Abstract
Results published in an article by Poore et al. (Nature. 2020;579:567–574) suggested that machine learning models can almost perfectly distinguish between tumour types based on their microbial composition using machine learning models. Whilst we believe that there is the potential for microbial composition to be used in this manner, we have concerns with the paper that make us question the certainty of the conclusions drawn. We believe there are issues in the areas of the contribution of contamination, handling of batch effects, false positive classifications and limitations in the machine learning approaches used. This makes it difficult to identify whether the authors have identified true biological signal and how robust these models would be in use as clinical biomarkers. We commend Poore et al. on their approach to open data and reproducibility that has enabled this analysis. We hope that this discourse assists the future development of machine learning models and hypothesis generation in microbiome research.
Keywords: cancer, bacteria, viruses, microbiome, machine learning, contamination
Most models do not perform any better than models constructed using no information
Poore et al. [1] detail the building of cancer type models based on microbial interrogation of TCGA (The Cancer Genome Atlas Program) cancer sequence data (which are predominantly RNA sequencing but with some whole genome sequences). Here, we evaluate these models within the framework of Whalen et al. [2] describing common modelling pitfalls, namely: (1) distributional differences, (2) confounding, (3) leaky preprocessing and (4) unbalanced classes.
Following their most stringent decontamination, only five of the 33 one-vs-all cancer type models examined were a statistically significantly improvement on models constructed using no information [at the 0.05 significance level, without false discovery correction for multiple models, ‘P-Value (Acc>NIR)’, available: http://cancermicrobiome.ucsd.edu/CancerMicrobiome_DataBrowser/] – this was not clear in the main text of their paper.
Models pronounce nonsensical genera are informative of tumour type
Even when the model does appear to identify samples better than the negative predictor, we have concerns that many of the key features used in the model are implausible. For example, the model predicting adrenocortical carcinoma is significantly better than a negative predictor (P=0.002) and boasts high sensitivity (0.9565), specificity (0.998) and positive predictive values (0.71). Therefore, this model should hold some features that truly distinguish it from the remaining cancer types. The top ten most important features for this model are Hepandensovirus (relative feature importance score: 9431, a virus that infects crustaceans [3]), Paeniclostridium (973), Comovirus (846), Thalassomonas (267, bacteria causing coral disease [4]), Simkania (160), Cronobacter (151), Simonsiella (148), Leucothrix (145, bacteria from marine macroalgae [5]), Phikmvlikevirus (128) and N4likevirus (88). It is unclear how Phikmvlikevirus and N4likevirus might be informative for adrenocortical carcinoma as they are bacteriophages and therefore would be dependent on the co-occurrence of their bacterial hosts in the adrenal glands (or alternatively the remaining anatomical locations [6, 7]). Many of the top performing features of other models under the most stringent decontamination approach also seem nonsensical (Table 1). This point is not covered by the Whalen pitfalls because it is generally presumed that the features being modelled exist to begin with, which in the case of taxonomic classification is not always true.
Table 1.
Top performing features for a selection of one-vs-all cancer type models in the most stringent decontamination approach as presented in Poore et al
These taxa include extremophiles that have not previously been isolated from humans. See Table S1, available in the online version of this article (bacteria), and Table S2 (viruses) for a full description as on NCBI of the sources for each representative species within these genera.
|
Genus |
Top feature in cancer type model |
Details |
|---|---|---|
|
Bladder cancer |
Bacteria from marine macroalgae [5] |
|
|
Uveal melanoma |
Bacteria causes disease in coral [4] |
|
|
Velarivirus |
Cervical cancer |
Grapevine is natural host [33] |
|
Tritimovirus |
Colon cancer |
Known to infect cereals [34] |
|
Dinovernavirus |
Renal clear cell carcinoma |
Contains insect viruses [35] |
|
Bacillarnavirus |
Lung squamous cell carcinoma |
Infects algae [36] |
|
Rymovirus |
Ovarian serous |
Infects species of grass [37] |
|
Prostate |
Identified in marine hydrothermal vents [19] |
|
|
Testicular cancer |
Halophilic genus identified from marine environments [38] |
Some models do demonstrate plausible and promising results. For example, in hepatocellular carcinoma, Orthohepadnavirus is known to have a causal relationship with cancer formation [8] and has been found to be specific to the liver in other datasets [9]. This is reflected well in Poore et al.’s model where the estimated variable importance score of Orthohepadnavirus in their model (2020.53) dwarfs the next most ‘important’ feature (Levivirus, 975.09). Despite this, the model is still not significantly better than a negative predictor [P-value (Acc>NIR)=1] and suffers a poor positive predictive value (0.4).
Potential for read misclassification
We believe that these nonsensical genera arise because the models produced by Poore et al. are built on many features that are likely to be taxonomically misclassified, from human reads or other contamination [10–14], and therefore do not originate from microbes in the sample. One possible reason for these misclassifications is that extra steps were not taken to remove human reads prior to model building. Poore et al. detail the extraction of reads unaligned to a human reference genome which are then the subject of taxonomic classification. The authors claim to have used ‘very stringent decontamination analyses that discarded up to 92.3 % of total sequence data’. This would suggest that 7.7 % of all sequencing reads were subject to taxonomic classification. This pool of reads will still contain human reads which have not aligned [15]. For example, this could be because the reads are of low quality or they are mutated in cancer genomes, or due to sequencing artefacts. In addition, the authors detail no human reference sequences in their taxonomic database, using 59 974 microbial genomes only. Therefore, it is highly likely that human sequences will have been misclassified as microbial. The subsequent application of SHOGUN alignment of Kraken-classified reads is more specific but may still involve the inappropriate classification of human reads to a database with no representation of the human genome. Additional human depletion filtering and steps to remove contamination such as those employed by the cancer microbiome atlas to distinguish tissue-resident microbiota from contaminants would have helped to remove misclassifications [16].
Normalization introduces variance and permits modelling
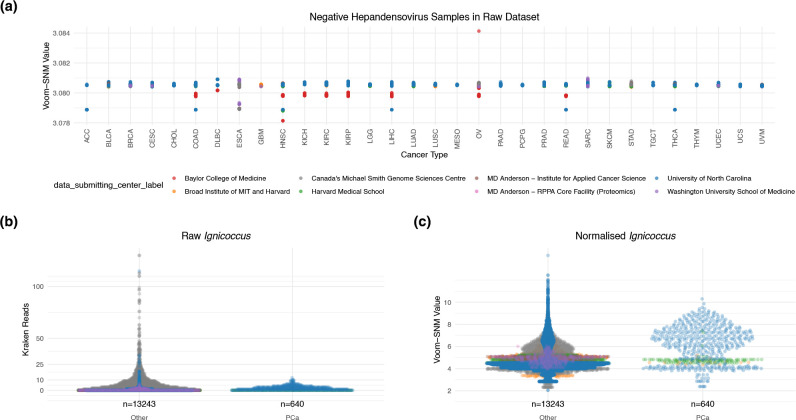
Another possible contributing factor to the issues with the models is in how the data were processed. Microbiome data are dynamic [17] (Whalen I: distributional differences), and are typically heteroskedastic (meaning that the variance of a variable is non-constant over values of an independent variable, i.e. the number of sequencing reads assigned to each of two genera) [18]. The authors resolve heteroskedasticity by applying a tool called Voom that is designed for RNA sequencing data of a single organism where the majority of genes have some level of expression. However, as applied by Poore et al. it suggests presence even when taxa are absent (Whalen III: leaky preprocessing). For example, for Hepandensovirus (genus of crustacean virus), the top feature for adrenocortical carcinoma, Voom transitions all zeros to non-zero values and untrue variation has been introduced by the global adjustment for technical variables including sequencing centre (Fig. 1a, batch correction relating to Whalen II: confounding). Therefore, this normalization appears beneficial on the global level but raises prominent concerns at the level of individual taxa.
Fig. 1.
(a) Voom-SNM normalized TCGA samples (n=17 624) that were negative for crustacean virus hepandensovirus with zero classified reads in the original Kraken dataset with the most stringent decontamination approach. One sample contained two sequencing reads for Hepandensovirus, which has been omitted from this figure to illustrate inappropriate variation introduced by SNM. The colour of each point indicates the centre where the sample was sequenced and from where the resulting data were submitted [University of North Carolina, Harvard Medical School, Canada’s Michael Smith Genome Sciences Centre, Broat Institute MIT and Harvard, Baylor College of Medicine, Washington University School of Medicine, MD Anderson – Institute for Applied Cancer Science, Johns Hopkins/University of Southern California, MD Anderson RPPA Core Facility (Proteomics)]. The x-axis demonstrates cancer types using TCGA abbreviations as in Poore et al. [1]. This is a prominent concern, especially given how closely linked sequencing centre and disease type are (Table S3). Raw (b) and Voom-SNM normalized (c) Ignicoccus values, which was deemed the most important feature for predicting prostate cancer (PCa) from all other cancer types (n=13 883 primary tumours). Median values are as follows: Kraken raw other 0, Kraken raw PCa 1, normalized other 4.49, normalized PCa 5.82. In both the raw and normalized cases, the distributions are significantly different (Wilcox signed rank-sum test P<2.2×10–16).
Another example of how the processing of data can be problematic is provided by the extremophile genus Ignicoccus in prostate cancer samples. Ignicoccus shows a statistically significant increase in prostate cancer samples compared to other cancers in the normalized dataset (Wilcox signed rank-sum test P<2.2×10–16, Fig. 1b, c). In the raw, unprocessed data no increase in prostate cancer samples is apparent. Indeed, most values are zero and the maximum number of reads found in the raw prostate cancer data for Ignicoccus is 12 (low evidence of detection). It is also highly likely that these are false taxonomic assignments given that Ignicoccus was identified in marine hydrothermal vents [19]. This taxon should have been filtered out prior to model building – the application of a minimum read threshold (i.e. 100 classified reads) would have assisted the removal of spurious taxa.
The models are trained on unbalanced data
The performance of the models may in part be due to the major imbalance in class size in the datasets (Whalen IV: unbalanced classes), meaning that before model construction, data in the cancer set under investigation are multiplied up many times (upsampling) so that patient numbers in the ‘cancer groups’ and in the ‘all other cancers group’ become similar. This approach may amplify the prominence of implausible artefactual data. Adrenocortical carcinoma for example has 79 associated samples (as per Metadata-TCGA-All-18116-Samples.csv provided by Poore et al.). This means that 18 037 are not adrenocortical carcinoma. Adrenocortical carcinoma therefore represents 0.44 % of the whole dataset and therefore data from adrenocortical carcinoma are amplified up to 230 times to equal the sample size of the rest of the dataset. The modelling is therefore overexposed to inappropriate variation in taxa such as Hepandensovirus (Fig. 1).
Discussion
The detection of microbial composition via machine learning is increasingly being used in disease-based research. Extreme caution must be taken to avoid coming to inaccurate conclusions. In this letter we have reviewed the paper of Poore et al. [1] and highlighted many problems. Ideally, the authors would have followed as closely as possible the RIDE criteria set out by Eisenhofer et al. (also authored by Knight) [20]. Where this is not possible, the conclusions drawn should be more cautious and the limitations made clear. Poore et al. use many good practices in machine learning [21] but there is the need to avoid the pitfalls of Whalen et al. and use more stringent methods regarding contamination, taxonomic misclassification and previous microbiological evidence.
The hypothesis that microbes (including those found in tumours) are dependent on anatomical location is well founded based on previous work [22], but the models produced by Poore et al. are at best suggestive and do not substantiate this observation. Additional care should be taken to include only taxa with strong evidence of presence based on computational evidence, consideration of the likelihood of contamination and prior biological evidence that the taxa are present in the biological sample of interest.
After we raised our concerns [23], Poore et al. published a response to our points [24]. Despite its considerable length, it focused on the technical details of statistical modelling and did not address the core concerns raised in this letter regarding contamination, nonsensical taxa appearing as important and the flawed batch correction.
Even in the technical areas where they did respond, they did not address these points. For example, Poore et al. defended the use of Voom prior to batch correction by claiming that it had been cited >4000 times, but Voom was not developed for metatranscriptomics. Microbial community matrices are typically much sparser than single-organism gene expression matrices. Voom transforms zero values into non-zero values, subsequently with additional false signal introduced (Fig. 1a), which makes the use case of Voom followed by SNM inappropriate. In their response, they suggest that a difference of 0.006 in the normalized values for Hepandensovirus in adrenocortical carcinoma (Fig. 1a) is not significant. This is not correct. The machine learning algorithms used in Poore et al. do not require large differences to build a rule and make classifications. This is reflected in the fact that this genus is by far the most important feature in their near perfect performing model for predicting adrenocortical carcinoma.
The overarching problem, however, is the prevalence of nonsensical taxa appearing as informative in Poore et al.’s models. This is a sure sign that something is going wrong. Poore et al. have given some attention to the issue of contamination but nonsensical taxa with limited evidence of true involvement are still prominent, suggesting this has not gone far enough. In their response, Poore et al. also state that they ‘extensively remove human reads from metagenomic data’, but sequencing reads that are not aligned to the human genome are not equivalent to non-human reads and there is no evidence that a human genome was present in their taxonomic database, which is best practice [25]. Poore et al. noted that the most stringent decontaminated dataset was only produced to address a reviewer’s concerns but that the structure of the data soon became unrecognizable. It is therefore alarming that performance metrics are still high and that nonsensical taxa are still reported as the best performing features in the models built on these ‘unrecognizable’ datasets with ‘stringent’ decontamination. Contamination is undoubtedly a major concern in microbiome research and has critically affected the results of a significant amount of research [10, 11, 13, 14, 20]. Examples include the claims of a brain or placental microbiome [11, 12].
It is our contention that there are critical flaws in the study by Poore et al. resulting in misclassifications and contamination being considered as important features to predict tumour type. Unless this issue is addressed, no matter how good the subsequent analysis, the results will still be questionable. Therefore, we believe that our central point of urging ‘caution’ to those interpreting the data and results of Poore et al. remains valid.
Finally, we would like to highlight the controversy surrounding the use of the term ‘cancer microbiome’ in this context. There are many definitions of ‘microbiome’ [26], but the commonly accepted use of the term could imply that microbes are ubiquitous in every single cancer sample, which they are not. There are many sites in the body with highly disproven ‘microbiomes’ such as the uterus and brain [11–14]. Given the methodological issues we raise, it is difficult to see whether any of the reported microbes are cancer type specific or whether they go beyond the known tissue-specific microbes (hepatitis etc.). Therefore, it should be considered whether these really constitute a ‘microbiome’ or whether they are related to infection.
Conclusion
We believe that the study of microbes in tumours is an exciting field, and that the use of large sequencing datasets with rich metadata can reveal much more about the nature of the interplay between microbes and cancer. Poore et al. have used machine learning models to describe the ‘tumour microbiome’ as being specific to tumour type, but we have serious concerns. Overwhelming contamination and inappropriate handling of the data do not support the claims in the original title: ‘Microbiome analyses of blood and tissues suggest cancer diagnostic approach’. A dataset with a less pronounced batch effect, more balanced class sizes and modelling all tumour types at once (not one-vs-all models) might help to better distinguish the pan-cancer microbial structure. There needs to be a better demonstration of microbial differences between tumour types and rigorous validation of models before we can be certain of these differences and illuminate any taxa underpinning these differences. We are a long way from proving the utility of cancer microbial structure in improving cancer patient care.
Methods
All analysis in this paper was conducted on the open-source data made available by Poore et al. [1] available at: http://ftp.microbio.me/pub/cancer_microbiome_analysis/. Files analysed include: Kraken-TCGA-Raw-Data-17625-Samples.csv (MD5 checksum: 6af81818f69bf56b79836e1c317c3e03), Metadata-TCGA-All-18116-Samples.csv (MD5 checksum: dbdd1f64d45973977fc8435db2eb8b3e), and Kraken-TCGA-Voom-SNM-Most-Stringent-Filtering-Data.csv (MD5 checksum: b7e50700b791b8881426aeb1fa12c3bb).
Model performance and feature importance was accessed: http://cancermicrobiome.ucsd.edu/CancerMicrobiome_DataBrowser/. All data were analysed in R (version 4.2.1). Packages used include tidyverse [27] (version 1.3.2), ggpubr [28] (version 0.5.0), ggbeeswarm [29] (version 0.7.1), cowplot [30] (version 1.1.1) and EnvStats [31] (version 2.7.0). Hypothesis testing was performed with the wilcox.test() function.
Representative species within top features (Table S1) were identified by browsing GTDB [32] (release version 207). Associated metadata regarding isolation sources were found by accessing links presented on the GTDB taxonomy browser.
Supplementary Data
Funding information
This work was funded by Big C Cancer Charity (ref 16-09R), Prostate Cancer UK (MA-ETNA19-003) and The Bob Champion Cancer Trust.
Conflicts of interest
Colin S. Cooper, Daniel S. Brewer and Abraham Gihawi are co-inventors on a patent application (UK Patent Application No. 2200682.9) from the University of East Anglia/UEA Enterprises Limited regarding the application of biomarker bacterial genera in prostate cancer.
Footnotes
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary tables are available with the online version of this article.
References
- 1.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whalen S, Schreiber J, Noble WS, Pollard KS. Navigating the pitfalls of applying machine learning in genomics. Nat Rev Genet. 2022;23:169–181. doi: 10.1038/s41576-021-00434-9. [DOI] [PubMed] [Google Scholar]
- 3.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, et al. The family Parvoviridae. Arch Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosoya S, Adachi K, Kasai H. Thalassomonas actiniarum sp. nov. and Thalassomonas haliotis sp. nov., isolated from marine animals. Int J Syst Evol Microbiol. 2009;59:686–690. doi: 10.1099/ijs.0.000539-0. [DOI] [PubMed] [Google Scholar]
- 5.Liu T, Zhang Y, Zhang X, Zhou L, Meng C, et al. Leucothrix sargassi sp. nov., isolated from a marine alga [Sargassum natans (L.) Gaillon] Int J Syst Evol Microbiol. 2019;69:3857–3862. doi: 10.1099/ijsem.0.003694. [DOI] [PubMed] [Google Scholar]
- 6.Wittmann J, Klumpp J, Moreno Switt AI, Yagubi A, Ackermann H-W, et al. Taxonomic reassessment of N4-like viruses using comparative genomics and proteomics suggests a new subfamily - “Enquartavirinae.”. Arch Virol. 2015;160:3053–3062. doi: 10.1007/s00705-015-2609-6. [DOI] [PubMed] [Google Scholar]
- 7.Merabishvili M, Vandenheuvel D, Kropinski AM, Mast J, De Vos D, et al. Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii . PLoS One. 2014;9:e104853. doi: 10.1371/journal.pone.0104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringelhan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160274. doi: 10.1098/rstb.2016.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;574:329–334. doi: 10.1038/s41586-019-1628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedarf JR, Beraza N, Khazneh H, Özkurt E, Baker D, et al. Much ado about nothing? Off-target amplification can lead to false-positive bacterial brain microbiome detection in healthy and Parkinson’s disease individuals. Microbiome. 2021;9:75. doi: 10.1186/s40168-021-01012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, et al. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 14.de Goffau MC, Charnock-Jones DS, Smith GCS, Parkhill J. Batch effects account for the main findings of an in utero human intestinal bacterial colonization study. Microbiome. 2021;9:6. doi: 10.1186/s40168-020-00949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gihawi A, Rallapalli G, Hurst R, Cooper CS, Leggett RM, et al. SEPATH: benchmarking the search for pathogens in human tissue whole genome sequence data leads to template pipelines. Genome Biol. 2019;20:208. doi: 10.1186/s13059-019-1819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe. 2021;29:281–298. doi: 10.1016/j.chom.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber GK. The dynamic microbiome. FEBS Lett. 2014;588:4131–4139. doi: 10.1016/j.febslet.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 18.McMurdie PJ. Normalization of microbiome profiling data. Methods Mol Biol. 2018;1849:143–168. doi: 10.1007/978-1-4939-8728-3_10. [DOI] [PubMed] [Google Scholar]
- 19.Paper W, Jahn U, Hohn MJ, Kronner M, Näther DJ, et al. Ignicoccus hospitalis sp. nov., the host of “Nanoarchaeum equitans.”. Int J Syst Evol Microbiol. 2007;57:803–808. doi: 10.1099/ijs.0.64721-0. [DOI] [PubMed] [Google Scholar]
- 20.Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R, et al. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 22.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gihawi A, Cooper CS, Brewer DS. Caution regarding the specificities of pan-cancer microbial structure. Bioinformatics. 2023 doi: 10.1101/2023.01.16.523562. [DOI] [PMC free article] [PubMed]
- 24.Sepich-Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, et al. Reply to: caution regarding the specificities of pan-cancer microbial structure. Bioinformatics. 2023 doi: 10.1101/2023.02.10.528049. [DOI]
- 25.Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019;20:1125–1136. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:119. doi: 10.1186/s40168-020-00905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham H, Averick M, Bryan J, Chang W, McGowan L, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 28.Kassambara A. ggpubr: “ggplot2” Based Publication Ready Plots. 2022.
- 29.Clarke E-M, ggbeeswarm C. Categorical Scatter (Violin Point) Plots. 2022.
- 30.Wilke C. cowplot: Streamlined Plot Theme and Plot Annotations for 'ggplot2. 2020.
- 31.Millard SP. EnvStats: An R Package for Environmental. New York, NY: Springer; 2013. [Google Scholar]
- 32.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil PA, et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Qi S, Chang Z, Rong Q, Akinyemi IA, et al. Complete genome sequence of a novel velarivirus infecting areca palm in China. Arch Virol. 2015;160:2367–2370. doi: 10.1007/s00705-015-2489-9. [DOI] [PubMed] [Google Scholar]
- 34.Rabenstein F, Seifers DL, Schubert J, French R, Stenger DC. Phylogenetic relationships, strain diversity and biogeography of tritimoviruses. J Gen Virol. 2002;83:895–906. doi: 10.1099/0022-1317-83-4-895. [DOI] [PubMed] [Google Scholar]
- 35.Roundy CM, Azar SR, Rossi SL, Weaver SC, Vasilakis N. Insect-specific viruses: a historical overview and recent developments. Adv Virus Res. 2017;98:119–146. doi: 10.1016/bs.aivir.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Short SM, Staniewski MA, Chaban YV, Long AM, Wang D. Diversity of viruses infecting eukaryotic algae. Curr Issues Mol Biol. 2020;39:29–62. doi: 10.21775/cimb.039.029. [DOI] [PubMed] [Google Scholar]
- 37.Webster DE, Beck DL, Rabenstein F, Forster RLS, Guy PL. An improved polyclonal antiserum for detecting Ryegrass mosaic rymovirus. Arch Virol. 2005;150:1921–1926. doi: 10.1007/s00705-005-0531-z. [DOI] [PubMed] [Google Scholar]
- 38.Nedashkovskaya OI, Vancanneyt M, Kim SB, Han J, Zhukova NV, et al. Salinimicrobium marinum sp. nov., a halophilic bacterium of the family Flavobacteriaceae, and emended descriptions of the genus Salinimicrobium and Salinimicrobium catena . Int J Syst Evol Microbiol. 2010;60:2303–2306. doi: 10.1099/ijs.0.019166-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.