Abstract
Small-cell lung cancers (SCLCs) are highly aggressive and currently there are no available targeted therapies. To identify clinically actionable drug combinations, we analyzed our previously reported chemogenomics screens and identified a synergistically cytotoxic combination of the topoisomerase I (TOP1) inhibitor topotecan and cycle-dependent kinase 7 (CDK7) inhibitor THZ1. Topotecan causes cell death by generating TOP1-induced DNA breaks and DNA-protein crosslinks (TOP1-DPCs) that require proteolysis by the ubiquitin-proteasome pathway for their repair. We found that inhibition of the transcriptional kinase CDK7 by THZ1 induces ubiquitin-mediated proteasomal degradation of RNA polymerase II (Pol II) and prevents the proteasomal degradation of TOP1-DPCs. We also provide a mechanistic basis for combinatorial targeting of transcription using selective inhibitors of CDK7 and TOP1 in clinical trials to advance SCLC therapeutics.
Introduction
Transcription and replication are two essential processes for cell viability and proliferation. DNA topoisomerase I (TOP1) is a nuclear enzyme that relieves the torsional stress generated during unwinding of the DNA molecule during transcription and replication, thereby preventing transcription–replication conflicts (1). TOP1 inhibitors are widely used as anticancer drugs and are particularly effective in rapidly replicating cells (2). Targeting transcription has generally been challenging, but recent studies have identified small-molecule inhibitors of the transcriptional machinery with selectivity for cancer cells (3). Understanding how cancer cells resolve the damage arising from drugs interfering with replication and transcription might enable their effective use as anti-cancer agents.
Small cell lung cancer (SCLC) is an exemplar cancer to understand and exploit the dependence of cancer cells on transcription and replication (4). SCLC is the most lethal type of lung cancer, comprising 15% of all lung cancers. It is characterized by rapid growth, and early and widespread metastases (5). Although SCLCs display a strikingly high rate of mutations, these mutations are not targetable for therapeutic purposes (6). Most SCLC recurrent somatic alterations affect transcription factors and chromatin modifiers including members of MYC family, SOX2, MLL1/2, CREBBP-EP300, RBL2, and P73. SCLC is one of the few cancers in which mutations in transcription regulators RB and P53, are the primary genetic cause (4). SCLCs also exhibit sustained high expression of lineage transcription factors, including genes encoding transcription factors that regulate neuroendocrine development (4,6). Consequently, SCLC cells are highly vulnerable to perturbation of the transcriptional state (7). The standard treatment of SCLC consists mostly of DNA replication inhibitors such as platinum, and TOP1 and TOP2 inhibitors (8).
TOP1 inhibitors kill cells by trapping the protein-DNA catalytic intermediate termed TOP1 cleavage complex (TOP1cc) on DNA. The plant alkaloid camptothecin (CPT) and its analogs including TPT and SN-38, bind at the interface of TOP1ccs thereby blocking their reversal (9). The trapped TOP1ccs [which we refer to as TOP1 DNA-protein crosslinks (TOP1-DPC)] intervene in DNA metabolic processes such as replication and transcription, eventually resulting in cell death if left unrepaired. It has been shown that TOP1ccs are located within the actively transcribed regions in eukaryotes and arrest transcription once they are trapped by TOP1 inhibitors (1). The confrontation between TOP1-DPCs and the elongating RNA polymerase II (Pol II) appears to evoke the ubiquitin (Ub)-proteasome pathway to degrade TOP-DPC for repair (10,11).
We hypothesized that SCLCs are under replicative and transcriptional stress and that inhibition of transcription may enhance the effect of drugs interfering with replication such as TOP1 inhibitors (2). In a series of high throughput chemogenomics screens, we discovered marked antiproliferative activity of transcription inhibitors with TOP1 inhibitors. We demonstrate that THZ1, a covalent inhibitor of cyclin-dependent kinase 7 (CDK7) (3), induces rapid degradation of the large subunit of Pol II (Rpb1) through K48 polyubiquitylation in SCLC cell lines. Pol II depletion precludes transcription-coupled ubiquitin-proteasome-mediated repair of topotecan-induced DNA lesions, leading to enhanced cell killing. These results have implications for our understanding of how interference with transcription may contribute to anti-cancer activity, and for targeted SCLC treatments.
Materials and Methods
Quantitively high-throughput and matrix screening
H446, H196, H524, DMS114, DMS79, H187, H889 Cells were dissociated with TrypLE as needed to remove attached cells. Cells were seeded in 5 μL of growth media using a Multidrop Combi dispenser into 1536 well white polystyrene tissue culture treated Corning plates at a density of (500–1000) cells/well, depending on the specific cell line, to allow for compounds to be present during exponential growth phase. After cell addition, 23 nL of MIPE 5.0 compounds were added to individual wells (11 dosing tested for all compounds in separate well) via a 1536 pin-tool. Bortezomib (final concentration 2.3 mM) was used as a positive control for cell cytotoxicity. For drug combination screening, 10 nL of compounds were acoustically dispensed into 1536 well white TC treated plates. Cells were then added to compound-containing plates at the same density used for single agent screening in 5 μL of media. A 5-point custom concentration, with constant 1:4 dilution between points was used for the primary 6 × 6 matrix screening, and a 9-point concentration range with 1:2 dilution between points was used for secondary 10 × 10 matrix screening. Plates were incubated for 48 hr at standard incubator conditions, covered by a stainless steel gasketed lid to prevent evaporation. 3 μL of Cell Titer Glo (Promega, Madison, WI, USA) were added to each well and plates were incubated at room temperature for 15 min with a stainless-steel lid in place. Luminescence readings were taken using a Viewlux (PerkinElmer, Waltham, MA, USA) with a 2 s exposure time per plate. Compound dose-response curves were normalized to DMSO and empty well controls on each plate. All combination screening data is publicly available at https://matrix.ncats.nih.gov/. Each unique screen is independently searchable, and a ‘help’ tab provides an overview of methods and a tutorial to aid users as they search this database.
Cell lines
Small Cell Lung Cancer cells DMS114, H146 and H446 were obtained from ATCC and cultured in RPM1–1640 medium (Life Technologies) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 1 × GlutaMax (Thermo Fisher) in tissue culture dishes at 37 °C in a humidified CO2 - regulated (5%) incubator. All the cell lines were authenticated using short tandem repeat DNA profiling, passaged 15 times and examined by MycoAlert Mycoplasma Detection Kit (Lonza).
Chemicals
Topotecan, THZ1, DRB (5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole) and bortezomib were acquired from the Developmental Therapeutics Program (DCTD, NCI).
Antibodies
Anti-ubiquitin, mouse monoclonal, Santa Cruz, sc-8017; anti-TOP1, mouse monoclonal, BD Biosciences, 556597; anti-dsDNA, mouse monoclonal, Abcam, ab27156; anti-FLAG, mouse monoclonal, Sigma Aldrich, F1804; anti-FLAG, rabbit polyclonal, Sigma Aldrich, F7425; anti-γH2AX, rabbit polyclonal, Cell Signaling, 66564; anti-Cdk7, mouse monoclonal, Santa Cruz technology, sc-7344; anti-Pol II, mouse monoclonal, Santa Cruz technology, sc-17798; anti-Pol II pSer 5, mouse monoclonal, Santa Cruz technology, sc-47701; anti-β-Actin (ACTB), mouse monoclonal, Santa Cruz technology, sc-47778; anti-α-Tubulin, rat monoclonal, Santa Cruz technology, sc-53029.
Expression plasmids
pRK5 HA-ubiquitin-K48, Addgene, 17605; FLAG-Pol II (Rpb 1), Addgene, 35175. 48 h transfection was performed using lipofectamine™ 3000 transfection reagent (Invitrogen) following manufacturer’s instructions.
Viability Assay
To measure the sensitivity of cells to drugs, SCLC cells were continuously exposed to various concentrations of the drugs. Ten thousand cells were seeded in 96-well white plates (Perkin Elmer Life Sciences, 6007680) in 100 μl of medium per well. Cells were incubated for 48 hours in triplicate. Cellular viability was determined using the ATPlite 1-step kits (PerkinElmer). Briefly, 50 μl ATPlite solution was added in 96-well plates per well, respectively. After 5 min, luminescence was measured with an EnVision 2104 Multilabel Reader (PerkinElmer). The ATP level in untreated cells was defined as 100%. Viability (%) of treated cells was defined as ATP treated cells/ATP untreated cells x 100. Combination synergy was calculated using Combenefit software (12).
In vivo of complex (ICE) assay
TOP1-DPCs were isolated and detected using in vivo complex of enzyme (ICE) assay as previously described (13). Briefly, cells were lysed in sarkosyl solution (1% w/v) after treatment. Cell lysates were sheared through a 25g 5/8 needle (10 strokes) to reduce the viscosity of DNA and layered onto CsCl solution (150% w/v), followed by centrifugation in NVT 65.2 rotor (Beckman coulter) at 42,000 RPM for 20 hours at 25 °C. The resulting pellet containing nucleic acids and TOP-DPCs was obtained and dissolved in TE buffer. The samples were quantitated for DNA concentration and subjected to slot-blot for immunoblotting with various antibodies as indicated. 2 μg of DNA is applied per sample. For mass spectrometric analysis, ICE samples were treated with RNase A to eliminate RNA contamination. Experiments were performed in triplicate and TOP-DPCs were quantified by densitometric analysis using ImageJ.
Detection of Ubiquitylated and SUMOylated Topoisomerase DNA-Protein Crosslinks (DUST)
Ubiquitylated TOP1-DPCs were isolated and detected using DUST assay as previously described (14). After treatments (drug reversal was not performed), 1 × 106 DMS114 cells in 35 mm dish per sample were washed with 1 × PBS and lysed with 600 μl DNAzol (Invitrogen), followed by precipitation with 300 μl 200 proof ethanol. The nucleic acids were collected, washed with 75% ethanol, resuspended in 200 μl TE buffer then heated at 65°C for 15 minutes, followed by shearing with sonication (40% output for 10 sec pulse and 10 sec rest for 4 times). The samples are centrifuged at 15,000 rpm for 5 min and the supernatant are collected and treated RNase A (100 μg/ml) for 1 h, followed by addition of 1/10 volume of 3 M sodium acetate sodium acetate and 2.5 volume of 200 proof ethanol. After 20 min full speed centrifugation, DNA pellet was retrieved and resuspended in 100 μl TE buffer for spectrophotometric measurement to quantitate DNA content. 10 μg of each sample was digested with 50 units micrococcal nuclease (Thermo Fisher Scientific, 100 units/μl) in presence of 5 mM CaCl2, followed by SDS-PAGE electrophoresis for immunodetection of total TOP-DPCs, SUMOylated TOP-DPCs as well as ubiquitylated TOP-DPCs using specific antibodies. In addition, 2 μg of each sample was subjected to slot-blot for immunoblotting with anti-dsDNA antibody to confirm equal DNA loading.
FLAG Immunoprecipitation (IP)
DMS114 cells are washed with 1 × PBS and incubated with 220 μl IP lysis buffer (5 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.2% Triton X-100, 5% glycerol, 1 mM DTT, 20 mM N-ethylmaleimide and protease inhibitor cocktail) on a shaker for 15 min at 4 °C, followed by sonication and centrifugation. Supernatant was collected and treated with 1 μl benzonase (250 units/μl) for 1h. An aliquot (20 μl) of the lysate of each treatment group was saved as input. Lysates were resuspended in 900 μl IP lysis buffer containing 2.5 μl anti-FLAG M2 or anti-Myc antibody and rotated overnight at 4°C. 50 μl Protein A/G PLUS-agarose slurry was added and incubated with the lysates for another 4 hrs. After centrifugation, immunoprecipitates were washed with RIPA buffer 2 times then resuspended in 2 × Laemmli buffer for SDS-PAGE and immunoblotting with various antibodies as indicated.
γH2AX Immunofluorescence
SCLC cells were seeded on chamber slides. After treatments, cells were washed with PBS and fixed for 15 min at 4°C in 4% paraformaldehyde in PBS and permeabilized with 0.25% Triton X-100 in PBS for 15 min at 4°C. The samples were blocked with PBSTT- 1% BSA, followed by overnight incubation with γH2AX antibody (Millipore Sigma) in PBSTT-BSA at 4°C, cells were rinsed with PBSTT and incubated with Alexa Fluor 4888-conjugated secondary antibody (Invitrogen) at 1:1000 in PBSTT-BSA for 1 h in subdued light; washed and mounted using mounting medium with DAPI (Vectashield). Images were captured on Zeiss LSM 880/Airyscan confocal microscope, processed using ImageJ and analyzed using Imaris.
Western Blotting (WB)
Cellular proteins were detected by lysing cells with RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 7.5, 1 mM DTT and protease inhibitor cocktail), followed by sonication and centrifugation. The supernatant was collected and boiled for 10 mins, analyzed by SDS-PAGE, and immunoblotted with various antibodies as indicated.
Statistical Analyses
Error bars on bar graphs represent standard deviation (SD) and p-value was calculated using paired student’s t-test for independent samples.
Data availability
The data generated in this study are available within the article and its supplementary data files.
Results
High throughput screens identify a cytotoxic combination of topotecan and THZ1 for SCLC
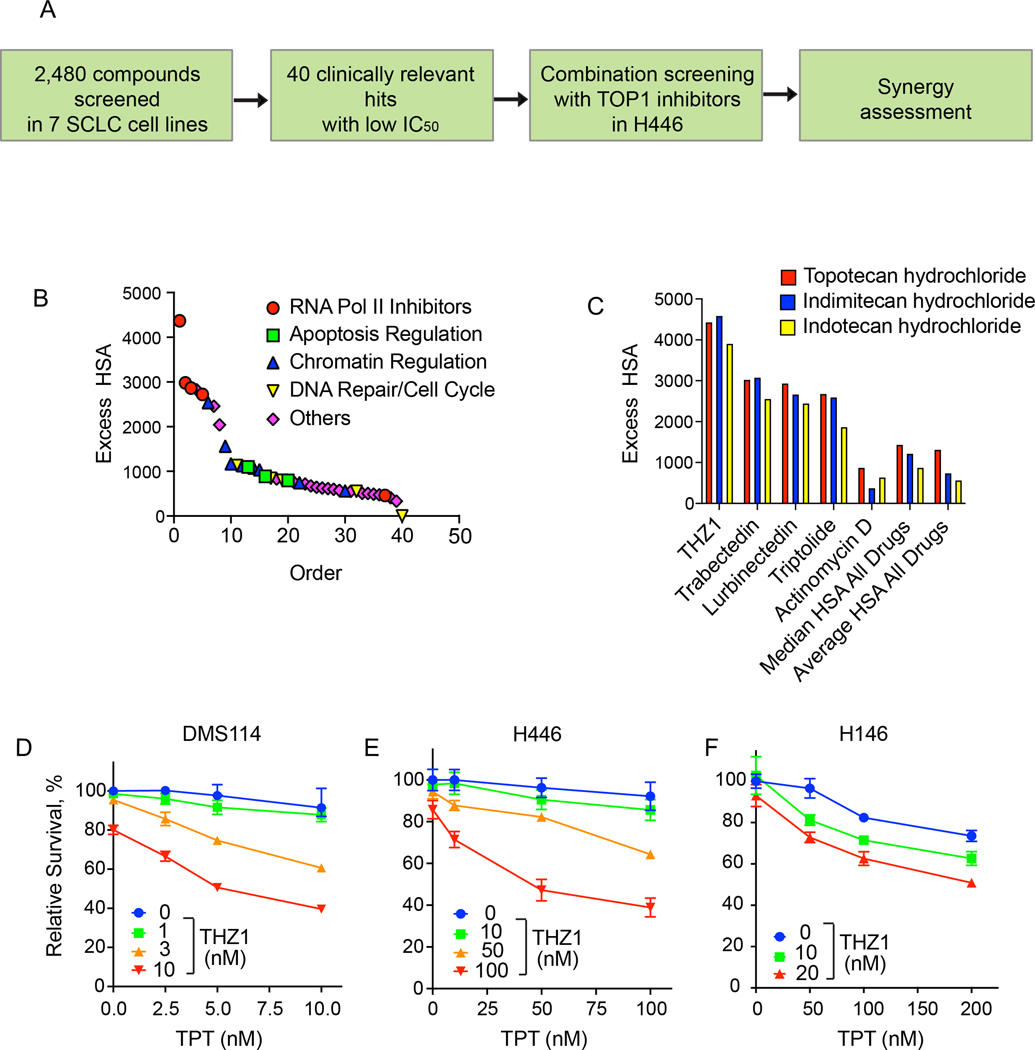
To identify small molecules that suppress SCLC cell growth in combination with TOP1 inhibitor topotecan (TPT), the standard care chemotherapeutic for recurrent SCLC (8), we re-analyzed our unbiased high-throughput screen in human SCLC cell lines using a library of over 2,000 annotated small-molecule inhibitors from NCATS drug library composed of both experimental compounds and early or advanced stage clinical candidates (15) (Figure 1A). Using TPT and indotecan (LMP400), a next-generation clinical TOP1 inhibitor (2,16), we explored combinations with the entire drug library, ranking combinations using the Excess over the Highest Single Agent (ExcessHSA) metric to quantitatively assess synergism and antagonism (Figure 1B and Supplementary Figure 1A). The top-ranked inhibitors fell broadly into three major putative categories of inhibitors targeting 1) cell cycle checkpoints (Chk1, ATR), 2) transcription (BRD4, CDK7, RNA polymerase II), 3) nuclear transport (XPO), and 4) anti-apoptotic pathways (Mcl1, Bcl2).
Figure 1. High-throughput chemogenomics screens identify an efficacious combination of topotecan and THZ1 in SCLCs.
A. Scheme of the different classes of drugs utilized in the NCATS screen. H446 cells were plated and then treated with drugs in 10 × 10 matrix format. Cells were analyzed at 8 and 16 hours for cleaved caspase activation. Synergy was assessed for the different therapeutics with TOP1 inhibitors.
B. Topotecan synergy assessed across different classes of drugs at 8 hours (to access the data, please refer to https://matrix.ncats.nih.gov/, small cell lung cancer project, ID 10073).
C. Synergy of all three TOP1 inhibitors (topotecan, Indimitecan [LMP776] and Indotecan [LMP400]) with transcription inhibitors at 8 hours.
D. Viability curves of DMS114 cells treated for 48 hours with TPT at the indicated concentrations (mean ± SD, n = 3) in presence or absence of THZ1 at the indicated concentrations.
E. Viability curves of H446 cells treated for 48 hours treatments with TPT at the indicated concentrations (mean ± SD, n = 3) in presence or absence of THZ1.
F. Viability curves of H146 cells treated for 48 hours treatments with TPT at the indicated concentrations (mean ± SD, n = 3) in presence or absence of THZ1.
The synergy of TOP1 inhibitors with cell cycle checkpoint inhibitors is well documented and combinations of these agents have demonstrated clinical efficacy in SCLC and high grade serous ovarian cancer (2). While targeting anti-apoptotic regulators such as Bcl-2 and Bcl-xL has shown efficacy in pre-clinical models, this has not been translated to clinical benefit (17). Nuclear transporter inhibitors are generally poorly tolerated in patients and a trial of selinexor in SCLC was terminated early (ClinicalTrials.gov Identifier: NCT02351505). BRD4 binds to acetylated histones and recruits chromatin modifiers and transcription factors. Although the bromodomain inhibitor JQ1 inhibits the growth of cancer cells with a significantly higher efficacy in MYC-amplified SCLC lines (18), targeting BRD4 in the clinic is yet to be successful. CDK7 orchestrates different phases of the transcription cycle of RNA polymerase II (RNAP II). Given the emerging interest in CDK7 targeting (19), with several drugs currently under clinical development, we focused our attention on THZ1, a covalent CDK7 inhibitor (3,7) (Figure 1C and Supplementary Figure 1B).
Next, we examined the synergistic effect of THZ1 and TPT in SCLC cell lines characterized by differential expression of key transcription regulators (6,20) achaete-scute homologue 1 (ASCL1), neurogenic differentiation factor 1 (NeuroD1), and yes-associated protein 1 (YAP1) using ATPlite luminescence assays. DMS114 cells (SCLC-YAP1) treated with THZ1 (3 and 10 nM) exhibited hypersensitivity to TPT (Figure 1D). By calculating the Loewe synergy score using Combenefit software (12), we confirmed the synergism of cotreatment with TPT and THZ1 in DMS114 cells (Supplementary Figure 1C). THZ1 (50 and 100 nM) also rendered H446 cells (SCLC-NEUROD1) hypersensitive to TPT (Figure 1E). The synergistic effect of the combination in H446 cells was further confirmed by Combenefit (Supplementary Figure 1D). Finally, we assessed the combination in H146 cells (SCLC-ASCL1) and confirmed the synergy (Figure 1F and Supplementary Figure 1E).
THZ1 blocks ubiquitin-dependent degradation of topotecan-induced TOP1-DPC in SCLC cells
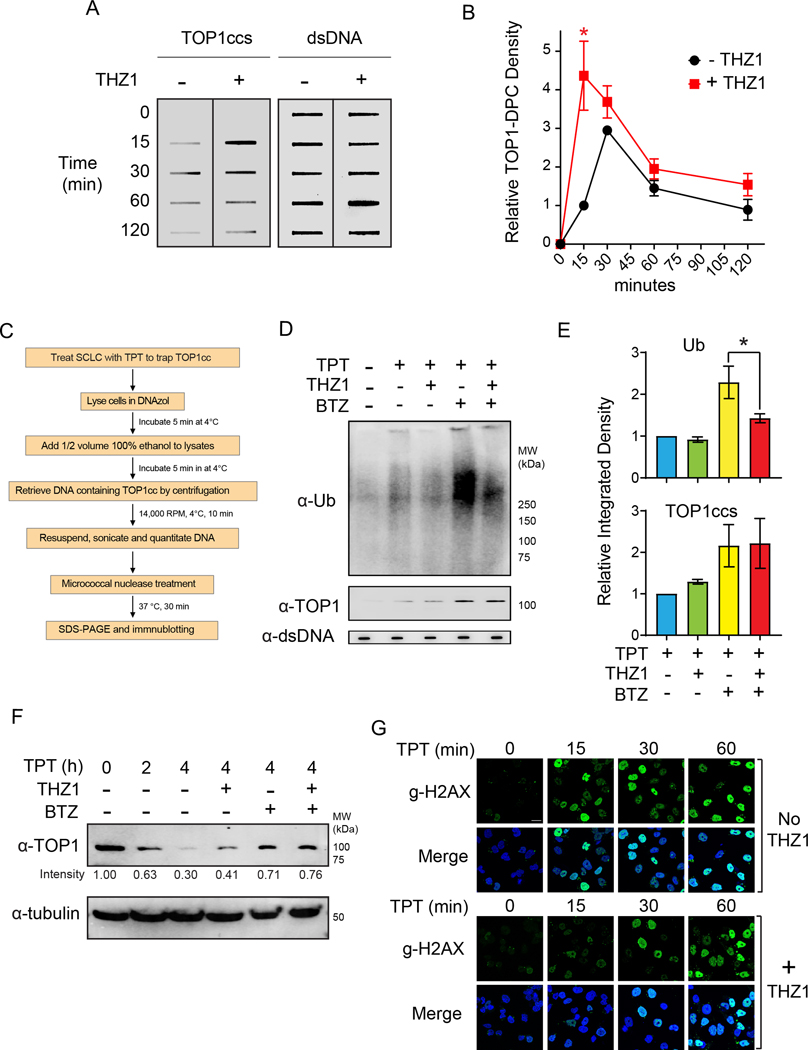
TPT and other camptothecin derivates target TOP1 by stabilizing TOP1ccs, and the resulting TOP1-DPCs can be converted to single-strand breaks (SSBs) and double-strand breaks (DSBs) that lead to cell death (2). Transcription has been implicated in the repair of TOP1-DPCs by triggering ubiquitylation and 26S proteasome-mediated degradation of the TOP1-DPC as well as RNA Pol II (10,11). To investigate whether THZ1 sensitizes SCLC cells to TPT by aggravating TPT-induced TOP1-DPCs, we examined whether CDK7 inhibition by THZ1 impairs the proteasomal degradation of TOP1-DPCs. We first carried out ICE (in vivo complex of enzyme) bioassay (13) to interrogate whether THZ1 impacts TPT-induced TOP1-DPCs in SCLC cells. We observed a 4.3-fold increase in the levels of TOP1-DPCs in THZ1-pretreated DMS114 cells 15 min after exposure to TPT (Figure 2A, B), suggesting a role of CDK7 in the repair of TOP1-DPCs. Yet, TOP1-DPCs in THZ1-pretreated cells were cleared as in control cells 60 minutes after TPT exposure (Figure 2A, B), suggesting that blocking transcription by inhibiting CDK7 delays does not completely block the repair of TOP1-DPCs, and that the delayed repair is independent of transcription (21,22).
Figure 2. THZ1 delays the ubiquitin-dependent degradation of topotecan-induced TOP1-DPC in DMS114 SCLC cells.
A. Cells treated with TPT (1 μM) for the indicated times in the presence and absence of THZ1 (1 μM) were collected for ICE assay to detect TOP1-DPC with anti-TOP1 antibody. THZ1 (1 μM) was added 2 h prior to the TPT treatment. Total DNA (dsDNA, right panel) was detected using anti-dsDNA antibody and served as loading control.
B. Densitometric analyses comparing the relative integrated densities of TOP1-DPCs from Independent experiments as shown in panel A. *: p < 0.05
C. Scheme for the Detection of Ubiquitylated and SUMOylated TOP-DPCs (DUST) assay.
D.Cells were pretreated with THZ1 (1 μM) or BTZ (1 μM) for 2 hours prior to cotreatment with TPT (20 μM) for 30 min and were subjected to DUST assay (upper panel) and total TOP1-DPCs (middle panel) using anti-ubiquitin and anti-TOP1 antibodies, respectively.
E. Quantitation of ubiquitylated TOP1-DPCs and total TOP1-DPCs. Upper panel: Densitometric analyses comparing relative integrated densities of Ub signals from independent experiments as shown in panel D. Lower panel: Densitometric analyses comparing relative integrated densities of TOP1-DPC signals from independent experiments as shown in D.
F. Cells were pretreated with THZ1 (1 μM) or BTZ for 2 hours prior to cotreatment with TPT (20 μM) for 1 or 4 hours. Cell were then lysed and digested with benzonase for Western blotting for immunodetection of TOP1 using anti-TOP1 antibody.
G. Cells were treated with TPT (1 μM) in the presence and absence of THZ1 (1 μM) and collected for immunofluorescence for detection of γH2AX.
To assess whether THZ1 impairs the repair of TOP1-DPCs by preventing their ubiquitin-mediated proteasomal degradation, we performed the DUST (Detection of Ubiquitylated and SUMOylated TOP-DPC) assay (Figure 2C) (23) in the presence of the proteasome inhibitor bortezomib (BTZ), which has been shown to reveal TOP1-DPCs by blocking their proteasomal degradation (23–25). We found that THZ1 decreases TPT-induced TOP1-DPC poly-ubiquitylation in (Figure 2D–E).
We next performed a time-course DUST assay to monitor the kinetics of TPT-induced ubiquitylation of TOP1-DPCs in the absence or presence of THZ1. TOP1-DPC ubiquitylation peaked at 1-hour TPT treatment and gradually diminished without THZ1 treatment whereas TOP1-DPC ubiquitylation levels remain low in cells treated with THZ1 (Supplementary Figure 2B). Coincidently, TOP1-DPCs remained unresolved in THZ1-treated cells even 4 hours after THZ1 treatment (Supplementary Figure 2B and Figure 2D and E). This finding is however inconsistent with the ICE assay in Figure 2A showing that TOP1-DPCs repair was delayed but not fully blocked by THZ1. This can be explained by the different TPT concentrations used in the two assays. We used 20 μM to detect TOP1-DPC ubiquitylation in the DUST assays whereas we only used 1 μM to detect total TOP1-DPCs in ICE assays. Such difference led to different levels of TOP1-DPCs and therefore suggests that the repair of high levels of TOP1-DPCs is largely dependent on transcription whereas low levels of TOP1-DPCs can be readily repaired by alternative mechanisms independently of transcription (23). Taken together, these findings indicate that the ubiquitin-dependent proteasomal degradation of TOP1-DPCs is partially dependent on transcription and CDK7 activity.
As SUMOylation plays a key role in the repair of TOP-DPCs and PARP trapping by recruiting the ubiquitin-proteasome pathway (23,26), we next assessed whether THZ1 impacts TOP1-DPC SUMOylation. Consistent with our previous finding that DRB did not affect CPT-induced TOP1-DPC SUMOylation (23), blocking transcription with THZ1 did not alter TOP1-DPC SUMO-1 modification or SUMO-2/3 modifications (Supplementary Figure 2C). This result shows that transcription-mediated TOP1-DPC ubiquitylation is independent of SUMOylation and confirms our prior results showing that SUMOylation of TOP1-DPCs does not require active transcription (23). Hence, we conclude that the SUMO-dependent ubiquitylation and transcription-dependent ubiquitylation are two parallel pathways.
By performing Western blotting, we found that, akin to inhibition of the proteasome by BTZ, CDK7 inhibition by THZ1 reduced the loss of cellular TOP1 in response to TPT (Figure 2F). Moreover, THZ1 did not further increase the levels of TOP1 in the presence of BTZ, further suggesting (10) the epistatic relationship between transcription (CDK7 activity) and the proteasome for the repair of TOP1-DPCs.
To investigate whether THZ1 sensitizes SCLC cells to TPT by aggravating TPT-induced DNA damage, we assessed the levels of γH2AX, a DSB marker readily activated by TOP1ccs (27–29). Pretreatment with THZ1 decreased the rapid induction of γH2AX upon exposure to TPT for 15 minutes (Figure 2G and Supplementary Figure 2). This finding is in line with independent studies showing that blocking the proteasome leads to a failure to expose the TOP1-concealed breaks and to activate DNA damage response (DDR) (10,21,23). Consistent with the ICE assays shown in Figure 2A, γH2AX accrual in THZ1-treated cells increased over time to the same levels as in control cells (Figure 2G and Supplementary Figure 2), suggesting that blocking transcription with THZ1 primarily delays the early debulking of the TOP1-concealed protein-associated-DPCs and their conversion to TOP1-free DNA breaks required for their repair (11,30).
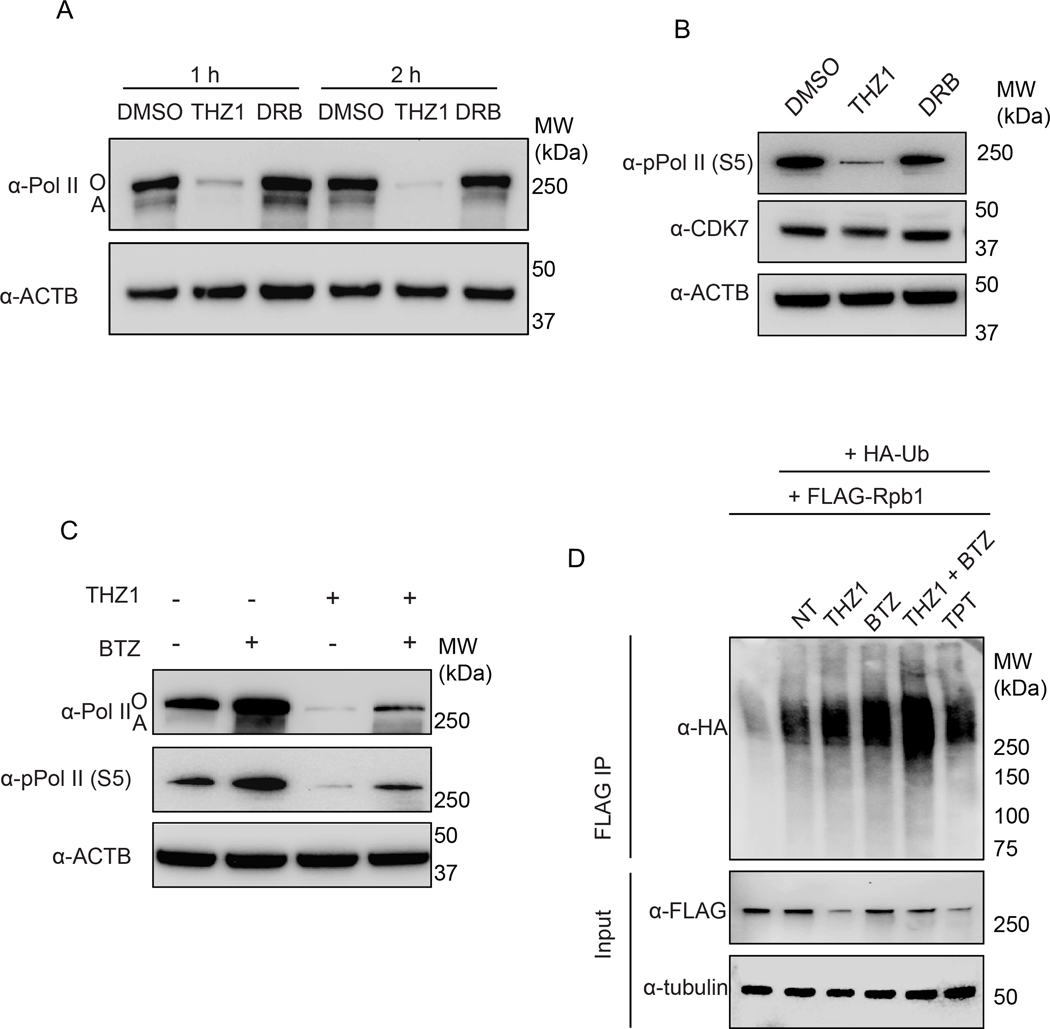
THZ1 induces ubiquitin-dependent degradation of the large subunit of RNA Pol II in SCLC cells
To elucidate the mechanism by which THZ1 prevents the ubiquitin-mediated degradation of TOP1-DPCs, we examined the levels of RNA polymerase II in SCLC cells treated with THZ1. Western blotting of whole cellular lysates showed that treatment with THZ1 reduced both phosphorylated and unphosphorylated Rpb1 (Pol IIo and Pol IIa, respectively), the largest subunit of RNA Pol II (31), in a time-dependent manner (Figure 3A). 5, 6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), another transcription inhibitor that acts by blocking CDK9 (32), did not affect the levels of total Pol II (Figure 3A). THZ1 also suppressed Serine 5-phosphorylation of Rbp1, which has been shown to be present during both transcription initiation and elongation (Figure 3B). Cellular levels of CDK7, on the contrary, were not significantly affected by THZ1 (Figure 3B).
Figure 3. THZ1 induces ubiquitin-dependent degradation of Rbp1 in DMS114 SCLC cells.
A. Cells were treated with THZ1 (1 μM) or DRB (50 μM) for 1 or 2 hours before Pol II Western blotting.
B. Cells were treated with THZ1 (1 μM) or DRB (50 μM) for 2 hours before Western blotting for immunodetection of Ser5-phosphorylated Pol II.
C. Cells were pretreated with BTZ (1 μM) for 1 hour before cotreatment with THZ1 (1 μM) for 2 hours and subjected to Western blotting for immunodetection of total Pol II and Ser5-phosphorylated Pol II.
D. FLAG-Rpb1-overexpressing DMS114 cells were transfected with HA-Ub K48 single lysine overexpression plasmid, followed by treatments with the indicated inhibitors. Immunoprecipitation (IP) using anti-FLAG tag antibody was performed after the treatments. IP samples and cell lysates (input) were subjected to immunoblotting (IB) with the indicated antibodies.
To test whether the decrease of Rbp1 protein in response to THZ1 treatment resulted from 26S proteasome-mediated degradation, we pretreated cells with the proteasome inhibitor BTZ prior to THZ1 treatment. BTZ blocked THZ1-induced Rbp1 reduction (Figure 3C and Supplementary Figure 3), indicating that THZ1 triggers the proteasomal degradation of Rbp1. To confirm this possibility, we transfected DMS114 cells with FLAG-Rbp1 and HA-Ub K48 expressing constructs and performed FLAG immunoprecipitation to examine whether THZ1 induces polyubiquitylation of Rbp1 through K48, the ubiquitin chain linkage for proteasomal degradation (11). Rbp1 K48 polyubiquitylation was significantly stimulated within 20 minutes in response to THZ1 and most clearly in the presence of BTZ to block proteasomal degradation (Figure 3D).
We also found that TPT increased Rbp1 polyubiquitylation (Figure 3D), implying that arrest of Pol II complexes by TOP1-DPCs triggers ubiquitin-dependent proteasomal destruction of both the TOP1-DPCs and blocked Pol II. Taken together, these data demonstrate that CDK7 inhibition by THZ1 induces K48 ubiquitylation-dependent proteasomal degradation of Rbp1, which precludes transcription elongation and processing of TOP1-DPCs.
Discussion
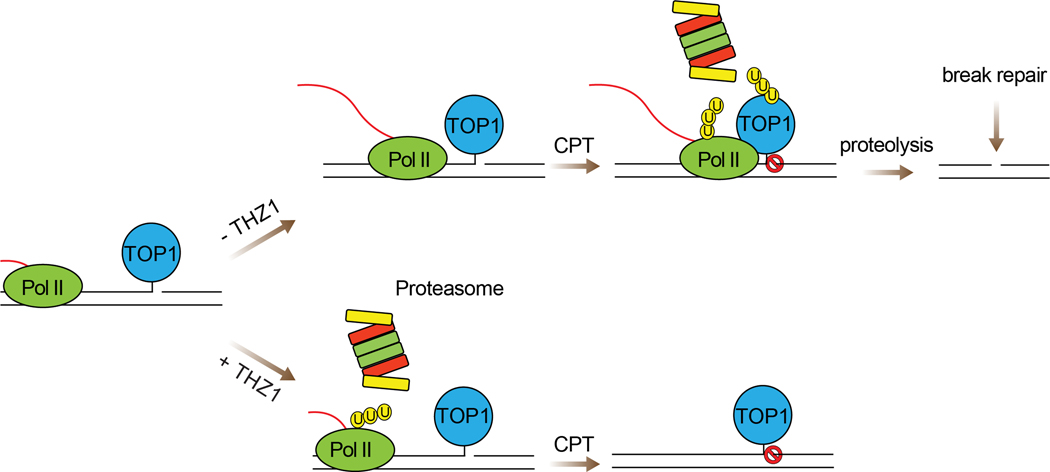
Here, we report combination treatment with the TOP1 inhibitor topotecan and CDK7 inhibition as a potential therapeutic paradigm for the treatment of SCLC. THZ1 renders SCLC cell lines hypersensitive to topotecan and warrants further evaluation in preclinical models. Mechanistically, THZ1 enhances topotecan-induced TOP1-DPC levels in SCLC cells. The enhancement results from the ubiquitin-mediated proteasomal degradation of Pol II large subunit Rpb1 induced by THZ1, which precludes the encounter between Pol II complexes and TOP1-DPCs, a step required to trigger proteasomal removal of the DPCs (Figure 4).
Figure 4. Model for the molecular mechanism by which THZ1 enhances TPT-mediated cytotoxicity in SCLC cells.
In absence of THZ1, TPT-stabilized TOP1cc arrests the elongating RNA Pol II complex, leading to polyubiquitylation of TOP1-DPCs for proteasomal degradation and conversion of TOP1-DPCs into DNA breaks for repair. In the presence of THZ1, the Pol II complex is polyubiquitylated and degraded by the proteasome, precluding its encounter with TOP1-DPC and the signaling for TOP1-DPC ubiquitylation. This leads to accumulation of unrepaired TOP1-DPCs that interfere with DNA metabolisms and cause cell death.
Recent studies have demonstrated the involvement of CDK7 functions throughout the Pol II transcription cycle, from promoter clearance and promoter-proximal pausing to co-transcriptional chromatin modification in gene bodies (31,33). As a crucial integrant of transcription factor II H (TFIIH), CDK7 primarily acts by promoting transcription initiation by phosphorylating serine 5 and 7 in the carboxy terminal domain (CTD) of RNA Pol II. Therefore, inhibition of CDK7 by THZ1 may represent a promising pharmacological approach for the treatment of SCLC (7), which features sustainedly high levels transcription of neuroendocrine genes that are coupled with high replication and low antigen-presenting characteristics (6,8,34,35). In this study, we used an unbiased small molecule screen approach and discovered that SCLC cells were highly vulnerable to combination treatment with the prototype covalent CDK7 inhibitor THZ1 (3,7) and the FDA-approved TOP1 inhibitor topotecan, as well as with the non-camptothecin TOP1 inhibitors Indimitecan (LMP776) and Indotecan (LMP400) (2). DMS114 cells were more sensitive to TPT and THZ1 than H146 and H446 cells (Figure 1D). This can be explained by the difference in their CDK7 dependency and SLF11 expressions.
CPT-induced TOP1-DPCs arrest the elongating RNA polymerase complexes (36–38) in vivo and in vitro, implying a transcription collision model where confrontation between elongating RNA polymerases and TOP1-DPCs on the template strands may engage DPC repair. Our data provide evidence that such collisions trigger the ubiquitylation and the subsequent proteasomal degradation of the TOP1-DPCs, leading to the debulking of the otherwise TOP1-concealed SSBs (11) (Figure 4, upper right). This possibility is consistent with previous studies suggesting the role of transcription in the repair of TOP1-DPCs. Those study showed that 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), a nucleoside analog that inhibits CDK9 kinase activity, impaired TOP1-DPC proteasomal degradation (10). CDK9 is a key constitute of P-TEFb (positive transcription elongation factor-b, or the CDK9/cyclin T1 complex) that facilitates transcription elongation by phosphorylating Ser2 in the CTD of Pol II (39,40). Here, to our knowledge, we provide the first evidence that THZ1 inhibits Pol II-mediated transcription by inducing the degradation of Pol II, and thereby preventing the transcription-driven UPP of TOP1-DPCs (32).
Transcription recovery studies have demonstrated that TOP1-DPC degradation is necessary for transcription recovery presumably by enabling transcription-coupled repair of TOP1-DPCs (10). According to our model shown in Figure 4, TOP1-DPCs are degraded through a Pol II-dependent ubiquitin-proteasome pathway. The tyrosyl-DNA phosphodiesterase TDP1, which removes tyrosine from the 3′ end of DNA, plays a key role in the repair of TOP1-DNA covalent complexes (1,11,30,41–43). In our model, TDP1 could potentially hydrolyze the otherwise occluded phosphotyrosyl bond after proteasomal debulking of the DPC to fully liberate the SSB termini and enable DNA repair (11,30,44).
Replication- and S-phase-specific cytotoxicity is critical for the antitumor activity of CPT derivatives, and is likely key to the activity of topotecan in SCLC (8). Yet, TOP1 inhibitors also inhibit gene expression by arresting Pol II complexes (32,36,45), which must transcribe the genome at a much higher frequency than DNA replicative polymerases. We propose that the timely repair of TOP1-DPCs by transcription-dependent proteolysis not only ensures the fidelity of transcription but also avoids replication collisions, thereby helping cells to survive TOP1-mediated DNA damage. Consistently, the 26S proteasome is commonly hyperactive in tumors (46) and may therefore contribute to the resistance to TOP1 inhibitors. Indeed, the proteasome inhibitor bortezomib has been shown to promote apoptosis in SCLC by preventing IkappaB degradation to decrease expression of the antiapoptotic protein Bcl-2 (47). BTZ was also reported to synergize with DNA damaging chemotherapeutics in SCLCs (48).
Our study not only reveals that the UPP repairs TOP1-DPCs in a Pol II-dependent manner, but also demonstrates that the CDK7 inhibitor THZ1 synergizes with topotecan in SCLCs by blocking the UPP-mediated TOP1-DPC degradation by inducing Pol II degradation, which is also carried out by the UPP (Figure 4, lower right). Further studies are warranted to identify the transcription-specific ubiquitin E3 ligases and their regulation for Pol II and TOP1-DPC degradation. Additionally, the studies presented here provide a mechanistic basis to explore in vivo selective CDK7 inhibitors that are currently in development and clinical trials (49) in combination with conventional TOP1 inhibitors such as topotecan and the novel antibody drug conjugates carrying TOP1 inhibitor payloads (2).
Supplementary Material
Acknowledgements
Our studies are supported by the Center for Cancer Research (CCR), the Intramural program of the National Cancer Institute, National Institutes of Health (Z01-BC006161 and Z01-BC006150)
Declaration of interests
A.T. and Y.P. report research funding to the institution from the following entities: EMD Serono Research & Development Institute Inc., Billerica, MA, USA; AstraZeneca; Tarveda Therapeutics; Immunomedics; Prolynx Inc.; and Taiho Pharmaceuticals Co.
References
- 1.Pommier Y, Nussenzweig A, Takeda S, Austin C. Human topoisomerases and their roles in genome stability and organization. Nat Rev Mol Cell Biol 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas A, Pommier Y. Targeting Topoisomerase I in the Era of Precision Medicine. Clin Cancer Res 2019;25:6581–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwiatkowski N, Zhang TH, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014;511:616−+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers 2021;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas A, Pattanayak P, Szabo E, Pinsky P. Characteristics and Outcomes of Small Cell Lung Cancer Detected by CT Screening. Chest 2018;154:1284–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tlemsani C, Pongor L, Elloumi F, Girard L, Huffman KE, Roper N, et al. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep 2020;33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang TH, et al. Targeting Transcriptional Addictions in Small Cell Lung Cancer with a Covalent CDK7 Inhibitor. Cancer Cell 2014;26:909–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas A, Pommier Y. Small cell lung cancer: Time to revisit DNA-damaging chemotherapy. Sci Transl Med 2016;8:346fs12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov 2011;11:25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, et al. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol 2003;23:2341–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Saha LK, Saha S, Jo U, Pommier Y. Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways. DNA Repair (Amst) 2020;94:102926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anand J, Sun Y, Zhao Y, Nitiss KC, Nitiss JL. Detection of Topoisomerase Covalent Complexes in Eukaryotic Cells. Methods Mol Biol 2018;1703:283–99 [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Jenkins LMM, Su YP, Nitiss KC, Nitiss JL, Pommier Y. A conserved SUMO-Ubiquitin pathway directed by RNF4/SLX5-SLX8 and PIAS4/SIZ1 drives proteasomal degradation of topoisomerase DNA-protein crosslinks. bioRxiv 2019:707661 [Google Scholar]
- 15.Thomas A, Takahashi N, Rajapakse VN, Zhang XH, Sun YL, Ceribelli M, et al. Therapeutic targeting of ATR yields durable regressions in small cell lung cancers with high replication stress. Cancer Cell 2021;39:566−+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coussy F, El-Botty R, Chateau-Joubert S, Dahmani A, Montaudon E, Leboucher S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Science Translational Medicine 2020;12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudin CM, Hann CL, Garon EB, de Oliveira MR, Bonomi PD, Camidge DR, et al. Phase II Study of Single-Agent Navitoclax (ABT-263) and Biomarker Correlates in Patients with Relapsed Small Cell Lung Cancer. Clinical Cancer Research 2012;18:3163–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, et al. Identification and Targeting of Long-Term Tumor-Propagating Cells in Small Cell Lung Cancer. Cell Rep 2016;16:644–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diab S, Yu M, Wang S. CDK7 Inhibitors in Cancer Therapy: The Sweet Smell of Success? J Med Chem 2020;63:7458–74 [DOI] [PubMed] [Google Scholar]
- 20.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J Biol Chem 2008;283:21074–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks. J Biol Chem 2009;284:28084–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Miller Jenkins LM, Su YP, Nitiss KC, Nitiss JL, Pommier Y. A conserved SUMO pathway repairs topoisomerase DNA-protein cross-links by engaging ubiquitin-mediated proteasomal degradation. Sci Adv 2020;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res 2001;61:5926–32 [PubMed] [Google Scholar]
- 25.Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem 1997;272:24159–64 [DOI] [PubMed] [Google Scholar]
- 26.Krastev DB, Li S, Sun Y, Wicks AJ, Hoslett G, Weekes D, et al. The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat Cell Biol 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, et al. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res 2010;16:4532–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA-double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem 2003;278:20303–12 [DOI] [PubMed] [Google Scholar]
- 29.Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep 2009;10:887–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Saha S, Wang W, Saha LK, Huang SN, Pommier Y. Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC). DNA Repair (Amst) 2020;89:102837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher RP. Cdk7: a kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription 2019;10:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sordet O, Larochelle S, Nicolas E, Stevens EV, Zhang C, Shokat KM, et al. Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J Mol Biol 2008;381:540–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci 2005;118:5171–80 [DOI] [PubMed] [Google Scholar]
- 34.Fisseler-Eckhoff A, Demes M. Neuroendocrine tumors of the lung. Cancers (Basel) 2012;4:777–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi N, Kim S, Rajapakse V, Schultz C, Zhang Y, Redon C, et al. Replication stress defines distinct molecular subtypes across cancers. Cell Rep 2021;Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bendixen C, Thomsen B, Alsner J, Westergaard O. Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry 1990;29:5613–9 [DOI] [PubMed] [Google Scholar]
- 37.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res 1989;49:5077–82 [PubMed] [Google Scholar]
- 38.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res 1997;25:4181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yankulov K, Yamashita K, Roy R, Egly JM, Bentley DL. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem 1995;270:23922–5 [DOI] [PubMed] [Google Scholar]
- 40.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2011;2:103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawale AS, Povirk LF. Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation. Nucleic Acids Res 2018;46:520–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem 2005;280:36518–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 1999;286:552–5 [DOI] [PubMed] [Google Scholar]
- 44.Sun Y, Chen J, Huang SN, Su YP, Wang W, Agama K, et al. PARylation prevents the proteasomal degradation of topoisomerase I DNA-protein crosslinks and induces their deubiquitylation. Nat Commun 2021;12:5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis 1996;17:31–5 [DOI] [PubMed] [Google Scholar]
- 46.Morozov AV, Karpov VL. Proteasomes and Several Aspects of Their Heterogeneity Relevant to Cancer. Front Oncol 2019;9:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortenson MM, Schlieman MG, Virudachalam S, Lara PN, Gandara DG, Davies AM, et al. Reduction in BCL-2 levels by 26S proteasome inhibition with bortezomib is associated with induction of apoptosis in small cell lung cancer. Lung Cancer 2005;49:163–70 [DOI] [PubMed] [Google Scholar]
- 48.Taromi S, Lewens F, Arsenic R, Sedding D, Sanger J, Kunze A, et al. Proteasome inhibitor bortezomib enhances the effect of standard chemotherapy in small cell lung cancer. Oncotarget 2017;8:97061–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galbraith MD, Bender H, Espinosa JM. Therapeutic targeting of transcriptional cyclin-dependent kinases. Transcription 2019;10:118–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files.