Abstract
The human gut microbiota can restrict the growth of pathogens to prevent them from colonizing the intestine (‘colonization resistance’). However, antibiotic treatment can kill members of the gut microbiota (‘gut commensals’) and reduce competition for nutrients, making these nutrients available to support the growth of pathogens. This disturbance can lead to the growth and expansion of pathogens within the intestine (including antibiotic-resistant pathogens), where these pathogens can exploit the absence of competitors and the nutrient-enriched gut environment. In this review, we discuss nutrient competition between the gut microbiota and pathogens. We also provide an overview of how nutrient competition can be harnessed to support the design of next-generation microbiome therapeutics to restrict the growth of pathogens and prevent the development of invasive infections.
Keywords: Gut microbiome, nutrient competition, colonisation resistance, antibiotics, pathogens
Introduction
What is the gut microbiota?
The human gut is colonized by a complex microbial community collectively referred to as the gut microbiota. The gut microbiota consists of a wide range of different micro-organisms, including bacteria, archaea, viruses, fungi and single-celled eukaryotes. The taxonomic composition of the gut microbiota changes throughout the gastrointestinal tract, according to nutrient availability, mucus structure, pH and oxygen availability [1–6]. In the adult faecal microbiota, 90 % of the commensal bacteria belong to the phyla Bacillota (formerly Firmicutes) and Bacteroidota (formerly Bacteroidetes), while the remaining 10 % belong to the phyla Actinomycetota (formerly Actinobacteria), Fusobacteriota (formerly Fusobacteria), Verrucomicrobiota (formerly Verrucomicrobia), and Pseudomonadota (formerly Proteobacteria) [7]. Of the bacteria within the adult gut microbiota, 99.9 % are obligate anaerobes and the remaining 0.1 % are facultative anaerobes [5, 8]. In infants, facultative anaerobes are more abundant, representing 30 % at 3 months, which decreases to just 1 % at 3 years [9].
Functional redundancy in the gut microbiota
There is high inter-individual variation in the taxonomic composition of the gut microbiota, and no universal species are present in all healthy individuals [10]. Despite these considerable differences, there is significant overlap in the microbial functional genes present between individuals [10, 11]. For example, the Human Microbiome Project demonstrated that healthy individuals can have large differences in the taxonomic composition of their faecal microbiota but share similar functional gene profiles [10]. This is because the gut microbiota has a high degree of functional redundancy, where phylogenetically unrelated taxa contain genes that perform similar functions [12]. Functional redundancy allows the host to maintain a healthy gut microbiota to preserve stability and resilience in response to perturbations [13, 14]. Therefore, identifying which microbial species are present in the gut microbiota is not necessarily sufficient to determine the functional output of the gut microbiota. In addition to asking ‘Who is there?’ we must also ask ‘What are they doing?’ and ‘How are they doing it?’ to answer important mechanistic questions in microbiome research and develop new microbiome therapeutics.
Role of the gut microbiota in the metabolism of dietary substrates
The gut microbiota plays a crucial role in the digestion of dietary and host substrates. The gut microbiota can metabolize dietary fibers that humans are otherwise unable to digest [15]. In the large intestine, the gut microbiota breaks down undigested fibers, such as resistant starch, cellulose, inulin and pectin, utilizing these nutrients as carbon sources to support their growth [15]. Many species within the gut microbiota possess carbohydrate-active enzymes, which are required to digest these dietary fibers into their constituent sugars (Table 1) [16]. Moreover, in the large intestine, the gut microbiota can also break down proteins to use as nitrogen sources to support their growth [17]. Host substrates, such as mucin and bile salts, can also be metabolized by the gut microbiota [18, 19].
Table 1.
Dietary polysaccharides and their breakdown products
|
Polysaccharide |
Degradation products |
Reference |
|---|---|---|
|
Starch |
Glucose, maltose, trehalose |
[130, 131] |
|
Inulin |
Fructose, glucose, sucrose |
[132] |
|
Pectin |
Glucose, arabinose, galactose, xylose, mannose, fructose |
[133] |
|
Arabinogalactan |
Arabinose, galactose |
[134] |
|
Mucin |
N-acetylglucosamine, galactose, fucose, amino acids |
[135] |
|
Xylan |
Xylose, arabinose |
[136] |
|
Cellulose |
Glucose |
[137] |
|
β-glucan |
Glucose |
[137] |
|
Xanthan |
Glucose, mannose, glucuronic acid |
[137] |
The gut microbiota can also participate in cross-feeding, where metabolic intermediates (such as acetate, lactate, succinate and formate) that are produced by gut commensals can act as nutrients to support the growth of other gut commensals [20, 21]. For example, members of the Bacteroidota and Negativicutes can convert succinate into propionate [22, 23]. Multiple commensal species within Pseudomonadota and Bacillota, such as Anaerobutyricum hallii , Desulfovibrio piger and Coprococcus catus , can convert lactate to acetate, propionate and butyrate [22, 24–27]. In co-culture A. hallii could deplete lactate (produced from the fermentation of starch by B. adolescentis ), which led to an increase in butyrate [25].
Gut microbiome-mediated colonization resistance
The gut microbiota protects against intestinal colonization by pathogens via colonization resistance through both direct and indirect mechanisms. Gut microbiota with high diversity have higher colonization resistance than gut microbiota with low diversity as a result of antibiotic treatment [28]. Perturbation of the gut microbiome can disrupt colonization resistance leading to intestinal colonization by pathogens.
Antibiotic usage is widely documented to cause a substantial shift in the composition and functionality of the gut microbiota [29]. Antibiotic treatment – especially treatment with broad-spectrum antibiotics – kills members of the gut microbiota and disrupts colonization resistance (Fig. 1). This promotes intestinal colonization (and even domination) by pathogens such as Clostridioides difficile , carbapenem-resistant Enterobacteriaceae (CRE) and vancomycin-resistant Enterococcus (VRE) [30–32]. An improved understanding of colonization resistance mechanisms against pathogens is vital for the development of novel microbiome therapeutics to prevent or reduce pathogen intestinal colonization.
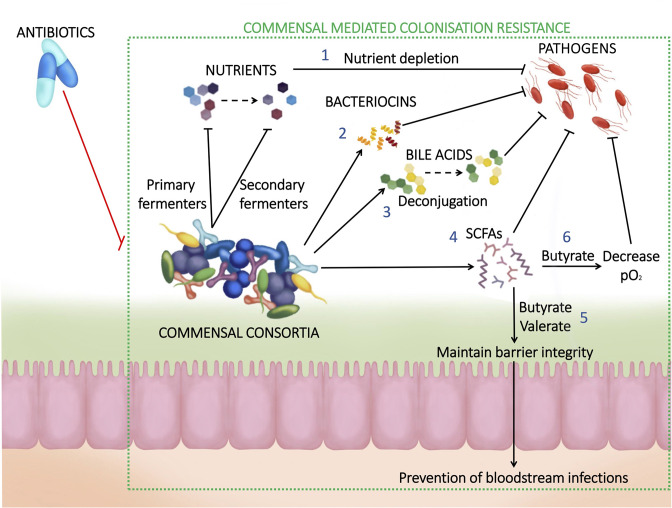
Fig. 1.
Commensal gut microbiota-mediated colonisation resistance. (1) Gut commensals compete with pathogens for nutrients that are essential to support their growth. (2) Some gut commensals produce small antimicrobial peptides (‘bacteriocins’), which can inhibit pathogen growth. (3) Metabolism of bile acids by gut commensals can reduce germination in spore-forming pathogens and inhibit vegetative growth. (4) Gut commensals produce SCFAs that inhibit pathogen growth through intracellular acidification. (5) Gut commensals produce butyrate and valerate, which maintain epithelial membrane barrier integrity and prevent pathogen translocation from the gut to the bloodstream. (6) Gut commensals produce butyrate, which is utilized by colonic epithelial cells as an energy source in a process that consumes oxygen. This creates an environment that is less supportive of the growth of facultatively anaerobic pathogens.
Direct versus indirect mechanisms of colonization resistance
Mechanisms of colonization resistance can be divided into both direct and indirect mechanisms. Direct mechanisms of colonization involve bacteria–bacteria interactions that are independent of host involvement, while indirect mechanisms of colonization resistance involve bacteria–host interactions that rely on host involvement. Examples of direct mechanisms of colonization resistance include nutrient competition between gut commensals and pathogens, production of inhibitory metabolites by gut commensals, production of bacteriocins by gut commensals, and the presence of bacteriophages [33, 34]. Indirect mechanisms of colonization resistance include interactions between the gut microbiota and the host immune response, changes in the gut microbiota that influence oxygen availability, modulation of the mucus layer by the gut microbiota, and promotion of cytokine release by the gut microbiota [34]. This review will focus on discussing direct mechanisms of colonization resistance.
Pathogen inhibition by short-chain fatty acids (SCFAs)
Metabolism by gut commensals drives colonization resistance not just by depleting nutrients, but also through the production of microbial metabolites. For example, the gut microbiota can ferment polysaccharides or proteins to produce short-chain fatty acids (SCFAs) [35]. Acetate, propionate and butyrate are the most abundant SCFAs in the gut, valerate is also present at lower concentrations [33, 36]. Microbial metabolites can inhibit the growth of some bacteria, including pathogens. For example, nonionized SCFAs can diffuse across the bacterial membrane and into the cytoplasm, where they dissociate into their ionized forms, lowering the intracellular pH and inhibiting the growth of susceptible bacteria, including antibiotic-resistant Enterobacteriaceae [37]. Administration of Lactobacillus to antibiotic-treated mice increased faecal butyrate levels and decreased intestinal colonization by Klebsiella pneumoniae [38]. Acetate production was associated with protection against Escherichia coli O157 in mice that were mono-colonized by Bifidobacterium [39]. Propionate produced by Bacteroides thetaiotaomicron negatively impacted the growth of Salmonella enterica serovar Typhimurium [40].
SCFA production can also reduce the pH of the intestine, which can influence the bacteria that can grow at that pH. For example, administering SCFA-producing commensals such as Bifidobacterium longum subspecies infantis to infants reduces intestinal pH from 5.97 to 5.15 and decreased virulence factor gene abundance [41–43]. Another study demonstrated that increasing the pH of ex vivo faecal cultures in a bioreactor system leads to an increase in Enterobacteriacae [44].
SCFAs also interact with the host to impact pathogen growth. Butyrate and valerate promote intestinal barrier function, which prevents the translocation of pathogens from the intestine into the bloodstream [45–48]. Butyrate acts as an energy source for colonic epithelial cells, which can improve gut barrier integrity [49]. In addition, butyrate interacts with gut epithelial cells to influence oxygen availability in the gut. When butyrate is reduced with antibiotic treatment oxygen availability increases, which promotes the growth of facultatively anaerobic pathogens [50].
Bile metabolism
Other microbial metabolites can promote or inhibit the growth of some pathogens that colonize the intestine. Bile salts play an important role in promoting the germination of C. difficile spores or the inhibition of vegetative C. difficile cells [51]. Bile salt hydrolase is an enzyme produced by some gut commensals (such as members of the Bacteroides, Bifidobacterium and Faecalibacterium genera) that deconjugate conjugated primary bile acids, including taurocholate. Taurocholate promotes the germination of C. difficile spores, which leads to C. difficile colonization in the intestine [52]. Some gut commensals (such as Clostridium scindens ) produce enzymes involved in the 7α-dehydroxylase pathway, which are involved in the conversion of unconjugated primary bile acids into secondary bile acids, such as deoxycholate and lithocholate. These secondary bile acids can inhibit vegetative C. difficile growth [53–55]. Additionally, while primary bile acids promote spore germination, secondary bile acids inhibit spore formation by C. difficille [56, 57]. Therefore, a healthy gut microbiota promotes colonization resistance against C. difficile through metabolism of bile acids to both reduce primary bile acids, which promote C. difficile spore germination, and increase secondary bile acids, which inhibit C. difficile vegetative growth.
Nutrient competition
Nutrient availability significantly impacts the diversity and abundance of the micro-organisms that colonize the intestine [58–63]. Each gut commensal strain has its own nutrient utilization ability and preference, and competition is high between bacteria with overlapping nutrient utilization abilities [64, 65]. Pathogens must compete with gut commensals for nutrients to colonize the intestine, and pathogens can more easily colonize an intestine with a low diversity gut microbiota that does not utilize all the available nutrients (e.g. following antibiotic treatment). Bacteria compete for carbon and nitrogen sources that are essential to support their growth within the intestine [66–68]. However, bacteria can also compete for other compounds that support their growth, such as iron and zinc reservoirs [69, 70].
The nutrient niche theory was first described by Rolf Freter in 1983 and proposed that the composition of the gut microbiota is dictated by nutrient availability. It also proposed that a micro-organism will only colonize the intestine if it can utilize one or more limiting nutrients with greater efficiency than its competitors [71]. A deeper understanding of the diversity of nutrient sources used by both pathogens and gut commensals (and the metabolites produced through the metabolism of these nutrients) is crucial to understand how antibiotics promote intestinal colonization of pathogens, and how we can restore colonization resistance by using this knowledge to develop new microbiome therapeutics.
Nutrient competition between similar bacterial taxa
Members of the same species have similar nutrient utilization profiles and therefore they are likely to occupy the same niche within the intestine. For example, Maldonado-Gómez and colleagues administered the probiotic strain Bifidobacterium longum AH1206 to healthy individuals and demonstrated that B. longum AH1206 was undetectable in the faeces from 64 % of individuals once administration was stopped, but that it persisted in the faeces of 27 % of individuals >166 days post-administration [72]. The baseline abundance of B. longum negatively correlated with the persistence of B. longum AH1206, suggesting these two B. longum strains occupied similar intestinal niches. Moreover, B. longum AH1206 persistence was associated with a reduced abundance of carbohydrate-utilizing genes detected in the probiotic-treated gut, suggesting that nutrient availability promoted colonization of this probiotic strain [72].
Another study by Lee et al. demonstrated that germ-free mice colonized by a single strain of Bacteroides species were resistant to colonization from the same, but not different, Bacteroides species [73]. The authors proposed that individual Bacteroides species occupy a unique niche within the intestine. They further discovered a class of polysaccharide utilisation loci (commensal colonization factors, or ‘ccf’), that was conserved amongst Bacteroides and required for colonization.
Several studies have demonstrated that commensal E. coli can compete with E. coli O157:H7 for nutrients. Maltby and colleagues demonstrated that two commensal E. coli strains ( E. coli HS and E. coli Nissle 1917) could prevent E. coli O157:H7 intestinal colonization via nutrient competition [74]. E. coli O157:H7 could use the mucus-associated monosaccharides arabinose, galactose, N-acetylglucosamine, ribose and mannose to support its growth. E. coli HS could use arabinose, galactose, N-acetylglucosamine and ribose (but not mannose). E. coli Nissle 1917 could use arabinose, galactose, N-acetylglucosamine and mannose (but not ribose). E. coli HS and E. coli Nissle 1917 could not restrict E. coli O157:H7 growth individually as neither strain fully occupied the nutrient niche that was occupied by E. coli O157:H7. However, when E. coli O157:H7, E. coli HS and E. coli Nissle 1917 were grown together, E. coli HS and E. coli Nissle 1917 could use all five sugars that E. coli O157:H7 could use, and these two commensal strains could fully occupy the nutrient niche of E. coli O157:H7 to restrict its growth. In another study, Momose et al. demonstrated that commensal E. coli compete with E. coli O157:H7 for proline [75]. They showed that high-proline-utilising E. coli strains depleted the proline pool in germ-free mouse caecal contents and inhibited the growth of E. coli O157:H7, but this was reversed by adding excess proline.
Previous studies have also investigated nutrient competition between Klebsiella oxytoca and Klebsiella michiganensis (a member of the K. oxytoca complex) against antibiotic resistant Enterobacteriaceae [76, 77]. Oliveira and colleagues demonstrated that antibiotic-treated mice that were colonized with K. michiganensis were resistant to colonization with E. coli [77]. They also showed that antibiotic-treated mice that were administered galactitol (a nutrient that is utilised by E. coli but not K. michiganensis ) eliminated colonization resistance mediated by K. michiganensis . This suggested that nutrient competition was the likely mechanism of colonization resistance conferred by K. michiganensis . In another study, Osbelt and colleagues screened faecal samples of healthy adults and children for their ability to inhibit multidrug-resistant K. pneumoniae growth in an ex vivo assay [76]. They found that K. pneumoniae growth varied by donor faecal microbiota, and that K. oxytoca was present at high levels in the two most protected faecal samples from children. They also showed that K. oxytoca accelerated clearance of K. pneumoniae in antibiotic-treated mice. Nutrient utilization assays demonstrated that K. oxytoca was able to utilize 100 carbon sources, while K. pneumoniae could only utilize 56 carbon sources, and there was an overlap in 55 of the carbon sources that K. oxytoca and K. pneumoniae could both utilize. Therefore, the authors suggested that the colonization resistance that K. oxytoca conferred against K. pneumoniae was due to competition for nutrients.
Pathogen exploitation of the altered nutrient environment following antibiotic treatment to overcome colonization resistance
Antibiotics disrupt microbiota-mediated colonization resistance, promoting the colonization and expansion of pathogens within the intestine. Broad-spectrum antibiotics cause significant decreases in the abundance and diversity of a wide range of gut commensals, thus reducing competition for nutrients [6, 78]. These nutrient-defined intestinal niches can then be exploited by pathogens, such as C. difficile , CRE and S. Typhimurium [37, 38, 57, 79, 80].
Healthy gut microbiota can also restrict the growth of specific gut commensals as well. For example, commensal Enterobacteriaceae are typically found at low abundances in the healthy gut microbiota, composing just 0.1–1 % relative abundance on average [10]. However, in the faecal microbiota of a patient treated with amoxicillin-clavulanic acid, abundance of Enterobacteriaceae increased from 2–34 % after 4 days of antibiotic treatment [81].
According to the nutrient niche theory, Enterobacteriaceae growth is limited in the healthy gut microbiota due to competition for nutrient sources with other gut commensals. Enterobacteriaceae can also be suppressed by production of SCFA and other metabolites produced by gut commensals [39, 79, 82]. However, antibiotics disrupt colonization resistance and allow for the expansion of pathogens (Fig. 1) [83].
Antibiotic treatment can also lead to the overgrowth of multidrug-resistant (MDR) pathogens in the intestine [3, 84]. Dense intestinal colonization with these MDR pathogens can lead to the development of invasive infections (such as bloodstream infections or urinary tract infections) and can promote the transmission of these pathogens between patients — a particular problem for immunocompromised patients [85]. Restoring colonization resistance (or preventing the initial loss of colonization resistance) is vital to restrict the growth of MDR pathogens in the intestine following antibiotic treatment [86]. We recently demonstrated that broad-spectrum antibiotics (which promote intestinal colonization by CRE) altered the nutrient landscape in the gut by increasing the availability of various monosaccharides, disaccharides and amino acids [79]. These enriched nutrients were used as carbon and nitrogen sources to support CRE growth both in vitro and in vivo. We also demonstrated that CRE isolates had preferences for specific nutrients over others when presented with a mixture of the nutrients that were enriched with antibiotic treatment. Moreover, CRE isolates were able to grow to higher levels on these nutrients in the presence of oxygen, which is increased in the gut following antibiotic treatment.
Previous studies demonstrated that C. difficile can also use nutrients that are increased with antibiotic treatment to support its growth. Theriot and colleagues demonstrated that several nutrients (sorbitol, mannitol, arabitol, xylitol, gluconate, sucrose, lactate, raffinose, stachyose, galactose and fructose) were increased in the caecal contents of antibiotic-treated mice susceptible to intestinal colonization with C. difficile [32]. They demonstrated that C. difficile could utilize mannitol, fructose, sorbitol, raffinose and stachyose as carbon sources to support its growth. Another study by Fletcher and colleagues demonstrated that amino acids (in particular, proline and branched-chain amino acids) and carbohydrates were decreased in antibiotic-treated mouse caecal content over time in C. difficile-colonized mice [87]. Gene expression data was consistent with the finding that C. difficile used these nutrients to support its growth.
Pathogens can also utilize mucin-derived sugars to support their growth. Ng et al. demonstrated that S. Typhimurium and C. difficile expansion was aided by elevated sugars released from mucin in the caecal contents of antibiotic-treated mice [80]. S. Typhimurium was able to utilize sialic acid and fucose, and mutants deficient in these sugar utilization pathways had impaired intestinal colonization. C. difficile was able to utilize sialic acid, while a mutant that was unable to utilize sialic acid had impaired intestinal colonization. Hudson and colleagues demonstrated that K. pneumoniae was also able to utilize mucin-derived fucose to support its growth in the mouse intestine [88]. They found that a K. pneumoniae mutant, that was unable to metabolize fucose (ΔfucI), showed significantly decreased faecal shedding in mice and decreased growth in filtered cecal contents compared to wild-type K. pneumoniae .
Antibiotic treatment affects the redox state in the gut by reducing competition for electron acceptors, leading to blooms of Enterobacteriaceae [89] Oxygen and nitrate availability was increased in the intestine following antibiotic treatment due to a depletion of butyrate-producing bacteria [90, 91]. Depletion of butyrate switches the metabolism of intestinal epithelial cells from butyrate metabolism (which consumes oxygen) to glycolysis (which does not consume oxygen), leading to increased levels of oxygen in the intestine [92]. Nitrate, which can serve as an alternative electron acceptor to oxygen, is important for the survival of E. coli under anaerobic conditions and can allow E. coli to outcompete bacteria using fermentation for energy generation [93, 94]. Nitrate availability is increased in the intestine following antibiotic treatment or in an inflamed intestine [89, 94]. S. Typhimurium intestinal colonization can trigger an increase in nitrate by inducing inflammation and can exploit this mechanism for intestinal expansion [95]. However, commensal E. coli are able to compete with S. Typhimurium for nitrate to restrict its growth [96]. In a mouse model, probiotic E. coli Nissle 1917 engineered to be deficient in nitrate-respiration were less efficient at restricting S. Typhimurium growth than strains, which could utilize nitrate, demonstrating the importance of nutrient competition in maintaining colonization resistance [96].
Harnessing nutrient competition to restrict intestinal growth by pathogens
We have demonstrated the important role that nutrient competition plays in gut microbiota-mediated colonization resistance against the intestinal colonization by pathogens. Using this information, new microbiome therapeutics need to be developed that modify or control the gut microbiota to re-establish colonization resistance through the restoration of nutrient competition (Figs 2 and 3).
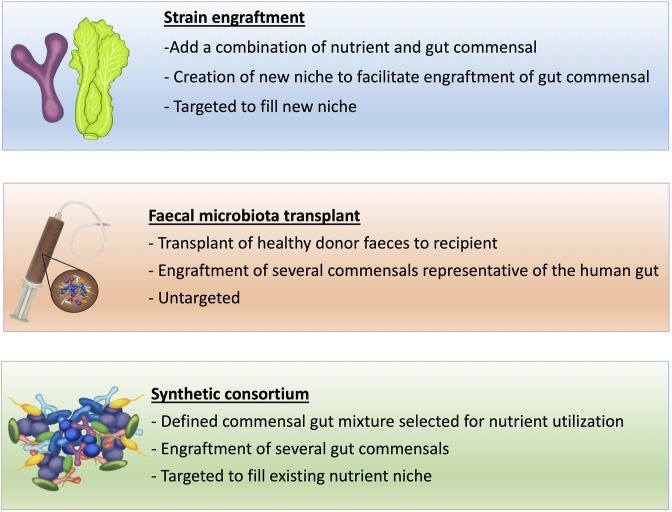
Fig. 2.
Methods to enhance the gut microbiome to restrict pathogen growth. Strain engraftment works through the creation of a new nutrient niche (introducing a new nutrient not utilized by other gut commensals) to allow engraftment of a target gut commensal strain by evading colonization resistance. Faecal microbiota transplant works by introducing an undefined mix of commensals to replenish the gut microbiota and restore colonization resistance. A synthetic consortium is designed to fill the existing nutrient niche to exclude a target pathogen.
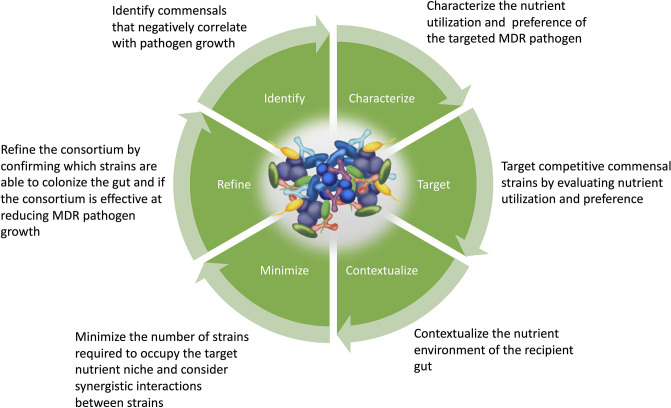
Fig. 3.
Stages in the design of a synthetic microbial consortium to target a specific pathogen.
Previous studies have demonstrated that it is possible to promote or restrict the growth of invading bacteria by altering the availability of nutrients, for example, by introducing nutrients to create a new nutrient-defined intestinal niche. In a study by Shepherd and colleagues, a Bacteroides ovatus strain was administered to human gut microbiota-associated mice with varying success depending on the donor microbiota [97]. This B. ovatus strain contained a rare porphyrin utilization locus, which allowed for the utilization of polysaccharides derived from seaweed. When mice were fed a diet high in seaweed after administering this B. ovatus strain, its abundance significantly increased, independent of the donor faecal microbiota [97]. This study demonstrated that the utilization of a specific carbon source can facilitate a bacterial strain to evade colonization resistance. Synergistic synbiotics have built upon a similar principle, as they contain a mixture of a substrate and a live micro-organism that can utilize that substrate to confer a health benefit to the host [98].
Diet can also influence pathogen growth in a context-dependent manner. For example, B. thetaiotaomicron was able to reduce intestinal growth of Citrobacter rodentium in mice fed a monosaccharide-rich diet, but not in a diet rich in monosaccharides and polysaccharides [99]. This was because B. thetaiotaomicron and C. rodentium competed for monosaccharides in the monosaccharide-only diet, but not in the monosaccharide and polysaccharide-rich diet that provided alternative nutrient sources for B. thetaiotaomicron , which could not be used by C. rodentium .
Faecal microbiota transplant (FMT) has been investigated to treat intestinal colonization with pathogens by reintroducing gut commensals into the intestine that are involved in both direct and indirect mechanisms of colonization resistance. For example, the success of FMT to treat C. difficile is associated with restoration of bile metabolism and SCFA production [100–106]. FMT has been used successfully to treat recurrent C. difficile infections with a higher success rate than antibiotic treatment [107]. C. difficile -associated diarrhoea was resolved in 81 % of patients treated with FMT compared to 31 % of patients treated with vancomycin alone [107]. A systematic review found that decolonization of MDR pathogens ranged from 20–90 % after FMT compared to between 11 and 66 % for controls [100]. A small trial found that FMT may be more successful at eradicating VRE carriage compared to CRE carriage [102]. However, FMT is not a risk-free procedure; whilst in most cases of FMT the adverse events are relatively minor, such as gastrointestinal discomfort [108], a much more serious consequence is the risk of transferring opportunistic pathogens, which may be carried asymptomatically by donors. Infections with Shiga toxin-producing E. coli and enteropathogenic E. coli have been reported following FMT pathogen screening, which impacts the viability of FMT by increasing costs and reducing the donor pool [109]. To bypass these issues, next-generation microbiome therapeutics should be donor-independent and contain a known composition of beneficial bacteria.
Previous work has demonstrated that synthetic microbial consortia (defined mixtures of gut commensals) can be effective at restricting the growth of pathogens in the intestine. Six phylogenetically distinct gut commensal strains were able to eradicate C. difficile colonization in mice [110]. A consortium of 33 strains isolated from human stool samples were used to prevent systemic infection of S. Typhimurium in antibiotic-treated mice [111]. The same consortium has been used to successfully eradicate C. difficile in two patients previously treated with multiple rounds of antibiotics, although there were differences in which commensal strains persisted in the faeces of the two patients following treatment [112].
The design of new microbiome therapeutics to restrict pathogen growth in the intestine requires the careful selection of gut commensals that are effective at restoring colonization resistance to promote pathogen clearance. One approach is to identify gut commensals that negatively correlate with pathogen growth. Isaac et al. treated mice with antibiotics with different spectra of activity and measured the amount of VRE in faecal samples [113]. Spearman correlation analysis was used to identify specific bacterial taxa that negatively correlated with VRE colonization. A synthetic bacterial consortium was developed consisting of strains from the genera Alistipes , Barnesiella , Olsenella , Oscillibacter and unclassified Ruminococcaceae that were isolated from mouse caecal content. This synthetic bacterial consortium restricted VRE intestinal colonization in antibiotic-treated mice. The effectiveness of this consortium was found to be due to depletion of fructose. Olsenella was capable of fructose utilization and reduced VRE colonization in mice both as a pre-treatment and when administered after VRE colonization [113]. Despite the restoration of colonization resistance being largely linked to Olsenella competing for fructose, the inhibition of VRE was greater with the whole consortium. This suggests that multiple mechanisms – and potential unidentified symbiotic interactions between members of the consortium – work together to reduce colonization of VRE.
Rational design of a microbial consortium can also be achieved by designing a mixture of commensals that targets the disruption of nutrient utilisation. As highlighted previously C. difficile is known to utilize host sugars following antibiotic treatment [80]. Pereira and colleagues designed a synthetic bacterial consortium that was capable of depleting N-acetylneuraminic acid and N-acetylglucosamine (mucus-derived sugars) that consisted of Akkermansia muciniphila , Ruthenibacterium lactatiformans , Alistipes timonensis , Muribaculum intestinale , and a Bacteroides sp. [114]. Depletion of N-acetylneuraminic acid and N-acetylglucosamine using this bacterial consortium restricted C. difficile growth both in vitro and in vivo.
Strategies to restore colonization resistance following antibiotic treatment should focus on introducing the minimal number of species possible to fill any unoccupied nutrient-defined intestinal niches to restrict pathogen growth. Due to the functional redundancy of the gut microbiota, it is possible to design a microbiome therapeutic that exhibits all the requisite functions that are required to have a therapeutic effect without administering a full and complex faecal microbiota. Defined microbial consortia must undergo regulatory approval, and the more complex the microbiome therapeutic, the more difficult it may be to pass this regulatory hurdle. However, no commensal strain is able to simultaneously use all nutrients which may be encountered in an antibiotic-treated gut microbiota [115, 116]. Bifidobacterium are present in almost all healthy human faecal samples and have genes that are predicted to be required for the degradation and internalization of a wide array of simple and complex carbohydrates [117–119]. Of the 47 sequenced Bifidobacterium strains investigated by Milani et al., all strains could ferment glucose, sucrose and raffinose. However, the fermentation capabilities of other sugars (such as lactose, galactose, maltose, melibiose, fructose, lactulose, maltodextrins, turanose, β-gentibiose and xylose) varied for most strains that were tested [117]. Bacteroides are primary fermenters, which break down complex dietary and host carbohydrates into monosaccharides and can ferment these to SCFAs and other metabolites [120]. Bacteroides exhibit substantial variation in the degredation profile of polysaccharides [121]. Bifidobacteria are secondary fermenters, which utilize the products of primary fermentation to produce SCFA [122]. A combination of both primary and secondary fermenters would be required for a minimal consortium to cover major intestinal niches and to restore colonization resistance through the production of inhibitory metabolites.
Previous studies have demonstrated that gut commensal strains can provide colonization resistance against intestinal pathogens in a context-dependent manner. Eberl and colleagues demonstrated that gnotobiotic mice colonized by 12 murine gut commensals (OMM) [12] and commensal E. coli Mt1B1 prevented S. Typhimurium intestinal colonization [123]. However, gnotobiotic mice colonized with only three gut commensals and E. coli Mt1B1 were not protective. This study demonstrated that E. coli Mt1B1 colonization depleted galactitol in OMM [12] colonized mice, and that galactitol supported S. Typhimurium growth in OMM [12] colonized mice that lacked E. coli Mt1B1. This study also demonstrated that Lachnospiraceae contributed to colonization resistance against S. Typhimurium by consuming C5 and C6 sugars. Therefore, when developing microbiome therapeutics, it is important to study nutrient competition in the context of a gut microbiome that is representative of patients that will receive this therapeutic to properly assess its effectiveness.
When designing synthetic microbial consortia, it is important to consider synergistic interactions between different species within the consortium, as gut commensals act together to alter the gut environment [124]. This is especially important when designing synthetic microbial consortia that target nutrient competition, as gut commensals work together to fully degrade food within the intestine [125, 126]. For example, Caballero and colleagues demonstrated that a synthetic bacterial consortium containing Blautia producta and Clostridium bolteae restricts VRE intestinal colonisation in mice [127]. They demonstrated that C. bolteae did not directly restrict VRE intestinal colonisation, but rather enabled intestinal colonization with B. producta , which directly restricted VRE intestinal colonization. Another study, by Djukovic and colleagues, established that Lactobacillus species promoted the recovery of Eubacteriales and an overall increase in microbiota density following antibiotic treatment in mice [38]. The increase in Eubacteriales was associated with an increase of SCFA, notably butyrate, which was inhibitory to multidrug-resistant Enterobacteriaceae.
It is also important to consider that there may be differences in the preferences that bacteria have for particular available nutrients, which will impact the development of microbiome therapeutics. Different species of Enterobacteriaceae have different nutrient utilization profiles in aerobic and anaerobic environments and also have a different order of nutrient preference [79, 128]. These results suggest that microbiome therapeutics should include multiple gut commensals that are able to outcompete pathogens for all available nutrients that could support pathogen growth to fully occupy all available nutrient niches in the gut and effectively restore colonization resistance. If nutrients available in the antibiotic-treated gut microbiota are only partially depleted, then the microbiome therapeutic will be ineffective as the pathogen could switch to utilizing alternative available nutrients to support its growth.
Conclusions
Exogenous micro-organisms can only colonize the intestine if they can occupy an available nutrient niche. The healthy gut microbiota fully occupies these niches and prevents pathogen intestinal colonization through multiple mechanisms, including nutrient competition. Antibiotic treatment causes significant disruption to the gut microbiota, which leads to the creation of new nutrient niches, which can become occupied by pathogens. With the urgent threat of antibiotic-resistant bacteria and the limitations of current treatment options, the development of novel microbiome therapeutics is vital to prevent this colonization or to promote decolonization, through introducing gut commensals that can outcompete pathogens for intestinal nutrients.
For microbiome therapeutics to be effective, a greater knowledge of certain areas of nutrient competition is required. Firstly, we must gain a better understanding of nutrient utilization and preferences by both pathogens and gut commensals. An improved understanding of how colonization resistance affects gut commensal colonization is essential to ensure the engraftment and persistence of a microbiome therapeutic. Further, knowledge of cross-feeding relationships is important to ensure the selected commensals can co-exist with commensals in the recipient’s gut microbiota. Development of microbiome therapeutics is also complicated by differences in the response of the gut microbiota to antibiotic treatment in different individuals due to differences in the composition and antibiotic resistance profiles of their baseline gut microbiota [129]. Individual bacterial taxa can show variable responses to antibiotics, which will impact the nutrient niches that are created in the intestine following antibiotic treatment [6]. Future studies should investigate how different antibiotics affect the nutrient and metabolite landscape encountered by pathogens in the intestine to inform selection of gut commensal strains that can occupy the available niches.
Funding information
This work was supported by funding awarded to JAKM: a Medical Research Council (MRC) New Investigator Research Grant (MR/W025655/1).
Author contributions
All authors wrote and edited the manuscript.
Conflicts of interest
J.A.K.M., A.Y.G.Y. and O.G.K. have filed a patent application related to nutrient competition (patent application number 2217266.2). V.H. and I.M.M. do not have any conflicts to disclose.
Footnotes
Abbreviations: CRE, carbapenem-resistant Enterobacteriaceae; FMT, faecal microbiota transplant; MDR, multidrug-resistant; SCFA, short chain fatty acid; VRE, vancomycin-resistant Enterococcus.
References
- 1.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–1032. doi: 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore WEC, Holdeman LV. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagpal R, Tsuji H, Takahashi T, Nomoto K, Kawashima K, et al. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird’s-eye view. Front Microbiol. 2017;8:1388. doi: 10.3389/fmicb.2017.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian L, Wang X-W, Wu A-K, Fan Y, Friedman J, et al. Deciphering functional redundancy in the human microbiome. Nat Commun. 2020;11:6217. doi: 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 15.Scott KP, Duncan SH, Flint HJ. Dietary fibre and the gut microbiota. Nutr Bull. 2008;33:201–211. doi: 10.1111/j.1467-3010.2008.00706.x. [DOI] [Google Scholar]
- 16.Wardman JF, Bains RK, Rahfeld P, Withers SG. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat Rev Microbiol. 2022;20:542–556. doi: 10.1038/s41579-022-00712-1. [DOI] [PubMed] [Google Scholar]
- 17.Davila A-M, Blachier F, Gotteland M, Andriamihaja M, Benetti P-H, et al. Re-print of “Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host.”. Pharmacol Res. 2013;69:114–126. doi: 10.1016/j.phrs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glover JS, Ticer TD, Engevik MA. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci Rep. 2022;12:8456. doi: 10.1038/s41598-022-11819-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziemer CJ. Newly cultured bacteria with broad diversity isolated from eight-week continuous culture enrichments of cow feces on complex polysaccharides. Appl Environ Microbiol. 2014;80:574–585. doi: 10.1128/AEM.03016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aranda-Díaz A, Willis L, Nguyen TH, Ho P-Y, Vila J, et al. Assembly of gut-derived bacterial communities follows “early-bird” resource utilization dynamics. bioRxiv. 2023:2023.01.13.523996. doi: 10.1101/2023.01.13.523996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rios-Covian D, Salazar N, Gueimonde M, de Los Reyes-Gavilan CG. Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front Microbiol. 2017;8:376. doi: 10.3389/fmicb.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquet P, Duncan SH, Chassard C, Bernalier-Donadille A, Flint HJ. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett. 2009;299:128–134. doi: 10.1111/j.1574-6968.2009.01750.x. [DOI] [PubMed] [Google Scholar]
- 25.Duncan SH, Louis P, Flint HJL-UB. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 27.Louis P, Duncan SH, Sheridan PO, Walker AW, Flint HJ. Microbial lactate utilisation and the stability of the gut microbiome. Gut Microb. 2022;3:e3. doi: 10.1017/gmb.2022.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, et al. Decreased diversity of the fecal microbiome in recurrent clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, et al. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;8:e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonten MJ, Willems R, Weinstein RA. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect Dis. 2001;1:314–325. doi: 10.1016/S1473-3099(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 31.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 32.Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, et al. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 2019;83:e00007–19. doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caballero-Flores G, Pickard JM, Núñez G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat Rev Microbiol. 2023;21:347–360. doi: 10.1038/s41579-022-00833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Repaske DR, Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981;145:1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216:84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djukovic A, Garzón MJ, Canlet C, Cabral V, Lalaoui R, et al. Lactobacillus supports Clostridiales to restrict gut colonization by multidrug-resistant Enterobacteriaceae . Nat Commun. 2022;13:5617. doi: 10.1038/s41467-022-33313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. 2018;24:296–307. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casaburi G, Frese SA. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Hum Microbiome J. 2018;9:7–10. doi: 10.1016/j.humic.2018.05.001. [DOI] [Google Scholar]
- 42.Duar RM, Kyle D, Casaburi G. Colonization resistance in the infant gut: the role of B. infantis in reducing pH and preventing pathogen growth. High Throughput. 2020;9:7. doi: 10.3390/ht9020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2 doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Firrman J, Liu L, Mahalak K, Tanes C, Bittinger K, et al. The impact of environmental pH on the gut microbiota community structure and short chain fatty acid production. FEMS Microbiol Ecol. 2022;98:fiac038. doi: 10.1093/femsec/fiac038. [DOI] [PubMed] [Google Scholar]
- 45.Gao G, Zhou J, Wang H, Ding Y, Zhou J, et al. Effects of valerate on intestinal barrier function in cultured Caco-2 epithelial cell monolayers. Mol Biol Rep. 2022;49:1817–1825. doi: 10.1007/s11033-021-06991-w. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. 2021;11:1463–1482. doi: 10.1016/j.jcmgh.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen DSG, Jensen BB, Theil PK, Nielsen TS, Knudsen KEB, et al. Effect of butyrate and fermentation products on epithelial integrity in a mucus-secreting human colon cell line. J Funct Food. 2018;40:9–17. doi: 10.1016/j.jff.2017.10.023. [DOI] [Google Scholar]
- 48.Fachi JL, Felipe J de S, Pral LP, da Silva BK, Corrêa RO, et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019;27:750–761. doi: 10.1016/j.celrep.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 49.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 50.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella . Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullish BH, McDonald JAK, Pechlivanis A, Allegretti JR, Kao D, et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut. 2019;68:1791–1800. doi: 10.1136/gutjnl-2018-317842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirano S, Nakama R, Tamaki M, Masuda N, Oda H. Isolation and characterization of thirteen intestinal microorganisms capable of 7 alpha-dehydroxylating bile acids. Appl Environ Microbiol. 1981;41:737–745. doi: 10.1128/aem.41.3.737-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66:1107–1113. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Theriot CM, Bowman AA, Young VB, Ellermeier CD. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salonen A, Salojärvi J, Lahti L, de Vos WM. The adult intestinal core microbiota is determined by analysis depth and health status. Clin Microbiol Infect. 2012;18 Suppl 4:16–20. doi: 10.1111/j.1469-0691.2012.03855.x. [DOI] [PubMed] [Google Scholar]
- 63.Donskey CJ, Hume ME, Callaway TR, Das SM, Hoyen CK, et al. Inhibition of vancomycin-resistant enterococci by an in vitro continuous-flow competitive exclusion culture containing human stool flora. J Infect Dis. 2001;184:1624–1627. doi: 10.1086/324533. [DOI] [PubMed] [Google Scholar]
- 64.Tramontano M, Andrejev S, Pruteanu M, Klünemann M, Kuhn M, et al. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat Microbiol. 2018;3:514–522. doi: 10.1038/s41564-018-0123-9. [DOI] [PubMed] [Google Scholar]
- 65.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio. 2015;6:e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmes AJ, Chew YV, Colakoglu F, Cliff JB, Klaassens E, et al. Diet-microbiome interactions in health are controlled by intestinal nitrogen source constraints. Cell Metab. 2017;25:140–151. doi: 10.1016/j.cmet.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 67.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29:R538–R544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keeney KM, Finlay BB. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr Opin Microbiol. 2011;14:92–98. doi: 10.1016/j.mib.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Behnsen J, Zhi H, Aron AT, Subramanian V, Santus W, et al. Siderophore-mediated zinc acquisition enhances enterobacterial colonization of the inflamed gut. Nat Commun. 2021;12:7016. doi: 10.1038/s41467-021-27297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie van Leeuwenhoek. 2008;94:165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- 76.Osbelt L, Wende M, Almási É, Derksen E, Muthukumarasamy U, et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021;29:1663–1679. doi: 10.1016/j.chom.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Oliveira RA, Ng KM, Correia MB, Cabral V, Shi H, et al. Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat Microbiol. 2020;5:630–641. doi: 10.1038/s41564-019-0658-4. [DOI] [PubMed] [Google Scholar]
- 78.Panda S, El khader I, Casellas F, López Vivancos J, García Cors M, et al. Short-term effect of antibiotics on human gut microbiota. PLoS One. 2014;9:e95476. doi: 10.1371/journal.pone.0095476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yip AYG, King OG, Omelchenko O, Kurkimat S, Horrocks V, et al. Antibiotics promote intestinal growth of carbapenem-resistant Enterobacteriaceae by enriching nutrients and depleting microbial metabolites. Microbiology. 2023 doi: 10.1101/2023.03.25.533086. [DOI] [PMC free article] [PubMed]
- 80.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roe AJ, O’Byrne C, McLaggan D, Booth IR. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology. 2002;148:2215–2222. doi: 10.1099/00221287-148-7-2215. [DOI] [PubMed] [Google Scholar]
- 83.Rivera-Chávez F, Lopez CA, Bäumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 84.Shimasaki T, Seekatz A, Bassis C, Rhee Y, Yelin RD, et al. Increased relative abundance of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin Infect Dis. 2019;68:2053–2059. doi: 10.1093/cid/ciy796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Correa-Martinez CL, Tönnies H, Froböse NJ, Mellmann A, Kampmeier S. Transmission of vancomycin-resistant enterococci in the hospital setting: uncovering the patient-environment interplay. Microorganisms. 2020;8:203. doi: 10.3390/microorganisms8020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ventola CL. The antibiotic resistance crisis. Pharm Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 87.Fletcher JR, Erwin S, Lanzas C, Theriot CM. Shifts in the gut metabolome and Clostridium difficile transcriptome throughout colonization and infection in a mouse model. mSphere. 2018;3:e00089-18. doi: 10.1128/mSphere.00089-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hudson AW, Barnes AJ, Bray AS, Ornelles DA, Zafar MA. Klebsiella pneumoniae l-Fucose metabolism promotes gastrointestinal colonization and modulates its virulence determinants. Infect Immun. 2022;90:e0020622. doi: 10.1128/iai.00206-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, et al. Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife. 2018;7:e35987. doi: 10.7554/eLife.35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cole J. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol Lett. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- 94.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McLaughlin PA, Bettke JA, Tam JW, Leeds J, Bliska JB, et al. Inflammatory monocytes provide a niche for Salmonella expansion in the lumen of the inflamed intestine. PLoS Pathog. 2019;15:e1007847. doi: 10.1371/journal.ppat.1007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liou MJ, Miller BM, Litvak Y, Nguyen H, Natwick DE, et al. Host cells subdivide nutrient niches into discrete biogeographical microhabitats for gut microbes. Cell Host Microbe. 2022;30:836–847. doi: 10.1016/j.chom.2022.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. 2020;17:687–701. doi: 10.1038/s41575-020-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kamada N, Kim Y-G, Sham HP, Vallance BA, Puente JL, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bilsen MP, Lambregts MMC, van Prehn J, Kuijper EJ. Faecal microbiota replacement to eradicate antimicrobial resistant bacteria in the intestinal tract - a systematic review. Curr Opin Gastroenterol. 2022;38:15–25. doi: 10.1097/MOG.0000000000000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109:1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dinh A, Fessi H, Duran C, Batista R, Michelon H, et al. Clearance of carbapenem-resistant Enterobacteriaceae vs vancomycin-resistant enterococci carriage after faecal microbiota transplant: a prospective comparative study. J Hosp Infect. 2018;99:481–486. doi: 10.1016/j.jhin.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 103.Saïdani N, Lagier J-C, Cassir N, Million M, Baron S, et al. Faecal microbiota transplantation shortens the colonisation period and allows re-entry of patients carrying carbapenamase-producing bacteria into medical care facilities. Int J Antimicrob Agents. 2019;53:355–361. doi: 10.1016/j.ijantimicag.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 104.Merrick B, Sergaki C, Edwards L, Moyes DL, Kertanegara M, et al. Modulation of the gut microbiota to control antimicrobial resistance (AMR)-a narrative review with a focus on Faecal Microbiota Transplantation (FMT) Infect Dis Rep. 2023;15:238–254. doi: 10.3390/idr15030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, et al. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology. 2018;155:1495–1507. doi: 10.1053/j.gastro.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seekatz AM, Theriot CM, Rao K, Chang Y-M, Freeman AE, et al. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe. 2018;53:64–73. doi: 10.1016/j.anaerobe.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile . N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 108.Rapoport EA, Baig M, Puli SR. Adverse events in fecal microbiota transplantation: a systematic review and meta-analysis. Ann Gastroenterol. 2022;35:150–163. doi: 10.20524/aog.2022.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta S, Mullish BH, Allegretti JR. Fecal microbiota transplantation: the evolving risk landscape. Am J Gastroenterol. 2021;116:647–656. doi: 10.14309/ajg.0000000000001075. [DOI] [PubMed] [Google Scholar]
- 110.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martz S-LE, McDonald JAK, Sun J, Zhang Y, Gloor GB, et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep. 2015;5:16094. doi: 10.1038/srep16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Isaac S, Flor-Duro A, Carruana G, Puchades-Carrasco L, Quirant A, et al. Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus. Nat Commun. 2022;13:7718. doi: 10.1038/s41467-022-35380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pereira FC, Wasmund K, Cobankovic I, Jehmlich N, Herbold CW, et al. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat Commun. 2020;11:5104. doi: 10.1038/s41467-020-18928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Palframan RJ, Gibson GR, Rastall RA. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol. 2003;4:71–75. [PubMed] [Google Scholar]
- 117.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.King CH, Desai H, Sylvetsky AC, LoTempio J, Ayanyan S, et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS One. 2019;14:e0206484. doi: 10.1371/journal.pone.0206484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10:e02566-18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eberl C, Weiss AS, Jochum LM, Durai Raj AC, Ring D, et al. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe. 2021;29:1680–1692. doi: 10.1016/j.chom.2021.09.004. [DOI] [PubMed] [Google Scholar]
- 124.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, et al. Glycan utilization and cross-feeding activities by Bifidobacteria. Trends Microbiol. 2018;26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 126.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, et al. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium . Cell Host Microbe. 2017;21:592–602. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chi Z, Liu J, Zhang W. Trehalose accumulation from soluble starch by Saccharomycopsis fibuligera sdu. Enzyme Microb Technol. 2001;28:240–245. doi: 10.1016/s0141-0229(00)00318-5. [DOI] [PubMed] [Google Scholar]
- 131.DeMartino P, Cockburn DW. Resistant starch: impact on the gut microbiome and health. Curr Opin Biotechnol. 2020;61:66–71. doi: 10.1016/j.copbio.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 132.Niness KR. Inulin and oligofructose: what are they? J Nutr. 1999;129:1402S–6S. doi: 10.1093/jn/129.7.1402S. [DOI] [PubMed] [Google Scholar]
- 133.Kaya M, Sousa AG, Crépeau M-J, Sørensen SO, Ralet M-C. Characterization of citrus pectin samples extracted under different conditions: influence of acid type and pH of extraction. Ann Bot. 2014;114:1319–1326. doi: 10.1093/aob/mcu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kelly GS. Larch arabinogalactan: clinical relevance of a novel immune-enhancing polysaccharide. Altern Med Rev. 1999;4:96–103. [PubMed] [Google Scholar]
- 135.Pultz NJ, Hoskins LC, Donskey CJ. Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb Drug Resist. 2006;12:63–67. doi: 10.1089/mdr.2006.12.63. [DOI] [PubMed] [Google Scholar]
- 136.Bastawde KB. Xylan structure, microbial xylanases, and their mode of action. World J Microbiol Biotechnol. 1992;8:353–368. doi: 10.1007/BF01198746. [DOI] [PubMed] [Google Scholar]
- 137.Ahmadi S, et al. Dietary polysaccharides in the amelioration of gut microbiome dysbiosis and metabolic diseases. Obes Control Ther Open Access. 2017;4 doi: 10.15226/2374-8354/4/2/00140. [DOI] [PMC free article] [PubMed] [Google Scholar]