Abstract
The present report describes an analysis of two virulence genes of Helicobacter pylori. Parts of the cagA gene, as well as parts from the signal (s) and middle (m) regions of the mosaic vacA gene, were amplified with biotin-labelled PCR primers and the products were subsequently analyzed by a single-step reverse hybridization line probe assay (LiPA). This assay comprises a strip containing multiple specific probes for the vacA s region (s1a, s1b, and s2 alleles), the vacA m region (m1 and m2 alleles), and the cagA gene. A total of 103 H. pylori-positive materials, including cultured isolates, gastric biopsy specimens, and surgical specimens from patients living in Portugal (n = 55) and The Netherlands (n = 48) were tested by the PCR-LiPA. cagA was detected in 84 and 73% of the Portuguese and Dutch patients, respectively. vacA typing results, as determined by reverse hybridization, were completely concordant with those of sequence analysis. Most Portuguese patients (72%) contained type s1b, whereas most Dutch patients (61%) contained type s1a (P < 0.001). The method is also very effective at detecting the presence of multiple genotypes in a single biopsy specimen. The prevalence of multiple strains in Portuguese patient samples was significantly higher (29%) than that in Dutch patient samples (8%) (P = 0.001). There was a significant association between the presence of ulcers or gastric carcinoma and the presence of vacA type s1 (s1a or s1b; P = 0.008) and cagA (P = 0.003) genes.
Helicobacter pylori is an important human pathogen that causes chronic gastritis and that is associated with the development of peptic ulcer disease, atrophic gastritis, and gastric malignancies (6). The establishment and maintenance of H. pylori colonization in the stomach depend on several host and bacterial factors. Expression of specific host antigens, such as the Lewis-type blood group antigens, may play an important role (2, 23). H. pylori itself secretes several enzymes, such as urease, that facilitate its survival in the acidic environment of the stomach (6).
The genetic variability among H. pylori strains is relatively high (16) and can be assessed by various methods, such as restriction fragment length polymorphism analysis and PCR fingerprinting (17, 19, 28, 29). Analysis of this overall genomic variability is useful for epidemiological studies, but this genomic variability is not related to the pathogenicity of H. pylori strains. Recently, specific bacterial genes that are associated with phenotypic strain virulence have been described (5, 7, 22). Approximately 50 to 60% of the H. pylori strains contain the cytotoxin-associated (cagA) gene and consequently produce the 128-kDa cagA protein (10). The presence of cagA is associated with duodenal ulceration, gastric mucosal atrophy, and gastric cancer (7). cagA is part of a larger genomic entity, designated the pathogenicity (cag) island (9), which contains multiple genes that are related to the virulence and pathogenicity of the H. pylori strain. Therefore, the presence of cagA can be considered a marker for this genomic pathogenicity island and is associated with more virulent H. pylori strains.
Another virulence factor, produced by approximately 50% of the H. pylori strains, is a cytotoxin that induces the formation of vacuoles in mammalian cells in vitro and that leads to cell death (20). This toxin is encoded by the vacA gene, which is present in virtually all H. pylori strains (11, 13, 25). Recently, the existence of different allelic variants in two parts of this gene has been described (3, 12) (Fig. 1). The N-terminal signal (s) region occurs as either an s1a, s1b, or s2 allele. The middle (m) region is present as an m1 or an m2 allele. This mosaic structure of the vacA gene (Fig. 1) accounts for the differences in the cytotoxin production between strains. The s1/m1 type strains produce high levels of toxin, s1/m2 type strains produce low to moderate levels of toxin, whereas s2/m2 strains do not produce any active toxin at all. Strains containing s2/m1 have not yet been found. The vacA s and m regions appear to have different clinical relevance (4). vacA s1a strains are associated with greater antral mucosal neutrophil and lymphocyte infiltrates than type s1b or s2 strains. vacA type m1 strains are associated with greater gastric epithelial damage than type m2 strains. Duodenal ulcer disease appears to be more prevalent in patients infected or colonized with type s1a strains than in patients colonized with type s1b and s2 strains.
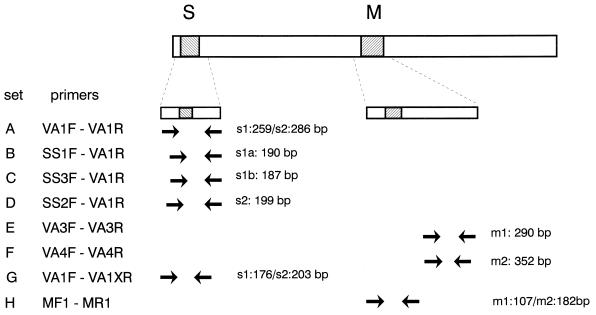
FIG. 1.
Mosaic structure of the vacA gene comprising the variable s and m regions. The locations of the various primer target sites are indicated by arrows, and the expected amplimer sizes are shown. Primer sets A through F were described by Atherton et al. (3). Primer sets G and H were described in the present study and permit general amplification of the vacA s and m regions with single primer sets.
The aim of the present study was to develop an efficient DNA-based detection and typing system for the virulence-associated cagA and vacA genes of H. pylori either in cultured isolates or directly in gastric biopsy specimens. The method was evaluated by comparison of the results obtained by the method with those obtained by direct sequence analysis of PCR products obtained from a number of patients from Portugal and The Netherlands. Finally, the clinical relevance of vacA and cagA genotyping was investigated with patients with functional dyspepsia, peptic ulcers, or gastric carcinomas.
MATERIALS AND METHODS
Patients.
A total of 103 Dutch (n = 48) and Portuguese (n = 55) patients were studied. Biopsy specimens were obtained from 48 consecutive patients who underwent standard gastroscopy procedures in a Dutch community hospital and were used directly for PCR. Of these patients, 23 had nonulcer dyspepsia (NUD), 18 had a duodenal ulcer, and 7 had a gastric ulcer.
Gastric biopsy specimens were also obtained from a total of 35 consecutive Portuguese patients. H. pylori was cultured from specimens from 20 of these patients, who had duodenal ulcer disease. The remaining 15 biopsy specimens were from patients with functional dyspepsia and were not used for culture but were used directly for PCR. Surgical specimens were obtained from 20 consecutive Portuguese patients with gastric carcinoma and were processed directly for PCR.
Biopsy specimens were transported in Portagerm medium (BioMérieux, Marcy l’Etoile, France) and were stored at −70°C. In all cases parallel biopsy specimens were processed for routine histopathological examination and/or the CLO test (Delta-West, Bentley, Australia).
Culture of H. pylori from gastric biopsy specimens.
Gastric biopsy specimens were collected from the transport medium and were plated onto agar medium (Gelose; BioMérieux), and the plates were incubated at 35°C under microaerobic conditions (5% O2, 15% CO2, 80% N2) for 5 days.
Isolates of H. pylori were identified by Gram staining as well as by oxidase, catalase, urease, gamma-glutamyltransferase, and nitrate reductase reactions (Rapidec pylori; BioMérieux) and were stored at −70°C in Trypticase soy broth containing 15% glycerol.
DNA isolation.
DNA was isolated from the cultured bacteria by harvesting cells from a plate in phosphate-buffered saline followed by centrifugation. The pelleted cells were resuspended in 200 μl of proteinase K solution (10 mM Tris-HCl [pH 7.8], 5 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 50 mg of proteinase K per ml), and the mixture was incubated at 55°C for 30 min. The DNA was extracted with phenol-chloroform-isoamyl alcohol by standard procedures (26). DNA was precipitated by the addition of 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 volumes of ethanol. After centrifugation, the DNA pellet was washed with 70% ethanol and was dissolved in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.3], 0.1 mM EDTA).
Gastric biopsy specimens were homogenized in 200 μl of proteinase K solution with a sterile micropestle (Eppendorf, Hamburg, Germany). DNA was isolated from the homogenate by the same protocol described above for the cultured isolates.
PCRs.
PCRs were performed in a volume of 100 μl containing 10 mM Tris-HCl [pH 9.0], 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, 0.1% Triton X-100, a 200 μM concentration of the deoxynucleoside triphosphates, 0.25 U of SuperTaq (SphaeroQ, Leiden, The Netherlands), and 10 to 50 pmol of both forward and reverse primers. The reaction mixtures were covered with mineral oil, and PCR was performed in a BioMed-60 thermocycler, comprising 2 min of preincubation at 95°C, followed by 40 cycles of 1 min at 95°C, 1 min at 50°C, and 1 min at 74°C. Final extension was performed for 5 min at 74°C. vacA typing with allele-specific PCR primers was performed as described earlier (3). PCR products were visualized by electrophoresis on 2% agarose gels by standard procedures (26).
PCR primers.
For analysis of the vacA s region, the following primers were used for PCR (see also Table 1 and Fig. 1). Primers VA1F and VA1R have been described earlier (3) and generate a fragment of 259 bp for s1a/b variants and a fragment of 286 bp for s2 variants. Primers VA1F and VA1XR generate a fragment of 176 bp for s1a/b variants and a fragment of 203 bp for s2 variants.
TABLE 1.
PCR primers for amplification of vacA and cagA sequences
| Gene and primer (polarity) | Sequence (5′-3′)a | Position |
|---|---|---|
| vacA s region | ||
| VA1F (+)b | ATGGAAATACAACAAACACAC | 1–21c,d |
| VA1R (−)b | CTGCTTGAATGCGCCAAAC | 241–259c and 268–286d |
| VA1XR (−) | CCTGARACCGTTCCTACAGC | 157–176c and 184–203d |
| vacA m region | ||
| HPMGF (+) | CACAGCCACTTTCAATAACGA | 1443–1463c and 1419–1439d |
| HPMGR (−) | CGTCAAAATAATTCCAAGGG | 1824–1843c and 1875–1894d |
| MF1 (+) | GTGGATGCYCATACRGCTWA | 1495–1514c and 1471–1490d |
| MR1 (−) | RTGAGCTTGTTGATATTGAC | 1582–1601c and 1633–1652d |
| cagA | ||
| cagAF (+) | TTGACCAACAACCACAAACCGAAG | 17–40e |
| cagAR (−) | CTTCCCTTAATTGCGAGATTCC | 178–199e |
R is A or G, W is A or T, and Y is = C or T.
VA1F and VA1R have been published earlier by Atherton et al. (3).
Nucleotide position numbers are according to the start codon of the vacA open reading frame in strain 60190 (GenBank accession no. U05676) for s1 and m1.
Nucleotide position numbers are according to the start codon of the vacA open reading frame in strain Tx30a (GenBank accession no. U29401) for s2 and m2.
For detection of cagA the positions are according to the start codon of the cagA open reading frame in the sequence of the strain with GenBank accession no. L11714.
For analysis of the m region of the vacA gene, initial analysis was performed with primers HPMGF and HPMGR, selected from the prototype sequences of m1 (GenBank U05676) and m2 (GenBank U29401), respectively. These primers generate a fragment of 401 bp for m1 variants and a fragment of 476 bp for m2 variants. These amplimers were used for sequence analysis. Primers MF1 and MR1 were deduced from the sequence alignments and were used for subsequent typing of the m region from patient samples. These biotinylated primers generate a fragment of 107 bp for m1 variants and a fragment of 182 bp for m2 variants.
For detection of the cagA gene, biotinylated primers cagAF and cagAR yield a fragment of 183 bp.
Sequence analysis.
The PCR products were used for direct sequence analysis as described previously (18). Briefly, PCR products, containing a biotin moiety on one of the primers, were immobilized onto streptavidin-coated paramagnetic particles (Dynabeads; Dynal, Oslo, Norway). The PCR product was denatured by alkaline treatment, and each strand was used in separate sequencing reactions with the T7 sequencing system (Pharmacia, Uppsala, Sweden). The sequences were read manually and were analyzed with PC-Gene software (Intelligenetics, Mountain View, Calif.).
The primer sequences were analyzed for specificity by using the Blast program at the National Institute of Health Data libraries (1).
LiPA.
Oligonucleotide probes (see Table 2) for specific identification of the different vacA types or subtypes were deduced from the sequence alignments. Probes were selected from those regions that showed consistent type or subtype-specific heterogeneity, as described earlier by Atherton et al. (3). For reverse hybridization, the probes were tailed with poly(dT) and were immobilized as parallel lines on nitrocellulose strips as described earlier (27).
TABLE 2.
LiPA probes for the vacA and cagA gene
| Gene and probe | Sequencea | Position | Specificity |
|---|---|---|---|
| vacA s region | |||
| P1S1a | GGAGCRTTRGTCAGCATCAC | 61–80b | s1a |
| P22S1a | GCTTTAGTAGGAGCRTTRGTC | 52–72b | s1a |
| P1S1b (S1b) | GGAGCGTTGATTAGYKCCAT | 61–80b | s1b |
| P2S1b | GTTTTAGCAGGAGCGTTGA | 52–72b | s1b |
| P1S2 (VAS2) | GCTAAYACGCCAAAYGATCC | 88–107c | s2 |
| P2S2 | GATCCCATACACAGCGAGAG | 103–122c | s2 |
| vacA m region | |||
| P1M1 | TTGATACGGGTAATGGTGG | 1526–1544b | m1 |
| P2M1 | GGGTAATGGTGGTTTCAACA | 1533–1552b | m1 |
| P1M2 | ACGAATTTAAGAGTGAATGGC | 1522–1542c | m2 |
| P2M2 | AGAGCGATAACGGGCTAAACA | 1577–1597c | m2 |
| cagA | |||
| cagApro1 | GTTGATAACGCTGTCGCTTC | 94–113d | cagA |
| cagApro2 | TAATCTTCARGTRGCTTTTCTT | 68–89d | cagA |
R is A or G, W is A or T, Y is C or T, and K is G or T.
Nucleotide position numbers are according to the start codon of the vacA open reading frame in strain 60190 (GenBank accession no. U05676) for s1 and m1.
Nucleotide position numbers are according to the start codon of the vacA open reading frame in strain Tx30a (GenBank accession no. U29401) for s2 and m2.
For cagA the positions are according to the start codon of the cagA open reading frame in the sequence of the strain with GenBank accession no. L11714.
Ten microliters of PCR product was denatured by the addition of 10 ml of 400 mM NaOH and 10 mM EDTA in a plastic trough. After incubation at ambient temperature for 10 min, 1 ml of prewarmed hybridization solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) was added and a reverse hybridization line probe assay (LiPA) strip was submerged. The troughs were incubated in a shaking water bath at 50 ± 0.5°C for 1 h. The strips were washed with 1 ml of 2× SSC–0.1% SDS for 30 min at 50°C. Subsequently, the strips were rinsed three times in phosphate buffer, and conjugate (streptavidin, alkaline phosphatase) was added. After 30 min of incubation at room temperature, the strips were rinsed again and 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate substrate was added. Positive hybridizations were visible as purple probe lines. Interpretation of the results was performed visually.
Statistical analysis.
Data were analyzed by the chi-square test.
RESULTS
Amplification and sequence analysis of vacA.
In order to investigate the variability of the vacA s region, amplification products from 34 isolates, comprising fragments of 259 bp (s1) or 286 bp (s2), generated by the primer set VA1F-VA1R as described by Atherton et al. (3) were directly sequenced. On the basis of the size of the PCR products and the homology to the prototype sequences, all sequences could be classified as either s1 or s2. Within the s1 group, s1a and s1b subtypes could be recognized. The sequences of representative vacA s1 and s2 variants are presented in Fig. 2. On the basis of sequence analysis a new conserved reverse primer (VA1XR) was chosen. Amplification with VA1F and VA1XR yielded PCR products of 176 and 203 bp for type s1 and type s2, respectively.
FIG. 2.
Nucleotide sequence alignment of vacA s1 and s2. The alignment shows nucleotides 36 to 108 and 36 to 135 of the s1 (GenBank accession no. U05676) and s2 (GenBank accession no. U29401) allelic types, respectively. Dots indicate the presence of a nucleotide identical to the nucleotide in the top sequence. A dash indicates a gap. U29401 and NL4600 represent s2 variants; U07145, PO30, and PO32 represent s1a variants; and NL4601 and PO12 represent s1b variants. The positions of the allele-specific probes are indicated by shading.
For analysis of the vacA m region, primers HPMGF and HPMFR were deduced from the alignment of the prototype sequences and were used to generate fragments from 18 isolates. On the basis of the sequence alignments, general primers for the amplification of both m1 and m2 were selected. These primers (MF1 and MR1) flank the highly variable insertion or deletion site of the m region and yield products of 107 and 182 bp for type m1 and type m2, respectively. The sequences of representative vacA m1 and m2 variants are presented in Fig. 3.
FIG. 3.
Nucleotide sequence alignment of vacA m1 and m2. The alignment shows nucleotides 1495 to 1607 and 1471 to 1658 of m1 (GenBank accession no. U5676) and m2 (GenBank accession no. U29401) allelic types, respectively. Dots indicate the presence of a nucleotide identical to the nucleotide in the top sequence. A dash indicates a gap. U29401 and NL4600 represent m2 variants, and U07145, PO30, PO32, NL4601, and PO12 represent m2 variants. The positions of the universal primers as well as those of the allele-specific probes are indicated by shading.
LiPA.
In order to confirm PCR amplification of fragments from the cagA and the vacA gene, a reverse hybridization assay that allows specific detection of the various PCR products in a single step was developed. Oligonucleotide probes (Table 2) were immobilized on a nitrocellulose strip as parallel lines. The outline and examples of the LiPA strip are presented in Fig. 4. For each of the allelic variants within the s region (s1a, s1b, and s2) and m region (m1 and m2), probes were developed and used on the strip. In order to exclude cross-reactivity or false-negative results due to unexpected mutations, two separate probes were used for each allelic variant. Samples from more than 50 individual patients were typed by LiPA as well as by direct sequence analysis. Identification of the vacA s1a, s1b, s2, m1, and m2 types was completely concordant between both methods. No cross-reactivity was observed between any of the different variants on the LiPA strip. All PCR fragments hybridized to both probes of a particular vacA subtype.
FIG. 4.
Outline and examples of the LiPA for the identification of the different vacA alleles and the cagA gene. Different representative examples are shown, including a sample containing multiple strains.
Clinical evaluation.
In order to evaluate the PCR-LiPA system further, a total of 103 H. pylori-positive clinical specimens were tested by the PCR-LiPA. The genotyping results are presented in Table 3. vacA typing of 20 Dutch strains was performed with allele-specific primers as described previously (3). This yielded results identical to those of sequencing and PCR-LiPA, although aspecific amplification products were observed, making interpretation of the results on the gel difficult in four cases.
TABLE 3.
vacA and cagA genotypes found in Portuguese and Dutch H. pylori isolates determined by PCR-LiPA
| Type | No. of isolatesa
|
||||
|---|---|---|---|---|---|
| Portugal
|
The Netherlands
|
||||
| NUD (15) | CA (20) | DU (20) | PU (25)b | NUD (23) | |
| s1a/m1 | 1 | 1 | 1 | 14 | 8 |
| s1a/m2 | 3 | 2 | |||
| s1b/m1 | 6 | 14 | 2 | 2 | 1 |
| s1b/m2 | 1 | 3 | 1 | ||
| s2/m2 | 4 | 3 | 4 | 9 | |
| Multiplec | 3 | 5 | 11 | 1 | 3 |
| cagA negative | 5 | 2 | 2 | 4 | 9 |
| cagA positive | 10 | 18 | 18 | 21 | 14 |
Abbreviations: CA, gastric carcinoma; DU, duodenal ulcers; PU, peptic ulcers. Numbers in parentheses represent number of patients.
One isolate did not yield a PCR fragment from the s region, but it contained vacA m1 and was cagA positive.
More than one vacA s or m type was detected by LiPA.
The cagA gene was detected in 46 of 55 (84%) of the Portuguese patients and in 35 of 48 (73%) of the Dutch patients (P > 0.05). Among the 103 patients, the prevalence of cagA-positive strains appeared to be higher among the patients with ulcers and gastric carcinoma than among patients with NUD (P = 0.003). Also, this association was similar in both countries.
In 4 of 48 (8%) of the Dutch biopsy specimens, multiple vacA genotypes were detected. Of the specimens with a single vacA genotype, 27 of 44 (61%) contained type s1a, 4 of 44 (9%) contained type s1b, and 13 of 44 (30%) contained type s2. Twenty-five samples (57%) contained type m1, and 19 samples (43%) contained type m2. One biopsy specimen repeatedly failed to yield a PCR product from the vacA s region, although PCR for the m region and the cagA gene was successful.
Overall, multiple vacA genotypes were observed in 19 of 55 (29%) of the Portuguese samples. This is significantly higher than the percentage observed in the Dutch samples (P = 0.001). Of the 36 Portuguese samples with a single genotype, 26 (72.3%) contained type s1b (72.3%), 3 (8.3%) contained type s1a, and 7 (19.4%) contained type s2. A total of 25 (69%) of the 36 samples contained type m1, whereas 11 of 36 (31%) contained type m2.
Among all Dutch and Portuguese patients with peptic ulcers and gastric carcinomas, the prevalence of vacA s1 strains was higher than that among patients with NUD (P = 0.008). This association was not significantly different between Portuguese and Dutch patients.
DISCUSSION
H. pylori infections can cause serious clinical problems, such as atrophic gastritis, gastric or duodenal ulcers, and gastric cancer. Therefore, it is important that accurate diagnostic methods are available.
There is increasing evidence that the genetic variability of H. pylori may also have clinical importance. Three different genes that are associated with bacterial pathogenicity have been described, i.e., cagA, vacA, and iceA (5, 8). The present study describes the development of a rapid method for analysis of the vacA and cagA genes. This method was evaluated by testing clinical samples from Portuguese and Dutch patients.
cagA detection.
PCR primers for the detection of the cagA gene were aimed at the 5′ part of the gene, which appears to be the most conserved region (10). There are indications of sequence variability of the cagA gene in isolates of different geographic origins (24), and this may lead to underestimation of the true prevalence of cagA-positive H. pylori strains. This requires more extensive sequence analysis of H. pylori strains of different geographic origins.
vacA detection and typing.
Analysis of the vacA gene is more complicated. The number of available vacA sequences in the data libraries is very limited. The PCR-based typing system of the s and m regions described by Atherton et al. (3) is based on a number of separate allele-specific PCRs for the detection of the s1/s2, s1a, s1b, s2, m1, and m2 alleles, respectively. This requires multiple PCRs for each strain, and in some cases, formation of aspecific PCR products made interpretation of the results on the gel difficult. To discriminate between types m1 and m2, two type-specific primer sets have been described (3). The target regions for these primer sets do not comprise the highly variable insertion or deletion site of the m region but are located more downstream in vacA. Since aberrant amplification products are often encountered or no amplification products are obtained, these primers were not satisfactory (21), which may be due to the fact that they were based on sequence information from H. pylori isolates obtained in North America.
We have deduced novel primers for general amplification of the s and m regions from sequences obtained from European H. pylori strains. These primers allowed effective amplification of the s and m region sequences. Amplification of the s region was not possible for only one sample from a Dutch patient, probably due to additional sequence variation in the primer target regions.
Reverse hybridization.
Since visual inspection of PCR products on agarose gels provides only limited reliability, hybridization to allele-specific probes is obligatory for confirming the typing results. Therefore, allele-specific probes were selected for the development of a reverse hybridization assay. The multiparameter design of the LiPA was aimed at specific determination of the vacA and cagA status in a single hybridization step. The complete concordance between direct sequencing and LiPA results confirmed the high degree of specificity of the reverse hybridization method. Despite the high degree of genetic diversity of H. pylori, the differences between vacA s- and m-region alleles appear to be highly consistent and in almost all cases permitted the use of general PCR primers and allele-specific probes.
The reverse hybridization format also permits rapid and sensitive detection of multiple genotypes in a clinical specimen. This is an important advantage of direct analysis of DNA from a gastric biopsy specimen by PCR and allele-specific hybridization. Investigation of cultured H. pylori strains may considerably underestimate the prevalence of simultaneous infections with multiple strains due to strain selection during in vitro culture.
Clinical evaluation.
In order to evaluate the PCR-LiPA system, a number of biopsy specimens and isolates from patients in Portugal and The Netherlands comprising patients with NUD, peptic ulcers, or gastric carcinomas were tested.
The prevalence of cagA-positive samples appears to be slightly higher among Portuguese patients than among Dutch patients. This difference was not statistically significant and may be due to the inclusion of patients with gastric carcinomas in the Portuguese group.
The majority of the Portuguese strains (72%) contained the s1b allele, whereas most of the Dutch strains (61%) contained type s1a. This highly significant difference is observed in both the NUD and the ulcer patients. Therefore, these results are likely to be indicative of the relative prevalence of vacA s types in both countries. The prevalence of the type s2 allele, as well as the type m1 and m2 alleles, does not appear to be different between strains from Portugal and those from The Netherlands. Since no gastric carcinoma patients from The Netherlands were studied, the prevalence of vacA types within this group could not be compared.
The geographic distribution of distinct H. pylori genotypes remains largely unknown. The prevalence of more pathogenic bacterial genotypes in certain areas may have important epidemiological consequences and may be associated with the severity of H. pylori-related diseases in such regions (15).
Since it has been demonstrated that H. pylori carries only a single copy of vacA (3, 11, 12), detection of multiple genotypes implies the presence of multiple strains in a clinical sample. The frequency of multiple genotypes in a single biopsy specimen appears to be much higher in Portugal than in The Netherlands (29 and 8%, respectively). This phenomenon may be related to the prevalence of H. pylori infections among the populations in each country. In Portugal, approximately 80% (14) of the adult population is infected, whereas in The Netherlands approximately 30% of the adult population is infected. The risk of coinfection or superinfection with multiple strains may be higher in countries with a high prevalence of H. pylori infection than in countries with a low prevalence.
The presence of multiple H. pylori strains, as determined by random amplification of polymorphic DNA from cultured isolates, has been reported earlier (17, 19, 28, 29). In the present study, we confirmed these findings by directly analyzing gastric biopsy specimens for the H. pylori vacA gene. It is unknown whether infection with multiple strains increases the risk of serious clinical implications, such as the development of ulcers and gastric cancer.
In conclusion, the present report describes the sequence analysis of the vacA s and m regions of Dutch and Portuguese H. pylori isolates. A rapid and simple PCR-reverse hybridization assay for vacA and cagA genotyping was developed. This assay may facilitate broader investigation of the clinical implications of the colonization with the various H. pylori genotypes. Furthermore, it offers an attractive tool for studying the variability of H. pylori isolates of different geographic origins.
ACKNOWLEDGMENTS
We thank Ger Scholte, An Lan Kam, Ricardo Sanna, Wink de Boer, and Peter Schneeberger for valuable contributions.
Part of this project was supported by PRAXIS XXI (project 1/2.1/SAU/1356/95).
REFERENCES
- 1.Altschull S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Med Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Appelmelk B J, Simoons-Smit I, Negrini R, Moran A P, Aspinall G O, Forte J G, de Vries T, Quan H, Verboom T, Maaskant J J, Ghiara P, Kuipers E J, Bloemena E, Tadema T M, Townsend R R, Tyagarajan K, Crothers J M, Monteiro M A, de Graaff J. Potential role of molecular mimicry between Helicobacter pylori lipopolysaccharide and host Lewis blood group antigens in autoimmunity. Infect Immun. 1996;64:2031–2040. doi: 10.1128/iai.64.6.2031-2040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atherton J C, Cao P, Peek R M, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 4.Atherton J C, Peek R M, Tham K T, Cover T L, Blaser M J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 5.Atherton J C. The clinical relevance of strains types of Helicobacter pylori. Gut. 1997;40:701–703. doi: 10.1136/gut.40.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser M J, Parsonnet J. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J Clin Invest. 1994;94:4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser M J. Intrastrain differences in Helicobacter pylori: a key question in mucosal damage? Ann Med. 1995;27:559–563. doi: 10.3109/07853899509002469. [DOI] [PubMed] [Google Scholar]
- 8.Blaser M J. Not all Helicobacter pylori strains are created equal: should all be eliminated? Lancet. 1997;349:1020–1022. doi: 10.1016/S0140-6736(96)09133-7. [DOI] [PubMed] [Google Scholar]
- 9.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1990;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 12.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 13.Cover T L, Glupczynski Y, Lage A P, Burette A, Tummuru M K R, Perez-Perez G I, Blaser M J. Serologic detection of infection with cagA+ Helicobacter pylori strains. J Clin Microbiol. 1995;33:1496–1500. doi: 10.1128/jcm.33.6.1496-1500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estevens J, Fidalgo P, Tendeiro T, Chagas C, Ferra A, Leitão C N, Mira F C. Anti-Helicobacter pylori antibodies prevalence and gastric adenocarcinoma in Portugal: report of a case-control study. Eur J Cancer Prev. 1993;2:377–380. doi: 10.1097/00008469-199309000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Genta R M, Gürer I E, Graham D Y. Geographical pathology of Helicobacter pylori infection: is there more than one gastritis? Ann Med. 1995;27:595–599. doi: 10.3109/07853899509002475. [DOI] [PubMed] [Google Scholar]
- 16.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic deversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschl A M, Richter M, Makristhatis A, Prückl P M, Willinger B, Schütze K, Rotter M L. Single and multiple strain colonization in patients with Helicobacter pylori-associated gastritis: detection by macrorestriction DNA analysis. J Infect Dis. 1994;170:473–475. doi: 10.1093/infdis/170.2.473. [DOI] [PubMed] [Google Scholar]
- 18.Hultman T, Bergh S, Moks T, Uhlen M. Bidirectional solid-phase sequencing of in vitro-amplified plasmid DNA. BioTechniques. 1991;10:84–93. [PubMed] [Google Scholar]
- 19.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazel S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 20.Leunk R D, Johnson P T, David B C, Kraft W G, Morgan D R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 21.Maeda S, Ogura K, Ishitobi M, Kanai F, Yoshida H, Ota S, Shiratori Y, Omata M. Diversity of Helicobacter pylori vacA gene in Japanese strains—high cytotoxin activity type s1 is dominant in Japan. Gastroenterology. 1996;110:A182. [Google Scholar]
- 22.Mobley H L T. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am J Med. 1996;100:2S–11S. doi: 10.1016/s0002-9343(96)80223-3. [DOI] [PubMed] [Google Scholar]
- 23.Negrini R, Savio A, Poesi C, Appelmelk B J, Buffoli F, Paterlini A, Cesari P, Graffeo M, Vaira D, Franzin G. Antigenic mimicry between Helicobacter pylori and gastric mucosa in the pathogenesis of body atrophic gastritis. Gastroenterology. 1996;111:655–665. doi: 10.1053/gast.1996.v111.pm8780570. [DOI] [PubMed] [Google Scholar]
- 24.Pan Z-J, van der Hulst R W M, Feller M, Xiao S-D, Tytgat G N J, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344–1347. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phadnis S H, Ilver D, Janzon L, Normark S, Westblom T U. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, van Heuverswyn H, Maertens G. Typing of HCV isolates and characterization of new (sub)types using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 28.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Ende A, Rauws E A J, Feller M, Mulder C J J, Tytgat G N J, Dankert J. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]