Abstract
Human cytomegalovirus (HCMV) primary infections are typically asymptomatic in healthy individuals yet can cause increased morbidity and mortality in organ transplant recipients, AIDS patients, neonates, and the elderly. The successful, widespread dissemination of this virus among the population can be attributed in part to its wide cellular tropism and ability to establish life-long latency. HCMV infection is a multi-step process that requires numerous cellular and viral factors. The viral envelope consists of envelope protein complexes that interact with cellular factors; such interactions dictate virus recognition and attachment to different cell types, followed by fusion either at the cell membrane or within an endocytic vesicle. Several HCMV entry factors, including neuropilin-2 (Nrp2), THBD, CD147, OR14I1, and CD46, are characterized as participating in HCMV pentamer-specific entry of non-fibroblast cells such as epithelial, trophoblast, and endothelial cells, respectively. This study focuses on characterizing the structural elements of CD46 that impact HCMV infection. Infectivity studies of wild-type and CD46 knockout epithelial cells demonstrated that levels of CD46 expressed on the cell surface were directly related to HCMV infectivity. Overexpression of CD46 isomers BC1, BC2, and C2 enhanced infection. Further, CD46 knockout epithelial cells expressing CD46 deletion and chimeric molecules identified that the intact ectodomain was critical for rescue of HCMV infection in CD46 knockout cells. Collectively, these data support a model that the extracellular domain of CD46 participates in HCMV infection due to its surface expression.
Keywords: human cytomegalovirus, CD46, epithelial cells, fibroblasts, virus entry
Impact Statement
Human cytomegalovirus (HCMV) can infect numerous cell types through interaction of the viral envelope protein complexes with diverse cellular factors. The multi-step process of HCMV entry is a tightly regulated and orchestrated event involving numerous cellular factors that ensures efficient entry and infection. The present study further characterizes the role of CD46 as an HCMV entry factor. Further, the surface expression levels of CD46 correlate linearly with HCMV infectivity in epithelial cells and is dependent on the CD46 ectodomain. Collectively, these findings imply that CD46 may function by directly interacting with the pentamer itself or with a complex consisting of viral and cellular factors to enhance virus infection. These findings suggest that HCMV infection utilizes multiple cellular factors for efficient entry into diverse cell types.
Introduction
Human cytomegalovirus (HCMV) is a β-herpesvirus that increases morbidity and mortality in immunocompromised individuals [1]. HCMV establishes latency following primary infection, which is typically asymptomatic in a healthy person. The major populations at high risk for HCMV-associated diseases are unborn children and transplant recipients. HCMV disease can manifest as neuronal defects in infants, and in fact is the leading infectious cause of birth defects affecting newborns worldwide. Further, HCMV causes gastrointestinal disorders, pneumonia, HCMV syndrome, and end-organ disease in transplant recipients [2, 3]. Primary HCMV infection of epithelial cells leads to viral propagation and an inflammatory response that recruits immune cells, including monocytes. These monocytes, when infected, disseminate virus to hematopoietic progenitor and myeloid cells (i.e. CD14+ monocytes) in the bone marrow and vascular endothelium. These cells can function as latent viral reservoirs maintaining viral genomes in the absence of virus replication [4]. Viral reactivation from latent reservoirs sustains viral persistence and fuels both dissemination [5–8] and immune-mediated injury [9–12].
HCMV is a successful pathogen due to its ability to infect and proliferate in diverse cell types of the nervous system, placenta, kidney, and lungs [2]. HCMV entry is exceedingly complex, requiring initial attachment to the cell surface followed by cell surface receptor clustering, likely through cholesterol-rich membrane microdomains, and then viral fusion with the membrane bilayer and capsid release in the cytosol [13]. The fusion step of virus entry may occur through a pH-independent process at the cell surface of fibroblasts, or a pH-dependent process within the endosome of non-fibroblast cells, including epithelial and endothelial cells [14, 15]. HCMV envelope proteins and complexes gM/gN, gB, and gH/gL/gO (referred to as the ‘trimer’) interact with membrane proteins in fibroblasts such as integrins, the platelet-derived growth factor-α receptor (PDGFRα), the epidermal growth factor receptor (EGFR), and/or CD90/THY-1 mediating virus tethering and binding to the cell surface and post-binding viral gene expression [1, 16–21]. Interestingly, the gH/gL/UL128/UL130/UL131a complex (referred to as the ‘pentamer’) is required for entry into non-fibroblast cells. CD46, CD147, neuropilin-2 (Nrp-2), thrombomodulin (THBD), and OR14I1 are relevant for infection of epithelial cells, endothelial cells, and trophoblasts, respectively [22–26]. Importantly, the mechanism(s) and potential synergistic activity of these proposed entry factors for HCMV infection has yet to be fully elucidated; thus warranting a comprehensive molecular analysis of interactions between viral and host factors.
CD46 was identified as an entry factor for HCMV infection of epithelial and trophoblast cells, but not fibroblasts [25]. Modulation of CD46 function using anti-CD46 antibodies, siRNA, and CD46-/- knockout cells all demonstrated a significant and consistent decrease of HCMV infectivity. Further, limiting virus entry in CD46-/- cells significantly decreased virus proliferation supporting the model of CD46 playing a role in the early steps of the virus life cycle. CD46 is a type-I membrane glycoprotein expressed on all nucleated cells whose major function is a complement regulatory protein [27]. CD46 is referred to as a ‘pathogens’ magnet’ due to its involvement in both bacterial and viral infections including S. pyogenes , N. meningitidis , HHV-6a, and measles virus [28]. CD46 functions as a receptor for the vaccine strain of measles virus in Jurkat and Hela cells, adenovirus (groups B and D) infection of epithelial cells, and HHV-6A infection of T cells [29]. CD46 also mediates adhesion of Neisseria gonorrhoeae to human epithelial cells [30] and group A streptococcus (GAS) to keratinocytes [31]. This work explores the domains of CD46 that participate in HCMV infection. Viral infection was examined in ARPE-19 epithelial cells that overexpress different CD46 isomers and CD46 mutants ultimately identifying that the extracellular domain of CD46 BC1 plays a role in HCMV infection.
Methods
Cells and viruses
Normal human dermal fibroblast cells (NHDFs, ATCC #PSC-201–010) and human lung fibroblasts (MRC-5, ATCC #CCL-171) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning #10–013-CV) supplemented with 10 % fetal bovine serum (FBS), 1 mM HEPES (Corning #25–060 CI), 100 U ml−1 penicillin and 100 µg ml−1 streptomycin (Corning, #30–002 CI). ARPE-19 human retinal epithelial cells (ATCC #CRL-2302) were cultured in DMEM/F-12 media (1 : 1) (Gibco, #11765–054) supplemented with FBS, HEPES and pen/strep as with DMEM. The HCMV strains TB40/E (gift from Dr Christian Sinzger, Ulm U., Germany) and a repaired AD169 (BADrUL131-C4) containing the UL131 open reading frame of the HCMV strain TR and expressing EGFP (AD169R [32], gift from Dr Thomas Shenk, Princeton U.), were propagated and titered as described [33]. Briefly, HCMV virus was propagated in NHDF cells. The virus was isolated from infected cell supernatant and lysates following sonification and then further isolated by ultracentrifugation (20 000 r.p.m., 1.5 h, 25 °C) over a 20 % sorbitol cushion (Fisher Scientific, S459-500) using a SW-28 rotor (Beckman Coulter). Resulting virus was resuspended in 3 % bovine serum albumin (BSA) in PBS and stored at −80 °C. Purified virus was titered in ARPE-19 and NHDF cells to determine the infectious units per millilitre (IU ml−1) for each cell type.
The antibody PY102 (anti-IAV) (gift from Dr Thomas Moran, Icahn School of Medicine), anti-gH antibody 15G11 [34], and anti-CD46 antibody [25] were purified from hybridoma supernatants using an ÄKTA pure FPLC system on protein G affinity columns (HiTrap-1ml, GE /Cytiva, #17-0404-01). The antibodies were dialysed against PBS and quantitated by both BCA and OD at 280 nm. The polyclonal rabbit anti-IE1 antibody was generated by Genscript against the peptide sequence N’-KRKMDPDNPDEGPS-C’. The polyclonal anti-CD46 immunoglobulins were generated in rabbits against the CD46 peptide corresponding to aa. 36–140 [25]. The mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EMD Millipore, MAB374), donkey anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (Invitrogen, A16035) and donkey anti-mouse-HRP (Invitrogen, A16017) antibodies were commercially available. The secondary antibodies used for detection using fluorescence were goat anti-mouse AlexaFluor 647 (AF647) (Invitrogen, A21236) and chicken anti-rabbit AF647 (Invitrogen, A21443). Anti-CD46 antibody GB24 [35] was a gift from Dr J.P. Atkinson (Washington University).
Virus infectivity assay
HCMV infection of ARPE-19 or MRC5 cells (10 000 cells/well in a 96-well plate) was typically analysed at 24 h post-infection (hpi), beginning with fixation using 4 % paraformaldehyde for 30 min at room temperature [34]. Infected cells were then permeabilized with 0.3 % Triton X-100 in PBS, followed by incubation with 4 % normal goat serum (NGS) in 0.3 % Triton X-100 in PBS for 1 h at 4 °C. Cells were incubated with the polyclonal anti-IE1 antibody (2 mg ml−1, 1 h at 4 °C) and washed 2× in PBS, followed by incubation with the chicken anti-rabbit Ig conjugated with Alexa647 (1 : 1000) for 1 h at 4 °C and then 2× PBS washes. Cells were then stained with Hoechst 33 342 (1 mg ml−1, Molecular Probes) in PBS for 15 min at room temperature before analysis by the Celigo Imaging Cytometer (Nexcellom Bioscience). Infection (%) was determined relative to non-binding mAb control PY102. All samples were tested in technical replicates of three.
Flow cytometry analysis
Cells were untreated or treated with CytoFix/CytoPerm (BD, 51-2090KZ) as recommended by the manufacturer and then resuspended in 1 % BSA (Akron Biotech, AK8905-0100)/PBS for intact cells or 1 % BSA/0.1 % Saponin/PBS for fixed/permeabilized cells. Cells were incubated with primary antibody (1 µg ml−1, 1 h at 4 °C) followed by incubation with goat anti-mouse IgG conjugated to AF647 (1 : 500, 1 h at 4 °C). Fluorescent data was collected on either an Intellicyte HTFC or an Attune NxT flow cytometer. The data was analysed using FlowJo software and graphed using Prism 9.0 software.
Pre/post-attachment infectivity assay
MRC5 or ARPE-19 cells (10 000 cells/well in 96-well plates) were pre-incubated with PY102, 2E7, or 15G11 antibody (0.016, 0.08, 0.4, 2, 10, and 50 mg ml−1) for 30 min on ice before washing 2× with media, followed by infection with HCMV-AD169R (MOI=0.2) for 1 h at 37 °C. The cells were then washed 2× with media, and infection was analysed 24 h post-infection (hpi). For virus pre-incubated with antibodies, AD169R (amount of virus that yields a MOI 0.2) was pre-incubated with PY102, 2E7, 15G11 antibody (0.016–50 mg ml−1) at 30 min on ice and then added to MRC5 or ARPE-19 cells times for 1 h at 37 °C. The cells were then washed 2× with media, and infection was analysed at 24hpi based on anti-IE1 immunostaining of cells. Percent infection was determined using infection levels of cells pretreated with irrelevant influenza antibody PY102 as 100 % infection.
Generation of wt ARPE-19 and ARPE CD46-/- cells that express CD46 constructs
The ARPE CD46-/- cells were generated as described [25]. cDNAs of CD46 isomers BC1 (variant c), BC2 (variant d), and C1 (variant e) were purchased from Genscript (BC1 [#OHu16915], BC2 [Ohu16805], and C1 [OHu16908]) and cloned into the retrovirus vector pLHCX (Clontech) using in InFusion cloning (Takara Bio). The deletion mutants BC1D1 (1-35, 100-377), BC1D2 (1-96, 158-377), and BC1D3 (1-160, 227-377) were synthesized by Genscript as a DNA fragment with restriction enzyme sites HindIII (5′ end) and ClaI (3′ end) for cloning directly into pLHCX. The chimeric CD46/CD4 constructs were generated by a two-step cloning process in which CD46 fragments were cloned with primers for amplification of nucleotide sequences corresponding to CD46-BC1 aa. 1–397 and aa. 1–366 and ligated with CD4 DNA fragments synthesized by Genscript corresponding to aa. 397–459 and aa. 419–458, followed by a PCR amplification with primers that amplify only the chimeric gene. The chimeric gene was then cloned into pLHCX using HindIII (5′ end) and ClaI (3′ end) restriction sites. The truncated CD46 1–360 was generated by amplifying the gene fragment using specific primers for InFusion cloning into pLHCX. The retrovirus vectors encoding the CD46 constructs were transiently transfected simultaneously with vector containing the VSV-G envelope into gp2-293 cells via a lipid-based protocol to generate pseudotyped Moloney Murine Leukaemia Virus amphotropic retrovirus, as described [36]. The transduced ARPE-19 cells were selected with hygromycin B (2.5 mg ml−1, Invitrogen #10687–010) for 7 days. Clones of the respective transduced cells were generated by plating 1 cell/well in a 24-well plate; clones were analysed and validated for CD46 construct expression by both flow cytometry and immunoblotting.
Results
CD46 mediates an early step of a HCMV infection
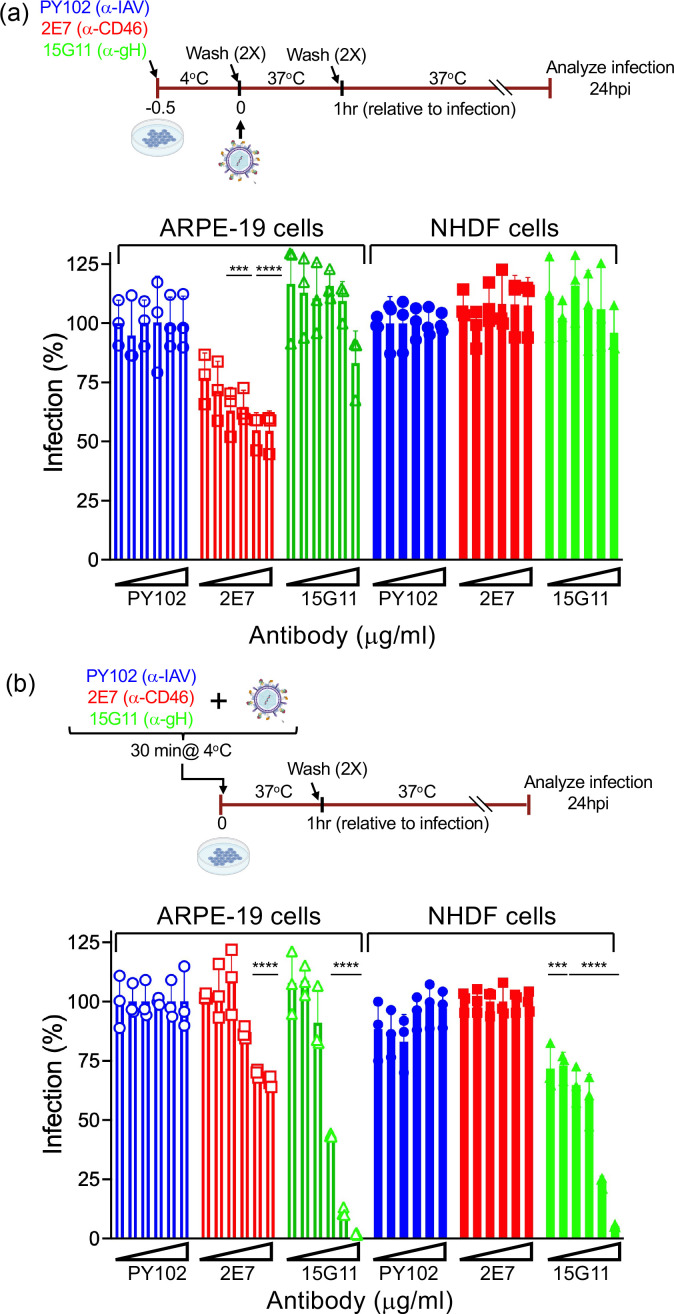
HCMV infection is mediated through interaction of viral and cellular factors, followed by virus fusion at the cell surface or within an endocytic vesicle [1]. CD46 was demonstrated to participate in HCMV infection during the early stages of infection of epithelial cells and trophoblasts, and not fibroblasts [25]. To further evaluate whether targeting CD46 prior to infection can limit subsequent HCMV infection, we examined virus infectivity in ARPE-19 and MRC5 cells pre-incubated with anti-CD46 monoclonal antibody (mAb) 2E7 [25], anti-gH mAb 15G11 [34], or an irrelevant mAb raised against influenza hemagglutinin (PY102) (Fig. 1). MRC5 and ARPE-19 cells were incubated with increasing concentrations of 2E7, 15G11, and PY102 for 30 min at 4 °C, followed by a wash step to remove unbound antibody. Cells were then infected with HCMV AD169R (MOI: 0.25) for 1 h at 37 °C (Fig. 1a). AD169R is a repaired AD169 (BADrUL131-C4) containing the UL131-UL128 open reading frame of HCMV strain TR and expresses EGFP [32]. Infectivity assay was used to determine percent infection with the corresponding PY102 incubation conditions as 100 % infection. Pre-incubation of ARPE-19 cells with anti-CD46 mAb 2E7 revealed a concentration-dependent decrease in virus infection with a ~50 % reduction in infection at the higher mAb concentrations (Fig. 1a). These results and the lack of 2E7-mediated inhibition of virus infection in fibroblasts was consistent with our previous findings [25] suggesting that CD46 functions with other cellular factors to mediate virus infection. As expected, the anti-gH mAb 15G11 had no impact on virus infection under these conditions due to its specificity for the gH viral envelope protein. However, AD169R pre-incubated with 15G11 reduced virus infection of both ARPE-19 and MRC5 cells [34] (Fig. 1b). Interestingly, the anti-CD46 2E7 mAb preincubated with HCMV limited infection of epithelial cells at highest mAb concentrations (Fig. 1b). These data imply that the anti-CD46 mAb functions more effectively in a pre-incubation step prior to virus binding, suggesting that the antibody may compete with the virus for binding to CD46. Alternatively, low levels of CD46 may be incorporated within the virion. However, a dilution of the 2E7 mAb did not block infection, while 15G11 continued to effectively limit virus infection (Fig. S1, available in the online version of this article), suggesting the anti-CD46 mAb functions by targeting cell surface CD46. Collectively, these studies further support a role for CD46 during the early stage of HCMV infection in epithelial cells.
Fig. 1.
Anti-CD46 mAb limits HCMV infection. (a) MRC5 and ARPE-19 cells pre-incubated with anti-CD46 mAb 2E7, anti-gH mAb 15G11, or anti-influenza mAb PY102 (0.016, 0.08, 0.4, 2, 10, and 50 mg ml−1) were infected with HCMV AD169R based on the schematic and analysed for infection at 24hpi. (b) HCMV AD169R pre-incubated with 2E7, 15G11, or PY102 prior to infection of MRC5 and ARPE-19 cells based on the schematic and analysed 24hpi. Percent infection was determined using the corresponding PY102 samples as 100 % infection. s.d. is depicted in the experiment. ***P<0.001, ****P<0.0001 (two-way ANOVA, Dunnett’s multiple comparison test).
Overexpression of CD46 expression enhances HCMV infection
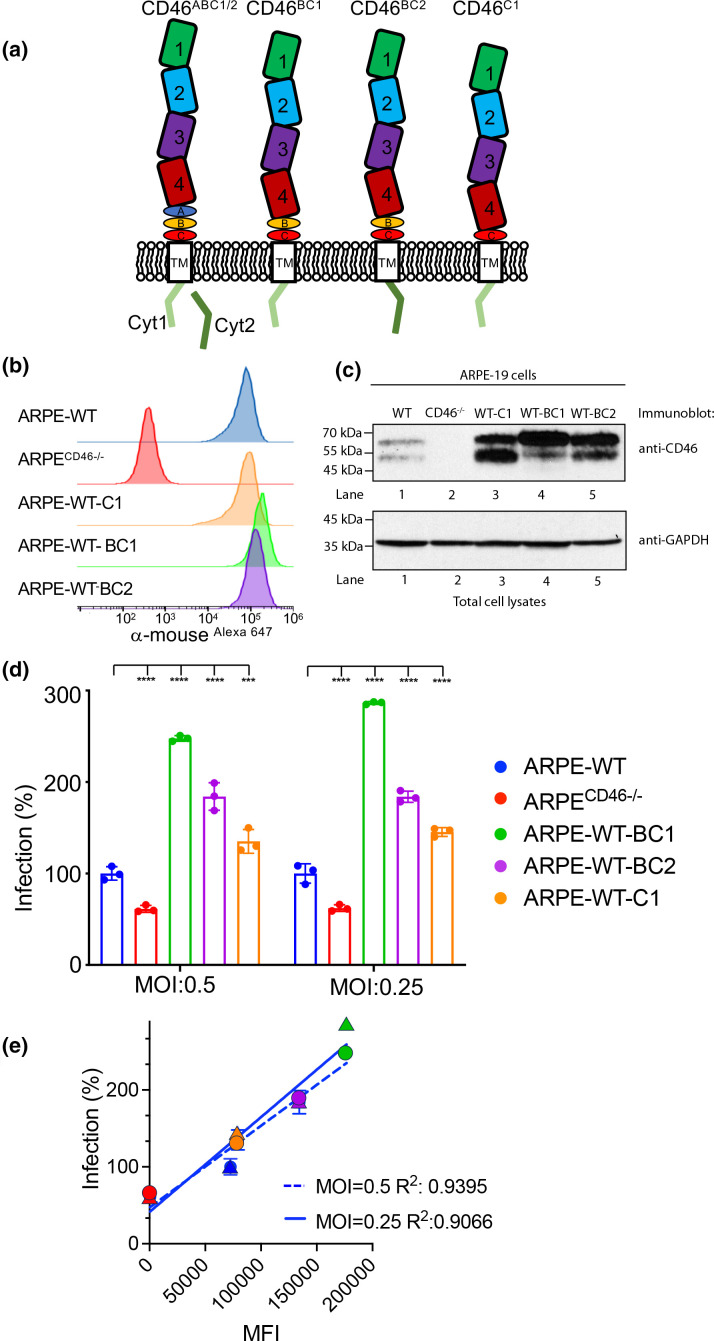
CD46 is a type I membrane protein consisting of four short consensus repeats (SCR)/complement control protein (CCP) domains, STP regions referred to as A, B, and C that are O-linked glycosylated, a transmembrane domain, and two alternatively spliced cytoplasmic tails designated Cyt-1 or Cyt-2 [37]. Due to alternative splicing events, CD46 exists as 16 isomers (A-N, 2, and 3) which are classified into alpha (isomer C and isomer D), beta (isomer E and isomer F), gamma (isomer A and isomer B), and delta (isomer N) isomers resulting in two prominent polypeptide species of 58 kDa–68kDa and 48 kDa–56kDa based on the presence of the STP region [38]. Further, the Cyt-1 or Cyt-2 CD46 species differ in the length and sequence of the carboxy-end of their cytoplasmic tails (Fig. 2a), which possess putative signals for phosphorylation by casein kinase two and protein kinase C [39]. The major CD46 isomers present in ARPE-19 cells are BC1, C1, BC2, and C2 and may vary in different cells and tissues [38, 40, 41] (Fig. 2a). To determine whether some CD46 isomers influence HCMV infection, we generated ARPE-19 cell lines that overexpress CD46 isomers BC1, BC2, and C1 by pseudoviral transduction. Unfortunately, cell lines that stably overexpress the CD46 isomer C2 could not be generated due to the instability of C2 protein and were excluded from the studies (unpublished data). Clones of ARPE-19 cells expressing endogenous CD46 and the CD46 isomers BC1 (ARPE-WT-BC1), BC2 (ARPE-WT-BC2), and C1 (ARPE-WT-C1) were analysed for CD46 protein levels on the surface of live cells by flow cytometry and total CD46 protein in cell lysates by Western blot (Fig. 2b, c). The analysis of these cells by flow cytometry (Fig. 2b) revealed higher expression levels of BC1 followed by C2 and BC2 in WT ARPE-19 cells possibly due to the number of cDNA integrations. Furthermore, immunoblot analysis demonstrated higher expression levels of the respective CD46 isomers (Fig. 2c, lanes 3–5). Together, the data confirm the expression of the specified CD46 isomers in ARPE-19 cells.
Fig. 2.
CD46 isomers enhance HCMV infection of ARPE-19 cells. (a) Schematic depicting the CD46 isomers. (b) Cell surface expression of CD46 in WT ARPE-19, ARPECD46-/- knockout cells, and WT ARPE-19 cells stably expressing CD46 C1 (ARPE-WT-C1), BC1 (ARPE-WT-BC1), and BC2 (ARPE-WT-BC2) was examined by fluorescence-based flow cytometry using anti-CD46 mAb GB24 followed by incubation with anti-mouse IgGAlexa647. The normalized cell number was plotted based on Alexa647 fluorescence intensity. (c) The cell lysates from the WT ARPE-19, ARPECD46-/-, ARPE-WT-C1, ARPE-WT-BC1, and ARPE-WT-BC2 cells were subjected to anti-CD46 and anti-GAPDH immunoblots, respectively. (d) WT ARPE-19, ARPECD46-/-, ARPE-WT-BC1, ARPE-WT-BC2, and ARPE-WT-C1 cells infected with AD169R (MOI:0.5 and 0.25) were assessed at 24hpi by a Celigo cytometer for infection. Percent infection was measured based on the anti-IE1 immunostain and WT ARPE-19 cells were used to normalize infection at 100 %. s.d. is depicted in the figure by errors bars. (e) A linear regression model was used to calculate the Goodness of Fit (R2 ) using infection (%) and mean fluorescence intensity (MFI) of CD46 surface expression of AD169R infection (MOI: 0.25 and 0.5). ***P<0.001, ****P<0.0001 (two-way ANOVA, Dunnett’s multiple comparison test) values are compared to ARPE-WT infection.
We next evaluated whether overexpression of CD46 isomers impacts virus infection (Fig. 2d). Wild-type ARPE, ARPECD46-/-, ARPE-WT-BC1, ARPE-WT-BC2, and ARPE-WT-C1 cells infected with HCMV-AD169R (MOI: 0.25 and 0.5) were evaluated using an infectivity assay. Percent infection was normalized to 100 % based on infection of wild-type ARPE-19 cells. Consistent with published data, there was a ~50 % reduction in infection of the ARPECD46-/- cells [25]. An enhanced HCMV infection was observed in ARPE-WT-BC1, ARPE-WT-BC2, and ARPE-WT-C1 cells independent of MOI and is consistent with CD46 isomer expression levels. A linear regression model comparing the mean fluorescent intensity (MFI) of CD46 isomers on the cell surface and HCMV infection at MOIs of 0.25 and 0.5 found R2 values of 0.91 and 0.94, respectively (Fig. 2e). The data demonstrate that HCMV infection is related to CD46 surface expression on epithelial cells. The expression levels of CD46 on the cell surface suggest that the increased levels of CD46 may enhance multivalent interaction with virus or act indirectly to enhance a virus entry step.
Expression of CD46 isomers rescues HCMV infection in ARPE-19 CD46 knock-out cells
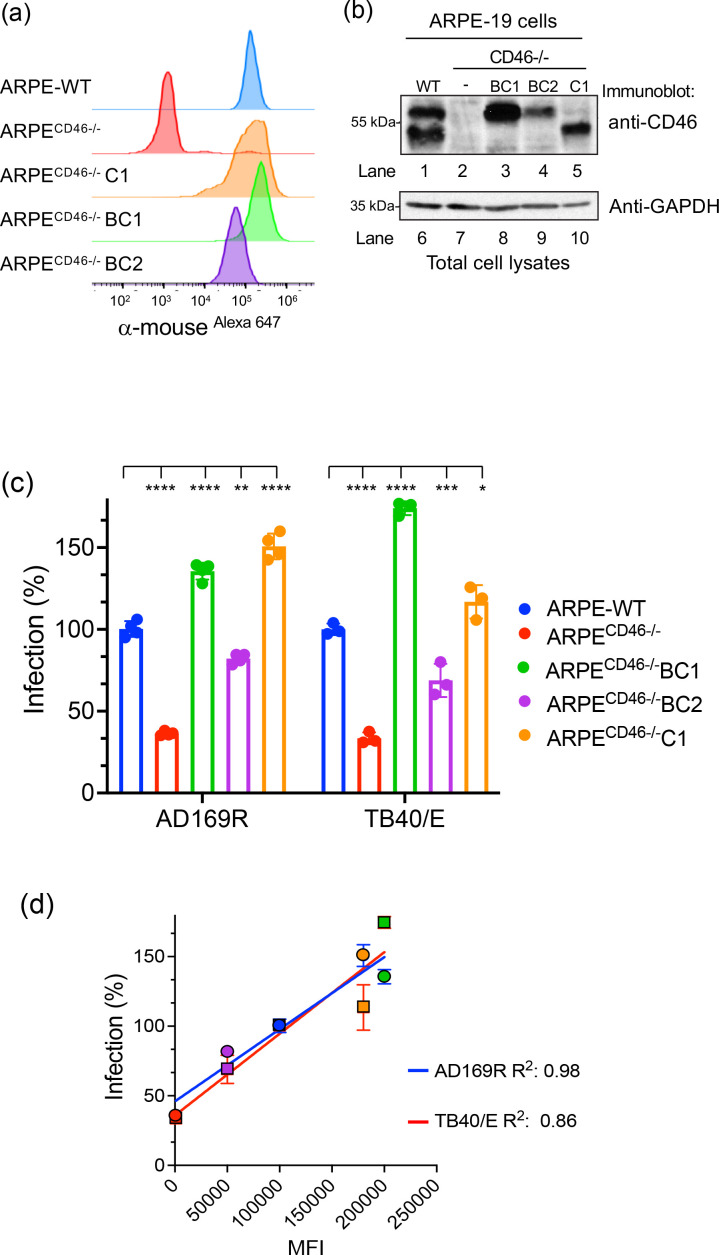
To investigate whether HCMV infection is dependent on the presence of specific isomers, we generated stable cell lines that overexpress the CD46 isomers BC1, BC2, and C1 in ARPECD46-/- cells and re-evaluated HCMV infection (Fig. 3). Again, we confirmed expression of the isomers by flow cytometry (Fig. 3a) and immunoblot analysis (Fig. 3b). Although the levels of CD46 isomer expression varied between the selected clones possibly due to the number of integration sites according to both flow cytometry and immunoblot analysis (Fig. 3b, lanes 3–5), the detection of the BC1, BC2, and C1 isomers by the anti-CD46 mAb GB24 indicates that these proteins are properly folded and capable of trafficking to the cell surface.
Fig. 3.
CD46 isomers rescue HCMV infection in ARPECD46-/- knockout cells. (a) Cell surface expression of WT ARPE-19, ARPECD46-/-, and ARPECD46-/- cells stably expressing CD46 C1, BC1, and BC2 (ARPECD46-/-C1, ARPECD46-/-BC1, and ARPECD46-/-BC2) was confirmed by fluorescence-based flow cytometry with anti-CD46 mAb GB24 followed by incubation with anti-mouse IgGAlexa647. The normalized cell number was plotted based on Alexa647 fluorescence intensity. (b) The cell lysates from the WT ARPE-19, ARPECD46-/-, ARPECD46-/-BC1, ARPECD46-/-BC2, and ARPECD46-/-C1 cells were subjected to anti-CD46 and GAPDH immunoblots, respectively. (c) HCMV AD169R and TB40/E infection of WT ARPE-19, ARPECD46-/-, ARPECD46-/-BC1, ARPECD46-/-BC2, and ARPECD46-/-C1 were assessed at 24hpi by Celigo cytometer. Percent infection was measured using an infectivity assay and WT ARPE-19 cells were used to normalize infection at 100 %. s.d. is depicted by errors bars. (d) A linear regression model was used to calculate the Goodness of Fit (R2 ) using infection (%) and mean fluorescence intensity (MFI) of CD46 surface expression of AD169R and TB40/E infected cells (c). *P<0.05, **P<0.01, ****P<0.0001 (two-way ANOVA, Sidak’s multiple comparison test) values are compared to ARPE-WT infection.
Next, we evaluated HCMV infection of ARPE-WT, ARPECD46-/-, ARPECD46-/-BC1, ARPECD46-/-BC2, and ARPECD46-/-C1 cells with both TB40/E and AD169R (MOI: 0.25) to demonstrate broad reactivity among strains (Fig. 3c). Infection was quantified based on the expression of the virally encoded IE1 protein at 18 hpi as detected by immunostaining. Again, we observed a >50 % reduction of infection in ARPECD46-/- cells for both the TB40/E and AD169R strains. A rescue of infection could be seen in cell lines that constitutively expressed the BC1, BC2 or C1 isomer relative to the parental CD46 KO cell line in both AD169R and TB40/E infection models. Further analysis of a linear regression model comparing cell surface expression levels of CD46 MFI and induction of IE1 following virus yielded a R2 value of 0.86 for TB40/E and a R2 value of 0.924 for AD169R (Fig. 3d) indicating a relationship between CD46 surface expression levels and HCMV infection, independent of the respective CD46 isomers. However, CD46 A and B regions and the C1 or C2 cytoplasmic tail may play a direct or indirect role in virus infection requiring further investigation. More broadly, the results demonstrate that levels of CD46 expression on the cell surface was related to HCMV infectivity further supporting the model that CD46 plays a role during virus infection.
The CD46 ectodomain plays a role in HCMV infection
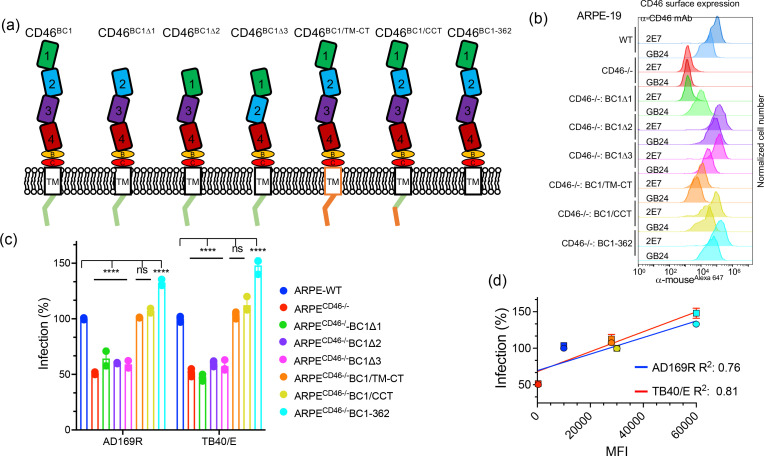
The SCR domain of CD46 mediate interactions with complement factors C3b and C4b and different pathogens [27, 42]. To determine if one of the CD46 SCR domains or CD46 transmembrane (TM)/cytoplasmic tail (CT) impact HCMV infection, we generated ARPECD46-/- constructs that express CD46 mutants lacking SCR 1, 2, or 3 domains (SCR1 region [BC1Δ1], SCR2 region [BC1Δ2], and SCR3 region [BC1Δ3]), CD46 chimaera in which the transmembrane and cytoplasmic tail regions of CD46 were replaced with the corresponding regions of CD4 (BC1/TM-CT and BC1/CCT), and a CD46 carboxy-terminal truncation mutant (CD46-362) (Fig. 4a). CD46 protein levels in ARPECD46-/-BC1Δ1, ARPECD46-/-BC1Δ2, ARPECD46-/-BC1Δ3, ARPECD46-/-BC1/TM-CT, ARPECD46-/-BC1/CCT, and ARPECD46-/-BC1-362 cells were initially evaluated by flow cytometry, using conformation-specific CD46 antibodies that target the SCR1 (2E7) or SCR4 (GB24) domains (Fig. 4b). Surface protein expression of CD46 mutants using 2E7 recognized CD46BC1Δ2, CD46BC1Δ3, CD46BC1/TM-CT, CD46BC1/CCT, and CD46BC1-362, but not CD46BC1Δ1 due to this construct lacking the 2E7-binding region. Flow cytometry analysis with GB24 confirmed varied surface expression levels among the different mutants (Fig. 4b). The expression levels of the CD46 BC1 mutants were validated by immunoblot analysis (Fig. S2). The difference in expression levels of the CD46 mutants may be due to variations in transduction efficiency, trafficking, and/or stability of the CD46 constructs. The recognition of the CD46 mutants on the cell surface using conformationally-specific antibodies validates the proper folding of the molecules.
Fig. 4.
The CD46 ectodomain plays a role in HCMV infection. (a) Schematic depicting CD46-BC1 mutant constructs: BC1 lacking SCR1 (Δ1), SCR2 (Δ2), SCR3 (Δ3), CD46-BC1 transmembrane and cytoplasmic tail (TM-CT) replaced with TM-CT of CD4 (BC1/TM-CT), CD46-BC1 whose carboxy-terminal end was replaced with the CD4 carboxy-terminal end (BC1/CCT), and CD46-BC1 carboxy-terminal truncation mutant (BC1-362). (b) Cell surface expression of CD46 molecules on WT ARPE-19, ARPECD46-/-, ARPECD46-/-BC1Δ1, ARPECD46-/-BC1Δ2, ARPECD46-/-BC1Δ3, ARPECD46-/-BC1/TM-CT, ARPECD46-/-BC1/CCT, and ARPECD46-/-BC1-362 was assessed by fluorescence-based flow cytometry with mAb 2E7 (anti-SCR1) and conformational mAb GB24 (anti-SCR4). The normalized cell number was plotted based on Alexa647 fluorescence intensity. (c) HCMV AD169R and TB40/E infection (MOI: 0.25) of ARPE-19 cells, ARPECD46-/- cells, and ARPECD46-/ cells expressing the CD46 BC1 mutants were assessed at 24hpi by Celigo cytometer. Percent infection was measured by an infectivity assay and WT ARPE-19 cells were used to normalize infection to 100 %. s.d. is depicted by errors bars. (d) A linear regression model was used to calculate the Goodness of Fit (R2 ) using infection (%) and mean fluorescence intensity (MFI) of CD46 surface expression of AD169R and TB40/E infected cells. ****P<0.0001 (two-way ANOVA, Sidak’s multiple comparison test) values are compared to ARPE-WT infection.
We next evaluated TB40/E and AD169R infection (MOI=0.25) of ARPE (WT), ARPECD46-/-, ARPECD46-/-BC1Δ1, ARPECD46-/-BC1Δ2, ARPECD46-/-BC1Δ3, ARPECD46-/-BC1/TM-CT, ARPECD46-/-BC1/CCT, and ARPECD46-/-BC1-362 cells using an infectivity assay (Fig. 4c). Percent infection was determined based on the number of IE1-positive cells and normalized to 100 % based on infection of ARPE-WT cells. Strikingly, expression of CD46BC1Δ1, CD46BC1Δ2, and CD46BC1Δ3 in ARPECD46-/- cells did not rescue virus infectivity suggesting these domains are important for virus binding or entry steps required for subsequent IE1 expression. On the other hand, CD46 BC1/TM-CT, CD46 BC1/CCT, and CD46 BC1-362 expression in ARPECD46-/- cells rescued virus infection with CD46 BC1-362 yielding increased infection levels than wild-type ARPE-19 cells. A linear regression model comparing expression levels of CD46 BC1/TM-CT, CD46 BC1/CCT, and CD46 BC1-362 to HCMV infection yielded R2 values of 0.81 for TB40/E and 0.76 for AD169R (Fig. 4d). Thus, the data suggest that the CD46 transmembrane domain and cytoplasmic tail do not directly impact HCMV infection. Given that the deletion of SCR domains 1, 2, or 3 did not rescue infection, a region that spans all three SCR domains may be involved in HCMV infection or a domain proximal to the bilayer is important. Collectively, the results support a model of the ectodomain of CD46-BC1 as an important structural element that is involved in HCMV infection.
Discussion
The function of CD46 as a pathogen entry factor likely occurs through the interaction of pathogen factors with one or several SCR domains of the extracellular region [27, 42]. SCR domains 1 and 2 interact with human adenovirus B PIV/FIBER protein and measles virus H protein, while domains 2 and 3 bind to HHV-6′s gH protein [43–45]. Also, SCR domain 3 is important for Neisseria binding, and domains 3 and 4 mediate interactions with Streptococcus pyogenes M protein [42]. Published findings concluded that a soluble version of CD46 interacted with recombinant gH/gL pentamer (KD ~10−5 M [23]) and in our current study we found several CD46 isomers and CD46 SCR deletion mutants to participate in HCMV infection (Figs 2–4), consistent with the paradigm that the SCR domains participate in virus infection. In the case of HCMV, the SCR 1, 2, and 3 domains together may form an interactive region that plays a role in HCMV infection. Another interesting observation was that CD46-BC1 isomers enhanced infection in the wild-type ARPE-19 and ARPECD46-/- cells suggesting this isomer may play a more dominant role in HCMV infection. It is interesting to speculate that the CD46 ectodomain may engage with viral proteins followed by engagement with other cellular factors that promote virus infection by triggering entry. This paradigm will require further studies to define CD46’s mechanism of action.
Nrp2 and THBD were identified, using cryo-electron microscopy (cryo-EM), as interacting with the HCMV pentamer through association with the UL128/UL130/UL131A subunits [26, 46]. In addition, a peptide of the OR14I1 entry factor limits HCMV infection through interactions with the pentamer [22]. These proteins likely function by directly interfacing with the HCMV pentamer as receptors on the cell surface. However, a monoclonal antibody that blocks HCMV infection of epithelial cells, but not NRP2 binding, suggests that the pentamer may engage with other factors at the cell surface that play a role in entry [46]. These other factors, which may include CD46, CD147, or OR14I1 may function through the activation of subsequential entry steps including endocytosis, fusion, and possibly activation of viral gene transcription. In the case of CD46, it was initially identified to interact with the pentamer using a proteomic screen [23]. Our study found that a linear relationship of CD46 cell surface expression levels and HCMV infectivity in epithelial cells was observed for several CD46 isoforms and CD46 transmembrane/cytoplasmic tail chimeric mutants (Figs. 2–4). Collectively, these findings imply that CD46 may function by directly interacting with the pentamer itself or a complex consisting of viral and cellular factors to enhance virus binding to cells or augment a post-binding step of infection. Thus, CD46 may act as an ‘efficiency factor’ ensuring the expeditious entry and infection of HCMV in non-fibroblast cell types.
The multi-step process of HCMV entry is a tightly regulated and orchestrated event involving numerous cellular factors that ensures efficient entry and infection of diverse cell types. The recent discovery of numerous HCMV entry factors involved in infection of non-fibroblast cells is consistent with the virus’s wide tropism, which may utilize myriad cellular factors for entry. Indeed, varying expression levels of these disparate entry factors across different cell types may dictate the cellular factors utilized for HCMV infection of non-fibroblast cells. The entry factors Nrp2, THBD, OR14I1, CD147, and CD46 are expressed at different levels [47] among cell types and tissues, supporting the ability of HCMV to infect diverse cell types by utilizing a wide variety of cellular entry factors and receptors. Alternatively, HCMV infection is known to require numerous viral envelope complexes that mediate virus tethering, binding, endocytosis (in non-fibroblast cells), and fusion. Given the complexity of HCMV infection, successful entry of the virus likely requires the engagement of multiple cellular proteins to mediate different steps of the entry process as well as multivalent interactions between host cell complexes and viral envelope complexes, including possible dimer formations of the pentamer, trimers of gB, and the heterodimer complex of gM/gN. The subsequent steps of virus entry as the activation of gB mediates fusion is suggested to occur through, in part, its interaction with gH/gL-complexes and possibly the cellular factor EGFR or PDGFRα [1]. In summary, the success of HCMV infection and spread is dependent on interactions with diverse cellular factors that function during the different steps of entry and dissemination.
Supplementary Data
Funding information
The NIH Institutional Research Training Award - T32-AI007647, R01AI139258, R21AI147632, and RF1AG059319, respectively, partially supported Andrea J. Parsons, Kathryn R. Stein, Kristina E. Atanasoff, Sabrina I. Ophir, Jailene Paredes Casado, and Domenico Tortorella.
Author contributions
A.J.P., K.R.S., and K.E.A. designed and performed the infectivity studies. S.I.O. and J.P.C. amplified H.C.M.V. strains. K.R.S., J.P.C., and D.T. generated the cell lines expressing the different CD46 constructs. A.J.P., K.R.S., K.E.A. and D.T. wrote and edited the manuscript. D.T. conceived and supervised the project.
Conflicts of interest
The author(s) declare that there are no conflicts of interest.
Footnotes
Two supplementary figures are available with the online version of this article.
Abbreviations: CCP, complement control protein; CD46, cluster of differentiation 46; HCMV, human cytomegalovirus; HHV6, human herpesvirus 6; MOI, multiplicity of infection; Nrp-2, neuropilin-2; OR14I1, olfactory receptor family 14 subfamily I member 1; pentamer, gH/gL/Ul128/Ul130/UL131a; SCR, short consensus repeats; STP, serines, threonines and prolines; THBD, thrombomodulin; trimer, gH/gL/gO.
References
- 1.Griffiths P, Reeves M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat Rev Microbiol. 2021;19:759–773. doi: 10.1038/s41579-021-00582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Söderberg-Nauclér C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med. 2006;259:219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 3.Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011;121:1673–1680. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrum F. Human cytomegalovirus latency: approaching the gordian knot. Annu Rev Virol. 2016;3:333–357. doi: 10.1146/annurev-virology-110615-042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benditt EP, Benditt JM. Evidence for a monoclonal origin of human atherosclerotic plaques. Proc Natl Acad Sci. 1973;70:1753–1756. doi: 10.1073/pnas.70.6.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabricant CG, Fabricant J, Minick CR, Litrenta MM. Herpesvirus-induced atherosclerosis in chickens. Fed Proc. 1983;42:2476–2479. [PubMed] [Google Scholar]
- 7.Gattone M, Iacoviello L, Colombo M, Castelnuovo AD, Soffiantino F, et al. Chlamydia pneumoniae and cytomegalovirus seropositivity, inflammatory markers, and the risk of myocardial infarction at a young age. Am Heart J. 2001;142:633–640. doi: 10.1067/mhj.2001.118118. [DOI] [PubMed] [Google Scholar]
- 8.Epstein SE, Zhou YF, Zhu J. Infection and atherosclerosis: emerging mechanistic paradigms. Circulation. 1999;100:e20–8. doi: 10.1161/01.cir.100.4.e20. [DOI] [PubMed] [Google Scholar]
- 9.Epstein SE, Zhu J, Burnett MS, Zhou YF, Vercellotti G, et al. Infection and atherosclerosis: potential roles of pathogen burden and molecular mimicry. Arterioscler Thromb Vasc Biol. 2000;20:1417–1420. doi: 10.1161/01.atv.20.6.1417. [DOI] [PubMed] [Google Scholar]
- 10.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 11.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 12.Ross R. Atherosclerosis is an inflammatory disease. Am Heart J. 1999;138:S419–20. doi: 10.1016/s0002-8703(99)70266-8. [DOI] [PubMed] [Google Scholar]
- 13.Compton T, Feire A. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. 2007. Early events in human cytomegalovirus infection. [DOI] [Google Scholar]
- 14.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton T, Nepomuceno RR, Nowlin DM. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 16.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/JVI.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kari B, Gehrz R. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J Gen Virol. 1993;74 (Pt 2):255–264. doi: 10.1099/0022-1317-74-2-255. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Huang DY, Huong SM, Huang ES. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat Med. 2005;11:515–521. doi: 10.1038/nm1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Wilkie AR, Weller M, Liu X, Cohen JI. THY-1 cell surface antigen (CD90) has an important role in the initial stage of human cytomegalovirus infection. PLoS Pathog. 2015;11:e1004999. doi: 10.1371/journal.ppat.1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu K, Oberstein A, Wang W, Shenk T. Role of PDGF receptor-α during human cytomegalovirus entry into fibroblasts. Proc Natl Acad Sci. 2018;115:E9889–E9898. doi: 10.1073/pnas.1806305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.E X, Meraner P, Lu P, Perreira JM, Aker AM, et al. OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc Natl Acad Sci. 2019;116:7043–7052. doi: 10.1073/pnas.1814850116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Martin N, Marcandalli J, Huang CS, Arthur CP, Perotti M, et al. An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell. 2018;174:1158–1171. doi: 10.1016/j.cell.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 24.Vanarsdall AL, Pritchard SR, Wisner TW, Liu J, Jardetzky TS, et al. CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells. mBio. 2018;9 doi: 10.1128/mBio.00781-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein KR, Gardner TJ, Hernandez RE, Kraus TA, Duty JA, et al. CD46 facilitates entry and dissemination of human cytomegalovirus. Nat Commun. 2019;10:2699. doi: 10.1038/s41467-019-10587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kschonsak M, Rougé L, Arthur CP, Hoangdung H, Patel N, et al. Structures of HCMV trimer reveal the basis for receptor recognition and cell entry. Cell. 2021;184:1232–1244. doi: 10.1016/j.cell.2021.01.036. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto H, Fara AF, Dasgupta P, Kemper C. CD46: the “multitasker” of complement proteins. Int J Biochem Cell Biol. 2013;45:2808–2820. doi: 10.1016/j.biocel.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Cattaneo R. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol. 2004;78:4385–4388. doi: 10.1128/jvi.78.9.4385-4388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liszewski MK, Kemper C, Price JD, Atkinson JP. Emerging roles and new functions of CD46. Springer Semin Immunopathol. 2005;27:345–358. doi: 10.1007/s00281-005-0002-3. [DOI] [PubMed] [Google Scholar]
- 30.Källström H, Blackmer Gill D, Albiger B, Liszewski MK, Atkinson JP, et al. Attachment of Neisseria gonorrhoeae to the cellular pilus receptor CD46: identification of domains important for bacterial adherence. Cell Microbiol. 2001;3:133–143. doi: 10.1046/j.1462-5822.2001.00095.x. [DOI] [PubMed] [Google Scholar]
- 31.Giannakis E, Jokiranta TS, Ormsby RJ, Duthy TG, Male DA, et al. Identification of the streptococcal M protein binding site on membrane cofactor protein (CD46) J Immunol. 2002;168:4585–4592. doi: 10.4049/jimmunol.168.9.4585. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner TJ, Bolovan-Fritts C, Teng MW, Redmann V, Kraus TA, et al. Development of a high-throughput assay to measure the neutralization capability of anti-cytomegalovirus antibodies. Clin Vaccine Immunol. 2013;20:540–550. doi: 10.1128/CVI.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons AJ, Ophir SI, Duty JA, Kraus TA, Stein KR, et al. Development of broadly neutralizing antibodies targeting the cytomegalovirus subdominant antigen gH. Commun Biol. 2022;5:387. doi: 10.1038/s42003-022-03294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsi B-L, Yeh C-JG, Fénichel P, Samson M, Grivaux C. Monoclonal antibody GB24 recognizes a trophoblast-lymphocyte cross-reactive antigen. Am J Reprod Immunol Microbiol. 1988;18:21–27. doi: 10.1111/j.1600-0897.1988.tb00228.x. [DOI] [PubMed] [Google Scholar]
- 36.Oresic K, Noriega V, Andrews L, Tortorella D. A structural determinant of human cytomegalovirus US2 dictates the down-regulation of class I major histocompatibility molecules. J Biol Chem. 2006;281:19395–19406. doi: 10.1074/jbc.M601026200. [DOI] [PubMed] [Google Scholar]
- 37.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 38.Post TW, Liszewski MK, Adams EM, Tedja I, Miller EA, et al. Membrane cofactor protein of the complement system: alternative splicing of serine/threonine/proline-rich exons and cytoplasmic tails produces multiple isoforms that correlate with protein phenotype. J Exp Med. 1991;174:93–102. doi: 10.1084/jem.174.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, Liszewski MK, Chan AC, Atkinson JP. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J Immunol. 2000;164:1839–1846. doi: 10.4049/jimmunol.164.4.1839. [DOI] [PubMed] [Google Scholar]
- 40.Hansen AS, Bundgaard BB, Møller BK, Höllsberg P. Non-random pairing of CD46 isoforms with skewing towards BC2 and C2 in activated and memory/effector T cells. Sci Rep. 2016;6:35406. doi: 10.1038/srep35406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnstone RW, Russell SM, Loveland BE, McKenzie IF. Polymorphic expression of CD46 protein isoforms due to tissue-specific RNA splicing. Mol Immunol. 1993;30:1231–1241. doi: 10.1016/0161-5890(93)90038-d. [DOI] [PubMed] [Google Scholar]
- 42.Liszewski MK, Atkinson JP. Membrane cofactor protein (MCP; CD46): deficiency states and pathogen connections. Curr Opin Immunol. 2021;72:126–134. doi: 10.1016/j.coi.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson BD, John L, Rafie K, Strebl M, Frängsmyr L, et al. Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proc Natl Acad Sci. 2021;118:e2020732118. doi: 10.1073/pnas.2020732118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchholz CJ, Koller D, Devaux P, Mumenthaler C, Schneider-Schaulies J, et al. Mapping of the primary binding site of measles virus to its receptor CD46. J Biol Chem. 1997;272:22072–22079. doi: 10.1074/jbc.272.35.22072. [DOI] [PubMed] [Google Scholar]
- 45.Greenstone HL, Santoro F, Lusso P, Berger EA. Human herpesvirus 6 and measles virus employ distinct CD46 domains for receptor function. J Biol Chem. 2002;277:39112–39118. doi: 10.1074/jbc.M206488200. [DOI] [PubMed] [Google Scholar]
- 46.Wrapp D, Ye X, Ku Z, Su H, Jones HG, et al. Structural basis for HCMV pentamer recognition by neuropilin 2 and neutralizing antibodies. Sci Adv. 2022;8:eabm2546. doi: 10.1126/sciadv.abm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Protein Atlas: CD46 and Nrp2. 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.