Abstract
Background
Accumulating evidence suggests that the upper airway bacterial microbiota is implicated in asthma inception, severity, and exacerbation. Unlike bacterial microbiota, the role of the upper airway fungal microbiome (mycobiome) in asthma control is poorly understood.
Research Question
What are the upper airway fungal colonization patterns among children with asthma and their relationship with subsequent loss of asthma control and exacerbation of asthma?
Study Design and Methods
The study was coupled with the Step Up Yellow Zone Inhaled Corticosteroids to Prevent Exacerbations (ClinicalTrials.gov Identifier: NCT02066129) clinical trial. The upper airway mycobiome was investigated using Internal transcribed spacer 1 (ITS1) sequencing of nasal blow samples collected from children with asthma when asthma was well controlled (baseline, n = 194) and during early signs of loss of asthma control (yellow zone [YZ], n = 107).
Results
At baseline, 499 fungal genera were detected in the upper airway samples, with two commensal fungal species, Malassezia globosa and Malassezia restricta, being most dominant. The relative abundance of Malassezia species varies by age, BMI, and race. Higher relative abundance of M globosa at baseline was associated with lower risk of future YZ episodes (P = .038) and longer time to development of first YZ episode (P = .022). Higher relative abundance of M globosa at YZ episode was associated with lower risk of progression from YZ episode to severe asthma exacerbation (P = .04). The upper airway mycobiome underwent significant changes from baseline to YZ episode, and increased fungal diversity was correlated highly with increased bacterial diversity (ρ = 0.41).
Interpretation
The upper airway commensal mycobiome is associated with future asthma control. This work highlights the importance of the mycobiota in asthma control and may contribute to the development of fungi-based markers to predict asthma exacerbation.
Key Words: asthma, fungi, Malassezia, mycobiome, upper airway
Take-home Points.
Study Questions: What is the composition of mycobiome in the upper airway of children with asthma? Is the composition of the mycobiome associated with future loss of asthma control or exacerbation of asthma? Does the mycobiome in the upper airways of children with asthma change from asymptomatic at baseline to time of loss of asthma control? Is the mycobiome correlated with bacterial microbiome in these children?
Results: We found that two Malassizia species, Malassezia globosa and Malassezia restricta, were the major mycobiome components in the upper airways of children with asthma. Relative abundance of M globosa at asymptomatic baseline was correlated inversely with future loss of asthma control. Higher relative abundance of M globosa at the time of loss of asthma control was correlated inversely with future severe asthma exacerbation. The upper airway mycobiome underwent significant changes from baseline to time of loss of asthma control. A complex correlation was found between fungal mycobiome and bacterial microbiome in the upper airways of children with asthma.
Interpretation: In children with asthma, the upper airway mycobiome is one important component of the entire microbiome that is associated with future asthma symptoms and exacerbations.
Asthma is the most common chronic disease in childhood and is characterized by recurrent episodes of cough, wheezing, and shortness of breath.1 Early signs of loss of asthma control, often termed yellow zone (YZ), represent a period when the patient is at risk of progressing to severe exacerbation that requires oral corticosteroid treatment.2 Prevention of asthma exacerbation is important because it is strongly associated with asthma mortality and potentially inhibits lung growth. Factors related to YZ development and to progression to acute exacerbation in children with asthma are not understood fully.
The human upper airway is a unique ecological niche that is colonized by a collection of bacteria, viruses, and fungi that interact with each other and with the host. Along with upper airway mucosa, upper airway microorganisms serve as the first line of defense against respiratory pathogens.3 Accumulating evidence strongly indicates that upper airway microbiota are associated with asthma inception, severity, and exacerbations.4,5 Our recent work involving a large cohort of school-aged children with asthma who were receiving low-dose inhaled corticosteroids demonstrated that higher relative abundance of upper airway Staphylococcus or Moraxella during periods of well-controlled asthma was associated with an increased risk of subsequent loss of asthma control (episodes of YZ), whereas higher relative abundance of the commensal bacteria Corynebacterium and Dolosigranulum was associated with decreased risk of subsequent loss of asthma control.6 However, the fungal component of the human upper airway microbiome and its role in asthma control have not been studied.
The collection of fungal species within the body, designated as mycobiota, is an important component of the airway microbiome in humans. Our understanding of mycobiota in asthma is restricted largely to several pathogenic fungi that grow at human body temperature and potentially infect the lungs. Aspergillus7 is one type of airborne fungus that causes allergic bronchopulmonary aspergillosis,8,9 which can contribute to poorly controlled asthma. Penicillium, Pneumocystis, and other indoor and outdoor fungal allergens also contribute to respiratory fungal infection and severe forms of allergic asthma.10 One study found that up to 30% of children with asthma and up to 60% of children with severe asthma were sensitized to these environmental fungi.11 In addition, higher Mucor exposure in the homes of children living in the inner city were associated with difficult-to-control asthma.12 With the development of high throughput sequencing, a comprehensive characterization of the commensal mycobiome in asthma is feasible. With a relatively small number of samples, a recent study showed that upper airway fungi are associated with respiratory illness in children with asthma.13 However, other than these few reports and despite the strong potential implications of fungi in respiratory infection and asthma severity, little is known about the characteristics of the commensal mycobiome in the upper airway in children with asthma and their relationship with loss of asthma control and exacerbation.
The overall goal of the study was to determine the relationship between the upper airway mycobiota and subsequent loss of asthma control and exacerbation of asthma. Leveraging a large, well-characterized prospective clinical trial,14 we analyzed the mycobiota from 301 nasal blow samples collected from children with asthma at baseline and at YZ episode and reported, for the first time to our knowledge, the composition and dynamics of the upper airway mycobiota in children with asthma. A specific fungal species, Malassezia globosa, was associated with future asthma activities.
Study Design and Methods
Study Design
This microbiome study was coupled to the Step Up Yellow Zone Inhaled Corticosteroids to Prevent Exacerbations (STICS) clinical trial,15 conducted by the National Heart, Lung, and Blood Institute’s AsthmaNet.14 The STICS clinical trial was approved by the AsthmaNet steering committee, protocol review committee, and data and safety monitoring board. The trial was approved independently by each of the institutional review boards of the clinical sites. We obtained informed consent from all participants.
The STICS clinical trial recruited school-aged children with mild asthma who were being treated with daily low-dose inhaled corticosteroids (ICS). Detailed of the STICS clinical trial study design, YZ criteria, clinical information of study participants, and the outcomes can be found in a previous publication.14 In brief, 254 children 5 to 11 years of age and from different sites across the United States were treated for 48 weeks with low-dose inhaled glucocorticoids. YZ episodes were identified by the occurrence of any of the following: the use of two doses (four inhalations) of rescue albuterol in 6 h, the use of three doses (six inhalations) of rescue albuterol in 24 h, or one night awakening that was the result of asthma that was treated with albuterol.14 When the children showed early signs of loss of asthma control (YZ episode), children were treated randomly with either the same dose of ICS (1 × ICS), or quintupled dose of ICS (5 × ICS) for 7 days.14
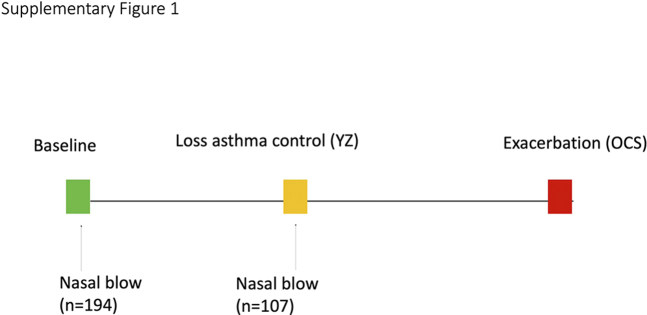
Nasal blow samples were collected in children at the randomization visit (baseline) when asthma symptoms were well controlled and at the time of the first episode of early signs of loss of asthma control (YZ episode) (e-Fig 1). The samples from the YZ time point were collected before ICS treatment to avoid potential influence of high-dose ICS to nasal microbiota. YZ samples collected by parents at home followed the sample collection protocol received at the randomization visit. Nasal samples were used for characterization of the bacterial microbiome previously6,14 and fungal microbiome in the current study.
ITS1 Sequencing and Data Processing
Total nucleic acid was extracted from nasal blow samples (200 μL) using the bioMerieux NucliSENS easyMAG automated extractor kit following standard protocol. Negative and positive controls were included in each sequence run. Internal transcribed spacer 1 (ITS1) region of fungal genomic DNA was amplified using primers: 18S-F—GTAAAAGTCGTAACAAGGTTTC and 5.8S-1R—GTTCAAAGAYTCGATGATTCAC, followed by sequencing on an Illumina Miseq (2 × 300 bp) platform. ITS1 was chosen because as it captures a more diverse fungal population than ITS2 and 18S rRNA.16,17
Raw sequencing reads were demultiplexed using Illumina software. Reads in each sample were processed using the Divisive Amplicon Denoising Algorithm (DADA) 2 pipeline (version 1.16) for taxonomy assignment to amplicon sequence variants (ASVs) based on the UNITE database (version 8.3).18 A classification with a confidence of < 0.5 was assigned as unclassified. To assign taxonomy of unclassified fungal genera additionally, we aligned all ASVs that were unclassified at the genus level to the Nucleotide database using the Basic Local Alignment Search Tool (BLAST+) (version 2.7.1). A given ASV was assigned to a specific genus based on Nucleotide taxonomy if the alignment of an ASV read was > 95% identity and 95% coverage to the Nucleotide database, as suggested previously.19
Samples with < 5,000 reads were removed from the analysis. The ITS1 sequencing reads from the remaining samples were rarefied to 5,000 reads/sample and were transformed to relative abundance. Taxa with mean relative abundance of > 0.1% were used for downstream analysis. Additionally, taxa identified as plant based on National Center for Biotechnology Information taxonomy were filtered out, including Solanum, Dioscorea, Pisum, Sinapis, Daucus, Cicer, Trifolium, Glycine, and Prunus. All the taxa found within negative controls (water and extraction controls) were < 0.1% in nasal wash samples; therefore, our analysis results were not impacted by potential contamination from negative controls.
Quantification of Fungal Load
We performed a FungiQuant assay (Thermo Fisher Scientific) to quantify the fungal 18S rRNA gene copy based on a previous study.20 Based on the quantitative polymerase chain reaction approach, a Candida albicans 18S rRNA gene clone was used as quantification standard.
Statistical Analysis of the Mycobiota
Hierarchical clustering using complete linkage was used to explore the mycobiota patterns among all participants. Fungal genera with relative abundance of > 0.1%, along with the remaining genera aggregated as “others,” were included in the construct of clustering and generating a heatmap. Clustering and a heatmap of Malassezia species at baseline also were produced. Pearson correlation analysis was used to examine associations between mycobiota characteristics (relative abundance, richness, Shannon diversity, fungal load) and continuous clinical variables. The Kruskal-Wallis test or Wilcoxon signed-rank test were used to test associations between mycobiota characteristics with categorical clinical variables. Linear regression analysis was performed for modelling the relationship between the annualized YZ episodes and the relative abundance of Malassezia species. In the model, annualized YZ episodes were treated as the response variable; the relative abundance of Malassezia species was the predictor; and age, sex, BMI, virus presence in nasal blow samples, or pet exposure were covariates.
Cox proportional hazards analysis assessed the association between time for YZ episode development (response) and log-transformed relative abundance of the fungal genera (predictor). The association was examined by likelihood ratio test and was visualized via Kaplan-Meier curves. Because the predictor (relative abundance of Malassezia species) was a continuous variable and Kaplan-Meier analysis handles only categorical predictors, we divided Malassezia species into the high-abundance group and the low-abundance group, with the best cutpoint that led to the largest difference in survival by log-rank test (each group has at least 30% of samples). The proportional hazards assumption was tested via scaled Schoenfeld residuals and was not rejected. The model stability was examined by removing two samples with slightly larger DFBETA, which led to consistent conclusions.
To test the robustness of our results on association between Malassezia species and future loss of asthma control or asthma exacerbation and to ensure that results are not driven by data from one geographical location or sample collection site, we performed sensitivity tests by excluding samples from one of nine geographic locations, so that each sensitivity test used samples from the eight remaining geographic locations.
To identify specific fungi (> 0.1% relative abundance) that statistically differ between baseline and YZ episode, we performed differential analyses based on the negative binomial distribution using the DESeq2 package version 1.34.0 (R Foundation for Statistical Computing). We further inspected the results by plotting raw and relative abundance of the fungal data and removed any differential fungal taxa that likely were driven by one or two samples.
To determine correlations between bacteria and fungi at baseline or YZ episode, we first performed centered log ratio transformation of relative abundance of fungi or bacteria. Bacterial genera with mean relative abundance of > 1% and fungal genera with mean relative abundance of > 0.5% were selected for Spearman correlation. All bacteria-fungi correlations were plotted using corrplot package in R software (R Foundation for Statistical Computing).
All of the above analyses were performed in R version 4.1.2. A P value of < .05 was considered statistically significant. P values were adjusted by false discovery rate when multiple comparisons were involved. These include association analysis between fungi and multiple clinical factors. An adjusted P value of < .1 was considered statistically significant.
Data Availability
Raw sequencing of this ITS1 dataset is available from the Sequence Read Archive (SRA) database with the accession number PRJNA830491.
Results
Mycobiota Composition in Upper Airway at Baseline
In total, 374 nasal blow samples from 214 children with asthma were collected and subjected to ITS1 sequencing. After data processing, 301 samples (194 at baseline and 107 at YZ episode) yielded > 5,000 reads/sample and were used for downstream analyses. The demographics and clinical characteristics of these children are shown in e-Table 1.
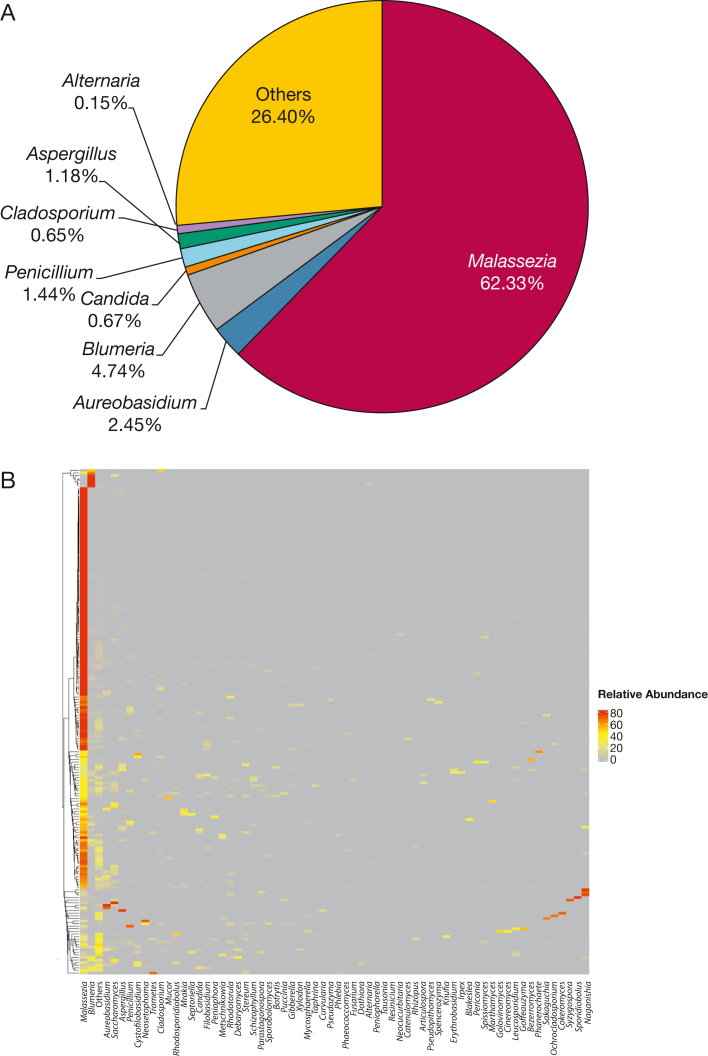
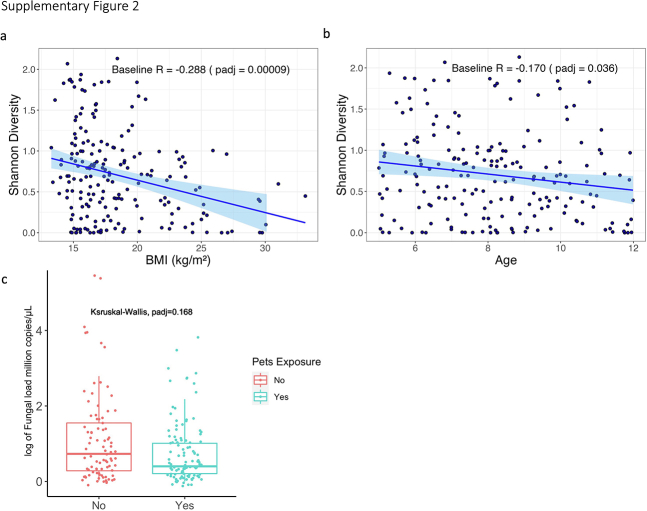
We first characterized the upper airway mycobiota composition at baseline. We identified 499 fungal genera and 4,704 ASVs from 194 baseline samples. The lipophilic skin-commensal Malassezia was the most dominant fungal genus in the samples (62.33% of relative abundance), followed by Blumeria (4.74%), Aureobasidium (2.45%), Penicillium (1.44%), Aspergillus (1.18%), Candida (0.67%), Cladosporium (0.65%), and Alternaria (0.15%) (Fig 1A). Unsupervised hierarchical clustering analysis revealed that most children (154/194 [79.4%]) harbored a single, highly abundant genus Malassezia (Fig 1B). The dominance by a single fungal genus in the mycobiota in the upper airway is different from the bacterial community of the same cohort in which several bacterial genera were dominant.6 Correlation analysis between Malassezia and baseline clinical factors showed that BMI and age were correlated inversely with fungal diversity, whereas pet exposure was associated with lower fungal load (e-Fig 2).
Figure 1.
Upper airway fungal composition, distribution, and association with clinical variables at baseline. A, Pie chart showing the relative abundance of upper airway fungal genera in nasal blow samples at baseline (n = 194). B, Heatmap generated to identify upper airway fungal pattern at baseline by hierarchical clustering and complete linkage approach. Fungal genera with relative abundance of > 0.1% are shown on the x-axis, and those with relative abundance of < 0.1% are assigned as “Others.”
Associations Between Malassezia Species and Clinical Factors at Baseline
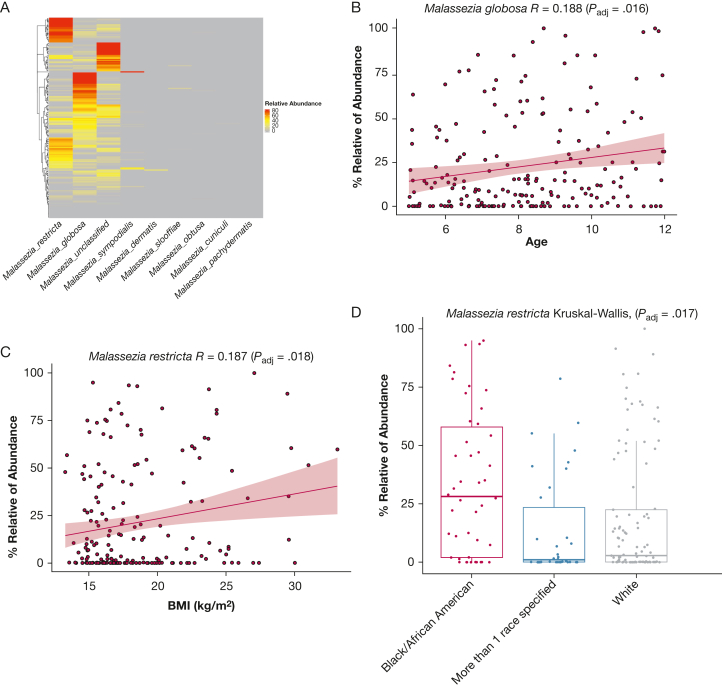
Because Malassezia was the most dominant genus at baseline, we further examined Malassezia species distribution patterns and their association with clinical variables. Eight different Malassezia species within the Malassezia genus were identified, with Malassezia globosa and Malassezia restricta being the most abundant (Fig 2A). M globosa and M restricta showed < 90% identity in their ITS1 sequences, indicating a confident distinction between the two species by ITS1 sequencing. M globosa was correlated positively with age (Padj = .016; ρglobosa = 0.1882) (Fig 2B). Alternatively, M restricta was correlated positively with BMI (Padj = .018; ρrestricta = 0.187) (Fig 2C). Additionally, relative abundance of M restricta differed by race, as demonstrated by higher relative abundance in Black or African American children (Fig 2D) (Padj = 0.017) compared with White children and children who identified more than one race. We did not find Malassezia species associated with other clinical variables (e-Table 1). These findings suggest that depending on specific species, relative abundance of Malassezia species vary by age, BMI, and race.
Figure 2.
Fungal Malassezia species identified from nasal blow samples at baseline and associations between Malassezia species and clinical variables. A, Heatmap showing the composition and abundance of Malassezia species in children with asthma at baseline (n = 194) as demonstrated by hierarchical clustering and complete linkage approach. B, C, Graphs showing Pearson correlations between relative abundance of M globosa and age at enrollment (B) and M restricta and BMI (C). D, Boxplot showing the relative abundance of M restricta compared across the following races: African American, White, and more than one race. Statistical significances of mycobiome richness or diversity among clusters were tested using the Kruskal-Wallis rank-based nonparametric test. Asian and Hispanic or Latino children were excluded in this analysis because of a limited number of samples (< 2 for both). Each dot represents one participant. The horizontal line in the middle of the boxplot represents the median value of relative abundance of the mycobiome; the top and bottom horizontal line of the boxplot represent the 25th and 75th percentiles of the data value.
M globosa Is Associated With Better Asthma Control
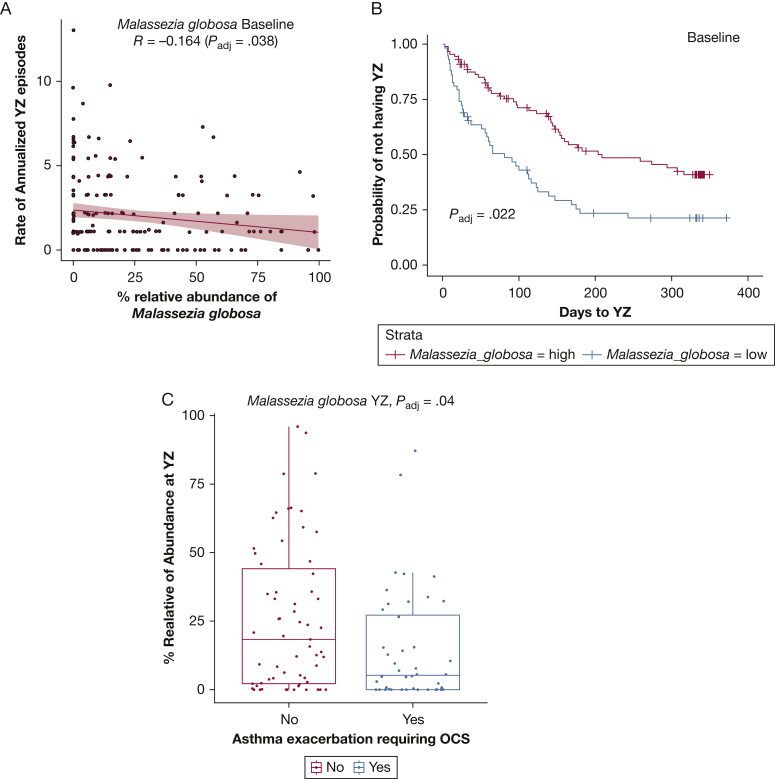
Among all children in the analysis, 69.6% experienced at least one episode of loss of asthma control (YZ episode). Linear regression analysis showed relative abundance of M globosa at baseline was correlated negatively with annualized YZ episodes after controlling for age, pet exposure, BMI, or sex (P = .038) (Fig 3A). These covariates are associated with the development of a YZ episode in this cohort.
Figure 3.
Associations between the relative abundance of M globosa at baseline and YZ episode and subsequent development of YZ episode or future asthma exacerbation. A, Graph showing the relative abundance of M globosa correlated with annualized rate of YZ episodes. B, Graph showing higher relative abundance of M globosa associated with longer time to YZ episode developing. C, Boxplot comparing relative abundance of M globosa at YZ episode between oral corticosteroid groups (yes = participants who have severe asthma exacerbation requiring OCS; no = participants have no severe asthma). Statistical significances of mycobiome relative abundance between groups was performed by Kruskal-Wallis test. OCS = oral corticosteroids; YZ = yellow zone.
Although we previously reported that the presence or absence of respiratory virus in nasal blow samples at baseline was not associated with future loss of asthma control or exacerbation of asthma,6 we still investigated whether the fungal findings were related to the presence of respiratory virus, given the important role of viruses in asthma development. After adjusting for the presence of respiratory virus, M globosa remained negatively associated with the annualized rate of YZ episodes (P = .0122). Children with higher relative abundance of M globosa at baseline also showed a longer time to development of the first YZ episode compared with patients with lower relative abundance of M globosa (Fig 3B) (P = .022), as shown by survival analysis with the Cox proportional hazards model.
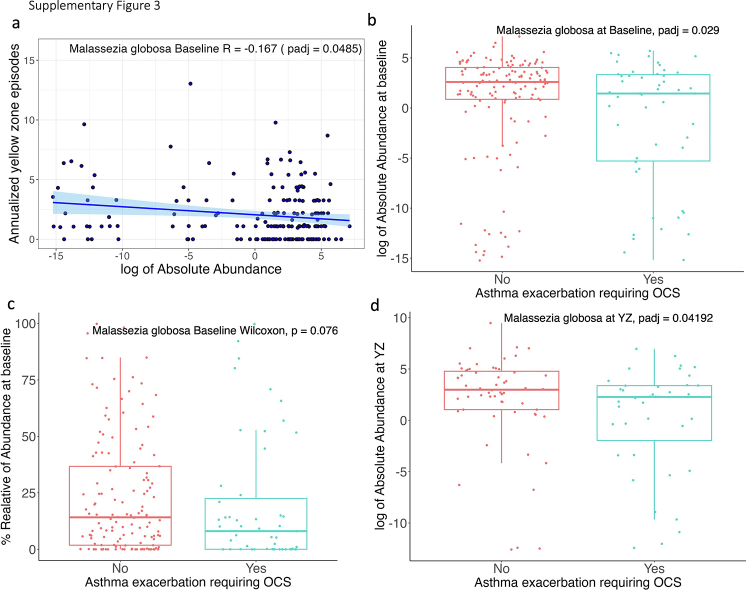
To determine whether the Malassezia species were associated with severe asthma exacerbation when oral corticosteroid therapy was required, we compared the relative abundance of Malassezia species at baseline or during YZ episodes in children with and without asthma exacerbation. Twenty-six of 214 participants (26.1%) experienced an asthma exacerbation. We found that the relative abundance of M globosa during the YZ episode was significantly lower in children requiring oral corticosteroid treatment after controlling for age, pets, BMI, virus presence, and sex in a linear regression analysis (Fig 3C).
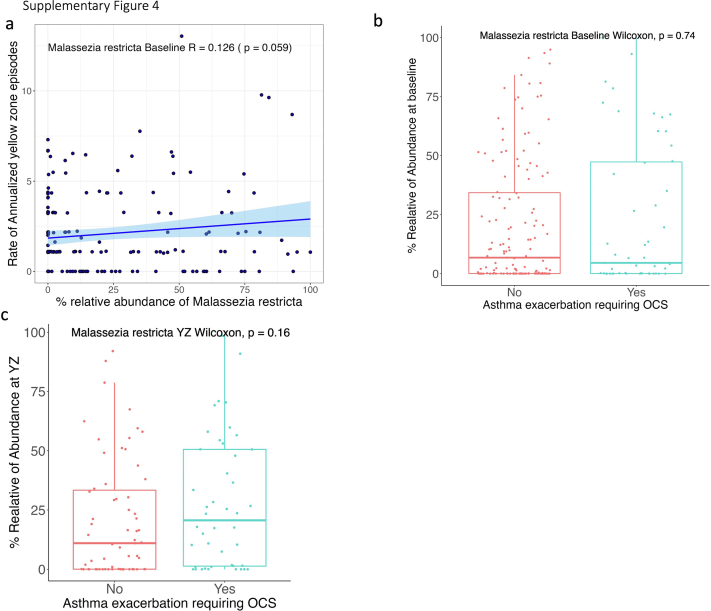
We further computed the absolute abundance of Malassezia species by multiplying their relative abundance with total fungal load. Results were consistent between the absolute abundance analysis and the relative abundance analysis (e-Fig 3). Additionally, the absolute abundance of M globosa at baseline was significantly lower in children with and without asthma exacerbation (P = .029) (e-Fig 3B), whereas relative abundance M globosa showed only marginal statistical difference between the two groups (P = .076) (e-Fig 3B). However, we did not find a significant association between M restricta and loss of asthma control or future exacerbation of asthma (e-Fig 4).
As a sensitivity analysis, we removed all samples from one geographic location (nine locations in total) (e-Table 2) and used the remaining samples to perform the above analyses. The sensitivity tests by geographic sites showed largely consistent results for M globosa (e-Table 2). Taken together, the findings suggest that M globosa in children is associated with a subsequently lower rate of YZ episodes and lower risk of asthma exacerbation.
Dynamic Changes of Upper Airway Mycobiota From Baseline to YZ Episode
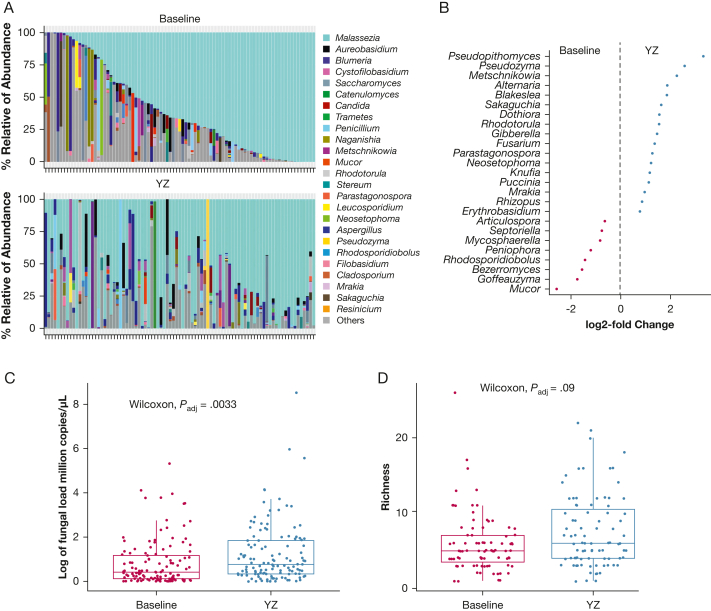
Leveraging the longitudinal design of the study, we determined the changes of upper airway mycobiota from baseline to YZ episode in the 87 children who contributed samples at both time points. We identified significant changes in these genera from baseline to YZ episode (Fig 4A). Differential taxa analysis using DESeq2 analysis identified 20 significantly increased and 10 significantly decreased fungal genera at YZ episode (Fig 4B) (Padj < .05). Pseudopithomyces, which is linked to facial eczema in animals,21 showed the highest log2-fold increase (3.32 folds) from baseline to YZ episode, followed by an environmental yeast Pseudozyma. Several previously reported asthma-associated fungi, Alternaria and Fusarium,11,22, 23, 24 also increased from baseline to YZ episode (Fig 4B) (Padj < .05). Alternatively, Mycosphaerella was decreased significantly at YZ episode in this cohort (Fig 4B) (Padj < .05). Mycosphaerella was reported to be correlated negatively with ICS use and enriched in patients with mild asthma or healthy control participants.24 Surprisingly, we found a mold, Mucor, decreased significantly at YZ episode.
Figure 4.
Dynamic changes of the upper airway mycobiota from baseline to YZ episode. A, Barplot showing changes in relative abundance of fungal genera in each child with asthma: the relative abundance of the top 25 fungi at baseline and YZ episode. B, Graph showing significantly different fungi from baseline to YZ episode by DEseq2 (Padj < . 05). C, D, Boxplots showing total fungal load (C) and fungal richness difference (D) between baseline and YZ episode. Statistical differences were tested by the Wilcoxon rank-sum test. YZ = yellow zone.
Our previous study showed that bacterial diversity and load were increased significantly from baseline to YZ episode.6 Total fungal load and fungal richness were significantly higher at YZ episode compared with baseline (Fig 4C, 4D) (Padj = .003 and Padj < .09, respectively, Wilcoxon rank-sum test). These results suggest an expansion of fungal microbiota from baseline to YZ episode.
Relationship Among Fungi, Bacteria, and Viruses in the Upper Airway
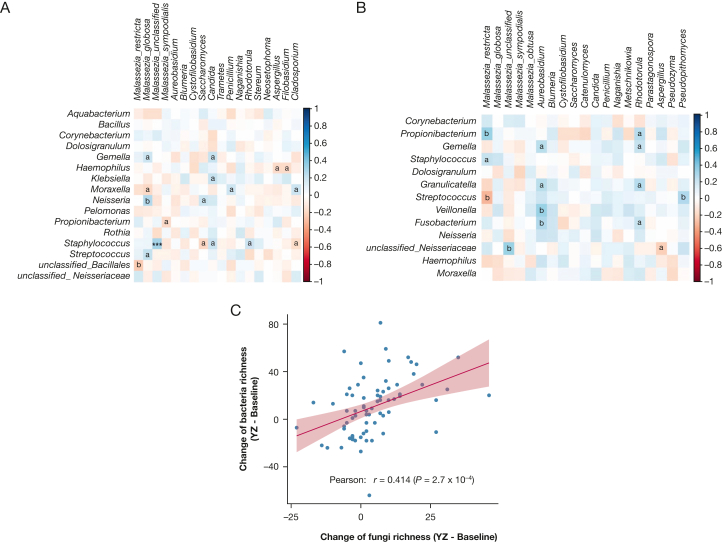
Pairwise correlations identified 288 and 247 bacteria-fungi correlations at baseline and YZ episode, respectively. At baseline, M globosa was correlated negatively with Moraxella (Fig 5A), which is a previously reported pathogenic bacteria that is associated with loss of asthma control.6 Other interactions include positive correlations between Candida and common bacteria pathogens Klebsiella, Staphylococcus, and Gemella, whereas Aspergillus showed a negative correlation with pathogenic Haemophilus.
Figure 5.
A, B, Fungi-bacteria correlation network at baseline (A) and YZ episode (B). Relationships between bacterial genera and relative abundance of Malassezia species and other major fungal genera were determined by Spearman correlation after central log ratio transformation of relative abundance of bacteria or fungi. Red and blue colors represent negative and positive correlations, respectively. aP < .05; bP < .01. C, Pearson correlation between changes of fungal diversity and changes of bacterial diversity from baseline to YZ episode. YZ = yellow zone.
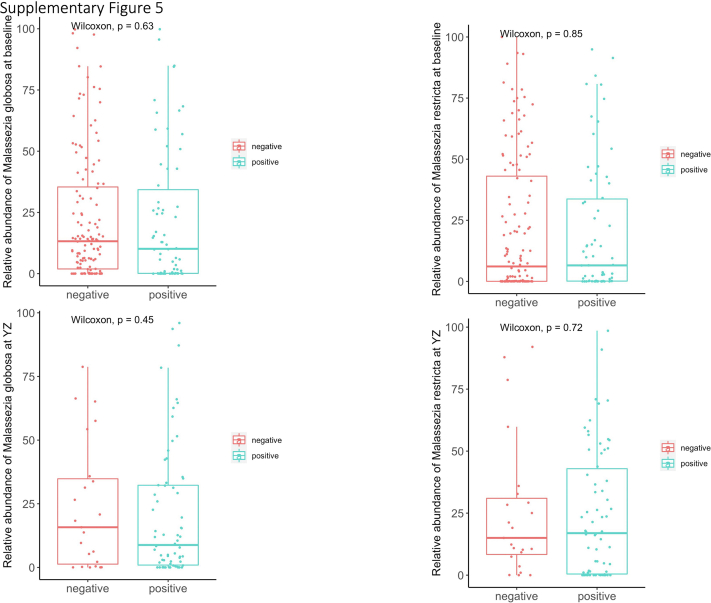
At YZ episode, we identified predominantly positive correlations between potentially pathogenic fungi and bacteria (60.3% correlations). For example, fungus Aureobasidium showed strong positive correlations with Fusobacterium and Veillonella. Fungal genus Pseudopithomyces, which also increased during YZ episode (Fig 5B), showed strong positive correlations with Streptococcus, which also was increased during YZ episode.6 Correlations between fungi and bacteria at YZ episode likely reflect concomitant increase of fungi and bacteria in response to YZ episode. Indeed, we found increased fungal diversity was correlated positively with increased bacterial diversity from baseline to YZ episode (Fig 5C) (P = 2.7 × 10–4; ρ = 0.414). These results suggest a complex co-occurrent and coexclusive relationships between fungi and bacteria and distinct correlation patterns between baseline and YZ episode in the upper airway. Fungal diversity and specific fungal compositions were not statistically different between virus-positive and virus-negative samples at either baseline or YZ episode (e-Fig 5).
Discussion
Our study defined the mycobiota characteristics, dynamics, and transkingdom interactions in the upper airway of school-aged children with asthma and provided the first novel evidence, to our knowledge, for associations between the airway fungi and asthma control. We found an association between abundance of M globosa and lower rate of annualized YZ episodes, along with a lower risk of progression from YZ episode to severe asthma exacerbation. The findings represent an important first step in studying the role of the commensal fungal mycobiome in asthma.
Malassezia is a lipid-dependent basidiomycetous yeast and is one of the most common fungi residing on human skin.25 We found Malassezia to be the most abundant fungal genera in nasal blow samples, which is consistent with previous studies.26, 27, 28, 29, 30 The anterior nares of the nasal passage are lined with keratinized squamous epithelium. Lipid-rich mucosa of the anterior nares can promote growth of lipophilic fungi such as Malassezia. Association between age and Malassizia also is evident from a study with a cohort of children from 6 to 17 years of age,13 which is likely because of increasing sebum production and sex hormone concentrations from early childhood to puberty.31,32 The positive correlation between Malassezia and BMI also may be related to higher sebum production in participants with higher BMI.33 Nevertheless, after controlling for age and BMI, the association between the abundance of Malassezia species during a YZ episode and asthma exacerbation remained significant, suggesting that Malassezia species may be related to asthma symptoms and exacerbation independent of age and BMI.
Malassezia is considered a commensal member of the human microbiome34 that has been understudied significantly.34,35 Its functional significance largely is derived from studies in skin diseases.25,36,37 For the first time, we reported that increased relative abundance of M globosa in the upper airway is associated with better control of asthma in children with asthma. The exact mechanisms underlying this potential benefit remain to be elucidated fully. However, one possible mechanism is through actively preventing or inhibiting pathogen colonization. For example, on skin, Malassezia can prevent colonization by pathogenic Staphylococcus aureus by secreting proteases.38 Interestingly, our correlation network analysis identified a coexclusion relationship between M globosa and bacterial pathogen Morexella that may be related to asthma exacerbation,39 supporting a competitive role of specific fungi and bacteria in asthma. Alternatively, M globosa can suppress inflammation indirectly, such as by inducing antiinflammatory cytokine IL-10 observed in keratinocytes after exposure to M globosa.40
Our study also identified cross-kingdom interactions between bacteria and fungi, but not between viruses and fungi, in the airways of children with asthma. Co-occurrence and a coexclusive relationship between fungi and bacteria suggest that these organisms may work cooperatively or competitively in driving asthma pathogenesis. However, these relationships may be a result of nutritional preference of these microorganisms. For example, co-occurrence of the organisms simply may reflect similar characteristics in nutrient use. Lack of correlations between viruses and fungi at baseline also may be the result of no active viral infection being present at a time when asthma symptoms are well controlled. More than 70% of children showed positive virus results at YZ episode, resulting in less statistical power in detecting any fungi difference in viral-positive and viral-negative groups. Subsequent in vitro and in vivo work are warranted to study cross-kingdom interactions and host immune-microbiome interactions, allowing for mechanistic understanding of how fungi influence the pathogenesis of asthma.
Our study has some limitations. First, the patient population included children with asthma who were treated with low doses of ICS, which may have an effect on the upper airway fungal profile. However, this is unlikely to affect the main findings of the study—the associations between M globosa abundance and loss of asthma control—because all study participants received this daily low dose ICS therapy. In addition, this study did not include healthy control participants. Therefore, we cannot exclude the possibility that changes of the mycobiome profile from the baseline to YZ episode were confounded by mycobiome changes in healthy conditions, if there are any. We also could not determine whether children with asthma have an altered mycobiome profile or mycobiome maturation trajectory compared with healthy control participants. Second, other limitations include lack of lower airway samples. However, the upper airways increasingly are recognized as an important gatekeeper to respiratory health.3,41 Upper airway samples are easy to access, rendering in wider clinical application than lower airway samples. Third, we cannot exclude the possibility that Malassizia detected from the nasal blow samples may include some microorganisms from the skin. However, a recent study reported that nasal passages and skin showed a similar fungal profile as the upper airway.32 Therefore, our data are informative in defining upper airway mycobiota profiles using nasal blow samples. Finally, our observations are based on associations; thus, further mechanistic studies are needed to demonstrate a causal relationship of M globosa in loss of asthma control and exacerbation of asthma.
Interpretation
Overall, a considerable knowledge gap exists regarding the commensal fungal microbiome in the upper airway in children with asthma. We took the initial step by establishing associations between airway mycobiota and loss of asthma control in children with asthma. These associations may allow for future mechanistic studies to understand better the role of commensal fungi in the pathogenesis of asthma and may open novel avenues to develop mycobiota-associated biomarkers to predict and monitor disease outcomes.
Funding/Support
L. B. B. is supported by the National Institute of Allergy and Infectious Diseases and the National Heart, Lung, and Blood Institute. D. J. is supported by the National Institute of Allery and Infectious Diseases, the National Heart, Lung, and Blood Institute, and the National Institutes of Health Office of the Director.
Financial/Nonfinancial Disclosures
The authors have reported to CHEST the following: L. B. B. reports personal fees from GlaxoSmithKline, Genentech/Novartis, DBV Technologies, Teva, Boehringer Ingelheim, AstraZeneca, WebMD/Medscape, Sanofi/Regeneron, Vectura, Circassia, Kinaset, Vertex, and OM Pharma and royalties from Elsevier outside the submitted work. D. J. reports grants from GlaxoSmithKline and Regeneron outside the submitted work and personal fees from AstraZeneca, GlaxoSmithKline, Genentech, Regeneron, Sanofi, OM Pharma, and Pfizer outside the submitted work. A. B. is a consultant to Sanofi and receives speaker fees from Sanofi. Y. Z. and G. M. W. hold shares in General Biomics, Inc. None declared (H. Y., Z. L., J. D., D. M., H. B., M. C., J. D., Y. J. H., R. F. L., G. A. S., K. W., R. C., A. M. F., W. P., R. G. R.).
Acknowledgments
Author contributions: H. Y. contributed to analysis, interpretation of data, and drafting the work. Z. L. contributed to analysis. J. D. contributed to analysis. L. B. B. contributed to acquisition and conception and design and substantively revised the paper. D. J. contributed to acquisition, conception and design, revision of the paper. D. M. contributed to acquisition, conception and design, revision of the paper. H. B. contributed to acquisition, conception and design, and revision of the paper. M. C. contributed to acquisition of data and conception and design and substantively revised the paper. J. D. contributed to data interpretation and substantively revised the paper. Y. J. H. contributed to revision of the paper. R. F. L. contributed to conception and design and revision of the paper. G. A. S. contributed to revision of the paper, acquisition, and conception and design. G. M. W. contributed to acquisition and revision of the paper. K. W. contributed to revision of the paper. R. C. contributed to conception and design, revision of the paper, participate in acquisition. A.M.F. contributed to conception and design, acquisition, and revision of the paper. W. P. contributed to conception and design, acquisition of data, and revision of the paper. R. G. R. contributed to acquisition of data and revision of the paper. A. B. contributed to design of the work, acquisition, and substantively revised the paper. Y. Z. contributed to design of the work and interpretation of data and drafted the work.
Other contributions: The authors thank Cassandra Suther for copyediting the manuscript.
Additional information: The e-Figures and e-Tables are available online under “Supplementary Data.”
Footnotes
A. B. and Y. Z. contributed equally to this work as co-corresponding authors.
Supplementary Data
Supplemental Figure 1.
Scheme of sample collection. Green rectangle represents the timepoints when children’s asthma is well controlled (baseline), yellow rectangle represents when children experience first episode of loss of asthma control (YZ), and red rectangle represents when children have asthma exacerbation requiring OCS treatment. The maximum follow-up time for children included in fungal analysis is up to 15 months.
Supplemental Figure 2.
Pearson’s Correlation between Shannon diversity and (A) BMI (Body Mass Index) and (B) Age of enrollment (5-11 years of age), both p value less than 0.05 after FDR adjustment. (c) Total fungal load difference between children with or without pet exposure at baseline. Y-axis represents 18S rRNA million copies per microliter estimated by qPCR. Children exposed to pets tended to have higher fungal load (p=0.028), but this became non-significant after FDR adjustment (padj=0.168).
Supplemental Figure 3.
(a) Correlation between log transformed absolute abundance of Malassezia globosa with annualized rate of YZ episodes. The results are controlled for age, pets, BMI, virus presence, and sex. (b) Comparison of log transformed absolute abundance of Malassezia globosa at baseline between OCS groups. (Yes: participants who have severe asthma exacerbation requiring OCS, No: participants have no severe asthma). (c) Comparison of relative abundance of Malassezia globosa at baseline between OCS groups. (d) Comparison of log transformed absolute abundance of Malassezia globosa at YZ between OCS groups.
Supplemental Figure 4.
(a) Relative abundance of Malassezia restricta is correlated with annualized rate of YZ episodes. Comparison of relative abundance of Malassezia restricta at baseline (b) and YZ (c) between OCS groups. Statistical significances of Malassezia restricta relative abundance between groups are performed by Kruskal Wallis test or Wilcoxon test.
Supplemental Figure 5.
Graphs showing no differences of relative abundance of Malassezia species in virus-positive and -negative samples at baseline and YZ.
References
- 1.Global Initiative for Asthma, Global strategy for asthma management and prevention, 2022. Accessed April 13, 2023. https://ginasthma.org/wp-content/uploads/2022/07/GINA-Main-Report-2022-FINAL-22-07-01-WMS.pdf
- 2.Dinakar C., Oppenheimer J., Portnoy J., et al. Management of acute loss of asthma control in the yellow zone: a practice parameter. Ann Allergy Asthma Immunol. 2014;113(2):143–159. doi: 10.1016/j.anai.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Fokkens W.J., Scheeren R.A. Upper airway defence mechanisms. Paediatr Respir Rev. 2000;1(4):336–341. doi: 10.1053/prrv.2000.0073. [DOI] [PubMed] [Google Scholar]
- 4.Boutin R.C., Petersen C., Woodward S.E., et al. Bacterial-fungal interactions in the neonatal gut influence asthma outcomes later in life. eLife. 2021;10 doi: 10.7554/eLife.67740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang R., Zhang Q., Ren Z., Li H., Ma Q. Different airway inflammatory phenotypes correlate with specific fungal and bacterial microbiota in asthma and chronic obstructive pulmonary disease. J Immunol Res. 2022;2022 doi: 10.1155/2022/2177884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y., Jackson D., Bacharier L.B., et al. The upper airway microbiota and loss of asthma control among asthmatic children. Nat Commun. 2019;10(1):5714. doi: 10.1038/s41467-019-13698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman D.L., Chen Z., Shankar V., Tyberg M., Vicencio A., Burk R. Lower airway microbiota and mycobiota in children with severe asthma. J Allergy Clin Immunol. 2018;141(2):808–811.e7. doi: 10.1016/j.jaci.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal R., Gupta D. Severe asthma and fungi: current evidence. Med Mycol. 2011;49(suppl 1):S150–S157. doi: 10.3109/13693786.2010.504752. [DOI] [PubMed] [Google Scholar]
- 9.Woolnough K., Fairs A., Pashley C.H., Wardlaw A.J. Allergic fungal airway disease: pathophysiologic and diagnostic considerations. Curr Opin Pulm Med. 2015;21(1):39–47. doi: 10.1097/MCP.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 10.Singh M., Paul N., Singh S., Nayak G.R. Asthma and fungus: role in allergic bronchopulmonary aspergillosis (ABPA) and other conditions. Indian J Pediatr. 2018;85(10):899–904. doi: 10.1007/s12098-018-2646-8. [DOI] [PubMed] [Google Scholar]
- 11.Baxi S.N., Portnoy J.M., Larenas-Linnemann D., Phipatanakul W., Environmental Allergens Workgroup Exposure and health effects of fungi on humans. J Allergy Clin Immunol Pract. 2016;4(3):396–404. doi: 10.1016/j.jaip.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vesper S., Wymer L., Kroner J., et al. Association of mold levels in urban children’s homes with difficult-to-control asthma. J Allergy Clin Immunol. 2022;149(4):1481–1485. doi: 10.1016/j.jaci.2021.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCauley K.E., Flynn K., Calatroni A., et al. Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J Allergy Clin Immunol. 2022;150(1):204–213. doi: 10.1016/j.jaci.2022.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Jackson D.J., Bacharier L.B., Mauger D.T., et al. Quintupling inhaled glucocorticoids to prevent childhood asthma exacerbations. N Engl J Med. 2018;378(10):891–901. doi: 10.1056/NEJMoa1710988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Institutes of Health Clinical Center, Step-up Yellow Zone Inhaled Corticosteroids to Prevent Exacerbations (STICS). NCT02066129. ClinicalTrials.gov. National Institutes of Health; 2014. https://clinicaltrials.gov/ct2/show/NCT02066129 Updated July 11, 2018.
- 16.Mbareche H., Veillette M., Bilodeau G., Duchaine C. Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ. 2020;8 doi: 10.7717/peerj.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Yu Y., Cai Z., Bartlam M., Wang Y. Comparison of ITS and 18S rDNA for estimating fungal diversity using PCR–DGGE. World J Microbiol Biotechnol. 2015;31(9):1387–1395. doi: 10.1007/s11274-015-1890-6. [DOI] [PubMed] [Google Scholar]
- 18.Abarenkov K, Zirk A, Piirmann T, et al. UNITE general FASTA release for fungi [published online ahead of print May 10, 2021]. Microorganisms, 10.15156/BIO/1280049 [DOI]
- 19.Vu D., Groenewald M., de Vries M., et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C.M., Kachur S., Dwan M.G., et al. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012;12(1):255. doi: 10.1186/1471-2180-12-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidhu JS, Suresh V, Baten A, et al. Genome sequence of Pseudopithomyces chartarum, causal agent of facial eczema (pithomycotoxicosis) in ruminants, and identification of the putative sporidesmin toxin gene cluster. bioRxiv. Published online April 30, 2021. 10.1101/2021.04.29.441555 [DOI]
- 22.Bush R.K., Prochnau J.J. Alternaria-induced asthma. J Allergy Clin Immunol. 2004;113(2):227–234. doi: 10.1016/j.jaci.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Nucci M., Anaissie E. Fusarium infections in immunocompromised patients. Clin Microbiol Rev. 2007;20(4):695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A., Laxman B., Naureckas E.T., et al. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol. 2019;144(5):1214–1227.e7. doi: 10.1016/j.jaci.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunte D.M.L., Gaitanis G., Hay R.J. Malassezia-associated skin diseases, the use of diagnostics and treatment. Front Cell Infect Microbiol. 2020;10:112. doi: 10.3389/fcimb.2020.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Baumgartner M., Li C., Kuntz T., et al. Differences of the nasal microbiome and mycobiome by clinical characteristics of COPD patients [published online ahead of print April 29, 2022], Chronic Obstr Pulm Dis. [DOI] [PMC free article] [PubMed]
- 27.Cleland E.J., Bassiouni A., Boase S., Dowd S., Vreugde S., Wormald P.J. The fungal microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes and patient outcomes. Int Forum Allergy Rhinol. 2014;4(4):259–265. doi: 10.1002/alr.21297. [DOI] [PubMed] [Google Scholar]
- 28.Jung W.H., Croll D., Cho J.H., Kim Y.R., Lee Y.W. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses. 2015;58(3):167–172. doi: 10.1111/myc.12296. [DOI] [PubMed] [Google Scholar]
- 29.Gelber J.T., Cope E.K., Goldberg A.N., Pletcher S.D. Evaluation of Malassezia and common fungal pathogens in subtypes of chronic rhinosinusitis. Int Forum Allergy Rhinol. 2016;6(9):950–955. doi: 10.1002/alr.21777. [DOI] [PubMed] [Google Scholar]
- 30.Zhang I., Pletcher S.D., Goldberg A.N., Barker B.M., Cope E.K. Fungal microbiota in chronic airway inflammatory disease and emerging relationships with the host immune response. Front Microbiol. 2017;8:2477. doi: 10.3389/fmicb.2017.02477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamloul G., Khachemoune A. An updated review of the sebaceous gland and its role in health and diseases. Part 1: embryology, evolution, structure, and function of sebaceous glands. Dermatol Ther. 2021;34(1):e14695. doi: 10.1111/dth.14695. [DOI] [PubMed] [Google Scholar]
- 32.Park J., Schwardt N.H., Jo J.H., et al. Shifts in the skin bacterial and fungal communities of healthy children transitioning through puberty. J Invest Dermatol. 2022;142(1):212–219. doi: 10.1016/j.jid.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai M.-C., Chen W., Cheng Y.-W., Wang C.-Y., Chen G.-Y., Hsu T.-J. Higher body mass index is a significant risk factor for acne formation in schoolchildren. Eur J Dermatol EJD. 2006;16(3):251–253. [PubMed] [Google Scholar]
- 34.Vijaya Chandra S.H., Srinivas R., Dawson T.L., Common J.E. Cutaneous Malassezia: commensal, pathogen, or protector? Front Cell Infect Microbiol. 2021;10:614446. doi: 10.3389/fcimb.2020.614446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawson T.L.J. Malassezia: the forbidden kingdom opens. Cell Host Microbe. 2019;25(3):345–347. doi: 10.1016/j.chom.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Jain C., Das S., Ramachandran V.G., Saha R., Bhattacharya S.N., Dar S. Malassezia yeast and cytokine gene polymorphism in atopic dermatitis. J Clin Diagn Res JCDR. 2017;11(3):DC01–DC05. doi: 10.7860/JCDR/2017/23948.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chng K.R., Tay A.S.L., Li C., et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol. 2016;1(9) doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- 38.Li H., Goh B.N., Teh W.K., et al. Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J Invest Dermatol. 2018;138(5):1137–1145. doi: 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 39.Kloepfer K.M., Lee W.M., Pappas T.E., et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133(5):1301–1307. doi: 10.1016/j.jaci.2014.02.030. 1307.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donnarumma G., Perfetto B., Paoletti I., et al. Analysis of the response of human keratinocytes to Malassezia globosa and restricta strains. Arch Dermatol Res. 2014;306(8):763–768. doi: 10.1007/s00403-014-1479-1. [DOI] [PubMed] [Google Scholar]
- 41.Lovato A., de Filippis C. Clinical presentation of COVID-19: a systematic review focusing on upper airway symptoms. Ear Nose Throat J. 2020;99(9):569–576. doi: 10.1177/0145561320920762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing of this ITS1 dataset is available from the Sequence Read Archive (SRA) database with the accession number PRJNA830491.