Abstract
Leptospirosis, the most widespread zoonotic disease in the world, is broadly understudied in multi-host wildlife systems. Knowledge gaps regarding Leptospira circulation in wildlife, particularly in densely populated areas, contribute to frequent misdiagnoses in humans and domestic animals. We assessed Leptospira prevalence levels and risk factors in five target wildlife species across the greater Los Angeles region: striped skunks (Mephitis mephitis), raccoons (Procyon lotor), coyotes (Canis latrans), Virginia opossums (Didelphis virginiana), and fox squirrels (Sciurus niger). We sampled more than 960 individual animals, including over 700 from target species in the greater Los Angeles region, and an additional 266 sampled opportunistically from other California regions and species. In the five target species seroprevalences ranged from 5 to 60%, and infection prevalences ranged from 0.8 to 15.2% in all except fox squirrels (0%). Leptospira phylogenomics and patterns of serologic reactivity suggest that mainland terrestrial wildlife, particularly mesocarnivores, could be the source of repeated observed introductions of Leptospira into local marine and island ecosystems. Overall, we found evidence of widespread Leptospira exposure in wildlife across Los Angeles and surrounding regions. This indicates exposure risk for humans and domestic animals and highlights that this pathogen can circulate endemically in many wildlife species even in densely populated urban areas.
Subject terms: Ecology, Urban ecology, Ecological epidemiology
Introduction
Leptospirosis, the disease caused by pathogenic bacteria from the genus Leptospira, is reported to be the most widespread zoonotic disease in the world1–3. Leptospira spp. are generalist pathogens capable of infecting a broad range of primarily mammalian hosts4,5. They are considered emerging pathogens of concern6, with one million human cases and 60,000 deaths estimated globally each year7,8, and many more cases likely undiagnosed9. Given the extensive global impact of Leptospira spp., it is surprising that their ecology and epidemiology remain understudied10. Though many pathogens, including all zoonoses, are capable of infecting multiple hosts, studying complex generalist pathogen systems remains a challenge, particularly in wildlife for which extensive, multi-host surveillance is required but resources and sample access are often limited11,12. Studying Leptospira poses additional challenges since host–pathogen epidemiology differs widely across host species and strains, and multiple Leptospira strains can co-circulate in communities. Consequently, extensive knowledge gaps remain regarding the role many hosts play in Leptospira transmission and the risks they pose to other host species, including humans. Epidemiological patterns also vary across climates and locations, with greater attention paid to the high incidence of human leptospirosis observed in wet tropical regions, and less concern historically placed on urban transmission, especially in more developed countries1,8. These complexities underscore the importance of broad surveillance efforts to inform veterinary and public health efforts, particularly in densely populated urban settings where the potential for human-wildlife contact is high and Leptospira transmission dynamics remain poorly understood.
Awareness of Leptospira genomic diversity has increased in recent years, with pathogenic Leptospira bacteria now categorized into 40 species9,13,14. Pathogenic Leptospira strains are classified into over 250 serovars based on cell surface antigen similarities and serologic reactivity4, with related serovars traditionally grouped into serogroups, though neither serovar nor serogroup are reliable predictors of Leptospira species15. Transmission usually occurs after leptospires are shed in the urine of infected hosts, leading to environmental contamination that can indirectly infect susceptible hosts via mucous membranes or abraded skin16–19. Following this contact, leptospires colonize the kidneys and other organs, which can cause pathology and associated clinical signs ranging from mild flu-like symptoms to fulminant multi-organ system failure and death17,20. Disease severity varies across Leptospira serovars and host species, with some broad host-serovar associations. Historically, the literature has categorized Leptospira epidemiology into: (i) maintenance hosts (and host-adapted serovars) that are likely to exhibit asymptomatic and/or chronic infections and maintain circulation of the pathogen, or (ii) accidental hosts (and non-host-adapted serovars) that exhibit symptomatic infections and do not maintain pathogen circulation1. These dichotomous classifications have been challenged in recent years, and there is clear evidence that clinical manifestations range widely irrespective of population-level circulation of the pathogen6,21,22. Evidence of infection is typically determined from urine or kidney samples using either polymerase chain reaction (PCR) to detect Leptospira DNA or culturing to detect live infectious leptospires. The most widely used diagnostic test for Leptospira spp. is the microscopic agglutination test (MAT)4,23, which tests serum for anti-Leptospira antibodies to assess past exposure. Serum MAT panels typically include serovars known or suspected to circulate in the geographic region of interest.
Despite increasing reports of Leptospira in urban areas, including dramatic outbreaks linked to flooding24, significant knowledge gaps remain regarding the prevalence and transmission risk across a variety of potential hosts in urban environments. Urbanization is increasing at a global scale25,26 and can influence wildlife pathogen dynamics through many mechanisms (e.g. altered community structure and contact rates27), with high densities of humans and domestic animals providing increased opportunity for cross-species contacts and possible spillover of infection28. Unhoused individuals in crowded urban areas may be at increased risk for zoonotic disease transmission due to poor sanitation and intensified contact with urban wildlife species, including in high-income countries where urban homelessness is a growing public health crisis29. Since changes to transmission risk are pathogen- and host-specific, investigations into pathogen dynamics and host diversity are critical for urban disease management and risk assessment. This is especially true for Leptospira, as most studies in cities have focused on rodents in high-density urban centers7,30–33, with much less attention placed on other urban-adapted wildlife that may influence the ecology of this important zoonosis1,34,35.
In the United States, many Leptospira infections in humans, domestic dogs, horses, and livestock are associated with spillover from wildlife36–38. Increasing reports of leptospirosis in domestic dogs39,40 and humans in some locations (e.g. Hawaii and California41–43) further highlight the need for expanded surveillance in wildlife. One recent study concluded that Leptospira exposure is common in a variety of wildlife across the country, and called for more studies investigating the relationship between serology and shedding to better understand the risk this poses to the health of humans, domestic pets, and livestock44. In California, a wildlife survey in the 1970s reported Leptospira exposure or active infections in multiple species, including coyotes (Canis latrans), raccoons (Procyon lotor), and striped skunks (Mephitis mephitis)45, and other surveys have detected Leptospira antibodies in black bears (Ursus americanus)46 and feral pigs (Sus scrofa)47. A recent survey in northern California reported Leptospira in multiple mesocarnivore and rodent species35, with both skunks and raccoons identified as potential reservoir hosts for Leptospira interrogans serovar Pomona48 – a serovar with a long history of circulation in terrestrial mammals on the California Channel Islands and California sea lions (Zalophus californianus) along the California coast21,22,49–53.
In contrast with northern and coastal regions, there has been little surveillance for Leptospira in inland southern California. The greater Los Angeles area features a range of landscapes, from natural and agricultural land to a dense urban center, as well as many wildlife species that are potential Leptospira carriers. There are scattered reports of Leptospira exposure in some local animals, including dogs54,55, deer (Odocoileus hemionus)56, mountain lions (Puma concolor) and bobcats (Lynx rufus)57, but prior studies have had limited host, geographic, and temporal ranges. The paucity of region-specific surveillance data in greater Los Angeles, the second largest metropolis in the United States58, represents a critical public health gap, and more comprehensive wildlife screening is needed to assess zoonotic spillover risk from the full range of susceptible wildlife hosts.
We conducted the first in-depth surveillance of Leptospira interrogans in wildlife in the greater Los Angeles area to shed light on the prevalence and circulation of this multi-host pathogen across a densely populated and complex urban landscape. We aimed to assess Leptospira exposure and active infections in common wildlife, harnessing novel tools for phylogenomic analysis of infecting Leptospira strains to additionally explore potential transmission links among host species. Serologic reactivity profiles across species provided additional insights into multi-host Leptospira ecology, and facilitated additional explorations into predictors of exposure and infections in local wildlife. Improved knowledge of Leptospira epidemiology in a greater range of susceptible urban host species, and the diversity of Leptospira strains they carry, will provide valuable context for wildlife management agencies and clinicians assessing illness in humans and domestic animals across the region.
Methods
Study animals
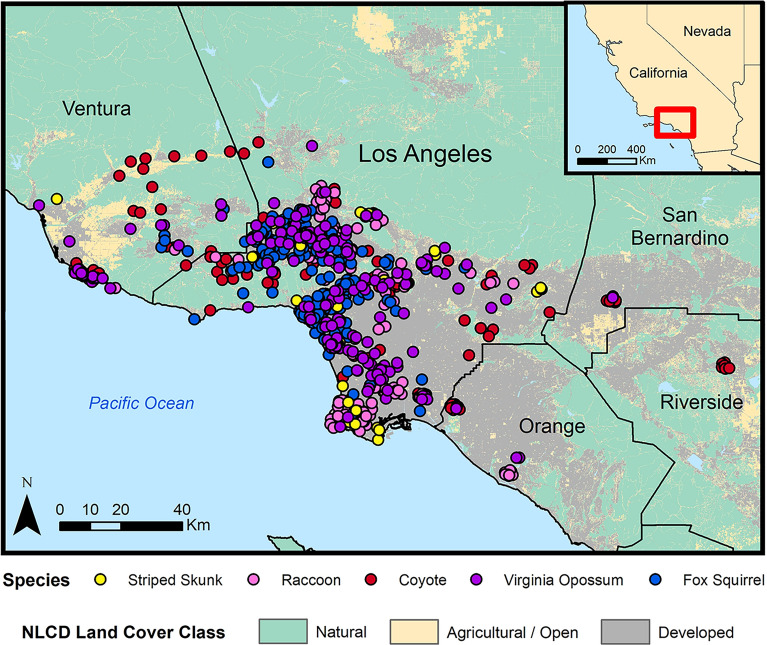
No live animals were captured or handled explicitly for the purposes of this study, and all wildlife sampling was carried out in accordance with relevant guidelines and regulations. This study focused primarily on wildlife from the greater Los Angeles region in southern California, with most results coming from opportunistically collected carcasses. Opportunistic carcass and sample collection was approved by the California Department of Fish and Wildlife (scientific collecting permits SC-13267 and SC-13700) and took place from September 2015 to June 2020. A small number of mountain lion (n = 11) and coyote (n = 19) samples were previously collected as part of National Park Service mountain lion and coyote studies, with capture and handling procedures permitted through the California Department of Fish and Wildlife (scientific collection permit SC-0005636) and the National Park Service Institutional Animal Care and Use Committee. Three additional serum samples were included from live-trapped striped skunks, with capture and handling procedures permitted for T. Stankowich through the California Department of Fish and Wildlife (scientific collection permit SC-006837) and the California State University Long Beach Institutional Animal Care and Use Committee (protocol #334). For the purposes of this study, the greater Los Angeles region refers to Los Angeles County and neighboring counties: Orange, Riverside, San Bernardino, and Ventura. Sample collection focused on our ‘target species’, five common mammals in the Los Angeles region: striped skunks, raccoons, coyotes, Virginia opossums (Didelphis virginiana) and fox squirrels (Sciurus niger; Table 1, Fig. 1). Collaborating wildlife agencies donated carcasses or existing samples, with the majority coming from animals killed by vehicle collisions or planned wildlife removal, or animals euthanized by animal control or rehabilitation agencies due to illness or injury. Carcasses were necropsied immediately or frozen at -20ºC and thawed in a refrigerator prior to necropsy. Animal measurements and demographic information were collected at the time of necropsy, with age class (adult or juvenile) determined using a combination of animal size and tooth wear59.
Table 1.
Descriptive characteristics for our five target wildlife species.
| Coyote (N = 137) |
Fox Squirrel (N = 187) |
Raccoon (N = 172) |
Striped Skunk (N = 40) |
Virginia Opossum (N = 171) |
Total (N = 707) |
|
|---|---|---|---|---|---|---|
| Age class | ||||||
| Adult | 100 (73.0%) | 154 (82.4%) | 136 (79.1%) | 22 (55.0%) | 133 (77.8%) | 545 (77.1%) |
| Juvenile | 32 (23.4%) | 26 (13.9%) | 32 (18.6%) | 13 (32.5%) | 27 (15.8%) | 130 (18.4%) |
| Unknown | 5 (3.6%) | 7 (3.7%) | 4 (2.3%) | 5 (12.5%) | 11 (6.4%) | 32 (4.5%) |
| Sex | ||||||
| Female | 63 (46.0%) | 84 (44.9%%) | 83 (48.3%) | 16 (40.0%) | 87 (50.9%) | 333 (47.1%) |
| Male | 67 (48.9%) | 79 (42.2%) | 80 (46.5%) | 16 (40.0%) | 71 (41.5%) | 313 (44.3%) |
| Unknown | 7 (5.1%) | 24 (12.8%) | 9 (5.2%) | 8 (20.0%) | 13 (7.6%) | 61 (8.6%) |
| County | ||||||
| Los Angeles | 82 (59.9%) | 182 (97.3%) | 152 (88.4%) | 38 (95.0%) | 147 (86.0%) | 601 (85.0%) |
| Orange | 15 (10.9%) | 0 (0%) | 9 (5.2%) | 0 (0%) | 3 (1.8%) | 27 (3.8%) |
| Riverside | 8 (5.8%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 8 (1.1%) |
| San Bernardino | 10 (7.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0.6%) | 11 (1.6%) |
| Ventura | 22 (16.1%) | 5 (2.7%) | 11 (6.4%) | 2 (5.0%) | 20 (11.7%) | 60 (8.5%) |
| Season | ||||||
| Dry | 60 (43.8%) | 81 (43.3%) | 53 (30.8%) | 27 (67.5%) | 73 (42.7%) | 294 (41.6%) |
| Wet | 77 (56.2%) | 106 (56.7%) | 119 (69.2%) | 13 (32.5%) | 98 (57.3%) | 413 (58.4%) |
Within-group percentages are proportions of column totals.
Figure 1.
Distribution of Leptospira sample locations for the five target wildlife species in the greater Los Angeles region (2015–2020). Land cover data were obtained from the National Land Cover Database (2019).
Additional species and regions
On an opportunistic basis, we also tested samples provided by other agencies that included non-target regions of California and non-target species, including ground squirrels (Otospermophilus beecheyi), desert cottontails (Sylvilagus audubonii), feral pigs, bobcats, mountain lions, gray foxes (Urocyon cinereoargenteus), and red foxes (Vulpes vulpes). A total of 266 additional samples were collected from 11 species. This included non-target species in Los Angeles County (n = 74) and Ventura County (n = 24), and all species from the following additional counties: Napa (n = 15), Monterey (n = 86), San Luis Obispo (n = 61) and Santa Barbara (n = 6; Supplementary Table S1).
Sample collection
Serum and urine samples collected at external agencies were analyzed and included when available. From fresh carcasses processed at UCLA, intracardiac blood was collected into serum separation tubes, then kept in a cooler with an ice pack until centrifugation (1350 × g for 10–15 min). Kidney samples were collected from all necropsies, and urine was collected when available using cystocentesis. The largest possible kidney sample that would fit in a 58 ml Whirl–PakⓇ was excised and homogenized in the sealed Whirl–PakⓇ using manual pressure (approximate size: entire kidney from smaller mammals such as squirrels or half a kidney from larger mammals such as coyotes). Serum and urine samples were transferred into cryovials prior to storage, and all cryovials and Whirl-PaksⓇ were stored at − 20 ºC or − 80 ºC prior to testing (− 80 ºC preferred and utilized when possible).
Leptospira PCR analysis
The presence of pathogenic Leptospira DNA was determined in urine and homogenized kidney samples. For all species except mountain lions, samples collected from 2015 to 2017 were analyzed at the Hollings Marine Laboratory (Charleston, South Carolina, USA) using a quantitative polymerase chain reaction (qPCR) assay targeting the lipL32 gene as detailed in Wu et al.60. Samples collected after 2017 were analyzed at Colorado State University Veterinary Diagnostic Laboratory (Denver, Colorado, USA) using the VetMAX™ qPCR Master Mix kit and the primers specified by Wu et al.60. Minor modifications to the protocol resulted in slightly higher sensitivity in the later samples, but results across laboratories were broadly consistent61. Samples that had a cycle threshold value less than 37 were considered PCR-positive and evidence of active infections. Samples from mountain lions were analyzed at the California Animal Health and Food Safety Laboratory (Davis, California, USA) using their standard protocol62.
Genetic sequencing and phylogenetic analyses
DNA capture and enrichment methods
To facilitate robust genomic level species identification and phylogenomic analyses, four Leptospira-positive DNA samples (CM-61, MM-3, PL-20, and PL-117) were subjected to pan pathogenic Leptospira DNA capture and enrichment as described in detail elsewhere63. Briefly, the sample DNAs were diluted separately to ~ 4 ng/µL in a volume of 40µL, sonicated to an average size of 225 bp using a Q800R2 sonicator (QSonica, Newtown, CT, USA), and then short-read next-generation libraries were prepared using Agilent Sure-Select methodology. The libraries were then subjected to one (MM-3 and PL-20) or two (CM-61 and PL-117) rounds of DNA capture and enrichment and then sequenced on an Illumina MiSeq instrument using a MiSeq v3 600 cycle kit (2 × 300 bp reads).
Read classifications
To estimate the percentage of Leptospira reads in the enriched sequences, reads were mapped against the standard Kraken database with Kraken v2.1.264.
Read mapping and phylogenomics
Single nucleotide polymorphisms (SNPs) were identified among the four enriched genomes and > 340 publicly available L. interrogans genomes (GenBank accession numbers provided in Supplementary Fig. S1 and Supplementary Fig. S2) by aligning reads against reference genome L. interrogans serovar Copenhageni strain Fiocruz L1-130 (GCA_000007685.1) using minimap2 v2.2265 and calling SNPs from the BAM file with GATK v4.2.266 using a depth of coverage ≥ 3 × and a read proportion of 0.9. SNPs that fell within duplicated regions, based on a reference self-alignment with MUMmer v3.167, were filtered from downstream analyses. All of these methods were wrapped by NASP v1.2.168. Maximum likelihood phylogenies were then inferred on the concatenated SNP alignments using IQ-TREE v2.2.0.3 with the “-fast” option, default parameters69, and the integrated ModelFinder method70; the phylogenies were midpoint rooted. To determine breadth of coverage across the reference, reads were aligned against the reference genome with minimap2 and the per base depth of coverage was calculated with Samtools v1.671.
Pomona reference phylogeny and WG-FAST placement of enriched samples
We then utilized the Whole Genome Focused Array SNP Typing (WG-FAST) tool, a publicly available pipeline designed for phylogenetically typing samples with only partial SNP profiles. Reads were simulated from genome assemblies with ART vMountRainier72 using the command “-p -na -ss MSv3 -l 250 -f 75 -m 300 -s 30”. Based on the position of the enriched reads in a complete L. interrogans phylogeny, simulated reads were aligned against L. interrogans serovar Pomona str. Pomona (GCA_000216355.3) with minimap2 v2.2465 and SNPs were called with GATK v4.2.6.166. A midpoint rooted maximum-likelihood phylogeny was inferred from a concatenated SNP alignment (529 positions out of a core genome size of 4,125,494 nts) with RAxML-NG v. 1.1.073. Enriched reads were inserted into this reference phylogeny with WG-FAST v1.274 using default settings.
Leptospira serology
Past exposure to Leptospira was assessed using serum microscopic agglutination testing (MAT). In this test, dark-field microscopy is used to assess the presence of anti-Leptospira antibodies in serum by evaluating agglutination (i.e. clumping) when samples are combined with live cultures of Leptospira species23. Serum samples are tested at doubling dilutions, beginning at 1:100 and continuing until endpoint, with the reported endpoint titers representing the highest dilution that achieved a 50% agglutination using the reference strain being tested. An antibody titer of 1:100 or higher to any serovar was considered positive for Leptospira exposure. Samples from 2018 to 2020 were analyzed at the California Animal Health and Food Safety Laboratory (CAHFS; Davis, California, USA) against a panel of 6 serovars that are common in the United States44,75: Bratislava, Canicola, Grippotyphosa, Hardjo, Icterohaemorrhagiae, and Pomona (Supplementary Table S2). The CAHFS lab uses L. interrogans serovar Copenhageni strain M20 as a representative member of the Icterohaemorrhagiae serogroup, and reports results as Icterohaemorrhagiae; it will henceforth be referred to as Icterohaemorrhagiae since they are used interchangeably on MAT testing76. Samples from 2015 to 2017 were analyzed at the Centers for Disease Control and Prevention (CDC; Atlanta, Georgia, USA) using an expanded panel of 20 serovars: Alexi, Australis, Autumnalis, Ballum, Bataviae, Borinca, Bratislava, Canicola, Celledoni, Cynopteri, Djasiman, Georgia, Grippotyphosa, Icterohaemorrhagiae, Javanica, Mankarso, Pomona, Pyrogenes, Tarassovi, and Wolffi (Supplementary Table S2). To assess consistency between the two laboratories, we compared the subset of wildlife samples analyzed at both laboratories (n = 469), demonstrating 98.3% agreement in seropositivity and minor quantitative differences between titers. Serovar cross-reactivity patterns were visualized using heatmaps of serovar titers from all seropositive animals using package ‘Heatmap’ in R version 4.3.077.
Analysis of surveillance data
Prevalence was estimated for Leptospira exposure and infections. For Leptospira serology, individuals could be reactive to multiple serovars, so seroprevalence was calculated for each host species in two ways: proportion positive against any serovar, and proportion positive against each specific serovar. All 95% binomial confidence intervals were estimated using package ‘PropCIs’ in R. Additional analyses were done in R, and maps were created using ArcGIS version 10.8.278.
Land cover analysis was conducted as detailed in Adducci II et al.79 using data from the National Land Cover Database80. Sample localities occurred across a diverse landscape gradient, ranging from natural vegetation and open space to the urban core of Los Angeles (Fig. 1). To account for land cover variation within home ranges, we extracted 2019 land cover data (30 m 30 m resolution) from home range buffers around each georeferenced sampling locality using the ‘raster’ and ‘rgdal’ packages in R. Buffer size varied by species based on home range estimates previously reported in the literature: 5 km2 for coyotes79, 2 km2 for raccoons and striped skunks81, 1 km2 for opossums82, and 0.5 km2 for fox squirrels83. As detailed in Adducci II et al.79, we grouped land cover classifications into three composite categories: urban/suburban (land with 20–100% impervious surface cover), agricultural/open (land used for pastures, crops and open development with < 20% impervious surface cover), and natural (shrubland, forest, grassland and wetland). We then calculated the relative proportions of these three land categories for each individual home range buffer, and used package ‘ggtern’ in R to make ternary plots showing Leptospira exposure (i.e. presence of antibodies) relative to land categories.
To explore potential predictors of Leptospira exposure (as indicated by an antibody titer of 1:100 or higher to any serovar), logistic regressions were conducted with the following covariates considered: age class, sex, season (wet Nov-April vs. dry May-Oct)35, and composite land classification (i.e. percentage of the home range that was developed vs. agricultural/open vs. natural). Since the correlation was high between developed and agricultural/open land (Spearman’s ), and developed and natural land (Spearman’s ), the developed category was excluded from these models.
Antibody titers have been identified as significant predictors of Leptospira shedding and PCR status (i.e. active infections) in California sea lions22,84. We therefore explored this association in raccoons, the only species with a sufficient number of paired antibody-PCR samples available (n = 81). Since these data exhibited complete separation (i.e. a clear distinction between the two outcomes), we applied a Firth’s bias-reduced logistic regression, a penalized maximum likelihood approach that is effective in the presence of data separation85, using the ‘logistf’ package in R to assess the association between antibody titers and PCR data.
Results
Broad patterns of Leptospira exposure and infection
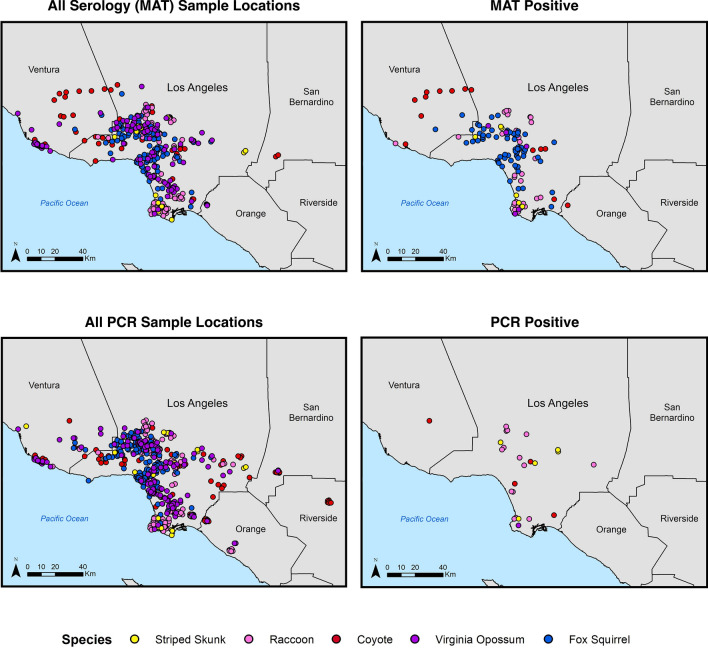
We detected evidence of Leptospira exposure in all five target species sampled in the greater Los Angeles region (Fig. 2, Table 2). Overall seroprevalence in each species was calculated as the proportion of samples that were positive against any serovar. Fox squirrels had the highest seroprevalence at 60.6% (Table 2), though most maximum titers in this species were low (Table 3). Seroprevalence was moderate in mesocarnivores (32.6% in raccoons, 28.6% in striped skunks, 25.9% in coyotes), and low (5.2%) in opossums.
Figure 2.
Locations of sample collection and associated Leptospira exposure and infection results for the five target wildlife species. Host sample locations are indicated in the left-hand panels, with the locations of animals with positive results shown in maps on the right. Samples tested by MAT for serum antibodies are shown in the top row, and those tested by PCR for pathogenic Leptospira DNA are shown on the bottom row.
Table 2.
Leptospira exposure and infection by species.
| Common name | Scientific name | Leptospira Exposure (MAT) | Leptospira Infections (PCR) | ||||
|---|---|---|---|---|---|---|---|
| POS | n | %POS (95% CI) | POS | n | %POS (95% CI) | ||
| Striped Skunk | Mephitis mephitis | 6 | 21 | 28.6 (11.3–52.2) | 5 | 33 | 15.2 (5.1–31.9) |
| Raccoon | Procyon lotor | 31 | 95 | 32.6 (23.4–43.0) | 14 | 162 | 8.6 (4.8–14.1) |
| Coyote | Canis latrans | 14 | 54 | 25.9 (15.0–39.7) | 4 | 108 | 3.7 (1.0–9.2) |
| Virginia Opossum | Didelphis virginiana | 5 | 97 | 5.2 (1.7–11.6) | 1 | 131 | 0.8 (0.0–4.2) |
| Fox Squirrel | Sciurus niger | 66 | 109 | 60.6 (50.7–69.8) | 0 | 148 | 0 (0.0–2.5) |
| Total | 122 | 376 | 32.4 (27.7–37.4) | 24 | 582 | 4.1 (2.7–6.1) | |
Leptospira antibody (MAT) and DNA (PCR) results in the five target host species sampled in the greater Los Angeles region. Antibody results include seropositives to all serovars tested.
Table 3.
Maximum antibody titers for our five target wildlife species.
| Common name | Scientific name | Serovar | 100 | 200 | 400 | 800 | 1600 | 3200 | ≥ 6400 | #Positive / Total | Percentage (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Striped Skunk | Mephitis mephitis | Pomona | 1 | 1 | 1 | 2 | 5/5 | 100 (47.8–100) | |||
| Raccoon | Procyon lotor | Bratislava | 2 | 1 | 3/30 | 10 (2.1–26.5) | |||||
| Icterohaemorrhagiae | 2 | 1 | 1 | 4/30 | 13.33 (3.8–30.7) | ||||||
| Pomona | 6 | 5 | 5 | 3 | 2 | 2 | 3 | 26/30 | 86.67 (69.3–96.2) | ||
| Coyote | Canis latrans | Bratislava | 2 | 2/7 | 28.57 (3.7–71) | ||||||
| Icterohaemorrhagiae | 1 | 1/7 | 14.29 (0.4–57.9) | ||||||||
| Pomona | 1 | 1 | 1 | 1 | 1 | 5/7 | 71.43 (29–96.3) | ||||
| Virginia Opossum | Didelphis virginiana | Icterohaemorrhagiae | 1 | 1/2 | 50 (1.3–98.7) | ||||||
| Pomona | 1 | 1/2 | 50 (1.3–98.7) | ||||||||
| Fox Squirrel | Sciurus niger | Bratislava | 5 | 12 | 8 | 4 | 2 | 31/60 | 51.67 (38.4-64.8) | ||
| Icterohaemorrhagiae | 25 | 11 | 6 | 4 | 1 | 1 | 48/60 | 80 (67.7–89.2) |
Maximum antibody titers and corresponding serovars for each individual from the five target species sampled in the greater Los Angeles region, reported for our five primary serovars (Bratislava, Canicola, Grippotyphosa, Icterohaemorrhagiae, and Pomona). Only individuals with a highest titer to one of these five primary serovars are included in the totals. In cases where there were ties for maximum titer, both serovars were counted in the table.
Of the 582 target animals tested by PCR in the greater Los Angeles region, 24 (4%) were PCR positive, with active infections detected in all species except fox squirrels (Table 2, Fig. 2). Infection prevalence ranged from 0.8% in opossums to 15% in skunks, with coyotes and raccoons intermediate at 3.7% and 8.6%, respectively. Infection prevalence was consistently lower than corresponding seroprevalence levels for each species, with no active infections detected in fox squirrels despite seroprevalence being highest in this species (Table 2).
Phylogenetic typing of Leptospira
We used DNA capture and enrichment to enable sequencing of Leptospira DNA present in DNA extracts obtained from kidney or urine samples from four of our PCR-positive animals: one coyote (CM-61), one striped skunk (MM-3), and two raccoons (PL-20 and PL-117). Two samples underwent two rounds of enrichment and had very high proportions of sequencing reads assigned to Leptospira: 90.52% for PL-117 (1,615,997 of 1,785,218 total reads) and 84.71% for CM-61 (1,194,246/1,409,734). The other two samples underwent just one round of enrichment and had lower proportions of reads assigned to Leptospira: 11.05% for PL-20 (141,648/1,281,984) and 6.98% for MM-3 (115,700/1,658,612).
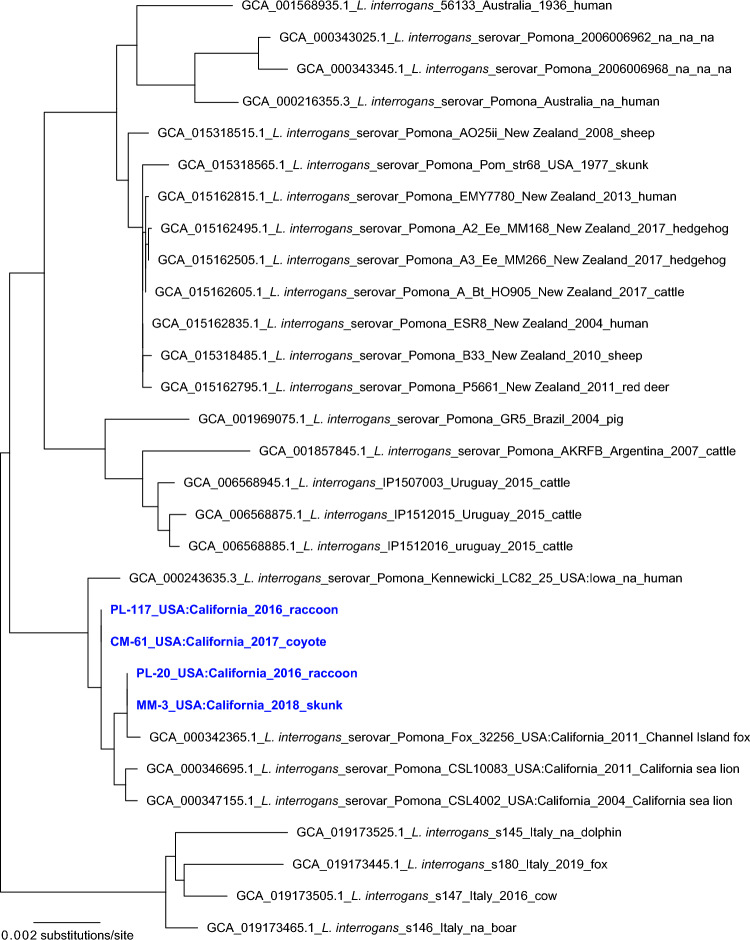
We used several approaches to phylogenetically characterize the Leptospira strains infecting these four animals based on the genomic sequence data we generated using targeted DNA capture and enrichment. First, we aligned the sequence reads from these four samples against reference genome L. interrogans serovar Copenhageni strain Fiocruz L1-130, which revealed coverages of 82.67% for PL-117 and 84.07% for CM-61, which both underwent two rounds of enrichment, and 53.17% for PL-20 and 54.61% for MM-3, which both underwent just one round of enrichment. We then constructed a phylogeny using the core genome shared by all four samples and the > 340 publicly available L. interrogans genomes, which corresponded to just 19.27% (891,916/4,627,366) of the reference genome. Within this phylogeny (Supplementary Fig. S1), the four enriched samples formed a single clade that was embedded within a larger clade containing published sequences from L. interrogans serovar Pomona isolates; the three most closely related isolates from outside our current study were collected within the last two decades in coastal California from two California sea lions (CSL4002 and CSL10083) and one Channel Island fox (Urocyon littoralis; Fox 32256). We then repeated this approach excluding the less-enriched samples PL-20 and MM-3, which increased the shared core genome among the analyzed samples/isolates to 41.53% (1,921,606/4,627,366) of the reference genome; the phylogenetic patterns remained unchanged (Supplementary Fig. S2). Finally, we used WG-FAST to map the Leptospira sequence reads from all four samples onto a reference phylogeny developed independently using only the published whole genome sequences from isolates, such that our new enriched genomes did not influence the tree topology. Again, the key patterns were preserved: our four new sequences were placed clearly within L. interrogans serovar Pomona and were clustered together along with recent isolates from California sea lions and a Channel Island fox (Fig. 3).
Figure 3.
L. interrogans serovar Pomona phylogeny. A maximum likelihood phylogeny of 26 L. interrogans serovar Pomona genomes together with enriched Leptospira genomic DNA from four PCR-positive animals in this study (highlighted with blue text). The tree was inferred from a concatenated SNP alignment (529 positions out of a core genome size of 4,125,494nts) with RAxML-NG v. 1.1.0. Leptospira from these four animals cluster in a single clade also containing isolates collected from two California sea lions sampled in 2011 and one Channel Island fox sampled in 2004.
Serological reactivity patterns
We analyzed patterns of serological reactivity to gain further insight into the Leptospira strains circulating in the Los Angeles region. We first investigated the five main serovars against which all samples were tested (serovars Bratislava, Canicola, Grippotyphosa, Icterohaemorrhagiae, and Pomona), and evaluated seroprevalence and titer magnitudes against each serovar (Supplementary Fig. S3, Supplementary Table S3). In skunks, raccoons, and coyotes, the maximum MAT titers in each individual were most frequently against serovar Pomona (100%, 87%, and 71%, respectively; Table 3); among individuals with higher titers (1:1600 or above), the maximum titer was always against serovar Pomona. Maximum MAT titer is an imperfect indicator of the infecting serovar, due to well-known challenges with MAT cross-reactivity74,86, but in our study the relationship was supported by the two animals that had both serologic and phylogenomic analyses conducted: the coyote (CM-61) and skunk (MM-3) samples both had highest titers to serovar Pomona (1:102,400 and 1:12,800, respectively), and both were phylogenomically clustered with isolates of L. interrogans serovar Pomona (Fig. 3). Fox squirrels exhibited a pattern that was distinct from the other hosts, with highest titers most often to serovar Icterohaemorrhagiae (80%), which was not highly reactive in any of the other host species. Of the two opossums that exhibited reactions against these 5 serovars, one individual (the only opossum in this study with an active infection) had a maximum titer to serovar Pomona (1:12,800).
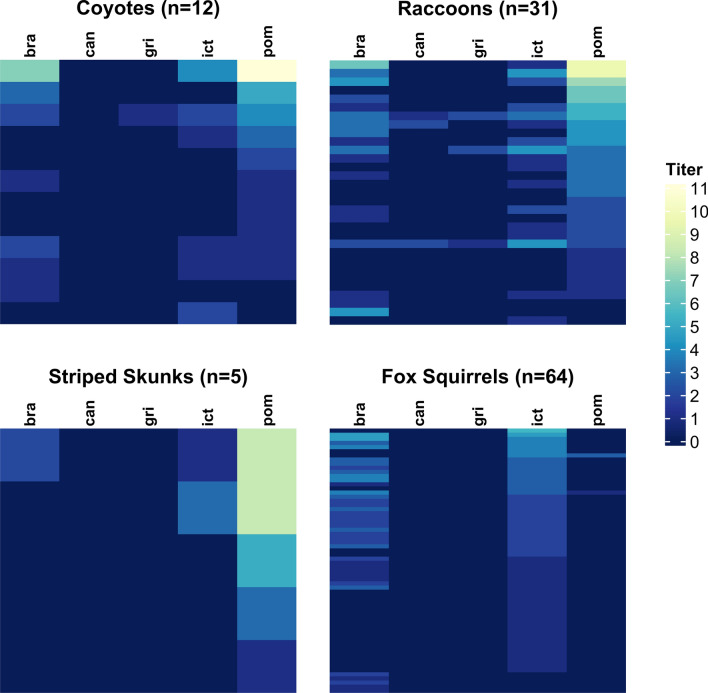
To gain insight into possible sources of reactivity against other serovars, we visualized individual seroreactivity profiles using heatmaps as a way to make sense of the multidimensional data from MAT panels (Fig. 4, Supplementary Fig. S4). The mesocarnivores show strongest reactivity against serovar Pomona. Reactions to other serovars tend to co-occur with strong reactions to serovar Pomona, consistent with the titers against other serovars being cross-reactions to an infection caused by serovar Pomona. A small number of individuals deviate from this pattern (e.g. one raccoon with high titer against serovar Bratislava and low titer against serovar Pomona, possibly indicating an isolated infection with another strain of Leptospira). Fox squirrels show strongest reactivity to serovar Icterohaemorrhagiae (Fig. 4, Supplementary Fig. S4), with other reactivities tending to co-occur.
Figure 4.
Serovar reactivity profiles for target species in the greater Los Angeles region. Data from all seropositive individuals are shown for the five serovars tested at both laboratories: Bratislava (‘bra’), Canicola (‘can’), Grippotyphosa (‘gri’), Icterohaemorrhagiae (‘ict’), and Pomona (‘pom’). Each row of colored bars represents MAT titers for an individual animal, with rows ordered by titer values for the serovar with the highest maximum titers in each species (serovar Pomona for coyotes, raccoons and skunks; serovar Icterohaemorrhagiae in fox squirrels). The colorbar legend shows antibody titer on a log2 scale (1:100 equivalent to 1, 1:200 equivalent to 2, 1:400 equivalent to 3, etc.).
We then considered MAT results for all 21 tested serovars in target species from all regions (Supplementary Table S3). Broad reactivity patterns in non-target regions were similar to patterns seen in greater Los Angeles with serovar Pomona highlighted in mesocarnivores (Supplementary Fig. S5). Serovar Pomona exhibited the highest seroprevalence in raccoons (40.7%), and was one of the highest in skunks (26.9%) and coyotes (18.9%). Mesocarnivore results from the CDC 20-serovar panel showed Pomona as dominant with strong cross-reactions to Autumnalis and Djasiman, with titers against those serovars higher than Pomona in some cases. Across all seropositive coyotes, maximum titers were most commonly observed to serovars Autumnalis (n = 9/22) and Pomona (n = 6/22), with 55% (n = 12/22) and 64% (n = 14/22) of coyotes positive to serovars Autumnalis and Pomona respectively. In coyotes with paired titers (n = 15), Pomona titers were most strongly correlated with titers against serovars Autumnalis (Spearman’s ) and Djasiman (Spearman’s ). In aggregate, the data from raccoons, coyotes and skunks are consistent with most infections being caused by serovar Pomona. In opossums, the broader dataset revealed several individuals with low titers against serovar Hardjo, making it the most frequently positive serovar (Supplementary Table S3), though Pomona was the highest titer observed in the one opossum with an active infection. Aside from the low-titer reactions against Hardjo that more than doubled the overall seroprevalence of opossums, the majority of our conclusions in target species did not change with consideration of the expanded serovar panel: Icterohaemorrhagiae is the dominant serovar in fox squirrels and Pomona is dominant in the other target species.
Associations between serology and PCR results
To analyze the association between active infections and the maximum antibody titer against any serovar, we used paired PCR and MAT results, for which only raccoons had a sufficient sample size (n = 81). Of those 81 animals, 29 were seropositive, and 86% of seropositive animals (n = 25/29) had maximum titers to serovar Pomona. We found that maximum titer level was a significant predictor of active infection in raccoons (Firth’s logistic regression; p-value = 2.09 × 10–9), which aligns with previous studies showing that maximum Leptospira MAT titers are effective predictors of infection in California sea lions22,84. Individual raccoons with titers above 1:1600 were highly likely to be PCR-positive, and therefore likely to present a transmission risk because of their potential to be actively shedding Leptospira (Fig. 5). Intriguingly, among host individuals of other target species that had paired PCR and MAT results, 80% (n = 4/5) of individuals with titers above 1:1600 were PCR-positive (1/1 coyotes, 2/2 skunks, 1/1 opossums and 0/1 fox squirrels).
Figure 5.
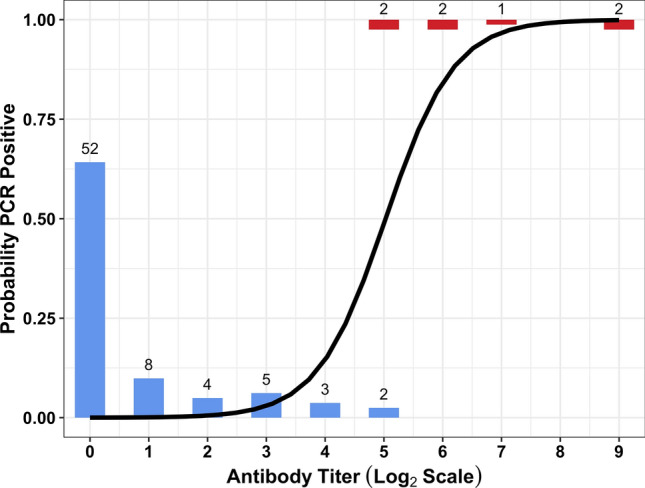
The predicted probability that raccoons are PCR-positive relative to maximum antibody titer to any serovar. The number of individuals with maximum MAT titer at a given level are represented by height of the bars and the corresponding number at the top of the bar. Animals with PCR negative results are shown in blue, and PCR positive individuals are shown in red. Antibody titer (x-axis) is shown on a log2 scale (1:100 equivalent to 1, 1:200 equivalent to 2, 1:400 equivalent to 3, etc.). The solid black line shows the best-fitting solution of a Firth’s logistic regression, which predicts that individuals with MAT titers greater than 1:1600 (5 on the log scale) are at least 80% likely to be PCR positive.
Spatial patterns of Leptospira exposure
We detected Leptospira exposure (i.e. MAT-positive individuals) throughout the sampled ranges of each host species (Fig. 2). When we evaluated sample location relative to the composite land cover classes, we found indications that different species use the landscape in different ways. For instance, fox squirrel and opossum samples were clustered around areas with higher levels of human development, providing evidence for increased use of urban and suburban regions in these non-native species (Supplementary Fig. S6). In contrast, coyotes and raccoons were found across all land classes. When we evaluated Leptospira exposure data in light of these land classes, no clear patterns emerged to distinguish the locations of positive and negative samples (Fig. 2, Supplementary Fig. S6), indicating that Leptospira circulates throughout the sampled range of each host species.
To probe how Leptospira exposure patterns might be influenced by land cover, season, and demographic factors, we used logistic regressions to assess possible correlates for each species. This was done in all species except skunks, which were excluded due to small overall sample size (n = 23). Of the covariates explored here (age class, sex, season, and land classification), none exhibited significant correlations with Leptospira exposure in any of the target species (all p-values > 0.09 in univariate analyses). This lack of correlation can be visualized as a lack of clustering of positive results in ternary plots (Supplementary Fig. S6), supporting that Leptospira is distributed throughout the sampled range of these species.
Additional species and regions
Overall prevalence levels in the non-target dataset were comparable to the target dataset: 4.4% (n = 4/91) of animals were PCR-positive (2 mountain lions, 1 feral pig and 1 bobcat) and antibodies were detected in 30.6% (n = 64/209), with species-specific seroprevalence levels ranging from 0 to 61% (Supplementary Table S1). Feral pigs had overall seroprevalence of 27% (n = 15/55) and were most frequently reactive to serovars Bratislava, Autumnalis, Djasiman, and Pomona (Supplementary Tables S1 and S3). Of the bobcats tested, 46% (n = 5/11) were seropositive (Supplementary Table S1), with titers to Bratislava and Pomona most frequently positive (Supplementary Table S3). Of the non-target species in the greater Los Angeles region, 10% (n = 3/30) of desert cottontails were seropositive, with the three seropositive individuals exhibiting titers against serovars Georgia, Icterohaemorrhagiae, and Pomona, respectively (Supplementary Table S3). Two mountain lions from the greater Los Angeles region were PCR-positive (18.2%; n = 2/11), with both animals showing clinical pathology consistent with leptospirosis (nephritis) upon necropsy.
Discussion
We conducted a large-scale survey of Leptospira in mainland terrestrial mammals in California, focusing on the understudied region of southern California. We had two goals: (1) to identify Leptospira prevalence and potential risk factors in the greater Los Angeles region, and (2) to inform our knowledge of broader multi-host circulation in coastal California wildlife. We identified Leptospira exposure in all target species (skunks, raccoons, coyotes, opossums, and fox squirrels), and detected active infections in all target species except fox squirrels. Widespread evidence of exposure, which was not correlated with specific landscape factors, highlights that Leptospira spp. are endemic and circulating throughout the ranges of these wildlife species in this major metropolitan area. Our findings extend the evidence that Leptospira are widespread in many wildlife species throughout California35,45 and potentially link transmission in mainland mammals to pathogen circulation in island and marine mammal species.
In our five target species, we detected low to moderate levels of Leptospira infection (0–15%) and markedly higher seroprevalence levels (5–60%). Skunks, raccoons and coyotes all exhibited moderate levels of infection (3.7–15.2%), whereas infection prevalence was low in opossums (0.8%) and fox squirrels (0%). Only one opossum was PCR positive, consistent with prior surveillance of opossums elsewhere in California, which have found no87 or low Leptospira infection prevalence35. Fox squirrels had the highest seroprevalence of all target hosts (60%) despite no infections being detected in this species. This could be due to a shorter duration of carriage, longer duration of titer decay (and hence seropositivity), or potentially an alternate route of transmission (e.g. sexual) and associated tissue distribution which could explain the lack of detection in the urinary tract. In addition to the widespread exposure detected in our target species, we detected exposure in all non-target species except gray foxes, and we provide the first evidence that desert cottontails are exposed to Leptospira in California.
In our target species, we detected lower infection prevalence and broadly comparable seroprevalence levels to those reported in more northern regions of California35 (Supplementary Table S4). This may reflect true regional differences, though comparisons across studies and laboratories should be considered carefully. Notably, the prior study used a PCR cycle threshold of 45 to define positivity35, resulting in higher sensitivity and lower specificity (i.e. a lower risk of false negatives, but a higher risk of false positives) than the cycle threshold of 37 used in this study. This could explain the slightly lower infection prevalence in our study. Regional differences in Leptospira infection incidence could also arise from environmental differences between northern and southern California, given that wetter environmental conditions facilitate the bacteria’s survival and transmission. For example, higher rainfall has been associated with higher Leptospira incidence in domestic dogs88, including in northern California89. Expanded analyses with broader sampling ranges and consistent protocols could elucidate possible environmental or seasonal patterns of Leptospira in California wildlife.
We used two broad approaches to characterize the Leptospira strains circulating in our system. For four individual animals (two raccoons, one coyote, and one skunk), we sequenced PCR-positive samples and used phylogenomic analysis to determine that they were infected with L. interrogans strains that exhibit little genetic distance from one another, and cluster within clades of L. interrogans serovar Pomona. For the broader set of individuals and species in our study, we analyzed patterns of seroreactivity to assess possible circulating strains, though we emphasize that serology cannot conclusively identify the Leptospira strain causing an infection86. Over half of the seropositive animals in this study were reactive to multiple serovars (67%; n = 124/186). Consideration of individual-level reactivity profiles indicated that most of these are likely cross-reactions since they co-occur with stronger reactions against other serovars (though infection by multiple strains of Leptospira is possible, as reported in cattle15). Trying to infer the infecting strain from serological data is challenging: this is sometimes assumed to be the serovar with the highest MAT titer, but this approach will fail if the infecting serovar is not on the test panel, or in cases of ‘paradoxical reactions’ where another serovar elicits a higher MAT titer than the true infecting serovar. In this study, coyotes, raccoons, and skunks typically had maximum antibody titers against serovar Pomona, which is consistent with our phylogenomic results, and aligns with findings from northern California where serovar Pomona predominated in mesocarnivores35. Conversely, squirrels showed minimal seroreactivity to serovar Pomona and typically had low maximum titers against serovar Icterohaemorrhagiae, which was not highly reactive in other species. In our two individual animals with both serology and genomic data (one raccoon and one coyote), we confirmed that the maximum titer against Pomona was indeed associated with infection by L. interrogans serovar Pomona. In a subset of samples tested against a broader array of serovars, some coyotes exhibited highest titers to serovar Autumnalis, though titers against serovar Pomona were also high, and titers against Pomona and Autumnalis were strongly correlated. We note that similar patterns have been reported in other canids (domestic dogs91 and Channel Island foxes61), including island foxes with culture-confirmed serovar Pomona infections that caused peak titers against serovar Autumnalis61. Though further genomic data are needed to identify all distinct strains circulating in Los Angeles wildlife, based on current evidence we propose that there are at least two: a strain of L. interrogans serovar Pomona circulating in mesocarnivores (and perhaps other unsampled species), and another strain (with serologic reactivity against serovar Icterohaemorrhagiae) circulating in fox squirrels.
Our finding of widespread Leptospira circulation, with clear indications that serovar Pomona may be predominant in multiple host species, has potential implications for ongoing research in marine and terrestrial island mammals in California21,49–53,61,90. Phylogenetic analyses of Leptospira genomes isolated from California sea lions, northern elephant seals (Mirounga angustirostris), Channel Island foxes and island spotted skunks (Spilogale gracilis amphiala) show evidence of repeated introductions of new strains of L. interrogans serovar Pomona into the broader coastal ecosystem52. The source of the introductions to the marine ecosystem is unknown, but mainland terrestrial mammals are one possibility. Our data are consistent with this hypothesis: our phylogenomic analysis of four PCR-positive samples from mainland mesocarnivores showed that they clustered with L. interrogans serovar Pomona genomes, and were nested closest to recent L. interrogans serovar Pomona isolates from two California sea lions and one Channel Island fox. More genomic data from mainland terrestrial mammals are needed to confirm the timing and direction of transmission links between these mainland and marine ecosystems91.
Information on wildlife disease occurrence is crucial to assessing wildlife and human health risks, but collecting samples to support broad-scale wildlife surveillance can be very challenging. New approaches to analyzing existing data and samples can provide valuable tools to scale up and make the most of surveillance efforts. Our work highlights the new insights that can be gained from emerging laboratory techniques like targeted DNA capture and enrichment63, which facilitate whole genome-based phylogenomic analyses in the absence of the cultured isolates that Leptospira genomics previously required. Novel insights can also be gained from more accessible samples when surveillance efforts are limited. For example, the relationship between Leptospira antibody titers and infection status, which has previously not been well characterized in wildlife44, may provide a proxy for determining infection prevalence from readily accessible serum samples. We showed that antibody titers are strongly predictive of infection status in raccoons, and similar results in California sea lions22,84 indicate a potentially robust general pattern. This could be a useful wildlife screening tool in situations where PCR results are not available or feasible to obtain, but more investigation is needed into the generality of this relationship given the known potential for Leptospira to exhibit species- or host-specific patterns. Consequently, using antibody titers as a proxy for infection status may not apply in some host species, such as squirrels, where antibody responses are variable92,93, or opossums where some individuals fail to mount an antibody response and seronegative shedding is possible35,94.
Baseline knowledge about the prevalence of zoonotic infections in urban wildlife has important utility for public health efforts. Diagnosing early cases of leptospirosis in humans and domestic animals can be challenging due to non-specific clinical signs. Raising clinical awareness about epidemiological risk factors (e.g. the prevalence in sympatric host species) is therefore critical for facilitating accurate diagnostics and disease risk assessments9. Broad-scale surveillance efforts also contribute to knowledge of circulating variants, which can be used to rapidly identify or exclude potential sources of transmission. For example, in 2021 there was a leptospirosis outbreak in Los Angeles dogs caused by L. interrogans serovar Canicola55. Very few individuals in our study showed any reactivity at all against serovar Canicola. This corroborates the conclusion that this outbreak did not originate from local wildlife55, highlighting the importance of longitudinal wildlife surveillance in determining (or ruling out) potential sources of outbreaks caused by multi-host pathogens.
This study has some limitations which suggest directions for additional surveillance efforts and future research. Since only five wildlife species could be sampled extensively, there could be unobserved host species contributing to Leptospira persistence and transmission in greater Los Angeles. Identifying these cryptic contributors, sometimes referred to as ‘epidemiological dark matter’12, remains a frontier in disease ecology and emphasizes the need for ongoing research to understand multi-host pathogen dynamics. We were primarily dependent on collaborating agencies for salvaged and opportunistically collected samples. This led to limited sample sizes with a degree of spatial clustering around collaborator facilities, which potentially reduced our ability to detect spatial patterns. Samples may also have been biased towards more developed areas, with higher road concentrations potentially increasing traffic-related deaths, and higher human densities increasing the likelihood of sick or dead animals being reported. Additionally, the composite land classification used here may have masked finer-scale spatial associations, and our use of circular home range buffers could overlook behavioral patterns in habitat use. Further investigations utilizing other metrics of urbanization (e.g. population density) or individual home ranges and land use patterns could reveal spatial relationships undetected in our analyses.
This study provides the first in-depth look at Leptospira ecology in terrestrial wildlife across the greater Los Angeles area. Expanded knowledge of this pathogen in southern California, including comparisons of prevalence levels and serological patterns across host species, provides key insights into multi-host pathogen dynamics and the potential for cross-species transmission, including from wildlife to humans and their pets. Evidence of Leptospira circulation in Los Angeles wildlife has been lacking, contributing to the perception that the pathogen does not pose a major risk in the area. Our study found evidence consistent with endemic circulation of at least two distinct strains of Leptospira among our five target species, and applied novel Leptospira DNA capture and enrichment techniques that yielded preliminary evidence linking mainland mammal transmission to nearby island and marine systems. High levels of exposure and wide geographic distribution indicate that Leptospira are ubiquitous across the region, with active infection rates substantial enough to warrant concern and to recommend that domestic dogs in the Los Angeles metropolitan area be vaccinated against this disease. Better understanding of Leptospira ecology and transmission dynamics in wildlife is critical to the management of this widely circulating pathogen, highlighting the need for more systematic, broad-scale research efforts to monitor this pathogen in wildlife, domestic animals, and humans.
Supplementary Information
Acknowledgements
The authors would like to thank all collaborating agencies and personnel that facilitated the collection of wildlife samples for this research, including the California Wildlife Center, the Los Angeles County Department of Animal Care and Control, the Los Angeles Department of Animal Services, the United States Department of Agriculture Animal and Plant Health Inspection Service (USDA APHIS), Dr. Ted Stankowich at California State University Long Beach, SPCA for Monterey County, Vandenberg Space Force Base, Naval Base Ventura County, the National Park Service, Dr. Duane Tom, Dr. Lorraine Barbosa, and all of our dedicated undergraduate research assistants who made the scale of this project possible. This research was supported by the following funding sources: UCLA Sustainable LA Grand Challenge, UCLA/La Kretz Center for California Conservation Science, Wildlife Disease Association Challenge Grant (Experiment.com), US National Science Foundation (DEB-1557022, OCE-1335657), US Cooperative Ecosystem Studies Unit (Cooperative Agreement #W9132T1920006). Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Author contributions
S.H., K.P. and J.L.S. conceived the study, and S.H., R.M., A.T., K.P., B.B., A.G., C.S. and J.L.S. contributed to methodology and data interpretation. S.H. conducted the wildlife sampling and testing with the support of A.T., M.G., R.M., C.C., J.N., I.T., D.H., R.G., J.M., J.B., S.R., J.S., A.F., J.L., K.P. and J.L.S. S.H. drafted the manuscript and made the maps. D.W., N.S. and J.S. performed the DNA amplification, genetic analyses and construction of all phylogenetic trees. S.H., R.M., A.T., B.B. and C.S. made all other figures and tables, and S.H., R.M., A.T., M.G., B.B. and J.M. performed the data analyses. All authors helped with revision of the manuscript.
Data availability
Data used for this paper are available at: https://github.com/SarahHelman/Lepto_in_SoCal_urban_wildlife. All sequence data were submitted to the NCBI sequence read archive under accession PRJNA979271. Source code for the Whole Genome Focused Array SNP Typing (WG-FAST) pipeline is available at: https://github.com/jasonsahl/wgfast.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sarah K. Helman, Email: sarahkh@ucla.edu
James O. Lloyd-Smith, Email: jlloydsmith@ucla.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-40322-2.
References
- 1.Levett PN. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, B. History of Leptospirosis and Leptospira. in Leptospira and Leptospirosis (ed. Adler, B.) vol. 387 1–9 (Springer Berlin Heidelberg, 2015). [DOI] [PubMed]
- 3.Torgerson PR, et al. Global burden of leptospirosis: Estimated in terms of disability adjusted life years. PLoS Negl. Trop. Dis. 2015;9:e0004122. doi: 10.1371/journal.pntd.0004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler B, de la Peña Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Cilia G, Bertelloni F, Albini S, Fratini F. Insight into the epidemiology of leptospirosis: A review of leptospira isolations from “unconventional” hosts. Animals. 2021;11:191. doi: 10.3390/ani11010191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko AI, Goarant C, Picardeau M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa F, et al. Patterns in Leptospira Shedding in Norway Rats (Rattus norvegicus) from Brazilian Slum Communities at High Risk of Disease Transmission. PLoS Negl. Trop. Dis. 2015;9:e0003819. doi: 10.1371/journal.pntd.0003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz-Zanzi C, et al. A systematic literature review of leptospirosis outbreaks worldwide, 1970–2012. Rev. Panam. Salud Pública. 2020;44:1. doi: 10.26633/RPSP.2020.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sykes JE, Reagan KL, Nally JE, Galloway RL, Haake DA. Role of diagnostics in epidemiology, management, surveillance, and control of leptospirosis. Pathogens. 2022;11:395. doi: 10.3390/pathogens11040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd-Smith JO, et al. Epidemic dynamics at the human–animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viana M, et al. Assembling evidence for identifying reservoirs of infection. Trends Ecol. Evol. 2014;29:270–279. doi: 10.1016/j.tree.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buhnerkempe MG, et al. Eight challenges in modelling disease ecology in multi-host, multi-agent systems. Epidemics. 2015;10:26–30. doi: 10.1016/j.epidem.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net). Genus Leptospira. https://lpsn.dsmz.de/genus/leptospira (2022). [DOI] [PubMed]
- 14.Vincent AT, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levett, P. N. Systematics of Leptospiraceae. in Leptospira and Leptospirosis (ed. Adler, B.) vol. 387 11–20 (Springer Berlin Heidelberg, 2015). [DOI] [PubMed]
- 16.Monahan AM, Callanan JJ, Nally JE. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 2008;76:4952–4958. doi: 10.1128/IAI.00511-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haake, D. A. & Levett, P. N. Leptospirosis in Humans. in Leptospira and Leptospirosis (ed. Adler, B.) vol. 387 65–97 (Springer Berlin Heidelberg, 2015). [DOI] [PMC free article] [PubMed]
- 18.Casanovas-Massana A, et al. Quantification of Leptospira interrogans survival in soil and water microcosms. Appl. Environ. Microbiol. 2018;84:e00507–e518. doi: 10.1128/AEM.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gostic KM, et al. Mechanistic dose-response modelling of animal challenge data shows that intact skin is a crucial barrier to leptospiral infection. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2019;374:20190367. doi: 10.1098/rstb.2019.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis, W. A. Animal Leptospirosis. in Leptospira and Leptospirosis (ed. Adler, B.) 99–137 (Springer, 2015). 10.1007/978-3-662-45059-8_6.
- 21.Lloyd-Smith JO, et al. Cyclical changes in seroprevalence of leptospirosis in California sea lions: Endemic and epidemic disease in one host species? BMC Infect. Dis. 2007;7:125–136. doi: 10.1186/1471-2334-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prager KC, et al. Linking longitudinal and cross-sectional biomarker data to understand host-pathogen dynamics: Leptospira in California sea lions (Zalophus californianus) as a case study. PLoS Negl. Trop. Dis. 2020;14:e0008407. doi: 10.1371/journal.pntd.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faine, S., Adler, B., Bolin, C. & Perolat, P. Leptospira and leptospirosis. (MediSci, 1999).
- 24.Lau CL, Smythe LD, Craig SB, Weinstein P. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 2010;104:631–638. doi: 10.1016/j.trstmh.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Bradley CA, Altizer S. Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 2007;22:95–102. doi: 10.1016/j.tree.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassell JM, Begon M, Ward MJ, Fèvre EM. Urbanization and disease emergence: Dynamics at the wildlife-livestock-human interface. Trends Ecol. Evol. 2017;32:55–67. doi: 10.1016/j.tree.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley, S. P. D., Serieys, L. E. K. & Moriarty, J. G. Infectious Disease and Contaminants in Urban Wildlife: Unseen and Often Overlooked Threats. in Urban Wildlife (eds. McCleery, R. A., Moorman, C. E. & Peterson, M. N.) 175–215 (Springer US, 2014). 10.1007/978-1-4899-7500-3_10.
- 28.Plowright RK, et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leibler JH, Zakhour CM, Gadhoke P, Gaeta JM. Zoonotic and vector-borne infections among urban homeless and marginalized people in the United States and Europe, 1990–2014. Vector Borne Zoonotic Dis. Larchmt. N. 2016;16:435–444. doi: 10.1089/vbz.2015.1863. [DOI] [PubMed] [Google Scholar]
- 30.Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC. Sporadic urban leptospirosis. Ann. Intern. Med. 1996;125:794–798. doi: 10.7326/0003-4819-125-10-199611150-00002. [DOI] [PubMed] [Google Scholar]
- 31.Dupouey J, et al. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. Dis. 2014;37:77–83. doi: 10.1016/j.cimid.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Minter A, et al. A model for leptospire dynamics and control in the Norway rat (Rattus norvegicus) the reservoir host in urban slum environments. Epidemics. 2018;25:26–34. doi: 10.1016/j.epidem.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Boey K, Shiokawa K, Rajeev S. Leptospira infection in rats: A literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 2019;13:e0007499. doi: 10.1371/journal.pntd.0007499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junge, R. E., Bauman, K., King, M. & Gompper, M. E. A serologic assessment of exposure to viral pathogens and Leptospira in an urban raccoon (Procyon lotor) population inhabiting a large zoological park. J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet.38, 18–26 (2007). [DOI] [PubMed]
- 35.Straub MH, Foley JE. Cross-sectional evaluation of multiple epidemiological cycles of Leptospira species in peri-urban wildlife in California. J. Am. Vet. Med. Assoc. 2020;257:840–848. doi: 10.2460/javma.257.8.840. [DOI] [PubMed] [Google Scholar]
- 36.Davis MA, et al. Serological Survey for Antibodies to Leptospira in Dogs and Raccoons in Washington State. Zoonoses Public Health. 2008 doi: 10.1111/j.1863-2378.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- 37.Gautam R, Wu C-C, Guptill LF, Potter A, Moore GE. Detection of antibodies against Leptospira serovars via microscopic agglutination tests in dogs in the United States, 2000–2007. J. Am. Vet. Med. Assoc. 2010;237:293–298. doi: 10.2460/javma.237.3.293. [DOI] [PubMed] [Google Scholar]
- 38.Blessington T, Schenck AP, Levine JF. Frequency of Animal Leptospirosis in the Southern United States and the Implications for Human Health. South. Med. J. 2020;113:240–249. doi: 10.14423/SMJ.0000000000001093. [DOI] [PubMed] [Google Scholar]
- 39.White AM, et al. Hotspots of canine leptospirosis in the United States of America. Vet. J. 2017;222:29–35. doi: 10.1016/j.tvjl.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Smith AM, Stull JW, Moore GE. Potential Drivers for the Re-Emergence of Canine Leptospirosis in the United States and Canada. Trop. Med. Infect. Dis. 2022;7:377. doi: 10.3390/tropicalmed7110377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meites E, et al. Reemerging Leptospirosis, California. Emerg. Infect. Dis. 2004;10:406–412. doi: 10.3201/eid1003.030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katz AR, Buchholz AE, Hinson K, Park SY, Effler PV. Leptospirosis in Hawaii, USA, 1999–2008. Emerg. Infect. Dis. 2011;17:221–226. doi: 10.3201/eid1702.101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerra MA. Leptospirosis: public health perspectives. Biol. J. Int. Assoc. Biol. Stand. 2013;41:295–297. doi: 10.1016/j.biologicals.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen K, et al. Leptospira antibodies detected in wildlife in the USA and the US Virgin Islands. J. Wildl. Dis. 2018;54:450–459. doi: 10.7589/2017-10-269. [DOI] [PubMed] [Google Scholar]
- 45.Cirone SM, Riemann HP, Ruppanner R, Behymer DE, Franti CE. Evaluation of the hemagglutination test for epidemiologic studies of leptospiral antibodies in wild mammals. J. Wildl. Dis. 1978;14:193–202. doi: 10.7589/0090-3558-14.2.193. [DOI] [PubMed] [Google Scholar]
- 46.Ruppanner R, Jessup DA, Ohishi I, Behymer DE, Franti CE. Serologic survey for certain zoonotic diseases in black bears in California. J. Am. Vet. Med. Assoc. 1982;181:1288–1291. [PubMed] [Google Scholar]
- 47.Clark RK, Jessup DA, Hird DW, Ruppanner R, Meyer ME. Serologic survey of California wild hogs for antibodies against selected zoonotic disease agents. J. Am. Vet. Med. Assoc. 1983;183:1248–1251. [PubMed] [Google Scholar]
- 48.Straub MH, Church M, Glueckert E, Foley JE. Raccoons ( Procyon lotor ) and Striped Skunks ( Mephitis mephitis ) as Potential Reservoirs of Leptospira spp. California. Vector-Borne Zoonotic Dis. 2020;20:418–426. doi: 10.1089/vbz.2019.2528. [DOI] [PubMed] [Google Scholar]
- 49.Gulland FMD, et al. Leptospirosis in California sea lions (Zalophus californianus) stranded along the central California coast, 1981–1994. J. Wildl. Dis. 1996;32:572–580. doi: 10.7589/0090-3558-32.4.572. [DOI] [PubMed] [Google Scholar]
- 50.Greig DJ, Gulland FMD, Kreuder C. A Decade of Live California Sea Lion (Zalophus californianus) Strandings Along the Central California Coast: Causes and Trends, 1991–2000. Aquat. Mamm. 2005;31:11–22. doi: 10.1578/AM.31.1.2005.11. [DOI] [Google Scholar]
- 51.Colagross-Schouten AM, Mazet JAK, Gulland FMD, Miller MA, Hietala S. Diagnosis and seroprevalence of leptospirosis in California sea lions from coastal California. J. Wildl. Dis. 2002;38:7–17. doi: 10.7589/0090-3558-38.1.7. [DOI] [PubMed] [Google Scholar]
- 52.Mummah, R. O. Leptospira in the coastal California ecosystem: Challenges and solutions for analyzing complex wildlife disease data. (University of California Los Angeles, 2021).
- 53.Zuerner RL, et al. Geographical dissemination of Leptospira interrogans serovar Pomona during seasonal migration of California sea lions. Vet. Microbiol. 2009;137:105–110. doi: 10.1016/j.vetmic.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Greene MR. A survey for leptospirosis in Southern California. Am. J. Epidemiol. 1941;34:87–90. doi: 10.1093/oxfordjournals.aje.a118739. [DOI] [Google Scholar]
- 55.LA County Department of Public Health (LADPH). Leptospirosis in Dogs in Los Angeles County in 2021. http://www.publichealth.lacounty.gov/vet/Leptospirosis2021.htm (2022).
- 56.Roug A, Swift P, Torres S, Jones K, Johnson CK. Serosurveillance for Livestock Pathogens in Free-Ranging Mule Deer (Odocoileus hemionus) PLoS ONE. 2012;7:e50600. doi: 10.1371/journal.pone.0050600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Straub MH, Rudd JL, Woods LW, Clifford DL, Foley JE. Leptospira prevalence and its association with renal pathology in mountain lions (Puma concolor) and bobcats (lynx rufus) in California, USA. J. Wildl. Dis. 2021;57:1. doi: 10.7589/JWD-D-20-00070. [DOI] [PubMed] [Google Scholar]
- 58.U.S. Census Bureau QuickFacts: Los Angeles city, California. https://www.census.gov/quickfacts/fact/table/losangelescountycalifornia,losangelescitycalifornia/PST045222,PST045221.
- 59.Grau GA, Sanderson GC, Rogers JP. Age Determination of Raccoons. J. Wildl. Manag. 1970;34:364. doi: 10.2307/3799023. [DOI] [Google Scholar]
- 60.Wu Q, et al. Development of a real-time PCR for the detection of pathogenic Leptospira spp. California sea lions. Dis. Aquat. Organ. 2014;110:165–172. doi: 10.3354/dao02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lloyd-Smith, J. O. & Prager, K. C. Leptospirosis in endangered island foxes and California sea lions: Outbreak prediction and prevention in a changing world. https://apps.dtic.mil/sti/citations/AD1189863 (2021).
- 62.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009;64:247–255. doi: 10.1016/j.diagmicrobio.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 63.Stone NE, et al. DNA capture and enrichment: A culture-independent approach for characterizing the genomic diversity of pathogenic Leptospira Species. Microorganisms. 2023;11:1282. doi: 10.3390/microorganisms11051282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delcher AL, Salzberg SL, Phillippy AM. Using mummer to Identify Similar Regions in Large Sequence Sets. Curr. Protoc. Bioinforma. 2003;1:10. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 68.Sahl JW, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2016;2:1. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang W, Li L, Myers JR, Marth GT. ART: A next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahl JW, et al. Phylogenetically typing bacterial strains from partial SNP genotypes observed from direct sequencing of clinical specimen metagenomic data. Genome Med. 2015;7:52. doi: 10.1186/s13073-015-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedersen K, et al. Widespread detection of antibodies to Leptospira in feral swine in the United States. Epidemiol. Infect. 2015;143:2131–2136. doi: 10.1017/S0950268814003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kmety, E. & Dikken, H. Classification of the species Leptospira interrogans and history of its serovars. (University Press Groningen, 1993).
- 77.R Core Team. R: A language and environment for statistical computing. (2023).
- 78.ArcGIS Desktop. (2011).
- 79.Adducci II, A. et al. Urban coyotes are genetically distinct from coyotes in natural habitats. J. Urban Ecol.6, juaa010 (2020).
- 80.US. Geological Survey (USGS). NLCD 2019 Land Cover Conterminous United States. (2019).
- 81.Šálek M, Drahníková L, Tkadlec E. Changes in home range sizes and population densities of carnivore species along the natural to urban habitat gradient. Mammal Rev. 2015;45:1–14. doi: 10.1111/mam.12027. [DOI] [Google Scholar]
- 82.Wright JD, Burt MS, Jackson VL. Influences of an Urban environment on home range and body mass of Virginia Opossums (Didelphis virginiana) Northeast. Nat. 2012;19:77–86. doi: 10.1656/045.019.0106. [DOI] [Google Scholar]
- 83.Prince A, DePerno CS, Gardner B, Moorman CE. Survival and home-range size of Southeastern Fox squirrels in North Carolina. Southeast. Nat. 2014;13:456. doi: 10.1656/058.013.0305. [DOI] [Google Scholar]
- 84.Helman, S. K. From California sea lions to urban coyotes: Maximizing insights from Leptospira surveillance in coastal California wildlife. (UCLA, 2022).
- 85.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. doi: 10.1093/biomet/80.1.27. [DOI] [Google Scholar]
- 86.André-Fontaine G, Triger L. MAT cross-reactions or vaccine cross-protection: retrospective study of 863 leptospirosis canine cases. Heliyon. 2018;4:e00869. doi: 10.1016/j.heliyon.2018.e00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krueger L, et al. Identification of Zoonotic and Vector-borne Infectious Agents Associated with Opossums (Didelphis virginiana) in Residential Neighborhoods of Orange County. California. Proc. Vertebr. Pest Conf. 2016;27:1. [Google Scholar]
- 88.Ward MP. Seasonality of canine leptospirosis in the United States and Canada and its association with rainfall. Prev. Vet. Med. 2002;56:203–213. doi: 10.1016/S0167-5877(02)00183-6. [DOI] [PubMed] [Google Scholar]
- 89.Adin CA, Cowgill LD. Treatment and outcome of dogs with leptospirosis: 36 cases (1990–1998) J. Am. Vet. Med. Assoc. 2000;216:371–375. doi: 10.2460/javma.2000.216.371. [DOI] [PubMed] [Google Scholar]
- 90.Buhnerkempe MG, et al. Detecting signals of chronic shedding to explain pathogen persistence: Leptospira interrogans in California sea lions. J. Anim. Ecol. 2017;86:460–472. doi: 10.1111/1365-2656.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Borremans, B., Faust, C., Manlove, K. R., Sokolow, S. H. & Lloyd-Smith, J. O. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Philos. Trans. R. Soc. B Biol. Sci.374, 20180344 (2019). [DOI] [PMC free article] [PubMed]
- 92.Diesch SL, Crawford RP, McCulloch WF, Top FH. Human Leptospirosis Acquired from Squirrels. N. Engl. J. Med. 1967;276:838–842. doi: 10.1056/NEJM196704132761504. [DOI] [PubMed] [Google Scholar]
- 93.Dirsmith K, et al. Leptospirosis in Fox Squirrels (Sciurus niger) of Larimer County, Colorado. USA. J. Wildl. Dis. 2013;49:641–645. doi: 10.7589/2012-10-265. [DOI] [PubMed] [Google Scholar]
- 94.Reilly JR. The susceptibility of five species of wild animals to experimental infection with Leptospira grippotyphosa. J. Wildl. Dis. 1970;6:289–294. doi: 10.7589/0090-3558-6.4.289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for this paper are available at: https://github.com/SarahHelman/Lepto_in_SoCal_urban_wildlife. All sequence data were submitted to the NCBI sequence read archive under accession PRJNA979271. Source code for the Whole Genome Focused Array SNP Typing (WG-FAST) pipeline is available at: https://github.com/jasonsahl/wgfast.