000), and mouse anti-lamin B1 monoclonal antibody (ab8982, 1:1000). Membranes were then washed in PBS and incubated with peroxydase conjugated anti-rabbit and anti-mouse secondary antibodies (Sigma), and finally revealed and imaged by chemiluminescence using the ChemiDoc (Bio-Rad). Quantification of luminescence intensity were performed with Image Lab software (Bio-Rad). References of reagents and antibodies are detailed in Supplementary Table 2.
Immunofluorescence microscopy of purified centrosomes
Isolated centrosomes were fixed with methanol for 6 min at −20°C and immune-stained with primary antibodies: mouse anti-TDP-43 (human specific) monoclonal antibody (60019-2-Ig, 1:2000), and rabbit anti-γ-tubulin polyclonal antibody.11 Finally, centrosomes were washed in PBS and incubated with Alexa Fluor conjugated secondary antibodies (Invitrogen, 1:1000) for 90 min at room temperature. Immunofluorescence microscopy was performed with an inverted motorized microscope (TCS SP8, Leica Microsystems) equipped with a UV diode (line 405) and two laser diodes (lines 488 and 552) for excitation, an oil immersion magnification 63×/numerical aperture 1.4 Plan-Apochromat CS2 objective lens and two PMT detectors. LAS X software (Leica Microsystems) was used for acquisition. Z-stacks were generated from optical sections taken at 0.3 µm intervals. Image stacks were processed using ImageJ software. References of reagents and antibodies are detailed in Supplementary Table 2.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee West II (DC-2011-146) and the Ethics Committee Île de France II (DC 2011-534). The study protocol was declared to the French commission for information technology and civil liberties (declaration number ar19-0012v0).
Statistical analysis
Statistical analyses were performed using PRISM software version 5.0 for Windows (GraphPad, La Jolla, CA). Comparisons of means were performed using the Mann-Whitney test for two-group analysis and the Kruskal-Wallis test for multiple-group analysis. Differences were considered significant at P < 0.01.
Results
TDP-43 is enriched at the centrosome
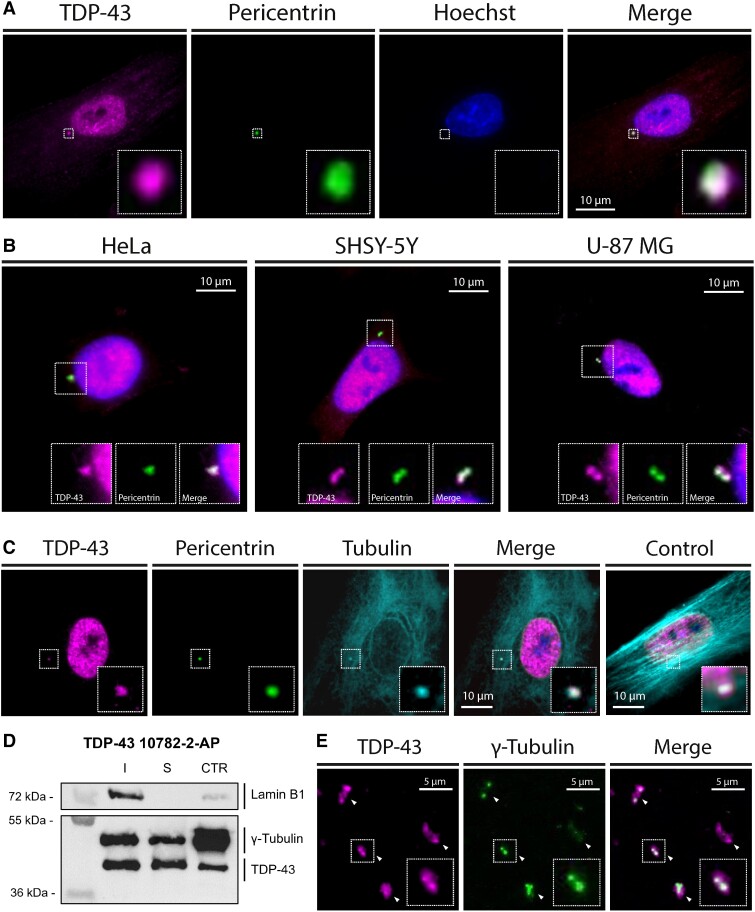
We first attempted to detect the presence of TDP-43 at the centrosome of cultured cells using TIRF microscopy at high magnification. Human skin fibroblasts (hSF), HeLa, U-87 MG and SH-SY5Y cells were fixed with cold methanol and immune-stained with antibodies against TDP-43 (10782-2-AP) and pericentrin, a centrosome marker. A co-localization was observed in all cells when the centrosome and the nucleus did not overlap, indicating a centrosomal enrichment of TDP-43 (Fig. 1A and B). Similar results were obtained with four other antibodies against distinct epitopes of TDP-43 (ab109535, 12892-1-AP, 60019-2 and ab57105, Supplementary Fig. 1A). The co-localization persisted after microtubule disruption with nocodazole (Fig. 1C), suggesting that TDP-43 does not require an active microtubule-dependent mechanism to be maintained at the centrosome, as for other specific centrosomal proteins.16,17 Indeed, centrosomes retain their distinctive morphology and their structural elements after microtubule depolymerization, whereas centrosomal microtubule-associated components tend to disperse.16,17 Furthermore, this result indicates that the detected signal was not aggresomal since pericentriolar aggresome formation requires an intact microtubule network.18,19
Figure 1.
Centrosomal enrichment of TDP-43 in human cells. (A and B) Total internal reflection fluorescence (TIRF) microscopy images of fixed healthy human skin fibroblasts (A), and pseudo-neuronal HeLa, U-87 MG and SH-SY5Y cells (B) immune-stained with antibodies against TDP-43 (10782-2-AP) and pericentrin, showing a co-localization of the two signals when merged. (C) Similar results were obtained in human skin fibroblasts after a 2-h treatment with nocodazole (5 mg/ml) inducing microtubule depolymerization. (D) Western blot performed on purified centrosome fractions from human T-lymphoblastoid KE-37 cells using primary antibodies against TDP-43 (10782-2-AP), γ-Tubulin and Lamin B1, showing an enrichment of TDP-43 at the centrosome. I = insoluble fraction; S = soluble fraction; CTR = centrosome fraction. (E) Immunofluorescence microscopy performed on purified centrosomes (arrowheads) fixed and immune-stained for TDP-43 (60019-2-Ig) and γ-Tubulin showing a co-localization of the two signals.
To confirm these results, we evaluated the presence of TDP-43 in isolated centrosomes. Western blots performed on purified centrosome fractions from KE-37 cells revealed a 43 kDa band when using a TDP-43 antibody (10782-2-AP) (Fig. 1D). Similar results were obtained using two other TDP-43 antibodies (12892-1-AP and 60019-2-Ig, Supplementary Fig. 1B and C). Since TDP-43 is mostly found in the nucleus, the nuclear fraction of the centrosome preparations was assessed by blotting Lamin B1 on the membranes. TDP-43 was more enriched in the centrosomal fractions than Lamin B1 compared to the insoluble fractions (Supplementary Fig. 1D), indicating minor nuclear contamination and confirming the presence of TDP-43 at the centrosome. Finally, fluorescence microscopy was performed on purified centrosomes immune-stained with antibodies against TDP-43 (60019-2-Ig) and γ-tubulin. All isolated centrosomes showed TDP-43 labelling (Fig. 1E). Together, these results demonstrate a centrosomal localization of TDP-43 in human cell lines.
TDP-43 centrosomal enrichment persists over time
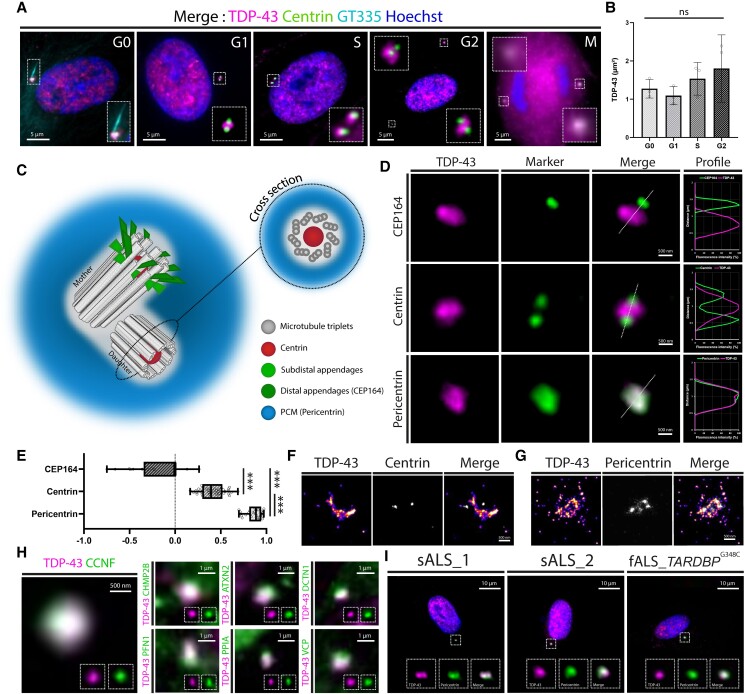
To further assess whether the centrosomal localization of TDP-43 varies over the cell cycle, we performed high magnification TIRF in cultured hSF. Each phase of the cell cycle was defined by the shape and position of the centrioles using a primary antibody against centrin, a centriole marker. The G0 phase was identified by immunostaining for polyglutamylated tubulin (GT335), a primary cilium marker. TDP-43 remained enriched at the centrosome in all phases of the cell cycle, including mitosis (Fig. 2A), further confirming that the centrosomal localization of TDP-43 persists in cells over time. Furthermore, we noticed that the surface area of TDP-43 centrosomal fraction tended to expand during the cell cycle (Fig. 2B), suggesting that the protein is part of the PCM.20
Figure 2.
Cell cycle independent pericentriolar enrichment of TDP-43 in physiological and pathological conditions. (A) Merged images of TIRF microscopy performed on human skin fibroblasts fixed and immune-stained for TDP-43, pericentrin and GT335, showing the presence of TDP-43 at the centrosome in all phases of the cell cycle, including mitosis. (B) Surface area of the TDP-43 centrosomal fraction (μm2) measured at each phase of the cell cycle (ns = not significant). (C) Schematic representation of the different parts of the centrosome, with corresponding protein markers. (D) High magnification TIRF microscopy images of the centrosome of human skin fibroblasts immune-stained with antibodies against TDP-43, centrin, CEP-164 and pericentrin. The co-localization profiles (fluorescence intensity of each channel) shown on the right and the Pearson coefficients of co-localization (E) indicate a pericentriolar enrichment of TDP-43 (P < 0.001). (F and G) Super resolution stochastic optical reconstruction microscopy (STORM) images of human skin fibroblasts immune-stained with antibodies against TDP-43 and centrin (F) or pericentrin (G). (H) TIRF microscopy images of TDP-43 and centrosomal TDP-43 partners associated to familial ALS/FTLD in fixed healthy human skin fibroblasts (magnification at the centrosome) disclosing a co-localization. (I) TIRF microscopy images of fixed human skin fibroblasts derived from sporadic and familial ALS (fALS) patients immune-stained with antibodies against TDP-43 and pericentrin, showing a co-localization of the two signals.
TDP-43 is located at the pericentriolar matrix
To define the precise localization of TDP-43 within the centrosome, we performed high magnification TIRF microscopy on hSF immune-stained with antibodies against TDP-43 and specific markers of three distinct parts of the centrosome: centrin (centrioles), CEP-164 (distal appendage) and pericentrin (PCM) (Fig. 2C). Co-localization profiles and Pearson coefficients (P < 0.0001) confirmed the pericentriolar localization of TDP-43 within the centrosome (Fig. 2D and E). Similar findings were obtained with super-resolutive STORM (Fig. 2F and G), supporting that TDP-43 is enriched at the PCM of the centrosome in human cells.
TDP-43 interacts with centrosomal RNAs and proteins
The pericentriolar enrichment of TDP-43 prompted us to evaluate whether the protein interacts with local mRNAs and proteins, as reported for other centrosomal RBPs.6–8 Interestingly, we found that four (PCNT, NUMA1, ASPM and CEP350) of the 11 conserved mRNAs recently detected at the centrosome7,8 were identified as TDP-43 targets by crosslinking and immunoprecipitation21,22 (Table 1). To identify centrosomal protein partners of TDP-43, we listed 518 proteins that directly interact with TDP-4323–25 and found that 16 of them are enriched at the centrosome (Table 2). Remarkably, 7 of these 16 proteins are encoded by genes whose variants are responsible for familial forms of ALS/FTLD (ATXN2, CCNF, CHMP2B, DCTN1, PFN1, PPIA and VCP) and three of them are kinases that phosphorylate TDP-43 and are associated with neurodegeneration (CDC7, CSNK1δ and TTBK1/2). The remaining six centrosomal TDP-43 interactors (BRCA1, DDX3X, ELAVL1/HuR, EWSR1, GSK3β and HDAC6) are also involved in neurodegenerative pathways in ALS/FTLD. Finally, we found that four proteins encoded by ALS/FTLD causative genes (ALSIN, CYLD, NEK1 and TBK1) are also enriched at the centrosome, but their direct interaction with TDP-43 has not yet been reported (Table 2). To further investigate the interaction of TDP-43 with the seven centrosomal proteins associated with familial ALS/FTLD and known to be TDP-43 partners (ATXN2, CCNF, CHMP2B, DCTN1, PFN1, PPIA and VCP), we performed high magnification TIRF microscopy on hSF. We disclosed a co-localization of TDP-43 with all the studied proteins at the PCM with similar distribution patterns (Fig. 2H). Taken together, these results support a role for TDP-43 at the PCM through local RNA binding and protein interactions and suggest that centrosomal dysfunction participates to TDP-43-related neurodegeneration.
Table 1.
Centrosomal mRNAs interacting with TDP-43
| mRNA | Protein | Centrosomal localization | TDP-43 interaction |
|---|---|---|---|
| ASPM | Abnormal spindle-like microcephaly-associated protein | Safieddine et al.,7 Chouaib et al.8 | Lang et al.,21 Van Nostrand et al.22 |
| NUMA1 | Nuclear mitotic apparatus prot1 | Safieddine et al.,7 Chouaib et al.8 | Lang et al.,21 Van Nostrand et al.22 |
| PCNT | Pericentrin | Safieddine et al.7 | Lang et al.,21 Van Nostrand et al.22 |
| CEP350 | Centrosomal protein 350 | Safieddine et al.7 | Lang et al.,21 Van Nostrand et al.22 |
Table 2.
Centrosomal proteins interacting with TDP-43
| Gene | Protein | Centrosomal localization | TDP-43 interaction | Neurodegeneration |
|---|---|---|---|---|
| ALSIN | Alsin | Millecamps et al.26 | Not reported | Yang et al.,27 Hadano et al.28 |
| ATXN2 | Ataxin 2 | Gnazzo et al.,29 Stubenvoll et al.30 | Elden et al.31 | Elden et al.,31 Hart et al.32 |
| BRCA1 | Breast cancer 1, early onset | Hsu and White33 | Hill et al.34 | Noristani et al.35 |
| CCNF | Cyclin F | D'Angiolella et al.36 | Rayner et al.37 | Williams et al.38 |
| CDC7 | Cell division cycle 7-related protein kinase | Müller-Taubenberger et al.39 | Liachko et al.40 | Liachko et al.,40 Vaca et al.41 |
| CHMP2B | Charged multivesicular body protein 2B | Ott et al.42 | Deng et al.43 | Skibinski et al.,44 Parkinson et al.45 |
| CSNK1D | Casein kinase I isoform delta | Greer et al.46 | Kametani et al.47 | Nonaka et al.,48 Alquezar et al.49 |
| CYLD | Ubiquitin carboxyl-terminal hydrolase CYLD | Eguether et al.50 | Not reported | Dobson-Stone et al.51 |
| DCTN1 | Dynactin 1 | Kodani et al.,52 Zhapparova et al.53 | Deshimaru et al.54 | Konno et al.55 |
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked | Chen et al.56 | Freibaum et al.57 | Cheng et al.58 |
| ELAVL1 | ELAV-like protein 1/Hu-antigen R (HuR) | Filippova et al.59 | Lu et al.60 | Matsye et al.61 |
| EWSR1 | Ewing sarcoma breakpoint region 1 | Leemann-Zakaryan et al.62 | Chi et al.63 | Couthouis et al.64 |
| GSK3β | Glycogen synthase kinase 3β | Yoshino and Ishioka65 | Moujalled et al.66 | Choi et al.67 |
| HDAC6 | Histone deacetylase 6 | Ran et al.68 | Hebron et al.69 | Fazal et al.70 |
| NEK1 | NIMA-related kinase 1 | White and Quarmby71 | Not reported | Brenner et al.72 |
| PFN1 | Profilin 1 | Nejedlá et al.73 | Tanaka et al.74 | Wu et al.75 |
| PPIA | Peptidyl-prolyl cis-trans Isomerase A or cyclophilin A | Bannon et al.76 | Lauranzano et al.77 | Pasetto et al.78 |
| TBK1 | TANK-binding kinase 1 | Pillai et al.79 | Not reported | Freischmidt et al.,80 Cirulli et al.81 |
| TTBK1/2 | Tau-tubulin kinase 1/2 | Čajánekand Nigg82 | Liachko et al.83 | Taylor et al.84 |
| VCP | Valosin containing protein | Balestra et al.,85 Madeo et al.86 | Freibaum et al.57 | Watts et al.87 |
ALS/FTLD causative genes are highlighted in bold.
Finally, we performed immune-fluorescence microscopy in hSF derived from a patient with familial ALS (TARDBPG348C, Supplementary Table 1) to assess mutant TDP-43 dynamics at the centrosome. In these cells, TDP-43 was still associated with the centrosome (Fig. 2I), demonstrating that the expression of the pathogenic TDP-43 mutation does not affect its localization to the organelle. Similar results were observed in hSF derived from patients with sporadic ALS (Fig. 2I and Supplementary Table 1).
Discussion
Here, we found that TDP-43 is physiologically enriched at the centrosome in cells. This TDP-43 centrosomal localization might have been unnoticed so far as it can only be detected by dedicated highly resolutive techniques. More specifically, our results evidence that TDP-43 is localized in the pericentriolar matrix, regardless of the cell-cycle, where it interacts with centrosomal specific mRNAs and proteins. This novel centrosomal localization of TDP-43 raises questions about the functions of the protein in this organelle.
The centrosome is the main microtubule organizing centre in human cells, thereby controlling cell shape, polarity, trafficking, motility, ciliogenesis and cell cycle. It can be assumed that the centrosomal fraction of TDP-43 is involved in these processes. By extension, the embryonic lethality and neurodevelopmental impairments caused by loss of TARDBP expression in animal models88–91 might be partly due to centrosome dysfunction, given the role of this organelle in embryogenesis and cell proliferation.
Although the precise role of TDP-43 at the PCM remains to be defined, the protein may be involved in the transport and translation of RNAs and interact with other proteins of the PCM, as for other centrosomal RBPs.6 Supporting this hypothesis, we found that 4 of the 11 RNAs recently detected at the centrosome were previously identified as TDP-43 targets, and that 16 TDP-43 interactors are also centrosomal proteins. More strikingly, all of these 16 proteins are involved in the pathophysiology of TDP-43 proteinopathies, seven of which are encoded by genes that cause familial ALS/FTLD, suggesting that local disruption participates to neurodegeneration in these diseases. Finally, we showed that TDP-43 was still associated with the centrosome in TARDBPG348C heterozygous hSF cells. Since this signal could originate from the restricted presence of the wild-type TDP-43, this result does not allow to prejudge whether the mutant TDP-43 is no longer associated to the centrosome with a possible local loss-of-function, or whether it is still targeted to the organelle with both toxic gain- and loss-of-function.
Taken together, our findings demonstrate that TDP-43 plays a role at the centrosome through RNA binding and protein interactions, and that centrosomal dysfunction could participate in TDP-43 related neurodegeneration. This first description of TDP-43 centrosomal localization in cells paves the way for a more comprehensive understanding of TDP-43 physiology and pathology.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Paul Guichard, University of Geneva, Section of Biology, Department of Molecular and Cellular Biology, Geneva, Switzerland for contribution to Fig. 2C.
Contributor Information
Alexia Bodin, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France; Neurobiology and neuropathology, University-Hospital of Angers, 49933 Angers, France.
Logan Greibill, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris Sud, Université Paris-Saclay, 91190 Gif sur Yvette, France.
Julien Gouju, Neurobiology and neuropathology, University-Hospital of Angers, 49933 Angers, France.
Franck Letournel, Neurobiology and neuropathology, University-Hospital of Angers, 49933 Angers, France.
Silvia Pozzi, Department of Psychiatry and Neuroscience, University of Laval, Québec City, Qc G1V 0A6, Canada; CERVO Brain Research Centre, Québec, Qc G1E 1T2, Canada.
Jean-Pierre Julien, Department of Psychiatry and Neuroscience, University of Laval, Québec City, Qc G1V 0A6, Canada; CERVO Brain Research Centre, Québec, Qc G1E 1T2, Canada.
Laurence Renaud, Département de Neurosciences, Université de Montréal, Montréal, Qc H3C 3J7, Canada; Groupe de recherche sur le système nerveux central, Université de Montréal, Montréal, Qc H3C 3J7, Canada.
Delphine Bohl, Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, 75013 Paris, France.
Stéphanie Millecamps, Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, 75013 Paris, France.
Christophe Verny, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France; Department of Neurology, Amyotrophic Lateral Sclerosis Center, University-Hospital of Angers, 49933 Angers, France.
Julien Cassereau, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France; Department of Neurology, Amyotrophic Lateral Sclerosis Center, University-Hospital of Angers, 49933 Angers, France.
Guy Lenaers, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France; Department of Neurology, Amyotrophic Lateral Sclerosis Center, University-Hospital of Angers, 49933 Angers, France.
Arnaud Chevrollier, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France.
Anne-Marie Tassin, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris Sud, Université Paris-Saclay, 91190 Gif sur Yvette, France.
Philippe Codron, Univ Angers, Equipe MitoLab, Unité MitoVasc, Inserm U1083, CNRS 6015, SFR ICAT, 49100 Angers, France; Neurobiology and neuropathology, University-Hospital of Angers, 49933 Angers, France; Department of Neurology, Amyotrophic Lateral Sclerosis Center, University-Hospital of Angers, 49933 Angers, France.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by the Conseil National de la Recherche Scientifique (CNRS), the Institut national de la santé et de la recherche médicale (Inserm), the European Regional Development Fund (ERDF), the Association pour la recherche sur la SLA (ARSLA) (2021 scientific grant), the University Hospital of Angers (grant 21_0266_1), and the University of Angers (PhD research fellowship to A.B.).
Competing interests
The authors report no competing interest.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. [DOI] [PubMed] [Google Scholar]
- 2. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. [DOI] [PubMed] [Google Scholar]
- 3. Nelson PT, Dickson DW, Trojanowski JQ, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): Consensus working group report. Brain. 2019;142:1503–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabashi E, Valdmanis PN, Dion P, et al. TARDBP Mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. [DOI] [PubMed] [Google Scholar]
- 5. Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zein-Sabatto H, Lerit DA. The identification and functional analysis of mRNA localizing to centrosomes. Front Cell Dev Biol. 2021;9:782802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Safieddine A, Coleno E, Salloum S, et al. A choreography of centrosomal mRNAs reveals a conserved localization mechanism involving active polysome transport. Nat Commun. 2021;12:1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chouaib R, Safieddine A, Pichon X, et al. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev Cell. 2020;54:773–791.e5. [DOI] [PubMed] [Google Scholar]
- 9. Codron P, Cassereau J, Vourc'h P, et al. Primary fibroblasts derived from sporadic amyotrophic lateral sclerosis patients do not show ALS cytological lesions. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19(5–6):446–456. [DOI] [PubMed] [Google Scholar]
- 10. Journe-Mallet I, Gouju J, Etcharry-Bouyx F, et al. Design and application of a customizable relational DataBase to assess clinicopathological correlations and concomitant pathology in neurodegenerative diseases. Brain Pathol. 2023;33:e13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast spc98p and its association with gamma-tubulin. J Cell Biol. 1998;141:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolff A, de Néchaud B, Chillet D, et al. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
- 13. Codron P, Letournel F, Marty S, et al. STochastic optical reconstruction microscopy (STORM) reveals the nanoscale organization of pathological aggregates in human brain. Neuropathol Appl Neurobiol. 2021;47:127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gogendeau D, Guichard P, Tassin AM. Purification of centrosomes from mammalian cell lines. Methods Cell Biol. 2015;129:171–189. [DOI] [PubMed] [Google Scholar]
- 15. Bailly E, Dorée M, Nurse P, Bornens M. P34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 1989;8:3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. [DOI] [PubMed] [Google Scholar]
- 17. Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. [DOI] [PubMed] [Google Scholar]
- 18. Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prosser SL, Tkach J, Gheiratmand L, et al. Aggresome assembly at the centrosome is driven by CP110-CEP97-CEP290 and centriolar satellites. Nat Cell Biol. 2022;24:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mennella V, Keszthelyi B, McDonald KL, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang B, Armaos A, Tartaglia GG. RNAct: Protein-RNA interaction predictions for model organisms with supporting experimental data. Nucleic Acids Res. 2019;47(D1):D601–D606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Nostrand EL, Pratt GA, Shishkin AA, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 2016;13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oughtred R, Rust J, Chang C, et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. UniProt Consortium . Uniprot: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 26. Millecamps S, Gentil BJ, Gros-Louis F, Rouleau G, Julien JP. Alsin is partially associated with centrosome in human cells. Biochim Biophys Acta. 2005;1745:84–100. [DOI] [PubMed] [Google Scholar]
- 27. Yang Y, Hentati A, Deng HX, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. [DOI] [PubMed] [Google Scholar]
- 28. Hadano S, Hand CK, Osuga H, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–173. [DOI] [PubMed] [Google Scholar]
- 29. Gnazzo MM, Uhlemann EME, Villarreal AR, Shirayama M, Dominguez EG, Skop AR. The RNA-binding protein ATX-2 regulates cytokinesis through PAR-5 and ZEN-4. Mol Biol Cell. 2016;27:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stubenvoll MD, Medley JC, Irwin M, Song MH. ATX-2, the C. elegans ortholog of human ataxin-2, regulates centrosome size and microtubule dynamics. PLoS Genet. 2016;12:e1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hart MP, Gitler AD. ALS-Associated Ataxin 2 PolyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hsu LC, White RL. BRCA1 Is associated with the centrosome during mitosis. Proc Natl Acad Sci U S A. 1998;95:12983–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hill SJ, Mordes DA, Cameron LA, et al. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci U S A. 2016;113:E7701–E7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noristani HN, Sabourin JC, Gerber YN, et al. Brca1 is expressed in human microglia and is dysregulated in human and animal model of ALS. Mol Neurodegeneration. 2015;10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. D’Angiolella V, Donato V, Vijayakumar S, et al. SCFCyclin F controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rayner SL, Yang S, Farrawell NE, et al. TDP-43 is a ubiquitylation substrate of the SCFcyclin F complex. Neurobiol Dis. 2022;167:105673. [DOI] [PubMed] [Google Scholar]
- 38. Williams KL, Topp S, Yang S, et al. CCNF Mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Müller-Taubenberger A, Ishikawa-Ankerhold HC, Kastner PM, Burghardt E, Gerisch G. The STE group kinase SepA controls cleavage furrow formation in Dictyostelium. Cell Motil Cytoskeleton. 2009;66:929–939. [DOI] [PubMed] [Google Scholar]
- 40. Liachko NF, McMillan PJ, Guthrie CR, Bird TD, Leverenz JB, Kraemer BC. CDC7 Inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration: CDC7 and TDP-43 pathology. Ann Neurol. 2013;74:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vaca G, Martinez-Gonzalez L, Fernandez A, et al. Therapeutic potential of novel cell division cycle kinase 7 inhibitors on TDP-43-related pathogenesis such as Frontotemporal Lobar Degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). J Neurochem. 2021;156:379–390. [DOI] [PubMed] [Google Scholar]
- 42. Ott C, Nachmias D, Adar S, et al. VPS4 Is a dynamic component of the centrosome that regulates centrosome localization of γ-tubulin, centriolar satellite stability and ciliogenesis. Sci Rep. 2018;8:3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deng X, Sun X, Yue W, et al. CHMP2B Regulates TDP-43 phosphorylation and cytotoxicity independent of autophagy via CK1. J Cell Biol. 2022;221:e202103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. [DOI] [PubMed] [Google Scholar]
- 45. Parkinson N, Ince PG, Smith MO, et al. ALS Phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology. 2006;67:1074–1077. [DOI] [PubMed] [Google Scholar]
- 46. Greer YE, Westlake CJ, Gao B, et al. Casein kinase 1δ functions at the centrosome and Golgi to promote ciliogenesis. Mol Biol Cell. 2014;25:1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kametani F, Nonaka T, Suzuki T, et al. Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem Biophys Res Commun. 2009;382:405–409. [DOI] [PubMed] [Google Scholar]
- 48. Nonaka T, Suzuki G, Tanaka Y, et al. Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1δ triggers mislocalization and accumulation of TDP-43. J Biol Chem. 2016;291:5473–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alquezar C, Salado IG, de la Encarnación A, et al. Targeting TDP-43 phosphorylation by casein kinase-1δ inhibitors: A novel strategy for the treatment of frontotemporal dementia. Mol Neurodegeneration. 2016;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eguether T, Ermolaeva MA, Zhao Y, et al. The deubiquitinating enzyme CYLD controls apical docking of basal bodies in ciliated epithelial cells. Nat Commun. 2014;5:4585. [DOI] [PubMed] [Google Scholar]
- 51. Dobson-Stone C, Hallupp M, Shahheydari H, et al. CYLD Is a causative gene for frontotemporal dementia—Amyotrophic lateral sclerosis. Brain. 2020;143:783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kodani A, Salomé Sirerol-Piquer M, Seol A, Manuel Garcia-Verdugo J, Reiter JF. Kif3a interacts with dynactin subunit p150 glued to organize centriole subdistal appendages. EMBO J. 2013;32:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhapparova ON, Fokin AI, Vorobyeva NE, Bryantseva SA, Nadezhdina ES. Ste20-like protein kinase SLK (LOSK) regulates microtubule organization by targeting dynactin to the centrosome. Mol Biol Cell. 2013;24:3205–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deshimaru M, Kinoshita-Kawada M, Kubota K, et al. DCTN1 Binds to TDP-43 and regulates TDP-43 aggregation. Int J Mol Sci. 2021;22:3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Konno T, Ross OA, Teive HAG, Sławek J, Dickson DW, Wszolek ZK. DCTN1-related Neurodegeneration: Perry syndrome and beyond. Parkinsonism Relat Disord. 2017;41:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen WJ, Wang WT, Tsai TY, Li HK, Lee YHW. DDX3 Localizes to the centrosome and prevents multipolar mitosis by epigenetically and translationally modulating p53 expression. Sci Rep. 2017;7:9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng W, Wang S, Zhang Z, et al. CRISPR-Cas9 Screens identify the RNA helicase DDX3X as a repressor of C9ORF72 (GGGGCC)n repeat-associated non-AUG translation. Neuron. 2019;104:885–898.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Filippova N, Yang X, King P, Nabors LB. Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function. J Biol Chem. 2012;287:32277–32287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lu L, Zheng L, Si Y, et al. Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS. J Biol Chem. 2014;289:31792–31804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matsye P, Zheng L, Si Y, et al. Hur promotes the molecular signature and phenotype of activated microglia: Implications for amyotrophic lateral sclerosis and other neurodegenerative diseases. Glia. 2017;65:945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leemann-Zakaryan RP, Pahlich S, Sedda MJ, Quero L, Grossenbacher D, Gehring H. Dynamic subcellular localization of the Ewing sarcoma proto-oncoprotein and its association with and stabilization of microtubules. J Mol Biol. 2009;386:1–13. [DOI] [PubMed] [Google Scholar]
- 63. Chi B, O’Connell JD, Yamazaki T, Gangopadhyay J, Gygi SP, Reed R. Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II machinery and contains multiple ALS/SMA-causative proteins. Sci Rep. 2018;8:8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Couthouis J, Hart MP, Erion R, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoshino Y, Ishioka C. Inhibition of glycogen synthase kinase-3 beta induces apoptosis and mitotic catastrophe by disrupting centrosome regulation in cancer cells. Sci Rep. 2015;5:13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moujalled D, James JL, Parker SJ, et al. Kinase inhibitor screening identifies cyclin-dependent kinases and glycogen synthase kinase 3 as potential modulators of TDP-43 cytosolic accumulation during cell stress. PLoS One. 2013;8:e67433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Choi HJ, Cha SJ, Lee JW, Kim HJ, Kim K. Recent advances on the role of GSK3β in the pathogenesis of amyotrophic lateral sclerosis. Brain Sci. 2020;10:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ran J, Yang Y, Li D, Liu M, Zhou J. Deacetylation of α-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci Rep. 2015;5:12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hebron ML, Lonskaya I, Sharpe K, et al. Parkin ubiquitinates tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J Biol Chem. 2013;288:4103–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fazal R, Boeynaems S, Swijsen A, et al. HDAC6 Inhibition restores TDP-43 pathology and axonal transport defects in human motor neurons with TARDBP mutations. EMBO J. 2021;40:e106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. White MC, Quarmby LM. The NIMA-family kinase, nek1 affects the stability of centrosomes and ciliogenesis. BMC Cell Biol. 2008;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brenner D, Müller K, Wieland T, et al. NEK1 Mutations in familial amyotrophic lateral sclerosis. Brain. 2016;139(Pt 5):e28. [DOI] [PubMed] [Google Scholar]
- 73. Nejedlá M, Klebanovych A, Sulimenko V, et al. The actin regulator profilin 1 is functionally associated with the mammalian centrosome. Life Sci Alliance. 2021;4:e202000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tanaka Y, Nonaka T, Suzuki G, Kametani F, Hasegawa M. Gain-of-function profilin 1 mutations linked to familial amyotrophic lateral sclerosis cause seed-dependent intracellular TDP-43 aggregation. Hum Mol Genet. 2016;25:1420–1433. [DOI] [PubMed] [Google Scholar]
- 75. Wu CH, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bannon JH, O’Donovan DS, Kennelly SME, Mc Gee MM. The peptidyl prolyl isomerase cyclophilin A localizes at the centrosome and the midbody and is required for cytokinesis. Cell Cycle. 2012;11:1340–1353. [DOI] [PubMed] [Google Scholar]
- 77. Lauranzano E, Pozzi S, Pasetto L, et al. Peptidylprolyl isomerase A governs TARDBP function and assembly in heterogeneous nuclear ribonucleoprotein complexes. Brain. 2015;138:974–991. [DOI] [PubMed] [Google Scholar]
- 78. Pasetto L, Grassano M, Pozzi S, et al. Defective cyclophilin A induces TDP-43 proteinopathy: Implications for amyotrophic lateral sclerosis and frontotemporal dementia. Brain. 2021;144:3710–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pillai S, Nguyen J, Johnson J, Haura E, Coppola D, Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun. 2015;6:10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Freischmidt A, Wieland T, Richter B, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. [DOI] [PubMed] [Google Scholar]
- 81. Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Čajánek L, Nigg EA. Cep164 triggers ciliogenesis by recruiting tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci U S A. 2014;111:E2841–E2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liachko NF, McMillan PJ, Strovas TJ, et al. The tau tubulin kinases TTBK1/2 promote accumulation of pathological TDP-43. PLoS Genet. 2014;10:e1004803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Taylor LM, McMillan PJ, Liachko NF, et al. Pathological phosphorylation of tau and TDP-43 by TTBK1 and TTBK2 drives neurodegeneration. Mol Neurodegener. 2018;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Balestra FR, Strnad P, Flückiger I, Gönczy P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev Cell. 2013;25:555–571. [DOI] [PubMed] [Google Scholar]
- 86. Madeo F, Schlauer J, Zischka H, Mecke D, Fröhlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell. 1998;9:131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Watts GDJ, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. [DOI] [PubMed] [Google Scholar]
- 88. Kraemer BC, Schuck T, Wheeler JM, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sephton CF, Good SK, Atkin S, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. [DOI] [PubMed] [Google Scholar]
- 91. Iguchi Y, Katsuno M, Niwa J, et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136(Pt 5):1371–1382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.