Abstract
PURPOSE:
Patients with EGFR-mutant non-small cell lung cancer (NSCLC) experience variable duration of benefit on EGFR tyrosine kinase inhibitors (TKI). The effect of concurrent genomic alterations on outcome has been incompletely described.
MATERIAL AND METHODS:
In this retrospective study, targeted next-generation sequencing data was collected from patients with EGFR-mutant lung cancer treated at the Dana-Farber Cancer Institute. Clinical data were collected and correlated with somatic mutation data. Associations between TP53 mutation status, genomic features, and mutational processes were analyzed.
RESULTS:
269 patients were identified for inclusion in the cohort. Among 185 response-evaluable patients with pre-treatment specimens, TP53 alterations were the most common event associated with decreased first-line progression-free survival (PFS), and associated with decreased overall survival along with DNMT3A, KEAP1 and ASXL1 alterations. Reduced PFS on later-line osimertinib in 33 patients was associated with MET, APC and ERBB4 alterations. Further investigation of the effect of TP53 alterations demonstrated an association with worse outcomes even in patients with good initial radiographic response, and faster acquisition of T790M and other resistance mechanisms. TP53 mutated tumors had higher mutational burdens and increased mutagenesis with exposure to therapy and tobacco. Cell cycle alterations were not independently predictive, but portended worse OS in conjunction with TP53 alterations.
CONCLUSIONS:
TP53 alterations associate with faster resistance evolution independent of mechanism in EGFR mutant NSCLC, and may cooperate with other genomic events to mediate acquisition of resistance mutations to EGFR TKIs.
Keywords: EGFR, non-small cell lung cancer, genomics
Introduction:
EGFR-directed tyrosine kinase inhibitors (TKI) have dramatically improved outcomes for patients whose tumors harbor sensitizing EGFR alterations1. However, long-term disease control remains elusive for most patients due to the inevitable development of resistance, typically within 8–18 months2,3. Genetic analyses to-date have largely focused on identifying acquired mechanisms of resistance by profiling specimens at time of progression; these analyses have identified EGFR-dependent mechanisms of resistance (e.g. EGFR T790M and C797S mutations)4–6, and EGFR-independent mechanisms that allow tumors to bypass EGFR pathway inhibition (e.g. small cell transformation or acquisition of other driver alterations such as MET amplification7–9). These studies helped define how cancers escape inhibition by EGFR TKIs, but do not explain what determines when and how a tumor will develop resistance. Understanding the biological basis of differential time to progression could help explain why some tumors evolve resistance within weeks and others remain suppressed for years, and further inform novel therapeutic strategies to delay resistance evolution.
Our study builds on previous work examining the hypothesis that the genomic context of the driver EGFR mutation plays a role in when and how resistance develops. Prior studies analyzing the impact of pre-treatment co-mutations have identified worse outcomes associated with alterations in TP53, PIK3CA, PTEN, and others2,10–13. However, these analyses have been limited by small sequencing panels, incomplete clinical annotations, or small cohort size, and it is likely that the diversity of co-occurring interactions remains underexplored. Here, we have assembled a cohort of patients treated with EGFR TKIs assessed by targeted next-generation sequencing (NGS) with comprehensive clinical annotations to further explore the role of concurrent alterations in mediating differential outcomes to EGFR TKI therapy.
Materials and Methods
Study population:
We retrospectively identified all patients with targetable EGFR-mutated metastatic NSCLC who had been treated at the Dana-Farber Cancer Institute (DFCI) between 2005 and 2019, had tumors assessed by targeted hybrid capture NGS, and had been treated with an EGFR TKI for metastatic or recurrent disease. For uniformity of outcome assessment, patients with historically non-targetable EGFR alterations, including exon 20 insertions, were excluded, as were patients with baseline T790M alterations. All patients included in this study had consented to institutional review board-approved protocols and the study was conducted in accordance with the declaration of Helsinki.
We collected clinical characteristics and detailed treatment histories for all patients, including smoking status (never smokers: patients who smoked <100 cigarettes; former smokers: patients who quit >12 months before diagnosis; current smokers: patients who quit <12 months or still smoked at diagnosis). Tumor measurements and response assessment were performed retrospectively by a thoracic radiologist (M.N.) on the baseline and follow-up scans during EGFR TKI therapy using Response Evaluation In Solid Tumors (RECIST) version 1.1, to determine best overall response and date of progression, as previously published.14–16 Progression-free survival was defined as time from the start of either first TKI therapy (PFS1), or start of later-line osimertinib (PFS-Osi) to the date of disease progression or death. Patients alive without disease progression were censored on the date of their last contact. Overall survival was defined as time from start of first-line EGFR TKI to death from any cause, with censoring also defined at the date of last contact. Resistance mechanisms were classified as EGFR mutation (T790M, C797S), small cell transformation, bypass pathway, or other, which includes cases with no identified mechanism. Mechanism was assigned based on clinical record or direct assessment of post-treatment specimens17, and development of T790M mutation or small cell mutation at any point after first TKI was annotated.
Mutational analysis:
Genetic sequencing and mutation calling were performed as previously described using the DFCI OncoPanel platform, which has been extensively validated for both mutation and copy number calling18–21. Only tissue derived sequencing was included. Version 1 of OncoPanel captures 287 genes; version 2 captures 323 genes; and version 3 captures 462 genes. Mutations were considered functionally significant if they were a loss-of-function alteration, including nonsense, frameshift, insertion/deletion or splice site alteration. Missense mutations were considered functionally significant if they were either: 1) present in the OncoKB hotspots database22, 2) present in the Catalogue of Somatic Mutations in Cancer23 > 3 times, or 3) deleterious based on in silico prediction from the PolyPhen-2 (Polymorphism Phenotyping v2) prediction tool24. Copy number events classified as ‘high amplification’ or homozygous deletions were considered functionally significant.
Tumor mutational burden (TMB) was calculated as the number of nonsynonymous alterations per megabase (Mb) of genome examined, specific to the OncoPanel version employed25. Because segment length and copy ratio were only available for a subset of samples, proportion copy number altered (CNA load) was estimated as the number of copy number altered genes over the number of genes included in each panel version. Mutational signature analysis was performed using SigMA26, a validated method for mutational signature analysis from targeted panels that utilizes likelihood-based measures and machine-learning to account for low mutation counts. We ran SigMA using pre-calculated OncoPanel weights and previously identified lung adenocarcinoma signatures (COSMIC signatures 1, 2, 3, 4, 5, 13, 17a, 17b, 18, and 28). We report results for signatures 1 (5’methylcytosine deamination), 3 (homologous recombination defect), 4 (tobacco mutagenesis), 5 (T>C substitution), 2 and 13 (APOBEC) due to low frequency of mutations in the other signatures. Mutual exclusivity testing was performed using WexT27. Genes were assigned to pathways based on previous annotations10,28,29.
TCGA Analysis:
Previously published lung adenocarcinoma sequencing and clinical metadata from The Cancer Genome Atlas (TCGA)30 was obtained from cBioPortal31. TMB was calculated as the sum of nonsynonymous mutations. Genome doubling as previously calculated using ABSOLUTE was obtained32. Proportion copy number altered was calculated as the sum of length of segments with |copy ratio| > 0.2 over the length of all segments. DeconstructSigs33 was used for mutational signature identification in the TCGA cohort.
Statistical analysis:
Categorical and continuous variables were summarized descriptively using percentages and medians. The Wilcoxon-Rank Sum test and Kruskal-Wallis test were used to test for differences between continuous variables, and Fisher’s exact test was used to test for associations between categorical variables. Pre- and post-treatment enrichment was performed via logistic regression with adjustment for TMB. Progression-free and overall survival were estimated using Kaplan-Meier methodology. Log-rank tests were used to test for differences in event-time distributions, and Cox proportional hazards models were fitted to obtain estimates of hazard ratios in univariate and multivariate models. Analyses were performed in sample subsets according to treatment time point as indicated in the text; for comutation or cohort-wide analyses, the earliest sample available for each patient was identified to ensure that patients with multiple biopsies did not bias the results (single-sample cohort). All P values are two-sided and confidence intervals are at the 95% level. Statistical significance is defined as P<0.05. Multiple hypothesis-test correction was not performed on these exploratory analyses. All analyses were performed using R version 4.0.3.
Results
Patient population:
A total of 269 patients were identified who had both received targeted therapy for an actionable EGFR alteration and who had at least one tumor specimen that had undergone genomic profiling (Table 1; Supp Fig 1A; Supp Table 1). Due to the historical nature of this cohort, most patients were initially treated with first-generation EGFR TKIs; 94 patients were treated with osimertinib after progression (Table 1). Pre-treatment specimens were available in 189 patients (pre-treatment cohort); 91 patients had sequencing on or after treatment with first TKI therapy (post-TKI1 cohort), of whom 37 were later treated with osimertinib (pre-Osi cohort); 31 were sequenced after later-line osimertinib (post-Osi cohort)(Supp Fig 1B; Supp Table 2). Patients treated with first-line osimertinib (n=2) are included in the TKI1 cohort. Thirty patients had a paired sample before and after a single TKI, 5 of whom had biopsies at each time point (Supp Fig 1C). Radiographic progression on first TKI therapy was evaluable in n=264 patients, and on later-line osimertinib in n=82. Median PFS was 10 months on first-line TKI (PFS1), and 6 months on later-line osimertinib (PFS-Osi). Median overall survival was 31.2 months (Supp Fig 2A-C). There was no statistically significant difference in outcome by EGFR driver alteration (Supp Fig 2D-F). Of 124 patients with post-TKI1 resistance annotations, 98 (79%) had a detectable T790M mutation, though direct detection rates of T790M mutation in the post-treatment specimens were only 55% (62/113). 5 patients developed small cell (SCLC) transformation after first-line therapy, and another 5 developed SCLC transformation later in their treatment course.
Table 1.
Clinical characteristics of patients in DFCI EGFR cohort.
| Cohort Characteristics | Pretreatment Cohort | |
|---|---|---|
| Characteristic | No. (%) | No. (%) |
| No of patients | 269 | 189 |
| Med age at diagnosis | 62 (29–93) | 64 (29–93) |
| Sex | ||
| Male | 79 (30) | 57 (30) |
| Female | 190 (70) | 132 (70) |
| Smoking status | ||
| Ever | 106 (40) | 80 (42) |
| Never | 163 (60) | 109 (58) |
| EGFR mutation | ||
| Exon 19 deletion | 137 (51) | 92 (49) |
| L858R | 103 (38) | 74 (39) |
| Other | 29 (11) | 23 (12) |
| Stage at diagnosis | ||
| I, II | 37 (14) | 29 (15) |
| III | 22 (8) | 15 (8) |
| IVa | 69 (26) | 52 (28) |
| IVb | 141 (52) | 93 (49) |
| Line of therapy, first TKI | ||
| First | 226 (84) | 162 (86) |
| Second | 39 (14) | 24 (13) |
| Third or higher | 4 (1) | 3 (1) |
| 1st TKI | ||
| Erlotinib | 255 (94) | 178 (94) |
| Afatinib | 9 (3) | 7 (4) |
| Gefitinib, Icotinib | 3 (1) | 2 (1) |
| Osimertinib | 2 (1) | 2 (1) |
| 1st-line Resistance mechanism assessed | 124 (46) | 67 (35) |
| T790M mutation | 98 | 55 |
| Bypass pathway | 7 | 5 |
| Small cell transformation | 5 | 2 |
| Other/not detected | 14 | 5 |
| Received osimertinib after TKI resistance | 94 (35) | 50 (26) |
| Osimertinib Resistance mechanism assessed | 8 | 3 |
| C797S mutation | 4 | |
| Small cell transformation | 4 | 3 |
| Other/not detected | 1 | |
Concurrent genomic alterations and predictors of outcome.
The most frequently co-occurring mutations and copy number events in pre and post-TKI samples are shown in Supp Fig 3. TP53 alterations were the most common, followed by alterations in PRKDC, RB1, KMT2D, and NKX2–1. TMB and CNA load increased with line of therapy (Supp Fig 4A-B), though in paired samples only CNA load trended toward statistical significance, suggesting that the change in TMB with therapy may be variable (Supp Fig 4C-D).
Focusing on putatively functional alterations (Methods), no gene was enriched in post-treatment tumors after correcting for TMB (Supp Fig 4E-F), though in these exploratory analyses there were non-significant trends toward increases in MET, BRD4, CDKN2A/B, and KEAP1 alterations. Post-osimertinib specimens showed non-significant trends toward PTEN, KMT2A, KRAS, NOTCH2, and MYC alterations compared to post-TKI1 specimens. As expected, EGFR T790M mutations were enriched in post-TKI1 specimens, and EGFR C797S was enriched in post-osimertinib specimens (Supp Fig 4 G-H). Aggregation of genes into pathways and all post-treatment samples into one group demonstrated increased post-treatment alterations in splicing genes (p=0.011), and non-significant increases in structural proteins (p=0.057), cell cycle (p=0.11), insulin signaling (p=0.110), and PI3K/AKT pathway genes (p=0.126) (Supp Fig 4I).
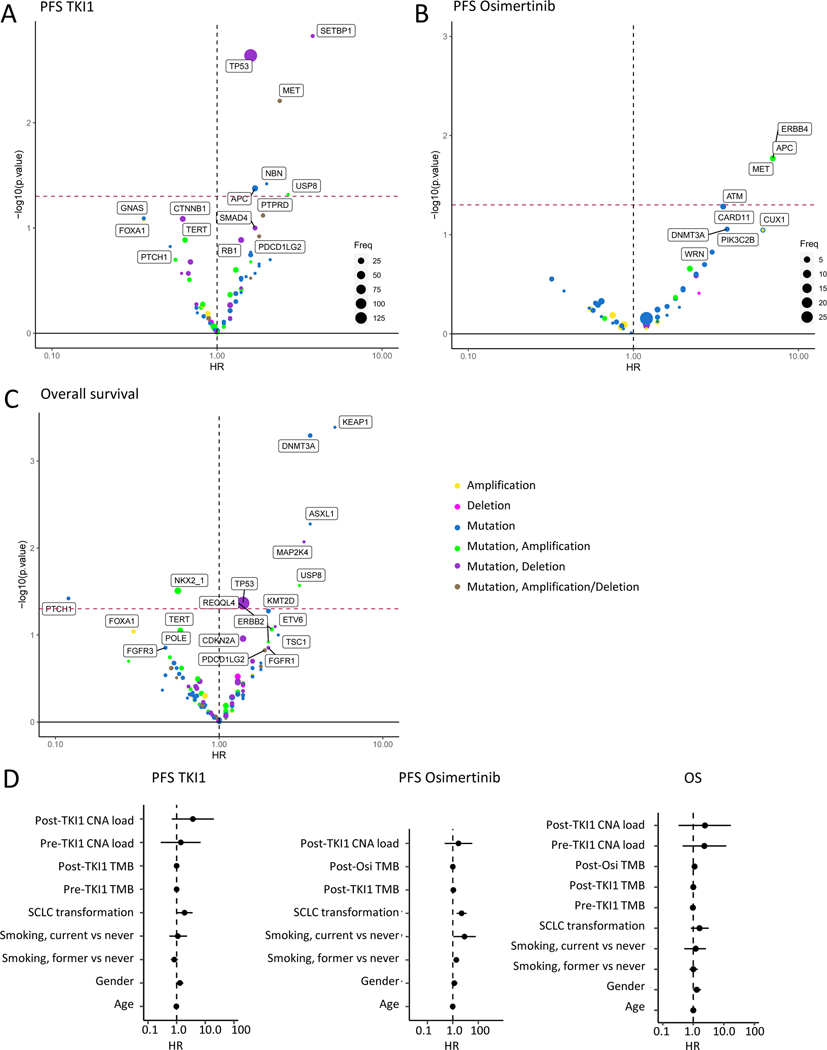
To identify genes whose concurrent alteration affects EGFR TKI outcomes, we performed cox-proportional hazards estimation of the effect of pre-treatment genetic changes on PFS1 (n=184). TP53 alterations were the most common event associated with reduced PFS1, but alterations in SETBP1 and MET also had a worse hazard ratio (Fig 1A). Focusing on patients with primary progressive disease, we observed that MET high amplifications were the only alteration enriched in these patients (Supp Fig 5A-B), though numbers were small. Other MET alterations, including splice site or low amplification, were not associated with PD (Supp Fig 5C). Analysis of post-progression, pre-osimertinib alterations associated with PFS-Osi (n=37) demonstrated worse outcomes in patients with MET, APC and ERBB4 alterations (Fig 1B). Considering samples across all treatment time points (single-sample cohort, n=269), alterations in TP53 were again the most frequent event associated with OS, but KEAP1 and DNMT3A alterations also corresponded with worse OS (Fig 1C). Forest plots for these analyses are shown in Supp Fig 6A-C. Analysis of clinicogenomic variables revealed no association between PFS1 and pre-treatment TMB or copy number burden, and a weak association with age (HR 0.98, 95% CI 0.97–0.99, p=0.0029). PFS-Osi associated with small cell transformation (HR 4.9, 95% CI 2–12, p=0.0007), smoking (Former vs never, HR 1.9, 95% CI 1.1–3.1, p=0.013), and post-Osi TMB (HR 1.1, 95% CI 1–1.2, p=0.023). No features were associated with OS (Fig 1D).
Figure 1. Genomic and clinical predictors of outcome to EGFR TKI therapy.
Association between co-occurring alterations and A. Progression-free survival (PFS) on first-line EGFR TKI (PFS1)(n=184), B. PFS on subsequent osimertinib (PFS-Osi)(n=37), and C. Overall survival (OS)(n=269). Hazard ratio is shown on the x-axis, -log10(p-value) from univariate cox-proportional hazards model is shown on the y-axis. Points are colored by the observed genetic events as indicated. Only genes altered in 5 or more samples are included in A & C, and in 2 or more samples in B. D. Forest plots of clinicogenomic variables and PFS1 (left), PFS-Osi (middle), and OS (right). CNA: copy number alteration; SCLC: small cell lung cancer; TKI: tyrosine kinase inhibitor; TMB: tumor mutational burden.
TP53 alterations associate with worse prognosis independent of resistance mechanism
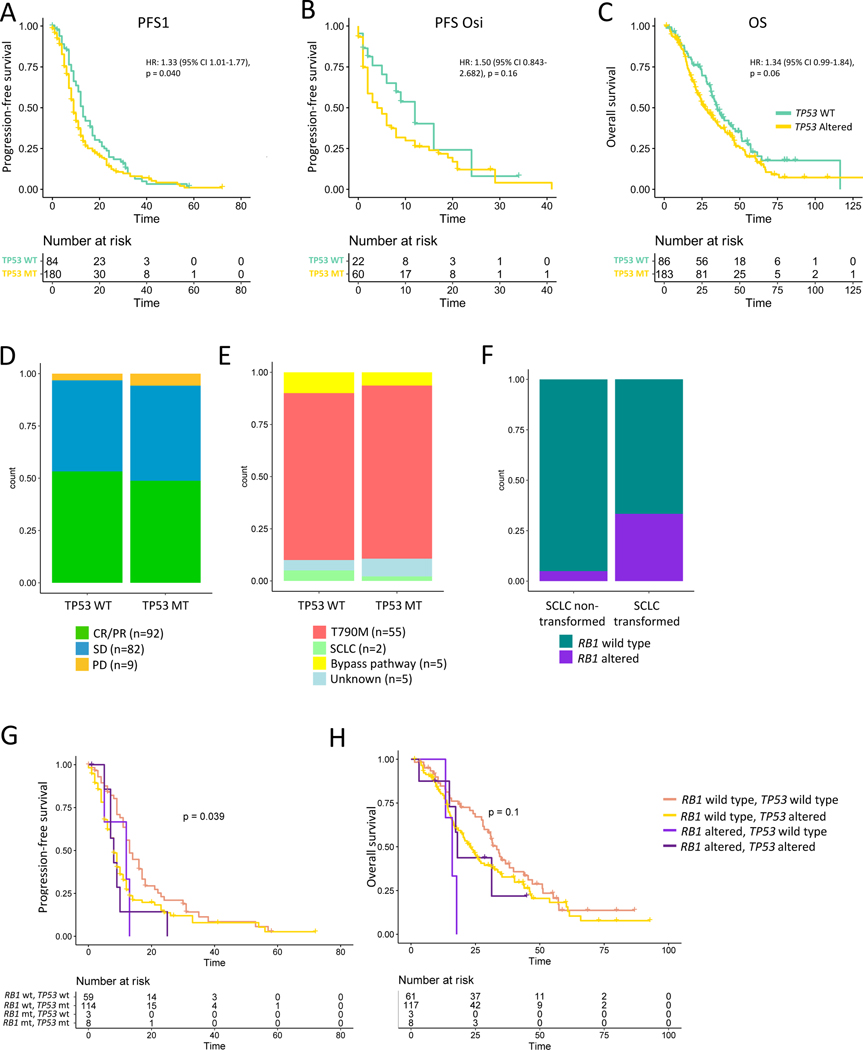
As TP53 alterations were present in significant numbers to allow further analysis, we decided to investigate this association further to better understand why TP53 associates with worse outcomes. We confirmed that, in addition to pre-treatment TP53 alterations (Fig 1A), patients with TP53 alterations detected at any time point had shorter PFS1 and trends toward reduced PFS-Osi and OS (Fig 2A-C). The same trends were present but less pronounced when analysis was restricted to disruptive TP53 mutations, defined as truncating mutations or nonsynonymous alterations affecting the L2-L3 region (Supp Fig 7A-C).34 Given prior reports associating TP53 exon 8 alterations and outcome13, we also investigated specific TP53 variants and exons; while analyses of specific mutational events are likely underpowered and confounded by differential event frequency, TP53 D281N mutations did associate with shorter PFS and OS (Supp Fig 7D-F). Exon-level events were not consistent across PFS1, PFS-Osi, and OS (Supp Fig 7G-I), and events in exon 8 were not statistically significant. While alterations in DNA binding domain exons (exons 5–8) trended toward worse outcomes, aggregated events in these regions did not associate with worse outcomes than those with TP53 mutations outside exons 5–8 (Supp Fig 7J-L).
Figure 2. Association between TP53 alteration, outcome, and resistance mechanism.
Association between TP53 alteration detected at any time point and A. First TKI progression-free survival (PFS1), B. PFS on subsequent osimertinib (PFS-Osi), and C. Overall survival (OS). D. Distribution of radiographic responses in patients with (MT) and without (WT) pre-treatment TP53 alteration (Fisher’s p-value=0.7541). E. Distribution of resistance mechanisms in pre-treatment TP53 MT vs WT patients (Fisher’s p-value=0.6483). F. Proportion of patients with pre-treatment Rb1 alterations in patients with and without small cell transformation at any time point. G. PFS1 and H. OS stratified by pre-treatment Rb1 and TP53 mutation status. TKI: tyrosine kinase inhibitor
We next asked whether TP53 alterations associate with worse outcomes by causing decreased therapeutic efficacy, manifest by higher rates of stable (SD) or progressive disease (PD). However, while almost all primary progressors harbored pre-treatment TP53 alterations (Supp Fig 5B), TP53 altered (MT) and wild type (WT) patients had similar RECIST distributions (Fig 2D; Supp Fig 8A). Even in patients with CR/PR as their best response, TP53-mutated patients had earlier progression (Supp Fig 8B-C), suggesting TP53 alterations affect the rate of resistance evolution rather than the likelihood or depth of initial response.
We next asked whether TP53 alterations associate with worse outcome by predisposing to small cell transformation. However, in both TP53 MT and WT patients the dominant mechanism of resistance was EGFR T790M (Fig 2E), and the negative effect of TP53 co-mutations was even more pronounced in the subset of patients that developed T790M (Supp Fig 8D-E), suggesting that TP53 alterations act independently of resistance mechanism. In contrast, pretreatment Rb1 loss significantly associated with SCLC transformation (Fig 2F) (Fisher’s p=0.04076), and patients with concurrent pre-treatment Rb1 loss and TP53 alteration had the worst outcomes, though numbers were small (Fig 2G-H). Among 13 patients who had a resistance biopsy and had TP53 and Rb1 alterations identified at any time point, 46% (6/13) had SCLC transformation, as did 2/4 (50%) patients with pre-treatment TP53 and Rb1 alterations.
While TP53 alterations were equally likely in samples harboring L858R vs exon 19 deletion, Rb1 loss was more likely to co-occur with exon 19 deletion, and, accordingly, these patients were more likely to develop small cell transformation (Supp Fig 9). These findings were largely recapitulated when TP53 mutations and Rb1 loss from any time point were considered (Supp Fig 10). Taken together, these analyses suggest that TP53 loss does not affect the initial efficacy of EGFR TKIs but does allows for the more rapid acquisition of resistance, independent of mechanism.
TP53 alterations associate with increased mutagenesis
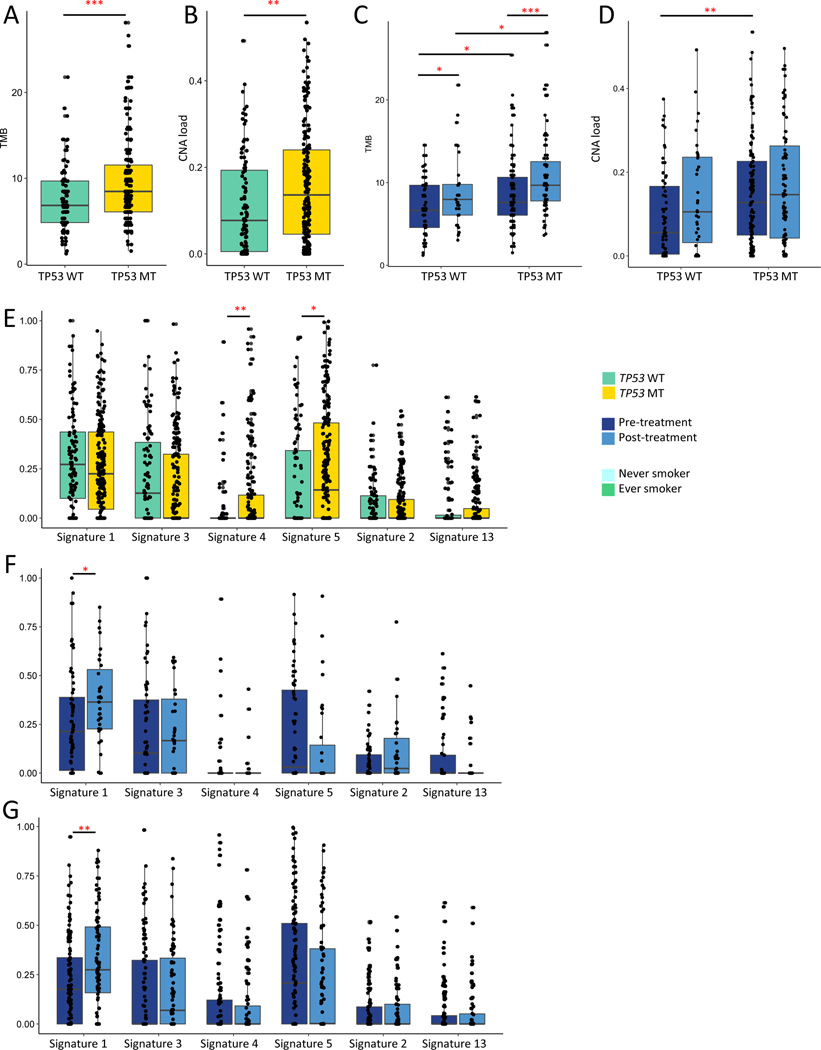
We next sought to understand how TP53 alterations promote resistance by examining whether TP53 alterations associate with increased genomic instability or specific mutagenesis patterns, as these processes might act as mechanism-independent engines for resistance evolution. Including all samples in the cohort to increase power (n=311), we observed that TP53 MT samples had a higher TMB (median 8.47 v 6.84, p=0.00051)(Fig 3A) and CNA load (median 0.136 v 0.077, p=0.0014)(Fig 3B). Because copy number measurement from panel data is imprecise, we validated these findings using published WES data from the TCGA30,32, which showed higher TMB, copy number load, and aneuploidy in TP53-mutated patients in general (Supp Fig 11A-C), and in EGFR-mutated samples more specifically (Supp Fig 11D-F). TP53 alterations associated with a more pronounced increase in TMB in post- vs pre-treatment samples (median 9.68 v 7.60, p=5.228e-05, Fig 3C); TKI treatment did not affect CNA load (Fig 3D).
Figure 3. TP53 alterations, mutation load, chromosomal instability and mutagenesis.
A. Tumor mutational burden (TMB) in TP53 wild type (WT) vs altered (MT) tumors (Wilcoxon p-value=0.00051). B. Proportion copy number altered (CNA) in TP53 WT vs MT (Wilcoxon p-value=0.0014). C. TMB in TP53 WT vs MT tumors stratified by treatment context (TP53 WT, pre vs post-treatment, p-value=0.042; TP53 MT, pre vs post-treatment, p-value=5.228e-05; pre-treatment, TP53 MT vs WT, p=0.02239; post-treatment, TP53 MT vs WT, p=0.01113). D. CNA load in TP53 WT vs MT tumors stratified by treatment context (TP53 WT, pre vs post-treatment, p-value=0.11; TP53 MT, pre vs post-treatment, p-value=0.45; pre-treatment, TP53 MT vs WT, p-value=0.00126). E. Proportion of mutations attributable to each signature in TP53 MT vs WT (Signature 4, p=0.0012; Signature 5, p=0.0282). F. Proportion of mutations attributable to each signature in pre vs post-treatment samples, TP53 WT samples (Signature 1, p=0.020. G. Proportion of mutations attributable each signature in pre vs post-treatment samples, TP53 MT samples; Signature 1, p=0.0024). All other comparisons, p > 0.05. *p<0.05; **p<0.01, ***p<0.001. N=311 samples, 212 with TP53 alterations, 99 without.
To further characterize the process by which TP53 MT tumors accumulate mutations, we performed mutational signature analysis of our DFCI cohort using SigMA26. Focusing on the most common signatures in this cohort, we observed that TP53 MT samples had a higher proportion of signature 4 (smoking) (p=0.0012) and signature 5 mutations (p=0.028)(Fig 3E). Validation in the TCGA dataset confirmed higher proportions of signature 4 alterations in TP53 MT samples, along with fewer signature 1 and more APOBEC-associated mutations (Supp Fig 11G). Stratification by disruptive vs non-disruptive TP53 mutations demonstrated highest mutation and copy number burden in tumors with disruptive TP53 mutations, and intermediate phenotypes in samples with non-disruptive TP53 mutations (Supp Fig 12).
To examine whether TP53 mutations promote resistance evolution through specific mutational processes, we stratified mutational signature proportions by treatment status. In both TP53 WT (Fig 3F) and mutated tumors (Fig 3G), the only significant change with treatment was an increased proportion of signature 1-related mutations (clock-like signature), with a non-significant trend toward decreased signature 5 mutations. These trends were present but not statistically significant in the subset of paired samples (Supp Fig 13A-C). Taken together, these findings suggest that TP53 alterations associate with increased genomic instability and mutagenic potential, but do not associate with distinct mutational processes.
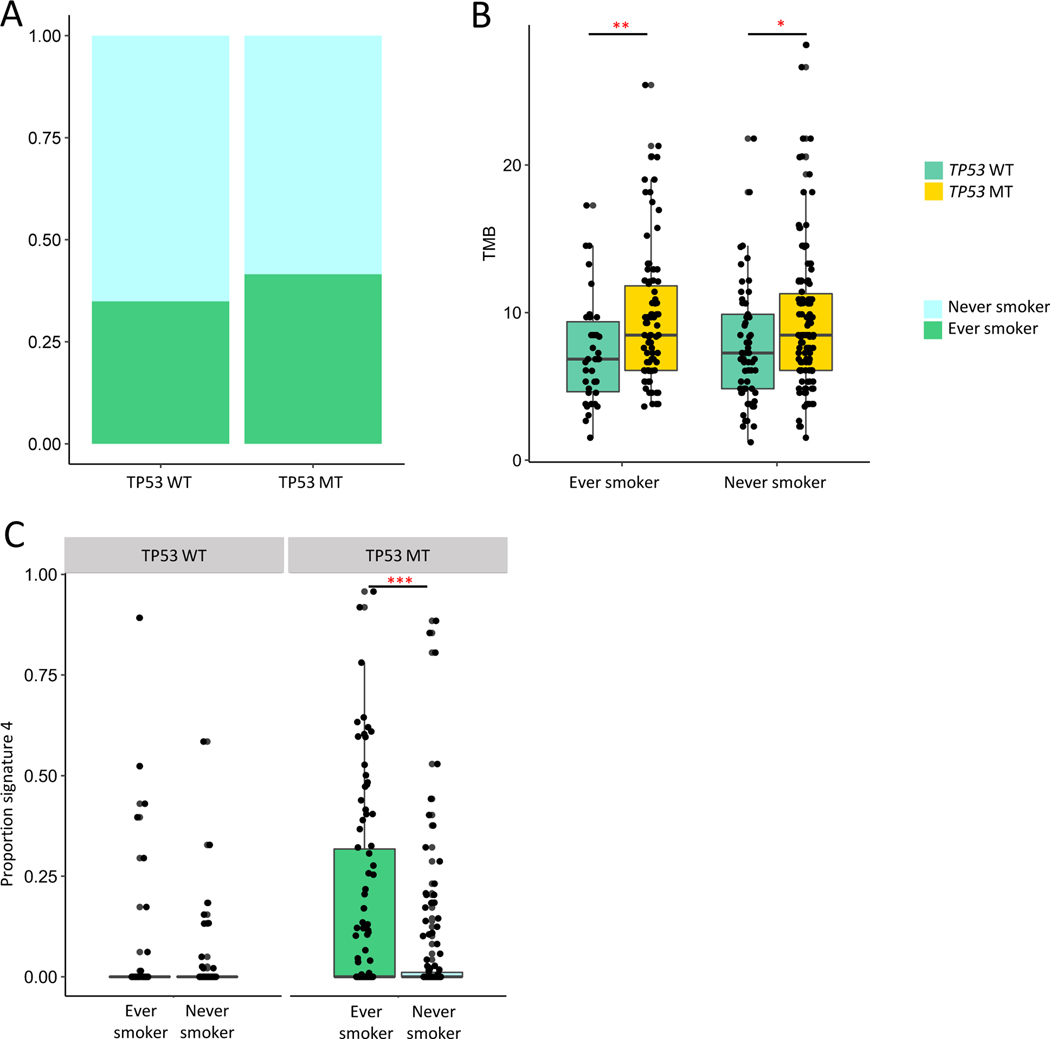
Focusing on the association with signature 4 alterations, despite more smoking-associated mutations, TP53 alterations were not more common in former/current smokers (Fig 4A; Table 2). To examine the joint effects of TP53 and smoking, we stratified patients by clinical smoking status. We observed that TP53 alterations associated with higher TMB regardless of smoking history (Fig 4B). As expected, current/former smokers with TP53 alterations had a higher proportion of signature-4 associated mutations (Fig 4C). However, among TP53 WT patients, even those patients with a clinical smoking history did not have a higher proportion of tobacco-attributable mutations than never smokers (Fig 4C). Joint analysis of signature 4, TP53 alteration status, and outcome demonstrated worse PFS1 in TP53 mutated patients both with and without signature 4 mutations. There was a non-significant trend toward worse outcomes in TP53 WT patients with signature 4 alterations, limited by the number of patients in this subgroup (Supp Fig 14).
Figure 4. TP53 alterations and smoking-associated mutagenesis.
A. Proportion of ever vs never smokers in TP53 WT vs MT patients (Chi-square p-value=0.3646). B. TMB in TP53 WT vs MT patients in ever (left) and never (right) smokers (Ever smoker, Wilcoxon p-value=0.006424; Never smoker, Wilcoxon p-value=0.0216). C. Proportion signature 4 mutations in ever vs never smokers, stratified by TP53 WT vs MT; TP53 WT, Wilcoxon p-value=0.46; TP53 MT, Wilcoxon p-value=0.0001. *p<0.05; **p<0.01, ***p<0.001. MT: altered; WT: wild type. N=269; 106 ever smokers, 163 never smokers.
Table 2.
Univariate logistic regression of clinical features associated with TP53 altered vs wild type status.
| Variable | Comparison | Beta | P-value |
|---|---|---|---|
| Gender | Male vs Female | 0.1888 | 0.517 |
| Smoking | Never vs ever | −0.2821 | 0.299 |
| Age | −0.03714 | 0.00189** | |
| Stage | Stage III v I,II | 0.3971 | 0.4723 |
| Stage IVA v I,II | 0.5306 | 0.2034 | |
| Stage IVB v I,II | 0.8346 | 0.0283* |
TP53 alterations define context specificity for effect of cell cycle alterations on outcome
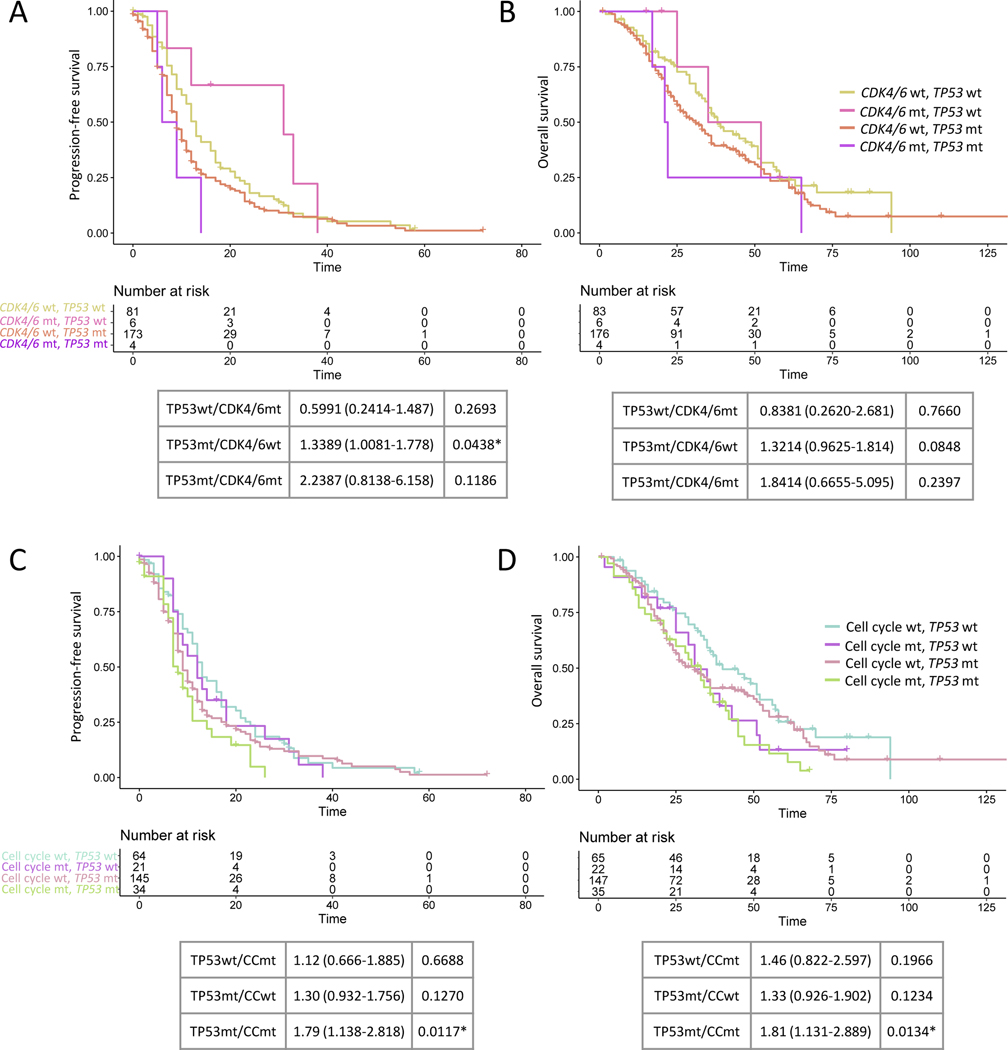
Finally, we sought to understand how TP53 interacts with other genes and pathways to situate these events in the context of other genomic events previously associated with outcome. Weighted mutual exclusivity assessment demonstrated strong mutual exclusivity between TP53 alterations and MDM2 amplification (weighted exclusivity, p= 4.79E-06)(Supp Fig 3; Supp Fig 15A). However, MDM2 amplification had no association with PFS or OS (Supp Fig 15B-D), suggesting MDM2 amplification alone does not recapitulate the effects of TP53 loss. Interestingly, despite multiple prior reports implicating cell cycle alterations in reduced PFS8,10, we also did not observe an independent effect of CDK4/6 amplifications specifically or cell cycle alterations more broadly on outcome (Supp Fig 15E-F). However, when considered in conjunction with TP53 alteration status, TP53/cell cycle co-mutated patients had worse outcomes (Fig 5A-D; Supp Fig 16A-B).
Figure 5. Cell cycle alterations and outcome.
A. First TKI progression-free survival (PFS1) and B. overall survival (OS) stratified by CDK4/6 and TP53 co-alteration status. Cox-proportional hazards for outcome relative to TP53 wild type/CDK4/6 wild type shown below. C. PFS1 and D. OS stratified by cell cycle and TP53 co-alteration status. Cox-proportional hazards for outcome relative to TP53 wild type/cell cycle wild type shown below. The most frequently altered cell cycle genes were considered and were: MDM2, CDK4, CDK6, CCND1, CCNE1, CDKN2A, CDKN2B, and EP300. MT: altered; TKI: tyrosine kinase inhibitor; WT: wild type.
Cox proportional hazards analysis of genes associated with outcome in TP53 WT tumors demonstrated shorter PFS in PIK3CA and ASXL1-mutated tumors, and shorter OS tumors harboring mutations in DNA epigenetic modifiers including ASXL1, DNMT3A, and KMT2D (Supp Fig 17A-B).
Discussion
In this study, we analyzed targeted sequencing data from a cohort of 269 patients with advanced, EGFR-mutated NSCLC treated with EGFR-directed TKIs. We found that pre-treatment alterations in TP53 were the most common concurrent genomic event associated with decreased first-line PFS and OS, and with a non-significant trend toward decreased PFS on later-line osimertinib. This finding is consistent with reports from several prior cohorts11–13,35–38, and indeed, TP53 alterations appear to be the genomic event most consistently associated with poor outcomes across different studies. However, the mechanism for why TP53 alterations associate with worse outcomes remains underexplored. While we cannot exclude an underlying prognostic effect, here we utilize our in-depth annotations to describe in more detail the clinical effects of TP53 alterations, demonstrating that TP53 alterations do not associate with decreased radiographic response or specific resistance mechanisms; rather, TP53 alterations associate with more rapid time to progression by any mechanism, even in those patients with the most favorable radiographic response.
Based on previous studies39, we hypothesized that TP53 alterations would facilitate more rapid resistance evolution by promoting cellular tolerance for genetic alterations. Consistent with this hypothesis, we found that TP53-altered tumors compared to wild type tumors had higher mutation burdens that became even more pronounced with TKI therapy, suggesting that TP53 alterations promote the acquisition of somatic mutations with therapy. While pre-treatment TMB did not associate with worse outcome, as had been previously suggested40, there was a statistically significant though small association between post-Osi TMB and PFS-Osi, suggesting that if TMB associates with outcome, it may be by facilitating a greater diversity of resistance mechanisms. Importantly, in our analysis of mutational signatures, we did not observe distinct mutational processes in TP53 MT compared to WT tumors41,42. Rather, it appeared that TP53 alterations facilitate the acquisition of mutations through those processes already underway, including tobacco-mediated mutagenesis in smokers, and spontaneous deamination of 5-methylcytosine over time. These trends were less apparent in the copy number space, where we observed higher pre-treatment copy number burden in TP53 MT tumors but more modest increases with therapy, suggesting acquisition of mutations rather than copy number events may be more important in this context. However, copy number calling from targeted NGS panels is less precise and these analyses would benefit from validation in paired samples assessed by WES.
Notably, the association between TP53 and outcome appeared independent of the location within the TP53 gene. While non-disruptive TP53 mutations34 associated with an intermediate TMB and CNA burden, suggestive of an intermediate phenotype or a more heterogeneous group, these patients had outcomes closer to patients with disruptive TP53 mutations than WT. Further studies will help determine whether this represents a thresholding effect of the TP53 phenotype on outcome, or distinct mechanisms driven by distinct TP53 genotypes. In contrast to prior reports implicating exon 813,38, we did not observe any robust associations between outcome and specific TP53 SNVs or exons; however, these subgroup analyses may be underpowered, and multiple exons demonstrated trends toward worse hazard ratios that might be significant in larger cohorts. Further exploration of the effect of specific TP53 alterations on chromosomal instability and treatment response to EGFR TKIs will help better characterize these associations.
We also note that TP53 alterations may interact with other genomic events to contribute to resistance evolution. In contrast to prior studies8,10,11, we did not identify an independent association between cell cycle events and worse outcomes, despite having an equal or greater number of events in our cohort. We did note, however, that patients with compound cell cycle and TP53 alterations had worse outcomes than dual wild type patients, suggesting that loss of cell cycle checkpoints in the context of increased tolerance of genetic changes may further accelerate cell turnover and resistance acquisition. These findings suggest that ongoing studies combining osimertinib with CDK4/6 inhibition (NCT03455829) may be particularly effective in TP53-mutated patients.
Our study also confirms several previous findings and identifies potential novel associations. The association between baseline MET amplification and reduced PFS has been previously demonstrated, and these findings together provide a strong rationale for investigating upfront therapy with dual EGFR and MET inhibition in these patients. The association between KEAP1 alterations and reduced OS validates recent experimental and limited patient data demonstrating decreased duration of EGFR TKI therapy in KEAP1/TP53 co-mutated patients43. Additionally, while most concurrent genomic events associated with reduced outcome occurred in the context of TP53 loss, our data also suggest that alterations in PIK3CA and epigenetic modifiers may be TP53-independent mediators of differential benefit. More work in larger cohorts will be necessary to validate and further explore these findings.
Our analysis has several limitations. Despite the large size of the cohort, many of the single-gene observations occurred in a limited number of samples. Consistent with prior studies35, we did not adjust our analyses for multiple comparisons in order to facilitate hypothesis generation, and consequently the findings reported here need additional validation in other cohorts and functional studies. Additionally, mutational signature analysis in panel-based data can be imprecise; we accounted for this limitation by using a validated algorithm that incorporates a panel-specific error model, and reassuringly observed similar trends in WES from TCGA. Validation of treatment effects on copy number and mutational signature is not currently possible due to limited WES cohorts of treated tumors, but will be important as such cohorts become available. Finally, as a retrospective analysis, this study has multiple intrinsic limitations, including variable response and resistance assessment, and historic treatment patterns with first- or second-generation TKIs as first TKI followed by osimertinib after resistance. We note that many of the trends we observed for PFS1 were present but underpowered in the pre-osimertinib specimens, and published studies of resistance to first-line osimertinib have implicated similar mechanisms in different distributions, suggesting that the same trends and overall biological pathways may be implicated8,44–46. Nonetheless, as cohorts of patients treated with first-line osimertinib mature, it will be important to validate that the same patterns hold.
In conclusion, our analysis further defines the effects of concurrent mutations on outcomes in EGFR mutated NSCLC, suggesting an important role for TP53 mutations in facilitating the acquisition of resistance. Our analyses further suggest that the deleterious effects of other concurrent mutations may be contingent on TP53 mutation status and should be studied in this context. These findings also have clear therapeutic implications; while compounds targeting or restoring TP53 function are of obvious interest47, these remain under investigation. However, in the short-term, these findings provide a clear rationale for trialing treatment intensification in TP53 mutant patients with chemotherapy or other therapies such as VEGF inhibitors, which are currently under active investigation (NCT04695925)48. Further exploration of these and other concurrent mutations may provide important prognostic information for patients and clinicians, and may facilitate combination therapeutic strategies to forestall resistance and prolong the duration of initial benefit from EGFR targeted therapy.
Supplementary Material
Acknowledgements and Funding:
This study was supported by the National Cancer Institute (R35 CA220497) (P.A.J.), the American Cancer Society (CRP-17-111-01-CDD) (P.A.J.), the Gohl Family Lung Cancer Research Fund (P.A.J.) and the Mock Family Fund for Lung Cancer Research (P.A.J.); NIH R01CA203636 (M.N.), U01CA209414 (M.N.); the Mark Foundation Damon Runyon Physician-Scientist Fellowship (N.I.V.), Conquer Cancer Foundation YIA (N.I.V.), Society for Immunotherapy of Cancer-Genentech Fellowship (N.I.V.); NIH/NCI U01-CA213273 (J.V.H.), University of Texas SPORE in Lung Cancer P5-CA070907 (J.V.H.); ASCO Conquer Cancer Foundation Career Development Award (X.L.), Rexanna’s Foundation for Fighting Lung Cancer (X.L., N.I.V.); R01CA227388 (E.M.V)
Conflict-of-interest statement:
Dr. Van Allen reports personal fees from Tango Therapeutics, personal fees from Genome Medical, personal fees from Invitae, personal fees from Enara Bio, personal fees from Janssen, personal fees from Manifold Bio, personal fees from Monte Rosa Therapeutics, grants from Novartis, grants from BMS IION, grants from Sanofi, personal fees from Foaley & Hoag, outside the submitted work; In addition, Dr. Van Allen has a patent Institutional patents filed on chromatin mutations and immunotherapy response, and methods for clinical interpretation pending. Dr. Heymach reports grants and personal fees from AstraZeneca, grants and personal fees from GlaxoSmithKline, personal fees from Boehringer Ingelheim, personal fees from Bristol Myers Squibb, personal fees from Merck, personal fees from Catalyst, personal fees from Guardant Health, personal fees from Foundation Medicine, personal fees from Hengrui, personal fees from Lilly, personal fees from Novartis, personal fees from EMD Serono, personal fees from Sanofi, personal fees from Biotree, personal fees from Takeda, outside the submitted work; In addition, Dr. Heymach has a patent PCT/US2019/022067 pending, and a patent PCT/US2017/062326 and U.S. Provisional Patent Application Nos. 62/423,732; 62/427,692 and 62/572,716, with royalties paid to The University of Texas System Board of Regents. L.M.S. reports consulting fees from EMD Serono, scientific advisory board roles for Loxo Oncology and Foghorn Therapeutics, and honorarium from Astra Zeneca. M.N. serves as a consultant to Daiichi Sankyo, AstraZeneca, and received institutional research grant from Merck, Canon Medical Systems, AstraZeneca, Daiichi Sankyo. Dr. Vokes reports consulting/advisory fees from Sanofi/Genzyme, Oncocyte, and Lilly. Dr Janne has received consulting fees from AstraZeneca, Boehringer-Ingelheim, Pfizer, Roche/Genentech, Takeda Oncology, ACEA Biosciences, Eli Lilly and Company, Araxes Pharma, Ignyta, Mirati Therapeutics, Novartis, LOXO Oncology, Daiichi Sankyo, Sanofi Oncology, Voronoi, SFJ Pharmaceuticals, Takeda Oncology, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Esai, Biocartis, Allorion Therapeutics, Accutar Biotech and Abbvie; receives post-marketing royalties from DFCI owned intellectual property on EGFR mutations licensed to Lab Corp; has sponsored research agreements with AstraZeneca, Daichi-Sankyo, PUMA, Boehringer Ingelheim, Eli Lilly and Company, Revolution Medicines and Astellas Pharmaceuticals; and has stock ownership in LOXO Oncology and Gatekeeper Pharmaceuticals. Dr. Le reports grants and personal fees from Eli Lilly, personal fees from AstraZeneca, grants and personal fees from Boehringer Ingelheim, personal fees from Spectrum Pharmaceutics, personal fees from Bristol Myers Squibb, personal fees from Celgene, personal fees from EMD Serono, personal fees from Janssen, personal fees from Hengrui Therapeutics, personal fees from Novartis, personal fees from Daiichi Sankyo, outside the submitted work. Emily Chambers reports personal fees from The Broad Institute of Harvard and MIT, personal fees from Takeda Pharmaceuticals, outside the submitted work.
References:
- 1.Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2020;382(1):41–50. doi: 10.1056/NEJMoa1913662 [DOI] [PubMed] [Google Scholar]
- 2.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17(6):1616–1622. doi: 10.1158/1078-0432.CCR-10-2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–562. doi: 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res. 2018;24(24):6195–6203. doi: 10.1158/1078-0432.CCR-18-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol. 2017;35(26):3065–3074. doi: 10.1200/JCO.2016.71.9096 [DOI] [PubMed] [Google Scholar]
- 10.Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–1704. doi: 10.1038/ng.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Lee B, Shim JH, et al. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J Thorac Oncol. 2019;14(2):193–202. doi: 10.1016/j.jtho.2018.10.150 [DOI] [PubMed] [Google Scholar]
- 12.VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer. 2017;106:17–21. doi: 10.1016/j.lungcan.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res. 2017;23(9):2195–2202. doi: 10.1158/1078-0432.CCR-16-0966 [DOI] [PubMed] [Google Scholar]
- 14.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 15.Nishino M, Cardarella S, Jackman DM, et al. RECIST 1.1 in NSCLC patients with EGFR mutations treated with EGFR tyrosine kinase inhibitors: comparison with RECIST 1.0. AJR Am J Roentgenol. 2013;201(1):W64–71. doi: 10.2214/ajr.12.9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino M, Cardarella S, Dahlberg SE, et al. Radiographic assessment and therapeutic decisions at RECIST progression in EGFR-mutant NSCLC treated with EGFR tyrosine kinase inhibitors. Lung Cancer. 2013;79(3):283–288. doi: 10.1016/j.lungcan.2012.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redig AJ, Taibi M, Oxnard GR, et al. A phase II trial of erlotinib for EGFR mutant NSCLC to prospectively assess biopsy feasibility and acquired resistance at disease progression. J Clin Oncol. 2015;33(15_suppl):8076–8076. doi: 10.1200/jco.2015.33.15_suppl.8076 [DOI] [Google Scholar]
- 18.Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med. 2017;141(6):751–758. doi: 10.5858/arpa.2016-0527-OA [DOI] [PubMed] [Google Scholar]
- 19.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19). doi: 10.1172/jci.insight.87062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cenaj O, Ligon AH, Hornick JL, Sholl LM. Detection of ERBB2 Amplification by Next-Generation Sequencing Predicts HER2 Expression in Colorectal Carcinoma. Am J Clin Pathol. 2019;152(1):97–108. doi: 10.1093/ajcp/aqz031 [DOI] [PubMed] [Google Scholar]
- 21.Robinson CL, Harrison BT, Ligon AH, et al. Detection of ERBB2 amplification in uterine serous carcinoma by next-generation sequencing: an approach highly concordant with standard assays. Mod Pathol Off J U S Can Acad Pathol Inc. 2021;34(3):603–612. doi: 10.1038/s41379-020-00695-5 [DOI] [PubMed] [Google Scholar]
- 22.Chakravarty D, Gao J, Phillips S, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;(1):1–16. doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed]
- 23.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vokes NI, Liu D, Ricciuti B, et al. Harmonization of Tumor Mutational Burden Quantification and Association With Response to Immune Checkpoint Blockade in Non–Small-Cell Lung Cancer. JCO Precis Oncol. 2019;(3):1–12. doi: 10.1200/PO.19.00171 [DOI] [PMC free article] [PubMed]
- 26.Gulhan DC, Lee JJK, Melloni GEM, Cortés-Ciriano I, Park PJ. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet. 2019;51(5):912–919. doi: 10.1038/s41588-019-0390-2 [DOI] [PubMed] [Google Scholar]
- 27.Leiserson MD, Reyna MA, Raphael BJ. A weighted exact test for mutually exclusive mutations in cancer. Bioinformatics. 2016;32(17):i736–i745. doi: 10.1093/bioinformatics/btw462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding L, Bailey MH, Porta-Pardo E, et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell. 2018;173(2):305–320.e10. doi: 10.1016/j.cell.2018.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–337.e10. doi: 10.1016/j.cell.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33(4):676–689.e3. doi: 10.1016/j.ccell.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17(1):31. doi: 10.1186/s13059-016-0893-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poeta ML, Manola J, Goldwasser MA, et al. TP53 Mutations and Survival in Squamous-Cell Carcinoma of the Head and Neck. 10.1056/NEJMoa073770. doi: 10.1056/NEJMoa073770 [DOI] [PMC free article] [PubMed]
- 35.Yu HA, Suzawa K, Jordan E, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res. 2018;24(13):3108–3118. doi: 10.1158/1078-0432.CCR-17-2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsui DWY, Murtaza M, Wong ASC, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR‐targeted therapies in non‐small cell lung cancer. EMBO Mol Med. 2018;10(6):e7945. doi: 10.15252/emmm.201707945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer Amst Neth. 2017;111:23–29. doi: 10.1016/j.lungcan.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 38.Molina-Vila MA, Bertran-Alamillo J, Gascó A, et al. Nondisruptive p53 Mutations Are Associated with Shorter Survival in Patients with Advanced Non–Small Cell Lung Cancer. Clin Cancer Res. 2014;20(17):4647–4659. doi: 10.1158/1078-0432.CCR-13-2391 [DOI] [PubMed] [Google Scholar]
- 39.Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR -mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9(1):216. doi: 10.1038/s41467-017-02584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Offin M, Rizvi H, Tenet M, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res. 2019;25(3):1063–1069. doi: 10.1158/1078-0432.CCR-18-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–256. doi: 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foggetti G, Li C, Cai H, et al. Genetic determinants of EGFR-Driven Lung Cancer Growth and Therapeutic Response In Vivo. Cancer Discov. Published online January 1, 2021. doi: 10.1158/2159-8290.CD-20-1385 [DOI] [PMC free article] [PubMed]
- 44.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018;4(11):1527–1534. doi: 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res. 2020;26(11):2654–2663. doi: 10.1158/1078-0432.CCR-19-3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramalingam SS, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29:viii740. doi: 10.1093/annonc/mdy424.063 [DOI] [Google Scholar]
- 47.Yang C, Lou G, Jin WL. The arsenal of TP53 mutants therapies: neoantigens and bispecific antibodies. Signal Transduct Target Ther. 2021;6(1):1–2. doi: 10.1038/s41392-021-00635-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore S, Wheatley-Price P. EGFR Combination Therapy Should Become the New Standard First-Line Treatment in Advanced EGFR-Mutant NSCLC. J Thorac Oncol. 2021;16(11):1788–1792. doi: 10.1016/j.jtho.2021.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.