Abstract
Trained immunity is a long-term memory of innate immune cells, generating an improved response upon reinfection. Shigella is an important human pathogen and inflammatory paradigm for which there is no effective vaccine. Using zebrafish larvae, we demonstrate that after Shigella training, neutrophils are more efficient at bacterial clearance. We observe that Shigella-induced protection is nonspecific and has differences with training by BCG and β-glucan. Analysis of histone ChIP-seq on trained neutrophils revealed that Shigella training deposits the active H3K4me3 mark on promoter regions of 1612 genes, dramatically changing the epigenetic landscape of neutrophils toward enhanced microbial recognition and mitochondrial ROS production. Last, we demonstrate that mitochondrial ROS plays a key role in enhanced antimicrobial activity of trained neutrophils. It is envisioned that signals and mechanisms we discover here can be used in other vertebrates, including humans, to suggest new therapeutic strategies involving neutrophils to control bacterial infection.
Shigella trains the zebrafish innate immune system to produce neutrophils with enhanced antimicrobial function via mtROS.
INTRODUCTION
Trained immunity is an immunological memory characterized by remodeling of the epigenetic landscape and metabolism of myeloid cells, conferring enhanced responses upon a new infection challenge (1). The most commonly used training stimuli are Mycobacterium bovis Bacille Calmette-Guérin (BCG) and the fungal wall component β-glucan (1); however, reports have described a large breadth of triggers that include metabolites, inflammatory cytokines, and danger signals (2–7). Although work in vitro, ex vivo, in mice and in humans has mostly been centered around monocytes/macrophages (1, 8), growing evidence shows that mechanisms of trained immunity can be observed in other immune and non-immune cells (such as endothelial and epithelial cells) (9–11). Recent reports have started to investigate training mechanisms by BCG and β-glucan in neutrophils (12, 13); however, the impact of trained neutrophils at a whole animal level is mostly unknown.
The zebrafish embryo has been widely used as a model for developmental biology, cell biology, and infectious diseases (14). Zebrafish are genetically tractable, share >80% of human genes associated with diseases, and strictly depend on innate immune responses until 4 weeks postfertilization when the adaptive immune system starts to develop (15, 16). Seminal studies have used zebrafish as a model to study hematopoietic stem and progenitor cell (HSPC) development and hematopoiesis (17–19), and the repertoire of innate immune cells in zebrafish is now well characterized (20, 21). Considering this, the zebrafish model presents itself as an attractive model to study trained immunity. The first evidence that training mechanisms may also be conserved in fish comes from decades of use of β-glucans in aquaculture, where their use improved the resistance of teleost fish to infections (22). Further work with carp macrophages (23), vaccine development (24–26), and pathogenic infections (19, 27, 28) support the use of fish as a model to dissect fundamental determinants of trained immunity.
Shigella is an important human pathogen, among the top 12 priority pathogens requiring urgent action by the World Health Organization (29), and for which there are no effective vaccines (30). Shigella infection is well known to induce inflammation, by its lipopolysaccharide (LPS) and type III secretion system (T3SS) (31, 32). We previously demonstrated that hallmarks of Shigella infection, including inflammation and macrophage cell death, can be observed using a zebrafish infection model (33, 34). Work has also shown that a nonlethal Shigella infection triggers immune responses that lead to emergency granulopoiesis and protects zebrafish larvae from secondary infection (28, 35).
Here, we introduce the zebrafish larvae infection model as a powerful system to study innate immune training. We show that Shigella training of zebrafish larvae generates protective neutrophils, and the training mechanism differs from that of BCG or β-glucan training. Genome-wide sequencing of epigenetic modifications in trimethylation at histone 3 lysine 4 (H3K4me3) in Shigella-trained neutrophils and downstream functional characterization shows that their epigenetic landscape is modified to increase mitochondrial reactive oxygen species (mtROS) production to better fight secondary infectious challenges.
RESULTS
Zebrafish training with a nonlethal dose of Shigella induces generation of protective neutrophils
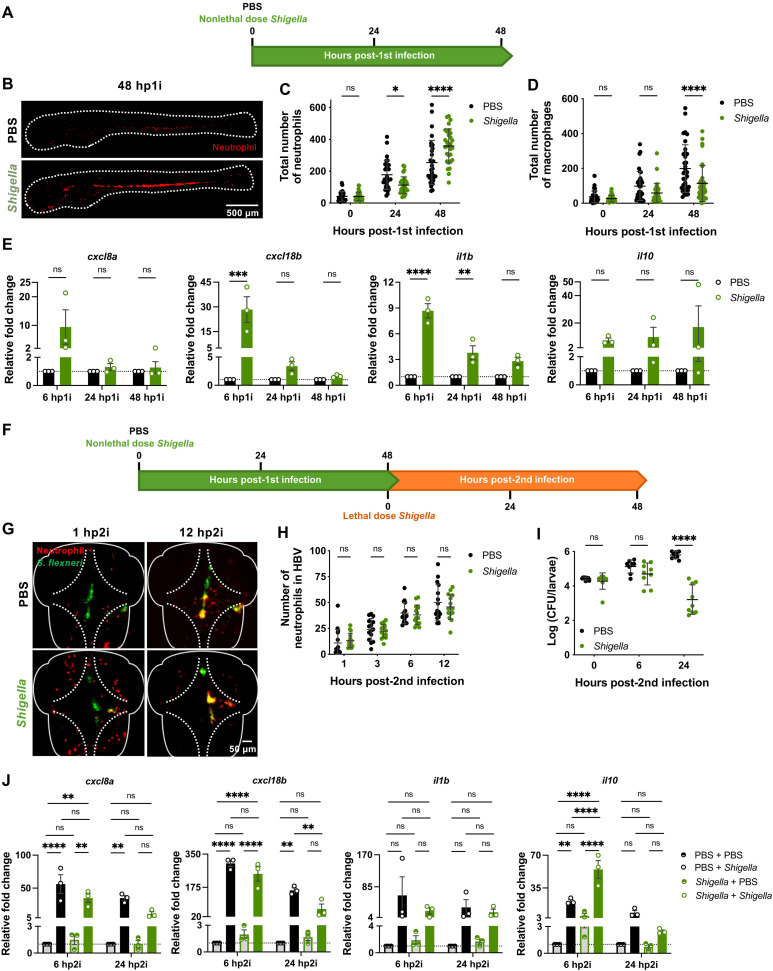
We previously demonstrated that a nonlethal dose of Shigella flexneri injected in the hindbrain ventricle (HBV) of zebrafish larvae at 2 days postfertilization (dpf) is rapidly cleared and induces HSPC proliferation and differentiation as well as emergency granulopoiesis in the aorta-gonad-mesonephros (AGM) region (28). To better understand the immune responses during a nonlethal Shigella infection, we quantified the total number of neutrophils and macrophages in larvae injected with ~2 × 103 colony-forming units (CFU) (Fig. 1A). Neutrophil numbers significantly decrease at 24 hours post–first infection (hp1i), and by 48 hp1i, the pool of neutrophils is replenished in excess, consistent with observed granulopoiesis in the AGM (Fig. 1, B and C). Although the macrophage population is also significantly reduced by 24 hp1i, it does not recover by 48 hp1i (Fig. 1D), suggesting that macrophage hematopoiesis is impaired to favor granulopoiesis, consistent with previous reports (3, 6, 36, 37).
Fig. 1. Zebrafish exposure to nonlethal Shigella dose induces generation of more resilient neutrophils.
(A) Training of 2 dpf zebrafish larvae. (B) Representative images of Tg(lyz::dsRed) larvae at 48 hp1i. Scale bar, 500 μm. (C) Quantification of neutrophils [Tg(lyz::dsRed)] in naïve and Shigella-trained (2 × 103 ± 1.4 × 103 CFUs) larvae. N = 3 with 10 larvae per time point (mean ± SD). (D) Quantification of macrophages [Tg(mpeg::mCherry)] in naïve and Shigella-injected (1.8 × 103 ± 8.6 × 102 CFUs) larvae. N = 3 with 11 larvae per time point (mean ± SD). (E) Expression of cxcl8a, cxcl18b, il1b, and il10 in Shigella-trained larvae (1.3 × 103 ± 5.5 × 102 CFUs) as compared to naïve larvae. N = 3 with >5 larvae per time point (mean ± SEM). (F) Reinfection of trained larvae with lethal dose injection of Shigella at 4 dpf. (G) Representative images of Tg(lyz::dsRed) naïve and Shigella-trained larvae reinfected with Shigella flexneri M90T. Scale bar, 50 μm. (H) Quantification of recruited neutrophils [Tg(lyz::dsRed)] to the HBV in naïve and Shigella-trained larvae following reinfection with Shigella (PBS, 2.6 × 104 ± 9 × 103 CFUs; Shigella, 2.7 × 104 ± 1 × 104 CFUs). N = 3 with 5 larvae per time point (mean ± SD). (I) Bacterial counts from naïve and Shigella-trained larvae reinfected with Shigella (PBS, 2.7 × 104 ± 4.5 × 103 CFUs; Shigella, 2.5 × 104 ± 1 × 104 CFUs). N = 3 with 3 larvae per time point (mean ± SD). (J) Expression of cxcl8a, cxcl18b, il1b, and il10 following reinfection with Shigella in naïve (2.5 × 104 ± 1 × 104 CFUs) and Shigella-trained (2.7 × 104 ± 4.5 × 103 CFUs) as compared to PBS-injected controls. N = 3 with >5 larvae per time point (mean ± SEM). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test (C, D, H, I, and J). ns, not significant.
Analysis of cytokine expression showed a significant increase in proinflammatory cytokines CXCL8a, CXCL18b, interleukin-1β (IL-1β), IL-6, and tumor necrosis factor–α (TNF-α) between 6 and 24 hp1i (Fig. 1E and fig. S1A), which correlates with neutrophil recruitment and bacterial clearance (fig. S1B). At 48 hp1i, the proinflammatory response induced by Shigella training returns to basal levels, but expression of the anti-inflammatory cytokine IL-10 remains elevated. These results are consistent with the requirement of inflammation to induce proliferation of HSPCs (18, 19, 28) and show that trained larvae (i.e., those injected with a nonlethal dose of Shigella) are at immunological homeostasis before reinfection.
Upon a second lethal Shigella challenge (Fig. 1F), trained larvae show significantly higher survival rates by 48 hours post–second infection (hp2i) as compared to naïve larvae [i.e., those injected with phosphate-buffered saline (PBS)]. Quantification of neutrophil recruitment to the HBV when infected with a lethal dose (>2 × 104 CFU) showed that recruitment in Shigella-trained larvae is not significantly different from naïve larvae (Fig. 1, G and H). At 6 hp2i, bacterial burden in Shigella-trained larvae starts to reduce, as compared to naïve larvae (Fig. 1I), suggesting that trained neutrophils are better at clearing bacteria. Similarities in neutrophil recruitment in both conditions during infection are consistent with expression of proinflammatory cytokines CXCL8a and CXCL18b, which are expressed to the same extent in naïve and trained larvae at 6 hp2i (Fig. 1J). In trained infected larvae, the anti-inflammatory cytokine IL-10 expression is significantly different compared to that in naïve larvae (both uninfected and infected), suggesting that trained larvae strongly induce anti-inflammatory responses. At 24 hp2i, all analyzed cytokines are less expressed in trained infected larvae compared to naïve infected larvae (Fig. 1J and fig. S1C). Together, these observations indicate that Shigella-trained larvae are protected against a secondary infectious challenge because of tighter control of immune responses and their neutrophils have increased killing capacity, which are important characteristics of innate immune training.
Shigella induces nonspecific and lasting training mechanisms in zebrafish larvae
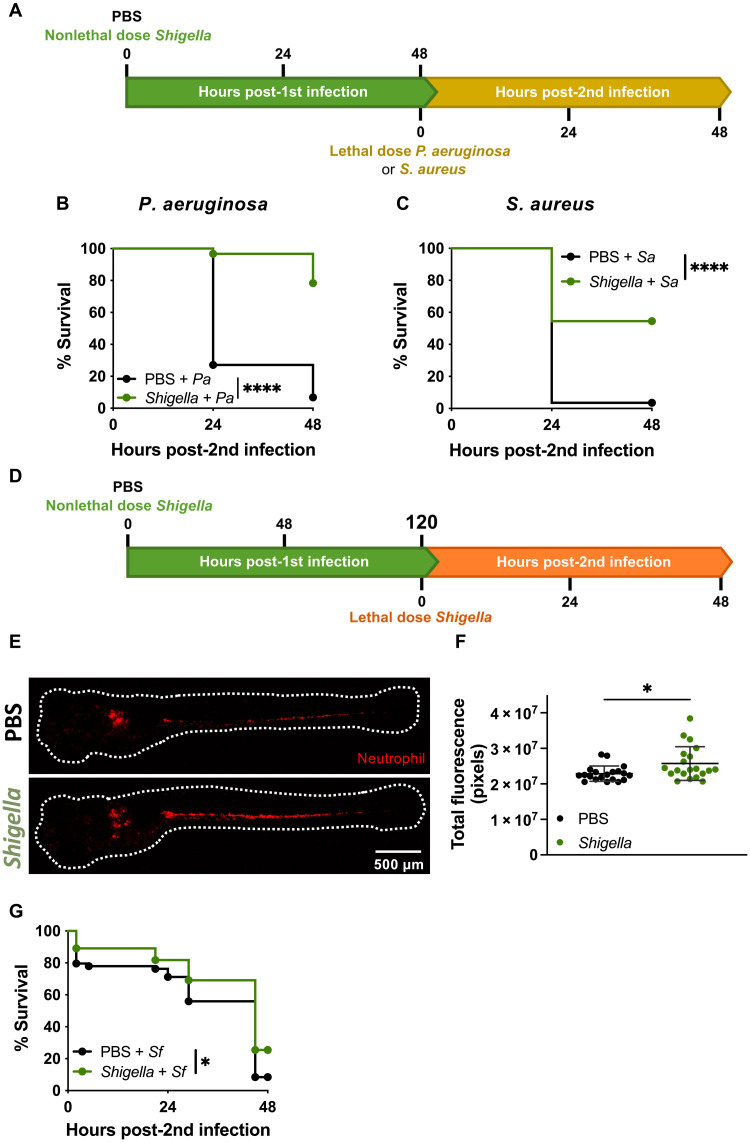
Work on trained innate immunity using BCG has demonstrated that induced protection is not specific to Mycobacterium tuberculosis and helps in controlling fungal infection by Candida albicans (38). In zebrafish, β-glucan and Salmonella enterica serovar Typhimurium have been shown to protect against Gram-negative and Gram-positive pathogens (27). Here, we tested our Shigella-trained larvae against lethal infections of Pseudomonas aeruginosa (a Gram-negative pathogen, 2 × 103 CFU) and Staphylococcus aureus (a Gram-positive pathogen, 2 × 104 CFU) (Fig. 2A). In both cases, the survival of Shigella-trained larvae is significantly higher as compared to the survival of naïve larvae (Fig. 2, B and C), and bacterial burden is controlled from 6 hp2i (fig. S2, A and B). These results demonstrate that Shigella-induced protection of zebrafish is nonspecific.
Fig. 2. Shigella induced protection is nonspecific.
(A) Reinfection of trained larvae with lethal dose injection of P. aeruginosa (Pa) or S. aureus (Sa) at 4 dpf. (B) Survival curves from naïve and Shigella-trained (2.2 × 103 ± 3.8 × 102 CFUs) larvae infected with P. aeruginosa (PBS, 2.6 × 103 ± 7.4 × 102 CFUs; Shigella, 2.3 × 103 ± 5.5 × 102 CFUs). N = 3 with 20 larvae per experiment. (C) Survival curves from naïve and Shigella-trained (1.4 × 103 ± 8.6 × 102 CFUs) larvae infected with S. aureus (PBS, 3.4 × 104 ± 1.1 × 104 CFUs; Shigella, 3.4 × 104 ± 6 × 103 CFUs). N = 3 with >20 larvae per experiment. (D) Reinfection of trained larvae with lethal dose injection of Shigella at 7 dpf (or 5 dp1i). (E) Representative images of naïve and Shigella-trained Tg(lyz::dsRed) larvae with at 5 dp1i. Scale bar, 500 μm. (F) Quantification of total fluorescence in Tg(lyz::dsRed) naïve and Shigella-trained (1.7 × 103 ± 5.7 × 102 CFUs) larvae at 5 dp1i. N = 3 with 10 larvae per time point (mean ± SD). (G) Survival curves from naïve and Shigella-trained larvae infected with Shigella at 5 dp1i. N = 3 with >16 larvae per experiment. *P < 0.05, ****P < 0.0001, log-rank (Mantel-Cox) test (B, C, and G) and unpaired Student’s t test (F).
To test the longevity of protection induced by Shigella training, we increased the resting interval between training and reinfection from 2 to 5 days (Fig. 2D). At this time point (7 dpf), we observed that neutrophil numbers remain higher in trained larvae as compared to naïve larvae (Fig. 2, E and F), but the differences are smaller than at 4 dpf (Fig. 1, A to C). Upon reinfection with a lethal dose of Shigella (2.5 × 104 CFU), survival of trained larvae is significantly higher compared to naïve larvae (Fig. 2G). However, differences in bacterial clearance are less detectable at 24 hp2i between groups (fig. S2C).
BCG and β-glucan induce protection in zebrafish larvae
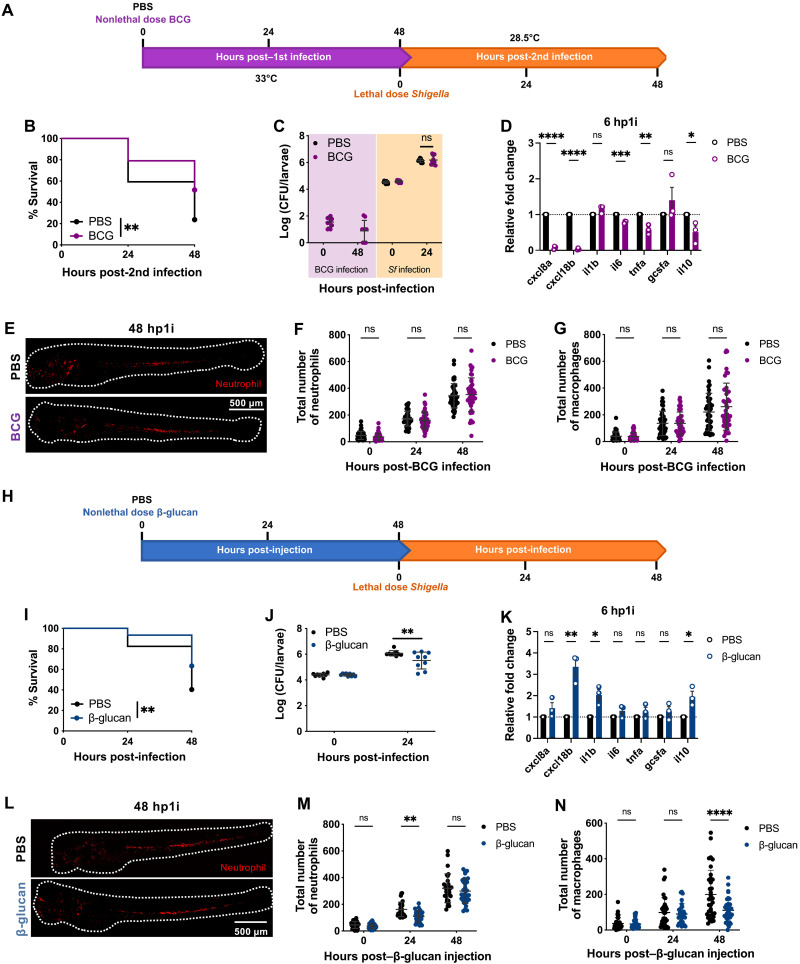
BCG and β-glucan have been widely studied as inducers of trained immunity (38, 39). To better understand the mechanisms by which Shigella can induce trained immunity, we developed zebrafish reinfection models with BCG and β-glucan for comparison. In the BCG reinfection model (Fig. 3A), a low dose of BCG (50 CFU) was injected in 2 dpf larvae that were then incubated at 33°C for 48 hours to stimulate BCG replication (40). Although BCG were not completely cleared from larvae (Fig. 3C), reinfection with a lethal dose of Shigella was done at 48 hp1i, and survival was followed for another 48 hours at 28.5°C. In this case, BCG-trained larvae had significantly greater survival than naïve larvae (Fig. 3B), although they presented a similar burden of Shigella at 24 hp2i (Fig. 3C). The difference between BCG and Shigella training may be associated with proinflammatory responses that are induced during training. Whereas Shigella strongly induces proinflammatory responses by 6 hp1i (Fig. 1E), BCG infection does not (Fig. 3D). Unexpectedly, the expressions of neutrophil chemokines CXCL8a and CXCL18b were repressed at 6 hp1i, which may explain the lack of neutrophil recruitment to the HBV by 24 hp1i (fig. S3A). Macrophages, in contrast, were recruited to the injection site and remained in the HBV until 24 hp1i (fig. S3B). Overall, no differences in neutrophil and macrophage numbers were observed during BCG infection (Fig. 3, E to G).
Fig. 3. BCG and β-glucan induce protection in zebrafish embryos.
(A) Reinfection of BCG-trained larvae with Shigella at 4 dpf. (B) Survival curves (N = 3 with >16 larvae per experiment) and (C) bacterial counts [N = 3 with 3 larvae per time point (mean ± SD)] from naïve and BCG-trained larvae infected with Shigella. (D) Expression of cxcl8a, cxcl18b, il1b, il6, tnfa, gcsfa, and il10 in BCG-trained larvae (4.7 × 101 ± 3.2 × 101 CFUs) as compared to naïve larvae. N = 3 with 10 larvae per time point (mean ± SEM). (E) Representative images of naïve and BCG-trained Tg(lyz::dsRed) larvae at 48 hp1i. Scale bar, 500 μm. (F) Quantification of neutrophils [Tg(lyz::dsRed)] and (G) macrophages [Tg(mpeg::mCherry)] in naïve and BCG-trained larvae [(F) 5.6 × 101 ± 4.3 × 101 CFUs, (G) 6.3 × 101 ± 3.7 × 101 CFUs]. N = 3 with >12 larvae per experiment (mean ± SD). (H) Reinfection of β-glucan–trained larvae with Shigella at 4 dpf. (I) Survival curves (N = 3 with >18 larvae per experiment) and (J) bacterial counts [N = 3 with 3 larvae per time point (mean ± SD)] from naïve and β-glucan–trained (20 ng) larvae infected with Shigella. (K) Expression of cxcl8a, cxcl18b, il1b, il6, tnfa, gcsfa, and il10 in β-glucan–trained larvae (20 ng) as compared to naïve larvae. N = 3 with 10 larvae per time point (mean ± SEM). (L) Representative images of naïve and β-glucan–trained Tg(lyz::dsRed) larvae at 48 hp1i. Scale bar, 500 μm. (M) Quantification of neutrophils [Tg(lyz::dsRed)] and (N) macrophages [Tg(mpeg::mCherry)] in naïve and β-glucan–trained larvae (20 ng). N = 3 with >8 larvae per experiment (mean ± SD). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, log-rank (Mantel-Cox) test (B and I), one-way ANOVA with Tukey’s multiple comparisons test (C and J), unpaired Student’s t test (D and K), and two-way ANOVA with Sidak’s multiple comparisons test (F, G, M, and N).
To establish a β-glucan (local) reinfection model (Fig. 3H), 20 ng of β-glucan was injected in the HBV of larvae and infection with Shigella occurred 48 hours after. Larvae trained with β-glucan had increased survival (Fig. 3I) and reduced bacterial burden by 24 hp2i (Fig. 3J). Analysis of cytokine expression at 6 hp1i demonstrated that cxcl18b, il1b, and il10 are significantly more expressed in β-glucan–trained larvae compared to naïve larvae (Fig. 3K), suggesting that β-glucan is recognized by the immune system. Similar to Shigella training, β-glucan induced changes in the neutrophil and macrophage populations. At 24 hp1i, the neutrophil numbers were significantly reduced, and by 48 hp1i, the population was restored to similar numbers as in naïve larvae (Fig. 3, L to M). The granulopoiesis observed was associated to expression of gcsfa expression at 24 hp1i (fig. S3F), but there was no significant increase in the number of neutrophils in the AGM as compared to naïve larvae (fig. S3G). Consequently, and similar to Shigella training, macrophage differentiation was blunted by 48 hp1i (Fig. 3N).
Considering differences across zebrafish models of trained innate immunity (Table 1), we conclude that the training mechanism induced by Shigella is different from that of BCG and β-glucan. However, the different training mechanisms share important protective phenotypes, such as the reduction of pathogen burden (independently of the bacterial species) and improvement of host survival.
Table 1. Comparison of training phenotypes between Shigella-, BCG-, and β-glucan–trained zebrafish larvae.
| Shigella | BCG | β-glucan | ||
|---|---|---|---|---|
| Training | Bacterial clearance | Yes | No | – |
| Neutropenia (24 hp1i) | Yes | No | Yes | |
| Emergency granulopoiesis (48 hp1i) | Yes | No | No | |
| Changes in macrophage population | Yes | No | Yes | |
| Induction proinflammatory responses | Yes | No | Yes | |
| Infection challenge | Survival protection | Yes | Yes | Yes |
| Infection control | Yes | No | Yes | |
| Nonspecific | Yes | Yes | Yes | |
Contribution of host factors, LPS, and Shigella effectors to training mechanisms
Gcsf signaling is crucial to induce emergency granulopoiesis in Shigella-trained larvae (28); however, it is not the only requirement to induce training mechanisms. To better understand triggers for Shigella-induced training in zebrafish larvae, we tested different factors important in Shigella infection of host cells (Fig. 3).
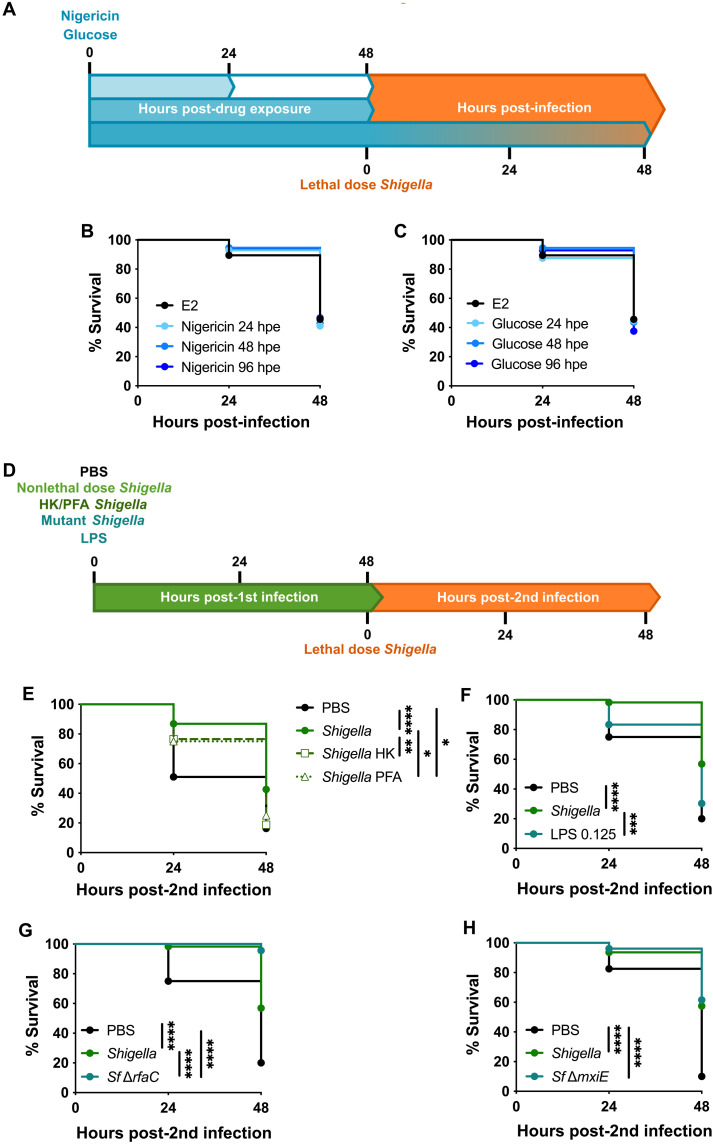
As previously described (18), inflammasome activation plays an important role in HSPCs production, and Shigella has been shown to be a potent activator of inflammasome pathways (31, 32). Therefore, we tested whether NOD-, LRR- and pyrin domain–containing protein 3 (NLRP3) inflammasome activation by nigericin and glucose in HSPCs is sufficient to induce training. Zebrafish larvae were bathed in either 0.1 μM nigericin or 1% glucose for at least 24 hours from 2 dpf, and at 4 dpf, larvae were injected with a lethal dose of Shigella (Fig. 4A). The survival assay showed that independently of exposure time to either nigericin or glucose, no differences in survival (Fig. 4, B and C) or bacterial burden (fig. S4, A and B) are observed. These results suggest that inflammasome and caspase-1 activation is not sufficient to induce training.
Fig. 4. Shigella stimulates inflammatory pathways to induce training.
(A) Exposure of zebrafish larvae to nigericin and glucose followed by Shigella infection. (B) Survival curves from E2- and nigericin (0.1 μM)–treated larvae infected with Shigella (PBS, 2.7 × 104 ± 8.5 × 103 CFUs; nigericin, 2.6 × 104 ± 4.1 × 103 CFUs). N = 3 with >14 larvae per experiment. hpe, hours post-drug exposure (C) Survival curves from E2- and glucose (1%)–treated larvae infected with Shigella (PBS, 2.7 × 104 ± 8.5 × 103 CFUs; glucose, 2.7 × 104 ± 1 × 104 CFUs). N = 3 with >14 larvae per experiment. (D) Reinfection of larvae trained with nonlethal dose of live or killed Shigella, Shigella mutants, or LPS followed by infection with Shigella at 4 dpf. (E) Survival curves from naïve, live-Shigella–trained (1.3 × 103 ± 6.6 × 102 CFUs), Shigella heat-killed (HK)–trained, and Shigella PFA–trained larvae infected with Shigella (PBS, 2.7 × 104 ± 6.1 × 103 CFUs; Shigella, 2.7 × 104 ± 1 × 104 CFUs; Shigella HK, 3.2 × 104 ± 3.3 × 104 CFUs; Shigella PFA, 3.2 × 104 ± 1.7 × 104 CFUs). N = 3 with >11 larvae per experiment. (F) Survival curves from naïve Shigella-trained (1.6 × 103 ± 6.5 × 102 CFUs) and LPS-primed (0.125 ng) larvae infected with Shigella (PBS, 1.9 × 104 ± 6.3 × 103 CFUs; Shigella, 2.8 × 104 ± 4.7 × 103 CFUs; LPS, 1.9 × 104 ± 9.1 × 103 CFUs). N = 3 with >18 larvae per experiment. (G) Survival curves from naïve, Shigella-trained (1.6 × 103 ± 6.5 × 102 CFUs), and Shigella ΔrfaC–trained (8.5 × 102 ± 7.5 × 102 CFUs) larvae infected with Shigella (PBS, 1.9 × 104 ± 6.3 × 103 CFUs; Shigella, 2.8 × 104 ± 4.7 × 103 CFUs; ΔrfaC, 2.4 × 104 ± 8.4 × 103 CFUs). N = 3 with >10 larvae per experiment. (H) Survival curves from naïve, Shigella-trained (7.9 × 102 ± 5.5 × 102 CFUs) and Shigella ΔmxiE–trained (8.8 × 102 ± 5.8 × 102 CFUs) larvae infected with Shigella (PBS, 1.9 × 104 ± 7.5 × 103 CFUs; Shigella, 2.5 × 104 ± 4.6 × 103 CFUs; ΔmxiE, 2.2 × 104 ± 7.6 × 103 CFUs). N = 3 with >10 larvae per experiment. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, log-rank (Mantel-Cox) test (B, C, E, F, G, and H).
Recently, studies with BCG and Salmonella have shown that inactivated or heat-killed bacteria can induce trained immunity mechanisms; however, the protection observed upon second challenge is weaker (27, 40, 41). In the case of Shigella (Fig. 4D), both heat-killed (HK) and paraformaldehyde (PFA)–killed Shigella failed to protect larvae survival upon a secondary lethal dose of Shigella (Fig. 4E and fig. S4C), strongly suggesting that Shigella needs to be alive to induce training.
To test the role of the LPS in training, LPS was used to prime larvae followed by secondary challenge with a lethal dose of Shigella (Fig. 4F and fig. S4, D to F). Larvae survival upon a lethal Shigella injection was not significantly affected by priming with LPS, highlighting that in our model LPS alone does not induce protection mechanisms. However, complete exposure of lipid A in a Shigella ∆rfaC mutant [which lacks the O-antigen, outer and inner core of LPS (42)] induced 100% protection upon reinfection with Shigella (Fig. 4G) and highly efficient bacterial clearance (fig. S4G). Virulence assays in zebrafish larvae showed that 1 × 104 CFU of Shigella ∆rfaC mutant can cause 100% lethality in 48 hours postinfection, whereas Shigella wild-type (WT) only kills 50% of the larvae (fig. S4H). The lethality was found to be caused by heighted proinflammatory responses and not due to bacterial burden (fig. S4, I and J). These results support work showing that stronger proinflammatory responses induced during training can result in better innate immune training (40).
Previous work has shown that the T3SS plays a role in training (28). Here, we tested the effect of T3SS effectors regulated by the master regulator MxiE (43). Larvae survival upon reinfection showed that MxiE-regulated effectors do not significantly affect how Shigella induces training (Fig. 4H and fig. S4K). We also tested a Shigella ∆ospF mutant, lacking the bacterial immunomodulin OspF (44, 45), where similar results to ∆mxiE mutant were obtained (fig. S4, L and M). These results are consistent with similar levels of virulence observed during infection of zebrafish larvae with Shigella ∆mxiE and ∆ospF mutants and Shigella WT (fig. S4, N to Q). Together, these data suggest that live Shigella, with a functional T3SS, is required to induce inflammatory responses that trigger training mechanisms.
Exposure to Shigella induces epigenetic reprogramming of neutrophils
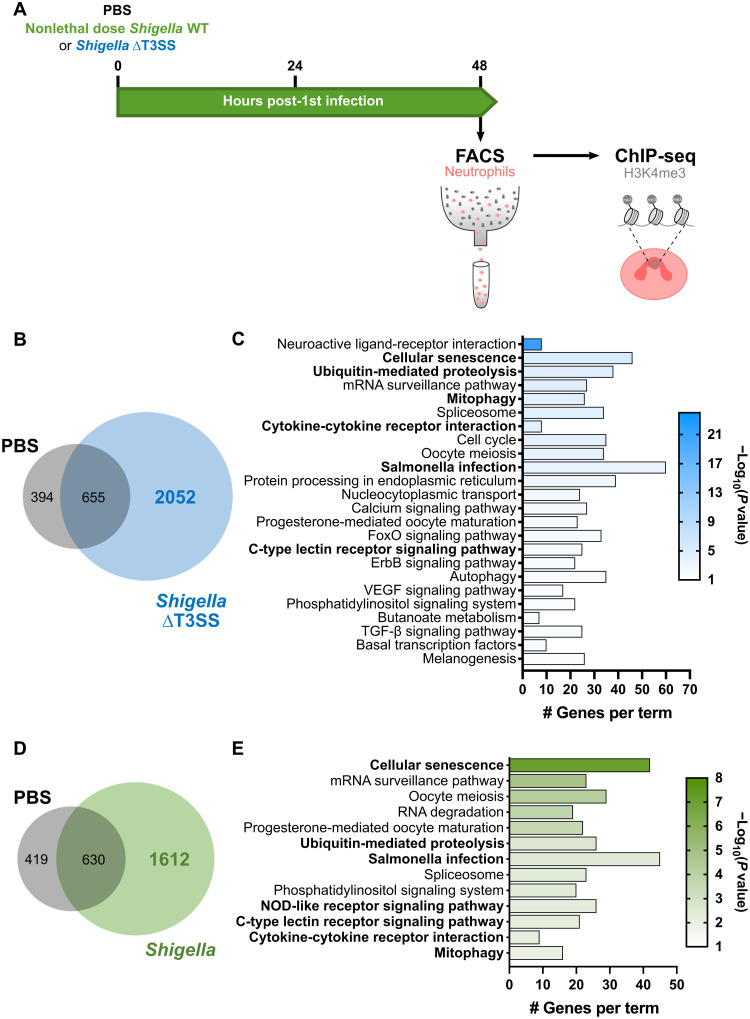
Epigenetic reprogramming is a defining characteristic underlying the innate immune training of myeloid cells, and the enrichment of H3K4me3 on gene promoter regions is typically associated to active/poised gene transcription (8, 38, 39, 46). Considering this, we investigated whether Shigella-trained neutrophils exhibit different H3K4me3 patterns that could explain their ability to better respond to reinfection. To test this, neutrophils were isolated by fluorescence-activated cell sorting (FACS) from naïve and trained [Shigella WT and Shigella ∆T3SS (i.e., mxiD mutant)] larvae at 48 hp1i (fig. S5A), and chromatin immunoprecipitation sequencing (ChIP-seq) was performed on the H3K4me3 mark (Fig. 5A). Genes were then annotated if they had a H3K4me3 mark in their promoter region [defined as ±3 kb from the transcription start site (TSS); data files S1 to S3]. Naïve neutrophils displayed H3K4me3 peaks at the promoter region of 1049 genes, which are associated with normal cell functioning (fig. S5, B and C), including surveying the quality of mRNA to increase the fidelity of RNA processing. Notably, neutrophil samples trained with Shigella WT or ΔT3SS showed an increase in the number of genes marked by H3K4me3 (2242 and 2707, respectively) as compared to naïve neutrophils, suggesting epigenetic remodeling of trained neutrophils (Fig. 5, B and D, and fig. S5, D and E).
Fig. 5. Exposure to Shigella induces epigenetic reprogramming of neutrophils.
(A) Isolation of neutrophils by FACS from naïve and Shigella-trained larvae for ChIP-seq on H3K4me3 modification. (B) Venn diagram showing the number of common and unique genes marked by H3K4me3 peaks in their promoter regions [±3 kb from transcription start site (TSS)] of PBS and Shigella ∆T3SS–trained larvae. (C) Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (P value < 0.01) associated with the 2052 unique Shigella ∆T3SS training genes marked by an H3K4me3 peak in their promoter regions (±3 kb from TSS). FoxO, Forkhead box O; ErbB, erythroblastic oncogene B; VEGF, vascular endothelial growth factor; TGF-β, transforming growth factor–β. (D) Venn diagram showing the number of common and unique genes marked by H3K4me3 peaks in their promoter regions (±3 kb from TSS) of PBS and Shigella WT trained larvae. (E) Enriched KEGG pathways (P value < 0.01) associated with the 1612 unique Shigella WT training genes marked by an H3K4me3 peak in their promoter regions (±3 kb from TSS). NOD, nucleotide binding oligomerization domain.
Upon Shigella ∆T3SS training, pathway analysis of marked gene promoter regions indicates that some modulated responses are common to Shigella WT training; however, many other different signaling pathways are affected (Fig. 5C and data file S2), highlighting that training with Shigella WT leads to a more precise and efficient immune response. Shigella WT–induced H3K4me3 deposition at promoter regions was observed for genes involved in cellular senescence and in infection signaling pathways, specifically associated to infections by Gram-negative pathogens (Fig. 5E). The analysis suggests that trained neutrophils have metabolic and gene expression alterations linked to senescence and also activation of innate immune signaling via mitogen-activated protein kinase (MAPK) signaling pathway to express proinflammatory cytokines, chemokines, and antimicrobial peptides. In addition, although most promoter regions of proinflammatory cytokines are not marked by H3K4me3, deposition of this mark was found in promoter regions of cytokine receptor genes il10rb, tgfbr1b, ifngr1l, cxcr3.2, and cxcr3.3 (data file S3). These results highlight the signaling pathways that may be activated and their importance for cell survival and immunomodulation.
Reprogrammed neutrophils produce more mtROS for enhanced antimicrobial function
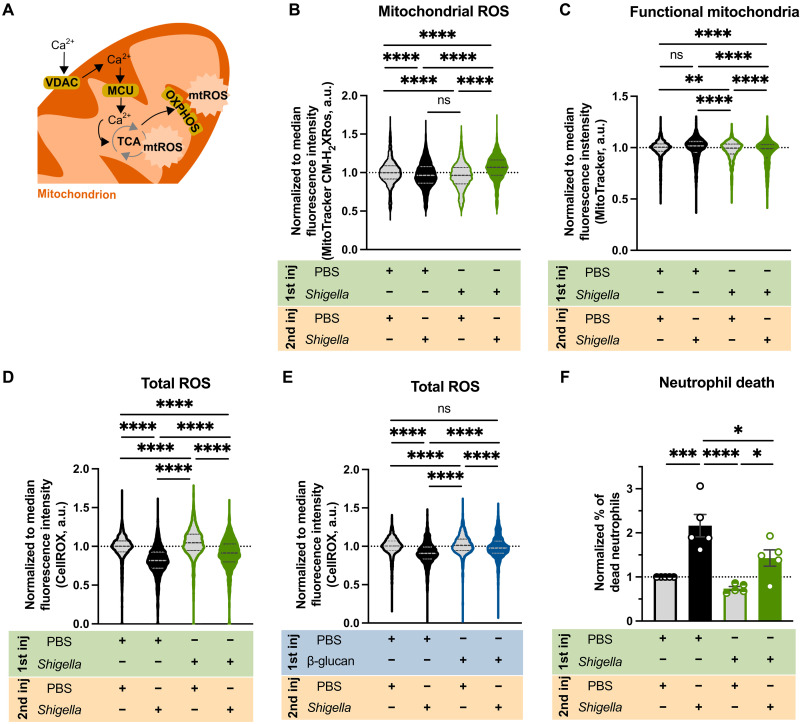
The ChIP-seq dataset strongly suggested increased mtROS production upon Shigella WT training (Fig. 5E). In agreement, we found that the promoter regions of the genes encoding mitochondrial calcium uniporter (MCU) and voltage-dependent anion channel (VDAC) transporters [mitochondrial permeability transition pore (mPTP)], as well as several genes involved in the tricarboxylic acid cycle and oxidative phosphorylation (Fig. 6A and data file S3), are marked by H3K4me3 in Shigella WT–trained but not in naïve larvae. To test whether mtROS production was increased in trained neutrophils, we used flow cytometry to quantify MitoTracker CM-H2XRos dye in naïve and Shigella-trained neutrophils in resting and restimulated state at 6 hp2i. Quantifications show that naïve neutrophils when infected with Shigella have reduced mtROS, compared to unstimulated neutrophils (Fig. 6B). Trained neutrophils also present lower levels of mtROS when unstimulated; however, after 6 hp2i, mtROS levels are significantly increased as compared to unstimulated naïve and trained neutrophils. To test for differences in mitochondrial mass between naïve and trained neutrophils, we measured functional mitochondria using MitoTracker staining (Fig. 6C). We found that naïve neutrophils exposed to infection (despite having more functional mitochondria) do not produce more mtROS to combat Shigella, as compared to unstimulated naïve neutrophils (Fig. 6, B and C, and fig. S6D). In contrast, trained neutrophils exposed to infection produce more mtROS without increasing the amount of functional mitochondria, as compared to unstimulated trained neutrophils.
Fig. 6. Trained neutrophils produce more mtROS.
(A) Increased calcium transportation via VDAC and MCU increases mtROS. (B) Normalized fluorescence of MitoTracker Red CM-H2XRos in neutrophils from naïve and Shigella-trained (1.2 × 103 ± 5.3 × 102 CFUs) larvae [Tg(mpx::GFP)] unstimulated or infected with Shigella (PBS, 2.6 × 104 ± 9.3 × 103 CFUs; Shigella, 3 × 104 ± 6.6 × 103 CFUs) at 6 hp2i. N = 3 with >10 larvae per experiment (mean ± SD). (C) Normalized fluorescence of MitoTracker Deep Red in neutrophils from naïve and Shigella-trained (1.8 × 103 ± 1 × 103 CFUs) larvae [Tg(mpx::GFP)] unstimulated or infected with Shigella (PBS, 2.1 × 104 ± 1.1 × 104 CFUs; Shigella, 1.9 × 104 ± 8.3 × 103 CFUs) at 6 hp2i. N = 3 with 30 larvae per experiment (mean ± SD). (D) Normalized fluorescence of CellROX Deep Red in neutrophils from naïve and Shigella-trained (1.4 × 103 ± 6.7 × 102 CFUs) larvae [Tg(mpx::GFP)] unstimulated or infected with Shigella (PBS, 2.6 × 104 ± 1 × 104 CFUs; Shigella, 2.6 × 104 ± 1 × 104 CFUs) at 6 hp2i. N = 3 with >10 larvae per experiment (mean ± SD). (E) Normalized fluorescence of CellROX Deep Red in neutrophils from naïve and β-glucan–trained larvae [Tg(mpx::GFP)] unstimulated or infected with a lethal dose of Shigella flexneri M90T (PBS, 2 × 104 ± 1.3 × 104 CFUs; β-glucan, 2.7 × 104 ± 9.8 × 103 CFUs) at 6 hp2i. N = 3 with >20 larvae per experiment (mean ± SD). (F) Normalized percentage of dead neutrophils (LIVE/DEAD Fixable Violet Stain) from naïve and Shigella-trained larvae [Tg(mpx::GFP)] unstimulated or infected with Shigella at 6 hp2i. N = 5 with >10 larvae per experiment (mean ± SD). *P < 0.05, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Tukey’s multiple comparisons test (B, C, D, and E). a.u., arbitrary units.
Given the redox cross-talk between mitochondria and NADPH (reduced form of NADP+) oxidases (47), we investigated total ROS production in neutrophils using CellROX dye. Quantifications showed that naïve neutrophils when infected (6 hp2i) have significantly lower total ROS levels as compared to unstimulated state (Fig. 6D). Shigella-trained neutrophils in resting phase showed higher levels of ROS, compared to naïve unstimulated neutrophils. However, when trained neutrophils became stimulated, total ROS levels decreased, but not to the same extent as in naïve infected neutrophils. The effect of both mtROS and ROS on bacterial killing was tested by adding specific inhibitors of mPTP [cyclosporin A (CsA)] and NADPH oxidase [diphenyleneiodonium chloride (DPI)] to naïve and Shigella-trained larvae bath during the 6 hp2i. Recovered CFU showed that inhibitors had more impact on bacterial burden control in Shigella-trained larvae than in naïve larvae (fig. S6, B, C, F, and G), suggesting that both mtROS and ROS are responsible for improved bacterial clearance in trained larvae. To understand differences between Shigella and β-glucan training, we tested whether ROS production in zebrafish neutrophils is affected by β-glucan training (Fig. 6E and fig. S6H). In this case, we observed that β-glucan–trained neutrophils exposed to Shigella are not significantly depleted of ROS, as observed for Shigella-trained neutrophils (Fig. 6D). However, we found that β-glucan–trained neutrophils can sustain enhanced ROS production following Shigella infection, highlighting that host defense mechanisms used by trained neutrophils are different between Shigella and β-glucan training.
Our ChIP-seq dataset also pointed to activation of cellular senescence, particularly prosurvival and anti-apoptotic pathways (PI3K-Akt signaling and mitochondrial pathways; Fig. 5, C and E, and data files S2 and S3) were marked (48). To test whether Shigella-trained neutrophils could survive longer than naïve neutrophils, we used flow cytometry and found that there was a significant increase in the percentage of dead naïve neutrophils when exposed to Shigella, as compared to Shigella-trained neutrophils (Fig. 6F). These data support the hypothesis that trained neutrophils are more resilient to death upon Shigella reinfection.
To test whether trained immunity can affect other aspects of host defense, we investigated levels of myeloperoxidase (MPO) in trained neutrophils. Using the transgenic zebrafish line Tg(mpx::GFP) in which GFP expression is driven by the MPO promoter, we quantified MPO production using FACS. In agreement with its absence from ChIP-seq analysis, we did not observe significant differences in MPO production between naïve and trained neutrophils when reinfected (fig. S6I), highlighting the specificity of the Shigella training toward mtROS production. Together, these findings suggest that Shigella-trained neutrophils sustain H3K4me3 modifications that result in enhanced antimicrobial function via mtROS in preparation for a second challenge.
DISCUSSION
Here, we introduce the use of zebrafish larvae to study innate immune training, demonstrating that local infection by Shigella, BCG, and β-glucan induces nonspecific protective effects. The zebrafish larval model is a highly attractive training model—as compared to in vitro (single-cell type system) and mouse models—as it is a vertebrate model with only innate immune responses during its first ~4 weeks of development.
Most work on trained immunity has been largely focused on monocytes and macrophages. However, there is growing evidence that neutrophils also play important roles upon training (12, 13). In the case of Shigella infection, macrophages are unable to control infection as bacteria induce cell death programs (31, 33, 49). Although neutrophils can also be killed by Shigella, they resist for longer than macrophages and can rapidly eliminate bacteria (31, 33). Here, we show that Shigella infection induces production of new neutrophils that are more protective and have better killing capacity than naïve neutrophils. We show that this enhanced infection control is linked to host control of its inflammatory state, which also contributes to increased survival upon a secondary infectious challenge. Considering that the immune status of zebrafish larvae returns to basal levels by 48 hp1i and that neutrophils retain epigenetic modifications following a first Shigella infection stimulus, we established that Shigella trains (and does not prime) the innate immune system of zebrafish. Shigella training protects against Gram-negative bacteria (as Shigella and P. aeruginosa) and also Gram-positive bacteria (as S. aureus), suggesting that it induces heterologous immunological memory. From our results showing that Shigella training increased survival and infection control, we conclude that the defense arm engaged by Shigella training contributes toward host resistance and not disease tolerance (50, 51). Although we observed that expression of the anti-inflammatory cytokine IL-10 remained elevated by 48 hp1i, future work is required to identify the precise source of IL-10 and investigate whether Shigella-trained neutrophils can adopt an anti-inflammatory state (52).
To test the hypothesis that Shigella induces lasting training mechanisms in zebrafish larvae, we extended the interval between infections to 5 days and observed protection against Shigella in those larvae. Although protection levels might wane, this is in agreement with the decline of BCG-induced protection after vaccination (53, 54). In the future, it will be of great interest to test adult zebrafish (>3 months) as well as investigate whether Shigella training mechanisms can be transmitted trans-generationally in zebrafish as shown using mice (55). We envision that with the practices of zebrafish laboratories to maintain adequate levels of heterozygosity (56), by consistently outbreeding their stocks (unlike mouse lines), the zebrafish model can be used to complement the mouse model aiming to elucidate transmission of trained immunity in humans.
Work in zebrafish has shown that BCG vaccination protects against Mycobacterium marinum and that β-glucan (systemic) protects against Salmonella Typhimurium (24, 27). Both BCG and β-glucan training induce protection against Shigella infection in zebrafish but rely on distinct training pathways for whole animal infection control. Consistent with this, work using mice has shown that BCG induces training via nucleotide binding oligomerization domain containing 2 (NOD2) receptor and β-glucan via dectin-1 receptor (38, 39, 57). Although Shigella training phenotypes in zebrafish share similarities with those from BCG and β-glucan training (such as nonspecific protection and enhanced survival upon new infection), the H3K4me3-enriched pathways following Shigella training differ from those found in BCG-trained human neutrophils (12), which supports our conclusion that both stimuli induce different training mechanisms in neutrophils. Our findings suggest that Shigella training is multifactorial, depending on Shigella recognition by innate immune receptors, strong cytokine production, and an active T3SS.
Enriched pathways from our ChIP-seq datasets indicate that mtROS is playing an important role in enhancing antimicrobial activity of Shigella-trained neutrophils. Historically, ROS production in neutrophils has been associated with NADPH oxidase, and therefore, the bactericidal role of mtROS in neutrophils has remained mostly elusive. However, recent work in human and mouse neutrophils suggested that mitochondria generated ROS enhanced bactericidal activity (58, 59). In agreement, our work highlights that mtROS in neutrophils plays a key role in bacterial killing, and we envision that future therapeutic strategies could manipulate mtROS by using pro-oxidants [such as SkQN (60) or MitoK3 (61)] specifically targeted to mitochondria in neutrophils.
Decades of research have tried to generate a vaccine with sufficient efficacy that could be used against different Shigella serotypes in humans (62). Our long-term goal is to guide vaccine studies in humans, exploiting innate immune memory to combat Shigella infections. A variety of animal models have significantly contributed to understanding shigellosis and feasibility of vaccines to combat it. We highlight zebrafish as an important animal model to investigate how innate immune training can be manipulated to control Shigella infection. It is next of great interest to train neutrophils using live Shigella in humans. In this way, we can discover fundamental components of immunity shared between zebrafish and humans that can be trained to promote a better immune response and combat Shigella infection.
MATERIALS AND METHODS
Ethics statements
Animal experiments were performed according to the Animals (Scientific Procedures) Act 1986 and approved by the Home Office (Project licenses: PPL P84A89400 and P4E664E3C).
Zebrafish husbandry
Embryos were obtained from naturally spawning zebrafish, and larvae were maintained at 28.5°C in embryo medium (0.5× E2 medium), unless specified otherwise. Transgenic zebrafish lines used here were Tg(lyz::dsRed)nz50 (63), Tg(mpx::GFP)i114 (64), and Tg(mpeg1::Gal4-FF)gl25/Tg(UAS-E1b::nfsB.mCherry)c264 (65). For injections and live microscopy, larvae were anesthetized with tricaine (200 μg/ml; Sigma-Aldrich) in embryo medium. For injections in larvae >5 dpf, lidocaine (2 μg/liter) was used as an analgesic and added to the embryo medium [together with tricaine (200 μg/ml; Sigma-Aldrich)] before injection. After checking for full recovery from the anesthetic, larvae were kept in E2 medium with lidocaine (2 μg/liter) for 18 to 24 hours at 28.5°C. Larvae were monitored for appearance, righting reflex, reactive reflex, and opercular movement from day 5 dpf for two to three times a day. Larvae were not fed during the course of the experiment.
Zebrafish injections
Bacterial strains used in this study can be found in table S1. Shigella, Pseudomonas, and Staphylococcus strains were grown on trypticase soy agar (TSA; Sigma-Aldrich) plates with the appropriate antibiotics—for Shigella growth, plates were supplemented with 0.01% Congo red (Sigma-Aldrich). Overnight cultures were grown from individual colonies at 37°C and 200 rpm, in 5 ml of trypticase soy broth (TSB; Sigma-Aldrich and Oxoid for Staphylococcus) supplemented with the appropriate antibiotics as above. For injections, bacteria were grown until an optical density (OD) of 0.55 to 0.65 at 600 nm (log phase) by diluting the overnight culture 50× in fresh TSB supplemented with the appropriate antibiotics. When in log phase, bacteria were spun down and washed in PBS (Sigma-Aldrich). The desired concentration was achieved by resuspension of bacteria in injection buffer [2% polyvinyl-pyrrolidone (PVP; Sigma-Aldrich) in PBS and 0.5% phenol red (Sigma-Aldrich)]. Control groups were injected with injection buffer (referred as PBS/naïve group).
For inactivated bacteria injections, suspensions were prepared as described above and, before injection, were incubated for 30 min at 60°C for heat killing or incubated in 4% PFA (Sigma-Aldrich) for 30 min at room temperature (RT) for PFA killing.
BCG was grown in 7H9 broth (supplemented with 10% albumin dextrose catalase (ADC) enrichment medium, 0.2% glycerol, and 0.02% Tween-80) until OD 0.5 to 0.6. The culture was then spun down and washed with PBS. The suspension was then centrifuged and resuspended in PVP to prevent large clumps. Single-cell suspensions were obtained by passing the bacteria through different gauge syringes (25G, 27G, and 29G) 10 times each. After achieving the desired concentration, bacteria were resuspended in injection buffer with higher concentration of PVP (4%).
A stock solution of β-glucan (Invivo Biosystems, catalog no. tlrl-bgp) was prepared at 20 mg/ml in PBS and stored at 4°C for a maximum of a month. For injections, indicated dilutions were prepared in injection buffer. Different amounts of β-glucan were first injected in the HBV of larvae to test for dose-dependent protection (fig. S3, C to E). The highest amount tested (20 μg/μl) was chosen as it suggested stronger protection responses.
LPS (Sigma-Aldrich, catalog no. L6143) was prepared to mimic the amount of bacteria injected in a nonlethal dose (2500 CFU). According to (66), 2500 bacterial cells in exponential phase would correspond to 0.125 ng of LPS. Therefore, a stock solution of LPS (0.25 mg/ml) in PBS was prepared and prior before injection was mixed 1:1 with 0.5% phenol red.
For injection, 1 to 2 nl of bacterial/β-glucan/LPS suspension or control solution was microinjected in the HBV of zebrafish larvae. For bacterial enumeration, larvae tissues were disrupted in 0.4% Triton X-100 (Sigma-Aldrich) with the aid of mechanical pestles, and serial dilutions were plated in the appropriate medium at the indicated time points. Plates were incubated at 37°C, and CFUs were enumerated when colonies were visible.
Design of S. flexneri ∆ospF mutant
S. flexneri ∆ospF mutant was created using a λ-Red–mediated recombination (67). Briefly, pKD4 plasmid was used as template to amplify a kanamycin resistance–encoding DNA cassette using primers containing 50-bp nucleotides homologous to the site of insertion (table S1). S. flexneri electrocompetent cells producing λ-Red recombinase were electroporated with resulting polymerase chain reaction (PCR) fragments. Electroporated cells were then plated in TSA plates supplemented with 0.01% of Congo red and kanamycin (50 μg/ml). To remove the kanamycin resistance–encoding DNA cassette, bacteria were transformed with pCP20 plasmid [that encodes the yeast flp recombinase (68)]. Deletions were verified by PCR using confirmation primers in table S1.
Microscopy and image analysis
Whole larvae and HBV images were acquired using stereo fluorescent microscope Leica M205FA (Leica, Germany) and by placing anesthetized larvae on 1% agarose-E2 plates.
Image files were processed using ImageJ/Fiji software. Whole larvae leukocyte quantification was performed according to (69) with modifications. Briefly, fluorescent images were converted to binary in ImageJ/Fiji, resulting in images in which fluorescence was converted into black pixels onto a white background. For each image, pixels of five individual cells were quantified. Total pixel count of each larva was divided by the average of five individual leukocytes to determine the total number of cells. Leukocyte quantification in the HBV was determined manually in the region highlighted in Fig. 1G.
Zebrafish chemical treatments
For inflammasome stimulation, larvae were kept at 28.5°C in E2 medium with 0.1 μM nigericin (Sigma-Aldrich, catalog no. 481990-5MG) or 1% d-glucose (Sigma-Aldrich, catalog no. G8270-100G) (18) for 24, 48, or 96 hours from 2 dpf. For NAPDH ROS and mtROS inhibition after the second infection, larvae were kept at 28.5°C in E2 medium with 100 μM DPI (Sigma-Aldrich, catalog no. D2926) (70) and 10 μM CsA (Enzo Life Sciences, catalog no. BML-A195-0100) (71), both in dimethyl sulfoxide (DMSO), for 6 hours until larvae were dissociated for flow cytometry. Control larvae were kept in E2 with 1% DMSO.
RNA extraction, cDNA synthesis, and quantitative reverse transcription–PCR
RNA was extracted from 5 to 10 snap-frozen larvae with the RNeasy Mini kit (Qiagen, catalog no. 74104) and reverse-transcribed using QuantiTect Reverse Transcription kit (Qiagen, catalog no 205311) according to manufacturer’s instructions. Quantitative PCR (qPCR) was performed using the 7500 Fast Real-Time PCR System machine and 7500 Fast Real-Time PCR software v2.3 (Applied Biosystems, Foster City, CA) and SYBR green master mix (Applied Biosystems, catalog no. 10187094). Template cDNA was subjected to PCR using primers described in table S1, and samples were run in technical duplicates. The comparative Ct method was used for gene expression quantification, and ef1a1l1 was used as the housekeeping gene.
Flow cytometry
Tg(lyz::dsRed)nz50 and Tg(mpx::GFP)i114 transgenic larvae were dissociated for flow cytometry following the protocol from (72), with some modifications. Briefly, 20 to 100 larvae were washed twice in calcium-free Ringer’s solution for 10 min with slow shaking at RT. After removal of Ringer’s solution, 2 ml of dissociation solution [0.5% Trypsin EDTA 10× no phenol (Thermo Fisher Scientific, catalog no.15400054)] was added. Larvae were incubated twice for 10 min at 28.5°C, with gentle pipetting in between incubations to help tissue digestion. The reaction was stopped by adding 10% fetal calf serum (Themo Fisher Scientific, catalog no. 11550356). The suspensions were sieved using 70-μm Nylon cell strainers (Merck, CLS431751-50EA) or cell strainer snap caps from test tubes (Corning, 352235) and then gently centrifuged for 3 min at 800g. Cells were then washed with 1 ml of PBS with 10 mM Hepes and 1 mM EDTA. For live/dead staining, cells were incubated with either LIVE/DEAD fixable near IR dead cell stain (1:2000; Thermo Fisher Scientific, catalog no. L34994) or LIVE/DEAD fixable violet dead cell stain (1:2000; Thermo Fisher Scientific, catalog no. L34964) dyes for 30 min at 28.5°C. Cells were fixed with 4% PFA for 10 min at RT for flow cytometry. For ROS staining, 5 μM CellROX Deep Red (Thermo Fisher Scientific, catalog no. C10422) was added to the E2 medium for 30 min before larvae dissociation, and the larvae were kept in the dark at 28.5°C. Single cells were measured on a LSRII (BD Biosciences) or on an Attune NxT Flow Cytometer (Thermo Fisher Scientific). For mtROS staining, 300 nM MitoTracker Red CM-H2XRos (Thermo Fisher Scientific, catalog no. M7513) was added to the dissociated cells for 30 min at 28.5°C. Single cells were measured on a FACSAria III cell sorter (BD Biosciences). Data were analyzed with FlowJo software v10.7.1. To prepare samples for ChIP-seq, cells were fixed with 1% formaldehyde for 8 min at RT with occasional mixing. To stop the reaction, 100 μl of glycine 1.25 M was added, and the samples were incubated for 5 min at RT. After centrifuging at 600g for 10 min at 4°C, cells were washed with ice-cold PBS containing protease inhibitor cocktail (Sigma-Aldrich, catalog no. 5892970001) and centrifuged again. Cells were then resuspended in PBS with 10 mM Hepes and 1 mM EDTA. FACS sorting was done on a FACSAria III cell sorter (BD Biosciences). Data were analyzed with BD FACSDiva software. Gating strategies can be found in fig. S5A for neutrophil sorting, in fig. S6A for mtROS measurement, and fig. S6E for CellROX measurement.
ChIP-seq and analysis
For ChIP experiments, a minimum of 20,000 sorted Tg(lyz:dsRed) neutrophils were collected per sample (PBS, Shigella, and Shigella ∆T3SS) and per replicate (each biological replicate corresponds to two experiments). Cells were kept at –80°C until sequencing. ChIP-seq was conducted by Diagenode ChIP-seq/ChIP-qPCR Profiling service (Diagenode, catalog no. G02010000). Chromatin was prepared for ChIP-seq using the True MicroChIP Kit (Diagenode, catalog no. C01010130) and was sheared using Bioruptor Pico sonication device (Diagenode, catalog no. B01060001) combined with the Bioruptor Water cooler for 5 cycles using a 30″ (on) 30″ (off) settings. Optimization of shearing conditions was done on chromatin corresponding to 10,000 cells. Immunoprecipitation was done on chromatin corresponding to the remaining 10,000 cells using antibodies against H3K4me3 (Diagenode, catalog no. C15410003)—10% of the chromatin was used for input. Libraries were prepared using the IP-Star Compact Automated System (Diagenode, catalog no. B03000002) from input and ChIP’d DNA using MicroPlex Library Preparation Kit v2 (12 indices) (Diagenode, catalog no. C05010013). Libraries were pooled and sequenced with Illumina Novaseq 6000, running NovaSeq Control Software 1.6.0, with paired-end reads of 50-bp length. Quality control of sequencing reads was performed using FastQC (73).
ChIP-seq reads were analyzed using the Galaxy platform (usegalaxy.eu). Reads were aligned to the reference genome GRCz10 using the Bowtie2 software (74), and PCR duplicates were removed with MarkDuplicates (75). Peak calling was performed using MACS2 callpeak (76), and the narrow peaks lists were used in Irreproducible Discovery Rate (IDR) (77) to identify reproducible peaks between replicates. Last, ChIPseeker (78) was used for peak annotation with a promoter defined as ±3 kb around the transcription start site (TSS). IGV v2.9.4 (79) was used for peak visualization.
For pathway annotation, the lists of genes associated with promoter regions highlighted in bold in Fig. 5 (B and D) and fig. S5 (B and D) (PBS, Shigella WT, and Shigella ΔT3SS) were analyzed against Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in Cytoscape v3.9.1 (80) using the plug-in ClueGO v2.5.9 (81).
Statistical analysis
Statistical significance was determined using GraphPad Prism v9. For differences in survival curves, the log-rank (Mantel-Cox) test was used. Data from bacterial burden and gene expression levels were log10- or log2-transformed, respectively. Pairwise comparisons were determined using a Student’s unpaired t test. For multiple comparisons, one-way or two-way analysis of variance (ANOVA) tests with Sidak’s or Tukey’s corrections were used, as indicated in the figure legend.
Acknowledgments
We thank Mostowy lab members, in particular V. Torraca, D. Lobato-Márquez, and A. T. López-Jiménez, for helpful discussions. We thank A. Filloux for providing P. aeruginosa. We thank T. Palmer for providing S. aureus. We thank M. Behr for providing BCG. We thank F. Randow for providing S. flexneri Ruby strains. We thank the LSHTM Biological Services Facility for the work and care of our fish stocks. We thank T. Prasjnar for troubleshooting embryo dissociation protocols. We thank Imperial College London Flow Cytometry Facility South Kensington, in particular L. Zarate-Garcia and J.Srivastava, for help with neutrophil sorting. We thank Diagenode for the work and helpful discussions on ChIP-seq.
Funding: D.B. is funded by the DFG Walter Benjamin Programme (BR 6637/1-1). Research in the F.C.W. laboratory is supported by the BBSRC (BB/R016283/). Research in S.M. laboratory is supported by a European Research Council Consolidator Grant (772853–ENTRAPMENT), Wellcome Trust Senior Research Fellowship (206444/Z/17/Z), and the Lister Institute of Preventive Medicine.
Author contributions: Conceptualization: M.C.G. and S.M. Methodology: M.C.G., D.B., and M.K.B. Software: M.C.G., D.B., M.K.B., and F.C.W. Validation: M.C.G. Formal analyses: M.C.G., D.B., and F.C.W. Investigation: M.C.G., D.B., and M.K.B. Data curation: M.C.G. Writing: M.C.G. and S.M. Supervision: S.M. Project administration: S.M. Funding acquisition: S.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Further information for resources and reagents should be directed to and will be fulfilled by S.M. at London School of Hygiene & Tropical Medicine, United Kingdom (serge.mostowy@lshtm.ac.uk). All unique/stable reagents generated in this study are available from S.M. without restriction. The ChIP-seq data of neutrophils 48 hours after Shigella training as reported in this paper can been found in the NCBI Gene Expression Omnibus under accession number GEO: GSE217063.
Supplementary Materials
This PDF file includes:
Table S1
Figs. S1 to S6
Legends for data files S1 to S3
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S3
REFERENCES AND NOTES
- 1.J. Domínguez-Andrés, J. C. Dos Santos, S. Bekkering, W. J. M. Mulder, J. W. M. van der Meer, N. P. Riksen, L. A. B. Joosten, M. G. Netea, Trained immunity: Adaptation within innate immune mechanisms. Physiol. Rev. 103, 313–346 (2023). [DOI] [PubMed] [Google Scholar]
- 2.R. J. W. Arts, A. Carvalho, C. La Rocca, C. Palma, F. Rodrigues, R. Silvestre, J. Kleinnijenhuis, E. Lachmandas, L. G. Gonçalves, A. Belinha, C. Cunha, M. Oosting, L. A. B. Joosten, G. Matarese, R. van Crevel, M. G. Netea, Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 17, 2562–2571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.E. Kaufmann, J. Sanz, J. L. Dunn, N. Khan, L. E. Mendonça, A. Pacis, F. Tzelepis, E. Pernet, A. Dumaine, J. C. Grenier, F. Mailhot-Léonard, E. Ahmed, J. Belle, R. Besla, B. Mazer, I. L. King, A. Nijnik, C. S. Robbins, L. B. Barreiro, M. Divangahi, BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 4.S. J. C. F. M. Moorlag, N. Khan, B. Novakovic, E. Kaufmann, T. Jansen, R. van Crevel, M. Divangahi, M. G. Netea, β-glucan induces protective trained immunity against Mycobacterium tuberculosis infection: A key role for IL-1. Cell Rep. 31, 107634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B. de Laval, J. Maurizio, P. K. Kandalla, G. Brisou, L. Simonnet, C. Huber, G. Gimenez, O. Matcovitch-Natan, S. Reinhardt, E. David, A. Mildner, A. Leutz, B. Nadel, C. Bordi, I. Amit, S. Srrazin, M. H. Sieweke, C/EBPβ-dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell 26, 657–674.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 6.A. Christ, P. Günther, M. A. R. Lauterbach, P. Duewell, D. Biswas, K. Pelka, C. J. Scholz, M. Oosting, K. Haendler, K. Baßler, K. Klee, J. Schulte-Schrepping, T. Ulas, S. J. C. F. M. Moorlag, V. Kumar, M. H. Park, L. A. B. Joosten, L. A. Groh, N. P. Riksen, T. Espevik, A. Schlitzer, Y. Li, M. L. Fitzgerald, M. G. Netea, J. L. Schultze, E. Latz, Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Bekkering, B. A. Blok, L. A. B. Joosten, N. P. Riksen, R. Van Crevel, M. G. Netea, In vitro experimental model of trained innate immunity in human primary monocytes. Clin. Vaccine Immunol. 23, 926–933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. G. Netea, J. Domínguez-Andrés, L. B. Barreiro, T. Chavakis, M. Divangahi, E. Fuchs, L. A. B. Joosten, J. W. M. van der Meer, M. M. Mhlanga, W. J. M. Mulder, N. P. Riksen, A. Schlitzer, J. L. Schultze, C. Stabell Benn, J. C. Sun, R. J. Xavier, E. Latz, Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. C. Sun, S. Madera, N. A. Bezman, J. N. Beilke, M. H. Kaplan, L. L. Lanier, Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 209, 947–954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Y. Sohrabi, S. M. M. Lagache, V. C. Voges, D. Semo, G. Sonntag, I. Hanemann, F. Kahles, J. Waltenberger, H. M. Findeisen, OxLDL-mediated immunologic memory in endothelial cells. J. Mol. Cell. Cardiol. 146, 121–132 (2020). [DOI] [PubMed] [Google Scholar]
- 11.S. Naik, S. B. Larsen, N. C. Gomez, K. Alaverdyan, A. Sendoel, S. Yuan, L. Polak, A. Kulukian, S. Chai, E. Fuchs, Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. J. C. F. M. Moorlag, Y. A. Rodriguez-Rosales, J. Gillard, S. Fanucchi, K. Theunissen, B. Novakovic, C. M. de Bont, Y. Negishi, E. T. Fok, L. Kalafati, P. Verginis, V. P. Mourits, V. A. C. M. Koeken, L. C. J. de Bree, G. J. M. Pruijn, C. Fenwick, R. van Crevel, L. A. B. Joosten, I. Joosten, H. Koenen, M. M. Mhlanga, D. A. Diavatopoulos, T. Chavakis, M. G. Netea, BCG vaccination induces long-term functional reprogramming of human neutrophils. Cell Rep. 33, 108387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L. Kalafati, I. Kourtzelis, J. Schulte-Schrepping, X. Li, A. Hatzioannou, T. Grinenko, E. Hagag, A. Sinha, C. Has, S. Dietz, A. M. de Jesus Domingues, M. Nati, S. Sormendi, A. Neuwirth, A. Chatzigeorgiou, A. Ziogas, M. Lesche, A. Dahl, I. Henry, P. Subramanian, B. Wielockx, P. Murray, P. Mirtschink, K. J. Chung, J. L. Schultze, M. G. Netea, G. Hajishengallis, P. Verginis, I. Mitroulis, T. Chavakis, Innate immune training of granulopoiesis promotes anti-tumor activity. Cell 183, 771–785.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. C. Gomes, S. Mostowy, The case for modeling human infection in zebrafish. Trends Microbiol. 28, 10–18 (2020). [DOI] [PubMed] [Google Scholar]
- 15.D. M. Page, V. Wittamer, J. Y. Bertrand, K. L. Lewis, D. N. Pratt, N. Delgado, S. E. Schale, C. McGue, B. H. Jacobsen, A. Doty, Y. Pao, H. Yang, N. C. Chi, B. G. Magor, D. Traver, An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood 122, e1–e11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. Howe, M. D. Clark, C. F. Torroja, J. Torrance, C. Berthelot, M. Muffato, J. E. Collins, S. Humphray, K. McLaren, L. Matthews, S. McLaren, I. Sealy, M. Caccamo, C. Churcher, C. Scott, J. C. Barrett, R. Koch, G. J. Rauch, S. White, W. Chow, B. Kilian, L. T. Quintais, J. A. Guerra-Assunção, Y. Zhou, Y. Gu, J. Yen, J. H. Vogel, T. Eyre, S. Redmond, R. Banerjee, J. Chi, B. Fu, E. Langley, S. F. Maguire, G. K. Laird, D. Lloyd, E. Kenyon, S. Donaldson, H. Sehra, J. Almeida-King, J. Loveland, S. Trevanion, M. Jones, M. Quail, D. Willey, A. Hunt, J. Burton, S. Sims, K. McLay, B. Plumb, J. Davis, C. Clee, K. Oliver, R. Clark, C. Riddle, D. Eliott, G. Threadgold, G. Harden, D. Ware, B. Mortimer, G. Kerry, P. Heath, B. Phillimore, A. Tracey, N. Corby, M. Dunn, C. Johnson, J. Wood, S. Clark, S. Pelan, G. Griffiths, M. Smith, R. Glithero, P. Howden, N. Barker, C. Stevens, J. Harley, K. Holt, G. Panagiotidis, J. Lovell, H. Beasley, C. Henderson, D. Gordon, K. Auger, D. Wright, J. Collins, C. Raisen, L. Dyer, K. Leung, L. Robertson, K. Ambridge, D. Leongamornlert, S. McGuire, R. Gilderthorp, C. Griffiths, D. Manthravadi, S. Nichol, G. Barker, S. Whitehead, M. Kay, J. Brown, C. Murnane, E. Gray, M. Humphries, N. Sycamore, D. Barker, D. Saunders, J. Wallis, A. Babbage, S. Hammond, M. Mashreghi-Mohammadi, L. Barr, S. Martin, P. Wray, A. Ellington, N. Matthews, M. Ellwood, R. Woodmansey, G. Clark, J. Cooper, A. Tromans, D. Grafham, C. Skuce, R. Pandian, R. Andrews, E. Harrison, A. Kimberley, J. Garnett, N. Fosker, R. Hall, P. Garner, D. Kelly, C. Bird, S. Palmer, I. Gehring, A. Berger, C. M. Dooley, Z. Ersan-Ürün, C. Eser, H. Geiger, M. Geisler, L. Karotki, A. Kirn, J. Konantz, M. Konantz, M. Oberländer, S. Rudolph-Geiger, M. Teucke, K. Osoegawa, B. Zhu, A. Rapp, S. Widaa, C. Langford, F. Yang, N. P. Carter, J. Harrow, Z. Ning, J. Herrero, S. M. J. Searle, A. Enright, R. Geisler, R. H. A. Plasterk, C. Lee, M. Westerfield, P. J. De Jong, L. I. Zon, J. H. Postlethwait, C. Nüsslein-Volhard, T. J. P. Hubbard, H. R. Crollius, J. Rogers, D. L. Stemple, The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y. Xue, D. Liu, G. Cui, Y. Ding, D. Ai, S. Gao, Y. Zhang, S. Suo, X. Wang, P. Lv, C. Zhou, Y. Li, X. Chen, G. Peng, N. Jing, J. D. J. Han, F. Liu, A 3D atlas of hematopoietic stem and progenitor cell expansion by multi-dimensional RNA-seq analysis. Cell Rep. 27, 1567–1578.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 18.J. M. Frame, C. Kubaczka, T. L. Long, V. Esain, R. A. Soto, M. Hachimi, R. Jing, A. Shwartz, W. Goessling, G. Q. Daley, T. E. North, Metabolic regulation of inflammasome activity controls embryonic hematopoietic stem and progenitor cell production. Dev. Cell 55, 133–149.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. J. Hall, M. V. Flores, S. H. Oehlers, L. E. Sanderson, E. Y. Lam, K. E. Crosier, P. S. Crosier, Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Stem Cell. 10, 198–209 (2012). [DOI] [PubMed] [Google Scholar]
- 20.P. Herbomel, B. Thisse, C. Thisse, Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745 (1999). [DOI] [PubMed] [Google Scholar]
- 21.P. P. Hernández, P. M. Strzelecka, E. I. Athanasiadis, D. Hall, A. F. Robalo, C. M. Collins, P. Boudinot, J.-P. Levraud, A. Cvejic, Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 3, eaau5265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Z. Zhang, H. Chi, R. A. Dalmo, Trained innate immunity of fish is a viable approach in larval aquaculture. Front. Immunol. 10, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. Petit, C. W. E. Embregts, M. Forlenza, G. F. Wiegertjes, Evidence of trained immunity in a fish: Conserved features in carp macrophages. J. Immunol. 203, 216–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. E. Oksanen, H. Myllymäki, M. J. Ahava, L. Mäkinen, M. Parikka, M. Rämet, DNA vaccination boosts Bacillus Calmette-Guérin protection against mycobacterial infection in zebrafish. Dev. Comp. Immunol. 54, 89–96 (2016). [DOI] [PubMed] [Google Scholar]
- 25.I. Sommerset, E. Lorenzen, N. Lorenzen, H. Bleie, A. H. Nerland, A DNA vaccine directed against a rainbow trout rhabdovirus induces early protection against a nodavirus challenge in turbot. Vaccine 21, 4661–4667 (2003). [DOI] [PubMed] [Google Scholar]
- 26.A. Martinez-Lopez, P. Garcia-Valtanen, M. Ortega-Villaizan, V. Chico, E. Gomez-Casado, J. M. Coll, A. Estepa, VHSV G glycoprotein major determinants implicated in triggering the host type I IFN antiviral response as DNA vaccine molecular adjuvants. Vaccine 32, 6012–6019 (2014). [DOI] [PubMed] [Google Scholar]
- 27.H. Darroch, J. W. Astin, C. J. Hall, Towards a new model of trained immunity: Exposure to bacteria and β-glucan protects larval zebrafish against subsequent infections. Dev. Comp. Immunol. 132, 104400 (2022). [DOI] [PubMed] [Google Scholar]
- 28.A. R. Willis, V. Torraca, M. C. Gomes, J. Shelley, M. Mazon-Moya, A. Filloux, C. Lo Celso, S. Mostowy, Shigella-induced emergency granulopoiesis protects zebrafish larvae from secondary infection. MBio 9, e00933-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher, N. Magrini; WHO Pathogens Priority List Working Group , Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 30.K. L. Kotloff, M. S. Riddle, J. A. Platts-Mills, P. Pavlinac, A. K. M. Zaidi, Shigellosis. Lancet 391, 801–812 (2018). [DOI] [PubMed] [Google Scholar]
- 31.P. Schnupf, P. J. Sansonetti, Shigella Pathogenesis: New insights through advanced methodologies. Microbiol. Spectr. 7, 10.1128/microbiolspec.bai-0023-2019, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.H. Ashida, M. Kim, C. Sasakawa, Manipulation of the host cell death pathway by Shigella. Cell. Microbiol. 16, 1757–1766 (2014). [DOI] [PubMed] [Google Scholar]
- 33.S. Mostowy, L. Boucontet, M. J. Mazon Moya, A. Sirianni, P. Boudinot, M. Hollinshead, P. Cossart, P. Herbomel, J. P. Levraud, E. Colucci-Guyon, The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLOS Pathog. 9, e1003588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. J. Mazon-Moya, A. R. Willis, V. Torraca, L. Boucontet, A. R. Shenoy, E. Colucci-Guyon, S. Mostowy, Septins restrict inflammation and protect zebrafish larvae from Shigella infection. PLOS Pathog. 13, e1006467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.V. Torraca, M. Kaforou, J. Watson, G. M. Duggan, H. Guerrero-Gutierrez, S. Krokowski, M. Hollinshead, T. B. Clarke, R. J. Mostowy, G. S. Tomlinson, V. Sancho-Shimizu, A. Clements, S. Mostowy, Shigella sonnei infection of zebrafish reveals that O-antigen mediates neutrophil tolerance and dysentery incidence. PLOS Pathog. 15, e1008006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. Cirovic, L. C. J. De Bree, L. Groh, N. Curtis, M. G. Netea, A. Schlitzer, Human Bacille Calmette-Guérin vaccination elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.I. Mitroulis, K. Ruppova, B. Wang, L. S. Chen, M. Grzybek, T. Grinenko, A. Eugster, M. Troullinaki, A. Palladini, I. Kourtzelis, A. Chatzigeorgiou, A. Schlitzer, M. Beyer, L. A. B. Joosten, B. Isermann, M. Lesche, A. Petzold, K. Simons, I. Henry, A. Dahl, J. L. Schultze, B. Wielockx, N. Zamboni, P. Mirtschink, Ü. Coskun, G. Hajishengallis, M. G. Netea, T. Chavakis, Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. Kleinnijenhuis, J. Quintin, F. Preijers, L. A. B. Joosten, D. C. Ifrim, S. Saeed, C. Jacobs, J. Van Loenhout, D. De Jong, S. Hendrik, R. J. Xavier, J. W. M. Van Der Meer, R. Van Crevel, M. G. Netea, Bacille Calmette-Guérin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 17537–17542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Quintin, S. Saeed, J. H. A. Martens, E. J. Giamarellos-Bourboulis, D. C. Ifrim, C. Logie, L. Jacobs, T. Jansen, B. J. Kullberg, C. Wijmenga, L. A. B. Joosten, R. J. Xavier, J. W. M. Van Der Meer, H. G. Stunnenberg, M. G. Netea, Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. J. W. Arts, B. A. Blok, P. Aaby, L. A. B. Joosten, D. de Jong, J. W. M. van der Meer, C. S. Benn, R. van Crevel, M. G. Netea, Long-term in vitro and in vivo effects of γ-irradiated BCG on innate and adaptive immunity. J. Leukoc. Biol. 98, 995–1001 (2015). [DOI] [PubMed] [Google Scholar]
- 41.R. A. Juste, E. Ferreras-Colino, J. de la Fuente, M. Domínguez, M. A. Risalde, L. Domínguez, A. Cabezas-Cruz, C. Gortázar, Heat inactivated mycobacteria, alpha-Gal and zebrafish: Insights gained from experiences with two promising trained immunity inductors and a validated animal model. Immunology 167, 139–153 (2022). [DOI] [PubMed] [Google Scholar]
- 42.D. Lobato-Márquez, J. Xu, G. Ö. Güler, A. Ojiakor, M. Pilhofer, S. Mostowy, Mechanistic insight into bacterial entrapment by septin cage reconstitution. Nat. Commun. 12, 4511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.C. D. Kane, R. Schuch, W. A. Day, A. T. Maurelli, MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184, 4409–4419 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L. Arbibe, D. W. Kim, E. Batsche, T. Pedron, B. Mateescu, C. Muchardt, C. Parsot, P. J. Sansonetti, An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8, 47–56 (2007). [DOI] [PubMed] [Google Scholar]
- 45.V. Reiterer, L. Grossniklaus, T. Tschon, C. A. Kasper, I. Sorg, C. Arrieumerlou, Shigella flexneri type III secreted effector OspF reveals new crosstalks of proinflammatory signaling pathways during bacterial infection. Cell. Signal. 23, 1188–1196 (2011). [DOI] [PubMed] [Google Scholar]
- 46.S. Saeed, J. Quintin, H. H. D. Kerstens, N. A. Rao, A. Aghajanirefah, F. Matarese, S. C. Cheng, J. Ratter, K. Berentsem, M. A. Van Der Ent, N. Sharifi, E. M. Jamssern-Megens, M. Ter Huurne, A. Mandoli, T. Van Schaik, A. Ng, F. Burden, K. Downes, M. Frontini, V. Kumar, E. J. Giamarellos-Bourboulis, W. H. Ouwehand, J. W. M. Van Der Meer, L. A. B. Joosten, C. Wijmenga, J. H. A. Martens, R. J. Xavier, C. Logie, M. G. Netea, H. G. Stunnenberg, Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. Kröller-Schön, S. Steven, S. Kossmann, A. Scholz, S. Daub, M. Oelze, N. Xia, M. Hausding, Y. Mikhed, E. Zinßius, M. Mader, P. Stamm, N. Treiber, K. Scharffetter-Kochanek, H. Li, E. Schulz, P. Wenzel, T. Münzel, A. Daiber, Molecular mechanisms of the crosstalk between mitochondria and NADPH oxidase through reactive oxygen species - Studies in white blood cells and in animal models. Antioxid. Redox Signal. 20, 247–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. Chaib, T. Tchkonia, J. L. Kirkland, Cellular senescence and senolytics: The path to the clinic. Nat. Med. 28, 1556–1568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. Brokatzky, S. Mostowy, Pyroptosis in host defence against bacterial infection. DMM Dis. Model. Mech. 15, dmm049414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J. L. McCarville, J. S. Ayres, Disease tolerance: Concept and mechanisms. Curr. Opin. Immunol. 50, 88–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. Rao, J. S. Ayres, Resistance and tolerance defenses in cancer: Lessons from infectious diseases. Semin. Immunol. 32, 54–61 (2017). [DOI] [PubMed] [Google Scholar]
- 52.E. Kolaczkowska, P. Kubes, Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 53.J. Kleinnijenhuis, J. Quintin, F. Preijers, C. S. Benn, L. A. B. Joosten, C. Jacobs, J. Van Loenhout, R. J. Xavier, P. Aaby, J. W. M. Van Der Meer, R. Van Crevel, M. G. Netea, Long-lasting effects of BCG vaccination on both heterologous TH1/TH17 responses and innate trained immunity. J. Innate Immun. 6, 152–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. Martinez, O. Cords, Q. Liu, C. Acuna-Villaorduna, M. Bonnet, G. J. Fox, A. C. C. Carvalho, P. C. Chan, J. Croda, P. C. Hill, E. Lopez-Varela, S. Donkor, K. Fielding, S. M. Graham, M. A. Espinal, B. Kampmann, A. Reingold, H. Huerga, J. A. Villalba, L. Grandjean, G. Sotgiu, U. Egere, S. Singh, L. Zhu, C. Lienhardt, J. T. Denholm, J. A. Seddon, C. C. Whalen, A. L. García-Basteiro, R. Triasih, C. Chen, J. Singh, L. M. Huang, S. Sharma, D. Hannoun, H. del Corral, A. M. Mandalakas, L. S. L. Malone, D. L. Ling, A. Kritski, C. M. Stein, R. Vashishtha, F. Boulahbal, C. T. Fang, W. H. Boom, E. M. Netto, A. C. Lemos, A. C. Hesseling, A. Kay, E. C. Jones-López, C. R. Horsburgh, C. Lange, J. R. Andrews, Infant BCG vaccination and risk of pulmonary and extrapulmonary tuberculosis throughout the life course: A systematic review and individual participant data meta-analysis. Lancet Glob. Heal. 10, e1307–e1316 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.N. Katzmarski, J. Domínguez-Andrés, B. Cirovic, G. Renieris, E. Ciarlo, D. Le Roy, K. Lepikhov, K. Kattler, G. Gasparoni, K. Händler, H. Theis, M. Beyer, J. W. M. van der Meer, L. A. B. Joosten, J. Walter, J. L. Schultze, T. Roger, E. J. Giamarellos-Bourboulis, A. Schlitzer, M. G. Netea, Reply to: ‘Lack of evidence for intergenerational inheritance of immune resistance to infections’. Nat. Immunol. 23, 208–209 (2022). [DOI] [PubMed] [Google Scholar]
- 56.M. J. Crim, C. Lawrence, A fish is not a mouse: Understanding differences in background genetics is critical for reproducibility. Lab Anim. (NY). 50, 19–25 (2021). [DOI] [PubMed] [Google Scholar]
- 57.B. Zhang, S. J. C. F. M. Moorlag, J. Domínguez-Andrés, Ö. Bulut, G. Kilic, Z. Liu, R. van Crevel, C.-J. Xu, L. A. B. Joosten, M. G. Netea, Y. Li, Single-cell RNA sequencing reveals induction of distinct trained immunity programs in human monocytes. J. Clin. Invest. 132, e147719 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. J. Dunham-Snary, B. G. Surewaard, J. D. Mewburn, R. E. Bentley, A. Y. Martin, O. Jones, R. Al-Qazazi, P. A. Lima, P. Kubes, S. L. Archer, Mitochondria in human neutrophils mediate killing of Staphylococcus aureus. Redox Biol. 49, 102225 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. E. Herring, S. Mao, M. Bhalla, E. Y. I. Tchalla, J. M. Kramer, E. N. Bou Ghanem, Mitochondrial ROS production by neutrophils is required for host antimicrobial function against Streptococcus pneumoniae and is controlled by A2B adenosine receptor signaling. PLOS Pathog. 18, e1010700 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.T. N. Goleva, K. G. Lyamzaev, A. G. Rogov, L. S. Khailova, K. K. Epremyan, G. P. Shumakovich, L. V. Domnina, O. Y. Ivanova, N. V. Marmiy, T. V. Zinevich, D. S. Esipov, R. A. Zvyagilskaya, V. P. Skulachev, B. V. Chernyak, Mitochondria-targeted 1,4-naphthoquinone (SkQN) is a powerful prooxidant and cytotoxic agent. Biochim. Biophys. Acta Bioenerg. 1861, 148210 (2020). [DOI] [PubMed] [Google Scholar]
- 61.J. Teixeira, R. Amorim, K. Santos, P. Soares, S. Datta, G. A. Cortopassi, T. L. Serafim, V. A. Sardão, J. Garrido, F. Borges, P. J. Oliveira, Disruption of mitochondrial function as mechanism for anti-cancer activity of a novel mitochondriotropic menadione derivative. Toxicology 393, 123–139 (2018). [DOI] [PubMed] [Google Scholar]
- 62.C. A. MacLennan, S. Grow, L. F. Ma, A. D. Steele, The Shigella vaccines pipeline. Vaccines 10, 1376 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.C. Hall, M. V. Flores, T. Storm, K. Crosier, P. Crosier, The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.S. A. Renshaw, C. A. Loynes, D. M. I. Trushell, S. Elworthy, P. W. Ingham, M. K. B. Whyte, A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978 (2006). [DOI] [PubMed] [Google Scholar]
- 65.F. Ellett, L. Pase, J. W. Hayman, A. Andrianopoulos, G. J. Lieschke, mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. W. Watson, T. J. Novitsky, H. L. Quinby, F. W. Valois, Determination of bacterial number and biomass in the marine environment. Appl. Environ. Microbiol. 33, 940–946 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.K. A. Datsenko, B. L. Wanner, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.P. P. Cherepanov, W. Wackernagel, Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995). [DOI] [PubMed] [Google Scholar]
- 69.F. Ellett, G. J. Lieschke, Computational quantification of fluorescent leukocyte numbers in zebrafish embryos. Methods Enzymol. 506, 425–435 (2012). [DOI] [PubMed] [Google Scholar]
- 70.P. Niethammer, C. Grabher, A. T. Look, T. J. Mitchison, A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D. E. Clift, R. J. Thorn, E. A. Passarelli, M. Kapoor, M. K. LoPiccolo, H. A. Richendrfer, R. M. Colwill, R. Creton, Effects of embryonic cyclosporine exposures on brain development and behavior. Behav. Brain Res. 282, 117–124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.J. Rougeot, A. Zakrzewska, Z. Kanwal, H. J. Jansen, H. P. Spaink, A. H. Meijer, "RNA sequencing of FACS-sorted immune cell populations from zebrafish infection models to identify cell specific responses to intracellular pathogens" in Host-Bacteria Interactions: Methods and Protocols, A. C. Vergunst, D. O’Callaghan, Eds. (Springer New York, New York, NY, 2014; 10.1007/978-1-4939-1261-2_15), pp. 261–274. [DOI] [PubMed]
- 73.S. Andrews, FastQC: A quality control tool for high throughput sequence data. Avaliable online at: https://bioinformatics.babraham.ac.uk/projects/fastqc/ (2010; https://bioinformatics.babraham.ac.uk/projects/fastqc/).
- 74.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Broad Institute, Picard toolkit. Available online at: http://broadinstitute.github.io/picard/ (2019; http://broadinstitute.github.io/picard/).
- 76.J. Feng, T. Liu, B. Qin, Y. Zhang, X. S. Liu, Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Q. Li, J. B. Brown, H. Huang, P. J. Bickel, Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011). [Google Scholar]
- 78.G. Yu, L. G. Wang, Q. Y. He, ChI Pseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 79.J. T. Robinson, H. Thorvaldsdóttir, W. Winckler, M. Guttman, E. S. Lander, G. Getz, J. P. Mesirov, Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D. Otasek, J. H. Morris, J. Bouças, A. R. Pico, B. Demchak, Cytoscape automation: Empowering workflow-based network analysis. Genome Biol. 20, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.G. Bindea, B. Mlecnik, H. Hackl, P. Charoentong, M. Tosolini, A. Kirilovsky, W. H. Fridman, F. Pagès, Z. Trajanoski, J. Galon, ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figs. S1 to S6
Legends for data files S1 to S3
Data files S1 to S3