Summary
Background
Efficient biomarker discovery and clinical translation depend on the fast and accurate analytical output from crucial technologies such as multiplex imaging. However, reliable cell classification often requires extensive annotations. Label-efficient strategies are urgently needed to reveal diverse cell distribution and spatial interactions in large-scale multiplex datasets.
Methods
This study proposed Self-supervised Learning for Antigen Detection (SANDI) for accurate cell phenotyping while mitigating the annotation burden. The model first learns intrinsic pairwise similarities in unlabelled cell images, followed by a classification step to map learnt features to cell labels using a small set of annotated references. We acquired four multiplex immunohistochemistry datasets and one imaging mass cytometry dataset, comprising 2825 to 15,258 single-cell images to train and test the model.
Findings
With 1% annotations (18–114 cells), SANDI achieved weighted F1-scores ranging from 0.82 to 0.98 across the five datasets, which was comparable to the fully supervised classifier trained on 1828–11,459 annotated cells (−0.002 to −0.053 of averaged weighted F1-score, Wilcoxon rank-sum test, P = 0.31). Leveraging the immune checkpoint markers stained in ovarian cancer slides, SANDI-based cell identification reveals spatial expulsion between PD1-expressing T helper cells and T regulatory cells, suggesting an interplay between PD1 expression and T regulatory cell-mediated immunosuppression.
Interpretation
By striking a fine balance between minimal expert guidance and the power of deep learning to learn similarity within abundant data, SANDI presents new opportunities for efficient, large-scale learning for histology multiplex imaging data.
Funding
This study was funded by the Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre.
Keywords: Deep learning, Self-supervised learning, Cell classification, Multiplex imaging, Multiplex immunohistochemistry, Imaging mass cytometry
Research in context.
Evidence before this study
We searched PubMed (https://pubmed.ncbi.nlm.nih.gov/) for studies using self-supervised learning or weakly-supervised learning to identify cell phenotypes in multiplex images. We found two relevant studies (PMID: 35758799; PMID: 35217454). In 2022, Murphy et al. trained a self-supervised model to learn features relevant to targeted genes from unlabelled immunohistochemistry images of kidney, and to predict the cell type specificity of images using single-cell transcriptomic data as references. The approach estimated the presence of cell types in a tissue region stained with a single marker, but was unable to locate and classify single cells defined by a combination of antibodies. In the same year, Daniel Jiménez-Sánchez et al. proposed a deep learning framework to associate clinical characteristics of patients to tumour microenvironment components, inferred from multiplex-stained cancer tissues. While the study encompassed cell phenotyping, its primary focus was on connecting cell types to clinical parameters, rather than classifying all cells targeted by the markers. To date, dedicated approaches for cell classification in multiplex images, especially in multiplex immunohistochemistry images where the intensities and combinations of staining are inferred from RGB images, has not yet been proposed.
Added value of this study
The current study introduces a self-supervised-based pipeline for label-efficient cell classification in multiplex immunohistochemistry and mass cytometry images. The method was evaluated across five datasets comprising slides from ovarian cancer, lung squamous cell carcinoma, ductal carcinoma in situ, myeloma and pancreas. The method dramatically reduced the annotation to 1%, equalling to 18–114 cells across five datasets, while achieving a performance comparable to the model trained on 1828–11,459 cells. Therefore, in the context of current research, this new study presents an efficient and accurate method with new functionalities to 1) classify single cells in multiplex stained tissue sections with a small set of user-specified examples. 2) be adopted to multiplex immunohistochemistry images without the need of estimating marker intensities based on prior knowledge of the colour spectrum of markers. 3) estimate the uncertainty of predicted cell classes based on the distance of predicted cells to user-defined examples, then automatically recommend the most uncertain cells for manual correction to efficiently improve model performance. 4) facilitate hypothesis-driven analysis of cellular spatial distributions on a large scale.
Implications of all the available evidence
By mitigating the annotation burden for accurate cell classification, the proposed pipeline demonstrats great potential to accelerate the multiplex imaging analysis, which would promote biomarker discovery and clinical applications.
Introduction
The abundance and spatial distribution of cell subsets are crucial to our understanding of disease progression and response to therapies.1 Rapid development of multiplex imaging techniques such as multiplex immunohistochemistry (mIHC) and imaging mass cytometry (IMC) has enabled the accurate quantitative localization of cellular markers in situ.2 However, the co-expression of antigens and the coexistence of abundant and rare cell types impose unique challenges for automated cell phenotyping in these images.3
The field of multiplex image analysis is currently dominated by fully supervised learning,4 a model training approach that requires annotation of every instance in order to guide the model in predicting labels for unseen instances.5 To train a model for cell classification in multiplex images, a substantial amount of annotations are crucial for capturing variations in marker intensities and cell morphologies within and across cell types. The annotation process is repeated for each panel with specific marker colours and cellular locations, resulting in dramatically increased annotation burden as the number of panels increases. Typically, fully supervised methods require more than 1000 annotations per cell type per panel,3,6 which sums to over 10 h of work from a pathologist to annotate for a panel with 3 markers. Also, existing methods could be sensitive to the class imbalance issue often observed in multiplex images.3 Unsupervised methods do not require manual annotations, but mainly rely on colour decomposition, which involves resolving intensities of determined colour vectors at each pixel. This method requires prior knowledge of the colour spectrum corresponding to each marker. Also, they are often limited to 4–6 colour channels7 and can be prone to background staining noise.8
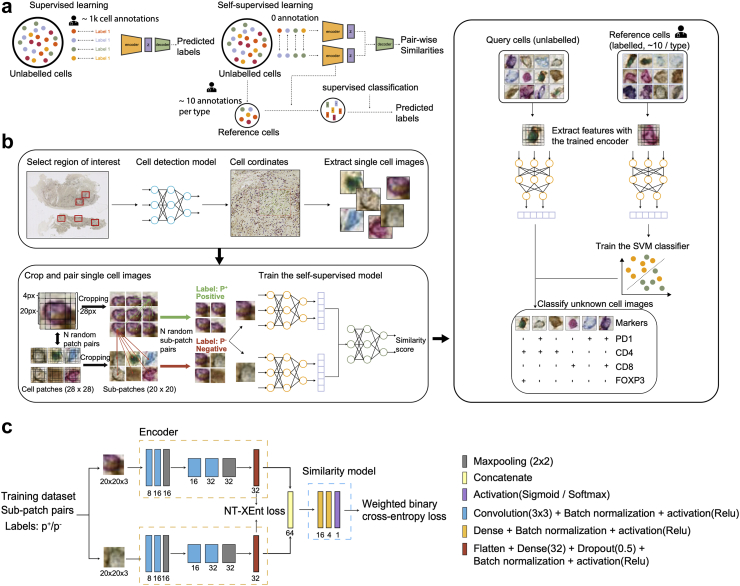
To leverage the latest advantages of deep learning and to minimise the annotation burden, we propose to apply a self-supervised deep learning-based approach that utilises intrinsic features from unlabelled data to facilitate cell phenotyping. Unlike supervised models trained using manual labels, self-supervised learning models can learn the relationship of unlabelled data without manual guidance (Fig. 1a). The first stage of self-supervised learning involves training a model on a pretext task. The data used for pretext tasks are assigned machine-generated pseudo labels denoting their similarities to other instances in the dataset. After being trained on a pretext task, the model can take an input sample and generate a vector that represents the intrinsic features of the input (Fig. 1a).9 Self-supervised learning has shown great promise in the classification of natural scene images,10, 11, 12 haematoxylin and eosin histology images,13,14 and microscope cell image data.15,16 Additionally, previous applications of self-supervised learning on immuno-stained tissue sections either aimed to estimate cell type compositions in a region,17 or reveal cell types associated with patient-level clinical characteristics.18 So far, dedicated approaches have not yet been developed for the classification of single cells in multiplex images with their unique experimental set-up, often consisting of multiple panels, therefore resulting in a particularly heavy annotation burden.
Fig. 1.
Overview of the SANDI pipeline. a, Illustration of label-based and pairwise comparison-based training strategies of supervised and self-supervised learning. Supervised training is based upon a large number of manual labels, whereas self-supervised learning first infers distinct features of cell types by learning from the pairwise similarities, and then classifies unlabelled cells using a small reference set. The reference set can be derived from unlabelled cells used for self-supervised learning, or from independent datasets stained by the same panel. b, Schematic representation of the SANDI pipeline. In the data preparation process, we selected multiple regions on the WSI that contain a variety of cell types. Then a pre-trained cell detection model was applied to the selected regions to map the coordinates of cells. Single-cell patches of 28 × 28 pixels were retrieved to constitute the training dataset. The patches were then randomly paired and cropped into 20 × 20 pixel sub-patches. Subpatch pairs that originated from the same patch were labelled as positive (), otherwise negative (). Pairs of input sub-patches were processed by two identical encoders to generate a feature vector of 32 dimensions. The encoded features were concatenated as inputs for the similarity model, which learnt to discriminate between and . The output score represents the pairwise similarities between a pair of sub-patches. A small set of cells was labelled by the pathologists as reference. Both the reference and the unknown cell image patches were cropped into 9 overlapping sub-patches of 20 × 20 pixels, which were then processed by the trained encoder to yield a feature vector of 9 × 32 dimensions. A support vector machine (SVM) classifier was trained on features extracted from the reference and to classify features extracted from unknown cells. c, Architecture of the self-supervised model.
Mitigating the annotation bottleneck of cell classification using self-supervised learning can produce a fast and precise mapping of cell phenotypes, thereby accelerating the biomarker discovery and the clinical translation of multiplex imaging.
Here we propose SANDI with a self-supervised learning framework, leading to a significant reduction in pathologists' time. By leveraging the intrinsic similarities in unlabelled cell images, SANDI was able to perform cell classification with a small reference set containing as few as 10 manual annotations per type, while achieving a comparable performance to that of the supervised model trained on thousands of cell annotations.
We validated the efficacy of SANDI by comparing its performance with that of the fully supervised model, and three state-of-the-art self-supervised frameworks, SimCLR,10 MoCo,11 and Debiased contrastive learning,12 across a range of annotation burdens. We also examined the performance of SANDI with automatically selected reference sets as an approach to further reduce the necessary annotations for desirable classification accuracy. We conducted the experiments on four mIHC datasets and one IMC dataset, which consisted of slides from ovarian cancer,19 lung squamous cell carcinoma (LUSC), ductal carcinoma in situ (DCIS),20 myeloma21 and pancreas22 (Table 1). We focused on the classification of immune cell types, whose distribution and abundance are known to have an impact on the disease progression and prognosis of different cancer types, and are therefore being targeted by a majority of multiplex imaging studies.
Table 1.
Composition of the 5 datasets used in the study.
| Dataset | Contributors | cell phenotypes | No. of annotations |
Total no. of annotations |
||
|---|---|---|---|---|---|---|
| Training + Validation | Testing | Training + Validation | Testing | |||
| Ovarian T cells | T.G and J.A.L19 | CD4+FOXP3+ | 292 | 197 | 1828 (4 slides) | 997 (5 slides) |
| CD4+FOXP3- | 596 | 168 | ||||
| PD1+CD8+ | 726 | 347 | ||||
| PD1-CD8+ | 139 | 203 | ||||
| PD1+CD4+ | 39 | 60 | ||||
| PD1+CD8-CD4- | 36 | 22 | ||||
| LUSC T cells | T.M, A.U.A, and J.L.Q | CD4+FOXP3+ | 746 | 228 | 2407 (2 slides) | 1383 (2 slides) |
| CD4+FOXP3- | 1225 | 696 | ||||
| CD8+ | 204 | 200 | ||||
| Haematoxylin-stained | 232 | 259 | ||||
| DCIS FOXP3 | F.S and E.S.H20 | FOXP3+ | 1030 | 576 | 11,459 (7 slides) | 3799 (5 slides) |
| FOXP3− | 10,429 | 3223 | ||||
| Myeloma | Y.H, C.S.Y.L, D.P, L.L, M.R-J and K.Y21 | CD8+ | 866 | 979 | 3269 (4 slides) | 1588 (2 slides) |
| CD4+FOXP3− | 2244 | 493 | ||||
| CD4+FOXP3+ | 159 | 116 | ||||
| IMC CD4-CD8 | Damond et al.22 | CD4+ | 987 | 828 | 3954 (80 slides) | 1085 (20 slides) |
| CD8+ | 2967 | 257 | ||||
Methods
Datasets
For experiments conducted in the study, the model was trained and validated on four mIHC datasets and one IMC dataset, including 9 ovarian cancer slides stained with CD8/CD4/FOXP3/PD1, 4 LUSC slides with CD8/CD4/FOXP3/haematoxylin, 12 DCIS slides with FOXP3, 6 Myeloma slides with CD8/CD4/FOXP3, and 100 IMC slides with CD4/CD8 channels extracted. Details of the five datasets are summarised in Table 1. Slides were assigned for model training, validation, and testing. Slides from the mIHC datasets were scanned at 40× magnification and were down-sampled to 20× (0.44 μm/px) before processing. We constrained the image resolution to test the model performance under the common resolution settings of highly multiplex images.23 The IMC dataset was scanned and processed at 1 μm/px resolution.22
Overview of the SANDI pipeline
The SANDI pipeline incorporated key strategies tailored for digital pathology to: (1) rapidly generate abundant examples of each cell type in regions of interest selected by pathologists, which can be achieved in minutes; (2) perform a series of operations to assign cell pairs as similar or dissimilar, extract features from unlabelled cell images that accurately represent cell identities, even when slight shifts in views are present; (3) convert learnt features into cell phenotyping based on a small set of references using a Support Vector Machine (SVM) classifier (Fig. 1b).
The self-supervised model of SANDI was built on a convolution neural network with two identical encoders24 (Fig. 1c). The model was trained to discriminate between pairs of subpatches that originated from the same cell image (), and different cell images (). Each subpatch was encoded into a vector of 32 features (Fig. 1c). The objective of the training step was to determine the optimal model parameters by minimising the loss function. This loss function was calculated as a combination of normalised temperature-scaled cross-entropy loss (NT-XEnt)10 and the weighted cross-entropy loss. By minimising the loss function, features originating from the same cell were positioned close to one another, while features derived from different cells were encouraged to be distant from each other within the feature space.
The trained self-supervised model of SANDI was able to extract distinct features for different cell phenotypes by learning to predict the pairwise similarities (Fig. 1c). To convert the encoded features into cell identities, we collected a small set of representative cell images as references, which showed clear staining, typical morphology and could be confidently assigned to a specific cell class. The encoder of the trained self-supervised model was used to extract features from both subpatches of references and unknown cells. A linear SVM trained on features of references was used to classify unknown cells.
Single-cell patches sampling
All slides were analysed for single-cell detection using a pre-trained deep learning model25 prior to the proposed pipeline. To build the dataset for self-learning purposes, the first step is typically to sample single-cell patches from the whole slide image (WSI).26 In an ideal situation where the percentage of each cell type present in the dataset is balanced, we can randomly sample from the pool of all detected cells and expect an equal chance of capturing each cell type of interest. However, in pathological data, cell type imbalance is common, which may cause some rare cell types to be missed out due to random sampling.
To tackle this problem and to investigate the impact of data imbalance on the model performance, we introduced a data sampling step to capture a variety of cell phenotypes and ensure the inclusion of rare cell types. First, small regions on the WSI enriched with diverse cell types were manually identified. The selection of regions should also consider excluding areas with low image quality, such as those exhibiting imperfect staining, artefacts, or being out-of-focus. Then, a pathologist will label the class of each cell within these regions by annotating the cell centre using different colours to denote different cell types. The selection of regions ensures that a considerable number of each cell types are included in the training dataset. Manual labels revealed the composition of cell types within the regions and provided ground truth for model evaluation. A 28 × 28 pixels patch around each dot annotation was retrieved to form the dataset. For cells present at the edge of a region, we applied mirror padding to expand the patch to 28 × 28 pixels. All image patches from slides used for model training and validation (Table 1) were pooled together and randomly allocated to the training or validation set with a 4:1 ratio. The validation patches were used for assessing the model performance during the training stage. The best model was chosen based on the lowest loss observed on the validation set. While patches on testing slides were employed to evaluate the best model's generalisation ability to unseen images after the completion of training.
Patch cropping and pairing
Given a dataset containing n 28 × 28 pixel (12.32 × 12.32 μm2) single-cell image patches , we first generated all possible combinations . For each batch, N pairs were randomly sampled from without replacement. For each pair of single-cell image patches, the acquired patches were each randomly cropped into 20 × 20 pixels (8.8 × 8.8 μm2) sub-patches , indicating the sub-patch cropped from the patch. Two sub-patches retrieved from the same patch and the paired patch were labelled as positive () and negative () respectively, indicating that they were from the same cell or different cells. These are described as follows:
| (1) |
| (2) |
where and denote the set of and . The total number of and in a batch is 2N with N set to 256 in the experiment. RGB-valued images were normalised to the range [0, 1] before being fed into the network. The sub-patches randomly cropped from a single-cell image represented different fields of view for the cell. By assigning sub-patches derived from the same cell to positive pairs, we encouraged the model to classify them as the same cell class. This approach aims to simulate the process of inspection performed by pathologists, where cell identification remains consistent regardless of the field of view.
To assess the effectiveness of the cropping strategy, we applied three additional methods to generate subpatches from the single-cell image patches. These methods include random flipping, blurring, and scaling. Random flipping randomly flips the image patch either horizontally or vertically. Blurring involves applying Gaussian blur to the image using a kernel size of 5 and a sigma value of 1. Scaling entails enlarging the image by a factor of 2 and then selecting the 28 × 28 pixels region at the centre of the rescaled image.
We trained the self-supervised model of SANDI using subpatches generated by these different methods, and compared the corresponding cell classification performance. It was observed that random cropping consistently resulted in the best or comparable classification performance across most datasets, except for the IMC CD4-CD8 dataset. In this particular dataset, the scaling method outperformed the cropping method at annotation budgets of 10% and 20% (Wilcoxon rank-sum test, P = 0.01, P = 0.01, Fig. S1a). These comparisons suggest that random cropping is generally more effective than the other augmentation methods tested for optimising the features learned by the self-supervised model.
Network architecture and training
As shown in Fig. 1c, the self-supervised model consists of two identical encoders conjoined at their last layers, followed by a single branch responsible for computing the pairwise similarity between the outputs of the two encoders. Each encoder contains a series of convolution, activation, batch normalisation, max-pooling, and dropout layers. These layers apply non-linear transformations to the input image, allowing high-dimensional inputs to be converted into a 32-dimension feature vector. This feature vector is a latent representation of an image in a low-dimensional space. The single branch concatenates feature vectors from two encoders and feeds them through a dense layer, followed by a series of activation layers to generate a value between 0 and 1. This output value corresponds to the predicted similarity score between the image pairs. A higher score indicates more similarity between the two images.
For cell phenotyping purposes, the network was expected to generate a high score for cells from the same class and a low score for cells from distinct classes. However, since the network was trained to identify similar or dissimilar pairs randomly sampled from the unlabelled dataset, two images from the same class might have been labelled as negative during the data preparation, which biased the network towards features that discriminate against images from the same class.12,27 To reduce the impact of uncertainty in negative labels, we modified the binary-entropy loss function by applying lower weights to the than to .
| (3) |
where denotes the similarity branch, N is the total number of or within a batch. denote the pre-defined weights applied to the entropy loss of positive pairs and negative pairs . The ratio of and determines the extent to which the model is encouraged to focus on positive pairs, which have been ascertained to be labelled correctly as of the same cell type. We tested various ratios of and , including 0.9:0.1, 0.8:0.2, 0.7:0.3, 0.6:0.3 and 1:1 across five datasets and a range of annotation budgets (Fig. S1b). The ratio of 0.7:0.3 demonstrated the highest performance in the Myeloma dataset across different annotation burdens (10%, 20%, 30%, 100%, Wilcoxon rank-sum test, P ≤ 0.043, Fig. S1b). This ratio also showed similar performance to other weight ratios in most of the other datasets when considering random sampling of annotations (Wilcoxon rank-sum test, P ≥ 0.074), except for the 20% annotations of the LUSC T cells dataset (P = 0.043, Fig. S1b). Therefore, we have chosen to set as 0.7 and as 0.3 for the remaining experiments.
To further constrain the latent representations to maximise the agreement between , we combined with the normalised temperature-scaled cross-entropy loss (NT-XEnt)8, which is expressed as
| (4) |
where and denote the normalised embedding of sub-patch and , denotes cosine similarity, equals to 1 if , otherwise 0. N is the total number of subpatch pairs within a batch. denotes the temperature parameter. We set the temperature parameters as 0.1 in the experiment, which was the optimal value proposed in its previous implementation.10 For a given sub-patch , the NT-XEnt loss treats the sub-patch originated from the same patch as positive samples, and all the other sub-patches within the batch as negative samples.
The combined loss is the combination of and , as given by
| (5) |
It is worth noting that a debiased variant of NT-XEnt has been proposed to account for the uncertainty in negative pairs.12 This Debiased loss is expressed as.
| (6) |
| (7) |
where the additional parameter denotes the probability of a negative pair to be of the same cell type. We compared the model performance of and to assess the effectiveness of different approaches in mitigating the negative impact of uncertainty in negative samples. In the experiment, and were set as 0.01 and 0.1 separately, which had been shown to produce the best performance in their previous implementations.10,12
For rigorous assessment of models, all training was performed on an Intel i7-9750H CPU for 100 epochs with a batch size of 256. The training was optimised using Adam with an initial learning rate of 10−3, along with other parameter settings suggested by the original paper.28 The models with the least validation loss were selected for evaluation.
Reference-based cell classification
Identification of cells from multiplex images is dependent on stain intensities and the morphology of the cell, which can be affected by experimental artefacts and out-of-focus regions. The noise in the data/label is a well-known issue affecting model performance in digital histology image analysis.29,30 Motivated by the need to reduce the annotation burden, we selected a set of reference images from the training dataset D as representations of each cell type. In practice, the reference can also be extracted from an independent subset of cells from images stained with the same panel as the training dataset. Each cell in a hold-out testing set is regarded as a query image . Both the reference image and query image were cropped into 9 20 × 20 pixel sub-patches and processed by the trained encoder to yield the feature vectors and of size 32 × 9. Assembling features of sub-patches allow the local regions adjacent to the cell to be incorporated for downstream classification, which has been shown to generate more accurate predictions.25
An SVM classifier with a linear kernel implemented in the libsvm library31 was trained on feature embeddings of references and predicted cell phenotypes for embeddings of unlabelled samples .
To evaluate the representative capability of features learned by the self-supervised model for cell categories, we conducted a comparative analysis with two alternative feature extraction methods: autoencoder and colour histogram. The autoencoder is a deep-learning model designed to reconstruct images from low-dimensional features. For a fair comparison, we constructed an autoencoder with the same encoder architecture as SANDI and a decoder that reverses the encoding process to reconstruct the original image. To extract features from the entire cell image, we used uncropped image patches of size 28 × 28 pixels as input. The autoencoder was trained using mean squared error as the loss function and optimised using Adam with an initial learning rate of 10−3. The batch size was set as 100 and the model was trained for 500 epochs. Unlike SANDI, the autoencoder was trained to reconstruct a cell image without contrastive learning from a pair of cells. The comparisons between features generated by SANDI and the autoencoder can therefore illustrate the effectiveness of the pairwise similarity learning strategy employed by SANDI. Another method for feature extraction is the colour histogram, which involves computing separate histograms for the red, green, and blue intensities, with a bin size of 50. The values of these three channels were concatenated into a vector, representing the colour distribution in an image patch.
Automatic expansion of the reference set
Although SANDI can obtain high accuracy using a limited number of labels, being trained on a small set of representatives may lead to an underestimation of the intra-cell-type variations in stain intensities, colour combinations, and morphologies.3,32,33 By contrast, a larger training set can expose the model to higher variability in the data but can also deteriorate model performance if poor-quality data is included.32,34 An ideal approach to capture a good level of variation while ensuring adequate data quality is to leverage information learnt by self-supervised training to inform the pathologist of cells that are prone to misclassification and thereby, create ground truth feedback to improve model performance. For this purpose, we proposed the automatic expansion method for iteratively adding human interpretation of the least confident instances as training events.
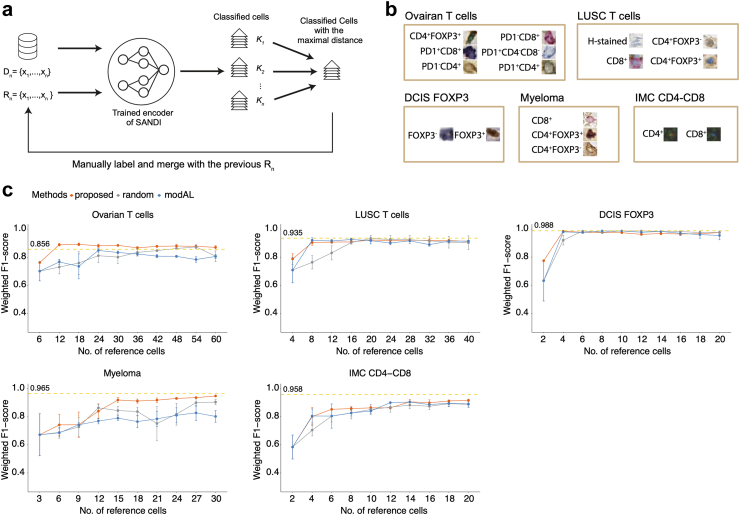
The flowchart illustrating the pipeline is shown in Fig. 2a. Firstly, we nominated 1 image for each cell phenotype as a representative, and then the minimal Euclidean distance between embeddings of unlabelled images and each reference image was used to determine the cell type of unlabelled cells. This distance-based classification method is described by:
| (8) |
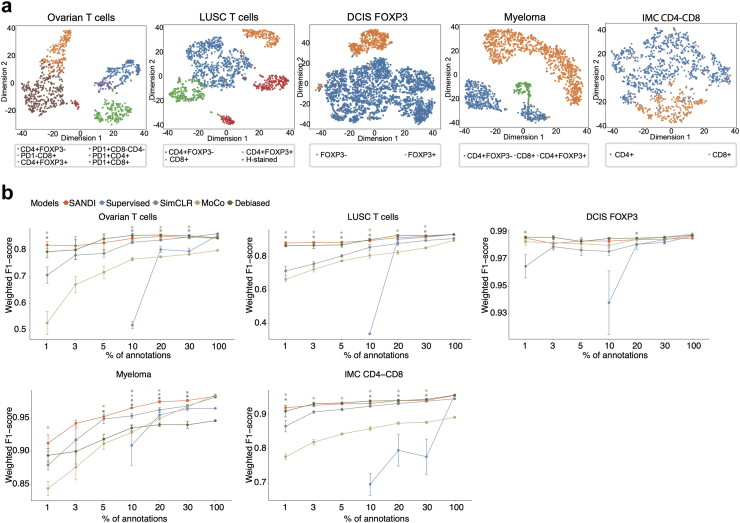
Fig. 2.
Performance of SANDI on five datasets. a, The t-SNE representation of test image embeddings. Cell labels are represented as colour codes. b, Comparison of the performance based on weighted F1-score of four self-supervised methods (SANDI, SimCLR, MoCo, Debiased contrastive learning) and the supervised classifier over increasing amounts of annotations. For annotations below 30%, the mean and standard error from five random samplings are shown. Asterisks above the curves indicate the statistical significance of comparisons between SANDI and the other four methods, with colours of asterisk representing the corresponding compared method. ∗, P < 0.05.
Second, as an automated reference set expansion, for each group of cells of class K, the cell with the maximum Euclidean distance to any of reference cells of the same class K was selected and manually labelled. These newly selected cells were then added to the previous reference set, while ignoring repeated instances. The two steps were repeated for 10 rounds and the weighted F1-score computed on the testing set was examined using the reference set from each round.
To assess the efficacy of the proposed automatic expansion method, we compared the classification performance based on reference selected by the proposed method, random selection, and an established active learning method in the modAL35 library. The random selection involved randomly sampling one cell per round from cells predicted as each category in the previous round. The modAL35 approach was configured using the entropy-based sampling strategy,36 which queries the cells with the least probability of being a certain class.
Assessment of model performance
To evaluate the model performance under various annotation budgets, we trained linear SVM classifiers on feature embeddings of randomly sampled training subsets containing 1%, 3%, 5%, 10%, 20%, and 30% of annotated samples from training slides of each dataset (Table 1). The random sampling was performed in a way that each subset contained approximately the same proportion of samples of each cell type as the complete set. The training of SVM was repeated five times on different randomly sampled training sets, and the model performance was tested on hold-out testing sets containing cells from slides excluded from training (Table 1). Results were compared against the performance of SVM-trained features generated by three state-of-the-art self-supervised methods SimCLR,10 MoCo,11 Debiased contrastive learning,12 and a supervised classifier trained on 10%, 20%, 30%, and 100% of the annotations.
For fair comparisons, the supervised classifier, SimCLR, MoCo, and Debiased contrastive learning were constructed with the same encoder as SANDI, and only random flipping was applied for data augmentation. As compared to SANDI, SimCLR and Debiased contrastive learning do not incorporate the similarity branch and were trained to optimise the NT-XEnt loss and the Debiased loss respectively. All methods were trained on the same training/validation set split, and were tested on the same hold-out testing set as SANDI.
Performance of the model was evaluated using the weighted F1-score, which is the average of F1-score for each class weighted by the number of their instances:
| (9) |
where n is the total number of instances, is the number of classes, and is the number of instances for class . TP, FP, and FN denote true positive, false positive and false negative respectively. The desired weighted f1-score can vary for different panels and study designs. In general, we expect a weighted f1-score of greater than 0.8 to be acceptable for generating robust results for downstream analysis.
Ethics
This research involved a retrospective review of deidentified medical images, and the requirement for informed consent was waived.
Statistics
Statistical significance of performance comparisons was determined by the Wilcoxon rank-sum test, with all P values adjusted using the Benjamini-Hochberg correction. Correlations among cell percentages in the Ovarian T cells dataset were evaluated using the Pearson correlation coefficient. All statistical analyses were performed in R (v4.2.2).
Role of funders
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The authors declare no competing non-financial interests that may have influenced the publication process.
Results
Evaluation of SANDI for cell classification across various annotation burdens
The effectiveness of SANDI in discriminating diverse cell types was first evaluated by visualising the embeddings of testing images in the latent space, which was performed using the t-distributed stochastic neighbour embedding (t-SNE). To capture the variability in cell appearance, each testing image was represented by the embeddings of nine sub-patches in its neighbourhood. The t-SNE plot revealed compact and distinguishable clusters corresponding to each cell type (Fig. 2a). In contrast, features extracted by an autoencoder or colour histogram (Methods) failed to separate different cell types into distinct clusters (Fig. S2a and b), suggesting that the self-supervised learning strategy of SANDI was crucial for capturing features representative of cell identities.
To investigate the size of reference set required for SANDI to achieve reasonable performance, we first trained linear SVM on feature embeddings of randomly sampled reference sets containing 1%, 3%, 5%, 10%, 20%, and 30% of annotated samples of each cell type. When the budget was limited to 1%, the number of annotations ranged from 1 for PD1+CD8-CD4-cells in the Ovarian T dataset to 104 for FOXP3- cells in the DCIS FOXP3 dataset (Table 1).
Across 5 datasets, SANDI achieved an impressive performance using only 1% of annotations (18–114 cells, Fig. 2b), comparable to a supervised classifier constructed using the same encoder trained on 1828–11,459 annotations (−0.002 to −0.053 of weighted F1-score, Wilcoxon rank-sum test, P = 0.31, Table 2, Table S1). Thus, SANDI can obtain adequate classification accuracy using 100 times fewer annotations than the conventional supervised training methods. With a budget of below 30% of annotation data (11–3129 cells per type), SANDI achieved superior or comparable performance than the supervised classifier, and the other three state-of-the-art self-supervised frameworks SimCLR,10 MoCo,11 and Debiased contrastive learning12 in all the five datasets (P ≥ 0.023, Fig. 2b, Table 2, Table S1). These comparisons demonstrate the effectiveness of the SANDI loss function and architecture for the task of cell classification in multiplex images. Importantly, the superiority of SANDI in the Ovarian T cells, LUSC T cells and Myeloma datasets with substantial data imbalance suggests a key advantage of SANDI in multiplex image analysis.
Table 2.
The weighted F1-score of the SVM classifier trained on features generated by different methods, with various percentages of annotations.
| Annotations | 1% | 3% | 5% | 10% | 20% | 30% | 100% |
|---|---|---|---|---|---|---|---|
| Ovarian T cells | |||||||
| No. of cells | 18 | 54 | 91 | 182 | 365 | 548 | 1828 |
| Supervised classifier | – | – | – | 0.518 (0.03) | 0.803 (0.026) | 0.797 (0.027) | 0.856 |
| SimCLR | 0.707 (0.069) | 0.782 (0.038) | 0.789 (0.032) | 0.831 (0.012) | 0.839 (0.007) | 0.850 (0.014) | 0.863 |
| MoCo | 0.527 (0.098) | 0.671 (0.068) | 0.718 (0.052) | 0.767 (0.014) | 0.776 (0.006) | 0.785 (0.007) | 0.800 |
| Debiased | 0.795 (0.053) | 0.802 (0.084) | 0.844 (0.042) | 0.857 (0.017) | 0.858 (0.008) | 0.855 (0.014) | 0.847 |
| SANDI | 0.820 (0.028) | 0.817 (0.077) | 0.829 (0.064) | 0.845 (0.014) | 0.853 (0.019) | 0.849 (0.012) | 0.846 |
| LUSC T cells | |||||||
| No. of cells | 24 | 72 | 120 | 240 | 481 | 722 | 2407 |
| Supervised classifier | – | – | – | 0.338 (0.005) | 0.905 (0.042) | 0.918 (0.016) | 0.935 |
| SimCLR | 0.716 (0.064) | 0.757 (0.03) | 0.806 (0.017) | 0.857 (0.041) | 0.880 (0.027) | 0.898 (0.005) | 0.910 |
| MoCo | 0.664 (0.031) | 0.725 (0.035) | 0.776 (0.012) | 0.808 (0.036) | 0.827 (0.03) | 0.854 (0.013) | 0.898 |
| Debiased | 0.867 (0.012) | 0.869 (0.045) | 0.872 (0.03) | 0.903 (0.02) | 0.929 (0.009) | 0.927 (0.007) | 0.935 |
| SANDI | 0.883 (0.019) | 0.886(0.013) | 0.887(0.014) | 0.898 (0.016) | 0.916 (0.022) | 0.922 (0.008) | 0.934 |
| DCIS FOXP3 | |||||||
| No. of cells | 114 | 343 | 572 | 1145 | 2291 | 3437 | 11,459 |
| Supervised classifier | – | – | – | 0.938 (0.052) | 0.981 (0.008) | 0.985(0.001) | 0.988 |
| SimCLR | 0.965 (0.019) | 0.979 (0.005) | 0.977 (0.009) | 0.976 (0.006) | 0.981 (0.001) | 0.982 (0.003) | 0.987 |
| MoCo | 0.983 (0.006) | 0.982 (0.005) | 0.982(0.006) | 0.980 (0.005) | 0.985(0.001) | 0.986(0.001) | 0.989 |
| Debiased | 0.986 (0.001) | 0.986 (0.003) | 0.983 (0.004) | 0.985 (0.002) | 0.986 (0.001) | 0.986 (0.002) | 0.988 |
| SANDI | 0.986(0.003) | 0.982 (0.008) | 0.984(0.006) | 0.984(0.003) | 0.985(0.001) | 0.985(0.001) | 0.986 |
| Myeloma | |||||||
| No. of cells | 32 | 98 | 163 | 326 | 653 | 980 | 3269 |
| Supervised classifier | – | – | – | 0.958 (0.003) | 0.961 (0.007) | 0.955 (0.011) | 0.965 |
| SimCLR | 0.879 (0.016) | 0.917 (0.04) | 0.949 (0.016) | 0.953 (0.008) | 0.962 (0.008) | 0.968 (0.005) | 0.982 |
| MoCo | 0.844 (0.024) | 0.876 (0.043) | 0.912 (0.02) | 0.928 (0.002) | 0.949 (0.006) | 0.967 (0.007) | 0.985 |
| Debiased | 0.894 (0.024) | 0.900 (0.042) | 0.918 (0.016) | 0.935 (0.010) | 0.940 (0.006) | 0.940 (0.012) | 0.946 |
| SANDI | 0.912(0.028) | 0.942(0.011) | 0.952(0.008) | 0.965(0.002) | 0.975(0.004) | 0.977(0.004) | 0.983 |
| IMC CD4-CD8 | |||||||
| No. of cells | 39 | 118 | 197 | 395 | 790 | 1186 | 3954 |
| Supervised classifier | – | – | – | 0.696 (0.073) | 0.795 (0.103) | 0.777 (0.115) | 0.958 |
| SimCLR | 0.867 (0.038) | 0.908 (0.008) | 0.916 (0.009) | 0.925 (0.006) | 0.933 (0.009) | 0.940 (0.003) | 0.947 |
| MoCo | 0.777 (0.018) | 0.820 (0.017) | 0.844 (0.007) | 0.859 (0.014) | 0.875 (0.009) | 0.878 (0.007) | 0.892 |
| Debiased | 0.910 (0.038) | 0.933 (0.011) | 0.935 (0.004) | 0.940 (0.007) | 0.942 (0.008) | 0.945 (0.006) | 0.958 |
| SANDI | 0.921(0.014) | 0.928 (0.01) | 0.932 (0.007) | 0.933 (0.005) | 0.942(0.005) | 0.942 (0.005) | 0.957 |
Results for annotation percentages ranging from 1% to 30% are the average over 5 trials with different random samplings. Standard deviations are shown inside the parentheses.
Bold values are within 0.003 lower than the best.
Notably, there is an inferior performance across all methods for the Ovarian T cells dataset compared to other datasets (Table 2). This dataset contains two cell types defined by the coexpression of two markers: PD1+CD8+ and PD1+CD4+ (Table 1). Cell types stained with a combination of markers displayed more variations in the staining compared to cells expressing a single marker. It is challenging for the model to distinguish between co-expressing cells and cells expressing each marker alone (i.e. PD1+CD8+ cells versus PD1+CD8- and PD1-CD8+ cells). This is illustrated in Fig. S3, where PD1-CD8+ cells with irregular staining intensities of CD8 were misclassified as PD1+CD8+ cells. Furthermore, we observed that the weighted cross-entropy loss improved over the non-weighted version; and when combined with the contrastive loss NT-XEnt to learn co-occurring modalities, resulted in the best overall performance regardless of image types (Table S2). Thus, SANDI is capable of boosting the performance of unbiased cell identification regardless of cell abundance, possibly due to its loss function design and independence of prior-defined labels.37
Performance with the automatic expansion of the reference set
To effectively select reference images that contribute the most to model performance improvement, we designed an automatic expansion of the reference set. This is achieved by iteratively estimating the confidence of cell phenotyping performed by the trained model, and recommending the least confident instances for manual labelling (Fig. 3a, Methods). The reference set was initialised with one arbitrarily selected image for each cell type (Fig. 3b). With 10 iterations, we gathered a reference set containing the 10 most diverse representations of the same cell type. It is worth noticing that we constrained the number of iterations to 10 for experiment purposes. In practice, the number of cells in the initial reference set, the number of iterations, and the number of cells recommended at each round can be modified for particular needs.
Fig. 3.
Automatic expansion of reference sets. a, The automatic expansion scheme of reference sets to effectively select reference images that contribute most to the improvement of model performance. The unlabelled cell images from the training set D and an initial reference set Rn containing 1 reference image for each cell type K were provided in the first round. Images in D and Rn were cropped to 9 20 × 20 pixel sub-patches and processed by the trained feature encoder. The unlabelled cells were assigned with cell type K based on the Euclidean distance between embeddings of reference and unlabelled cells. The instance with the maximal Euclidean distance was selected for manual labelling and merged with Rn from the previous round to form the new reference set. In the experiment, the process was repeated 10 times. b, Examples of initial reference sets for each of the five datasets. c, Weighted F1-score on testing sets for the linear SVM classifier trained on the reference set generated by 3 different methods at each round of automatic expansion. The process was repeated 3 times with different initial reference images. The error bar indicates the standard error. The yellow horizontal line denotes the weighted F1-score achieved by the supervised classifier trained on 100% annotations.
Initial references and example cells classified at the 10th iteration were shown in Fig. S3. As the number of references increases, we have observed an inconsistent improvement in the weighted F1-score, with the highest value achieved prior to the 10th iteration in the Ovarian T cells, LUSC T cells, and DCIS FOXP3 datasets (Fig. 3c). The decrease in accuracy can potentially be attributed to the inclusion of references located near the decision boundary. These references may have neighbouring cells from a mixture of classes, leading to confusion and difficulty in distinguishing adjacent cell classes within the feature space. Despite the fluctuation in performance, references at the 10th iteration yielded comparable weighted F1-scores than the supervised model trained on 100% of annotations (+0.014 to −0.041 of weighted F1-score, Wilcoxon rank-sum test, P = 0.55, Table 2, Table 3). These results suggest that the confidence-based reference selection scheme can effectively boost classification accuracy using as few as 10 annotations per cell type.
Table 3.
Weighted F1-score on the testing set for the linear SVM classifier trained on reference sets generated from each round of automatic expansion.
| Datasets | Rounds | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ovarian T cells | Ref. Size | 6 | 12 | 18 | 24 | 30 | 36 | 42 | 48 | 54 | 60 |
| Weighted F1-score | 0.762 (0.007) | 0.889(0.015) | 0.891(0.016) | 0.881 (0.02) | 0.884 (0.008) | 0.866 (0.016) | 0.877 (0.012) | 0.879 (0.019) | 0.877 (0.019) | 0.87 (0.02) | |
| LUSC T cells | Ref. Size | 4 | 8 | 12 | 16 | 20 | 24 | 28 | 32 | 36 | 40 |
| Weighted F1-score | 0.787 (0.064) | 0.904 (0.033) | 0.907 (0.017) | 0.908 (0.015) | 0.922(0.011) | 0.918 (0.027) | 0.921(0.021) | 0.918 (0.02) | 0.918 (0.014) | 0.912 (0.004) | |
| DCIS FOXP3 | Ref. Size | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 |
| Weighted F1-score | 0.775 (0.008) | 0.984(0.003) | 0.977 (0.008) | 0.975 (0.001) | 0.974 (0.002) | 0.962 (0.011) | 0.968 (0.015) | 0.965 (0.017) | 0.967 (0.016) | 0.975 (0.008) | |
| Myeloma | Ref. Size | 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | 27 | 30 |
| Weighted F1-score | 0.672 (0.2590) | 0.742 (0.131) | 0.743 (0.156) | 0.839 (0.091) | 0.919 (0.028) | 0.912 (0.025) | 0.918 (0.026) | 0.93 (0.012) | 0.936 (0.006) | 0.948(0.001) | |
| IMC CD4-CD8 | Ref. Size | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 |
| Weighted F1-score | 0.584 (0.147) | 0.801 (0.11) | 0.853 (0.005) | 0.857 (0.024) | 0.863 (0.021) | 0.863 (0.015) | 0.906 (0.029) | 0.899 (0.036) | 0.912 (0.012) | 0.917(0.015) |
The average of 3 repeats with different initial reference sets is shown, with standard deviations shown inside the parentheses.
Bold values are within 0.003 lower than the best.
To further evaluate the efficacy of the proposed automatic expansion approach, we compared the classification performance using reference images selected by three approaches: the proposed automatic expansion method, random selection, and the entropy-based sampling strategy36 in the modAL library,35 which is an established active learning method for querying uncertain instances to be labelled (Methods). All approaches were initialised with the same reference set and evaluated on the same whole-out testing set. Although the three approaches did not show significant differences in the classification performance (Wilcoxon rank-sum test, P ≥ 0.191, Fig. 3c, Table S3), the analysis of the standard deviation of the weighted F1-score revealed noteworthy findings. Specifically, in the Ovarian T cells, LUSC T cells, and IMC CD4-CD8 datasets, the proposed method's selected references resulted in significantly lower standard deviation in weighted F1-scores compared to randomly selected references (Wilcoxon rank-sum test, P = 0.043, P = 0.016, P = 0.085, Table S3), indicating a higher level of performance consistency. Furthermore, in the Ovarian T cells dataset, the proposed method's selected references also demonstrated significantly lower standard deviation compared to references selected by modAL (Wilcoxon rank-sum test, P = 0.039, Table S3). Notably, a significant difference in standard deviation between modAL and randomly selected references was observed solely in the LUSC T cells dataset (Wilcoxon rank-sum test, P = 0.041, Table S3). These findings suggest that the proposed automatic expansion method has the potential to select references that lead to more reliable and stable results and contribute to robust enhancements in classification accuracy.
SANDI reveals the association between PD1 expression and T regulatory cell proximity in the Ovarian T cells dataset
To examine the capability of SANDI in identifying biological meaningful cellular distributions, we performed it on cells auto-detected by a pre-trained neural network6 on 9 slides from the Ovarian T cells dataset. It is worth noting that the auto-detected dataset contains tissue backgrounds that were over-detected as cells (Fig. S4). Despite such noise within the data, SANDI trained on 4431 auto-detected cells from 19 regions achieved a weighted F1-score of 0.855 with the linear SVM classifier trained on 20% of randomly selected training samples and evaluated on the same testing set as previously described, suggesting its robustness against incorrect detection of cells. Additionally, SANDI is capable of correcting over-detected cells using patches of tissue background as references (Fig. S4).
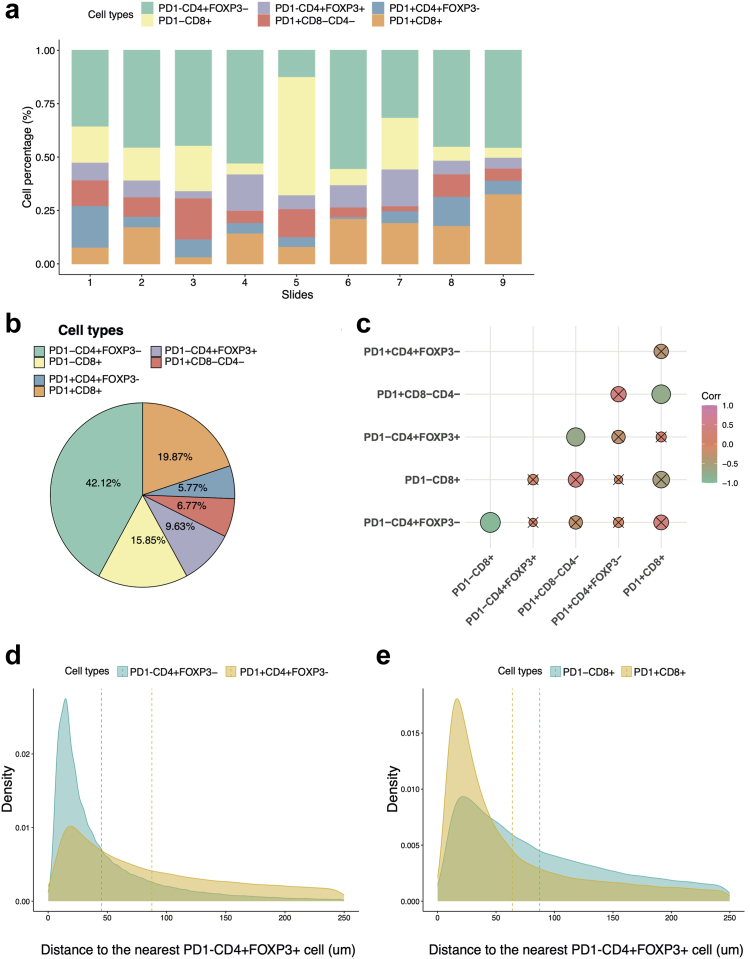
We applied SNADI to classify six immune cell subsets using references from the 10th iteration of the automatic expansion scheme. The classified cells exhibit a diverse composition across the 9 samples (Fig. 4a), with PD1-CD4+FOXP3-, PD1+CD8+ and PD1-CD8+ being the top three abundant cell types (Fig. 4b). We observed negative associations between percentages of PD1-CD4+FOXP3- T helper cells (Th) and PD1-CD8+ cells (Rho = −0.922, P = 0.0004, 95% confidence interval (CI) = −0.984 to −0.665), PD1-CD4+FOXP3+ T regulatory cells (Treg) and PD1+CD8-CD4-cells (Rho = −0.720, P = 0.029, 95% CI = −0.936 to −0.107), and between PD1+CD8+ cells and PD1+CD8-CD4-cells (Rho = −0.759, P = 0.018, 95% CI = −0.946 to −0.191, Fig. 4c). PD1 expression has been associated with activation and exhaustion of CD8+ and CD4+ T cells.38 To quantify the impact of PD1 expression on the T regulatory cell (Treg) mediated immunosuppression, we measured the distance of PD1+ and PD1- T cells to the nearest PD1-CD4+FOXP3+ Treg cell. We constrained the analysis to distance within 250 μm, which was shown to be the maximal distance of effective cell–cell interactions.39 This approach showed that PD1-CD4+FOXP3- Th cells were nearer to Treg cells than PD1+CD4+FOXP3- Th cells (Fig. 4d), whereas PD1+CD8+ cells were closer to Treg cells compared to PD1-CD8+ cells (Fig. 4e). It has been documented that CD4+ cells with low PD1 expression displayed reduced cytokine production and was associated with poor overall survival in follicular lymphoma.40 By contrast, high expression of PD1 is known to characterise the dysfunctional CD8+ T cells,38 and the irreversible exhaustion is partly attributed to the Treg interaction.41 These findings raised the possibility that high PD1 expression on CD8+ T cytotoxic cells may be linked to increased interaction with Treg cells and co-orchestrate immunosuppression while having an opposite effect on Th cells. Overall, these results demonstrated the potential of SANDI not only to classify cellular components but also to facilitate hypothesis-driven analysis of cell–cell interactions in complex tissues.
Fig. 4.
Cellular composition and cell–cell distance in the Ovarian T cells dataset revealed by SANDI. a. The percentage of immune cell subsets in each of the 9 ovarian slides. b. Overall compositions of six immune cell subsets. c, Correlation heatmap to illustrate the association between percentages of six immune cell subsets. d. Density plots showing the distribution of PD1-CD4+FOXP3-, PD1+CD4+FOXP3- within 250 μm to the nearest PD1-CD4+FOXP3+ Treg cells. e. Density plot showing the distribution of PD1-CD8+ and PD1+CD8+ T cells within 250 μm to the nearest PD1-CD4+FOXP3+ Treg cells. The mean distance is shown as the horizontal line.
Discussion
In this work, we developed and demonstrated the performance of a self-supervised framework SANDI for cell classification in multiplex images to minimise the workload for pathologists. Results obtained from five datasets showed that, with an average of 10 labels per cell type, the performance of SANDI was comparable to that of the fully supervised classifier trained on more than 1800 single-cell annotations. Specifically, SANDI achieved a mean weighted F1-score 0.002 below that of the fully supervised classifier in the DCIS FOXP3 dataset. We also showed that SANDI achieved superior or comparable performance to other self-supervised frameworks when the annotation budget was below 30%, indicating that our proposed framework is highly effective at reducing the number of annotations required for accurate classification. We achieved these results by using a self-supervised model that learns the distinct features of cell classes using pairwise similarities between subpatches of the same cell and different cells as labels. Notably, the model demonstrated the ability to generalise effectively to datasets containing various compositions of cell types (Table 1, Table 2), suggesting that the model was robust against varying distributions of cell types, which can potentially be introduced by different choices of regions of interest. Additionally, we showed that the trained encoders can help identify cells that are prone to misclassification, thus guiding the annotation efforts towards cells that can effectively improve classification accuracy. With SANDI applied to the Ovarian T cell slides, we revealed a distinct association of PD1 expression on CD8+ and CD4+ T cells with the Treg-mediated immunosuppression. These results demonstrate the capability of SANDI in deconstructing cellular spatial organisation at scale and suggest its potential application in biomarker discovery and clinical translations.
This work has several limitations. First, the pipeline still requires manual selection of regions that contain a variety of cell phenotypes and are of high image quality to ensure that a considerable number of cells of interest are included in the training. Future work to evaluate automated methods to guide region selection will help address this issue. Second, the training images of the self-supervised model is currently limited to cell-containing images, which involves a pre-trained detection model applied prior to the pipeline to locate image patches of single cells. Classification on automatically detected cells showed that SANDI was capable of distinguishing cell-containing images from the tissue background when representative images of background were provided as references. It would be of interest to identify background patches using the self-supervised model trained on randomly cropped image patches to reduce false positives in cell detection. Also, the performance of SANDI was contingent on the selection of the weight ratio in the loss function. An intriguing direction for future research is to automatically learn the weight ratio along with the weights in layers during model training. Additionally, the increase in classification performance as the automatic expansion of the reference set was inconsistent, which might be due to the varied quality of references. A quantitative measure of image quality is required to evaluate the sustainability of model performance to different choices of references. Lastly, future work should tailor this method to other multiplex imaging techniques, such as phenocycler42 and multiplexed ion beam imaging43 to aid the cell phenotyping in the context of a large number of antibodies.
In conclusion, SANDI enables labour-efficient cell phenotyping in multiplex images with minimal manual inputs, which facilitates the analysis of large-scale datasets and paves the way for translating multiplex image analysis into clinical practice. By employing the prediction of pairwise similarity as the pretext task, self-supervised learning leverages intrinsic information from the rich amount of unlabelled data independent of prior knowledge of cell phenotypes. This strategy greatly reduces the expert annotations required for desired classification performance and establishes self-supervised learning as a promising new technology in medical artificial intelligence.
Contributors
H.Z and Y.Y conceived and designed the study. Y.Y supervised the work. H.Z conducted the validation experiments and analysed the results. K.A provided insights for the pipeline. H.Z and K.A verified the underlying data. T.G and J.A.L provided the Ovarian T cells dataset. J.L.Q, T.M, and A.U.A provided the LUSC T cell dataset. F.S and E.S.H provided the DCIS FOXP3 dataset. Y.H, C.S.Y.L, D.P, L.L, M.R-J and K.Y provided the Myeloma dataset. H.Z, K.A, Y.Y wrote the manuscript with input from all the authors. All authors have read and approved the final version of the manuscript.
Data sharing statement
The images and annotations of the DCIS FOXP3, Myeloma, and the IMC CD4-CD8 datasets can be obtained from the corresponding publication. The Ovarian T cells dataset is available at https://doi.org/10.5281/zenodo.8219131. The LUSC T cells dataset is available at https://doi.org/10.5281/zenodo.8219177.
Code availability
The scripts for training and implementing SANDI are available at https://doi.org/10.5281/zenodo.8063718.
Declaration of interests
The authors declare the following competing financial interests: a patent has been filed for the methodology reported in the paper (applicant, the Institute of Cancer Research; inventors, H.Z and Y.Y; application number, UK patent GB 2106397.9 and PCT/EP2022/061941). Other authors have no conflict of interest to disclose.
Acknowledgements
This study was funded by an anonymous private donation in support of H.Z. Y.Y acknowledges funding from Cancer Research UK Career Establishment Award (CRUK C45982/A21808), CRUK Early Detection Program Award (C9203/A28770), CRUK Sarcoma Accelerator (C56167/A29363), CRUK Brain Tumour Award (C25858/A28592), Breast Cancer Now (2015NovPR638), Rosetrees Trust (A2714), Children's Cancer and Leukaemia Group (CCLGA201906), NIH U54 CA217376, NIH R01 CA185138, CDMRP Breast Cancer Research Program Award BC132057, European Commission ITN (H2020-MSCA-ITN-2019), and The Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre. K.W, L.L and M.R-J are partly funded by the UCL/UCLH NIHR Biomedical Research Centre.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104769.
Appendix A. Supplementary data
References
- 1.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan W.C.C., Nerurkar S.N., Cai H.Y., et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020;40:135–153. doi: 10.1002/cac2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fassler D.J., Abousamra S., Gupta R., et al. Deep learning-based image analysis methods for brightfield-acquired multiplex immunohistochemistry images. Diagn Pathol. 2020;15:100. doi: 10.1186/s13000-020-01003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serag A., Ion-Margineanu A., Qureshi H., et al. Translational AI and deep learning in diagnostic pathology. Front Med. 2019;6:185. doi: 10.3389/fmed.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greener J.G., Kandathil S.M., Moffat L., Jones D.T. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23:40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 6.AbdulJabbar K., Raza S.E.A., Rosenthal R., et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med. 2020;26:1–9. doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankhead P., Loughrey M.B., Fernández J.A., et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geread R.S., Morreale P., Dony R.D., et al. IHC color histograms for unsupervised Ki67 proliferation index calculation. Front Bioeng Biotechnol. 2019;7:226. doi: 10.3389/fbioe.2019.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan R., Rajpurkar P., Topol E.J. Self-supervised learning in medicine and healthcare. Nat Biomed Eng. 2022;6:1346–1352. doi: 10.1038/s41551-022-00914-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen T., Kornblith S., Norouzi M., Hinton G. 2020. A simple framework for contrastive learning of visual representations. [Google Scholar]
- 11.He K., Fan H., Wu Y., Xie S., Girshick R. Proceedings of the IEEE computer society conference on computer vision and pattern recognition. 2020. Momentum contrast for unsupervised visual representation learning; pp. 9726–9735. [DOI] [Google Scholar]
- 12.Chuang C.-Y., Robinson J., Lin Y.-C., Torralba A., Jegelka S. Debiased contrastive learning. Adv Neural Inf Process Syst. 2020;33:8765–8775. Curran Associates, Inc. [Google Scholar]
- 13.Koohbanani N.A., Unnikrishnan B., Khurram S.A., Krishnaswamy P., Rajpoot N. Self-path: self-supervision for classification of pathology images with limited annotations. IEEE Trans Med Imag. 2021;40:2845–2856. doi: 10.1109/TMI.2021.3056023. [DOI] [PubMed] [Google Scholar]
- 14.Ciga O., Xu T., Martel A.L. Self supervised contrastive learning for digital histopathology. Mach Learn Appl. 2022;7 doi: 10.1016/J.MLWA.2021.100198. [DOI] [Google Scholar]
- 15.Kobayashi H., Cheveralls K.C., Leonetti M.D., Royer L.A. Self-supervised deep learning encodes high-resolution features of protein subcellular localization. Nat Methods. 2022;19:995–1003. doi: 10.1038/s41592-022-01541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong K.S., Zhong X., Low C.S.L., Kanchanawong P. Self-supervised classification of subcellular morphometric phenotypes reveals extracellular matrix-specific morphological responses. Sci Rep. 2022;12 doi: 10.1038/s41598-022-19472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy M., Jegelka S., Fraenkel E. Self-supervised learning of cell type specificity from immunohistochemical images. Bioinformatics. 2022;38:i395–i403. doi: 10.1093/bioinformatics/btac263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiménez-Sánchez D., Ariz M., Chang H., Matias-Guiu X., de Andrea C.E., Ortiz-de-Solórzano C. NaroNet: discovery of tumor microenvironment elements from highly multiplexed images. Med Image Anal. 2022;78 doi: 10.1016/j.media.2022.102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., Grunewald T., Akarca A.U., et al. Vol. 156. 2021. Symmetric dense inception network for simultaneous cell detection and classification in multiplex immunohistochemistry images; pp. 246–257. (MICCAI Computational Pathlogy (COMPAY) Workshop). [Google Scholar]
- 20.Sobhani F., Muralidhar S., Hamidinekoo A., et al. Spatial interplay of tissue hypoxia and T-cell regulation in ductal carcinoma in situ. NPJ Breast Cancer. 2022;8:1–11. doi: 10.1038/s41523-022-00419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagos Y.B., Lecat C.S., Patel D., et al. Proceedings–International Symposium on Biomedical Imaging 2021. 2021. Cell abundance aware deep learning for cell detection on highly imbalanced pathological data; pp. 1438–1442. [Google Scholar]
- 22.Damond N., Engler S., Zanotelli V.R.T., et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019;29:755–768.e5. doi: 10.1016/j.cmet.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenwald N.F., Miller G., Moen E., et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat Biotechnol. 2022;40:555–565. doi: 10.1038/s41587-021-01094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak D., Krahenbuhl P., Donahue J., Darrell T., Efros A.A. Proceedings of the IEEE computer society conference on computer vision and pattern recognition 2016. 2016. Context encoders: feature learning by inpainting; pp. 2536–2544. [DOI] [Google Scholar]
- 25.Sirinukunwattana K., Raza S.E.A., Tsang Y.W., Snead D.R.J., Cree I.A., Rajpoot N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imag. 2016;35:1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 26.Falk T., Mai D., Bensch R., et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 27.Khosla P., Teterwak P., Wang C., et al. In: Larochelle H., Ranzato M., Hadsell R., Balcan M.F., Lin H., editors. vol. 33. Curran Associates, Inc.; 2020. Supervised contrastive learning; pp. 18661–18673. (Advances in neural information processing systems). [Google Scholar]
- 28.Kingma D.P., Ba J.L. 3rd international conference on learning representations, ICLR 2015–conference track proceedings, international conference on learning representations. ICLR; 2015. Adam: a method for stochastic optimization. [Google Scholar]
- 29.Janowczyk A., Zuo R., Gilmore H., Feldman M., Madabhushi A. JCO Clinical Cancer Informatics; 2019. HistoQC: an open-source quality control tool for digital pathology slides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jm T., Akturk G., Angelo M., et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 2020;8 doi: 10.1136/JITC-2019-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C.C., Lin C.J. LIBSVM: a Library for support vector machines. ACM Trans Intell Syst Technol. 2011;2 doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 32.Nalepa J., Kawulok M. Selecting training sets for support vector machines: a review. Artif Intell Rev. 2019;52:857–900. doi: 10.1007/s10462-017-9611-1. [DOI] [Google Scholar]
- 33.Frénay B., Verleysen M. Classification in the presence of label noise: a survey. IEEE Trans Neural Netw Learn Syst. 2014;25:845–869. doi: 10.1109/TNNLS.2013.2292894. [DOI] [PubMed] [Google Scholar]
- 34.Tsyurmasto P., Zabarankin M., Uryasev S. Value-at-risk support vector machine: stability to outliers. J Combin Optim. 2014;28:218–232. doi: 10.1007/s10878-013-9678-9. [DOI] [Google Scholar]
- 35.Danka T., Horvath P. ArXivOrg; 2018. modAL: a modular active learning framework for Python.https://arxiv.org/abs/1805.00979v2 [Google Scholar]
- 36.Lewis D.D., Catlett J. Machine Learning Proceedings 1994. Elsevier; 1994. Heterogeneous uncertainty sampling for supervised learning; pp. 148–156. [DOI] [Google Scholar]
- 37.Liu H., HaoChen J.Z., Gaidon A., Ma T. 2021. Self-supervised learning is more robust to dataset imbalance. [Google Scholar]
- 38.Hashimoto M., Kamphorst A.O., Im S.J., et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med. 2018;69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 39.Francis K., Palsson B.O. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc Natl Acad Sci U S A. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z.-Z., Grote D.M., Ziesmer S.C., Xiu B., Novak A.J., Ansell S.M. PD-1 expression defines two distinct T-cell sub-populations in follicular lymphoma that differentially impact patient survival. Blood Cancer J. 2015;5:e281. doi: 10.1038/bcj.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngiow S.F., Young A., Jacquelot N., et al. A threshold level of intratumor CD8+ T-cell PD1 expression dictates therapeutic response to anti-PD1. Cancer Res. 2015;75:3800–3811. doi: 10.1158/0008-5472.CAN-15-1082. [DOI] [PubMed] [Google Scholar]
- 42.Goltsev Y., Samusik N., Kennedy-Darling J., et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/J.CELL.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keren L., Bosse M., Marquez D., et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174:1373–1387.e19. doi: 10.1016/J.CELL.2018.08.039/ATTACHMENT/DB711C24-528F-47F0-B379-F3DB037CA6BD/MMC3.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.