Abstract
Recent advances in genome sequencing have led to the identification of new ion and metabolite transporters, many of which have not been characterized. Due to the variety of subcellular localizations, cargo and transport mechanisms, such characterization is a daunting task, and predictive approaches focused on the functional context of transporters are very much needed. Here we present a case for identifying a transporter localization using evolutionary rate covariation (ERC), a computational approach based on pairwise correlations of amino acid sequence evolutionary rates across the mammalian phylogeny. As a case study, we find that poorly characterized transporter SLC30A9 (ZnT9) coevolves with several components of the mitochondrial oxidative phosphorylation chain, suggesting mitochondrial localization. We confirmed this computational finding experimentally using recombinant human SLC30A9. SLC30A9 loss caused zinc mishandling in the mitochondria, suggesting that under normal conditions it acts as a zinc exporter. We therefore propose that ERC can be used to predict the functional context of novel transporters and other poorly characterized proteins.
Introduction
Localization in a specific organelle or subset of organelles defines the functional context for many molecules, especially ion and metabolite transporters. For example, there are ion channels and transporters that localize to the plasma membrane or endo/sarcoplasmic reticulum, and even though they might conduct the same ions, they clearly have a different impact on cellular function [1–3]. Similarly, a large cohort of zinc transporters, responsible for pumping ionic zinc into distinct organelles or in or out of cells, subserve distinct functions, including synaptic modulation [4] and co-secretion with milk, in the latter case providing an essential dietary supplement and shaping mammary gland development [5]. The recent advances in genomic sequencing have resulted in identification of many putative transporters. While some aspects of their function can be inferred based on sequence homology or structural similarity to previously characterized molecules, important details, such as subcellular localization, are usually beyond the reach of these approaches. The main objective of the present studies is to evaluate whether a computational approach based on molecular evolution can accurately predict cellular localization of poorly characterized cellular proteins. Here, this idea was tested using zinc transporters as a model.
Zinc is an essential component of cellular function, as a key cofactor in enzymatic reactions, transcription and synaptic transmission [6]. Moreover, it has been estimated that ~10% of human proteome binds zinc [7]. Zinc deficit is a factor in growth delays, hair and skin pathologies and behavioral defects [8]. On the other hand, excess zinc promotes the production of reactive oxygen species and many forms of cell damage [9–12]. Because of zinc’s duality, cellular zinc concentrations are tightly regulated by a system of transporters and metal-binding proteins. In particular, zinc entry into the cytoplasm is managed by the SLC39A (Zip) family of transporters, while its expulsion or sequestration into organelles is driven by the SLC30A family. Metallothioneins, in addition to other zinc-binding proteins, provide an important intracellular buffer for the metal, leading to extremely low (sub nanomolar), or perhaps non-existent, levels of free zinc within the cytoplasm [13]. The zinc transporter families contain two dozen of structurally related members, which were discovered as a result of advances in sequencing technology and gene annotation [6]. However, many of these molecules remain under-characterized at the level of subcellular localization or transport mechanisms. The full extent of annotations supported by several lines of experimental evidence indicates that, for example, SLC30A1 and SLC39A1 function in the plasma membrane [14,15], SLC30A3 functions in synaptic vesicles [16] and SLC30A2 and SLC30A4 function in lysosomes and related organelles [17,18]. For many of the remaining SLC30A and SLC39A transporters, however, functional annotations, including cellular localizations, are fragmentary. Importantly, some organelles appear to contain several zinc transporters with overlapping functions, and the reason for this is unclear [6,19].
At the same time, some key aspects of organellar zinc transport remain poorly understood and/or lack an assigned transporter. This is especially the case for mitochondrial zinc, which, as a cofactor of several mitochondrial enzymes, is a critical component of mitochondrial physiology [20,21]. In parallel to full-cell consequences of zinc dysregulation, mitochondrial zinc overload is toxic and has been linked to traumatic brain injury and stroke [12]. The mechanisms of mitochondrial zinc regulation are poorly understood and neither SLC30A nor SLC39A members have been directly shown to localize in the mitochondria. Moreover, while the mitochondrial calcium uniporter MCU was proposed to be responsible for zinc uptake into the mitochondria from the cytoplasm [21,22], the pathway that dissipates the mitochondrial zinc has not been described.
These considerations illustrate a need for a predictive approach to match zinc transporters to specific organelles and focus experimental hypotheses to a reasonable set of targeted studies. We propose Evolutionary Rate Covariation (ERC) as the link between functional genomic information about zinc transporters and their associated organelles. ERC is based on the central hypothesis that proteins with shared functional context will share evolutionary pressure and thus evolve at similar rates, even as their rates change over time and between species [23,24]. This evolutionary rate covariation occurs in physically interacting proteins that coevolve as well as functionally related proteins such as gene regulatory elements, membrane traffic adaptors, and transporters. It is also important to note that many, but not all, recognized functional relationships between genes are reflected by high ERC correlations. Because of its predictive capability in some pathways, ERC has been used to identify the functions of uncharacterized genes, including novel DNA damage-induced apoptosis suppressor, sex peptide network components, regulators of cell adhesion, mitochondrial components and ion and aminoacid transporters [23,25–30]. ERC is calculated as correlation between evolutionary rates throughout a phylogeny. In fact, the work presented here will focus on values calculated across 33 mammal species. Genes are inferred to be functionally related if they have a high degree of correlation in evolutionary rates, or, in other words, if the genes in question have similar evolutionary rates across species.
We present ERC as a powerful tool for de novo prediction of molecular function with characterization of the zinc transporter SLC30A family, specifically SLC30A9, its most poorly characterized member. Although there are previously reported links to zinc transport [31,32], this transporter had previously not been linked to an organellar function in mammalian cells. We show here that ERC analysis suggests a strong and specific signal for SLC30A9 coevolution with several components of the mitochondrial oxidative phosphorylation chain including complex I, and the mitochondrial H+ driven ATP synthase (complex V [33,34]). The ERC signal between SLC30A9 and mitochondrial complexes was significantly higher than the signal between SLC30A9 and the vacuolar/vesicular/lysosomal H+ pump. We also find that recombinant SLC30A9 co-localizes with the mitochondrial protein marker TOM20, strongly suggesting mitochondrial localization. This is consistent with prior evidence contained in MitoCarta, which integrates multiple lines of evidence for mitochondrial localization and function of proteins, including APEX_matrix, targetP signal, yeast mitochondrial homolog, mitochondrial protein domain, induction, and MS/MS [35,36]. Furthermore, SLC30A9 knockdown in HeLa cells using siRNA suppresses the dissipation of mitochondrial zinc after zinc overload. We therefore propose that SLC30A9 is a mitochondrial zinc exporter and thus possesses a unique functional profile unique for members of the SLC30A family. Of interest, further inquiry of SLC30A sequence revealed that SLC30A9 has followed a unique evolutionary trajectory — it is deeply and highly conserved from mammals through archaea and proteobacteria, while other SLC30As are likely a result of more recent gene duplication events. These findings both illuminate the function of SLC30A9 and distinguish it as a unique molecule among SLC30A family members.
Materials and methods
Discovery and categorization of SLC30 sequences
Mammalian and other multi-cellular eukaryotic amino acid sequences of SLC30 family members were obtained using Pubmed search and aligned using CLUSTALW [37]. The ExxHxxxD…(IVL)(IVL)xED and DxxHxxxD… HxxxD tempates were used as wildcard searches of single-celled eukaryotic and prokaryotic sequences by means of BLASTp. The resulting sequences were verified for the presence of six full or partial transmembrane domains using TMHMM server.
Phylogenetic analysis
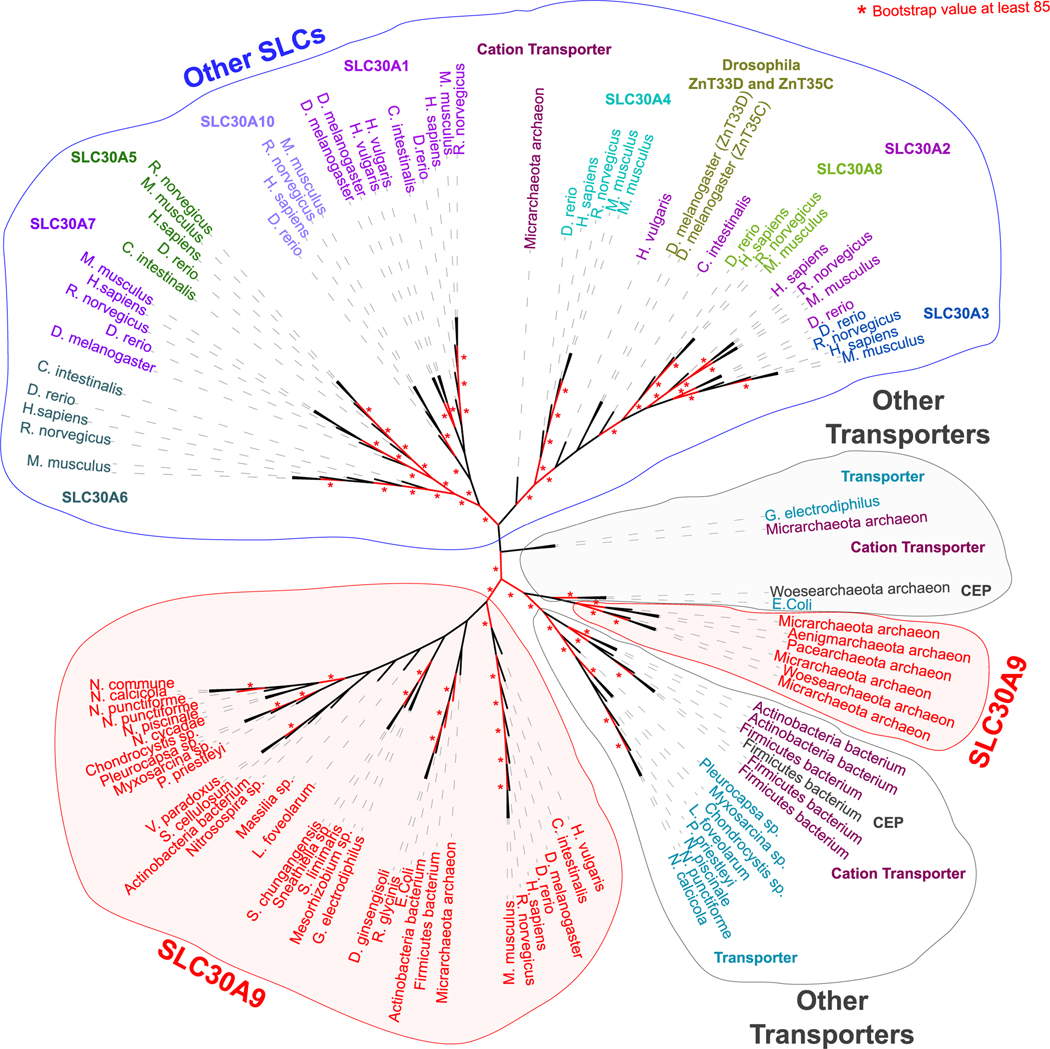
SLC30 family proteins and related homologs collected in the previous step were analyzed in terms of their divergence and phylogeny. Amino acid sequences were aligned using CLUSTALW [38]. The resulting alignment was used in PhyML 3.0 to infer their phylogenetic relationships and perform branch support analysis using 100 bootstrap replicates [39]. The ‘LG’ amino acid substitution model was used and rate heterogeneity was modeled using a class of invariable sites with freely estimated size and a Gamma rate shape parameter discretized as four rate categories. The tree figure in Figure 2 was made using Interactive Tree of Life (iTOL) [40,41].
Figure 2. SLC30A dendrogram suggests ancient divergence of SLC30A9 and other zinc transporters.
The branch leading to all SLC30A9s, both mammalian and bacterial, as well as some other transporters, is supported with high bootstrap confidence. Other SLC30As in animals and some other transporters branch separately.
Evolutionary rate covariation
The calculation of ERC values was performed as in previous publications [23,24]. In this case, orthologous gene sequences from 33 mammalian species were obtained from the 100-way alignment at the University of California Santa Cruz Genome Browser [42]. The species chosen were: Homo sapiens (human), Pongo pygmaeus abelii (orang-utan), Macaca mulatta (rhesus macaque), Callithrix jacchus (marmoset), Tarsius syrichta (tarsier), Microcebus murinus (mouse lemur), Otolemur garnettii (bushbaby), Tupaia belangeri (tree shrew), Cavia porcellus (guinea pig), Dipodomys ordii (kangaroo rat), Mus musculus (mouse), Rattus norvegicus (rat), Spermophilus tridecemlineatus (squirrel), Oryctolagus cuniculus (rab- bit), Ochotona princeps (pika), Vicugna pacos (alpaca), Sorex araneus (shrew), Bos taurus (cow), Tursiops truncatus (dolphin), Pteropus vampyrus (megabat), Myotis lucifugus (micro- bat), Erinaceus europaeus (hedgehog), Equus caballus (horse), Canis lupus familiaris (dog), Felis catus (cat), Choloepus hoffmanni (sloth), Echinops telfairi (tenrec), Loxodonta africana (elephant), Procavia capensis (rock hyrax), Dasypus novemcinctus (armadillo), Monodelphis domestica (opossum), Macropus eugenii (wallaby), and Ornithorhynchus anatinus (platypus). Those 17 486 coding sequence alignments were used to calculate gene-specific branch lengths over the mammalian species tree topology using codeml of the PAML package [43]. For each orthologous gene group/tree, the branch lengths were normalized into relative evolutionary rates through a projection operator [44]. Those relative rates were then used to calculate the Pearson correlation coefficient, the ERC value, between each pair of genes.
Enrichment analysis of organellar proteomes
Pairwise ERC values between SLC30 genes and all other genes were extracted from the ERC website: https://csb.pitt.edu/erc_analysis/. Values for gene subsets of interest were used to generate visualizations and calculate enrichment statistics. Shown in Figure 3A are ERC values using proteomes included in Supplementary File S1 represent key elements of mitochondrial and lysosomal proteomes that would interact with zinc transporters. Enrichment statistics were calculated using two sets of annotations to map genes to functions. First, search terms were used (shown in Figure 3B) to search gene descriptors for each gene taken from the UCSC Genome Browser gene track. If a descriptor contained the term, the gene was mapped to that search term annotation. Also included were MitoCarta annotations [35,36] describing mitochondrial genes. After extracting terms, enrichment was calculated in two ways. First, a one-tailed Fisher’s exact test was performed to detect an increased proportion of annotated genes with ERC values above 0.3. This test indicates that a gene being included in a particular annotation and the gene having a high ERC value are related. Second, a Wilcoxon rank-sum test was performed to look for shifts in distributions of ERC values for genes in a particular annotation compared with genes not in that annotation. AUC values, which range from 0 to 1, were calculated directly from the Wilcoxon rank-sum W statistic as W/(number of annotated genes × number of unannotated genes). Small AUC values indicate that annotated genes have lower ERC values than unannotated genes and large AUC values indicate that annotated genes have higher ERC values than unannotated genes.
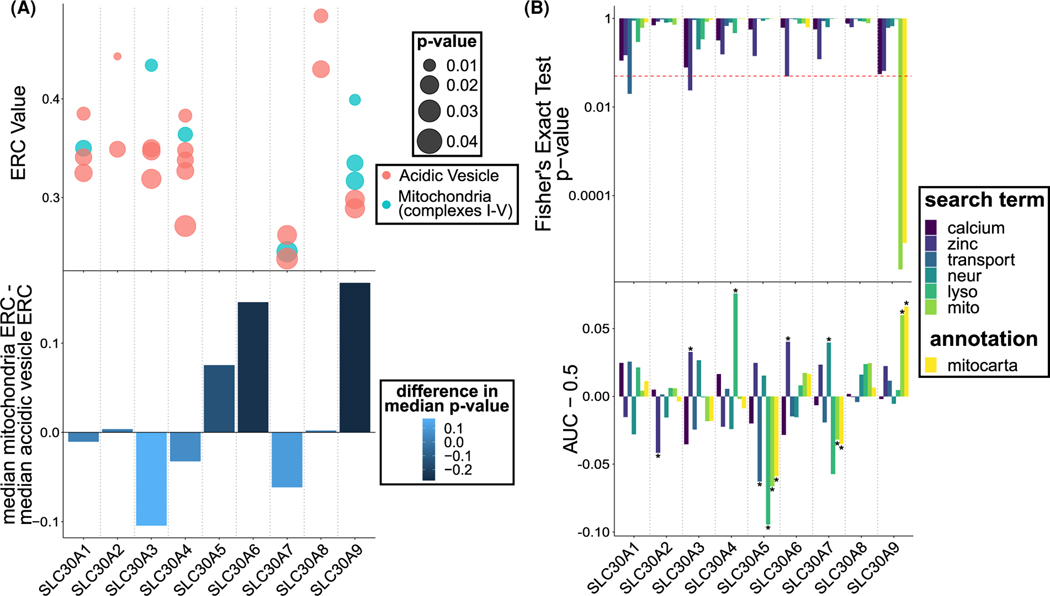
Figure 3. ERC analysis of SLC30 proteins reveals that SLC30A9 (ZnT9) shows unique patterns of correlated evolutionary rates with mitochondrial proteins.
(A) ERC between SLC30A9 and mitochondrial proteins shows higher correlation and lower P-value compared with ERC between SLC30A9 and lysosomal proteins. This contrast is stronger for SLC30A9 than for any other SLC30. Dots represent genes with positive ERC values and P-values less than 0.05. Size of dots are continuously scaled with the P-value. Bars heights show difference in median ERC value between mitochondrial and lysosomal proteins for all genes and bar colors show difference in median P-value between mitochondrial and lysosomal proteins for all genes. (B) SLC30A9 genes with high ERC values are uniquely enriched for mitochondrial function. Search terms represent words identified in gene descriptions to assign them to that term. Also included are Mitocarta annotations. One-tailed Fisher’s Exact Tests were performed to test for significant enrichment of search terms and annotations with ERC values above 0.05, and notably only SLC30A9 shows a significant P-value for mitochondria (bar beyond the red dashed line). A Wilcoxon rank-sum test also showed a uniquely significant positive shift of ERC values for mitochondrial terms and SLC30A9 (asterisks show significance). Centered AUC values demonstrate whether the term was enriched (positive values) or depleted (negative values).
Clustering
Clusters depicted in Figure 4A were constructed using igraph [45]. Genes included were selected from genes listed in Supplementary File S2 that represent mitochondrial complexes and ATP transporters. Lines were drawn to connect genes with ERC values greater than or equal to 0.3 and singleton genes that did not connect to other genes were removed. The optimal clustering algorithm implemented in igraph was used to define clusters.
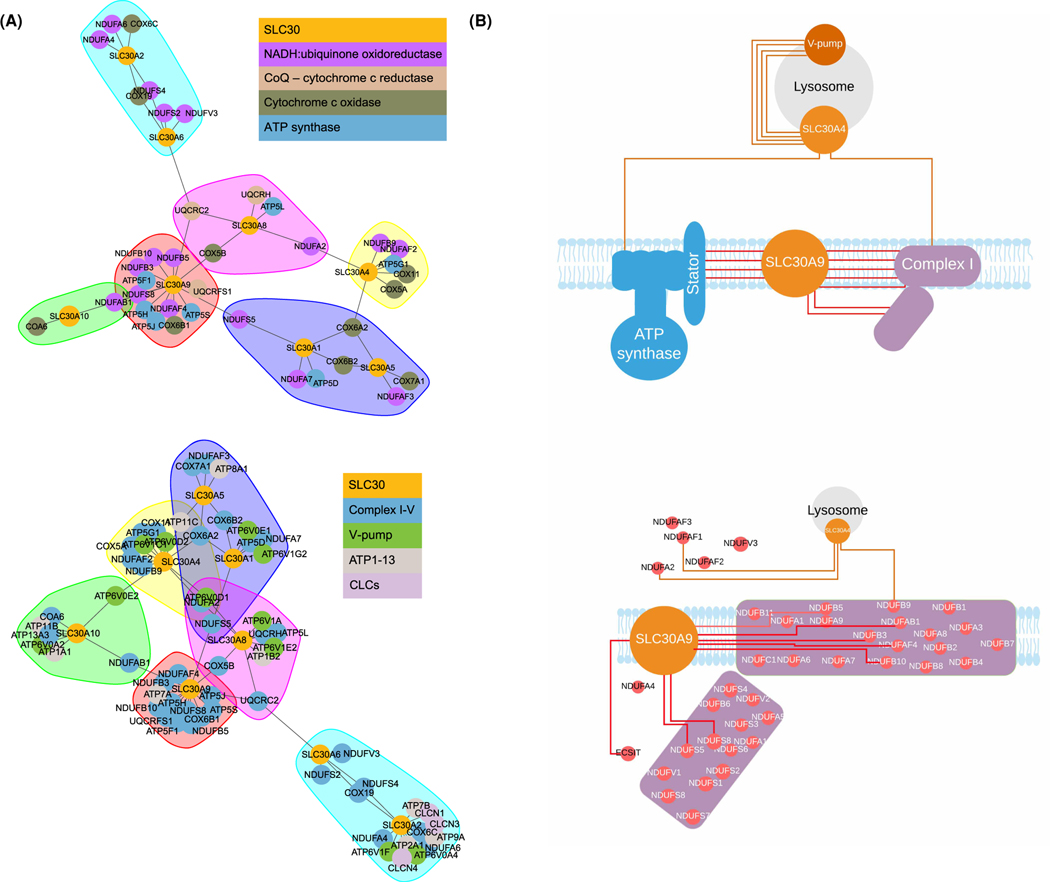
Figure 4. SLC30 evolutionary network clusters and diagrams of SLC30A9 interaction with mitochondrial components.
(A) Clustering of SLC30A transporters with mitochondrial proteins. Clusters highlighted with arbitrary colors indicate communities identified during clustering. Top: SLC30A9 shows robust clustering with numerous mitochondria-related proteins, including NDUF and ATP proteins. Note the cluster including SLC30A9 (in red) with many NADH proteins (purple dots) and ATP synthase proteins (blue dots). Bottom: Clustering using an expanded geneset shows distinctive clustering of SLC30A9 (red cluster) with numerous mitochondria complex I–V proteins (blue circles). (B) Wire diagram comparing ERC-positive interactions (at R > 0.3) between SLC30A9 and the mitochondrial OxPhos components. SLC30A4 is shown as a control.
Protein expression and widefield microscopy
YFP-tagged C- and N-terminal in-frame fusions of human SLC30A9 sequence (NP_006336.3) were synthesized by Vectorbuilder (Chicago, IL, U.S.A.) and transfected in HeLa cells using Lipofectamine 3000 (ThermoFisher, Waltham, MA, U.S.A.). Before transfection, the cells were grown in plastic culture dishes in DMEM supplemented with 10% FBS, at 37°C, 5% CO2, in a humidified incubator and plated on glass coverslips at ~70% confluency. The cells were fixed in 4% formaldehyde (in phosphate-based solution, 5 min incubation) 16–24 h post-transfection, permeabilized with 0.1% Triton X100 in phosphate-based solution, 5 min incubation, transferred to blocking buffer (1% bovine serum albumin and 1% goat serum in phosphate-based solution, 1–24 h) and then treated with primary and secondary (fluorescent) antibodies. T0M20 antibodies (polyclonal, cat number PA5–52843) and Alexa 568-tagged secondary antibodies were from ThermoFisher. The imaging was performed using Nikon SP-1 microscope and analyzed using FIJI [46].
Zinc imaging and qPCR
HeLa cells cultured as above and grown in 12-well culture plates or 35 mm Mattek dishes (MatTek Corp) were transfected with SLC30A9 esiRNA (Sigma, St Louis, MO, U.S.A.) and used 16–48 h post-transfection. qPCR analysis was performed as before [47], and the data were analyzed using the DDCt method. ACTB was used as a housekeeping gene; primers were optimized as described before [47]. For zinc-loading experiments, the cells were exposed to 100–300 μM ZnCl2 in DMEM/FBS for 16 h. Next, the cells were washed, loaded with 5 mM Rhod-2,am (ThermoFisher) in a zinc-containing HEPES-based salt solution (HBSS, in mM: 140 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES (pH 7.4) and 1 g/l glucose, supplemented with 100–300 μM ZnCl2) and washed again. The dishes were inserted into a closed, thermo-controlled (37°C) stage top incubator (Tokai Hit Co.) above the motorized stage of an inverted Nikon TiE fluorescent microscope equipped with a 60× optic (Nikon, CFI Plan Fluor, NA 1.4), a diode-pumped light engine (SPECTRA X, Lumencor). Emissions were detected using an ORCA-Flash 4.0 sCMOS camera (Hamamatsu) and excitation and emission filters were from Chroma. Zinc removal was performed by manual aspiration and gravity-fed application of nominally zinc-free HBSS. The recorded image stacks were analyzed using FIJI. Specifically, discrete regions of interest containing clearly identifiable mitochondria were outlined by hand and the dynamics of average fluorescence intensity in these images was analyzed and compared. Data are presented as the mean ± standard error of the mean.
Results
SLC30A9 phylogeny
We sought to investigate SLC30A9 function by analyzing its evolution throughout the tree of life. Although SLC30A9 variants have been previously identified in some species, including rat, mouse, zebrafish, and fruit fly (Pubmed search), we performed a more rigorous, targeted search. Due to heterogeneity of the SLC30A/SLC39A superfamily, we began by establishing defining features of SLC30A9 and other SLC30As. Zinc permeation by SLC30A is coordinated by aspartate and histidine residues contributed by the HxxxD motif (flanked by D at position 39 of human SLC30A1, forming DxxHxxxD signature) and by the HxxxD motif located downstream of the first motif (at position 250 of human SLC30A1) (Figure 1A; full alignment in Supplementary Figure S1). Previously published crystal structures and homology modeling show that these motifs are located in the transmembrane domains 2 and 5 of human SLC30As [48–50]. Amino acid alignment of human and other organisms’ SLC30A9 with other SLC30As, coupled with homology modeling, reveals a distinct and conserved arrangement (Figure 1A and Supplementary Figure S1). In human SLC30A9, the HxxxD sequence is flanked by a glutamate residue (ExxHxxxD at position 268 of human SLC30A9) and the downstream sequence is ILLED.
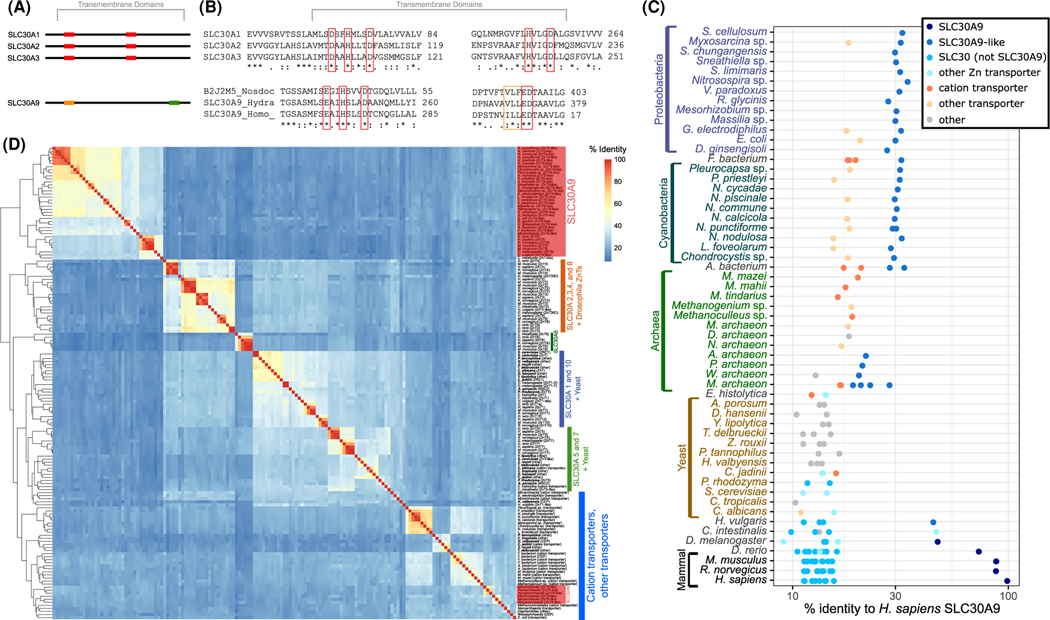
Figure 1. Defining features of SLC30A9.
(A) Cartoon indicating defining features of SLC30A9 and other SLC30As. (B) Partial amino acid alignment of select human SLC30A (top) and SLC30A9 from humans and other organisms (bottom), specifically focused on the conserved differences in the putative permeation center depicted in part A. (C) Bacterial SLC30A9s show higher percent identity with human SLC30A9 than do other animal SLC30As. This suggests potential divergence of SLC30A9 before animal duplication events give rise to other SLC30As. (D) Hierarchical clustering based on percent sequence identity demonstrates that bacterial and animal SLC30A9s cluster together separately from other animal SLC30As, yeast transporters, cation transporters, and other ion transporters, further supporting ancient divergence of SLC30A9 prior to duplication of other SLC30As.
These substitutions provide a unique signature that identifies SLC30A9 and distinguishes it from other SLC30A family members. The signatures are surprisingly well conserved: homologous sequences containing these substitutions and transmembrane domain arrangements reminiscent of human SLC30A9, are found throughout the tree of life (Figure 1A and Supplementary Figure S1). Critically, a detailed phylogenetic analysis of SLC30A9 candidates and other members of the SLC30A family identifies SLC30A9 as a distinct phylogenetic entity, components of which are more related to each other across the tree of life, than to other SLC30A members.
The substitutions specific to SLC30A9 appear to be exceptionally well-conserved as we can detect SLC30A9 homologous sequences containing the invariant ExxHxxxD…(I/V/L)(I/V/L)xED in every clade except for the yeast and other fungi (Table 1, Figures 1C, 2, see additional trees in Supplementary Figures S2, S3). This includes archaea, proteobacteria, cyanobacteria, firmicutes, and actinobacteria, as well as every animal species tested. Organisms on which SLC30A9 is present typically contain a single SLC30A9 homolog, with the exception of Nostoc punctiforme (2 copies), Actinobacteria bacterium (2 copies), and Micrarchaeota archaeon (3 copies). The SLC30A superfamily increases in complexity in eukaryotes, with ten members (including SLC30A9) in mammals, six members in Ciona intestinalis and Drosophila melanogaster, and 4 members in Hydra vulgaris. However, of important note, the numerous duplications of SLC30As are not paralleled by increasing numbers of SLC30A9 homologs (Figures 1C, 2). This distinction points toward a unique evolutionary trajectory for SLC30A9 compared with other SLC30s, potentially because of a unique molecular function for this transporter.
Table 1.
Expression of SLC30A9, MCU, MICU, MMT and their homologs in various organisms (x shows expression)
| Clade | Species | ZNT9 | MCU | MICU | MMT |
|---|---|---|---|---|---|
|
| |||||
| Animals | Homo sapiens | x | x | x | |
| Mus musculus | x | x | x | ||
| Rattus norvegicus | x | x | x | ||
| Danio rerio | x | x | x | ||
| Ciona intestinalis | x | x | x | ||
| Hydra vulgaris | x | x | x | ||
| Drosophila melanogaster | x | x | x | ||
| Fungi | Yarrowia lipolytica | x | |||
| Saccharomyces cerivisiae | x | ||||
| Debaryomyces hansenii | x | ||||
| Pachysolen tannophilus | x | ||||
|
Williopsis saturnus
Ustilago maydis 521 |
x | x | |||
| Cryptococcus neoformans | x | x | |||
| Laccaria bicolor S238N-H82 | x | ||||
| Magnaporthe oryzae 70–15 | x | x | |||
| Penicillium chrysogenum 54–1255 | x | x | |||
| Uncinocarpus reesii 1704 | x | ||||
| Green Algae | Ostreococcus lucimarinus CCE9901 | x | x | x | |
| Chlamydomonas reinhardtii | x | x | x | ||
| Volvox carteri f. nagareinsis | x | x | x | ||
| Protists |
Trichomonas vaginalis G3 Leishmania infantum JPCM5 |
x | x | ||
| Trypanosoma brucei TREU927 | x | x | |||
| Tetrahymena thermophila SB210 | x | x | |||
| Paramecium tetraurelia strain d4–2 | x | ||||
|
Phaeodactylum tricornutum CCAP 1055/1 Plasmodium chabaudi chabaudi |
x | ||||
| Archaea | Candidatus Pacearchaeota archaeon | x | |||
| Candidatus Woesearchaeota archaeon | x | ||||
|
Candidatus Aenigmarchaeota archaeon ex4484_52 Candidatus Diapherotrites archaeon Methanohalophilus mahii Methanolobus tindarius Methanosarcina mazei |
x | ||||
| Bacteria | Sneathiella chungangensis | x | |||
| Massilia sp. LC238 | x | ||||
| Geopsychrobacter electrodiphilus | x | ||||
| Escherichia coli | x | ||||
| Chondrocystis sp. NIES-4102 | x | ||||
| Nosdoc punctiforme | x | ||||
| Leptolyngbya foveolarum | x | ||||
| Nostoc cycadae | x | ||||
As further evidence of SLC30A9’s unique evolution, the human SLC30A9 is more closely related to bacterial SLC30A9 than to other human SLC30As based both on sequence identity and on clustering through sequencebased phylogenetic tree construction (Figures 1D, 2). The SLC30A9 sequences appear to be an ancient clade and have remained a single-copy orthologous group during eukaryotic and bacterial evolution. This monophyletic relationship of the ancient SLC30A9 subtree is supported by strong branch support (100% bootstrap support). The phylogenetic tree also shows a clade of SLC30A9 sequences from archaea that may not be monophyletic with the rest of the SLC30A9 clade, making their membership in this family uncertain. Together these findings show that SLC30A9 is an ancient, deeply conserved protein that likely arose via ancient duplication followed by neofunctionalization, meaning that it should be considered functionally distinct from the rest of the eukaryotic SLC30A family. As such, it appears that the ancestor of the SLC30A family was present early in prokaryote evolution. Indeed, SLC30A9 has remained relatively static, not undergoing further duplications. In contrast, the remaining SLC30A family members have gone through multiple gene duplications and deletions, leading to the SLC30A1–8 and 10 paralogs in eukaryotes.
Yeast and other fungi do not appear to have SLC30A9. MMT1/2 yeast transporters, which have been suggested to be the yeast analog of SLC30A9, do not have the ExxHxxxD…(I/V/L)(I/V/L)ED substitution, and thus likely have a different function. Nonetheless, there appears to be mutual exclusivity between SLC30A9 and MMT1/2 expression, as organisms containing SLC30A9 do not seem to have MMT1/2 (Table 1, Figure 2), suggesting that MMT1/2 may replace SLC30A9 in those species, although our sequence identity and phylogeny construction analyses suggest that MMT1/2 would not have originated from substitutions in SLC30A9. Although we did not further pursue this avenue of inquiry, mechanisms by which yeast species compensated for the loss of such a deeply conserved protein is of great interest. MMT1/2 are found in the yeast mitochondria. This, together with the correlation between the presence of SLC30A9 and MCU in many species (Table 1) strongly suggested that SLC30A9 would be present in the mitochondria.
SLC30A9 coevolution with the mitochondrial components
Since we found that SLC30A9 is an evolutionarily distinct and ancient protein, we next applied another evolutionary method to generate a hypothesis about its function and localization. To gain an insight into the cellular localization of SLC30A9 we performed an ERC analysis between SLC30A9 and a battery of other organellar transporters. Because some SLC30A transporters have been shown or proposed to use a proton gradient as a driving force for zinc transport [49–51], our rationale for this evolutionary approach was as follows: if SLC30A9 uses an ionic gradient to pump zinc, then its evolution could be influenced by the same evolutionary pressures acting on the transporter that establishes the ionic gradient. Hence, the two transporters will have correlated evolutionary rates, which will be uncovered by ERC. We can, therefore, use well established ion transporters, whose localization is known, as baits to search for SLC30As that have coevolved with them, and, as such, probably located within the same organelle.
The analysis was performed using a publicly available ERC portal (https://csb.pitt.edu/erc_analysis/) [52]. This approach utilizes gene-specific evolutionary rates from 33 mammalian species and their ancestral branches calculated for more than 19 000 protein-coding genes. A gene pair’s ERC value is calculated as the correlation coefficient between their rates in those species. Analysis of SLC30A9 focused on several organellar transporters including the vacuolar/vesicular/lysosomal H+ pump (multimolecular complex comprising structurally unrelated proteins that are coded by the ATP6 family of genes [53,54]), the endolysosomal ClC transporters [55,56], and the components of the mitochondrial oxidative phosphorylation chain [57], which either establish or utilize the mitochondrial H+ gradient (see Materials and Methods and Supplementary Figure S2 for the list). We calculated ERC values between SLC30A9 and each of the genes annotated to those batteries of transporters. MCU coevolution with MICU1 [58,59] was used as a positive control for coevolution of functionally linked proteins (R value 0.584, P = 0.002).
SLC30A3 and SLC30A4, known to localize in the acidic organelles containing high levels of ATP6, namely synaptic vesicles [16,60] and lysosomes [18], respectively, were used as controls for specificity of the observed effects. Both of these transporters were shown, or proposed, to utilize H+ gradient established by ATP6 pumps in order to move zinc. As predicted in this control analysis, SLC30A3 and SLC30A4 show several significant ERC values with the ATP6 components (Figure 3A, top, red points; see original plots of ERC values in Supplementary Figure S4), which is consistent with their cellular localization and H+ requirements for transport. In contrast, their ERC scores with the mitochondrial components (blue points) were lower, and, in both cases, ERC was skewed towards the vacuolar/vesicular/lysosomal H+ pump relative to the mitochondrial signal (Figure 3A, bottom).
In contrast, we found that SLC30A9 shows a strong coevolutionary signal with several components of the mitochondrial oxidative phosphorylation chain, which is a very unique pattern when compared with other members of the SLC30A family (Figure 3A, top). Specifically, it has significantly correlated rates with three individual mitochondrial components, belonging to mitochondrial ATP synthase and complex I. In addition, the full distribution of SLC30A9’s scores with the mitochondrial complex encoding genes was much higher than its scores with lysosomal components (Figure 3A, bottom).
We next examined the full set of scores for each SLC30A in mammalian genomes to learn with which functions high-scoring genes were associated with it. This study also shows that SLC30A9 is uniquely enriched for high ERC values with genes encoding proteins that are associated with the mitochondria (Figure 3B). First, we searched the human RefSeq gene descriptions of the high-scoring genes (ERC value >0.3) for specific search terms related to calcium, zinc, lysosomes, and mitochondria [61]. SLC30A9’s high-scoring genes were strongly associated with the ‘mito’ search term and with genes annotated as mitochondrial by MitoCarta 2.0 (Figure 3B, upper) [35,36]. Similarly, we studied the entire distribution of genes as ranked by ERC values with SLC30As. That distribution for SLC30A9 was significantly enriched at high ERC scores for genes with mitochondrial annotations (RefSeq gene description and MitoCarta status) (Figure 3B, lower). In fact, out of all SLC30As, only SLC30A9’s ERC scores showed such associations with mitochondrial annotations.
We also examined the networks of connections drawn based on ERC scores between SLC30As and components of mitochondrial oxidative phosphorylation complexes (Figure 4A, upper) and organellar ion transporters, including the vesicular ClC chloride/proton transporters (Figure 4A, lower). See also Supplementary Figure S5 for more detail. It is clear that SLC30A9 uniquely co-evolves with many mitochondrial components, involving several connections to them, when compared with other SLC30As, as well as other organellar markers. The models in Figure 4B summarize the findings. Based on the totality of these evolutionary studies, we propose that SLC30A9 localizes to the mitochondria. We set out next to test this hypothesis.
SLC30A9 localization and effect on mitochondrial zinc
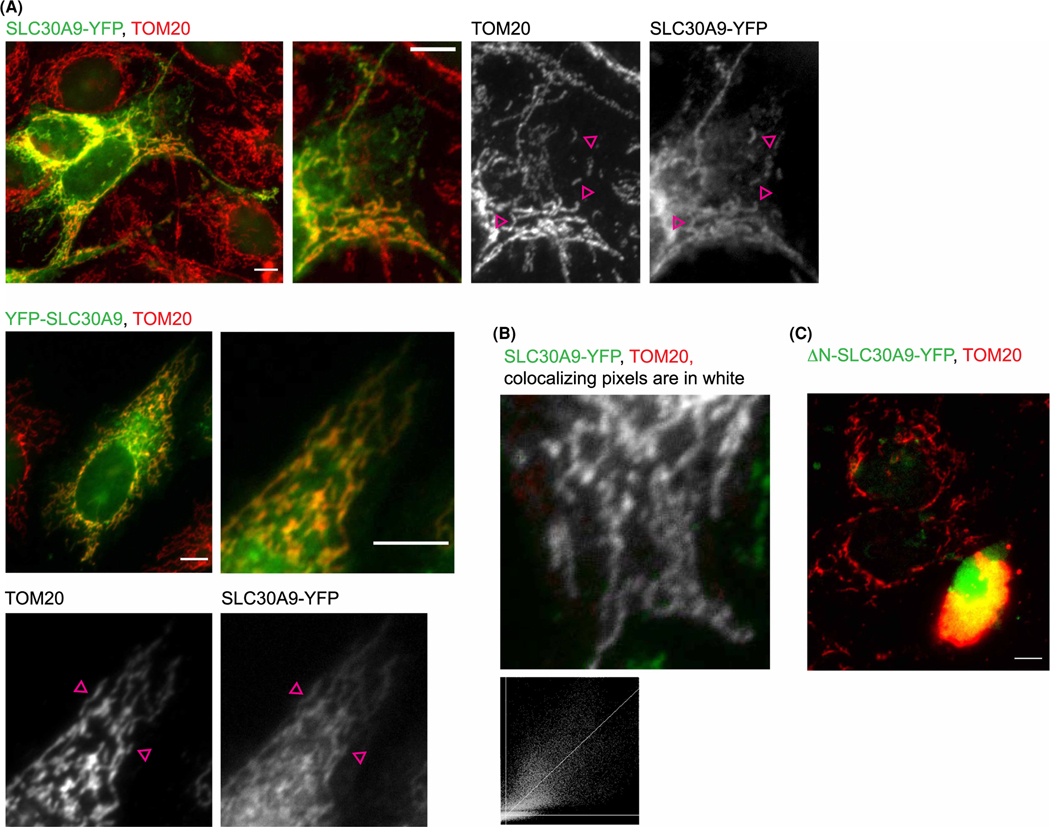
Although MitoCarta predicts the mitochondrial localization of SLC30A9, it has not been directly demonstrated. We did not find commercially available antibodies that produce a convincing organellar stain. Therefore, we transfected HeLa cells with recombinant cDNA constructs coding for human SLC30A9 with in-frame C- or N-terminal YFP fusions. Figure 5A,B shows that both constructs produced a reticular stain overlapping with the mitochondrial marker TOM20 [62]. The stretch of positively charged amino acids in the extreme N-terminus of human SLC30A9 is indicative of mitochondrial localization signal, which is supported by Mitoprot II prediction [63]. Deleting this sequence (MLPGLAAAAAHRCSWSSLCRLRLRCR, aminoacids 1–26) resulted in SLC30A9 mislocalization and significant cell toxicity following transfection, indicating that the mitochondrial localization of SLC30A9 is essential (Figure 5C). Based on this evidence, we conclude that SLC30A9 localizes in the mitochondria. This is the first direct evidence of SLC30A9 localization to this or any other organelle.
Figure 5. SLC30A9 localization.
(A) Widefield fluorescence image of N-terminal (top) and C-terminal (bottom) YFP fusion of human SLC30A9 (green) expressed in HeLa cells and co-stained with antibodies against TOM20 (red). In addition, zoom-in images are shown for clarity. Pink arrows point to examples of overlapping stains. Scale bars are 10 μm. (B) An example of colocalization analysis of SLC30A9 and TOM20 obtained using the Colocalization threshold Plugin of FIJI. Histogram below shows linear regression of intensities of the overlapping pixels in the red and green channels. (C) Evidence of cell death in cells overexpressing N-terminal deletion of SLC30A9. A representative widefiled image of HeLa cells transfected with GFP-tagged N-terminal deletion construct of human SLC30A9 stained as above. Note the swelling and detachment of the transfected cell as well as swelling and compacting at the nucleus of the mitochondria, indicating cell death.
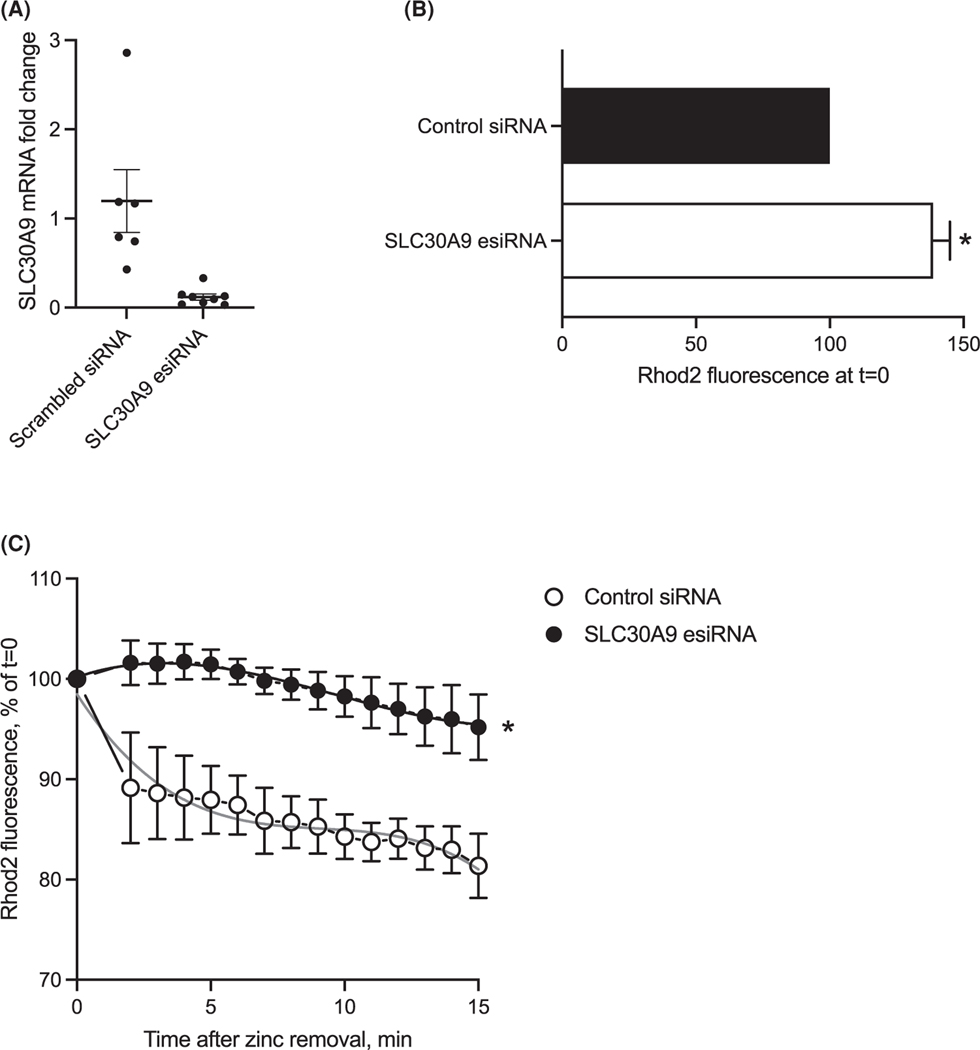
siRNA-driven SLC30A9 deletion in HeLa cells produced significant changes in mitochondrial zinc handling. Mitochondrial zinc content was analyzed using fluorescent divalent-cation sensitive dye Rhod-2,am, which was loaded into the cells for 15 min, followed by a washout. Fluorescence intensity was analyzed using time-lapse fluorescent imaging; the intensity was measured in the regions of interest which were drawn manually. In zinc-loaded cells, the pattern of the fluorescent signals seemed to conform to the mitochondrial shape. When exposed to 300 ¼M zinc overnight, SLC30A9 siRNA-transfected cells (knockdown confirmed by qPCR, Figure 6A) accumulated significantly more zinc, which is evident from statistically higher fluorescence in these cells, compared with control-transfected cells (Figure 6B). Next, we measured the dynamics of the mitochondrial zinc dissipation by returning zinc-exposed cells to a nominally zinc-free medium. Under such conditions, SLC30A9 siRNA-transfected cells lost mitochondria fluorescence signal at a significantly slower rate, indicating slower dissipation of zinc (Figure 6C). These data suggest that SLC30A9 is a functional mitochondrial zinc transporter, which, under normal conditions, is responsible for zinc extrusion from the mitochondria.
Figure 6. Mitochondrial zinc handling deficits in SLC30A9-knockdown HeLa cells.
(A) qPCR analysis of HeLa cells transfected with SLC30A9 esiRNA and harvested 24 h post-transfection. (B) Mitochondrial zinc content in control and SLC30A9-transfected HeLa cells was evaluated using Rhod-2,am. The cells were exposed to zinc for 24 h. Average fluorescence in each region of interest containing clearly identifiable mitochondria was recorded and normalized to the values in the cells that were transfected with scrambled shRNA. (C) The delayed loss of mitochondrial zinc in SLC30A9-deficient cells exposed to nominally zinc-tree medium after 24 h long zinc load. The cells were treated as above; at t = 0 the zinc-containing medium was replaced with nominally zinc free medium and images were taken every 60 s. The fluorescence intensity was normalized to the values recorded at t = 0, which was taken as 100%. In Panels A and B, data represent three independent trials involving 7–10 cells. Data are shown ± SEM. Asterisks represent statistical significance at P < 0.05, for Student’s test in Panel A. In Panel B, the curves were fitted using a third order polynomial curve (smooth gray lines) and an extra sum-of-squares test was performed to answer whether one curve can adequately fit both datasets. That hypothesis was rejected at P < 0.0001 level. Here and below, statistical analysis and data plotting were performed using Prism 9.
Discussion
In the course of the present studies, we used ERC, a bioinformatics assay, to predict the localization of a poorly understood ion transporter. While ERC has been described and tested before [23,25–30], our approach utilized a novel idea that tracking evolutionary histories of functionally linked proteins may inform their functional context, such as localization. Therefore, using this approach we were able to predict protein localization in the absence of obvious clearly defined functional domains. We suggest that this approach can have broad applications in diverse biological fields.
Little is known about the SLC30A9 function, although it has recently been linked to Covid-19 because of strong co-purification of SLC30A9 with SARS-CoV-2 proteins (Supplementary Data in [64]). Furthermore, SLC30A9 has recently been identified as a master regulator of Parkinson’s pathogenesis [65]. Given the central role of mitochondria in Parkinson’s pathology [66] and the emerging role of zinc in this process [67], the evidence of SLC30A9 in the mitochondria suggests a novel contributing factor to Parkinson’s disease.
While SLC30A9 is considered to be a member of the zinc family of transporters, the evidence of its involvement in zinc handling is scarce. Interestingly, polymorphisms in SLC30A9 in African populations have been linked to environmental zinc availability [32], and dysregulation of zinc handling has been detected in fibroblasts from human patients a novel autosomal recessive cerebro-renal syndrome, which has been linked to mutations in SLC30A9 [31]. Our data suggest that under normal conditions, SLC30A9 is a mitochondrial zinc exporter. While this seems to be counteractive to the known function of other members of the SLC30A family, namely, moving zinc into organelles, it is important to note that the mitochondrial proton gradient operates in the opposite direction in mitochondria, when compared with other membranous organelles such as lysosomes and the Golgi apparatus. Given that some zinc transporters have been shown or proposed to be proton-driven, the ‘reversed’ mitochondrial proton gradient may explain the outward direction of SLC30A9-driven cation transport detected in our system and attributed to SLC30A9 activity.
Mitochondrial zinc transport mechanisms are poorly understood. Several components of the mitochondrial respiratory chain require zinc [20,21]. Furthermore, proteostasis, protein insertion, enzymatic activities and other functions of the mitochondria are regulated by zinc [68–71]. With this in mind, the SLC30A9 coevolution with the mitochondrial oxidative phosphorylation chain components is informative. However, excess zinc has adverse impacts on the mitochondrial respiratory chain and has been implicated in several pathologies including Alzheimer’s disease and stroke [11,72–76]. Importantly, a mechanism of zinc efflux from the mitochondria had, heretofore, not been identified. Ascribing this role to SLC30A9 is likely to have far-reaching implications in a more complete understanding of zinc management by the mitochondria.
At the moment, the mechanistic reasons for coevolution between SLC30A9 and mitochondrial oxidative phosphorylation chain components are not completely clear. ERC suggests functional, but not necessarily physical interaction between these proteins. The coevolution may reflect the shared function of zinc flux and utilization in the mitochondria, as discussed above. Furthermore, SLC30A9 appears to specifically coevolve with the components of the peripheral stalk of the mitochondrial ATP synthase (Figure 4B) suggesting a shared role in proton transport, as the membrane components of the peripheral stalk were suggested to contribute to the formation of the proton channel [77]. Further investigation would help uncover the mechanism behind the coevolution between SLC30A9 and mitochondrial respiratory chain components and further validate ERC as a cutting-edge tool of molecular discovery and characterization.
Supplementary Material
Acknowledgements
This work was supported by the NIH grant 1R21NS111944 to K.K. and E.A., and by NIH grant 1R01HG009299 to M.C. and N.L.C.
Abbreviations
- ERC
evolutionary rate covariation
- HBSS
HEPES-based salt solution
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
CRediT Author Contribution
Kirill Kiselyov: Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing — original draft, Project administration, Writing — review and editing. Amanda Kowalczyk: Conceptualization, Resources, Data curation, Formal analysis, Investigation, Visualization, Writing — original draft, Writing — review and editing. Omotola Gbadamosi: Investigation. Kathryn Kolor: Investigation, Visualization. Jahree Sosa: Investigation. Livia Andrzejczuk: Investigation. Gregory A. Gibson: Investigation. Claudette M. St. Croix: Resources, Supervision, Investigation. Maria Chikina: Conceptualization, Resources, Funding acquisition. Elias Aizenman: Conceptualization, Resources, Supervision, Funding acquisition, Writing — original draft, Project administration, Writing — review and editing. Nathan Clark: Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Writing — original draft, Project administration, Writing — review and editing.
Data Availability
All supporting data are included within the main article and its Supplementary Files.
References
- 1.Lemos FO, Bultynck G. and Parys JB (2021) A comprehensive overview of the complex world of the endo- and sarcoplasmic reticulum Ca2+-leak channels. Biochim. Biophys. Acta Mol. Cell Res 1868, 119020 10.1016/j.bbamcr.2021.119020 [DOI] [PubMed] [Google Scholar]
- 2.Lopez JJ, Jardin I, Albarrán L, Sanchez-Collado J, Cantonero C, Salido GM et al. (2020) Molecular basis and regulation of store-operated calcium entry. Adv. Exp. Med. Biol 1131, 445–469 10.1007/978-3-030-12457-1_17 [DOI] [PubMed] [Google Scholar]
- 3.Gandini MA and Zamponi GW (2021) Voltage-gated calcium channel nanodomains: molecular composition and function. FEBS J. Online ahead of print 10.1111/febs.15759 [DOI] [PubMed] [Google Scholar]
- 4.Krall RF, Tzounopoulos T. and Aizenman E. (2021) The function and regulation of zinc in the brain. Neuroscience 457, 235–258 10.1016/j.neuroscience.2021.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCormick NH, Hennigar SR, Kiselyov K. and Kelleher SL (2014) The biology of zinc transport in mammary epithelial cells: implications for mammary gland development, lactation, and involution. J. Mammary Gland Biol. Neoplasia 19, 59–71 10.1007/s10911-013-9314-4 [DOI] [PubMed] [Google Scholar]
- 6.Kambe T, Tsuji T, Hashimoto A. and Itsumura N. (2015) The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev 95, 749–784 10.1152/physrev.00035.2014 [DOI] [PubMed] [Google Scholar]
- 7.Andreini C, Banci L, Bertini I. and Rosato A. (2006) Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 5, 196–201 10.1021/pr050361j [DOI] [PubMed] [Google Scholar]
- 8.Prasad AS (2013) Discovery of human zinc deficiency: its impact on human health and disease. Adv. Nutr 4, 176–190 10.3945/an.112.003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galasso SL and Dyck RH (2007) The role of zinc in cerebral ischemia. Mol. Med 13, 380–387 10.2119/2007-00044.Galasso [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sensi SL, Paoletti P, Koh J-Y, Aizenman E, Bush AI and Hershfinkel M. (2011) The neurophysiology and pathology of brain zinc. J. Neurosci 31, 16076–16085 10.1523/JNEUROSCI.3454-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granzotto A, Canzoniero LMT and Sensi SL (2020) A neurotoxic ménage-à-trois: glutamate, calcium, and zinc in the excitotoxic cascade. Front. Mol. Neurosci 13, 600089 10.3389/fnmol.2020.600089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji SG, Medvedeva YV, Wang H-L, Yin HZ and Weiss JH (2019) Mitochondrial zn2+ accumulation: A potential trigger of hippocampal ischemic injury. Neuroscientist 25, 126–138 10.1177/1073858418772548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thirumoorthy N, Shyam Sunder A, Manisenthil Kumar K, Senthil Kumar M, Ganesh G. and Chatterjee M. (2011) A review of metallothionein isoforms and their role in pathophysiology. World J. Surg. Oncol 9, 54 10.1186/1477-7819-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmiter RD and Findley SD (1995) Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 14, 639–649 10.1002/j.1460-2075.1995.tb07042.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milon B, Wu Q, Zou J, Costello LC and Franklin RB (2006) Histidine residues in the region between transmembrane domains III and IV of hZip1 are required for zinc transport across the plasma membrane in PC-3 cells. Biochim. Biophys. Acta 1758, 1696–1701 10.1016/j.bbamem.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 16.Palmiter RD, Cole TB, Quaife CJ and Findley SD (1996) ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc. Natl Acad. Sci. U.S.A 93, 14934–14939 10.1073/pnas.93.25.14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez V. and Kelleher SL (2009) Zinc transporter-2 (ZnT2) variants are localized to distinct subcellular compartments and functionally transport zinc. Biochem. J 422, 43–52 10.1042/BJ20081189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukic I, Lee JK, Coblentz J, Kelleher SL and Kiselyov K. (2013) Zinc-dependent lysosomal enlargement in TRPML1-deficient cells involves MTF-1 transcription factor and ZnT4 (Slc30a4) transporter. Biochem. J 451, 155–163 10.1042/BJ20121506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kambe T, Matsunaga M. and Takeda TA (2017) Understanding the contribution of zinc transporters in the function of the early secretory pathway. Int. J. Mol. Sci 18, 2179 10.3390/ijms18102179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turan B. and Tuncay E. (2021) The role of labile Zn2+ and Zn2+-transporters in the pathophysiology of mitochondria dysfunction in cardiomyocytes. Mol. Cell. Biochem 476, 971–989 10.1007/s11010-020-03964-8 [DOI] [PubMed] [Google Scholar]
- 21.Ji SG, Medvedeva YV and Weiss JH (2020) Zn2+ entry through the mitochondrial calcium uniporter is a critical contributor to mitochondrial dysfunction and neurodegeneration. Exp. Neurol 325, 113161 10.1016/j.expneurol.2019.113161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medvedeva YV and Weiss JH (2014) Intramitochondrial Zn2+ accumulation via the Ca2+ uniporter contributes to acute ischemic neurodegeneration. Neurobiol. Dis 68, 137–144 10.1016/j.nbd.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark NL, Alani E. and Aquadro CF (2012) Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 22, 714–720 10.1101/gr.132647.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priedigkeit N, Wolfe N. and Clark NL (2015) Evolutionary signatures amongst disease genes permit novel methods for gene prioritization and construction of informative gene-based networks. PLoS Genet. 11, e1004967 10.1371/journal.pgen.1004967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talsness DM, Owings KG, Coelho E, Mercenne G, Pleinis JM, Partha R. et al. (2020) A drosophila screen identifies NKCC1 as a modifier of NGLY1 deficiency. elife 9, e57831 10.7554/eLife.57831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunette GJ, Jamalruddin MA, Baldock RA, Clark NL and Bernstein KA (2019) Evolution-based screening enables genome-wide prioritization and discovery of DNA repair genes. Proc. Natl Acad. Sci. U.S.A. 116, 19593–19599 10.1073/pnas.1906559116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raza Q, Choi JY, Li Y, O’Dowd RM, Watkins SC, Chikina M. et al. (2019) Evolutionary rate covariation analysis of E-cadherin identifies raskol as a regulator of cell adhesion and actin dynamics in Drosophila. PLoS Genet 15, e1007720 10.1371/journal.pgen.1007720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegler AB, Augustin H, Clark NL, Berthelot-Grosjean M, Simonnet MM, Steinert JR et al. (2016) The amino acid transporter JhI-21 coevolves with glutamate receptors, impacts NMJ physiology, and influences locomotor activity in drosophila larvae. Sci. Rep 6, 19692 10.1038/srep19692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL and Wolfner MF (2014) Evolutionary rate covariation identifies new members of a protein network required for drosophila melanogaster female post-mating responses. PLoS Genet. 10, e1004108 10.1371/journal.pgen.1004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Z, Ye G. and Werren JH (2019) Evolutionary rate correlation between mitochondrial-encoded and mitochondria-associated nuclear-encoded proteins in insects. Mol. Biol. Evol 36, 1022–1036 10.1093/molbev/msz036 [DOI] [PubMed] [Google Scholar]
- 31.Perez Y, Shorer Z, Liani-Leibson K, Chabosseau P, Kadir R, Volodarsky M. et al. (2017) SLC30A9 mutation affecting intracellular zinc homeostasis causes a novel cerebro-renal syndrome. Brain 140, 928–939 10.1093/brain/awx013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Li J, Tian L, Lu D, Yuan K, Yuan Y. et al. (2015) Differential natural selection of human zinc transporter genes between african and non-African populations. Sci. Rep 5, 9658 10.1038/srep09658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sazanov LA (2015) A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388 10.1038/nrm3997 [DOI] [PubMed] [Google Scholar]
- 34.Neupane P, Bhuju S, Thapa N. and Bhattarai HK (2019) ATP synthase: structure, function and inhibition. Biomol. Concepts 10, 1–10 10.1515/bmc-2019-0001 [DOI] [PubMed] [Google Scholar]
- 35.Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ et al. (2021) Mitocarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 10.1093/nar/gkaa1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvo SE, Clauser KR and Mootha VK (2016) Mitocarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–7 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N. et al. (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG et al. (2003) Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31, 3497–3500 10.1093/nar/gkg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W. and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 40.Letunic I. and Bork P. (2007) Interactive tree Of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 41.Letunic I. and Bork P. (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol 24, 1586–1591 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 44.Sato T, Yamanishi Y, Kanehisa M. and Toh H. (2005) The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21, 3482–3489 10.1093/bioinformatics/bti564 [DOI] [PubMed] [Google Scholar]
- 45.Gustavsen JA, Pai S, Isserlin R, Demchak B. and Pico AR (2019) RCy3: Network biology using cytoscape from within R. [version 3; peer review: 3 approved]. F1000Res. 8, 1774 10.12688/f1000research.20887.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peña KA and Kiselyov K. (2015) Transition metals activate TFEB in overexpressing cells. Biochem. J 470, 65–76 10.1042/BJ20140645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoch E, Lin W, Chai J, Hershfinkel M, Fu D. and Sekler I. (2012) Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc. Natl Acad. Sci. U.S.A 109, 7202–7207 10.1073/pnas.1200362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohana E, Hoch E, Keasar C, Kambe T, Yifrach O, Hershfinkel M. et al. (2009) Identification of the Zn2+ binding site and mode of operation of a mammalian Zn2+ transporter. J. Biol. Chem 284, 17677–17686 10.1074/jbc.M109.007203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoch E, Levy M, Hershfinkel M. and Sekler I. (2020) Elucidating the H+ coupled zn2+ transport mechanism of ZIP4; implications in acrodermatitis enteropathica. Int. J. Mol. Sci 21, 734 10.3390/ijms21030734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golan Y, Alhadeff R, Warshel A. and Assaraf YG (2019) Znt2 is an electroneutral proton-coupled vesicular antiporter displaying an apparent stoichiometry of two protons per zinc ion. PLoS Comput. Biol 15, e1006882 10.1371/journal.pcbi.1006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfe NW and Clark NL (2015) ERC analysis: web-based inference of gene function via evolutionary rate covariation. Bioinformatics 31, 3835–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beyenbach KW and Wieczorek H. (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol 209, 577–589 10.1242/jeb.02014 [DOI] [PubMed] [Google Scholar]
- 54.Grabe M, Wang H. and Oster G. (2000) The mechanochemistry of V-ATPase proton pumps. Biophys. J 78, 2798–2813 10.1016/S0006-3495(00)76823-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jentsch TJ (2007) Chloride and the endosomal-lysosomal pathway: emerging roles of CLC chloride transporters. J. Physiol 578, 633–640 10.1113/jphysiol.2006.124719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poroca DR, Pelis RM and Chappe VM (2017) Clc channels and transporters: structure, physiological functions, and implications in human chloride channelopathies. Front. Pharmacol 8, 151 10.3389/fphar.2017.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kühlbrandt W. (2015) Structure and function of mitochondrial membrane protein complexes. BMC Biol. 13, 89 10.1186/s12915-015-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patron M, Checchetto V, Raffaello A, Teardo E, Vecellio Reane D, Mantoan M. et al. (2014) MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell 53, 726–737 10.1016/j.molcel.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallilankaraman K, Doonan P, Cárdenas C, Chandramoorthy HC, Müller M, Miller R. et al. (2012) MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644 10.1016/j.cell.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole TB, Wenzel HJ, Kafer KE, Schwartzkroin PA and Palmiter RD (1999) Elimination of zinc from synaptic vesicles in the intact mouse brain by disruption of the ZnT3 gene. Proc. Natl Acad. Sci. U.S.A. 96, 1716–1721 10.1073/pnas.96.4.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruitt KD and Maglott DR (2001) Refseq and locusLink: NCBI gene-centered resources. Nucleic Acids Res. 29, 137–140 10.1093/nar/29.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S. et al. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 10.1016/S0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- 63.Claros MG and Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem 241, 779–786 10.1111/j.1432-1033.1996.00779.x [DOI] [PubMed] [Google Scholar]
- 64.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM et al. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vargas DM, De Bastiani MA, Parsons RB and Klamt F. (2021) Parkinson’s disease master regulators on substantia nigra and frontal cortex and their use for drug repositioning. Mol. Neurobiol 58, 1517–1534 10.1007/s12035-020-02203-x [DOI] [PubMed] [Google Scholar]
- 66.Trinh D, Israwi AR, Arathoon LR, Gleave JA and Nash JE (2021) The multi-faceted role of mitochondria in the pathology of Parkinson’s disease. J. Neurochem 156, 715–752 10.1111/jnc.15154 [DOI] [PubMed] [Google Scholar]
- 67.Park J-S, Koentjoro B, Veivers D, Mackay-Sim A. and Sue CM (2014) Parkinson’s disease-associated human ATP13A2 (PARK9) deficiency causes zinc dyshomeostasis and mitochondrial dysfunction. Hum. Mol. Genet 23, 2802–2815 10.1093/hmg/ddt623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Audano M, Pedretti S, Cermenati G, Brioschi E, Diaferia GR, Ghisletti S. et al. (2018) Zc3h10 is a novel mitochondrial regulator. EMBO Rep. 19, e45531 10.15252/embr.201745531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costello LC, Liu Y, Franklin RB and Kennedy MC (1997) Zinc inhibition of mitochondrial aconitase and its importance in citrate metabolism of prostate epithelial cells. J. Biol. Chem 272, 28875–28881 10.1074/jbc.272.46.28875 [DOI] [PubMed] [Google Scholar]
- 70.Atkinson A, Khalimonchuk O, Smith P, Sabic H, Eide D. and Winge DR (2010) Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J. Biol. Chem 285, 19450–19459 10.1074/jbc.M110.109793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye B, Maret W. and Vallee BL (2001) Zinc metallothionein imported into liver mitochondria modulates respiration. Proc. Natl Acad. Sci. U.S.A 98, 2317–2322 10.1073/pnas.041619198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin HZ, Wang H-L, Ji SG, Medvedeva YV, Tian G, Bazrafkan AK et al. (2019) Rapid intramitochondrial zn2+ accumulation in CA1 hippocampal pyramidal neurons after transient global ischemia: A possible contributor to mitochondrial disruption and cell death. J. Neuropathol. Exp. Neurol 78, 655–664 10.1093/jnen/nlz042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nolte C, Gore A, Sekler I, Kresse W, Hershfinkel M, Hoffmann A. et al. (2004) ZnT-1 expression in astroglial cells protects against zinc toxicity and slows the accumulation of intracellular zinc. Glia 48, 145–155 10.1002/glia.20065 [DOI] [PubMed] [Google Scholar]
- 74.Isaev NK, Stelmashook EV and Genrikhs EE (2020) Role of zinc and copper ions in the pathogenetic mechanisms of traumatic brain injury and Alzheimer’s disease. Rev. Neurosci 31, 233–243 10.1515/revneuro-2019-0052 [DOI] [PubMed] [Google Scholar]
- 75.Lee J-Y, Cho E, Seo J-W, Hwang JJ and Koh J-Y (2012) Alteration of the cerebral zinc pool in a mouse model of Alzheimer disease. J. Neuropathol. Exp. Neurol 71, 211–222 10.1097/NEN.0b013e3182417387 [DOI] [PubMed] [Google Scholar]
- 76.Cherny RA, Atwood CS, Xilinas ME, Gray DN, Jones WD, McLean CA et al. (2001) Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer’s disease transgenic mice. Neuron 30, 665–676 10.1016/S0896-6273(01)00317-8 [DOI] [PubMed] [Google Scholar]
- 77.Colina-Tenorio L, Dautant A, Miranda-Astudillo H, Giraud M-F and González-Halphen D. (2018) The peripheral stalk of rotary atpases. Front. Physiol 9, 1243 10.3389/fphys.2018.01243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supporting data are included within the main article and its Supplementary Files.