The tumor suppressor gene CDKN2A encodes p16INK4a and p14ARF which regulate cell cycle progression, and is frequently deleted in cancer. Homozygous deletion (homdel) of CDKN2A has been incorporated as a Grade 4 defining criterion for IDH-mutant gliomas without 1p-/19q-codeletion (IDHmut-noncodel) in the WHO CNS5 classification.1 However, the prognostic value of hemizygous CDKN2A deletions (hemidel) is unclear. We assessed copy number variation profiles of initial (n = 1256) and recurrent (n = 494) IDHmut gliomas to evaluate the impact of CDKN2A hemidel on overall survival (OS) outcomes (Figure 1).
Figure 1.
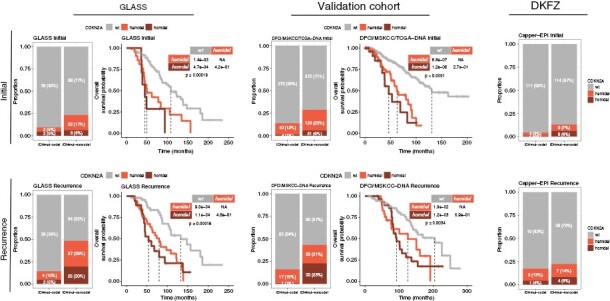
Distribution of CDKN2A homozygous deletion, hemizygous deletion and wild type (wt) across six IDHmut glioma cohorts. The discovery cohort consists of the GLASS DNAseq and DNA methylation profiling datasets. The validation cohort consists of the DFCI, MSKCC, and TCGA datasets, with further validation in DKFZ methylation profiling dataset. Survival analyses are specific to IDHmut-noncodel gliomas. Upper panel depicts initial cases; lower panel depicts recurrent cases. Stacked bar plots showing the total number and relative proportion of cases with CDKN2A homozygous deletion, hemizygous deletion and wt, separated by molecular subtype into columns representing IDHmut-codel and IDHmut-noncodel. Fisher’s exact test was applied as a statistical test to compare initial and recurrent gliomas. Kaplan–Meier survival plots depict overall survival probability (y-axis) and survival time (x-axis). Overall survival indicates the time from diagnosis to death or last date of follow-up as censoring. Global log-rank test was applied for comparison of 3 groups, and pairwise log-rank test was applied for comparison of 2 groups. Note that the TCGA cohort included only initial glioma cases. No survival data was available for the DKFZ cohort. GLASS, Glioma Longitudinal Analysis Consortium; DFCI, Dana-Farber Cancer Institute; MSKCC, Memorial Sloan Kettering Cancer Center; TCGA, The Cancer Genome Atlas; DKFZ, Deutsches Krebsforschungszentrum.
Using the GLASS primary/recurrent glioma dataset,2,3 we inferred CDKN2A status using DNAseq (n = 240) and DNA methylation array data (n = 100). Longitudinal analyses of IDH-mutant gliomas without 1p-/19q-codeletion (IDHmut-noncodels) revealed a significant increase in CDKN2A homdel at recurrence, as previously described,4 from n = 8 to n = 25 (6% to 20%, P = .0002, Fisher’s-exact test), but also in CDKN2A hemidel from n = 22 to n = 37 (17% to 29%, P = .0037). Overlapping DNAseq and DNA methylation profiles, available for n = 29 cases, illustrated strong concordance of log2-CDKN2A values between these platforms (R = 0.82, P = 4.4e-13, Pearson correlation).
We inferred tumor purity from DNAseq and DNA methylation array data. Neither the CDKN2A status derived from DNA methylation array (P = .27) nor from DNAseq (P = .61) was affected by tumor purity, indicating a robust assessment of CDKN2A status independent of platform or purity. Importantly, CDKN2A hemidels were present in a large proportion of cancer cells (median cancer cell fraction = 0.98), indicating a high degree of clonality. These data reflect the observation that CDKN2A hemidels are not misclassifications or merely subcloncal homozygous deletions. Similar distribution analyses of IDHmut-codels demonstrated that the increase in CDKN2A homdel and hemidel at recurrence is specific to IDHmut-noncodels.
These findings prompted us to characterize the impact of CDKN2A hemidel on overall survival, defined as time from diagnosis to death or last follow-up, in IDHmut-noncodel gliomas. The presence of CDKN2A hemidel was significantly associated with poor OS at initial diagnosis (P = .00019, log-rank test) and recurrence (P = 00018). This association remained significant in a multivariable Cox regression model adjusting for age and treatment with radiotherapy and/or alkylating agents: CDKN2A hemidel HR 2.15 (95% CI: 1.15–4.03, P = .02) and homdel HR 3.99 (95% CI: 1.83–8.70, P < .001).
We then sought to validate our observations using the DFCI,5 MSKCC,6 and TCGA7 datasets, which were combined to form a discovery cohort. For the TCGA dataset,7 cases included in GLASS were excluded, and only initial cases were included. In the DFCI dataset,5 we noted a higher rate of CDKN2A hemidel among IDHmut-noncodels recurrences compared to non-matched primaries (14% to 23%, P = .025, Fishers’ exact test). Analyses of the MSKCC dataset6 also revealed a similar distribution of CDKN2A status and an increase of CDKN2A homdel/hemidel at recurrence, particularly in IDHmut-noncodels. Clinical outcome analyses of the discovery cohort demonstrated worse OS in initial (P < .0001) and recurrent (P < .0034) CDKN2A deleted IDHmut-noncodel gliomas. This association remained significant in a multivariable Cox regression model that accounted for age, treatment and center: CDKN2A hemidel HR 2.08 (95% CI: 1.41–3.08, P < .001) and homdel HR 2.45 (95% CI: 1.54–3.88, P < .001).
Taken together, we have confirmed in four independent datasets4–7 that CDKN2A hemidel was associated with worse OS in IDHmut-noncodel gliomas independent of time point. These results highlight the clinical relevance of assessing CDKN2A status at initial diagnosis and at recurrence.
CDKN2A status can be accurately assessed by DNAseq, including the panel sequencing used in the DFCI and MSKCC datasets. DNA methylation array profiling is also increasingly being integrated into clinical workflows8 and can be used to derive DNA copy number profiles, enabling assessment of the CDKN2A status in gliomas. We analyzed the DKFZ DNA methylation dataset8 and found that the frequency of CDKN2A hemidels more than doubled in recurrent cases (P = .016), suggesting that DNA methylation profiles are amenable to detection of CDKN2A hemidel.
As a technical note, the CDKN2A copy number status was obtained from previous publications when available.2,4,5,7 For the MSKCC6 and the DNA methylation array8 datasets, the distinction between homozygous and hemizygous deletions was not available, and we applied conservative cutoffs defining a log2 copy ratio ≤ −1-1 as homozygous deletion and −1.1 < log2 copy ratio ≤ −0.4 as hemizygous deletion, consistent with the stringent criteria of the other datasets.2,4,5,7
In conclusion, our analysis shows that the presence of CDKN2A hemidel at initial diagnosis and recurrence is associated with significantly worse OS in IDHmut-noncodel gliomas. Similar to CDKN2A homdel,4CDKN2A hemidel is enriched in post-treatment, recurrent IDHmut-noncodel gliomas, confirming the value of CDKN2A status (re-) assessment in recurrences. Our results highlight the importance of incorporating CDKN2A status into the diagnostic work-up to inform prognosis and treatment strategies.
Contributor Information
Emre Kocakavuk, Department of Neurosurgery, Yale School of Medicine, New Haven, Connecticut, USA; Department of Hematology and Stem Cell Transplantation, West German Cancer Center (WTZ), National Center for Tumor Diseases (NCT) West, University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Kevin C Johnson, Department of Neurosurgery, Yale School of Medicine, New Haven, Connecticut, USA.
Thais S Sabedot, Department of Neurosurgery, Hermelin Brain Tumor Center, Henry Ford Hospital, Detroit, Michigan, USA.
Hans Christian Reinhardt, Department of Hematology and Stem Cell Transplantation, West German Cancer Center (WTZ), National Center for Tumor Diseases (NCT) West, University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Houtan Noushmehr, Department of Neurosurgery, Hermelin Brain Tumor Center, Henry Ford Hospital, Detroit, Michigan, USA.
Roel G W Verhaak, Department of Neurosurgery, Yale School of Medicine, New Haven, Connecticut, USA; Department of Neurosurgery, Amsterdam University Medical Center, Amsterdam, The Netherlands.
Funding
This work is supported by National Institutes of Health grants, R01CA237208, R21NS114873, R21CA256575, U2CCA252979, and P30CA034196 (RGWV). E.K. is supported by the Clinician Scientist Fellowship (FU 356/12-1) funded by DFG (Deutsche Forschungsgemeinschaft) and UMEA (University Medicine Clinician Scientist Academy) and a recipient of the Forbeck Scholar Award by the William Guy Forbeck Research Foundation.
Conflict of interests statement
RGWV is a co-founder of Boundless Bio.
References
- 1. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barthel FP, Johnson KC, Varn FS, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576(7785):112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varn FS, Johnson KC, Martinek J, et al. Glioma progression is shaped by genetic evolution and microenvironment interactions. Cell. 2022;185(12):2184–2199.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kocakavuk E, Anderson KJ, Varn FS, et al. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat Genet. 2021;53(7):1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jonsson P, Lin AL, Young RJ, et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res. 2019;25(18):5537–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]


