Summary
SARS-CoV-2 Omicron quickly spread globally, also in regions with high vaccination coverage, emphasizing the importance of exploring the immunological requirements for protection against Omicron breakthrough infection.
The test-negative matched case-control study (N = 964) characterized Omicron breakthrough infections in triple-vaccinated individuals from the ENFORCE cohort. Within 60 days before a PCR test spike-specific IgG levels were significantly lower in cases compared to controls (GMR [95% CI] for BA.2: 0.83 [0.73–0.95], p = 0.006). Multivariable logistic regression showed significant associations between high antibody levels and lower odds of infection (aOR [95% CI] for BA.2 spike-specific IgG: 0.65 [0.48–0.88], p = 0.006 and BA.2 ACE2-blocking antibodies: 0.46 [0.30–0.69], p = 0.0002). A sex-stratified analysis showed more pronounced associations for females than males.
High levels of vaccine-induced antibodies provide partial protection against Omicron breakthrough infections. This is important knowledge to further characterize a threshold for protection against new variants and to estimate the necessity and timing of booster vaccination.
Subject areas: Immunology, Molecular medicine, Immune response
Graphical abstract
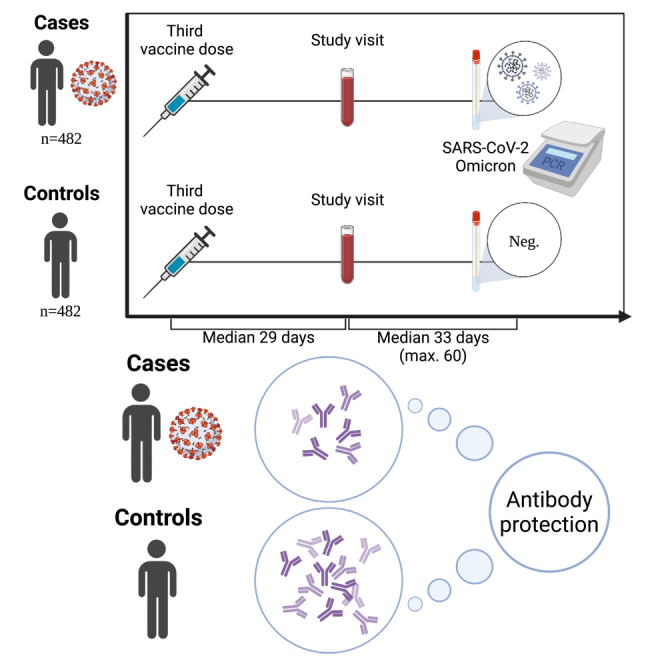
Highlights
-
•
High antibody levels are associated with low odds of Omicron breakthrough infection
-
•
Vaccine-induced antibodies are an immune marker of protection against infection
-
•
Sex stratification revealed a clear association for females in particular
Immunology; Molecular medicine; Immune response
Introduction
In late 2019, the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged and caused the outbreak of the coronavirus disease 2019 (COVID-19) pandemic. Global administration of expeditiously developed vaccines has been of significant importance in controlling the health impact of the pandemic.1 The SARS-CoV-2 Omicron variant (B.1.1.529) was discovered in South Africa in November 2021 and quickly spread, creating a new global challenge for pandemic control. The Omicron variant has more than 30 mutations in the spike protein, increasing evasion from existing neutralizing antibodies and exhibiting higher transmissibility compared to previous variants.2 Thus, while vaccine-induced immunity continues to prevent severe disease, breakthrough infections are more likely to occur with Omicron compared to previous viral variants.2,3,4,5 We and others have shown that high levels of vaccine-induced antibodies provide protection against infection with past variants.6,7,8,9,10,11 However, the association between vaccine-induced antibodies and breakthrough infection is less clear for Omicron.12,13,14,15,16,17,18 Consequently, there is a critical need for a better understanding of the protection elicited by vaccine-induced antibodies against Omicron variants.
The objective of the present study was to characterize vaccine-induced immunity among monovalent triple-vaccinated individuals from the Danish National Cohort Study of Effectiveness and Safety of SARS-CoV-2 vaccines (ENFORCE) and to evaluate how vaccine-induced immunity may provide protection against Omicron breakthrough infection. Omicron spike-specific immunoglobulin G (IgG) levels and angiotensin-converting enzyme 2 (ACE2)-blocking antibody titers were evaluated in samples collected prior to a positive (case) or negative (control) polymerase chain reaction (PCR) test. Lastly, pre-to post-infection nucleocapsid IgG levels among cases were determined to investigate seroconversion.
Results
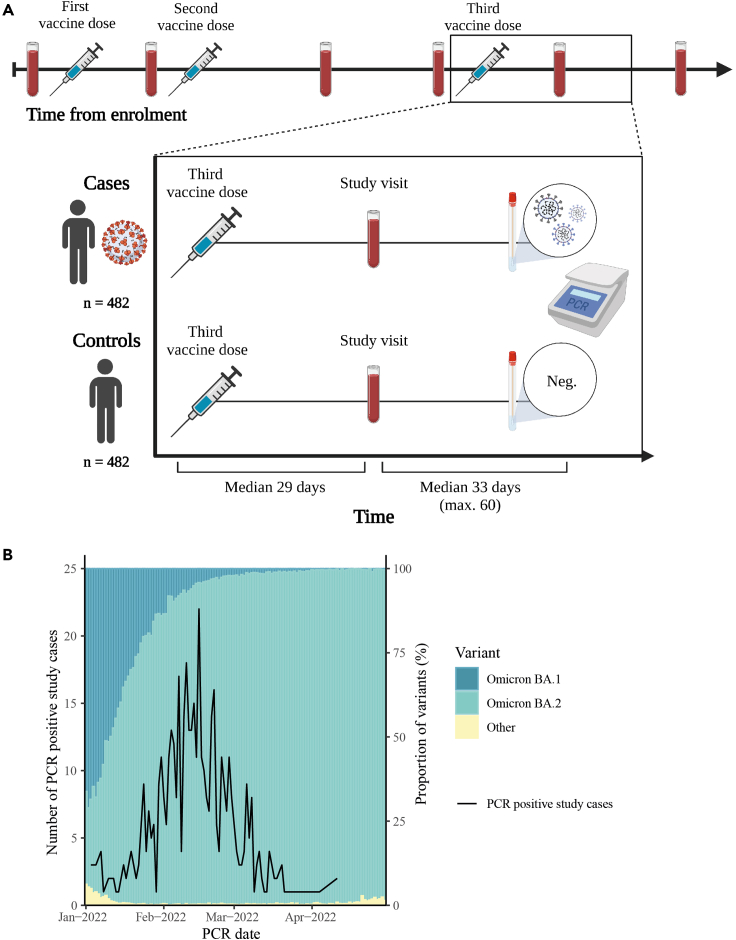
From the ENFORCE cohort, a total of 482 breakthrough infection cases were selected and matched with 482 uninfected controls (Figures 1A and S1). Both cases and controls had a median age of 64 years (IQR: [56–73] and [57–74], respectively) with 197 males (40.9%) and 285 females (59.1%) in each group. The demographic characteristics of the study participants are shown in Table 1. The prevalence of comorbidities was higher among controls compared with cases (28.3% in controls and 17.2% in cases) and the timing of the study visits relative to the PCR tests also differed slightly between cases and controls (median [IQR]: 36 days [25–50] and 29 days [18–42], respectively). All participants had received three monovalent vaccine doses (three doses of Pfizer-BioNTech [n = 240, 49.8%], Moderna [n = 207, 42.9%], or one dose of AstraZeneca and two doses of an mRNA vaccine [n = 35, 7.3%]). All participants had blood sampled at a study visit median 29 days after the third vaccine dose (IQR: [27–33] and [27–32], respectively) which was used to measure antibody levels. The frequency of PCR testing for cases and controls was comparable from first vaccination until May 1st, 2022 (median [IQR]: 6 [3–12] and 5 [3–10], respectively). Based on PCR test date, 9.3% of cases were estimated to be Omicron BA.1 breakthrough infections and 90.2% were estimated to be BA.2 (Figure 1B).
Figure 1.
Study design and inclusion of participants
(A) Schematic overview of the ENFORCE study design with focus on inclusion of study participants for evaluation of Omicron breakthrough infections. All study participants had received a third vaccine dose and completed a subsequent study visit maximum 60 days prior to a positive (case) or negative (control) PCR test.
(B) Illustration of the proportion of SARS-CoV-2 variants (Omicron BA.1 and Omicron BA.2) in Denmark from January 1st to May 1st 2022 and the number of PCR positive study cases included by PCR date.
Table 1.
Demographics of study participants
| Cases (n = 482) | Controls (n = 482) | p-value | |
|---|---|---|---|
| Age at enrollment (median, IQR) | 64 (56–73) | 64 (57–74) | 0.40 |
| Age group (n, %)∗ | NA | ||
| <55 | 100 (20.7) | 100 (20.7) | . |
| 55–65 | 152 (31.5) | 152 (31.5) | . |
| >65 | 230 (47.7) | 230 (47.7) | . |
| Sex (n, %)∗ | NA | ||
| Male | 197 (40.9) | 197 (40.9) | . |
| Female | 285 (59.1) | 285 (59.1) | . |
| Vaccine (n, %)∗ | NA | ||
| Pfizer-BioNTech | 240 (49.8) | 240 (49.8) | . |
| Moderna | 207 (42.9) | 207 (42.9) | . |
| AstraZeneca/mRNA | 35 (7.3) | 35 (7.3) | . |
| Study visit (n, %)∗ | NA | ||
| 28 days after 3rd dose | 470 (97.5) | 470 (97.5) | . |
| Up to 170 days after 3rd dose | 12 (2.5) | 12 (2.5) | . |
| Charlson Comorbidity Index (n, %) | <0.0001 | ||
| 0 | 399 (82.8) | 346 (71.8) | . |
| 1–2 | 78 (16.2) | 115 (23.9) | . |
| >2 | 5 (1.0) | 21 (4.4) | . |
| Days from 3rd vaccine to study visit (median, IQR) | 29 (27–33) | 29 (27–32) | 0.045 |
| Days from study visit to PCR test (median, IQR) | 36 (25–50) | 29 (18–42) | <0.0001 |
The demographic characteristics of the cases (n = 482) and controls (n = 482). The categorical variables are described as number of participants (n) and the percentage (%), while the continuous variables are described as the median and the interquartile range (IQR). The matched variables are indicated with an asterisk (∗).
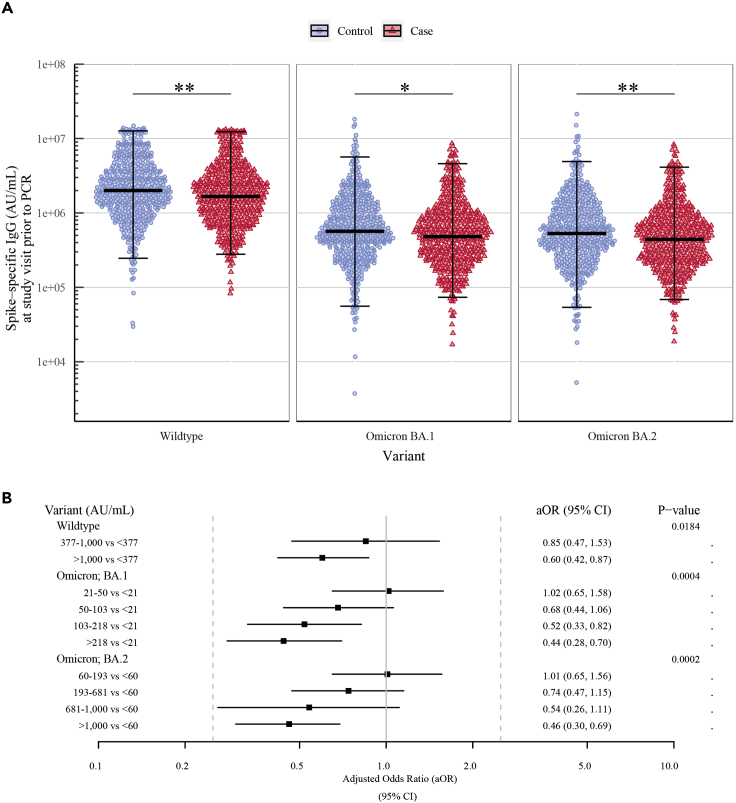
High levels of spike-specific IgG and ACE2-blocking antibody titers were associated with lower odds of Omicron breakthrough infection
Significantly lower spike-specific IgG levels were observed among cases compared to controls for SARS-CoV-2 wildtype and Omicron variants BA.1 and BA.2 (GM ratio cases-to-controls [95% CI] for wildtype: 0.84 [0.75–0.95], BA.1: 0.84 [0.74–0.96], and BA.2: 0.83 [0.73–0.95]) (Figure 2A and Table 2). Multivariable logistic regression was used to analyze the association between antibody levels and the risk of Omicron breakthrough infection after adjusting for the matched variables: age group, sex, vaccine, and study visit, and the unmatched variables: vaccine priority group, CCI, visit year, time from study visit to PCR test, and time from third vaccine dose to PCR test. Higher levels of wildtype spike-specific IgG were associated with 40% lower odds of breakthrough infection (aOR [95% CI]: 0.60 [0.43–0.85] per log10-fold increase). Similar associations were observed for spike-specific Omicron BA.1 and BA.2 IgG levels (0.69 [0.51–0.93] and 0.65 [0.48–0.88], respectively) (Table 2).
Figure 2.
High levels of spike-specific IgG and ACE2-blocking antibody titers were associated with lower odds of Omicron breakthrough infection
(A) Comparison of spike-specific IgG levels (AU/mL) quantified at a study visit prior to a positive (cases, n = 482) or negative PCR test (controls, n = 482) showing the geometric mean and 95% CI. SARS-CoV-2 variants from left to right: Wildtype (Wuhan-Hu-1), Omicron BA.1, and Omicron BA.2. ∗ = p ≤ 0.05 and ∗∗ = p < 0.01.
(B) Multivariable logistic regression showing the adjusted odds ratios (aORs) and 95% CI for breakthrough infection for ACE2-blocking antibody titers in tertiles (wildtype) or quintiles (Omicron variants). The analysis adjusts for the matched variables: age group, sex, vaccine, and study visit, and for the unmatched variables: vaccine priority group, Charlson comorbidity index (CCI), visit year, days from study visit to PCR test, and days from third vaccination to PCR test.
Table 2.
High levels of spike-specific IgG were associated with lower odds of Omicron breakthrough infection
| Variant | n | Geometric Mean |
Adjusted Odds Ratio |
||||
|---|---|---|---|---|---|---|---|
| Cases 95% CI |
Controls 95% CI |
GM ratio 95% CI |
p-value | aOR 95% CI |
p-value | ||
| Wildtype | 481 | 1,744,920 1585978–1919791 |
2,075,195 1897717–2269271 |
0.84 0.75–0.95 |
0.005 | 0.60 0.43–0.85 |
0.003 |
| Omicron BA.1 | 482 | 507,775 459492–561132 |
601,740 542144–667888 |
0.84 0.74–0.96 |
0.012 | 0.69 0.51–0.93 |
0.015 |
| Omicron BA.2 | 481 | 464,534 421169–512365 |
558,175 503889–618309 |
0.83 0.73–0.95 |
0.006 | 0.65 0.48–0.88 |
0.006 |
Quantification of spike-specific IgG levels prior to a positive (case) or negative PCR test (control) showing the geometric mean (GM) with 95% CI and GM ratio cases-to-controls. Multivariable logistic regression showing the adjusted odds ratios (aORs) and 95% CI for breakthrough infection for a log10-fold increase of spike-specific IgG levels. The analysis adjusts for the matched variables: age group, sex, vaccine, and study visit, and for the unmatched variables: vaccine priority group, Charlson comorbidity index (CCI), visit year, days from study visit to PCR test, and days from third vaccination to PCR test.
ACE2 data were not treated as continuous, due to many samples reaching the assay’s detection limit, particularly for wildtype. Thus, the data were split into quintiles. The highest quintiles that reached the assay’s upper detection limit were subsequently grouped together (Table S1). In multivariate logistic regression, we observed significant associations with both wildtype and Omicron BA.1 and BA.2 ACE2-blocking antibodies, and risk of Omicron breakthrough infection. Participants with moderate titers (377–1000 AU/mL) of wildtype antibodies had 15% lower odds of breakthrough infection compared to participants with the lowest antibody titers (<377 AU/mL) (aOR [95% CI]: 0.85 [0.47–1.53]), while those with the highest titers (>1000 AU/mL) were 40% less likely to have a breakthrough infection (0.60 [0.42–0.87], p value: 0.0184). Similarly, for Omicron-specific ACE2-blocking antibodies, participants with higher antibody titers were less likely to have a breakthrough infection compared to participants with the lowest titers (BA.1: 0.68 [0.44–1.06], 0.52 [0.33–0.82], and 0.44 [0.28–0.70] for 50–103 AU/mL, 103–218 AU/mL, and >218 AU/mL, respectively, p value: 0.0004. BA.2: 0.74 [0.47–1.15], 0.54 [0.26–1.11], and 0.46 [0.30–0.69] for 193–681 AU/mL, 681–1000 AU/mL, and >1000 AU/mL, respectively, p value: 0.0002). However, there was no difference between low antibody titers (BA.1: 21–50 AU/mL and BA.2: 60–193 AU/mL) and the lowest titers (BA.1: <21 AU/mL and BA.2: <60 AU/mL) (BA.1: 1.02 [0.65–1.58] and BA.2: 1.01 [0.65–1.56]) (Figure 2B). Results were consistent in sensitivity analysis using conditional logistic regression and only adjusting for the unmatched variables (data not shown).
For an unmatched subpopulation of the study cohort (Table S2), we assessed spike-specific CD4+ and CD8+ T cell frequencies. No significant differences between cases and controls were observed (Figure S2A).
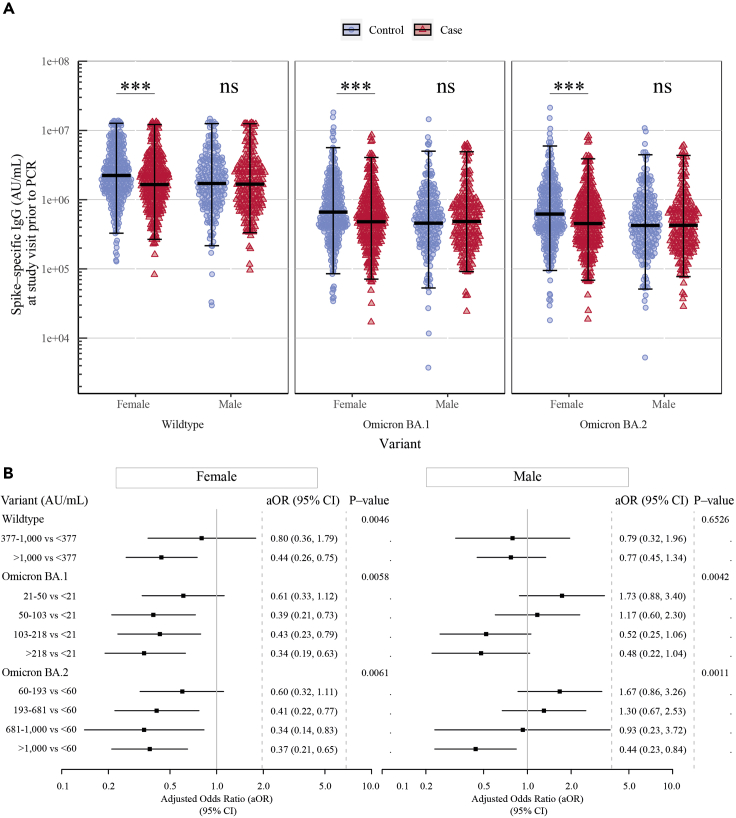
Association between high levels of vaccine-induced antibodies and lower odds of Omicron breakthrough infection were more pronounced in females than males
To investigate whether sex had an influence on protection from breakthrough infection, the comparison of spike-specific IgG levels and ACE2-blocking antibody titers was stratified by sex (Table S3). Compared with female controls, female cases had significantly lower spike-specific IgG levels (GM ratio cases-to-controls [95% CI] for wildtype: 0.76 [0.65–0.89]), BA.1 (0.84 [0.74–0.96]), and BA.2 (0.83 [0.73–0.95])). In contrast, no significant differences were observed for either variant between male cases and male controls (wildtype: 0.98 [0.82–1.18], BA.1: 1.02 [0.83–1.26], and BA.2: 0.99 [0.81–1.21]) (Figure 3A and Table S4). Multivariable logistic regression also revealed that higher levels of spike-specific IgG were associated with significantly lower odds of breakthrough infection among females (aOR [95% CI] for wildtype: 0.49 [0.31–0.77], BA.1: 0.52 [0.34–0.79], and BA.2: 0.53 [0.35–0.79]), but not among males (wildtype: 0.84 [0.49–1.45], BA.1: 1.00 [0.63–1.58], and BA.2: 0.88 [0.55–1.43]) (Table S4).
Figure 3.
Association between high levels of vaccine-induced antibodies and lower odds of Omicron breakthrough infection were more pronounced in females than to males
(A) Comparison of spike-specific IgG levels (AU/mL) quantified at a study visit prior to a positive (case) or negative (control) PCR test stratified by sex showing the geometric mean and 95% CI (n = 285 females and n = 197 males). SARS-CoV-2 variants from left to right: Wildtype (Wuhan-Hu-1), Omicron BA.1, and Omicron BA.2. ∗∗∗ = p < 0.001.
(B) Multivariable logistic regression showing the adjusted odds ratios (aORs) and 95% CI for breakthrough infection for ACE2-blocking antibody titers in tertiles (wildtype) or quintiles (Omicron variants) stratified by sex. The analysis adjusts for the matched variables: age group, sex, vaccine, and study visit, and for the unmatched variables: vaccine priority group, Charlson comorbidity index (CCI), visit year, days from study visit to PCR test, and days from third vaccination to PCR test.
Multivariable logistic regression was also used to investigate the association between ACE2-blocking antibodies and the risk of Omicron breakthrough infection, stratified by sex. For females, an association was found for both wildtype and Omicron variants, consistent with spike-specific IgG levels. For males, an association between ACE2-blocking antibodies and lower odds of breakthrough infection was only found for participants with the highest antibody titers compared to participants with the lowest antibody titers for Omicron BA.1 and BA.2 (Figure 3B).
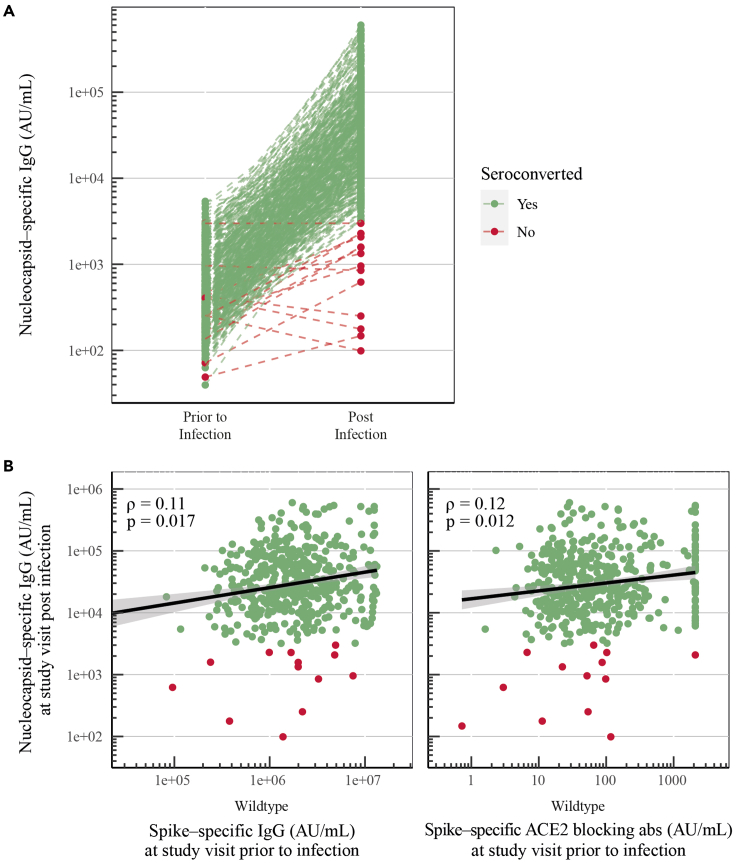
Nucleocapsid-specific IgG seroconversion following breakthrough infection was not correlated with vaccine-induced immunity
To investigate whether nucleocapsid seroconversion after viral infection was impacted by vaccine-induced immunity, nucleocapsid-specific IgG levels were quantified post infection (median of 114 days after positive PCR test [IQR: 101–126]). Samples were available for 457 cases of which 96.9% (n = 443) seroconverted post breakthrough infection (Figure 4A).
Figure 4.
Nucleocapsid-specific IgG seroconversion following breakthrough infection was not correlated with vaccine-induced immunity
(A) Nucleocapsid-specific IgG levels (AU/mL) prior to and post breakthrough infection for cases (n = 457). Seroconversion is defined by nucleocapsid-specific antibodies >3,000 AU/mL and a 2-fold increase.
(B) Spearman correlations between infection-induced nucleocapsid-specific IgG (AU/mL), and vaccine-induced spike-specific IgG (AU/mL) (Wuhan-Hu-1) and ACE2-blocking antibodies (AU/mL) (Wuhan-Hu-1).
Disease severity has been shown to increase the levels of humoral and cellular immunity induced by infection and thereby leading to higher levels of nucleocapsid seroconversion. Therefore, we evaluated whether nucleocapsid-specific antibody levels post infection correlated with vaccine-induced immunity.19,20 No clear correlations were found between nucleocapsid-specific IgG levels post infection, and spike-specific IgG levels and ACE2-blocking antibody titers prior to infection (ρ = 0.11 and ρ = 0.12, respectively) (Figure 4B). Nucleocapsid-specific IgG levels post infection did also not correlate with spike-specific CD4+ or CD8+ T cells prior to infection (Figure S2).
Discussion
In this test-negative matched case-control study, we evaluated humoral immune responses in a large triple-vaccinated cohort to investigate the immunological requirements for protection against Omicron breakthrough infection. Higher vaccine-induced antibody levels were found to be associated with lower odds of breakthrough infection. A sex-stratified analysis revealed that this association was consistent and clear for females while the association was unclear or absent for males. In addition, we demonstrated nucleocapsid seroconversion in almost all cases and we found no correlation between vaccine-induced immunity and nucleocapsid seroconversion.
In this study, we report the protection of humoral vaccine-induced immunity against Omicron breakthrough infection in a large test-negative matched case-control study with antibody levels measured within 60 days before infection. Identification of an immune correlate of protection against SARS-CoV-2 infection is of major importance to navigate the effects of waning immunity, the necessity and timing of booster vaccination, and whether vaccinated individuals remain protected from infection. Several studies have shown associations between vaccine-induced antibodies and protection from breakthrough infection with previous SARS-CoV-2 variants, including wildtype, Alpha, and Delta,7,8,9,10,11,12,13,14,15,16 but only very few studies have reported similar association for Omicron variants.17,18 We observed significantly higher spike-specific IgG levels and ACE2-blocking antibody titers in controls compared to cases for both wildtype and Omicron variants, that remained after adjusting for demographic differences, demonstrating a significant impact of vaccine-induced immunity against Omicron breakthrough infection. Still, there was not a defining protective threshold, indicating confounding factors, such as the combined impact of the immune system, gene variations, the level of societal infections or lifestyles, which would need additional investigations. Nonetheless, the data support the general assumption of high antibody titers eliciting protection against SARS-CoV-2 infection and suggest an inverse correlation between antibody levels and odds of breakthrough infection for all viral variants.
High levels of SARS-CoV-2-specific CD4+ and CD8+ T cells have been correlated to a mild COVID-19 disease course.21 The cellular data presented in this study did not show a direct correlation between T cell levels and protection against breakthrough infection. Still, others have shown a rapid end extensive recall of SARS-CoV-2-specific T cells upon breakthrough infection which may aid in protection of viral replication.22
The study included participants infected from January 1st to May 1st 2022, where Omicron variants BA.1 and BA.2 were predominant in Denmark and worldwide. During the inclusion period, societal transmission levels in Denmark were soaring. Especially in February 2022, where around 7,500 PCR-confirmed cases per 1 million people were registered daily, as well as a PCR test positivity rate of ∼30%.23 Increasing transmission of SARS-CoV-2 in society is highly correlated with increasing risk of breakthrough infection.14 The high antibody levels associated with lower odds of breakthrough infection in this study, may therefore be higher than the antibody levels needed to stay protected from infection during periods with lower societal transmission levels of SARS-CoV-2.
Many studies have shown sex-based differences in immunity following vaccination and infection. One such difference is the observation of increased antibody production elicited by vaccination in females compared to males.24 Susceptibility to SARS-CoV-2 infection seems to be similar in males and females, while higher severity and mortality rates are observed for males.25,26,27 ACE2 is the receptor utilized by SARS-CoV-2 to enter cells, and different expression patterns of ACE2 in the respiratory tract in males and females may add to the sex-based differences in susceptibility to infection and mortality, interplaying with the sex-based differences in the immune system.28,29,30 In this study, a sex-stratified analysis showed that both spike-specific IgG levels and ACE2-blocking antibody titers act as an immune marker of protection against Omicron breakthrough infection for females, while only high ACE2-blocking antibody titers showed protection against infection for males. This may partially be explained by small differences in age compositions of our study cohort, with a higher percentage of male participants >65 years of age (56.3% males compared to 41.8% females). Additionally, this may link to the sex-based difference in ACE2 expression.28,29,30 A study has reported that males maintain innate immunity, but have a more rapid decline of adaptive immunity with increasing age compared to females.31 Another consideration is the type of antibodies quantified. In this study, a sex-based difference in vaccine-induced immunity and protection from infection was discovered with antibody levels as a strong immune marker of protection for females, while males may have additional immunological components impacting protection against infection. These observations further emphasize the need of sex-stratifying analyses when investigating vaccine-induced immunity.
Previous studies have shown that disease severity and high viral loads were associated with increased humoral and cellular immune responses.19,20,32 SARS-CoV-2 vaccination has been demonstrated to provide substantial protection against severe disease and to reduce the infectious viral load significantly, bringing into question whether vaccine-induced immunity reduces seroconversion following infection.33,34 In this study, we did not find a negative correlation between vaccine-induced immunity and infection-induced seroconversion. In contrast, we observed a very weak positive correlation between vaccine-induced antibodies and infection-induced nucleocapsid-specific antibodies, which may reflect a general correlation between the ability to produce antibodies following vaccination and seroconversion upon infection.
Our study had several strengths. The matched case-control study design eliminated many demographic discrepancies and focused exclusively on vaccine-induced immunity. The risk of including participants with an undetected prior infection was reduced by examination of nucleocapsid-specific IgG seroconversion prior to first vaccination and at the study visit. Additionally, the criteria of a documented negative PCR test for controls secured a similar behavioral testing pattern during the pandemic for both groups. The quantification of antibody levels, within 60 days prior to breakthrough infection, placed a suitable time frame to display clinical outcomes. Furthermore, the study was able to measure both wildtype- and Omicron-specific antibodies.
In conclusion, our study described an inverse correlation between vaccine-induced antibodies and risk of Omicron breakthrough infection. This supports the general assumption of vaccine-induced antibodies as an immune marker of protection against SARS-CoV-2 infection for all viral variants, including Omicron BA.1 and BA.2. The inverse correlation may extend to other sub-lineages of Omicron, such as BA.5, BA.2.75, BQ.1, and XBB, although the required protective antibody levels may be higher, since these sub-lineages have more enhanced immune-evasion capacities than previous Omicron variants. Overall, further research into vaccine-induced antibodies and breakthrough infections is important to characterize a threshold for protection against infection and to estimate the necessity and timing of booster vaccination to remain protected.
Limitation of the study
This study focused on Omicron-specific antibody levels to protect against Omicron breakthrough infection. We utilized a platform measuring multiple variant-specific antibodies in a single well, requiring one dilution of the sample. This introduced a saturation of the signal for wildtype antibodies. This limitation can be observed for both MSD assays, but particularly the ACE2 assay. Neutralizing antibody titers were quantified by the MSD ACE2 assay, which was preferred over the usage of the more laborious pseudovirus neutralization assay, as we have previously shown a very strong correlation between ACE2-blocking antibodies measured by MSD assays and neutralizing titers measured by pseudovirus neutralization assays.6 Our study also had a few other limitations. Even with the matched study design, some variation was still present in the cohort, including the number of individuals with comorbidities. Individuals with comorbidities are generally considered to be at a higher risk of infection, since a diminished humoral response has been associated with comorbidities.24 Even so, in our study cohort, there were more controls with comorbidities compared to cases, indicative of a preventive behavioral pattern among high-risk populations. Another drawback of the study was the limited number of participants who donated PBMCs for the evaluation of T cell immunity. T cell-mediated immunity may be particularly important for providing protection against infection over time.
Consortia
ENFORCE Study Group: J. Lundgren, L. Østergaard, T. Benfield, L. Krohn-Dehli, D. K. Petersen, K. Fogh, E. Højmark, K. K. Iversen, P. Bek, V. Klastrup, F. Larsen, S. H. Rasmussen, M. H. Schleimann, S. Schieber, N. B. Stærke, A. Søndergaard, B. Tarp, M. Tousgaard, Y. Yehdego, J. Bodilsen, H. Nielsen, K. T. Petersen, M. Ruwald, R. K. Thisted, S. F. Caspersen, M. Iversen, L. S. Knudsen, J. L. Meyerhoff, L. G. Sander, L. Wiese, C. Abildgaard, I. K. Holden, N. E. Johansen, I. S. Johansen, L. Larsen, S. O. Lindvig, L. W. Madsen, A. Øvrehus, N. A. Kruse, H. Lomholdt, T. G. Krause, P. Valentiner-Branth, B. Søborg, T. K. Fischer, C. Erikstrup, S. R. Ostrowski, M. Tolstrup, O. S. Søgaard, D. Raben, E. Jylling, D. Hougaard, S.D. Andersen, K. Lykkegaard, S. R. Andreasen, E. Baerends, L. L. Dietz, A. K. Hvidt, A. K. Juhl, R. Olesen, K. K. Andersen, W. Bannister, C. Bjernved, T. W. Elsing, F. V. Esmann, M. A. Ghafari, E. Gravholdt, S. F. Jakobsen, M. L. Jakobsen, C. M. Jensen, T. Ø. Jensen, D. Kristensen, L. R. Kumar, C. Matthews, N. Normand, C. Olsson, J. Reekie, A. Traytel, T. Weide, A. M. Hvas, H. Støvring.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3- PerCP-Cy5.5 | Biolegend | CAT#BL344808; RRID: AB_10640736 |
| CD4-BV650 | Biolegend | CAT#BL300536; RRID: AB_2632791 |
| CD8-BV605 | Biolegend | CAT#BL301040; RRID: AB_2563185 |
| CD69-APC | Biolegend | CAT#BL310910; RRID: AB_314845 |
| OX40-BV421 | Biolegend | CAT#BL350014; RRID: AB_2564184 |
| 41BB-PE | Biolegend | CAT#BL309804; RRID: AB_314783 |
| Chemicals, peptides, and recombinant proteins | ||
| LIVE/DEAD-APC-H7 | Invitrogen | CAT#L34976 |
| Brilliant Stain Buffer | BD | CAT#566349 |
| PepMixTM SARS-CoV-2 (Swiss-Prot ID: P0DTC2) | JPT | CAT#PM-WCPV-S-2 |
| Critical commercial assays | ||
| V-PLEX SARS-CoV-2 Panel 25 (IgG) kit | Meso Scale Discovery | CAT#K15583U-2 |
| V-PLEX SARS-CoV-2 Panel 25 (ACE2) kit | Meso Scale Discovery | CAT#K15586U-2 |
| V-PLEX SARS-CoV-2 Panel 2 (IgG) kit | Meso Scale Discovery | CAT#K15383U-2 |
| Software and algorithms | ||
| MESO QuickPlex SQ 120MM | Meso Scale Discovery | https://www.mesoscale.com/en/products_and_services/instrumentation/quickplex_sq_120mm |
| Discovery Workbench Software (version 4.0) | Meso Scale Discovery | https://www.mesoscale.com/en/products_and_services/software |
| MACSQuant Analyzer 16 Flow Cytometer | Miltenyi Biotec | https://www.miltenyibiotec.com/DK-en/products/macsquant-analyzer-16.html |
| FlowJo™ Software (version 10.8.1) | BD Biosciences | https://www.flowjo.com/ |
| SAS Studio Software (version 9.4) | SAS Studio | https://www.sas.com/ |
| R Software (version 4.2.2) | R CRAN | https://www.r-project.org/ |
| RStudio Desktop (version 2022.07.2) | Posit | https://posit.co/download/rstudio-desktop/ |
| BioRender | BioRender | https://www.biorender.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Astrid Hvidt (asthvi@rm.dk).
Materials availability
Samples from the ENFORCE cohort may be made available to researchers upon approval of an application to the ENFORCE scientific steering committee and further approval by relevant authorities. Applications for material must be sent to enforce.rigshospitalet@regionh.dk. The ENFORCE protocol is available at www.enforce.dk and more detailed information about material access may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Governance.
Experimental model and study participant details
The Danish National Cohort Study of Effectiveness and Safety of SARS-CoV-2 vaccines (ENFORCE) was designed as an open-label, non-randomized, parallel group, phase IV study.35 The study enrolled adults prior to their first vaccination offered through the Danish vaccination program (clinicaltrails.gov, identifier: NCT04760132). From February to August 2021, 6,943 individuals were enrolled at seven study sites that covered all five Danish regions. The study enrolled 3,014 males (43.4%) and 3,929 females (56.6%) with a median age of 64 years (IQR: 53, 75). Information on ancestry, race, and/or ethnicity has not been considered in this study. The study protocol was approved by the Danish Medicines Agency (#2020-006003-42) and the National Committee on Health Research Ethics (#1-10-72-337-20). All participants provided written informed consent.
Study design and study population
In this test-negative matched case-control study, participants were selected from the ENFORCE cohort. Cases were defined as individuals that had: 1) a positive SARS-CoV-2 PCR test between January 1st and May 1st 2022, 2) a study visit within 60 days prior to the PCR test, and 3) received a third vaccination prior to the PCR test and >20 days prior to the study visit. Controls were defined by the same criteria but with the requirement of a negative PCR test (Figure 1A). Individuals with a previous SARS-CoV-2 infection were excluded. Previous infection was defined as: 1) previously PCR- or antigen-test positive, 2) spike Ig positive prior to first vaccination, or 3) seroconversion prior to first vaccination (nucleocapsid-specific IgG >3,000 AU/mL). To ensure exclusion of previous infections, nucleocapsid-specific IgG at the study visit had to be less than double the value prior to first vaccination and ≤3,000 AU/mL. Recipients of a fourth vaccine dose during the inclusion period were also excluded (Figure S1). Controls and cases were matched 1:1 by age, sex, vaccine, and study visit.
Data collection
Information on age, sex, medical history, vaccination dates, and vaccine were collected prior to first vaccination and confirmed by the Danish National Patient Registry and the Danish Vaccination Registry. SARS-CoV-2 infections were confirmed by positive PCR (data from the Key Infectious Diseases System database and the Danish National Microbiology database). In Denmark, the capacity of standard variant sequencing of viral isolates was saturated by the end of December 2021, due to the extensive increase in positive PCR tests. Consequently, less than 10% of the viral isolates were sequenced during the following time period. This study investigated breakthrough infections from January 1st to May 1st 2022, where Omicron BA.1 was the predominant variant during the first weeks of January, after which the BA.2 variant became predominant.36
Blood samples collected at the last study visit prior to the PCR test were used to profile the humoral immune response by quantifying spike-specific IgG levels and ACE2-blocking antibody titers.
Method details
Profiling of the humoral immune response
Serum levels of spike-specific IgG and ACE2-blocking antibodies against SARS-CoV-2 wildtype (Wuhan-Hu-1) and Omicron variants BA.1 and BA.2 were quantified by the Meso Scale Discovery (MSD) platform (Meso Scale Diagnostics LLC, Maryland, USA) using panel 25: IgG kit (K15583U-2) and ACE2 kit (K15586U-2). The assays were performed according to the manufacturer’s protocol. Unspecific antibody binding was blocked by MSD Blocker A (Cat. No. R93AA-2). Serum samples were diluted 1:100,000 in MSD Diluent 100 (Cat. No. R50-AA3) for IgG assays and 1:10 for ACE2 assays and run as singlets. For IgG assays, serum samples were incubated along with MSD Reference Standard 1 (Cat. No. C00ADK-2) to establish a calibration curve. At the time of analysis, MSD did not provide a calibration reagent for the ACE2 assays including Omicron variants. An internal monoclonal antibody calibrator, that bound both wildtype and Omicron spike proteins, was included for all ACE2 assays. For IgG assays, bound IgG was detected by MSD SULFO-TAG anti-human IgG antibody (Cat. No. D21ADF-3). For ACE2 assays, MSD SULFO-TAG human ACE2 protein (Cat. No. D21ADG-3) was added to the wells before washing to allow ACE2 to compete with antibody binding to spike. GOLD Read Buffer B (Cat. No. R60AM-2) was added and plates were read on a MESO QuickPlex SQ 120 reader. Raw data were processed by Discovery Workbench 4.0 Software. Quantifications were reported in arbitrary units per mL (AU/mL).
Serum levels of nucleocapsid-specific IgG against SARS-CoV-2 wildtype (Wuhan-Hu-1) were also quantified by MSD using panel 2: IgG kit (K15383U-2). The assays were performed according to the manufacturer’s protocol with a serum dilution of 1:5,000. Seroconversion was identified by: 1) nucleocapsid-specific IgG >3,000 AU/mL and 2) a two-fold increase in nucleocapsid-specific antibodies compared to a previous study visit.
Profiling of the cellular immune response
For a small subset of participants (controls: n=40, cases: n=33), peripheral blood mononuclear cells (PBMCs) were donated and used to profile the cellular immune response by quantifying spike-specific (Wuhan-Hu-1) CD4+ and CD8+ T cells by the activation induced marker (AIM) assay.37 Antigen specific T cells were defined as cells that express 2 or 3 activation induced markers. The AIMs used for this study were CD69, OX40 (CD134), and 41BB (CD137).
Purified PBMCs were stimulated with PepMix™ SARS-CoV-2 (JPT peptides product code PM-WCPV-S-2, Swiss-Prot ID: P0DTC2) at 2μg/ml or Dimethyl sulfoxide (negative control) for 20 h. Following stimulation, cells were washed, stained and run on MACSQuant16 (Miltenyi Biotec). Data was analyzed using FlowJoTM v10.8.1 Software (BD Biosciences).
For viability staining 0.1 μl LIVE/DEAD-APC-H7 (cat# L34976) was diluted in 99.9 μl PBS. The staining master mix contained CD3- PerCP-Cy5.5 (1 μl, cat# BL344808), CD4-BV650 (2 μl, cat# BL300536), CD8-BV605 (1μl, cat# BL301040), CD69-APC (1μl, cat# BL310910), OX40-BV421 (5 μl, cat# BL 350014), and 41BB-PE (2.5 μl, cat# BL309804) in 52.5 μl Brilliant Stain Buffer (cat# 566349).
Quantification and statistical analysis
Demographic characteristics were analyzed by Chi-squared tests (categorical variables) and Wilcoxon rank sum tests (continuous variables). Spike-specific IgG data were presented with the geometric mean (GM) and error bars showing 95% confidence interval (CI). A paired t-test was used to compare the log-transformed titers. The mean differences and 95% CI were subsequently exponentiated to give the GM ratios and 95% CI. Multivariate logistic regression analysis investigated whether antibody levels were associated with breakthrough infection after adjusting for the matched variables: age group (<55, 55-65, or >65), sex (female or male), vaccine (Pfizer-BioNTech, Moderna, or AstraZeneca/mRNA), and study visit (∼28 days or up to 170 days after third vaccination), and adjusting for the unmatched variables: vaccine priority group (general population, individuals at increased risk, or health care workers), Charlson comorbidity index38 (CCI) (0, 1-2, or >2), visit year (2021 or 2022), days from study visit to PCR test, and days from third vaccination to PCR test. Data was presented as a forest plot showing the adjusted odds ratios (aORs) and 95% CI. ACE2 data were not normally distributed and many samples reached the assay’s detection limit. The ACE2 data were therefore split into quintiles. The assay had an upper detection limit of 1,000 AU/mL and the samples reaching this limit were designated to the highest quintile. This resulted in the data being presented in five groups for Omicron variants and in three groups for wildtype (Table S1). ACE2 data were also analyzed by multivariable logistic regression, as described for spike-specific IgG data. To investigate the robustness of the results, sensitivity analysis using multivariable conditional logistic regression, adjusting for the unmatched variables was performed. To investigate if the findings were consistent among males and females, sensitivity analysis was conducted to analyze the data stratified by sex. Spearman’s rank correlation test was used to assess a correlation between infection-induced nucleocapsid-specific antibodies and vaccine-induced spike-specific antibodies and ACE2-blocking antibodies.
P-values ≤0.05 were considered statistically significant. P-values were denoted as following: ∗=P≤0.05, ∗∗=P<0.01, ∗∗∗=P<0.001, and ∗∗∗∗=P<0.0001. Data analysis and visualization was conducted in SAS Studio (version 9.4), R (version 4.2.2), and RStudio Desktop (version 2022.07.2). Schematic overviews of the study were created using BioRender.com.
Acknowledgments
ENFORCE has received a grant from the Danish Ministry of Health (SUM).
The ENFORCE study group members all contributed substantially to the study. A full list of members of the ENFORCE study group may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Group.
We would like to thank C. Hansen, M. Gram, and P. Valentiner-Branth, who provided a complete dataset of viral variant sequencing data in Denmark through personal communications (November 16th 2022). These data were utilized in Figure 1B.
Author contributions
J.L., L.Ø., O.S.S., N.B.S., J.R., D.R., T.K.F., C.E., P.V.B., and M.T. conceptualized the work. H.N., K.T.P., M.R.J., I.S.J., S.O.L., L.W.M., L.W., L.S.K., M.B.I., T.L.B., and K.K.I. did the clinical visits and collected the samples. E.B., A.K.H., S.D.A., S.R.A., L.L.D., and A.K.J. performed the laboratory analyses. E.B., A.K.H., and J.R. performed the data analysis and visualization. M.T. supervised and led the study. E.B., A.K.H., J.R., and M.T. drafted the manuscript and have accessed and verified the underlying data. All authors read and approved the final draft. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
N.B.S. declares to have served as an investigator in clinical trials sponsored by Pfizer, Gilead, and Bavarian Nordic. H.N. declares to have been on advisory boards for GSK and MSD. T.B. declares receipt of unrestricted grants from Novo Nordisk Foundation, Simonsen Foundation, Lundbeck Foundation, Kai Foundation, Erik and Susanna Olesen’s Charitable Fund, GSK, Pfizer, Gilead Sciences, and MSD; and being advisory board member for GSK, Pfizer, Gilead Sciences, MSD, Janssen, and Astra Zeneca; and being principal investigator on clinical trials conducted by Pfizer, Boehringer Ingelheim, Gilead Sciences, MSD, Roche, Novartis, Kancera AB, Bavarian Nordic, and Janssen; and being board member on Pentabase; and receiving consulting fees from GSK and Pfizer; and receiving honorarium for lectures from GSK, Pfizer, Gilead Sciences, Boehringer Ingelheim, AbbVie, Astra Zeneca, and Bavarian Nordic; and receiving donation of trial medication (baricitinib) from Eli Lilly. MT declares to be on a Data Safety Monitoring Board for Immunocore. The remaining authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 14, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107621.
Contributor Information
Astrid K. Hvidt, Email: asthvi@rm.dk.
on behalf of the ENFORCE Study Group:
J. Lundgren, L. Østergaard, T. Benfield, L. Krohn-Dehli, D.K. Petersen, K. Fogh, E. Højmark, K.K. Iversen, P. Bek, V. Klastrup, F. Larsen, S.H. Rasmussen, M.H. Schleimann, S. Schieber, N.B. Stærke, A. Søndergaard, B. Tarp, M. Tousgaard, Y. Yehdego, J. Bodilsen, H. Nielsen, K.T. Petersen, M. Ruwald, R.K. Thisted, S.F. Caspersen, M. Iversen, L.S. Knudsen, J.L. Meyerhoff, L.G. Sander, L. Wiese, C. Abildgaard, I.K. Holden, N.E. Johansen, I.S. Johansen, L. Larsen, S.O. Lindvig, L.W. Madsen, A. Øvrehus, N.A. Kruse, H. Lomholdt, T.G. Krause, P. Valentiner-Branth, B. Søborg, T.K. Fischer, C. Erikstrup, S.R. Ostrowski, M. Tolstrup, O.S. Søgaard, D. Raben, E. Jylling, D. Hougaard, S.D. Andersen, K. Lykkegaard, S.R. Andreasen, E. Baerends, L.L. Dietz, A.K. Hvidt, A.K. Juhl, R. Olesen, K.K. Andersen, W. Bannister, C. Bjernved, T.W. Elsing, F.V. Esmann, M.A. Ghafari, E. Gravholdt, S.F. Jakobsen, M.L. Jakobsen, C.M. Jensen, T.Ø. Jensen, D. Kristensen, L.R. Kumar, C. Matthews, N. Normand, C. Olsson, J. Reekie, A. Traytel, T. Weide, A.M. Hvas, and H. Støvring
Supplemental information
Data and code availability
-
•
The data reported in this study cannot be deposited in a public repository. Data is restricted to protect the privacy of the study participants in this cohort. Data from the ENFORCE cohort may be made available to researchers upon approval of an application to the ENFORCE scientific steering committee and further approval by relevant authorities. Applications for data must be sent to enforce.rigshospitalet@regionh.dk. If approval is granted data will be provided as deidentified data. The ENFORCE protocol is available at www.enforce.dk and more detailed information about data access may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Governance.
-
•
This paper does not report original code. Requests for code should be directed to the lead contact.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tseng H.F., Ackerson B.K., Luo Y., Sy L.S., Talarico C.A., Tian Y., Bruxvoort K.J., Tubert J.E., Florea A., Ku J.H., et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and Delta variants. Nat. Med. 2022;28:1063–1071. doi: 10.1038/s41591-022-01753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Zhi H., Teng Y. The outbreak of SARS-CoV-2 Omicron lineages, immune escape, and vaccine effectivity. J. Med. Virol. 2023;95:e28138. doi: 10.1002/jmv.28138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 6.Hvidt A.K., Baerends E.A.M., Søgaard O.S., Stærke N.B., Raben D., Reekie J., Nielsen H., Johansen I.S., Wiese L., Benfield T.L., et al. Comparison of vaccine-induced antibody neutralization against SARS-CoV-2 variants of concern following primary and booster doses of COVID-19 vaccines. Front. Med. 2022;9:994160. doi: 10.3389/fmed.2022.994160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldridge R.W., Yavlinsky A., Nguyen V., Eyre M.T., Shrotri M., Navaratnam A.M.D., Beale S., Braithwaite I., Byrne T., Kovar J., et al. SARS-CoV-2 antibodies and breakthrough infections in the Virus Watch cohort. Nat. Commun. 2022;13:4869. doi: 10.1038/s41467-022-32265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei J., Pouwels K.B., Stoesser N., Matthews P.C., Diamond I., Studley R., Rourke E., Cook D., Bell J.I., Newton J.N., et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat. Med. 2022;28:1072–1082. doi: 10.1038/s41591-022-01721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lumley S.F., O'Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Accorsi E.K., Britton A., Fleming-Dutra K.E., Smith Z.R., Shang N., Derado G., Miller J., Schrag S.J., Verani J.R. Association Between 3 Doses of mRNA COVID-19 Vaccine and Symptomatic Infection Caused by the SARS-CoV-2 Omicron and Delta Variants. JAMA. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres I., Giménez E., Albert E., Zulaica J., Álvarez-Rodríguez B., Burgos J.S., Peiró S., Limón R., Vanaclocha H., Rodado C., et al. SARS-CoV-2 Omicron BA.1 variant breakthrough infections in nursing home residents after an homologous third dose of the Comirnaty(R) COVID-19 vaccine: Looking for correlates of protection. J. Med. Virol. 2022;94:4216–4223. doi: 10.1002/jmv.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams E., Colson J., Valiathan R., Carreño J.M., Krammer F., Hoffer M., Pallikkuth S., Pahwa S., Andrews D. Permissive omicron breakthrough infections in individuals with binding or neutralizing antibodies to ancestral SARS-CoV-2. Vaccine. 2022;40:5868–5872. doi: 10.1016/j.vaccine.2022.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stærke N.B., Reekie J., Nielsen H., Benfield T., Wiese L., Knudsen L.S., Iversen M.B., Iversen K., Fogh K., Bodilsen J., et al. Levels of SARS-CoV-2 antibodies among fully vaccinated individuals with Delta or Omicron variant breakthrough infections. Nat. Commun. 2022;13:4466. doi: 10.1038/s41467-022-32254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smoot K., Yang J., Tacker D.H., Welch S., Khodaverdi M., Kimble W., Wen S., Amjad A., Marsh C., Perrotta P.L., Hodder S. Persistence and Protective Potential of SARS-CoV-2 Antibody Levels After COVID-19 Vaccination in a West Virginia Nursing Home Cohort. JAMA Netw. Open. 2022;5:e2231334. doi: 10.1001/jamanetworkopen.2022.31334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Park S., Kim J.Y., Lim S.Y., Chang E., Bae S., Jung J., Kim M.J., Chong Y.P., Lee S.O., et al. No correlation of neutralizing antibody titers against the Omicron variant after a booster dose of COVID-19 vaccines with subsequent breakthrough Omicron infections among healthcare workers. J. Infect. 2022;85:e177–e180. doi: 10.1016/j.jinf.2022.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carazo S., Skowronski D.M., Brisson M., Barkati S., Sauvageau C., Brousseau N., Gilca R., Fafard J., Talbot D., Ouakki M., et al. Protection against omicron (B.1.1.529) BA.2 reinfection conferred by primary omicron BA.1 or pre-omicron SARS-CoV-2 infection among health-care workers with and without mRNA vaccination: a test-negative case-control study. Lancet Infect. Dis. 2023;23:45–55. doi: 10.1016/S1473-3099(22)00578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R., Liu N., Li X., Peng Q., Yiu C.-K., Huang H., Yang D., Du Z., Kwok H.-Y., Au K.-K., et al. Three-dose vaccination-induced immune responses protect against SARS-CoV-2 Omicron BA.2: a population-based study in Hong Kong. Lancet Reg. Health. West. Pac. 2023;32:100660. doi: 10.1016/j.lanwpc.2022.100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen S.S., Vibholm L.K., Monrad I., Olesen R., Frattari G.S., Pahus M.H., Højen J.F., Gunst J.D., Erikstrup C., Holleufer A., et al. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine. 2021;68:103410. doi: 10.1016/j.ebiom.2021.103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vibholm L.K., Nielsen S.S.F., Pahus M.H., Frattari G.S., Olesen R., Andersen R., Monrad I., Andersen A.H.F., Thomsen M.M., Konrad C.V., et al. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine. 2021;64:103230. doi: 10.1016/j.ebiom.2021.103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koutsakos M., Reynaldi A., Lee W.S., Nguyen J., Amarasena T., Taiaroa G., Kinsella P., Liew K.C., Tran T., Kent H.E., et al. SARS-CoV-2 breakthrough infection induces rapid memory and de novo T cell responses. Immunity. 2023;56:879–892.e4. doi: 10.1016/j.immuni.2023.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bæk K.T. COVID-19 in Denmark - Data from SSI (Statens Serum Institut) https://covid19danmark.dk/
- 24.Søgaard O.S., Reekie J., Johansen I.S., Nielsen H., Benfield T., Wiese L., Stærke N.B., Iversen K., Fogh K., Bodilsen J., et al. Characteristics associated with serological COVID-19 vaccine response and durability in an older population with significant comorbidity: the Danish Nationwide ENFORCE Study. Clin. Microbiol. Infect. 2022;28:1126–1133. doi: 10.1016/j.cmi.2022.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., Liu S., Yang J.K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortolan A., Lorenzin M., Felicetti M., Doria A., Ramonda R. Does gender influence clinical expression and disease outcomes in COVID-19? A systematic review and meta-analysis. Int. J. Infect. Dis. 2020;99:496–504. doi: 10.1016/j.ijid.2020.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foresta C., Rocca M.S., Di Nisio A. Gender susceptibility to COVID-19: a review of the putative role of sex hormones and X chromosome. J. Endocrinol. Invest. 2021;44:951–956. doi: 10.1007/s40618-020-01383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagliardi M.C., Tieri P., Ortona E., Ruggieri A. ACE2 expression and sex disparity in COVID-19. Cell Death Dis. 2020;6:37. doi: 10.1038/s41420-020-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majdic G. Could Sex/Gender Differences in ACE2 Expression in the Lungs Contribute to the Large Gender Disparity in the Morbidity and Mortality of Patients Infected With the SARS-CoV-2 Virus? Front. Cell. Infect. Microbiol. 2020;10:327. doi: 10.3389/fcimb.2020.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beyerstedt S., Casaro E.B., Rangel É.B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:905–919. doi: 10.1007/s10096-020-04138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Márquez E.J., Chung C.H., Marches R., Rossi R.J., Nehar-Belaid D., Eroglu A., Mellert D.J., Kuchel G.A., Banchereau J., Ucar D. Sexual-dimorphism in human immune system aging. Nat. Commun. 2020;11:751. doi: 10.1038/s41467-020-14396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Follmann D., Janes H.E., Buhule O.D., Zhou H., Girard B., Marks K., Kotloff K., Desjardins M., Corey L., Neuzil K.M., et al. Antinucleocapsid Antibodies After SARS-CoV-2 Infection in the Blinded Phase of the Randomized, Placebo-Controlled mRNA-1273 COVID-19 Vaccine Efficacy Clinical Trial. Ann. Intern. Med. 2022;175:1258–1265. doi: 10.7326/M22-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodbridge Y., Amit S., Huppert A., Kopelman N.M. Viral load dynamics of SARS-CoV-2 Delta and Omicron variants following multiple vaccine doses and previous infection. Nat. Commun. 2022;13:6706. doi: 10.1038/s41467-022-33096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puhach O., Adea K., Hulo N., Sattonnet P., Genecand C., Iten A., Jacquérioz F., Kaiser L., Vetter P., Eckerle I., Meyer B. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat. Med. 2022;28:1491–1500. doi: 10.1038/s41591-022-01816-0. [DOI] [PubMed] [Google Scholar]
- 35.Stærke N.B., Reekie J., Johansen I.S., Nielsen H., Benfield T., Wiese L., Søgaard O.S., Tolstrup M., Iversen K.K., Tarp B., et al. Cohort Profile: The Danish National Cohort Study of Effectiveness and Safety of SARS-CoV-2 vaccines (ENFORCE) BMJ Open. 2022;12:e069065. doi: 10.1136/bmjopen-2022-069065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen C.H., Friis N.U., Bager P., Stegger M., Fonager J., Fomsgaard A., Gram M.A., Christiansen L.E., Ethelberg S., Legarth R., et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a nation-wide population-based study in Denmark. Lancet Infect. Dis. 2023;23:167–176. doi: 10.1016/S1473-3099(22)00595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietz L.L., Juhl A.K., Søgaard O.S., Reekie J., Nielsen H., Johansen I.S., Benfield T., Wiese L., Stærke N.B., Jensen T.Ø., et al. Impact of age and comorbidities on SARS-CoV-2 vaccine-induced T cell immunity. Commun. Med. 2023;3:58. doi: 10.1038/s43856-023-00277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charlson M., Szatrowski T.P., Peterson J., Gold J. Validation of a combined comorbidity index. J. Clin. Epidemiol. 1994;47:1245–1251. doi: 10.1016/0895-4356(94)90129-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data reported in this study cannot be deposited in a public repository. Data is restricted to protect the privacy of the study participants in this cohort. Data from the ENFORCE cohort may be made available to researchers upon approval of an application to the ENFORCE scientific steering committee and further approval by relevant authorities. Applications for data must be sent to enforce.rigshospitalet@regionh.dk. If approval is granted data will be provided as deidentified data. The ENFORCE protocol is available at www.enforce.dk and more detailed information about data access may be found at https://chip.dk/Research/Studies/ENFORCE/Study-Governance.
-
•
This paper does not report original code. Requests for code should be directed to the lead contact.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.