Abstract
Decma fissa is the most widely distributed species of the genus Decma occuring in southern China. This study presents the first phylogeographic work of D. fissa based on COI, Cytb and ITS sequence. We examined genetic diversity with ITS and mitochondrial sequence respectively, and phylogenetic work was based on the mitochondrial data. A high-level genetic diversity was revealed based on mitochondrial data but a low-level diversity was shown with ITS sequence. For the mitochondrial data, divergence time analysis displayed five lineages. Based on the Mantel test, geographic and genetic distances among D. fissa populations revealed a significant positive correlation. Bayesian skyline plot (BSP) analyses implied that none of three major lineages of D. fissa was seemingly affected by the last glacial maximum (LGM, 0.015–0.025 Mya). Ecological niche modeling was used to predict the distribution of D. fissa in four periods (LGM, Mid-Holocene, current and 2070) in China. Analysis of the ancestral area reconstruction indicated that D. fissa occurred in the South China area.
Keywords: Decma fissa, Phylogeography, Ecological niche models, Population structure, Demographic history
BACKGROUND
The distribution and dispersing of insects are affected by many factors such as mutation, geographic isolation, natural selection, climate change and gene flow (Hewitt 2004; Garrido et al. 2012; Saeb and Al-Naqeb 2016; Tang et al. 2022). Understanding the historical processes of species can help us reveal their ability to confront environmental changes (Porretta et al. 2007; Lyons et al. 2012; Wei et al. 2013 2015). In particular, the repeated changes of the Quaternary climate (Clark et al. 2009) had a profound impact on the distribution pattern of existing species. Species had been reduced to refugia during the ice period, and then expanded again during the warm climate period (Hewitt 2000; Song et al. 2016). In addition, different species have different historical distributions and responses to environmental changes, so they would be affected differently (Liu et al. 2018). Unlike the climate of Europe and North America, due to the uplift of the Qinghai Tibet Plateau, the climate was warmer in eastern Asia in the LGM (Dai et al. 2011; Wei et al. 2013). Such a climate led to the absence of ice cover in low altitude areas. Although phylogeographic studies were carried out in many vertebrates (Dufresnes et al. 2016; Qiu et al. 2016; Liu et al. 2019), the distribution and genetic diversity of insects have been only partially studied in China (Ye et al. 2018; Zhou et al. 2021; Li et al. 2022; Wang et al. 2022; Tang et al. 2022). These studies show that insect species in eastern Asia might often survive in more than one refugia.
Decma fissa (Orthoptera: Tettigoniidae: Meconematinae) is a long-wing species of Meconematinae, which is a subfamily containing a high number of species (Cigliano et al. 2022). It was described as Xiphidiopsis fissa beloning to the genus Xiphidiopsis (Xia and Liu 1993). The genus Decma was established by Gorochov (1993) for Decma (Decma) stshelkanovtzevi. Gorochov et al (2005) moved Xiphidiopsis fissa into the genus Decma, and the new combination, Decma fissa, continues to be used today. Compared to the narrow distributions of other species of this genus (Liu and Yin 2004; Gorochov et al. 2005; Liu and Zhou 2007; Shi et al. 2013), the distribution of D. fissa ranges across several provinces in southern China (Xia and Liu 1993; Wei et al. 2016). In the present study, we predicted the potential distribution based on climate data and ecological modelling methods, and explored the dispersal pathway by ancestral area reconstruction. Our aims are to (a) determine the genetic structure and diversity; (b) study the demographic history among populations; and (c) identify the potential locations.
MATERIALS AND METHODS
Specimen Collection
A total of 232 specimens were collected from 15 locations (Fig. 1). The location details are shown in table S1. All the individuals have been identified by morphological method and preserved in absolute ethanol and stored at -20°C until DNA extraction. The key that we used for species identification is from Wang (2020).
Fig. 1.
The geographic distribution collection points.
DNA extraction, amplification and sequencing
Genomic DNA was extracted using TIANamp Genomic DNA Kit (Tiangen, Beijing) following the manufacturer’s protocol from muscle tissue of each adult individual. PCR was performed with Premix TaqTM (Takara, Beijing). A fragment of COI was amplified and sequenced using the primers COBU/COBL (Huang et al. 2013), the primers Cytb-N/Cytb-J (Simon et al. 2006) were used for a fragment of Cytb, and the primers ITS-F/ITS-R (Weeker et al. 2001) were used for a fragment of ITS. PCR cycle profiles are: for COI, an initial denaturing of 3 min at 94°C followed by 35 cycles of: 30s at 94°C, 30s at 49°C and 90s at 72°C, then a step of 8 min at 72°C, finally forever at 4°C, for CYTB, an initial denaturing of 5 min at 95°C followed by35cyclesof:30sat95°C,30sat50°Cand1minat 72°C, then a step of 8 min at 72°C, finally forever at 4°C. For ITS, an initial denaturing of 3 min at 94°C followed by 35 cycles of: 30s at 94°C, 30s at 46°C and 90s min at 72°C, then a step of 8 min at 72°C, finally forever at 4°C. The information about primers is given in table 1. The PCR products were sent to Azenta (Tianjin, China) for sequencing in both directions after being analyzed with 1% agarose gels with ethidium bromide following electrophoresis. All sequences were edited with Seqman (DNASTAR, Lasergene 7.1) and aligned with BioEdit (Hall 1999). The combined data was concatenated with PhyloSuite v1.2.2 (Zhang et al. 2020). At last, a total of 232 COI, 232 Cytb and 229 ITS sequences were used in this work.
Table 1.
Information about primers used in this study
Ecological niche models
MaxEnt 3.3.3 (Phillips et al. 2006; Phillips and Dudík 2008; Warren et al. 2013) was used to predict the potential distribution with 23 distribution records including 15 collection points and 8 points from literature (Xia and Liu 1993; Liu and Yin 2004; Gorochov et al. 2005; Liu and Zhou 2007; Shi et al. 2013; Wei et al. 2016). All the distribution data from literature are provided in table S2. The ecological factors of the Last Glacial Maximum (LGM, about 22000 years ago), Mid-Holocene (about 6000 years ago), present (1960) and the future (2070) were downloaded from WorldClim (http://www.world clim. org) at 30s arc-min resolution and RCP4.5 (Thomson et al. 2011; Zhou et al. 2014). Firstly, we extracted the data of ecological factors with Arcgis 10.7 (Esri, Redlands, CA, USA) and counted the correlation between factors with IBM SPSS Statistics 20.0 (IBM Corp, Armonk, NY, USA). Then, we used ecological niche modeling (ENM) for current climate conditions using all factors. The model was performed with 10 replicate runs, 10000 maximum iterations and 25% for model training. The importance of factors was determined by percentage contribution in the result. The factors with high-relation (r > 0.8) and low percentage contribution were removed (Yang et al. 2013). Finally, nine factors were used to predict the distribution area of D. fissa in China: Bio2 (Mean Diurnal Range), Bio3 (Isothermality), Bio6 (Min Temperature of Coldest Month), Bio7 (Temperature Annual Range), Bio10 (Mean Temperature of Warmest Quarter), Bio12 (Annual Precipitation), Bio16 (Precipitation of Wettest Quarter), Bio17 (Precipitation of Driest Quarter), Bio18 (Precipitation of Warmest Quarter).
We used the ENMeval package to optimize the regularization multiplier and feature class parameters in the R version 4.2.2 software (Muscarella et al. 2014; Kass et al. 2021). Based on the AICc calculated in the ENMeval, the best model setting for MaxEnt was the feature class (FC): linear (L), quadratic (Q), hinge (H), and regularization multiplier (RM) equal to 2.
The accuracy of each model prediction was determined based on AUC scores. The score ranges from 0.5 to 1, and indicates excellent power of predicting when it is above 0.9 (Ye et al. 2014).
Genetic diversity
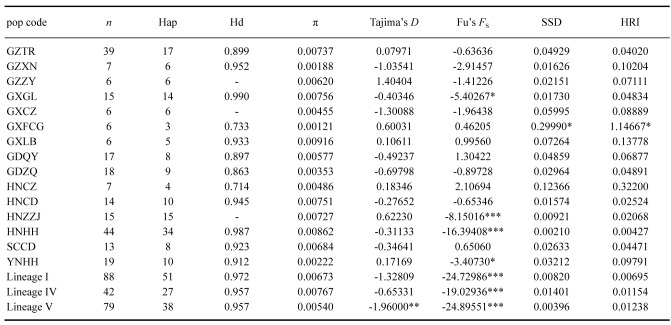
The number of haplotypes (Hap), haplotype diversity (Hd), nucleotide diversity (π), and variable sites were analyzed using DnaSP 5.10 (Librado and Rozas 2009). Population differentiation (FST) was implemented in Arlequin 3.5 (Excoffier and Lischer 2010) conducted with 10,000 permutations. FST value ranges 0 to 1, and the 0 implies complete panmixia (Hudson et al. 1992; Hudson and Richard 2000; Chabot et al. 2015).
Mitochondrial haplotype network and divergence times
Haplotype data of combined sequences were edited with DnaSP v5.10. The haplotype network was constructed in PopArt 1.7 with Median Joining (Leigh and Bryant 2015).
Divergence time was estimated with Beast v1.10.4 (Suchard et al. 2018). Due to the lack of fossil record, a uniform calibration prior, which is based on prior estimated divergence time, was set for the species Xizicus fascipes (GenBank code: NC018765.1) and Decma fissa at 42.84 Ma (Zhou et al. 2017). The analysis was run 100 million generations with TN + F + G4 model, uncorrelated relaxed clock, lognormal distribution, random starting tree, the Yule process, and sampling every 10,000 steps. The first 10% of the trees were discarded as burn-in. Tracer v1.7.1 (Rambaut et al. 2018) was used to assess convergence (all ESS parameters were > 200). The maximum clade credibility tree was produced in TreeAnnotator v1.10.4 and visualized with Figtree v4.1.3 (Rambaut 2012).
Demographic history and population structure of mitochondrial data
To trace the demographic changes of each population and three major lineages of D. fissa, neutrality tests and mismatch distribution analyses were conducted in Arlequin 3.5 (Excoffier and Lischer 2010). Two neutrality tests, Fu’s FS and Tajima’s D were used to evaluate historical demographic expansion with 1,000 simulated samples. Negative values of these two tests indicate that the population had undergone expansion, while positive values indicate the population experienced a bottleneck (Tajima 1989; Fu 1997). Mismatch distribution for three major lineages were conducted for the sum of squared deviations (SSD) and Harpending’s raggedness index (HRI) (Rogers and Harpending 1992; Harpending 1994; Excoffier 2004) by 1000 bootstrap replications.
Genetic distances between populations were calculated with Mega X (Kumar et al. 2018) using Kimura’s 2-parameter and Tamura 3-parameter models.
AMOVA (analysis of molecular variance) was implemented in Arlequin 3.5 conducted with 10,000 permutations. The spatial analysis of molecular variance (SAMOVA) was implemented by SAMOVA 2.0 (Dupanloup et al. 2002) with K = 2 to 15, and 10,000 iterations. The final K was determined by FCT (Heuertz et al. 2004).
The population size of three major lineages over time was assessed using Bayesian skyline plots (BSP) analysis with concatenated mitochondrial genes (Drummond et al. 2005; Minin et al. 2008). This analysis was performed using BEAST v1.10.4 (Suchard et al. 2018). We focused on three major lineages (I, IV, and V) because other lineages (II and III) contained too few haplotypes. TN + F + G4 for lineage I and lineage V, HKY + F + G4 for lineage IV were determined to be the best substitution model with PhyloSuite (Zhang et al. 2020). Analyses were run using a strict molecular clock, assuming a divergence rate of 3.54% per million years for COI (Clock.rate parameter was set as a normal prior with an initial value = 0.0177, mean = 0.0177, and SD = 0.004) (Papadopoulou et al. 2010), and 4.22% per million years for Cytb (Clock.rate parameter was set as a normal prior with an initial value = 0.0211, mean = 0.0211, and SD = 0.004) (Pons and Vogler 2005). Analyses were run for 100 million generations and sampling every 10000 steps for lineages IV and V, and 200 million generations and sampling every 20000 steps for lineage I. Tracer v1.7.1 (Rambaut et al. 2018) was used to assess convergence (all ESS parameters were > 200) and visualization of median and 95% highest posterior probability density intervals (95% HPD).
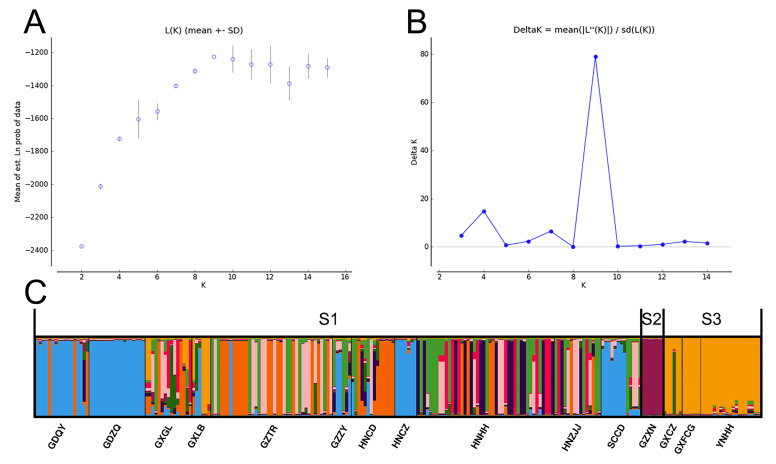
The population genetic structure of D. fissa was analyzed by Structure v. 2.3.4 (Pritchard et al. 2000). The number of clusters (K) was set as 2 to 15. Ten runs for K = 2 to 15 were analyzed, and a burn-in of 200000 followed by 1000000 Markov chain Monte Carlo (MCMC) iterations. The optimal K-value was determined with Structure Harvester 0.6.93 (Earl and Vonholdt 2012) based on the Delta K method. The results of 10 replicates for the chosen K-value were uploaded to Clumpak server (http://clumpak.tau.ac.il/index) (Kopelman et al. 2015) to generate the final plots.
Mantel test
A Mantel test was conducted by Arlequin 3.5 with 1000 permutations to ensure that there was a relationship between the population differentiation (FST) based on mitochondrial combined sequence and geographic distance.
Ancestral area reconstruction
To trace the origin and diffusion process of D. fissa, Bayesian binary MCMC (BBM) analysis was implemented in RASP v4.2 (Yu et al. 2015). Five areas were considered based on the environmental conditions in the distribution range of D. fissa: (A) Central China, (B) South China, (C) Yunnan-Guizhou Plateau, (D) Southeast coastal area, and (E) Sichuan Basin. As input data, 20000 trees were produced by Beast 1.10.4 based on the mitochondrial dataset with HKY + F + G4 model (chosen with PhyloSuite). Clock rate and MCMC parameters were the same with BSP. 10000 random trees were selected to use for analysis after 4000 trees (20%) were discarded as burn-in. BBM analysis was implemented with default settings, except the number of maximum areas, which was set to two because of the limited dispersal ability of the Meconematinae (Wang et al. 2019).
RESULTS
Distribution modeling
Ecological niche models showed a high mean model fit (AUC > 0.9) indicating excellent performance of the models. Figure 2 shows the predicted areas of different periods. As for the present, the predicted areas were mainly the mountain regions including existing records. During the LGM, one high-suitability area is most obvious in the Sichuan basin. During the Mid-Holocene, a large low-suitability area existed in Yunnan province for the first time and the higher suitability areas were similar to the present-day predicted areas. Under RCP 4.5 in the year 2070, the high-suitability areas were slightly expanded, and other predicted areas were quite similar to the present-day predicted areas.
Fig. 2.
Potential distribution areas of D. fissa in different periods. Potential areas for (A) Current day; (B), Last Glacial Maximum; (C), Mid-Holocene; (D), year 2070 (RCP 4.5).
Population genetic diversity and structure
ITS sequences were 846 bp long, and 27 haplotypes were obtained based on 70 polymorphic sites. The haplotype diversity (Hd) and nucleotide diversity (π) are shown in table 2.
Table 2.
Diversity parameter based on ITS sequences
For mitochondrial data, the combined sequences were 1215 bp long (COI 624 bp, and Cytb 591 bp). A total of 134 haplotypes were identified based on 165 polymorphic sites (91 sites from COI, and 74 sites from Cytb). The numbers of haplotypes, haplotype diversity (Hd), and nucleotide diversity (π) are shown in table 3. The genetic distances between populations based on Kimura’s 2-parameter and Tamura 3-parameter models are shown as a heatmap in figure 3 (details information in Tables S3, S4). Pairwise FST between populations ranged from 0.02125 to 0.92767. The highest FST occurred between GXFCG and GZXN. The lowest FST occurred between GXFCG and YNHH (detailed information in Table S5, S6).
Table 3.
Diversity parameter based on combined mitochondrial sequences
Fig. 3.
The genetic distances among populations. A: based on Kimura’s 2-parameter; B: based on Tamura 3-parameter.
The SAMOVA showed that the FCT value clearly increased from K = 2 to 3. Three groups (FCT = 0.427, p < .001) were delimited (S1: GDQY + GDZQ + GXGL+GXLB+GZTR+GZZY+HNCZ+HNCD + HNHH + HNZJJ + SCCD; S2: GZXN; S3: GXCZ + GXFCG + YNHH). AMOVA revealed that 59.68% genetic variation was found within populations, whereas 40.32% genetic variation was explained by differences among populations. Based on SAMOVA results, the most genetic variation was found within populations (45.71%), followed by the genetic variance among 3 groups (42.96%) and among populations within groups (11.60%). Results of the SAMOVA and AMOVA test are shown in table 4.
Table 4.
Results of SAMOVA and AMOVA analysis based on mitochondrial data
We determined the population genetic structure of D. fissa. Plots of ΔK showed multiple peaks at K = 9 (Fig. 4). We found genetic similarities among populations GDQY, GDZQ, HNCZ and SCCD, and the individuals collected from populations GXCZ, GXFCG and YNHH demonstrate similarities.
Fig. 4.
Structure clustering results. (A) the posterior probability of each K; (B) the distribution of Delta K values; (C) Bayesian clustering results at K = 9; S1–3 was the groups defined by SOMOVA.
The geographic distance among populations ranged from 25.60 km (GDZQ vs. GDQY) to 1111.99 km (GDZQ vs. SCCD). Mantel tests indicated a significant correlation between genetic differentiation (FST) and geographic distance (r = 0.3924, p < 0.001).
Haplotype network and demographic history
We obtained 134 haplotypes of combined sequences (designated as H1-H134). Among the 134 haplotypes, 14 haplotypes (H1, H3, H12, H17, H22, H24, H26, H42, H48, H49, H50, H76, H103 and H105) were shared by different population. H3 and H44 were both the most frequent haplotype shared by 10 samples, which suggests they are likely to be the ancestral haplotypes (Castelloe and Templeton 1994). H4 was only seen in population GZTR and H44 was shared by GZTR (n = 1), GDQY (n = 4) and GDZQ (n = 5).
Divergence time analysis (MCC tree) revealed five mitochondrial lineages. The primary divergence within D. fissa commenced 1.31 Mya when two lineages (lineage IV and lineage V) split from the rest. Lineage IV and lineage V split around 1.01 Mya. The time of lineage III split from lineage I and two haplotypes (H73 and H74) was around 0.99 Mya (Fig. 5). The haplotype network (Fig. 6) showed phylogeographic relationships. H3 was the core of the “star-like” topology in lineage V.
Fig. 5.
Divergence time estimation based on combined mitochondrial data of D. fissa.
Fig. 6.
The mitochondrial haplotype network of concatenated sequences. The dotted box represents the five lineages based on divergence time analysis.
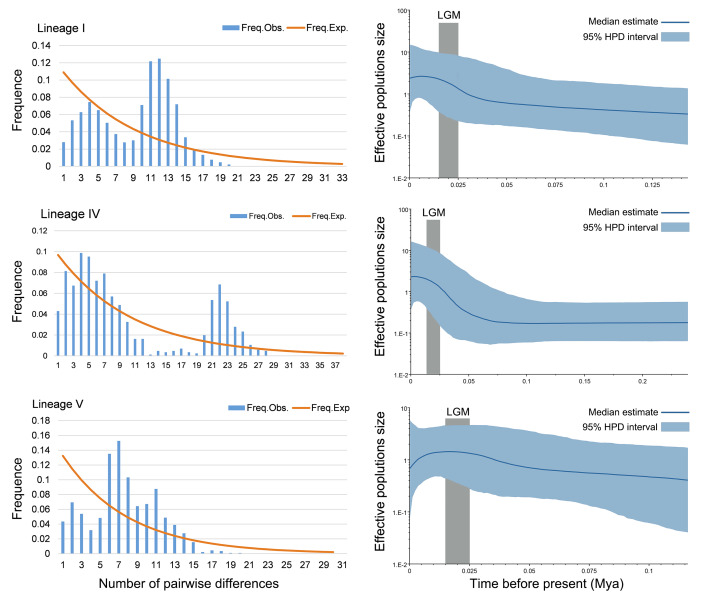
Tajima’s D test for lineage V (p < 0.01) was negative, while lineage I and lineage IV were negative but not significant. Fu’s FS test had significantly negative values (p < 0.001) for all three major lineages. Significantly negative values of Fu’s FS and Tajima’s D tests indicate a recent population expansion. The mismatch distribution analysis suggests that three lineages remained relatively stable. According to the BSP results, lineage I and lineage IV showed a population expansion in recent times (from around 0.025 and 0.05 Mya). Lineage V showed a constant population size for a long time, which suddenly decreased beginning around 0.01 Mya. Moreover, the BSP results showed that no lineage was affected by LGM (Fig. 7).
Fig. 7.
Mismatch distributions (left) and Bayesian skyline plots (right) for lineages I, IV, and V of D. fissa based on mitochondrial data.
Ancestral area reconstruction
The ancestral area reconstructions of D. fissa by BBM are shown in figure 8. BBM analyses indicated that D. fissa originated from the South China (area B) and dispersed to Central China (area A). Then, D. fissa dispersed to Yunnan-Guizhou Plateau (area C), Southeast coastal area (area D). The population of Sichuan Basin (area E) was from Central China and Yunnan-Guizhou Plateau (Fig. 9).
Fig. 8.
The result of the BBM with RASP
Fig. 9.
The dispersed path of D. fissa. A: Central China; B: South China; C: Yunnan-Guizhou Plateau; D: Southeast coastal area; E: Sichuan Basin.
DISCUSSION
The genetic diversity of the nuclear gene (ITS) is significantly lower than that of mitochondrial data. Because there is no significant sex-bias in migration capacity or adaptability in D. fissa, the reason for this condition may be the highly elevated mutation rate of mitochondrial DNA in natural populations or subsequent secondary contact after geographic isolation (Toews and Brelsford 2012). The low diversity also makes the ITS sequence unable to provide sufficient information in phylogeny.
The 134 mtDNA haplotypes formed 5 haplotype lineages (lineage II only consisted of two haplotypes). Most haplotypes (120 of 134) were unique to one population, which indicated high level genetic differentiation among D. fissa populations. SAMOVA analysis showed that S2 (GZXN) and S3 (YNHH, GXFCG and GXCZ) formed a separate group respectively. The STRUCTURE analysis based on concatenated data also implied that S2 and S3 groups have unique genetic structures. The GZXN population had six unique haplotypes (H58 to H63), which connected with adjacent haplotype by more than 5 mutational steps. We also checked the difference between the results of SAMOVA and MCC tree. The S2 group defined by SOMAVA corresponds to a part of lineage I, and is connected with adjacent haplotype by more than 10 mutational steps. All haplotypes of the S2 group belong to lineage I. Lineages II, III, V and the rest of lineages I, IV correspond to the S1 group.
High level haplotype diversity was observed in most populations, which may be a result of rapid population growth of ancestral populations (Yu et al. 2014). High genetic diversity often relates to the species with long evolutionary history and wide distribution (Li et al. 2019; Zhou et al. 2021). D. fissa is a long-winged Meconematinae species and its distribution is wider than most other species of Meconematinae in China. The Mantel test revealed a significant correlation between genetic differentiation (FST) and geographic distance. Dispersal ability is an important factor that affects the correlation between geographic distances and genetic diversity of the species (Wu et al. 2019).
Significant FST values indicated that the D. fissa populations had undergone obvious genetic differentiation. In east Asia, the gene flow of remarkable array insects was restricted by topography such as mountains and rivers (Liu et al. 2018). Significant negative values of Fu’s FS may be the result of population expansion. Fu’s FS test is more sensitive than Tajima’s D (Pilkington et al. 2008), which explains why the values of three Fu’s FS tests were significant (p < 0.001) while just one value of Tajima’s D had statistical p-value < 0.01. Three major lineages of D. fissa were all formed before the LGM. According to BSP results, the expansion of lineage I (0.035 Mya) and lineage IV (0.05 Mya) occurred earlier than the LGM, and both lineages were not adversely affected by LGM. Lineage V showed a stable population size during the LGM.
Because the AUC value was higher than 0.9, the predictions generated by models are assumed to be reliable. We believe that D. fissa had a large middle-level suitable area as LGM refugia in south and central China that might have been caused by the Tibetan Plateau blocking cold snaps (Yang et al. 2016). The predicted distribution of D. fissa displayed that although the high-level suitable areas had been reduced, there was still a middle-level suitable area connecting them to prevent them from being completely isolated in south and central China during LGM. This pattern could be the reason that none of the three major lineages were adversely affected by LGM. Based on the predicted distribution in 2070, the habitat of D. fissa will not be seriously affected in the future. However, the habitat has shown a trend of moving towards high-altitude and high-latitude areas. If global warming continues to intensify, the high-altitude populations in southern China may be separated from each other.
Despite the results of BBM, D. fissa may occur from South China. First, the population dispersed to Central China and Sichuan Basin, and then the population of Central China dispersed in two directions: 1. from Central China to Sichuan Basin; 2. from Central to Southeast Coastal area. It is unclear why D. fissa did not disperse to Southeast Coastal area from South China directly, but by way of Central China.
In summary, our results indicated that the populations of D. fissa in China are highly genetically diverse. Although our present study made some contributions regarding D. fissa genetic patterns and occurrence location, the study for D. fissa is not complete. Systematic research based on nuclear genes has not been done yet. More studies are needed to identify accurate migration paths and evolutionary history, especially based on more extensive sampling and more types of data.
CONCLUSIONS
In this study, we examined the genetic diversity and demographic history of Decma fissa, which has a high-level diversity based on mitochondrial data. The 134 mitochondrial haplotypes formed 5 haplotype lineages. The BSP analysis suggested that all three main lineages were not adversely affected by LGM, and this matches the results of MaxEnt model that most of the suitable living areas were reserved during LGM. By reconstructing the ancestral area, we revealed the migration path and the possible origin of D. fissa in China.
Supplementary materials
Information of the collection point and haplotype distribution.
Distribution point used in MaxEnt from literature.
Genetic distance among populations based on Kimura’s 2-parameter.
Genetic distance among populations based on Tamura 3-parameter.
Genetic difference (FST) and p-value among populations.
p-value of Genetic difference (FST) among populations.
Acknowledgments
The study was supported by National Natural Science Foundation of China (Grant Number: 31672259 and 31872268). We thank Dr Jianhua Huang and Dr Haijian Wang for collecting the research material, Mr Jianyu Chen and Dr Tong Liu for their instructions on processing the molecular and climate data.
Footnotes
Authors’ contributions: Qi Guo: Contributed to the study design, collected specimens, data analysis, and wrote the manuscript. Qi-Di Zhu: Conceived and designed the study, collected specimens. Zhi-Jun Zhou: Methodology and visualization. Shi-Fu Ming: Resources, data curation and supervision.
Competing interests: The authors declare that they have no conflicts of interest.
Availability of data and materials: All sequences are available at GenBank (COI: OP799863-OP800094, Cytb: OP818506-OP818742, ITS: OP805649-OP805877). The following data are in supplementary materials.
Consent for publication: We agree on having our work published by Zoological Studies.
Ethics approval consent to participate: Not applicable.
References
- Castelloe J, Templeton AR. 1994. Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol 3(2):102–113. doi:10.1006/mpev.1994.1013. [DOI] [PubMed]
- Chabot CL, Espinoza M, Mascarenas-Osorio I, Rocha-Olivares A. 2015. The effect of biogeographic and phylogeographic barriers on gene flow in the brown smoothhound shark, Mustelus henlei, in the northeastern Pacific. Ecol Evol 5(8):1585–1600. doi:10.1002/ece3.1458. [DOI] [PMC free article] [PubMed]
- Cigliano MM, H Braun, DC Eades, D Otte. 2022. Orthoptera Species File. Version 5.0/5.0. Available at: http://Orthoptera.SpeciesFile. org. Accessed 23 Oct. 2022.
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The Last Glacial Maximum. Science 325(5941):710–714. doi:10.1126/science.1172873. [DOI] [PubMed]
- Dai CY, Zhao N, Wang WJ, Lin CT, Gao B, Yang XJ, Zhang ZW, Lei FM. 2011. Profound climatic effects on two East Asian Black Throated Tits (Ave: Aegithalidae), revealed by ecological niche models and phylogeographic analysis. PLoS ONE 6(12):e29329. doi:10.1371/journal.pone.0029329. [DOI] [PMC free article] [PubMed]
- Dufresnes C, Litvinchuk SN, Borzée A, Jang Y, Li J-T, Miura I, Perrin N, Stöck M. 2016. Phylogeography reveals an ancient cryptic radiation in East-Asian tree frogs (Hyla japonica group) and complex relationships between continental and island lineages. BMC Evol Biol 16(1):253. doi:10.1186/s12862-016-0814-x. [DOI] [PMC free article] [PubMed]
- Dupanloup I, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11(12):2571–2581. doi:10.1046/j.1365-294X.2002.01650.x. [DOI] [PubMed]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22(5):1185–1192. doi:10.1093/molbev/msi103. [DOI] [PubMed]
- Earl DA, Vonholdt BM. 2012. STRUCTURE HARVESTER: A web-site and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361. doi:10.1007/s12686-011-9548-7.
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567. doi:10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed]
- Excoffier L. 2004. Patterns of DNA sequence diversity and genetic structure after a range expansion: Lessons from the infinite-island model. Mol Ecol 13(4):853–864. doi:10.1046/j.1365-294X.2003.02004.x. [DOI] [PubMed]
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925. doi:10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed]
- Garrido JL, Fenu G, Mattana E, Bacchetta G. 2012. Spatial genetic structure of Aquilegia taxa endemic to the Island of Sardinia. Ann Bot 109:953–964. doi:10.1093/aob/mcs011. [DOI] [PMC free article] [PubMed]
- Gorochov AV. 1993. A contribution to the knowledge of the tribe Meconematini (Orthoptera: Tettigoniidae). Zoosystematica Rossica 2(1):63–92.
- Gorochov AV, Liu CX, Kang L. 2005. Studies on the tribe Meconematini (Orthoptera: Tettigoniidae: Meconematinae) from China. Orient Insects 39:63–88. doi:10.1080/00305316.2005.10 417418.
- Harpending HC. 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66(4):591–600. [PubMed]
- Heuertz M, Fineschi S, Anzidei M, Pastorelli R, Salvini D, Paule L, Frascaria-Lacoste N, Hardy OJ, Vekemans X, Vendramin GG. 2004. Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol Ecol 13(11):3437–3452. doi:10.1111/j.1365-294X.2004.02333.x. [DOI] [PubMed]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. doi:10.1038/35016000. [DOI] [PubMed]
- Hewitt G. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos Soc B 2004(359):183–195. doi:10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed]
- Huang JH, Zhang AB, Mao SL. 2013. DNA barcoding and species boundary delimitation of selected species of Chinese Acridoidea (Orthoptera: Caelifera). PLoS ONE 8:e82400. doi:10.1371/journal.pone.0068688. [DOI] [PMC free article] [PubMed]
- Hudson RR, Boos DD, Kaplan NL. 1992. A statistical test for detecting geographic subdivision. Mol Biol Evol 9(1):138–151. doi:10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed]
- Hudson RR, Richard R. 2000. A new statistic for detecting genetic differentiation. Genetics 155(4):2011–2014. doi:10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15(5):1179–1191. doi:10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed]
- Kumar S, Steche G, Li M. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. doi:10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed]
- Leigh JW, Bryant D. 2015. POPART: Full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116. doi:10.1111/2041-210X.12410.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. doi:10.1093/bioinformatics/btp187. [DOI] [PubMed]
- Liu JL, Guo XG, Chen DL, Li J, Yue BS, Zeng XM. 2019. Diversification and historical demography of the rapid racerunner (Eremias velox) in relation to geological history and Pleistocene climatic oscillations in arid Central Asia. Mol Phylogenet Evol 130:244–258. doi:10.1016/j.ympev.2018.10.029. [DOI] [PubMed]
- Li J, Jin Q, Zhu GP, Jiang C, Zhang AB. 2019. Phylogeography of Dendrolimus punctatus (Lepidoptera: Lasiocampidae): Population differentiation and last glacial maximum survival. Ecol Evol 9(13):7480–7496. doi:10.1002/ece3.5278. [DOI] [PMC free article] [PubMed]
- Li X, Wu S, Xu Y, Liu Y, Wang J. 2022. Population Genetic Structure of Chlorops oryzae (Diptera, Chloropidae) in China. Insects 13:327. doi:10.3390/insects13040327. [DOI] [PMC free article] [PubMed]
- Liu XW, Yin HS. 2004. Orthoptera: Tettigonioidea and Stenopelmatoidea. In: Yang, X.K. (Ed.), Insects from Mt. Shiwandashan Area of Guangxi. China. Forestry Publishing House, Beijing, p. 98.
- Liu XW, Zhou M. 2007. A taxonomic study on the genus Decma Gorochov from China (Orthoptera: Tettigonioidea: Meconematidae). Acta Entomol Sinica 50(6):610–615. doi:10.16380/J.KCXB.2007.06.010.
- Liu YX, Qiu Y, Wang X, Yang H, Hayashi M, Wei C. 2018. Morphological variation, genetic differentiation and phylogeography of the East Asia cicada Hyalessa maculaticollis (Hemiptera: Cicadidae). Syst Entomol 43(2):308–329. doi:10.1111/syen.12276.
- Kass JM, Muscarella R, Galante PJ, Bohl CL, Pinilla‐Buitrago GE, Boria RA, Anderson RP. 2021. ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol Evol 12(9):1602–1608. doi:10.1111/2041-210X.13628.
- Lyons JI, Pierce AA, Barribeau SM. 2012. Lack of genetic differentiation between monarch butterflies with divergent migration destinations. Mol Ecol 21:3433–3444. doi:10.1111/j.1365-294X.2012.05613.x. [DOI] [PubMed]
- Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25(7):1459–1471. doi:10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed]
- Muscarella R, Galante PJ, Soley‐Guardia M, Boria RA, Kass JM, Uriarte M, Anderson RP. 2014. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol Evol 5(11):1198–1205. doi:10.1111/2041-210X. 12261.
- Papadopoulou A, Anastasiou I, Vogler AP. 2010. Revisiting the insect mitochondrial molecular clock: The mid-aegean trench calibration. Mol Biol Evol 27(7):1659–1672. doi:10.1093/molbev/msq051. [DOI] [PubMed]
- Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259. doi:10.1016/j.ecolmodel.2005.03.026.
- Phillips SJ, Dudík M. 2008. Modeling of species distributions with Maxent, new extensions and a com-prehensive evaluation. Ecography 31:161–175. doi:10.1111/j.0906-7590.2008.5203.x.
- Pilkington MM, Wilder JA, Mendez FL, Cox MP, Woerner A, Angui T, Kingan S, Mobasher Z, Batini C, Destro-Bisol G, Soodyall H, Strassmann BI, Hammer MF. 2008. Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol Biol Evol 25(3):517–525. doi:10.1093/molbev/msm279. [DOI] [PubMed]
- Pons J, Vogler AP. 2005. Complex pattern of coalescence and fast evolution of a mitochondrial rRNA pseudogene in a recent radiation of tiger beetles. Mol Biol Evol 22:991–1000. doi:10.1093/molbev/msi085. [DOI] [PubMed]
- Porretta D, Canestrelli D, Bellini R, Celli G, Urbanelli S. 2007. Improving insect pest management through population genetic data: a case study of the mosquito Ochlerotatus caspius (Pallas). J Appl Ecol 44:682–691. doi:10.1111/j.1365-2664.2007.01301.x.
- Pritchard J, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genetype data. Genetics 155:945–959. doi:10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed]
- Qiu F, Li H, Lin H, Ding S, Miyamoto MM. 2016. Phylogeography of the inshore fish, Bostrychus sinensis, along the Pacific coastline of China. Mol Phylogenet Evol 96:112–117. doi:10.1016/j.ympev. 2015.11.020. [DOI] [PubMed]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol 67(5):901–904. doi:10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed]
- Rambaut A. 2012. FigTree v 1.4. Molecular evolution, phylogenetics and epidemiology. University of Edinburgh, Institute of Evolutionary Biology, Edinburgh, UK.
- Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9(3):552–569. doi:10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed]
- Saeb A, Al-Naqeb D. 2016. The impact of evolutionary driving forces on human complex diseases: a population genetics approach. Scientifica 2016:1–10. doi:10.1155/2016/2079704. [DOI] [PMC free article] [PubMed]
- Shi FM, Bai JR, Zhang Y, Chang YL. 2013. Notes on a collection of Meconematinae (Orthoptera: Tettigoniidae) from Damingshan, Guangxi, China with the description of a new species. Zootaxa 3717(4): 593–597. doi:10.11646/zootaxa.3717.4.8. [DOI] [PubMed]
- Simon C, Buckley TR, Frati F. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol S 37:545–579. doi:10.1146/annurev.ecolsys.37.091305.110018.
- Song W, Cao LJ, Li BY, Gong YJ, Hoffmann AA, Wei SJ. 2016. Multiple refugia from penultimate glaciations in East Asia demonstrated by phylogeography and ecological modelling of an insect pest. BMC Evol Biol 18:152. doi:10.1186/s12862-018-1269-z. [DOI] [PMC free article] [PubMed]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus. Evolution 4(1):vey016. doi:10.1093/ve/vey016. [DOI] [PMC free article] [PubMed]
- Tang XT, Lu MX, Du YZ. 2022. Molecular phylogeography and evolutionary history of the pink rice borer (Lepidoptera: Noctuidae): Implications for refugia identification and pest management. Syst Entomol 47(2):371–383. doi:10.1111/syen. 12535.
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. doi:10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed]
- Toews DPL, Brelsford A. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21(16):3907–3930. doi:10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed]
- Thomson AM, Calvin KV, Smith SJ, Kyle GP, Volke A, Patel P, Delgado-Arias S, Bond-Lamberty B, Wise MA, Clarke LE, Edmonds JA. 2011. RCP4.5: a pathway for stabilization of radiative forcing by 2100. Climate Change 109(1):77–94. doi:10.1007/s10584-011-0151-4.
- Wang F, Li M, Zheng H, Dong T, Zhang XA. 2022. Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China. Insects 13:145. doi:10.3390/insects 13020145. [DOI] [PMC free article] [PubMed]
- Wang HQ. 2020. Meconematinae in China. Shanghai Science Popularization Press, Shanghai, pp. 159–160.
- Wang T, Zhu QD, Klaus-Gerhard H, Zhou ZJ, Shi FM. 2019. Phylogenetic relationships and phylogeography of the genus Sinocyrtaspis Liu, 2000 (Orthoptera: Tettigoniidae: Meconematinae) reveal speciation processes related to climate change. Syst Entomol 45(1):144–159. doi:10.1111/syen.12384.
- Warren R, Vanderwal J, Price J. 2013. Quantifying the benefit of early climate change mitigation in avoiding biodiversity loss. Nat Clim Change 3:678–682. doi:10.1038/NCLIMATE1887.
- Wei SJ, Cao LJ, Gong YJ, Shi BC, Wang S, Zhang F, Guo XJ, Wang YM, Chen XX. 2015. Population genetic structure and approximate Bayesian computation analyses reveal the southern origin and northward dispersal of the oriental fruit moth Grapholita molesta (Lepidoptera: Tortricidae) in its native range. Mol Ecol 24:4094–4111. doi:10.1111/mec.13300. [DOI] [PubMed]
- Wei SJ, Shi BC, Gong YJ. 2013. Genetic structure and demographic history reveal migration of the diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) from the southern to northern regions of China. PLoS ONE 8:e59654. doi:10.1371/journal.pone.0059654. [DOI] [PMC free article] [PubMed]
- Wei X, Mao ML, Wang JF, Huang JH. 2016. Tettigoniidae (Orthoptera: Tettigonioidea) from Hunan Province, CHINA. PART 2. Subfamilies Conocephalinae, Hexacentrinae and Meconematinae. Far Eastern Entomologist 305:13–24.
- Wu JX, Wang WP, Deng DG, Zhang K, Peng SX, Xu XX, Zhang YN, Zhou ZZ. 2019. Genetic diversity and phylogeography of Daphnia similoides sinensis located in the middle and lower reaches of the Yangtze River. Ecol Evol 9(8):4362–4372. doi:10.1002/ece3.4880. [DOI] [PMC free article] [PubMed]
- Xia KL, Liu XW. 1993. Orthoptera: Tettigonioidea and Grylloidea. In: Huang, F.S. (Ed.), Insects of Wuling Mountains Aera, Southwestern China. Science Press, Beijing, p. 97.
- Yang H, Lin CP, Liang AP. 2016. Phylogeography of the rice spittle bug (Callitettix versicolor) implies two long-term mountain barriers in South China. Zool Sci 33(6):592–602. doi:10.2108/zs160042. [DOI] [PubMed]
- Yang XQ, Kushwaha SPS, Saran S, Xu J, Roy PS. 2013. Maxent modeling for predicting the potential distribution of medicinal plant, Justicia adhatoda L. in Lesser Himalayan foothills. Ecol Eng 51:83–87. doi:10.1016/j.ecoleng.2012.12.004.
- Ye Z, Zhen YH, Damgaard J, Chen PP, Zhu L, Zheng CG, Bu WJ. 2018. Biogeography and diversification of Holarctic water striders: Cenozoic temperature variation, habitat shifting and multiple intercontinental dispersals. Syst Entomol 43(1):19–30. doi:10.1111/syen.12274.
- Ye Z, Zhu G, Chen P, Zhang D, Bu W. 2014. Molecular data and ecological niche modelling reveal the Pleistocene history of a semi-aquatic bug (Microvelia douglasi douglasi) in East Asia. Mol Ecol 23(12):3080–3096. doi:10.1111/mec.12797. [DOI] [PubMed]
- Yu TL, Lin HD, Weng CF. 2014. A new phylogeographic pattern of endemic Bufo bankorensis in Taiwan Island is attributed to the genetic variation of populations. PLoS ONE 9(5):e98029. doi:10.1371/journal.pone.0098029. [DOI] [PMC free article] [PubMed]
- Yu Y, Harris AJ, Blair C, He X. 2015. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol Phylogenet Evol 87:46–49. doi:10.1016/j.ympev.2015.03.008. [DOI] [PubMed]
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20(1):348–355. doi:10.1111/1755-0998.13096. [DOI] [PubMed]
- Zhou ZJ, Zhao L, Liu N, Guo HF, Guan B, Di JX, Shi FM. 2017. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol Phylogenet Evol 108:22–33. doi:10.1016/j.ympev.2017.01.014. [DOI] [PubMed]
- Zhou ZJ, Zhen YX, Guan B, Ma L, Wang WJ. 2021. Phylogeography and genetic diversity of the widespread katydid Ducetia japonica (Thunberg, 1815) across China. Ecol Evol 11(9):4276–4294. doi:10.1002/ece3.7324. [DOI] [PMC free article] [PubMed]
- Zhou TJ, Zou LW, Wu B, Jin CX, Song FF, Chen XL, Zhang LX. 2014. Development of earth/climate system models in China: A review from the Coupled Model Intercomparison Project perspectove. Acta Meteorol Sin 72(5):892–907. doi:10.1007/s13351-014-4501-9.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of the collection point and haplotype distribution.
Distribution point used in MaxEnt from literature.
Genetic distance among populations based on Kimura’s 2-parameter.
Genetic distance among populations based on Tamura 3-parameter.
Genetic difference (FST) and p-value among populations.
p-value of Genetic difference (FST) among populations.