Highlights
-
•
The concentration of MIF in peripheral blood and the expression of MIF in tumor microenvironment were negatively correlated with clinical efficacy.
-
•
Intratumoral MIF levels correlate with changes in the immune cell population in the tumor microenvironment.
-
•
Serum MIF concentration level can be used as a marker to predict the efficacy of immunotherapy combined with chemotherapy in the treatment of ESCC.
Keywords: Macrophage migration inhibitory factor (MIF), Neoadjuvant therapy, Immunotherapy, Esophageal squamous cell carcinoma, Biomarker
Abstract
Purpose
This study aimed to screen biomarkers to predict the efficacy of programmed cell death 1 (PD-1) blockade immunotherapy combined with chemotherapy as neoadjuvant therapy for esophageal squamous cell carcinoma (ESCC).
Methods
In the first stage of the study, the baseline concentrations of 40 tumor-related chemokines in the serum samples of 50 patients were measured to screen for possible biomarkers. We investigated whether the baseline concentration of the selected chemokine was related to the therapeutic outcomes and tumor microenvironment states of patients treated with the therapy. In the second stage, the reliability of the selected biomarkers was retested in 34 patients.
Results
The baseline concentration of macrophage migration inhibitory factor (MIF) was negatively correlated with disease-free survival (DFS) and overall survival (OS) in patients treated with the therapy. In addition, a low baseline expression level of MIF is related to a better tumor microenvironment for the treatment of ESCC. A secondary finding was that effective treatment decreased the serum concentration of MIF.
Conclusion
Baseline MIF levels were negatively correlated with neoadjuvant therapy efficacy. Thus, MIF may serve as a predictive biomarker for this therapy. The accuracy of the prediction could be improved if the serum concentration of MIF is measured again after the patient received several weeks of treatment.
Graphical abstract
Introduction
Esophageal carcinoma, which ranks seventh in cancer incidence and sixth in overall cancer-related mortality, is a severe malignancy. Based on its histopathological features, esophageal carcinoma can be classified as squamous cell carcinoma or adenocarcinoma. Esophageal squamous cell carcinoma (ESCC) is the predominant cancer subtype in Asia [1,2]. The traditional treatments for ESCC include surgery, chemotherapy, and radiotherapy [3,4]. Although surgical resection is the most common treatment, the R0 resection rate is only approximately 50% in patients with locally advanced tumors, leading to a high recurrence rate [5]. To improve clinical outcomes, neoadjuvant therapies using preoperative chemotherapy or chemoradiotherapy have been developed and have significantly increased the R0 resection and survival rates of patients with ESCC [6]. However, a low response rate to chemotherapy has become a new problem [7]. In this case, a method using programmed cell death protein 1 (PD-1) blockade immunotherapy combined with chemotherapy as a neoadjuvant therapy was discovered.
PD-1 can elicit the immune checkpoint response of T cells in the tumor microenvironment (TME), resulting in the escape of tumor cells from immune surveillance. PD-1 blockade immunotherapy provides a way to overcome immune escape and generate immune responses against tumor cells. The National Comprehensive Cancer Network guidelines recommend the use of PD-1 blockade immunotherapy for advanced esophageal cancer. The combination of PD-1 blockade immunotherapy and chemotherapy can generate stronger anti-tumor responses [8]. The use of such a combination therapy as a neoadjuvant therapy can lead to better clinical outcomes and a higher response rate than chemotherapy alone. When treating patients with resectable non-small cell lung cancer and esophageal cancer, PD-1 blockade immunotherapy combined with chemotherapy as neoadjuvant therapy has successfully achieved tumor regression and improved the survival rate of patients [9,10]. Additionally, when camrelizumab is combined with carboplatin and nab-paclitaxel to treat ESCC, patients have an objective response rate of 90.5%, confirming the high response rate of combination therapy [9]. Recently, in patients with advanced ESCC, compared to those who were treated with chemotherapy alone, it improved progression-free survival (PFS) and overall survival (OS) and increased the objective response rate by 69.3% in patients treated with a combination of toripalimab and chemotherapy [7]. Despite the promising outcomes, not all patients showed complete responses, and disease progression was noted, suggesting that a clinically applicable biomarker is needed to predict the patient response and treatment efficacy of this neoadjuvant therapy.
Several predictive biomarkers for gastroesophageal carcinoma immunotherapy have been validated, including programmed death ligand-1 (PD-L1), mismatch repair, microsatellite instability (MMR/MSI), and tumor mutation burden (TMB). However, these biomarkers have limited sensitivity or specificity in predicting treatment efficacy [11]. Chemokines control cell migration, localization, and interactions within tissues. They guide immune cells to the TME and mediate local anti-tumor immune responses [12]. The peripheral circulating chemokines CXCL8, CXCL9, and CXCL10 can be used as predictive biomarkers for PD-1 blockade immunotherapy in the treatment of melanoma and lung cancer [13], [14], [15]. However, few studies have been conducted to identify predictive biomarkers for PD-1 blockade immunotherapy combined with chemotherapy as a neoadjuvant therapy for ESCC.
Our purpose was to screen for a biomarker for predicting the efficacy of PD-1 blockade immunotherapy combined with chemotherapy as a neoadjuvant therapy for ESCC. The baseline levels of Macrophage Migration Inhibitory Factor (MIF) in patients’ peripheral blood and TME were negatively correlated with the clinical outcomes of neoadjuvant therapy. Thus, MIF may serve as a clinically applicable predictive biomarker for neoadjuvant therapy.
Materials and methods
Patients
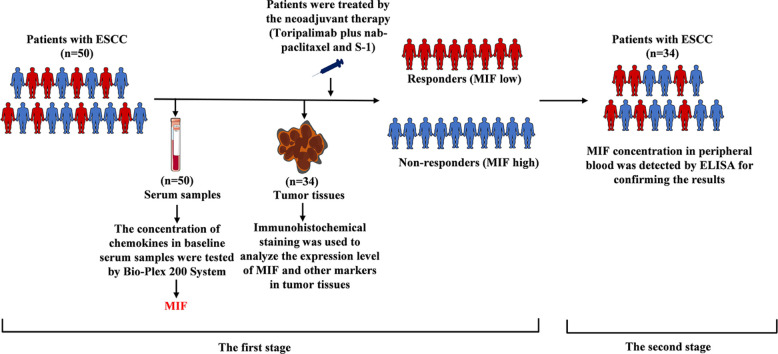
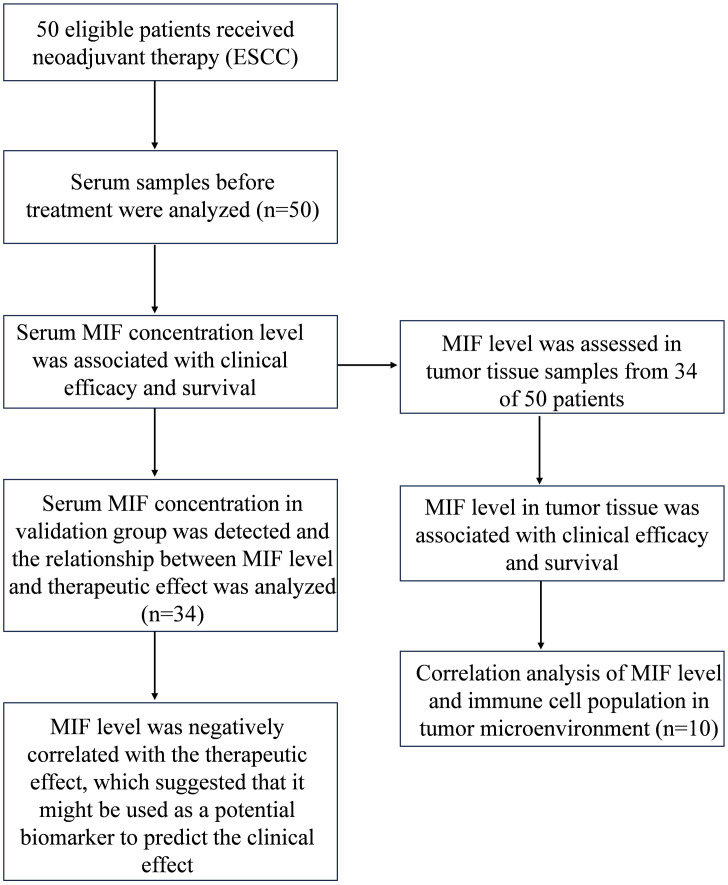
Fig. 1 shows an overview of this study. All samples were acquired from a single-arm, single-center, open-label phase II clinical trial. Fifty patients newly diagnosed with resectable ESCC were included in the first stage of this study, samples were provided by 50 patients with newly diagnosed resectable ESCC. Patient characteristics are summarized in Supplementary Table 1. In the second stage of the study, samples were obtained from 34 patients with newly diagnosed resectable ESCC, and their characteristics are summarized in Supplementary Table 2. The patients received PD-1 blockade immunotherapy (teriprizumab) combined with chemotherapy as a neoadjuvant therapy before surgical resection. Baseline serum and tumor tissue samples were obtained before treatment initiation. Details of the neoadjuvant therapy are presented in the supplementary materials. All the patients enrolled in the trial underwent the entire therapeutic procedure and provided written informed consent. The study protocol followed the Declaration of Helsinki, GCP ethical guidelines, and the Chinese regulations for clinical trial research. The study was approved by the Ethics Committee of the PLA General Hospital.
Fig. 1.
Flow chart.
Clinical outcomes evaluation
The efficacy of neoadjuvant therapy was evaluated according to the criteria from RECIST 1.1. The survival of patients after treatment was evaluated using disease-free survival (DFS; the time from the initiation of treatment to the onset of tumor recurrence or death caused by any cause) and OS. The therapeutic effects were evaluated as complete response (CR), partial response (PR), stable disease (SD), and disease progression (PD). Patients who achieved CR or PR were classified into the response (R) group, and those who achieved SD or underwent PD were classified into the non-response (NR) group. Treatment responses were evaluated in the absence of corticosteroids. Overall Response Rate (ORR) and Disease Control Rate (DCR) were used to evaluate pathological responses after treatment.
Quantitative analysis of chemokines
The chemokine concentrations in baseline serum samples were tested using the Bio-Plex 200 System. The accuracy of the system was verified using the Bio-Plex Validation Kit and Bio-Plex Calibration Kit (Bio-Rad, USA). Patient sera were sampled before the initiation of neoadjuvant therapy. After being pretreated by centrifugation (13000 rpm, 10 min at 4 °C), serum samples were tested by Bio-Plex ProTM Human Chemokine Panel (Bio-Rad, USA), which could detect 40 common tumor-related chemokines listed in supplementary materials. The concentrations of chemokines in the serum samples were obtained and calculated as concentration scores using Bio-plex Manager 5.0 Software in Luminex Analysis System.
Immunohistochemistry and image assessment
Tumor tissue samples were obtained from patients before the initiation of therapy. Samples were immobilized by 10% formalin and embedded with paraffin before being sliced into thin sections of 3 μm. Sections were deparaffinized with xylene and rehydrated using a series of descending alcohol concentrations. After using citrate buffer to achieve Antigen retrieval, sections were stained by the primary antibody against human MIF (dilution, 1:50; Abcam, UK) and incubated overnight at 4 °C. The secondary antibody (Gene Tech, China) was added and the sections were incubated at 37°C for 20 min.. After washing with PBS, 3, 3-Diaminobezidine (Gene Tech, China) was used for color rendering. Immunohistochemistry (IHC) staining of six immunological markers, PD-L1 (dilution, 1:200; Cell Signaling Technology, USA), CD4 (dilution,1:400; Gene Tech, China), CD8 (dilution, 1:200; Abcam, UK), CD20 (dilution, 1:200; Abcam, UK), CD163 (dilution, 1:800; Gene Tech, China), and Foxp3 (dilution, 1:300; Abcam, UK), was performed using the same method. IHC images were visualized using cellSens Platform (Olympus, Japan). The images were examined using a semi-quantitative score. The parenchyma and stroma were evaluated as previously described [16]. The average percentage of positive cells among all cells was calculated by averaging the results from five randomly selected high-magnification ( × 200) fields. Images were analyzed using image analysis software (ImageJ, USA).
ELISAs
Serum MIF concentration was quantified using a Human MIF Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA). The procedures were conducted according to the manufacturer's instructions. Serum samples were diluted five times with sample diluent buffer (Calibrator Diluent RD5-20). After adding 100 μl of Assay Diluent RD1-53 to the detection plate, 50 μl of samples and standards (0, 0.156, 0.313, 0.625, 1.25, 2.5, 5, and 10 ng/ml) were added and incubated in the plate for 2 hours at room temperature. After incubation, the plate was washed four times with the Wash Buffer (400 μl/well). Next, the Human MIF Conjugate (200 μl/well) was added, and the plate was incubated again for 2 h at room temperature. After incubation, the plate was washed four times with the Wash Buffer (400 μl/well). The substrate Solution (200 μl/well) was added, and the plate was incubated for 30 min. in the dark. After incubation, the Stop Solution was added (50 μl/well). The absorbance was read at 450 nm using a microplate reader (Multiskan Go, Thermo Fisher Scientific, USA). The MIF concentration in each test sample was quantified using a standard curve plotted according to the standards. Serum samples were measured in duplicate. The coefficient of variation was less than 15%.
Statistical methods
GraphPad Prism (version 8.0, USA), IBM SPSS Statistics (IBM Corp., USA), and software R 3.6.3. The R package of ggplot2 was used for visualization [17]. The R package of Complex Heatmap was used to make a heatmap for chemokine visualization. The R package of pROC was used to make ROC curves. The R package of Survival was used for DFS and OS survival analysis, and Kaplan–Meier curves were statistically analyzed by log-rank test. The categorical variables of different groups were compared using the chi-square test and Fisher's exact probability test. The continuous variables of different groups were compared using a t-test and Wilcoxon rank sum test. The Cox proportional hazard regression model was used to identify independent prognostic factors associated with DFS and OS. The RNAseq data of pan-cancer in TCGA database was collected in FPKM format, and the differences in chemokine and chemotaxis-relevant analyte expressions between tumor tissues and normal tissues were compared after log2 transformation. The R package of GSVA was used to analyze the associations between the expression levels of chemokines and chemotaxis-relevant analytes and their immune infiltrations [18,19]. GraphPad Prism 8 was used for calculating the immunohistochemical statistics of each immunological marker. Significant difference terms were indicated as follows: CI, confidence interval; ns, no significance; *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Selection of MIF to be a predictive biomarker for testing the efficacy of the neoadjuvant therapy for ESCC
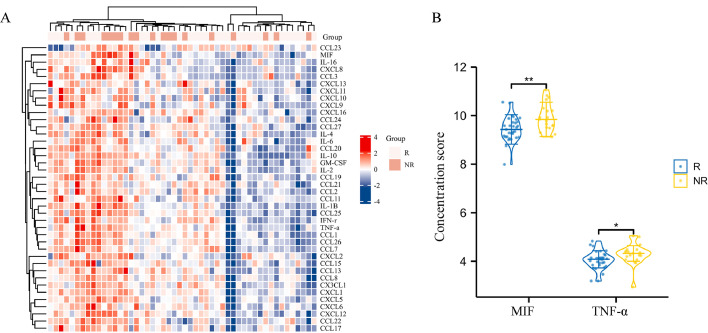
The concentrations of 40 common tumor-related chemokines in the baseline serum of the R and NR groups were measured and compared to screen for a biomarker for predicting the efficacy of PD-1 blockade immunotherapy combined with chemotherapy as a neoadjuvant therapy for ESCC. First, unsupervised hierarchical clustering was performed by comparing the concentrations of the chemokines tested in the baseline serum of each patient (Fig. 2A), with each column representing a patient and each row representing a chemokine tested. In the heat map, distinct colors in the clusters indicate that there were concentration differences between individuals, suggesting that detailed evaluations were worthwhile. Next, the median concentration of each chemokine in the R and NR groups was calculated using the scores (Supplementary Table 3) and compared (Supplementary Fig. 1). Only the concentrations of MIF and tumor necrosis factor-α (TNF-α) in the sera of the R and NR groups had statistically significant differences (Fig. 2B). The median concentration score of MIF in the sera of the NR group was 0.475 higher than that in those of the R group (95% CI: 0.176–0.812, p = 0.004), while the score difference of TNF-α between two groups was 0.266 (95% CI: 0.093-0.48, p = 0.014). The greater distinction between the baseline MIF levels of the two groups suggests that the potential of MIF as a predictive biomarker for testing the efficacy of neoadjuvant therapy is worth investigating. Therefore, whether the baseline level of MIF in both the serum and TME is associated with the efficacy of neoadjuvant therapy should be verified.
Fig. 2.
Comparisons between the baseline concentration of 40 chemokines in responders and non-responders. (A) Unsupervised hierarchical clustering of baseline concentrations of 40 chemokines in patients (n = 50). High concentration levels are shown in red, while low concentration levels are shown in blue. Each column represents a patient and each row represents a chemokine. (B) Baseline serum concentration score of MIF and TNF-α in R and NR groups. R, responder; NR, non-responder. *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to color in the text, the reader is referred to the web version of this article.)
Baseline serum MIF level was negatively correlated with the clinical outcomes of the neoadjuvant therapy
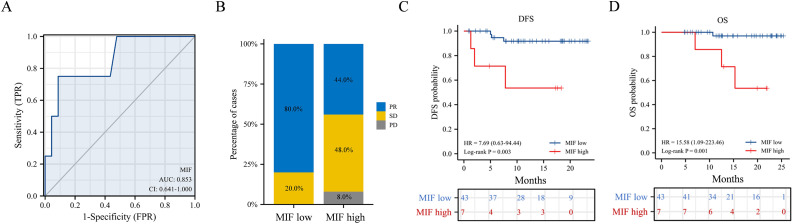
We studied whether the baseline concentration of MIF in the serum was associated with the clinical outcomes of neoadjuvant therapy for ESCC. Based on baseline serum MIF levels and patient survival (n = 50), a receiver operating characteristic (ROC) curve was plotted to investigate the ability of baseline serum MIF levels to predict the efficacy of neoadjuvant therapy (Fig. 3A). The area under the curve (AUC) is 0.853, indicating that the baseline serum MIF level had high specificity and sensitivity for predicting therapeutic outcomes. A ROC cut-off value of 10.254 was used to classify patients into the low-MIF-level group (n = 43) and high-MIF-level group (n = 7). In the low-MIF-level group, 80% of the patients showed PR, which was much higher than that in the high-MIF-level group (44 %). None of the patients in the low-MIF-level group underwent PD (Fig. 3B). Patients in the low-MIF-level group had better DFS and OS than those in the high-MIF-level group (p = 0.003 and p = 0.001, respectively; Fig. 3C,D). Thus, baseline serum MIF levels negatively correlated with the clinical outcomes of neoadjuvant therapy.
Fig. 3.
Comparisons of the clinical outcomes between patients in the low- and high-MIF-level groups. (A) ROC curve for evaluating the ability of serum MIF levels to predict the therapeutic outcomes of the neoadjuvant therapy. AUC, area under the curve; TPR, true positive rate; FPR, false positive rate. (B) Percentages of patients that had different clinical responses in the low-MIF-level group (n = 43) and the high-MIF-level group (n = 7). PR, partial response; SD, stable disease; PD, progression of the disease. Kaplan–Meier (C) DFS and (D) OS curves of patients in the low- and high-MIF-level groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Using DFS and OS as endpoints, we conducted univariate and multivariate COX regression analyses on patient age, sex, degree of tissue differentiation, initial diagnosis stage, postoperative stage, smoking history, Eastern Cooperative Oncology Group (ECOG) score, and MIF expression. Univariate and multivariate analyses of DFS showed that elevated baseline serum levels of MIF significantly increased the risk of tumor recurrence in patients with ESCC (HR = 4.365, 95% CI:1.115–17.088, p = 0.034; Supplementary Table 4). Moreover, univariate and multivariate analyses of OS demonstrated that elevated baseline serum MIF levels significantly increased the risk of death in patients with ESCC (HR = 7.78, 95% CI: 1.340–45.190, p = 0.022; Supplementary Table 5). In conclusion, the baseline MIF concentration in the peripheral blood was an independent risk factor for DFS and OS in patients with ESCC treated with neoadjuvant therapy.
The baseline level of MIF in the TME was negatively correlated with the clinical outcomes of the neoadjuvant therapy
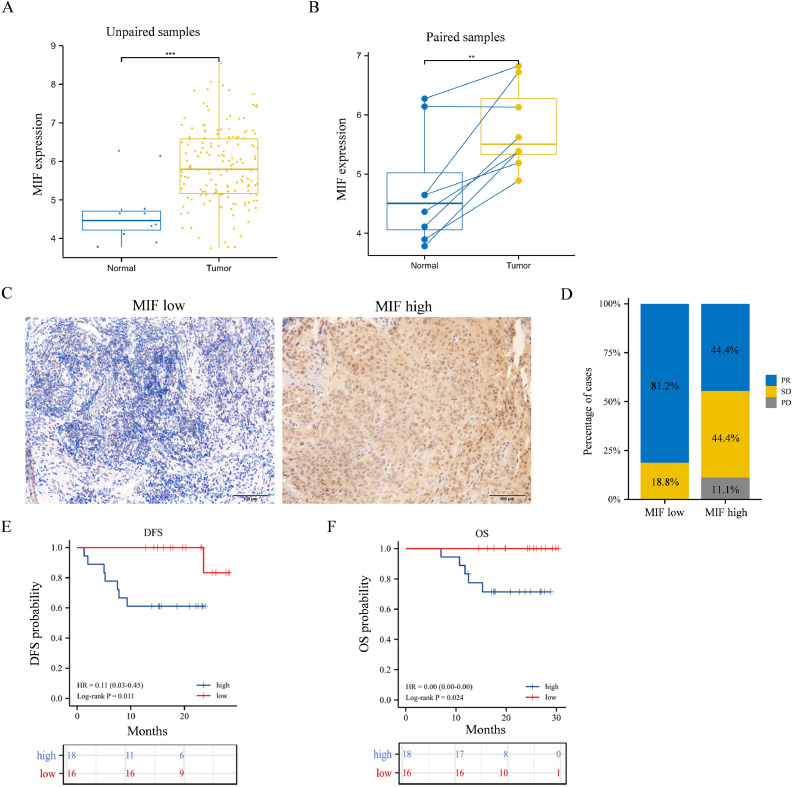
RNAseq data obtained from TCGA database showed that elevated MIF expression in the TME was observed in many types of tumors (Supplementary Fig. 2). Analysis of unpaired samples from TCGA database indicated that the average expression level of MIF in ESCC tumor tissues was significantly higher than that in normal tissues (95% CI: 0.833%–2.057, p< 0.001; Fig. 4A). Similar results were obtained by analyzing paired ESCC samples from TCGA database (95% CI: 0.609–1.891, p = 0.002; Fig. 4B). Hence, increased levels of MIF in the TME were related to tumor growth.
Fig. 4.
Comparisons of the clinical outcomes between patients in the low- and high-MIF-expression groups. (A) Comparisons between the expression levels of MIF in unpaired ESCC tissues and normal tissues. Data were obtained from TCGA database. (B) Comparisons between the expression levels of MIF in paired ESCC tissues and normal tissues. Data were obtained from TCGA database. (C) IHC images of samples from the low- and high-MIF-expression groups. Original magnification: × 200. (D) Percentages of patients (n= 34 out of 50) that had different clinical responses in the low- (n =16) and high- (n = 18) expression groups. (E) Kaplan–Meier DFS and (F) OS curves of patients in the low- and high-MIF-expression groups. *p < 0.05, **p < 0.01, ***p < 0.001.
We further investigated whether the baseline expression levels of MIF in the TME were associated with the clinical outcomes of neoadjuvant therapy. Pre-treatment tumor tissue samples were obtained from 34 patients among the 50 patients involved in the first stage of the study. Patients were classified into low- (n = 16) and high- (n = 18) MIF-expression groups according to the staining intensity on IHC images. If the average percentage of positive cells was more than 50%, the patient was assigned to the high-MIF-expression group, and vice versa. Since MIFs are expressed in the cell membrane, cytoplasm, and tumor stroma, the standing intensity in IHC images was higher in the high-MIF-expression group (Fig. 4C). In the low-MIF-expression group, 81.2% of patients showed PR, whereas only 44% showed PR in the high-MIF-expression group. Moreover, none of the patients in the low-MIF-expression group underwent PD (Fig. 4D). Patients in the low-MIF-expression group had better DFS and OS than those in the high-MIF-expression group (p < 0.011 and p< 0.024, respectively; Fig. 4E,F). Therefore, baseline MIF levels in the TME were negatively correlated with the clinical outcomes of neoadjuvant therapy.
Immune cell infiltrations were related to MIF expression levels in the TME
We investigated whether the expression level of MIF in tumor tissues was related to the infiltration of immune cells into the TME. Based on the RNA-seq data obtained from TCGA database, 162 samples in the database were separated by their median MIF expression levels in the TME into low- and high-expression groups. Next, the infiltration of different types of immune cells into the TME in the two groups was compared (Fig. 5A). More immune cells infiltrated the low-MIF-expression group, including T cells, B cells, eosinophils, mast cells, neutrophils, T helper cells, Th1 cells, Th17 cells, CD8 T cells, Tem cells, TFH cells, Treg cells, and pDC cells.
Fig. 5.
IHC images of immunological markers in the low- and high-MIF-expression groups. (A) Baseline infiltration levels of different immune cells in the low- and high-MIF-expression groups from TCGA database. The image samples and expression levels of (B) CD4, (C) CD8, (D) CD163, (E) Foxp3, (F) CD20, and (G) PD-L1 in the low- and high-MIF-expression groups. (H) IHC images of TLS. Original magnification: × 200. TLS, Tertiary lymphoid structures. *p < 0.05, **p < 0.01, ***p < 0.001. ns, non-significant.
Subsequently, IHC was conducted to detect the expression levels of six immunological markers: CD4, CD8, CD163, Foxp3, CD20, and PD-L1. We randomly selected ten MIF staining samples for immunological marker staining, of which five were in the high-expression group and five were in the low-expression group. The average densities of immunological marker-stained cells and IHC images of the two groups were compared (Fig. 5B–G). The numbers of CD8+ cells and CD163+ M2 macrophages in the low-MIF-expression group were significantly higher than those in the high-MIF-expression group (p = 0.0348 and p = 0.0415, respectively). Although no statistical differences were observed in other markers, the number of CD4+ T cells and CD20+ B cells tended to increase in the low-expression group. Meanwhile, the number of Foxp3+ Treg cells was higher in the high-expression group. Furthermore, tertiary lymphoid structures (TLSs) were found in the TME of the two responders in the low-MIF-expression group (Fig. 5H and Supplementary Fig. 3). A group of CD20+ B cells was surrounded by CD4+ T cells, CD8+ T cells, and CD163+ M2 macrophages, whereas only a limited number of Foxp3+ Treg cells and PD-L1 expression were observed (Fig. 5H). Both TCGA database analysis and IHC results showed that the expression level of MIF was related to immune cell infiltration in the TME, which may explain why the expression level of MIF is related to the efficacy of neoadjuvant therapy.
Decreased serum MIF level was related to effective treatment by the neoadjuvant therapy
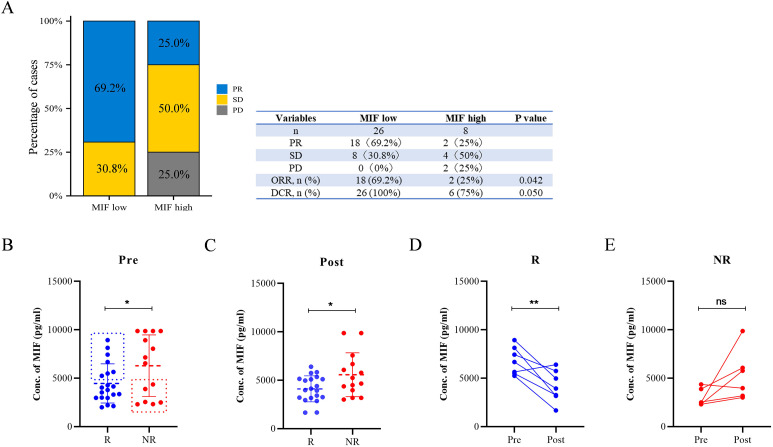
New samples (n = 34) were used to validate the feasibility of using serum MIF levels as predictive biomarkers for neoadjuvant therapy. The baseline concentration of MIF in serum samples was measured using ELISA. Patients were separated into low- (n = 26) and high- (n = 8) MIF-level groups based on their ROC cut-off value of 12.917. In the low-level group, 69.2% of the patients showed PR, and none of the patients underwent PD. In comparison, only 25% of patients showed a PR, and 25% of patients underwent PD in the high-level group (Fig. 6A). The objective response rate (ORR) for neoadjuvant therapy in the low-level group was 69.2%, whereas that in the high-level group was 25%. Additionally, the disease control rate (DCR) of neoadjuvant therapy was higher in the low-level group than in the high-level group (Fig. 6A). Overall, the serum MIF level was closely related to the efficacy of neoadjuvant therapy, suggesting that it might be a feasible predictor for clinical use.
Fig. 6.
Baseline serum concentration of MIF and clinical outcomes from another group of patients (n=34). (A) Percentages of patients who had different clinical responses in the low- (n = 26) and high- (n = 8) MIF-level groups. ORR, objective response rate; DCR, disease control rate. Serum MIF levels in R (n = 20) and NR (n = 14) groups (B) before the initiation of the treatment and (C) after two cycles of the treatment. The changes in the serum MIF level in exceptional cases in the (D) R group (n = 7) and (E) NR group (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001.
A secondary result was that effective neoadjuvant therapy resulted in a decrease in serum MIF concentration. Patients involved in the second stage of the study provided serum samples before the initiation of therapy and after receiving two cycles of treatment. Serum MIF concentrations in the R and NR groups before treatment are plotted in Fig. 6B, demonstrating that the MIF levels in the R group were significantly lower than those in the NR group (p = 0.0487). The serum MIF concentrations in the R and NR groups after two cycles of treatment are plotted in Fig. 6C, and the significant difference between the two groups was maintained (p = 0.025). Despite the differences between the two groups, we observed some exceptions: 7 out of 20 responders had relatively high serum MIF concentrations, and 6 out of 14 non-responders had low serum MIF concentrations, which are highlighted by dashed boxes. The concentration changes in these exceptions before and after the two treatment cycles are plotted in Fig. 6D and E. In the R group, although seven responders had high serum MIF levels before treatment, six out of seven showed significant decreases in MIF levels after treatment (p = 0.0077). At the same time, four out of six non-responders with low serum MIF levels had increased MIF levels after treatment (p = 0.0634). Thus, effective treatment with neoadjuvant therapy could decrease serum MIF levels, even in patients with high serum MIF levels before treatment, demonstrating that there is a close relationship between the efficacy of neoadjuvant therapy and serum MIF concentration.
Discussion
The purpose of this study was to screen for a biomarker for predicting the efficacy of PD-1 blockade immunotherapy combined with chemotherapy as neoadjuvant therapy for ESCC. Here, MIF was found to be negatively correlated with the clinical outcomes of neoadjuvant therapy and could be used as a predictive biomarker. Our research starts with measuring the baseline serum concentrations of 40 common tumor-related chemokines, MIF and TNF-α show statistically significant differences between responders and non-responders of the neoadjuvant therapy. Since the concentration difference of MIF is more obvious compared to TNF-α, MIF is selected for further investigations. MIF was first reported in 1966 and is associated with delayed-type hypersensitivity [20]. It is expressed in various immune cells, including macrophages, monocytes, T cells, B cells, mast cells, and neutrophils [21]. MIF interacts with CD74 and initiates the downstream PI3K and/or MAPK pathways [22]. MIF also facilitates tumorigenesis by inhibiting the typical tumor suppressor gene p53 [23]. Moreover, it promotes the differentiation and proliferation of myeloid-derived suppressor cells (MDSCs) and drives them to become more suppressive. Thus, MIF has inhibitory effects on both innate and adaptive immunity against tumors. Our analyses of RNA-seq data from TCGA and GTEx databases match previous research showing that MIF overexpression exists in many tumor types, including ESCC. Serum MIF levels can be used as diagnostic or prognostic markers for hepatocellular carcinoma [24], gastric cancer [25], and resectable pancreatic ductal adenocarcinoma [26]. Therefore, the potential of MIF as a predictive biomarker for neoadjuvant therapy in ESCC is worth studying.
Baseline levels of MIF in both the serum and TME were negatively correlated with the survival and clinical outcomes of patients with ESCC treated with neoadjuvant therapy. The relationship between the baseline MIF level and treatment outcomes was first measured using a chemokine quantitative analysis kit and confirmed using ELISA to measure samples provided by another group of patients. Patients in the low serum MIF group had better DFS and OS, which is consistent with a previous finding that patients with stage I-III ESCC had lower serum MIF levels than those with stage IV ESCC [27]. Moreover, IHC imaging supports the finding that higher expression of MIF in the TME results in more non-responders to neoadjuvant therapy and worse treatment outcomes. These results are consistent with a review that mentioned that high MIF expression is associated with poor OS and DFS in patients with many cancer types, including ESCC [28]. Thus, the close relationship between the level of MIF and the efficacy of neoadjuvant therapy supports the feasibility of using MIF as a predictor.
A possible explanation for why MIF could be a predictive biomarker for neoadjuvant therapy was investigated by IHC imaging of six immunological markers in the TME. The infiltration levels of immune cells are related to MIF expression levels in the TME. First, CD8+ T cells obviously infiltrate the baseline TME under conditions of low MIF expression. Although the infiltration of CD4+ T cells was not statistically significant between the low- and high-MIF-expression groups, probably because of the small sample size, a trend of increasing infiltration in the low-MIF-expression group still exists. Our results are consistent with previous ones showing that MIF inhibits the activity of intratumoral CD8+ T cells. Additionally, anti-MIF treatments successfully increased the infiltration of CD4+ and CD8+ T cells into the TME in animal experiments [22,29]. Since valid immune checkpoint blockade (ICB) treatment is highly related to an elevated number of T cells in the TME, the close link between MIF levels and T-cell infiltration suggests that MIF is a useful biomarker for ICB immunotherapy. Second, an increased number of CD163+ M2 macrophages were observed in the high MIF expression group. MIF promotes the transformation of anti-tumor M1 macrophages into M2 macrophages, which can promote the occurrence and metastasis of tumor cells [22]. This result is not shown in Fig. 5A, possibly because the macrophages measured in TCGA database were not divided into different subtypes. Furthermore, when the expression of MIF in the TME increases, Foxp3+ Tregs, which can inhibit anti-tumor immunity, also increase in number in the TME. This result is consistent with previous findings that MIF promotes the differentiation and proliferation of Treg cells in a mouse model [30]. Finally, baseline PD-L1 expression levels were similar in the low and high MIF expression groups. Since patients in the two groups showed a great difference in treatment outcomes, the baseline PD-L1 expression level was rechecked as a poor predictor of neoadjuvant therapy, as previously reported [11]. Surprisingly, TLSs with a large number of CD20+ B cells were observed in the two responders belonging to the low-MIF-expression group. TLSs are local immune cell aggregates formed by chronic inflammatory stimulation. They are composed of B cells, T cells, DC cells, and other immune cells. TLSs are related to strong anti-tumor immune responses and enhanced efficacy of ICB therapy in several types of cancers [31]. The increased number of CD20+ B cells in the TME, as shown by IHC imaging, may support the formation of TLSs. Although whether the low expression level of MIF in the TME is directly associated with the formation of TLSs requires further investigation, it still indicates that a low baseline MIF level is associated with a more antitumor baseline TME for neoadjuvant therapy against ESCC. Overall, a low baseline level of MIF is related to the formation of antitumor state TME, which may explain why MIF is a feasible predictor of neoadjuvant therapy.
A secondary finding was that effective neoadjuvant therapy resulted in a decrease in serum MIF concentration. This finding supports the hypothesis that lower MIF levels are related to better clinical outcomes; however, it raises the problem that using only baseline serum MIF levels cannot ensure that the patient is a responder. Patients with low baseline serum MIF levels may not always respond to neoadjuvant therapy and vice versa. Therefore, although baseline serum MIF can predict treatment outcomes for most patients, exceptions still exist. The analysis of these exceptions showed that after two cycles of treatment, the levels of MIF decreased in responders and increased in non-responders. Therefore, the prediction can be more accurate if the serum MIF level can be tested again after several weeks of treatment. This secondary finding provides a more accurate and reliable method for using MIF as a predictive biomarker for testing the efficacy of PD-1 blockade immunotherapy combined with chemotherapy as neoadjuvant therapy for ESCC.
Conclusions
The key finding of this study was that the level of MIF negatively correlated with the efficacy of PD-1 blockade immunotherapy combined with chemotherapy as a neoadjuvant therapy for ESCC. Hence, MIF could serve as a clinically applicable predictive biomarker for neoadjuvant therapy. The prediction can be made more accurately by retesting serum MIF levels after several weeks of treatment. A limitation of this study lies in the limited number of participants; therefore, the exact concentration of MIF as a classification standard cannot be provided. Further studies should focus on identifying the best criteria for using the MIF level as a predictor. Furthermore, as TNF-α is also related to the efficacy of the neoadjuvant therapy, its potential to be a predictive biomarker can be studied in the future.
Data availability
The datasets used and/or analyzed during the study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Chinese PLA General Hospital (Beijing, China; approval no. S2019-213-02). The clinical trial registration date is 2/11/2019 and registration number is ChiCTR1900027160. All the patients enrolled signed written, informed consents.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Liangliang Wu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Yiming Gao: Methodology, Validation, Visualization, Resources. Shengzhi Xie: Data curation, Resources. Wan Ye: Writing – review & editing. Yasushi Uemura: Investigation. Rong Zhang: Investigation. Yanju Yu: Methodology. Jinfeng Li: Methodology. Man Chen: Software. Qiyan Wu: Methodology. Pengfei Cui: Investigation. Hongyu Liu: Software. Shuai Mu: Visualization. Yilan Li: Methodology. Lingxiong Wang: Methodology. Chunxi Liu: Resources. Jiahui Li: Methodology. Lijun Zhang: Resources. Shunchang Jiao: Resources, Investigation, Project administration. Guoqing Zhang: Conceptualization, Resources, Investigation, Project administration. Tianyi Liu: Conceptualization, Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft.
Declaration of Competing Interest
The authors declare no potential conflicts of interest regarding this study.
Acknowledgements
We thank all staff in the Department of Oncology of PLA General Hospital for their support to this research project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101775.
Contributor Information
Shunchang Jiao, Email: jiaoshunchang301@126.com.
Guoqing Zhang, Email: zhanggq_301@sina.com.
Tianyi Liu, Email: ht514@126.com.
Appendix. Supplementary materials
References
- 1.Lagergren J., Smyth E., Cunningham D., Lagergren P. Oesophageal cancer. The Lancet. 2017;390(10110):2383–2396. doi: 10.1016/s0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Cowie A., Noble F., Underwood T. Strategies to improve outcomes in esophageal adenocarcinoma. Expert. Rev. Anticancer Ther. 2014;14(6):677–687. doi: 10.1586/14737140.2014.895668. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Medical research council oesophageal cancer working party. Surgical resection with or without preoperative chemotherapy in oesophageal cancer: a randomised controlled trial. The Lancet. 2002;359(9319):1727–1733. doi: 10.1016/s0140-6736(02)08651-8. [DOI] [PubMed] [Google Scholar]
- 6.Leng X.F., Daiko H., Han Y.T., Mao Y.S. Optimal preoperative neoadjuvant therapy for resectable locally advanced esophageal squamous cell carcinoma. Ann. N Y Acad. Sci. 2020;1482(1):213–224. doi: 10.1111/nyas.14508. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z.X., Cui C., Yao J., Zhang Y., Li M., Feng J., et al. Toripalimab plus chemotherapy in treatment-naive, advanced esophageal squamous cell carcinoma (JUPITER-06): a multi-center phase 3 trial. Cancer Cell. 2022;40(3):277–288. doi: 10.1016/j.ccell.2022.02.007. e273. [DOI] [PubMed] [Google Scholar]
- 8.Emens L.A., Middleton G. The interplay of immunotherapy and chemotherapy: harnessing potential synergies. Cancer Immunol. Res. 2015;3(5):436–443. doi: 10.1158/2326-6066.CIR-15-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W., Xing X., Yeung S.J., Wang S., Chen W., Bao Y., et al. Neoadjuvant programmed cell death 1 blockade combined with chemotherapy for resectable esophageal squamous cell carcinoma. J. Immunother. Cancer. 2022;10(1) doi: 10.1136/jitc-2021-003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Provencio M., Nadal E., Insa A., García-Campelo M.R., Casal-Rubio J., Dómine M., et al. Neoadjuvant chemotherapy and nivolumab in resectable non-small-cell lung cancer (NADIM): an open-label, multicentre, single-arm, phase 2 trial. The Lancet Oncolo. 2020;21(11):1413–1422. doi: 10.1016/s1470-2045(20)30453-8. [DOI] [PubMed] [Google Scholar]
- 11.Park R., Da Silva L.L., Saeed A. Immunotherapy predictive molecular markers in advanced gastroesophageal cancer: MSI and beyond. Cancers (Basel) 2021;13(7) doi: 10.3390/cancers13071715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow M.T., Luster A.D. Chemokines in cancer. Cancer Immunol. Res. 2014;2(12):1125–1131. doi: 10.1158/2326-6066.CIR-14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow M.T., Ozga A.J., Servis R.L., Frederick D.T., Lo J.A., Fisher D.E., et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 2019;50(6):1498–1512. doi: 10.1016/j.immuni.2019.04.010. e1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanmamed M.F., Perez-Gracia J.L., Schalper K.A., Fusco J.P., Gonzalez A., Rodriguez-Ruiz M.E., et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann. Oncol. 2017;28(8):1988–1995. doi: 10.1093/annonc/mdx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Xie S., Wang L., Li J., Han L., Qin B., et al. The ratio of IP10 to IL-8 in plasma reflects and predicts the response of patients with lung cancer to anti-PD-1 immunotherapy combined with chemotherapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.665147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Chang N., Wu L., Li J., Zhang L., Chen Y., et al. A nomogram-based immunoprofile predicts clinical outcomes for stage II and III human colorectal cancer. Mol. Clin. Oncol. 2021;15(6):257. doi: 10.3892/mco.2021.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxford, England) 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 18.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom B.R., Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science (New York, NY) 1966;153(3731):80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 21.Calandra T. Macrophage migration inhibitory factor and host innate immune responses to microbes. Scand. J. Infect. Dis. 2003;35(9):573–576. doi: 10.1080/00365540310016277. [DOI] [PubMed] [Google Scholar]
- 22.Noe J.T., Mitchell R.A. MIF-dependent control of tumor immunity. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.609948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conroy H., Mawhinney L., Donnelly S.C. Inflammation and cancer: macrophage migration inhibitory factor (MIF)–the potential missing link. QJM: monthly j. Assoc. Physic. 2010;103(11):831–836. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y.M., Wang L., Dai Z., Wang D.D., Hei Z.Y., Zhang N., et al. Validity of plasma macrophage migration inhibitory factor for diagnosis and prognosis of hepatocellular carcinoma. Int. J. Cancer. 2011;129(10):2463–2472. doi: 10.1002/ijc.25918. [DOI] [PubMed] [Google Scholar]
- 25.He X.X., Yang J., Ding Y.W., Liu W., Shen Q.Y., Xia H.H. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 2006;55(6):797–802. doi: 10.1136/gut.2005.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed A., Kohler S., Klotz R., Giese N., Lasitschka F., Hackert T., et al. Peripheral blood and tissue assessment highlights differential tumor-circulatory gradients of IL2 and MIF with prognostic significance in resectable pancreatic ductal adenocarcinoma. Oncoimmunology. 2021;10(1) doi: 10.1080/2162402X.2021.1962135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R.M., Sun D.N., Jiao Y.L., Wang P., Zhang J., Wang M., et al. Macrophage migration inhibitory factor promotes tumor aggressiveness of esophageal squamous cell carcinoma via activation of Akt and inactivation of GSK3beta. Cancer Lett. 2018;412:289–296. doi: 10.1016/j.canlet.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Koh H.M., Kim D.C. Prognostic significance of macrophage migration inhibitory factor expression in cancer patients: a systematic review and meta-analysis. Medicine (Baltimore) 2020;99(32):e21575. doi: 10.1097/MD.0000000000021575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe R., Peng T., Sailors J., Bucala R., Metz C.N. Regulation of the CTL response by macrophage migration inhibitory factor. J. Immunol. 2001;166(2):747–753. doi: 10.4049/jimmunol.166.2.747. [DOI] [PubMed] [Google Scholar]
- 30.Choi S., Kim H.R., Leng L., Kang I., Jorgensen W.L., Cho C.S., et al. Role of macrophage migration inhibitory factor in the regulatory T cell response of tumor-bearing mice. J. Immunol. 2012;189(8):3905–3913. doi: 10.4049/jimmunol.1102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sautes-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer. 2019;19(6):307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the study are available from the corresponding author upon reasonable request.