Abstract
Antimicrobial resistance (AMR) is a growing threat to public health, global food security and animal welfare. Despite efforts in antibiotic stewardship, AMR continues to rise worldwide. Anthropogenic activities, particularly intensive agriculture, play an integral role in the dissemination of AMR genes within natural microbial communities – which current antibiotic stewardship typically overlooks. In this review, we examine the impact of anthropogenically induced temperature fluctuations, increased soil salinity, soil fertility loss, and contaminants such as metals and pesticides on the de novo evolution and dissemination of AMR in the environment. These stressors can select for AMR – even in the absence of antibiotics – via mechanisms such as cross-resistance, co-resistance and co-regulation. Moreover, anthropogenic stressors can prime bacterial physiology against stress, potentially widening the window of opportunity for the de novo evolution of AMR. However, research to date is typically limited to the study of single isolated bacterial species – we lack data on how intensive agricultural practices drive AMR over evolutionary timescales in more complex microbial communities. Furthermore, a multidisciplinary approach to fighting AMR is urgently needed, as it is clear that the drivers of AMR extend far beyond the clinical environment.
Keywords: Anthropocene, AMR, antibiotics, evolution, cross-resistance, agriculture, soil
Introduction
Antimicrobial resistance (AMR) is a pressing global issue expected to cause more than 10 million deaths annually in 2050 [1, 2]. While responsible antibiotic stewardship has been advocated as crucial for controlling AMR, emerging evidence suggests that reducing antibiotic use alone may not be sufficient for curbing [3–5] or reversing AMR [6]. The prevailing paradigm suggesting that antibiotic resistance is metabolically costly is being increasingly challenged because compensatory mutations and genetic co-selection can negate the cost of resistance in natural bacterial populations [6]. Moreover, environmental stressors such as pesticides [7], increasing temperatures [8] and heavy metal contamination [9] could make AMR genes beneficial – even in the absence of antibiotics. Worryingly, this suggests that increasingly intensive agricultural practices (Fig. 1) could drive selection for AMR, potentially compromising the efforts of antibiotic stewardship programmes [10]. In this review, we discuss how anthropogenic activities, particularly intensive agricultural practices, could be important yet overlooked drivers of AMR.
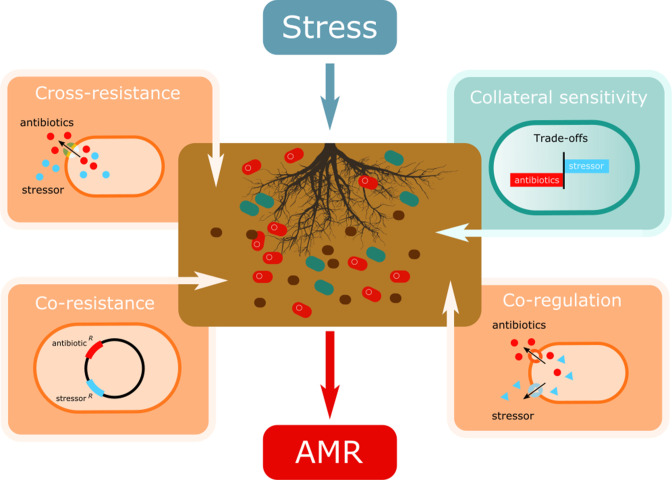
Fig. 1.
Illustration of the eco-evolutionary mechanisms at play in intensively farmed soils which can shape the dissemination of antimicrobial resistance (AMR) in microbial communities. Cross-resistance and co-regulation occurs when stress-resistance mechanisms inadvertently upregulate AMR genes either directly (cross-resistance) or indirectly (co-regulation). The spread of mobile genetic elements (MGEs) in soil bacterial communities exposed to stress can also cause AMR dissemination since stress-resistance and AMR genes are often present on MGEs (co-resistance) [15]. Finally, trade-offs between resistance mechanisms could instead constrain AMR evolution when selection for resistance against one stressor increases sensitivity to a second stressor [91].
Intensive agricultural practices have been highlighted as significant drivers of AMR [11], with conventionally farmed sites harbouring more AMR genes than organically farmed sites [12]. Intensively farmed soils and their constituent microbial communities are frequently exposed to anthropogenically induced stressors, such as agricultural chemicals and pollutants [11]. Such stressors can drive AMR in three key ways (Fig. 1). First, stressors can select for stress-resistance mechanisms in bacteria that cause cross-resistance to antibiotics (Fig. 1). For instance, efflux pumps are often upregulated in response to heavy metals but can also expel a wide range of clinical antibiotics [13]. Second, environmental stress can select for mutations in global stress regulators, which can have myriad effects on the cell, including enhanced AMR [14]. Third, stressors can accelerate the spread of AMR mobile genetic elements (MGEs), when stress and antibiotic resistance genes are located on the same MGE (co-resistance; Fig. 1) [15]. Together, this suggests that agricultural fields may serve as reservoirs of AMR which could spread through the food chain and into clinical settings (Fig. 2). In this review, we highlight five key ways in which intensive agricultural practices amid climate change could drive AMR. We discuss how, even without antibiotic exposure, intensive agricultural practices could independently drive the emergence of AMR through physiological responses, de novo evolution, species sorting toward resistant taxa and enhanced horizontal transfer of AMR genes.
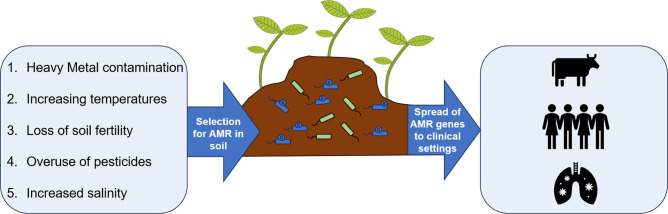
Fig. 2.
Anthropogenic stressors such as heavy metal contamination, pesticide application, increased salinity, loss of soil fertility and increasing temperatures could be key drivers of AMR dissemination in intensively farmed agricultural sites. Intensive agriculture could act as a reservoir of AMR genes, which can then subsequently spread to animals and humans.
Metal contamination
Mining is a major cause of land degradation, having left a substantial historical and contemporary global footprint [16]. For example, it has been estimated that an area of ~1 million km2 globally is covered by mine waste, a large proportion of which is located in populated areas [17]. Other important sources of environmental metal pollution include agriculture and atmospheric deposition resulting from industrial processes [18]. In particular, practices associated with intensive agriculture [19], such as the application of biocides (e.g. copper), manure and sewage sludge [20], are often a local source of environmental metal pollution.
While many metals are essential to biological functions, most are toxic at high concentrations [20]. Hence, the release and remobilization of non-degradable metals into the environment can pose a serious long-term threat to ecosystem- and human-health [21]. For example, consumption of crops and other food products harvested from metal-polluted environments can negatively impact human health via trophic cascades in the food chain (e.g. arsenic accumulation in paddy rice [22]; mercury accumulation in freshwater fish [23]). Metal contamination can also indirectly impact human health by co-selecting microorganisms that are resistant to antibiotics (AMR) [24], even in the absence of antibiotics themselves [25]. For example, using pot experiments, a recent study found enhanced levels of antibiotic resistance genes (ARGs) in soil communities fertilized with swine slurry compared to non-treated soils. Importantly, metals were a key factor in driving the observed increase in ARGs in the resident soil communities [26]. A recent metagenomic study confirms these findings: the addition of zinc oxide (a growth promotor in animal husbandry) to pig feed led to increased co-occurrence of metal and antibiotic resistance genes in pig faeces and caused the dissemination of these genes into the wider agricultural environment [27]. Similarly, in terrestrial subsurface soils, heavy metal content was found to be a better predictor of ARG content than bacterial community composition, MGEs or physico-chemical factors [28]. Because of its clinical and environmental importance, there is a large body of work on the co-selection of metal and antibiotic resistance [9, 19, 29, 30]. These studies have revealed that different mechanisms can underpin the co-selection process, having important implications for the acquisition and spread of AMR in natural environments.
Bacteria have evolved a wide range of mechanisms to cope with toxic metals [31–35]. Some of these mechanisms confer cross-resistance to other toxic compounds. Examples of cross-resistance mechanisms include multi-drug resistant (MDR) efflux pumps [13] and several factors reducing cell permeability and the influx of compounds (e.g. biofilm formation [31]; downregulation or deletion of outer-membrane porins [36, 37]). For instance, bacteria commonly encode (sometimes multiple) MDR efflux pumps that can expel a wide range of compounds, including clinical antibiotics. However, efflux of clinical antibiotics is often a side-effect of the primary role of MDR efflux pumps [24] – protection against natural toxins (e.g. metals, bile salts, aromatic compounds and quorum-sensing molecules) and host colonization [38]. The presence of xenobiotics in agricultural soils (e.g. metals, pesticides) can therefore selectively favour bacteria that harbour, or overexpress, MDR efflux pumps, thereby inadvertently selecting for increased resistance to clinical antibiotics. Indeed, the expression of cross-resistance mechanisms, such as efflux systems and biofilms, often changes in response to abiotic stress [9, 24, 39].
Bacteria can also harbour resistance mechanisms that specifically target toxic metals, including metal reduction and extracellular sequestration. For example, most bacteria produce metal-chelating siderophores [40, 41]. Bacteria release these compounds into the environment, where they bind to toxic metals, preventing them from being taken up and killing bacterial cells [42, 43]. The production of siderophores is typically upregulated in response to toxic metal stress, both at the species level [44] and community level (i.e. ecological species sorting favouring siderophore-producing taxa [45, 46]). Crucially, while siderophores do not confer increased antibiotic resistance, their production can increase pathogen virulence. The fact that siderophores are potent iron chelators enables bacteria to scavenge poorly soluble iron from their host and be taken up by cells carrying specific outer-membrane receptors [47]. A recent study confirms this notion showing that copper-mediated selection favours higher levels of siderophore production in the opportunistic pathogen Pseudomonas aeruginosa , thereby increasing pathogen virulence [48].
Some metal resistance genes do not provide cross-resistance to other toxic compounds but are associated with AMR because they are co-located on the same chromosome or MGE (i.e. co-resistance [9]). Importantly, resistance genes are often enriched on MGEs [33], including plasmids that spread horizontally by conjugation. Such horizontal gene transfer (HGT) is considered to be a major evolutionary force [49, 50] and is thought to play a vital role in the spread of AMR genes, both within and across bacterial pathogen species [24, 51, 52]. Previous work has shown that the presence of metals in the environment can enhance the horizontal spread of plasmids [53–55], including those that harbour both AMR and metal-resistance genes. For example, genes encoding aerobactin – a metal-chelating siderophore – and AMR are located on the same plasmid in Escherichia coli strains isolated from sewage [56]. Co-location could then subsequently enhance the dissemination of AMR genes in environments where the production of siderophores is selectively favoured (i.e. in iron-deficient or metal-polluted environments).
Heavy metals continue to be used routinely for both crop and livestock production, although the use of some heavy metals as antimicrobials (e.g. zinc oxide) was banned in the EU in June 2022 [Regulation (EU) 2019/6 on Veterinary Medicinal Products]. A recent scoping review article by Anedda et al. [57] examined 73 studies addressing the impact of heavy metal contamination on ARG dissemination [57]. Despite differences in objectives, sample types, locations and methods between these studies, they all asserted a clear link between heavy metals and AMR in the primary food production environment. Together, this is strong evidence to support better management of heavy metal use in agriculture, including legislation that supresses the use of heavy metals or heavy-metal-containing fertilizers in routine farm management.
Increasing temperatures
Anthropogenic activity is warming the climate at an unprecedented rate, with an average projected increase of 1.5–2 °C in the coming decades [58]. Extreme temperature events, warmer nights and more variable precipitation will significantly reduce agricultural crop yield and expand the potential habitable range of some insect and disease pests [59]. Increasing global soil temperatures and associated drought can similarly shape soil microbial community composition, diversity and functioning, disrupting microbe–plant feedback [60–62]. However, far less is known about how increasing temperatures can alter the evolutionary trajectory of soil microbial populations per se, and the impact such evolved changes can have on microbe–plant interactions. There is solid evidence to suggest that the global burden of AMR is positively correlated with increased local temperature, suggesting that even minor temperature increases could influence the evolution and spread of AMR in soil microbial communities [63].
Since the origin of life, microbes have been faced with temperature stress. In fact, microbial pathogens may have been the driving force behind the evolution of warm-blooded animals, as fever would be more effective at repelling infections in warm-blooded versus cold-blooded animals [64]. Temperatures exceeding the optimum temperature for bacteria (T opt) can cause cellular proteins to misfold, damage DNA and RNA, and increase membrane fluidity [65]. Bacteria can cope with short-term temperature extremes via transient heat shock responses (HSRs) [65, 66]. In E. coli, the HSR is regulated by the alternative sigma factor σ32 [67], characterized by reduced growth rates and upregulation of heat shock proteins, such as (i) chaperones to prevent protein misfolding (e.g. ClpB, DnaK, DnaI and GroEL/ES); (ii) proteases to degrade misfolded proteins (e.g. ClpP and ClpX); (iii) DNA/RNA repair enzymes; (iv) metabolic enzymes; (v) outer membrane stability proteins; and (vi) membrane transport proteins [68]. This HSR is highly conserved and allows cells to temporarily counteract the effects of short-term stress by slowing down growth and re-directing resources into preventing DNA damage.
It has been recently suggested that mechanisms of AMR are co-opted from such stress responses to temperature [8, 69]. Different classes of antibiotics can simulate heat stress, or cold stress, depending on the class of antibiotic. For example, using 2D gel electrophoresis, it has been shown that treating E. coli with aminoglycosides (antibiotics that target the 30S ribosomal subunit) results in protein expression changes that are ‘virtually indistinguishable from that produced by a shift in temperature’ from 28 °C to 42 °C [70]. More recently, Cruz-Loya et al. [8] used stressor interaction networks to reveal that E. coli physiological responses to low or high temperatures are clearly separated, and each is grouped with particular antibiotics that have similar effects to cold or heat respectively. For example, aminoglycosides, as well as nitrofurantoin and trimethoprim, all have similar physiological effects to heat stress (44–46 °C), while macrolides, tetracycline and fluoroquinolones emulate cold stress (22–37 °C). While the exact causes of this interaction similarity are unclear [71] it was hypothesized that translational misreading caused by both heat and aminoglycoside antibiotics warrant a similar protective response by the cell. For example, heat shock chaperones (DnaK and GroEL) protect cells against aminoglycoside antibiotics by preventing protein misfolding and aggregation in Acinetobacter baumanii and E. coli [49]. Moreover, the deletion of cspB – associated with the cold-shock response – can lead to an enhanced HSR and consequently increased resistance to heat-similar aminoglycosides and trimethoprim [50]. Together, this suggests that as temperatures increase under climate change, particular classes of antibiotics may be more vulnerable to resistance evolution (i.e. heat-similar antibiotics, such as aminoglycosides). Conversely, resistance toward cold-similar antibiotics (e.g. tetracycline and chloramphenicol) could become more costly [72].
Despite the potential for temperature stress to influence AMR via activation of the HSR, it is less clear whether temperature-similarity profiles feed into selection over evolutionary time. The HSR is generally transient (in the order of magnitude of minutes) in E. coli , and hence is fitting for short-term rather than long-term temperature extremes. However, activation of the HSR could ‘prime’ bacterial populations for resistance before a heat-similar antibiotic is applied [73]. In this case, cells with an active HSR would gain a slight advantage over non-expressing cells since they would be better equipped to deal with misfolded DNA. Even such a short-term physiological advantage could expand the window of opportunity for resistance mutations to evolve.
A tour de force by Rodríguez-Verdugo and colleagues [8, 71, 74] provides unique insights into how bacteria might adapt to heat stress over evolutionary time and how this could impact AMR. Rodríguez-Verdugo et al. [74] compared the mid-exponential phase gene-expression profile of E. coli growing at 37 and 42 °C, revealing differential expression of 1737 genes between the two temperature treatments. Downregulated genes at 42 °C included ribosomal constituents (rpl, rpm and rps) involved in translation, amino acid biosynthesis, flagellum motility and ribonucleoside biosynthesis. Perhaps most surprisingly, however, is that growth at 42 °C resulted in the downregulation of heat shock proteins involved in the HSR, including subunits of core RNA polymerase (RNAP) rpoA, rpoB and rpoC. Similarly, most HSR encoding chaperones were also downregulated (clpB, dnaJ, groEL and groES) at 42 °C. This result was explained by the fact that the gene-expression profiles were assayed at the mid-exponential phase, at which point the HSR was already turned off in this particular experimental setup. This suggests that the transient HSR might play a minimal role in priming populations for AMR. However, further experiments are needed to test if a short-term advantage could enhance selection for resistance over longer timescales.
Rodríguez and colleagues also experimentally evolved 114 E. coli populations under thermal stress (42.2 °C for 2000 generations) [74]. While initially, the general stress response system was activated (slowing down growth and reducing gene expression of RNAP, increasing transcriptional efficiency at 42 °C), this heightened stress response was not maintained throughout the experiment. Conversely, after 2000 generations, mutations in the β-subunit of RNAP (rpoB) instead restored growth and allowed the cell to revert to a gene expression profile similar to the ancestral (pre-stressed) state [71]. In 12 populations, resistance was mediated by single SNPs in codon 572, causing amino acid substitutions in rpoB and incidentally conferring rifampicin resistance ranging from ×10 to ×320 higher than susceptible cells. Using constructed rpoB mutants with SNPs in codon 572 they confirmed that this particular rpoB mutation (i) conferred high levels of rifampicin resistance via alterations in RNAP, and (ii) increased growth relative to the wild-type at 42 °C. Their work shows that the interplay between the costs and benefits of bacterial stress responses across multiple timescales can dictate the spread of AMR, at least in E. coli.
While the HSR could allow bacteria to tolerate short-term temperature extremes, more moderate increases in temperature could drive AMR evolution in multiple ways. Higher temperatures can have mutagenic effects by increasing replication errors and causing DNA damage [75]. Increased mutation rates could enhance the standing genetic diversity of populations, which, on exposure to antibiotics, could accelerate selection for resistance. Indeed, the evolutionary speed hypothesis (ESH) demonstrates that higher temperatures lead to faster evolutionary processes by increasing mutation rates and accelerating natural selection [76]. Temperature could also modulate HGT of AMR genes. MacFadden et al. [63] found that in the USA, an increase in daily minimum temperature of 10 °C (which is conceivable for some parts of the world by the end of the century) correlated with increased levels of AMR in E. coli, Klebsiella pneumoniae, and Staphylococcus aureus . Although the mechanism was not elucidated, it was speculated that increased temperatures accelerate HGT of AMR genes. Similarly, Reverter et al. [77] analysed data from 40 countries to pinpoint the key predictors of ARG frequencies in aquaculture [77]. They found a negative relationship between a country's vulnerability index (CVI; lower scores=higher vulnerability) and ARG abundance. Notably, this association was underpinned by the physical component of the CVI score, which encompassed mean temperature, water availability and frequency of extreme weather events. Together, these findings emphasize the deadly combination of AMR and climate change, where increasing temperatures could worsen the already growing crisis of AMR.
Loss of soil fertility
The nutrient content of natural soils is declining on a global scale [78]. Conversion of native vegetation and land to intensive agricultural fields results in substantial losses of C, N, P and S, which in turn reduce the nutrient content of food crops [78]. The causes of nutrient loss via conventional farming are multifaceted, including intensive and continuous crop cultivation, soil erosion, leaching, and removing or burning stubble [79]. Crop production is now heavily dependent on artificial and organic fertilizers to replenish the low nutrient content of soils.
While the impact of soil fertility loss on plants is now evident [79], we know far less about how nutrient loss might influence soil microbes directly. In soil, scarcity of resources as well as competition with other species can heavily limit bacterial growth. Soil bacteria spend most of their lives under nutrient limitation in long-term stationary phase [80], with short pulses of nutrient input (feast/famine dynamics) [81, 82]. However, as levels of C and N are depleted in intensively farmed soils, it is likely that bacteria will spend increasingly longer proportions of their lives in stationary phase, with selection to enhance long-term survival in an increasingly nutrient-limited environment. Worryingly, studies examining short-term microbial responses to starvation and long-term adaptation under nutrient limitation suggest that AMR and loss of soil fertility could be inextricably linked [83–85].
When nutrients are scarce, physiological responses by bacterial cells can permit long-term survival. Activation of the stationary phase sigma factor S (σ s) stimulates key physiological changes, including (i) genome compaction, (ii) reduced membrane permeability, (iii) increased production of osmoprotectants and (iv) decreased expression of growth-promoting genes [85]. In E. coli , rpoS mutations conferring σ s overexpression can also have consequences for AMR – for example, a mutation in rpoS (87 D-6) can cause resistance to nalidixic acid [14]. Studies examining the starvation response of E. coli through prolonged incubation on agar plates also reveal the predictable emergence of rifampicin-resistant mutants, despite incubation taking place in the absence of antibiotics [14]. This finding was classically explained by stress-induced mutagenesis – where resistance emerges as a by-product of an increased mutation rate [86]. However, it is now acknowledged that rifampicin resistance is beneficial per se under starvation due to the pleiotropic effects of mutations in RNAP β-subunit (rpoB) that enhance fitness under starvation while incidentally conferring rifampicin resistance [83]. Mutations in rpoB are also the cause of high-level β-lactam resistance in methicillin-resistant Staphylococcus aureus [87]. Similarly, under nutrient-limitation, Salmonella enterica rspL mutants fail to induce the stationary phase sigma factor σs, causing them to outcompete the slow-growing wild-type. These rspL mutations also enhance resistance to streptomycin [84]. Interestingly, these dynamics are reversed under nutrient-rich conditions, so the antibiotic-susceptible wild-type outcompetes rspL mutants. This result is presumably due to poorer translational fidelity of the mutant relative to the wild-type when nutrients are rich [84].
While stress response systems may drive AMR under nutrient limitation in the short term, experimental evolution approaches can reveal whether such responses will probably feed into long-term selection dynamics. One experimental evolution study of E. coli in laboratory media (in the absence of antibiotics) reported reduced susceptibility to erythromycin (2–4× MIC), fosfomycin (2–6× MIC), rifampicin (2–32× MIC) and streptomycin (2–3× MIC) after 500–1000 generations of evolution [83]. AMR genes evolved more frequently in low-nutrient versus high-nutrient media, reaching high frequencies and exhibiting extensive parallelism within the same treatment, indicating a strong selective advantage under nutrient limitation. Similarly, experimental evolution of P. aeruginosa in M9 minimal media found that under nutrient-limiting (but not nutrient-rich) conditions, lasR mutations evolved that caused enhanced levels of antibiotic resistance (possibly via increased expression of efflux pumps) [88, 89]. Interestingly, P. aeruginosa lasR mutants are common in clinical settings, and typically display enhanced resistance to tobramycin [90]. While such studies suggest that manipulating soil nutrient content could be a potential avenue for AMR control, we lack experiments that test how soil nutrients drive AMR in natural communities. In natural communities, resource competition can impose further constraints on the allocation of resources towards different traits/functions. This constraint can lead to trade-offs where adaptations evolved to optimize one trait are accompanied by a reduction in another [91]. Trade-offs can select against costly resistance mechanisms (e.g. porins keep out antibiotics but potentially also necessary nutrients) and constrain the evolution of multi-drug resistance [92]. However, the impact of soil fertility loss on AMR in natural microbial communities remains unexplored.
The loss of soil fertility is a complex problem, but solutions are relatively easy to implement. The EU Green Deal is encouraging the farming sector to adopt more sustainable methods, such as crop rotation, cover cropping, reduced tillage and better irrigation [93]. These approaches will have the dual effect of reducing emissions while restoring soil fertility. Still, further research is needed into the long-term benefits of such practices on the dissemination of ARGs in soil microbial communities.
Overuse of pesticides
Intensive agricultural practices have led to cultivating densely packed crop monocultures to make the most economical use of space. Unfortunately, these conditions also facilitate the success of ‘pests’ such as fungi, bacteria, viruses, insects, arachnids and rodents, as well as undesirable vegetation (i.e. weeds) [94]. To mitigate crop death and prevent famine, it has become routine to apply pesticides. Pesticides are biocidal or biostatic compounds used to prevent and treat agricultural pest infestations. These pesticides are grouped into classes based on their target organism; for example, fungicides target fungi, and herbicides target undesirable vegetation.
Despite being designed to target specific pests (i.e. fungicides ‘only’ target fungi), many pesticides have been shown to have harmful effects on humans and the broader ecosystem [95]. Policies are now being implemented to rectify this emerging issue; for example, the European Union has recently banned some neonicotinoid pesticides due to their harmful effects on bees [96]. However, the impact of pesticides is not constrained to larger organisms; some fungicides have been recently found to inhibit the growth of soil bacteria [97, 98]. For example, pesticides, including the fungicides azoxystrobin and flutriafol, have been linked with the decreased abundance of nitrifiers which can reduce soil fertility and health [99]. Furthermore, the herbicide bromoxynil has been shown to inhibit the growth of soil bacterial populations, in turn reducing bromoxynil biodegradation and increasing fungicide persistence in soils [100]. It is now clear that pesticides can drive loss of soil microbial diversity, alter microbial soil community composition and even drive the rapid evolution of AMR in soil bacteria [7, 99, 100]. Despite the acknowledged non-target effects of pesticides and their potentially harmful implications on public health, they are still widely overused, further contributing towards the emerging AMR crisis.
The first line of defence by bacteria against pesticides is the downregulation of membrane porins or upregulation of efflux pumps to remove the chemicals from the cell [101, 102]. For example, the herbicide Dicamba has been shown to induce soxRS in E. coli [103]. The soxRS system can upregulate AcrAB efflux pumps which have been attributed to both pesticide and antibiotic resistances, including fluoroquinolones [104]. Copper-based fungicides can similarly drive the de novo evolution of mutations in the AcrAB-TolC multi-drug export pump, causing cross-resistance to tetracycline and chloramphenicol [105]. Mutations that cause overexpression of efflux pumps are a common early adaptation in response to both antibiotics and pesticides [15, 104, 106, 107] (cross-resistance; Fig. 1). This evidence suggests that pesticide-treated soils could act as a reservoir for AMR genes, which can ultimately return to the food chain (Fig. 2) [102].
Many bacteria can also degrade pesticides into less harmful residuals [108]. This biodegradation process can protect both the bacterium and other organisms from the stress of the pesticide. Therefore, such microbes and their enzymes have become areas of interest for bioremediation. One such group of enzymes are hydrolases (e.g. esterases, organophosphorus hydrolase and lipase) which can degrade certain chemicals through reactions with water. For example, organophosphorus insecticides are potent acetylcholinesterase inhibitors that are harmful to human health and other organisms [109, 110]. Bacterial-derived organophosphorus hydrolases are an effective means of bioremediation of these pesticides, as they reduce toxicity via the hydrolysis of phosphodiester bonds [109, 111–113]. The organophosphate degradation genes (opd) have been identified in a plethora of soil-dwelling bacteria such as Flavobacterium , Pseudomonas , Bacillus and Agrobacterium [114–116]. However, enzymatic modelling studies by Rangasamy et al. [117–119] have suggested that organophosphorus hydrolase from Geobacillus could dock with and hydrolyse streptomycin, ampicillin, chloramphenicol and cefotaxime. Furthermore, a plasmid-bound α–β hydrolase known to degrade organophosphate-degrading α–β hydrolases has been shown to reside on plasmids and provide resistance to a range of antibiotics [119].
Another group of enzymes involved in detoxification are glutathione S-transferases (GSTs). These isozymes are widely used by plants and insects as pesticide resistance mechanisms [120, 121]. GTSs have also been widely found in prokaryotes, and are associated with pesticide degradation in the rhizosphere [122, 123]. However, GTSs have also been associated with the degradation of antibiotics such as tetracycline, sulfathiazole and ampicillin [124]. Interestingly, some GTSs have also been shown to have peroxidase activity – an important family of enzymes which can degrade phenylamide herbicides [125]. Peroxidases and other pesticide-resistance mechanisms such as soxR can protect against oxidative (redox) stress. Since many antibiotics partly inhibit bacteria via the formation of reactive oxygen species (ROS) [126], it is likely that such pesticide-resistance mechanisms will similarly protect against antibiotics.
Finally, the co-occurrence of resistance genes to pesticides and antibiotics on MGEs poses a significant risk to public health, enabling the rapid dissemination of antibiotic resistance in bacterial populations. Studies have shown that exposure to certain pesticides can facilitate the transmission of AMR genes on MGEs in natural populations (i.e. co-resistance; Fig. 1). For instance, Liao et al. demonstrated that soils treated with glyphosate herbicides show an increased abundance of ARGs and MGEs compared to the non-herbicide-treated control [15]. These ARGs included genes associated with aminoglycoside, vancomycin, chloramphenicol and tetracycline resistance [15]. Similarly, azoxystrobin and carbendazim fungicides increase the expression of conjugation-related genes on plasmids, thereby increasing the spread of MGEs containing ARGs [127]. Pyrethroid-insecticides such as permethrin can also increase conjugation and mutation rates in E. coli [128]. Furthermore, a study of E. coli grown in a lab-based medium containing a cocktail of 23 common pesticides revealed selection for mutations conferring streptomycin resistance [129]. In the same study, co-exposure of E. coli to pesticides and ampicillin selects for cross-resistance to ciprofloxacin, tetracycline and chloramphenicol, possibly due to mutations in transcriptional regulators responsible for oxidative stress defence or biofilm formation [129].
While it is clear that pesticides have the potential to directly drive AMR evolution, it is also possible that pesticides could shape AMR indirectly, through disrupting soil microbial communities. By reducing community diversity, pesticides can leave communities open to invasion by pesticide- (and antibiotic-) resistant species. These dynamics are frequently reported in the gut microbiota, where antibiotic treatment leaves the gut microbiota vulnerable to invasion by antibiotic-resistant Clostridium difficile [130]. Hence, the non-targeted biocidal effect of pesticides, alongside selection for AMR, could create a perfect recipe for a rapid sweep of ARGs through soil communities.
Given the accumulating evidence of pesticide use on the dissemination of AMR, it is important that policy is implemented to prevent further AMR spread and evolution. To address this issue, several solutions can be implemented. First, promoting integrated pest management practices that combine various eco-friendly methods, such as crop rotation, biological control and mechanical pest control, can help minimize pesticide reliance. Second, research into the development of pesticides targeted to a specific pest could significantly lessen the indirect impact on soil microbiota while still effectively controlling pests. Insect viruses, such as baculoviruses, are highly specific in their host range and could be used to target a single insect pest [131]. Plant pathogenic fungi can be controlled by adding soil that contains microbes antagonistic to fungal pathogens. For example, species of Trichoderma have been used to control fungal plant diseases caused by Fusarium and Rhizoctonia [132]. Finally, implementing buffer zones around agricultural fields can also act as a safeguard, preventing pesticide runoff into adjacent ecosystems. By adopting these proactive strategies, we can mitigate the adverse effects of pesticide use on soil microbial communities and reduce the risk of AMR emergence, fostering a more resilient and sustainable agricultural landscape.
Increased salinity
Climate change brings increased levels of evaporation, low rainfall and global sea level rise [133]. The influx of salts into soils through flooding, alongside a reduction of natural irrigation to remove existing salt, has led to the salinization of one-third of the world’s arable land [134–136]. Bacteria respond to high salinity via (i) increased expression of inorganic ion membrane transporters, porins, and efflux pumps associated with Na+, Cl−, and K+ influx and efflux; and (ii) increased biosynthesis or uptake of compatible solutes – which are compounds that mimic the osmotic properties of salts [137, 138]. Both mechanisms ultimately balance the osmotic gradient across the cell membrane, preventing an influx of salt, which can damage cellular machinery.
High-saline environments have been associated with increased antibiotic production by resident microbes. For example, studies on rhizobia in mangroves show gradients of increasing salinity are associated with increased abundance of antibiotic biosynthesis genes encoding streptomycin (rffG, rffH and ISYNA1); monobactams (met3, asf, LysC); carbapenem (proA, proB); and penicillin (pac) [139]. Conversely, AMR genes similarly increased in abundance under high versus low salt. For example, high salinity levels increased abundance of cusS and copS (causing reduced membrane permeability), as well as AbcA and BmrA efflux pumps conferring β-lactam resistance [139]. One study confirmed a direct link between increased salinity and the maintenance of AMR genes in soil communities: by performing quantitative PCR on cattle manure microbial communities treated with increasing salt concentrations, Li et al. [140] found that high salinity can prevent the loss of AMR genes tetM, sul1, ermB and intI1, but not tetV, mexk or bacA [140]. Hence, AMR genes can persist in high-salinity environments, despite the energetic cost of maintaining AMR genes in the absence of antibiotics [140].
Cross-resistance or co-regulation of salt resistance and AMR genes can enhance AMR under high salinity. For example, the EnvZ/OmpR membrane-bound histidine kinases are chemoreceptors which can detect changes in external osmolarity [141]. These receptors regulate the expression of cell permeability porins, OmpC and OmpF, as well as the AcrAB–TolC multi-drug-resistance efflux pump [107, 141–145], enhancing both salt tolerance and AMR [146]. In Listeria monocytogenes, salt stress similarly increases the expression of LiaR – a response regulator involved in cell envelope stress and toxic ion responses [147, 148]. Deleting the liaR gene increases the sensitivity of L. monoctytogenes to nisin [147]. While such studies do not go as far as demonstrating that salt stress drives AMR evolution, they suggest that elevated salt stress could potentially select for de novo mutants that have reduced antibiotic susceptibility. An experimental evolution approach could reveal how physiological responses to salt stress feed into selection for AMR over evolutionary time.
MGEs can co-select for ARG in high-salinity environments. Metagenomic analysis of manure samples treated with increasing salt concentrations revealed that high-salinity environments maintain AMR genes tetM, sul1 and ermB, because they resided on MGEs containing salt stress resistance mechanisms [140]. Similarly, other salt-resistant mechanisms, such as the biosynthesis of the compatible solute ectoine and the expression of K+ transporters [124, 125], have been found on plasmids isolated from halophilic (‘salt-loving’) strains. The loss of such plasmids can alter the sensitivity of halophiles to antibiotics, including ampicillin and vancomycin [144]. Conversely, studies on saline soils have revealed a reduction in ARGs and MGEs under high- compared to low-salinity soils [149]. This was probably caused by a fitness cost of plasmid carriage under salt stress, as well as the loss of important ARG carriers such as Actinobacteria in highly saline soils [149]. Hence, the impact of increasing salinity on AMR depends on opportunities for co-resistance (i.e. whether salt-resistance and AMR genes are located on the same MGE) as well as the costs of MGE carriage in a particular environment.
The salinization of arable land is a growing concern, as land use intensifies amid the growing impact of climate change. In order to reduce or maintain soil salinity levels and prevent AMR dissemination, improved irrigation practices are required. This could be achieved by using efficient irrigation methods such as drip irrigation or precision agriculture; farmers can reduce water wastage and prevent the build-up of salts in the soil [150]. However, this approach may not be suitable for arid regions (where the problem is most prevalent) or may be cost-prohibitive for some farmers. Furthermore, promoting soil conservation practices, such as mulching and cover cropping, can help retain soil moisture and reduce salinity levels. However, the effectiveness of these practices depends on local climatic conditions and farming traditions, making adoption challenging in some areas. Salinization is regarded as a serious form of soil degradation and is estimated to be a major challenge for sustaining plant and animal life in the coming decades [133, 151]. Hence, the myriad benefits of employing mitigation measures to protect soils against salinization range from tackling AMR to safeguarding the future production of food.
Perspectives and future work
Anthropogenic activities have increasingly led to accelerated climate change, environmental pollution and the disruption of natural ecosystems. These events have been identified as potential contributors to the proliferation of AMR [152]. Emerging evidence suggests that anthropogenic actions resulting in increased temperatures, salinity and chemical influx could reduce or negate the cost of AMR in soil microbes. This finding is important because it becomes more difficult to control or reverse AMR if resistance mechanisms do not carry a cost (or are beneficial) in the absence of antibiotics. Worryingly, although antimicrobial stewardship programmes are becoming more successful, global population numbers and food production are becoming more intense. Together, this suggests that without a one-health approach to fighting AMR, intensive agriculture in the face of climate change could negate the positive efforts of antimicrobial stewardship programmes. Below, we highlight three areas of research that should be prioritized:
A myriad of studies linking bacterial stress responses to AMR stem from gene expression studies of knockout mutant strains lacking stress response gene(s) (e.g. sigma factor σ32 in E. coli ). The question remains about how relevant short-term stress responses are (which often temporarily enhance tolerance or antibiotic resistance) to long-term AMR selection. Moreover, the knowledge we have gained by studying stress responses in a single species (typically E. coli ) should now be expanded to gauge the role of stress response systems in natural microbial communities, where the cost and benefits of stress responses are likely to change [153]. Semi-natural experimental systems (e.g. [45]) are useful for testing the responses of a focal species (or community) to a stressor over both short and long timescales in a controlled laboratory setting.
While our review focuses on how anthropogenic stress can drive AMR, trade-offs could equally influence resistance evolution. However, few studies have investigated trade-offs between mechanisms other than those targeting different clinical antibiotics (i.e. collateral sensitivity [154–156]). Using an experimental evolution approach, Vasse et al. [157] demonstrated that antibiotic resistance evolution in P. aeruginosa comes at a population-level cost by selectively favouring siderophore 'cheats' that do not bear the cost of siderophore production but reap the metal-chelating benefits of siderophores produced by others [157]. These results indirectly imply that AMR might trade-off with metal chelation (and detoxification). Although there is strong evidence for trade-offs in experimental populations of bacteria [91], whether such trade-offs occur in metal-polluted communities – and influence the spread of AMR genes – remains unclear and is a worthwhile avenue for future research.
Our review highlights the profound role of natural environments in shaping AMR, and follows several excellent reviews in this area [72, 158]. However, the role anthropogenic stressors (other than antibiotic overuse) could have in driving AMR typically does not reach policy – where the focus is on managing antibiotic use, mainly in the clinic. A recent report by the World Health Organization [159] outlined 40 key research topics for informing policy on AMR – however, agriculture was not mentioned anywhere in this report. There remains a worrying assumption that tackling AMR begins and ends in the clinic, a perception that should now be challenged with translatory research that transcends from science into policy. Opportunities to second or partner research scientists into the public service (such as Science foundation Ireland’s Public Service Fellowship Programme) could allow researchers to bring their expertise to policy-making and innovation at a national and international level.
Funding information
The authors would like to acknowledge the financial support of NERC (MK: NE/S00713X/1), BBSRC Discovery (SOB: BB/T009446/1), IRC New Foundations and Community Foundation Ireland (SOB: NF/2022/39250777) and a UKRI Future Leaders Fellowship (EH: MR/V022482/1).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ARG, antimicrobial resistance gene; CVI, country vulnerability index; EU, European Union; GST, glutathione S-transferase; HGT, horizontal gene transfer; HSR, heat shock responses; MDR, multi-drug resistant; MGE, mobile genetic element; RNAP, RNA polymerase; Topt, optimum temperature.
References
- 1.O’Neill J. Review on Antimicrobial Resistance - Wellcome Collection; 2014. [ June 1; 2023 ]. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations.https://wellcomecollection.org/works/rdpck35v accessed. [Google Scholar]
- 2.de Kraker MEA, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13:e1002184. doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charani E, Holmes A. Antibiotic stewardship—twenty years in the making. Antibiotics. 2019;8:7. doi: 10.3390/antibiotics8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mudenda S, Daka V, Matafwali SK. World Health Organization AWaRe framework for antibiotic stewardship: Where are we now and where do we need to go? An expert viewpoint. Antimicrob Steward Healthc Epidemiol. 2023;3:e84. doi: 10.1017/ash.2023.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charani E, Cooke J, Holmes A. Antibiotic stewardship programmes--what’s missing? J Antimicrob Chemother. 2010;65:2275–2277. doi: 10.1093/jac/dkq357. [DOI] [PubMed] [Google Scholar]
- 6.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 7.Qiu D, Ke M, Zhang Q, Zhang F, Lu T, et al. Response of microbial antibiotic resistance to pesticides: an emerging health threat. Sci Total Environ. 2022;850:158057. doi: 10.1016/j.scitotenv.2022.158057. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Loya M, Tekin E, Kang TM, Cardona N, Lozano-Huntelman N, et al. Antibiotics shift the temperature response curve of Escherichia coli growth. mSystems. 2021;6:e0022821. doi: 10.1128/mSystems.00228-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Al-Omari A, Al Mutair A, Alhumaid S, Salih S, Alanazi A, et al. The impact of antimicrobial stewardship program implementation at four tertiary private hospitals: results of a five-years pre-post analysis. Antimicrob Resist Infect Control. 2020;9:95. doi: 10.1186/s13756-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaviani Rad A, Astaykina A, Streletskii R, Afsharyzad Y, Etesami H, et al. An overview of antibiotic resistance and abiotic stresses affecting antimicrobial resistance in agricultural soils. Int J Environ Res Public Health. 2022;19:4666. doi: 10.3390/ijerph19084666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassitta R, Nottensteiner A, Bauer J, Straubinger RK, Hölzel CS. Spread of antimicrobial resistance genes via pig manure from organic and conventional farms in the presence or absence of antibiotic use. J Appl Microbiol. 2022;133:2457–2465. doi: 10.1111/jam.15717. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz S, Hershberg R. Elevated mutagenesis does not explain the increased frequency of antibiotic resistant mutants in starved aging colonies. PLoS Genet. 2013;9:e1003968. doi: 10.1371/journal.pgen.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao H, Li X, Yang Q, Bai Y, Cui P, et al. Herbicide selection promotes antibiotic resistance in soil microbiomes. Mol Biol Evol. 2021;38:2337–2350. doi: 10.1093/molbev/msab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson-Edwards K. Tackling mine wastes. Science. 2016;352:288–290. doi: 10.1126/science.aaf3354. [DOI] [PubMed] [Google Scholar]
- 17.Lottermoser BG. Characterization, Treatment and Environmental Impacts. third edition. Vol. 352. Berlin Heidelberg: Springer; 2010. Mine wastes; pp. 288–290. vol. [Google Scholar]
- 18.Bradl HB. Heavy Metals in the Environment Origin, Interaction and Remediation. Amsterdam: Elsevier Academic Press; 2005. [Google Scholar]
- 19.Seiler C, Berendonk TU. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali H, Khan E, Ilahi I. Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. J Chemist. 2019;2019:1–14. doi: 10.1155/2019/6730305. [DOI] [Google Scholar]
- 21.Hudson-Edwards KA, Jamieson HE, Lottermoser BG. Mine wastes: past, present, future. Elements. 2011;7:375–380. doi: 10.2113/gselements.7.6.375. [DOI] [Google Scholar]
- 22.Abedin MJ, Cresser MS, Meharg AA, Feldmann J, Cotter-Howells J. Arsenic accumulation and metabolism in rice (Oryza sativa L.) Environ Sci Technol. 2002;36:962–968. doi: 10.1021/es0101678. [DOI] [PubMed] [Google Scholar]
- 23.Castro-González MI, Méndez-Armenta M. Heavy metals: implications associated to fish consumption. Environ Toxicol Pharmacol. 2008;26:263–271. doi: 10.1016/j.etap.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Alonso A, Sánchez P, Martínez JL. Environmental selection of antibiotic resistance genes. Environ Microbiol. 2001;3:1–9. doi: 10.1046/j.1462-2920.2001.00161.x. [DOI] [PubMed] [Google Scholar]
- 25.Dickinson AW, Power A, Hansen MG, Brandt KK, Piliposian G, et al. Heavy metal pollution and co-selection for antibiotic resistance: a microbial palaeontology approach. Environ Int. 2019;132:105117. doi: 10.1016/j.envint.2019.105117. [DOI] [PubMed] [Google Scholar]
- 26.Sui Q, Zhang J, Chen M, Wang R, Wang Y, et al. Fate of microbial pollutants and evolution of antibiotic resistance in three types of soil amended with swine slurry. Environ Pollut. 2019;245:353–362. doi: 10.1016/j.envpol.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Peng S, Zheng H, Herrero-Fresno A, Olsen JE, Dalsgaard A, et al. Co-occurrence of antimicrobial and metal resistance genes in pig feces and agricultural fields fertilized with slurry. Sci Total Environ. 2021;792:148259. doi: 10.1016/j.scitotenv.2021.148259. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Lan B, Fei H, Wang S, Zhu G. Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils. J Hazard Mater. 2021;411:124848. doi: 10.1016/j.jhazmat.2020.124848. [DOI] [PubMed] [Google Scholar]
- 29.Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, et al. Metal resistance and its association with antibiotic resistance. Adv Microb. 2017;70:261–313. doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 32.Valls M, de Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev. 2002;26:327–338. doi: 10.1111/j.1574-6976.2002.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 33.Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14:262–269. doi: 10.1016/s0958-1669(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien S, Buckling A. The sociality of bioremediation. EMBO Reports. 2015;16:1241–1245. doi: 10.15252/embr.201541064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruins MR, Kapil S, Oehme FW. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45:198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 36.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 37.Caille O, Rossier C, Perron K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa . J Bacteriol. 2007;189:4561–4568. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piddock LJV. Multidrug-resistance efflux pumps ? not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 40.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 41.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 42.Hesse E, O’Brien S, Luján AM, Sanders D, Bayer F, et al. Stress causes interspecific facilitation within a compost community. Ecol Lett. 2021;24:2169–2177. doi: 10.1111/ele.13847. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien S, Hesse E, Luján A, Hodgson DJ, Gardner A, et al. No effect of intraspecific relatedness on public goods cooperation in a complex community. Evolution. 2018;72:1165–1173. doi: 10.1111/evo.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13:2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 45.Hesse E, O’Brien S, Tromas N, Bayer F, Luján AM, et al. Ecological selection of siderophore-producing microbial taxa in response to heavy metal contamination. Ecol Lett. 2018;21:117–127. doi: 10.1111/ele.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hesse E, Padfield D, Bayer F, van Veen EM, Bryan CG, et al. Anthropogenic remediation of heavy metals selects against natural microbial remediation. Proc Biol Sci. 2019;286:20190804. doi: 10.1098/rspb.2019.0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer J, Özkaya Ö, Kümmerli R. Bacterial siderophores in community and host interactions. Nat Rev Microbiol. 2020;18:152–163. doi: 10.1038/s41579-019-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lear L, Hesse E, Buckling A, Vos M. Copper selects for siderophore-mediated virulence in Pseudomonas aeruginosa . BMC Microbiol. 2022;22:303. doi: 10.1186/s12866-022-02720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardoso K, Gandra RF, Wisniewski ES, Osaku CA, Kadowaki MK, et al. DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii . J Med Microbiol. 2010;59:1061–1068. doi: 10.1099/jmm.0.020339-0. [DOI] [PubMed] [Google Scholar]
- 50.Duval BD, Mathew A, Satola SW, Shafer WM. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob Agents Chemother. 2010;54:2283–2290. doi: 10.1128/AAC.01786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 52.Dimitriu T. Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiology. 2022;168:001214. doi: 10.1099/mic.0.001214. [DOI] [PubMed] [Google Scholar]
- 53.Klümper U, Dechesne A, Riber L, Brandt KK, Gülay A, et al. Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 2017;11:152–165. doi: 10.1038/ismej.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Yu Z, Ding P, Lu J, Klümper U, et al. Non-antibiotic pharmaceuticals promote conjugative plasmid transfer at a community-wide level. Microbiome. 2022;10:1–5. doi: 10.1186/s40168-022-01314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen LD, Sørensen SJ. The effect of longterm exposure to mercury on the bacterial community in marine sediment. Curr Microbiol. 1998;36:291–297. doi: 10.1007/s002849900312. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalo MP, Martínez JL, Baquero F, Gómez-Lus R, Pérez-Díaz JC. Aerobactin production linked to transferable antibiotic resistance in Escherichia coli strains isolated from sewage. FEMS Microbiol Lett. 1988;50:57–59. doi: 10.1111/j.1574-6968.1988.tb02911.x. [DOI] [Google Scholar]
- 57.Anedda E, Farrell ML, Morris D, Burgess CM. Evaluating the impact of heavy metals on antimicrobial resistance in the primary food production environment: A scoping review. Environ Pollut. 2023;320:121035. doi: 10.1016/j.envpol.2023.121035. [DOI] [PubMed] [Google Scholar]
- 58.Intergovernmental Panel on Climate Change (IPCC) Cambridge: Cambridge University Press; 2018. An IPCC Special Report on the impacts of global warming of 1.5 °C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty. Global warming of 1.5 °C. [Google Scholar]
- 59.Skendžić S, Zovko M, Živković IP, Lešić V, Lemić D. The impact of climate change on agricultural insect pests. Insects. 2021;12:440. doi: 10.3390/insects12050440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang X, Luo S, Lyu S. Observed soil temperature trends associated with climate change in the Tibetan Plateau, 1960–2014. Theor Appl Climatol. 2019;135:169–181. doi: 10.1007/s00704-017-2337-9. [DOI] [Google Scholar]
- 61.Rasmussen PU, Bennett AE, Tack AJM, Singh B. The impact of elevated temperature and drought on the ecology and evolution of plant–soil microbe interactions. J Ecol. 2020;108:337–352. doi: 10.1111/1365-2745.13292. [DOI] [Google Scholar]
- 62.Rasmussen PU, Bennett AE, Tack AJM, Singh B. The impact of elevated temperature and drought on the ecology and evolution of plant–soil microbe interactions. J Ecol. 2020;108:337–352. doi: 10.1111/1365-2745.13292. [DOI] [Google Scholar]
- 63.MacFadden DR, McGough SF, Fisman D, Santillana M, Brownstein JS. Antibiotic resistance increases with local temperature. Nat Clim Chang. 2018;8:510–514. doi: 10.1038/s41558-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Logan ML. Did pathogens facilitate the rise of endothermy? Ideas in Ecol Evol. 2019;12 [Google Scholar]
- 65.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 66.Roncarati D, Scarlato V. Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol Rev. 2017;41:549–574. doi: 10.1093/femsre/fux015. [DOI] [PubMed] [Google Scholar]
- 67.Grossman AD, Erickson JW, Gross CA. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 68.Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodríguez-Verdugo A, Lozano-Huntelman N, Cruz-Loya M, Savage V, Yeh P. Compounding effects of climate warming and antibiotic resistance. iScience. 2020;23:101024. doi: 10.1016/j.isci.2020.101024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.VanBogelen RA, Neidhardt FC. Ribosomes as sensors of heat and cold shock in Escherichia coli . Proc Natl Acad Sci. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodríguez-Verdugo A, Tenaillon O, Gaut BS. First-step mutations during adaptation restore the expression of hundreds of genes. Mol Biol Evol. 2016;33:25–39. doi: 10.1093/molbev/msv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodríguez-Verdugo A, Lozano-Huntelman N, Cruz-Loya M, Savage V, Yeh P. Compounding effects of climate warming and antibiotic resistance. iScience. 2020;23:101024. doi: 10.1016/j.isci.2020.101024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hilker M, Schwachtje J, Baier M, Balazadeh S, Bäurle I, et al. Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev Camb Philos Soc. 2016;91:1118–1133. doi: 10.1111/brv.12215. [DOI] [PubMed] [Google Scholar]
- 74.Rodríguez-Verdugo A, Gaut BS, Tenaillon O. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol. 2013;13:50. doi: 10.1186/1471-2148-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu XL, Zhang BW, Zhang QG, Zhu BR, Lin K, et al. Temperature responses of mutation rate and mutational spectrum in an Escherichia coli strain and the correlation with metabolic rate. BMC Evol Biol. 2016;16:1–24. doi: 10.1186/s12862-018-1252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohde K. Latitudinal gradients in species diversity: the search for the primary cause. Oikos. 1992;65:514. doi: 10.2307/3545569. [DOI] [Google Scholar]
- 77.Reverter M, Sarter S, Caruso D, Avarre J-C, Combe M, et al. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat Commun. 2020;11:1870. doi: 10.1038/s41467-020-15735-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kopittke PM, Dalal RC, Finn D, Menzies NW. Global changes in soil stocks of carbon, nitrogen, phosphorus, and sulphur as influenced by long-term agricultural production. Glob Chang Biol. 2017;23:2509–2519. doi: 10.1111/gcb.13513. [DOI] [PubMed] [Google Scholar]
- 79.Kurothe RS, Kumar G, Singh R, Singh HB, Tiwari SP, et al. Effect of tillage and cropping systems on runoff, soil loss and crop yields under semiarid rainfed agriculture in India. Soil Tillage Res. 2014;140:126–134. doi: 10.1016/j.still.2014.03.005. [DOI] [Google Scholar]
- 80.Gefen O, Fridman O, Ronin I, Balaban NQ. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci. 2014;111:556–561. doi: 10.1073/pnas.1314114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Himeoka Y, Mitarai N. Dynamics of bacterial populations under the feast-famine cycles. Phys Rev Res. 2020;2:013372. doi: 10.1103/PhysRevResearch.2.013372. [DOI] [Google Scholar]
- 82.Hazan R, Schoemann M, Klutstein M. Endurance of extremely prolonged nutrient prevention across kingdoms of life. iScience. 2021;24:102745. doi: 10.1016/j.isci.2021.102745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knöppel A, Näsvall J, Andersson DI. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob Agents Chemother. 2017;61:e01495-17. doi: 10.1128/AAC.01495-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paulander W, Maisnier-Patin S, Andersson DI. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (sigmaS) Genetics. 2009;183:539–546. doi: 10.1534/genetics.109.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schellhorn HE. Function, evolution, and composition of the RpoS regulon in Escherichia coli . Front Microbiol. 2020;11:560099. doi: 10.3389/fmicb.2020.560099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panchal VV, Griffiths C, Mosaei H, Bilyk B, Sutton JAF, et al. Evolving MRSA: High-level β-lactam resistance in Staphylococcus aureus is associated with RNA Polymerase alterations and fine tuning of gene expression. PLoS Pathog. 2020;16:e1008672. doi: 10.1371/journal.ppat.1008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding F, Han L, Xue Y, Yang IT, Fan X, et al. Multidrug-resistant Pseudomonas aeruginosa is predisposed to lasR mutation through up-regulated activity of efflux pumps in non-cystic fibrosis bronchiectasis patients. Front Cell Infect Microbiol. 2022;12:934439. doi: 10.3389/fcimb.2022.934439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang QG, Buckling A. Resource-dependent antagonistic coevolution leads to a new paradox of enrichment. Ecology. 2016;97:1319–1328. doi: 10.1890/15-1408.1. [DOI] [PubMed] [Google Scholar]
- 90.Jean-Pierre F, Hampton TH, Schultz D, Hogan DA, Groleau M-C, et al. Community composition shapes microbial-specific phenotypes in a cystic fibrosis polymicrobial model system. Elife. 2023;12:e81604. doi: 10.7554/eLife.81604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferenci T. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 2016;24:209–223. doi: 10.1016/j.tim.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 92.Phan K, Ferenci T. A design-constraint trade-off underpins the diversity in ecologically important traits in species Escherichia coli . ISME J. 2013;7:2034–2043. doi: 10.1038/ismej.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Montanarella L, Panagos P. The relevance of sustainable soil management within the European Green Deal. Land Use Policy. 2021;100:104950. doi: 10.1016/j.landusepol.2020.104950. [DOI] [Google Scholar]
- 94.Miller SA, Ferreira JP, LeJeune JT. Antimicrobial use and resistance in plant agriculture: a one health perspective. Agriculture. 2022;12:289. doi: 10.3390/agriculture12020289. [DOI] [Google Scholar]
- 95.Rani L, Thapa K, Kanojia N, Sharma N, Singh S, et al. An extensive review on the consequences of chemical pesticides on human health and environment. J Clean Product. 2021;283:124657. doi: 10.1016/j.jclepro.2020.124657. [DOI] [Google Scholar]
- 96.Blacquière T, van der Steen JJ. Three years of banning neonicotinoid insecticides based on sub-lethal effects: can we expect to see effects on bees? Pest Manag Sci. 2017;73:1299–1304. doi: 10.1002/ps.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marinho M da C, Diogo BS, Lage OM, Antunes SC. Ecotoxicological evaluation of fungicides used in viticulture in non-target organisms. Environ Sci Pollut Res. 2020;27:43958–43969. doi: 10.1007/s11356-020-10245-w. [DOI] [PubMed] [Google Scholar]
- 98.Roman DL, Voiculescu DI, Filip M, Ostafe V, Isvoran A. Effects of triazole fungicides on soil microbiota and on the activities of enzymes found in soil: a review. Agriculture. 2021;11:893. doi: 10.3390/agriculture11090893. [DOI] [Google Scholar]
- 99.Sim JXF, Doolette CL, Vasileiadis S, Drigo B, Wyrsch ER, et al. Pesticide effects on nitrogen cycle related microbial functions and community composition. Sci Total Environ. 2022;807:150734. doi: 10.1016/j.scitotenv.2021.150734. [DOI] [PubMed] [Google Scholar]
- 100.Baxter J, Cummings SP. The degradation of the herbicide bromoxynil and its impact on bacterial diversity in a top soil. J Appl Microbiol. 2008;104:1605–1616. doi: 10.1111/j.1365-2672.2007.03709.x. [DOI] [PubMed] [Google Scholar]
- 101.Medvedeva SE. Transfer of xenobiotics through cell membranes of luminous bacteria. Luminescence. 1999;14:267–270. doi: 10.1002/(SICI)1522-7243(199909/10)14:5<267::AID-BIO545>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 102.Nogacka AM, Gómez-Martín M, Suárez A, González-Bernardo O, de Los Reyes-Gavilán CG, et al. Xenobiotics formed during food processing: their relation with the intestinal microbiota and colorectal cancer. Int J Mol Sci. 2019;20:2051. doi: 10.3390/ijms20082051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurenbach B, Marjoshi D, Amábile-Cuevas CF, Ferguson GC, Godsoe W, et al. Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. mBio. 2015;6:e00009-15. doi: 10.1128/mBio.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sato T, Yokota S, Okubo T, Ishihara K, Ueno H, et al. Contribution of the AcrAB-TolC efflux pump to high-level fluoroquinolone resistance in Escherichia coli isolated from dogs and humans. J Vet Med Sci. 2013;75:407–414. doi: 10.1292/jvms.12-0186. [DOI] [PubMed] [Google Scholar]
- 105.Yu S, Wang Y, Shen F, Fang H, Yu Y. Copper-based fungicide copper hydroxide accelerates the evolution of antibiotic resistance via gene mutations in Escherichia coli . Sci Total Environ. 2022;815:152885. doi: 10.1016/j.scitotenv.2021.152885. [DOI] [PubMed] [Google Scholar]
- 106.Fernández L, Hancock REW. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raczkowska A, Trzos J, Lewandowska O, Nieckarz M, Brzostek K, et al. Expression of the AcrAB components of the AcrAB-TolC multidrug efflux pump of Yersinia enterocolitica is subject to dual regulation by OmpR. PLoS One. 2015;10:e0124248. doi: 10.1371/journal.pone.0124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumar M, Yadav AN, Saxena R, Paul D, Tomar RS. Biodiversity of pesticides degrading microbial communities and their environmental impact. Biocat Agri Biotechnol. 2021;31:101883. doi: 10.1016/j.bcab.2020.101883. [DOI] [Google Scholar]
- 109.Paidi MK, Satapute P, Haider MS, Udikeri SS, Ramachandra YL, et al. Mitigation of organophosphorus insecticides from environment: Residual detoxification by bioweapon catalytic scavengers. Environ Res. 2021;200:111368. doi: 10.1016/j.envres.2021.111368. [DOI] [PubMed] [Google Scholar]
- 110.Eddleston M. Novel clinical toxicology and pharmacology of organophosphorus insecticide self-poisoning. Annu Rev Pharmacol Toxicol. 2019;59:341–360. doi: 10.1146/annurev-pharmtox-010818-021842. [DOI] [PubMed] [Google Scholar]
- 111.Barman DN, Haque MA, Islam SMA, Yun HD, Kim MK. Cloning and expression of ophB gene encoding organophosphorus hydrolase from endophytic Pseudomonas sp. BF1-3 degrades organophosphorus pesticide chlorpyrifos. Ecotoxicol Environ Saf. 2014;108:135–141. doi: 10.1016/j.ecoenv.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 112.Serdar CM, Gibson DT. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta . Nat Biotechnol. 1985;3:567–571. doi: 10.1038/nbt0685-567. [DOI] [Google Scholar]
- 113.Zhao S, Xu W, Zhang W, Wu H, Guang C, et al. Overview of a bioremediation tool: organophosphorus hydrolase and its significant application in the food, environmental, and therapy fields. Appl Microbiol Biotechnol. 2021;105:8241–8253. doi: 10.1007/s00253-021-11633-z. [DOI] [PubMed] [Google Scholar]
- 114.Horne I, Sutherland TD, Harcourt RL, Russell RJ, Oakeshott JG. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl Environ Microbiol. 2002;68:3371–3376. doi: 10.1128/AEM.68.7.3371-3376.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ali M, Naqvi TA, Kanwal M, Rasheed F, Hameed A, et al. Detection of the organophosphate degrading gene opdA in the newly isolated bacterial strain Bacillus pumilus W1. Ann Microbiol. 2012;62:233–239. doi: 10.1007/s13213-011-0251-4. [DOI] [Google Scholar]
- 116.Chaudhry GR, Ali AN, Wheeler WB. Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Appl Environ Microbiol. 1988;54:288–293. doi: 10.1128/aem.54.2.288-293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rangasamy K, Athiappan M, Devarajan N, Parray JA, Shameem N, et al. Cloning and expression of the organophosphate pesticide-degrading α-β hydrolase gene in plasmid pMK-07 to confer cross-resistance to antibiotics. Biomed Res Int. 2018;2018:1535209. doi: 10.1155/2018/1535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rangasamy K, Athiappan M, Devarajan N, Parray JA. Emergence of multi drug resistance among soil bacteria exposing to insecticides. Microb Pathog. 2017;105:153–165. doi: 10.1016/j.micpath.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 119.Rangasamy K, Athiappan M, Devarajan N, Samykannu G, Parray JA, et al. Pesticide degrading natural multidrug resistance bacterial flora. Microbial Pathogen. 2018;114:304–310. doi: 10.1016/j.micpath.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 120.Schröder P. Significance of Glutathione to Plant Adaptation to the Environment 2001;155-183. Dordrecht: Springer Netherlands; The role of glutathione and glutathione S-transferases in plant reaction and adaptation to xenobiotics. [DOI] [Google Scholar]
- 121.Koirala B K S, Moural T, Zhu F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int J Biol Sci. 2022;18:5713–5723. doi: 10.7150/ijbs.77141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zablotowicz RM, Hoagland RE, Locke MA, Hickey WJ. Glutathione-S-transferase activity and metabolism of glutathione conjugates by rhizosphere bacteria. Appl Environ Microbiol. 1995;61:1054–1060. doi: 10.1128/aem.61.3.1054-1060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Park H, Choung YK. Degradation of antibiotics (tetracycline, sulfathiazole, ampicillin) using enzymes of glutathion S-transferase. HERA. 2007;13:1147–1155. doi: 10.1080/10807030701506223. [DOI] [Google Scholar]
- 125.Bordeleau LM, Rosen JD, Bartha R. Herbicide-derived chloroazobenzene residues. Pathway of formation. J Agric Food Chem. 1972;20:573–578. doi: 10.1021/jf60181a001. [DOI] [PubMed] [Google Scholar]
- 126.Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012;20:227–234. doi: 10.1016/j.tim.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 127.Zhang H, Song J, Zheng Z, Li T, Shi N, et al. Fungicide exposure accelerated horizontal transfer of antibiotic resistance genes via plasmid-mediated conjugation. Water Res. 2023;233:119789. doi: 10.1016/j.watres.2023.119789. [DOI] [PubMed] [Google Scholar]
- 128.Schmidt SBI, Rodríguez-Rojas A, Rolff J, Schreiber F. Biocides used as material preservatives modify rates of de novo mutation and horizontal gene transfer in bacteria. J Hazard Mater. 2022;437:129280. doi: 10.1016/j.jhazmat.2022.129280. [DOI] [PubMed] [Google Scholar]
- 129.Xing Y, Wu S, Men Y. Exposure to environmental levels of pesticides stimulates and diversifies evolution in Escherichia coli toward higher antibiotic resistance. Environ Sci Technol. 2020;54:8770–8778. doi: 10.1021/acs.est.0c01155. [DOI] [PubMed] [Google Scholar]
- 130.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 131.Wang M, Hu Z. Cross-talking between baculoviruses and host insects towards a successful infection. Phil Trans R Soc B. 2019;374:20180324. doi: 10.1098/rstb.2018.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Harman GE. Overview of mechanisms and uses of Trichoderma spp. Phytopathology. 2006;96:190–194. doi: 10.1094/PHYTO-96-0190. [DOI] [PubMed] [Google Scholar]
- 133.Corwin DL. Climate change impacts on soil salinity in agricultural areas. Eur J Soil Sci. 2021;72:842–862. doi: 10.1111/ejss.13010. [DOI] [Google Scholar]
- 134.Shao H, Chu L, Lu H, Qi W, Chen X, et al. Towards sustainable agriculture for the salt-affected soil. Land Degrad Dev. 2019;30:574–579. doi: 10.1002/ldr.3218. [DOI] [Google Scholar]
- 135.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fan X, Pedroli B, Liu G, Liu Q, Liu H, et al. Soil salinity development in the yellow river delta in relation to groundwater dynamics. Land Degrad Dev. 2012;23:175–189. doi: 10.1002/ldr.1071. [DOI] [Google Scholar]
- 137.Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4:12176–12181. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- 139.Sepúlveda-Correa A, Daza-Giraldo LV, Polanía J, Arenas NE, Muñoz-García A, et al. Genes associated with antibiotic tolerance and synthesis of antimicrobial compounds in a mangrove with contrasting salinities. Mar Pollut Bull. 2021;171:112740. doi: 10.1016/j.marpolbul.2021.112740. [DOI] [PubMed] [Google Scholar]
- 140.Li B, Jeon MK, Li X, Yan T. Differential impacts of salinity on antibiotic resistance genes during cattle manure stockpiling are linked to mobility potentials revealed by metagenomic sequencing. J Hazard Mater. 2023;445:130590. doi: 10.1016/j.jhazmat.2022.130590. [DOI] [PubMed] [Google Scholar]
- 141.Dawan J, Ahn J. Bacterial stress responses as potential targets in overcoming antibiotic resistance. Microorganisms. 2022;10:1385. doi: 10.3390/microorganisms10071385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brindangnanam P, Sawant AR, Prashanth K, Coumar MS. Bacterial effluxome as a barrier against antimicrobial agents: structural biology aspects and drug targeting. Tissue Barriers. 2022;10:2013695. doi: 10.1080/21688370.2021.2013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chetri S, Singha M, Bhowmik D, Nath K, Chanda DD, et al. Transcriptional response of OmpC and OmpF in Escherichia coli against differential gradient of carbapenem stress. BMC Res Notes. 2019;12:138. doi: 10.1186/s13104-019-4177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mukhtar S, Ahmad S, Bashir A, Mehnaz S, Mirza MS, et al. Identification of plasmid encoded osmoregulatory genes from halophilic bacteria isolated from the rhizosphere of halophytes. Microbiol Res. 2019;228:126307. doi: 10.1016/j.micres.2019.126307. [DOI] [PubMed] [Google Scholar]
- 145.Sun S, Berg OG, Roth JR, Andersson DI. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics. 2009;182:1183–1195. doi: 10.1534/genetics.109.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thiolas A, Bornet C, Davin-Régli A, Pagès JM, Bollet C. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes . Biochem Biophys Res Commun. 2004;317:851–856. doi: 10.1016/j.bbrc.2004.03.130. [DOI] [PubMed] [Google Scholar]
- 147.Bergholz TM, Tang S, Wiedmann M, Boor KJ. Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl Environ Microbiol. 2013;79:5682–5688. doi: 10.1128/AEM.01797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bergholz TM, Bowen B, Wiedmann M, Boor KJ. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl Environ Microbiol. 2012;78:2602–2612. doi: 10.1128/AEM.07658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan L, Wang F, Liang M, Wang X, Das R, et al. Antibiotic resistance genes attenuated with salt accumulation in saline soil. J Hazard Mater. 2019;374:35–42. doi: 10.1016/j.jhazmat.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 150.Machado RM, Serralheiro RP. Soil salinity: effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae. 2017;3:30. doi: 10.3390/horticulturae3020030. [DOI] [Google Scholar]
- 151.Shrivastava P, Kumar R. Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci. 2015;22:123–131. doi: 10.1016/j.sjbs.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.de Jongh EJ, Harper SL, Yamamoto SS, Wright CJ, Wilkinson CW, et al. One health, one hive: a scoping review of honey bees, climate change, pollutants, and antimicrobial resistance. PLoS One. 2022;17:e0242393. doi: 10.1371/journal.pone.0242393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barraclough TG. How do species interactions affect evolutionary dynamics across whole communities? Annu Rev Ecol Evol Syst. 2015;46:25–48. doi: 10.1146/annurev-ecolsys-112414-054030. [DOI] [Google Scholar]
- 154.Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pál C, Papp B, Lázár V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015;23:401–407. doi: 10.1016/j.tim.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 157.Vasse M, Noble RJ, Akhmetzhanov AR, Torres-Barceló C, Gurney J, et al. Antibiotic stress selects against cooperation in the pathogenic bacterium Pseudomonas aeruginosa . Proc Natl Acad Sci. 2017;114:546–551. doi: 10.1073/pnas.1612522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Malagón-Rojas JN, Parra Barrera EL, Lagos L. From environment to clinic: the role of pesticides in antimicrobial resistance. Revista Panamericana de Salud Pública. 2020;44:1. doi: 10.26633/RPSP.2020.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.World Health Organisation (WHO) Global research agenda for antimicrobial resistance in human health. 2023. [ July 1; 2023 ]. https://www.who.int/publications/m/item/global-research-agenda-for-antimicrobial-resistance-in-human-health accessed.