Abstract
RNA viruses play an important role in Leishmania biology and virulence. Their presence was documented in three (out of four) Leishmania subgenera. Sauroleishmania of reptiles remained the only underinvestigated group. In this work, we analyzed the viral occurrence in Sauroleishmania spp. and detected RNA viruses in three out of seven isolates under study. These viruses were of two families—Narnaviridae and Totiviridae. Phylogenetic inferences demonstrated that totiviruses from L. adleri and L. tarentolae group together within a larger cluster of LRV2s, while a narnavirus of L. gymnodactyli appeared as a phylogenetic relative of narnaviruses of Blechomonas spp. Taken together, our work not only expanded the range of trypanosomatids that can host RNA viruses but also shed new light on the evolution and potential routes of viral transmission in these flagellates.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00436-023-07928-x.
Keywords: Leishmania (Sauroleishmania) adleri, L. (S.) gymnodactyli, L. (S.) hoogstraali, L. (S.) tarentolae, Narnaviridae, LRV2
Introduction
Leishmaniasis is a neglected parasitic disease threatening millions of people worldwide (WHO 2023). It is caused by flagellated protists of the genus Leishmania, which are mainly transmitted to vertebrate hosts by blood-feeding female phlebotomine sand flies (Diptera: Psychodidae) (Bruschi and Gradoni 2018). The genus comprises four subgenera, from which Leishmania, Viannia, and Mundinia are associated with human diseases, while members of the subgenus Sauroleishmania are restricted to reptiles (Espinosa et al. 2018; Kostygov et al. 2021b).
Although Leishmania in reptiles was first described over a century ago (Wenyon 1920), information about their life cycle is still limited, likely because they were not considered pathogenic to humans. Nevertheless, several historical and recent reports documented the presence of Sauroleishmania spp. in mammals, including dogs and humans (Adler 1962; Coughlan et al. 2017; Latrofa et al. 2021; Mendoza-Roldan et al. 2022b; Pombi et al. 2020).
Sauroleishmania has been isolated from a range of reptiles, mostly lizards and geckos of the families Agamidae, Gekkonidae, Lacertidae, Scincidae, and Varanidae (Belova 1971; Wilson and Southgate 1979), and sand flies of the genus Sergentomyia, which are considered the main vectors as they preferentially feed on cold-blooded vertebrates (Killick-Kendrick 1990). However, sand flies of the genus Phlebotomus are also susceptible to Sauroleishmania infection in vitro and in vivo (Tichá et al. 2021; Tichá et al. 2022).
It is now generally accepted that Sauroleishmania has evolved from the mammal-infecting parasites and all its 21 described species form a monophyletic group within the genus Leishmania (Akhoundi et al. 2016; Lukeš et al. 2018). The type species, L. (S.) tarentolae, has been extensively studied and is commonly used as a laboratory model in many fields, including biotechnology (Breitling et al. 2002; Klatt et al. 2019; Mendoza-Roldan et al. 2022a).
Numerous studies have reported presence of RNA viruses in representatives of the family Trypanosomatidae. To date, five different families of viruses were reported to infect these flagellates, of which Leishbuviridae, Narnaviridae, and Totiviridae are the most frequent (Grybchuk et al. 2018a; Grybchuk et al. 2018c).
Narnaviruses (naked RNA viruses) are capsid-less positive-strand RNA cytoplasmic elements encoding a single RNA-dependent RNA polymerase (RDRP) protein, although a multi-segmented narna-like virus has been described from the trypanosomatid Leptomonas seymouri (Kraeva et al. 2015; Lye et al. 2016). Narnaviridae, along with RNA bacteriophages (Leviviridae), mitochondrial capsidless RNA elements of eukaryotes (Mitoviridae), and plant viruses (Botourmiaviridae), belong to the phylum Lenarviricota. Based on phylogenetic inferences, it has been postulated that capsid-less mito- and narnaviruses diverged upon eukaryogenesis from an RNA phage that infected alphaproteobacteria, the ancestors of mitochondria (Koonin et al. 2015; Sadiq et al. 2022; Wolf et al. 2018). Narnaviruses of Trypanosomatidae are not monophyletic, and their evolution was likely shaped by several horizontal transfers (Grybchuk et al. 2018c).
Leishmania RNA virus (LRV) is a double-stranded RNA virus of the family Totiviridae infecting trypanosomatids from two genera: Leishmania (LRV1/2) (Scheffter et al. 1995; Stuart et al. 1992) and Blechomonas (LRV3/4) (Grybchuk et al. 2018c). These viruses form non-enveloped icosahedral virus particles about 40 nm in diameter (Procházková et al. 2021). No cellular receptors have been identified for LRVs. Thus, vertical inheritance is thought to be the predominant mode of viral transmission resulting in general co-evolution of LRVs and Leishmania spp. (Cantanhêde et al. 2021; Widmer and Dooley 1995). Notably, occasional horizontal transfers (both intra- and interspecific) have been also reported (Kostygov et al. 2021a). Such transfers are possible owing to the exploitation of host’s exosomes as vehicles for transmission between flagellates (Atayde et al. 2019; Lafleur and Olivier 2022; Olivier and Zamboni 2020). Infrequent mating events may also contribute to horizontal transmission of viruses in trypanosomatids (Akopyants et al. 2009; Rougeron et al. 2010; Sádlová et al. 2011).
There is strong evidence that LRVs provide a survival advantage to Leishmania guyanensis and L. aethiopica in vertebrate hosts through upregulation of pro-inflammatory cytokines facilitating the spread of parasites from the initial infection site (de Carvalho et al. 2019; Ives et al. 2011; Zangger et al. 2014). The LRV presence also downregulates apoptotic pathways and promotes parasite persistence (Eren et al. 2016). Of note, the molecular mechanisms governing viral maintenance may differ between LRV species (Saura et al. 2022).
In this work, we analyzed the viral occurrence in Sauroleishmania spp. and detected RNA viruses in three out of seven analyzed isolates. These viruses belong to two families—Narnaviridae and Totiviridae. Phylogenetic analyses showed totiviruses from L. adleri LV30 and L. tarentolae LV108 group together within a larger cluster of LRV2s, while a narnavirus of L. gymnodactyli LV247 appeared to be a phylogenetic relative of narnaviruses of Blechomonas spp. Taken together, our work expands the range of trypanosomatids that can host RNA viruses to include Sauroleishmania, the only Leishmania subgenus that has not been scrutinized in this respect so far.
Materials and methods
Isolates and cultivation
Seven cultures of Leishmania (Sauroleishmania) were examined: L. (S.) adleri RLIZ/KE/1954/1433 (LV30) isolated from the common long-tailed lizard Latastia longicaudata in Kenya in 1954 (Heisch 1958); L. (S.) gymnodactyli RGEC/SU/1964/Ag (LV247) isolated from the agamid lizard Trapelus (Agama) sanguinolenta in Turkmenistan in 1964 (Saf'janova 1966); L. (S.) hoogstraali RHEM/SD/1963/NG-26 (LV31) isolated from the Mediterranean house gecko Hemidactylus turcicus in Sudan in 1963 (McMillan 1965); L. (S.) tarentolae RTAR/IT/1981/ISS21-G6c (ISS21) isolated from the common wall gecko Tarentola mauritanica in Italy in 1981 (Pozio et al. 1986); L. (S.) tarentolae RCYR/IT/1981/ISS24-CK3 (ISS24) isolated from the Kotschy's gecko Mediodactylus (Cyrtodactylus) kotschyi in Italy in 1981 (Pozio et al. 1983); L. (S.) tarentolae IMIN/IT/2017/ISS3200RM-5 (ISS3200) isolated from the sand fly Sergentomyia minuta in Italy in 2017 (Di Muccio et al. 2015); and L. (S.) tarentolae RTAR/SE/67/G10 (LV108) isolated from the white-spotted wall gecko Tarentola annularis in Senegal in 1967 (Ranque 1973). This is one of the most comprehensive collections of the currently available Sauroleishmania isolates. Cells were cultivated as described previously (Tichá et al. 2022) and their identities were confirmed as in (Yurchenko et al. 2006).
Screening for dsRNA and next-generation sequencing
Trypanosomatid cultures were screened as described previously (Grybchuk et al. 2020). In short, 50 μg of total RNA from each strain was digested with DNase I (Thermo Fisher Scientific, Carlsbad, USA) and S1 nuclease (Sigma-Aldrich, St. Louis, USA) and analyzed by gel electrophoresis. The three gel-positive samples were sequenced at Macrogen (Seoul, South Korea) following the protocol established before (Kleschenko et al. 2019).
Sequence data processing and phylogenetic inferences
The raw sequence reads were trimmed with Trimmomatic v. 0.40 (Bolger et al. 2014), assembled de novo in Trinity v. 2.13.2 (Haas et al. 2013), and mapped back to the obtained contigs using Bowtie2 v. 2.4.4 (Langmead and Salzberg 2012) and SAMtools v. 1.17 (Danecek et al. 2021). BEDTools v. 2.30.0 software was used to estimate read coverage (Quinlan 2014). The identity of each contig was determined by running BLASTN (BLAST+ v. 2.13.0 (Camacho et al. 2009)) against a custom database of publicly available trypanosomatid genomes and BLASTX (DIAMOND v. 2.0.2 (Buchfink et al. 2021)) against UniClust50 protein database (Mirdita et al. 2017). All viral contigs were found by BLASTX. In addition, unmatched contigs were checked for the presence of long ORFs (as it usually the case for viral genomes); however, none was found.
The LRV phylogeny was inferred from concatenated protein sequence alignment of capsid and RDRP genes. The dataset was taken from previous work (Kostygov et al. 2021a). Each gene was aligned iteratively in MAFFT v. 7.490 (Katoh and Standley 2013) with G-INS-i algorithm and trimmed with a range of gap thresholds in TrimAl v. 1.4 (Capella-Gutiérrez et al. 2009). The optimal alignment length (665 positions at 0.7 gap threshold for capsid and 828 positions at 0.6 gap threshold for RDRP) and substitution model (LG + I + F + G4 for both capsid and RDRP) were selected based on the average bootstrap support value (ultra-fast bootstraps in IQ-TREE 2 v. 2.2.2.6 (Minh et al. 2020)). The respective alignments were concatenated and subject to maximum likelihood (ML) phylogenetic inference with 1,000 thorough bootstrap replicas in IQ-TREE 2 without partitioning. Bayesian tree was inferred in MrBayes v. 3.2.7. (Ronquist et al. 2012) with default settings and the same model as in the ML analysis. The tree was rooted at the midpoint.
The relationships of LRV2 viruses were inferred based on nucleotide sequences obtained here and all those available in the GenBank. The alignment was performed in MAFFT as above, but no trimming was applied. The ML analysis in IQ-TREE followed the same strategy, but the best automatically selected model was TIM2 + F + I + G4 and the branch support was estimated using 1,000 thorough bootstrap replicates as above.
The narnavirus phylogeny was inferred from the protein sequence of the RDRP gene. The dataset contained representatives of three families of Lenarviricota: Mitoviridae, Narnaviridae, and Botourmiaviridae. The alignment and trimming were done as above resulting in 496 positions long alignment obtained at 0.8 gap threshold and the best-fit model LG + I + F + G4. The ML and Bayesian phylogenies were inferred as above. The tree was rooted with Mitoviridae as an outgroup based on previous studies (Sadiq et al. 2022; Wolf et al. 2018).
Results
Three more species of Leishmania revealed to host RNA viruses
The gel analysis revealed that three out of seven isolates were positive for dsRNA (Fig. 1). Leishmania (S.) adleri LV30 displayed a single dsRNA band with similar mobility of approximately 6 kb, but at least by one order of magnitude lower intensity, as compared to that of the LRV1-4 from L. (V.) guyanensis M4147 that was used here as a positive control (Zakharova et al. 2022). On the contrary, a single dsRNA band of nearly the same size detected in L. (S.) tarentolae LV108 was severalfold brighter than that in the control. BLAST searches identified LRV2 in these two isolates of Sauroleishmania.
Fig. 1.
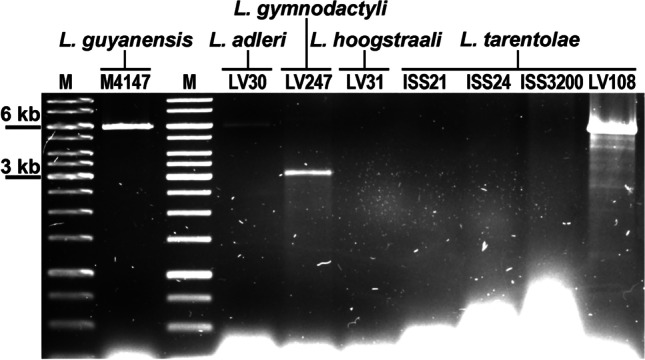
dsRNA agarose gel with screened Sauroleishmania isolates. L. guyanensis Lg-M4147 was used as a positive control. M - 1 kb DNA ladder
Leishmania (S.) gymnodactyli LV247 had a prominent major band of approximately 3 kb along with a fainter band that migrated at about 6 kb. The global BLASTX (all contigs versus clustered UniProt DB) detected only a narnavirus, which corresponded to the lower bright band, while the identity of the faint upper band could not be reliably established. One assembled contig of about 5.4 kb had no detectable ORFs and, as such, returned no BLASTX or BLASTN hits. Other contigs ranging between 5.5 and 7 kb had hits to the Leishmania genome. No dsRNA bands were documented in L. (S.) hoogstraali LV31 or three L. (S.) tarentolae isolates (ISS21, ISS24, and ISS3200).
Phylogenetic position of the viruses from Sauroleishmania
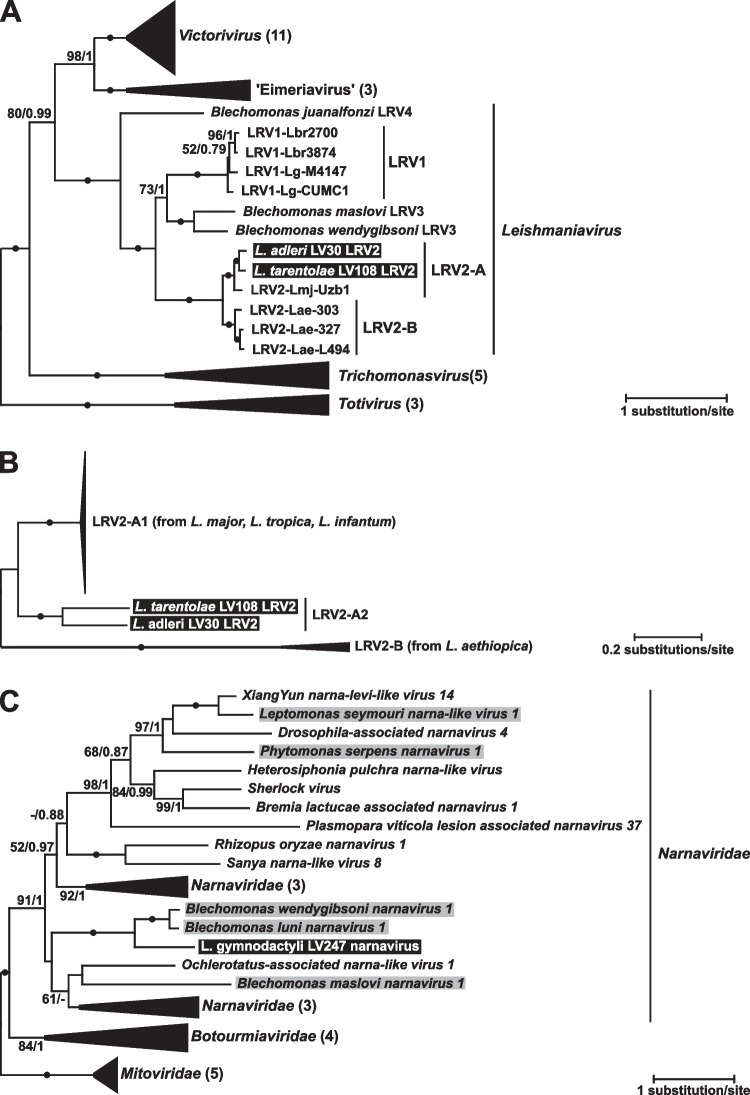
The phylogenetic analyses unambiguously (as judged by absolute statistical supports) demonstrated that the viruses from L. (S.) adleri and L. (S.) tarentolae LV108 belong to the LRV2, specifically to the clade associated with L. (L.) major. We denoted this clade as LRV2-A, as opposed to LRV2-B, containing viral sequences from L. (L.) aethiopica (Fig. 2A). To understand the relationships of the new viruses with their relatives from the LRV2-A clade, we performed an additional phylogenetic analysis using all nucleotide RDRP sequences of LRV2 available in the GenBank (Figs. 2B and S1). Although most of these sequences were rather short (ranging between 263 and 520 nt) preventing detailed resolution of their relationships (Fig. S1), it became obvious that the LRV2-A clade has a well-supported deep split between the viruses from L. (Sauroleishmania) [LRV2-A2] and those from L. (Leishmania) [LRV2-A1]. The latter subclade mostly contains viral sequences from L. (L.) major and only a few (nine sequences of 4 distinct haplotypes from L. (L.) tropica and one from L. (L.) infantum). We could not document host species-specific phylogroups in the LRV2-A1 clade.
Fig. 2.
Phylogenetic position of viruses from Sauroleishmania. A Midpoint-rooted maximum likelihood tree based on concatenated capsid and RDRP amino acid sequences of selected Totiviridae. B Maximum likelihood tree of LRV2 based on nucleotide sequences of RDRP rooted with LRVs from L. aethiopica. C Maximum likelihood tree based on RDRP amino acid sequences of selected Narnaviridae and rooted with Mitoviridae. A−B Clades were collapsed for better visibility with a number of samples indicated in brackets. Viruses reported in this study are highlighted in black, narnaviruses previously found in other trypanosomatids are highlighted in gray. Numbers at branches represent bootstrap supports and Bayesian posterior probabilities; circles indicate absolute support (100/1); values below 50% or 0.5 are replaced with dashes or not shown
The virus from L. (S.) gymnodactyli nested within the speciose family Narnaviridae with the closest relatives being the two sister viruses from trypanosomatids of the genus Blechomonas, namely, Blechomonas wendygibsoni narnavirus 1 and Blechomonas luni narnavirus 1 (Grybchuk et al. 2018c) (Fig. 2C). Interestingly, while statistical supports varied across the tree, the two internal branches determining such relationships between these three species showed maximal values for bootstrap percentage and posterior probability.
Discussion
Our screening of Sauroleishmania isolates revealed viruses in three species. Out of these, the first two, namely, L. adleri and L. tarentolae, harbored the same viral species, which has been characterized earlier—Leishmania RNA virus 2 (LRV2), while a new virus has been discovered in L. gymnodactyli. Although being new, it belongs to Narnaviridae, a group, the members of which has been repeatedly recorded in various trypanosomatids (Grybchuk et al. 2018a; Grybchuk et al. 2018c; Lye et al. 2016). Interestingly, only one L. tarentolae isolate (that from Senegal) harbored LRV2, while those three obtained in two different regions of Italy from three distinct animal species (one sand fly and two lizards) tested negative. This suggests that the distribution of viruses may be region-specific. However, it cannot be excluded that such a result is biased because of the small sample size.
Our findings shed a new light on evolution of viruses in trypanosomatids, which represents a quaint mixture of co-evolution and horizontal transfers. It was previously known that LRV2 is subdivided into two clades designated here as LRV2-A and LRV-2B for viruses from (mainly) L. major and L. aethiopica, respectively (Kostygov et al. 2021a). The divergence of these two groups is so ancient that the viruses belonging to them already acquired structural differences in their genome (Grybchuk et al. 2018b). In the current work, we revealed that the LRVs of Sauroleishmania fall into the LRV2-A clade, and this result can be explained by occasional co-infections of a common sand fly vector by parasites from these two subgenera (das Chagas et al. 2022; Latrofa et al. 2021; Mendoza-Roldan et al. 2022b; Pombi et al. 2020; Saf'janova 1991; Saf'janova et al. 1976). However, the additional phylogenetic analysis with all available LRV2 sequences suggests that such a viral transfer is not recent as judged by a relatively deep split between the LRV2-A1 of L. (Leishmania) and LRV2-A2 of L. (Sauroleishmania). Considering the limited number of analyzed isolates, we cannot judge whether this transition was unique. However, we argue that establishment of the infection in a phylogenetically distant and, therefore, physiologically different host is challenging and such events should be rare. Conversely, all documented transitions of LRV2-A1s from L. major to phylogenetically closer L. tropica and L. infantum appear quite recent (Hajjaran et al. 2016; Nalçacı et al. 2019; Saberi et al. 2020; Yurchenko et al. 2023) and occurred independently in different lineages of this viral clade, as we demonstrated here for the first time by combining all the available sequences of these viruses.
The new narnavirus discovered in L. gymnodactyli provides another example of an interesting link between Leishmania and Blechomonas. The sister relationship of this virus to those from B. wendygibsoni and B. luni is reminiscent of the situation with leishmania viruses: LRVs from these two trypanosomatid genera represent a monophyletic group, but it comprises more lineages (Grybchuk et al. 2018c). Previously, we hypothesized that all narnaviruses in Blechomonas spp. could be acquired independently from other trypanosomatids. However, the phylogenetic position of the virus from L. gymnodactyli suggests that, at least in this case, the initial acquisition was followed by a horizontal transfer to another trypanosomatid. As it was argued in our previous study, the direction of this transfer was from Leishmania to Blechomonas, since a contact between trypanosomatids of these two genera could occur only in the gut of a flea, the host of Blechomonas spp. (Grybchuk et al. 2018c). Out of the two variants possible for LRVs, i.e., transfer in the gut of either adult fleas or larvae, the second one appears more plausible. Indeed, the presence of Sauroleishmania (reptilian parasites) in the gut of adult fleas, which feed on mammalian blood, is unlikely. However, the larvae developing in the dens of mammals are scavengers that can consume bodies of dead insects, including sand flies, frequently appearing in such biotopes. Interestingly, this intergeneric transfer of viruses became possible because of the ability of some sand flies (e.g., Sergentomyia spp.) to feed on both reptiles and mammals (Polanská et al. 2020; Tichá et al. 2023). The repeated events of viral transition between Blechomonas and Leishmania (one for narnaviruses and at least two for leishmania viruses) suggest that these might also involve Leishbuviridae (LBVs), which have been documented in the flea-infecting Blechomonas. To date, only a single representative of this viral family (Leishmania martiniquensis LBV1) was found in Leishmania (Grybchuk et al. 2020).
In conclusion, detection of RNA viruses in Sauroleishmania not only closed the gap in our knowledge about viral presence in different groups of Leishmania but also shed light on the viral evolution in trypanosomatids. At the same time, our study raises new questions requiring screening of more samples and functional studies to reveal the significance of these viruses in Sauroleishmania biology.
Supplementary information
Author contribution
The study was designed by V.Y. Data collection and analyses were carried out by D.K., D.G., L.T., J.V., P.V., and A.Y.K. V.Y. and A.Y.K. drafted the manuscript. All authors contributed to the final draft and editing, giving their approval for publication and agreeing to be held accountable for the work performed herein.
Funding
Open access publishing supported by the National Technical Library in Prague. This work was supported by the Grant Agency of Czech Republic (20-22689) to V.Y. Computational resources were partially funded by the European Regional Funds (CZ.02.1.01/16_019/ 0000759) to V.Y. and P.V; L.T. and P.V. were supported by the project LX22NPO5103, Next Generation EU of the National Institute of Virology and Bacteriology.
Data availability
All sequence data obtained in this work were submitted to GenBank with the following accession numbers: OR192165 (narnavirus of L. gymnodactyli LV247), OR192166 (LRV2 of L. adleri LV30), and OR192167 (LRV2 of L. tarentolae LV108).
Declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
V.Y. is an Editor for Parasitology Research. Other authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Donnamae Klocek and Danyil Grybchuk contributed equally.
Contributor Information
Alexei Yu. Kostygov, Email: kostygov@gmail.com
Vyacheslav Yurchenko, Email: vyacheslav.yurchenko@osu.cz.
References
- Adler S. The behaviour of a lizard Leishmania in hamsters and baby mice. Rev Inst Med Trop Sao Paulo. 1962;4:61–64. [PubMed] [Google Scholar]
- Akhoundi M, et al. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis. 2016;10(3):e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants NS, et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324(5924):265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atayde VD, et al. Exploitation of the Leishmania exosomal pathway by Leishmania RNA virus 1. Nat Microbiol. 2019;4:714–723. doi: 10.1038/s41564-018-0352-y. [DOI] [PubMed] [Google Scholar]
- Belova EM. Reptiles and their importance in the epidemiology of leishmaniasis. Bull World Health Organ. 1971;44(4):553–560. [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, et al. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr Purif. 2002;25(2):209–218. doi: 10.1016/S1046-5928(02)00001-3. [DOI] [PubMed] [Google Scholar]
- Bruschi F, Gradoni L. The leishmaniases: old neglected tropical diseases. Cham, Switzerland: Springer; 2018. [Google Scholar]
- Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantanhêde LM, et al. The maze pathway of coevolution: a critical review over the Leishmania and its endosymbiotic history. Genes. 2021;12(5):657. doi: 10.3390/genes12050657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan S, Mulhair P, Sanders M, Schönian G, Cotton JA, Downing T. The genome of Leishmania adleri from a mammalian host highlights chromosome fission in Sauroleishmania. Sci Rep. 2017;7:43747. doi: 10.1038/srep43747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):1–4. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Chagas BD, et al. Interspecies and intrastrain interplay among Leishmania spp. parasites. Microorganisms. 2022;10(10):1883. doi: 10.3390/microorganisms10101883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho RVH, et al. Leishmania RNA virus exacerbates leishmaniasis by subverting innate immunity via TLR3-mediated NLRP3 inflammasome inhibition. Nat Commun. 2019;10(1):5273. doi: 10.1038/s41467-019-13356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Muccio T, et al. Epidemiology of imported leishmaniasis in Italy: implications for a European endemic country. PLoS One. 2015;10(6):e0129418. doi: 10.1371/journal.pone.0129418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren RO, et al. Mammalian innate immune response to a Leishmania-resident RNA virus increases macrophage survival to promote parasite persistence. Cell Host Microbe. 2016;20(3):318–328. doi: 10.1016/j.chom.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa OA, Serrano MG, Camargo EP, Teixeira MMG, Shaw JJ. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology. 2018;145(4):430–442. doi: 10.1017/S0031182016002092. [DOI] [PubMed] [Google Scholar]
- Grybchuk D, et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc Natl Acad Sci U S A. 2018;115(3):E506–E515. doi: 10.1073/pnas.1717806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grybchuk D, Kostygov AY, Macedo DH, d'Avila-Levy CM, Yurchenko V. RNA viruses in trypanosomatid parasites: a historical overview. Mem Inst Oswaldo Cruz. 2018;113(4):e170487. doi: 10.1590/0074-02760170487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grybchuk D, Kostygov AY, Macedo DH, Votýpka J, Lukeš J, Yurchenko V. RNA viruses in Blechomonas (Trypanosomatidae) and evolution of Leishmaniavirus. mBio. 2018;9(5):e01932–e01918. doi: 10.1128/mBio.01932-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grybchuk D, et al. The first non-LRV RNA virus in Leishmania. Viruses. 2020;12(2):168. doi: 10.3390/v12020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjaran H, et al. Detection and molecular identification of Leishmania RNA virus (LRV) in Iranian Leishmania species. Arch Virol. 2016;161(12):3385–3390. doi: 10.1007/s00705-016-3044-z. [DOI] [PubMed] [Google Scholar]
- Heisch RB. On Leishmania adleri sp. nov. from lacertid lizards (Latastia sp.) in Kenya. Ann Trop Med Parasitol. 1958;52(1):68–71. doi: 10.1080/00034983.1958.11685846. [DOI] [PubMed] [Google Scholar]
- Ives A, et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331(6018):775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med Vet Entomol. 1990;4(1):1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Klatt S, Simpson L, Maslov DA, Konthur Z. Leishmania tarentolae: taxonomic classification and its application as a promising biotechnological expression host. PLoS Negl Trop Dis. 2019;13(7):e0007424. doi: 10.1371/journal.pntd.0007424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleschenko Y, et al. Molecular characterization of Leishmania RNA virus 2 in Leishmania major from Uzbekistan. Genes. 2019;10:e830. doi: 10.3390/genes10100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479-480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov AY, et al. Analyses of Leishmania-LRV co-phylogenetic patterns and evolutionary variability of viral proteins. Viruses. 2021;13:2305. doi: 10.3390/v13112305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov AY, et al. Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses. Open Biol. 2021;11:200407. doi: 10.1098/rsob.200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraeva N, et al. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 2015;11(8):e1005127. doi: 10.1371/journal.ppat.1005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur A, Olivier M. Viral endosymbiotic infection of protozoan parasites: how it influences the development of cutaneous leishmaniasis. PLoS Pathog. 2022;18(11):e1010910. doi: 10.1371/journal.ppat.1010910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrofa MS, Mendoza-Roldan JA, Manoj R, Dantas-Torres F, Otranto D. A duplex real-time PCR assay for the detection and differentiation of Leishmania infantum and Leishmania tarentolae in vectors and potential reservoir hosts. Entomol Gen. 2021;41(5):543–551. doi: 10.1127/entomologia/2021/1178. [DOI] [Google Scholar]
- Lukeš J, Butenko A, Hashimi H, Maslov DA, Votýpka J, Yurchenko V. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol. 2018;34(6):466–480. doi: 10.1016/j.pt.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Lye LF, Akopyants NS, Dobson DE, Beverley SM. A Narnavirus-like element from the trypanosomatid protozoan parasite Leptomonas seymouri. Genome Announc. 2016;4(4):e00713–e00716. doi: 10.1128/genomeA.00713-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan B. Leishmaniasis in the Sudan Republic. 22. Leishmania hoogstraali sp. n. in the gecko. J Parasitol. 1965;51(3):336–339. doi: 10.2307/3275947. [DOI] [PubMed] [Google Scholar]
- Mendoza-Roldan JA, et al. Leishmania tarentolae: a new frontier in the epidemiology and control of the leishmaniases. Transbound Emerg Dis. 2022;69(5):e1326–e1337. doi: 10.1111/tbed.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Roldan JA, et al. Leishmania (Sauroleishmania) tarentolae isolation and sympatric occurrence with Leishmania (Leishmania) infantum in geckoes, dogs and sand flies. PLoS Negl Trop Dis. 2022;16(8):e0010650. doi: 10.1371/journal.pntd.0010650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, von den Driesch L, Galiez C, Martin MJ, Söding J, Steinegger M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017;45(D1):D170–D176. doi: 10.1093/nar/gkw1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalçacı M, et al. Detection of Leishmania RNA virus 2 in Leishmania species from Turkey. Trans R Soc Trop Med Hyg. 2019;113(7):410–417. doi: 10.1093/trstmh/trz023. [DOI] [PubMed] [Google Scholar]
- Olivier M, Zamboni DS. Leishmania (Viannia) guyanensis, LRV1 virus and extracellular vesicles: a dangerous trio influencing the faith of immune response during muco-cutaneous leishmaniasis. Curr Opin Immunol. 2020;66:108–113. doi: 10.1016/j.coi.2020.08.004. [DOI] [PubMed] [Google Scholar]
- Polanská N, et al. Sergentomyia schwetzi: salivary gland transcriptome, proteome and enzymatic activities in two lineages adapted to different blood sources. PLoS One. 2020;15(3):e0230537. doi: 10.1371/journal.pone.0230537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, et al. Molecular detection of Leishmania (Sauroleishmania) tarentolae in human blood and Leishmania (Leishmania) infantum in Sergentomyia minuta: unexpected host-parasite contacts. Med Vet Entomol. 2020;34(4):470–475. doi: 10.1111/mve.12464. [DOI] [PubMed] [Google Scholar]
- Pozio E, Gramiccia M, Gradoni L, Maroli M. Hemoflagellates in Cyrtodactylus kotschyi (Steindachner, 1870) (Reptilia, Gekkonidae) in Italy. Acta Trop. 1983;40(4):399–400. [PubMed] [Google Scholar]
- Pozio E, Gramiccia M, Gradoni L, Maroli M. Hémoflagellés de Tarentola mauritanica L., 1758 (Reptilia, Gekkonidae) In: Rioux JA, editor. Leishmania Taxonomie et phylogenèse. Montpellier: IMEEE; 1986. pp. 149–155. [Google Scholar]
- Procházková M, et al. Capsid structure of Leishmania RNA Virus 1. J Virol. 2021;95(3):e01957–e01920. doi: 10.1128/JVI.01957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranque P. Etude morphologique et biologique de quelques Trypanosomatides récoltés au Sénégal. Ph.D. thesis, Université de Aix-Marseilles; 1973. [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V, De Meeûs T, Kako Ouraga S, Hide M, Bañuls AL. "Everything you always wanted to know about sex (but were afraid to ask)" in Leishmania after two decades of laboratory and field analyses. PLoS Pathog. 2010;6(8):e1001004. doi: 10.1371/journal.ppat.1001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi R, et al. Presence and diversity of Leishmania RNA virus in an old zoonotic cutaneous leishmaniasis focus, northeastern Iran: haplotype and phylogenetic based approach. Int J Infect Dis. 2020;101:6–13. doi: 10.1016/j.ijid.2020.08.033. [DOI] [PubMed] [Google Scholar]
- Sadiq S, Chen YM, Zhang YZ, Holmes EC. Resolving deep evolutionary relationships within the RNA virus phylum Lenarviricota. Virus Evol. 2022;8(1):veac055. doi: 10.1093/ve/veac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sádlová J, et al. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One. 2011;6(5):e19851. doi: 10.1371/journal.pone.0019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saf'janova VM. Serological comparison of leptomonad strains isolated from sandflies with Leishmania tropica and leptomonads of reptiles. Med Parazitol (Mosk) 1966;6:686–695. [Google Scholar]
- Saf'janova VM. Phlebotominae sandflies as the key link of Leishmania parasitic systems. Parassitologia. 1991;33:505–511. [PubMed] [Google Scholar]
- Saf'janova VM, Alekseev AN, Karapet'ian AB. The fate of the promastigotes of Leishmania tropica major and L. gymnodactyli in the body of Phlebotomus papatasi under conditions of a mixed infection. Parazitologiia. 1976;10(1):78–83. [PubMed] [Google Scholar]
- Saura A, et al. Elimination of LRVs elicits different responses in Leishmania spp. mSphere. 2022;7(4):e0033522. doi: 10.1128/msphere.00335-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffter SM, Ro YT, Chung IK, Patterson JL. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology. 1995;212(1):84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- Stuart KD, Weeks R, Guilbride L, Myler PJ. Molecular organization of Leishmania RNA virus 1. Proc Natl Acad Sci U S A. 1992;89(18):8596–8600. doi: 10.1073/pnas.89.18.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichá L, Kykalová B, Sádlová J, Gramiccia M, Gradoni L, Volf P. Development of various Leishmania (Sauroleishmania) tarentolae strains in three Phlebotomus species. Microorganisms. 2021;9(11):2256. doi: 10.3390/microorganisms9112256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichá L, Sádlová J, Bates P, Volf P. Experimental infections of sand flies and geckos with Leishmania (Sauroleishmania) adleri and Leishmania (S.) hoogstraali. Parasit Vectors. 2022;15(1):289. doi: 10.1186/s13071-022-05417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichá L, et al. Experimental feeding of Sergentomyia minuta on reptiles and mammals: comparison with Phlebotomus papatasi. Parasit Vectors. 2023;16(1):126. doi: 10.1186/s13071-023-05758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenyon CM. Observations on the intestinal protozoa of three Egyptian lizards, with a note on a cell-invading fungus. Parasitology. 1920;12(4):350–365. doi: 10.1017/S0031182000014347. [DOI] [Google Scholar]
- WHO (2023) Leishmaniasis. https://www.who.int/en/news-room/fact-sheets/detail/leishmaniasis. Accessed 1 Jun 2023
- Widmer G, Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995;23(12):2300–2304. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Southgate B. Lizard Leishmania. In: Lumsden W, Evans DA, editors. Biology of Kinetoplastida. New York: Academic Press; 1979. pp. 242–268. [Google Scholar]
- Wolf YI, et al. Origins and evolution of the global RNA virome. mBio. 2018;9(6):e02329–e02318. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko V, Chistyakov DS, Akhmadishina LV, Lukashev AN, Sádlová J, Strelkova MV. Revisiting epidemiology of leishmaniasis in Central Asia: lessons learnt. Parasitology. 2023;150(2):129–136. doi: 10.1017/S0031182022001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko V, Lukeš J, Xu X, Maslov DA. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae) J Eukaryot Microbiol. 2006;53(2):103–111. doi: 10.1111/j.1550-7408.2005.00078.x. [DOI] [PubMed] [Google Scholar]
- Zakharova A, et al. Leishmania guyanensis M4147 as a new LRV1-bearing model parasite: phosphatidate phosphatase 2-like protein controls cell cycle progression and intracellular lipid content. PLoS Negl Trop Dis. 2022;16(6):e0010510. doi: 10.1371/journal.pntd.0010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangger H, et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2014;8(4):e2836. doi: 10.1371/journal.pntd.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data obtained in this work were submitted to GenBank with the following accession numbers: OR192165 (narnavirus of L. gymnodactyli LV247), OR192166 (LRV2 of L. adleri LV30), and OR192167 (LRV2 of L. tarentolae LV108).