Abstract
Background and hypothesis
Psychiatric disorders impose a huge health and economic burden on modern society. However, there is currently no proven completely effective treatment available, partly owing to the inefficiency of drug target identification and validation. We aim to identify therapeutic targets relevant to psychiatric disorders by conducting Mendelian randomization (MR) analysis.
Study design
We performed genome-wide MR analysis by integrating expression quantitative trait loci (eQTL) of 4479 actionable genes that encode druggable proteins and genetic summary statistics from genome-wide association studies of psychiatric disorders. After conducting colocalization analysis on the brain MR findings, we employed protein quantitative trait loci (pQTL) data as genetic proposed instruments for intersecting the colocalized genes to provide further genetic evidence.
Study results
By performing MR and colocalization analysis with eQTL genetic instruments, we obtained 31 promising drug targets for psychiatric disorders, including 21 significant genes for schizophrenia, 7 for bipolar disorder, 2 for depression, 1 for attention deficit and hyperactivity (ADHD) and none for autism spectrum disorder. Combining MR results using pQTL genetic instruments, we finally proposed 8 drug-targeting genes supported by the strongest MR evidence, including gene ACE, BTN3A3, HAPLN4, MAPK3 and NEK4 for schizophrenia, gene NEK4 and HAPLN4 for bipolar disorder, and gene TIE1 for ADHD.
Conclusions
Our findings with genetic support were more likely to be to succeed in clinical trials. In addition, our study prioritizes approved drug targets for the development of new therapies and provides critical drug reuse opportunities for psychiatric disorders.
Keywords: drug targets, Mendelian randomization, psychiatric disorders, GWAS, eQTL, pQTL
Introduction
Psychiatric disorders are one of the leading causes of disability worldwide and impose an enormous health and economic burden on human society.1,2 Emerging evidence suggests that the current coronavirus disease 2019 (COVID-19) pandemic is worsening the prevalence of psychiatric disorders globally.3,4 To date, no effective treatments able to prevent the recurrence of psychiatric disorder symptoms are available, although available therapies offer symptomatic relief. In spite of considerable efforts in drug discovery and development, over 90% of proposed therapeutics fail in the course of clinical trials due to poor therapeutic efficacy and unacceptable safety.5,6 This contributes to the costs for the development of a novel drug and bringing it to the market can be ranging from $314 million to $2.8 billion.7 There is a compelling need for the discovery of novel drugs for the management of psychiatric disorders, which is time-consuming and expensive.
Human genetic studies are now widely adopted in drug development for many complex diseases. Drug target pairs supported by human genetic evidence are more likely to be clinically successful.8 For example, genetic loci identified in genome-wide association studies (GWASs) of type 2 diabetes, which contain genes encoding targets for the sulphonylurea and glitazone drug classes, have been used to treat diabetes.9,10 In the past decade, GWASs have identified multiple genetic loci harboring associated single nucleotide polymorphisms (SNPs) for psychiatric disorders. Nevertheless, the genetic associations from GWAS cannot reliably pinpoint causal genes and directly inform future drug design and development efforts for disease.
Well-designed randomized clinical trials (RCTs) are generally accepted as the gold standard approaches for assessing whether drug treatment is efficacious (figure 1).11,12 However, RCTs are expensive to undertake and not always feasible.13 Mendelian randomization (MR) is now widely used to infer causal relationships between exposures and outcomes by using genetic factors as instrumental variables,14–18 providing opportunities for informing therapeutic targeting. Specifically, genetic variants acting in “cis” and “trans” on the druggable gene (ie, gene encoding the approved and clinical-phase drugs target proteins or drug-like small-molecule binding partners as annotated by Finan et al.10) expression, also referred to as expression quantitative trait loci (eQTL), can be used as MR instruments for drug exposure. The association between the same genetic variants and complex disease (ie, the outcome) can then be derived from GWAS for the outcome. Thus, integration of GWAS (ie, SNP-disease associations) and eQTL data (ie, SNP-gene-expression) using MR will help to infer the causal effect of the exposure on the outcome (ie, identifying the potentially druggable genes), as they mimic the on-target beneficial or harmful effects observed by pharmacological modification. Recent studies have adopted such an approach through a combination of 2 sample MR analysis19 to investigate the association between druggable gene expression and disease outcomes including Parkinson’s disease20 and COVID-19.21
Fig. 1.
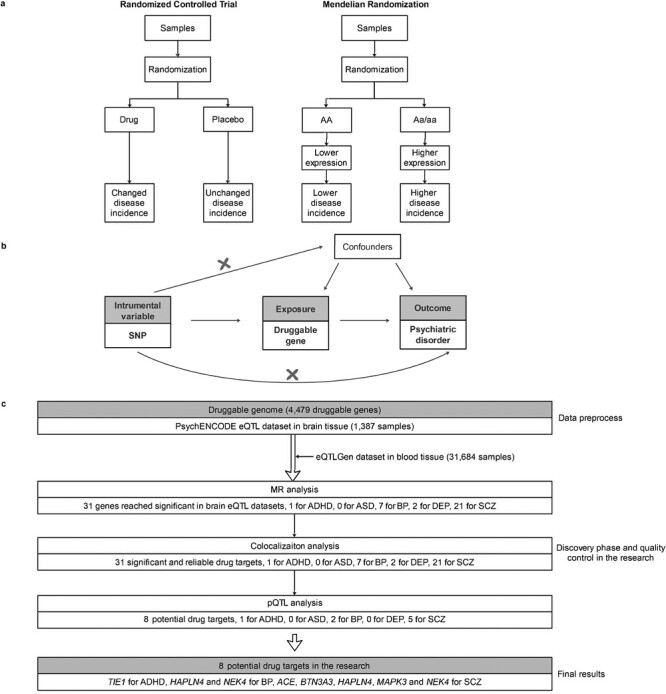
Overview of MR and the study design. (a) The comparison between MR and randomized controlled trial. MR is similar to the randomized controlled trial. (b) Diagram of MR analysis and assumptions. (c) Workflow and statistical results of this research. MR: Mendelian randomization.
In this study, to identify the most promising drug targets and to seek drug repurposing candidates for psychiatric disorders, we performed a 2 sample MR analysis by integrating summary statistics from the most recent and largest GWAS of psychiatric disorders (including schizophrenia (SCZ), bipolar disorder (BP), depression (DEP), autism spectrum disorder (ASD), and attention deficit and hyperactivity (ADHD)) and publicly available eQTL data in brain and blood tissue of 4479 actionable druggable genes (including protein targets or drug-like small-molecule binding partners that encode approved) (figure 1). By combining evidence of colocalization of effects, we identified 31 promising drug targets for psychiatric disorders. Of these, we proposed 8 genetically supported promising actionable novel drug targets with strong evidence for psychiatric disorders by protein quantitative trait loci (pQTL) verification, including gene ACE, BTN3A3, HAPLN4, MAPK3 and NEK4 for SCZ, gene NEK4 and HAPLN4 for BP, and gene TIE1 for ADHD but no drug-targeting gene with DEP and ASD. Our analysis provides a road map for the identification of potential therapeutic targets with genetically supported evidence in psychiatric disorders.
Materials and Methods
Genome-Wide Association Studies Summary Statistics Used in This Study
To incorporate human genetic studies in drug discovery and development, the most recent and largest-scale GWASs summary statistics that identified the genetic risk variants across various psychiatric disorders, including SCZ, BP, DEP, ASD, and ADHD, were used in this study. For SCZ, we used transancestry SCZ meta-analysis results (67 390 cases and 94 015 controls) reported by Trubetskoy et al.22For BP, we used meta-analysis results of Europe, North America and Australia cohorts (41 917 cases and 371 549 controls) reported by Mullins et al.23 For DEP, we used meta-analysis results (246 363 cases and 561 190 controls) reported by Howard et al. which samples collected from the 3 largest GWAS of depression.24 For ASD, we used meta-analysis results from the Danish population (18 381 cases and 27 969 controls) reported by Grove et al.25 For ADHD, we used ADHD meta-analysis results on samples from multi cohorts (20 183 cases and 35 191 controls) reported by Demontis et al.26 We obtained these GWAS summary datasets from the Psychiatric Genomics Consortium (PGC) (https://www.med.unc.edu/pgc/). Details on the processing of these GWAS summary statistics are provided in the original publications.
Quantitative Trait LociDatasets Used for Genetic Instrumental Variables Selection
The eQTL dataset has been extensively utilized for identifying genetic variants that regulate gene expression.27 The eQTL data can be an indispensable connection between instrumental variables and exposures in MR analysis. Therefore, eQTL datasets from brain and blood tissues were included in the MR analysis. We downloaded publicly available data from the PsychENCODE Consortium (http://resource.psychencode.org/) as eQTL data for brain tissue (1387 samples, mostly from European cohorts) which results reached significant FDR < 0.05.28 Moreover, we obtained publicly available eQTL data for blood tissues (31 684 samples, mostly from European cohorts) from the eQTLGen consortium (https://eqtlgen.org/) selecting those that reached significance with false discovery rate (FDR) < 0.05.29 The detailed descriptions of these eQTL datasets can be found in the original publications.
Actionable Drug Targets
In order to ensure that final experimental results are usable and reliable for drug target genes, the dataset of druggable genes required a stringent verification process. The druggable genome dataset in the public supplementary materials of the original publication10 was downloaded to ensure that the results of our experiments were corresponding to drug targets. This druggable genome dataset is straight set into diverse classes for different stages of the drug development procedure. It includes the approved and clinical-phase drugs target proteins, proteins analogous to approved drug targets and proteins accessible to drug-like small molecules. See the Supplementary Methods for details.10 We took the intersection of the druggable genome and eQTL datasets on the gene level and then obtained the input dataset of druggable genes with corresponding SNPs as exposures and instrumental variables. After potential drug targets were inferred, the drug information can be retrieved through DrugBank,30 with the purpose of highlighting novel repurposing opportunities for existing drugs.
Mendelian Randomization
GWAS has identified a large number of risk SNPs associated with diseases.31 However, GWAS cannot be utilized directly for drug discovery and development because druggable genes encode proteins rather than SNPs. MR analysis used genetic variants as instrumental variables to assess whether an association between an exposure and an outcome is consistent with a causal effect.32,33 This is because the natural, random assortment of genetic variants during meiosis yields a random distribution of genetic variants in a population32,34 (figure 1a). Two-sample MR analysis is a function based on MR analysis which is used to estimate the causal effect of an exposure on an outcome35 (figure 1b). Two-sample MR analysis could be conducted with the R package TwoSampleMR (version 0.5.4) (https://mrcieu.github.io/TwoSampleMR/). Different in-built functions were integrated when we performed a 2 sample MR analysis through the R package. If the druggable gene only contained 1 available SNP after the data processing, the Wald ratio was used to estimate the causal effect between druggable genes and diseases. Inverse-Variance Weighted (IVW) was utilized to assess the effect between the druggable gene with 2 available SNPs and psychiatric disorders. Moreover, a weighted median was conducted in the analysis as well for druggable genes with the number of available SNPs > 2. More specifically, IVW combines the Wald ratio estimates of each of the instrumental variables (ie, SNP) into one causal estimate for each risk factor. The weighted median estimate is the median of the weighted empirical distribution function of individual SNP Wald ratio estimates.
Details on data preprocessing before MR analysis are provided in the Supplementary Methods. By conducting a 2 sample MR analysis, the effect of druggable genes on psychiatric disorders can be estimated to predict the efficient drug targets for diseases. The MR analysis calculated the P-value of each druggable gene which indicated the probability of causal association between druggable gene and psychiatric disorder. To avoid the type 1 error, Bonferroni correction for multiple testing (ie, Bonferroni-corrected P-value cutoff of 0.05/27 421 total significant eQTLs in brain and blood = 1.82 × 10–6) was used to identify significant associations.36
Colocalization Analysis
For 2 sample MR analysis, SNP-druggable gene association and SNP-psychiatric disorder association should be rooted in the shared SNP. However, there were certain occasions when 2 different SNPs can influence druggable genes and psychiatric disorders independently. To estimate whether the druggable gene and psychiatric disorder association are consistent with an overlap causal variant, we conducted the colocalization analysis (based on the Bayesian framework37) to adjust such spurious results and posterior probabilities (PP) for 5 hypotheses (H0, H1, H2, H3, and H4) were calculated. Specifically, H4 indicates that one shared SNP associates with the druggable genes and the psychiatric disorders. The correct hypothesis above is H4 and the PP of H4 approximately probes the probability of one shared causal variant influencing druggable gene and psychiatric disorder. SNPs with strong evidence (PP.H4 > 0.8) and their corresponding genes were kept. Those druggable genes that reached significance in MR analysis and pass colocalization analysis were more likely to be drug targets for the corresponding psychiatric disorders.17 The R package coloc (Version 5.1.0) was used to conduct the colocalization analysis (https://www.rdocumentation.org/packages/coloc/versions/5.1.0).
Protein Quantitative Trait Loci Analysis
The pQTL study has identified genetic variants that regulate protein expression in various tissues. The proteins play an indispensable role in disease pathogenesis38 and drug target genes are based on protein. The pQTL dataset was selected based on 2 criteria, namely that the pQTL should be significant and provide all the information required for 2 sample MR analysis. Therefore, we obtained 4 public pQTL datasets in blood tissues39–42 (see the Supplementary Methods for details) and performed MR analysis on each pQTL dataset. We then conducted MR analysis again with pQTLs and GWASs of 5 psychiatric disorders. If druggable genes remained significant and reliable in a series of experiments of pQTL, we considered that these druggable genes were more likely to be drug targets for the corresponding diseases.
Mendelian Randomization Analysis in Non-European Study Participants
We further examined whether our MR findings are informative for non-European study populations using eQTL and GWAS data of non-European populations. By searching the PGC website for GWAS statistics in 5 psychiatric disorders, we found that the publicly downloadable non-European GWAS on the PGC website were only from East Asian (EAS) populations, including schizophrenia,43 and depression.44 Because there is no publicly available EAS brain eQTL data available for analysis, we used EAS blood eQTL data (N = 162) from Stranger et al.45 for MR analysis of non-European (ie, EAS) participants. The threshold for significant associations with MR evidence was set at P< 1.12 × 10–5 (ie, Bonferroni-corrected P-value cutoff of 0.05/ 4479 druggable gene). See Supplementary Material for details.
Results
Genetic Proposed Instruments for Actionable Druggable Genes
To identify potential drug repurposing opportunities for psychiatric disorders, we collected the most comprehensive druggable genome library to date from Finan et al.,10 covering a total of 4479 human genes that encode druggable proteins (Supplementary table 1). Furthermore, we identified all significant cis-eQTLs with FDR < 0.05 in the brain (1387 prefrontal cortex samples of mostly European ancestry from PsychENCODE Consortium), which acted in cis within 5 kb on either side of that encoded druggable gene and blood (31 684 samples from eQTLGen, mostly European ancestry individuals), which acted in cis within 5 kb either side of that encoded druggable gene. Moreover, only autosomal genes were kept for further research. Eventually, eQTL data for 2300 and 2580 druggable genes in brain and blood tissue entered the further MR analyses, respectively. For all selected druggable genes, we kept eQTLs after clumping at r2 = 0.001 based on the 1000 genomes European reference panel to avoid LD bias.
Mendelian Randomization Identifies Potential Drug Targets for Psychiatric Disease
Using brain and blood eQTL proposed instruments, we performed 2 sample MR analysis19on summary statistics from the 5 largest-scale GWAS published to date for psychiatric diseases (including SCZ, BP, DEP, ASD, and ADHD) (figure 1). The threshold for significant associations with MR evidence was set at P< 1.82 × 10–6 (ie, 0.05 Bonferroni-corrected for 27 421 total significant eQTLs of brain and blood) and full significant MR results are shown in Supplementary table 2. Using cis-eQTLs fromPsychENCODE dataset as proposed instruments, we identified a significant relationship between 21 genes and SCZ, 7 genes and BP, 2 genes and DEP, and a single gene for ADHD. Using cis-eQTLs from the eQTLGen dataset as proposed instruments, we discovered a significant relationship between 33 genes and SCZ, 15 genes and BP, 7 genes and DEP, 3 genes and ADHD, and none of the genes associated with ASD risk. Among these significant genes, the treatment of the target gene may be associated with disease risk because it has an odds ratio (OR) of greater than 1.00 (ie, OR > 1.00 indicates a relationship between increased gene expression and increased disease risk).
Furthermore, we found that a set of replicated genes reached significance in both 2 eQTL datasets (Supplementary table 3), including 11 genes for SCZ, 3 genes for BP, and 1 gene for DEP and ADHD. However, 12 of the 16 replicated genes had different directions of the MR effect between tissues (Supplementary figure 1 and Supplementary table 3). For example, genetically raised gene BTN3A2 expression was associated with reduced DEP risk in brain tissue (OR = 0.95; 95% CI, 0.94–0.97; P = 1.65 × 10–12) and increased DEP risk in blood tissue (OR = 1.06; 95% CI, 1.03–1.08; P = 5.09 × 10–7), and this pattern was consistent with SCZ. Caution is required in interpreting MR results with opposite directions of effect.
Taking into account the priority given to brain tissue probably most relevant to psychiatric disorders and the different directions of the MR effect between tissues, we adapted a potentially useful approach to screening for potentially druggable genes as follows: prioritizing genes associated with brain eQTL, even if they are not in the blood. Therefore, a total of 31 significant genes identified in MR analysis might be used as potential drug targets for psychiatric disease. Twenty-six of these 31 potential targets identified were nominated by the psychiatric disease GWAS previously and the rest five additional potential targets were reported here, which suggests that integrating eQTL evidence may help substantiate GWAS findings. Intriguingly, we found that several candidate genes with consistent directions of effect in the brain and blood, including one SCZ gene (FURIN), 2 BP genes (FURIN, MCHR1) and 1 ADHD gene (TIE1), strongly suggest that these genes represent potential drug targets for brain disorders (Supplementary figure 1). Moreover, we examined whether the significant MR results were overrepresented in a particular tier (Supplementary figure 2). Chi-squared tests assessed differences between the groups (ie, tiers) and we founded no significant relationship between any of the significant MR results and particular tier (P > 0.05, chi-squared test, Supplementary table 4).
Mendelian Randomization Quality Control Further Identifies That Promising Drug Targets Have the Most Robust Mendelian Randomization Evidence for Psychiatric Disease Risk
Given the low number of instruments used in this study (N = 1–3, Supplementary tables), MR–Egger and heterogeneity analyses may be less reliable, thus we did not perform sensitivity analyses. For potential associations that reached the significance threshold in brain MR analysis, we undertook an important quality control step to prioritize the MR significant results: colocalization analysis (Online Methods). Due to the presence of LD, there may be 2 distinct pathogenic SNPs in the association region of SNP exposure and SNP outcome, resulting in a spurious MR result.46 To further investigate whether the genetic associations with both gene expression and disease trait shared the same causal variant in the region, we performed colocalization analysis on MR preliminary results of the brain (Supplementary table 2) for psychiatric diseases. As summarized in Supplementary table 5, all of these 31 signals showed strong evidence of colocalization between gene expression and disease (posterior probability of shared causal variant across 2 traits, hypothesis 4 (PP.H4) > 0.8), indicating that both eQTL instrument (ie, gene expression) and outcome (ie, disease trait) are associated with the same region and share a same causal variant. Evidence shows that genes with strong evidence of MR and colocalization are more likely to become promising drug targets.17 In total, we prioritized 31 significant genes as promising drug targets for psychiatric disorders after MR quality control, including 21 significant genes for SCZ, 7 significant genes for BP, 2 significant genes for DEP and 1 significant gene for ADHD (figure 2 andSupplementary table 5).
Fig. 2.
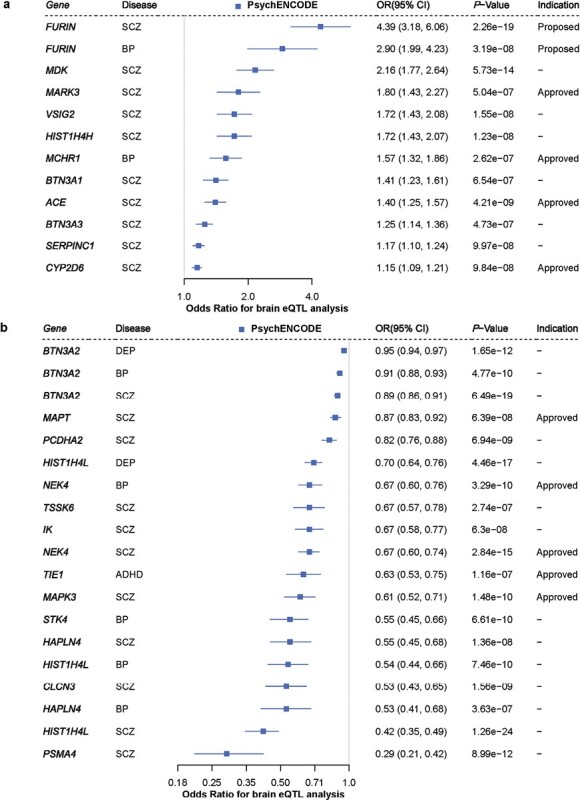
Thirty-one promising drug targets were proposed for psychiatric disorders. (a) Forest plots showing the results that drug targets as an antagonist reached significance and pass stringent quality tests in the brain eQTL dataset (blue =PsychENCODE). (b) Forest plots showing the results that drug targets as an agonist reached significance and pass stringent quality tests in the brain eQTL dataset (blue =PsychENCODE). The “Indication” column indicates whether the corresponding drug is approved or proposed (the drug information can be retrieved through DrugBank). The term “NA” used in the plot represents encoded druggable genes that have not been included in the DrugBank. Replicated significant genes with concordant effect direction between blood and brain tissues are distinguished and bolded. Additionally, we calculated a 95% CI for the OR. SCZ: schizophrenia, BP: bipolar disorder, DEP: depressive disorder, ADHD: attention deficit hyperactivity disorder.
Protein Quantitative Trait Loci Data Provide Further Genetic Evidence for Identifying Actionable Therapeutic Targets for Psychiatric Disorders
Considering that most clinical use of proteins as drug targets rather than gene expression and the nonlinear relationship between protein and mRNA levels,47 it is necessary and important to utilize pQTLs to model drug target effects (ie, defined as MR instruments for drug exposure).48 Using pQTLs in the MR analyses for the 31 proposed targets (figure 2), we identified 8 protein-trait associations (figure 3 andSupplementary table 6) on three diseases with strong evidence of MR (P< 1.12 × 10-5 at a Bonferroni-corrected threshold; after applying colocalization), including 5 (ACE, NEK4, BTN3A3, HAPLN4, and MAPK3) significant proteins for SCZ, 2 (NEK4 and HAPLN4) significant protein for BP and 1 (TIE1) significant protein for ADHD. By integrating drug-target information from the DrugBank database,30 4 of the 8 significant genes mentioned above encode targets of approved or clinical-phase drugs, presenting a possible repurposing opportunity: NEK4, MAPK3, ACE, and TIE1 (table 1). If the drug target is used as an inhibitor, there would need to be an OR > 1 in the MR, otherwise, it could be a safety concern and may not be a good drug target. For instance, by integrating summary data from brain eQTL/blood pQTL and SCZ GWAS in the MR analysis, we found that ACE with OR > 1 in both datasets (Supplementary table 2 and figure 3), indicating that a higher gene ACE expression level corresponded to a higher risk of SCZ. As shown in Table 1, perindopril is an ACE inhibitor prodrug with an appropriate direction of effect, possibly representing a repurposing opportunity. In addition, we observed that several identified potential druggable genes corresponding to the same drug (ie, fostamatinib), including TIE1 and NEK4 (table 1). Interestingly, another MARK3 gene that was significant only in brain tissue MR analysis also corresponded to the fostamatinib drug (Supplementary table 7). We found that the direction of effect of the fostamatinib (action as inhibition) on MARK3 expression was consistent with the effect seen in MR Analysis (MARK3 with OR > 1 in brain MR results, Supplementary table 2) and inconsistent with the other 2 gene effects (TIE1 and NEK4 with OR < 1 in brain MR results, Supplementary table 2), thus warranting further safety investigation in follow-up studies. Similar to the above, MAPK3, encodes the target of an approved-phase medication (ie, Arsenic trioxide) with an appropriate direction of effect (ie, inducer with negative MR effect), possibly representing an actionable therapeutic target for SCZ in future clinical studies (table 1).
Fig. 3.
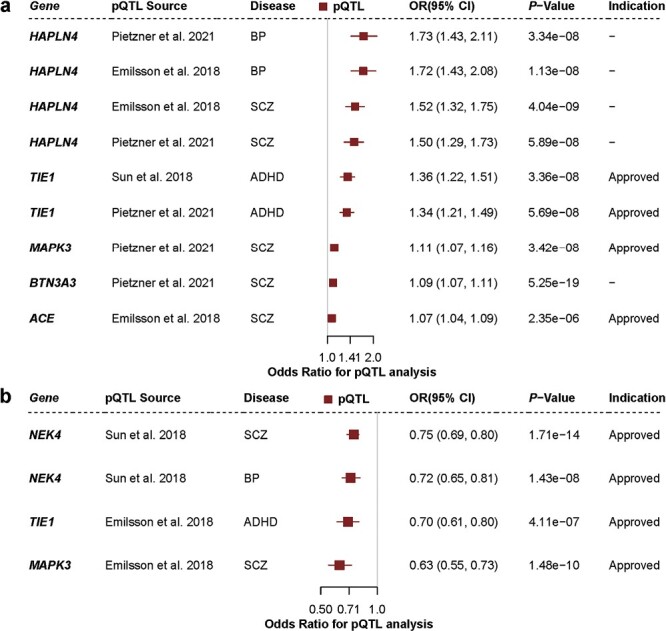
Protein quantitative trait loci in blood provide further genetic evidence. (a) Forest plots showing the results that all proteins as an antagonist and outcomes where a pQTL was available. (b) Forest plots showing the results that all proteins as an agonist and outcomes where a pQTL was available. The term “NA” used in the plot represents encoded druggable genes that have not been included in the DrugBank. The “pQTL Source” column indicates which pQTL study the SNPs were derived from. The “Indication” column indicates whether the corresponding drug is approved or proposed (the drug information can be retrieved through DrugBank). 95% CI, OR, SCZ: schizophrenia, BP: bipolar disorder, ADHD: attention deficit hyperactivity disorder.
Table 1.
Four Potential Drug Targets Offer Possible Drug Repurposing Opportunities as Clinical Drug Candidates for Psychiatric Disorders
| Gene | Outcome | Drug | Clinical phase | Action | Indication |
|---|---|---|---|---|---|
| ACE | SCZ | Perindopril | Approved | Inhibitor | Perindopril is an ACE inhibitor prodrug used to treat hypertension, mild to moderate congestive heart failure, and to reduce cardiovascular risk in patients with hypertension or postmyocardial infarction. |
| MAPK3 | SCZ | Arsenic trioxide | Approved | Inducer | Arsenic trioxide is a chemotherapeutic agent used in the treatment of refractory or relapsed acute Promyelocytic leukemia in patients with prior retinoid and anthracycline chemotherapy. |
| NEK4 | SCZ | Fostamatinib | Approved | Inhibitor | Fostamatinib is a spleen tyrosine kinase inhibitor used to treat chronic immune Thrombocytopenia after attempting one other treatment. |
| NEK4 | BP | Fostamatinib | Approved | Inhibitor | Fostamatinib is a spleen tyrosine kinase inhibitor used to treat chronic immune Thrombocytopenia after attempting one other treatment. |
| TIE1 | ADHD | Fostamatinib | Approved | Inhibitor | Fostamatinib is a spleen tyrosine kinase inhibitor used to treat chronic immune Thrombocytopenia after attempting one other treatment. |
Note: These drugs are approved, clinical phase, and drug indication based on https://go.drugbank.com.
Gene: The name of the drug target gene.
Outcome: The psychiatric disorder in which the drug target gene associates.
Drug: The name of the drug to the corresponding disorder.
Clinical phase: The phase of drug discovery procession.
Action: Pharmacological action of the drug.
Indication: Brief introduction to the drug.
Discussion
To identify drug repurposing opportunities and find novel drug targets for psychiatric disorders (including SCZ, BP, DEP, ASD, and ADHD), we undertook a large-scale 2 sample MR analysis by integrating eQTL/pQTL datasets for druggable genes with genetic summary statistics from the largest currently available GWAS datasets. By performing a colocalization analysis of the MR significant results in the brain, we further identified 31 promising drug targets with robust evidence (figure 2 and Supplementary table 5). By combining pQTL MR results, we proposed 8 drug targets with the strongest MR evidence. Among them, 4 of which (ACE, MAPK3, NEK4, and TIE1) offer possible drug repurposing opportunities as clinical drug candidates for psychiatric disorders (table 1). The potential contributions of the above 4 drug-target genes to psychiatric disorders are discussed in detail in the Supplementary Discussion. Overall, we highlight the importance and opportunities of eQTL MR analyses for drug repositioning and emphasize the key analytical methods to support such inference.
Several previous similar MR studies49,50 have been performed to provide valuable information for brain-related traits by integrating eQTL and GWAS from multiple sources. Detailed discussions on comparison with previous similar MR studies are provided in the Supplementary Discussion.
Due to the relatively small number of non-European study participants in the psychiatric GWASs and eQTL dataset, it is uncertain whether the MR results from this study are applicable to other non-European study populations. Therefore, we performed MR analysis using blood eQTL and GWAS data of schizophrenia and depression in EAS populations and full significant MR results are shown in Supplementary table 8. Considering that differences in LD patterns between populations tend to affect the MR and colocalization results, we have repeated the MR analysis using brain eQTL data and European-specific GWAS summary statistics of SCZ (including 53 386 cases and 77 258 controls) to compare the EAS MR results (the detailed European-specific MR results are summarized in Supplementary table 9). Using cis-eQTL SNPs from the blood eQTL dataset of the EAS population as genetic instruments, we identified 3 significant drug targets for schizophrenia (including MAPK3, BTN3A2, and ITIH4) but no significant drug-targeting gene with depression. Of note, we noticed that MAPK3 and BTN3A2 also showed significant associations in the European-specific MR study (Supplementary table 2). However, we found that both 2 significant genes (MAPK3 and BTN3A2) had different directions of effect between tissues, possibly due to the discrepancy in effect direction and the heterogeneity of different eQTL tissues.
There are also several potential limitations to this study. First, although we used SNP instruments derived from different data sources in MR analysis, they still limit the generalizability of the results because the genetic instruments obtained did not cover the whole actionable druggable genome. Second, most previous GWAS and eQTL studies of psychiatric disorders have used data from individuals of European descent. This limits the understanding of the underlying biology of psychiatric disorders and raises questions about the transferability of findings between populations. Our study also suggests ancestry-specific MR findings diverge and converge across modalities in schizophrenia and depression. Third, we mainly focused on the effect of SNPs (ie, variants) on gene expression and protein levels. This work did not take into account other factors that may affect disease, including chromatin accessibility, histone modification, and DNA methylation. Also, we overlooked the effect of variants on expression levels of gene isoforms. Finally, MR cannot fully reproduce clinical trials (ie, the MR results do not yet directly reflect the practical effect size) and does not perfectly predict a drug effect, considering that standard MR studies typically determine lifespan and low-dose exposure, whereas RCTs only report the effects of relatively shorter and high-dose exposures.
In conclusion, we performed comprehensive 2 sample MR analyses covering all actionable druggable genes to identify potentially causal genes for psychiatric disorders. By focusing on brain eQTL to mimic exposure to medications, we prioritized 31 promising causal genes based on genetic data (Supplementary table 10). Eight of these genes with the strongest pQTL MR evidence served as actionable drug targets for common psychiatric disorders (figure 2). Our work provides both an analytical framework for prioritizing potential new targets and provides actionable and promising drug candidates for drug repurposing in psychiatric disorders. The strategies proposed in this study can also be applied to other complex diseases, like cancer, that have extensive eQTL data available but still have many cases without well-defined, therapeutic targets with genetically supported evidence.
Supplementary Material
Acknowledgments
We thank the participants and investigators of the Working Group of the Psychiatric Genomics Consortium, the eQTLGen Consortium and the PsychENCODE Consortium for generating and making the summary statistics available for us and making this work possible.
Contributor Information
Xiaoyan Li, Key Laboratory of Intelligent Computing and Signal Processing of Ministry of Education and Information Materials and Intelligent Sensing Laboratory of Anhui Province, Institutes of Physical Science and Information Technology, Anhui University, Hefei, Anhui 230601, China.
Aotian Shen, Key Laboratory of Intelligent Computing and Signal Processing of Ministry of Education and Information Materials and Intelligent Sensing Laboratory of Anhui Province, Institutes of Physical Science and Information Technology, Anhui University, Hefei, Anhui 230601, China.
Yiran Zhao, Key Laboratory of Intelligent Computing and Signal Processing of Ministry of Education and Information Materials and Intelligent Sensing Laboratory of Anhui Province, Institutes of Physical Science and Information Technology, Anhui University, Hefei, Anhui 230601, China.
Junfeng Xia, Key Laboratory of Intelligent Computing and Signal Processing of Ministry of Education and Information Materials and Intelligent Sensing Laboratory of Anhui Province, Institutes of Physical Science and Information Technology, Anhui University, Hefei, Anhui 230601, China.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (U22A2038 and 82101611).
References
- 1. Collaborators GBDCRD. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 2020;8(6):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doran CM, Kinchin I.. A review of the economic impact of mental illness. Aust Health Rev. 2019;43(1):43–48. [DOI] [PubMed] [Google Scholar]
- 3. Guessoum SB, Lachal J, Radjack R, et al. Adolescent psychiatric disorders during the COVID-19 pandemic and lockdown. Psychiatry Res. 2020;291:113264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collaborators C-MD. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet. 2021;398(10312):1700–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smietana K, Siatkowski M, Moller M.. Trends in clinical success rates. Nat Rev Drug Discov. 2016;15(6):379–380. [DOI] [PubMed] [Google Scholar]
- 6. Harrison RK. Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov. 2016;15(12):817–818. [DOI] [PubMed] [Google Scholar]
- 7. Wouters OJ, McKee M, Luyten J.. Estimated research and development investment needed to bring a new medicine to market, 2009–2018. JAMA. 2020;323(9):844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856–860. [DOI] [PubMed] [Google Scholar]
- 9. Mit LU, et al. ; Diabetes Genetics Initiative of Broad Institute of H, Novartis Institutes of BioMedical R. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. [DOI] [PubMed] [Google Scholar]
- 10. Finan C, Gaulton A, Kruger FA, et al.The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9(383):eaag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okuda PMM, Klaiman C, Bradshaw J, Reid M, Cogo-Moreira H.. Assessing risk of bias in randomized controlled trials for autism spectrum disorder. Front Psychiatry. 2017;8:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fralick M, Kesselheim AS, Avorn J, Schneeweiss S.. Use of health care databases to support supplemental indications of approved medications. JAMA Intern Med. 2018;178(1):55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Escala-Garcia M, Morra A, Canisius S, et al. Breast cancer risk factors and their effects on survival: a Mendelian randomisation study. BMC Med. 2020;18(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y, Zeng J, Zhang F, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun. 2018;9(1):918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 16. Qi T, Wu Y, Zeng J, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9(1):2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng J, Haberland V, Baird D, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. 2020;52(10):1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diogo D, Tian C, Franklin CS, et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9(1):4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Storm CS, Kia DA, Almramhi MM, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC). Finding genetically-supported drug targets for Parkinson’s disease using Mendelian randomization of the druggable genome. Nat Commun. 2021;12(1):7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gaziano L, Giambartolomei C, Pereira AC, et al. ; VA Million Veteran Program COVID-19 Science Initiative. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. 2021;27(4):668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trubetskoy V, Pardinas AF, Qi T, et al. ; Indonesia Schizophrenia Consortium. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604(7906):502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mullins N, Forstner AJ, O’Connell KS, et al. ; HUNT All-In Psychiatry. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53(6):817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Howard DM, Adams MJ, Clarke TK, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grove J, Ripke S, Als TD, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilad Y, Rifkin SA, Pritchard JK.. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24(8):408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li M, Santpere G, Imamura Kawasawa Y, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362(6420):eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vosa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horwitz T, Lam K, Chen Y, Xia Y, Liu C.. A decade in psychiatric GWAS research. Mol Psychiatry. 2019;24(3):378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith GD, Ebrahim S.. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. [DOI] [PubMed] [Google Scholar]
- 33. Emdin CA, Khera AV, Kathiresan S.. Mendelian randomization. JAMA. 2017;318(19):1925–1926. [DOI] [PubMed] [Google Scholar]
- 34. Hingorani A, Humphries S.. Nature’s randomised trials. Lancet 2005;366(9501):1906–1908. [DOI] [PubMed] [Google Scholar]
- 35. Davey Smith G, Hemani G.. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chauquet S, Zhu Z, O’Donovan MC, Walters JTR, Wray NR, Shah S.. Association of antihypertensive drug target genes with psychiatric disorders: a Mendelian randomization study. JAMA Psychiatry. 2021;78(6):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Williams SM, Giambartolomei C, Vukcevic D, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suhre K, McCarthy MI, Schwenk JM.. Genetics meets proteomics: perspectives for large population-based studies. Nat Rev Genet. 2021;22(1):19–37. [DOI] [PubMed] [Google Scholar]
- 39. Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374(6569):eabj1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lam M, Chen CY, Li Z, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giannakopoulou O, Lin K, Meng X, et al. The genetic architecture of depression in individuals of East Asian ancestry: a genome-wide association study. JAMA Psychiatry 2021;78(11):1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stranger BE, Montgomery SB, Dimas AS, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8(4):e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hemani G, Bowden J, Davey Smith G.. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195–R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Albert FW, Treusch S, Shockley AH, Bloom JS, Kruglyak L.. Genetics of single-cell protein abundance variation in large yeast populations. Nature. 2014;506(7489):494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt AF, Finan C, Gordillo-Maranon M, et al. Genetic drug target validation using Mendelian randomisation. Nat Commun. 2020;11(1):3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baird DA, Liu JZ, Zheng J, et al. Identifying drug targets for neurological and psychiatric disease via genetics and the brain transcriptome. PLoS Genet. 2021;17(1):e1009224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Klein N, Tsai EA, Vochteloo M, et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat Genet. 2023;55(3):377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


