Summary
In this study we examined how genetic risk for asthma associates with different features of the disease and with other medical conditions and traits. Using summary statistics from two multi-ancestry genome-wide association studies of asthma, we modeled polygenic risk scores (PRSs) and validated their predictive performance in the UK Biobank. We then performed phenome-wide association studies of the asthma PRSs with 371 heritable traits in the UK Biobank. We identified 228 total significant associations across a variety of organ systems, including associations that varied by PRS model, sex, age of asthma onset, ancestry, and human leukocyte antigen region alleles. Our results highlight pervasive pleiotropy between asthma and numerous other traits and conditions and elucidate pathways that contribute to asthma and its comorbidities.
Keywords: asthma, phenome-wide association study, PheWAS, polygenic risk score, PRS, UK Biobank, UKB, HLA, pleiotropy, Global Biobank Meta-analysis Initative, GBMI
We performed phenome-wide association studies of asthma polygenic risk scores in the UK Biobank, identifying significant associations across a variety of organ systems, including associations that varied by genetic risk model, sex, age of asthma onset, ancestry, and human leukocyte antigen region alleles.
Introduction
Asthma is a common respiratory disorder, affecting 3%–4% of people globally, with prevalences exceeding 15% in many regions among different groups.1,2,3 The disease is characterized by episodic airflow obstruction caused by airway narrowing from bronchial constriction and inflammation, which can be induced by inhaled exposures, such as viruses, allergens, and pollutants.4 Genetic factors account for a substantial portion of overall disease liability,5 with over 200 different genetic risk loci reported for asthma in large genome-wide association studies (GWASs).6,7 These GWAS findings have implicated many genes and pathways in the pathogenesis of asthma,8 but the significant GWAS alleles contribute only modestly to asthma heritability9 and individually paint a limited picture of the overall genetic risk for asthma.10 By instead considering the cumulative contribution from all common genetic variants and modeling a comprehensive polygenic risk score (PRS), we can measure how genetic risk for asthma differs by sex, age of disease onset, and ancestry and the extent to which it is associated with other traits and diseases, generating new insights into asthma etiology and its shared biology with other traits.
Previous studies that have developed PRSs for asthma have focused on predictive performance11,12,13,14,15 or associations with specific traits,15,16,17,18,19,20,21 while broader studies of cross-trait genetic correlations between asthma and other conditions offer less interpretable findings than PRS associations in terms of relative disease risk.7,22,23,24,25,26,27,28,29 In this study, we aimed to provide a quantitative overview of the genetic landscape for asthma in terms of other heritable traits and conditions and their associations with genetic asthma risk. We first modeled PRSs of asthma using GWAS summary statistics from the Trans-National Asthma Genetic Consortium (TAGC; 23,948 cases, 118,538 controls)30 and the Global Biobank Meta-analysis Initiative (GBMI; 153,763 cases, 1,647,022 controls)7 and then derived scores for individuals in the UK Biobank (UKB; n = 398,744).31 We assessed the predictive performance of our models in several UKB populations stratified by age of disease onset, sex, and ancestry. We then performed phenome-wide association studies (PheWASs) of genetic asthma risk with 371 heritable traits reported in the UKB using both GBMI and TAGC PRS models and analyzed sensitivities to asthma status, age of disease onset, ancestry, and human leukocyte antigen (HLA) region alleles. Our results highlight shared genetic architecture between asthma and numerous other traits and conditions, including some with sex, age, and/or ancestry dependencies, elucidating the affected pathways that contribute to asthma and its comorbidities.
Subjects and methods
Study populations and asthma phenotyping
The polygenic risk models used in this study were derived from summary statistics of the TAGC and GBMI meta-analyses of asthma.7,30,32 The TAGC meta-analysis integrated data from 66 GWASs from multiple populations: European ancestry (19,954 cases, 107,715 controls), African ancestry (2,149 cases, 6,055 controls), Japanese (1,239 cases, 3,976 controls), and Latino (606 cases, 792 controls). Summary statistics from TAGC were available for the entire multi-ancestry sample (23,948 cases, 118,538 controls) and for the European-ancestry subset (19,954 cases, 107,715 controls). Twenty-seven of the 66 studies included in TAGC only included cases of asthma with age of onset ≤16 years (8,976 cases, 18,399 controls). Cases of asthma were based on doctor’s diagnosis and/or standardized questionnaires.
The GBMI includes combined health and genomic data from over 2.2 million participants across 23 biobanks.33 The discovery GWAS dataset used to model the GBMI asthma PRS consisted of 197,342 cases and 1,903,937 controls from eight biobanks (BioBank Japan,34 BioVU,35 Lifelines,36 Ontario Health Study,37 Estonian Biobank,38 FinnGen,39 Michigan Genomics Initiative,40 and the HUNT Study41). The cohort was 66.3% European (141,957/1,251,642 cases/controls), 27.7% East Asian (33,312/549,116 cases/controls), 2.4% African (8,661/42,279 cases/controls), 2.1% South Asian (6,162/38,103 cases/controls), and 1.4% admixed American (7,250/22,797 cases/controls).32 Asthma case and control statuses were assigned according to specific International Classification of Diseases (ICD)-9 and ICD-10 codes in participant electronic health records,33 except for BioBank Japan, which used physician’s diagnosis or past medical history (including self-report) to assign asthma status, and the Lifelines study, which used doctor’s diagnosis or current symptoms/treatment.
To evaluate the TAGC and GBMI polygenic risk models, PRSs were derived for genotyped individuals in the UKB.31 We tested our model in three different population samples defined by ancestry: white British (n = 376,237), white non-British (n = 27,026), and African (n = 7,196). The white British and white non-British populations were as defined by the UKB,31 while the African-ancestry population combined samples with African ancestries from multiple UKB cohorts. For this African-ancestry population, we included individuals from the “Black or Black British,” “African,” “Caribbean,” and “Any other Black background” cohorts with a first principal component (PC) of ancestry value >150 (UKB data field 22009), which indicated significant African admixture and greater genetic dissimilarity from the white UKB cohorts. UKB data are available following a process described at https://www.ukbiobank.ac.uk/enable-your-research.
We used quality-control metrics derived centrally by the UKB to process the genotype data, including ancestry PC values and ancestry group designation.31 We excluded individuals with poor-quality genotypes based on excess heterozygosity, missingness, and/or ambiguous genetic sex assignment. For pairs of individuals who were first- or second-degree relatives, only the individual with the smallest number of missing genotypes was included. We performed additional quality control of genotypes, requiring an imputation quality score (INFO) >0.8, a minor allele frequency (MAF) >0.1%, variant call rate >95%, and a Hardy-Weinberg equilibrium p value >0.1 × 10−10.
We defined asthma in the UKB according to age of asthma diagnosis fields (UKB data fields 3786 and 22147), with individuals with a diagnosis before age 66 years considered to have had asthma. As in our previous study,9 we defined childhood-onset asthma (COA) and adult-onset asthma (AOA) in the UKB using strict age-of-onset criteria to minimize the likelihood of misclassification; COA was defined as an asthma diagnosis before 12 years of age (n = 9,611) and AOA was defined as asthma onset after age 25 years and before 66 years (n = 21,427). Participants older than 38 years of age without an asthma diagnosis (self-reported or ICD-10 codes) were included as controls for these fields. Individuals with chronic obstructive pulmonary disease (COPD), emphysema, or chronic bronchitis (self-reported or ICD-10 codes) were excluded from the AOA and control groups.
PRS modeling
Polygenic risk models were generated from each of the TAGC and GBMI results, separately. For TAGC, we derived polygenic risk models from the GWAS summary statistics. For GBMI, we adopted the model published by Wang and colleagues32 from the GBMI leave-UKB-out multi-ancestry GWAS deposited on the Polygenic Score Catalog (PGS001787).42 All polygenic risk models were generated using PRS-CS,43 a Python-based program that applies a high-dimensional Bayesian regression framework with continuous shrinkage priors placed on single-nucleotide polymorphism (SNP) effect sizes. The default half-Cauchy priors, α = 1 and β = 0.5, were used for determining the local shrinkage parameters in the PRS-CS model (ψj). To determine the global shrinkage parameter (φ), the “PRS-CS-auto” algorithm was applied to estimate the value directly from the GWAS summary statistics. Each autosomal chromosome was modeled separately, producing different parameters for each. For the Gibbs sampling used to derive the global shrinkage parameter and posterior SNP effect sizes, we ran 10,000 Markov Chain Monte Carlo iterations with 5,000 burn-in steps for the TAGC models, and the GBMI models used the default parameters of 1,000 and 500, respectively.32 For each set of TAGC summary statistics, we trained two different models, incorporating European linkage disequilibrium (LD) panels from either the 1000 Genomes (1KG) Project44 or from the UKB, as prepared by the PRS-CS developers.43 The GBMI model utilized the European 1KG LD panel. To improve computational efficiency and standardize the reference sets, the LD panels were created for common (minor allele frequency >1%), non-ambiguous (no A/T and C/G) HapMap3 SNPs with INFO >0.8 (UKB = 1,117,425 SNPs; 1KG = 1,120,696 SNPs).43 LD block boundaries were determined using LDetect on European samples from 1KG (n = 1,703 blocks).45 Previous studies have shown that European-based LD panels can adequately approximate LD in multi-ancestry GWAS with majority European ancestry.32,46 Posterior SNP effect sizes were combined into aggregate scores for individuals in the UKB using the Plink (v2.00a2 AFX2) “score” function.47 Effects for missing genotypes were imputed as the posterior SNP effect size multiplied by the effect allele frequency. We converted the PRSs to Z scores for analysis.
Asthma prediction in the UKB
We evaluated scores from five different polygenic risk models, four from TAGC utilizing either the 1KG or UKB LD reference panels with each of the European- and multi-ancestry summary statistics and one from GBMI excluding UKB participants and utilizing the 1KG LD panel.7,32 We assessed the predictive performance of the five PRSs in the white British, white non-British, and African-ancestry populations in the UKB using receiver-operating characteristic curves.48 We estimated the variance explained by each PRS using R2 on liability scale, using bootstrap sampling (n = 1,000) to derive 95% confidence intervals (CIs).49 We selected the PRS with the highest area under the receiver-operating-characteristic curve (AUC) for all subsequent primary statistical analyses. PRS associations with asthma were evaluated using logistic regressions, adjusting for sex and the first 10 PCs of ancestry (first 20 PCs in African-ancestry samples). In assessing relative risks between PRS quantiles, we set individuals with scores between the 40th and 60th percentiles as the reference group. Interaction effects with sex for PRS associations were tested by adding a PRS-by-sex interaction term to the logistic regression models.
PheWAS
Traits included in the PheWAS were selected from 4,178 UKB phenotypes with additive SNP-heritability estimates from the Neale Lab UKB SNP-Heritability Browser (http://www.nealelab.is/uk-biobank/), as updated on October 19, 2019. SNP heritability was estimated using partitioned LD score regression50 and assigned confidence ratings based on effective sample sizes, standard errors, sex biases, and ordinal coding. We considered traits with estimated SNP heritability >0.10 with low, medium, or high confidence estimates (n = 519). We then omitted those that were not interpretable as biomedical phenotypes or laboratory values, such as occupations or food preferences. We extracted phenotypes from the UKB using the ukbREST tool on a local data server.10 Phenotypes derived from ICD-10 codes in participant hospital inpatient records correspond to primary diagnoses (UKB data field 41202).
Some phenotypes were refined to improve interpretability. We removed the self-reported (UKB data field 20002) and doctor-diagnosed diabetes (UKB data field 2443) fields, which did not distinguish between type 1 and type 2 and instead used primary and secondary ICD-10 codes (UKB data fields 41202 and 41204) to indicate either disease. For forced expiratory volume in 1 s (FEV1), we only considered the percentage predicted phenotype (UKB data field 20154). For COPD, we expanded the definition to account for all self-reported or doctor-diagnosed COPD, emphysema, or chronic bronchitis (UKB data fields 20002, 41202, 41204, 22128, 22129, 22130, 22148, 22149, 22150, 3992). Following filtering and refinement, 371 traits remained (208 binary phenotypes, 163 quantitative traits) for association testing with the PRS. Twenty-nine of the traits were measured in only one sex, including traits that could apply to either sex but were assessed in sex-specific survey questions (e.g., UKB data fields 6153 and 6177). Traits were manually categorized into 13 groups according to their affiliated biological system: anthropometric (n = 45), asthma and allergic disease (n = 23); cardiovascular (n = 39), gastrointestinal (n = 12); hematologic and blood chemistry (n = 46); musculoskeletal and skin (n = 34); neoplasms (n = 14); neurologic and behavioral (n = 48); ocular (n = 32); other (n = 14); pulmonary (n = 17); renal and urologic (n = 9); and reproductive, endocrine, and metabolic (n = 38).
We performed separate PheWAS in the following UKB cohorts: white British, white British excluding individuals with diagnosis or self-report of asthma, white non-British, and African ancestry. We tested associations between each of the PheWAS traits and the asthma PRS using logistic regressions for binary phenotypes and linear regressions for quantitative traits, adjusting for sex and the first 10 PCs of ancestry (first 20 PCs in African-ancestry samples). Quantitative traits were standardized using a Z score transformation but were not normalized prior to association testing, as it was assumed sample sizes were sufficiently large to invoke the central limit theorem. Phenome-wide significance was determined using Bonferroni multiple-test correction (p < 0.05/371), which was conservative considering the high correlations between many pairs of traits (e.g., left arm fat mass, right arm fat mass). For display purposes, we labeled highly correlated trait groupings as single traits, such as for anthropometric measures with values from both the left and right sides, resulting in 253 non-redundant traits, and displayed the strongest trait association statistics from within each grouping. Trait associations with p values less than the minimum subnormal double-precision floating-point number, 2−1074, were labeled as p < 4.9 × 10−324.
In defining the non-asthma PheWAS population in the UKB, we excluded individuals with a report of asthma in any of the following UKB fields: 3786, 22147, 6152, 20002, 22127, and 41202. We did not further exclude individuals based on COPD, emphysema, or chronic bronchitis, as we did in defining the AOA and control groups, so that we would compare the PRS association effects for these traits across PheWAS cohorts.
When comparing individual trait associations between different PRS models, we determined bias toward one PRS or the other based on the absolute differences between regression effect sizes. Therefore, for quantitative traits, the biases equated to the total differences in standardized effect sizes [Δβz] between PRS associations, whereas, for binary traits, the biases were equivalent to the relative change in odds ratio (OR) (OR1/OR2). To evaluate whether there were differences in association strengths between PRSs for specific phenotypic categories, we regressed the effect sizes or ORs of all nominally significant (p < 0.05) PheWAS associations of one PRS against the other with a fixed intercept at zero for each category and then performed one-sample t tests of the resultant slopes against a value of 1. When applicable, we combined the binary and quantitative results into one p value for each category using Stouffer’s method weighted by the number of traits in each category by variable type (binary or quantitative).
To test for significant differences in individual PRS-trait association effects, whether between different populations for the same PRS or between different PRSs for the same population, we used the Wald statistic:
where β1 and β2 are the estimated effect sizes for each PRS association, and W asymptotically follows a z distribution for binary traits and a t distribution for quantitative traits. For the PheWAS in non-asthmatic individuals, we used inverse probability weighting to mitigate potential selection bias induced from conditioning on asthma status.51 Regression weights were set as the inverse predicted probability of never having asthma, as determined from a logistic regression of asthma (union of all asthma phenotypes) against the PRS, sex, ancestry PCs 1–10, and four additional environmental confounders: body mass index (BMI), the Townsend deprivation index, disability allowance, and pack years smoking.
HLA effects
To evaluate the extent to which the HLA region contributed to PRS-trait associations, we calculated modified PRSs for individuals in the white British cohort, in which all contributions from SNPs in the extended HLA region (chr6:28477797-33448354, GRCh37) were removed. We then performed a PheWAS using the HLA-removed scores and compared the association effect sizes with those generated from the initial PRS PheWAS in the white British population. Statistical probabilities for differences in AUCs between models were estimated using 1,000 bootstrap permutations.
Trait mediation analysis
To quantify the relative contributions of eosinophilia, lung function, and allergic disease on the overall genetic risk of asthma, we performed mediation analyses52 with each factor modeled as a mediator between the PRS and asthma. We performed separate tests for asthma, COA, and AOA. We used blood eosinophil count (UKB field 30150), ratio of FEV1 to forced vital capacity (FVC) (UKB field 20258), and age of hay fever diagnosis (UKB field 3761) as the mediators in our models, denoting eosinophilia, lung function, and allergic disease, respectively. Z score transformation was first applied to each trait within the white British UKB population. The mediation analysis consisted of three regression models:
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
where Y is the dependent asthma phenotype; X is the PRS; M is the mediating trait; c, a, c′, and b are coefficients relating the independent variables to the dependent variables; Z is a matrix of baseline covariates with β coefficients; i is the intercept; and e are the residuals. We included sex and the first 10 PCs of ancestry as fixed covariates. We also included BMI (UKB field 21001), the Townsend deprivation index (UKB field 189), and estimated pack years of smoking (UKB fields 2867, 2887, 2897, 2907, 3436, 3456, 3486, 6183, 6194, 20160, 21022) as fixed covariates to mitigate against unmeasured confounding. The proportion of the PRS effect on asthma mediated by each mediating trait was determined using the standardized logistic solution proposed by MacKinnon et al.53:
| (Equation 4) |
| (Equation 5) |
where cs is the standardized form of the c coefficient from Equation 1 and σ2MX is the residual variance from Equation 2. We performed bootstrap sampling (n = 1,000) to derive 95% CIs.
Results
Genetic risk prediction of asthma
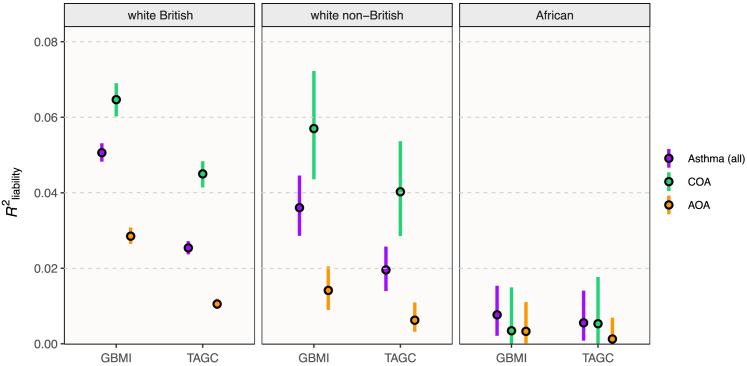
We evaluated the predictive performance of five genetic risk models for asthma, derived using the PRS-CS polygenic prediction method,43 on summary statistics from either the TAGC30 or GBMI7 meta-analyses of asthma GWASs. The four TAGC scores featured between 694,173 SNPs and 699,291 SNPs, depending on the LD panel. The GBMI-based PRS consisted of 637,438 SNPs.7 We compared the performance of these five scores in predicting asthma in white British, white non-British, and African-ancestry population samples in the UKB (Table S1).
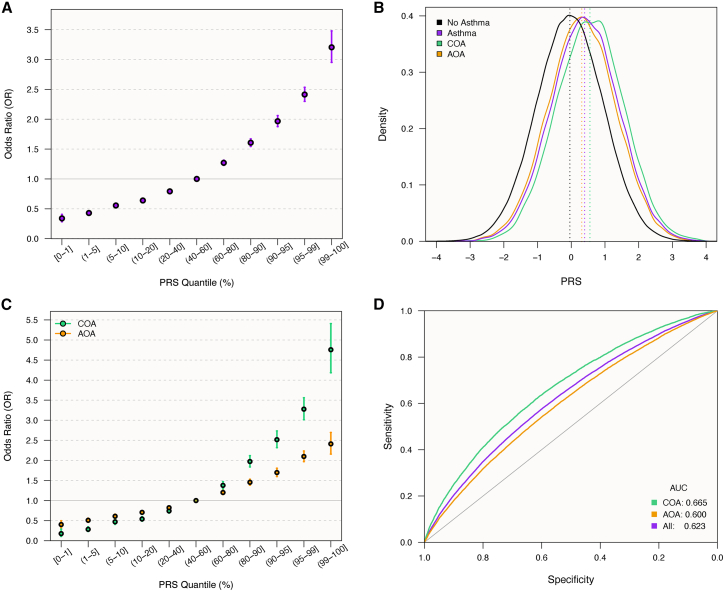
The GBMI PRS was the most accurate in all populations, with an AUC of 0.623 in white British, 0.608 in white non-British, and 0.545 in African-ancestry individuals. Because the GBMI model was the best performing in each group (Figure 1), we used the individual risk scores generated from that model for the remainder of the study, except where we further compare trait associations between models below (see section “trait association differences by PRS model”). The OR for asthma per standard deviation (SD) increase in the PRS was 1.56 (95% CI = 1.54–1.58). Individuals with a PRS in the top decile were 4.69 times (95% CI = 4.43–4.95) more likely to have had asthma than those in the lowest decile (Figure 2A).
Figure 1.
Asthma liability explained by GBMI and TAGC polygenic risk scores
R2 on the liability scale with 95% CIs are shown for the GBMI PRS and best-performing TAGC PRS for all asthma, COA (<12 years old), and AOA (≥25 and <66 years old) in three different UKB populations according to ancestry. AUC, area under the curve.
Figure 2.
PRS prediction of asthma in white British UKB participants
(A) Odds ratios (ORs) with 95% CIs of asthma risk for individuals in different GBMI PRS quantiles relative to those with scores in the 40th–60th quantile.
(B) Density plot showing distributions of PRS scores by asthma status, with mean scores indicated by vertical lines.
(C) ORs with 95% CIs of COA and AOA risk for individuals in different PRS quantiles relative to those with scores in the 40th–60th quantile.
(D) Receiver-operating-characteristic curves for the PRS as a predictor of asthma, COA, and AOA.
Genetic risk by sex and age of onset
Many features of asthma differ by sex and age at disease onset. For example, COA is more prevalent in males and has a high rate of remission, while AOA is more prevalent in females and is associated with more severe symptoms.54,55 Moreover, genetic risk factors contribute more to the development of COA relative to AOA.9,56 We therefore tested PRS associations with asthma by age of onset and by sex to quantify these differences and identify potential interaction effects. The mean PRS was significantly higher in individuals with COA (μ = 0.55) than in individuals with AOA (μ = 0.31; p = 3.4 × 10−87; Figure 2B). The OR for each SD increase in the PRS was 1.83 (95% CI = 1.79–1.87) for COA and 1.43 (95% CI = 1.41–1.45) for AOA (Figure 2C). Individuals with a PRS in the top decile were 8.3 times (95% CI = 7.4–9.3) more likely to have had COA and 3.5 times (95% CI = 3.3–3.8) more likely to have had AOA than those in the lowest decile. Accordingly, the asthma PRS was better at predicting COA (AUC = 0.665) than AOA (AUC = 0.600; Figure 2D). These results reinforce how genetic risk factors contribute more to COA. Indeed, AOA was more prevalent than COA in individuals with a lower asthma PRS, and COA was more common in individuals with a higher PRS (Figure 3A).
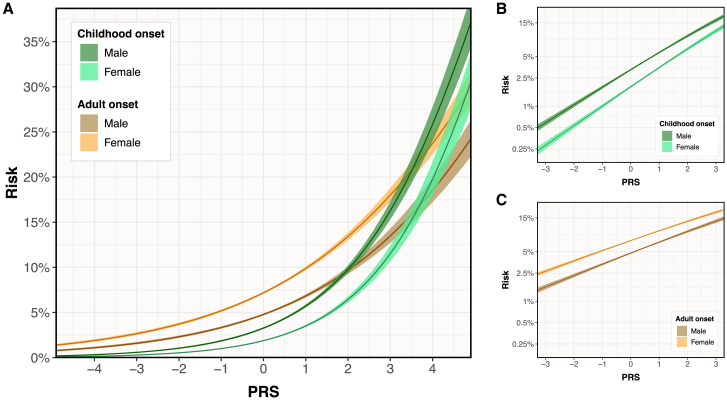
Figure 3.
Asthma risk vs. GBMI PRS by sex and age of onset
(A) Predicted risks of COA and AOA are plotted against the PRS score separately for males and females in the white British UKB population with 95% CIs.
(B) Predicted risks of COA are plotted on a log scale with 95% CIs, with the different slopes demonstrating the PRS-sex interaction for COA.
(C) Predicted risks of AOA are plotted on a log scale with 95% CIs, demonstrating the PRS-sex interaction for AOA.
We also observed significant interaction effects between the PRS and sex, depending on age of onset. While the overall risk of COA was higher in males than in females, regardless of genetic risk, the PRS was slightly more correlated with disease risk in females (interaction p = 1.0 × 10−2; ratio of ORs = 1.06; Figure 3B). We observed the opposite effects in AOA, with a higher overall risk in females but a greater PRS correlation in males (interaction p = 4.6 × 10−2; ratio of ORs = 1.03; Figure 3C). Overall, these results showed that genetic risk is higher in COA than AOA and that sex interacted with the age of asthma onset.
PheWAS of asthma PRS
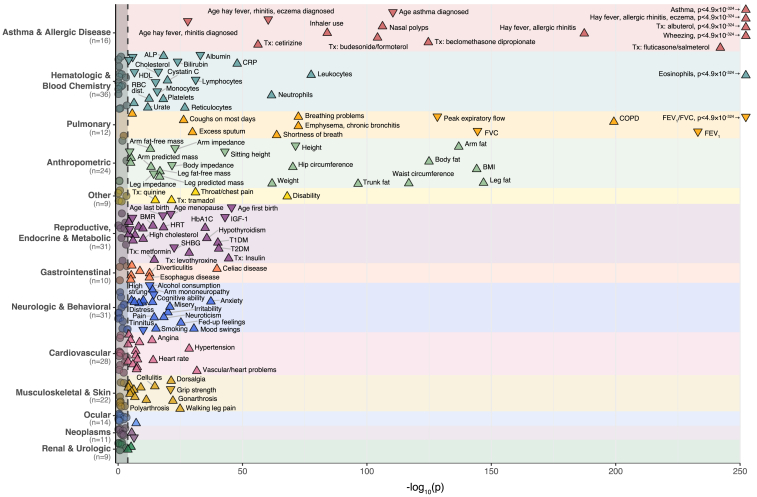
To quantify the polygenic pleiotropy between asthma and other traits and conditions, we performed a PheWAS of the GBMI asthma PRS with 371 heritable biomedical traits in the UKB white British population (n = 376,237). After applying a conservative Bonferroni correction for multiple testing, 214 traits were significantly associated with the asthma PRS across a variety of biological systems (Figure 4; Table S2). As expected, asthma and its related traits and comorbidities were the most strongly associated phenotypes, including wheezing (p < 4.9 × 10−324, OR = 1.30), treatment with albuterol (p < 4.9 × 10−324, OR = 1.59), FEV1/FVC (p < 4.9 × 10−324, βz = −0.08), diagnosis of hay fever/allergic rhinitis or eczema (p < 4.9 × 10−324, OR = 1.24), and eosinophil count (p < 4.9 × 10−324, standardized effect of 1 SD increase in PRS [βz] = 0.11). In line with our earlier GWAS findings,9 the PRS was significantly associated with younger age of onset as a quantitative trait (p = 3.7 × 10−111, βz = −0.12), where each SD increase in the PRS corresponded to 2.1 years (95% CI = 1.9–2.3) younger age of onset. Other significant disease associations of note include several immune-mediated diseases, such as celiac disease (p = 1.5 × 10−40, OR = 1.38), type 1 diabetes (T1DM; p = 8.2 × 10−41, OR = 1.26), and hypothyroidism (p = 2.3 × 10−36, OR = 1.10).
Figure 4.
Phenome-wide association test of GBMI asthma PRS in the UKB
The p values for associations with the asthma PRS are shown for 257 non-redundant traits out of 371 heritable phenotypes tested in the UKB. Traits to the right of the dashed line were significantly associated after correcting for multiple testing (p < 1.35 × 10−4). Traits with association p values <1 × 10−10 are labeled. Traits are grouped by corresponding organ system. The direction of each arrow corresponds to the direction of the association effect. ALP, alkaline phosphatase; AST, aspartate aminotransferase; BMI, body mass index; CRP, C-reactive protein; COPD, chronic obstructive pulmonary disease; DED, diabetes-related eye disease; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; GGT, gamma-glutamyl transferase; HbA1C, glycated hemoglobin; Hgb, hemoglobin; HDL, high-density lipoprotein; HRT, hormone-replacement therapy; MCH, mean corpuscular hemoglobin; RBC, red blood cell; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus; Tx, treatment.
Twenty-nine of the UKB traits included in our study were measured in only one sex (Table S2), and 17 of those were significantly associated with the asthma PRS (Figure 4). In females, age at first live birth (p = 2.5 × 10−46, βz = −0.04, β = −0.17 years), age at menopause (p = 7.1 × 10−22, βz = −0.03, β = −0.14 years), and hormone-replacement therapy (p = 5.6 × 10−19, OR = 1.26) were the top three sex-specific associations with the asthma PRS. In males, insulin treatment (p = 3.0 × 10−29, OR = 1.26), hypertension treatment (p = 1.1 × 10−9, OR = 1.03), and male pattern baldness (p = 1.7 × 10−7, OR = 1.03) were the top three sex-specific associations.
Trait association differences by PRS model
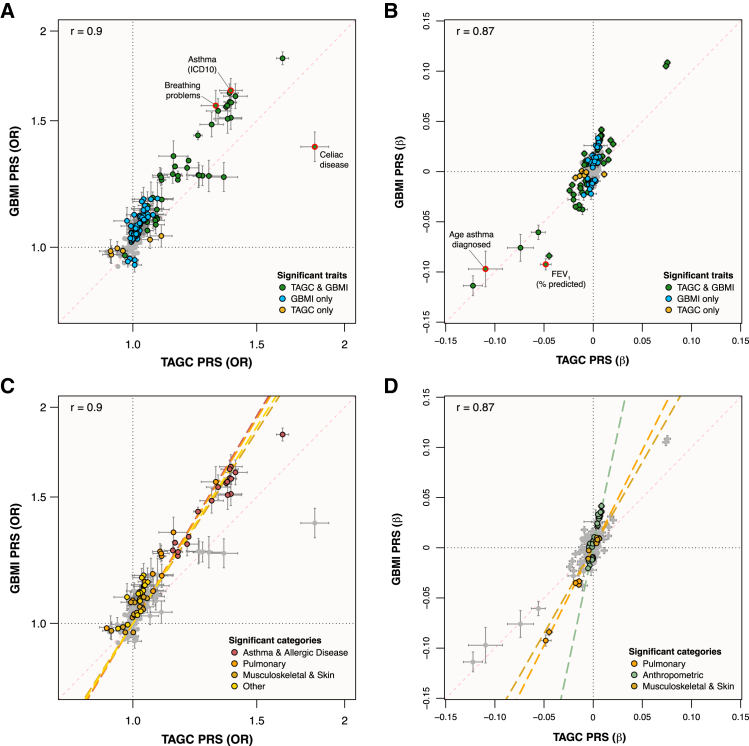
The GBMI and TAGC PRSs were derived from different cohorts that used different phenotype definitions for asthma. The GBMI meta-analysis defined asthma predominantly according to ICD codes from health records of adult participants,7,33 whereas TAGC relied more on physician diagnosis and over 40% of its studies were limited to COA.30 To explore how these differences might affect pleiotropic associations with asthma, we repeated the PheWAS using the best-performing TAGC PRS (multi-ancestry cohort, UKB LD panel), and compared the association results with those from the GBMI PRS PheWAS. The association effect size and OR correlations between PheWASs were r = 0.87 and r = 0.90, respectively (Figure 5).
Figure 5.
Scatterplots of GBMI PRS vs. TAGC PRS-trait associations in the UKB
(A and B) Binary (A) and quantitative (B) traits association effects are shown with 95% CIs, colored according to which PRSs had significant associations. Traits highlighted in red had the largest effect bias toward one or the other PRS models.
(C and D) Binary (C) and quantitative (D) traits association effects are shown with 95% CIs, colored according to which trait categories were significantly biased toward one of the PRS models. ICD-10, International Classification of Diseases, 10th revision.
The TAGC PRS was significantly associated with 110 traits after Bonferroni correction (Figure S1; Table S3), including 14 that were uniquely associated with the TAGC PRS. The TAGC PRS was not significantly associated with 118 of the traits significantly associated with the GBMI PRS. Of the 45 binary traits that were significantly associated with both PRSs, celiac disease (UKB field 20002) had the largest relative effect size bias toward the TAGC PRS (ORTAGC = 1.81, ORGBMI = 1.38), and breathing problems at work (UKB field 22616) had the largest effect size bias toward the GBMI PRS (ORTAGC = 1.31, ORGBMI = 1.57; Figure 5A). Of the 51 quantitative phenotypes significantly associated with both PRSs, age of asthma diagnosis (UKB field 22147) had the largest difference in association effect sizes biased toward the TAGC PRS (βz,TAGC = −0.11, βz,GBMI = −0.10), whereas FEV1 (percentage predicted; UKB field 20154) had the largest bias toward the GBMI PRS (βz,TAGC = −0.05, βz,GBMI = −0.09; Figure 5B).
Categorically, anthropometric trait associations were the most biased toward the GBMI PRS (p = 4.4 × 10−14), followed by asthma and allergic disease (p = 1.0 × 10−11), pulmonary (p = 1.1 × 10−6), musculoskeletal and skin (p = 5.4 × 10−8), and other (p = 1.8 × 10−3) traits (Figures 5C and 5D). Notably, the TAGC PRS was not significantly associated with any of the 24 neurologic and behavioral traits that were significantly associated with the GBMI PRS, including five measures of smoking. Most of the remaining traits that were significantly associated with the GBMI PRS and not the TAGC PRS were related to cardiovascular and metabolic health (Tables S2 and S3).
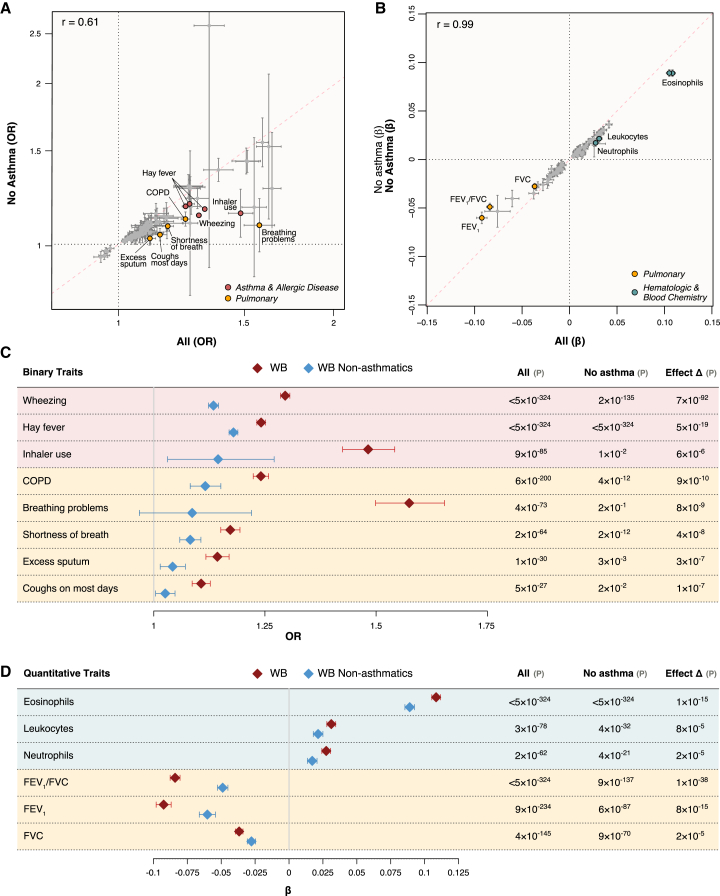
Trait associations in a non-asthma population
Trait associations with asthma genetic risk scores do not necessarily arise from shared genetic architecture but could be secondary to disease-induced physiological and/or behavioral changes. To evaluate the degree to which the trait associations with asthma genetic risk were independent of asthma, we repeated the PheWAS of the GBMI asthma PRS in the white British population, excluding all individuals with a report or diagnosis of asthma (remaining n = 328,353). We used inverse probability weighting to mitigate selection bias induced from conditioning on asthma (see section “subjects and methods”). Of the 205 non-asthma-defining significant traits in the initial PheWAS, 54 (26.3%) were no longer significantly associated with the asthma PRS after correcting for multiple testing, and 17 had significantly weaker association effects in the non-asthmatic cohort (Figure 6; Table S4). Of the 17 traits with weaker association effects, five were related to asthma and allergic disease, eight were related to pulmonary function, and four were related to leukocyte counts. PRS associations with FEV1 (Δβ = −35%, pΔβ = 8.4 × 10−15), FVC (Δβ = −25%, pΔβ = 2.0 × 10−5), FEV1/FVC (Δβ = −42%, pΔβ = 1.4 × 10−38), and COPD prevalence (Δβ = −49%, pΔβ = 8.6 × 10−10) were all significantly weaker in the non-asthmatic cohort. The association with total leukocyte counts was also significantly weaker (Δβ = −31%, pΔβ = 7.9 × 10−5), including significant reductions in association strength for eosinophil counts (Δβ = −18%, pΔβ = 1.1 × 10−15) and neutrophil counts (Δβ = −38%, pΔβ = 2.4 × 10−5), specifically. These results highlight specific trait associations with genetic asthma risk that arise largely due to disease-induced changes, but most of the traits with significantly reduced effect sizes (n = 13/17, 76%) nonetheless remained significantly associated with the asthma PRS. Importantly, for the vast majority of traits, their association effect sizes did not change significantly in the non-asthmatic cohort relative to the cohort with asthmatics, indicating that most trait associations with genetic asthma risk are due to pleiotropic effects arising from shared biology.
Figure 6.
GBMI PRS-trait associations in non-asthmatics
(A and B) Scatterplots of binary (A) and quantitative (B) trait associations with the GBMI PRS in UKB white British for individuals with no history of asthma vs. all individuals are shown with 95% CIs. Traits with significant differences in association effect sizes are labeled and colored according to trait category.
(C and D) Forest plots of binary (C) and quantitative (D) trait associations with the GBMI PRS with significant effect size differences between the non-asthmatic population and the entire white British cohort are shown with 95% CIs. COPD, chronic obstructive pulmonary disease; WB, white British.
Replication in subjects with different ancestries
To assess the robustness of the asthma PRS-trait associations, we performed PheWASs of the GBMI PRS in two other UKB cohorts, a white non-British population (n = 27,026; Table S5) and a population of individuals with African ancestries (n = 7,196; Table S6). In the white non-British population, 208 (97%) of the trait associations had consistent directions of effect with the white British cohort, of which 139 (65%) replicated at p < 0.05. Considering asthma prediction was similar in the white British and white non-British cohorts (Table S1), the non-replicated traits were likely due to the loss of power in the smaller cohort.
In the population with African ancestries, 109 (52%) of the significant trait associations from the white British PheWAS yielded the same direction of effect, with 20 (11%) at p < 0.05. Although this cohort was relatively small, the PRS was also less predictive of asthma in this cohort relative to the others (Table S1), demonstrating the limited portability of PRSs across populations,57 even when using a PRS based on multi-ancestry GWASs. For the traits that replicated at p < 0.05 in the white non-British and African-ancestry cohorts (n = 18), we tested for differences in association effect sizes between African ancestry and the white British and white non-British cohorts. We identified four traits with significantly different effects, including wheezing in the past year (p = 9.7 × 10−5), and three measures of self-reported asthma (UKB data fields 20002, p = 8.8 × 10−4; 6152 [asthma], p = 4.5 × 10−4; 6152 [none], p = 7.2 × 10−4). In each case, the PRS association effect sizes were significantly smaller in the African-ancestries population (Table S6) and likely reflect the lower predictive accuracy of the PRS in the population with African ancestries.
Influence of the HLA region
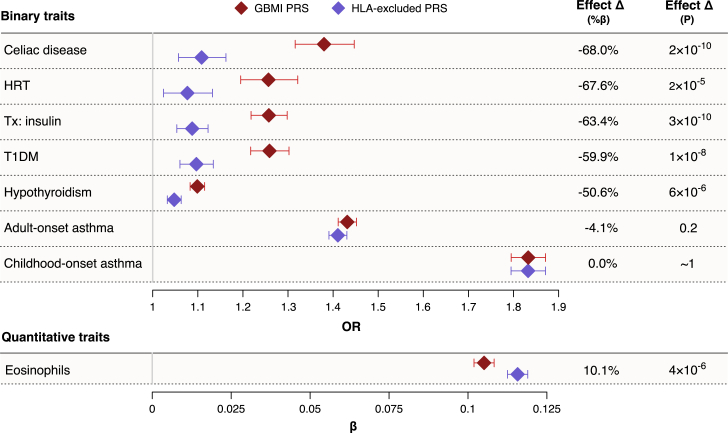
The HLA region on chromosome 6p21.3 is the most gene-dense and polymorphic region in the human genome,58 encoding proteins with critical roles in immune responses. Genetic variants in the HLA region have been robustly associated with risk for both autoimmune and allergic diseases.59,60 To investigate the influence of HLA region alleles on asthma PRS associations, we measured GBMI PRS-trait associations after removing HLA region variants from the asthma PRS (Table S7). The region contained only 0.7% (n = 4,694) of the total number of modeled SNPs but included 39 of the top 100 by scoring weight. The correlation between the whole-genome PRS and the HLA-removed PRS was 0.88.
Removal of the HLA region had a negligible effect on the PRS association and predictive performance with asthma (Δβ=0%, ΔAUC = −0.003). We observed no difference for COA (Δβ=0%, ΔAUC = 0.000). The HLA-removed PRS was marginally worse at predicting AOA (Δβ = −4%, ΔAUC = −0.004) compared to the whole-genome PRS, but none of these differences were statistically significant.
In contrast, PRS associations with autoimmune conditions were dramatically reduced after removing the HLA region variants. Associations with celiac disease (Δβ = −68%, pΔβ = 1.5 × 10−10), T1DM (Δβ = −60%, pΔβ = 1.5 × 10−8), hypothyroidism (Δβ = −51%, pΔβ = 6.5 × 10−6), and their corresponding treatments were significantly weaker with the HLA-removed PRS (Figure 7). The only trait with a significantly stronger association with the asthma PRS after removing the HLA region was eosinophil levels (Δβ = 10%, pΔβ = 4.2 × 10−6), suggesting an opposite effect of the HLA region compared to the rest of the genome’s shared architecture with blood eosinophil levels. No other traits demonstrated significantly different association effect sizes with the HLA-removed PRS. Overall, these results reveal that the pleiotropy between asthma and immune-mediated traits is largely concentrated within the HLA region.
Figure 7.
Changes in asthma PRS associations after removing HLA region
Changes in PRS association effect sizes after removing the HLA region from the PRS (Δβ) are shown with 95% confidence intervals for traits with significant effect reductions and for COA and AOA. Traits with a negative Δβ had a weaker association with the asthma PRS, and traits with a positive Δβ had a stronger association with the asthma PRS after removing HLA region alleles from consideration.
Mediation of cardinal asthma-associated traits on genetic risk for asthma
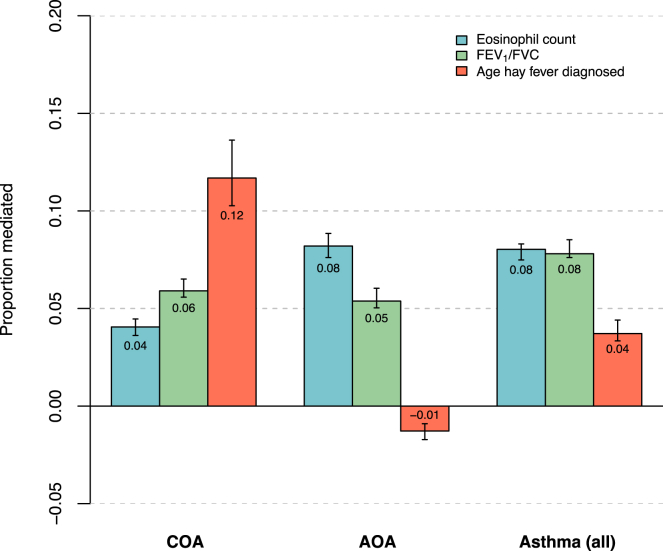
Allergic disease, lung function, and eosinophils are all highly correlated with asthma, yet there is considerable heterogeneity among these traits in asthmatic patients. To quantify the relative extent to which each of these traits mediate the genetic risk of asthma, we performed mediation analyses52 with each trait modeled as a mediator between the GBMI PRS and asthma, stratified by COA and AOA. Blood eosinophil count, FEV1/FVC, and age of hay fever diagnosis were used as the metrics for eosinophilia, lung function, and allergic disease, respectively (Figure 8). For COA, age of hay fever diagnosis mediated the largest proportion of genetic risk (12%) relative to eosinophil count (6%) and FEV1/FVC (4%). In AOA, however, age of hay fever diagnosis did not mediate any of the genetic risk (−1%), while eosinophils mediated the greatest proportion (8%), followed by FEV1/FVC (5%). These results are consistent with clinical observations of allergic disease being a prominent comorbidity with COA and further indicates that this may be due to shared genetics.
Figure 8.
Mediation of cardinal asthma-associated traits on genetic risk for asthma
For the PRS associations with asthma, the estimated proportions mediated by eosinophil count (blue), FEV1/FVC (green), and age of hay fever diagnosis (orange) are shown with 95% confidence intervals. Mediation analysis was conducted separately for COA, AOA, and all asthma. In each instance, the asthma PRS was the independent variable, asthma status was the dependent variable, and one of the three traits was the mediator.
Discussion
This study provides a quantitative overview of the genetic landscape for asthma, revealing polygenic pleiotropy between asthma and hundreds of other traits and conditions across a variety of biological systems and highlights differences by sex and age of onset. Previous studies have developed PRSs for asthma,6,7,11,12,13,14,15,16,17,18,19,20,21 with comparable AUC values when reported,12,14,21 but most were more limited by just focusing on predictive performance11,12,13,14,15 or by testing associations with specific traits.15,16,17,18,19,20,21 Our study features a more comprehensive evaluation of the traits and conditions associated with genetic asthma risk, including dependencies on age of onset, sex, ancestry, disease status, and HLA alleles, thereby revealing novel features of asthma and its associated traits.
Utilization of a PRS enabled us to perform direct quantification of relative disease risks and facilitated additional analyses that can account for genetic risk strata, disease status, mediating effects, and/or confounding factors. In contrast, previous studies of cross-trait genetic correlations with asthma provide estimates of shared genetic effects among common variants but are less interpretable in terms of relative disease risk.7,22,23,24,25,26,27,28,29 The relative risks between different disease pairs with similar genetic correlation estimates can vary dramatically.61 PRS-based associations therefore offer a different view of multimorbidity with additional utility than cross-trait genetic correlation estimates alone.
Age and sex both significantly alter genetic effects on asthma risk. Asthma that begins in childhood (COA) is more often diagnosed in males and is associated with different risk factors compared to asthma that develops in adulthood (AOA), which is more commonly diagnosed in females and is associated with increased respiratory symptoms and a lower frequency of quiescent disease.54,55,62 In fact, clinically, COA and AOA are often considered different subtypes of asthma. Recent studies have revealed that the genetic etiologies of COA and AOA are indeed partly distinct,9,56 with an estimated genetic correlation of rg = 0.67.56 We previously reported that the genetic risks for AOA were largely a subset of those for COA but with overall smaller effect sizes.9 Consistent with this, SNP-based heritability estimates for COA were approximately three times greater than those estimated for AOA.9,56 These combined findings indicate that genetics plays a more prominent role in asthma that presents earlier in life and, conversely, risk of AOA is more environmentally mediated. In the present study, the genetic risk for asthma was significantly associated with a younger age of onset and was a better predictor of COA than AOA.
Our results further reinforced that genetic risk for asthma acts through different mechanisms depending on the age of onset. The mediation analyses revealed that genetic risk for COA is more related to allergic disease, whereas genetic risk for AOA is more related to eosinophilia. These findings align with observations that allergic asthma is the predominant endotype in children.54,62,63
Mechanistic differences between COA and AOA may also be contributing to the trait association differences we observed between the GBMI PRS PheWAS and the TAGC PRS PheWAS. The GBMI PRS was modeled primarily on ICD codes in adults and is therefore likely identifying more of an adult-onset phenotype than the PRS from TAGC, in which the cases in 40% of the studies were COA. Indeed, the binary UKB trait with the second largest association bias toward the GBMI PRS was the primary asthma ICD-10 code. Of the 1,667 individuals in the UKB with a primary asthma ICD-10 code and information on age of asthma diagnosis, only 471 (28%) reported an asthma diagnosis in childhood, whereas 945 (57%) reported a first asthma diagnosis in adulthood (>25 and <66 years old). Anthropometric traits, consisting almost entirely of body-fat measures, were the most biased toward the GBMI PRS, and the TAGC PRS was not associated with any of the 24 neurologic and behavioral traits associated with the GBMI PRS, including five different measures of smoking behavior. These findings accord with genetic correlation results reported by Ferreira et al.,56 in which COA had a larger allergic component than AOA, whereas only AOA had significant genetic overlap with obesity-related traits and smoking history. Of the traits more strongly associated with the TAGC PRS, celiac disease stood out as having the greatest effect size difference. Previous studies have reported correlations between COA and celiac disease incidence,64,65,66,67,68 including a study by Hemminki et al.65 that found the risk of celiac disease following an asthma-related hospitalization was greater only in individuals that were diagnosed with asthma before 20 years of age. Importantly, while the relative composition of COA and AOA cases in GBMI and TAGC likely contributed to trait association differences between their respective PRS PheWASs, other factors, such as relative training data sample size, overall age distribution, ancestry composition, phenotype accuracy, and/or overlap between asthma cases and COPD, could also have contributed. Therefore, we cannot be certain what underlies the differences between the GBMI and TAGC PRS PheWASs. Nonetheless, our results demonstrate how compositional differences in PRS training datasets inevitably produce models with different genetic architectures for the same disease.
In addition to allergic diseases (hay fever/allergic rhinitis, eczema), pulmonary traits were among those most strongly associated with the asthma PRSs. Multiple studies have previously reported significant genetic overlap between asthma and various measures of lung function,7,21,24,56,69,70 but their causal relationships are complex.71 In our study, most asthma PRS associations with pulmonary traits remained significant after excluding individuals with an asthma diagnosis, indicating that asthma and lung function share a genetic architecture independent of asthma diagnosis. However, the corresponding effect sizes upon excluding asthmatics were greatly reduced, and half of the traits that were no longer significantly associated with the asthma PRS were pulmonary traits, suggesting that asthma itself leads to lower lung function. For example, after removing the asthma cases, the PRS association with COPD was still statistically significant, but its effect was substantially weaker in the non-asthmatic cohort (Δβ = −48%, pΔβ = 2.5 × 10−9). This combination of shared genetics and disease-induced changes could explain the contradictory results regarding the causal order of the association between lung function and asthma.71
Many risk loci for asthma and allergy are also associated with one or more autoimmune diseases.57,59,72 We observed significant associations with the GBMI asthma PRS and celiac disease, hypothyroidism, and T1DM, and additionally for ulcerative colitis with the TAGC PRS. Our results showed that the genetic overlap between asthma and immune-mediated conditions was primarily due to genetic variation at the HLA locus, as their PRS associations were significantly reduced after removing HLA region variants from the PRS. Interestingly, genetic risk of asthma, according to the TAGC PRS, had a protective effect on risk for ulcerative colitis, which is consistent with reports that many alleles associated with both asthma and autoimmune diseases have opposite effects on pathogenesis.59,60
The asthma PRSs were also significantly associated with immune cell types. Eosinophils, in particular, had the most significant quantitative trait associations with both the GBMI (p < 4.9 × 10−324, βz = 0.11) and TAGC (p < 4.9 × 10−324, βz = 0.07) PRSs. Eosinophils are granulocytes that contribute to immunity during multicellular parasitic infection, but, in geographical regions where parasites are less abundant, eosinophils can contribute to type 2 inflammation and subtypes of severe asthma.73 Notably, the PRS association with eosinophil levels was the only association that was significantly stronger after removing the HLA region from the PRS (Δβ = 10%, pΔβ = 4.2 × 10−6), suggesting that the shared architecture between asthma and blood eosinophil levels resides outside of the HLA region.
We also found links between asthma, obesity, and insulin resistance, an observation that is well established in both children and adults.74 Mendelian randomization studies indicated that BMI causally increases the risk of asthma,28,75,76 but findings from prospective studies suggest that asthma is also a risk factor for subsequent obesity.77,78,79,80 Our study suggested that the PRS associations with body fat were not due to asthma onset: most of the associations with body-fat-related traits remained significant after removing the asthma cases from the PheWAS. The GBMI PRS had greater association effects with obesity-related traits than the TAGC PRS did, which could be due to significant genetic correlations between obesity-related traits and AOA but not COA.28,56 Insulin resistance has been reported to modify the association between obesity and asthma in adults,81 suggesting that features of metabolic syndrome may contribute to asthma risk independent of obesity. A recent study found that asthma was associated with metabolic syndrome independent of BMI, but only in adults.82 In our study, body-fat traits had stronger associations than T2DM with the GBMI PRS but weaker associations than T2DM for the TAGC PRS. Together these findings showed that both insulin and obesity are associated with asthma risk, but the strength of their associations depend on the population and/or how asthma is defined.
Another notable finding was the significant association between the asthma PRS and bilirubin, a metabolite of heme. A study by Turi et al.83 identified dose-dependent protective associations between unconjugated bilirubin measured in plasma at age 1 year and subsequent diagnoses of recurrent wheeze and COA.83 The study found that higher concentrations of bilirubin within the normal physiological range were associated with protection from asthma. Our study extends the link between asthma and bilirubin by showing that higher genetic risk for asthma is significantly correlated with lower total bilirubin levels and further demonstrates that this relationship is due at least in part to shared genetics.
There are some important caveats and limitations of our study. First, the genetic risk for asthma captured by the PRSs were limited to HapMap3 SNPs genotyped in the training data and the UKB. The genetic risk modeled in this study was therefore limited to that captured by common tag SNPs and could not account for independent contributions from lower-frequency variants or from a more comprehensive SNP coverage. Second, the PRSs were modeled using summary statistics from multi-ancestry GWASs,30,32 but most samples nonetheless consisted of individuals of European ancestries. Residual population stratification in GWASs can lead to biased PRS associations in test cohorts with overlapping population structure.84 Therefore, any stratification biases within the TAGC or GBMI GWASs could have confounded certain PRS associations, and we did not model this potential effect. Moreover, performances of PRSs suffer when applied to populations with different ancestries, due to differences in LD structure, allele frequencies, effect sizes, and heritability.85 Unsurprisingly, the accuracy of the PRSs was lowest in the African-ancestry cohort. We had less power to detect PRS-trait associations in this cohort as well, due to its smaller sample size. Larger non-European training samples and the development of new methods to improve bias correction and cross-population PRS accuracy are needed to prevent PRS use from exacerbating health disparities.86 Third, the PRSs applied in this study were modeled on asthma phenotypes that included both COA and AOA cases. The models therefore likely best represent genetic risk common to both COA and AOA. PRSs modeled on COA or AOA alone may associate with different sets of traits than those identified in this study and better capture genetic differences between COA and AOA. Fourth, we did not account for potential assortative mating effects, which can inflate genetic correlation estimates for etiologically independent traits,87 although the impact of assortative mating on PRS-trait associations has not yet been thoroughly investigated.88 Fifth, the phenotypes based on ICD codes included in our PheWAS correspond to principal diagnoses responsible for hospital admission and do not include secondary diagnoses that coexist at admission or subsequently develop. Use of ICD codes to denote disease phenotypes is also subject to misclassification.89,90 Consequently, the associations reported in our study for such traits may not be representative of overall risk. Sixth, the UKB captured all phenotype data from adults. Therefore, for traits measured at the time of study, such as BMI or lung function or leukocyte proportions, we are unable to make conclusions about potential associations earlier in life. For example, eosinophil count measured in childhood would likely mediate more of the genetic risk of COA than would the eosinophil counts recorded in the UKB. Furthermore, although our previous sensitivity analysis on COA in the UKB suggested that recall bias did not contribute to our GWAS results,9 we cannot exclude recall bias among participants for early-life diagnoses, such as COA, which could have inflated associations in the non-asthmatic cohort if a proportion of COA cases were mis-specified as controls. Seventh, estimation of direct and indirect effects within our mediation analyses inherently assumed no unmeasured mediator-outcome confounders affected by the PRS. We modeled for confounding with BMI, the Townsend deprivation index, disability, and smoking history, but, considering the polygenic pleiotropy demonstrated by the PRS, it is unlikely that this assumption was strictly met for all models. Therefore, our mediation results serve to illustrate relative correlations rather than provide strict causal inferences. Finally, associations with the asthma PRS do not necessarily represent causal biological relationships. We removed traits that were not interpretable as biomedical phenotypes or laboratory values, but any number of intermediate or correlated factors could contribute to associations observed with the asthma PRS.
In summary, we quantified the probabilities of developing asthma based on relative genetic risk and identified numerous traits across a wide spectrum of biological systems that were significantly associated with genetic risk for asthma. We identified trait associations that were limited to specific PRS models, demonstrating how different PRS training data produce models with different architectures. Most PRS-trait associations remained significant after controlling for asthma status, indicating shared genetics, but lung function traits and inflammatory markers were significantly less associated with genetic asthma risk in non-asthmatics, suggesting disease-state-induced changes of these traits. We found the genetic overlap with autoimmune-related phenotypes was largely limited to the HLA region and showed etiological differences between COA and AOA, with genetic risk of COA mediated more by allergic disease and AOA more closely tied to eosinophilia. Overall, this study sheds light on how asthma is related to other traits and conditions and opens new avenues for investigation, where further research may guide improved risk management and more effective therapies.
Data and code availability
The TAGC GWAS summary statistics are publicly available via the GWAS catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics). GBMI PRSs are publicly available via the Polygenic Score Catalog (https://www.pgscatalog.org/publication/PGP000262/). UKB data are available following a process described at https://www.ukbiobank.ac.uk/enable-your-research. The TAGC PRS allele weights and all original code are available on GitHub (https://github.com/mdapas/Asthma_PRS_PheWAS_UKB) and were published on Zenodo (https://doi.org/10.5281/zenodo.8237286).
Acknowledgments
This research was conducted using the UKB resource under application number 44300. This research was supported by NIH grants U19 AI62310 and P30 DK20595. M.D. was supported by NIH TL1 TR002388 and T32 HL007605; N.S. was supported by NIH K08 HL153955.
Author contributions
M.D., C.O., and N.S. conceived and designed the study. M.D. and Y.L.L. performed all analyses. W.W.S. managed all biobank data and performed phenotype derivations. H.K.I. facilitated biobank data access and contributed to the interpretation of results with M.D., C.O., and N.S. M.D. wrote the manuscript. All authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100233.
Contributor Information
Carole Ober, Email: cober@uchicago.edu.
Nathan Schoettler, Email: nschoettle@bsd.uchicago.edu.
Supplemental information
The p values for associations with the TAGC asthma PRS are shown for 257 non-redundant traits out of 371 heritable phenotypes tested in the UKB. Labeled traits to the right of the dashed line were significantly associated after correcting for multiple testing (p < 1.35 × 10−4). Traits are grouped by corresponding biological system. The direction of each arrow corresponds to the direction of the association effect.
References
- 1.GBD Chronic Respiratory Disease Collaborators Prevalence and attributable health burden of chronic respiratory diseases, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020;8:585–596. doi: 10.1016/S2213-2600(20)30105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta R.S., Zhang X., Sharp L.K., Shannon J.J., Weiss K.B. Geographic variability in childhood asthma prevalence in Chicago. J. Allergy Clin. Immunol. 2008;121:639–645.e1. doi: 10.1016/j.jaci.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Pate C.A., Zahran H.S., Qin X., Johnson C., Hummelman E., Malilay J. Asthma Surveillance - United States, 2006-2018. MMWR. Surveill. Summ. 2021;70:1–32. doi: 10.15585/mmwr.ss7005a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castillo J.R., Peters S.P., Busse W.W. Asthma Exacerbations: Pathogenesis, Prevention, and Treatment. J. Allergy Clin. Immunol. Pract. 2017;5:918–927. doi: 10.1016/j.jaip.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polderman T.J.C., Benyamin B., de Leeuw C.A., Sullivan P.F., van Bochoven A., Visscher P.M., Posthuma D. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 2015;47:702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 6.Han Y., Jia Q., Jahani P.S., Hurrell B.P., Pan C., Huang P., Gukasyan J., Woodward N.C., Eskin E., Gilliland F.D., et al. Genome-wide analysis highlights contribution of immune system pathways to the genetic architecture of asthma. Nat. Commun. 2020;11:1776. doi: 10.1038/s41467-020-15649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuo K., Zhou W., Wang Y., Kanai M., Namba S., Gupta R., Majara L., Nkambule L.L., Morisaki T., Okada Y., et al. Multi-ancestry meta-analysis of asthma identifies novel associations and highlights the value of increased power and diversity. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K.W., Ober C. Lessons Learned From GWAS of Asthma. Allergy Asthma Immunol. Res. 2019;11:170–187. doi: 10.4168/aair.2019.11.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pividori M., Schoettler N., Nicolae D.L., Ober C., Im H.K. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir. Med. 2019;7:509–522. doi: 10.1016/S2213-2600(19)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pividori M., Im H.K. ukbREST: efficient and streamlined data access for reproducible research in large biobanks. Bioinformatics. 2019;35:1971–1973. doi: 10.1093/bioinformatics/bty925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavazos T.B., Witte J.S. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. HGG Adv. 2021;2 doi: 10.1016/j.xhgg.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sordillo J.E., Lutz S.M., Jorgenson E., Iribarren C., McGeachie M., Dahlin A., Tantisira K., Kelly R., Lasky-Su J., Sakornsakolpat P., et al. A polygenic risk score for asthma in a large racially diverse population. Clin. Exp. Allergy. 2021;51:1410–1420. doi: 10.1111/cea.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijk F.N., Folkersma C., Gruzieva O., Kumar A., Wijga A.H., Gehring U., Kull I., Postma D.S., Vonk J.M., Melén E., Koppelman G.H. Genetic risk scores do not improve asthma prediction in childhood. J. Allergy Clin. Immunol. 2019;144:857–860.e7. doi: 10.1016/j.jaci.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Kothalawala D.M., Kadalayil L., Curtin J.A., Murray C.S., Simpson A., Custovic A., Tapper W.J., Arshad S.H., Rezwan F.I., Holloway J.W., On Behalf Of Stelar/Unicorn Investigators Integration of Genomic Risk Scores to Improve the Prediction of Childhood Asthma Diagnosis. J. Personalized Med. 2022;12 doi: 10.3390/jpm12010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belsky D.W., Sears M.R., Hancox R.J., Harrington H., Houts R., Moffitt T.E., Sugden K., Williams B., Poulton R., Caspi A. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir. Med. 2013;1:453–461. doi: 10.1016/S2213-2600(13)70101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Z., Hasegawa K., Ma B., Fujiogi M., Camargo C.A., Jr., Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J. Allergy Clin. Immunol. 2020;146:327–329.e4. doi: 10.1016/j.jaci.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang T.T., Sikdar S., Xu C.J., Lee M.K., Cardwell J., Forno E., Imboden M., Jeong A., Madore A.M., Qi C., et al. Epigenome-wide association study of DNA methylation and adult asthma in the Agricultural Lung Health Study. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00217-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leffa D.T., Horta B., Barros F.C., Menezes A.M.B., Martins-Silva T., Hutz M.H., Bau C.H.D., Grevet E.H., Rohde L.A., Tovo-Rodrigues L. Association between Polygenic Risk Scores for ADHD and Asthma: A Birth Cohort Investigation. J. Atten. Disord. 2022;26:685–695. doi: 10.1177/10870547211020111. [DOI] [PubMed] [Google Scholar]
- 19.Lareau C.A., DeWeese C.F., Adrianto I., Lessard C.J., Gaffney P.M., Iannuzzi M.C., Rybicki B.A., Levin A.M., Montgomery C.G. Polygenic risk assessment reveals pleiotropy between sarcoidosis and inflammatory disorders in the context of genetic ancestry. Gene Immun. 2017;18:88–94. doi: 10.1038/gene.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hüls A., Vanker A., Gray D., Koen N., MacIsaac J.L., Lin D.T.S., Ramadori K.E., Sly P.D., Stein D.J., Kobor M.S., Zar H.J. Genetic susceptibility to asthma increases the vulnerability to indoor air pollution. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.01831-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namjou B., Lape M., Malolepsza E., DeVore S.B., Weirauch M.T., Dikilitas O., Jarvik G.P., Kiryluk K., Kullo I.J., Liu C., et al. Multiancestral polygenic risk score for pediatric asthma. J. Allergy Clin. Immunol. 2022;150:1086–1096. doi: 10.1016/j.jaci.2022.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z., Lee P.H., Chaffin M.D., Chung W., Loh P.R., Lu Q., Christiani D.C., Liang L. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat. Genet. 2018;50:857–864. doi: 10.1038/s41588-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H., An J., Yu Z. Identifying Shared Risk Genes for Asthma, Hay Fever, and Eczema by Multi-Trait and Multiomic Association Analyses. Front. Genet. 2020;11:270. doi: 10.3389/fgene.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John C., Guyatt A.L., Shrine N., Packer R., Olafsdottir T.A., Liu J., Hayden L.P., Chu S.H., Koskela J.T., Luan J., et al. Genetic Associations and Architecture of Asthma-COPD Overlap. Chest. 2022;161:1155–1166. doi: 10.1016/j.chest.2021.12.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballard J.L., O'Connor L.J. Shared components of heritability across genetically correlated traits. Am. J. Hum. Genet. 2022;109:989–1006. doi: 10.1016/j.ajhg.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehto K., Pedersen N.L., Almqvist C., Lu Y., Brew B.K. Asthma and affective traits in adults: a genetically informative study. Eur. Respir. J. 2019;53 doi: 10.1183/13993003.02142-2018. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Z., Zhu X., Liu C.L., Shi H., Shen S., Yang Y., Hasegawa K., Camargo C.A., Jr., Liang L. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur. Respir. J. 2019;54 doi: 10.1183/13993003.01507-2019. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Z., Guo Y., Shi H., Liu C.L., Panganiban R.A., Chung W., O'Connor L.J., Himes B.E., Gazal S., Hasegawa K., et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2020;145:537–549. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y., Liang Z.S., Jin Y., Ding J., Huang T., Moore J.H., Zheng Z.J., Huang J. Shared Genetic Architecture and Causal Relationship Between Asthma and Cardiovascular Diseases: A Large-Scale Cross-Trait Analysis. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.775591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demenais F., Margaritte-Jeannin P., Barnes K.C., Cookson W.O.C., Altmüller J., Ang W., Barr R.G., Beaty T.H., Becker A.B., Beilby J., et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Namba S., Lopera E., Kerminen S., Tsuo K., Läll K., Kanai M., Zhou W., Wu K.H., Favé M.J., et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2022.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W., Kanai M., Wu K.H.H., Rasheed H., Tsuo K., Hirbo J.B., Wang Y., Bhattacharya A., Zhao H., Namba S., et al. Global Biobank Meta-analysis Initiative: Powering genetic discovery across human disease. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagai A., Hirata M., Kamatani Y., Muto K., Matsuda K., Kiyohara Y., Ninomiya T., Tamakoshi A., Yamagata Z., Mushiroda T., et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol. 2017;27:S2–S8. doi: 10.1016/j.je.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowton E.A., Collier S.P., Wang X., Sutcliffe C.B., Van Driest S.L., Couch L.J., Herrera M., Jerome R.N., Slebos R.J.C., Alborn W.E., et al. Phenotype-Driven Plasma Biobanking Strategies and Methods. J. Personalized Med. 2015;5:140–152. doi: 10.3390/jpm5020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sijtsma A., Rienks J., van der Harst P., Navis G., Rosmalen J.G.M., Dotinga A. Cohort Profile Update: Lifelines, a three-generation cohort study and biobank. Int. J. Epidemiol. 2022;51:e295–e302. doi: 10.1093/ije/dyab257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dummer T.J.B., Awadalla P., Boileau C., Craig C., Fortier I., Goel V., Hicks J.M.T., Jacquemont S., Knoppers B.M., Le N., et al. The Canadian Partnership for Tomorrow Project: a pan-Canadian platform for research on chronic disease prevention. CMAJ (Can. Med. Assoc. J.) 2018;190:E710–E717. doi: 10.1503/cmaj.170292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leitsalu L., Haller T., Esko T., Tammesoo M.L., Alavere H., Snieder H., Perola M., Ng P.C., Mägi R., Milani L., et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 39.Kurki M.I., Karjalainen J., Palta P., Sipilä T.P., Kristiansson K., Donner K.M., Reeve M.P., Laivuori H., Aavikko M., Kaunisto M.A., et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zawistowski M., Fritsche L.G., Pandit A., Vanderwerff B., Patil S., Schmidt E.M., VandeHaar P., Willer C.J., Brummett C.M., Kheterpal S., et al. The Michigan Genomics Initiative: A biobank linking genotypes and electronic clinical records in Michigan Medicine patients. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2023.100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Åsvold B.O., Langhammer A., Rehn T.A., Kjelvik G., Grøntvedt T.V., Sørgjerd E.P., Fenstad J.S., Heggland J., Holmen O., Stuifbergen M.C., et al. Cohort Profile Update: The HUNT Study, Norway. Int. J. Epidemiol. 2023;52:e80–e91. doi: 10.1093/ije/dyac095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert S.A., Gil L., Jupp S., Ritchie S.C., Xu Y., Buniello A., McMahon A., Abraham G., Chapman M., Parkinson H., et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021;53:420–425. doi: 10.1038/s41588-021-00783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge T., Chen C.Y., Ni Y., Feng Y.C.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yengo L., Vedantam S., Marouli E., Sidorenko J., Bartell E., Sakaue S., Graff M., Eliasen A.U., Jiang Y., Raghavan S., et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S.H., Goddard M.E., Wray N.R., Visscher P.M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 50.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monsees G.M., Tamimi R.M., Kraft P. Genome-Wide Association Scans for Secondary Traits Using Case-Control Samples. Genet. Epidemiol. 2009;33:717–728. doi: 10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacKinnon D.P. Lawrence Erlbaum Associates; 2008. Introduction to Statistical Mediation Analysis. [Google Scholar]
- 53.MacKinnon D.P., Lockwood C.M., Brown C.H., Wang W., Hoffman J.M. The intermediate endpoint effect in logistic and probit regression. Clin. Trials. 2007;4:499–513. doi: 10.1177/1740774507083434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trivedi M., Denton E. Asthma in Children and Adults-What Are the Differences and What Can They Tell us About Asthma? Front. Pediatr. 2019;7:256. doi: 10.3389/fped.2019.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naeem A., Silveyra P. Sex Differences in Paediatric and Adult Asthma. Eur. Med. (Edicion Espanola) J. 2019;4:27–35. [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreira M.A.R., Mathur R., Vonk J.M., Szwajda A., Brumpton B., Granell R., Brew B.K., Ullemar V., Lu Y., Jiang Y., et al. Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct. Am. J. Hum. Genet. 2019;104:665–684. doi: 10.1016/j.ajhg.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carapito R., Radosavljevic M., Bahram S. Next-Generation Sequencing of the HLA locus: Methods and impacts on HLA typing, population genetics and disease association studies. Hum. Immunol. 2016;77:1016–1023. doi: 10.1016/j.humimm.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Kreiner E., Waage J., Standl M., Brix S., Pers T.H., Couto Alves A., Warrington N.M., Tiesler C.M.T., Fuertes E., Franke L., et al. Shared genetic variants suggest common pathways in allergy and autoimmune diseases. J. Allergy Clin. Immunol. 2017;140:771–781. doi: 10.1016/j.jaci.2016.10.055. [DOI] [PubMed] [Google Scholar]
- 60.Li X., Ampleford E.J., Howard T.D., Moore W.C., Torgerson D.G., Li H., Busse W.W., Castro M., Erzurum S.C., Israel E., et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J. Allergy Clin. Immunol. 2012;130:861–868.e7. doi: 10.1016/j.jaci.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong G., Feng J., Sun F., Chen J., Zhao X.M. A global overview of genetically interpretable multimorbidities among common diseases in the UK Biobank. Genome Med. 2021;13:110. doi: 10.1186/s13073-021-00927-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sood A., Qualls C., Schuyler M., Arynchyn A., Alvarado J.H., Smith L.J., Jacobs D.R., Jr. Adult-onset asthma becomes the dominant phenotype among women by age 40 years. the longitudinal CARDIA study. Ann. Am. Thorac. Soc. 2013;10:188–197. doi: 10.1513/AnnalsATS.201212-115OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaur R., Chupp G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019;144:1–12. doi: 10.1016/j.jaci.2019.05.031. [DOI] [PubMed] [Google Scholar]
- 64.Kero J., Gissler M., Hemminki E., Isolauri E. Could TH1 and TH2 diseases coexist? Evaluation of asthma incidence in children with coeliac disease, type 1 diabetes, or rheumatoid arthritis: a register study. J. Allergy Clin. Immunol. 2001;108:781–783. doi: 10.1067/mai.2001.119557. [DOI] [PubMed] [Google Scholar]
- 65.Hemminki K., Li X., Sundquist J., Sundquist K. Subsequent autoimmune or related disease in asthma patients: clustering of diseases or medical care? Ann. Epidemiol. 2010;20:217–222. doi: 10.1016/j.annepidem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Canova C., Pitter G., Ludvigsson J.F., Romor P., Zanier L., Zanotti R., Simonato L. Coeliac disease and asthma association in children: the role of antibiotic consumption. Eur. Respir. J. 2015;46:115–122. doi: 10.1183/09031936.00185714. [DOI] [PubMed] [Google Scholar]
- 67.Assa A., Frenkel-Nir Y., Tzur D., Katz L.H., Shamir R. Large population study shows that adolescents with celiac disease have an increased risk of multiple autoimmune and nonautoimmune comorbidities. Acta Paediatr. 2017;106:967–972. doi: 10.1111/apa.13808. [DOI] [PubMed] [Google Scholar]
- 68.Patel B., Wi C.I., Hasassri M.E., Divekar R., Absah I., Almallouhi E., Ryu E., King K., Juhn Y.J. Heterogeneity of asthma and the risk of celiac disease in children. Allergy Asthma Proc. 2018;39:51–58. doi: 10.2500/aap.2018.39.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hobbs B.D., de Jong K., Lamontagne M., Bossé Y., Shrine N., Artigas M.S., Wain L.V., Hall I.P., Jackson V.E., Wyss A.B., et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat. Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang D., Hunkapiller J., Bhangale T., Reeder J., Mukhyala K., Tom J., Cowgill A., Vogel J., Forrest W.F., Khan Z., et al. A whole genome sequencing study of moderate to severe asthma identifies a lung function locus associated with asthma risk. Sci. Rep. 2022;12:5574. doi: 10.1038/s41598-022-09447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sánchez-Solís M. Early Lung Function and Future Asthma. Front. Pediatr. 2019;7:253. doi: 10.3389/fped.2019.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferreira M.A., Vonk J.M., Baurecht H., Marenholz I., Tian C., Hoffman J.D., Helmer Q., Tillander A., Ullemar V., van Dongen J., et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat. Genet. 2017;49:1752–1757. doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Busse W.W., Sedgwick J.B. Eosinophils in asthma. Ann. Allergy. 1992;68:286–290. [PubMed] [Google Scholar]
- 74.Peters U., Dixon A.E., Forno E. Obesity and asthma. J. Allergy Clin. Immunol. 2018;141:1169–1179. doi: 10.1016/j.jaci.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Granell R., Henderson A.J., Evans D.M., Smith G.D., Ness A.R., Lewis S., Palmer T.M., Sterne J.A.C. Effects of BMI, fat mass, and lean mass on asthma in childhood: a Mendelian randomization study. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skaaby T., Taylor A.E., Thuesen B.H., Jacobsen R.K., Friedrich N., Møllehave L.T., Hansen S., Larsen S.C., Völker U., Nauck M., et al. Estimating the causal effect of body mass index on hay fever, asthma and lung function using Mendelian randomization. Allergy. 2018;73:153–164. doi: 10.1111/all.13242. [DOI] [PubMed] [Google Scholar]
- 77.Chen Z., Salam M.T., Alderete T.L., Habre R., Bastain T.M., Berhane K., Gilliland F.D. Effects of Childhood Asthma on the Development of Obesity among School-aged Children. Am. J. Respir. Crit. Care Med. 2017;195:1181–1188. doi: 10.1164/rccm.201608-1691OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Contreras Z.A., Chen Z., Roumeliotaki T., Annesi-Maesano I., Baïz N., von Berg A., Bergström A., Crozier S., Duijts L., Ekström S., et al. Does early onset asthma increase childhood obesity risk? A pooled analysis of 16 European cohorts. Eur. Respir. J. 2018;52 doi: 10.1183/13993003.00504-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasler G., Gergen P.J., Ajdacic V., Gamma A., Eich D., Rössler W., Angst J. Asthma and body weight change: a 20-year prospective community study of young adults. Int. J. Obes. 2006;30:1111–1118. doi: 10.1038/sj.ijo.0803215. [DOI] [PubMed] [Google Scholar]
- 80.Moitra S., Carsin A.E., Abramson M.J., Accordini S., Amaral A.F.S., Anto J., Bono R., Casas Ruiz L., Cerveri I., Chatzi L., et al. Long-term effect of asthma on the development of obesity among adults: an international cohort study. Thorax. 2023;78:128–135. doi: 10.1136/thoraxjnl-2021-217867. [DOI] [PubMed] [Google Scholar]
- 81.Cardet J.C., Ash S., Kusa T., Camargo C.A., Jr., Israel E. Insulin resistance modifies the association between obesity and current asthma in adults. Eur. Respir. J. 2016;48:403–410. doi: 10.1183/13993003.00246-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Boer G.M., Tramper-Stranders G.A., Houweling L., van Zelst C.M., Pouw N., Verhoeven G.T., Boxma-de Klerk B.M., Braunstahl G.J., In 't Veen J.C.C.M., van Rossum E.F.C., Hendriks R.W. Adult but not childhood onset asthma is associated with the metabolic syndrome, independent from body mass index. Respir. Med. 2021;188 doi: 10.1016/j.rmed.2021.106603. [DOI] [PubMed] [Google Scholar]
- 83.Turi K.N., McKennan C., Gebretsadik T., Snyder B., Seroogy C.M., Lemanske R.F., Jr., Zoratti E., Havstad S., Ober C., Lynch S., et al. Unconjugated bilirubin is associated with protection from early-life wheeze and childhood asthma. J. Allergy Clin. Immunol. 2021;148:128–138. doi: 10.1016/j.jaci.2020.12.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blanc J., Berg J.J. Testing for differences in polygenic scores in the presence of confounding. bioRxiv. 2023 doi: 10.1101/2023.03.12.532301. Preprint at. [DOI] [Google Scholar]
- 85.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W.J., Khera A.V., Okada Y., Biobank Japan Project. Martin A.R., Finucane H.K., Price A.L. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54:450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Border R., Athanasiadis G., Buil A., Schork A.J., Cai N., Young A.I., Werge T., Flint J., Kendler K.S., Sankararaman S., et al. Cross-trait assortative mating is widespread and inflates genetic correlation estimates. Science. 2022;378:754–761. doi: 10.1126/science.abo2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Border R., O'Rourke S., de Candia T., Goddard M.E., Visscher P.M., Yengo L., Jones M., Keller M.C. Assortative mating biases marker-based heritability estimators. Nat. Commun. 2022;13:660. doi: 10.1038/s41467-022-28294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joo J., Hobbs B.D., Cho M.H., Himes B.E. Trait Insights Gained by Comparing Genome-Wide Association Study Results using Different Chronic Obstructive Pulmonary Disease Definitions. AMIA Jt. Summits Transl. Sci. Proc. 2020;2020:278–287. [PMC free article] [PubMed] [Google Scholar]
- 90.Prieto-Centurion V., Rolle A.J., Au D.H., Carson S.S., Henderson A.G., Lee T.A., Lindenauer P.K., McBurnie M.A., Mularski R.A., Naureckas E.T., et al. Multicenter study comparing case definitions used to identify patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2014;190:989–995. doi: 10.1164/rccm.201406-1166OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The p values for associations with the TAGC asthma PRS are shown for 257 non-redundant traits out of 371 heritable phenotypes tested in the UKB. Labeled traits to the right of the dashed line were significantly associated after correcting for multiple testing (p < 1.35 × 10−4). Traits are grouped by corresponding biological system. The direction of each arrow corresponds to the direction of the association effect.
Data Availability Statement
The TAGC GWAS summary statistics are publicly available via the GWAS catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics). GBMI PRSs are publicly available via the Polygenic Score Catalog (https://www.pgscatalog.org/publication/PGP000262/). UKB data are available following a process described at https://www.ukbiobank.ac.uk/enable-your-research. The TAGC PRS allele weights and all original code are available on GitHub (https://github.com/mdapas/Asthma_PRS_PheWAS_UKB) and were published on Zenodo (https://doi.org/10.5281/zenodo.8237286).