Abstract
The decision of whether cells are activated or not is controlled through dynamic intracellular molecular networks. However, the low population of cells during the transition state of activation renders the analysis of the transcriptome of this state technically challenging. To address this issue, we have developed the Time-Dependent Cell-State Selection (TDCSS) technique, which employs live-cell imaging of secretion activity to detect an index of the transition state, followed by the simultaneous recovery of indexed cells for subsequent transcriptome analysis. In this study, we used the TDCSS technique to investigate the transition state of group 2 innate lymphoid cells (ILC2s) activation, which is indexed by the onset of interleukin (IL)-13 secretion. The TDCSS approach allowed us to identify time-dependent genes, including transiently induced genes (TIGs). Our findings of IL4 and MIR155HG as TIGs have shown a regulatory function in ILC2s activation.
Subject terms: Immune cell isolation, Time-lapse imaging, ELISPOT
Time-Dependent Cell-State Selection combines live cell imaging and single-cell RNA sequencing to characterize and analyse ILC2s during their activation, revealing a set of genes that are transitionally upregulated (TIGs) in ILC2s during this phase.
Introduction
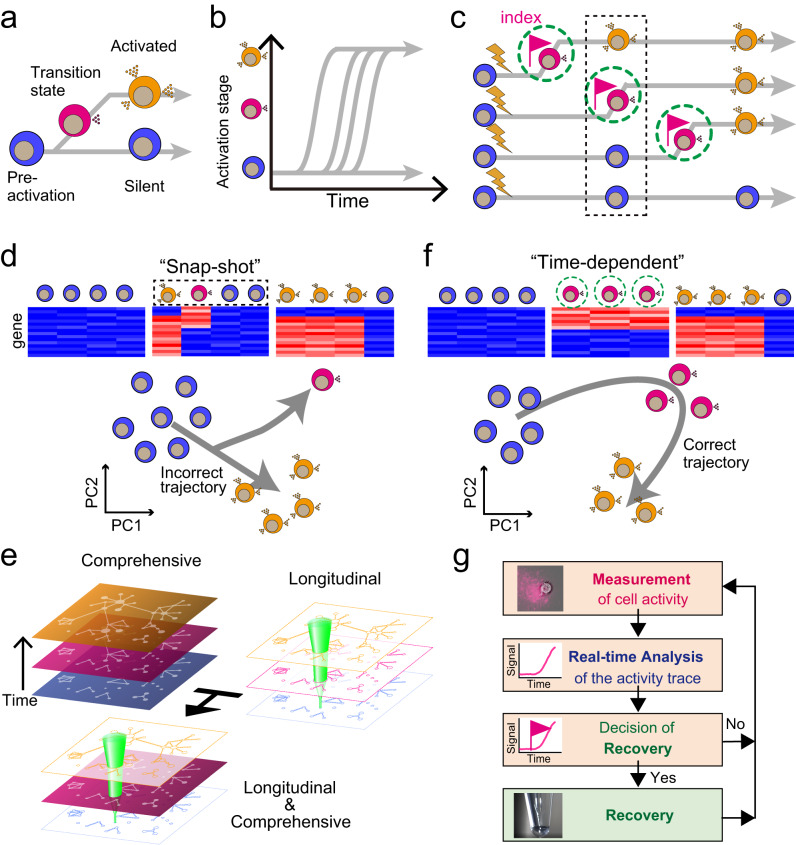
Cells determine their fate, such as differentiation1, cell death2, and activation during stress response3 and immune response4, through dynamic intracellular molecular mechanisms5,6. In order to comprehend the mechanisms by which cells determine their fate, it is necessary to identify the molecular signatures of the early stages of the fate determination process, which control gene expression and ultimately determine the cell’s fate (Fig. 1a). In general, as fate determination is asynchronous7–9 (Fig. 1b), a population of cells collected by a “snap-shot” manner at a specific time point would contain cells in various stages of the fate determination process (Fig. 1c, d).
Fig. 1. Concept of time-dependent cell-state selection, TDCSS.
a Model depicting the transition of cellular states during the process of cell fate determination. b Variation in the timing of fate determination among cells. c Comparison of two cell recovery methods during the process of cell fate determination. The conventional strategy involves collecting cells in a “snapshot” manner (black dashed rectangle), while the “time-dependent” method involves collecting cells at each cell’s individual transition timing (green dashed circle). d Single-cell transcriptomes obtained through snapshot cell recovery methods and inferred trajectories of fate determination. e TDCSS methodology combines longitudinal and comprehensive analysis. f Single-cell transcriptomes obtained through time-dependent cell recovery methods and deduced trajectories of fate determination. g Schematic representation of the TDCSS technique.
There are two traditional methods for studying cell fate determination processes. Single-cell transcriptome analysis allows us to computationally infer the fate determination process by ordering cells based on similarities in their gene expression patterns10,11 (Fig. 1d). This approach is effective for slow fate determination processes, such as differentiation, but it is not effective for rapid fate determination processes with a short transition period, such as inflammation. Additionally, this approach can sometimes inaccurately estimate the cell fate trajectory due to the scarcity of cells in a transition state within a collected population of cells (Fig. 1d). This scarcity is caused by both the low frequency of cells entering a transition state and the short duration of the state itself12–14.
Another approach is a longitudinal analysis, such as live-cell imaging (LCI) techniques, that can reliably capture a transition state even if it is infrequent and short by visualizing the molecules associated with the fate determination process2,9,12,15,16. However, LCI analysis may not provide a comprehensive view of potential factors as it is limited to the predetermined molecular dynamics that are being visualized.
Here we report the development of the Time-Dependent Cell-State Selection (TDCSS) technique which combines the advantages of single-cell transcriptome analyses and LCI (Fig. 1e). TDCSS is a time-dependent cell recovery method that utilizes longitudinal cellular state information recorded using an LCI technique to identify the molecular signatures at the targeted transition state of the fate determination process (Fig. 1c, f), instead of relying on computational inference for time-dependent analysis. We demonstrate the effectiveness of the TDCSS technique by applying it to mouse group 2 innate lymphoid cells (mILC2s)17,18 that select their fate to induce a rapid type 2 immune response by producing a large amount of type 2 cytokines. We further demonstrate the power of the TDCSS technique by applying it to the rare human ILC2s (hILC2s)19–21 and obtaining the signature of expression in the transition state of the fate determination process.
Results
Development of the TDCSS technique
The key technical challenge of our method is integrating the destructive single-cell transcriptome analysis and the non-invasive LCI without compromising the benefits of either one (Fig. 1g). To overcome this challenge, we have designed and developed the TDCSS technique, which employs LCI to detect an index of the transition state, followed by the simultaneous recovery of indexed cells for subsequent transcriptome analysis. The TDCSS technique is composed of four steps: (1) Measurement of individual cell activity using LCI, (2) Real-time analysis of the activity trace, (3) Decisions on cell recovery, and (4) Recovery of the target cells. In the measurement step, images of individual cells are acquired using LCI. In the subsequent analysis step, the acquired images are analyzed in real-time to identify cells in the targeted activation state. In the third step, the activity of the identified cells is evaluated in a time-series manner to determine whether to flag them for recovery. Finally, in the recovery step, flagged cells are selectively harvested for transcriptome analysis.
Evaluating the effectiveness of the TDCSS
As a proof of concept and to evaluate the effectiveness of the TDCSS technique, we selected the cytokine secretion response of mouse ILC2s (mILC2s) for the following reasons. First, mILC2s rapidly produce a large amount of type 2 cytokines, such as interleukin (IL)-5 and IL-13, during the activation process17, which makes the initiation of the transition state easily detectable by the live-cell imaging (LCI) technique. Second, mILC2s are activated by humoral factors, such as IL-3317, which facilitates the reconstitution of their activation response in microscopy experiments. Third, mILC2s are non-adherent cells, thus the flagged mILC2s can be easily recovered using a glass capillary.
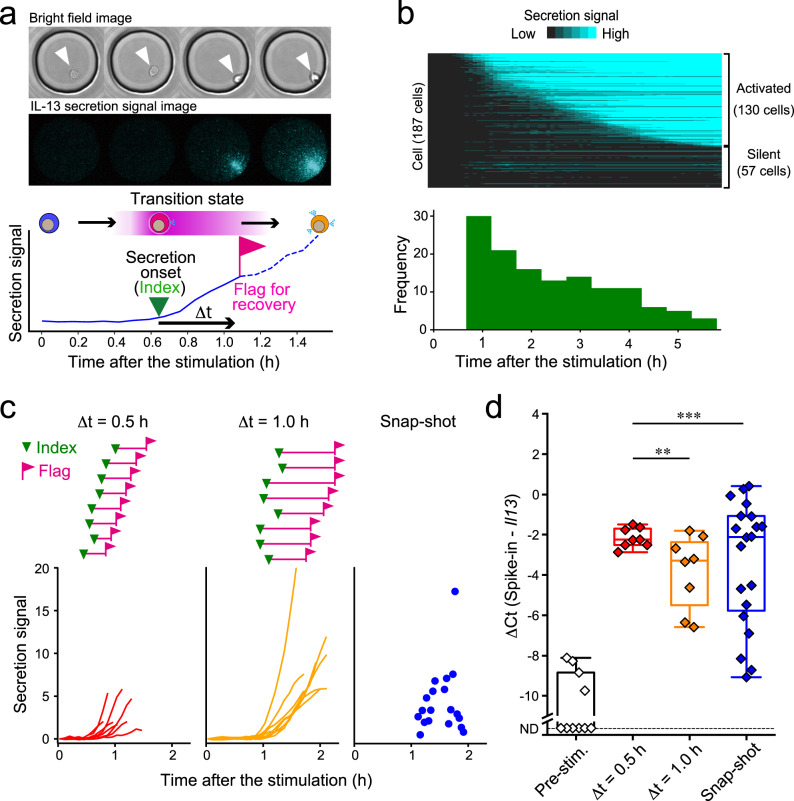
To perform the TDCSS technique, we observed the IL-13 secretion activities of 187 individual ILC2s from mouse fat tissues stimulated by recombinant IL-33 using real-time single-cell imaging of protein secretion12 (LCI of secretion activity, LCI-S) (Fig. 2a). The traces of the secretion signal showed that cells started secretion after various latencies over several hours (Fig. 2b). We found that a delay time (⊿t) over 0.5 h from the onset of secretion (index) is needed to reliably flag the target cells (Supplementary Fig. 1).
Fig. 2. The TDCSS technique found a uniform Il13 mRNA level of mILC2 in transition state.
a Time-lapse images and intensity trace of IL-13 secretion signal from a stimulated mILC2 acquired by LCI-S. b Heatmap of IL-13 secretion signal from individual mILC2s and a histogram of their secretion onset times. c IL-13 secretion onset (inverted triangle), recovery timing (flag), and signal trace (bottom) of each mILC2 subjected to qRT-PCR analysis. d Box plots showing the median and interquartile range of Il13 mRNA levels after stimulation. Whiskers represent minimum and maximum values. ND: not detected. **P < 0.01, ***P < 0.001, F-test.
To validate the TDCSS technique, we compared mRNA levels of Il13, which encodes IL-13, between “time-dependent” recovered cells using the TDCSS technique at ⊿t = 0.5 or 1.0 h (Supplementary Fig. 1) and “snap-shot” recovered cells only from secretion-positive mILC2s (Fig. 2c) using single-cell quantitative RT-PCR (qRT-PCR) (Fig. 2d, Supplementary Fig. 2). The variation of Il13 expression in mILC2s recovered in the “time-dependent” manner at ⊿t = 0.5 h was significantly reduced compared to that of mILC2s recovered in a “snap-shot” manner. Additionally, the variation in mILC2s at at ⊿t = 1.0 h was greater than the variation in mILC2s at ⊿t = 0.5 h. These results indicate homogeneity in the Il13 mRNA level in the transition state of mILC2’s activation process among cells, which diversified within 1.0 h. In conclusion, we found that using the onset of IL-13 secretion as an index was effective in obtaining the intracellular molecular information of mILC2 in the transition state using the TDCSS technique.
The transcriptome of mILC2 in the transition state
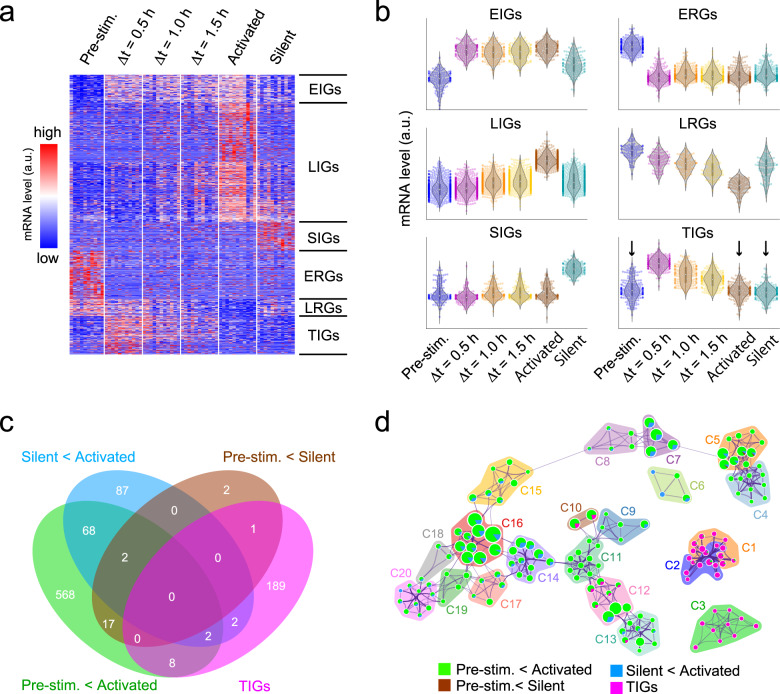
In order to explore the activation process of mILC2s comprehensively, we subjected mILC2s in the transition state (⊿t = 0.5, 1.0, and 1.5 h) and in other states (pre-stimulation: the state before the stimulation, activated: the state in which secretory activity was detected at 8 h of stimulation, when the majority of cells that had undergone the transition as shown in Fig. 2b, and silent: the state in which no secretion activity was detected at 8 h of stimulation) to single-cell RNA-seq (scRNA-seq). Based on the principal component analysis (PCA) or the trajectory inference (TI) results, we found that the transition cells were in a cluster/branch different from that of pre-stimulation cells or activated cells (Supplementary Fig. 3a, b). The TI method, which uses similarity between elements as an indicator, provides little analogy for abrupt changes in gene expression. Therefore, it is likely that the cells immediately after the change were displayed as the ends of different branches, due to the estimation of a pseudo-time different from the original time course (Supplementary Fig. 3b, c). To understand cell fate determination in rapid cell state transitions such as inflammatory responses, it is necessary to introduce real-time information obtained by our TDCSS method. To classify genes with variable expression during the activation process, we clustered differentially expressed genes (DEGs) between the activation states (one-way ANOVA p < 0.05). We classified 1,471 DEGs based on the transition modes in their expression levels into six classes: early induced genes (EIGs: 147), early reduced genes (ERGs: 254), late induced genes (LIGs: 627), late reduced genes (LRGs: 90), silent induced genes (SIGs: 151), and transiently induced genes (TIGs: 202) (Fig. 3a). Importantly, the TIGs could only be identified using the TDCSS technique because their expression levels were indistinguishable between the pre-stimulation and the activated / silent states (Fig. 3b, arrows). The difference in the dynamic nature of these transition modes explained why the pseudo-time estimated by the TI method differed from the actual elapsed time (Supplementary Fig. 3d). We further confirmed that the TIGs provided by the TDCSS technique were distinctive from the DEGs obtained by snapshot collection of pre- and post-stimulation cellular states (Fig. 3c). Interestingly, several genes reported to be involved in ILC2 activation were included in the TIGs we newly identified in TDCSS (Supplementary Data 1), for example, the growth inducers; Rel (cRel gene)22 and Bcl2l1 (BCL-XL gene)23, the immune checkpoint factors; Relb24, Klrg1, Lilrb4a and Cd274 (PD-L1 gene)25, the post-transcriptionally regulators; Mir155hg (host gene for miR-155)26,27 and Zc3h12a (Regnase-1 gene)28, the transcription factors; Egr2, Nfkbiz and Nr4a129 and the anti-apoptotic protein; Bcl330. The TIGs also contained Il4 (type 2 cytokine IL-4 gene), Il7rb (IL-25 receptor gene), and Tnfaip3 (A20 gene which is a negative regulator of IL-25 signals31). To study TIGs’ functions, we performed gene ontology (GO) analysis and found the relationships with immune activation-related terms such as “leukocyte activation” and “regulation of cytokine production” (Fig. 3d and Supplementary Data 2). The result that TIGs had a uniquely enriched ontology cluster indicates that the TDCSS technique succeeded in revealing the context of the transition state of mILC2’s activation process previously unavailable.
Fig. 3. Unbiased characterisation of gene expression during ILC2’s activation process.
a Heat map depicting the expression levels of 1471 differentially expressed genes (DEGs). b The trend of relative expression levels of genes in six transition classes. Additional information about each class can be found in Supplementary Data 1. c Overlap of the TIGs with the specified DEGs. d Network visualization of enriched ontology clusters of the TIGs and specified DEGs by using Metascape. Each node represents an enriched ontology cluster. The size of the node indicates the number of hit genes, while the pie chart illustrates the proportion of hit genes in the list. Clusters C1–C20, classified by similarity of enriched terms, are depicted in each coloured area. Additional information about each cluster can be found in Supplementary Data 2.
To investigate the influence of the index on the identification of TIGs, we reassigned the index to the onset of IL-5 secretion and re-evaluated the TIGs (Supplementary Fig. 4, Supplementary Data 3). Despite some shared GO terms between the IL-5 indexed TIGs and IL-13 indexed TIGs, these two groups of TIGs were found to be distinct (Supplementary Fig. 4e, f, Supplementary Data 4 and 5). We explored the relationship between the initiation of IL-13 and IL-5 secretion from individual cells during the activation process using two-colour LCI-S (Supplementary Fig. 5). The results indicate that under the experimental conditions described in this study, most mILC2s initiate secretion of both IL-5 and IL-13 simultaneously or IL-13 first. This suggests that the index based on IL-13 is able to detect an earlier transition state in the activation process. These findings demonstrate that the TDCSS technique can be employed to comprehensively characterize the gene expression in the transition state of the mILC2 activation process.
Identification of TIGs of human ILC2
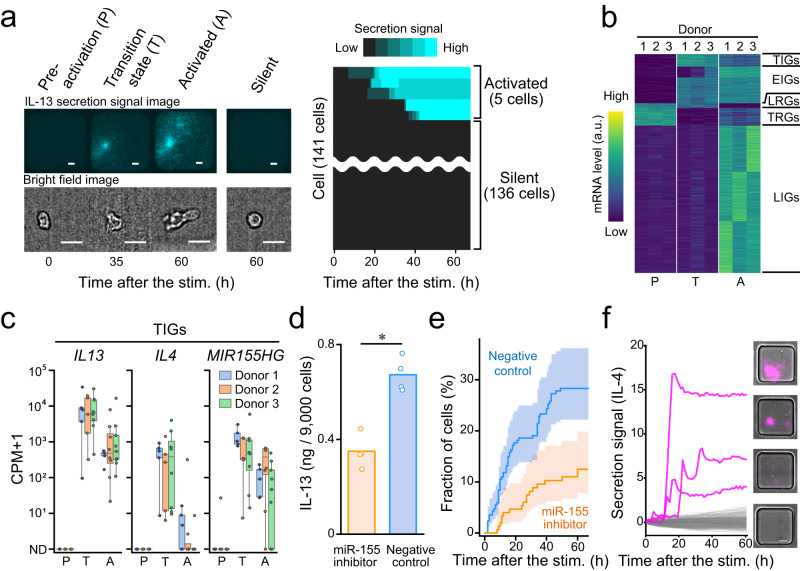
Given the demonstration of the efficacy of the TDCSS technique through experiments utilizing expanded culture mILC2s, we subsequently analysed the expression signature of human ILC2s (hILC2s)20,21,32 during the transition state. Besides being a rare cell type in peripheral blood (50–150/mL)20, we confirmed that the population of hILC2s that initiated secretion (2–4%) exhibited a wide range of latency within 60 h after stimulation (Fig. 4a, Supplementary Fig. 6a, b), as previously reported (Supplementary Fig. 6c)33. Given the populational rarity and temporal heterogeneity, it is challenging to efficiently collect cells in the transition state of activation using a “snapshot” approach (Supplementary Fig. 6b).
Fig. 4. Identification of genes expressed in different activation steps from human specimens.
a Representative time-lapse images of secretion signal and cell morphology (left) and heatmap of IL-13 secretion signal (right) of individual hILC2s. Scale bar, 25 µm. b Heatmap showing the average mRNA levels in each activation state of each specimen in standard scores (z-score). Details of each class are provided in Supplementary Data 6. c Box plot illustrating the expression levels of representative TIGs; IL13, IL4 and MIR155HG in each activation state. Each colour represents a different specimen. The centre line, box limits, whiskers, and dots indicate the median, upper and lower quartiles, 1.5x interquartile range and individual cells, respectively. ND: not detected. d Comparison of IL-13 production in cultured human ILC2 with and without miR-155 inhibitor by ELISA of bulk culture supernatants. Inhibition was performed by electroporation of a synthetic miRNA inhibitor with miR-155 specific or control sequence. e Comparison of temporal changes in the estimated proportion of IL-13-secreting cells in cultured human ILC2s with and without miR-155 inhibitor using the Nelson-Aalen estimator with their respective 95% confidence intervals (coloured bands). The number of cells in negative control and inhibition were 259 and 153, respectively. The difference between them was compared with the log-rank test (observed p value = 0.0013). f Secretion signal traces and images of IL-4 producing individual hILC2s. Three secretion-positive traces are shown in magenta and other traces are shown in grey (388 traces).
We applied the TDCSS technique to hILC2 by indexing the onset of IL-13 secretion and recovered the hILC2s in the transition state (⊿t within 3 h, Supplementary Fig. 6d). We compared the transcriptome among hILC2s of three donors in the transition state and other states (pre-stimulation and activated at 60 h of stimulation or later). As a result, we identified 91 TIGs as well as genes in four other classes of transition patterns (EIGs:270, LIGs:1081, LEGs:35 and transiently reduced genes: TRGs 128) from 1605 DEGs (ANOVA p < 0.01) (Fig. 4b and Supplementary Data 6). Another approach was to identify TIGs for genes whose expression levels transiently increase and subsequently decrease, and the robustness of five of these TIGs (IL17RB, CYTIP, IL13, IL4, MIR155HG) was confirmed (Supplementary Fig. 7). As with the results in mILC2s, the dynamic nature of the transition patterns, especially TIGs, explained why the pseudo-time estimated by the TI method differed from the actual elapsed time (Supplementary Fig. 8).
Next, to confirm the functional involvement of TIGs in the activation process of ILC2, we investigated the roles of IL4 and MIR155HG (Fig. 4c). Using LCI-S, we verified the contribution of miR-155 to ILC2 activation and found that the inhibition of miR-155 attenuated the stimulus-dependent activation of expanded culture hILC2s (Fig. 4d, e). IL-4 is a key inducer of type 2 immune response34 and plays an important role in hILC2 function, including proliferation and maintenance of the CRTH2 expression35. Although hILC2s have been reported to produce IL-419, it was difficult to characterize the IL-4 secretion dynamics of hILC2s due to their low production of IL-4. Therefore, we examined IL-4 secretion dynamics from individual hILC2s by LCI-S and detected their transient secretion for several hours (Fig. 4f). Although we could propose the plausibility of transient IL-4 production, further exploration of experimental conditions is required to reproduce the protein-level secretion of IL-4.
Discussion
We succeeded in a comprehensive exploration of the potential regulators of fate determination by developing and utilizing the TDCSS technique to recover individual cells at the transition state, even though such potential regulators existed in a small population of cells and only for a short duration. The discovery of a uniquely enriched gene ontology cluster of the transiently induced genes (Fig. 3d) indicates that the TDCSS technique is able to exploit the IL-13 secretion onset to identify the characteristic state during the activation process of ILC2.
The process by which activated ILC2s determine their fate to produce a tremendous amount of cytokine17,36 was previously unknown. IL4 and MIR155HG, whose respective products IL-4 and miR-155 are two positive regulators of the cytokine production and proliferation of ILC226,35, are upregulated in the transition state of the activation process, suggesting that they are key regulators of the subsequent activation process. MiR-155 is known to target and repress IL-33 signalling repressors, including INPP5D36–38 and DUSP1039,40. Although these genes were not expressed immediately after the secretion onset of IL-13, they were expressed at the late stage of the activation process under IL-33 stimulation (Supplementary Fig. 7c). MIR155HG may be prepared to silence these negative regulators from the transition state of the activation process to maintain ILC2s’ proliferation and their cytokine production activity. Furthermore, miR-155 also targets SOCS141,42 and DUSP443,44, both encoding inhibitors of IL-2 signalling that support ILC2’s activation45,46, and IFNGR147, encoding a receptor of IFN-γ that suppresses ILC2’s activation48. This indicates that miR-155 contributes to the fate determination in several ways, however, more detailed observations are required to understand whether miR-155 regulates by a transcriptional or a translational repression. IL-435, the other positive regulator we focused on, was confirmed to be transiently produced at the protein level. Our data show a constant expression of IL4R, which encodes an IL-4 receptor. This suggests that IL-4 produced by ILC2 early in its activation process may contribute to the positive regulation of activation in an autocrine manner, although further research is needed to confirm this hypothesis. The genes expressed in the transition state of the activation process that currently have an unknown function in the ILC2 should be studied in the future in order to gain a deeper understanding of the mechanism of fate determination in ILC2.
The selection of surface antigens for human ILC2 (hILC2) remains a topic of ongoing research, and the TDCSS technique has the potential to provide new insights not only in identifying transiently induced genes, but also in selecting surface antigens that index the activation of hILC2. Liu et al. have reported that CD30, TNFR2, and ICOS are newly proposed surface antigens that define ILCs producing type 2 cytokines49. In contrast, our TDCSS results revealed that CD30, TNFR2, and ICOS were stably expressed on ILC2 after activation, while CRTH2 (PTGDR2) and IL7R were downregulated on ILC2 after activation (Supplementary Fig. 9). These data suggest that the newly proposed surface antigen markers identified by Liu et al. are markers of activated ILC2. This activation-dependent change in surface antigens is likely to be observed not only in ILC2, but also in various immune cells, and thus the TDCSS technique will greatly contribute to the identification of surface antigen markers that reflect not only the cell type, but also the stage of activation.
While we have succeeded in exploring the activation process in ILC2, the TDCSS technique has yet to reveal the full picture of the fate-determination mechanism. Towards this future goal, TDCSS still faces a number of technical challenges. Firstly, TDCSS must increase its current throughput, which is currently hindered by the time-consuming manual recovery steps and scanning of cellular images. Secondly, the information obtained by TDCSS is dependent on the choice of index used to define the transition state. In this study, different TIGs were extracted when IL-13 and IL-5 were used as indexes (Supplementary Fig. 4), indicating that different indexes capture different aspects of the transition state. Therefore, it will be necessary to use multimodal information based on LCI, such as cell morphology, motility, and secretion, to achieve a comprehensive understanding of the fate-determination process.
While techniques exist to record the transcriptional histories of cells50–52, it is difficult to analyse rare clinical specimens as TDCSS has achieved, due to the need for genetic modification or aggregation of information from a large number of cells. While we have focused on the activation process of ILC2 in this study, many LCI techniques have been developed to trace various fate determination processes, including immune response15, cell differentiation53,54, cell death2, and stimulus response9,55. By utilizing these LCI techniques, the TDCSS technique can aid in the elucidation of the context of these fate determination processes. Additionally, it is known that in some immune responses, secretion is initiated immediately after the initiation of the transition state, via post-transcriptional or post-translational regulation56–59. Using an index that does not include delays due to transcription, allows for exploration of earlier stages of the fate-determining process.
Methods
Reagents
For cytokine secretion imaging using the LCI-S, we used anti-human IL-13 (MAB213 and BAF 213) and IL-4 (MAB604 and BAF204), anti-mouse IL-13 (MAB413 and BAF413) and IL-5 (MAB405 and BAM705) antibodies (R&D Systems, Minneapolis, MN, USA). For the measurement of IL-13 in culture supernatant, we used enzyme-linked immuno-sorbent assay (ELISA) kit (Human IL-13 Quantikine ELISA Kit, R&D Systems). For cell sorting of hILC2s, we used lineage antibody cocktails of human CD3 (UCHT1), CD14 (HCD14), CD16 (3G8), CD19 (HIB19), CD20 (2H7), CD56 (HCD56), PE-conjugated anti-human CD127 (A019D5), PE/Cy7-conjugated anti-human CD161 (HP-3G10) antibodies (BioLegend, San Diego, CA, USA), and Alexa Fluor 647-conjugated anti-human CRTH2 (BM16) antibody (BD Biosciences, San Jose, CA, USA).
For hILC2 stimulation, we used recombinant human IL-2 (Imunace35, Shionogi & Co., Ltd, Japan), IL-33, and TSLP (3625-IL/CF and 1398-TS/CF, R&D Systems, Minneapolis, MN, USA). For mILC2 stimulation, we used recombinant mouse IL-2 and IL-33 (402-ML and 3626-IL, R&D Systems).
For the miR-155 functional assay for hILC2 activation, we used miR-155 inhibitor (MISSION synthetic miRNA inhibitor, Sigma-Aldrich) and control (MISSION synthetic miRNA inhibitor negative control #1).
Isolation of hILC2s
Peripheral blood was obtained from three volunteers at Keio University School of Medicine. The study protocol was approved by the Institutional Review Board (IRB) of the Keio University School of Medicine (IRB approval number: 20090009). All subjects provided their written informed consent. For isolation of ILC2s from human peripheral blood, mononuclear cells were obtained using LymphoprepTM (Axis-Shield, Dundee, UK), according to the manufacturer’s protocols. Lineage-negative (CD3, CD14, CD16, CD19, CD20 and CD56) and CD45-, CD127-, CRTH2- and CD161-positive cells were sorted using a MoFloTM XDP flow cytometer (Beckman Coulter, Brea, CA, USA).
Preparation of mILC2
We isolated mILC2s from mouse fat tissues using a previously described protocol60. Briefly, we obtained pooled mesentery from 20 wildtype C57BL/6 mice. Lineage-negative cells were selected by negative selection using Microbeads and AutoMACS (Miltenyi Biotec GmbH). Then, c-Kit+Sca-1+ cells were sorted as ILC2s using a FACSAria flow cytometer (BD Bioscience). The acquired mILC2s were cultured for several months in RPMI-1640 media (R8758, Sigma-Aldrich, St. Louis, MO, USA) containing 10% foetal calf serum (FCS) (S1820, Japan Bioserum, Hiroshima, Japan), 10 mM HEPES buffer (H3537, Sigma-Aldrich), 1 mM sodium pyruvate (11360-070, Thermo Fisher Scientific, Waltham, MA, USA), 1x non-essential amino acids (M7145, Sigma-Aldrich), 100 U/mL penicillin and 100 µg/mL streptomycin (15140-122, Thermo Fisher Scientific), 14.3 mM 2-mercaptoethanol (21985023, Thermo Fisher Scientific), and 10 ng/mL recombinant mouse IL-2 (1399-IL, R&D Systems) in 96-well round-bottom plates. ILC2 cells were usually isolated from retired female breeder mice unless stated otherwise. All experiments were approved by the Animal Care and Use Committee of RIKEN or Keio University and performed according to institutional guidelines.
LCI of secretion activity of ILC2s
A fully automated inverted microscope (ECLIPSE Ti-E; Nikon, Tokyo, Japan) was used for time-lapse imaging using total internal reflection fluorescence (TIRF) illumination of LED light (X-Cite XLED1, mounted with RLX: 615 -655 nm, Excelitas Technologies Corp., Waltham, MA) through white-light TIRF optics (high-performance Epi-fl illuminator module TI-SFL). The optical configurations used were the following: excitation filter = FF01-635/18; emission filter = FF01-692/40; and dichroic mirror = FF560/659-Di01. These optical filters were purchased from Semrock (Rochester, NY, USA). We introduced pre-stimulation ILC2s into a microfabricated well array dish, which composed with a polydimethylsiloxane (PDMS), amorphous fluorocarbon polymer CYTOPTM, and glass with the immobilised capture antibody. We selected wells that contained single cells and immediately started scanning after replacing the medium with detection medium containing CF660R-labelled detection antibody and cytokine stimuli.
In the hILC2s experiment, we used a detection medium containing 10 U/mL IL-2, 25 ng/mL IL-33, and 25 ng/mL TSLP. For mILC2s, we used detection medium containing 10 ng/mL IL-2 and 10 ng/mL IL-33. Scan intervals were 15 min for hILC2s experiments and 4 min for mILC2s experiments.
When the secretion dynamics of the two types of cytokines were measured separately, a mixture of IL-13 and IL-5 capture antibodies was fixed on the bottom of the chip, and IL-13 and IL-5 detection antibodies are labelled with fluorescent substances of different wavelengths (CF660R or Cy3). Then, we perform live-cell imaging by switching between two sets of filters and lighthouse of different wavelengths to obtain separate signal increases at each wavelength.
Functional assay using miR-155 inhibitor
We obtained hILC2 from peripheral blood, proliferated by culturing in the presence of IL-2 and IL-33, followed by 5 days of culture with IL-2 alone. Electroporation was performed using Neon Electroporation System (Thermo Fisher Scientific) at 1600 V, 10 ms, 3 pulses in T buffer. miR-155 inhibitor or control was introduced at a final concentration of 0.1 uM. After electroporation, the cells were incubated overnight and introduced into the LCI-S platform to measure the secretion of IL-13 under the same conditions as in other experiments. In the bulk experiment, 9000 cells were incubated with IL-2 (10 U/mL), IL-33 (25 ng/mL), and TSLP (25 ng/mL) for 2 days, following which the IL-13 concentration in the supernatant was measured by ELISA.
Detection of the secretion onset
For detection of increased secretion signals from each cell, acquired images were sequentially analysed using Python 3 software developed in-house (https://github.com/TanaYumi/TDCSS). The program calculated the mean intensity of each well and compared the intensity to the first image intensity. The resulting value was used as the secretion signal. When the last three secretion signals were higher than the value estimated using linear regression of the past signal by one standard deviation, the program notified the operator of potential secretion onset.
After the experiments, we reanalysed the secretion signals to determine the accurate time between secretion onset and cell recovery (⊿t). The secretion onset time was determined by fitting the values to the following formula (1):
| 1 |
In the formula (1), represents drift, represents the slope of the secretion signal increase, represents the time after stimulation and represents the time of secretion onset. The initial value of was set to the visually determined onset on the automated image-analysis system. ⊿t was obtained by calculating the interval between secretion onset and cell recovery.
Cell recovery
Single cells or single colonies were recovered using a glass capillary (L-Tip 15 µm 60° 15 mm, Yodaka Co., Ltd., Kawasaki, Japan) and a pneumatic microinjector (IM-11-2, NARISHIGE, Tokyo, Japan). The glass capillary was positioned using a micromanipulator (TransferMan 4r, Eppendorf, Hamburg, Germany, or Quick Pro, Micro Support Co. Ltd., Shizuoka, Japan). The aspirated cell was ejected to 2.5 μL RNase-free water (06442-95, Nacalai tesque, Kyoto, Japan) for RNA-seq or 2.5 μL of the qRT-PCR reaction mixture (CellsDirect One-Step qRT-PCR Kits, Thermo Fisher Scientific) for qRT-PCR in a PCR tube.
The recovered cells were immediately frozen in liquid nitrogen and stored at -80°C for subsequent gene expression analysis. The pre-stimulation cells were recovered prior to the addition of the stimulus from the LCI-S platform. In the experiments involving mILC2s, activated and silent cells were randomly recovered from cells displaying positive and negative secretion signals 8 h after the initiation of stimulation. In the experiments involving hILC2s, activated and silent cells were randomly recovered 36 or 60 h (from 2 specimens) and 120 h (from 1 specimen) after the initiation of stimulation.
qRT-PCR
To increase the accuracy of single-cell mRNA quantification, we added the same amount of ERCC Spike-In RNA (622,819 copies per cell) to each sample and used it as a reference. cDNA was synthesised using CellsDirect One-Step qRT-PCR Kits (Thermo Fisher Scientific) using TaqMan probes targeting four genes (Il13, Il5, Gapdh and Rplp0) and four spike-in RNA sequences (ID130, ID136, ID131 and ID85). The TaqMan probes used were: Il13 (Mm.PT.58.31366752), Il5 (Mm.PT.58.41498972), Gapdh (Mm_GAPDH), Rplp0 (Mm.PT.58.43894205) purchased from IDT Integrated DNA Technologies (Coralville, IA, USA), ID130 (Ac03459943_a1), ID136 (Ac03459946_a1), ID131 (Ac03459944_a1) and ID85(Ac03459923_a1) purchased from Thermo Fisher Scientific. The PCR product was diluted ten-fold in RNase-free water, and 1 μL of the PCR product was analysed using qPCR using TaqMan Fast Universal PCR Master Mix (Thermo Fisher Scientific). Relative gene expression was calculated using the ΔΔCt method using ID136 or Gapdh as a reference. Samples, where Gapdh or Rplp0 could not be detected, were omitted from the analysis.
RNA sequencing
We synthesised cDNA libraries using SMART-Seq v4 3’-DE Kits (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s instructions, with some modifications, adding the synthetic oligo RNA/DNA to suppress undesirable concatemers. A total of 3,892 copies of ERCC spike-in RNA were added to each sample. The cDNA was purified using Agencourt AMPure XP magnetic beads (Beckman Coulter). Library quality check was performed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and Agilent High Sensitivity DNA Kits (5067-4626). Degraded or low-yield samples were removed. Qubit High Sensitivity assays (Thermo Fisher Scientific) were performed to quantify the cDNA in each library, and 5 or 6 libraries with different indexed primers were evenly pooled. Pooled cDNA (400 pg) was tagmented using Nextera XT DNA Library Prep Kits (Illumina, San Diego, CA, USA), as described in the protocol. Library size and cDNA amount were quantified using an Agilent High Sensitivity DNA Kit and Qubit High Sensitivity assays, respectively. Pooled libraries were sequenced using 91-bp paired-end sequencing on a MiSeq instrument (Illumina). After library demultiplexing and adaptor trimming, we aligned read 1 to reference sequences (human: GRCh38.90, mouse: GRCm38) using TopHat61, and the read counts were calculated using HTSeq62.
In mILC2 experiment, total 103 transition cells were collected using TDCSS. We reanalysed the secretory signals of these cells, determined the exact ⊿t, and classified them into cells with ⊿t = 0.5 h, 1.0 h, and 1.5 h. The collected cells were sequentially prepared into cDNA libraries, and library preparation was repeated until 11 libraries of each Δt were obtained. As a result, we obtained 33 cDNA libraries (11 libraries for each ⊿t = 0.5 h, 1.0 h, and 1.5 h), all passed QC at the post sequencing analysis. In hILC2 experiment, 13, 9, 13 transition cells were collected using TDCSS from each donor and library preparation was conducted for all cells, resulting in 5, 5, 8 transition cells passed QC at the post sequencing analysis.
Principal component analysis
PCA was performed using the Python 3 module “Scikit-learn63”. All cells’ expression data were used for the analyses. Count data of all genes were normalized by count per million (CPM). We used all genes for the PCA, except for genes with log2(CPM + 1) > 1 under 9 cells.
Pseudo-time analyses
The pseudo-time analysis was performed using Monocle64(2.20.0). All cells’ expression data were used for the analyses. Genes with an average expression level of less than 1 across cells were removed from further analysis. After that, highly variable genes were obtained by PCA and used for the trajectory inference. Used software versions and parameters of each processes can be found in the scripts at (https://github.com/TanaYumi/TDCSS).
Enrichment analysis
We used Metascape65 for enrichment analysis using default settings.
Statistics and reproducibility
Variety of the expression levels acquired by qRT-PCR experiment was tested by F-test.
To identify the differentially expressed genes among activation cell states (pre stimulation, transition state, activated and Silent for mouse ILC2s), one-way ANOVA was performed. In the analysis of mILC2, the earliest transition state (Δt = 0.5 h) was used for comparison. In the analysis of hILC2, we used expression data from three donors. The expression average among the same states in individuals were used for the clustering to avoid the bias caused by the difference in the number of cells among the states. Genes with P < 0.05 (mILC2) or < 0.01 (hILC2) were classified as highly variable. We used these highly variable genes for subsequent analyses.
Differential expression analysis of activated > pre-stimulation, silent > pre-stimulation and activated > silent states were performed using the Mann-Whitney rank test by SciPy 1.066 on genes that were expressed in more than four cells and had a fold-change 4 or above. Genes with an adjusted p value < 0.05 (adjusted using the Benjamini-Hochberg method with FDR < 0.1) were selected as differentially expressed genes.
K-Means clustering
K-Means clustering was performed with the Python 3 module “Scikit-learn63”. First, we classified the genes into 14 classes for mILC2 and 8 classes for hILC2 by the K-Means method. We grouped clusters with similar variation patterns into the same class, resulting in six or five classes, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank Mr. Hirotaka Matsumoto, Ms. Kaede Miyata, and Mr. Kazuki Yoda for their scientific advice and Yodaka Co. Ltd (Kawasaki, Japan) for supporting the cell recovery system, and for Mr. Mitsutaka Shirasaki who corrected the English text. This study was supported by JST, PRESTO (grant no. JPMJPR17H6), AMED (grant no. JP16ek0410031), Cabinet Office, Government of Japan (the ImPACT Program of the Council for Science, Technology, and Innovation) and JSPS (grant no. 18J20143 to Y.T. and 20H04512 to Y.S). This study was partially supported by JST, CREST (grant no. JPMJCR1412 to T.T and JPMJCR18H1 to YS), AMED (grant no. JP20hm0102082 and JP23ek0410110), JSPS (Core-to-Core Program), and White Rock Foundation. This manuscript was edited by Life Science Editors.
Author contributions
Y.T., Y.S. and M.Y. conceived the study and designed the experiments. Y.T., Y.S., M.Y., Y.O. and N.S. established the measurement equipment and environment and the analysis environment. Y.T., M.I. and Y.S. performed and analysed the experiments. Y.M., Ts.K. and K.M. provided the murine sample. Ta.K., M.I. and H.K. prepared the human sample. Y.S., T.T., K.G., O.O., T.F., K.F., K.M. and S.U. supervised the experiments. Y.T., Y.S., K.G., O.O., T.F., K.F., K.M. and S.U. contributed to acquisition of the financial support for the project leading to this publication. Y.T., Y.S., M.Y., Y.M.,Ts.K, Ta.K, M.I,, H.K., K.F., K.M. and S.U. interpreted the biological significance of the results. Y.T., Y.S., Ta.K., Y.O., T.T. and F.M. examined the statistical adequacy of the analysis. Y.T., Y.S. and M.Y. interpreted the data and wrote the paper.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Anam Akhtar and Christina Karlsson Rosenthal.
Data availability
RNA-sequencing data will be available through the GEO database; mILC2 expression data (GSE222762), hILC2 expression data (GSE213810, GSE222761). Source data for main figures are provided with the supplementary data 7. Other datasets generated during and/or analysed during the current study are available from the corresponding authors on reasonable request.
Code availability
Code is available at: https://github.com/TanaYumi/TDCSS. Environments for gene expression analysis are available at: https://hub.docker.com/r/yumikotanaka/tdcss.
Competing interests
The University of Tokyo has filed a patent application covering the methodology of cell recovery system described in this paper. M.Y. has a financial interest in Live Cell Diagnosis, Ltd., the company that was launched for the development of LCI-S. K.G. is a shareholder of CYBO and Cupido. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Koichi Fukunaga, Kazuyo Moro, Sotaro Uemura, Yoshitaka Shirasaki.
Contributor Information
Sotaro Uemura, Email: uemura@bs.s.u-tokyo.ac.jp.
Yoshitaka Shirasaki, Email: shirasaki@g.ecc.u-tokyo.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05297-w.
References
- 1.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu T, et al. Single-cell imaging of Caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Rep. 2013;8:974–982. doi: 10.1016/j.celrep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Clerk A, et al. Signaling pathways mediating cardiac myocyte gene expression in physiological and stress responses. J. Cell. Physiol. 2007;212:311–322. doi: 10.1002/jcp.21094. [DOI] [PubMed] [Google Scholar]
- 4.Best JA, et al. Transcriptional insights into the CD8 + T cell response to infection and memory T cell formation. Nat. Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiller DG, Wood CD, Rand DA, White MRH. Measurement of single-cell dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 6.Jeknić S, Kudo T, Covert MW. Techniques for studying decoding of single cell dynamics. Front. Immunol. 2019;10:755. doi: 10.3389/fimmu.2019.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 8.Polonsky M, Chain B, Friedman N. Clonal expansion under the microscope: studying lymphocyte activation and differentiation using live‐cell imaging. Immunol. Cell Biol. 2016;94:242–249. doi: 10.1038/icb.2015.104. [DOI] [PubMed] [Google Scholar]
- 9.Park MC, Hur JY, Cho HS, Park SH, Suh KY. High-throughput single-cell quantification using simple microwell-based cell docking and programmable time-course live-cell imaging. Lab Chip. 2011;11:79–86. doi: 10.1039/c0lc00114g. [DOI] [PubMed] [Google Scholar]
- 10.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirasaki Y, et al. Real-time single-cell imaging of protein secretion. Sci. Rep. 2014;4:4736. doi: 10.1038/srep04736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitrophanov AY, Groisman EA. Positive feedback in cellular control systems. BioEssays. 2008;30:542–555. doi: 10.1002/bies.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno S, et al. Kinetic trans-omic analysis reveals key regulatory mechanisms for insulin-regulated glucose metabolism in adipocytes. iScience. 2020;23:101479. doi: 10.1016/j.isci.2020.101479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chyan W, Raines RT. Enzyme-activated fluorogenic probes for live-cell and in vivo imaging. ACS Chem. Biol. 2018;13:1810–1823. doi: 10.1021/acschembio.8b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moro K, et al. Innate production of TH 2 cytokines by adipose tissue-associated c-Kit+ Sca-1+ lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 18.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camelo A, et al. IL-33, IL-25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv. 2017;1:577–589. doi: 10.1182/bloodadvances.2016002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J. Allergy Clin. Immunol. 2014;134:671. doi: 10.1016/j.jaci.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mjösberg JM, et al. Human IL-25-and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, et al. The transcription factor RelB restrains group 2 innate lymphoid cells and type 2 immune pathology in vivo. Cell. Mol. Immunol. 2020;18:230–242. doi: 10.1038/s41423-020-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nat. 2017;549:351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz C, et al. ILC2s regulate adaptive Th2 cell functions via PD-L1 checkpoint control. J. Exp. Med. 2017;214:2507–2521. doi: 10.1084/jem.20170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson K, Malmhäll C, Ramos-Ramírez P, Rådinger M. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. J. Allergy Clin. Immunol. 2017;139:1007–1016.e9. doi: 10.1016/j.jaci.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 27.Knolle MD, et al. MicroRNA-155 protects group 2 innate lymphoid cells from apoptosis to promote type-2 immunity. Front. Immunol. 2018;9:2232. doi: 10.3389/fimmu.2018.02232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushita K, et al. Regnase-1 degradation is crucial for IL-33– and IL-25–mediated ILC2 activation. JCI Insight. 2020;5:e131480. doi: 10.1172/jci.insight.131480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricardo-Gonzalez RR, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 2018;19:1093–1099. doi: 10.1038/s41590-018-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner JE, et al. IL-9–mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J. Exp. Med. 2013;210:2951–2965. doi: 10.1084/jem.20130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C, et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell. 2018;174:271–284.e14. doi: 10.1016/j.cell.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim AI, et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 2016;213:569–583. doi: 10.1084/jem.20151750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba R, et al. Upregulation of IL-4 receptor signaling pathway in circulating ILC2s from asthma patients. J. Allergy Clin. Immunol. Glob. 2022;1:299–304. doi: 10.1016/j.jacig.2022.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fallon PG, et al. IL-4 induces characteristic Th2 responses even in the combined absence of IL-5, IL-9, and IL-13. Immunity. 2002;17:7–17. doi: 10.1016/s1074-7613(02)00332-1. [DOI] [PubMed] [Google Scholar]
- 35.Bal SM, et al. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 2016;17:636–645. doi: 10.1038/ni.3444. [DOI] [PubMed] [Google Scholar]
- 36.Furusawa J, et al. Critical role of p38 and GATA3 in natural helper cell function. J. Immunol. 2013;191:1818–1826. doi: 10.4049/jimmunol.1300379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrova T, Pesic J, Pardali K, Gaestel M, Arthur JSC. p38 MAPK signalling regulates cytokine production in IL-33 stimulated Type 2 Innate Lymphoid cells. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-60089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. Usa. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto T, et al. DUSP10 constrains innate IL-33-mediated cytokine production in ST2hi memory-type pathogenic Th2 cells. Nat. Commun. 2018;9:2–11. doi: 10.1038/s41467-018-06468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlad G, Suciu-Foca N. Induction of antigen-specific human T suppressor cells by membrane and soluble ILT3. Exp. Mol. Pathol. 2012;93:294–301. doi: 10.1016/j.yexmp.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ. JAB/SOCS1/SSI-1 is an interleukin-2-induced inhibitor of IL-2 signaling. Blood. 2001;97:221–226. doi: 10.1182/blood.v97.1.221. [DOI] [PubMed] [Google Scholar]
- 42.Ye J, et al. MiR-155 Regulated Inflammation Response by the SOCS1-STAT3-PDCD4 Axis in Atherogenesis. Mediat. Inflamm. 2016;2016:8060182. doi: 10.1155/2016/8060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CY, et al. DUSP4 deficiency enhances CD25 expression and CD4 + T-cell proliferation without impeding T-cell development. Eur. J. Immunol. 2012;42:476–488. doi: 10.1002/eji.201041295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandhu SK, et al. MiR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eμ-miR-155 transgenic mouse model. Proc. Natl Acad. Sci. USA. 2012;109:20047–20052. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roediger B, et al. IL-2 is a critical regulator of group 2 innate lymphoid cell function during pulmonary inflammation. J. Allergy Clin. Immunol. 2015;136:1653–1663.e7. doi: 10.1016/j.jaci.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 46.Mirchandani AS, et al. Type 2 innate lymphoid cells drive CD4 + Th2 cell responses. J. Immunol. 2014;192:2442–2448. doi: 10.4049/jimmunol.1300974. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee A, Schambach F, Dejong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-γ signaling in CD4+ T cells. Eur. J. Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moro K, et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, et al. Optimal identification of human conventional and nonconventional (CRTH2–IL7Rα–) ILC2s using additional surface markers. J. Allergy Clin. Immunol. 2020;146:390–405. doi: 10.1016/j.jaci.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanna T, Schmidt F, Cherepkova MY, Okoniewski M, Platt RJ. Recording transcriptional histories using Record-seq. Nat. Protoc. 2020;15:513–539. doi: 10.1038/s41596-019-0253-4. [DOI] [PubMed] [Google Scholar]
- 51.Tang W, Liu DR. Rewritable multi-event analog recording in bacterial and mammalian cells. Science (80-.) 2018;360:eaap899. doi: 10.1126/science.aap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lane K, et al. Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst. 2017;4:458–469.e5. doi: 10.1016/j.cels.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W, et al. Live-cell imaging and analysis reveal cell phenotypic transition dynamics inherently missing in snapshot data. Sci. Adv. 2020;6:eaba9319. doi: 10.1126/sciadv.aba9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishimura K, et al. Live-cell imaging of subcellular structures for quantitative evaluation of pluripotent stem cells. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-018-37779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davey, A. M., Liu, W., Sohn, H. W., Brzostowski, J. & Pierce, S. K. Understanding the initiation of B cell signaling through live cell imaging. in Methods in Enzymology506, 265–290 (Academic Press Inc., 2012). [DOI] [PMC free article] [PubMed]
- 56.Jurgens AP, Popović B, Wolkers MC. T cells at work: How post‐transcriptional mechanisms control T cell homeostasis and activation. Eur. J. Immunol. 2021;51:2178. doi: 10.1002/eji.202049055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kouzaki H, Tojima I, Kita H, Shimizu T. Transcription of Interleukin-25 and extracellular release of the protein is regulated by allergen proteases in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2013;49:741–750. doi: 10.1165/rcmb.2012-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Travers J, et al. Chromatin regulates IL-33 release and extracellular cytokine activity. Nat. Commun. 2018;9:1–15. doi: 10.1038/s41467-018-05485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franciszkiewicz K, et al. Synaptic release of CCL5 storage vesicles triggers CXCR4 surface expression promoting CTL migration in response to CXCL12. J. Immunol. 2014;193:4952–4961. doi: 10.4049/jimmunol.1401184. [DOI] [PubMed] [Google Scholar]
- 60.Moro K, Ealey KN, Kabata H, Koyasu S. Isolation and analysis of group 2 innate lymphoid cells in mice. Nat. Protoc. 2015;10:792–806. doi: 10.1038/nprot.2015.047. [DOI] [PubMed] [Google Scholar]
- 61.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedregosa F. et al. Scikit-learn: Machine Learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res.12, 2825–2830 (2011).
- 64.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Virtanen P, et al. SciPy 1.0-fundamental algorithms for scientific computing in python. Nat. Methods. 2019;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
RNA-sequencing data will be available through the GEO database; mILC2 expression data (GSE222762), hILC2 expression data (GSE213810, GSE222761). Source data for main figures are provided with the supplementary data 7. Other datasets generated during and/or analysed during the current study are available from the corresponding authors on reasonable request.
Code is available at: https://github.com/TanaYumi/TDCSS. Environments for gene expression analysis are available at: https://hub.docker.com/r/yumikotanaka/tdcss.