Significance
Neutrophils are critical first-line responders to infection forming a potent line of innate immune defense. While we know that neutrophils are armed with highly glycosylated microbicidal proteins essential for combatting invading pathogens, little is known about how, when, and where the microbicidal proteins are glycosylated during neutrophil maturation. Using systems glycobiology to investigate blood neutrophils and their progenitors, we find that profound glycoproteome remodeling accompanies the early neutrophil maturation phase when microbicidal glycoproteins are produced. The study is significant as it provides spatiotemporal insights into the glycophenotype of key neutrophil compartments and the peculiar glycosylation that decorates microbicidal proteins. The study forms a resource to guide future explorations targeting the biological roles and dysregulation of glycosylation in neutrophil-related disorders.
Keywords: granulopoiesis, neutrophil, granule, glycoproteome, remodeling
Abstract
Neutrophils store microbicidal glycoproteins in cytosolic granules to fight intruding pathogens, but their granule distribution and formation mechanism(s) during granulopoiesis remain unmapped. Herein, we comprehensively profile the neutrophil N-glycoproteome with spatiotemporal resolution by analyzing four key types of intracellular organelles isolated from blood-derived neutrophils and during their maturation from bone marrow–derived progenitors using a glycomics-guided glycoproteomics approach. Interestingly, the organelles of resting neutrophils exhibited distinctive glycophenotypes including, most strikingly, highly truncated N-glycans low in α2,6-sialylation and Lewis fucosylation decorating a diverse set of microbicidal proteins (e.g., myeloperoxidase, azurocidin, neutrophil elastase) in the azurophilic granules. Excitingly, proteomics and transcriptomics data from discrete myeloid progenitor stages revealed that profound glycoproteome remodeling underpins the promyelocytic-to-metamyelocyte transition and that the glycophenotypic differences are driven primarily by dynamic changes in protein expression and less by changes within the glycosylation machinery. Notable exceptions were the oligosaccharyltransferase subunits responsible for initiation of N-glycoprotein biosynthesis that were strongly expressed in early myeloid progenitors correlating with relatively high levels of glycosylation of the microbicidal proteins in the azurophilic granules. Our study provides spatiotemporal insights into the complex neutrophil N-glycoproteome featuring intriguing organelle-specific N-glycosylation patterns formed by dynamic glycoproteome remodeling during the early maturation stages of the myeloid progenitors.
Neutrophils are critical first-line responders to infection and form a potent line of innate immune defense against invading pathogens (1, 2). Circulating in their resting state in blood, neutrophils are equipped with an arsenal of prepacked cytosolic granules (3). The carefully timed mobilization of these intracellular granules released upon specific environmental cues enables neutrophils to extravasate and traffic to inflammatory sites to elicit an effective, timely, and well-balanced response against invading pathogens, which they fight by means of phagocytosis, degranulation of granule-resident microbicidal glycoproteins, and neutrophil extracellular traps (NETs) (3, 4).
Discrete granule populations are formed during the week-long maturation of neutrophils (granulopoiesis) in the bone marrow (3, 5). Azurophil (Az) granules are formed during the early myeloblast-to-promyelocyte (MB-to-PM) maturation stages, while specific (Sp) granules are formed during the PM-to-myelocyte/metamyelocyte (MC/MM) stages. Finally, gelatinase (Ge) granules form during the late MM-to-band cell (BC) maturation stages. Secretory (Se) vesicles, although not considered granules per se, form another type of membrane-enclosed intracellular compartments within neutrophils that arise from endocytosis and plasma membrane invagination during the final BC-to-mature neutrophil maturation (3). Now well established, the granule populations harbor distinct proteome signatures; the temporal granule formation during granulopoiesis is, through a mechanism known as “targeting-by-timing,” a recognized contributor to the granule-specific protein repertoires of neutrophils (6, 7).
Protein N-glycosylation, the covalent attachment of N-glycans to motif-restricted asparagine residues (Asn-X-Ser/Thr, X ≠ Pro), adds important structural and functional heterogeneity to proteins. Both the neutrophil cell surface and granule-resident proteins are richly decorated with N-glycans that facilitate and modulate key immune and trafficking functions of neutrophils (4). For example, neutrophil proteins carry N-glycans containing Lewis X (Lex) and sialyl-Lewis X (sLex) glycoepitopes (8), which interact with endothelial P- and E-selectins to mediate neutrophil tethering, rolling, and extravasation to peripheral tissues (9, 10).
Following our identification of paucimannosidic N-glycans (Man1-3GlcNAc2Fuc0-1) in the neutrophil-rich sputum from cystic fibrosis-affected individuals (11, 12), we and others have demonstrated for a few specific proteins that neutrophils indeed produce paucimannosylation, which abundantly decorates microbicidal glycoproteins residing in the Az granules, e.g., cathepsin G (CTSG), neutrophil elastase (ELANE), and myeloperoxidase (MPO) (12–18). The biological roles of paucimannosidic proteins in neutrophils, however, remain poorly understood despite emerging evidence pointing to their involvement in key innate immune processes (19, 20).
While protein glycosylation is widely recognized to shape neutrophil function (4), the spatial N-glycoproteome distribution across the neutrophil compartments and the dynamic glycoproteome changes (remodeling) that accompany the dramatic metamorphosis associated with the differentiation of myeloid progenitor cells to mature polymorphonuclear (PMN) blood cells through the granulopoiesis process remain unknown.
Herein, we comprehensively profile the dynamic and complex neutrophil N-glycoproteome with spatial (across granules/vesicles) and temporal (across neutrophil maturation stages of developing myeloid cells) information focusing specifically on characterizing the luminal (soluble) organelle fractions hosting most of the microbicidal glycoproteins involved in neutrophil defense. Enabled by a glycomics-guided glycoproteomics approach applied to isolated neutrophil compartments complemented with multiomics data interrogation of myeloid progenitors still developing in the bone marrow, we provide spatiotemporal insights into neutrophil N-glycosylation processes. We find that the neutrophil N-glycoproteome exhibits fascinating site-, protein-, and organelle-specific N-glycosylation features and undergoes profound changes in the protein repertoire and remodeling of the glycosylation machinery during the early stages of neutrophil maturation in the bone marrow. Our study offers important clues to the mechanisms driving the organelle-specific glycosylation of neutrophils, notably the formation of the unique glycophenotype of the Az granules packed with potent microbicidal glycoproteins and provides a valuable resource to guide future studies that seek to explore the biological roles and dysregulation of neutrophil glycosylation.
Results
Glycoproteome Profiling of Resting and Maturing Neutrophils.
The N-glycoproteome of resting neutrophils was profiled with spatial resolution by applying an integrated glycomics-assisted glycoproteomics approach (21–23) to isolated granule and vesicular populations (Fig. 1A). The temporal N-glycoproteome remodeling accompanying granulopoiesis was explored by interrogating robust proteomics and transcriptomics resources generated from discrete myeloid progenitor subsets (24, 25) for previously overlooked glycoprotein and glycoenzyme expression patterns (Fig. 1B).
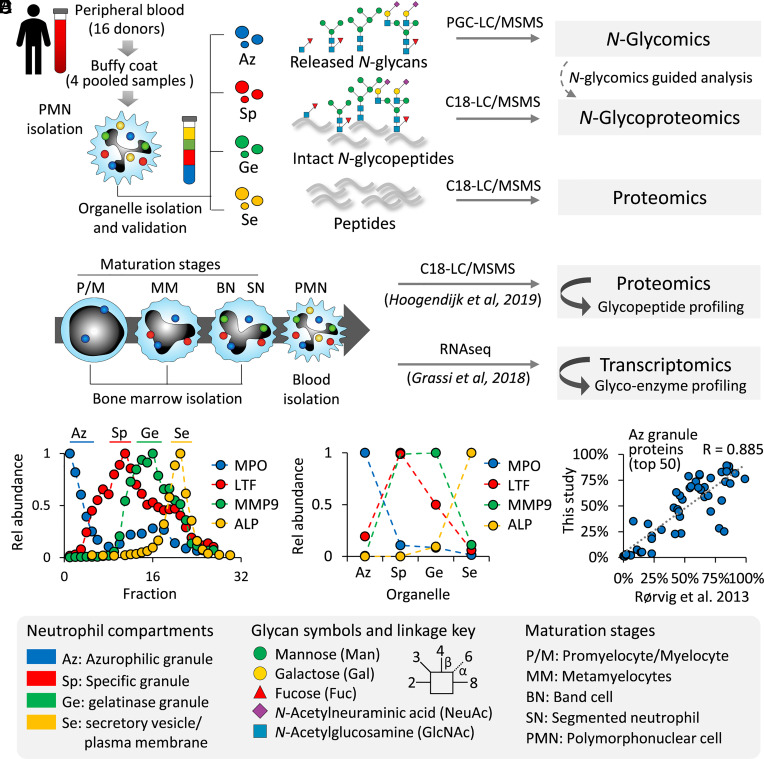
Fig. 1.
Spatiotemporal profiling of the N-glycoproteome of the neutrophil organelles and myeloid maturation stages using systems glycobiology approaches. (A) Spatial profiling of the N-glycoproteome using quantitative glycomics–assisted glycoproteomics of four intracellular compartments isolated from resting neutrophils. (B) Temporal profiling of the N-glycoproteome and the underpinning glycosylation machinery of discrete neutrophil maturation stages from bone marrow–derived myeloid progenitor cells through interrogation of available proteomics and transcriptomics resources (24, 25). The neutrophil compartments were separated using density gradients, and their isolation was guided by (C) ELISA (LTF, MMP9) and enzyme activity assays (MPO, ALP) of established organelle markers and further confirmed by (D) quantitative proteomics of the same marker proteins. (E) The effective separation of the neutrophil compartments was validated by an observed concordance of the 50 most abundant proteins identified in each of the compartments against a reference study also performing quantitative proteomics of separated neutrophil organelles (7). See Dataset S1 for the correlation of the other compartments. See Inset for key to the glycan symbols, linkage nomenclature, and the neutrophil organelles and maturation stages.
The separation of four intracellular compartments of neutrophils was guided by established immunoblot and enzyme activity assays against known organelle markers across the Percoll density fractions (Fig. 1C) (14, 26). The enrichment of discrete subsets of granules and vesicles was orthogonally validated using the acquired proteomics data by plotting the protein expression profile of the organelle markers (Fig. 1D). Further supporting the precision of the organelle separation, concordance was observed between our compartment-specific proteomics data and a landmark study also performing quantitative proteomics on separated neutrophil compartments (7) as illustrated by the correlation of proteins identified in the Az granules (R = 0.885) (Fig. 1E) and in other compartments (Sp: 0.854; Ge: 0.610; Se: 0.890) (Dataset S1). The relatively low correlation for the Ge granules is likely a result of the different Percoll density gradients (three- vs. four-layered) used in the two studies.
Distinct N-glycome Signatures across the Neutrophil Granules.
First, quantitative glycomics performed using PGC-LC-MS/MS revealed a total of 71 protein-linked N-glycan structures spanning 43 N-glycan compositions across the neutrophil compartments (Datasets S2 and S3).
Principal component analysis (PCA) of the N-glycomics data indicated that the neutrophil compartments carry distinct N-glycome signatures (Fig. 2A). The Az granules appeared particularly well separated from the other organelles based on the N-glycome data. With respect to the N-glycan type distribution, the Az granules displayed abundant paucimannosylation (~55% of the total Az N-glycome) (Fig. 2B). The Sp granules displayed mainly sialofucosylated complex-type N-glycans (~75%), while oligomannosidic- and less decorated complex-type structures were characteristic N-glycan types of both the Ge and Se compartments.
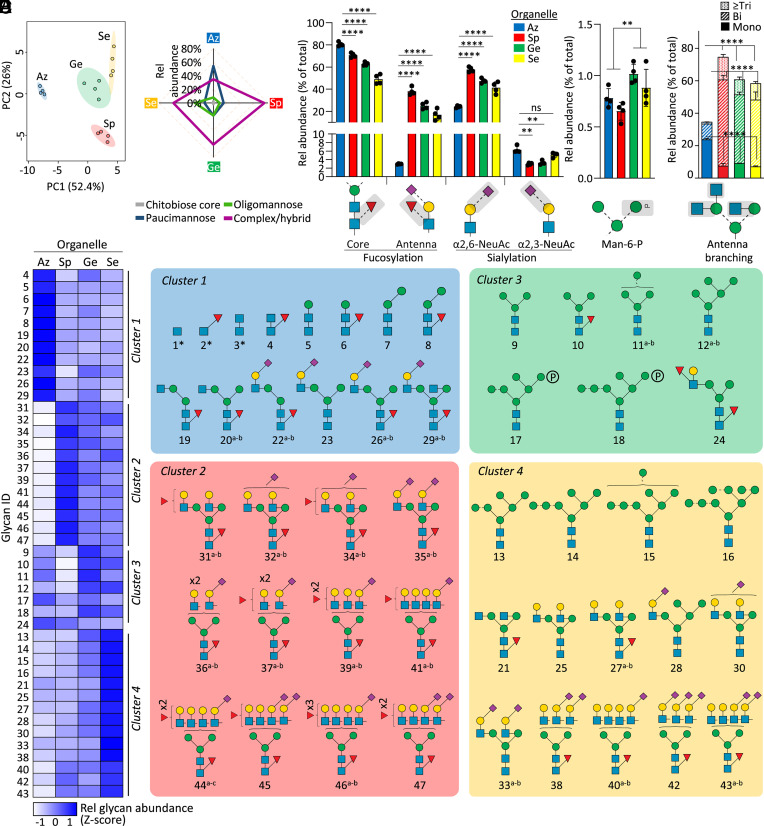
Fig. 2.
Organelle-specific N-glycome signatures in resting neutrophils. (A) PCA of quantitative N-glycomics data of the four cytosolic compartments (Az, Sp, Ge, and Se) isolated from resting neutrophils. (B) N-glycan class distribution across compartments. Key glycofeatures across organelles including (C) core/antenna fucosylation and α2,3-/α2,6-sialylation, (D) mannose-6-phosphate (M6P) modifications, and (E) antennary branching patterns (relative abundance of each glycofeature against the total N-glycome). For panel B−E, data plotted as mean and SD (n = 4 biological replicates, SD left out in panel B for simplicity, ANOVA test, **P < 0.01, ****P < 0.0001, ns, not significant). Organelle-specific differences were also observed for fucosylation, sialylation, M6P, and antennary branching when measured as a proportion of identified structures having the potential to be modified by the specific glycofeature rather than against the total N-glycome, an informative measure when exploring biosynthetic relationships associated with observed N-glycome differences (SI Appendix, Fig. S1 A and B). (F, Left) Unsupervised clustering (see SI Appendix, Fig. S1C for dendrogram) and heatmap of N-glycomics data illustrating four major clusters representing the neutrophil organelles. Data plotted as average of four biological replicates in each compartment at the composition level (relative abundances of N-glycan isomers were summed) and Z-score transformed. (Right) N-glycan structures contributing to each cluster. The most common structure is depicted for glycan compositions exhibiting multiple isomers. *chitobiose core-type N-glycans were prevalently observed at the glycopeptide level and, thus, for balance, were added to this map. See Fig. 1 for key. See Dataset S2 for glycan identifiers consistently used throughout the study.
Profound organelle-specific differences were also observed for the fine structural elements of the complex-type N-glycans in each neutrophil compartment. While core fucosylation (α1,6-fucosylation) was a prevalent feature across all organelles, the Sp, Ge, and Se compartments exhibited significantly lower core fucosylation (~45-70%) compared to the Az granules (~80%, all P < 0.0001, ANOVA test) (Fig. 2C). In contrast, antenna fucosylation forming functionally important Lewis-type glycoepitopes was near-absent in the Az granules (~3%) while being prominent features in other compartments (~20 to 40%, all P < 0.0001, ANOVA test). Interestingly, sialyl linkage switching was also observed across the granule populations; the Az granules exhibited less α2,6-sialylation and high α2,3-sialylation relative to the other compartments (all P < 0.01 except for Az vs. Se α2,3-sialylation, ANOVA test). Albeit of generally low abundance (0.5 to 1% of N-glycome), two mannose-6-phosphate (M6P) containing oligomannosidic N-glycans (Man6-7GlcNAc2) were additionally found to be elevated in the Ge granules and Se vesicles relative to the Az and Sp granules (P < 0.01, Student’s t test) (Fig. 2D). Last, dramatic differences in the antennary branching pattern were observed across the organelles; unusual monoantennary complex-type N-glycans were features of the Az granules contrasting the bi-, tri- and higher branching patterns observed in the other compartments (all P < 0.0001, ANOVA test) (Fig. 2E). Organelle-specific differences were also observed for these N-glycan features (core/antenna fucosylation, α2,3-/α2,6-sialylation, M6P, antennary branching) when measured as a proportion of identified structures having the potential to be modified by the specific glycofeature (SI Appendix, Fig. S1 A and B). This plotting method is informative when exploring biosynthetic mechanisms underpinning the observed organelle-specific N-glycome differences (27).
Recapitulating the organelle-specific N-glycome differences, unsupervised clustering analysis of the glycomics data indicated four major glycan clusters (cluster 1-4), one for each compartment type, and revealed individual N-glycan structures contributing to the organelle differences (Fig. 2D and SI Appendix, Fig. S1C). In short, cluster 1 (Az granules) was rich in peculiar chitobiose core-, paucimannosidic-, and monoantennary complex-type N-glycans, cluster 2 (Sp granules) featured highly sialylated and core/antenna fucosylated complex-type branched N-glycans, cluster 3 (Ge granules) exhibited M6P-oligomannosylation, and cluster 4 (Se vesicles) was enriched in oligomannose and less sialofucosylated complex-type N-glycans. Taken together, our comprehensive N-glycome map of luminal (soluble) proteins from the neutrophil compartments, the most detailed to date, demonstrates prominent organelle-specific N-glycan differences across resting neutrophils.
Glycoproteomics Reveals Granule-, Protein-, and Site-Specific N-glycosylation in Neutrophils.
Organelle-specific glycoproteomics data of the neutrophil compartments were generated and searched for intact N-glycopeptides using a tailored glycan database comprising only N-glycans already identified by glycomics and a few ultratruncated N-glycans (Glycan #1-3) known to be expressed by neutrophils yet commonly overlooked by our PGC-LC-MS/MS-based glycomics approach (15, 17). Glycomics-guided glycoproteomics (21–23, 28) is beneficial relative to uninformed searches as the prior information reduces the glycopeptide mis-identification rate and the search time, two bottlenecks in glycoproteomics data analysis (29, 30), while it, in parallel, provides quantitative details of the N-glycans in the N-glycoproteome.
Collectively, a total of 27,663 HCD-based N-glycoPSMs from 4,772 unique N-glycopeptides (unique protein, site, glycan) spanning 627 N-glycosites and mapping to 352 N-glycoproteins were identified across the neutrophil compartments (SI Appendix, Fig. S2A and Dataset S4). Despite using conservative confidence thresholds and a tailored glycan database to ensure accurate glycopeptide identification, our analysis provides the, to date, deepest coverage of the neutrophil N-glycoproteome, expanding on findings from a recent neutrophil glycoproteomics study performed without granule fractionation (13) (SI Appendix, Fig. S2 B and C). Our glycomics-informed glycoproteomics approach therefore facilitated both an extensive and accurate view of the glycoproteome as supported by correlations between our glycomics and glycoproteomics data (Az: R = 0.588, Sp: 0.505, Ge: 0.529, Se: 0.419) and the enriched and unenriched glycopeptide fractions (Az: R = 0.875, Sp: 0.649, Ge: 0.789, Se: 0.906) across the investigated organelles (SI Appendix, Fig. S3). The performance of our glycomics-guided glycoproteomics approach hinges on the assumption that glycomics captures a representative snapshot of the N-glycans present in a sample; we therefore compared the glycoprofile of the top 10% most abundant glycoproteins in each compartment (those expected to contribute mostly to the N-glycome) and the remaining lower 90% glycoproteins in each compartment. Pleasingly, these analyses revealed that similar N-glycosylation patterns decorate the high and low abundant proteins in each neutrophil organelle (Az: R = 0.876, Sp: 0.674, Ge: 0.912, Se: 0.876) thereby validating the approach used (SI Appendix, Fig. S4).
Interestingly, the glycopeptide spectral prevalence in the Az granules (averaging 2,558 N-glycoPSMs/LC-MS/MS run) far outnumbered the glycopeptide spectral counts in other compartments (700 to 1,501 N-glycoPSMs/LC-MS/MS run) (Fig. 3A). Despite the higher proportion of glycopeptide spectra in the Az datasets, a similar number of unique glycopeptides but mapping to less source N-glycoproteins was observed for the Az granules (94 proteins) relative to the other organelle populations (139 to 199 proteins/compartment). Since the same amount of protein starting material was used for the organelle-specific glycoproteomics experiments, the uneven glycoPSM distribution across compartments suggests a higher glycosylation “efficiency” of the proteins residing in the Az granules. Supporting these findings (which we further expand on below), a significantly higher overall presence of a diagnostic glycopeptide ion (m/z 204.0867 HexNAc oxonium ion) was found in the LC-MS/MS data for the Az granule (25.9%) relative to the other compartments (8.9 to 21.1%, P < 0.05, Az vs. other organelles, Student’s t tests) (SI Appendix, Fig. S5). Annotated HCD-MS/MS spectra of representative glycopeptides carrying different classes of N-glycans (e.g., paucimannosidic-, oligomannosidic-, and complex-type glycans) characteristic for each neutrophil compartment showed that the HexNAc oxonium ion signature is an abundant feature across all glycopeptide classes validating the oxonium ion counting method as a useful proxy for the prevalence of glycosylation in the peptide mixtures from the neutrophil organelles.
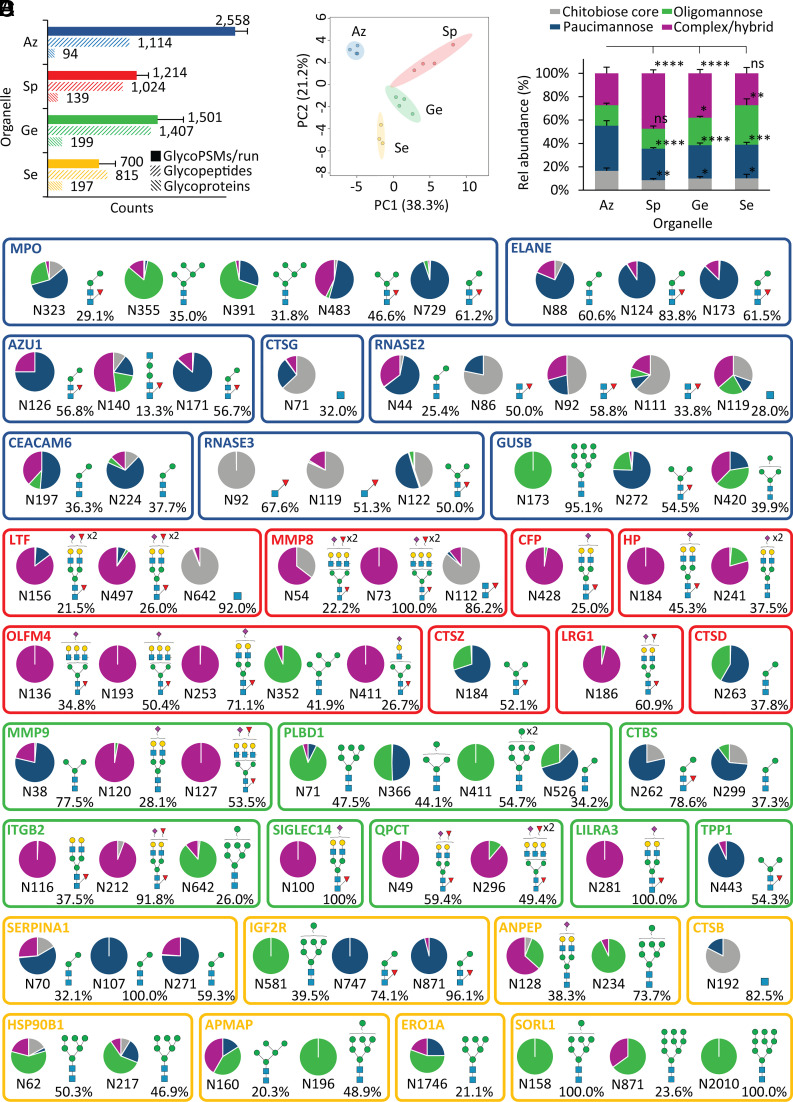
Fig. 3.
Glycoproteomics reveals site-, protein-, and organelle-specific N-glycosylation across the neutrophil compartments. (A) Glycoproteome coverage across the neutrophil compartments as measured by N-glycoPSM counts, unique N-glycopeptides (unique protein, site, glycan), and source N-glycoproteins identified by quantitative glycoproteomics. GlycoPSM data are plotted as mean ± SD, while glycopeptide/glycoprotein data are plotted as total counts from four biological replicates (n = 4). (B) Glycoproteomics-informed PCA of the neutrophil compartments (Az, Sp, Ge, and Se). (C) N-glycan type distribution across the neutrophil compartments based on glycoPSM spectral counting. Data plotted as mean ± SD (n = 4 biological replicates, *P < 0.05, **P < 0.01, ****P < 0.0001, ns, not significant, ANOVA test). (D) Extensive N-glycan diversity across sites and proteins as illustrated for the eight most abundant N-glycoproteins (out of 100 to 200 glycoproteins/granule) across each neutrophil compartment. The quantification of the site-specific glycoform distribution was performed by dividing the area-under-the-curve (AUC) of each glycoform by the summed AUC of all glycoforms identified within the same site from the same glycoprotein. The most abundant glycoform (and its relative abundance) is indicated for each site. An additional N-glycosylation site (N139), not part of the mature polypeptide chain, was observed for MPO, but left out of the map due to a low abundance (< 5%) relative to the other sites. See Fig. 1 for key.
PCA of the glycoproteomics data recapitulated the glycomics-informed separation of the four neutrophil compartments supporting their distinctive glycophenotypes and highlighted again the unique glycosylation features of the Az granules (Fig. 3B). In line with the glycomics data, the organelle-specific glycoproteomics data indeed showed that the Az granules exhibit higher levels of chitobiose core- and paucimannosidic-type N-glycans and lower levels of oligomannosidic- and complex-type N-glycans relative to the other compartments (Fig. 3C).
The information-rich glycoproteomics data revealed a profound microheterogeneity featuring an extensive diversity and variation of the N-glycan distribution across sites and proteins as illustrated for the eight most abundant glycoproteins (of ~100 to 200 glycoproteins/granule) in each compartment (Fig. 3D and Dataset S5). Importantly, while each glycoprotein (and each site) exhibited distinct glycosylation patterns, the organelle-specific N-glycosylation features documented by glycomics were recapitulated in the glycoproteomics data as illustrated by similar glycan-based clustering patterns across the neutrophil compartments (SI Appendix, Fig. S6).
Taken together, our comprehensive glycomics-assisted glycoproteomics analysis of the four cytosolic compartments of resting neutrophils provides spatial insight into the heterogeneous neutrophil N-glycoproteome that exhibits fascinating organelle-, protein- and site-specific N-glycosylation features.
Profound Glycoproteome Remodeling during Neutrophil Granulopoiesis.
To begin to unpick mechanisms driving the intriguing compartment-specific N-glycosylation in resting neutrophils, we investigated longitudinally the N-glycoproteome remodeling accompanying the granulopoiesis process in the bone marrow. Specifically, we mined robust proteomics and transcriptomics data collected from discrete myeloid progenitor stages for previously overlooked N-glycopeptide and glycoenzyme expression patterns (24, 25). Owing to the high data quality, a total of 11,928 N-glycoPSMs from 2,276 unique N-glycopeptides mapping to 1,079 N-glycoproteins (SI Appendix, Fig. S2A and Datasets S6 and S7) and 45 glycoenzymes driving diverse N-glycosylation biosynthetic processes (Dataset S8) were longitudinally profiled from these available resources.
In early granulopoiesis (P/M stage), the myeloid progenitors displayed prominent paucimannosylation (~45%) and chitobiose core-type (~25%) N-glycosylation, which were dramatically reduced in the subsequent maturation stage (MM stage, ~20% and ~15%, respectively), which instead featured elevated complex- (~15% → ~35%) and oligomannosidic-type (~15% → ~30%) N-glycans as assessed using glycoPSM-based quantitation of newly expressed glycoproteins at each stage (Fig. 4A and Dataset S6). The glycoproteome remodeling was notably reduced in mid- (MM) and late- (BN, SN) stage granulopoiesis.
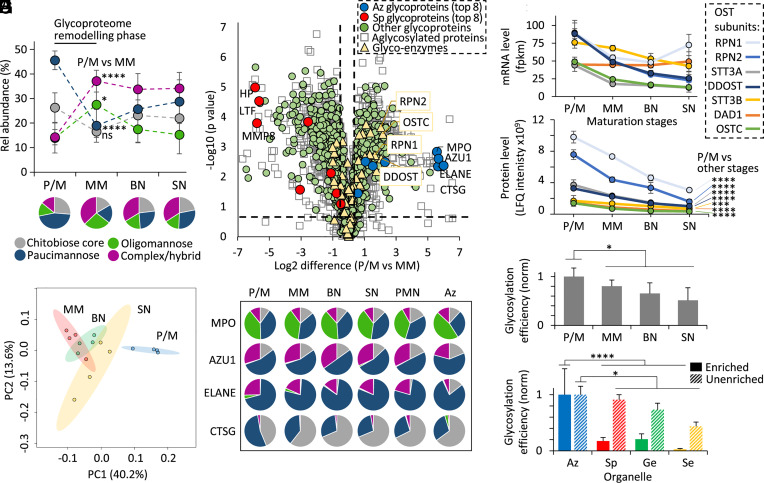
Fig. 4.
Dramatic glycoproteome remodeling in early granulopoiesis is accompanied by changes in proteome expression and the glycosylation initiation machinery. (A) N-glycan class distribution during granulopoiesis as established by glycopeptide profiling from proteomics of key myeloid progenitor cell types (P/M, MM, BN, SN) (25). For this analysis, glycoPSM-based quantitation was performed using an approach involving subtraction of glycoproteins identified in the maturation stage(s) prior to the specific maturation stage being assessed (hence considering only newly expressed glycoproteins at each stage). (B) N-glycopeptide-informed PCA of the four populations of myeloid progenitors. (C) Differential transcript expression between P/M and MM progenitors. Permutation-based FDRs < 0.05 and S0 > 0.1 were calculated using Perseus. Dashed x/y lines represent significance thresholds. Proteins were assigned with organelle location/glycosylation status based on glycoproteomics data (Dataset S5). Key N-glycosylation enzymes (yellow) including OST subunits (boxed, yellow) and the eight most abundant Az (blue) and Sp (red) granule glycoproteins among other glycosylated (green) and nonglycosylated (white) granule proteins are highlighted. (D) Uniform N-glycosylation of key Az granule glycoproteins during granulopoiesis, in blood neutrophils (PMN) and in their principal compartment (“Az”) of resting neutrophils. For simplicity, multiple sites were combined for the glycoprofile of MPO, AZU1, and ELANE. (E) Transcript (mRNA) and (F) protein expression profile of the OST subunits during myeloid differentiation. Data plotted as mean ± SD (n = 4 biological replicates, ***P < 0.001, ****P < 0.0001, Student’s t tests, P/M vs. other maturation stages combined). Global protein N-glycosylation efficiency (G) during granulopoiesis and (H) across the corresponding neutrophil compartments mapped and compared separately for enriched (full bars) and unenriched (broken bars) glycoproteomics data. Data plotted as mean ± SD and normalized against P/M stage or Az granule (n = 4 biological replicates. *P < 0.05, ****P < 0.0001, Student’s t tests, P/M or Az vs. other maturation stages or other compartments combined). See Fig. 1 for key.
Early myeloid progenitors (P/M stage) indeed showed a distinctly different glycophenotype compared to progenitors in later maturation stages as illustrated by the separation of P/M cells by PCA, further supporting that strong N-glycoproteome remodeling accompanies early neutrophil maturation (Fig. 4B). We therefore used transcriptomics to monitor glycoprotein and glycoenzyme expression during the P/M-to-MM transition (Fig. 4C). Interestingly, the analysis revealed that prominent changes in the proteome expression as opposed to dramatic glycoenzyme regulation are main contributors to the N-glycoproteome remodeling. Expectedly, the Az granule proteins expressed at the P/M stage (e.g., MPO, AZU1, ELANE) and Sp granule proteins expressed at the MM stage (e.g., HP, LTF, MMP8) showed strong and opposite expression differences (up to 5 to 30-fold) between the two stages. In fact, ~75% of the transcriptome dataset (2,579/3,484 transcripts) demonstrated expression differences between the P/M and MM stages (Student’s t test, FDR < 0.05, Dataset S8). Supporting these findings, a correlation was observed between the glycoprofile of the P/M stage of myeloid cells undergoing granulopoiesis and the glycoprofile of the Az granules (R = 0.865), suggesting that the glycosylation characteristics of the granules already form during granulopoiesis in the bone marrow as opposed to after the genesis of the neutrophil granules during storage and circulation (SI Appendix, Fig. S7) in line with previous findings (17). However, to unequivocally confirm this relationship, one would need to map the glycosylation of the cytosolic compartments isolated from maturing bone marrow myeloid cells, which was out of the scope of this study.
In line with the prominent protein expression differences and comparably minor changes to the glycosylation machinery during granulopoiesis, the glycosylation of key proteins enriched in the Az granules but found throughout all neutrophil compartments (MPO, AZU1, ELANE, CTSG) remained relatively unchanged during neutrophil maturation (Fig. 4D and Dataset S6). Taken together, these results indicate that the organelle-specific N-glycosylation in resting neutrophils is driven primarily by glycoprotein repertoire changes during granulopoiesis as opposed to prominent global changes in the cellular glycosylation machinery.
As a notable exception, the global expression analysis indicated altered P/M-to-MM expression of the oligosaccharyltransferase (OST) enzyme complex responsible for the initiation of N-glycoprotein biosynthesis. We therefore used already mentioned -omics resources (24, 25) to monitor the transcript- and protein-level expression profile of the subunits forming the OST enzyme complex (STT3A/B, RPN1/2, DDOST, DAD1, OSTC) during granulopoiesis and found considerably reduced OST mRNA (Fig. 4E) and protein expression (all P < 0.001, Student’s t test, P/M vs. other maturation stages combined) (Fig. 4F) over the course of neutrophil development. Correlating with the observed OST dynamics, proteins expressed at the P/M stage and those identified in the corresponding Az granules displayed a higher glycosylation efficiency compared to proteins expressed later during differentiation and those residing in other corresponding granules (all P < 0.05, Student’s t tests, P/M or Az vs. other maturation stages or other organelles combined) (Fig. 4 G and H). Strengthening further the notion of high glycosylation efficiency of the Az granule proteins formed early during granulopoiesis, the LC-MS/MS data of the Az granules and the P/M myeloid cells featured higher levels of glycopeptide signature ions relative to other granules and maturation stages, respectively (both P < 0.05, Student’s t tests, Az or P/M vs. other granules or other maturation stages combined) (SI Appendix, Figs. S5 and S8). Collectively, the temporal profiling of the N-glycoproteome and the N-glycosylation machinery during myeloid differentiation has revealed that profound glycoproteome remodeling accompanies early granulopoiesis, which provides important system-wide molecular-level clues to mechanisms driving the organelle-specific N-glycosylation observed in circulating blood neutrophils.
Discussion
Neutrophils store cytosolic granules and vesicles packed with highly glycosylated and potent hydrolytic enzymes, membrane receptors, and microbicidal glycoproteins that can be mobilized in a timely and coordinated manner to allow neutrophils to traffic to sites of infection and fight invading pathogens (31). While the literature has documented diverse roles for protein N-glycosylation in neutrophil function (4), the structural diversity and spatial distribution of the N-glycoproteome across the neutrophil compartments and the temporal dynamics during the profound metamorphosis underpinning neutrophil granulopoiesis remain unmapped. Closing this knowledge gap is important since while under normal physiology and homeostasis, neutrophils play protective roles throughout the body, it is becoming increasingly apparent that neutrophils, under certain conditions, play disease-promoting roles upon dysregulation of processes directly or indirectly involving protein glycosylation including impaired neutrophil maturation, recruitment, activation, and turnover (4, 32, 33).
In this study, we employed an integrated glycomics-assisted glycoproteomics method (22, 23, 28) to comprehensively profile the N-glycoproteome of four key intracellular compartments from resting neutrophils (Az, Sp, Ge, and Se). The initial N-glycome profiling facilitated a quantitative view of the glycan fine structures and aided the informative [yet challenging (29, 30, 34)] N-glycoproteomics data analysis enabling us to establish with precision the, to date, most detailed quantitative map of the heterogeneous N-glycoproteome across the neutrophil compartments. Further, mining of N-glycopeptide and glycoenzyme expression patterns using robust proteomics (25) and transcriptomics (24) resources obtained from discrete myeloid progenitor populations unveiled, with temporal resolution, the dynamic proteome expression and non-template-driven glycosylation processes accompanying neutrophil granulopoiesis.
Expanding significantly on previous efforts aiming to globally map neutrophil N-glycosylation (12–14, 35), we here provide direct evidence for the distinctive glycophenotypes exhibited by the neutrophil compartments and report on site-specific N-glycan features and carrier protein repertoires contributing to the intriguing organelle-specific N-glycosylation observed in blood neutrophils.
The Az granules, which consistently displayed the most distinctive N-glycosylation signatures, exhibited elevated levels of unconventional paucimannosidic-, chitobiose core-, and monoantennary complex-type N-glycans as well as low α2,6-sialylation and Lewis fucosylation. Recapitulating observations from structure–function studies of key neutrophil glycoproteins (12, 15, 17, 18), these unusual N-glycosylation features were found to be carried by MPO, ELANE, and CTSG among ~100 other less studied Az granule glycoproteins. The identification of peculiar N-glycosylation in the Az granules is important as this compartment is regarded as the microbicidal granule that neutrophils mobilize both intra- and extracellularly when combating pathogens at infection sites (3, 31). While the observation that unconventional N-glycans decorate a diverse repertoire of potent Az granule proteins is intriguing, their role(s) in shaping the microbicidal potential of the Az granules remains largely unstudied (4, 20). Our report of a bacteriostatic potential of the paucimannosidic N-glycans carried by ELANE toward clinical strains of Pseudomonas aeruginosa is, to our knowledge, the only literature on this topic (15), but we anticipate that future efforts will seek to uncover the biological roles that paucimannosylation and other truncated N-glycans play in neutrophil defense and communication.
In addition to phagosomal fusion, a subset of the Az granules degranulate their content into the extracellular environment upon neutrophil activation (36). Thus, there is also the potential for the unconventional glycosylation of the Az granule proteins to mediate cell–cell communication through putative signaling receptors toward paucimannose (19) and induce production of autoantibodies in neutrophil-related autoimmune diseases, such as antineutrophil cytoplasmic autoantibody–associated vasculitis (18). Supporting the former, we and others have demonstrated that mannan-binding lectin (MBL2) (12) and the macrophage mannose receptor (CD206) (37) display affinities to paucimannosidic glycoepitopes, but lectin receptors with high affinity/avidity to paucimannose and any cell–cell communication and other functional consequences resulting from their interaction require further exploration. Since neutrophil-derived MPO reportedly is internalized via endocytosis by CD206 on macrophage cell surfaces (38), paucimannosylation may proposedly also function as danger-associated molecular patterns (39) or alternatively serve as “removal tags” of the potent (thus potentially harmful) Az granule glycoproteins to prevent damage to host tissues following excessive release and pathogen killing.
Our finding that biologically relevant glycoepitopes (sLe/Le) decorate a considerable subset of the ~140 glycoproteins, e.g., LTF, MMP8, and LRG1 identified in the Sp granules is in line with (but considerably expands on) the existing literature (14, 40, 41). Lewis-type epitopes are known to mediate neutrophil tethering and rolling, and, in turn, facilitate extravasation to inflammatory sites through interactions with endothelial selectins (42). However, the literature points primarily to membrane-embedded glycoproteins residing in readily mobile secretory vesicles, e.g., PSGL-1, ESL-1, and CD44 being responsible for this trafficking process (4). Our study reveals that many luminal Sp granule glycoproteins also carry s/Le glycoepitopes. The extracellular roles of these glycoproteins remain essentially unstudied apart from a few reports suggesting that Lex on luminal Sp granule glycoproteins (e.g., LTF) mediates binding to the scavenger receptor C-type lectin (SRCL) expressed on endothelial cells to facilitate internalization and clearance of such bioactive (and potentially harmful) glycoproteins (40, 43).
Finally, the Ge granules and Se vesicles each hosted ~200 N-glycoproteins carrying predominantly oligomannosidic- and less sialylated/fucosylated N-glycans. While traditionally regarded as an intracellular glycofeature pertaining to incompletely processed N-glycoproteins trafficking the secretory machinery, oligomannosylation is increasingly reported i) on mature (fully processed) neutrophil glycoproteins (4, 19), ii) to decorate the surface of neutrophils (44) and other blood (45) as well as epithelial (46, 47) cells, and iii) to be elevated in cancer (48, 49) and other neutrophil-related diseases (50). Our study supports that oligomannosylation is a prominent feature of the Ge and Se compartments. This is of potential significance since neutrophils readily expose, in a context-dependent manner, the granule content from these highly mobile compartments to the extracellular environment to enable (possibly through the oligomannosidic glycoepitopes) communication with surrounding tissues or removal from circulation (4, 19).
Excitingly, our investigation of myeloid progenitors across different maturation stages revealed considerable plasticity in key N-glycosylation processes and detailed a global remodeling of the N-glycoproteome during neutrophil granulopoiesis. Particularly dramatic N-glycoproteome remodeling was found to accompany early myeloid maturation (P/M-to-MM transition). Notably, the Az and Sp granules formed during the P/M and MM differentiation stages, respectively, exhibited striking glycoproteome differences providing important mechanistic clues to the formation of the distinct glycoproteome characteristics found across these two granule populations. The P/M-to-MM transition is important as it marks the change from self-replicating promyelocytes to nondividing and committed metamyelocytes, a transition that is accompanied by notable phenotypic (morphological, molecular, functional) alterations (25). This study demonstrates glycoproteome remodeling in developing myeloid progenitor cells. In fact, maturation stage–specific glycosylation appears scarcely investigated with recently available and highly informative glycoproteomics approaches as used herein. Examples from the glycobiological literature include our own report on dynamic glycosylation underpinning myogenesis and muscle development (51), and N-glycoproteome changes during neuronal (52), and cardiomyocyte (53) differentiation reported by other groups.
Importantly, our multiomics interrogations established that the N-glycoproteome remodeling occurring during granulopoiesis is driven primarily by temporal changes in the proteome expression patterns leading to different glycoprotein repertoires across the individual neutrophil organelles. Less prominent were changes within the glycosylation machinery responsible for the enzymatic processing of nascent N-glycoproteins trafficking the secretory pathway. This observation is important as it illustrates alternative factors (other than the glycosylation machinery) mediating glycoproteome remodeling in neutrophils and serves to explain how some N-glycoproteins expressed throughout granulopoiesis (e.g., MPO, ELANE) can receive relatively uniform glycosylation regardless of the timing of their expression and their organelle location (Fig. 4D). To this end, both the protein repertoire and the glycosylation machinery can be considered determinants for the distinct glycophenotypes of the neutrophil compartments. However, beyond relatively simple solvent accessibility><glycan processing relationships (54, 55), a molecular-level understanding of how neutrophil proteins carry information that enables their individualized modification patterns across the neutrophil compartments is still lacking. Finally, we acknowledge that other dynamic factors such as Golgi restructuring, altered microtrafficking routes, and changes to nonenzymatic components of the glycosylation machinery (i.e., nucleotide-sugar levels and transporters), all of which were not explored in this study, may also accompany granulopoiesis consequently adding another level of complexity to consider when attempting to decode mechanisms governing the dynamics of the neutrophil glycoproteome.
As a notable exception to the relatively stable glycosylation machinery during neutrophil granulopoiesis, the expression of the OST enzyme complex responsible for the initiation of N-glycoprotein biosynthesis was significantly reduced during neutrophil maturation, which correlated with a relatively high glycosylation efficiency of proteins in the Az granules relative to other compartments (Fig. 4 E–H and SI Appendix, Figs. S5 and S8). We are not aware of similar reports of OST dynamics during the maturation of neutrophils (or other cell types), but a growing body of evidence points to context-dependent OST regulation and interesting links to cancer, immune evasion, and ER stress (56, 57). It is tempting to speculate that the Az granule proteins require a high level of glycosylation to prevent premature proteolytic degradation during the extended storage in these hydrolytic compartments hosting many potent proteases (e.g., ELANE, CTSG). Although energetically expensive, the installation of voluminous and flexible glycans on protein surfaces is, in fact, shown to confer effective protection against proteolysis (58, 59).
In steady-state conditions, neutrophils form from hematopoietic stem and progenitor cells in the bone marrow (1). However, during systemic infection and inflammation involving a prolonged demand for neutrophils (e.g., cancer and chronic infections), the hematopoietic system switches from steady-state to “emergency granulopoiesis” (60, 61). This so-called “left-shift” results in the appearance of neutrophil precursors in circulation featuring immature phenotypes including aberrant (progenitor-like) morphology and, presumably, an incomplete repertoire of granules and/or granule content. Considering our identification of maturation stage-specific glycosylation, we hypothesize that immature neutrophils arising from emergency granulopoiesis display an “immature” (not fully remodeled) N-glycoproteome (4). Impaired granulopoiesis is also central to acute myeloid leukemia (AML) (62), in which morphologically immature blood neutrophils, expectedly exhibiting aberrant glycosylation, were found to show decreased capacity for NET formation (63). The link (if any) between poor NET formation and aberrant glycosylation of AML neutrophils remains unexplored. Guided by our high-precision N-glycoproteome map of healthy neutrophils, future glycoprofiling studies of immature neutrophils from individuals exhibiting emergency granulopoiesis and AML patients are now well positioned to substantiate these speculations and establish causal links to neutrophil dysfunction.
In conclusion, our detailed N-glycoproteome profiling of developing myeloid progenitors and mature neutrophils has painted the, to-date, most comprehensive picture of the intriguingly complex neutrophil N-glycoproteome notably with spatiotemporal information and has laid an important foundation to explain its formation. Our systems glycobiology (multiomics) approach revealed that strong N-glycoproteome remodeling underpins neutrophil granulopoiesis, which, when considered with the widely accepted “targeting-by-timing” model (6, 7), provides mechanistic clues to the distinctive glycophenotypes exhibited by the four intracellular compartments of neutrophils as detailed herein. Importantly, our data-rich omics-centric study serves as a resource to guide future explorations targeting the biological roles and dysregulation of neutrophil glycosylation that holds an untapped potential for the discovery of glycoprotein-related markers for and therapeutics against a spectrum of neutrophil-related disorders.
Materials and Methods
Donors, Neutrophil Isolation, and Organelle Separation.
Neutrophils were isolated from buffy coats from healthy donors at The Blood Center, Sahlgrenska University Hospital, Gothenburg, Sweden. According to Swedish law on ethical conduct in human research, ethics approval was not needed because the buffy coats were provided anonymously. Resting neutrophils were isolated to >95% purity in four biological replicates each comprising pooled buffy coats from four individuals as described (14). Neutrophil granules and vesicles were separated using an established three-layered Percoll method (26), monitored using established immunoblot and enzyme activity assays (14), and validated using proteomics. The isolated organelles were lysed and the luminal (soluble) protein extract collected after ultracentrifugation. See Extended methods for details of all experimental procedures.
Glycome Profiling.
N-glycans were prepared for glycomics as described (28). Briefly, N-glycans were released using N-glycosidase F, hydroxylated, and reduced prior to N-glycan desalting. The N-glycans were profiled in negative polarity mode on a Velos Pro linear ion trap connected to a Dionex Ultimate-3000 HPLC (Thermo Fisher Scientific) using an established PGC-LC-MS/MS method (14). GlycoMod and GlycoWorkBench v2.1 (64) assisted the otherwise manual de novo glycan annotation as described (65). Briefly, N-glycans were identified based on molecular mass, MS/MS fragmentation pattern, and relative and absolute PGC-LC retention time. AUC-based glycan quantification was performed using Skyline v.19.1 (66, 67).
Glycoproteome Profiling.
Proteins were precipitated, reduced, alkylated, and digested using trypsin. Peptides were desalted using C18-SPE and either analyzed directly by LC-MS/MS or enriched for glycopeptides using hydrophilic interaction liquid chromatography SPE as described (15).
Unenriched peptides were analyzed in positive ion polarity using a Q-Exactive HF-X Hybrid Quadrupole-Orbitrap coupled to an Easy nLC-1200 HPLC (Thermo Scientific). Peptides were separated using reversed-phase C18 chromatography and fragmented using higher-energy collisional dissociation (HCD)-MS/MS.
Enriched glycopeptides were analyzed in positive ion polarity using an Orbitrap Fusion coupled to a Dionex 3500RS HPLC (Thermo Scientific). Glycopeptides were separated using reversed-phase C18 chromatography and fragmented using HCD-, ETD-, and CID-MS/MS.
Employing the Andromeda search engine and the generated HCD-MS/MS data, MaxQuant v1.6.10.43 (68) was used for protein identification and quantification. Identifications were filtered to <1% protein FDR. Label-free AUC-based quantification was performed, and protein abundance was calculated based on the normalized protein intensity (LFQ intensity) (69).
Glycopeptides were identified and quantified from the HCD-MS/MS data using Byonic v4.5.2 (Protein Metrics) in Proteome Discoverer v.2.5. Glycopeptide identifications were filtered to FDR <1%. CID- and ETD-MS/MS data were used to manually confirm select glycopeptides. (Glyco)peptides were quantified based on AUC of precursor ions using the Minora Feature Detector or glycoPSMs as indicated.
Interrogation of Available Proteomics and Transcriptomics Resources.
Proteomics data of developing and mature neutrophils (PXD013785) (25) were interrogated for intact glycopeptides (see above). Matching transcriptomics data (https://blueprint.haem.cam.ac.uk/neutrodiff) (24) were interrogated for glycoenzymes (e.g., OST subunits) and granule protein expression patterns.
Statistical Analyses.
Significance was assessed using unpaired two-tailed Student's t tests or ANOVA followed by the Tukey test for multiple comparison with FDR < 0.05. For transcriptomics and glycomics data analyses, significance was corrected for multiple test comparisons. P < 0.05 was considered significant. GraphPad Prism v9.4.1 (Dotmatics) and Perseus v2.0.7.0 (70) were used for statistical analyses. Correlation analyses were performed using the Pearson correlation coefficient using Microsoft Excel. PCA was performed using Metaboanalyst v. 5.0 (71).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (PDF)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Acknowledgments
We thank Dr. Nathan Croft and Joshua Ferring for assistance with glycopeptide data mining. R.K. was supported by the Cancer Institute of New South Wales (ECF181259). J.U. was supported by a Macquarie Research Excellence Scholarship. S.C. was supported by an International Macquarie Research Excellence Scholarship (iMQRES 2017152). I.L. was supported by an International Research Training Program Scholarship funded by the Australian Government. H.C.T. was supported by a Macquarie University COVID-19 fellowship. Z.S.-B. was supported by a JSPS fellowship. J.B. was supported by grants from the Swedish Research Council (2019-01123) and TUA Research Funding; The Sahlgrenska Academy at University of Gothenburg/Region Västra Götaland, Sweden (TUAGBG-917531). A.K.-B. is supported by the Swedish Research Council (2018-03077). M.T.-A. is supported by an Australian Research Council Future Fellowship (FT210100455).
Author contributions
R.K., J.U., I.L., and M.T.-A. designed research; R.K., S.C., H.C.T., I.L., B.L.P., and V.V. performed research; R.K., B.L.P., V.V., R.D., Z.S.-B., A.K.-B., J.B., and M.T.-A. contributed new reagents/analytic tools; R.K., J.U., S.C., H.C.T., I.L., and M.T.-A. analyzed data; and R.K., J.U., and M.T.-A. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Preprint server: https://www.biorxiv.org/content/10.1101/2023.01.18.524318v2.
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All LC-MS/MS data and metadata have been deposited in public repositories. Specifically, glyco/proteomics LC-MS/MS data were deposited to PRIDE (PXD039387 and PXD021131) (72, 73). Glycomics LC-MS/MS raw data were deposited to GlycoPOST (GPST000315) (74). Previously published data were used for this work (PRIDE: PXD013785 and https://blueprint.haem.cam.ac.uk/neutrodiff) (75, 76).
Supporting Information
References
- 1.Liew P. X., Kubes P., The Neutrophil’s role during health and disease. Physiol. Rev. 99, 1223–1248 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Rosales C., Neutrophil: A cell with many roles in inflammation or several cell types? Front. Physiol. 9, 113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowland J. B., Borregaard N., Granulopoiesis and granules of human neutrophils. Immunol. Rev. 273, 11–28 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ugonotti J., Chatterjee S., Thaysen-Andersen M., Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol. Aspects Med. 79, 100882 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Borregaard N., Cowland J. B., Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 89, 3503–3521 (1997). [PubMed] [Google Scholar]
- 6.Le Cabec V., Cowland J. B., Calafat J., Borregaard N., Targeting of proteins to granule subsets is determined by timing and not by sorting: The specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc. Natl. Acad. Sci. U.S.A. 93, 6454–6457 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rorvig S., Ostergaard O., Heegaard N. H., Borregaard N., Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: Correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 94, 711–721 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Babu P., et al. , Structural characterisation of neutrophil glycans by ultra sensitive mass spectrometric glycomics methodology. Glycoconj. J. 26, 975–986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foxall C., et al. , The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewis(x) oligosaccharide. J. Cell Biol. 117, 895–902 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimrichter L., et al. , E-selectin receptors on human leukocytes. Blood 112, 3744–3752 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatakrishnan V., Thaysen-Andersen M., Chen S. C., Nevalainen H., Packer N. H., Cystic fibrosis and bacterial colonization define the sputum N-glycosylation phenotype. Glycobiology 25, 88–100 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Thaysen-Andersen M., et al. , Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290, 8789–8802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiding K. R., Lin Y. H., van Alphen F. P. J., Meijer A. B., Heck A. J. R., Neutrophil azurophilic granule glycoproteins are distinctively decorated by atypical pauci- and phosphomannose glycans. Commun. Biol. 4, 1012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatakrishnan V., et al. , Glycan analysis of human neutrophil granules implicates a maturation-dependent glycosylation machinery. J. Biol. Chem. 295, 12648–12660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loke I., Ostergaard O., Heegaard N. H. H., Packer N. H., Thaysen-Andersen M., Paucimannose-Rich N-glycosylation of Spatiotemporally Regulated Human Neutrophil Elastase Modulates Its Immune Functions. Mol. Cell Proteomics 16, 1507–1527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loke I., Packer N. H., Thaysen-Andersen M., Complementary LC-MS/MS-based N-Glycan, N-Glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-Glycosylation of human neutrophil cathepsin G. Biomolecules 5, 1832–1854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tjondro H. C., et al. , Hyper-truncated Asn355- and Asn391-glycans modulate the activity of neutrophil granule myeloperoxidase. J. Biol. Chem. 296, 100144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiding K. R., et al. , Neutrophil myeloperoxidase harbors distinct site-specific peculiarities in its glycosylation. J. Biol. Chem. 294, 20233–20245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loke I., Kolarich D., Packer N. H., Thaysen-Andersen M., Emerging roles of protein mannosylation in inflammation and infection. Mol. Aspects Med. 51, 31–55 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Tjondro H. C., Loke I., Chatterjee S., Thaysen-Andersen M., Human protein paucimannosylation: Cues from the eukaryotic kingdoms. Biol. Rev. Camb Philos Soc. 94, 2068–2100 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Parker B. L., et al. , Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol. Cell Proteomics 15, 141–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara R., et al. , The complexity and dynamics of the tissue glycoproteome associated with prostate cancer progression. Mol. Cell Proteomics 20, 100026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnielli C. M., et al. , Comprehensive glycoprofiling of oral tumours associates N-glycosylation with lymph node metastasis and patient survival. Mol. Cell Proteomics 22, 100586 (2023), 10.1016/j.mcpro.2023.100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grassi L., et al. , Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoogendijk A. J., et al. , Dynamic transcriptome-proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. Cell Rep. 29, 2505–2519.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Borregaard N., Heiple J. M., Simons E. R., Clark R. A., Subcellular localization of the b-cytochrome component of the human neutrophil microbicidal oxidase: Translocation during activation. J. Cell Biol. 97, 52–61 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moh E. S., Thaysen-Andersen M., Packer N. H., Relative versus absolute quantitation in disease glycomics. Proteomics Clin. Appl. 9, 368–382 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Chau T. H., et al. , Glycomics-assisted glycoproteomics enables deep and unbiased N-glycoproteome profiling of complex biological specimens. Methods Mol. Biol. 2628, 235–263 (2023). [DOI] [PubMed] [Google Scholar]
- 29.Kawahara R., et al. , Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies for serum glycopeptide analysis. Nat. Methods 18, 1304–1316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chau T. H., Chernykh A., Kawahara R., Thaysen-Andersen M., Critical considerations in N-glycoproteomics. Curr. Opin. Chem. Biol. 73, 102272 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Rosales C., Neutrophils at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 108, 377–396 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Kaplan M. J., Role of neutrophils in systemic autoimmune diseases. Arthritis Res. Ther. 15, 219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonego F., et al. , Paradoxical roles of the neutrophil in sepsis: protective and deleterious. Front. Immunol. 7, 155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polasky D. A., Nesvizhskii A. I., Recent advances in computational algorithms and software for large-scale glycoproteomics. Curr. Opin. Chem. Biol. 72, 102238 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee S., et al. , Oxonium ion-guided optimization of ion mobility-assisted glycoproteomics on the timsTOF Pro. Mol. Cell Proteomics 22, 100486 (2022). 10.1016/j.mcpro.2022.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia R. C., Peterson C. G., Segal A. W., Venge P., Elastase in the different primary granules of the human neutrophil. Biochem Biophys Res Commun 132, 1130–1136 (1985). [DOI] [PubMed] [Google Scholar]
- 37.Stavenhagen K., et al. , Tumor cells express pauci- and oligomannosidic N-glycans in glycoproteins recognized by the mannose receptor (CD206). Cell Mol. Life Sci. 78, 5569–5585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepherd V. L., Hoidal J. R., Clearance of neutrophil-derived myeloperoxidase by the macrophage mannose receptor. Am. J. Respir Cell Mol. Biol. 2, 335–340 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Gong T., Liu L., Jiang W., Zhou R., DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20, 95–112 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Graham S. A., et al. , Identification of neutrophil granule glycoproteins as Lewis(x)-containing ligands cleared by the scavenger receptor C-type lectin. J. Biol. Chem. 286, 24336–24349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudd P. M., et al. , Glycosylation of natural human neutrophil gelatinase B and neutrophil gelatinase B-associated lipocalin. Biochemistry 38, 13937–13950 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Wright R. D., Cooper D., Glycobiology of leukocyte trafficking in inflammation. Glycobiology 24, 1242–1251 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Taylor M. E., Drickamer K., Mammalian sugar-binding receptors: Known functions and unexplored roles. FEBS J. 286, 1800–1814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinbaum D. L., Sullivan J. A., Mandell G. L., Receptors for concanavalin A cluster at the front of polarized neutrophils. Nature 286, 725–727 (1980). [DOI] [PubMed] [Google Scholar]
- 45.Cao H., et al. , Measurement of erythrocyte membrane mannoses to assess splenic function. Br. J. Haematol. 198, 155–164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park D., et al. , Characteristic changes in cell surface glycosylation accompany intestinal epithelial cell (IEC) differentiation: High mannose structures dominate the cell surface glycome of undifferentiated enterocytes. Mol. Cell Proteomics 14, 2910–2921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heindel D. W., et al. , Glycomic analysis of host response reveals high mannose as a key mediator of influenza severity. Proc. Natl. Acad. Sci. U.S.A. 117, 26926–26935 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee S., et al. , Trends in oligomannosylation and alpha1,2-mannosidase expression in human cancers. Oncotarget 12, 2188–2205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park D. D., et al. , Metastasis of cholangiocarcinoma is promoted by extended high-mannose glycans. Proc. Natl. Acad. Sci. U.S.A. 117, 7633–7644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kissel T., Toes R. E. M., Huizinga T. W. J., Wuhrer M., Glycobiology of rheumatic diseases. Nat. Rev. Rheumatol. 19, 28–43 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blazev R., et al. , Integrated glycoproteomics identifies a role of N-glycosylation and galectin-1 on myogenesis and muscle development. Mol. Cell Proteomics 20, 100030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura K., et al. , Glycoproteomic analysis of the changes in protein N-glycosylation during neuronal differentiation in human-induced pluripotent stem cells and derived neuronal cells. Sci. Rep. 11, 11169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashwood C., Waas M., Weerasekera R., Gundry R. L., Reference glycan structure libraries of primary human cardiomyocytes and pluripotent stem cell-derived cardiomyocytes reveal cell-type and culture stage-specific glycan phenotypes. J. Mol. Cell Cardiol. 139, 33–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thaysen-Andersen M., Packer N. H., Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lee L. Y., Lin C. H., Fanayan S., Packer N. H., Thaysen-Andersen M., Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harada Y., Ohkawa Y., Kizuka Y., Taniguchi N., Oligosaccharyltransferase: A gatekeeper of health and tumor progression. Int. J. Mol. Sci. 20, 6074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevens K. L. P., et al. , Diminished Ost3-dependent N-glycosylation of the BiP nucleotide exchange factor Sil1 is an adaptive response to reductive ER stress. Proc. Natl. Acad. Sci. U.S.A. 114, 12489–12494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Q., et al. , Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife 9, e61552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.King S. L., et al. , TAILS N-terminomics and proteomics reveal complex regulation of proteolytic cleavage by O-glycosylation. J. Biol. Chem. 293, 7629–7644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malengier-Devlies B., Metzemaekers M., Wouters C., Proost P., Matthys P., Neutrophil homeostasis and emergency granulopoiesis: The example of systemic juvenile idiopathic arthritis. Front. Immunol. 12, 766620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manz M. G., Boettcher S., Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Mackey J. B. G., Coffelt S. B., Carlin L. M., Neutrophil maturity in cancer. Front. Immunol. 10, 1912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lukasova E., et al. , Granulocyte maturation determines ability to release chromatin NETs and loss of DNA damage response; these properties are absent in immature AML granulocytes. Biochim. Biophys. Acta 1833, 767–779 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Ceroni A., et al. , GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome. Res. 7, 1650–1659 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Everest-Dass A. V., Kolarich D., Campbell M. P., Packer N. H., Tandem mass spectra of glycan substructures enable the multistage mass spectrometric identification of determinants on oligosaccharides. Rapid. Commun. Mass. Spectrom. 27, 931–939 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Adams K. J., et al. , Skyline for small molecules: A unifying software package for quantitative metabolomics. J. Proteome Res. 19, 1447–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashwood C., Lin C. H., Thaysen-Andersen M., Packer N. H., Discrimination of isomers of released N- and O-glycans using diagnostic product ions in negative ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass Spectrom. 29, 1194–1209 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Cox J., et al. , Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Xia J., Psychogios N., Young N., Wishart D. S., MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawahara R., Thaysen-Andersen M., Spatiotemporal glycoproteome remodelling in neutrophils. PRIDE - Proteomics Identification Database. https://www.ebi.ac.uk/pride/archive/projects/PXD039387. Deposited 13 January 2023.
- 73.Kawahara R., Thaysen-Andersen M., The bimodal sugar code of neutrophil myeloperoxidase. PRIDE - Proteomics Identification Database. https://www.ebi.ac.uk/pride/archive/projects/PXD021131. Deposited 26 August 2020.
- 74.Kawahara R., Thaysen-Andersen M., Glycoproteome remodelling and organelle-specific N-glycosylation accompany myeloid progenitor-to-neutrophil metamorphosis. GlycoPOST. https://glycopost.glycosmos.org/entry/GPST000315. Deposited 9 January 2023.
- 75.van den Biggelaar M., Dynamic transcriptome-proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. PRIDE - Proteomics Identification Database. https://www.ebi.ac.uk/pride/archive/projects/PXD013785. Deposited 9 May 2019. [DOI] [PubMed]
- 76.van den Biggelaar M., Epigenomic and functional dynamics of human bone marrow myeloid differentiation to mature blood neutrophils. BLUEPRINT-EPIGENOME. https://blueprint.haem.cam.ac.uk/neutrodiff. Deposited 5 April 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (PDF)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Data Availability Statement
All LC-MS/MS data and metadata have been deposited in public repositories. Specifically, glyco/proteomics LC-MS/MS data were deposited to PRIDE (PXD039387 and PXD021131) (72, 73). Glycomics LC-MS/MS raw data were deposited to GlycoPOST (GPST000315) (74). Previously published data were used for this work (PRIDE: PXD013785 and https://blueprint.haem.cam.ac.uk/neutrodiff) (75, 76).